Abstract
Lactobacillus delbrueckii represents a technologically relevant member of lactic acid bacteria, since the two subspecies bulgaricus and lactis are widely associated with fermented dairy products. In the present work, we report the characterization of two commercial strains belonging to L. delbrueckii subspecies bulgaricus, lactis and a novel strain previously isolated from a traditional fermented fresh cheese. A phenomic approach was performed by combining metabolomic and proteomic analysis of the three strains, which were subsequently supplemented as food source to the model organism Caenorhabditis elegans, with the final aim to evaluate their possible probiotic effects. Restriction analysis of 16S ribosomal DNA revealed that the novel foodborne strain belonged to L. delbrueckii subspecies lactis. Proteomic and metabolomic approaches showed differences in folate, aminoacid and sugar metabolic pathways among the three strains. Moreover, evaluation of C. elegans lifespan, larval development, brood size, and bacterial colonization capacity demonstrated that L. delbrueckii subsp. bulgaricus diet exerted beneficial effects on nematodes. On the other hand, both L. delbrueckii subsp. lactis strains affected lifespan and larval development. We have characterized three strains belonging to L. delbrueckii subspecies bulgaricus and lactis highlighting their divergent origin. In particular, the two closely related isolates L. delbrueckii subspecies lactis display different galactose metabolic capabilities. Moreover, the L. delbrueckii subspecies bulgaricus strain demonstrated potential probiotic features. Combination of omic platforms coupled with in vivo screening in the simple model organism C. elegans is a powerful tool to characterize industrially relevant bacterial isolates.
Keywords: Lactobacillus delbrueckii metabolism, bacterial folate biosynthesis, bacterial galactose metabolism, tagatose pathway, lactic acid bacteria, foodborne bacteria
Introduction
The species Lactobacillus delbrueckii comprises three main subspecies, namely lactis, bulgaricus and delbrueckii, classified in 1983 on the basis of metabolic and phenotypic properties (Weiss et al., 1983). L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis are usually associated with dairy products, such as yogurt or cheeses, while L. delbrueckii subsp. delbrueckii is typical of fermented vegetables. More recently, a novel subspecies, namely indicus, has been described for L. delbrueckii isolated from Indian dairy products (Dellaglio et al., 2005). Lactobacillus delbrueckii is a member of the acidophilus complex, a heterogeneous group of lactobacilli related to L. acidophilus, comprising several bacterial species, many of which of gastrointestinal origin (i.e., L. acidophilus, L. johnsonii, and L. gasseri), and characterized by potential probiotic properties, particularly related to immune modulation. On the other hand, the species L. delbrueckii is rather known for its dairy applications in yogurt and cheesemaking (El Kafsi et al., 2014), and a deeper understanding of the mechanisms underlying its beneficial effects on human health represents a challenging goal. Traditional fermented dairy products are obtained by manufacturing procedures that often employ raw material, relying on the natural microflora preexisting in such ingredients, whose species composition reflects local environments, therefore characterized by a high biodiversity in terms of numbers of strains, whose origin is prevalently environmental (Morea et al., 1999; Tamang et al., 2016).
Several food-associated Lactobacillus species are also found as normal inhabitants of the human gut microbiota (Liu, 2003). The interplay between food and gut microbial consortia can affect human health status, since several foodborne strains also display probiotic features (Devirgiliis et al., 2011). Characterization of novel foodborne isolates of environmental origin is therefore crucial to predict their potential beneficial effects on human health. A broad range of different “omics” technologies are now available to allow a deep characterization of microbial physiology (Calvani et al., 2014). In particular, metabolomics and proteomics are emerging as powerful tools in the field of probiotics (Rebollar et al., 2016; Ruiz et al., 2016).
Evaluation of probiotic properties requires, along with molecular techniques, suitable in vivo models. The nematode Caenorhabditis elegans represents a valuable tool to test the effects of ingested bacteria on host physiology. C. elegans is a differentiated multicellular organism with a nervous system, reproductive organs, and digestive apparatus. Furthermore, it has a simple structure and a short life cycle (less than 3 days). Dietary sources, such as bacteria, play an important role in the control of C. elegans lifespan (So et al., 2011). A recent review elegantly discusses an important aspect concerning the relationship between bacteria and C. elegans: bacterial biomass indeed represents the worm food, and in this sense their trophic relationship is different from the synergistic one between mammals and gut microbiota. However, since live bacteria can influence the nematode physiology through their metabolites, the bacterial component of the C. elegans model can represent both direct and indirect aspects of a diet (Yilmaz and Walhout, 2014).
The nematode has been employed as a useful host for a wide variety of microbes relevant for human health (Clark and Hodgkin, 2014), including foodborne bacteria. It was recently reported that lactic acid bacteria (LAB) can exert protective functions on C. elegans by acting on intestinal permeability (Zhao et al., 2015). Foodborne strains of Weissella koreensis and Weissella cibaria significantly extend the lifespan of C. elegans (Lee et al., 2015). Moreover, in the recent past a growing body of literature demonstrated that the nematode can be successfully used to screen the probiotic features of several bacterial strains. Park et al. (2015) analyzed potentially probiotic Bacillus licheniformis strains isolated from traditional Korean food sources in terms of protection against Staphylococcus aureus infection (Yun et al., 2014), as well as of ability to enhance longevity of nematodes. Recently, Lactobacillus gasseri SBT2055, which had been previously demonstrated to exert beneficial effects in mice and humans, in terms of improvement of the intestinal environment and prevention of infection by influenza A virus (Nakayama et al., 2014), showed a positive impact on longevity and/or aging in the nematode through the modulation of skn-1 gene (Nakagawa et al., 2016).
In the present work we have applied comprehensive analytical approaches based on the combination of metabolomics and proteomics as well as in vivo screening to compare different strains belonging to L. delbrueckii bulgaricus and lactis subspecies, in order to characterize their metabolic activities and to investigate on their potential probiotic properties.
Materials and Methods
Bacterial Strains and Growth Conditions
Lactobacillus delbrueckii subspecies used in this study were L. delbrueckii bulgaricus ATCC11842, L. delbrueckii lactis LMG6401 and the isolate L. delbrueckii 23 originating from Mozzarella di Bufala Campana (MBC) (Zanni et al., 2015). Lactobacillus rhamnosus GG (LGG, ATCC53103) was also used. All the strains were routinely maintained in Elliker broth (DIFCO) and grown at 37°C overnight under anaerobic conditions. Escherichia coli OP50 was grown on LB broth at 37°C overnight.
Lactobacillus delbrueckii Subspecies Identification by ARDRA
Total bacterial DNA extraction and 16S rDNA amplification were performed as previously described (Zanni et al., 2015). ARDRA analysis was conducted by digestion of amplified 16S rDNA with EcoRI or Tru9I restriction endonucleases (Promega Italia, Milan, Italy), according to manufacturer’s instructions.
Cellular Extraction Procedure
Cultured bacterial cells grown as described above were washed three times with cold H2Odd and suspended in 900 μL of cold methanol (-20°C) to quench intracellular metabolism. In the meantime an aliquot of the cultures was washed and the wet weight calculated. To extract the metabolites and proteins the method reported in (Miccheli et al., 1988) was followed. The method allowed the separation of polar, organic phases and protein pellets, which were separately dried under N2 flux and stored at -80°C until further analysis.
NMR Analysis
The freeze-dried polar samples were re-dissolved in 600 μL of D2O phosphate buffer solution (pH 7.4) containing 2 mM sodium 3-(tri-methylsilyl)propionate-2,2,3,3-d4 (TSP) as 1H NMR reference, and transferred to 5 mm NMR glass tubes for analysis. The organic phases were re-dissolved in 600 μL of CDCl3 containing 2 mM hexamethyl-di-siloxane (HMDSO) as a 1H NMR reference.
NMR Spectroscopy and Data Analysis
1H NMR spectra were acquired at 25°C using a Bruker Avance III 400 spectrometer (Bruker BioSpin GmbH, Germany) equipped with a magnet operating at 9.4 Tesla, where the 1H nucleus resonates at 400.13 MHz. The probe-head was a 5 mm diameter multinuclear PABBO BB-1H/D (Z108618/0044) equipped with z-gradient.
The pulse sequence adopted for spectra acquisition was a presaturation–single 90 detection pulse–acquire–delay sequence where the D1 relaxation delay was optimized to 2.5 s to allow the acquisition of 64 k data point in about 5.5 s, satisfying full relaxation conditions.
The length of the detection pulse was calibrated previously to the acquisition of each spectrum, the spectral width was set to 6009.62 Hz (15 ppm) and 64 scans were collected for each spectrum.
1H NMR spectra were processed using the 1D-NMR Manager ver. 12.0 software (Advanced Chemistry Development, Inc., Toronto, ON, Canada).
The assignment of the peaks to specific metabolites was achieved by standard two-dimensional (2D)1H-1H total correlation spectroscopy (TOCSY), 1H-13C heteronuclear single quantum correlation (HSQC), and heteronuclear multiple bond correlation (HMBC) and confirmed using an internal library of compounds, in comparison with literature data (Fan and Lane, 2008; Brasili et al., 2013; Gorietti et al., 2014).
The acquired NMR spectra were manually phased and baseline corrected; polar and organic spectra were referenced to the chemical shift of the TSP or HMDSO methyl resonance at δ 0.00 and 0.055 ppm, respectively. The quantification of metabolites was obtained by comparison of the integrals of specific signals to the internal standard (TSP or HMDSO) integral.
Proteomic Analysis
Total extract of each Lactobacillus samples was resuspended in 200 μl of Ammonium Bicarbonate 0.1 M pH 8 and then homogenized.
After the addiction of Rapigest 0.2% v/v (Waters Corporation, Milford, MA, United States) and after incubation at 100°C for 15 min, protein content was quantified using SPN-Protein Assay (G-Biosciences, United States) and 50 μg of each sample was digested with trypsin (1:50) at 37°C o/n (Sequencing Grade Modifier Trypsin, Promega). A second aliquot of trypsin was added to samples and after 4h the digestion was stopped with TFA 0.5% (v/v). Finally, samples were incubated for 40 min at 37°C, centrifuged at 13 000 × g for 10 min in order to precipitate Rapigest and desalted by PepClean C-18 spin column (Pierce Biotechnology, Rockford, IL, United States).
Trypsin digested samples were analyzed by means of two-dimensional micro liquid chromatography coupled to linear ion trap mass spectrometer LTQ (Thermo Fisher Scientific, Waltham, MA, United States) as described in Comunian et al. (2011) with some modifications. Briefly, the separation of peptides was obtained through an acetonitrile gradient (eluent A, 0.1% formic acid in water; eluent B, 0.1% formic acid in acetonitrile) and the gradient profile was 5–10% eluent B in 5 min, 10–40% B in 40 min, 40–80% B in 8 min, and 80–95% in 3 min.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD006551.
C. elegans Strains and Growth Conditions
The wild-type C. elegans strain N2 was used in all experiments and propagated on nematode growth medium (NGM) modified to be peptone-free (mNGM) (Zanni et al., 2015). Worms were fed with bacterial cultures, daily prepared as follows: aliquots of frozen bacteria stock were inoculated in Elliker broth (DIFCO) and grown at 37°C overnight under anaerobic conditions. Afterward, 25 μL of each bacterial suspension in M9 buffer, corresponding to 10 mg of bacterial cells, was spread on 3.5 cm diameter mNGM plates.
Lifespan Assays
Synchronized N2 adults were allowed to lay embryos for 2 h directly on mNGM, covered with the indicated bacterial lawns, and then sacrificed. All lifespan assays started when the progeny became fertile (t0). Animals were transferred to new plates spread with fresh lawns and monitored daily. They were scored as dead when they no longer responded to gentle prodding with a platinum wire. Worms that crawled off the plates were not included in the analysis.
Brood Size Measurement
Progeny production was evaluated according to (Zanni et al., 2015), with some modifications. Briefly, synchronized worms obtained as above were grown on mNGM plates seeded with bacteria and then were allowed to lay embryos at 16°C. Next, animals were transferred onto a fresh bacteria plate every day, and the number of progeny was counted with a Zeiss Axiovert 25 microscope. The procedure was repeated for 4 days until the mother worms stopped laying eggs. Each day the progeny production was recorded.
Body Size Measurement
Individual animals were photographed after 3, 4, 5, and 6 days from egg hatching using a Leica MZ10F stereomicroscope connected to Jenoptik CCD camera. Length of worm body was determined by using the Delta Sistemi IAS software. At least 30 nematodes were imaged on at least three independent experiments.
Estimation of Bacterial CFU within the Nematode Gut
For each experiment, 10 animals at L4 stage and at 5 days of adulthood were washed and lysed according to (Uccelletti et al., 2010). Whole worm lysates were plated onto MRS-agar plates. The number of CFU was counted after 48 h of incubation at 37°C, anaerobically.
Statistical Analysis
Experiments were performed at least in triplicate. Data are presented as mean ± SD, and Student’s-test or one-way ANOVA analysis coupled with a Bonferroni post test (GraphPad Prism 4.0 software) was used to determine the statistical significance between experimental groups. Statistical significance was defined as ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
In the case of metabolomics analysis, multivariate data analysis was carried out using Unscrambler 9.8 Software (CAMO, Oslo, Norway). Spectral data were mean-centered and autoscaled before analysis. Principal components analysis (PCA) was used to explore inherent clustering, to identify outliers and significant metabolites in the separation between sample groups.
U Mann–Whitney test was also applied; a P-value < 0.05 was considered for a statistically significant difference between sample groups.
For proteomic analysis, the experimental mass spectra produced by MudPIT analyses were correlated to tryptic peptide sequences by comparing with theoretical mass spectra, obtained by in silico digestion of a protein database downloaded from the NCBI website1 containing Lactobacillus delbrueckii subsp. bulgaricus and Lactobacillus delbrueckii subsp. lactis sequences. Data processing was performed using the 3.3.1. Bioworks version, based on SEQUEST algorithm (University of Washington, licensed to Thermo Finnigan Corp., San Josè, CA, United States), and the following parameters: Xcorr scores greater than 1.5 for singly charged peptide ions and 2.0 and 2.5 for doubly and triply charged ions, respectively, the peptide probability ≤ 0.001 and the protein consensus score value ≥ 10. These filters guaranteed that the resulting proteins have a probability value p ≤ 0.001. Data were treated with an in-house algorithm called MAProMa (Mauri and Deho, 2008) (Multidimensional Algorithm Protein Map), in particular a tool of MAProMa permits the comparison of the protein list obtain from the analysis of the samples. Proteins with significant differences in level, were identified by other two tools of MAProMA: DAve (Differential Average) and DCI (Differential Coefficient Index) (Mauri et al., 2005). These two algorithms are based on score values assigned by SEQUEST software to each identified protein in samples to be compared. Specifically, DAve is an index of the relative ratio between control and mutant and DCI is an index to evaluate the absolute variation of score value of each protein. Briefly, using MAProMA each identified protein in the two samples were aligned and then DAve and DCI indexes were calculated for all proteins. The threshold values imposed were very stringent: DAve > 0.4 and DAve < –0.4, DCI > 400 and DCI < –400. To increase the confidence, it is necessary that both indexes, DAve and DCI, satisfy these thresholds.
Hierarchical clustering was performed applying Ward’s method and Euclidean’s distance metric (Zhao and Karypis, 2005).
Results
Identification of the Foodborne Lactobacillus delbrueckii Strain Subspecies
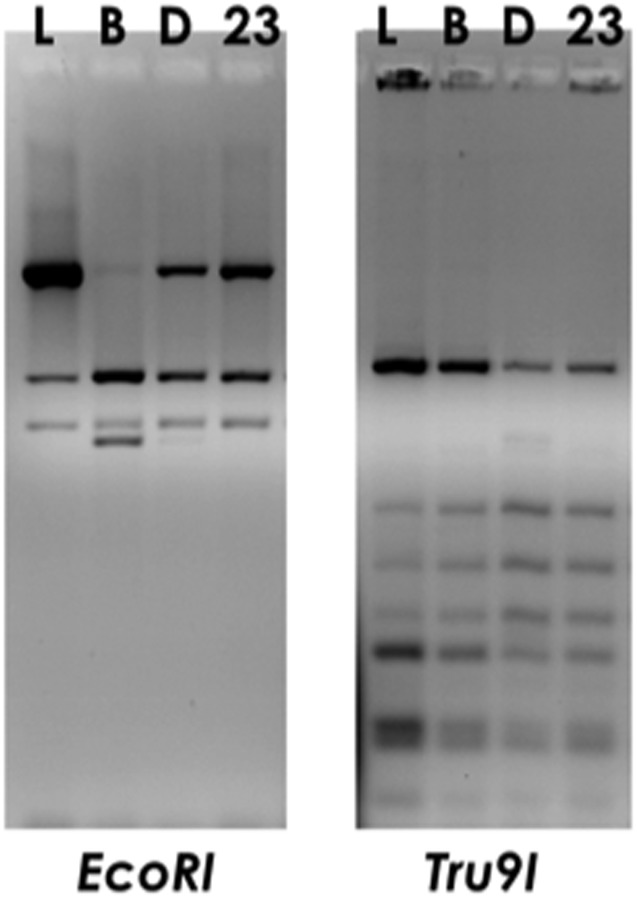
The strain Lactobacillus delbrueckii 23 used in this work was originally isolated from the fermented cheese MBC (Devirgiliis et al., 2008; Zanni et al., 2015), however, it had not been characterized at the subspecies level. To this aim, restriction analysis of amplified 16S ribosomal DNA (ARDRA) was performed by using endonucleases EcoRI, which allowed to differentiate subspecies bulgaricus from subspecies lactis/delbrueckii (Miteva et al., 2001), and Tru9I, which was used to discriminate between subspecies delbrueckii from subspecies lactis/bulgaricus. The results shown in Figure 1 demonstrated that the strain L. delbrueckii 23 belongs to lactis subspecies, as revealed by the comparison of restriction patterns with reference L. delbrueckii subspecies.
FIGURE 1.
ARDRA profiles of foodborne isolate Lactobacillus delbrueckii 23 (23) obtained by digestion of PCR-amplified 16S rDNA with EcoRI or Tru9I restriction enzymes. Commercial isolates L. delbrueckii subsp. lactis LMG6401 (L), subsp. bulgaricus (B) or subsp. delbrueckii LMG6412T (D) were used as reference subspecies controls.
Proteomic Analysis of L. delbrueckii Subspecies
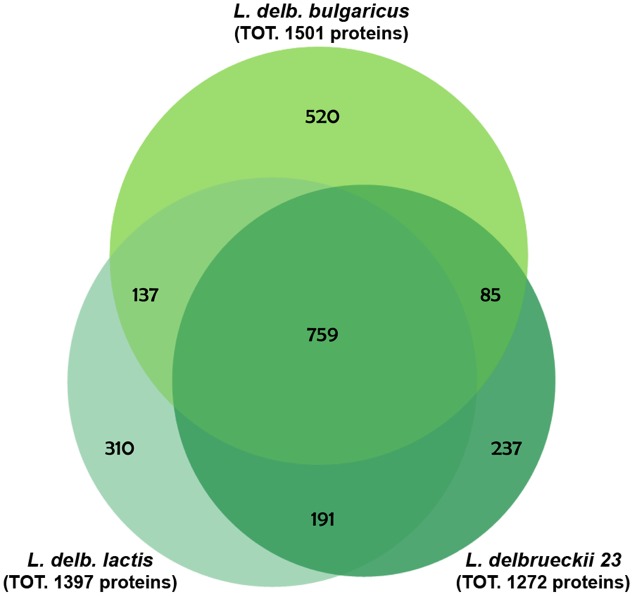
To highlight differences among the three strains belonging to L. delbrueckii subspecies relevant in dairy technologies, a proteomic analysis of L. delbrueckii subsp. bulgaricus, lactis and L. delbrueckii 23 was performed, using protein pellets from bacterial cells grown on Elliker’s broth, as specified in “Materials and Methods.” A total of 2239 proteins were identified in all samples (Supplementary Table S1), whose distribution across strains was analyzed by Venn diagram (Figure 2). L. delbrueckii subsp. lactis and L. delbrueckii 23 shared 191 proteins, while 137 and 85 proteins were shared by these strains with L. delbrueckii bulgaricus, respectively. A total of 759 proteins resulted common to all the strains, whereas 520, 310, and 237 proteins were specific to L. delbrueckii subsp. bulgaricus, lactis and L. delbrueckii 23, respectively (Figure 2).
FIGURE 2.
Venn diagram of protein distribution across the analyzed strains. Proteins are identified from total cell extracts derived from lactobacilli grown overnight in Elliker broth.
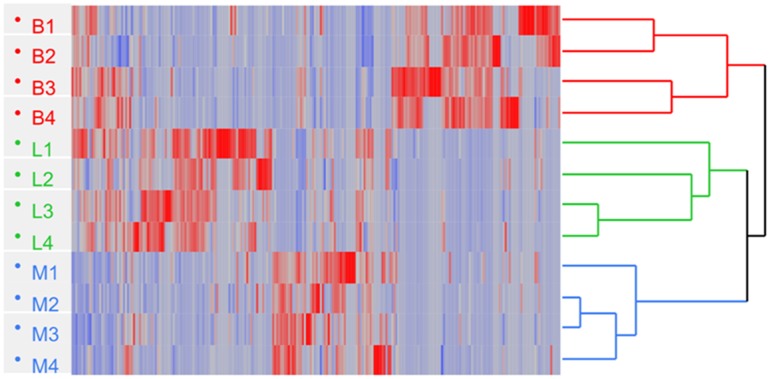
To further analyze the abundance profiles of the significantly changed proteins that were commonly identified in the three strains, hierarchical clustering analysis was performed. The two strains of L. delbrueckii subsp. lactis resulted closer with respect to L. delbrueckii subsp. bulgaricus, although differences among them were detected (Figure 3).
FIGURE 3.
Hierarchical clustering of L. delbrueckii subsp. bulgaricus (B; n = 4), lactis (L; n = 4), and L. delbrueckii 23 (M; n = 4) obtained by processing the SpC (Spectral Count) of proteins; Euclidean’s distance and Ward method were applied. Heat map shows the SpC and indicates down (blue) and up-regulated (red) proteins, respectively.
The majority of differentially expressed proteins were related to bacterial metabolism. In particular, proteins exclusively expressed by L. delbrueckii subsp. bulgaricus appeared to be related to aminoacid metabolism, those exclusively expressed by L. delbrueckii subsp. lactis were related to glutathione and maltose metabolism, while sugar metabolism and fatty acid biosynthesis were specific to the strain L. delbrueckii 23 (Supplementary Table S2). In particular, enzymes involved in galactose metabolism through the tagatose pathway were specifically identified in such strain (Supplementary Figure S1). It is worth noting that dihydropteroate synthase as well as GTP cyclohydrolase, key enzymes of folate biosynthetic pathway, were upregulated in L. delbrueckii subsp. bulgaricus (Supplementary Figure S2).
NMR Analysis of L. delbrueckii Subspecies
In order to further explore the differences among the three strains in terms of metabolites, a 1H-NMR spectroscopy analysis was performed. To this aim, an NMR-based metabolic profiling from aqueous as well as organic cellular extract phases was generated from the same bacterial cell samples used for proteomic experiments.
In Supplementary Table S3, the resonance assignments of the metabolites measured on polar and organic extracts are depicted. A representative 1H NMR spectrum of cell polar extracts was reported in Supplementary Figure S3. The analysis of intracellular metabolites reported in Table 1 revealed the presence of higher amounts of several amino acids in L. delbrueckii subsp. bulgaricus.
Table 1.
Amount of intracellular metabolites expressed as μmol/g calculated as the average of three samples +/- standard deviation.
| L. delbrueckii bulgaricus | L. delbrueckiilactis | L. delbrueckii 23 | Significativity | |
|---|---|---|---|---|
| Aminoacids | ||||
| Valine | 0.28 ± 0.07 | 0.25 ± 0.02 | 0.47 ± 0.05 | C |
| Isoleucine | 0.22 ± 0.05 | 0.16 ± 0.03 | 0.30 ± 0.04 | C |
| Leucine | 0.34 ± 0.06 | 0.35 ± 0.04 | 0.75 ± 0.08 | BC |
| Threonine | 0.46 ± 0.00 | 1.30 ± 0.28 | 0.33 ± 0.05 | AC |
| Alanine | 0.59 ± 0.00 | 0.40 ± 0.06 | 0.64 ± 0.16 | |
| Glutamate | 9.17 ± 0.77 | 1.70 ± 0.28 | 2.26 ± 0.56 | AB |
| Glutamine | 2.42 ± 0.05 | 0.02 ± 0.00 | 1.34 ± 0.51 | AC |
| Glutathione | 0.00 ± 0.00 | 0.16 ± 0.02 | 0.54 ± 0.10 | ABC |
| Aspartate | 5.18 ± 0.16 | 1.07 ± 0.19 | 1.35 ± 0.64 | AB |
| Asparagine | 2.47 ± 0.41 | 0.34 ± 0.08 | 0.94 ± 0.39 | ABC |
| Lysine | 2.88 ± 0.03 | 1.27 ± 0.33 | 5.23 ± 0.40 | ABC |
| Arginine | 0.11 ± 0.07 | 7.25 ± 0.93 | 0.39 ± 0.09 | ABC |
| Serine | 6.42 ± 0.08 | 0.00 ± 0.00 | 0.00 ± 0.00 | AB |
| Glycine | 0.61 ± 0.00 | 0.21 ± 0.02 | 0.35 ± 0.05 | AB |
| Tyrosine | 0.17 ± 0.05 | 0.04 ± 0.01 | 1.79 ± 1.39 | BC |
| Histidine | 0.19 ± 0.11 | 0.29 ± 0.16 | 0.14 ± 0.05 | |
| Phenylalanine | 0.04 ± 0.02 | 0.08 ± 0.02 | 0.16 ± 0.02 | |
| Tryptophan | 0.08 ± 0.05 | 0.05 ± 0.02 | 0.08 ± 0.03 | |
| Organic acids | ||||
| Lactate | 0.74 ± 0.07 | 0.25 ± 0.02 | 15.83 ± 1.82 | ABC |
| Acetate | 0.53 ± 0.03 | 0.97 ± 0.19 | 1.32 ± 0.08 | AB |
| Formate | 0.19 ± 0.00 | 0.36 ± 0.19 | 0.29 ± 0.04 | |
| Carbohydrates | ||||
| Glucose | 1.53 ± 0.81 | 0.00 ± 0.00 | 0.00 ± 0.00 | AB |
| Fatty Acids | ||||
| Saturated fatty acid | 1.32 ± 0.14 | 2.77 ± 0.18 | 2.51 ± 0.21 | AB |
| Monoinsaturated fatty acid | 7.82 ± 1.08 | 5.97 ± 1.17 | 2.18 ± 0.17 | BC |
| Monoacyl glycerol | 2.26 ± 0.05 | 2.73 ± 0.11 | 2.07 ± 0.12 | |
| Miscellaneous | ||||
| Dilactate | 0.26 ± 0.00 | 0.07 ± 0.01 | 0.06 ± 0.02 | AB |
| Choline | 0.00 ± 0.00 | 3.72 ± 0.53 | 0.09 ± 0.02 | AC |
| Uridine Phosphate | 1.42 ± 0.16 | 2.89 ± 0.29 | 0.59 ± 0.11 | ABC |
| Cytosine Phosphate | 0.73 ± 0.05 | 0.64 ± 0.09 | 0.34 ± 0.05 | |
| Guanidine Phosphate | 0.56 ± 0.08 | 0.69 ± 0.13 | 0.90 ± 0.10 | |
| Adenine Phosphate | 0.31 ± 0.05 | 0.55 ± 0.07 | 0.12 ± 0.03 | C |
| NAD | 0.50 ± 0.05 | 0.74 ± 0.08 | 0.90 ± 0.12 | |
Univariate statistical analysis was performed employing the non-parametrical U Mann–Whitney test. The letters A, B, and C indicate significant statistical differences (p < 0.05) with the code A indicating a discrimination between L. delbrueckii bulgaricus and L. delbrueckii lactis, B between L. delbrueckii bulgaricus and L. delbrueckii 23, and C between L. delbrueckii lactis and L. delbrueckii 23.
Concerning fat content, saturated fatty acids (SFA) resulted lower in L. delbrueckii subsp. bulgaricus, while the monounsaturated fatty acids were higher. Intracellular glucose was detected only in this subspecies, whereas glutathione was present only in L. delbrueckii subsp. lactis, although at different amounts in the two strains. Finally, dilactic acid showed a higher intracellular concentration in L. delbrueckii subsp. bulgaricus (Table 1).
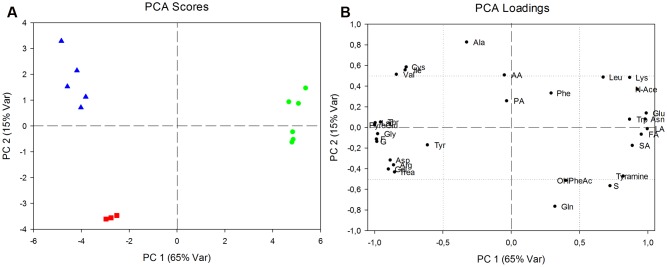
The analysis of synthesis and consumption of metabolites provided more information on specific metabolism of the single subspecies than intracellular content. Principal component analysis was performed to explore the data field (Figure 4).
FIGURE 4.
Analysis of production and consumption of metabolites, expressed by net balances. (A) Principal components analysis (PCA) score plot relative to L. delbrueckii 23 (green circles), L. delbrueckii bulgaricus (red squares) and L. delbrueckii lactis (blue triangles). Eighty percent of total variability represented in two components (PC1: 65%; PC2: 15%). (B) PCA loadings plot indicating statistically significant differences of excreted or consumed metabolites between the two L. delbrueckii 23 and L. delbrueckii lactis with respect to L. delbrueckii bulgaricus along PC1, or between both L. delbrueckii lactis subspecies and L. delbrueckii bulgaricus along PC2.
Component scores plot showed along PC1 a net separation between L. delbrueckii 23 and L. delbrueckii subsp. lactis together with L. delbrueckii subsp. bulgaricus; whereas a separation between L. delbrueckii subsp. lactis strains and L. delbrueckii subsp. bulgaricus was showed along PC2.
The data, reported in Table 2, are representative of net balances, with positive and negative values being considered as an estimate of net fluxes of production and utilization of metabolites, respectively. Both aminoacid and sugar metabolism revealed differences among the three isolates, and in particular between the two strains of L. delbrueckii subsp. lactis. Interestingly, all the three strains produced aminoacids such as leucine, alanine, aspartate, arginine and aromatic aminoacids and consumed glutamate. Conversely, threonine, glutamine, pyroglutamate, cysteine, and asparagine displayed opposite delta values between the two L. delbrueckii lactis strains, and in the case of valine, glutamic acid and glycine the amounts of produced/consumed metabolites, although displaying the same trend, were significantly different between the two strains. However, the sugar metabolism resulted to be particularly strain-specific. L. delbrueckii 23 showed a strong ability to metabolize lactose, galactose, fructose, and trealose with a parallel production of high amounts of lactic acid. It is worth noting that L. delbrueckii subsp. lactis and L. delbrueckii subsp. bulgaricus produced glucose and galactose, although this latter component was detected at lower levels in the case of lactis strain. These sugars derived from the incomplete catabolism of lactose by these two lactobacilli being not able to further metabolize galactose. As a consequence, the sugars were secreted out of the cell, probably through lactose-galactose antiporter system. Sucrose was consumed only by L. delbrueckii subsp. lactis, while glucose was consumed only by L. delbrueckii 23.
Table 2.
Medium balances expressed as mM and calculated as the average of three samples +/- standard deviation.
| L. delbrueckii bulgaricus | L. delbrueckii lactis | L. delbrueckii 23 | Significativity | |
|---|---|---|---|---|
| Aminoacids | ||||
| Valine | 0.55 ± 0.02 | 1.31 ± 0.10 | 0.26 ± 0.06 | A∗BC∗ |
| Isoleucine | -1.00 ± 0.01 | -0.64 ± 0.03 | -1.11 ± 0.06 | C |
| Leucine | 1.46 ± 0.04 | 1.97 ± 0.18 | 3.07 ± 0.40 | B |
| Threonine | 1.18 ± 0.16 | 1.61 ± 0.13 | -0.30 ± 0.11 | B∗C∗ |
| Alanine | 0.44 ± 0.01 | 0.99 ± 0.02 | 0.65 ± 0.11 | A |
| Lysine | -3.23 ± 0.03 | -1.97 ± 0.12 | 0.16 ± 0.19 | A∗B∗C∗ |
| Arginine | 10.93 ± 0.11 | 9.63 ± 0.04 | 7.46 ± 0.21 | A∗B∗C∗ |
| Pyroglutamate | 6.89 ± 0.01 | 8.53 ± 0.09 | -0.02 ± 0.21 | B∗C∗ |
| Glutamate | -8.15 ± 0.01 | -8.14 ± 0.01 | -1.67 ± 0.09 | B∗C∗ |
| Glutamine | 1.97 ± 0.04 | -0.43 ± 0.13 | 0.83 ± 0.19 | A∗BC |
| Aspartate | 12.92 ± 0.34 | 11.78 ± 0.37 | 8.56 ± 0.21 | AB∗C∗ |
| Asparagine | -1.52 ± 0.09 | -1.67 ± 0.12 | 0.62 ± 0.11 | B∗C∗ |
| Cysteine | -1.45 ± 0.34 | 1.98 ± 0.27 | -1.84 ± 0.17 | A∗C∗ |
| Glycine | -1.46 ± 0.16 | -1.41 ± 0.53 | -8.57 ± 0.19 | B∗C∗ |
| Tyrosine | 1.09 ± 0.05 | 1.07 ± 0.09 | 0.86 ± 0.03 | |
| Phenylalanine | 0.92 ± 0.07 | 1.09 ± 0.14 | 1.18 ± 0.13 | |
| Tryptophan | 0.60 ± 0.06 | 0.64 ± 0.19 | 1.46 ± 0.06 | B∗C∗ |
| Organic Acids | ||||
| Lactate | 50.62 ± 0.34 | 37.26 ± 2.48 | 106.12 ± 1.55 | A∗B∗C∗ |
| Acetate | 4.54 ± 0.06 | 5.21 ± 0.73 | 4.93 ± 0.32 | |
| N-Acetyl | 1.47 ± 0.03 | 1.73 ± 0.02 | 2.52 ± 0.06 | B |
| Pyruvate | 0.84 ± 0.01 | 1.09 ± 0.16 | 0.98 ± 0.02 | |
| Succinate | 0.55 ± 0.01 | 0.39 ± 0.09 | 0.85 ± 0.02 | BC∗ |
| Formate | 0.51 ± 0.02 | 0.05 ± 0.21 | 1.75 ± 0.05 | C |
| Carbohydrates | ||||
| Sucrose | 0.63 ± 0.19 | -2.62 ± 0.18 | 0.63 ± 0.22 | A∗C∗ |
| Fructose | -3.23 ± 0.23 | -3.11 ± 0.14 | -8.67 ± 0.01 | A |
| Lactose | -4.33 ± 0.13 | -0.71 ± 0.23 | -16.11 ± 0.02 | C |
| Galactose | 3.91 ± 0.18 | 0.86 ± 0.42 | -5.65 ± 0.01 | B |
| Trealose | -0.26 ± 0.09 | -0.62 ± 0.11 | -1.37 ± 0.01 | B |
| Glucose | 5.79 ± 0.29 | 5.77 ± 0.18 | -0.89 ± 0.01 | BC∗ |
| Miscellaneous | ||||
| Tyramine | 0.37 ± 0.01 | 0.16 ± 0.04 | 0.44 ± 0.02 | A∗C∗ |
| 4-Hydroxyphenylacetate | 0.19 ± 0.02 | 0.02 ± 0.10 | 0.18 ± 0.01 | AC |
A positive value corresponds to the production of a metabolite compared to the starting medium, while a negative one indicates consumption. The letters A, B, and C indicate significant statistical (p < 0.05) differences according to ANOVA on ranks analysis with the code A discriminating between L. delbrueckii bulgaricus and L. delbrueckii lactis, B between L. delbrueckii bulgaricus and L. delbrueckii 23, and C between L. delbrueckii lactis and L. delbrueckii 23. Asterisks indicate the discriminations with a oxp-value < 0.01.
Evaluation of Potential Probiotic Effects of L. delbrueckii Subspecies in C. elegans
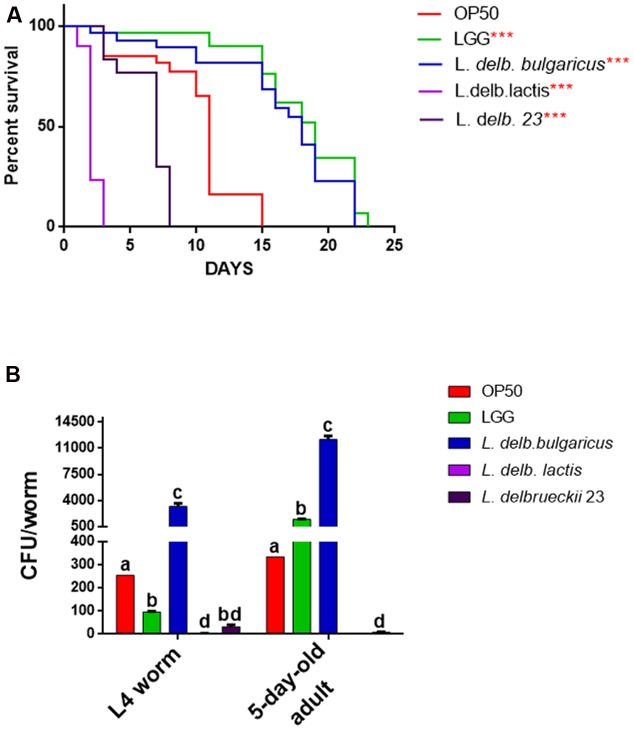
We next aimed at understanding possible probiotic effects exerted by these isolates by using the model organism C. elegans. To this purpose, phenotypical analysis was performed on animals grown on the L. delbrueckii strains. Notably, the dietary administration of L. delbrueckii subsp. bulgaricus led to an increased worm longevity with respect to both the commercial and the foodborne isolate L. delbrueckii subsp. lactis (Figure 5A).
FIGURE 5.
Effect of L. delbrueckii subspecies on nematode lifespan and gut colonization capacity. (A) Kaplan–Mèier survival plot of N2 worms fed the three L. delbrueckii strains or controls (LGG and Escherichia coli OP50); n = 60 for each data point of single experiments. Asterisks indicate the P-values (log-rank test) with respect to the control E. coli OP50. Different letters indicate statistically significant differences (P < 0.05). (B) Bacterial colony forming units (CFU) recovered from nematodes, obtained by plating whole lysates of L4 and 5-days adults fed the three L. delbrueckii strains or controls (LGG and E. coli OP50). Bars represent the mean of three independent experiments.
Indeed, a median survival of 18 days was found in animals fed with L. delbrueckii subsp. bulgaricus, while a median survival of 11 days was observed in worms fed with the standard food source OP50. Moreover, the use of a claimed probiotic strain, namely LGG, as a food source, resulted in an almost identical effect on worm longevity. On the other hand, median survival values obtained for nematodes fed with L. delbrueckii subsp. lactis and L. delbrueckii 23 strains were recorded at 2 and 7 days, respectively. In addition, bacterial colonization capacity, a feature strongly related to probiotic activity, was assessed at L4 and 5-days adulthood stages in animals fed with the different bacteria (Figure 5B). In L4 worms, L. delbrueckii subsp. bulgaricus showed the maximum colonization capacity, expressed as CFU/worm; about 30-fold higher with respect of the probiotic control LGG. By contrast, the two strains of L. delbrueckii subsp. lactis exhibited almost undetectable CFU/worm values, which resulted even lower with respect to OP50 control (Figure 5B). In 5-days adult nematodes the same trend was observed, with L. delbrueckii subsp. bulgaricus and LGG showing the maximum colonization capacity, although with a less marked relative difference. Moreover, L. delbrueckii 23 CFU/worm decreased by four times with respect to the earlier animal stage (Figure 5B).
In order to evaluate other physiological effects exerted by L. delbrueckii strains, analysis of larval development was conducted. L. delbrueckii subsp. bulgaricus did not influence the worm length, while reduction in measures was observed in animals fed with both L. delbrueckii lactis strains (Figure 6A).
FIGURE 6.
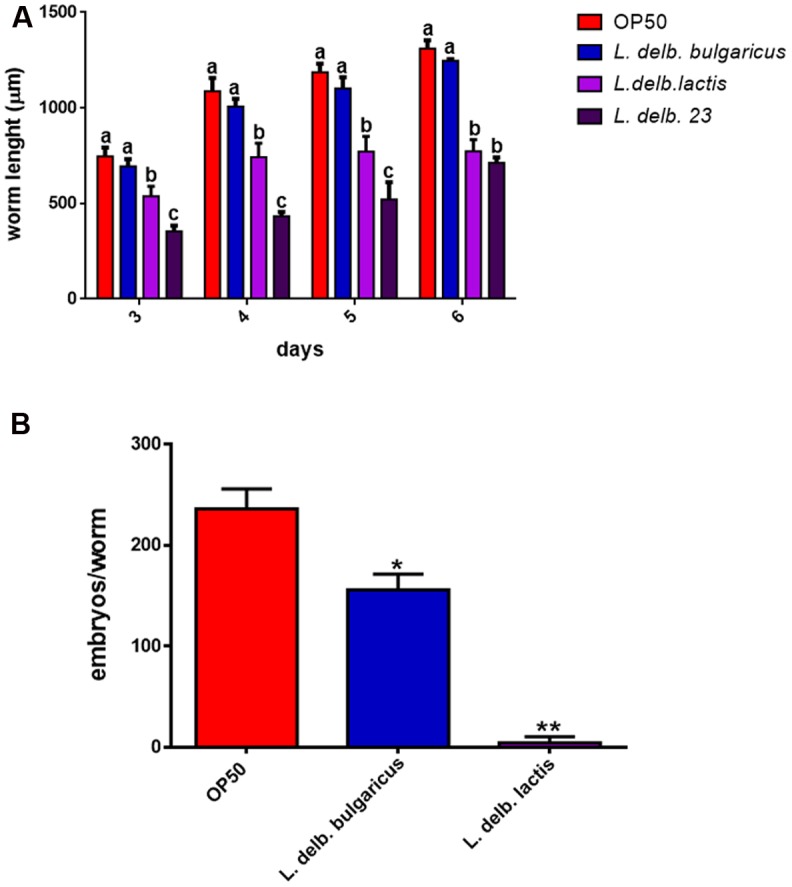
Analysis of body size and fertility rate in worms fed L. delbrueckii subspecies. (A) Effect on Caenorhabditis elegans body size. Worms were grown in the presence of the three L. delbrueckii strains or OP50, their length was measured from head to tail at the indicated time points. Different letters indicate statistically significant differences (P < 0.05). (B) Average embryos production per worm of L. delbrueckii strains-fed animals. (∗P < 0.05; ∗∗P < 0.01).
Finally, nematode fertility was also evaluated, through brood size analysis, expressed as number of embryos/worm. A slight reduction of progeny production was observed in worms fed with L. delbrueckii subsp. bulgaricus with respect to those fed with OP50, whereas a very low number of embryos laid by animals grown on L. delbrueckii subsp. lactis was recorded (Figure 6B). L. delbrueckii 23 diet induced a sterile phenotype in the nematodes (data not shown).
Discussion
In the present work a multidisciplinary approach was applied to characterize three strains belonging to L. delbrueckii subspecies bulgaricus and lactis, by combining metabolomic and proteomic analysis as well as in vivo screening in the model organism C. elegans, with the final aim to evaluate their possible probiotic effects.
In particular, one of the strains, namely L. delbrueckii 23, was newly isolated from the fermenting microbiota of a traditional fermented fresh cheese (Zanni et al., 2015), while the other two strains, belonging to the subspecies bulgaricus and lactis, were acquired from bacterial culture collections ATCC and LMG, respectively. ARDRA analysis performed on L. delbrueckii 23 revealed that this strain belonged to subspecies lactis.
Both proteomic and metabolomic approaches showed differences among the three isolates. In particular, hierarchical cluster analysis of proteomic data highlighted that the two strains of L. delbrueckii subsp. lactis resulted closer with respect to L. delbrueckii subsp. bulgaricus, even if differences among them were detected. NMR spectroscopy-metabolomic profiles obtained for the L. delbrueckii subspecies highlighted different metabolome profiles among the three isolates. We have considered both the intracellular metabolites as well as the specific metabolism of the single subspecies, this latter through the analysis of synthesis and consumption of metabolites, expressed by net balances of extracellular metabolites. In particular, the obtained results showed that metabolism of L. delbrueckii subsp. bulgaricus and lactis are mainly sustained from a protein-based source, on the contrary, the L. delbrueckii 23 strain appeared to be mainly dependent on carbohydrate metabolism. Analysis of sugar metabolism showed differences between the two strains of L. delbrueckii subsp. lactis, in particular concerning galactose metabolism, in agreement with the proteomic experiments. Intriguingly, the newly isolated strain L. delbrueckii 23 resulted capable of metabolizing galactose through the tagatose pathway, a trait that could reflect its origin from MBC. In fact, most strains of L. delbrueckii subsp. bulgaricus, used as starter cultures for Mozzarella cheese, ferment only the glucose portion of lactose and release the galactose portion into the medium or curd, whereas L. delbrueckii subsp. lactis strains can metabolize, in addition, galactose (Suzzi et al., 2000; Germond et al., 2003). This avoids accumulation of the galactose in dairy products, which may lead to several undesirable effects, such as browning of Mozzarella cheese (Vaillancourt et al., 2004).
Furthermore, we took advantage of the simple model organism C. elegans to perform in vivo analysis in order to evaluate the potential probiotic effects exerted by the three L. delbrueckii strains. Our findings demonstrated that feeding worms with different L. delbrueckii subspecies results in opposite effects on host metabolism. In particular, L. delbrueckii subsp. bulgaricus exerted probiotic features in terms of life-span extension and gut colonization capacity, whereas the two strains belonging to L. delbrueckii subsp. lactis impacted more severely on lifespan and larval development. One of the key aspects in determining these different behaviors could be represented by the gut colonization capacity, since L. delbrueckii subsp. bulgaricus resulted capable of colonizing the worm’s gut more efficiently with respect to the subsp. lactis. According to our hypothesis, Park et al. (2014) observed that newly isolated strains of L. plantarum, which were more efficient in colonizing the nematode gut with respect to LGG, were able to extend the worm’s lifespan. However, this aspect is still controversial, since other authors reported a negative correlation between colonization capacity and lifespan extension (Brokate-Llanos et al., 2014; Park et al., 2015), or suggested that gut colonization does not directly influence the host defense response (Yun et al., 2014). It must be pointed out that in the case of E. coli as food source, the reduction of colonization capacity could lower bacterial pathogenicity resulting in an increased worm’s longevity (Brokate-Llanos et al., 2014). Moreover, it has been observed that the effects of bacterial feeding on worm longevity are often strain-specific (Park et al., 2015).
It has been demonstrated that bacterial metabolism plays a key role in affecting nematode longevity (Nguyen and Clarke, 2012; Brokate-Llanos et al., 2014; Yu et al., 2015). A trait characterizing L. delbrueckii subsp. bulgaricus strain was represented by its ability to synthesize folate, as indicated by proteomic analysis. Bacterial folate metabolism has received great consideration in terms of influencing C. elegans life span, in particular correlating bacterial folate synthesis inhibition with reduction of worm longevity (Virk et al., 2012). A recent paper clarified that bacterial folate status acts on E. coli physiology in a way that influences worm aging (Virk et al., 2016). To the best of our knowledge, however, this kind of studies have never been conducted in alternative worm’s food sources, such as LAB.
Analysis of amino acid metabolism revealed that serine, glycine, aspartic acid, asparagine, and glutamic acid accumulated in L. delbrueckii subsp. bulgaricus cells, whereas their levels were lower in the two lactis strains. Aminoacid metabolism has been shown to influence lifespan, in particular tryptophan and proline, among others, were associated with longevity, while aspartic acid and phenylalanine seem to have anti-longevity effects (Edwards et al., 2015). It could be likely that the differentially accumulated aminoacids in the two subspecies can contribute to the effects exerted on host lifespan. Concerning fat metabolism, both strains of L. delbrueckii subsp. lactis exhibited higher intracellular levels of SFA than L. delbrueckii subsp. bulgaricus. According to our results, Kudo et al. (2012) found that the content of several SFA, including palmitic (C:16) and stearic (C:18) acids, was increased in L. delbrueckii subsp. lactis with respect to L. delbrueckii subsp. bulgaricus. Moreover, it has been reported that C. elegans lifespan was slightly reduced by supplementation of palmitic acid in the diet (Shmookler Reis et al., 2011). We can thus not exclude that the increased levels of SFA may in part contribute to the effects exerted by L. delbrueckii subsp. lactis on worm longevity.
Sugar metabolism can also be correlated to lifespan, since evidences suggest that the presence of several carbohydrates, such as glucose, galactose or lactose, derived from bacterial metabolism, increase the nematode longevity (Brokate-Llanos et al., 2014). The ability of L. delbrueckii subsp. bulgaricus to secrete galactose could be related to its beneficial effects observed in C. elegans.
Conclusion
In the present work three strains belonging to L. delbrueckii subspecies bulgaricus and lactis were characterized applying a phenomic approach based on proteomic and metabolomic analysis as well as in vivo evaluation of their potential probiotic effects. Metabolic profiling highlighted the different origin of the three strains. Moreover, the L. delbrueckii subspecies bulgaricus strain demonstrated potential probiotic features. Overall, our results demonstrate that combination of omic platforms coupled with in vivo screening in the model organism C. elegans represents a powerful tool to characterize industrially relevant bacterial isolates.
Author Contributions
Conceived and designed the experiments: DU, CD, and AM. Wrote the paper: CD, DU, and EZ. Critical revision of manuscript: GP and CP. Did animal experiments/treatments: EZ and ES. Performed metabolomics experiments: FS. Performed proteomics experiments: SM. Analyzed and supervised proteomics data: PM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Part of the present study was supported by FEASR, Reg.CE n.1698/2005, PSR Regione Abruzzo 2007/2013: Misura 1.2.4.: G.A.P.INNO project (2013–2015) to AM, Ateneo 2015 C26A15Z54W to DU and Avvio alla ricerca 2016 to EZ. We thank CNR Project ‘FaReBio di Qualita,’ CNR-Regione Lombardia AMANDA Project to PM, as well as Valeria Bellettato for technical support.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01206/full#supplementary-material
References
- Brasili E., Mengheri E., Tomassini A., Capuani G., Roselli M., Finamore A., et al. (2013). Lactobacillus acidophilus La5 and Bifidobacterium lactis Bb12 induce different age-related metabolic profiles revealed by 1H-NMR spectroscopy in urine and feces of mice. J. Nutr. 143 1549–1557. 10.3945/jn.113.177105 [DOI] [PubMed] [Google Scholar]
- Brokate-Llanos A. M., Garzon A., Munoz M. J. (2014). Escherichia coli carbon source metabolism affects longevity of its predator Caenorhabditis elegans. Mech. Ageing Dev. 14 22–25. 10.1016/j.mad.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Calvani R., Brasili E., Pratico G., Sciubba F., Roselli M., Finamore A., et al. (2014). Application of NMR-based metabolomics to the study of gut microbiota in obesity. J. Clin. Gastroenterol. 48(Suppl. 1), S5–S7. 10.1097/MCG.0000000000000236 [DOI] [PubMed] [Google Scholar]
- Clark L. C., Hodgkin J. (2014). Commensals, probiotics and pathogens in the Caenorhabditis elegans model. Cell Microbiol. 16 27–38. 10.1111/cmi.12234 [DOI] [PubMed] [Google Scholar]
- Comunian C., Rusconi F., De Palma A., Brunetti P., Catalucci D., Mauri P. L. (2011). A comparative MudPIT analysis identifies different expression profiles in heart compartments. Proteomics 11 2320–2328. 10.1002/pmic.201000479 [DOI] [PubMed] [Google Scholar]
- Dellaglio F., Felis G. E., Castioni A., Torriani S., Germond J. E. (2005). Lactobacillus delbrueckii subsp. indicus subsp. nov., isolated from Indian dairy products. Int. J. Syst. Evol. Microbiol. 55 401–404. 10.1099/ijs.0.63067-0 [DOI] [PubMed] [Google Scholar]
- Devirgiliis C., Barile S., Perozzi G. (2011). Antibiotic resistance determinants in the interplay between food and gut microbiota. Genes Nutr. 6 275–284. 10.1007/s12263-011-0226-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devirgiliis C., Caravelli A., Coppola D., Barile S., Perozzi G. (2008). Antibiotic resistance and microbial composition along the manufacturing process of Mozzarella di Bufala Campana. Int. J. Food Microbiol. 128 378–384. 10.1016/j.ijfoodmicro.2008.09.021 [DOI] [PubMed] [Google Scholar]
- Edwards C., Canfield J., Copes N., Brito A., Rehan M., Lipps D., et al. (2015). Mechanisms of amino acid-mediated lifespan extension in Caenorhabditis elegans. BMC Genet. 16:8 10.1186/s12863-015-0167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kafsi H., Binesse J., Loux V., Buratti J., Boudebbouze S., Dervyn R., et al. (2014). Lactobacillus delbrueckii ssp. lactis and ssp. bulgaricus: a chronicle of evolution in action. BMC Genomics 15:407 10.1186/1471-2164-15-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W. M. T., Lane A. N. (2008). Lane Structure-based profiling of metabolites and isotopomers by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 52 69–117. 10.1007/s10858-011-9484-6 [DOI] [Google Scholar]
- Germond J. E., Lapierre L., Delley M., Mollet B., Felis G. E., Dellaglio F. (2003). Evolution of the bacterial species Lactobacillus delbrueckii: a partial genomic study with reflections on prokaryotic species concept. Mol. Biol. Evol. 20 93–104. 10.1093/molbev/msg012 [DOI] [PubMed] [Google Scholar]
- Gorietti D., Zanni E., Palleschi C., Delfini M., Uccelletti D., Saliola M., et al. (2014). Depletion of casein kinase I leads to a NAD(P)(+)/NAD(P)H balance-dependent metabolic adaptation as determined by NMR spectroscopy-metabolomic profile in Kluyveromyces lactis. Biochim. Biophys. Acta 1840 556–564. 10.1016/j.bbagen.2013.10.020 [DOI] [PubMed] [Google Scholar]
- Kudo Y., Oki K., Watanabe K. (2012). Lactobacillus delbrueckii subsp. sunkii subsp. nov., isolated from sunki, a traditional Japanese pickle. Int. J. Syst. Evol. Microbiol. 62 2643–2649. 10.1099/ijs.0.037051-0 [DOI] [PubMed] [Google Scholar]
- Lee J., Kwon G., Lim Y. H. (2015). Elucidating the mechanism of weissella-dependent lifespan extension in Caenorhabditis elegans. Sci. Rep. 5:17128 10.1038/srep17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. Q. (2003). Practical implications of lactate and pyruvate metabolism by lactic acid bacteria in food and beverage fermentations. Int. J. Food Microbiol. 83 115–131. 10.1016/S0168-1605(02)00366-5 [DOI] [PubMed] [Google Scholar]
- Mauri P., Deho G. (2008). A proteomic approach to the analysis of RNA degradosome composition in Escherichia coli. Methods Enzymol. 447 99–117. 10.1016/S0076-6879(08)02206-4 [DOI] [PubMed] [Google Scholar]
- Mauri P., Scarpa A., Nascimbeni A. C., Benazzi L., Parmagnani E., Mafficini A., et al. (2005). Identification of proteins released by pancreatic cancer cells by multidimensional protein identification technology: a strategy for identification of novel cancer markers. FASEB J. 19 1125–1127. 10.1096/fj.04-3000fje [DOI] [PubMed] [Google Scholar]
- Miccheli A., Aureli T., Delfini M., Di Cocco M. E., Viola P., Gobetto R., et al. (1988). Study on influence of inactivation enzyme techniques and extraction procedures on cerebral phosphorylated metabolite levels by 31P NMR spectroscopy. Cell Mol. Biol. 34 591–603. [PubMed] [Google Scholar]
- Miteva V., Boudakov I., Ivanova-Stoyancheva G., Marinova B., Mitev V., Mengaud J. (2001). Differentiation of Lactobacillus delbrueckii subspecies by ribotyping and amplified ribosomal DNA restriction analysis (ARDRA). J. Appl. Microbiol. 90 909–918. 10.1046/j.1365-2672.2001.01320.x [DOI] [PubMed] [Google Scholar]
- Morea M., Baruzzi F., Cocconcelli P. S. (1999). Molecular and physiological characterization of dominant bacterial populations in traditional mozzarella cheese processing. J. Appl. Microbiol. 87 574–582. 10.1046/j.1365-2672.1999.00855.x [DOI] [PubMed] [Google Scholar]
- Nakagawa H., Shiozaki T., Kobatake E., Hosoya T., Moriya T., Sakai F., et al. (2016). Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in Caenorhabditis elegans. Aging Cell 15 227–236. 10.1111/acel.12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y., Moriya T., Sakai F., Ikeda N., Shiozaki T., Hosoya T., et al. (2014). Oral administration of Lactobacillus gasseri SBT2055 is effective for preventing influenza in mice. Sci. Rep. 4:4638 10.1038/srep04638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. P., Clarke C. F. (2012). Folate status of gut microbiome affects Caenorhabditis elegans lifespan. BMC Biol. 10:66 10.1186/1741-7007-10-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. R., Oh S., Son S. J., Park D. J., Kim S. H., Jeong D. Y., et al. (2015). Bacillus licheniformis isolated from traditional korean food resources enhances the longevity of Caenorhabditis elegans through serotonin signaling. J. Agric. Food Chem. 63 10227–10233. 10.1021/acs.jafc.5b03730 [DOI] [PubMed] [Google Scholar]
- Park M. R., Yun H. S., Son S. J., Oh S., Kim Y. (2014). Short communication: development of a direct in vivo screening model to identify potential probiotic bacteria using Caenorhabditis elegans. J. Dairy Sci. 97 6828–6834. 10.3168/jds.2014-8561 [DOI] [PubMed] [Google Scholar]
- Rebollar E. A., Antwis R. E., Becker M. H., Belden L. K., Bletz M. C., Brucker R. M., et al. (2016). Using “Omics” and integrated multi-omics approaches to guide probiotic selection to mitigate chytridiomycosis and other emerging infectious diseases. Front. Microbiol. 7:68 10.3389/fmicb.2016.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz L., Hidalgo C., Blanco-Miguez A., Lourenco A., Sanchez B., Margolles A. (2016). Tackling probiotic and gut microbiota functionality through proteomics. J. Proteomics 147 28–39. 10.1016/j.jprot.2016.03.023 [DOI] [PubMed] [Google Scholar]
- Shmookler Reis R. J., Xu L., Lee H., Chae M., Thaden J. J., Bharill P., et al. (2011). Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging (Albany NY) 3 125–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So S., Tokumaru T., Miyahara K., Ohshima Y. (2011). Control of lifespan by food bacteria, nutrient limitation and pathogenicity of food in C. elegans. Mech. Ageing Dev. 132 210–212. 10.1016/j.mad.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Suzzi G., Lombardi A., Lanorte M. T., Caruso M., Andrighetto C., Gardini F. (2000). Phenotypic and genotypic diversity of yeasts isolated from water-buffalo Mozzarella cheese. J. Appl. Microbiol. 88 117–123. 10.1046/j.1365-2672.2000.00926.x [DOI] [PubMed] [Google Scholar]
- Tamang J. P., Watanabe K., Holzapfel W. H. (2016). Review: diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7:377 10.3389/fmicb.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uccelletti D., Zanni E., Marcellini L., Palleschi C., Barra D., Mangoni M. L. (2010). Anti-Pseudomonas activity of frog skin antimicrobial peptides in a Caenorhabditis elegans infection model: a plausible mode of action in vitro and in vivo. Antimicrob. Agents Chemother. 54 3853–3860. 10.1128/AAC.00154-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt K., LeMay J. D., Lamoureux M., Frenette M., Moineau S., Vadeboncoeur C. (2004). Characterization of a galactokinase-positive recombinant strain of Streptococcus thermophilus. Appl. Environ. Microbiol. 70 4596–4603. 10.1128/AEM.70.8.4596-4603.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virk B., Correia G., Dixon D. P., Feyst I., Jia J., Oberleitner N., et al. (2012). Excessive folate synthesis limits lifespan in the C. elegans: E. coli aging model. BMC Biol. 10:67 10.1186/1741-7007-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virk B., Jia J., Maynard C. A., Raimundo A., Lefebvre J., Richards S. A., et al. (2016). Folate Acts in E. coli to Accelerate C. elegans Aging Independently of Bacterial Biosynthesis. Cell Rep. 14 1611–1620. 10.1016/j.celrep.2016.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., et al. (2016). 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 44 D447–D456. 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss N., Schillinger U., Kandler O. (1983). Lactobacillus lactis, Lactobacillus leichmannii and Lactobacillus bulgaricus, subjective synonyms of Lactobacillus delbrueckii, and description of Lactobacillus delbrueckii subsp. lactis comb. nov. and Lactobacillus delbrueckii subsp. bulgaricus comb. nov. Syst. Appl. Microbiol. 4 552–557. 10.1016/S0723-2020(83)80012-5 [DOI] [PubMed] [Google Scholar]
- Yilmaz L. S., Walhout A. J. (2014). Worms, bacteria, and micronutrients: an elegant model of our diet. Trends Genet. 30 496–503. 10.1016/j.tig.2014.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Yan X., Ye C., Zhao H., Chen X., Hu F., et al. (2015). Bacterial respiration and growth rates affect the feeding preferences, brood size and lifespan of Caenorhabditis elegans. PLoS ONE 10:e0134401 10.1371/journal.pone.0134401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun H. S., Heo J. H., Son S. J., Park M. R., Oh S., Song M. H., et al. (2014). Bacillus licheniformis isolated from Korean traditional food sources enhances the resistance of Caenorhabditis elegans to infection by Staphylococcus aureus. J. Microbiol. Biotechnol. 24 1105–1108. 10.4014/jmb.1406.06008 [DOI] [PubMed] [Google Scholar]
- Zanni E., Laudenzi C., Schifano E., Palleschi C., Perozzi G., Uccelletti D., et al. (2015). Impact of a complex food microbiota on energy metabolism in the model organism Caenorhabditis elegans. Biomed. Res. Int. 2015:621709 10.1155/2015/621709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Karypis G. (2005). Data clustering in life sciences. Mol. Biotechnol. 31 55–80. 10.1385/MB:31:1:055 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Yu X., Jia R., Yang R., Rui Q., Wang D. (2015). Lactic acid bacteria protects Caenorhabditis elegans from toxicity of graphene oxide by maintaining normal intestinal permeability under different genetic backgrounds. Sci. Rep. 5:17233 10.1038/srep17233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.