ABSTRACT
Kaposi's sarcoma herpesvirus (KSHV) establishes lifelong latency. The viral latency-associated nuclear antigen (LANA) promotes viral persistence by tethering the viral genome to cellular chromosomes and by participating in latent DNA replication. Recently, the structure of the LANA C-terminal DNA binding domain was solved and new cytoplasmic variants of LANA were discovered. We discuss how these findings contribute to our current view of LANA structure and assembly and of its role during viral persistence.
KEYWORDS: Kaposi's sarcoma-associated herpesvirus, LANA speckles, cytoplasmic DNA sensors, cytoplasmic variants, structure, virus persistence
INTRODUCTION
Kaposi's sarcoma herpesvirus (KSHV) is the cause of three human malignancies—Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and the plasma cell variant of multicentric Castleman's disease (MCD). Like other herpesviruses, it establishes a lifelong infection of the host. KSHV persistence involves latency, during which no viral particles are produced, only a limited set of viral genes is expressed, and the circular latent genome is replicated and segregated to daughter cells as an episome by the host replication and cell division machinery. However, this mechanism of persistence is probably not very efficient as illustrated by the fact that, in cell culture, most KSHV-infected cells lose the viral genome rapidly (1). Notable exceptions to this rule are cultured cell lines established from PEL samples, which retain the viral genome indefinitely (2, 3). It is therefore possible that occasional low-level virus production and infection of new cells may be required for the persistence of KSHV in the infected host (1).
LANA DISCOVERY AND CHARACTERIZATION
One of the few proteins expressed during the latent phase of the viral life cycle, as well as in all KSHV-infected tumor cells, is the latency-associated nuclear antigen (LANA), which is essential for viral persistence. In this review, we focus on recently identified features of LANA that relate to its role in both the latent and lytic replication cycles during viral persistence. For a more comprehensive overview of LANA and its functions, we refer the reader to several excellent recent reviews (4–7).
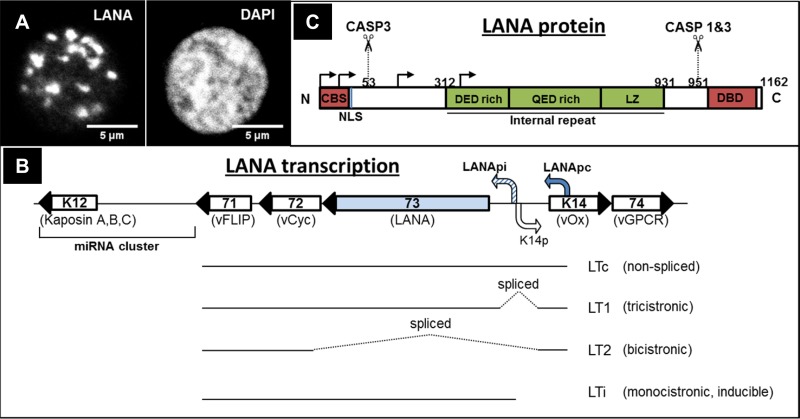
LANA was initially identified as a “speckled” nuclear fluorescence staining pattern (see Fig. 1A) recognized by serum antibodies from KSHV-infected patients (8–11). The size of the LANA protein is heterogeneous, and multiple protein bands in the range of 150 to 230 kDa, thought to be mostly the result of posttranslational modifications, can be seen on Western blots of lysates of infected cells (12–17). LANA is encoded by KSHV ORF73, and ORF73 and the neighboring genes ORF72/vcyc (viral cyclin), ORF71/vFLIP (viral FLICE inhibitory protein), and K12/kaposin and a transcript encoding 12 microRNAs (miRNAs) make up the major latency locus of KSHV (16, 18–21). LANA is translated from a spliced mRNA (LT1), whose transcription is directed by the constitutively active latency promoter (Fig. 1B) (22–25). In addition, a bidirectional promoter, located in the intron of the latent LANA transcript and activated by the KSHV lytic regulator RTA (replication and transcription activator), directs the expression of a “lytic” or “inducible” LANA transcript following the activation of the lytic replication cycle, as well as the expression of a bicistronic mRNA for the neighboring ORFK14/vOX2 (viral homolog of CD200 glycoprotein) and ORF74/vGPCR (viral G protein-coupled receptor) genes (Fig. 1B) (20, 26, 27). This transcriptional arrangement suggests a role for LANA during both latency and lytic reactivation.
FIG 1.
(A) Immunofluorescence staining of LANA speckles in PEL cells. (Left panel) LANA staining. (Right panel) DAPI (4′,6-diamidino-2-phenylindole) staining of the nucleus. (B) Simplified structure of KSHV latency locus, indicating the LANA constitutive promoter (Lana pc) and the bidirectional lytic LANA inducible (LANA pi)/K14 promoter, as well as transcripts produced from the latency locus (20, 22, 26, 27). Viral open reading frame (ORF) numbers are indicated inside the respective genes, and the customary names are given below. ORF71 (vFLIP—viral FLICE inhibitory protein), ORF72 (vCyc—viral cyclin), ORF73 (LANA), ORF K14 (vOx—viral homologue of OX2), ORF74 (vGPCR—viral G protein coupled receptor) are indicated. (C) Schematic representation of LANA domains and important motifs. Caspase 1 and 3 cleavage sites (CASP1/3) (13) are also indicated. Arrows indicate canonical and alternative translation initiation sites (17). CBS, chromatin binding site; NLS, nuclear localization signal; DBD, DNA binding domain; LZ, leucine zipper.
ROLE OF LANA IN VIRAL PERSISTENCE
LANA speckles (Fig. 1A) contain latent viral DNA and are attached to mitotic chromosomes during cell division (28–30). As mentioned above, LANA is essential for latent persistence: experiments using small interfering RNA (siRNA) to silence LANA expression in PEL cells harboring latent KSHV, or deleting LANA from a recombinant KSHV genome, showed that LANA is necessary for persistence of the viral genome in an episomal state (31–35). The presence of LANA is also sufficient to mediate the replication and maintenance of a plasmid containing the KSHV latent origin of replication in transfected cells (28, 36–38). These observations suggested a model of LANA replicating and tethering the KSHV episome to host chromosomes during cell division.
In order to perform these two functions—latent replication and tethering of viral episomes to mitotic chromosomes—LANA associates with cellular histones H2A and H2B via a domain at its N-terminal end (chromatin binding sequence [CBS; Fig. 1C]), binds to the viral latent origin of replication, which is located in each of the multiple terminal repeat (TR) subunits flanking the viral genome (29, 36, 39, 40), and recruits the cellular replication machinery. LANA was shown to colocalize and interact with ORC (origin recognition complex) (41, 42) and RFC (replication factor C) (43). Additionally, the MCM (minichromosome maintenance) complex, TopoIIβ (topoisomerase 2 β), and PCNA were shown to be recruited to the TR (41, 43, 44). LANA is also known to recruit a member of the replication fork protection complex, the Tim protein, to TR in order to regulate the formation of recombination structures that arise at the TR during replication and to promote the stability of TR elements (45).
LANA-MEDIATED CHROMATIN ASSOCIATION AND TRANSCRIPTIONAL REGULATION
LANA binds to the viral genome directly at the terminal repeat region (see below) but can also associate with it indirectly through protein-protein interactions. On the other hand, LANA associates with cellular DNA or chromatin, mostly through protein-protein interactions, but has also been suggested to bind directly to LANA binding site (LBS)-like sequences in the human genome (46–48). In the cellular chromatin/DNA, LANA associates preferentially with active promoters and was found to bind at locations close to transcriptional start sites (TSS) of H3K4me3-decorated promoters (46–48). Specific LBSs within the human genome seem to differ in different cell types (46–48). LANA was previously observed to associate with transcriptional activators and repressors (49–55) and may regulate the transcription of both viral and cellular genes. LANA activates its own promoter and represses the promoter of the lytic switch protein—RTA (replication and transcription activator) (47, 49, 56, 57). With regard to cellular genes, ectopic expression of LANA leads to both activation and repression of transcription (54, 57–59). However, the mechanism of LANA-mediated regulation of promoter activity has been identified only in the case of a few cellular genes (46, 60, 61). Additionally, a recent study in LEC (lymphatic endothelial cells) showed no correlation between LANA binding sites in the cellular promoters and their transcriptional activity (48). This suggests that mere binding of LANA to host regulatory sequences is, at least in the majority of cases, not sufficient for the control of cellular gene transcription.
LANA was also observed to associate with chromatin-modifying complexes, including the H3K9 methyltransferase SUV39H, H3K4 methyltransferase hSET1, a H3K9 demethylase, the histone acetyltransferase CBP, the histone deacetylase mSin3, and chromatin remodelers (FACT, CBP, and BET proteins) (46, 50, 62–65). Therefore, a role for LANA in the epigenetic modification of the KSHV or cellular genome has recently been suggested. The deposition of histones on the viral genome and their epigenetic modification, as well as the methylation of KSHV DNA, occur after entry of the virus into the cell and upon circularization of the viral genome (66–69). KSHV latency is associated with deposition of H3K27me3, a bivalent mark representing “poised” heterochromatin that can repress transcription despite the presence of activating modifications, on the promoters of lytic genes (67). LANA has been shown to be involved in the establishment of this epigenetic modification by directly recruiting PRC2 (polycomb repressive complex 2), which contains EZH2 (enhancer of zeste homolog 2), an H3K27me3 histone methyltransferase, or by mediating the redistribution of Sp100, a negative regulator of PRC2 recruitment, into different chromatin compartments (69, 70). LANA was also found to interact with hSET1, an H3K4-specific lysine methyltransferase, thereby inducing transcriptional activation of H3K4me3-decorated promoters (46). LANA has also been reported to promote lytic gene expression by recruiting JMJD1A (Jumonji domain-containing protein 1A)/KDM3 (lysine demethylase 3), an H3K9me1/2 histone demethylase, to the viral genome, resulting in an increase of viral gene expression (63). The balance between repressive H3K27me3 and activating H3K4me3 marks at both KSHV latent promoters as well as at promoters essential for reactivation may affect the switch between latency and lytic replication.
LANA STRUCTURE AND BINDING TO THE VIRAL LATENT ORIGIN OF REPLICATION
The prototypical LANA protein in the KSHV BC-1 strain consists of 1,162 amino acids (aa) (71) and can be divided into an N-terminal domain, a C-terminal domain, and the internal repeat region (Fig. 1C). The N-terminal domain contains the nuclear localization signal (NLS) responsible for the nuclear localization of the full-length LANA protein (72), as well as the motif responsible for the tethering of LANA speckles to mitotic chromatin (the CBS described above) (29, 39, 73). The N-terminal domain of LANA was also shown to play a role in replication and transcriptional regulation (29, 74, 75) and contains a motif for the recruitment of E3-type ubiquitin ligases (76). The internal repeat region consists of three sections, two of which are of a very acidic nature (DED rich and QED rich) and one that contains a leucine zipper (16, 18, 19, 71). This internal repeat region is required for KSHV genome persistence and contributes to immune evasion by preventing the presentation of antigenic peptides on MHC-I (major histocompatibility complex class I) molecules (31, 32, 77). The C-terminal domain contains a region responsible for the binding to the latent replication origin in the terminal repeat (TR) region of the viral genome (DNA binding domain [DBD]), contributes to the association of LANA with interphase chromatin, and is essential for latent replication (40, 78–82). The interaction with the interphase chromatin may involve binding to one or more candidates from a long list of cellular proteins, including, e.g., MeCP2 (methyl CpG binding protein 2), DEK, and Brd2/4 (bromodomain- and ET domain-containing protein 2/4) (53, 55, 82–86).
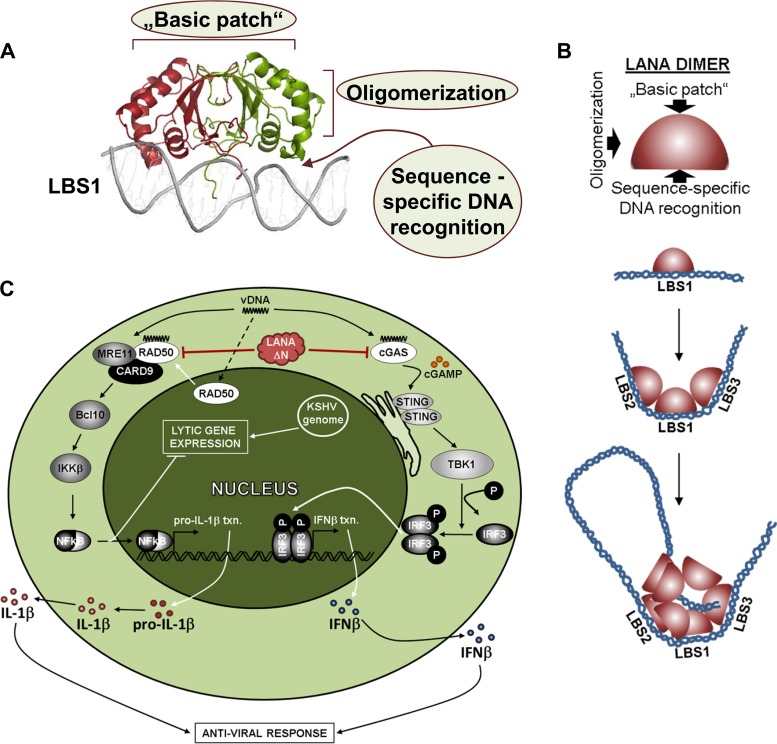
We and others have determined the structure of the LANA DNA binding domain (DBD) by X-ray crystallography (83, 87, 88). The purified LANA DBD forms dimers and multimers of dimers in solution. Some of the crystals obtained consisted of LANA DBD tetramers or pentamers of dimers arranged in a ring; in one crystal form, the pentamers appeared to be arranged in what looked like the beginning of a right-handed spiral of dimers (83, 88).
The basic LANA DBD dimer is formed by an antiparallel β-barrel structure at the dimerization interface and has three α-helices on each side of the β-barrel (Fig. 2A) (83, 87, 88). The bottom face of the dimer contains a specific binding site for the three LANA binding sites (LBS1 to LBS3) that are located in each of the 801-bp terminal repeat (TR) subunits flanking the KSHV genome in various numbers (40, 78–80, 87, 88). We recently also solved the structure of the LANA DBD dimer in complex with LBS1 (88). Unlike the latent origin binding proteins of Epstein-Barr virus (EBNA-1 [Epstein-Barr nuclear antigen 1]) and bovine papillomavirus (E2), the LANA DBD dimer binds asymmetrically to LBS1.
FIG 2.
(A) Structure of KSHV LANA dimer bound to LBS1. Functionally important surfaces are indicated. (B) Model showing how sequence-specific binding of LANA dimers to LBS1, LBS2, and LBS3 in combination with oligomerization and sequence-independent binding of LANA to the long unique region of the viral genome may lead to the assembly of larger structures that could form the basis of LANA speckles (Fig. 1A). (C) Diagram of N-terminally truncated LANA (LANAΔN) antagonizing viral DNA sensing by cGAS and the double-strand break repair machinery in the cytoplasm. A simplified version of the affected signaling pathways is shown. IL-1β, interleukin-1 beta; vDNA, viral DNA; IFNβ, beta interferon.
A LANA DBD dimer binds to LBS1 with approximately 100-fold-higher affinity than to LBS2 and LBS3, and the binding to LBS1, LBS2, and LBS3 is cooperative (88). The three LBSs are spaced apart by 22 bp and are therefore positioned on the same face of DNA, with one (LBS3) being located on the DNA strand opposite LBS1 and LBS2. This allows the three LANA DBD dimers binding to these three adjacent LBSs to interact with each other via a lateral alpha helical domain (88). Taking the data together, the asymmetric binding of three LANA DBD dimers to three adjacent LBS motifs was shown to induce a bend in the TR subunit DNA (Fig. 2B) (88–90).
The LANA DBD dimer face located opposite the sequence-specific DNA binding site features a characteristic “basic patch,” which was shown to be the binding site for the bromodomain and ET domain (BET) proteins but also to represent a sequence-independent DNA binding site (Fig. 2B) (83, 87, 88). We found that, in vitro, a LANA DBD mutant devoid of its specific binding site for LBS1 to LBS3 is able to coat DNA molecules, regardless of their sequence, by arranging a spiral of dimers around them (88). We postulated that a combination of these features, i.e., the sequence-specific interaction of LANA with the three LBSs (via the bottom face of the LANA DBD), together with the sequence-independent interaction with any DNA (host or viral) through the basic patch, as well as the ability of LANA to form different oligomer arrangements and to interact with other proteins, provides the basis for the assembly of the typical LANA speckles (Fig. 1A and 2B). These features may also allow LANA to assemble viral DNA structures as a first step in viral latent DNA replication (Fig. 2B). Latent replication could initiate at the TR region but also in other areas of the viral genome, including the long unique region (LUR) of the viral genome (91).
LANA ISOFORMS
As mentioned above, LANA is detectable on SDS-PAGE/Western blots as several bands with different molecular weights. While posttranslational modifications explain some of these multiple protein bands, there is currently also evidence of alternative translation initiation, alternative stop codons, internal frameshifting, and proteolytic cleavage contributing to the complexity of LANA isoforms (12, 13, 15, 17). Transfection of LANA expression vectors encoding a mutated start codon generates LANA variants by initiating translation at downstream translation initiation sites (dTIS); consequently, these lack the main nuclear localization signal located within the first 30 amino acids of full-length LANA (Fig. 1C) and are therefore located in the cytoplasm rather than the nucleus (17). In addition, frameshifting within the LANA internal repeat region results in shortened LANA variants with an alternative C-terminal domain translated from a different reading frame (15). Furthermore, the presence of proteolytic cleavage sites targeted by caspase 1 and 3 has been noted in the N- and C-terminal LANA domains (Fig. 1C) and caspase-mediated cleavage could therefore conceivably contribute to the generation of novel LANA isoforms (13).
ROLE OF CYTOPLASMIC LANA IN ANTAGONIZING INNATE IMMUNITY
The existence of N-terminally truncated LANA variants in the cytoplasm can be observed in KSHV-infected cells (17, 92). We recently showed that these play a role in the regulation of latency and in counteracting innate immune responses triggered by the presence of viral DNA in the cytosol. In particular, cytoplasmic LANA variants interact with and antagonize the innate DNA sensor cGAS (cyclic GMP-AMP synthase) (Fig. 2C) (92). The physiological role of cGAS is to synthesize the cyclic dinucleotide cGAMP upon recognizing cytoplasmic foreign DNA. cGAMP then binds to and activates STING (stimulator of interferon [IFN] genes), which, following relocation to the endoplasmic reticulum, induces the phosphorylation and activation of the cellular kinase TBK1 (TANK binding kinase 1) and thereby TBK1-mediated phosphorylation of IRF3 and subsequent increased expression from type I interferon gene promoters (93–98). Like other herpesviruses, KSHV is known to activate the cGAS-IRF3-IFN pathway during lytic replication (92, 99), presumably in response to viral DNA that may be inappropriately released from leaky capsids in the cytoplasm, as shown for herpes simplex virus 1 (HSV1) (100). For both HSV and KSHV, cGAS-dependent activation of interferon gene expression has been shown to restrict viral lytic replication (92, 101, 102). By binding to cGAS, cytoplasmic LANA variants inhibit the phosphorylation of TBK1 and IRF3 that occurs in response to the presence of transfected or viral DNA and may therefore antagonize the restriction on lytic KSHV replication imposed by cGAS and thus promote KSHV reactivation from latency, as well as—in an experimental heterologous system—lytic HSV1 replication (92). Thus, by promoting KSHV lytic replication, cytoplasmic LANA variants appear to exert a role that is opposed to that of full-length LANA, which, as reviewed above, promotes the establishment and maintenance of latency.
Antagonizing the cGAS-dependent activation of the IFN pathway clearly plays an important role in the KSHV lytic replication cycle, as illustrated by the fact that several other KSHV proteins antagonize either cGAS directly (e.g., orf52) (103) or the downstream STING protein (vIRF1) (99). In addition, multiple other KSHV proteins have been shown to downregulate the IFN response induced by transfected DNA (99), but their mode of action has not yet been clarified. The relative importance of cytoplasmic LANA versus these other KSHV proteins in antagonizing cGAS also needs to be investigated in more depth.
The role of cytoplasmic LANA as an antagonist of the innate immune response may go beyond its ability to inhibit cGAS-dependent IFN activation. By the use of coimmunoprecipitation and mass spectrometry, we and others have shown the interaction of LANA with Mre11 and RAD50, members of the MRN (Mre11, RAD50, NBS1) complex involved in the repair of the DNA double-strand breaks (92, 104). We found that, in concert with RAD50 and CARD9 (caspase recruitment domain-containing protein 9), cytoplasmic LANA variants can inhibit the previously reported (105) ability of Mre11 to activate the NF-κB pathway in response to foreign DNA (Fig. 2C) (106). The NF-κB pathway plays an important role in suppressing the lytic replication of KSHV and MHV68 and promoting latency (107–109). Several latent KSHV proteins, including full-length LANA and vFLIP, are potent activators of NF-κB (109–113). The ability of cytoplasmic LANA variants to antagonize Mre11-dependent NF-κB activation reinforces the notion that cytoplasmic LANA can oppose the effects of full-length LANA. It also provides an interesting example of the connection between DNA damage response (DDR) pathways and the innate immune response to foreign or damaged “self” DNA and the need for KSHV to restrain these cellular responses.
OUTLOOK
Our model of how LANA may assemble latent viral DNA into the typical nuclear speckles (Fig. 1A and 2B) and the observation that cytoplasmic LANA isoforms may play a role in the modulation of innate immune response pathways (Fig. 2C) suggest several questions:
If cytoplasmic LANA isoforms do indeed play a role in promoting the reactivation from latency by antagonizing the innate immune response, is their generation linked to the use of the alternative lytic LANA promoter (Fig. 1B)?
If so, does the shorter 5′ untranslated region (UTR) of the lytic LANA transcript (Fig. 1B) favor the use of alternative translation initiation codons?
If LANA has a role in antagonizing the innate immune response, initiated by either cGAS or the double-strand-break (DSB) repair machinery, is it conceivable that not just latent replication but also “shielding” of nuclear viral DNA from the innate immune response is an important role of LANA speckles?
If so, where does latent viral replication happen and could it be mediated by LANA molecules that are not packaged in the typical speckles?
Answering these and other questions will help our understanding of how KSHV, in spite of not being very efficient with regard to the latent replication/maintenance of its episomes, manages to persist in infected individuals throughout their lives.
ACKNOWLEDGMENTS
This work was supported by a grant to T.F.S. from the Deutsche Forschungsgemeinschaft (DFG) (Collaborative Research Centre SFB900 “Chronic Infections: Microbial Persistence and its Control,” Project C1). G.M. is a recipient of PhD fellowship IRTG 1273, funded by the DFG.
REFERENCES
- 1.Grundhoff A, Ganem D. 2004. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J Clin Invest 113:124–136. doi: 10.1172/JCI200417803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Mesri EA, Nador RG, Said JW, Asch AS, Knowles DM, Cesarman E. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648–2654. [PubMed] [Google Scholar]
- 3.Cesarman E, Moore PS, Rao PH, Inghirami G, Knowles DM, Chang Y. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708–2714. [PubMed] [Google Scholar]
- 4.Juillard F, Tan M, Li S, Kaye KM. 2016. Kaposi's sarcoma herpesvirus genome persistence. Front Microbiol 7:1149. doi: 10.3389/fmicb.2016.01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purushothaman P, Dabral P, Gupta N, Sarkar R, Verma SC. 2016. KSHV genome replication and maintenance. Front Microbiol 7:54. doi: 10.3389/fmicb.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uppal T, Banerjee S, Sun Z, Verma SC, Robertson ES. 2014. KSHV LANA–the master regulator of KSHV latency. Viruses 6:4961–4998. doi: 10.3390/v6124961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei F, Gan J, Wang C, Zhu C, Cai Q. 2016. Cell cycle regulatory functions of the KSHV oncoprotein LANA. Front Microbiol 7:334. doi: 10.3389/fmicb.2016.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao SJ, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo CR, Saah A, Phair J, Detels R, Chang Y, Moore PS. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med 2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 9.Kedes DH, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med 2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 10.Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcomae. J Virol 70:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson GR, Schulz TF, Whitby D, Cook PM, Boshoff C, Rainbow L, Howard MR, Gao SJ, Bohenzky RA, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder RS, Weller IV, Weiss RA, Moore PS. 1996. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet 348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 12.Canham M, Talbot SJ. 2004. A naturally occurring C-terminal truncated isoform of the latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus does not associate with viral episomal DNA. J Gen Virol 85:1363–1369. doi: 10.1099/vir.0.79802-0. [DOI] [PubMed] [Google Scholar]
- 13.Davis DA, Naiman NE, Wang V, Shrestha P, Haque M, Hu D, Anagho HA, Carey RF, Davidoff KS, Yarchoan R. 2015. Identification of caspase cleavage sites in KSHV latency-associated nuclear antigen and their effects on caspase-related host defense responses. PLoS Pathog 11:e1005064. doi: 10.1371/journal.ppat.1005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao SJ, Kingsley L, Hoover DR, Spira TJ, Rinaldo CR, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore PS. 1996. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N Engl J Med 335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 15.Kwun HJ, Toptan T, Ramos da Silva S, Atkins JF, Moore PS, Chang Y. 2014. Human DNA tumor viruses generate alternative reading frame proteins through repeat sequence recoding. Proc Natl Acad Sci U S A 111:E4342–E4349. doi: 10.1073/pnas.1416122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rainbow L, Platt GM, Simpson GR, Sarid R, Gao SJ, Stoiber H, Herrington CS, Moore PS, Schulz TF. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol 71:5915–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toptan T, Fonseca L, Kwun HJ, Chang Y, Moore PS. 2013. Complex alternative cytoplasmic protein isoforms of the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 generated through noncanonical translation initiation. J Virol 87:2744–2755. doi: 10.1128/JVI.03061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kedes DH, Lagunoff M, Renne R, Ganem D. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Invest 100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss RA, Talbot SJ. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J Hum Virol 1:19–29. [PubMed] [Google Scholar]
- 20.Pearce M, Matsumura S, Wilson AC. 2005. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J Virol 79:14457–14464. doi: 10.1128/JVI.79.22.14457-14464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadler R, Wu L, Forghani B, Renne R, Zhong W, Herndier B, Ganem D. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J Virol 73:5722–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J Virol 72:1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol 72:8309–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarid R, Wiezorek JS, Moore PS, Chang Y. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol 73:1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talbot SJ, Weiss RA, Kellam P, Boshoff C. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 26.Hilton IB, Dittmer DP. 2012. Quantitative analysis of the bidirectional viral G-protein-coupled receptor and lytic latency-associated nuclear antigen promoter of Kaposi's sarcoma-associated herpesvirus. J Virol 86:9683–9695. doi: 10.1128/JVI.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumura S, Fujita Y, Gomez E, Tanese N, Wilson AC. 2005. Activation of the Kaposi's sarcoma-associated herpesvirus major latency locus by the lytic switch protein RTA (ORF50). J Virol 79:8493–8505. doi: 10.1128/JVI.79.13.8493-8505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballestas ME, Chatis PA, Kaye KM. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 29.Barbera AJ, Ballestas ME, Kaye KM. 2004. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J Virol 78:294–301. doi: 10.1128/JVI.78.1.294-301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cotter MA II, Robertson ES. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 31.Alkharsah KR, Schulz TF. 2012. A role for the internal repeat of the Kaposi's sarcoma-associated herpesvirus latent nuclear antigen in the persistence of an episomal viral genome. J Virol 86:1883–1887. doi: 10.1128/JVI.06029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De León Vázquez E, Kaye KM. 2011. The internal Kaposi's sarcoma-associated herpesvirus LANA regions exert a critical role on episome persistence. J Virol 85:7622–7633. doi: 10.1128/JVI.00304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Godfrey A, Anderson J, Papanastasiou A, Takeuchi Y, Boshoff C. 2005. Inhibiting primary effusion lymphoma by lentiviral vectors encoding short hairpin RNA. Blood 105:2510–2518. doi: 10.1182/blood-2004-08-3052. [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Zhou F, Ye F, Gao SJ. 2008. Genetic disruption of KSHV major latent nuclear antigen LANA enhances viral lytic transcriptional program. Virology 379:234–244. doi: 10.1016/j.virol.2008.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang YJ, Wang KY, Stein DA, Patel D, Watkins R, Moulton HM, Iversen PL, Matson DO. 2007. Inhibition of replication and transcription activator and latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus by morpholino oligomers. Antiviral Res 73:12–23. doi: 10.1016/j.antiviral.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballestas ME, Kaye KM. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J Virol 75:3250–3258. doi: 10.1128/JVI.75.7.3250-3258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grundhoff A, Ganem D. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J Virol 77:2779–2783. doi: 10.1128/JVI.77.4.2779-2783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu J, Garber AC, Renne R. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J Virol 76:11677–11687. doi: 10.1128/JVI.76.22.11677-11687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 40.Garber AC, Shu MA, Hu J, Renne R. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Virol 75:7882–7892. doi: 10.1128/JVI.75.17.7882-7892.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stedman W, Deng Z, Lu F, Lieberman PM. 2004. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J Virol 78:12566–12575. doi: 10.1128/JVI.78.22.12566-12575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verma SC, Choudhuri T, Kaul R, Robertson ES. 2006. Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J Virol 80:2243–2256. doi: 10.1128/JVI.80.5.2243-2256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Q, Tsurimoto T, Juillard F, Li L, Li S, De Leon Vazquez E, Chen S, Kaye K. 2014. Kaposi's sarcoma-associated herpesvirus LANA recruits the DNA polymerase clamp loader to mediate efficient replication and virus persistence. Proc Natl Acad Sci U S A 111:11816–11821. doi: 10.1073/pnas.1404219111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purushothaman P, McDowell ME, McGuinness J, Salas R, Rumjahn SM, Verma SC. 2012. Kaposi's sarcoma-associated herpesvirus-encoded LANA recruits topoisomerase IIbeta for latent DNA replication of the terminal repeats. J Virol 86:9983–9994. doi: 10.1128/JVI.00839-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dheekollu J, Chen HS, Kaye KM, Lieberman PM. 2013. Timeless-dependent DNA replication-coupled recombination promotes Kaposi's sarcoma-associated herpesvirus episome maintenance and terminal repeat stability. J Virol 87:3699–3709. doi: 10.1128/JVI.02211-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J, Yang Y, Turner PC, Jain V, McIntyre LM, Renne R. 2014. LANA binds to multiple active viral and cellular promoters and associates with the H3K4methyltransferase hSET1 complex. PLoS Pathog 10:e1004240. doi: 10.1371/journal.ppat.1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu J, Verma SC, Cai Q, Saha A, Dzeng RK, Robertson ES. 2012. The RBP-Jkappa binding sites within the RTA promoter regulate KSHV latent infection and cell proliferation. PLoS Pathog 8:e1002479. doi: 10.1371/journal.ppat.1002479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mercier A, Arias C, Madrid AS, Holdorf MM, Ganem D. 2014. Site-specific association with host and viral chromatin by Kaposi's sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation. J Virol 88:6762–6777. doi: 10.1128/JVI.00268-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong JH, Orvis J, Kim JW, McMurtrey CP, Renne R, Dittmer DP. 2004. Regulation and autoregulation of the promoter for the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Biol Chem 279:16822–16831. doi: 10.1074/jbc.M312801200. [DOI] [PubMed] [Google Scholar]
- 50.Krithivas A, Young DB, Liao G, Greene D, Hayward SD. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J Virol 74:9637–9645. doi: 10.1128/JVI.74.20.9637-9645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim C, Lee D, Seo T, Choi C, Choe J. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J Biol Chem 278:7397–7405. doi: 10.1074/jbc.M211912200. [DOI] [PubMed] [Google Scholar]
- 52.Ottinger M, Christalla T, Nathan K, Brinkmann MM, Viejo-Borbolla A, Schulz TF. 2006. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J Virol 80:10772–10786. doi: 10.1128/JVI.00804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platt GM, Simpson GR, Mittnacht S, Schulz TF. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol 73:9789–9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shamay M, Krithivas A, Zhang J, Hayward SD. 2006. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc Natl Acad Sci U S A 103:14554–14559. doi: 10.1073/pnas.0604469103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viejo-Borbolla A, Ottinger M, Bruning E, Burger A, Konig R, Kati E, Sheldon JA, Schulz TF. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J Virol 79:13618–13629. doi: 10.1128/JVI.79.21.13618-13629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeong J, Papin J, Dittmer D. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J Virol 75:1798–1807. doi: 10.1128/JVI.75.4.1798-1807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renne R, Barry C, Dittmer D, Compitello N, Brown PO, Ganem D. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Virol 75:458–468. doi: 10.1128/JVI.75.1.458-468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.An FQ, Compitello N, Horwitz E, Sramkoski M, Knudsen ES, Renne R. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J Biol Chem 280:3862–3874. doi: 10.1074/jbc.M407435200. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe T, Sugaya M, Atkins AM, Aquilino EA, Yang A, Borris DL, Brady J, Blauvelt A. 2003. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen prolongs the life span of primary human umbilical vein endothelial cells. J Virol 77:6188–6196. doi: 10.1128/JVI.77.11.6188-6196.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu F, Tsai K, Chen HS, Wikramasinghe P, Davuluri RV, Showe L, Domsic J, Marmorstein R, Lieberman PM. 2012. Identification of host-chromosome binding sites and candidate gene targets for Kaposi's sarcoma-associated herpesvirus LANA. J Virol 86:5752–5762. doi: 10.1128/JVI.07216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su L, Liao Q, Wu Y, Chen X. 2011. Kaposi's sarcoma-associated herpesvirus-encoded LANA down-regulates IL-22R1 expression through a cis-acting element within the promoter region. PLoS One 6:e19106. doi: 10.1371/journal.pone.0019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu J, Liu E, Renne R. 2009. Involvement of SSRP1 in latent replication of Kaposi's sarcoma-associated herpesvirus. J Virol 83:11051–11063. doi: 10.1128/JVI.00907-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim KY, Huerta SB, Izumiya C, Wang DH, Martinez A, Shevchenko B, Kung HJ, Campbell M, Izumiya Y. 2013. Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen regulates the KSHV epigenome by association with the histone demethylase KDM3A. J Virol 87:6782–6793. doi: 10.1128/JVI.00011-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim C, Gwack Y, Hwang S, Kim S, Choe J. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Biol Chem 276:31016–31022. doi: 10.1074/jbc.M102431200. [DOI] [PubMed] [Google Scholar]
- 65.Sakakibara S, Ueda K, Nishimura K, Do E, Ohsaki E, Okuno T, Yamanishi K. 2004. Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen. J Virol 78:7299–7310. doi: 10.1128/JVI.78.14.7299-7310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bechtel JT, Winant RC, Ganem D. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J Virol 79:4952–4964. doi: 10.1128/JVI.79.8.4952-4964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Günther T, Grundhoff A. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog 6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, Jung JU. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog 6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toth Z, Papp B, Brulois K, Choi YJ, Gao SJ, Jung JU. 2016. LANA-mediated recruitment of host polycomb repressive complexes onto the KSHV genome during de novo infection. PLoS Pathog 12:e1005878. doi: 10.1371/journal.ppat.1005878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Günther T, Schreiner S, Dobner T, Tessmer U, Grundhoff A. 2014. Influence of ND10 components on epigenetic determinants of early KSHV latency establishment. PLoS Pathog 10:e1004274. doi: 10.1371/journal.ppat.1004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A 93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cherezova L, Burnside KL, Rose TM. 2011. Conservation of complex nuclear localization signals utilizing classical and non-classical nuclear import pathways in LANA homologs of KSHV and RFHV. PLoS One 6:e18920. doi: 10.1371/journal.pone.0018920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Piolot T, Tramier M, Coppey M, Nicolas JC, Marechal V. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J Virol 75:3948–3959. doi: 10.1128/JVI.75.8.3948-3959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lim C, Choi C, Choe J. 2004. Mitotic chromosome-binding activity of latency-associated nuclear antigen 1 is required for DNA replication from terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus. J Virol 78:7248–7256. doi: 10.1128/JVI.78.13.7248-7256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong LY, Matchett GA, Wilson AC. 2004. Transcriptional activation by the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen is facilitated by an N-terminal chromatin-binding motif. J Virol 78:10074–10085. doi: 10.1128/JVI.78.18.10074-10085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. 2006. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog 2:e116. doi: 10.1371/journal.ppat.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwun HJ, da Silva SR, Qin H, Ferris RL, Tan R, Chang Y, Moore PS. 2011. The central repeat domain 1 of Kaposi's sarcoma-associated herpesvirus (KSHV) latency associated-nuclear antigen 1 (LANA1) prevents cis MHC class I peptide presentation. Virology 412:357–365. doi: 10.1016/j.virol.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fejér G, Medveczky MM, Horvath E, Lane B, Chang Y, Medveczky PG. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts preferentially with the terminal repeats of the genome in vivo and this complex is sufficient for episomal DNA replication. J Gen Virol 84:1451–1462. doi: 10.1099/vir.0.18940-0. [DOI] [PubMed] [Google Scholar]
- 79.Garber AC, Hu J, Renne R. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J Biol Chem 277:27401–27411. doi: 10.1074/jbc.M203489200. [DOI] [PubMed] [Google Scholar]
- 80.Kelley-Clarke B, Ballestas ME, Srinivasan V, Barbera AJ, Komatsu T, Harris TA, Kazanjian M, Kaye KM. 2007. Determination of Kaposi's sarcoma-associated herpesvirus C-terminal latency-associated nuclear antigen residues mediating chromosome association and DNA binding. J Virol 81:4348–4356. doi: 10.1128/JVI.01289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelley-Clarke B, De Leon-Vazquez E, Slain K, Barbera AJ, Kaye KM. 2009. Role of Kaposi's sarcoma-associated herpesvirus C-terminal LANA chromosome binding in episome persistence. J Virol 83:4326–4337. doi: 10.1128/JVI.02395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krithivas A, Fujimuro M, Weidner M, Young DB, Hayward SD. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J Virol 76:11596–11604. doi: 10.1128/JVI.76.22.11596-11604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hellert J, Weidner-Glunde M, Krausze J, Richter U, Adler H, Fedorov R, Pietrek M, Ruckert J, Ritter C, Schulz TF, Luhrs T. 2013. A structural basis for BRD2/4-mediated host chromatin interaction and oligomer assembly of Kaposi sarcoma-associated herpesvirus and murine gammaherpesvirus LANA proteins. PLoS Pathog 9:e1003640. doi: 10.1371/journal.ppat.1003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matsumura S, Persson LM, Wong L, Wilson AC. 2010. The latency-associated nuclear antigen interacts with MeCP2 and nucleosomes through separate domains. J Virol 84:2318–2330. doi: 10.1128/JVI.01097-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Si H, Verma SC, Lampson MA, Cai Q, Robertson ES. 2008. Kaposi's sarcoma-associated herpesvirus-encoded LANA can interact with the nuclear mitotic apparatus protein to regulate genome maintenance and segregation. J Virol 82:6734–6746. doi: 10.1128/JVI.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.You J, Srinivasan V, Denis GV, Harrington WJ Jr, Ballestas ME, Kaye KM, Howley PM. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J Virol 80:8909–8919. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Domsic JF, Chen HS, Lu F, Marmorstein R, Lieberman PM. 2013. Molecular basis for oligomeric-DNA binding and episome maintenance by KSHV LANA. PLoS Pathog 9:e1003672. doi: 10.1371/journal.ppat.1003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hellert J, Weidner-Glunde M, Krausze J, Lunsdorf H, Ritter C, Schulz TF, Luhrs T. 2015. The 3D structure of Kaposi sarcoma herpesvirus LANA C-terminal domain bound to DNA. Proc Natl Acad Sci U S A 112:6694–6699. doi: 10.1073/pnas.1421804112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ponnusamy R, Petoukhov MV, Correia B, Custodio TF, Juillard F, Tan M, Pires de Miranda M, Carrondo MA, Simas JP, Kaye KM, Svergun DI, McVey CE. 2015. KSHV but not MHV-68 LANA induces a strong bend upon binding to terminal repeat viral DNA. Nucleic Acids Res 43:10039–10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wong LY, Wilson AC. 2005. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen induces a strong bend on binding to terminal repeat DNA. J Virol 79:13829–13836. doi: 10.1128/JVI.79.21.13829-13836.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Verma SC, Lu J, Cai Q, Kosiyatrakul S, McDowell ME, Schildkraut CL, Robertson ES. 2011. Single molecule analysis of replicated DNA reveals the usage of multiple KSHV genome regions for latent replication. PLoS Pathog 7:e1002365. doi: 10.1371/journal.ppat.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang G, Chan B, Samarina N, Abere B, Weidner-Glunde M, Buch A, Pich A, Brinkmann MM, Schulz TF. 2016. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc Natl Acad Sci U S A 113:E1034–E1043. doi: 10.1073/pnas.1516812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. 2013. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. 2013. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep 3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. 2013. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V. 2014. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J 33:2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell 51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. 2015. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A 112:E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Horan KA, Hansen K, Jakobsen MR, Holm CK, Soby S, Unterholzner L, Thompson M, West JA, Iversen MB, Rasmussen SB, Ellermann-Eriksen S, Kurt-Jones E, Landolfo S, Damania B, Melchjorsen J, Bowie AG, Fitzgerald KA, Paludan SR. 2013. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol 190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, Neipel F, Sturzl M, Whitby D, Li H, Zhu F. 2015. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, Von Dollen J, Maher MC, Johnson T, Newton W, Jager S, Shales M, Horner J, Hernandez RD, Krogan NJ, Glaunsinger BA. 2015. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol Cell 57:349–360. doi: 10.1016/j.molcel.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roth S, Rottach A, Lotz-Havla AS, Laux V, Muschaweckh A, Gersting SW, Muntau AC, Hopfner KP, Jin L, Vanness K, Petrini JH, Drexler I, Leonhardt H, Ruland J. 2014. Rad50-CARD9 interactions link cytosolic DNA sensing to IL-1beta production. Nat Immunol 15:538–545. doi: 10.1038/ni.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mariggiò G, Koch S, Zhang G, Weidner-Glunde M, Rückert J, Kati S, Santag S, Schulz TF. 21 April 2017. Kaposi sarcoma herpesvirus (KSHV) latency-associated nuclear antigen (LANA) recruits components of the MRN (Mre11-Rad50-NBS1) repair complex to modulate an innate immune signaling pathway and viral latency. PLoS Pathog doi: 10.1371/journal.ppat.1006335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bussey KA, Reimer E, Todt H, Denker B, Gallo A, Konrad A, Ottinger M, Adler H, Sturzl M, Brune W, Brinkmann MM. 2014. The gammaherpesviruses Kaposi's sarcoma-associated herpesvirus and murine gammaherpesvirus 68 modulate the Toll-like receptor-induced proinflammatory cytokine response. J Virol 88:9245–9259. doi: 10.1128/JVI.00841-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ehrlich ES, Chmura JC, Smith JC, Kalu NN, Hayward GS. 2014. KSHV RTA abolishes NFkappaB responsive gene expression during lytic reactivation by targeting vFLIP for degradation via the proteasome. PLoS One 9:e91359. doi: 10.1371/journal.pone.0091359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Graham C, Matta H, Yang Y, Yi H, Suo Y, Tolani B, Chaudhary PM. 2013. Kaposi's sarcoma-associated herpesvirus oncoprotein K13 protects against B cell receptor-induced growth arrest and apoptosis through NF-kappaB activation. J Virol 87:2242–2252. doi: 10.1128/JVI.01393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Field N, Low W, Daniels M, Howell S, Daviet L, Boshoff C, Collins M. 2003. KSHV vFLIP binds to IKK-gamma to activate IKK. J Cell Sci 116:3721–3728. doi: 10.1242/jcs.00691. [DOI] [PubMed] [Google Scholar]
- 111.Konrad A, Wies E, Thurau M, Marquardt G, Naschberger E, Hentschel S, Jochmann R, Schulz TF, Erfle H, Brors B, Lausen B, Neipel F, Sturzl M. 2009. A systems biology approach to identify the combination effects of human herpesvirus 8 genes on NF-kappaB activation. J Virol 83:2563–2574. doi: 10.1128/JVI.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu L, Eby MT, Rathore N, Sinha SK, Kumar A, Chaudhary PM. 2002. The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the Ikappa B kinase complex. J Biol Chem 277:13745–13751. doi: 10.1074/jbc.M110480200. [DOI] [PubMed] [Google Scholar]
- 113.Matta H, Chaudhary PM. 2004. Activation of alternative NF-kappa B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1 beta-converting enzyme inhibitory protein (vFLIP). Proc Natl Acad Sci U S A 101:9399–9404. doi: 10.1073/pnas.0308016101. [DOI] [PMC free article] [PubMed] [Google Scholar]