Abstract
In genetic screens for ribosomal export mutants, we identified CFD1, NBP35 and NAR1 as factors involved in ribosome biogenesis. Notably, these components were recently reported to function in extramitochondrial iron–sulfur (Fe–S) cluster biosynthesis. In particular, Nar1 was implicated to generate the Fe–S clusters within Rli1, a potential substrate protein of unknown function. We tested whether the Fe–S protein Rli1 functions in ribosome formation. We report that rli1 mutants are impaired in pre-rRNA processing and defective in the export of both ribosomal subunits. In addition, Rli1p is associated with both pre-40S particles and mature 40S subunits, and with the eIF3 translation initiation factor complex. Our data reveal an unexpected link between ribosome biogenesis and the biosynthetic pathway of cytoplasmic Fe–S proteins.
Keywords: eIF3, iron–sulfur cluster, Nar1, ribosome export, Rli1
Introduction
The ribosome is a complex ribonucleoprotein assembly that carries out the protein synthesis. The eukaryotic ribosome consists of a 60S subunit, which contains 25S, 5.8S and 5S rRNAs and ribosomal L-proteins, and a 40S subunit composed of the 18S rRNA and ribosomal S-proteins. The formation of ribosomal particles has been extensively studied in the yeast Saccharomyces cerevisiae by genetic and biochemical analyses. This yielded considerable insight into a most complicated process of pre-rRNA processing and pre-ribosome assembly (Kressler et al, 1999; Venema and Tollervey, 1999; Tschochner and Hurt, 2003). The complexity of ribosome formation is underscored by the observations that more than 150 non-ribosomal proteins and myriad of small nucleolar RNAs (snoRNAs) participate in pre-rRNA formation and processing, and formation of pre-ribosomal particles (Kressler et al, 1999; Samarsky and Fournier, 1999; Tschochner and Hurt, 2003). However, the function of the vast majority of these proteins remains unknown.
In a first step, the 35S pre-rRNA is synthesized by RNA polymerase I, and an early precursor particle, the 90S pre-ribosome, is formed in the nucleolus, which carries many rRNA processing and assembly factors. This 90S pre-ribosomal particle then undergoes a series of processing and maturation steps, of which the cleavage of the 35S pre-rRNA at sites A2 and A3 initiates the separation of pre-40S and pre-60S particles (Venema and Tollervey, 1999). Subsequently, these two branches follow a separate biogenesis pathway, ending with the export of 60S and 40S subunits to the cytoplasm. Nuclear export of the 60S pre-ribosome to the cytoplasm requires the Nmd3 adaptor protein, which exhibits a leucine-rich nuclear export sequence (NES) that is recognized by the general export receptor Xpo1/Crm1 (Fornerod et al, 1997; Fukuda et al, 1997; Ho et al, 2000; Gadal et al, 2001b; Thomas and Kutay, 2003; Trotta et al, 2003). The mechanism of 40S subunit export is unknown, but Xpo1/Crm1 and the Ran cycle have been implicated in this process (Moy and Silver, 2002). Another Ran-binding protein, Rrp12, has also been implicated in the export of both ribosomal subunits (Oeffinger et al, 2004).
Insight into the coupling between ribosome biogenesis and export of subunits to the cytoplasm was derived from various in vivo and in situ hybridization approaches including a GFP-based assay that monitors intracellular location of GFP-labeled large and small subunit ribosomal proteins (Hurt et al, 1999; Stage-Zimmermann et al, 2000; Gleizes et al, 2001; Milkereit et al, 2003; Schäfer et al, 2003). Using the GFP-based assay for the 60S subunit export, a screen of randomly generated temperature-sensitive (ts) mutants yielded many ribosomal export (rix) mutants (Gadal et al, 2001a, 2001b, 2002).
We report that one of these rix mutants, rix19-1, is complemented by the YIL003W gene (also known as CFD1 or DRE3) that encodes an essential and highly conserved putative P-loop ATPase. Notably, Cfd1p was recently shown to play an essential role in the biogenesis of cytoplasmic iron–sulfur (Fe–S) proteins (Roy et al, 2003). Fe–S proteins are an ancient group of proteins found in all life forms. As a characteristic feature they contain different types of iron–sulfur clusters (ISCs) (Lill and Kispal, 2000). Fe–S proteins are involved in a variety of cellular processes, including intermediary metabolism, electron transport and gene expression.
Moreover, we have shown that two additional factors, Nbp35 and Nar1, which are functionally linked to Cfd1, have also roles in ribosome export (this work) and in extramitochondrial ISC biogenesis (Kispal et al, 2005). Importantly, we have found that the cytoplasmically located Fe–S protein Rli1, which requires Nar1 for its ISC biogenesis, is associated with the eIF3 translation initiation complex, of which eIF3j is also involved in 20S to 18S rRNA processing (Valasek et al, 2001). Thus, our data revealed a novel link between ribosome biogenesis and formation of ISCs in extramitchondrial proteins. During the course of this work, Hinnebusch and co-workers (Dong et al, 2004) reported that Rli1 functions in translation by promoting preinitiation complex assembly.
Results
In a genetic screen, which detects ribosome export defects (Gadal et al, 2001b), we identified the ts rix19-1 mutant (Figure 1A), which showed a significant nuclear accumulation of the 60S subunit reporter Rpl25-GFP after shift to the restrictive temperature (Figure 1B). Moreover, rix19-1 also exhibited a 40S subunit export defect (Figure 1C). The gene complementing the ts phenotype and ribosomal export defect of rix19-1 was cloned by complementation and shown to be the CFD1/YIL003W gene (Figure 1A and B). Surprisingly, CFD1 was previously reported to be involved in the assembly of ISCs in non-mitochondrial cytoplasmic proteins (Roy et al, 2003). Rix19 is highly homologous to Nbp35 (YGL091c), which is also an essential putative ATPase of the ParA/MinD protein family (Vitale et al, 1996). The major difference between these two proteins is the presence of an additional cysteine-rich N-terminal region in Nbp35 that has the potential to coordinate ISCs (R Lill, personal communication) and was shown to be essential for cell viability (Vitale et al, 1996). Interestingly, similar pairs of nucleotide binding proteins (Cfd1-Nbp35) can be found in all eukaryotic organisms suggesting a conserved function for this family of putative ATPases.
Figure 1.
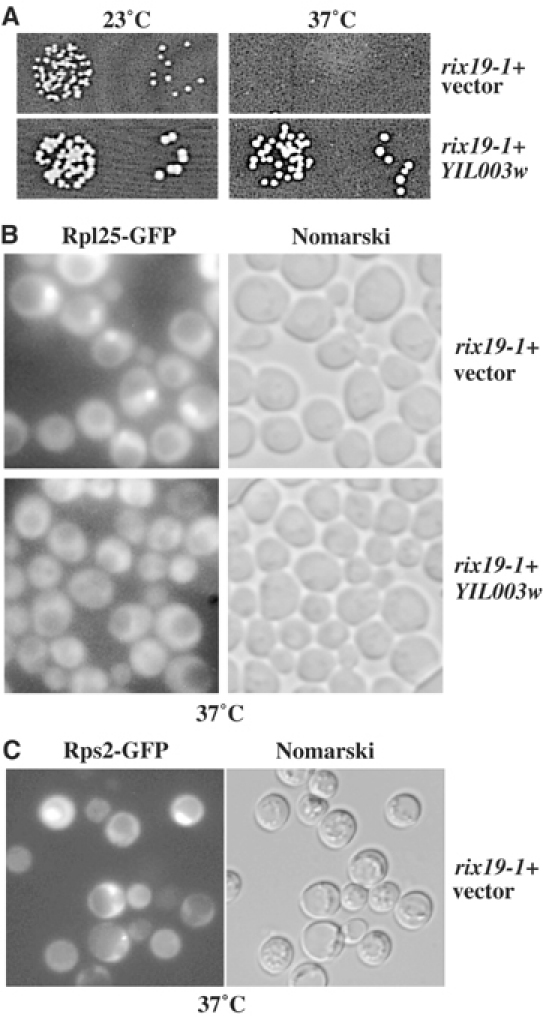
Cloning of RIX19. YIL003w can complement the ts (A) and rix phenotype (B) of the rix19-1 allele. The strain rix19-1 was transformed with pRS315 plasmid either without insert or containing YIL003w. (B, C) Rps2-GFP and Rpl25-GFP accumulation in rix19-1. Cells with the indicated reporter plasmids were examined in the fluorescence microscope 2 h after shift from 23 to 37°C.
To find out whether Nbp35 is also involved in ribosomal export, we generated ts nbp35 mutants and tested them for defects in ribosomal subunit export. Strains carrying the nbp35-1 ts allele (Figure 2A) accumulated both 60S and 40S subunits in the nucleolus/nucleus after shift to the nonpermissive temperature (Figure 2B). To test whether CFD1 and NBP35 functionally interact, we combined rix19-1 and nbp35-1 alleles in a haploid cell. The double mutant was not viable, indicating a synthetic lethal (sl) interaction (Figure 2C). We conclude that Rix19 and Nbp35 have both unique and overlapping functions required for export of both ribosomal subunits.
Figure 2.
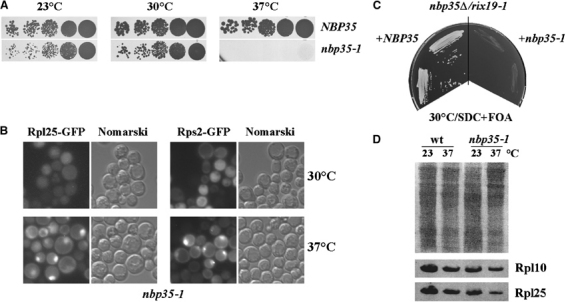
NBP35 is genetically linked to RIX19. (A) The NBP35 shuffle strain (nbp35∷KAN-MX4+pRS316-NBP35) was transformed with pRS315 plasmid containing nbp35-1 allele or the wild-type NBP35. After FOA selection, cells were grown on YPD plates for 5 days. (B) Rps2-GFP and Rpl25-GFP accumulation in the nbp35-1 ts mutant, which was transformed with indicated reporter plasmids. Exponentially growing cells were shifted from 30 to 37°C and examined after 4 h in the fluorescence microscope. (C) rix19-1 strain was crossed with the NBP35 shuffle strain. After tetrad analysis, a resulting rix19-1/nb35Δ haploid was transformed with NBP35 or nbp35-1. Two individual transformants were grown on the SDC plate containing 5-FOA for 5 days at 30°C. (D) The levels of ribosomal proteins Rpl10 and Rpl25 are not elevated in nbp35-1 strains. Overnight cultures of indicated stains were diluted to OD600 0.2 and grown at 23 or 37°C for 4 h. Whole cell lysates were separated by SDS–PAGE and transferred to nitrocellulose. Ponceau S staining (upper panel) revealed the protein loading, and the amount of Rpl10 and Rpl25 was determined by Western using specific antibodies (lower panel).
CFD1/RIX91 was recently identified in a screen for mutations that derepressed synthesis of certain ribosomal proteins (J Woolford, personal communication). To rule out that the observed nuclear accumulation of ribosomal proteins is due to overproduction, we analyzed by Western the level of Rpl10 and Rpl25 in rix19-1 and nbp35-1 cells. However, the levels of these ribosomal proteins were rather decreased than increased after shift to the restrictive temperature in both rix19-1 (not shown) and nbp35-1 strains (Figure 2D). This could be due to downregulation of ribosomal protein expression in response to a mild heat shock (Warner, 1999).
To characterize the function of RIX19/CFD1 in ribosome biogenesis, we performed an sl screen with the rix19-1 allele. Among 105 colonies screened, we found two sl mutants, which were complemented by NBP35 (data not shown) and NAR1/YNL240C, respectively (Figure 3A).
Figure 3.
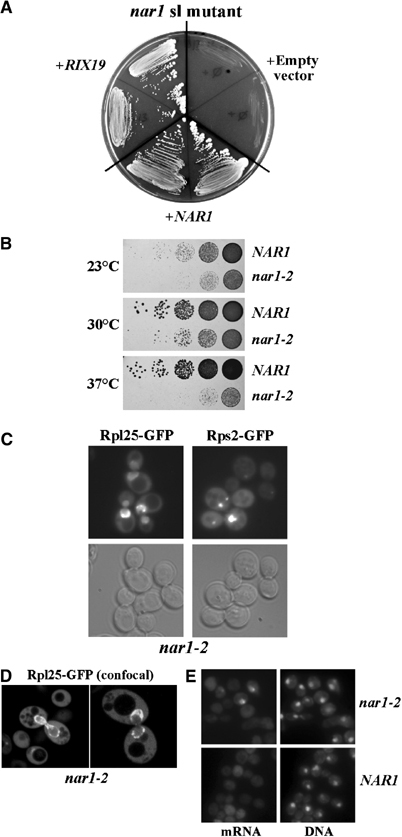
NAR1 is genetically linked to RIX19 and is required for nuclear export of ribosomes. (A) The rix19/nar sl mutant was transformed with pRS315 plasmid without insert or containing NAR1 or RIX19. Cells were grown on 5-FOA containing SDC plates for 5 days at 30°C. (B) The NAR1 shuffle strain was transformed with pRS315 plasmids containing either NAR1 or nar1-2. After plasmid shuffling on 5-FOA containing SDC plates, the cells were grown in serial dilutions on YPD plates at the indicated temperatures for 3 days. (C, D) nar1-2 strain was transformed with Rps2-GFP and Rpl25-GFP plasmids. Cells growing in liquid SDC medium at 30°C were observed in the fluorescence (C) and confocal (D) microscope. (E) Poly(A)+ RNA localization in nar1-2 cells, grown at 30°C, was analyzed by in situ hybridization.
NAR1 (nuclear architecture related) is an essential gene that is highly homologous to bacterial Fe-only hydrogenases, in which six ISCs form the active site and are involved in electron transport reactions (Peters et al, 1998). Since Nar1 lacks two of the six ISCs, it is, however, unlikely that it has enzymatic hydrogenase activity (Barton and Worman, 1999). There are two human homologs of yeast Nar1, NARFL and NARF. One of them, NARF (nuclear prelamin A recognition factor), was found to interact with the prenylated form of the lamin A precursor and was localized to the nuclear envelope and nucleus (Barton and Worman, 1999). Since lamins are absent from yeast, the function of Nar1 was unclear.
To assess whether Nar1 has a role in nuclear export of ribosomes, we generated nar1 mutant alleles by in vitro mutagenesis (Figure 3B) and tested them for defects in nuclear export mechanisms (Figure 3C–E). As shown in Figure 3C and D, nar1-2 mutant cells exhibited a strong nuclear accumulation of the large subunit reporter Rpl25-GFP in the nucleus with a significant concentration at the nuclear periphery both at 23 and 37°C (note that nar1-2 cells are also impaired in growth at permissive temperatures, for example, 23 and 30°C; see Figure 3B). Such a striking accumulation of Rpl25-GFP around the nuclear envelope has not been observed in other rix mutants. The significance of this perinuclear accumulation is unclear, but this finding is consistent with a model in which Nar1 is required for a late step in ribosomal biogenesis and/or nuclear export (see below). In addition, we observed nuclear accumulation of poly(A)+ RNA in nar1-2 cells (Figure 3E). However, a smaller fraction of the nar1-2 cells accumulated poly(A)+ RNA (10%), whereas accumulation of ribosomal subunits was observed in >50% of the cells. In contrast, cfd1 and nbp35 mutant cells did not exhibit an mRNA export defect (data not shown). We assume that nuclear accumulation of mRNA in nar1 mutant cells could be a secondary effect. It is possible that accumulation of pre-ribosomes at the nuclear periphery in nar1-2 cells competes with mRNA export.
In summary, our data have shown that three essential and conserved proteins, Rix19, Nbp35 and Nar1, are required for the nuclear export of ribosomal subunits. Surprisingly, recent reports suggested that these proteins are required for the assembly of ISCs in non-mitochondrial proteins (Roy et al, 2003; Kispal et al, 2005). Thus, it seemed possible that an essential factor involved in ribosome biogenesis is a Fe–S protein, whose biogenesis depends on the function of the Rix19–Nbp35–Nar1 machinery.
We speculated that the uncharacterized protein Rli1 could be this factor, based on the observations that (i) Rli1 is an essential cytoplasmic Fe–S protein whose ISC formation is inhibited in Rix19/Cfd1- and Nar1-depleted cells (Kispal et al, 2005) and (ii) Rli1 was predicted to be linked to translation initiation (Letovsky and Kasif, 2003). In addition, Rli1 belongs to the ATP-binding cassette (ABC) superfamily (Decottignies and Goffeau, 1997), and its human homolog, RLI (RNase L inhibitor), serves as a regulator of the RNAse L-dependent antiviral activity (Bisbal et al, 1995). However, yeast does not contain an RNAse L ortholog, leaving the function of Rli1 unclear.
To investigate the link between Rli1 and ribosome export, we analyzed a yeast strain in which RLI1 was under the control of a tetracycline-regulated promoter (Hughes et al, 2000). Following doxycycline treatment of tet:rli1 cells, which represses RLI1, we observed a strong accumulation of the Rpl25-GFP reporter in the nucleus (Figure 4A). Moreover, nuclear export of the small subunit reporter Rps2-GFP was inhibited in these cells (Figure 4A). This shows that nuclear export of both 60S and 40S subunits is impaired in RLI1-repressed cells. In contrast to nar1 mutant cells, no mRNA export defect was observed in rli1-depleted cells (data not shown).
Figure 4.
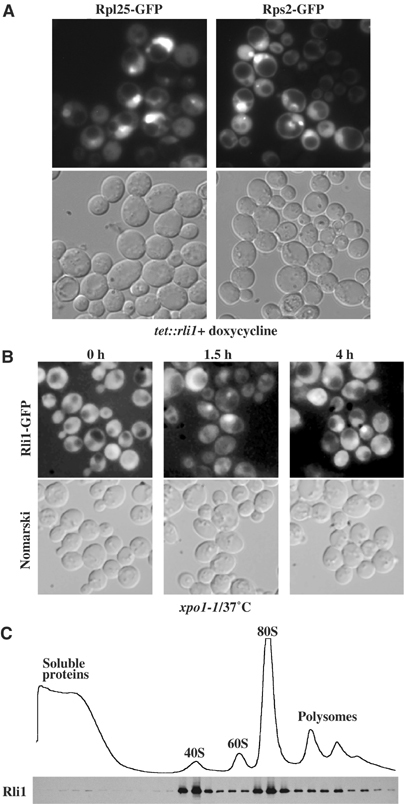
Rli1 is a shuttling protein involved in ribosomal export. (A) A yeast strain, in which the endogenous RLI1 promoter was replaced by the TetOFF promoter, was transformed with Rpl25-GFP and Rps2-GFP plasmids. The cells were grown in liquid SDC-leu medium at 30°C. At early exponential growth (OD 0.2), doxycycline was added to a final concentration of 5 μg/ml. Cells were examined after 12 h of incubation. (B) The RLI1-GFP/xpo1-1 cells were grown in YPD at 30°C to OD600 0.2 before shift to 37°C. After 1.5 and 4 h of incubation at 37°C, cells were examined in the fluorescence microscope. (C) Rli1 mainly cosediments with 40S subunits and 80S ribosomes. Rli1-TAP was visualized in the fractions derived from the sucrose gradient by Western using anti-protein A antibodies.
Consistent with the apparent role of Rli1 in ribosomal subunit export, Hinnebusch and co-workers (Dong et al, 2004) reported that Rli1-GFP has both a cytoplasmic and nuclear location. To find out whether Rli1 can shuttle between the nucleus and the cytoplasm, Rli1-GFP was expressed in the ts xpo1-1 mutant, which is impaired in nuclear export of NES-containing cargo proteins (Stade et al, 1997). As shown in Figure 4B, after shift to the nonpermissive temperature, Rli1-GFP accumulates in xpo1-1 cells, suggesting that Rli1 is able to shuttle between the nucleus and the cytoplasm.
To assess whether Rli1 is directly associated with (pre-) ribosomal particles, the sedimentation of tagged Rli1-TAP was analyzed on sucrose gradients. This showed that Rli1 is associated with 40S subunits, 80S ribosomes and possibly polysomes under low-salt conditions (Figure 4C). A minor pool of Rli1p was also found in the soluble fractions. In contrast, Rix19, Nbp35 and Nar1 were predominantly found in the low molecular weight fractions of the sucrose gradient (data not shown). When the salt concentration in the sucrose gradient was increased to 150 mM, significant amount of Rli1 was shifted to the soluble fractions (not shown).
Northern hybridization and pulse-chase labeling were used to determine whether Rli1 depletion inhibits rRNA maturation (see Figure 5A for scheme of rRNA processing). Accumulation of the early 35S pre-rRNA was seen by 4 h after doxycycline addition, together with reduced levels of the 27SA2 and 20S pre-rRNAs, demonstrating the inhibition of processing steps on the pathway of 18S rRNA synthesis (Figure 5B). Defects in 60S synthesis were also apparent from elevated levels of the 27SB pre-rRNA (Figure 5B). At later time points, the mature 25S and 5.8S rRNAs were also reduced (Figure 5B and C). The mature rRNAs are stable and must be depleted by growth, resulting in slower alterations in their abundance than are seen for the pre-rRNA. To confirm this apparent reduction in rRNA synthesis, metabolic labeling was performed with [5,6-3H]uracil (Figure 5D). In the absence of doxycycline, no clear differences in rRNA synthesis between the wild-type and tet-rli1 strains were detected (data not shown). Following treatment with doxycycline for 6 h (Figure 5D), the 35S and 32S pre-rRNAs were more prominent in the Rli1-depleted strain, clearly showing impaired pre-rRNA processing. In contrast, synthesis of the mature 25S and 18S rRNA was strongly reduced.
Figure 5.
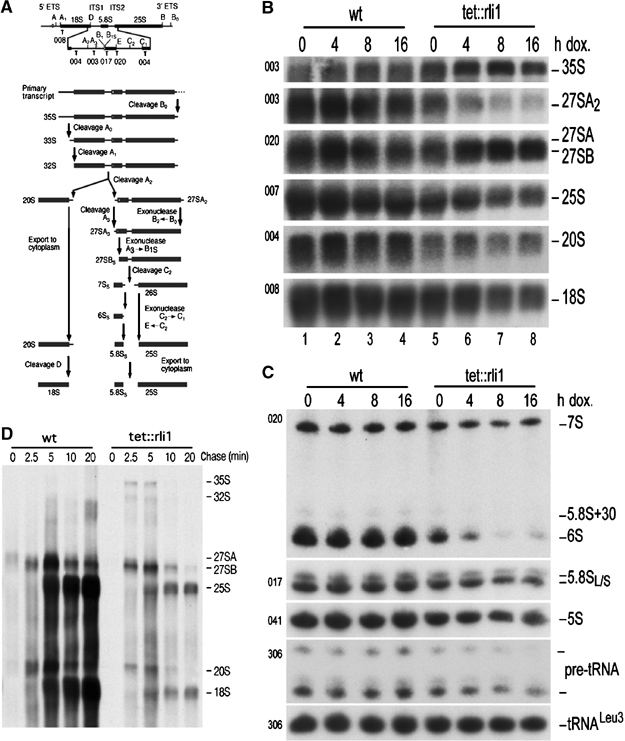
Rli1 is involved in pre-rRNA maturation. (A) Schematic of the pre-rRNA showing the positions of the oligonucleotide probes used for hybridization. Total RNA was extracted from strains expressing wild-type or Tet-regulated RLI1 (tet∷rli1) during growth in the absence of doxycycline (0 h samples), and at time points following doxycycline addition. (B) Northern analyses of high molecular weight RNAs separated on a 1.2% agarose/formaldehyde gel. (C) Northern analyses of low molecular weight RNAs separated on an 8% polyacrylamide/urea gel. Pre-rRNAs and rRNAs detected are indicated on the right of the figure. Oligonucleotides used are listed on the left. tRNALeu was used as a loading control (oligonucleotide 306). (D) Pulse-chase labeling of high molecular weight RNAs. Wild-type and tet∷rli1 strains were pregrown in minimal medium and treated with doxycycline for 6 h prior to pulse labeling with [5,6-3H]uracil for 2 min. A large excess of unlabeled uracil was added and samples were taken immediately (0 min) and at the time points indicated. RNA was extracted, separated on a 1.2% agarose/formaldehyde gel, transferred to a nylon membrane and visualized by fluorography.
Analysis of low molecular weight RNAs (Figure 5C) revealed the striking loss of the 6S pre-rRNA, which is a late precursor in the synthesis of mature 5.8S rRNA. Notably, this phenotype has been observed in several other strains with defects in the export of pre-60S ribosomes (Gadal et al, 2001a, 2001b; Milkereit et al, 2001; Neumann et al, 2003; Galani et al, 2005). We conclude that depletion of Rli1 impairs the processing of pre-rRNAs on both the 40S and 60S subunit synthesis pathways.
In order to determine which pre-rRNA species are associated with Rli1, rRNAs co-precipitated with Rli1-TAP were analyzed by Northern hybridization (Figure 6A). As a control, the precipitation was performed in parallel with TAP-tagged Ded1, a DEAD box ‘RNA helicase' that functions as a translation factor and associates with both pre-40S particles and mature 40S subunits (de la Cruz et al, 1997; Schäfer et al, 2003; Berthelot et al, 2004; Fairman et al, 2004). Comparison with the otherwise isogenic nontagged strains clearly supported the association of both Ded1 and Rli1 with the 20S pre-rRNA and therefore with pre-40S ribosomes (Figure 6A). The 7S pre-rRNA was also recovered in both the Rli1-TAP and Ded1-TAP precipitates at higher levels than in the control strains, suggesting an association with late pre-60S ribosomes. Consistent with the gradient analyses, co-precipitation with Rli1-TAP was seen for the mature rRNAs from both the 40S subunit (18S rRNA) and the 60S subunit (25S, 5.8S and 5S rRNAs). In contrast, no co-precipitation was detected for pre-rRNAs that are earlier in the synthesis pathway (35S, 27SA and 27SB pre-rRNAs), for the snoRNAs U3 and snR30, which are associated with the early 90S pre-ribosomes, or for tRNALeu.
Figure 6.
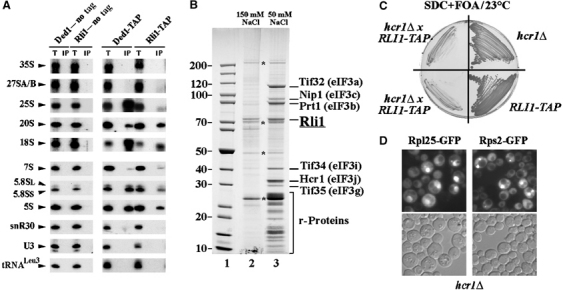
Rli1 is associated with pre-rRNA, rRNA and eIF3. (A) Immunoprecipitation of TAP-tagged Rli1 and Ded1 was performed in the presence of 150 mM NaCl using IgG-Sepharose. RNAs were extracted from the pellets obtained after precipitation (lanes IP) or from an amount of total extract corresponding to 1/30 of that used for the precipitation (lanes T). Co-precipitated RNAs were identified by Northern hybridization. (B) TAP purification of Rli1 was performed in the presence of 150 mM NaCl (lane 2) or 50 mM NaCl (lane 3). Lane 1 depicts a protein standard. Bands marked with asterisks are the commonly found contaminants in TAP purifications. (C) RLI1 and HCR1 are genetically linked. The hcr1Δ strain pretransformed with pRS315-RLI1 plasmid was crossed with RLI1-TAP strain. After subsequent tetrad analysis, two haploid strains from one tetrad carrying both mutations were selected and plated on FOA-containing media together with parental strains. (D) Ribosomal export is impaired in hcr1Δ cells. hcr1Δ strain was transformed with Rpl25-GFP or Rps2-GFP plasmids. The cells were grown in liquid SDC-leu medium at 30°C. Cells were examined in the fluorescence microscope.
To identify proteins that might interact physically with Rli1, two-step affinity purification of a TAP-tagged version of Rli1 was performed. When the protein analysis was performed following isolation in the presence of 150 mM salt, insufficient proteins were specifically co-precipitated with Rli1-TAP to allow their identification (Figure 6B, lane 2). However, when the Rli1-TAP purification was performed in the presence of 50 mM salt, several strongly copurifying proteins were seen (Figure 6B, lane 3). Notably, these associated proteins were the subunits of eIF3 (identified by mass spectrometry), which constitutes a eukaryotic translation initiation factor complex (Browning et al, 2001) and ribosomal S-proteins (rps; see also Supplementary Figure 1). The Hcr1/eIF3j band appeared to be particularly strongly co-precipitated with Rli1-TAP in comparison with other eIF3 subunits (Figure 6B, lane 3). However, a differential staining could also account for the apparent higher amount of Hcr1 in the Rli1-TAP preparation. Notably, eIF3j was previously reported to be a substoichiometric component of eIF3 complex (Valasek et al, 2001). Visible enrichment of Hcr1/eIF3j in Rli1-TAP purification suggests that these proteins may directly bind to each other, a possibility that is supported by two-hybrid data (Ito et al, 2001; Kispal et al, 2005). Consistent with our previous data (Figures 4C, 5D and 6A), small ribosomal subunit proteins were also copurified with Rli1.
The direct interaction between Rli1 and Hcr1 could suggest that Hcr1 links Rli1 to eIF3. An experiment to test this hypothesis would be a TAP purification of Rli1 in the absence of Hcr1. However, an attempt to construct the strain, in which TAP-tagged Rli1 would be combined with the deletion of eIF3j/Hcr1, was unsuccessful. The resulting RLI1-TAP/hcr1Δ haploid strains were viable only in the presence of wild-type, nontagged copy of Rli1 (Figure 6C). Thus, the TAP tag on Rli1 is unfavorable in a yeast strain lacking HCR1. These genetic data suggest that Rli1 and Hcr1 exhibit a functional interaction.
Since HCR1 is a nonessential gene, we tested whether an hcr1Δ strain exhibits ribosomal export defects. This analysis showed that logarithmically growing hcr1Δ cells strongly accumulated both ribosomal 60S and 40S subunits in the nucleus (Figure 6D). In contrast, neither mutations in another subunit of eIF3, eIF3c/Rip1 (T Schäfer, unpublished data), or in subunits of eIF2B, eIF2Bγ/Gcd1 and eIF2Bα/Sui2 (Senger et al, 2001), cause pronounced defects in synthesis and export of ribosomes. Therefore, we conclude that involvement of Rli1 and Hcr1 in biogenesis of ribosomes is specific, which is not observed for other initiation factors.
Discussion
The key finding of this work is that the predominantly cytoplasmic protein Rli1, which contains essential ISCs in its N-domain is required for normal rRNA maturation and nuclear export of 60S and 40S subunits. In addition, mutations in the components of extramitochondrial ISC biogenesis machinery, Rix19/Cfd1, Nbp35 and Nar1, also induce pronounced defects in ribosome biogenesis and export. Altogether, the data suggest that the rix phenotype observed in rix19, nbp35 and nar1 mutants is indirect and could be caused by a failure to assemble the ISCs in Rli1. As a consequence, Rli1 can no longer perform its role in rRNA processing and ribosome export (this work). The essential role of ISCs in Rli1 function was confirmed by mutational analysis (Kispal et al, 2005).
Northern analysis of an Rli1-depleted strain revealed a defect in processing of the 7S pre-rRNA to the mature 5.8S rRNA, which is one of the last steps in 60S subunit biogenesis (Venema and Tollervey, 1999). Co-precipitation of the 7S pre-rRNA with Rli1-TAP was weak, but clearly above the background in an otherwise isogenic, nontagged strain, suggesting a direct interaction. This late maturation step is performed by the concerted action of a number of 3′>5′ exonucleases, many of which are part of the exosome (Allmang et al, 1999; Faber et al, 2002). Interestingly, several analyses have indicated a connection between 5.8S rRNA maturation and 60S subunit export. Examples include the late 60S biogenesis factors Rrp12 (Oeffinger et al, 2004) and Rix1 complex members (Galani et al, 2005), the 60S nuclear export factor Nmd3 (Ho and Johnson, 1999) and the RanGTPase Gsp1, which regulates nucleocytoplasmic transport in general (Suzuki et al, 2001). The precise role of Rli1 in RNA processing and export remains unknown, but its homology to the human ribonuclease L inhibitor RLI suggests that Rli1 might regulate 7S pre-rRNA processing. In addition, Rli1-TAP was associated with 20S pre-rRNA (Figure 6A), showing its association with pre-40S particles. In principle, this could occur in either the nucleus or the cytoplasm, but the evidence for nuclear–cytoplasmic shuttling of Rli1 and for a defect in the export of pre-40S subunits in the rli1 mutant strain both suggest that it interacts with nuclear pre-40S particles, perhaps remaining associated during export. In the Rli1-depleted strain, the level of 20S pre-rRNA was reduced, indicating that the nuclear-restricted 20S pre-rRNA is relatively unstable in this background.
In addition to its role in ribosome synthesis, we have found that Rli1 is physically associated with the translation initiation factor eIF3 and with 40S and 80S ribosomes. Using co-immunoprecipitation, which is less stringent method than TAP, Hinnebusch and co-workers (Dong et al, 2004) identified by Western blotting that Rli1 is associated with eIF2, eIF3 and eIF5. Our results from Rli1-TAP purification and subsequent mass spectrometry analysis suggest that Rli1 directly interacts with eIF3, and interaction of eIF2 and eIF5 might be indirect (Figure 6B).
The interaction of Rli1 with initiation factors suggests a role in the initiation of protein translation. The recent experimental data confirmed that Rli1 is required for efficient formation and stabilization of 43S and 48S preinitiation complexes (Dong et al, 2004). The eIF3j/Hcr1 subunit of eIF3, a substoichiometric component of eIF3 (Valasek et al, 2003), was specifically enriched as compared to other eIF3 subunits copurified with Rli1-TAP (Figure 6B). Therefore, Hcr1 might be the direct interaction partner of Rli1 within eIF3. This conclusion is consistent with two-hybrid data (Ito et al, 2001; Kispal et al, 2005) and co-immunoprecipitation data, which show relatively more efficient association between Rli1 and Hcr1 compared to other translation initiation factors (Dong et al, 2004).
Notably, eIF3j/Hcr1 functions in the maturation of 20S pre-rRNA to 18S rRNA, the terminal step in 40S subunit maturation (Valasek et al, 2001). The existence of a functional link between Rli1 and Hcr1 is supported by their genetic interaction (Figure 6C). Thus, Rli1 could also be involved in 40S biogenesis, consistent with the co-precipitation data (Figure 6A) and the finding that rli1 mutants are impaired in 40S subunit export (Figure 4A) and maturation (Figure 5D).
We can suggest the following model. In actively growing cells, the major pool of Rli1 is localized in the cytoplasm and required for efficient formation of 43S and 48S preinitiation complexes (Dong et al, 2004). At the same time, a minor pool of Rli1 is able to enter the nucleus, where it plays an essential role in 7S to 5.8S pre-rRNA processing and also participates in the export of the pre-40S particle. Notably, another translation initiation factor, Ded1, was also found to associate with both pre-40S and pre-60S subunits (Figure 6A) (de la Cruz et al, 1997; Schäfer et al, 2003; Berthelot et al, 2004) indicating that this dual localization is not unique to Rli1.
Due to their remarkable ability for structural rearrangements induced by electron uptake/donation (Beinert and Kiley, 1999), ISCs within Rli1 could serve as sensors for the concentration of reactive oxygen species (ROS) in an immediate environment and control the biological activity of the protein. The concentration of ROS inside of the cell is a very important parameter, which can be altered directly or indirectly according to many vital external and internal stimuli (Morel and Barouki, 1999). In response to the oxidative stress, Rli1 could efficiently downregulate both ribosomal biogenesis and initiation of translation, adapting the cell to changing environmental and intercellular conditions.
In conclusion, these results demonstrate for the first time the essential role of the cytoplasmic Fe–S protein Rli1 and components of its biogenesis pathway in the maturation and export of ribosomal subunits. In addition, physical and genetic interactions between Rli1 and the eIF3j/Hcr1 subunit of translation initiation factor 3 suggest presence of a new functional link between ribosomal biogenesis, initiation of translation and maturation of ISCs.
Materials and methods
Yeast strains and plasmids
Yeast strains and plasmids used in this study are shown in Supplementary data.
Sucrose density gradient centrifugation
Sedimentation analysis was performed essentially as described (Baßler et al, 2001). Briefly, cells were lysed in cold buffer A (20 mM Tris–HCl pH 7.5, 10 mM KCl, 5 mM MgCl2 and 1 mM dithiothreitol (DTT)). In all, 5 OD (260 nm) of the lysate was loaded on a 10–50% sucrose gradient prepared in buffer A.
rRNA analysis
Northern hybridization was performed as described using whole cell extracts (Beltrame and Tollervey, 1992; Tollervey et al, 1993). Oligonucleotides used were: 003: TGT TAC CTC TGG GCC C; 004: CGG TTT TAA TTG TCC TA; 007: CTC CGC TTA TTG ATA TGC; 008: CAT GGC TTA ATC TTT GAG AC; 017: GCG TTG TTC ATC GAT GC; 020: TGA GAA GGA AAT GAC GCT; 041: CTA CTC GGT CAG GCT C; 306: GCA TCT TAC GAT ACC TG. Analysis of rRNA species associated with Rli1-TAP was performed after immunoprecipitation with IgG-Sepharose essentially as described (Dez et al, 2004) in the presence of 150 mM of salt. The oligonucleotides indicated above were used for detection of pre-rRNA species.
Affinity purification of Rli1-TAP
The purification was performed essentially as described using 2–6 l of yeast cultures (Puig et al, 2001). For lysis, buffer A supplemented with 50 or 150 mM NaCl was used. Protein was visualized using Novex SDS–4–12% gradient polyacrylamide gels (Invitrogen), stained with colloidal Coomassie (Sigma).
Miscellaneous
DNA recombinant work was performed according to Maniatis et al (1982). sl screen with rix19-1 was performed according to Wimmer et al (1992). SDS–PAGE and immunoblotting were performed according to Siniossoglou et al (1996). Mass spectrometry using tryptic digests of bands excised from the Coomassie stained SDS–polyacrylamide gel was performed according to Baßler et al (2001). The generation of ts mutants was performed according to Baßler et al (2001). The in vivo assay for ribosome export using the Rpl25-GFP and Rps2-GFP reporters was performed according to Gadal et al (2001b) and Schäfer et al (2003). Poly(A)+ RNA export was analyzed according to Santos-Rosa et al (1998).
Supplementary Material
Supplementary data
Acknowledgments
We are grateful to Dr Olivier Gadal and Daniela Strauss for the initial isolation of the rix19-1 mutant, Sabine Merker and Dr Lechner (BZH, University of Heidelberg) for performing mass spectrometry, to Dr Tracy Nissan, Dr Dieter Kressler and Professor Roland Lill (University of Marburg, Germany) for helpful discussions and to Professor Karsten Weis for xpo1-1 strain. EH was supported by grants from the Deutsche Forschungsgemeinschaft (Gottfried Wilhelm Leibniz Program) and Fonds der Chemischen Industrie. VP was supported by HFSP long-term fellowship. CD was supported by an EMBO long-term fellowship. EP and DT were supported by the Wellcome Trust. VP was recipient of a fellowship from the Human Frontier Science Program.
References
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baßler J, Grandi P, Gadal O, Leßmann T, Tollervey D, Lechner J, Hurt EC (2001) Identification of a 60S pre-ribosomal particle that is closely linked to nuclear export. Mol Cell 8: 517–529 [DOI] [PubMed] [Google Scholar]
- Barton RM, Worman HJ (1999) Prenylated prelamin A interacts with Narf, a novel nuclear protein. J Biol Chem 274: 30008–30018 [DOI] [PubMed] [Google Scholar]
- Beinert H, Kiley PJ (1999) Fe–S proteins in sensing and regulatory functions. Curr Opin Chem Biol 3: 152–157 [DOI] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D (1992) Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J 11: 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot K, Muldoon M, Rajkowitsch L, Hughes J, McCarthy JE (2004) Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol Microbiol 51: 987–1001 [DOI] [PubMed] [Google Scholar]
- Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T (1995) Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2–5A pathway. J Biol Chem 270: 13308–13317 [DOI] [PubMed] [Google Scholar]
- Browning KS, Gallie DR, Hershey JW, Hinnebusch AG, Maitra U, Merrick WC, Norbury C (2001) Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem Sci 26: 284. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Iost I, Kressler D, Linder P (1997) The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 94: 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decottignies A, Goffeau A (1997) Complete inventory of the yeast ABC proteins. Nat Genet 15: 137–145 [DOI] [PubMed] [Google Scholar]
- Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y (2004) Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol Cell Biol 24: 6324–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG (2004) The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem 279: 42157–42168 [DOI] [PubMed] [Google Scholar]
- Faber AW, van Dijk M, Raue A, Vos JC (2002) Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA 8: 1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E (2004) Protein displacement by DExH/D ‘RNA helicases' without duplex unwinding. Science 304: 730–734 [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E (1997) CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390: 308–311 [DOI] [PubMed] [Google Scholar]
- Gadal O, Strauß D, Braspenning J, Hoepfner D, Petfalski E, Philippsen P, Tollervey D, Hurt EC (2001a) A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J 20: 3695–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauß D, Kessl J, Trumpower B, Tollervey D, Hurt E (2001b) Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a NES-containing factor Nmd3p that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O, Strauss D, Petfalski E, Gleizes PE, Gas N, Tollervey D, Hurt E (2002) Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J Cell Biol 157: 941–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E (2005) Rea1, a dynein related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60S subunits. J Biol Chem 279: 55411–55418 [DOI] [PubMed] [Google Scholar]
- Gleizes PE, Noaillac-Depeyre J, Leger-Silvestre I, Teulieres F, Dauxois JY, Pommet D, Azum-Gelade MC, Gas N (2001) Ultrastructural localization of rRNA shows defective nuclear export of preribosomes in mutants of the Nup82p complex. J Cell Biol 155: 923–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JHN, Johnson AW (1999) NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol 19: 2389–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JHN, Kallstrom G, Johnson AW (2000) Nmd3p is a Crm1p dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151: 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH (2000) Functional discovery via a compendium of expression profiles. Cell 102: 109–126 [DOI] [PubMed] [Google Scholar]
- Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G (1999) A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J Cell Biol 144: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janáky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, Lill R (2005) Biogenesis of cytosolic ribosomes requires the essential iron–sulfur protein Rli1p and mitochondria. EMBO J 2005, 20 January [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Linder P, De La Cruz J (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 19: 7897–7912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letovsky S, Kasif S (2003) Predicting protein function from protein/protein interaction data: a probabilistic approach. Bioinformatics 19 (Suppl 1): i197–i204 [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G (2000) Maturation of cellular Fe–S proteins: an essential function of mitochondria. Trends Biochem Sci 25: 352–356 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch ET, Sambrook J (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H (2001) Maturation and intranuclear transport of pre-ribosomes requires Noc-proteins. Cell 105: 499–509 [DOI] [PubMed] [Google Scholar]
- Milkereit P, Strauss D, Bassler J, Gadal O, Kuhn H, Schutz S, Gas N, Lechner J, Hurt E, Tschochner H (2003) A Noc complex specifically involved in the formation and nuclear export of ribosomal 40S subunits. J Biol Chem 278: 4072–4081 [DOI] [PubMed] [Google Scholar]
- Morel Y, Barouki R (1999) Repression of gene expression by oxidative stress. Biochem J 342: 481–496 [PMC free article] [PubMed] [Google Scholar]
- Moy TI, Silver PA (2002) Requirements for the nuclear export of the small ribosomal subunit. J Cell Sci 115: 2985–2995 [DOI] [PubMed] [Google Scholar]
- Neumann S, Petfalski E, Brügger B, Großhans H, Wieland F, Tollervey D, Hurt E (2003) Formation and nuclear export of tRNA, rRNA and mRNA is regulated by the ubiquitin ligase Rsp5p. EMBO Rep 4: 1156–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Dlakic M, Tollervey D (2004) A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev 18: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC (1998) X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science 282: 1853–1858 [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Roy A, Solodovnikova N, Nicholson T, Antholine W, Walden WE (2003) A novel eukaryotic factor for cytosolic Fe–S cluster assembly. EMBO J 22: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky DA, Fournier MJ (1999) A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res 27: 161–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E (1998) Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol 18: 6826–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T, Strauß D, Petfalski E, Tollervey D, Hurt EC (2003) The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J 22: 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Lafontaine D, Graindorge J-S, Gadal O, Camasses A, Sanni A, Garnier J-M, Bteitenbach M, Hurt E, Fasiolo F (2001) The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 8: 1363–1373 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC (1996) A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275 [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K (1997) Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T, Schmidt U, Silver PA (2000) Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol Biol Cell 11: 3777–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Noguchi E, Nakashima N, Oki M, Ohba T, Tartakoff A, Ohishi M, Nishimoto T (2001) The Saccharomyces cerevisiae small GTPase, Gsp1p/Ran, is involved in 3′ processing of 7S to 5.8S rRNA and in degradation of the excised 5′-A0 fragment of 35S pre-rRNA, both of which are carried out by the exosome. Genetics 158: 613–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F, Kutay U (2003) Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116: 2409–2419 [DOI] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Jansen RP, Kern H, Hurt EC (1993) Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72: 443–457 [DOI] [PubMed] [Google Scholar]
- Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE (2003) Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J 22: 2841–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Valasek L, Hasek J, Nielsen KH, Hinnebusch AG (2001) Dual function of eIF3j/Hcr1p in processing 20S pre-rRNA and translation initiation. J Biol Chem 276: 43351–43360 [DOI] [PubMed] [Google Scholar]
- Valasek L, Mathew A, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG (2003) The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 33: 261–311 [DOI] [PubMed] [Google Scholar]
- Vitale G, Fabre E, Hurt EC (1996) NBP35 encodes an essential and evolutionary conserved protein in Saccharomyces cerevisiae with homology to a superfamily of bacterial ATPases. Gene 178: 97–106 [DOI] [PubMed] [Google Scholar]
- Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440 [DOI] [PubMed] [Google Scholar]
- Wimmer C, Doye V, Grandi P, Nehrbass U, Hurt E (1992) A new subclass of nucleoporins that functionally interacts with nuclear pore protein NSP1. EMBO J 11: 5051–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


