Abstract
Piwi-interacting RNAs (piRNAs) are central components of the piRNA pathway, which directs transposon silencing and guarantees genome integrity in the germ cells of several metazoans. In Drosophila, piRNAs are produced from discrete regions of the genome termed piRNA clusters, whose expression relies on the RDC complex comprised of the core proteins Rhino, Deadlock, and Cutoff. To date, the RDC complex has been exclusively implicated in the regulation of the piRNA loci. Here we further elucidate the function of Cutoff and the RDC complex by performing genome-wide ChIP-seq and RNA-seq assays in the Drosophila ovaries and analyzing these data together with other publicly available data sets. In agreement with previous studies, we confirm that Cutoff is involved in the transcriptional regulation of piRNA clusters and in the repression of transposable elements in germ cells. Surprisingly, however, we find that Cutoff is enriched at and affects the expression of other noncoding RNAs, including spliceosomal RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). At least in some instances, Cutoff appears to act at a transcriptional level in concert with Rhino and perhaps Deadlock. Finally, we show that mutations in Cutoff result in the deregulation of hundreds of protein-coding genes in germ cells. Our study uncovers a broader function for the RDC complex in the Drosophila germline development.
Keywords: Drosophila, germline, piRNAs, noncoding RNAs, Cutoff
INTRODUCTION
Noncoding RNAs (ncRNAs) exert essential roles in a variety of developmental events, where they regulate mechanisms ranging from transcription to RNA processing and translation. In recent years, a specific class of small ncRNAs termed Piwi-interacting RNAs or piRNAs have emerged as key components of a defense mechanism that protects the genome against transposable and repetitive sequences. In Drosophila, piRNAs are mostly produced from regions of the genome referred to as piRNA clusters (Brennecke et al. 2007). Both in the germline and in the somatic tissues of the ovary, the processing of cluster transcripts leads to the production of mature piRNAs, which interact with the Piwi protein to silence transposable elements (TEs) dispersed in the genome (Brennecke et al. 2007; Malone et al. 2009). In the germ cells, however, two additional members of the Piwi-clade Argonaute family, namely, Aubergine (Aub) and Argonaute3 (Ago3), sustain the production of piRNAs by promoting a feed-forward amplification loop known as the “ping–pong” mechanism (Brennecke et al. 2007; Gunawardane et al. 2007; Li et al. 2009a). piRNA biogenesis in the germline also requires the RNA helicase Vasa and a number of cytoplasmic proteins, which generally accumulate together with Aub and Ago3 in a perinuclear organelle known as the “nuage” (Pek et al. 2012; Xiol et al. 2014; Nishida et al. 2015). While Aub and Ago3 are cytoplasmic germline-specific factors, Piwi is mostly nuclear and is found both in the germ cells and in the somatic follicle cells of the ovary. Piwi bound to antisense piRNAs imposes the transcriptional silencing of TEs by regulating the levels of Histone H3 tri-methyl Lysine 9 (H3K9trimeth) and the recruitment of RNA Pol2 at the target element (Sienski et al. 2012).
Genomic regions that have the capacity of generating piRNAs have been identified in distantly related organisms, including mouse and human (Girard et al. 2006; Aravin et al. 2007). In Drosophila, piRNA clusters are predominantly composed of fragmented and mostly inactive transposable elements and are commonly divided into two classes. The uni-strand clusters, like the flamenco locus (flam), are populated by polarized transposon relics and produce piRNAs from one strand of the genome (Sarot et al. 2004; Mével-Ninio et al. 2007; Malone et al. 2009; Dennis et al. 2013). The majority of the piRNA clusters in this species, however, belong to the so-called dual-strand class, where transposon remnants are harbored both on the plus and minus strand of the genome. Paradigmatic of this class is Cluster 1/42AB (Cl1/42AB), which spans ∼250 kb in the pericentromeric region of the Chromosome II right arm. Like all bidirectional loci, the piRNA pool produced from Cl1/42AB is composed of both sense and antisense piRNAs. Dual-strand cluster expression in germ cells is regulated by the Cutoff (Cuff) protein in a complex with the HP1 variant Rhino (Rhi) (Klattenhoff et al. 2009; Pane et al. 2011). It was recently shown that these two factors also interact with the Deadlock (Del) protein to form the so-called “RDC” complex (Mohn et al. 2014). While Del does not display obvious catalytic domains, Cuff belongs to the Rai1-like family of transcription termination cofactors (Chen et al. 2007). In accordance with this, it was proposed that the RDC complex is essential at piRNA clusters to suppress transcription termination and promote the production of the long piRNA precursor transcripts necessary to generate a full complement of mature piRNAs (Mohn et al. 2014; Chen et al. 2016). Furthermore, the activity of the complex also prevents the splicing of cluster transcripts, which would otherwise eliminate sequences that are potential sources of piRNAs (Mohn et al. 2014; Zhang et al. 2014). This regulatory mechanism requires the activity of the DEAD-box RNA helicase UAP56, an evolutionarily conserved splicing and export factor (Gatfield et al. 2001; Zhang et al. 2012, 2014). UAP56 was shown to colocalize with Rhi in nurse cell nuclear speckles, suppress the splicing of cluster-derived transcripts, and mediate their transport to the cytoplasmic nuage (Zhang et al. 2012, 2014). Furthermore, UAP56 is a component of the THO/TREX complex, whose component Thoc5 interacts with UAP56 and is necessary for the expression of the piRNA clusters (Hur et al. 2016). While UAP56 has been reported to control the splicing and export of a variety of transcripts, to date the components of the RDC complex have been exclusively connected to the regulation of the piRNA clusters and the biogenesis of piRNAs in Drosophila germline tissues.
In the current study, we further investigate the function of the Cuff protein. We perform genome-wide RNA-seq and ChIP-seq assays in Drosophila ovaries and combine them with publicly available data sets produced by other laboratories (Zhang et al. 2012; Mohn et al. 2014). In order to obtain a more accurate understanding of the function of Cuff during oogenesis, we used high sequencing depth for each library, adopted 100-bp read length protocols, and always produced biological replicates for each assay (Supplemental Table S1). Our integrative computational analysis of these data—consisting of newly generated Cuff ChIP-seq data, RNA Pol2 ChIP-seq data, and RNA-seq data in wild-type and Cuff mutant ovaries, and analogous existing data for Rhi and Del—confirms that Cuff extensively associates with piRNA cluster sequences and is required for their transcription in germ cells, in agreement with previous reports. Strikingly, our analysis reveals that the Cuff protein is not specific to the piRNA pathway, but also contributes to the expression of several small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). snRNAs and snoRNAs have been implicated in various regulatory mechanisms, but they have been best characterized for their role in splicing and ribosomal RNA processing, respectively (Will and Lührmann 2011; Dupuis-Sandoval et al. 2015). We show that Cuff and Rhi ChIP-seq binding profiles often overlap at snRNA and snoRNA loci and that the UAP56 factor associates with snoRNA precursor transcripts. These observations strongly point to a model whereby the RDC complex and the UAP56 factor regulate the biogenesis of different classes of noncoding RNAs in the Drosophila germline.
RESULTS
Computational analysis of ChIP-seq data shows significant enrichment of Cuff, Rhi, and Del proteins in noncoding RNA gene loci
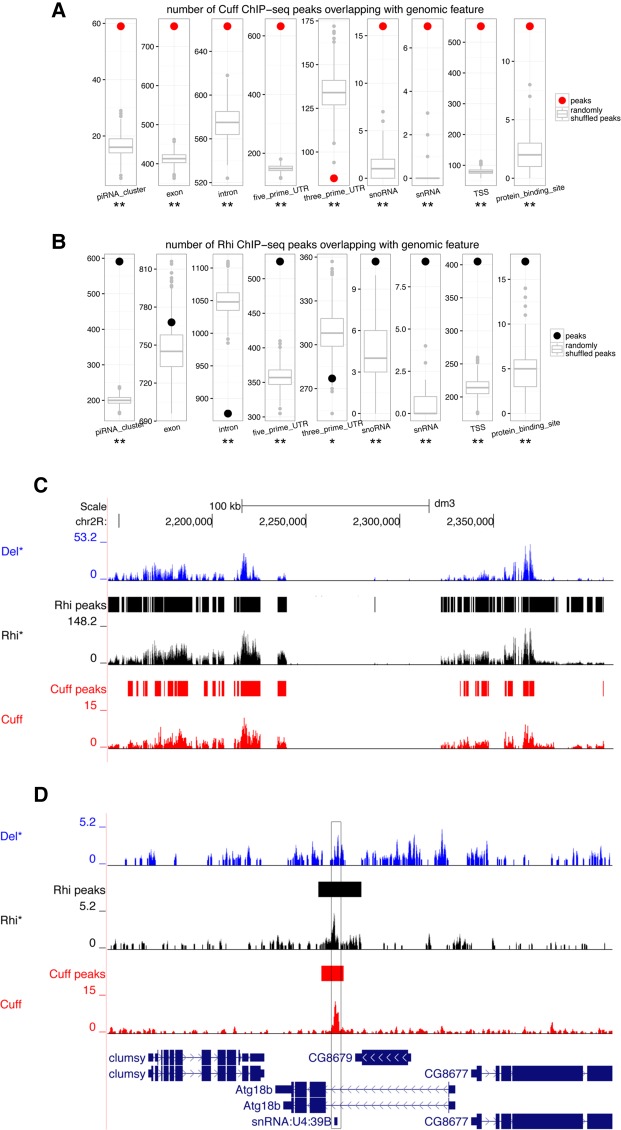
Using chromatin immunoprecipitation (ChIP) assays, we have previously shown that Cuff is enriched at specific regions within the germline dual-strand piRNA cluster 1/42AB, while it does not bind the uni-strand clusters flam and Cluster 2 (Pane et al. 2011). In order to expand our analysis and identify all the Cuff binding sites in the genome, we performed ChIP followed by deep sequencing (ChIP-seq) in ovaries expressing a Cuff-EGFP fusion protein (Pane et al. 2011). DNA fragments from Cuff-EGFP ChIP and control experiments were isolated and used to generate ChIP-seq libraries. We mapped the sequencing reads back to the Drosophila genome and pooled all the reads uniquely mapping to the genome. This approach allowed us to identify 937 regions in the Drosophila genome that are reproducibly bound by the Cuff protein (IDR threshold 0.05, see Materials and Methods for details). Since Cuff acts in a complex with Rhi and Del, we also analyzed previously published ChIP-seq data for Rhi and Del (Mohn et al. 2014). We assessed the enrichment of Cuff peaks at certain genomic features using a randomization procedure (see Materials and Methods for details). As expected, we could identify a significant enrichment for Cuff at sequences belonging to piRNA clusters (Fig. 1A; Supplemental Figs. S1, S2). Surprisingly, Cuff appears to strongly associate with other loci that express noncoding RNAs, like snRNA and snoRNA genes. Since Cuff is found in a complex with Rhi, one would expect to observe an enrichment of Rhi at the same genomic features. In accordance with this hypothesis, we can detect a significant overlap between Cuff and Rhi peaks genome-wide and an enrichment of Rhi binding at snRNAs and snoRNAs (Fig. 1B; Supplemental Figs. S1, S2). The recruitment of the RDC complex at the piRNA clusters relies on the interaction between Rhi and H3K9me3-marked chromatin (Mohn et al. 2014). However, we do not observe a significant enrichment of this histone mark at snRNA and snoRNA loci, suggesting the binding of the RDC complex or some of its components at these ncRNA loci might occur through a different and yet unknown mechanism (Supplemental Fig. S2). Consistent with our previous study, we find a robust enrichment of Cuff at Cl1/42AB (Fig. 1C), while we did not detect significant binding at the uni-strand flam and Cl2. Cuff peaks could not be readily detected at several dual-strand clusters, like the major cluster Cl13, although Rhi peaks are evident (Supplemental Fig. S3), and the piRNAs produced from this locus are lost in the cuff mutant ovaries (Pane et al. 2011) indicating that Cuff is regulating their expression. This lack of measurable binding might reflect in part the fact that Cuff does not directly interact with the DNA, rather it is recruited by Rhi and Del (Mohn et al. 2014). Intriguingly, our analysis reveals that Cuff and Rhi peaks also overlap at certain snoRNA and snRNA loci. For instance, they are enriched at the snRNA:U4 variant hosted in the intron of the Atg18b gene (Fig. 1D). These observations pointed to a role for the RDC complex in the regulation of other genomic loci in addition to piRNA clusters. We therefore set to systematically investigate the genome-wide distribution and function of Cuff in germline tissues.
FIGURE 1.
Genome-wide analysis of Cuff activity in Drosophila germ cells. (A,B) Association of Cuff and Rhi binding sites with prominent genomic features. A dot (Cuff, red; Rhi, black) shows the number of peaks overlapping with a particular genomic feature (e.g., exons or snRNAs). A boxplot shows the distribution, from 1000 random trials, of the number of overlaps when peaks are randomly shuffled while preserving their size and chromosome (see Supplemental Data for details). (*) P < 0.05, (**) P < 0.001. (C,D) Del (blue), Rhi (black), and Cuff (red) background subtracted ChIP-seq profiles along the Cl1/42AB cluster and the genomic region encompassing a snRNA:U4 gene. Loci reproducibly bound by Cuff (red) and Rhi (black) across replicates are marked with solid lines in separate tracks (not reported for Del due to lack of biological replicates). The black box highlights the Cuff and Rhi peaks at the snRNA:U4 locus. The Rhi and Del ChIP-seq data sets were produced by Mohn et al. (2014).
RNA-seq and Pol2 ChIP-seq analysis confirms the role of Cuff in regulating piRNA cluster expression
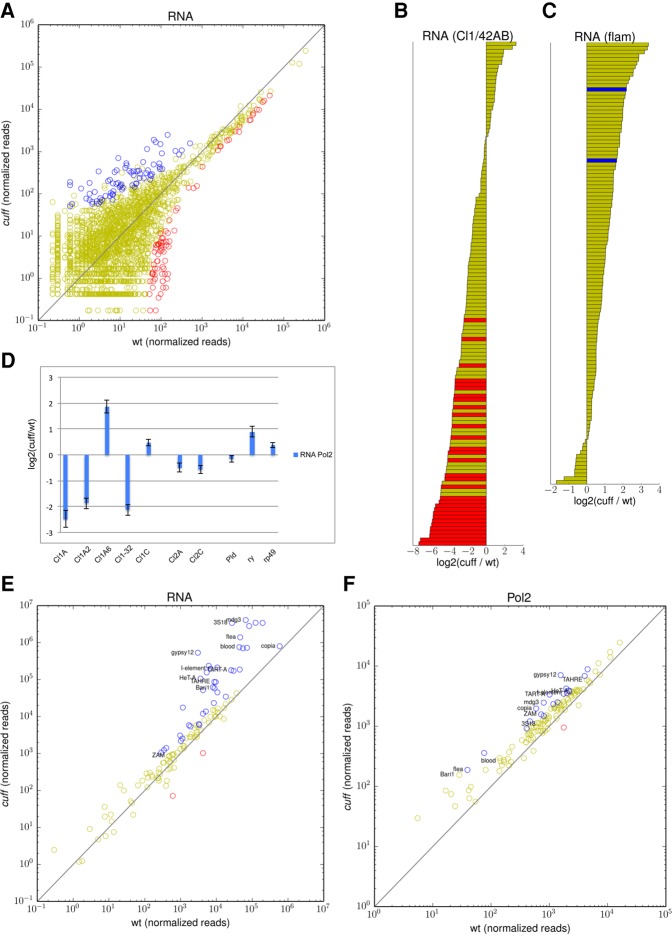
First, we evaluated the impact of mutations in cuff on piRNA cluster expression. We used RNA-seq assays and compared the expression levels of all the known piRNA loci in wild-type and cuff mutant ovaries. For this study, rRNA-depleted total RNA was used to produce cDNA libraries suitable for massive parallel sequencing. We mapped the reads obtained from each assay back to the genome. We then focused on reads that mapped to the genome uniquely. In order to obtain an accurate comparison of different clusters in the wild-type and cuff ovaries, we split each piRNA cluster into 500-bp fragments, and computed for each of them the fold difference between mutant and wild-type and estimated the differential expression between the two genetic backgrounds (Fig. 2A). This approach yielded 9933 unique fragments belonging to the 140 reported piRNA clusters. Despite the depth of our RNA-seq data sets, we could only find uniquely aligned reads for 3737 fragments in at least one of the two genotypes. Of these, 116 displayed significant down-regulation (FDR < 0.05) by at least twofold in the absence of the Cuff protein, while 86 fragments displayed a significant up-regulation (FDR < 0.05) by at least twofold in the cuff ovaries (false discovery rate calculated using the Benjamini–Hochberg procedure to correct for multiple testing; see Materials and Methods for details). The remaining regions did not show significant changes. We then narrowed our analyses to the major clusters Cl1/42AB and flam. We could determine that Cl1/42AB harbors ∼130 unique fragments with aligned reads in at least one of the genotypes. None of these fragments is significantly up-regulated in the cuff mutant, while 30 showed a significant down-regulation (at least twofold, FDR < 0.05) (Fig. 2B). For the flam locus, we could identify ∼120 unique fragments with aligned reads in at least one of the genotypes (Fig. 2C). Of these, two fragments displayed a significant up-regulation in the cuff mutant (three- to fourfold), while none of the regions was significantly down-regulated. Although limited to a set of unique fragments, our analysis confirms that Cuff is required for the expression of the piRNA cluster Cl1/42AB, while it is not involved in the regulation of the somatic flam locus. It is noteworthy that a substantial number of fragments from various clusters displayed increased expression levels in the cuff mutant ovaries. It is tempting to speculate that piRNAs might themselves contribute to the expression of the clusters by down-regulating certain regions. Alternatively, the transcripts produced from the clusters might, in some cases, fail to be processed into mature piRNAs and might therefore accumulate in germline tissues.
FIGURE 2.
Mutations in Cuff alter the transcription of piRNA clusters and transposable elements. (A) Steady-state RNA levels at piRNA clusters. Scatter plot displays 500-bp fragments of piRNA clusters that are up-regulated (blue dots), down-regulated (red dots), or not significantly affected (yellow dots) in the cuff ovaries (normalized read counts). (B,C) Barplot displaying the fold difference between cuff and wild-type ovaries [log2 (cuff/wt)] at 500-bp fragments of Cl1/42AB and flam. Fragments with significantly higher (blue bars) and lower (red bars) expression levels in the cuff mutant are shown. Yellow bars indicate regions that were not significantly affected by mutations in Cuff. (D) RNA Pol2 occupancy was analyzed by ChIP-qPCR at specific positions at Cl1/42AB, Cl2, and the control Pld, ry, and rp49 genes. Values are reported as fold difference between cuff and wt [log2 (cuff/wt)]. Error bars represent standard deviation. (E) Scatter plot displaying the distribution of the RNA-seq reads mapping to canonical transposon families in cuff versus wild-type ovaries. Transposons that are up-regulated (blue dots), down-regulated (red dots), or not significantly altered (yellow dots) in the cuff ovaries are reported. Shown on the x- and y-axes are consensus normalized read coverage values computed over replicates. (F) Scatter plot displaying the distribution of RNA Pol2 ChIP-seq reads mapping to canonical transposons in cuff versus wild-type ovaries. Transposons showing increased (blue dots), reduced (red dots), or unaltered (yellow dots) Pol2 occupancy in the cuff mutant are indicated. Shown on the x- and y-axes of scatter plots are consensus normalized read coverage values computed over replicates.
The observation that the steady-state RNA levels of cluster transcripts are sometimes altered in the absence of a functional Cuff protein led us to investigate whether these changes are caused by a transcriptional or post-transcriptional mechanism. To test these alternatives, we attempted to determine the impact of mutations in Cuff on the occupancy of RNA-Pol2 over the piRNA clusters by ChIP-seq assays (Supplemental Fig. S4). In accordance with the RNA-seq profiles, Pol2 occupancy does not display uniformly reduced levels at the piRNA clusters in the absence of a functional Cuff protein. Rather, we could identify 49 fragments displaying a substantial increase in Pol2 when we compared the cuff mutant versus the wild-type ovaries, while 15 fragments displayed a significant decrease of Pol2 in the cuff mutant. Because of the very limited number of regions that displayed significantly altered Pol2 levels in the mutant ovaries, we turned to Pol2 ChIP assays followed by qPCR on selected regions of the dual-strand Cl1/42AB and the uni-strand Cl2 to validate the conclusions of high-throughput data analysis (Fig. 2D). As control for this experiment, we analyzed the Pol2 occupancy in wild-type and cuff ovaries at the Pld, ry, and rp49 genes (Fig. 2D). This assay revealed a clear loss of Pol2 in the mutant ovaries at regions Cl1A, Cl1A2, and Cl1-32 within the Cl1/42AB cluster. In agreement with our previous results, these were the same regions that we had found to be expressed at a lower level in the absence of a functional Cuff protein (Pane et al. 2011). We also found that region Cl1A6 displayed a clear enrichment of Pol2 in the mutant ovaries, while the levels of Pol2 at region Cl1C appeared unaltered. Similarly, the levels of Pol2 at Cl2A, Cl2C, Pld, ry, and rp49 were comparable in wild-type and mutant ovaries. These observations strongly suggest that mutations in Cuff perturb the expression pattern of the clusters by affecting the levels of Pol2 along their sequences.
Transposable elements display increased RNA levels and Pol2 occupancy in the cuff mutant ovaries
A primary role of the piRNA pathway in Drosophila is to silence transposable and repetitive sequences either in germline or in somatic tissues (Senti and Brennecke 2010; Guzzardo et al. 2013). For instance, we reported earlier that cuff mutant ovaries display elevated levels of the Het-A and TART telomere-specific retrotransposons (Chen et al. 2007). We used our RNA-seq data sets to expand this analysis by comparing the expression levels of all the canonical transposable elements in wild-type versus cuff ovaries. Drosophila transposons have been divided into 122 families. Our RNA-seq approach allowed us to determine that 40 transposon families are significantly up-regulated in the cuff mutant (FDR < 0.05), 80 are apparently not significantly affected, and only two seem to be significantly down-regulated (Fig. 2E). Among the transposons that are up-regulated in cuff, the values of the fold difference between the mutant and the wild-type vary from ∼1.4-fold difference for copia to more than 120-fold for gypsy12 and 3S18. Consistent with our previous study, the Het-A transposon appears to be up-regulated by more than 30-fold and TART-A by approximately sevenfold in the mutant ovaries. The TAHRE element, which is involved in telomere regulation together with Het-A and TART, is also significantly de-repressed in the absence of Cuff.
We then asked whether the mutations in cuff cause transposon deregulation through a transcriptional versus a post-transcriptional mechanism. To answer this question, we compared the RNA-Pol2 occupancy over the transposable elements in wild-type and cuff ovaries (Fig. 2F). Interestingly, we observed a significant increase in RNA-Pol2 over 19 transposon families in the absence of a functional Cuff protein with fold differences varying from approximately twofold to more than fourfold. These include the Het-A, TART-B, and TAHRE elements (Supplemental Fig. S5). The gypsy12 element is the most sensitive to mutations in cuff both in terms of transcript levels as well as RNA Pol2 occupancy. When comparing cuff over wild-type log-fold-change values of RNA level with cuff over wild-type log-fold-change values of RNA-Pol2 level across all transposons detectable in both assays, we observed them to be significantly positively correlated (Spearman's correlation 0.3615, P-value < 4 × 10−5), which confirms massive transposon deregulation at the transcriptional level.
Intriguingly, Cuff binding events can be detected at several transposon families including the Het-A, TART, and TAHRE elements (Supplemental Fig. S5). These telomere-specific transposons are at the same time the sources and the targets of piRNAs. Hence, Cuff might control transposon expression not only indirectly, by its role in piRNA cluster regulation and piRNA production, but also directly, by associating with sequences within the transposable elements.
Cuff regulates spliceosomal RNA expression
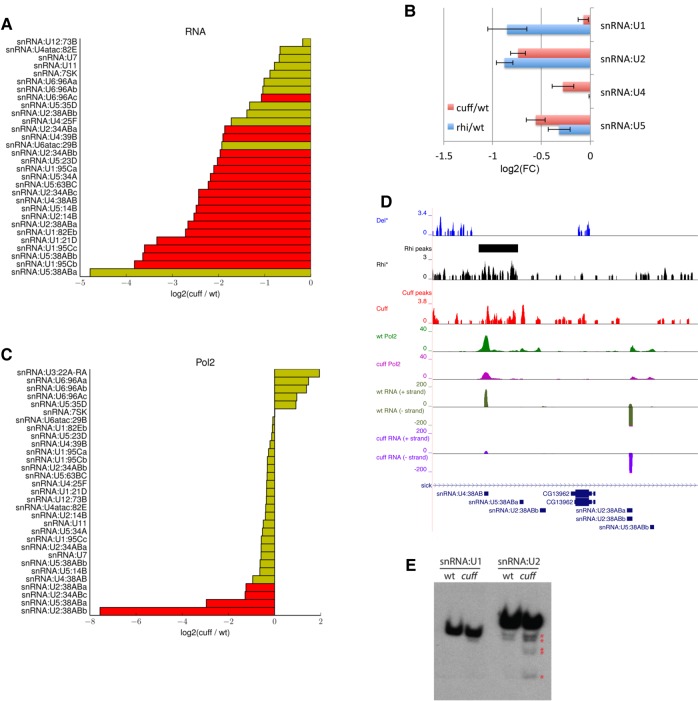
To date, the activity of Cuff and of the RDC complex has been connected only to piRNA cluster regulation in Drosophila germ cells. However, our ChIP-seq assays showed that Cuff is enriched at hundreds of positions in the genome including snRNA and snoRNA loci. The Drosophila genome harbors 31 snRNAs, which are generally divided into two classes: the U2-type, which catalyzes 99% of the splicing events in eukaryotic cells, and the U12-type (Alioto 2007; Lu and Matera 2014). The major U1, U2, U4, U5, and U6 snRNAs belong to the first class and can display multiple paralogs in the genome, while the U12-type are single-copy genes. We find that mutations in Cuff significantly affect the levels of 18 snRNAs belonging to the U2-type, which drop from two- to more than eightfold as per RNA-seq in the mutant ovaries, while U12-type snRNAs are not significantly affected (Fig. 3A). In order to lend support to our high-throughput analyses, we performed qRT-PCR assays with oligonucleotides specific for the major spliceosomal RNAs in wild-type, cuff, and rhi mutant ovaries (Fig. 3B). We could observe a reduction of the U2, U4, and U5 snRNA levels in the cuff mutant compared to the wild-type ovaries. In agreement with these observations, a consistent reduction of the snRNA levels can be detected also in the rhi mutant ovaries (Fig. 3B). The U2 and U5 snRNAs are significantly down-regulated by up to 1.8-fold both in the cuff and in the rhi mutants, thus suggesting that the RDC complex might exert a prominent role in the regulation of these snRNAs.
FIGURE 3.
Cuff controls the expression of spliceosomal RNAs. (A) Barplot of RNA-seq fold difference between wild-type and cuff ovaries for the small nuclear RNAs [log2 (cuff/wt)]. snRNAs with significantly lower (red bars) expression levels in the cuff mutant are shown. Yellow bars indicate snRNAs that are not significantly affected by mutations in Cuff. (B) Impact of mutations in cuff and rhi on the expression levels of the snRNAs U1, U2, U4, and U5 as per qRt-PCR. Y-axis displays the fold difference [log2 (FC)] between cuff and wt (blue bars) and between rhi and wt (red bars). Error bars represent standard deviation. (C) RNA Pol2 ChIP-seq levels at small nuclear RNAs. Barplot shows snRNAs characterized by reduced (red) or not significantly changed (yellow) RNA Pol2 occupancy in cuff versus wild-type (normalized read counts) [log2 (cuff/wt)]. (D) Del (blue), Rhi (black), Cuff (red), RNA Pol2 in wild-type (green), RNA Pol2 in cuff mutant (magenta) ChIP-seq profiles along selected snRNAs. Loci reproducibly bound by Rhi are marked with solid line in separate track (not reported for Del due to lack of biological replicates). RNA-seq profiles in wild-type (dark green) and cuff mutant (purple) are also shown. Rhi and Del ChIP-seq data sets were produced by Mohn et al. (2014). (E) Northern blotting analysis of total RNA extracted from wild-type and cuff mutant ovaries. Oligonucleotide probes specific to sequences in the snRNA:U1 and snRNA:U2 were used to analyze snRNA expression levels in wild-type and cuff ovaries. Red asterisks mark partial transcript of the snRNA:U2, which accumulate in the cuff mutant background.
The comparison of the Pol2 occupancy at these loci by ChIP-seq reveals that in some cases the reduced expression levels are likely caused by loss of Pol2 over the gene region (Fig. 3C,D). For instance, mutations in Cuff cause a partial loss of Pol2 over the genomic region encompassing the snRNAs U4:38Ab, U5:38Aba, and U2:38ABb and a concomitant decrease in their RNA levels (Fig. 3D). The region spanning these snRNAs is enriched in the Rhi protein. Although Cuff peaks cannot be clearly detected in this case, the data suggest that sets of neighboring snRNAs can be coregulated by the RDC complex at a transcriptional level.
Northern blotting on total RNA extracted from ovaries shows a clear accumulation of partial transcripts for the snRNA:U2 in the cuff mutant compared to the wild-type (Fig. 3E). While we cannot rule out that these fragments might be the result of partial degradation of the snRNA:U2, it is tempting to speculate that they might reflect incomplete transcription of the locus in the absence of Cuff. Different from the U2, partial or fragmented transcripts are not detected for U1, U4, and U5 (Fig. 3E and data not shown). Nevertheless, our data strongly point to a direct role for Cuff and Rhi in the regulation of certain snRNA loci in the Drosophila germline.
Small nucleolar RNA expression is controlled by the Cuff protein
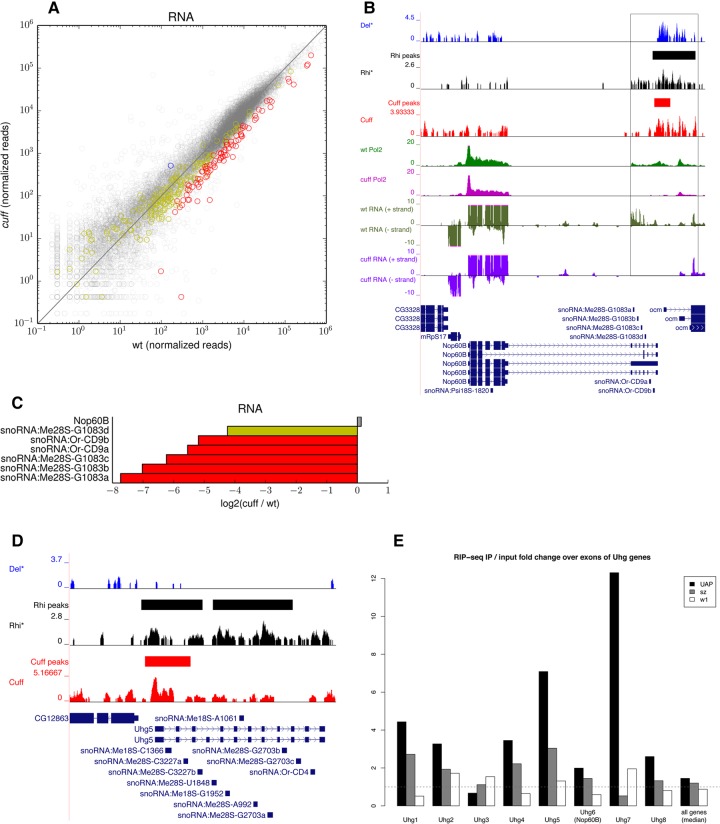
SnoRNAs are more numerous and heterogeneous than snRNAs and their functions are still largely unclear. These ncRNAs are generally hosted in the intronic sequences of protein-coding genes, and the release of the mature snoRNAs relies on the regulated splicing of the host precursor RNAs. We find that mutations in cuff significantly affect 92 of the 281 reported snoRNAs, whose expression levels drop by two- to ninefold in the mutant ovaries (Fig. 4A). One remarkable example is represented by an array of snoRNAs residing within the 3′UTR of the Nop60B gene. The expression of the snoRNAs is coupled to the transcription of Nop60B, whereby the usage of alternative transcription termination sites in the 3′UTR of this gene allows the production of transcripts encompassing the snoRNA set (Riccardo et al. 2007). Intriguingly, combining our data with that of Mohn et al. (2014), we can clearly detect partially overlapping peaks for the Cuff, Rhi, and probably Del proteins at the 3′ end of Nop60B (Fig. 4B). Mutations in cuff cause a significant loss of Pol2 over the Nop60B 3′UTR containing the snoRNAs without affecting the Pol2 occupancy along the coding region. This observation is substantiated by the significant down-regulation of the snoRNAs belonging to this cluster in the cuff ovaries as per RNA-seq, whereas the expression levels of the Nop60B gene are not significantly altered (Fig. 4B,C). Thus, Cuff, probably in a complex with Rhi and perhaps Del, appears to suppress an alternative transcription termination site at this locus and, in doing so, it couples the expression of the mRNA encoding the Nop60B protein to the production of the snoRNAs in germline tissues.
FIGURE 4.
Small nucleolar RNAs are down-regulated in cuff mutant ovaries. (A) Scatter plot displaying the snoRNAs that are up-regulated (blue), down-regulated (red), and not significantly affected (yellow) in the cuff mutant as per RNA-seq. Expression levels of protein-coding genes are displayed in the background (black). Shown on x- and y-axes are consensus normalized read coverage values computed over replicates. (B) Del (blue), Rhi (black), Cuff (red), RNA Pol2 in wild-type (green), and RNA Pol2 in cuff mutant (magenta) ChIP-seq profiles along Nop60B locus. Loci reproducibly bound by Cuff (red) and Rhi (black) are marked with solid lines in separate tracks (not reported for Del due to lack of biological replicates). RNA-seq profiles in wild-type (dark green) and cuff mutant (purple) are also shown. Black frame box highlights the snoRNA cluster in the Nop60B 3′UTR and downstream region that is bound by the RDC complex components. (C) Barplot of the fold difference between wild-type and cuff ovaries for the Nop60B coding region and for the snoRNAs hosted within its 3′UTR as per RNA-seq [log2 (cuff/wt)]. (D) Del (blue), Rhi (black), and Cuff (red) ChIP-seq profiles along the Uhg5 gene. Regions reproducibly bound by Cuff (red) and Rhi (black) as per ChIP-seq are marked with solid lines. (E) UAP56 RIP-seq at Uhg transcripts. Barplot shows the fold enrichment of the RIP-seq signal over corresponding input for the exons of the Uhg genes. Barplot for the wild-type UAP56 (UAP), the mutated UAP56sz15″ variant (sz), and for the control (w1) are shown. Only the last seven exons were used for Uhg6 (Nop60B), and only exons not overlapping the RpL23A locus were used for Uhg7, to avoid contamination from protein-coding sequence. Median of the values for all genes is shown in the last column. This analysis was performed based on data sets from Zhang et al. (2012).
snoRNAs are mostly found within the intronic sequences of protein-coding genes. In Drosophila, however, snoRNAs are also generated by specific loci termed noncoding host genes (Uhg). Uhg genes do not encode for proteins, but serve as carriers of snoRNAs, which are hosted in their intronic sequences (Tycowski and Steitz 2001). The release of mature snoRNAs proceeds through regulated splicing and trimming of the Uhg transcripts. Interestingly, Cuff and Rhi appear to be enriched at Uhg5 (Fig. 4D), thus suggesting that these proteins might directly regulate the expression of the Uhg5 snoRNA set. In order to strengthen these observations, we turned to genome-wide studies independently performed by other laboratories. It was recently shown that the processing and transport of the piRNA cluster transcripts from the nucleus to the cytoplasm is mediated by the DEAD-box helicase UAP56, which is found to colocalize with Rhi in the nurse cell nuclei (Zhang et al. 2012). Zhang and coworkers performed UAP56 RNA immunoprecipitation assays followed by deep sequencing (RIP-seq) and unveiled a significant enrichment of the piRNA precursors in the UAP56 immunoprecipitates. We reasoned that if UAP56 is implicated in the regulation of the Uhg loci together with Cuff and Rhi, we would expect to find it associated with the Uhg transcripts. In agreement with this hypothesis, we could determine that Uhg RNAs are strongly associated with the wild-type UAP56 protein, while we did not detect a preferential binding of this factor to protein-coding genes (Fig. 4E). The Uhg5 and Uhg7 transcripts show the strongest enrichment in the wild-type UAP56 protein precipitates (Fig. 4E). Transcripts from the Nop60B also seem to be enriched in UAP56 complexes albeit to a lesser extent, whereas Uhg3 RNAs are not (Fig. 4E). RIP-seq assays in germ cells were performed expressing the tagged wild-type UAP56 or the mutated UAP56sz15′ variant, where the mutated protein is characterized by an E245K substitution affecting a conserved residue (Zhang et al. 2012). Intriguingly, the binding of UAP56 at Uhg transcripts is significantly compromised by the E245K substitution harbored in UAP56sz15′ (Fig. 4E). This is particularly evident for Uhg5 and Uhg7 (Fig. 4E) and suggests that this conserved residue is critical for the capacity of UAP56 to specifically recognize and associate with Uhg transcripts. Our study implicates the components of the RDC complex and the UAP56 splicing/export factor in the regulation of snoRNA loci in germline tissues.
RNA-seq and ChIP-seq analysis reveals mutations in Cuff affect the expression of hundreds of protein-coding genes
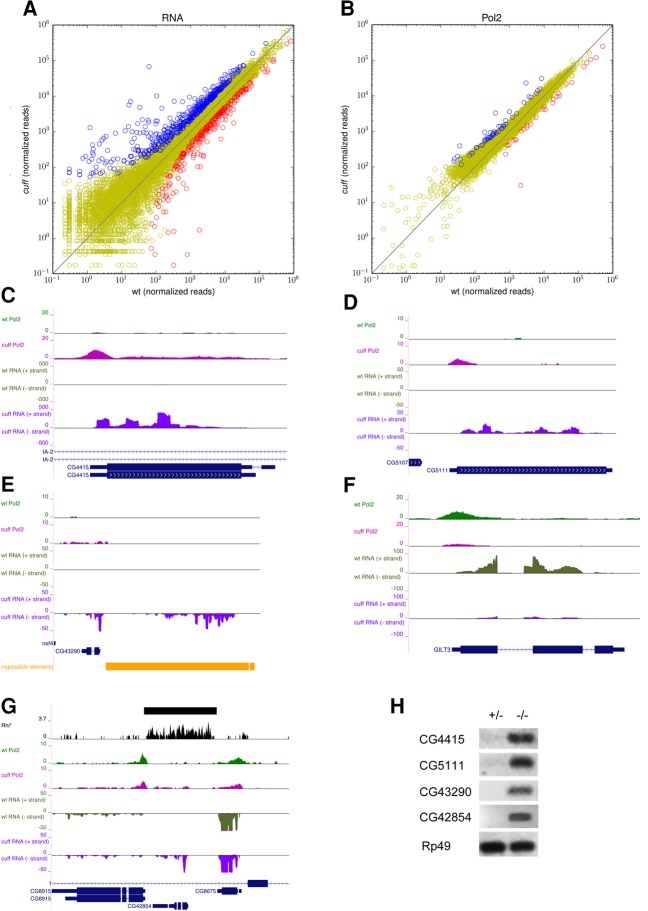
Previous studies reported that mutations in piRNA pathway components do not generally impact the expression levels of protein-coding genes (Klattenhoff et al. 2009; Mohn et al. 2014). We explored our genome-wide data sets to determine whether we reach the same conclusions for the cuff mutant. To this aim, we searched our RNA-Pol2 ChIP-seq and RNA-seq data sets for genes that were differentially expressed in wild-type versus cuff ovaries. We could identify 526 genes that displayed a significant up-regulation and 313 genes displaying a significant reduction (at least twofold, FDR < 0.05) in the absence of Cuff (Fig. 5A). The analysis of the Pol2 occupancy at protein-coding genes in wild-type versus cuff mutant ovaries suggests that in several cases the observed changes in the expression levels are the result of transcriptional, rather than post-transcriptional, events (Fig. 5B). Consistent with this, we observe a significant positive correlation (Spearman correlation 0.25, P-value < 6 × 10−104) when we compare the log-fold changes in the steady-state RNA levels and Pol2 occupancy in cuff over wt across all genes detectable in both assays. When restricting this comparison to genes with significant over- or under-expression of RNA levels, we observe even stronger correlation (Spearman's correlation 0.57, P-value < 2 × 10−50). For instance, the steady-state RNA levels for the uncharacterized CG4415 and CG5111 are approximately 1000-fold and 360-fold higher, respectively, in cuff versus wild-type ovaries. Concomitantly, a higher RNA-Pol2 occupancy is observed in the mutant either at the promoter or over the body of the gene, thus pointing to a transcriptional control mechanism (Fig. 5C,D). The alignment of the RNA-Pol2 ChIP-seq sequence reads to the reference Drosophila genome excluded the presence of novel transposon insertions in the vicinity of or within CG4415 and CG5111 in the genome of the cuff homozygous mutant flies (Supplemental Bioinformatic Methods). In contrast, the CG43290 lies in close proximity of the micropia element on the minus strand of the genome (Fig. 5E). This uncharacterized gene is expressed below detectable levels in wild-type ovaries, while it displays elevated expression in the cuff mutant. The tandem arrangement suggests that the deregulation of the mobile element in cuff might be affecting the expression of the downstream CG43290. A similar mechanism is induced upon loss of Piwi in Drosophila OSC cells, where Pol2 spreading from transposon sequences was shown to affect the expression of neighboring loci (Sienski et al. 2012). In addition to genes that are expressed at substantially higher levels, we also find a group of genes with reduced RNA levels in the cuff ovaries. For instance, mutations in Cuff appear to cause a loss of RNA-Pol2 at the GILT3 gene and a concomitant reduction of the corresponding mRNA levels (Fig. 5F). In all the cases we have illustrated so far, Cuff and Rhi peaks are apparently not present within the genes or in the neighboring regions. However, according to our Cuff ChIP-seq assays, Cuff is enriched at many protein-coding genes (Fig. 1A). In addition, Cuff-bound and Rhi-bound loci display a mild, but significant up-regulation in the absence of a functional Cuff protein (Supplemental Fig. S6). Thus we cannot rule out that Cuff and other components of the RDC complex might also contribute directly to the regulation of certain protein-coding loci. The analysis of the CG42854 lends some support to these conclusions (Fig. 5G). Rhi binding is apparent at this uncharacterized CG, while Cuff peaks are not detected. Despite the absence of clear Cuff binding, however, CG42854 is significantly up-regulated in the cuff mutant ovaries as per RNA-seq.
FIGURE 5.
Protein-coding genes are differentially expressed in the absence of a functional Cuff protein. (A) Scatter plot displaying the impact of mutations in cuff on the expression levels of protein-coding genes as per RNA-seq. Genes that are up-regulated (blue dots), down-regulated (red dots), and not significantly affected (yellow dots) in the cuff mutant are displayed. Shown on x- and y-axes are consensus normalized read counts over replicates. (B) Scatter plot displaying changes in RNA Pol2 occupancy at protein-coding genes as per ChIP-seq assays. Genes characterized by increased (blue dots) and decreased (red dots) RNA Pol2 occupancy in the cuff mutant are reported. Yellow dots indicate genes that display apparently unaffected RNA Pol2 occupancy. Shown on x- and y-axes are normalized consensus read counts over replicates. (C–F) RNA Pol2 in wild-type (green) and RNA Pol2 in cuff mutant (magenta) ChIP-seq profiles along selected protein-coding genes. RNA-seq profiles in wild-type (dark green) and cuff mutant (purple) are also shown. (G) Rhi (black), RNA Pol2 in wild-type (green), and RNA Pol2 in cuff mutant (magenta) ChIP-seq profiles along CG42854. Reproducible Rhi binding is shown by a solid line in a separate track. RNA-seq profiles in wild-type (dark green) and cuff mutant (purple) are also shown. (H) RT-PCR in wild-type and cuff mutant ovaries with primers specific for the CG4415, CG5111, CG43290, CG42854, and the control rp49.
In order to confirm the RNA-seq data, we performed RT-PCR assays on total RNA extracted from wild-type and cuff mutant ovaries with oligonucleotides specific for CG4415, CG5111, CG43290, and the Rhi-bound CG42854. Oligonucleotides specific for rp49 were used as a control for this experiment. Consistent with the RNA-seq data, all the genes analyzed are expressed at very low or undetectable levels in wild-type ovaries, while a clear signal is detectable for each of them in the cuff mutant ovaries (Fig. 5H). We did not observe significant differences in the signals produced by rp49 in wild-type and cuff ovaries (Fig. 5H). The absence of any Cuff, Rhi, or Del peaks at or in the vicinity of the CG4415, CG5111, and CG43290 loci suggests that their deregulation is likely an indirect effect of mutations in the Cuff protein. However, the analysis of the CG42854 locus strongly points to a direct role for the RDC complex or at least some of its components in the direct regulation of certain protein-coding genes.
DISCUSSION
In distantly related organisms, piRNAs generated by genomic regions termed piRNA clusters are essential to guarantee transposon silencing and genome integrity during germ cell development. In Drosophila, piRNA cluster expression is controlled by the RDC complex, which is centered on the core proteins Cuff, Rhi, and Del (Klattenhoff et al. 2009; Pane et al. 2011; Le Thomas et al. 2014; Mohn et al. 2014). To date, this complex has been exclusively implicated in the early steps of piRNA biogenesis in the Drosophila germ cells. Surprisingly, our genome-wide analyses unveil a broader function for Cuff in the regulation of different classes of noncoding RNAs. We find that Cuff is not only enriched at piRNA clusters, but it also binds snRNA and snoRNA loci, and it is necessary for their expression in germline tissues. Similar to the piRNA clusters, snoRNAs and snRNAs appear to be often enriched for the Rhi protein, suggesting that the underlying regulatory mechanism might be controlled by the RDC complex, rather than by Cuff alone. The role of Cuff in Drosophila shares some analogies with the function of the SNAPc complex subunit SNPC-4, which was recently shown to bind and regulate piRNA clusters, snRNAs, and snoRNAs in the germline of the nematode worm C. elegans (Kasper et al. 2014).
In support of our conclusions, the transcripts produced from snoRNA sources, like the Uhg loci, associate with the UAP56 protein. This splicing/export factor colocalizes with Rhi in nuclear foci and is crucial for the suppression of splicing events at piRNA clusters and the export of the piRNA precursors to the cytoplasmic nuage compartment (Zhang et al. 2012, 2014). Interestingly, the UAP56sz mutant form of the protein is unable to efficiently bind the Ugh transcripts, while the same mutation did not seem to substantially affect the binding to piRNA cluster transcripts (Zhang et al. 2012). It is possible that the amino acid residue that is substituted in the mutated form of UAP56 is essential for the specific interaction between UAP56 and Uhg RNAs. The UAP56 protein was recently shown to interact with Thoc5, a subunit of the THO/TREX complex (Hur et al. 2016). Interestingly, when we analyzed the RNA-seq data sets produced by Hur and coworkers (Hur et al. 2016), we found that mutations in Thoc5 result in increased snRNA and snoRNA expression levels (data not shown). The coordinated activity of the RDC complex components, the UAP56 protein and THO/TREX complex might be critical to ensure that different noncoding RNA precursors are expressed and sorted into the appropriate biogenesis pathway.
Recently, Cuff was proposed to prevent transcription termination at piRNA clusters by binding the CAP structure of nascent transcripts and protecting them from degradation (Mohn et al. 2014). Together with the suppression of splicing and polyadenylation events, this mechanism elicits the expression of long piRNA precursors from the clusters, an essential prerequisite for the production of a proper piRNA complement (Mohn et al. 2014; Zhang et al. 2014; Chen et al. 2016). Our study suggests that Cuff, Rhi, and Del might also control transcription termination at loci other than the piRNA clusters. This is illustrated by the regulation of the snoRNA set hosted in the 3′UTR of the Nop60B gene, where Cuff, probably in a complex with Rhi and Del, appears to be enriched at the 3′ end of the locus and suppresses an alternative transcription termination site in the Nop60B gene. This regulatory mechanism promotes the inclusion of the snoRNAs in the nascent Nop60B transcripts and likely couples the expression of the Nop60B protein to the production of the snoRNAs in the germ cells. In addition, our study seems to add new layers of complexity to the repertoire of functions exerted by the Cuff protein. For instance, the loss of Cuff at the snRNA:U4:39B does not appear to affect the overall Pol2 occupancy, although the expression levels of this snRNA are severely diminished in the cuff mutant ovaries. Thus, Cuff might act cotranscriptionally to control the production or the stability of the nascent transcript at this locus. Differently, the snRNA variants U4, U5, and U2 hosted in the intron of the sickle gene seem to be coregulated at a transcriptional level. Loss of Cuff causes a clear reduction of the Pol2 levels over the region encompassing these loci. It is conceivable that reduction of the snRNA levels might also contribute to the general down-regulation of the snoRNA and piRNA populations given that the biogenesis of both these ncRNA species is based on the regulation or suppression of splicing events.
Importantly, we also find that Cuff appears to control the expression of some protein-coding genes. For example, the uncharacterized gene CG42854 appears to be bound along its length by the Rhi protein and is expressed at very low levels in the wild-type, while its expression is greatly increased in the cuff ovaries. This observation reveals that the RDC complex or at least some of its components can function as negative regulators of gene expression in germ cells and that the genomic loci regulated by this complex are not limited to noncoding RNAs, but can include certain protein-coding genes. It is possible that the interaction with additional factors together with inherent structural motifs in the nascent RNAs or cis-acting elements at the regulated loci might exert an essential role in defining the specific activity of Cuff. This functional flexibility is probably crucial at piRNA clusters to bypass cis-regulatory elements (i.e., transcription termination and splice sites) carried by the transposon sequences and guarantee the production of long piRNA precursors that can be efficiently converted into a full set of piRNAs.
To date, mutations in piRNA pathway components have not been found to affect the transcript levels of protein-coding genes in the Drosophila germ cells. However, we could identify a few hundred loci with dramatically altered steady-state RNA levels in the cuff mutant ovaries. In some cases, the Pol2 ChIP-seq assays support a model whereby loss of Cuff causes a change in the transcriptional activity at certain loci. For example, higher expression levels of CG4415 and CG5111 are consistent with a significant accumulation of Pol2 at their promoter regions and over the gene body in the cuff mutant. The absence of Cuff peaks at these CGs as well as at other genes suggests that Cuff is only indirectly involved in their transcriptional regulation. We and others have reported that the loss of the piRNA pathway activities leads to the activation of a meiotic checkpoint system, centered on the Checkpoint kinase Chk2, in germline tissues (Chen et al. 2007; Klattenhoff et al. 2007). This checkpoint senses the DNA damage induced by massive transposon mobilization and translates it into the loss of germ and stem cells, the defective assembly of the oocyte nucleus (i.e., karyosome phenotype), and the disruption of the Dorsal–Ventral axis of the egg and future embryo, which characterize cuff as well as other piRNA pathway mutants. Although the details of the signaling mechanism are not known, it is conceivable that Chk2 might act by regulating the expression of yet-unknown downstream genes. In this scenario, the deregulation of certain protein-coding loci observed in cuff mutant ovaries might at least in part depend upon the activation of checkpoint mechanisms in the germline. In accordance with this, we find that the expression levels of CG4415 and CG5111, which are dramatically increased in the cuff mutant, are restored to wild-type levels in ovaries doubly mutant for cuff and chk2 (A Pane, unpubl.). It is therefore tempting to speculate that these CGs might belong to a set of genes that form a Chk2-mediated cellular response to the transposon-induced DNA damage.
Our data strongly implicate Cuff and the RDC complex in the biogenesis of piRNAs, snRNAs, and snoRNAs and reveal a previously unknown degree of crosstalk between different classes of ncRNAs in the Drosophila germline. In line with our findings, Klenov and coworkers have recently shown that loss of nuclear Piwi affects the levels of certain spliceosomal RNAs, including the snRNA:U2:Aba, snRNA:U5:38ABa, and snRNA:U4:39B, that we find to be down-regulated in cuff ovaries (Klenov et al. 2014). The discovery that the initial steps in the production of different classes of noncoding RNAs relies on a shared set of proteins will help shed light on the biogenesis of piRNAs, snRNAs, and snoRNAs in the Drosophila germ cells.
MATERIALS AND METHODS
Fly stocks
The cuffWM25 allele used in this study was isolated in a screening for female sterile mutations (Schüpbach and Wieschaus 1989). The rhiKG00910 allele originates from a P-element insertion in the rhi locus (Volpe et al. 2001) (Bloomington Stock Center). rhiKG00910 was used in combination with the deficiency Df(2R)Exel17149 (Bloomington Stock Center) that spans the rhi gene. Heterozygous cuffWM25/CyO or rhiKG00910/CyO flies were used as wild-type control.
RNA-seq and ChIP-seq assays
RNA-seq and ChIP-seq assays were performed in triplicate, and for each biological replicate we dissected ovaries from approximately 30 female flies (2–4 d old). Total RNA was extracted using TRIzol reagents (Life Technologies) and treated with the turbo DNase-free kit (Ambion) to eliminate traces of genomic DNA. DNA-free total RNA was subjected to two rounds of rRNA depletion using the Ribo-Zero rRNA removal kit (Epicentre) as per the manufacturer's instructions. The RNA-seq library preparation was obtained with the ScriptSeq v2 kit (Epicentre) and the libraries were sequenced on the Illumina platforms.
The conditions and the oligonucleotides used for the Pol2 chromatin immunoprecipitation followed by qPCR (ChIP-qPCR) have been previously described (Blythe et al. 2009; Pane et al. 2011). Pol2 ChIP-qPCR assays were performed using anti Pol2 antibodies (clone 8WG16, Millipore). Pol2 enrichment at specific regions of the piRNA cluster 1/42AB, Cl2, ry, Pld, and rp49 was calculated as percent of the Input chromatin and displayed as fold change [log2(cuff/wt)]. The same ChIP protocol was used for the ChIP-seq assays, where chromatin fragments were used to generate DNA libraries with the ChIP-seq DNA Sample Prep kit (Illumina). The Cuff-EGFP expressing transgenic line, GFP antibodies, and the conditions used for the Cuff ChIP-seq experiment have been previously described (Blythe et al. 2009; Pane et al. 2011).
Bioinformatic analyses
For ChIP-seq analysis, reads were aligned uniquely to the genome using Bowtie (Langmead and Salzberg 2012). Cuff and Rhi peaks were called using MACS2 (Zhang et al. 2008), and reproducible peaks were detected using IDR (Zhang et al. 2008), threshold 0.05 (for Rhi, publicly available ChIP-seq data from Mohn et al. 2014 was used). Peaks were randomly shuffled in the genome (preserving chromosome and peak length) in order to assess the significance of overlap of peaks with genomic features. For visualization in the UCSC Genome Browser (Kent et al. 2002), bigWig files with genome-wide background subtracted ChIP signal were produced using BEDTools (Quinlan and Hall 2010). For RNA-seq analysis, reads were aligned uniquely to the genome using Tophat (Kim et al. 2013), and reads aligned to each genomic feature of interest (protein-coding gene, snRNA, snoRNA, piRNA cluster 500 bp fragment) counted with custom scripts utilizing the python interface to SAMtools (package pysam) (Li et al. 2009b). Differential expression analysis was run using DESeq (Anders and Huber 2010). The Benjamini–Hochberg procedure was used to calculate false discovery rates (FDR) for multiple hypotheses testing corrections. Genomic features with zero read count in all replicates were excluded from the analysis. Differential Pol2 occupancy was analyzed in the same manner. Analysis for canonical transposons was run similarly with the exception that reads aligned multiple times to the same transposon were also taken into account. We also used publicly available RIP-seq data (Zhang et al. 2012) to analyze the RNA binding profile for UAP56. For this we used bedGraph files with genome-wide normalized RIP-seq signal at single-nucleotide resolution produced by the authors and available at Gene Expression Omnibus. A detailed description of the bioinformatic analyses is provided in the Supplemental Data. The total number of raw and aligned reads for each library is shown in Supplemental Table 1.
Northern blotting, RT-PCR, and qRT-PCR
Fifteen micrograms total RNA extracted from cuffWM25 and cuffWM25/Cyo flies were separated on 6% Urea gels (Life Technologies) using the SureLock System (Life Technologies). The RNA was then transferred onto Nylon membranes (Genescreen Plus) and hybridized with P32-labeled antisense oligonucleotide probes corresponding to sequences in snRNA:U1 and snRNA:U2. Hybridization conditions have been previously described (Pane et al. 2007). Probe sequences are reported in Supplemental Table 2.
Rt-PCR and qRt-PCR reagents, conditions, and oligonucleotides specific for rp49, Pld, ry, and for selected regions within piRNA clusters have been previously described (Chen et al. 2007; Pane et al. 2011). Oligonucleotides specific for protein-coding genes and for snRNAs, which were designed for the current study, are described in Supplemental Table 2.
DATA DEPOSITION
ChIP-seq and RNA-seq raw sequencing data are available at NCBI GEO, accession number GSE98635.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lance Parsons, Donna Storton, and Jessica Buckles in the Microarray Facility at Princeton University for help with RNA-seq and ChIP-seq library sequencing; Gail Barcelo, Natalia Ferreira, and Aline de Graça Sampaio for assistance with molecular techniques. We are grateful to Paul Schedl, Michael Sammeth, and Helena Araujo for critical reading of the manuscript and members of the Schupbach, Singh, Wieschaus, and Araujo laboratories for helpful discussions and suggestions. This work was supported in part by US Public Health Service Grants NIH-RO1 GM077620 (T.S. and A.P.) and NIH-RO1 GM076275 (M.S. and Y.P.), National Science Foundation 1458457 (M.S.), the Howard Hughes Medical Institute (T.S.), the CNPq Ciencia Sem Fronteiras Program 410594/2012-2 (A.P.), the INCT (A.P.), and FAPERJ 218071 (A.P.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.058594.116.
REFERENCES
- Alioto TS. 2007. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res 35: D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kessler DS, Klein PS. 2009. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev Dyn 238: 1422–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. [DOI] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schüpbach T. 2007. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol 17: 637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YCA, Stuwe E, Luo Y, Ninova M, Le Thomas A, Rozhavskaya E, Li S, Vempati S, Laver JD, Patel DJ, et al. 2016. Cutoff suppresses RNA polymerase II termination to ensure expression of piRNA precursors. Mol Cell 63: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis C, Zanni V, Brasset E, Eymery A, Zhang L, Mteirek R, Jensen S, Rong YS, Vaury C. 2013. “Dot COM”, a nuclear transit center for the primary piRNA pathway in Drosophila. PLoS One 8: e72752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis-Sandoval F, Poirier M, Scott MS. 2015. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA 6: 381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Hir H, Schmitt C, Braun IC, Köcher T, Wilm M, Izaurralde E. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol 11: 1716–1721. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590. [DOI] [PubMed] [Google Scholar]
- Guzzardo PM, Muerdter F, Hannon GJ. 2013. The piRNA pathway in flies: highlights and future directions. Curr Opin Genet Dev 23: 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Luo Y, Moon S, Ninova M, Marinov GK, Chung YD, Aravin AA. 2016. Splicing-independent loading of TREX on nascent RNA is required for efficient expression of dual-strand piRNA clusters in Drosophila. Genes Dev 30: 840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper DM, Wang G, Gardner KE, Johnstone TG, Reinke V. 2014. The C. elegans SNAPc component SNPC-4 coats piRNA domains and is globally required for piRNA abundance. Dev Cell 31: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler AD. 2002. The Human Genome Browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. 2007. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 12: 45–55. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Korbut AP, Stolyarenko AD, Yakushev EY, Reuter M, Pillai RS, Gvozdev VA. 2014. Impact of nuclear Piwi elimination on chromatin state in Drosophila melanogaster ovaries. Nucleic Acids Res 42: 6208–6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen YCA, Luo Y, Sachidanandam R, Toth KF, et al. 2014. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev 28: 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009a. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009b. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Matera AG. 2014. Developmental analysis of spliceosomal snRNA isoform expression. G3 (Bethesda) 5: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mével-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A. 2007. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175: 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. 2014. The Rhino-Deadlock-Cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157: 1364–1379. [DOI] [PubMed] [Google Scholar]
- Nishida KM, Iwasaki YW, Murota Y, Nagao A, Mannen T, Kato Y, Siomi H, Siomi MC. 2015. Respective functions of two distinct siwi complexes assembled during PIWI-interacting RNA biogenesis in bombyx germ cells. Cell Rep 10: 193–203. [DOI] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T. 2007. Zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Jiang P, Zhao DY, Singh M, Schüpbach T. 2011. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J 30: 4601–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, Patil VS, Kai T. 2012. piRNA pathway and the potential processing site, the nuage, in the Drosophila germline. Dev Growth Differ 54: 66–77. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardo S, Tortoriello G, Giordano E, Turano M, Furia M. 2007. The coding/non-coding overlapping architecture of the gene encoding the Drosophila pseudouridine synthase. BMC Mol Biol 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschêne G, Bucheton A, Pélisson A. 2004. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. 1989. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics 121: 101–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Brennecke J. 2010. The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet 26: 499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Steitz JA. 2001. Non-coding snoRNA host genes in Drosophila: expression strategies for modification guide snoRNAs. Eur J Cell Biol 80: 119–125. [DOI] [PubMed] [Google Scholar]
- Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA. 2001. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics 159: 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Couté Y, Conn S, Kadlec J, Sachidanandam R, et al. 2014. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 157: 1698–1711. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. 2012. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 151: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. 2014. The HP1 homolog Rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 157: 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.