Significance
(Bacterio)chlorophylls harvest and convert the solar energy that powers the biosphere. The absorption properties of these pigments are determined in part by formation of the isocyclic ring, which confers their characteristic green color. This last remaining uncharacterized biosynthetic step has remained enigmatic for more than 60 y, and only a single subunit of the O2-dependent cyclase, AcsF, has been identified. Here we demonstrate that Ycf54 is a bona fide subunit of this enzyme in oxygenic phototrophs; identify a new cyclase subunit, BciE, in Alphaproteobacteria; and confirm that the AcsF found in other classes of bacterial phototrophs is the principal form of the cyclase, requiring neither Ycf54 nor BciE for activity.
Keywords: photosynthesis, chlorophyll, bacteriochlorophyll, cyclase
Abstract
The biosynthesis of (bacterio)chlorophyll pigments is among the most productive biological pathways on Earth. Photosynthesis relies on these modified tetrapyrroles for the capture of solar radiation and its conversion to chemical energy. (Bacterio)chlorophylls have an isocyclic fifth ring, the formation of which has remained enigmatic for more than 60 y. This reaction is catalyzed by two unrelated cyclase enzymes using different chemistries. The majority of anoxygenic phototrophic bacteria use BchE, an O2-sensitive [4Fe-4S] cluster protein, whereas plants, cyanobacteria, and some phototrophic bacteria possess an O2-dependent enzyme, the major catalytic component of which is a diiron protein, AcsF. Plant and cyanobacterial mutants in ycf54 display impaired function of the O2-dependent enzyme, accumulating the reaction substrate. Swapping cyclases between cyanobacteria and purple phototrophic bacteria reveals three classes of the O2-dependent enzyme. AcsF from the purple betaproteobacterium Rubrivivax (Rvi.) gelatinosus rescues the loss not only of its cyanobacterial ortholog, cycI, in Synechocystis sp. PCC 6803, but also of ycf54; conversely, coexpression of cyanobacterial cycI and ycf54 is required to complement the loss of acsF in Rvi. gelatinosus. These results indicate that Ycf54 is a cyclase subunit in oxygenic phototrophs, and that different classes of the enzyme exist based on their requirement for an additional subunit. AcsF is the cyclase in Rvi. gelatinosus, whereas alphaproteobacterial cyclases require a newly discovered protein that we term BciE, encoded by a gene conserved in these organisms. These data delineate three classes of O2-dependent cyclase in chlorophototrophic organisms from higher plants to bacteria, and their evolution is discussed herein.
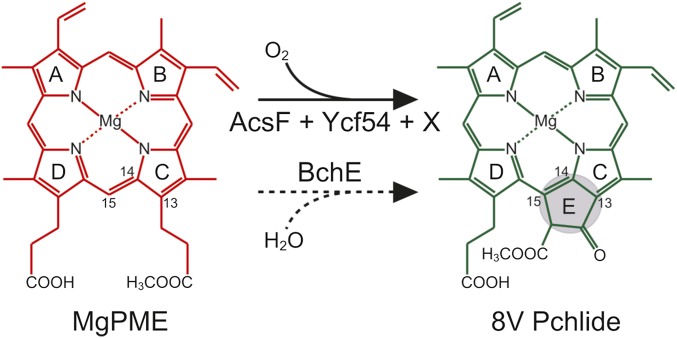
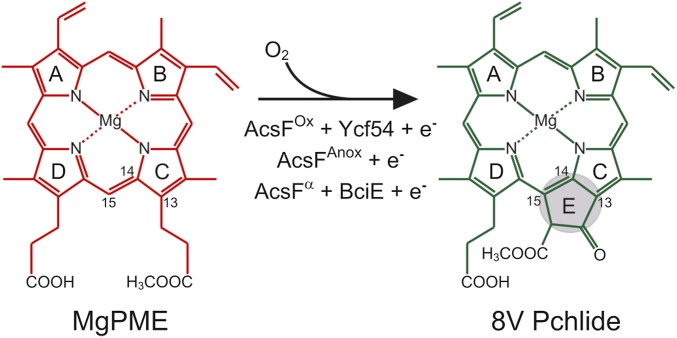
The (bacterio)chlorophylls, or (B)Chls, are ubiquitous pigments used by chlorophototrophic organisms for light harvesting and photochemistry, so elucidation of their biosynthetic pathways is of great importance. The least well-characterized step in the common pathway for all (B)Chls is formation of the isocyclic E ring, occurring via oxidation and cyclization of the C13 propionate group of magnesium protoporphyrin IX monomethyl ester (MgPME), producing 8-vinyl protochlorophyllide (8V Pchlide) (Fig. 1). The reaction is catalyzed by two distinct enzymes using different chemistries: an O2-sensitive protein containing [4Fe-4S] and cobalamin prosthetic groups (1) that derives oxygen from water (2), and an oxidative diiron enzyme that requires molecular oxygen (3). The O2-independent MgPME cyclase [EC:1.21.98.3] is believed to be encoded by a single gene, bchE (4), that is essential for BChl biosynthesis in bacterial phototrophs inhabiting anoxic environments. The O2-dependent MgPME cyclase [EC:1.14.13.81] catalyzes this reaction in plants and cyanobacteria (5, 6), and is believed to be composed of multiple subunits (7).
Fig. 1.
Cyclization reactions involved in (B)Chl biosynthesis. Shown is isocyclic ring formation via O2-dependent and -independent routes, catalyzed by AcsF and Ycf54 (solid arrow) and BchE (dashed arrow), respectively. Here x denotes the as-yet unassigned subunit required for the O2-dependent reaction. International Union of Pure and Applied Chemistry numbering of the relevant macrocycle carbons is indicated, and formation of the ring E is highlighted. The oxygen sources for the O2-dependent and -independent enzymes are molecular oxygen and water, respectively.
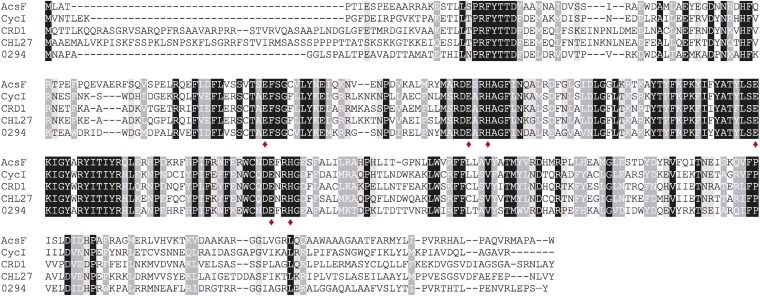
The first subunit assigned to the O2-dependent reaction was identified in the purple betaproteobacterium Rubrivivax (Rvi.) gelatinosus and was named AcsF (aerobic cyclization system Fe-containing subunit) (8). Subsequently, it was demonstrated that Rvi. gelatinosus contains both BchE and AcsF cyclases, conferring the ability to synthesize BChl under varying O2 regimes (9). Orthologs of acsF are widely distributed in phototrophs and have been studied in higher plants (10, 11), algae (12) and cyanobacteria (13), the green nonsulfur bacterium Chloroflexus aurantiacus (14), and the purple alphaproteobacterium Rhodobacter (Rba.) sphaeroides (15) (Fig. S1).
Fig. S1.
Amino acid sequence alignments of known AcsF proteins. Sequences are those from Rvi. gelatinosus (AcsF), Synechocystis (CycI), C. reinhardtii (CRD1), A. thaliana (CHL27), and Rba. sphaeroides (Rsp_0294; abbreviated as 0294). Conserved, highly similar, and similar residues are highlighted in black, dark gray, and light gray, respectively. The putative diiron center ligands are marked by red diamonds.
Two isoforms of AcsF in the unicellular alga Chlamydomonas reinhardtii, CRD1 and CTH1, catalyze E ring formation under copper-deficient and -replete conditions, respectively (12, 16). These proteins are localized to both the thylakoid membrane and chloroplast envelope (17), a pattern shared with the single AcsF in Arabidopsis thaliana, CHL27 (10, 18). The cyanobacterium Synechocystis sp. PCC 6803 (hereinafter Synechocystis) also contains two isoforms of AcsF, designated CycI and CycII (19). Constitutively expressed cycI encodes the sole AcsF protein responsible for cyclase activity under oxic conditions, with cycII expressed only under microoxic conditions (13); overexpression of cycII cannot rescue cyclase function under a range of O2 tensions when CycI levels are depleted (19).
The O2-dependent enzyme has been resolved into membrane-bound and soluble fractions from cucumber chloroplasts and Synechocystis, suggesting that more than one subunit is required for a functional enzyme (7, 20). Later in vivo pulldown experiments in Synechocystis with tagged CycI and CycII identified Ycf54 as an interaction partner of both AcsF isoforms; tagged Ycf54 produced in the same manner was shown to interact with CycI (21). A mutant depleted in Ycf54 had a significantly reduced Chl content, accumulated MgPME, was only able to tolerate low light intensities and was unable to grow under photoautotrophic conditions (21, 22). Antisense mutants of the ortholog of ycf54 in tobacco, LCAA, also demonstrated impaired cyclase activity, and the dimeric protein was shown to interact with CHL27 in the chloroplast (23). The foregoing findings suggest that Ycf54/LCAA, conserved among oxygenic phototrophs, is the “missing” soluble component of the O2-dependent cyclase. The subunit requirement for this enzyme across phototrophic organisms has not been resolved, however.
In the present study, we demonstrate that acsF from Rvi. gelatinosus corrects the loss of both cycI and ycf54 in Synechocystis, suggesting that this AcsF protein does not require a Ycf54 component. Reciprocally, CycI substitutes for AcsF in Rvi. gelatinosus only in the presence of Ycf54, providing validation of this protein as a subunit of the O2-dependent cyclase. Furthermore, we identify BciE as a cyclase subunit conserved among AcsF-containing Alphaproteobacteria. This work delineates three distinct classes of O2-dependent cyclase; phylogenetic analysis identifies defined clades, the evolution of which is discussed.
Results
Rvi. gelatinosus acsF Complements the Loss of cycI in Synechocystis, Regardless of the Presence of ycf54.
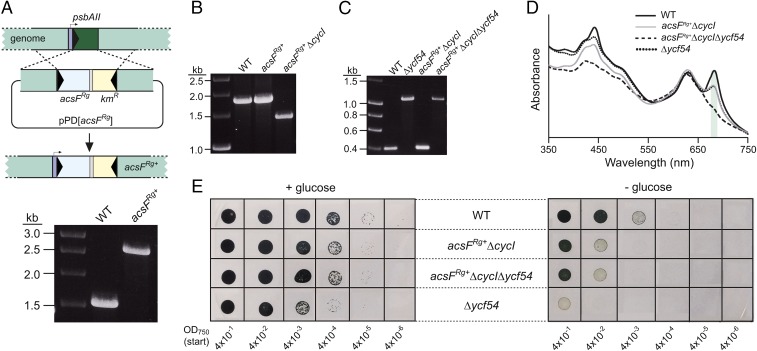
The apparent absence of ycf54 orthologs in phototrophic bacteria containing orthologs of acsF suggests that the Ycf54 component of the O2-dependent cyclase either is not required for function of the bacterial enzyme or that an unrelated protein performs the same function in its place. To determine which of these possibilities is the case, we integrated acsF from Rvi. gelatinosus into the genome of the ycf54-containing model cyanobacterium Synechocystis in place of the nonessential, light-responsive psbAII as described previously (24) (Fig. 2A). Deletion of the native CycI-encoding gene (sll1214) was attempted in this acsFRg+ background; a previous attempt to delete cycI in the wild type (WT) under oxic conditions proved unsuccessful (19). Full segregation of ΔcycI in acsFRg+ was achieved (Fig. 2B), indicating that acsF complements the loss of cycI in Synechocystis. Subsequently, deletion of ycf54 (slr1780) in acsFRg+ ΔcycI was achieved by replacement of the native gene with a zeocin resistance cassette as described previously (22), yielding acsFRg+ ΔcycIΔycf54 (Fig. 2C).
Fig. 2.
Construction and phenotypic analyses of Synechocystis cyclase mutants. (A) Diagram depicting replacement of the psbAII gene with acsFRg via pPD[acsFRg] (Upper), and construction of the fully segregated strain confirmed by colony PCR (Lower). (B and C) Inactivation of cycI (B) and ycf54 (C) genes via replacement with chloramphenicol and zeocin resistance cassettes, respectively, confirmed by colony PCR. (D) Whole-cell absorption spectra of strains grown mixotrophically under low light conditions. The peaks for Chl-containing complexes are marked with a green shadow. (E) Drop growth assays of strains on solid agar, supplemented with or lacking glucose. Photographs were taken after incubation for 12 d.
We performed phenotypic analyses of the acsFRg+ strains lacking cycI and both cycI and ycf54, along with WT and Δycf54 controls. Liquid cultures were grown photomixotrophically under low light to an OD750 of ∼0.4. Absorption spectra of these suspensions indicate that deletion of ycf54 almost abolishes the assembly of Chl-containing photosystems, as judged by the near absence of a peak at ∼680 nm (Fig. 2D). The restoration of a 680-nm absorption band by the introduction of acsFRg into strains lacking cycI, irrespective of the presence of ycf54, shows that acsFRg is necessary and sufficient for Chl a biosynthesis in Synechocystis. This conclusion is further reinforced by the Chl content of these strains grown under moderate light, calculated when all apart from Δycf54 were grown without glucose (mg·L−1·OD750-1, % relative to WT): WT, 3.22 ± 0.05, 100%; Δycf54, 0.24 ± 0.01, 7.5%; acsFRg+ΔcycI, 3.08 ± 0.07, 96%; acsFRg+ ΔcycIΔycf54, 3.08 ± 0.01, 96%. These data indicate that the O2-dependent cyclase of Rvi. gelatinosus integrates into a cyanobacterial Chl pathway and dispenses with the requirement for Ycf54 normally exhibited by its native partner, CycI.
In addition, we performed drop growth assays on solid agar with and without 5 mM glucose (Fig. 2E). As expected, supplementation with glucose resulted in improved growth at identical dilutions for each strain. Strains containing acsFRg showed the same pattern as seen in WT and grew under photoautotrophic conditions; they also were able to grow at higher dilutions than the Δycf54 mutant under photomixotrophic conditions. These data suggest that AcsFRg restores Chl biosynthesis and photoautotrophic growth to Synechocystis in the absence of Ycf54.
Ycf54 Is a Catalytic Component of the O2-Dependent Cyclase Enzyme in Oxygenic Phototrophs.
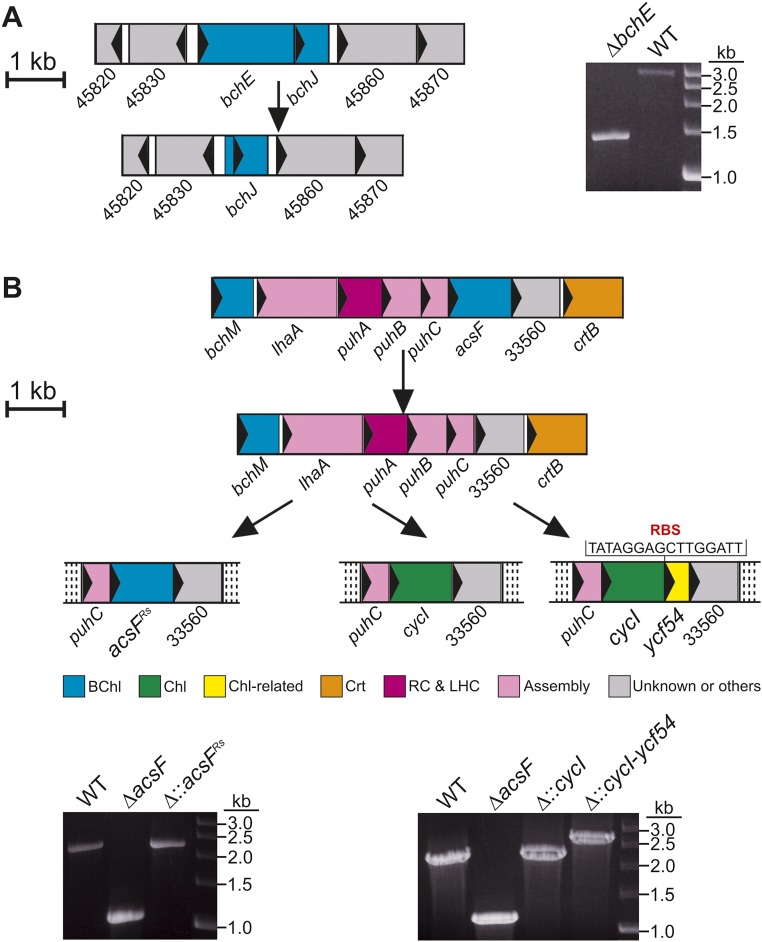
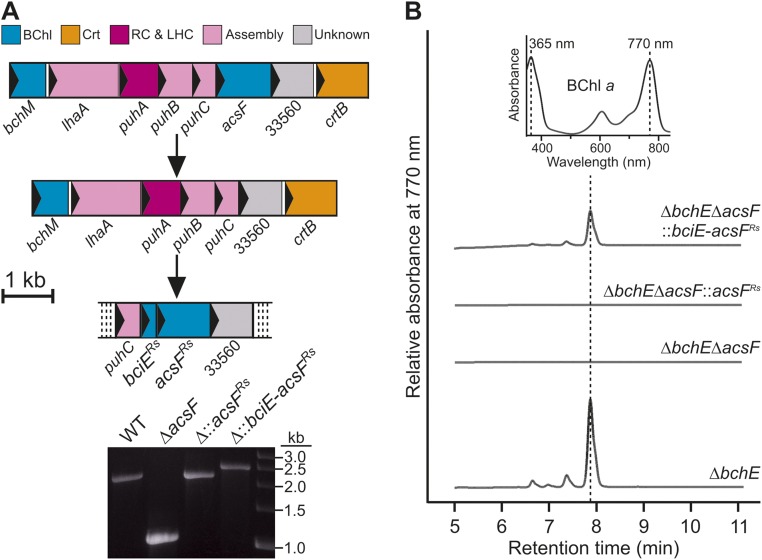
Previous work has shown that Synechocystis Δycf54 makes a small amount of Chl, ∼13% of WT levels (21, 22); thus, Ycf54 appears to be important, but not essential, for cyclase activity. To assess the contribution of Ycf54 more precisely, we developed a reciprocal system for the heterologous expression of Synechocystis genes in Rvi. gelatinosus, which synthesizes BChl a under conditions ranging from oxic to anoxic using O2-dependent and -independent cyclase enzymes, respectively (8, 9). Genes encoding the known components of these enzymes were removed using an in-frame, markerless deletion method to avoid the polar effects often encountered with resistance cassette-mediated gene disruption. The O2-independent cyclase was inactivated by deletion of bchE (Fig. S2A), and BChl biosynthesis was completely inactivated by the subsequent deletion of acsF (Fig. S2B). This ΔbchEΔacsF strain, which accumulates the cyclase substrate MgPME, provides a background for testing components of the O2-dependent enzyme. The cycI gene from Synechocystis was integrated at the original acsF locus both alone and in combination with ycf54 from the same organism encoded downstream of cycI (Fig. S2B). A third complemented strain used acsF from Rba. sphaeroides, which was recently shown to be essential for O2-dependent cyclase activity in this model anoxygenic phototroph (15) (Fig. S2B).
Fig. S2.
Genetic knockouts and replacements in Rvi. gelatinosus. (A) Depiction of the deletion of bchE (Left), confirmed by colony PCR (Right). (B) Depiction of deletion of acsF, and subsequent integration of foreign genes at the acsF locus, under control of the native promoter (Upper), confirmed by colony PCR (Lower). The regions subjected to genetic manipulation are depicted in proportion to the scale bar. ORFs are represented as colored filled rectangles, within which the arrow indicates the direction of transcription. Crt, carotenoid biosynthesis; RC&LHC, reaction center and light-harvesting complexes.
The three resulting strains, ∆bchE∆acsF::cycI, ∆bchE∆acsF::cycI-ycf54, and ∆bchE∆acsF::acsFRs, along with positive and negative control strains ∆bchE and ∆bchE∆acsF, respectively, were cultured under oxic conditions in the dark in liquid medium, standardized by OD680, and pelleted. The pigments accumulated by these strains were extracted, and BChl a content was analyzed by HPLC (Fig. 3). As expected, BChl a accumulated to a high level in ∆bchE (Fig. 3A), but ∆bchE∆acsF was unable to synthesize BChl (Fig. 3B). The presence of cycI in this background did not restore BChl biosynthesis (Fig. 3C), whereas BChl was detected in the strain complemented with both cycI and ycf54 (Fig. 3D). These data confirm that Ycf54 is essential for activity of the the O2-dependent cyclase from oxygenic phototrophs. Surprisingly, although a cyanobacterial cyclase was functional, acsF from the more closely related Rba. sphaeroides was unable to restore BChl biosynthesis to this strain (Fig. 3E).
Fig. 3.
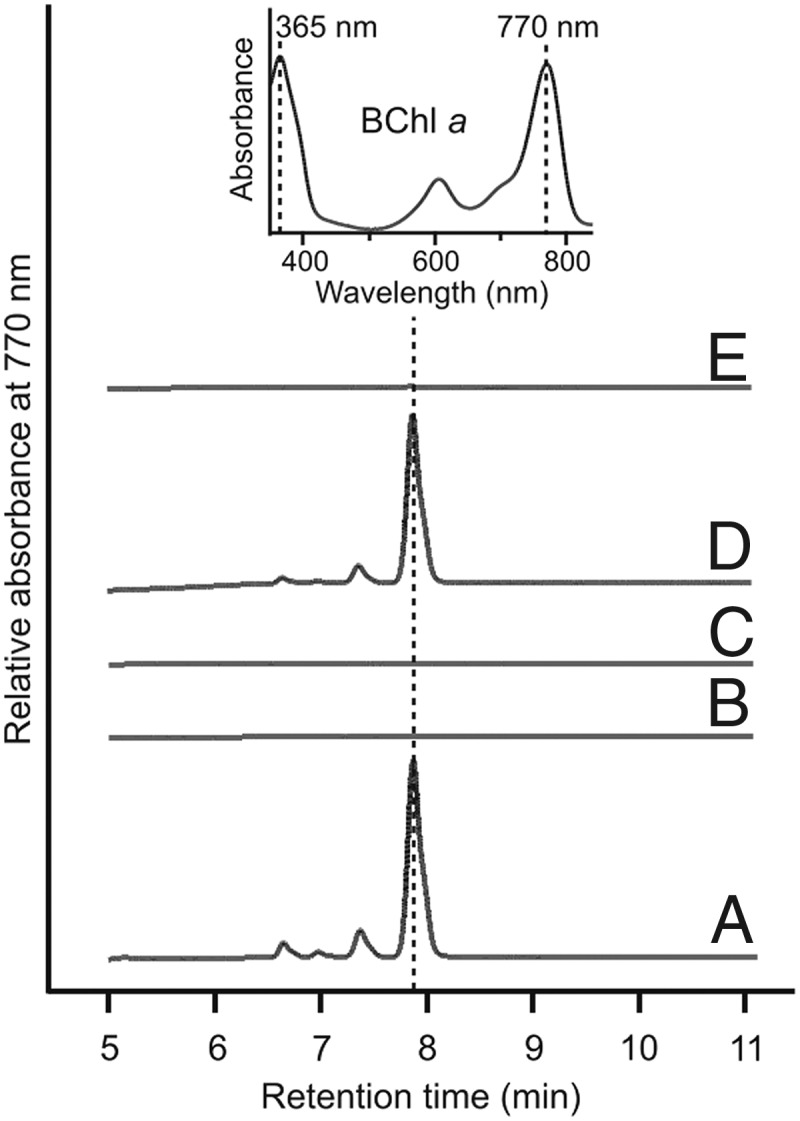
HPLC analysis of pigments extracted from Rvi. gelatinosus strains. Pigments were extracted from the same number of cells of each strain except for the ∆bchE strain, which had a much greater BChl a content than the other strains. (A) ∆bchE. (B) ∆bchE∆acsF. (C) ∆bchE∆acsF::cycI. (D) ∆bchE∆acsF::cycI-ycf54. (E) ∆bchE∆acsF::acsFRs. (Inset) Retention times and Soret/Qy maxima of peaks were used to identify BChl a.
Bioinformatic Analysis of a Conserved ORF Upstream of acsF in Alphaproteobacteria Reveals a Cyclase Subunit.
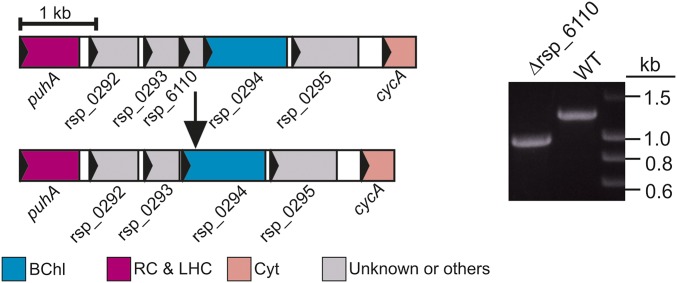
The unexpected lack of activity of acsFRs in Rvi. gelatinosus led us to postulate that the functional enzyme from Rba. sphaeroides may require a “Ycf54-like” protein. The genome of Rba. sphaeroides does not contain an ortholog of ycf54, so there was no obvious candidate encoding a missing cyclase component. All known genes essential for BChl biosynthesis in this model organism are contained within a 40.7-kb photosynthesis gene cluster (PGC) (25). PGCs are conserved throughout bacterial phototrophs and are a likely location for a “purple bacterial ycf54.” Analysis of the PGC of Rba. sphaeroides revealed a small ORF (rsp_6110) found directly upstream of, and overlapping with, acsF; orthologs of this gene are conserved upstream of acsF in all studied Alphaproteobacteria containing acsF but are absent in all other taxonomic groups (26), including the betaproteobacterium Rvi. gelatinosus (Table S1). To determine whether rsp_6110 encodes an essential component of the alphaproteobacterial O2-dependent cyclase, the ORF was deleted from Rba. sphaeroides ∆bchE∆ccoP (Fig. S3), in which native AcsF function has been detected via accumulation of BChl a chelated with either Mg or Zn (15). The resulting Rba. sphaeroides strain, ∆bchE∆ccoP∆6110, and its parent were cultured under oxic conditions in the dark, and pigments were extracted from pellets and analyzed as described previously (15).
Table S1.
Distribution of acsF and bchE genes among sequenced phototrophic Proteobacteria, along with the presence of orthologs of rsp_6110
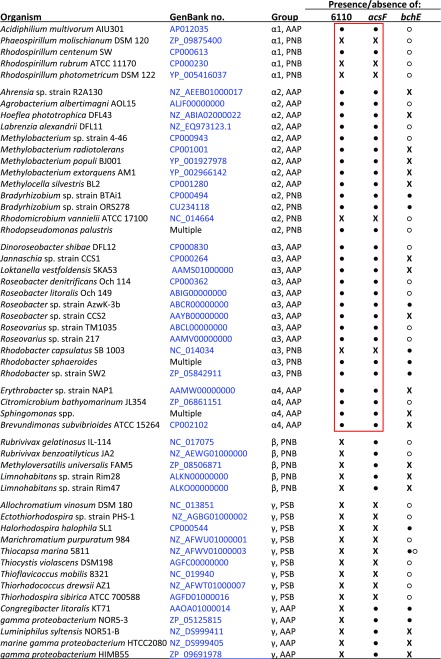 |
Modified from (26). ●, gene present in PGC; ○, gene present outside PGC; X, gene absent. The red box indicates an identical pattern of presence/absence of orthologs of rsp_6110 and acsF among Alphaproteobacteria. AAP, aerobic anoxygenic phototroph; PNB, purple nonsulfur bacterium; PSB, purple sulfur bacterium.
Fig. S3.
Deletion of rsp_6110 in Rba. sphaeroides. Diagram depicting deletion of rsp_6110 (Left), and confirmation by colony PCR (Right).
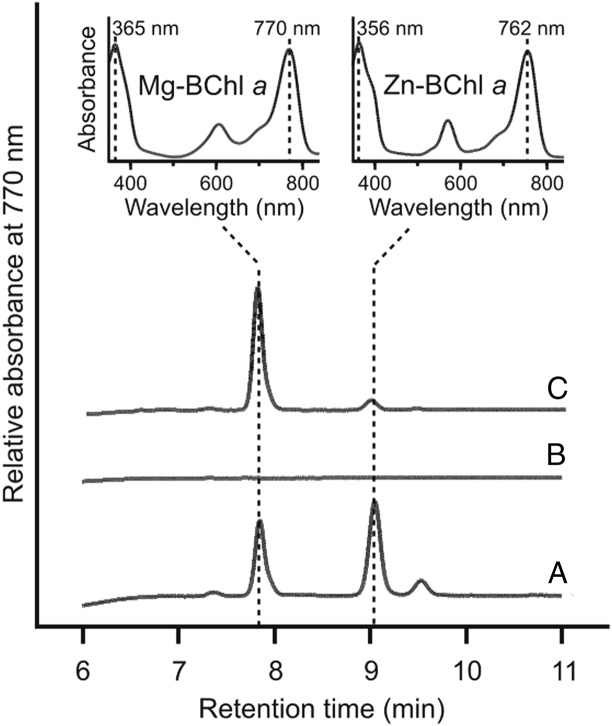
Both Mg-BChl a and Zn-BChl a were detected in ∆bchE∆ccoP (Fig. 4A), whereas these pigments were absent in ∆bchE∆ccoP∆6110 (Fig. 4B). To ensure that this result was due to deletion of rsp_6110, rather than a polar effect on acsF, rsp_6110 was expressed in trans in ∆bchE∆ccoP∆6110 from pBBRBB-Ppuf843–1200 (27). Pigments from this strain were analyzed as above, and Mg-BChl a and Zn-BChl a were detected in this extract (Fig. 4C), confirming that the O2-dependent cyclase from Rba. sphaeroides requires Rsp_6110 to function. Therefore, we propose, according to the Demerec nomenclature, that rsp_6110 be renamed bciE.
Fig. 4.
HPLC analysis of pigments extracted from Rba. sphaeroides strains. Pigments were extracted from strains standardized by cell number. (A) ∆bchE∆ccoP. (B) ∆bchE∆ccoP∆6110. (C) ∆bchE∆ccoP∆6110 + pBB[6110]. (Insets) Retention times and Soret/Qy maxima of peaks were used to identify Mg- and Zn-chelated species of BChl a.
To determine whether heterologous expression of bciE from Rba. sphaeroides, along with acsF from the same organism in Rvi. gelatinosus ∆bchE∆acsF, is able to restore BChl biosynthesis, where acsFRs alone is not, we amplified the overlapping bciE and acsF genes directly from the genome of Rba. sphaeroides and integrated at the acsF locus of Rvi. gelatinosus, as described earlier (Fig. S4A). This strain, ∆bchE∆acsF::bciE-acsFRs, was grown, and its pigments were extracted and analyzed as described above. Coexpression of bciE and acsFRs resulted in accumulation of BChl (Fig. S4B).
Fig. S4.
Construction and phenotypic analysis of Rvi. gelatinosus mutant expressing bciE and acsF from Rba. sphaeroides. (A) Diagram depicting integration of bciE and acsF from Rba. sphaeroides in place of the native acsF in Rvi. gelatinosus (Upper), and confirmation by colony PCR (Lower). (B) HPLC analysis of pigments extracted from Rvi. gelatinosus strains, extracted from the same number of cells of each strain except for the ∆bchE strain, which had a much greater BChl a content compared with the other strains. (Inset) Retention times and Soret/Qy maxima of peaks were used to identify BChl a.klj.
Phylogenetic Analysis of AcsF Proteins.
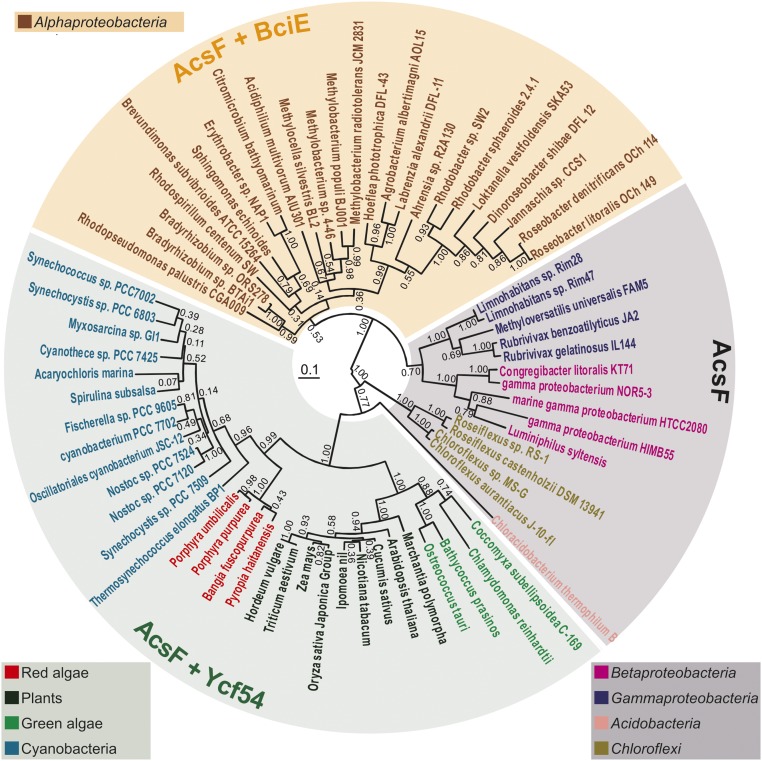
To investigate the evolutionary history of AcsF orthologs, we conducted phylogenetic analysis with example protein sequences from plants, algae, and acsF-containing phyla of phototrophic (cyano)bacteria, as described in Materials and Methods (Fig. 5). AcsF proteins from species belonging to the same group cluster in the same clade and the topology of the tree correspond closely with the evolutionary relationships among the species being analyzed (28) (Fig. 5). In addition, we checked for the presence or absence of BciE and Ycf54 orthologs in the 69 studied species by performing DELTA-BLAST searches using either Rba. sphaeroides BciE (WP_002720458) or Synechocystis Ycf54 (P72777) as a query. Distribution patterns of BciE and Ycf54 orthologs are seemingly related to the phylogeny of AcsF proteins.
Fig. 5.
Phylogenetic analysis of AcsF proteins. Evolutionary analysis via a phylogenetic tree was conducted in MEGA6 using the maximum likelihood method based on the JTT matrix-based model. The analysis involved 69 protein sequences. The tree with the highest log-likelihood (−17,513.1099) is shown. Numbers next to each node indicate bootstrap values (1,000 replicates) as percentages. Phyla are distinguished by color of species name. The length of each branch represents the number of amino acid substitutions per site in proportion to the scale bar at the center of the tree. The presence/absence of BciE/Ycf54 is indicated by shading over the species names: gray, no BciE or Ycf54; orange, BciE present; green, Ycf54 present. Note that orthologs of both bciE and ycf54 are not found together in the genome of any organism sequenced to date.
Discussion
The O2-dependent cyclase is the “missing link” in (B)Chl biosynthesis, and it has remained enigmatic for more than 60 y. Before the present study, only AcsF had been identified as a bona fide cyclase subunit (8, 13). Ycf54 and its ortholog LCAA were subsequently discovered to be required for the normal activity of cyanobacterial and plant enzymes, respectively (21, 22), but the absence of genes encoding Ycf54 from acsF-containing anoxygenic bacterial phototrophs suggested that it might not be a catalytic subunit. Our demonstration that only the Ycf54-CycI combination replaces the native cyclase function of AcsF in Rvi. gelatinosus shows that, irrespective of the other roles of Ycf54 in Synechocystis, it can be considered a subunit of this enzyme. In the case of Synechocystis Δycf54, it may be that the small amount of Chl a produced by this strain is due to the presence of three orthologs of bchE in the genome of this organism. It once was believed that proteins encoded by these genes did not contribute to Chl biosynthesis (13); however, the recent finding that cyanobacterial bchE orthologs from two strains of Cyanothece restore BChl a biosynthesis in a bchE mutant of Rba. capsulatus demonstrates the activity of O2-independent BchE orthologs from oxygenic phototrophs (29).
In the other half of the reciprocal experiment, expression of acsFRg in Synechocystis was found to complement the loss of either the native cycI or ycf54. The step occurring after the formation of ring E in (B)Chl biosynthesis, conversion of Pchlide to chlorophyllide, can be catalyzed by two unrelated enzymes, light-activated Pchlide oxidoreductase (POR) and dark-operative POR (DPOR), both of which are present in Synechocystis. The light-activated POR is dominant under the conditions that we used for culturing the complemented strains of Synechocystis (30). This enzyme is absent from all anoxygenic bacterial phototrophs, so the recombinant bacterial AcsF is able to functionally integrate into the Chl pathway in an oxygenic phototroph; this tallies with observations that other recombinant (B)Chl enzymes expressed in cyanobacterial or purple phototrophic hosts can function in nonnative pathways (31–33). These results suggest that there is much promise for combining pigment biosynthetic pathways with the aim of producing novel (B)Chls with unique spectral properties.
The existence of cyclase enzymes that require an extra subunit in plants and cyanobacteria is reminiscent of the role played by Gun4 in the activity of magnesium chelatase (MgCH), the enzyme catalyzing the first committed step in (B)Chl biosynthesis. Plant and cyanobacterial mutants in gun4 display reduced Chl content and impaired growth, and they accumulate the substrate for MgCH, whereas phototrophic bacteria lack orthologs of gun4 (34). Gun4 was found to stimulate cyanobacterial MgCH activity in vitro (35) and to be involved in increasing flux into the Chl biosynthesis pathway in vivo (36). Similarly, it is conceivable that Ycf54 may play a role in substrate channeling; pull-down experiments identified protein–protein interactions between Ycf54 and the cyanobacterial AcsF, CycI (21), and CycI with other Chl biosynthesis enzymes (22). These interactions are abrogated in the absence of ycf54, and the level of CycI is reduced, suggesting that Ycf54 may stabilize the CycI protein (22).
We have identified and validated a subunit of the O2-dependent cyclase in Alphaproteobacteria, which we have named BciE. The ORF encoding this protein is found directly upstream of acsF in this bacterial class. Deletion in Rba. sphaeroides led to the abolition of O2-dependent cyclase activity, reinstatement of bciE in trans restored activity, and AcsFRs was found to require BciE to function in the heterologous Rvi. gelatinosus system. No conserved domain can be identified in BciE based on the National Center for Biotechnology Information’s Conserved Domain Database (37). Thus, BciE may represent a novel protein family; its precise role in the alphaproteobacterial enzyme is unclear, but it may play a role similar to that of Ycf54 or Gun4 in stabilization of the major subunit and/or stimulation of the forward reaction.
A cyanobacterial progenitor of algal and plant chloroplasts (38) is also the likely origin of the acsF and ycf54 genes common to all oxygenic phototrophs. Our phylogenetic analysis is consistent with this theory, with Ycf54-requiring AcsF sequences clustering in a well-defined clade. In addition, it is believed that acsF was horizontally transferred from cyanobacteria to Proteobacteria before the divergence of Alphaproteobacterial, Betaproteobacterial, and Gammaproteobacterial lineages, because cyanobacteria were initially oxygenating the atmosphere, conferring an advantage to anoxygenic phototrophs previously reliant solely on O2-sensitive BchE (26). A newly discovered bacterial phototroph belonging to the phylum Gemmatimonadetes is believed to have acquired an acsF-containing purple bacterial PGC via horizontal gene transfer (39), and thus is more recent than the acquisition of acsF by the Proteobacteria. The acsF gene is also found in Chloroflexi and the single phototrophic member of the phylum Acidobacteria discovered to date (28). Although Chloroflexi are more closely related to cyanobacteria than to other phyla mentioned here, their AcsF proteins more closely resemble the proteobacterial enzyme, whereas the acidobacterial AcsF is more similar to those from cyanobacteria, yet it does not require Ycf54. These observations imply that multiple horizontal transfers have occurred, possibly to a common ancestor of Acidobacteria and Proteobacteria, followed by transfer from a proteobacterium to the Chloroflexi. Because acsF and ycf54 are not clustered in cyanobacteria, it follows that they would not be readily cotransferred to another organism. Mutations in the transferred acsF might have relieved the requirement for Ycf54, explaining the absence of ycf54 in anoxygenic bacteria. The bciE gene may have appeared after the divergence of Alphaproteobacteria from other subgroups, and the emergence of BciE may be necessary for the function of the enzyme under as-yet unidentified cellular conditions.
It has long been believed that the O2-dependent cyclase reaction requires NAD(P)H as a reductant (5), with dependence observed in plant, algal, and cyanobacterial systems (20, 40). The barley xantha-l and viridis-k mutants that accumulate MgPME are deficient in the membrane components of the O2-dependent enzyme, with all xantha-l mutations mapping to the single acsF ortholog that is intact in the viridis-k mutants (11). Subsequently, Bollivar et al. (41) concluded that viridis-k mutants do not lack Ycf54; thus, it is possible that the missing subunit of the enzyme, disrupted in viridis-k mutants, is a membrane-associated NAD(P)H-binding protein. Recently it was proposed that the O2-dependent cyclase may derive electrons from reduced quinone, as in the case for other diiron enzymes, plastid terminal oxidase and the mitochondrial alternative oxidase (42). The authors suggested that NAD(P)H may maintain the redox state of the plastoquinone pool, via NAD(P)H dehydrogenase, which they proposed to be directly coupled to efficient cyclase activity. Those studies indicated that the last remaining hurdle in the study of this enzyme, and thus in the pathway common to all (B)Chl pigments, is to determine the electron donor to the diiron center of AcsF (Fig. S5).
Fig. S5.
Current status of known components of the oxygen-dependent cyclase. AcsFα, AcsFAnox, and AcsFOx represent AcsF proteins from Alphaproteobacteria, anoxygenic phototrophs other than the Alphaproteobacteria, and oxygenic phototrophs, respectively. e− denotes the electron donor to the diiron center of AcsF.
Conclusion
We have used genetic techniques to test and identify components of the O2-dependent cyclase in three unrelated model organisms. Bioinformatic analyses coupled with heterologous gene expression (i) show that Ycf54 is essential for cyclase function in oxygenic phototrophs; (ii) identify a previously unknown protein component of the alphaproteobacterial enzyme, BciE; and (iii) indicate that the AcsF from other taxonomic groups of anoxygenic phototrophs requires neither Ycf54 nor BciE for activity.
Materials and Methods
Bacterial Strains and Growth Conditions.
Rvi. gelatinosus strains were grown in a rotary shaker in the dark at 30 °C in PYS medium (43). Microoxic and oxic growth regimes were achieved as described previously (15). When required, kanamycin was added at 50 μg·mL−1. Synechocystis strains were grown photoautotrophically in a rotary shaker at 30 °C under low light (5 μmol photons m−2·s−1) or moderate light (30 μmol photons m−2·s−1) as described previously (35). Photomixotrophic growth was achieved by the addition of glucose to a final concentration of 5 mM. Rba. sphaeroides strains were grown in the dark at 30 °C as described previously (15). Escherichia coli strains were grown in LB medium, supplemented with 30 μg·mL−1 kanamycin when required. The bacterial strains and plasmids used in this study are listed in Table S2.
Table S2.
Strains and plasmids described in this study
| Strain/plasmid | Genotype/characteristics | Source |
| E. coli | ||
| JM109 | Cloning strain for plasmid constructs | Promega |
| S17-1 | Conjugation strain for pK18mobsacB constructs | (48) |
| Rvi. gelatinosus | ||
| WT | IL144 | S. Nagashima* |
| ∆bchE | Unmarked deletion mutant of bchE in WT | This study |
| ∆bchE∆acsF | Unmarked deletion mutant of acsF in ∆bchE | This study |
| ∆bchE∆acsF::acsFRs | acsFRs replacement of acsF in ∆bchE | This study |
| ∆bchE∆acsF::bciE-acsFRs | acsF replaced with rsp_6110-acsFRs in ∆bchE | This study |
| ∆bchE∆acsF::cycI | cycI replacement of acsF in ∆bchE | This study |
| ∆bchE∆acsF::cycI-ycf54 | cycI-ycf54 replacement of acsF in ∆bchE | This study |
| Synechocystis | ||
| WT | sp. PCC6803 | R. Sobotka† |
| acsFRg+ | acsFRg and KmR replacement of psbAII in WT | This study |
| acsFRg+ ∆cycI | CmR replacement of cycI in acsFRg+ | This study |
| acsFRg+ ∆cycI∆ycf54 | ZeoR replacement of central portion of ycf54 in acsFRg+∆cycI | This study |
| ∆ycf54 | ZeoR replacement of central portion of ycf54 in WT | (22) |
| Rba. sphaeroides | ||
| WT | 2.4.1 | S. Kaplan‡ |
| ∆bchE∆ccoP | Unmarked deletion mutant of bchE and ccoP in WT | (15) |
| ∆bchE∆ccoP∆acsF | Unmarked deletion mutant of acsF in ∆bchE∆ccoP | (15) |
| ∆bchE∆ccoP∆6110 | Unmarked deletion mutant of rsp_6110 in ∆bchE∆ccoP | This study |
| Plasmids | ||
| pK18mobsacB | Allelic exchange vector, KmR | J. Armitage§ |
| pK18∆bchERg | Upstream-NdeI-downstream of bchERg cloned into BamHI/HindIII sites of pK18mobsacB | This study |
| pK18∆acsFRg | Upstream-NdeI-downstream of acsFRg cloned into BamHI/HindIII sites of pK18mobsacB | This study |
| pK18∆6110 | Upstream-downstream of rsp_6110 cloned into XbaI/HindIII sites of pK18mobsacB | This study |
| pK18[acsFRs] | acsFRs cloned into the NdeI site of pK18∆acsFRg | This study |
| pK18[6110-acsFRs] | rsp_6110-acsFRs cloned into the NdeI site of pK18∆acsFRg | This study |
| pK18[cycI] | cycI cloned into the NdeI site of pK18∆acsFRg | This study |
| pK18[cycI-ycf54] | cycI-ycf54 cloned into the NdeI site of pK18∆acsFRg | This study |
| pPD-FLAG | Cloning site, KmR, flanked by psbAII upstream and downstream regions, AmpR | (21) |
| pPD[acsFRg] | acsFRg cloned into NdeI/BglII sites of pPD-FLAG | This study |
| pBBRBB-Ppuf843–1200 | Expression vector carrying the 843–1,200 region of puf promoter of Rba. sphaeroides, KmR | (27) |
| pBB[6110] | rsp_6110 cloned into the BglII/NotI sites of pBBRBB-Ppuf843–1200 | This study |
Research Institute for Photosynthetic Hydrogen Production, Kanagawa University, Yokohama, Japan.
Institute of Microbiology, Department of Phototrophic Microorganisms, Třeboň, Czech Republic.
Department of Microbiology and Molecular Genetics, University of Texas Medical School, Austin, TX.
Department of Biochemistry, University of Oxford, Oxford, United Kingdom.
Construction of Mutants.
Heterologous expression acsFRg in Synechocystis was achieved by cloning the ORF between NdeI/BglII sites in pPD-FLAG (21). The resulting pPD[acsFRg] was transformed into WT Synechocystis, and transformants were isolated as described previously (21). The regions upstream and downstream of cycI were amplified with 1214UpF and 1214UpR and with 1214DownF and 1214DownR primers, respectively. The chloramphenicol (Cm) resistance cassette from pACYC184 was amplified with 1214UpCmF and 1214DownCmR. The three PCR products were fused by overlap extension PCR. The resulting fragment was used to disrupt cycI, and segregation was achieved by sequentially doubling the concentration of Cm from 5 to 80 μg·mL−1. A ycf54 disruption fragment used in this study was constructed as described previously (22). Markerless deletion mutants in Rba. sphaeroides and Rvi. gelatinosus were constructed as described previously (15). Gene replacements were achieved by cloning ORFs into the NdeI site in the relevant pK18∆gene construct. The pBB[gene] plasmid was used to express foreign genes in Rba. sphaeroides and was constructed by cloning the relevant ORF between BglII/NotI sites in pBBRBB-Ppuf843–1200 (27). Rvi. gelatinosus was transformed using a method described by Nagashima et al. (43). Sequences of primers used in this study are listed in Table S3.
Table S3.
Primers used in this study
| Primer | Sequence (5′-3′) |
| 6110UpF | GCTCTAGAGGAGCTGATCCCGCCCTTCC |
| 6110UpR | GGAGAGCCCTCCGGCCGGCGCGTTCATGGGGGTTCCCTTCTCTTGG |
| 6110DownF | CCAAGAGAAGGGAACCCCCATGAACGCGCCGGCCGGAGGGCTCTCC |
| 6110DownR | GCAAGCTTCCCAGGTTCACCGCCACGCC |
| 6110CheckF | GCCCCGGAGCGACAAGGAC |
| 6110CheckR | GTATTTCTTGGCCTTGGTCAGG |
| 6110F_NdeI | GAGTCTCATATGGGTCTGTTCACGAAACAAGCG |
| 6110F_BglII | GGCAGATCTATGGGTCTGTTCACGAAACAAGCGGAA |
| 6110R_NotI | TCTGCGGCCGCTCACAGCGTCACCTGCTCGGAGAA |
| 0294F_NdeI | CCAGTACATATGTGAACGCGCCGGCCGGAGG |
| 0294R_NdeI | CCAGTACATATGTCAATAGCTCGGCTCCAGTCGG |
| 45840UpF | CTAGGTCAAGTAGGATCCTCATGCCGGCGGCGATCATG |
| 45840UpR | CTAGGTCAAGTACATATGGGAAACGGCTCCTCGCGATTC |
| 45840DownF | CTAGGTCAAGTACATATGCGACGGCTGGGTCACGATGC |
| 45840DownR | CTAGGTCAAGTAAAGCTTTGCCGGTGTAGAAGTCGCACGC |
| 45840CheckF | TAGCCGCCGACCATGCCGA |
| 45840CheckR | GCGGTGCACCAGCACCGTGA |
| 33550UpF | GAGTCTGGATCCCTGCATGAGCGACAACGCGTC |
| 33550UpR | GAGTCTCATATGGAGGGTCTCCGTGGTGTGTCA |
| 33550DownF | GAGTCTCATATGAAGCGAGGACAGGATGCTGAGC |
| 33550DownR | GAGTCTAAGCTTGGAACTCCTCGCTCAGGTTGCG |
| 33550CheckF | GAACGTTTGCCGGACACGGT |
| 33550CheckR | ACGAGGTACTTCAGGTGCTCC |
| 33550F_NdeI | GAGTCTCATATGCTCGCGACCCCGACGATCG |
| 33550R_BamHI | GAGTCTGGATCCTCACCATGCCGGGGCCATG |
| 1214UpF | GCCGATCCGGTTAACCTAGGCA |
| 1214UpR | ATATCCAGTGATTTTTTTCTCCATAGAGTTGTTTAAAATAGTTTCC |
| 1214UpCmF | GGAAACTATTTTAAACAACTCTATGGAGAAAAAAATCACTGGATAT |
| 1214DownCmR | GGTGATCCAGCGGAAGACAACCTTACGCCCCGCCCTGC |
| 1214DownF | GCAGGGCGGGGCGTAAGGTTGTCTTCCGCTGGATCACC |
| 1214DownR | TGGAGTTGTTGGGAGAGTTCGGTC |
| 1214F_NdeI | GGAATTCCATATGGTTAATACCCTCGAAAAGCCCG |
| 1214R_NdeI | GGAATTCCATATGTTAGCGCACAGCTCCAGCCA |
| 1214RBS1780F | GTTGGCTGGAGCTGTGCGCTAATATAGGAGCTTGGATTGTGGAAAGTTGGGCATTGACGA |
| 1214RBS1780R | TCGTCAATGCCCAACTTTCCACAATCCAAGCTCCTATATTAGCGCACAGCTCCAGCCAAC |
| 1780F | GTGGAAAGTTGGGCATTGACG |
| 1780R | CTAATCCAGGGATGCAAGGGG |
| 1780R_NdeI | GAGTCTCATATGCTAATCCAGGGATGCAAGGGG |
Whole-Cell Absorption Spectroscopy and Determination of Chl Content.
Absorption spectra were obtained as detailed previously (15). Pigments were extracted from 4 mL of Synechocystis culture (OD750 = 0.4) with methanol, and Chl concentrations were determined spectroscopically from extracts of biological triplicates using a published method (44).
Pigment Extraction and Analysis by HPLC.
Pigment extraction and HPLC analysis were carried out as described previously (15).
Plate-Based Growth Assays.
Synechocystis drop growth assays were conducted using liquid cultures of Synechocystis grown under low light, adjusted to OD750 = 0.4, and serially diluted. Dilutions were spotted on solid medium ±5 mM glucose and incubated at low light.
Phylogenetic Tree Construction.
The phylogenetic tree was built with MEGA6 (45). AcsF sequences from 24 Alphaproteobacteria, 5 Betaproteobacteria, 5 Gammaproteobacteria, 13 Cyanobacteria, 1 Acidobacterium sp., 4 Chloroflexi, 9 algae, and 8 plants were retrieved via DELTA-BLAST using Rvi. gelatinosus AcsF (RGE_33550) as a query. The protein sequences thus obtained (Table S1) were aligned using the ClustalW algorithm provided in MEGA6. All positions containing gaps and missing data in the alignment were removed. Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using the Jones–Taylor–Thornton (JTT) amino acid substitution model (46). Evolutionary history was inferred by using the maximum likelihood method with the JTT model and was tested by the bootstrap method with 1,000 replicates. The tree with the highest log-likelihood (−17,513.1099) was adopted and visualized using Interactive Tree of Life v2 (47).
Acknowledgments
We thank Dr. Sakiko Nagashima (Kanagawa University) for supplying the Rvi. gelatinosus IL144. G.E.C. was supported by a University of Sheffield doctoral studentship. D.P.C. and C.N.H. were supported by a grant from the Biotechnology and Biological Sciences Research Council (BB/M000265/1). C.N.H. is supported by the European Research Council (Advanced Award 338895). D.P.C. is supported by the European Commission (Marie Skłodowska-Curie Global Fellowship 660652).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701687114/-/DCSupplemental.
References
- 1.Gough SP, Petersen BO, Duus JØ. Anaerobic chlorophyll isocyclic ring formation in Rhodobacter capsulatus requires a cobalamin cofactor. Proc Natl Acad Sci USA. 2000;97:6908–6913. doi: 10.1073/pnas.97.12.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porra RJ, et al. Origin of the two carbonyl oxygens of bacteriochlorophyll a: Demonstration of two different pathways for the formation of ring E in Rhodobacter sphaeroides and Roseobacter denitrificans, and a common hydratase mechanism for 3-acetyl group formation. Eur J Biochem. 1996;239:85–92. doi: 10.1111/j.1432-1033.1996.0085u.x. [DOI] [PubMed] [Google Scholar]
- 3.Walker CJ, Mansfield KE, Smith KM, Castelfranco PA. Incorporation of atmospheric oxygen into the carbonyl functionality of the protochlorophyllide isocyclic ring. Biochem J. 1989;257:599–602. doi: 10.1042/bj2570599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter CN, Coomber SA. Cloning and oxygen-regulated expression of the bacteriochlorophyll biosynthesis genes bch E, B, A, and C of Rhodobacter sphaeroides. J Gen Microbiol. 1988;134:1491–1497. [Google Scholar]
- 5.Chereskin BM, Wong YS, Castelfranco PA. In vitro synthesis of the chlorophyll isocyclic ring: Transformation of magnesium-protoporphyrin IX and magnesium-protoporphyrin IX monomethyl ester into magnesium-2,4-divinyl pheoporphyrin A(5) Plant Physiol. 1982;70:987–993. doi: 10.1104/pp.70.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beale SI. Enzymes of chlorophyll biosynthesis. Photosynth Res. 1999;60:43–73. [Google Scholar]
- 7.Wong YS, Castelfranco PA. Resolution and reconstitution of mg-protoporphyrin-IX monomethyl ester (oxidative) cyclase, the enzyme system responsible for the formation of the chlorophyll isocyclic ring. Plant Physiol. 1984;75:658–661. doi: 10.1104/pp.75.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinta V, Picaud M, Reiss-Husson F, Astier C. Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear iron cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J Bacteriol. 2002;184:746–753. doi: 10.1128/JB.184.3.746-753.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouchane S, Steunou AS, Picaud M, Astier C. Aerobic and anaerobic Mg-protoporphyrin monomethyl ester cyclases in purple bacteria: A strategy adopted to bypass the repressive oxygen control system. J Biol Chem. 2004;279:6385–6394. doi: 10.1074/jbc.M309851200. [DOI] [PubMed] [Google Scholar]
- 10.Tottey S, et al. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA. 2003;100:16119–16124. doi: 10.1073/pnas.2136793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rzeznicka K, et al. Xantha-l encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc Natl Acad Sci USA. 2005;102:5886–5891. doi: 10.1073/pnas.0501784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moseley JL, et al. Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell. 2002;14:673–688. doi: 10.1105/tpc.010420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minamizaki K, Mizoguchi T, Goto T, Tamiaki H, Fujita Y. Identification of two homologous genes, chlAI and chlAII, that are differentially involved in isocyclic ring formation of chlorophyll a in the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem. 2008;283:2684–2692. doi: 10.1074/jbc.M708954200. [DOI] [PubMed] [Google Scholar]
- 14.Tang K-H, Wen J, Li X, Blankenship RE. Role of the AcsF protein in Chloroflexus aurantiacus. J Bacteriol. 2009;191:3580–3587. doi: 10.1128/JB.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen GE, Canniffe DP, Martin EC, Hunter CN. Absence of the cbb3 terminal oxidase reveals an active oxygen-dependent cyclase involved in bacteriochlorophyll biosynthesis in Rhodobacter sphaeroides. J Bacteriol. 2016;198:2056–2063. doi: 10.1128/JB.00121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moseley J, Quinn J, Eriksson M, Merchant S. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 2000;19:2139–2151. doi: 10.1093/emboj/19.10.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen MD, Kropat J, Merchant SS. Regulation and localization of isoforms of the aerobic oxidative cyclase in Chlamydomonas reinhardtii. Photochem Photobiol. 2008;84:1336–1342. doi: 10.1111/j.1751-1097.2008.00440.x. [DOI] [PubMed] [Google Scholar]
- 18.Bang WY, et al. Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development, and gene expression profiling. Plant Cell Physiol. 2008;49:1350–1363. doi: 10.1093/pcp/pcn111. [DOI] [PubMed] [Google Scholar]
- 19.Peter E, et al. Differential requirement of two homologous proteins encoded by sll1214 and sll1874 for the reaction of Mg protoporphyrin monomethylester oxidative cyclase under aerobic and micro-oxic growth conditions. Biochim Biophys Acta. 2009;1787:1458–1467. doi: 10.1016/j.bbabio.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Bollivar DW, Beale SI. The chlorophyll biosynthetic enzyme Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase: Characterization and partial purification from Chlamydomonas reinhardtii and Synechocystis sp PCC 6803. Plant Physiol. 1996;112:105–114. doi: 10.1104/pp.112.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollingshead S, et al. Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J Biol Chem. 2012;287:27823–27833. doi: 10.1074/jbc.M112.352526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollingshead S, et al. Synthesis of chlorophyll-binding proteins in a fully-segregated ∆ycf54 strain of the cyanobacterium Synechocystis PCC 6803. Front Plant Sci. 2016;7:292. doi: 10.3389/fpls.2016.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albus CA, et al. LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. 2012;160:1923–1939. doi: 10.1104/pp.112.206045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canniffe DP, Jackson PJ, Hollingshead S, Dickman MJ, Hunter CN. Identification of an 8-vinyl reductase involved in bacteriochlorophyll biosynthesis in Rhodobacter sphaeroides and evidence for the existence of a third distinct class of the enzyme. Biochem J. 2013;450:397–405. doi: 10.1042/BJ20121723. [DOI] [PubMed] [Google Scholar]
- 25.Naylor GW, Addlesee HA, Gibson LCD, Hunter CN. The photosynthesis gene cluster of Rhodobacter sphaeroides. Photosynth Res. 1999;62:121–139. [Google Scholar]
- 26.Boldareva-Nuianzina EN, Bláhová Z, Sobotka R, Koblízek M. Distribution and origin of oxygen-dependent and oxygen-independent forms of Mg-protoporphyrin monomethylester cyclase among phototrophic proteobacteria. Appl Environ Microbiol. 2013;79:2596–2604. doi: 10.1128/AEM.00104-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tikh IB, Held M, Schmidt-Dannert C. BioBrick™-compatible vector system for protein expression in Rhodobacter sphaeroides. Appl Microbiol Biotechnol. 2014;98:3111–3119. doi: 10.1007/s00253-014-5527-8. [DOI] [PubMed] [Google Scholar]
- 28.Bryant DA, et al. Comparative and functional genomics of anoxygenic green bacteria from the taxa Chlorobi, Chloroflexi, and Acidobacteria. In: Burnap R, Vermaas W, editors. Functional Genomics and Evolution of Photosynthetic Systems. Springer; Amsterdam, The Netherlands: 2012. pp. 47–102. [Google Scholar]
- 29.Yamanashi K, Minamizaki K, Fujita Y. Identification of the chlE gene encoding oxygen-independent Mg-protoporphyrin IX monomethyl ester cyclase in cyanobacteria. Biochem Biophys Res Commun. 2015;463:1328–1333. doi: 10.1016/j.bbrc.2015.06.124. [DOI] [PubMed] [Google Scholar]
- 30.Kopečná J, Sobotka R, Komenda J. Inhibition of chlorophyll biosynthesis at the protochlorophyllide reduction step results in the parallel depletion of Photosystem I and Photosystem II in the cyanobacterium Synechocystis PCC 6803. Planta. 2013;237:497–508. doi: 10.1007/s00425-012-1761-4. [DOI] [PubMed] [Google Scholar]
- 31.Wilks HM, Timko MP. A light-dependent complementation system for analysis of NADPH:protochlorophyllide oxidoreductase: Identification and mutagenesis of two conserved residues that are essential for enzyme activity. Proc Natl Acad Sci USA. 1995;92:724–728. doi: 10.1073/pnas.92.3.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canniffe DP, Chidgey JW, Hunter CN. Elucidation of the preferred routes of C8-vinyl reduction in chlorophyll and bacteriochlorophyll biosynthesis. Biochem J. 2014;462:433–440. doi: 10.1042/BJ20140163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hitchcock A, et al. Biosynthesis of chlorophyll a in a purple bacterial phototroph and assembly into a plant chlorophyll-protein complex. ACS Synth Biol. 2016;5:948–954. doi: 10.1021/acssynbio.6b00069. [DOI] [PubMed] [Google Scholar]
- 34.Wilde A, Mikolajczyk S, Alawady A, Lokstein H, Grimm B. The gun4 gene is essential for cyanobacterial porphyrin metabolism. FEBS Lett. 2004;571:119–123. doi: 10.1016/j.febslet.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 35.Davison PA, et al. Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry. 2005;44:7603–7612. doi: 10.1021/bi050240x. [DOI] [PubMed] [Google Scholar]
- 36.Kopečná J, et al. Porphyrin binding to Gun4 protein, facilitated by a flexible loop, controls metabolite flow through the chlorophyll biosynthetic pathway. J Biol Chem. 2015;290:28477–28488. doi: 10.1074/jbc.M115.664987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFadden GI, van Dooren GG. Evolution: Red algal genome affirms a common origin of all plastids. Curr Biol. 2004;14:R514–R516. doi: 10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 39.Zeng Y, Feng F, Medová H, Dean J, Koblížek M. Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. Proc Natl Acad Sci USA. 2014;111:7795–7800. doi: 10.1073/pnas.1400295111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nasrulhaq-Boyce A, Griffiths WT, Jones OT. The use of continuous assays to characterize the oxidative cyclase that synthesizes the chlorophyll isocyclic ring. Biochem J. 1987;243:23–29. doi: 10.1042/bj2430023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bollivar D, Braumann I, Berendt K, Gough SP, Hansson M. The Ycf54 protein is part of the membrane component of Mg-protoporphyrin IX monomethyl ester cyclase from barley (Hordeum vulgare L.) FEBS J. 2014;281:2377–2386. doi: 10.1111/febs.12790. [DOI] [PubMed] [Google Scholar]
- 42.Steccanella V, Hansson M, Jensen PE. Linking chlorophyll biosynthesis to a dynamic plastoquinone pool. Plant Physiol Biochem. 2015;97:207–216. doi: 10.1016/j.plaphy.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Nagashima KV, Shimada K, Matsuura K. Shortcut of the photosynthetic electron transfer in a mutant lacking the reaction center-bound cytochrome subunit by gene disruption in a purple bacterium, Rubrivivax gelatinosus. FEBS Lett. 1996;385:209–213. doi: 10.1016/0014-5793(96)00382-1. [DOI] [PubMed] [Google Scholar]
- 44.Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- 45.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 47.Letunic I, Bork P. Interactive Tree of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475-8. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]