Abstract
Background:
Despite being a high burden disorder, the pathogenesis of severe refractory asthma (SRA) is poorly understood. There are some evidences for the involvement of members of the signal transducer and activator of transcription (STAT) family, including STAT3 and STAT5a. Our study aimed to evaluate the gene expression of STAT3 and STAT5a in asthma and SRA to establish if there is an association.
Materials and Methods:
Using quantitative real-time polymerase chain reactions (qRT-PCR), the transcript levels of STAT3 and STAT5a were evaluated in peripheral blood mononuclear lymphocytes (PBML) isolated from 13 patients with SRA, 14 with mild asthma, and 30 healthy volunteers.
Results:
There were no significant differences in STAT3 transcript levels between study groups. There was however a significant difference in STAT5a transcript levels between cases and controls (p-value=0.03). In comparison to healthy controls, the levels of STAT5a were notably lower in patients with mild asthma and significantly least in those with SRA.
Conclusion:
Our study found no appreciable association between STAT3 gene expression and either mild asthma or SRA. However, the STAT5a down regulation in asthmatics and especially SRA is a notable finding which denotes on association between STAT5a and different level of asthma.
Keywords: Asthma, Severe Asthma, STAT3, STAT5a, Gene Expression
INTRODUCTION
Chronic asthma is a non-communicable inflammatory airway disorder in which patients present with recurring bouts of breathlessness and wheezing. Although the exact pathogenesis of asthma is not fully understood, numerous causative environmental and/or triggering agents have been described. These include allergens, tobacco smoke, chemical irritants, and microorganisms. These factors interact with an individual’s genetic and epigenetic background, leading to the development of asthma or the triggering of attacks (1).
Asthma is suspected to cause approximately 250,000 premature deaths annually and the World Health Organization (WHO) estimates that more than 300 million individuals are affected worldwide. This number is predicted to increase to 400 million by 2025. Due to the fact that asthma is a major cause of disability, poor quality of life, increased health resource utilization, and is a public health concern, it is essential to fully understand the genetic basis of the disease (2, 3).
During previous decades, there has been some controversy concerning the definition and classification of severe refractory asthma (SRA), found in approximately in 5–10% of cases (4). In 2011, an international consensus statement was published by the Innovative Medicine Initiative (IMI) that aimed to clarify a unique definition, classification, and diagnostic algorithm for SRA. This stated that “the term ‘severe refractory asthma’ should be reserved for patients with asthma in whom alternative diagnoses have been excluded, comorbidities have been treated, trigger factors have been removed (if possible) and compliance with treatment has been checked, but still have poor asthma control or frequent (≥2) severe exacerbations per year despite the prescription of high-intensity treatment or can only maintain adequate control when taking systemic corticosteroids and are thereby at risk of serious adverse effects of treatment” (2).
In order to understand the fundamental mechanisms of asthma pathogenesis, efforts have been made to identify genetic associations with asthma and SRA (5,6). Among the numerous genes known to associate with asthma, members of the signal transducer and activator of transcription (STAT) pathway appear to have an important function (7). Seven proteins of this family have been shown to have roles in signal transduction pathways and/or gene transcription (8) and may be involved with the mechanisms that underlie asthma and SRA.
The gene coding for STAT3 is located in chromosomal region 17q21.2 and consists of 24 exons. Through alternative splicing, STAT3 can be expressed as three different splice variants. STAT3 is an essential protein involved in cell growth and apoptosis, and is expressed in all tissue types. Furthermore, it can act as a transcription factor and may also be a co-activator of signal transduction by glucocorticoid receptors (9, 10).
Like STAT3, STAT5 is related to glucocorticoid receptors and acts as a transcription activator in the immune system (11). There are two isoforms of STAT5 (STAT5a and STAT5b), coded by two separate genes located at inverted positions within the 17q21.2 or 17q11.2 genomic regions (12,13). Several important cellular processes are influenced by STAT5 isoforms, including replication, apoptosis, differentiation, and inflammation. It has also been shown that STAT5a/b are important for lymphocyte proliferation, apoptosis, and have been used for targeted gene therapy and therapeutics (e.g., for asthma and cancer) (14–17).
There is some evidence supporting the involvement STAT3 and STAT5 in the development of asthma (18, 19). STAT3 has been demonstrated to be involved in airway inflammation, allergy, and asthma through several proposed mechanisms. These include the Th2/Th17 immune responses and epidermal growth factor receptor (EGFR) signaling (20–22). Furthermore, it has been proposed that STAT3 is a potential target for asthma and SRA therapeutics (23). However, there are also several studies that discount a role for STAT3 in asthma (24, 25).
STAT5 is an important regulator of mast cell activity and mediates their proliferation, survival, and homeostasis. Mast cells have been shown to have a key role in the development of asthma, and are implicated in SRA. This suggests that STAT5 may be involved in asthma through mast cell pathogenesis (8, 26) and there have been several studies supporting such a link (27–31). The association between asthma and the STAT5b isoform has been the most well studied relationship to date but a potential role for the STAT5a isoform is unclear (14). A previous study by Tsitsiou et al. that evaluated gene expression in patients with severe asthma reported that STAT3 and STAT5b expression levels were 1.59 and 1.62 times higher, respectively in these patients (3). Our study, therefore, aimed to investigate STAT3 and STAT5 gene expression in asthma and SRA.
MATERIALS AND METHODS
Our study was a joint investigation by the National Research Institute of Tuberculosis and Lung Diseases (NRITLD) and the Tarbiat Modares University (TMU) of Tehran-Iran. The cross-sectional study was conducted from 2012–2014 using 13 patients with SRA, 14 non-severe asthma cases, and 30 healthy volunteers. The 2011 international consensus for the definition of severe asthma (2) was used for diagnosis and inclusion of SRA patients. These cases were selected sequentially from the NRITLD asthma clinic. Patients with non-severe asthma were enrolled from the same clinic using criteria outlined by the Global Initiative for Asthma (GINA) (32). Patient involvement was approved by certified pulmonologists. Healthy participants had no history of any compounding disorders at the time of the study. The enrollment of participants was voluntary and signed informed consent forms were collected for each participant. Patient data were kept confidentially and no intervention was applied throughout their clinical management. All stages of the study were approved by the ethical committee of the NRITLD under the code sbmu1.REC.1391.1, dated May 14, 2012.
Following demographic and clinical data gathering, 5 mL samples of peripheral venous blood were obtained from each patient and immediately stored at 4°C. Peripheral blood mononuclear lymphocytes (PBMLs) were separated using a Lympholyte-H (Cedarlane Co., Ontario, Canada) solution during 2 hours. Following the manufacturer’s protocols, RNA was isolated using RNXPlus solution (CinnaGen Co., Tehran, Iran) and the quality and quantity verified using agarose gel electrophoresis and spectrophotometry, respectively.
For each sample, 3 μg of isolated RNA was used to synthezise cDNA using Oligo-dT, random hexamers, and reverse transcriptase enzyme (Fermentas, Thermo Fisher Scientific Co., Waltham, Massachusetts, USA). This was validated using PCR specific to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene. qRT-PCR was performed using an Applied Biosystems 7500 sequence detection system (Applied Biosystems, Foster City, CA, USA) and Power SYBR Green I PCR Master Mix (Takara, Japan), according to the manufacturer’s protocol. Primers used for STAT3 (including all splice variants) and STAT5A are indicated in Table 1. The conditions for each qRT-PCR were a preliminary denaturing stage at 95°C for 15 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 20 seconds, and extension at 72°C for 20 seconds. The housekeeping gene of GAPDH was used as an endogenous control to normalize STAT3 and STAT5A expression (Table 1).
Table 1.
The used primers in the study
| Gene | Dir | Primer Sequence | Length (bp) | Product length (bp) | Efficiency (%) |
|---|---|---|---|---|---|
| GAPDH | F | 5′-CCATGAGAAGTATGACAAC-3′ | 19 | 115 | 98.3 |
| R | 5′-GAGTCCTTCCACGATACC-3′ | 18 | |||
| STAT3 (Var. 1,2,3) | F | 5′-AGCAGGAGGGCAGTTTGAGTC-3′ | 21 | 241 | 99.1 |
| R | 5′-TTTAAAAGTGCCCAGATTGCTC-3′ | 22 | |||
| STAT5A | F | 5′- ACATGTACCCACAGAACCCTGACC-3′ | 24 | 239 | 98.2 |
| R | 5′- CACAACACGACCGCTTCACATTGC-3′ | 24 |
A comparative analysis of the transcript expressioin of STAT3, STAT5A, and GAPDH was performed, using the mean Ct of at least two replicates for each sample. Delta Ct (ΔCt), defined as the difference between the mean Cts of each gene (STAT3 or STAT5A) and the endogenous control (GAPDH), was used for further analysis. Statistical differences between the mean ΔCts of the mild asthma, SRA, and control groups were assessed by independent Student’s t-tests and one way ANOVA with a Tamhane’s post hoc test. The primers used for amplification were found to have high efficacy (99.1% for STAT3 and 98.2% for STAT5A), allowing the fold change in transcript levels to be calculated using 2−ΔΔCt methodology (33). Any putative correlation between transcript levels were evaluated using Pearson’s tests. Statistical analyses were performed using the Statistical Package for Social Sciences (SPSS) Version 21.0 (Microsoft, Chicago, IL, USA), with a threshold of significance set at a P-value of 0.05.
RESULTS
Participant Characteristics
Table 2 summarizes the demographics of the participants. The mean age was significantly different between healthy individuals and both asthma groups, mainly due to the voluntary nature of the study. Additionally, 90% of the controls were male, while 61% were male for the mild asthma group and 64% for the SRA group. Patient body mass indices (BMIs) were used as a general gauge of the nutritional condition and physical health of participants. This was found to be approximately 26.40 across the three studied groups (P-value = 0.99). Finally, the ethnicities of participants were found to be similar in each of the three groups (60% Fars, 8% Turks, 12% Lores, and 20% Kurds).
Table 2.
Demographic findings for the participants
| Severe asthma | Asthma | Healthy | Total | P-value | |
|---|---|---|---|---|---|
| Gender | |||||
| Male | 8 | 9 | 27 | 44 | |
| Female | 5 | 5 | 3 | 13 | |
| Total | 13 | 14 | 30 | 57 | |
| Age | |||||
| Mean (SE) | 50.23 (3.80) | 54.71 (4.31) | 39.17 (1.54) | 45.51 (1.80) | <0.001 |
| 95% CI | 41.95–58.51 | 45.39–64.4 | 36.01–42.33 | 41.89–49.13 | |
| Min.–Max. | 21–76 | 24–80 | 23–60 | 21–80 | |
| BMI | |||||
| Mean (SE) | 26.39 (1.25) | 26.41 (0.81) | 26.49 (0.70) | 26.45 (0.50) | 0.99 |
| 95% CI | 23.65–29.14 | 24.65–28.16 | 25.04–27.93 | 25.44–27.45 | |
| Min.–Max. | 19.53–35.49 | 20.05–30.12 | 18.65–34.09 | 18.65–35.49 |
Comparison of Transcript Abundance
Student’s t-tests comparing the qRT-PCR data revealed that the transcript expressions of STAT3 and STAT5A were not significantly different between genders (Tables 3 and 4). STAT3 and STAT5a transcript expressions also did not correlate with age of all participants (r=0.42, P-value=0.35 for STAT3 and r=0.31, P-value=0.67 for STAT5a). Furthermore the gene expression (mean ΔCts) of STAT3 and STAT5A were compared by ANOVA statistical tests between cases and control groups.
Table 3.
The expression difference (ΔCts) of STAT3 in genders and disease groups
| Mean ± SE | 95% CI | Min.–Max. | P-value | |
|---|---|---|---|---|
| Gender | ||||
| Male | 0.303 ± 0.16 | 0.02 – 0.66 | −2.03 – 2.89 | 0.47 |
| Female | 0.011 ± 0.50 | −1.09 – 1.11 | −3.00 – 2.55 | |
| Case/control | ||||
| Severe asthma | 0.254 ± 0.47 | −0.78 – 1.29 | −3.00 – 2.89 | 0.95 |
| Asthma | 0.141 ± 0.40 | −0.73 – 1.01 | −1.86 – 2.55 | |
| Healthy | 0.274 ± 0.16 | −0.06 – 0.61 | −2.37 – 1.65 | |
| Total | 0.237 ± 0.16 | −0.96 – 0.57 | −3.00 – 2.89 |
Table 4.
The expression difference (ΔCts) of STAT5A in genders and disease groups
| Mean ± SE | 95% CI | Min.–Max. | P-value | |
|---|---|---|---|---|
| Gender | ||||
| Male | 2.88 ± 0.24 | 2.39 – 3.37 | −0.43 – 7.15 | 0.72 |
| Female | 2.72 ± 0.57 | 1.46 – 3.98 | −1.44 – 5.34 | |
| Case/control | ||||
| Severe asthma | 3.64 ± 0.64 | 2.24 – 5.05 | −1.44 – 7.15 | 0.03 |
| Asthma | 3.29 ± 0.35 | 2.52 – 4.06 | 1.85 – 5.52 | |
| Healthy | 2.33 ± 0.23 | 1.85 – 2.81 | −0.43 – 4.03 | |
| Total | 2.86 ± 0.22 | 2.42 – 3.31 | −1.44 – 7.15 |
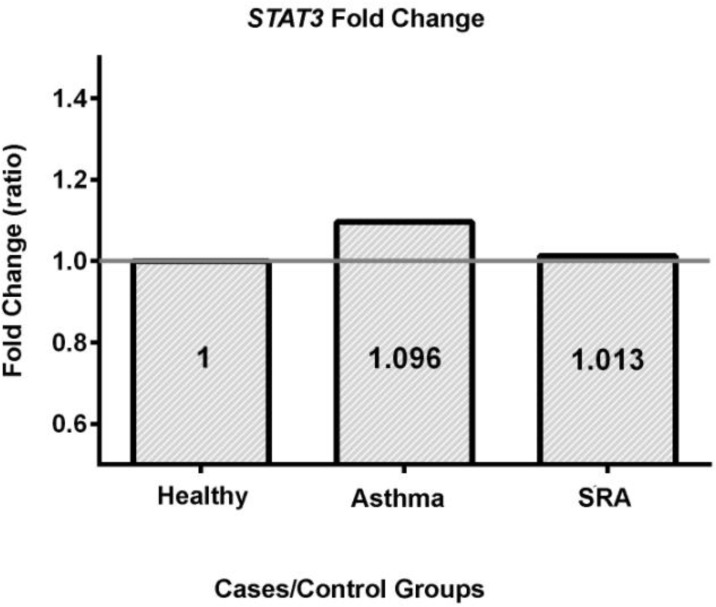
Table 3 indicates that there was no significant different between the disease groups in terms of STAT3 gene expression. Meanwhile, the fold change analysis (2−ΔΔCt) showed the expression ratios of asthma and SRA groups against the control group are 1.096 and 1.013 respectively, when the control adjusted to 1 (Figure 1). These findings show the level of STAT3 gene expression in asthma groupis followed by SRA and control groups.
Figure 1.
The fold change of gene expression for STAT3
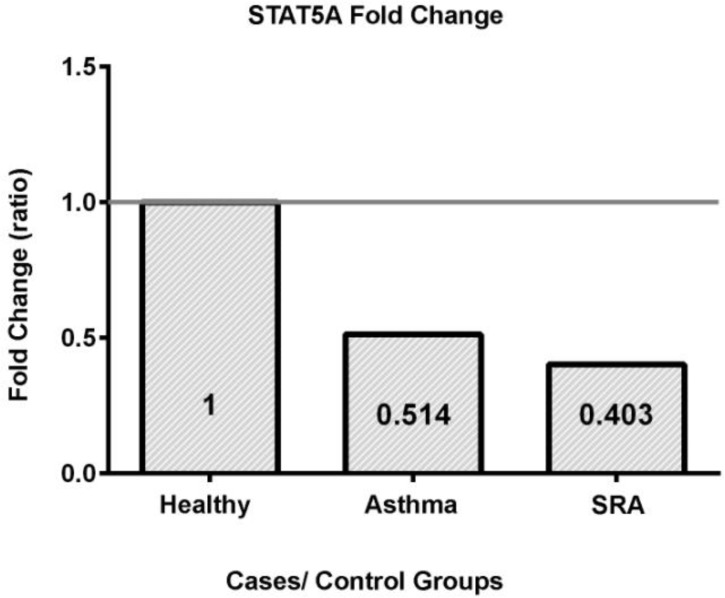
When examining STAT5a transcript levels, we found a significant difference between the disease groups (P-value=0.03) (Table 4). A Tukey’s post hoc test suggested that the only significant difference was between healthy and SRA groups (P-value=0.04). The P-value for a putative difference between the mild asthma versus SRA groups, and the asthma versus healthy groups, were 0.83 and 0.15, respectively. Fold change analysis (2−ΔΔCt) revealed the highest STAT5a transcript levels were in healthy controls, followed by the mild asthma and then SRA groups (the control group was set to 1.0) (Figure 2).
Figure 2.
The fold change of gene expression for STAT5A
DISCUSSION
Considering all participants, the STAT3 and STAT5a gene expressions were not associated with age and significantly different in both sexes. Thus, significant differences of age and gender between the study groups are not confounding factors. Also, similarity in BMI and ethnicity of the groups shows that participants’ physical conditions and genetic backgrounds do not affect the results.
As the steroid regimens and pulmonary function tests of all participants were variable, there was possibility that these factors interfere with our results. To reduce these effects, we applied certain standards restrictedly to prevent bias. For example, the cases were selected if they had diagnostic criteria of GINA (for asthma) and IMI (for SRA) for more than 2 years and had no experience of exacerbation in the 6 months prior to sampling. Furthermore, they were controlled by inhaled corticosteroids which have least systemic effects (34).
STAT3
As previously mentioned, the role of STAT3 in the pathogenesis of asthma and SRA is somewhat controversial. Our study revealed that STAT3 gene expression was not significantly different between the three groups of healthy controls, asthma and severe asthma. This finding is compatible with some reports. For example, Chiba et al. found that STAT3 had no notable role in the pathogenesis of bronchial allergic asthma (24, 25). Furthermore, the polymorphic relation of asthma and STAT3 has been denied previously (35) and intracellular flow cytometry of CD4(+)CD161(+) T cells found no differences in phosphorylated STAT3 levels between patients with asthma and controls (36). On the other hand, many studies signify the STAT3 role in pathogenesis of asthma and SRA (18–23). It is also believed that airway remodeling may be influenced by STAT3 (37–40).
Based on our results and previous studies, we hypothesize that, STAT3 may have some role in the metabolic pathways of asthma, however, it does not seem to be directly involved in the pathogenesis of asthma and SRA. Meanwhile, the controversies in studies may be due to different methods used in each study, the individual role of the three STAT3 isoforms, an inadequate sample size, or even unknown confounding factors.
STAT5a
In contrast to STAT3, we found a significant difference in the transcript levels of STAT5a between the study groups. The STAT5a expression in healthy controls was nearly twice of patients with asthma. The groups of patients with SRA had even less transcript, approximately 20% lower than patients with mild asthma. This demonstrates that there is STAT5a down regulation in asthma and SRA cases, suggesting that this isoform has a role in the pathogenesis of asthma, particularly its severe form.
The genome wide association studies (GWAS) have found that severe asthma is associated with the 17q21 chromosomal region; the region where codes some proteins like STAT5a (6). Furthermore, many evidences signified the role of STAT5a in asthma (18, 27), such as that by Stefanowicz et al, who found that STAT5a gene expression is decreased in the epithelial cells of airways (41).
Although the exact mechanisms of asthma pathogenesis remain unclear, some studies have suggested possible mechanisms for how STAT5a may be involved. These include roles for STAT5a in controlling IL-9 expression, the differentiation of Th2, Th9, and Th17 cells (19), the activity of the CD69 receptor and its regulatory role in Th17 cells (42), several mast cell pathways (8, 26), the activity of glucocorticoid receptors (11), lymphocyte proliferation (28), nitric oxide-mediated STAT5 dephosphorylation (29, 43), and induction of IL-4 producing eosinophils by IL-5 (30). In addition to their individual roles, pathogenesis may be due to a complicated combination of any or all of these factors, or even factors not yet identified. More directly, Burnham et al. found evidence for down regulation of STAT5 in eosinophil cells during allergic asthma (44), although at least one study found the opposite result. Gernez et al. also showed that there was no difference in intracellular phosphorylated STAT5 levels in CD4(+)CD161(+) T cells between asthmatics and healthy controls (36). As an assumption, the non-different expression of STAT5 may be due to summation of STAT5b up regulation (1.62 times; as Tsitsiou et. al. showed) (3) and STAT5a down regulation (>2 times; as we found).
The evidences that STAT5, and particularly STAT5a, is involved in the pathogenesis of asthma and SRA may present us a new diagnostic or therapeutic horizon. We believe that further study is required to evaluate the importance of STAT5a expression as a potential diagnostic tool for SRA. Kabata et al. showed that STAT5 inhibitors (e.g., Pimozide) can be used to overcome resistance to corticosteroid therapy in patients with asthma (45). The STAT5 metabolic pathway is therefore a potentially a new target for SRA treatment.
CONCLUSION
In conclusion, we found no evidence to support the suggestion that STAT3 is involved in asthma and SRA. Further investigation may provide more information to elucidate its role in respiratory inflammation disorders. Meanwhile, down regulation of STAT5a in asthma, and especially SRA, is a notable finding which worth to be considered more in future studies. Using transcriptomic and proteomic methods with higher sample size, the expression study of STAT5a, STAT5b and total STAT5 may provide considerable results on their roles in asthma pathogenesis.
Acknowledgements
The authors gratefully thank the kind cooperation of Dr. Alireza Eslaminejad and Dr. Guitti Pourdowlat (pulmonologists) for their clinical support, Dr. Seyed Alireza Nadji and his staff for their assistance in laboratory sample preparation, the patients and volunteers who participated, and the Iran National Science Foundation and Department of Research Affairs of Tarbiat Modares University for funding the study.
REFERENCES
- 1.Masjedi MR. Asthma. In: Azizi F, Hatami H, Janghorbani M. Epidemiology and control of common diseases in Iran. Tehran: Eshtiagh Publications; 2000;P:342–62. [Google Scholar]
- 2.Bel EH, Sousa A, Fleming L, Bush A, Chung KF, Versnel J, et al. Diagnosis and definition of severe refractory asthma: an international consensus statement from the Innovative Medicine Initiative (IMI). Thorax 2011;66(10):910–7. [DOI] [PubMed] [Google Scholar]
- 3.Tsitsiou E, Williams AE, Moschos SA, Patel K, Rossios C, Jiang X, et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol. 2012;129(1):95–103. [DOI] [PubMed] [Google Scholar]
- 4.Barnes PJ. Severe asthma: advances in current management and future therapy. J Allergy Clin Immunol 2012;129(1):48–59. [DOI] [PubMed] [Google Scholar]
- 5.Thomsen SF. Genetics of asthma: an introduction for the clinician. Eur Clin Respir J 2015;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones BL, Rosenwasser LJ. Linkage and Genetic Association in Severe Asthma. Immunol Allergy Clin North Am 2016;36(3):439–47. [DOI] [PubMed] [Google Scholar]
- 7.Sampath D, Castro M, Look DC, Holtzman MJ. Constitutive activation of an epithelial signal transducer and activator of transcription (STAT) pathway in asthma. J Clin Invest 1999;103(9):1353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morales JK, Falanga YT, Depcrynski A, Fernando J, Ryan JJ. Mast cell homeostasis and the JAK-STAT pathway. Genes Immun 2010;11(8):599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Jones S, Hagood JS, Fuentes NL, Fuller GM. STAT3 acts as a co-activator of glucocorticoid receptor signaling. J Biol Chem 1997;272(49):30607–10. [DOI] [PubMed] [Google Scholar]
- 10.Lerner L, Henriksen MA, Zhang X, Darnell JE., Jr STAT3-dependent enhanceosome assembly and disassembly: synergy with GR for full transcriptional increase of the alpha 2-macroglobulin gene. Genes Dev 2003;17(20):2564–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyszomierski SL, Yeh J, Rosen JM. Glucocorticoid receptor/signal transducer and activator of transcription 5 (STAT5) interactions enhance STAT5 activation by prolonging STAT5 DNA binding and tyrosine phosphorylation. Mol Endocrinol 1999;13(2):330–43. [DOI] [PubMed] [Google Scholar]
- 12.Ambrosio R, Fimiani G, Monfregola J, Sanzari E, De Felice N, Salerno MC, et al. The structure of human STAT5A and B genes reveals two regions of nearly identical sequence and an alternative tissue specific STAT5B promoter. Gene 2002; 285(1–2):311–8. [DOI] [PubMed] [Google Scholar]
- 13.Crispi S, Sanzari E, Monfregola J, De Felice N, Fimiani G, Ambrosio R, et al. Characterization of the human STAT5A and STAT5B promoters: evidence of a positive and negative mechanism of transcriptional regulation. FEBS Lett 2004;562(1–3):27–34. [DOI] [PubMed] [Google Scholar]
- 14.Qiu C, Peng WK, Shi F, Zhang T. Bottom-up assembly of RNA nanoparticles containing phi29 motor pRNA to silence the asthma STAT5b gene. Genet Mol Res 2012;11(3):3236–45. [DOI] [PubMed] [Google Scholar]
- 15.Rani A, Murphy JJ. STAT5 in Cancer and Immunity. J Interferon Cytokine Res 2016;36(4):226–37. [DOI] [PubMed] [Google Scholar]
- 16.Xu C, Zhang L, Li H, Liu Z, Duan L, Lu C. MiRNA-1469 promotes lung cancer cells apoptosis through targeting STAT5a. Am J Cancer Res 2015;5(3):1180–9. [PMC free article] [PubMed] [Google Scholar]
- 17.Szelag M, Wesoly J, Bluyssen HA. Advances in peptidic and peptidomimetic-based approaches to inhibit STAT signaling in human diseases. Curr Protein Pept Sci 2016;17(2):135–46. [DOI] [PubMed] [Google Scholar]
- 18.Hausding M, Tepe M, Ubel C, Lehr HA, Röhrig B, Höhn Y, et al. Induction of tolerogenic lung CD4+ T cells by local treatment with a pSTAT-3 and pSTAT-5 inhibitor ameliorated experimental allergic asthma. Int Immunol 2011;23(1):1–15. [DOI] [PubMed] [Google Scholar]
- 19.Yang XO, Zhang H, Kim BS, Niu X, Peng J, Chen Y, et al. The signaling suppressor CIS controls proallergic T cell development and allergic airway inflammation. Nat Immunol 2013;14(7):732–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim H, Cho M, Choi G, Na H, Chung Y. Dynamic control of Th2 cell responses by STAT3 during allergic lung inflammation in mice. Int Immunopharmacol 2015;28(2):846–53. [DOI] [PubMed] [Google Scholar]
- 21.Lührmann A, Tschernig T, von der Leyen H, Hecker M, Pabst R, Wagner AH. Decoy oligodeoxynucleotide against STAT transcription factors decreases allergic inflammation in a rat asthma model. Exp Lung Res 2010;36(2):85–93. [DOI] [PubMed] [Google Scholar]
- 22.Cao D, Tal TL, Graves LM, Gilmour I, Linak W, Reed W, et al. Diesel exhaust particulate-induced activation of Stat3 requires activities of EGFR and Src in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 2007;292(2):L422–9. [DOI] [PubMed] [Google Scholar]
- 23.Simeone-Penney MC, Severgnini M, Rozo L, Takahashi S, Cochran BH, Simon AR. PDGF-induced human airway smooth muscle cell proliferation requires STAT3 and the small GTPase Rac1. Am J Physiol Lung Cell Mol Physiol 2008;294(4):L698–704. [DOI] [PubMed] [Google Scholar]
- 24.Chiba Y, Todoroki M, Misawa M. Antigen exposure causes activations of signal transducer and activator of transcription 6 (STAT6) and STAT1, but not STAT3, in lungs of sensitized mice. Immunopharmacol Immunotoxicol 2011;33(1):43–8. [DOI] [PubMed] [Google Scholar]
- 25.Chiba Y, Todoroki M, Misawa M. Phosphorylation of signal transducer and activator of transcription 6 (STAT6) and STAT1, but not STAT3, induced by antigen inhalation in bronchial smooth muscles of sensitized mice. Biol Pharm Bull 2010;33(1):146–9. [DOI] [PubMed] [Google Scholar]
- 26.Pullen NA, Falanga YT, Morales JK, Ryan JJ. The Fyn-STAT5 Pathway: A New Frontier in IgE- and IgG-Mediated Mast Cell Signaling. Front Immunol 2012;3:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu C, Zhang T, Qi H, Peng WK, Shi F. Effect of STAT5 gene silencing on the proliferation of T lymphocytes in a mouse model of asthma. Zhonghua Jie He He Hu Xi Za Zhi 2012;35(1):50–4. [PubMed] [Google Scholar]
- 28.Li G, Liu Z, Ran P, Qiu J, Zhong N. Activation of signal transducer and activator of transcription 5 (STAT5) in splenocyte proliferation of asthma mice induced by ovalbumin. Cell Mol Immunol 2004;1(6):471–4. [PubMed] [Google Scholar]
- 29.Eriksson U, Egermann U, Bihl MP, Gambazzi F, Tamm M, Holt PG, et al. Human bronchial epithelium controls TH2 responses by TH1-induced, nitric oxide-mediated STAT5 dephosphorylation: implications for the pathogenesis of asthma. J Immunol 2005;175(4):2715–20. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Chen L, Huang Z, Alkan S, Bunting KD, Wen R, et al. Cutting edge: IL-5 primes Th2 cytokine-producing capacity in eosinophils through a STAT5-dependent mechanism. J Immunol 2004;173(5):2918–22. [DOI] [PubMed] [Google Scholar]
- 31.Turlej RK, Fiévez L, Sandersen CF, Dogné S, Kirschvink N, Lekeux P, et al. Enhanced survival of lung granulocytes in an animal model of asthma: evidence for a role of GM-CSF activated STAT5 signalling pathway. Thorax 2001;56(9):696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Global strategy for asthma management and prevention: Global Initiative for Asthma (GINA); 2012. Available from: http://www.ginasthma.org/.
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 34.Barnes PJ. Inhaled corticosteroids. Pharmaceuticals 2010;3(3):514–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wjst M, Lichtner P, Meitinger T, Grimbacher B. STAT3 single-nucleotide polymorphisms and STAT3 mutations associated with hyper-IgE syndrome are not responsible for increased serum IgE serum levels in asthma families. Eur J Hum Genet 2009;17(3):352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gernez Y, Tirouvanziam R, Nguyen KD, Herzenberg LA, Krensky AM, Nadeau KC. Altered phosphorylated signal transducer and activator of transcription profile of CD4+CD161+ T cells in asthma: modulation by allergic status and oral corticosteroids. J Allergy Clin Immunol 2007;120(6):1441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie M, Mustovich AT, Jiang Y, Trudeau JB, Ray A, Ray P, et al. IL-27 and type 2 immunity in asthmatic patients: association with severity, CXCL9, and signal transducer and activator of transcription signaling. J Allergy Clin Immunol 2015;135(2):386–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Liu F, Zhao J, Wei Y, Lv J, Dong F, et al. Thymic stromal lymphopoietin promotes asthmatic airway remodelling in human lung fibroblast cells through STAT3 signalling pathway. Cell Biochem Funct 2013;31(6):496–503. [DOI] [PubMed] [Google Scholar]
- 39.Doganci A, Eigenbrod T, Krug N, De Sanctis GT, Hausding M, Erpenbeck VJ, et al. The IL-6R alpha chain controls lung CD4+CD25+ Treg development and function during allergic airway inflammation in vivo. J Clin Invest 2005;115(2):313–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagahama KY, Togo S, Holz O, Magnussen H, Liu X, Seyama K, et al. Oncostatin M modulates fibroblast function via signal transducers and activators of transcription proteins-3. Am J Respir Cell Mol Biol 2013;49(4):582–91. [DOI] [PubMed] [Google Scholar]
- 41.Stefanowicz D, Hackett TL, Garmaroudi FS, Günther OP, Neumann S, Sutanto EN, et al. DNA methylation profiles of airway epithelial cells and PBMCs from healthy, atopic and asthmatic children. PLoS One 2012;7(9):e44213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martín P, Sánchez-Madrid F. CD69: an unexpected regulator of TH17 cell-driven inflammatory responses. Sci Signal 2011;4(165):pe14. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Moilanen E, Lahti A, Hämäläinen M, Giembycz MA, Barnes PJ, et al. Regulation of eosinophil apoptosis by nitric oxide: Role of c-Jun-N-terminal kinase and signal transducer and activator of transcription 5. J Allergy Clin Immunol 2003;112(1):93–101. [DOI] [PubMed] [Google Scholar]
- 44.Burnham ME, Koziol-White CJ, Esnault S, Bates ME, Evans MD, Bertics PJ, et al. Human airway eosinophils exhibit preferential reduction in STAT signaling capacity and increased CISH expression. J Immunol 2013;191(6):2900–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabata H, Moro K, Fukunaga K, Suzuki Y, Miyata J, Masaki K, et al. Thymic stromal lymphopoietin induces corticosteroid resistance in natural helper cells during airway inflammation. Nat Commun 2013;4:2675. [DOI] [PubMed] [Google Scholar]