ABSTRACT
Pollen, fungi, and bacteria are the main microscopic biological entities present in outdoor air, causing allergy symptoms and disease transmission and having a significant role in atmosphere dynamics. Despite their relevance, a method for monitoring simultaneously these biological particles in metropolitan environments has not yet been developed. Here, we assessed the use of the Hirst-type spore trap to characterize the global airborne biota by high-throughput DNA sequencing, selecting regions of the 16S rRNA gene and internal transcribed spacer for the taxonomic assignment. We showed that aerobiological communities are well represented by this approach. The operational taxonomic units (OTUs) of two traps working synchronically compiled >87% of the total relative abundance for bacterial diversity collected in each sampler, >89% for fungi, and >97% for pollen. We found a good correspondence between traditional characterization by microscopy and genetic identification, obtaining more-accurate taxonomic assignments and detecting a greater diversity using the latter. We also demonstrated that DNA sequencing accurately detects differences in biodiversity between samples. We concluded that high-throughput DNA sequencing applied to aerobiological samples obtained with Hirst spore traps provides reliable results and can be easily implemented for monitoring prokaryotic and eukaryotic entities present in the air of urban areas.
IMPORTANCE Detection, monitoring, and characterization of the wide diversity of biological entities present in the air are difficult tasks that require time and expertise in different disciplines. We have evaluated the use of the Hirst spore trap (an instrument broadly employed in aerobiological studies) to detect and identify these organisms by DNA-based analyses. Our results showed a consistent collection of DNA and a good concordance with traditional methods for identification, suggesting that these devices can be used as a tool for continuous monitoring of the airborne biodiversity, improving taxonomic resolution and characterization together. They are also suitable for acquiring novel DNA amplicon-based information in order to gain a better understanding of the biological particles present in a scarcely known environment such as the air.
KEYWORDS: airborne biodiversity, Hirst-type spore trap, bioaerosol monitoring, methods in aerobiology, next-generation sequencing (NGS)
INTRODUCTION
The outdoor air carries many biological particles such as viruses, archaea, bacteria, fungi, and pollen coming from soil, vegetation, water sources, and other origins (1). Several studies have proposed that they play a significant role in weather conditions and atmospheric dynamics by triggering ice nucleation and producing oxidizing compounds (2–5). Moreover, some of these airborne organisms or parts thereof can cause allergy symptoms and relevant diseases in humans such as influenza, tuberculosis, and aspergillosis (6–8) and can also affect animals and plants, with critical consequences for health and the economy (6, 9, 10). Despite their importance, a real-time system for monitoring all this biodiversity has not yet been developed, partly because of technical factors such as different chemical and physical properties, low concentrations, and variable residence time in the atmosphere that make difficult an integrated analysis (7). Impingers, filters, and impactors are conventionally employed to collect these airborne biological entities for subsequent characterization by culture and microscopy, although new approaches such as mass spectrometry, immunological techniques, and DNA sequencing have been alternatively tested (1, 11–13).
Inside cities, where human population is congregated and more extensively exposed to airborne disease transmission, aerobiological networks perform an inestimable labor. With a worldwide distribution, these stations constantly monitor urban atmosphere to provide valuable information about the presence, abundance, and peak season of allergenic pollen and fungal propagules. These aerobiological stations mostly operate with Hirst-type spore traps to collect these biological particles (14, 15), and further identification is performed by microscopy, recognizing distinctive characters such as size, apertures, and furrows for pollen and color, scars, and uni- or multicelled spores for fungi (17, 18). While this traditional characterization is extremely time-consuming and requires considerable expertise, the results are widely accepted and have been consistent across the years. Unfortunately, precise taxonomic discrimination (especially to the genus and species levels) is not always accomplished, and some groups are frequently clustered during annotation as pollen or spore types with different taxonomic correspondences. For instance, pollen grains from all the grass species are usually included within the pollen type “Poaceae” because of their matching morphologies, and grains from the families Amaranthaceae and Chenopodiaceae cannot be distinguished. Fungal spores are also difficult to classify because the spores look very similar under a light microscope. Therefore, propagules from the genera Mucor, Histoplasma, and Rhizopus may be easily misplaced into the group “Aspergillus/Penicillium.” As a result, information about the global biodiversity present in the air is limited and is not always supplemented with a rigorous taxonomic classification, which is valuable knowledge in regard to allergens and pathogenic agents.
On the other hand, the current taxonomy is strongly supported by DNA sequence information in comparison to the traditional classification based on morphological features. 16S rRNA data for bacteria (19, 20), internal transcribed spacer (ITS) data for fungi (21), mitochondrial cytrochrome oxidase I (COI) gene data for animals (22), and data corresponding to the plastid RuBisCO (rbcL) and maturase K (matK) genes complemented with ITS2 for land plants (23, 24) represent the most popular gene markers employed for phylogenetic classification in individual isolates. Recent advances in sequencing technologies have boosted new disciplines such as metagenomics. Thus, the different next-generation sequencing (NGS) platforms have become powerful tools for identification of any organism present in heterogeneous microbiomes such as those represented by soil, water, or human specimens (25–29). Although NGS is fairly established for the study of bacterial communities in different environments, there is a shortage of surveys that apply this technology to air samples in urban spaces. Moreover, the existing publications are frequently focused on one particular group of organisms (usually bacteria or fungi) and there is no general consensus about sampling methodology or sequencing proceedings (30).
Here we evaluate the use of the Hirst-type spore trap for simultaneous characterization of the main groups of microscopic biological particles (bacteria, fungi, and pollen) present in the urban atmosphere, applying high-throughput DNA sequencing techniques. We also compare the results with traditional identification by microscopy for pollen and fungi and analyze the concordance between the two approaches. We propose to use the samples collected in aerobiological stations worldwide to provide additional metagenomic information about the airborne organisms in urban areas to uncover their dynamics, relationships, and distribution.
RESULTS
Analysis of DNA concentrations between replicates.
Our main goal was to validate the Hirst-type spore trap as a sampler device for high-throughput DNA sequencing analysis applied to bioaerosols in urban airborne studies. In order to collect a representative sample of the biodiversity of the metropolitan atmosphere, a weekly sampling approach was chosen to reduce the daily variability described in previous studies (31–33). Accordingly, three independent 7-day sampling assays (labeled A, B, and C; Table 1) were performed using two volumetric spore samplers running synchronously and acting as biological replicates.
TABLE 1.
Sampling information and DNA concentrations of the samples determined by PicoGreen assay
| Assay | Sampling dates | Sample IDa | Airflow (liters/min) (start–end) | [DNA] (pg/m3) | Mean | SD (% relative SD) |
|---|---|---|---|---|---|---|
| A | 2–9 December 2015 | A1 | 10.00–10.00 | 34.78 | 19.19 | 15.59 (81.24) |
| A2 | 10.00–8.50 | 3.60 | ||||
| B | 9–16 December 2015 | B1 | 9.70–9.20 | 59.98 | 48.46 | 11.52 (23.77) |
| B2 | 9.70–9.70 | 36.94 | ||||
| C | 16–23 December 2015 | C1 | 9.50–9.40 | 54.80 | 67.33 | 12.53 (18.61) |
| C2 | 9.50–9.60 | 79.85 | ||||
| M | 20–27 July 2015 | M0 | 9.50–9.60 | 271.20 |
ID, identifier.
First, we checked the DNA concentration obtained from the replicates. As shown in Table 1, the amounts of DNA recovered from the surveys were small (less than 1 ng/m3 but within the limit of detection of the measurement method), supporting previous studies that demonstrated the low concentration of DNA and organisms present in the air compared to other biomes (34–36). Additionally, in our study, some biological replicates showed significant variations in the final DNA concentration obtained. For instance, the relative standard deviation (SD) in assay A represented 81.24%. Although several factors can interfere with the sampling and DNA extraction, the only remarkable difference that we detected was a depletion in the airflow rate in sample A2 during collection (∼8.5 liters/min [checked at the end of the sampling] instead of 10 liters/min; Table 1), which may partly explain this divergence. Significantly lower values were obtained for the other two surveys, surveys B and C (relative SD = 23.77% and 18.61%, respectively), suggesting a more consistent collection.
Analysis of pollen diversity.
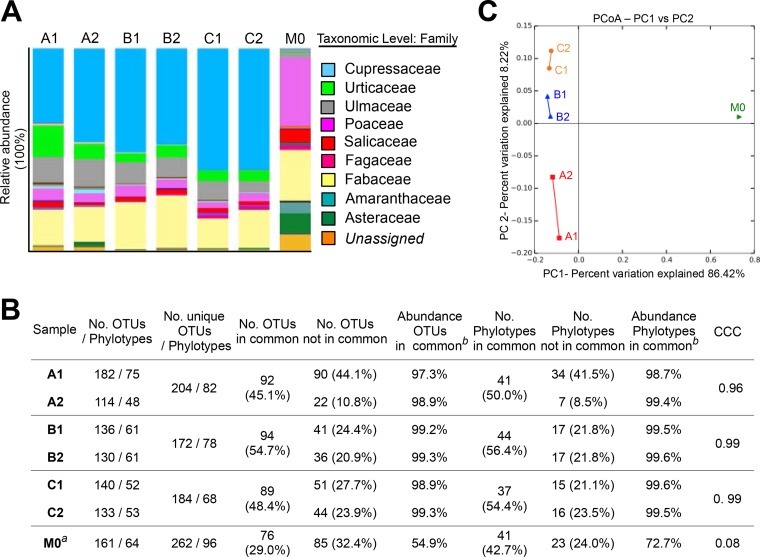
Next, we submitted the DNA samples to high-throughput sequencing in a targeted amplicon approach, assuming similar taxon identifications between replicates. For the analysis of the pollen diversity, the taxonomic assignment was performed using a customized database for the 5.8S-ITS2 region present in the plant genome (see Materials and Methods). Representatives of Cupressaceae, Urticaceae, Ulmaceae, and Fabaceae were the dominant taxa detected at that time of the year by NGS (Fig. 1A). An additional sample collected during the summer season taken in the same location was included as a contrast (M0), showing remarkable differences in the relative abundances of the taxa (Fig. 1A).
FIG 1.
Diversity of plants determined by NGS, statistics, and beta diversity analysis. (A) Relative abundances of the taxonomic assignments. Only the taxa representing ≥2% are identified in the legend. (B) Statistical analyses for OTUs and phylogenetic taxa. CCC, concordance correlation coefficient. The superscript “a” indicates a comparison with sample A1; the superscript “b” indicates relative abundances compiled by analysis of the OTUs/phylotypes in common. (C) Beta diversity analysis. Principal-coordinate analysis (PCoA) was performed to determine the diversity of plants from a Bray-Curtis distance matrix comprising the replicates and the divergence sample M0.
The counts from the total number of sequences assigned to each operational taxonomic unit (OTU) were used to perform statistical analyses. The concordance correlation coefficient (CCC) described by Lin (37) that was used for evaluation of the agreement between two measures of the same variable was determined to analyze the consistency of the sampling method. Almost perfect concordances between the replicates were found for assays B and C (CCC ≥ 0.99). In assay A, with a high difference in DNA concentrations between replicates, a lower CCC value was determined (0.96, representing substantial concordance). A remarkably poor coefficient value was obtained in comparisons with sample M0 (taken in summer season) (for A1 versus M0, CCC = 0.08) (Fig. 1B).
The OTUs in common within the replicates ranged from 45.1% to 54.7% of the total number of OTUs for each assay, corresponding to 50.0% to 56.4% of the total number of phylotypes. Nonetheless, these values represented >97% of the total relative abundance for each sample, indicating that the pollen diversity was fairly well represented in both samplers. Moreover, although the number of noncommon OTUs/phylotypes was apparently high, it represented only a minor fraction of the total relative abundance (<3%; Fig. 1B). Interestingly, only ca. 30% of the OTUs were shared between samples A1 (late fall) and M0 (summer), representing 54.9% of the abundance in M0 (Fig. 1B). This result suggests an important change in the relative abundance of the pollen diversity in the cold season compared to summer, confirmed by the pollen calendar in the region (38), supporting the sensitivity of the NGS analysis for detection of changes in the pollen community following this procedure.
Subsequently, we studied the dissimilarity between the replicates (beta diversity), assuming that the samples from the same assay clustered together. Principal-coordinate analysis (PCoA) of the Bray-Curtis distance matrix showed that replicates tended to group and that the largest divergence was found between samples in assay A (Fig. 1C). This result was anticipated since there was an important number of OTUs not shared between A1 and A2 (Fig. 1B). However, the three replicates clustered far apart from a divergent sample (M0), suggesting that these internal differences were minor and resulting a global R value of 0.777 (P < 0.01; determined by analysis of similarities [ANOSIM], considering 4 groups constituted by each replicate and M0) (Fig. 1C). Globally, these results suggest that real differences in pollen diversity and abundance can be graphically and statistically detected by analyzing DNA sequence information.
As additional validation of the Hirst spore trap for DNA sequencing studies, we compared the NGS outcome (for samples A1, B1, and C1) with the results obtained from traditional morphological identification of the same set of samples by microscopy (see Materials and Methods for details). We would expect that sequencing would detect at least the same taxa that microscopy revealed and, ideally, that the proportions of the relative abundances of each taxa would be maintained between the two methods. As shown in Table 2, the most abundant pollen type determined by morphology was Cupressaceae/Taxaceae (81% to 89% across the samples), followed distantly by Urticaceae (2% to 4%) and by other types with less representation. Remarkably, NGS results confirmed a major presence of the Cupressaceae/Taxaceae pollen type (37% to 60%), although some divergences from other types were found. Moreover, NGS identified more plant diversity (with a minor contribution to total abundance), as represented in the group “Others,” which contained additional families and genera that are not visualized by microscopy (data not shown).
TABLE 2.
Comparison between morphological identification and NGS results for pollena
| Taxonomic level: order | Pollen type | A1 |
B1 |
C1 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Morphology |
NGS (% DNA sequences) | Morphology |
NGS (% DNA sequences) | Morphology |
NGS (% DNA sequences) | |||||
| No. of pollen grains/m3/wk | % total grains | No. of pollen grains/m3/wk | % total grains | No. of pollen grains/m3/wk | % total grains | |||||
| Apiales | Apiaceae | 0 | 0.00 | 0.05 | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.01 |
| Asterales | Artemisia | 1 | 0.33 | 0.06 | 2 | 0.67 | 0.20 | 2 | 0.36 | 0.4 |
| Compositae (others) | 2 | 0.66 | 0.24 | 1 | 0.34 | 0.10 | 1 | 0.18 | 0.1 | |
| Caryophyliales | Caryophyllaceae | 0 | 0.00 | 0.20 | 0 | 0.00 | 0.09 | 0 | 0.00 | 0.01 |
| Chenopodiaceae/Amaranthaceae | 0 | 0.00 | 0.01 | 4 | 1.35 | 0.01 | 3 | 0.54 | 0.00 | |
| Ericales | Ericaceae | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.00 |
| Fabales | Sophora/Styphnolobium | 0 | 0.00 | 16.00 | 0 | 0.00 | 22.00 | 0 | 0.00 | 14.00 |
| Fabaceae (others) | 2 | 0.66 | 2.00 | 2 | 0.67 | 1.00 | 3 | 0.54 | 1.00 | |
| Fagales | Alnus | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.06 | 1 | 0.18 | 0.04 |
| Corylus | 0 | 0.00 | 0.01 | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.00 | |
| Quercus | 3 | 0.99 | 0.30 | 8 | 2.69 | 0.40 | 12 | 2.14 | 2.00 | |
| Lamiales | Fraxinus | 2 | 0.66 | 0.50 | 3 | 1.01 | 0.10 | 9 | 1.61 | 1.00 |
| Ligustrum | 0 | 0.00 | 0.07 | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.03 | |
| Olea | 0 | 0.00 | 0.00 | 3 | 1.01 | 0.00 | 0 | 0.00 | 0.00 | |
| Plantago | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.05 | 0 | 0.00 | 0.00 | |
| Malpighiales | Populus | 0 | 0.00 | 3.00 | 1 | 0.34 | 2.00 | 0 | 0.00 | 2.00 |
| Myrtales | Eucalyptus | 1 | 0.33 | 0.00 | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.00 |
| Pinales | Cupressaceae/Taxaceae | 272 | 89.77 | 37.00 | 243 | 81.82 | 52.00 | 501 | 89.46 | 60.00 |
| Pinaceae | 1 | 0.33 | 0.00 | 4 | 1.35 | 0.00 | 4 | 0.71 | 0.00 | |
| Poales | Poaceae | 5 | 1.65 | 6.00 | 9 | 3.03 | 5.00 | 6 | 1.07 | 3.00 |
| Proteales | Platanus | 2 | 0.66 | 1.00 | 0 | 0.00 | 0.40 | 0 | 0.00 | 0.60 |
| Rosales | Celtis | 0 | 0.00 | 0.50 | 0 | 0.00 | 0.20 | 0 | 0.00 | 0.20 |
| Ulmus | 0 | 0.00 | 13.00 | 0 | 0.00 | 11.00 | 0 | 0.00 | 9.00 | |
| Urticaceae | 7 | 2.31 | 16.00 | 13 | 4.38 | 4.00 | 16 | 2.86 | 5.00 | |
| Sapindales | Acer | 0 | 0.00 | 0.20 | 0 | 0.00 | 0.05 | 0 | 0.00 | 0.01 |
| Unidentified | 5 | 1.65 | 1.91 | 4 | 1.35 | 0.44 | 2 | 0.36 | 0.79 | |
| Others (only NGS) | 0 | 0.00 | 1.95 | 0 | 0.00 | 0.90 | 0 | 0.00 | 0.80 | |
| Total | 303 | 100.00 | 100.00 | 297 | 100.00 | 100.00 | 560 | 100.00 | 100.00 | |
Pollen types with no representatives in either of the two analyses have been omitted.
To compare the results from the two methods statistically, we focused on the pollen types detected by morphology and the percentages of NGS results were corrected (see Materials and Methods [see also Table S2 in the supplemental material]). CCC values determined a substantial concordance between the two approaches (CCC ≥ 0.96), except for sample A1 (CCC = 0.84 [poor concordance]).
Overall, these results confirmed that NGS analyses of samples collected with a Hirst spore trap are suitable for airborne pollen characterization (detecting greater diversity than microscopy) and that the results were in good agreement with the traditional identification.
Comparison of fungal communities.
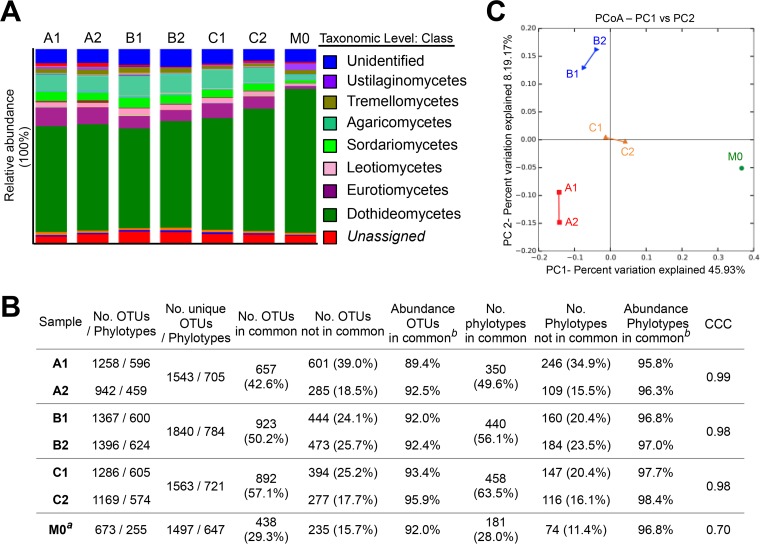
Next, we compared the communities of fungi between replicates. Sequences assigned to the class Dothideomycetes were highly represented in all the samples (Fig. 2A) and consisted mostly of sequences from Davidiella/Cladosporium (range, 33.48% to 50.35%; data not shown), in agreement with other works that analyzed airborne fungal communities in urban spaces by NGS and microscopy identification (39–43). CCC values of ≥0.98 confirmed a good concordance between the replicates (Fig. 2B). Sample M0 obtained a value for CCC of 0.70 (which indicates poor agreement between samples A1 and M0). The replicates shared 42.6% to 57.1% of the OTUs, representing 89.4% to 95.9% of the total relative abundance in each sample. As in the pollen analysis, these values showed a good representation of the fungal population in the air, although the values were slightly lower, likely because of the greater diversity of fungi that exists in the air (supported by the higher number of unique OTUs detected; Fig. 2B). Interestingly, samples A1 and M0 had only 29.3% of OTUs in common (28% of phylotypes) but represented 92.4% of the total abundance in M0 (96.8% with respect to the phylotypes). These values suggested a higher diversity in summer season, as described by other authors (41–43), but a relative steady abundance of the main and common taxa.
FIG 2.
Diversity of fungi determined by NGS, statistics, and beta diversity analysis. (A) Relative abundances of the taxonomic assignments. Only the taxa representing ≥2% are identified in the legend. (B) Statistical analyses for OTUs and phylogenetic taxa. CCC, concordance correlation coefficient. The superscript “a” indicates a comparison with sample A1; the superscript “b” indicates relative abundances compiled by analysis of the OTUs/phylotypes in common. (C) Beta diversity analysis. Principal-coordinate analysis (PCoA) was performed to determine the diversity of fungi from a Bray-Curtis distance matrix comprising the replicates and the divergence sample M0.
Subsequently, we checked the beta diversity graphically by PCoA. As shown in Fig. 2C, the replicates were clustered together, suggesting high similarity between the samples in fungal composition (also for assay A), and were placed at a significant distance from sample M0 (global R = 1.000, P < 0.01 [determined by ANOSIM]), indicating that the global differences in abundance and those divergent taxa present in M0 (representing <8% of the total relative abundance compared with A1) are enough to distinguish dissimilar samples.
Similarly to the pollen analysis, we compared the results from NGS with microscopy identification (see Materials and Methods). A taxonomic adjustment for comparisons with NGS results is shown in Table 3 (for complete morphology classification results, see Table S4 in the supplemental material). Results of classification of traditional fungal propagules and spore types by morphological characterization do not always correlate with the results of a strict taxonomic analysis, and specific recognition of the organisms cannot always be achieved. Thus, for the sake of comparison, we selected the morphological spore types that can be assigned to a unique taxon. The rest of the morphological spore and propagule types were included in the category “Others” (ranging from 38% to 65% across the samples), hampering the comparison with NGS results (see Discussion). Nonetheless, morphology confirmed the dominant presence of Cladosporium detected by DNA sequencing and of other allergens such as Alternaria or Epicoccum. Next, the results were corrected to compare NGS outcome with morphological identification (see Materials and Methods for details; see also Table S3 in the supplemental material). Moderate concordance between NGS and morphological identification was found for samples A1 and B1 (CCC = 0.94 in both cases), while a substantial concordance (CCC = 0.98) was determined for sample C1.
TABLE 3.
Comparison between morphological identification of fungi and NGS resultsa
| Taxonomic level: class | Morphological type | A1 |
B1 |
C1 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Morphology |
NGS (% DNA sequences) | Morphology |
NGS (% DNA sequences) | Morphology |
NGS (% DNA sequences) | |||||
| No. of fungal particles/m3/wk | % fungal particles | No. of fungal particles/m3/wk | % fungal particles | No. of fungal particles/m3/wk | % fungal particles | |||||
| Dothideomycetes | Davidiella = Cladosporium (C. herbarum + C. cladosporoides) | 696 | 26.83 | 35.00 | 2,464 | 46.02 | 35.00 | 3,581 | 55.13 | 45.00 |
| Polythrincium (= Mycosphaerella) | 2 | 0.08 | 0.02 | 3 | 0.06 | 0.00 | 7 | 0.11 | 0.00 | |
| Epicoccum | 12 | 0.46 | 2.00 | 6 | 0.11 | 5.00 | 19 | 0.29 | 2.00 | |
| Alternaria | 34 | 1.31 | 12.00 | 30 | 0.56 | 8.00 | 27 | 0.42 | 7.00 | |
| Drechslerab | 18 | 0.69 | 0.50 | 9 | 0.17 | 0.20 | 23 | 0.35 | 0.40 | |
| Curvularia | 0 | 0.00 | 0.08 | 0 | 0.00 | 0.10 | 2 | 0.03 | 0.01 | |
| Tetraploa | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.00 | 2 | 0.03 | 0.00 | |
| Spegazzinia | 0 | 0.00 | 0.00 | 24 | 0.45 | 0.06 | 6 | 0.09 | 0.01 | |
| Sordariomycetes | Chaetomium | 18 | 0.69 | 1.00 | 18 | 0.34 | 0.70 | 19 | 0.29 | 0.70 |
| Fusarium | 0 | 0.00 | 0.06 | 30 | 0.56 | 0.30 | 0 | 0.00 | 0.20 | |
| Xylariales | 12 | 0.46 | 0.20 | 15 | 0.28 | 0.20 | 2 | 0.03 | 0.20 | |
| Arthrinium | 18 | 0.69 | 0.10 | 18 | 0.34 | 0.01 | 40 | 0.62 | 0.06 | |
| Khuskia | 18 | 0.69 | 0.01 | 39 | 0.73 | 0.03 | 25 | 0.38 | 0.01 | |
| (Ascomycota) Incertae sedis | Beltrania | 0 | 0.00 | 0.00 | 0 | 0.00 | 0.00 | 2 | 0.03 | 0.01 |
| Agaricomycetes | Psathyrellaceae | 30 | 1.16 | 0.50 | 57 | 1.06 | 0.50 | 56 | 0.86 | 0.90 |
| Bovista | 14 | 0.54 | 0.80 | 9 | 0.17 | 0.50 | 66 | 1.02 | 0.60 | |
| Ganoderma | 4 | 0.15 | 0.03 | 15 | 0.28 | 0.06 | 21 | 0.32 | 0.05 | |
| Boletales | 6 | 0.23 | 0.30 | 0 | 0.00 | 0.20 | 6 | 0.09 | 0.30 | |
| Thelephoraceae | 10 | 0.39 | 0.09 | 9 | 0.17 | 0.07 | 22 | 0.34 | 0.20 | |
| Pucciniomycetes | Pucciniomycetes | 4 | 0.15 | 0.40 | 9 | 0.17 | 0.80 | 7 | 0.11 | 0.30 |
| Ustilaginomycetes | Ustilaginomycetes | 14 | 0.54 | 1.00 | 39 | 0.73 | 0.30 | 73 | 1.12 | 0.50 |
| Subtotal | 910 | 35.08 | 54.09 | 2,794 | 52.19 | 52.03 | 4,006 | 61.67 | 58.45 | |
| Others | 1,684 | 64.92 | 45.91 | 2,560 | 47.81 | 47.97 | 2,490 | 38.33 | 41.55 | |
| Total | 2,594 | 100.00 | 100.00 | 5,354 | 100.00 | 100.00 | 6,496 | 100.00 | 100.00 | |
Morphological spore types with no representatives in either of the two analyses have been omitted.
Data from the teleomorphs Pyrenophora, Cochliobolus, and Bipolaris and the anamorph Helminthosporium are also included.
In general, the differences between the samples of each replicate were due to minor representatives of the total fungal diversity (relative abundances of ≤11% for OTUs and <5% for phylotypes) and the NGS results correlated with the morphological identification, validating the employment of the Hirst-type sampler for fungal characterization by DNA sequencing analyses.
Comparison of bacterial communities.
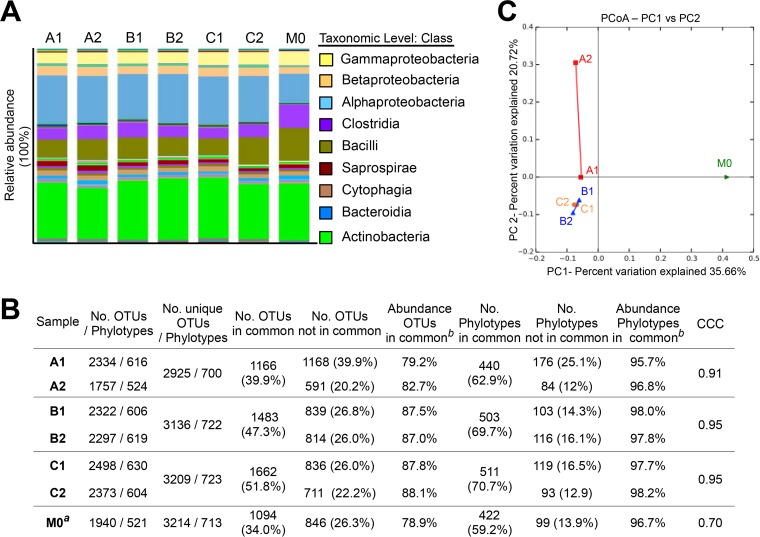
The similarity in bacterial diversity between replicates was also determined (see Fig. S3 in the supplemental material for details). As shown in Fig. 3A, OTUs assigned to the phyla Actinobacteria and Proteobacteria were the dominant taxa in all the samples, in accordance with previous studies (44–46). Statistical analyses determined substantial concordance of the results for replicates B and C (CCC = 0.95) and moderate concordance for assay A (CCC = 0.91). The diversity of bacteria collected by both samplers (measured as observed OTUs) ranged from 39.9% to 51.8%, correlating with 79.2% to 88.1% of the total relative abundance. Although these values were moderately lower than those for fungal diversity, they represented similar percentages of abundance with respect to phylotypes (62.9% to 70.7% of phylotypes in common, corresponding to 95.7% to 98.2% of the total relative abundance). Thus, despite the fact that the device is not primary designed for collecting bacteria, a representative sample of prokaryotic diversity can be reliably obtained.
FIG 3.
Diversity of bacteria determined by NGS, statistics, and beta diversity analysis. (A) Relative abundances of the taxonomic assignments. Only the taxa representing ≥2% are identified in the legend. (B) Statistical analyses for OTUs and phylogenetic taxa. CCC, concordance correlation coefficient. The superscript “a” indicates a comparison with sample A1; the superscript “b” indicates relative abundances compiled by analysis of the OTUs/phylotypes in common. (C) Beta diversity analysis. Principal-coordinate analysis (PCoA) was performed to determine bacterial diversity from a Bray-Curtis distance matrix comprising the replicates and the divergence sample M0.
Remarkably, a lower value of CCC was obtained for the comparison of samples A1 and M0: CCC = 0.70 (poor concordance) (Fig. 3B). Interestingly, 59.2% of the phylotypes were shared between the two samples (34.0% of the total OTUs), representing 96.7% of the total relative abundance in M0 (78.9% in assessing the abundance compiled by the OTUs in common). These values were similar to those obtained with the other replicates, suggesting lower variation of the bacterial diversity in the air than for the other biological entities analyzed.
Next, we studied the beta diversity graphically by PCoA. While replicates B and C clearly clustered together (small differences between the paired samples), an apparently big disparity was found between sample A1 and sample A2 (Fig. 3C), in accordance with the statistical analyses. Even though the value obtained for this clustering was lower than that found for fungi or pollen (R = 0.630, P < 0.01), it was still possible clearly to distinguish the replicates (containing a similar diversity) from sample M0. Moreover, we noticed that the largest variation between the A1 and A2 replicates fell along the second component (explaining 20% variance) whereas it clustered together with replicates in the B and C samples, all far apart from the M0 sample, along the main component.
Altogether, these results suggested that the diversity of airborne bacteria can be described by employing the Hirst spore trap and subsequent DNA analysis and that it is also possible to identify differences in composition by combining graphical and statistical analyses.
DISCUSSION
The Hirst spore trap is the most widely used sampler in aerobiological surveys for monitoring fungal propagules and pollen grains. Recent studies have taken advantage of this device to perform DNA-based studies for pollen identification as described by Kraaijeveld and coworkers (47), who obtained promising results, especially for grass pollen. Here we assessed the use of the Hirst spore trap for monitoring not only pollen but also fungal propagules and airborne bacteria by high-throughput sequencing.
Previous studies analyzing the Hirst spore sampler established that there are no significant differences in counts or diversity among samples collected by two devices placed in the same location, although error rates of around 20% to 30% during visual identification must be assumed (48). Our results from DNA sequence analyses showed a consistent sampling of the air biodiversity between the two samplers working synchronically, excluding assay A because the technical issues observed might have been interfering. Differences in the proportions of OTUs detected by the two devices ranged from 17% to 27% for all the biological particles studied (13% to 23% in analysis of diversity by phylotypes). The OTUs in common (47% to 57%) composed ≥92% of the total relative abundance in each sampler for fungal and pollen (≥87% in the case of bacteria), indicating that the microscopic biological diversity is well represented in each sample and that the differences are due to minor representatives with a small contribution from each one. It is worthwhile to point out that the OTU level set at 97% similarity does not always correlate with different species. In fact, in the analyses of phylotypes in contrast to OTUs, the percentages in common were increased to 54% to 71%, corresponding to ≥95% of the total relative abundance. Accordingly, simpler organisms (bacteria and fungi) showed higher diversity (with respect to numbers of OTUs), likely because of their potential for genetic diversification, but they also showed higher percentages of phylotypes in common. Additionally, these scores correlated with almost perfect concordance between the devices for pollen (CCC ≥ 0.99) and substantial concordance for fungi (CCC ≥ 0.98), confirming previous studies that investigated sampling consistency by microscopy (49, 50). Lower values were determined for bacteria (CCC ≥ 0.95 [moderate concordance]), probably because of greater diversity, although these devices had never been evaluated previously for prokaryotes by NGS, and more studies may be required. Overall, these results suggest high consistency during collection of the airborne biological particles by DNA sequencing technologies employing the Hirst spore trap for global characterization.
We also compared NGS results with traditional identification of fungal and pollen diversity performed by light microscopy since it is essential to validate the results obtained from DNA sequencing. Qualitative (detected/nondetected) differences can be expected since the two methods analyze different items (DNA versus complete pollen grains or fungal spores), with the expectation that NGS would be more sensitive. Moreover, we used the DNA extracted from the whole half-tape used in the sampler, whereas the standard procedure for identification by microscopy analyzes only 10% to 15% of the tape (51). Therefore, some divergences were observed during the analyses. For instance, in the pollen diversity analysis, 9% to 13% of the sequences were assigned to Ulmus and 14% to 22% to Sophora (= Styphnolobium), whereas these pollen types were not detected by microscopy and are not expected to be found in that region according to their phenological calendar (38). Because NGS detects the DNA not only from inside the pollen grains but also from any other plant residues or fragments carrying nucleic acids, this type of disagreement may be found.
With regard to quantitative differences, this is a problem that has yet to be solved for the targeted amplicon sequencing (TAS) strategy. The NGS data may be biased by the number of copies of the targeted region present in the genome. Although quantitative PCR (qPCR) may be applied, performance of additional analyses that focus on a few species is required, as Kraaijeveld and colleagues did for grass pollen (47). Our results using a nuclear marker for plants (5.8S-ITS2) worked fairly well, with good agreement with morphological identification (CCC ≥ 0.96), while CCC values of ≥0.94 (moderate concordance) were determined in comparisons of microscopy and NGS for fungi. Taxonomic classification by visual identification is challenging because of the great diversity of fungi in the air. Comparison with the results from NGS is as yet also hampered because of poor knowledge of the teleomorph and anamorph stages with respect to fungal diversity. Nonetheless, performing visual and DNA sequencing analyses in parallel enriches the information for both approaches, as Pashley and coworkers had also demonstrated (52).
On the other hand, DNA sequencing has very interesting advantages in aerobiological studies. All types of biological entities (bacteria, fungi, pollen, archaea, etc.) can be analyzed using the same sample, and the relationships among them, including groups from different taxonomic kingdoms, can be studied. Moreover, NGS facilitates the identification for those cryptic groups of plants and fungi that are difficult to identify by morphology. For instance, in our analyses, 17 different genera of the family Poaceae were identified across the samples (data not shown) whereas pollen features do not allow such distinction, and other members of 11 orders that are not studied routinely were also detected. Furthermore, the procedure that we have described is sensitive enough to detect not only significant divergences between samples (comparisons with sample M0) but also subtler differences, as between the replicates, which were taken across consecutive weeks and for which great dissimilarities were not expected (see Fig. S4 in the supplemental material).
Our study evaluated the use of the Hirst spore trap for simultaneous monitoring of pollen, fungi, and bacteria by high-throughput DNA sequencing. Sampling in aerobiological stations can easily be adapted to perform both traditional and DNA-based analyses, therefore providing additional information for comprehending the complexity of the bioaresols in urban environments and their connections with meteorological parameters, airborne disease transmission, and physical and chemical pollutants.
MATERIALS AND METHODS
Sampling methodology.
Two volumetric spore traps (Burkard Manufacturing Co., United Kingdom) were placed on the roof (height, 23 m) of the building Escuela Técnica Superior de Ingenieros Industriales (Universidad Politécnica de Madrid, Madrid, Spain [40.439881°N, 3.689409°W, ∼700 m above sea level]), with a 2-m separation. All the material that was to be in contact with the sample was sterilized in advance. The drums employed in the spore trap device, Melinex tape, boxes to carry the drums, and metallic stuff for manipulation were cleaned with 10% sodium hypochlorite solution, rinsed thoroughly with Milli-Q (MQ) water, sprayed with ethanol (70%), and allowed to dry before autoclaving was performed (121°C, 20 min). Further steps after sterilization were performed in a Telstar AV-100 biosafety cabinet, with the use of sterile gloves to prevent DNA contamination. The Melinex tape was cut longitudinally into two equal parts and attached to the drums. Next, it was covered with pharmaceutical sterile petroleum jelly (Vaseline; Interapothek) as an adherence collection surface. One half of the tape was used for DNA extraction and the other half for microscopy analyses. The two spore traps were run synchronously, and three 7-day samples were taken in December 2015 (see Table 1; see also Table S1 in the supplemental material for meteorological conditions). Airflow generated by the vacuum pump of the samplers was checked before and after sampling to verify that the flow rate (∼10 liters/min) was constant across the time period and between devices. An additional 7-day sample was taken in summer in the same location for use as a comparative-divergence sample for NGS analysis and statistics. A negative control was set as a 7-day sample, keeping the vacuum of the sampler off. An amount of DNA insufficient for performing any analysis was obtained, and none of the PCR product was amplified during the preparation of the amplicon libraries using this specimen.
DNA extraction and quantitation.
After sampling, the petroleum jelly from one half of the Melinex tape was collected for analysis in the biosafety cabinet by using a sterilized razor and the DNA was extracted with a PowerSoil DNA isolation kit (Mo Bio Laboratories, CA, USA) following the manufacturer's instructions. Purified DNA was eluted in a final volume of 60 μl and quantified with a Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Invitrogen, Molecular Probes) using a QuantiFluor Fluorometer (Promega). Aliquots from the extracted DNA were used for next-generation DNA sequencing analyses.
Next-generation sequencing.
High-throughput sequencing analyses were performed using the purified DNA from each sample. Universal primers attached to adaptors and multiplex identifier sequences were used to amplify specific regions from 16S rRNA for bacteria [for Bakt_341 (F), 5′-CCTACGGGNGGCWGCAG-3′; for Bakt_805 (R), 5′-GACTACHVGGGTATCTAATCC-3′ (53)], from 5.8S-ITS2 for fungi [for ITS86 (F), 5′-GTGAATCATCGAATCTTTGAA-3′ (54); for ITS-4 (R), 5′-TCCTCCGCTTATTGATATGC-3′ (55)], and for plants [for ITS-D (F), 5′-YGACTCTCGGCAACGGATA-3′ (56); for ITS-4 (R), 5′-TCCTCCGCTTATTGATATGC-3′ (55)], in a targeted amplicon sequencing (TAS) approach. Purified-amplicon libraries were sequenced using an Illumina MiSeq platform (2 × 300 reads) at Parque Científico de Madrid (Madrid, Spain).
Bioinformatic analysis.
Data from NGS were first submitted to a general checking with FastQC software (version 0.11.3; Babraham Bioinformatics Group, Babraham Institute, United Kingdom [www.bioinformatics.babraham.ac.uk/projects/]). Paired-end sequences were assembled with PANDAseq (57) (version 2.8; https://github.com/neufeld/pandaseq/wiki/PANDAseq-Assembler), removing primer sequences and filtering by quality. For the fungal ITS2 library, as the sequencing protocol exceeded the length of the amplicon, we employed “read_fastq” from Biopieces (version 2.0; http://maasha.github.io/biopieces/) to remove the primer sequence at the end of the amplicon followed by “fastq-join” (59) (https://github.com/brwnj/fastq-join) to pair the reads. Global processing was carried out in the Qiime suite environment (60) (version 1.9.1; http://qiime.org). Taxonomic assignments were performed using the Greengenes database (61) for bacteria (version gg_13_8 implemented in Qiime software by default) and UNITE (62) (version no. 7.0; https://unite.ut.ee/) for fungi. A customized database was created for plant assignment. Briefly, 5.8S-ITS2 sequences from local species (based on the work of Gavilán and coworkers [63]) were extracted from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) and formatted for use within the Qiime workflow. The unidentified sequences found during the preliminary tests were subjected to BLAST analysis using Web BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and the database was manually upgraded until the “unassigned” proportion was <3% for the three assays (806 sequences; ∼764 species in the final database).
For 16S analysis of bacteria, chloroplast and mitochondrion sequences were removed. During fungal ITS2 analysis, sequences from other eukaryotic organisms were discarded. Supplementary filtering was carried out in all analyses to remove OTUs with 5 or fewer counts (n ≤ 5) in any sample. OTUs were defined at 97% sequence similarity. Every unique taxonomic assignment was considered a phylotype, independenly of the taxonomic level. For additional information and the rarefaction curves of each library, see Fig. S1 to S3 in the supplemental material.
Fungal and pollen identification and quantification.
One half of the Melinex tape from the samples A1, B1, and C1 was used for morphological determination and quantification by microscopy following the procedures of the Spanish Aerobiological Network (15). The longitudinal half of the Melinex tape was cut into seven equal portions (48 mm in length) which were mounted on the microscopic slide using fuchsin-stained glycerin gelatin as a mounting medium. Samples were observed using a light microscope (Nikon Eclipse 50i and Nikon Eclipse E200) at a magnification of ×400. One and four continuous nonoverlapping horizontal sweeps over the whole slide were analyzed for fungal spores and pollen grains, respectively. Every spore and pollen particle was counted and identified as one spore and pollen type, based on morphological features (17, 18). The number of airborne particles counted was multiplied by a factor that takes into account the volume of air sampled, the area analyzed, and the size of the microscope field of vision used (15). Finally, total pollen or fungal spore counts per week were expressed as the sum of daily mean counts per cubic meter of air (see Table 2 and 3; see also Table S4 in the supplemental material).
Statistics and graphical representation.
Calculations of statistics were performed using the software packages in the R software environment (version V3.2.1; https://www.r-project.org/). The statistical agreement between paired samples (replicate) was determined by calculating the concordance correlation coefficient (CCC) described by Lin (37) using rarefied OTU data filtered (n ≥ 5) for each pair (with the rarefaction depth adjusted to each amplicon; see Fig. S1 to S3 in the supplemental material), and the calculations were performed with the “epiR” package in R (function “epi.ccc” with a confidence level at 95%). The scale of agreement for the test interpretation was set as follows (64): almost perfect (>0.99), substantial (0.99 to 0.95), moderate (0.95 to 0.90), and poor (<0.90). In order to compare the NGS results to the morphological data from microscopy, counts of pollen grains and fungal spores were transformed to relative abundance data in reference to the total count of the sample. For statistical analyses, only pollen or fungal types identified by morphology were selected, the relative abundances of NGS were corrected to 100% (keeping the proportions constant), and subsequent CCC was performed as described above (see Table S2 and S3 in the supplemental material).
Distance matrix analysis, principal-coordinate analysis (PCoA) graph calculations, and analysis of similarities (ANOSIM [which compares the mean of ranked dissimilarities between groups to the mean of ranked dissimilarities within groups]) were performed using Qiime scripts with the same rarefied OTU data used for statistics.
Accession number(s).
Raw sequence data obtained in this study are available in the National Center for Biotechnology Information Sequence Read Archive under accession no. SRP095022.
Supplementary Material
ACKNOWLEDGMENTS
All of us contributed to conceive this study. A.N. performed the experimental design, sample collection, DNA sequence processing, data analysis, and main redaction of the manuscript; G.A.D.P. and Z.F. performed the fungal and pollen identification, helped to analyze the data, and wrote some parts of the manuscript. A.R. and R.G. helped with data analyses and redaction. A.M.G., A.A., A.M.G.-B., and D.A.M. (principal investigator) contributed to redaction.
This study was funded by the Community of Madrid (Spain), under the AIRBIOTA-CM Program (S2013/MAE-2874).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00472-17.
REFERENCES
- 1.Després VR, Huffman JA, Burrows SM, Hoose C, Safatov AS, Buryak G, Fröhlich-Nowoisky J, Elbert W, Andreae MO, Pöschl U, Jaenicke R. 2012. Primary biological aerosol particles in the atmosphere: a review. Tellus B Chem Phys Meteorol 64:15598. doi: 10.3402/tellusb.v64i0.15598. [DOI] [Google Scholar]
- 2.DeLeon-Rodriguez N, Lathem TL, Rodriguez-R LM, Barazesh JM, Anderson BE, Beyersdorf AJ, Ziemba LD, Bergin M, Nenes A, Konstantinidis KT. 2013. Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc Natl Acad Sci U S A 110:2575–2580. doi: 10.1073/pnas.1212089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris CE, Sands DC, Bardin M, Jaenicke R, Vogel B, Leyronas C, Ariya PA, Psenner R. 2011. Microbiology and atmospheric processes: research challenges concerning the impact of airborne micro-organisms on the atmosphere and climate. Biogeosciences 8:17–25. doi: 10.5194/bg-8-17-2011. [DOI] [Google Scholar]
- 4.Šantl-Temkiv T, Sahyoun M, Finster K, Hartmann S, Augustin-Bauditz S, Stratmann F, Wex H, Clauss T, Nielsen NW, Sørensen JH, Korsholmd US, Wickf LY, Karlsona UG. 2015. Characterization of airborne ice-nucleation-active bacteria and bacterial fragments. Atmos Environ 109:105–117. doi: 10.1016/j.atmosenv.2015.02.060. [DOI] [Google Scholar]
- 5.Steiner AL, Brooks SD, Deng C, Thornton DCO, Pendleton MW, Bryant V. 2015. Pollen as atmospheric cloud condensation nuclei. Geophys Res Lett 42:3596–3602. doi: 10.1002/2015GL064060. [DOI] [Google Scholar]
- 6.Fernstrom A, Goldblatt M. 2013. Aerobiology and its role in the transmission of infectious diseases. J Pathog 2013:1–13. doi: 10.1155/2013/493960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fronczek CF, Yoon J-Y. 2015. Biosensors for monitoring airborne pathogens. J Lab Autom 20:390–410. doi: 10.1177/2211068215580935. [DOI] [PubMed] [Google Scholar]
- 8.Simon-Nobbe B, Denk U, Pöll V, Rid R, Breitenbach M. 2008. The spectrum of fungal allergy. Int Arch Allergy Immunol 145:58–86. doi: 10.1159/000107578. [DOI] [PubMed] [Google Scholar]
- 9.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. 2012. Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folloni S, Kagkli D-M, Rajcevic B, Guimarães NCC, Van Droogenbroeck B, Valicente FH, Van den Eede G, Van den Bulcke M. 2012. Detection of airborne genetically modified maize pollen by real-time PCR. Mol Ecol Resour 12:810–821. doi: 10.1111/j.1755-0998.2012.03168.x. [DOI] [PubMed] [Google Scholar]
- 11.Georgakopoulos DG, Després V, Fröhlich-Nowoisky J, Psenner R, Ariya PA, Pósfai M, Ahern HE, Moffett BF, Hill TCJ. 2009. Microbiology and atmospheric processes: biological, physical and chemical characterization of aerosol particles. Biogeosciences 6:721–737. doi: 10.5194/bg-6-721-2009. [DOI] [Google Scholar]
- 12.Ghosh B, Lal H, Srivastava A. 2015. A review of bioaerosols in indoor environment with special reference to sampling, analysis and control mechanisms. Environ Int 85:254–272. doi: 10.1016/j.envint.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez K, Rao C, Burton N. 2004. Exposure assessment and analysis for biological agents. Grana 43:193–208. doi: 10.1080/00173130410000794. [DOI] [Google Scholar]
- 14.Hirst JM. 1952. An automatic volumetric spore-trap. Ann Appl Biol 39:257–265. doi: 10.1111/j.1744-7348.1952.tb00904.x. [DOI] [Google Scholar]
- 15.Galán C, González PC, Teno PA, Vilches ED. 2007. Spanish Aerobiology Network (REA): management and quality manual. Serv. Publicaciones Univ; Córdoba, Córdoba, Spain. [Google Scholar]
- 16.Reference deleted.
- 17.Smith EG. 1990. Sampling and identifying allergenic pollens and molds: an illustrated identification manual for air samplers. Blewstone Press, San Antonio, TX. [Google Scholar]
- 18.Weber RW. 1998. Pollen identification. Ann Allergy Asthma Immunol 80:141–148. doi: 10.1016/S1081-1206(10)62947-X. [DOI] [PubMed] [Google Scholar]
- 19.Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig W, Schleifer KH. 1994. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol Rev 15:155–173. doi: 10.1111/j.1574-6976.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 21.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. 2012. Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc B Biol Sci 270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proc Natl Acad Sci U S A 106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollingsworth PM, Graham SW, Little DP. 2011. Choosing and using a plant DNA barcode. PLoS One 6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coelho SM, Simon N, Ahmed S, Cock JM, Partensky F. 2013. Ecological and evolutionary genomics of marine photosynthetic organisms. Mol Ecol 22:867–907. doi: 10.1111/mec.12000. [DOI] [PubMed] [Google Scholar]
- 26.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A 109:21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López-Bueno A, Tamames J, Velázquez D, Moya A, Quesada A, Alcamí A. 2009. High diversity of the viral community from an Antarctic lake. Science 326:858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- 29.Yoccoz NG, Bråthen KA, Gielly L, Haile J, Edwards ME, Goslar T, Von Stedingk H, Brysting AK, Coissac E, Pompanon F, Sønstebø JH, Miquel C, Valentini A, De Bello F, Chave J, Thuiller W, Wincker P, Cruaud C, Gavory F, Rasmussen M, Gilbert MT, Orlando L, Brochmann C, Willerslev E, Taberlet P. 2012. DNA from soil mirrors plant taxonomic and growth form diversity. Mol Ecol 21:3647–3655. doi: 10.1111/j.1365-294X.2012.05545.x. [DOI] [PubMed] [Google Scholar]
- 30.Núñez A, Amo de Paz G, Rastrojo A, García AM, Alcamí A, Gutiérrez-Bustillo AM, Moreno DA. 2016. Monitoring of airborne biological particles in outdoor atmosphere. Part 2: metagenomics applied to urban environments. Int Microbiol 19:69–80. doi: 10.2436/20.1501.01.265. [DOI] [PubMed] [Google Scholar]
- 31.Fierer N, Liu Z, Rodríguez-Hernández M, Knight R, Henn M, Hernandez MT. 2008. Short-term temporal variability in airborne bacterial and fungal populations. Appl Environ Microbiol 74:200–207. doi: 10.1128/AEM.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lighthart B, Shaffer BT. 1995. Airborne bacteria in the atmospheric surface layer: temporal distribution above a grass seed field. Appl Environ Microbiol 61:1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabariego S, Díaz de la Guardia C, Alba F. 2000. The effect of meteorological factors on the daily variation of airborne fungal spores in Granada (southern Spain). Int J Biometeorol 44:1–5. doi: 10.1007/s004840050131. [DOI] [PubMed] [Google Scholar]
- 34.Burrows SM, Elbert W, Lawrence MG, Pöschl U. 2009. Bacteria in the global atmosphere – part 1: review and synthesis of literature data for different ecosystems. Atmos Chem Phys 9:9263–9280. doi: 10.5194/acp-9-9263-2009. [DOI] [Google Scholar]
- 35.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A 95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin LI-K. 1989. A concordance correlation coefficient to evaluate reproducibility. Biometrics 45:255–268. doi: 10.2307/2532051. [DOI] [PubMed] [Google Scholar]
- 38.Gutiérrez M, Sabariego S, Cervigón P. 2006. Calendario polínico de Madrid (Ciudad Universitaria). Periodo 1994–2004. Lazaroa 27:21–27. http://revistas.ucm.es/index.php/LAZA/article/view/LAZA0606110021A/8927. [Google Scholar]
- 39.De Antoni Zoppas BC, Valencia-Barrera RM, Vergamini Duso SM, Fernández-González D. 2006. Fungal spores prevalent in the aerosol of the city of Caxias do Sul, Rio Grande do Sul, Brazil, over a 2-year period (2001–2002). Aerobiologia 22:117–124. doi: 10.1007/s10453-006-9022-2. [DOI] [Google Scholar]
- 40.Fröhlich-Nowoisky J, Pickersgill DA, Després VR, Pöschl U. 2009. High diversity of fungi in air particulate matter. Proc Natl Acad Sci U S A 106:12814–12819. doi: 10.1073/pnas.0811003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrero AD, Ruiz SS, Bustillo MG, Morales PC. 2006. Study of airborne fungal spores in Madrid, Spain. Aerobiologia 22:133–140. doi: 10.1007/s10453-006-9025-z. [DOI] [Google Scholar]
- 42.Oliveira M, Ribeiro H, Delgado JL, Abreu I. 2009. The effects of meteorological factors on airborne fungal spore concentration in two areas differing in urbanisation level. Int J Biometeorol 53:61–73. doi: 10.1007/s00484-008-0191-2. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto N, Bibby K, Qian J, Hospodsky D, Rismani-Yazdi H, Nazaroff WW, Peccia J. 2012. Particle-size distributions and seasonal diversity of allergenic and pathogenic fungi in outdoor air. ISME J 6:1801–1811. doi: 10.1038/ismej.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertolini V, Gandolfi I, Ambrosini R, Bestetti G, Innocente E, Rampazzo G, Franzetti A. 2013. Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of northern Italy. Appl Microbiol Biotechnol 97:6561–6570. doi: 10.1007/s00253-012-4450-0. [DOI] [PubMed] [Google Scholar]
- 45.Bowers RM, McLetchie S, Knight R, Fierer N. 2011. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J 5:601–612. doi: 10.1038/ismej.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franzetti A, Gandolfi I, Gaspari E, Ambrosini R, Bestetti G. 2011. Seasonal variability of bacteria in fine and coarse urban air particulate matter. Appl Microbiol Biotechnol 90:745–753. doi: 10.1007/s00253-010-3048-7. [DOI] [PubMed] [Google Scholar]
- 47.Kraaijeveld K, de Weger LA, Ventayol García M, Buermans H, Frank J, Hiemstra PS, den Dunnen JT. 2015. Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Mol Ecol Resour 15:8–16. doi: 10.1111/1755-0998.12288. [DOI] [PubMed] [Google Scholar]
- 48.Comtois P, Alcazar P, Néron D. 1999. Pollen counts statistics and its relevance to precision. Aerobiologia 15:19–28. doi: 10.1023/A:1007501017470. [DOI] [Google Scholar]
- 49.Irdi GA, Jones JR, White CM. 2002. Pollen and fungal spore sampling and analysis. Statistical evaluations. Grana 41:44–47. doi: 10.1080/00173130260045495. [DOI] [Google Scholar]
- 50.Tormo Molina R, Maya Manzano JM, Fernández Rodríguez S, Gonzalo Garijo Á, Silva Palacios I. 2013. Influence of environmental factors on measurements with Hirst spore traps. Grana 52:59–70. doi: 10.1080/00173134.2012.718359. [DOI] [Google Scholar]
- 51.Galán C, Smith M, Thibaudon M, Frenguelli G, Oteros J, Gehrig R, Berger U, Clot B, Brandao R. 2014. Pollen monitoring: minimum requirements and reproducibility of analysis. Aerobiologia 30:385–395. doi: 10.1007/s10453-014-9335-5. [DOI] [Google Scholar]
- 52.Pashley CH, Fairs A, Free RC, Wardlaw AJ. 2012. DNA analysis of outdoor air reveals a high degree of fungal diversity, temporal variability, and genera not seen by spore morphology. Fungal Biol 116:214–224. doi: 10.1016/j.funbio.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 53.Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turenne CY, Sanche SE, Hoban DJ, Karlowsky JA, Kabani AM. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J Clin Microbiol 37:1846–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White T, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand D, Shinsky J, White T (ed), PCR protocols: a guide to methods and applications. Academic Press, London, United Kingdom. doi: 10.1016/0307-4412(91)90165-5. [DOI] [Google Scholar]
- 56.Cheng T, Xu C, Lei L, Li C, Zhang Y, Zhou S. 2016. Barcoding the kingdom Plantae: new PCR primers for ITS regions of plants with improved universality and specificity. Mol Ecol Resour 16:138–149. doi: 10.1111/1755-0998.12438. [DOI] [PubMed] [Google Scholar]
- 57.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. 14 February 2012 PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reference deleted.
- 59.Aronesty E. 2013. Comparison of sequencing utility programs. Open Bioinform J 7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 60.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeSantis TZ, Dubosarskiy I, Murray SR, Andersen GL. 2003. Comprehensive aligned sequence construction for automated design of effective probes (CASCADE-P) using 16S rDNA. Bioinformatics 19:1461–1468. doi: 10.1093/bioinformatics/btg200. [DOI] [PubMed] [Google Scholar]
- 62.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH. 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 63.Gavilán R, Echevarría JE, Casas I. 1993. Catálogo de la flora vascular de la Ciudad Universitaria de Madrid (España). Bot Complut 18:175–202. http://revistas.ucm.es/index.php/BOCM/article/view/BOCM9393110175A. [Google Scholar]
- 64.McBride G. 2005. A proposal for strength-of-agreement criteria for Lin.S.concordance correlation coefficient. NIWA client report: HAM2005-062. National Institute of Water & Atmospheric Research Ltd. https://www.medcalc.org/download/pdf/McBride2005.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.