Abstract
In a plant cell, a subset of actin filaments function as a scaffold that positions the endomembrane system and acts as a substrate on which organelle motility occurs. Other actin filament arrays appear to be more dynamic and reorganize in response to growth signals and external cues. The distorted group of trichome morphology mutants provides powerful genetic tools to study the control of actin filament nucleation in the context of morphogenesis. In this article, we report that DISTORTED3 (DIS3) encodes a plant-specific SCAR/WAVE homolog. Null alleles of DIS3, like those of other Arabidopsis thaliana WAVE and Actin-Related Protein (ARP) 2/3 subunit genes, cause trichome distortion, defects in cell–cell adhesion, and reduced hypocotyl growth in etiolated seedlings. DIS3 efficiently activates the actin filament nucleation and branching activity of vertebrate Arp2/3 and functions within a WAVE-ARP2/3 pathway in vivo. DIS3 may assemble into a WAVE complex via a physical interaction with a highly diverged Arabidopsis Abi-1–like bridging protein. These results demonstrate the utility of the Arabidopsis trichome system to understand how the WAVE and ARP2/3 complexes translate signaling inputs into a coordinated morphogenetic response.
INTRODUCTION
Growing plant cells contain several types of actin arrays that are likely to conduct different functions in the cell. Heavily bundled actin filaments are commonly observed throughout the cytoplasm and in transvacuolar strands in growing plant cells. They provide a scaffold that positions the endoplasmic reticulum (Boevink et al., 1998) and maintain conduits for long distance organelle transport. Organelles such as chloroplasts (Sheahan et al., 2004), peroxisomes (Collings et al., 2003), and Golgi stacks (Nebenfuhr et al., 1999) traffic on actin bundles. In highly vacuolated cells, long-distance transport may be important for organelle inheritance during cell division or may satisfy local metabolic demand. Actin filaments mediate short-range recycling of endosomal compartments such as those in which the auxin efflux carrier resides (Geldner et al., 2001). Plant cells also contain cortical actin filaments that are in close proximity to the plasma membrane. In many cell types, regions of active cell expansion correlate with the presence of fine networks of actin filaments; however, the in vivo organization and function of cortical actin remains a mystery. In tip-growing pollen tubes, cortical actin may regulate the directed transport of vesicles to the tube tip, facilitate endocytotic recovery of excess membrane and integral membrane proteins, or maintain the cytoplasmic architecture of the tip zone (Gibbon et al., 1999; Fu et al., 2001). In non-plant cells, the energy of actin polymerization itself provides essential functions that drive plasma membrane protrusion, organelle motility, and endocytosis (Welch and Mullins, 2002; Pollard and Borisy, 2003). Although plants employ many conserved actin binding proteins (Staiger and Hussey, 2004), the signaling pathways and protein complexes that control actin filament nucleation are not well characterized.
Genetic approaches to plant morphogenesis have provided many new clues about the cytoskeletal basis of growth (Wasteneys and Galway, 2003). The “distorted group” is a particular class of trichome morphology mutants that define a WAVE–Actin-Related Protein 2/3 (ARP2/3) morphogenesis pathway (Basu et al., 2004; Brembu et al., 2004; Deeks et al., 2004; El-Assal et al., 2004b). Arp2/3 is an evolutionarily conserved seven-subunit multiprotein complex that was originally isolated from Acanthamoeba (Machesky et al., 1994). Arp2/3 is an actin filament-nucleating machine that binds to the sides of existing actin filaments and nucleates new filaments at a distinctive 70° angle (Blanchoin et al., 2000). In non-plant cells, Arp2/3 provides essential driving forces for plasma membrane protrusion during cell crawling (Svitkina and Borisy, 1999). Mutations in Arp2/3 subunit genes in flies and worms cause embryonic lethality (Hudson and Cooley, 2002; Stevenson et al., 2002; Sawa et al., 2003). Arp2/3-mediated cell migration is dynamic and allows cells to alter their shape and reorient their motility in response to extracellular signals. In Drosophila, signaling through the WAVE complex mediates growth cone guidance in developing neurons (Bogdan and Klambt, 2003; Bogdan et al., 2004).
The nearly identical phenotypes of DISTORTED2 (DIS2 [ARPC2]), WURM (ARP2), DISTORTED1 (DIS1 [ARP3]), and CROOKED (CRK [ARPC5]) mutants strongly suggest that an Arabidopsis thaliana ARP2/3 complex exists. The ability of human ARPC5 to rescue crk (arpc5) phenotypes (Mathur et al., 2003b) and the ability of DIS1 (ARP3) (Le et al., 2003) and DIS2 (ARPC2) (El-Assal et al., 2004a) to rescue the severe growth defects of budding yeast cells that lack the corresponding Arp2/3 subunit gene demonstrate the potential of the plant genes to function in the context of an Arp2/3 complex. The binding of ARPC2 to the ARPC4 subunit defines the core dimer that is necessary for Arp2/3 assembly (Gournier et al., 2001). The physical interaction between DIS2 (ARPC2) and ARPC4 subunits is a retained and essential aspect of Arabidopsis ARP2/3 function (El-Assal et al., 2004a). However, despite strong indirect evidence for the presence of a plant ARP2/3 complex, direct biochemical data for the existence and activity of a seven-subunit complex is absent.
In the absence of activators (also called nucleation promoting factors) Arp2/3 has little or no activity. The WASP (for Wiskott-Aldrich syndrome protein)/Scar (for suppressor of cAMP receptor defects)/WAVE (for WASP family verprolin homologous protein) family of Arp2/3 activators encodes a composite WA domain (also referred to as VCA) at the C terminus. WA binds G-actin and Arp2/3 as part of its activation mechanism (Welch and Mullins, 2002; Stradal et al., 2004). WASP proteins are autoinhibited. Binding of ligands such as Cdc42 and PtdIns(4,5)P2 unleashes the potential of WASP to activate Arp2/3 (Rohatgi et al., 2000). SCAR/WAVE activity is regulated in trans by physical interaction with the heterotetrameric WAVE complex (Eden et al., 2002). Assembly of SCAR/WAVE into a WAVE complex occurs through the N-terminal SCAR homology domain, which is the defining feature of SCAR/WAVE proteins. The five subunits of the WAVE complex have many names: Sra-1/PIR121/GEX-2/CYFIP, NAP1/NAP125/HEM/GEX-3/KETTE, Abi-1 or Abi-2, Scar/WAVE, and HSPC300/Brick1. The WAVE-ARP2/3 pathway is an important pathway for actin filament nucleation in multicellular organisms, but unifying principles of RHO-GTPase-WAVE-ARP2/3 pathway functions have not yet emerged (Blagg and Insall, 2004).
The distorted group genes also encode WAVE complex subunits. Mutations in the WAVE complex subunit genes PIROGI (ATSRA-1/PIRP [SRA-1]) and GNARLED (ATNAP125/ATNAP1/NAPP [NAP1]), like those in ARP2/3 subunit genes, reduce shoot fresh weight, decrease hypocotyl length in the dark, and alter pavement cell shape as well as epidermal cell–cell adhesion (Basu et al., 2004; Brembu et al., 2004; Deeks et al., 2004; El-Assal et al., 2004b). WAVE and ARP2/3 subunit mutants also have very similar actin cytoskeleton defects. The simplest explanation for these observations is that Arabidopsis WAVE subunit genes positively regulate ARP2/3. Arabidopsis WAVE may be a Rho-of-Plants (ROP) effector complex. PIR (SRA-1) and human SRA-1 bind interchangeably and with a high degree of specificity to active forms of RAC1 and ROP2 small GTPases (Basu et al., 2004). Given the conserved functions of PIR (SRA-1) and GRL (NAP1) (Basu et al., 2004; El-Assal et al., 2004b), it seems likely, despite the absence of clear homologs in the plant sequence databases, that additional WAVE subunits exist and would cause trichome distortion when mutated. Because the rules for WAVE complex assembly are well established (Echarri et al., 2004; Gautreau et al., 2004; Innocenti et al., 2004), we and others have noted the possibility that Arabidopsis proteins have retained the structural domains that mediate WAVE complex assembly. In the case of DISTORTED3 (DIS3), bioinformatic and forward genetic approaches converged on the discovery of a putative WAVE complex subunit that directly activates ARP2/3.
In this article, we show that DIS3 encodes a Scar/WAVE-like subunit of a putative plant WAVE complex. DIS3 has a domain organization that resembles Scar/WAVE proteins. The DIS3 WA domain efficiently activates Arp2/3. Moreover, the evolutionarily conserved SCAR homology domain of DIS3 physically interacts with a highly diverged Arabidopsis Abi-1–like protein. By several genetic and biochemical criteria, DIS3 functions as a redundant positive regulator of ARP2/3 in a WAVE-ARP2/3 pathway. The functional analysis of DIS3 provides important new knowledge about the composition and activity of the putative plant WAVE complex.
RESULTS
DIS3 Affects Multiple Morphogenetic Processes
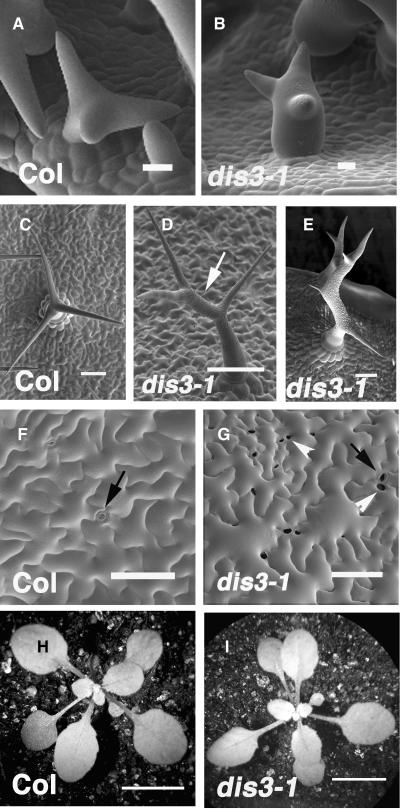
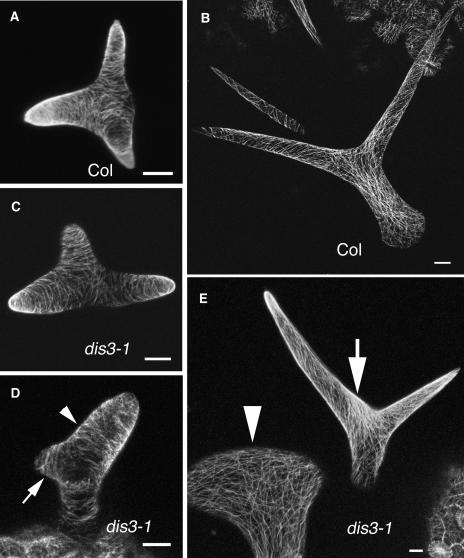
We isolated several recessive dis3 alleles in a mutant screen for plants that had trichomes with a normal branch number but with a swollen shape. After branch initiation, wild-type stalks and branches elongate in a highly predictable and coordinated manner (Szymanski et al., 1999), and the tip transitions from a hemispherical dome to a progressively refined pointed morphology (Figure 1A). Stage 4 trichomes have a blunt tip (Szymanski et al., 1998). Similarly staged dis3 trichomes displayed abnormal cell swelling and reduced branch length (Figure 1B), and 20% (n = 25) of the dis3 trichomes swelled to a diameter exceeding 30 μm, a value that is never observed in the wild type. This percentage of severely swollen cells was less than the values reported for similarly staged dis1 (arp3, 40%), dis2 (arpc2, 30%), pir (sra-1, 36%), and grl (nap1, 32%) mutants (Le et al., 2003; Basu et al., 2004; El-Assal et al., 2004a, 2004b). The decreased severity of the dis3 phenotype compared with other WAVE and ARP2/3 subunit mutants was also apparent in mature trichomes. Of the dis3-1 (n = 105), dis3-2 (n = 89), dis3-4 (n = 91), and dis3-T1 (n = 65) trichomes that were measured on the first leaf pair, between 5 and 10% had elongated stalks and tapered branches that closely resembled those of the wild type. The fraction of trichomes with a wild-type appearance was 28% (n = 97) for dis3-3. As a standard for comparison, 100% of the dis2 (arpc2) trichomes (n = 103) had a strong distorted phenotype. For all of the dis3 alleles, trichomes displaying distortion fell into two categories. Approximately half of them had a mild phenotype in which the interbranch zone, which is defined as the stalk-like cell volume that connects the most basal branch and the adjacent proximal branch, was abnormally expanded (Figure 1D, arrow). In most wild-type trichomes, the interbranch zone was minimal (Table 1) and the branches emanated from a confined apical region of the stalk (Figure 1C). The remaining fraction of dis3 trichomes had short and twisted branches and cell swelling at multiple locations (Figure 1E).
Figure 1.
Mutation of DISTORTED3 Causes Trichome Distortion and Defective Cell–Cell Adhesion in the Cotyledon Epidermis.
(A) Scanning electron micrograph of a wild-type stage 4 trichome that contains young branches with a blunt tip morphology.
(B) A representative stage 4 dis3-1 trichome with a blunt tip morphology and a swollen stalk.
(C) Mature wild-type (Columbia-0 [Col-0]) trichome displaying three highly elongated and tapered branches.
(D) An example of the mild dis3-1 mature trichome phenotype. Trichomes in this class display an abnormally elongated interbranch zone (arrow) and mild branch swelling.
(E) An example of the severe dis3-1 mature trichome phenotype. Branch lengths are reduced and slightly swollen. The interbranch zone is abnormally elongated and swollen.
(F) Upper surface of a representative 12-DAG wild-type cotyledon. Pavement cells are highly lobed. Image was taken from the most highly expanded cells in the apical third of the cotyledon.
(G) Upper surface of representative 12-DAG dis3-1 cotyledon pavement cells. Image was taken from the most highly expanded cells in the apical third of the cotyledon.
(H) Low magnification image of a representative 14-DAG wild-type seedling.
(I) Low magnification image of a representative 14-DAG dis3-1 seedling.
The white arrowheads label the gaps between adjacent pavement cells, black arrows label stomatal pores, and white arrows label the interbranch zone of a dis3 trichome. Bars = 10 μm in (A) and (B), 100 μm in (C) to (H), and 1 cm in (I) and (J).
Table 1.
Quantitation of the Trichome Phenotypes of Col, dis3, dis2, and dis3 dis2 Plants
| Wild Type | dis3-1 | dis2-1 | dis3-1 dis2-1 | |
|---|---|---|---|---|
| Trichome branch length (μm) | ||||
| Branch 1 | 229 ± 80a (n = 9) | 187 ± 54 (n = 10) | 58 ± 51* (n = 13) | 63 ± 46* (n = 17) |
| Branch 2 | 198 ± 44 (n = 9) | 115 ± 50* (n = 10) | 23 ± 12* (n = 13) | 20 ± 14* (n = 17) |
| Branch 3 | 137 ± 43 (n = 9) | 92 ± 48* (n = 10) | 18 ± 7* (n = 13) | 13 ± 5* (n = 17) |
| Interbranch zone length (μm) | 15 ± 5 (n = 9) | 58 ± 30* (n = 10) | 59 ± 48* (n = 13) | 43 ± 37* (n = 17) |
| Maximum diameter (μm) | 40 ± 9 (n = 9) | 41 ± 8 (n = 10) | 39 ± 12 (n = 13) | 44 ± 15 (n = 17) |
Mean value ± sd; (*), significantly different than the wild type according to the Student's t test (P value < 0.05).
To better understand the shape defects of dis3 trichomes, we compared directly several parameters of wild-type, dis3, and dis2 (arpc2) trichomes. Wild-type trichomes usually contain three branches of progressively shorter lengths. The longest had a mean length of ∼230 μm, and the shortest branch had a mean length of ∼140 μm (Table 1). For dis3-1 trichomes, the mean length of two of three branches was significantly less than the wild type. However, the reduced branch length of dis3 was less severe than dis2 (arpc2) (Table 1) and published mean branch lengths of pir (sra-1) and grl (nap1) (Basu et al., 2004; El-Assal et al., 2004b). dis3-1 and dis2 (arpc2) trichomes contained an abnormally expanded interbranch zone that was significantly longer than the wild type (Table 1). Although distorted mutants are abnormally swollen and occasionally acquire large cell diameters that are never observed in the wild type, the mean maximum diameters of dis2-1 (arpc2) and dis3-1 trichomes were not significantly different from the wild type (Table 1). The branch lengths of dis3-1, dis3-2, dis3-3, dis3-4, and dis3-T1 also were measured from digital images acquired on a dissecting microscope. For each of these alleles, at least two of the three trichome branches were significantly shorter than the wild type, and there were no clear differences among the dis3 alleles (Table 2).
Table 2.
Quantitation of the Branch Lengths from Trichomes on the First Leaf Pair of Wild-Type, dis3-1, dis3-3, dis3-4, and dis3-T1 Plants
| Wild Type | dis3-1 | dis3-3 | dis3-4 | dis3-T1 | |
|---|---|---|---|---|---|
| Trichome branch length (μm) | |||||
| Branch 1 | 236 ± 22a (n = 11) | 198 ± 52 (n = 10) | 163 ± 40* (n = 10) | 165 ± 50* (n = 14) | 210 ± 51 (n = 12) |
| Branch 2 | 202 ± 48 (n = 11) | 148 ± 54* (n = 10) | 122 ± 40* (n = 10) | 112 ± 50* (n = 14) | 145 ± 34* (n = 12) |
| Branch 3 | 178 ± 43 (n = 11) | 97 ± 48* (n = 10) | 62 ± 31* (n = 10) | 62 ± 26* (n = 14) | 102 ± 28* (n = 12) |
Mean length in μm ± sd; (*), significantly different compared to the wild type according to a Student's t test (P value < 0.05).
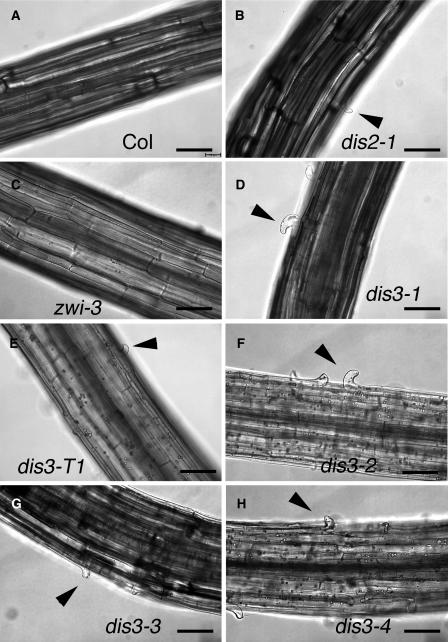
We examined all of the dis3 alleles for additional cotyledon and hypocotyl phenotypes. At 12 d after germination (DAG), the wild-type cotyledon epidermis consisted of a population of highly lobed, interdigitated pavement cells (Figure 1F). In the wild type, adjacent cells tightly adhere, and the only gaps we have observed correspond to stomatal pores. As reported for several ARP2/3 and WAVE complex subunit mutants (Le et al., 2003; Basu et al., 2004; El-Assal et al., 2004a, 2004b), dis3 seedlings displayed aberrant connections between pavement cells (Figure 1G). The severity of the cell–cell adhesion defects was quite variable within and between genotypes, but for each of the dis3 alleles, almost all tissue culture–grown cotyledons had obvious gaps between pavement cells. The mean number of gaps/mm2 ranged from 13 to 52 gaps/mm2 (Table 3). Gaps were commonly observed adjacent to guard cells and at seemingly random locations along the pavement cell perimeter. The presence of gaps does not correlate with a decreased cell size or altered shape (Basu et al., 2004). ARP2/3 and WAVE subunit mutants consistently display pavement cell–cell adhesion defects in both soil- and tissue culture–grown plants. However, in soil-grown dis3 seedlings, gaps were extremely rare. Under these growth conditions, using scanning electron microscopy of cryo-fixed samples and reflected light in a light microscope, we could detect gaps, but the frequency was <1/whole cotyledon (data not shown). Defective cell–cell contacts were also observed in the hypocotyls of etiolated dis3 seedlings (Figure 2). Like other distorted group mutants, etiolated dis3 seedlings had a reduced hypocotyl length, and the mean cell size was reduced in the hypocotyl epidermis (Table 3). The failure of cells to adhere was not the result of reduced hypocotyl elongation alone because the zwichel (zwi) mutant, which presumably affects microtubule-based processes (Oppenheimer et al., 1997), had a reduced hypocotyl length but had no apparent cell–cell adhesion defects (Table 3, Figure 2C).
Table 3.
Quantitation of the Hypocotyl Size and Cotyledon Cell–Cell Adhesion Defects of Wild-Type and dis3 Plants
| Wild Type | dis3-1 | dis3-2 | dis3-3 | dis3-4 | dis3-T1 | dis2-1 | zwi-3 | |
|---|---|---|---|---|---|---|---|---|
| Hypocotyl length (mm)a | 16.8 ± 1.1b (n = 8) | 11.3 ± 1.6* (n = 10) | 11.8 ± 1.8* (n = 8) | 9.1 ± 0.9* (n = 8) | 8.1 ± 0.6* (n = 8) | 10.7 ± 1.4* (n = 8) | 10 ± 1.0* (n = 9) | 11.9 ± 1.6* (n = 8) |
| Hypocotyl cell length (μm)a | 693 ± 100 (n = 8) | 437 ± 90* (n = 15) | 434 ± 98* (n = 12) | 415 ± 57* (n = 11) | 375 ± 76* (n = 9) | 465 ± 105* (n = 11) | 416 ± 49* (n = 9) | 540 ± 49* (n = 14) |
| Gaps/mm2a | 0 ± 0 (n = 3) | 13 ± 7* (n = 3) | 21 ± 9* (n = 3) | 15 ± 12* (n = 3) | 52 ± 85* (n = 5) | 32 ± 31* (n = 3) | 14 ± 11* (n = 3) | n.d. |
Plants grown in vitro on half-strength MS media plus 1% sucrose.
Mean value ± sd.
(*), Significantly different than the wild type according to a Student's t test (P value < 0.05). n.d., not determined.
Figure 2.
Cell–Cell Adhesion Defects in Etiolated Hypocotyls of dis3 Alleles.
(A) Wild-type hypocotyls with adherent epidermal cells.
(B) Epidermal cell–cell adhesion defects in dis2-1 (arpc2).
(C) zwi-3 hypocotyls with adherent epidermal cells.
(D) Epidermal cell–cell adhesion defects in dis3-1.
(E) Epidermal cell–cell adhesion defects in dis3-T1.
(F) Epidermal cell–cell adhesion defects in dis3-2.
(G) Epidermal cell–cell adhesion defects in dis3-3.
(H) Epidermal cell–cell adhesion defects in dis3-4.
Differential interference contrast images of whole-mounted, 7-DAG, dark-grown seedlings. Arrowheads indicate the location of cell–cell adhesion defects. Bar = 100 μm in all panels.
A severely simplified cotyledon and leaf pavement cell shape has been reported for a subset of ARP2/3 subunit mutants (Li et al., 2003; Mathur et al., 2003a, 2003b). Mutants of the WAVE complex homologs SRA1 and NAP1 have a highly lobed pavement cell morphology (Basu et al., 2004; Brembu et al., 2004; El-Assal et al., 2004b). We focused our analyses of pavement cell shape on the apical region of healthy 12-DAG cotyledons. This minimizes the contribution of cell division and allows us to examine cell shape in a population of cells that are, to a large extent, fully expanded and synchronized in their development (Qiu et al., 2002). In this population of pavement cells, we reported subtle defects in lobe formation for dis1 (arp3) (Le et al., 2003) but no consistent effects on cell shape for other ARP2/3 and WAVE complex subunit mutants (Basu et al., 2004; El-Assal et al., 2004a, 2004b). Similarly staged dis3-1 cotyledons contained highly lobed pavement cells (Figure 1G) that were not noticeably different than the wild type (Figure 1F). However, like other ARP2/3 and WAVE subunit mutants, lobe formation was occasionally reduced in regions where cell–cell adhesion defects were the most severe (data not shown).
Although the overall plant architecture of ARP2/3 and WAVE complex subunit mutants is normal, hypocotyl length is reduced in etiolated seedlings, and shoot fresh weight is also reduced. The organ level defects of etiolated hypocotyls have been correlated with reduced cell size and actin localization defects (Mathur et al., 2003a; El-Assal et al., 2004a). Etiolated dis3-1 seedlings had a reduced hypocotyl length (Table 3). Unlike several other distorted group mutants that reduce fresh weight by ∼30%, we failed to detect a difference in fresh weight between dis3-1 seedlings and segregating wild-type control plants. Taken as a whole, DIS3, WAVE, and ARP2/3 subunit mutants displayed the same array of phenotypes, but in many aspects those of dis3 were the least severe.
DIS3 Encodes a Scar/WAVE Homolog
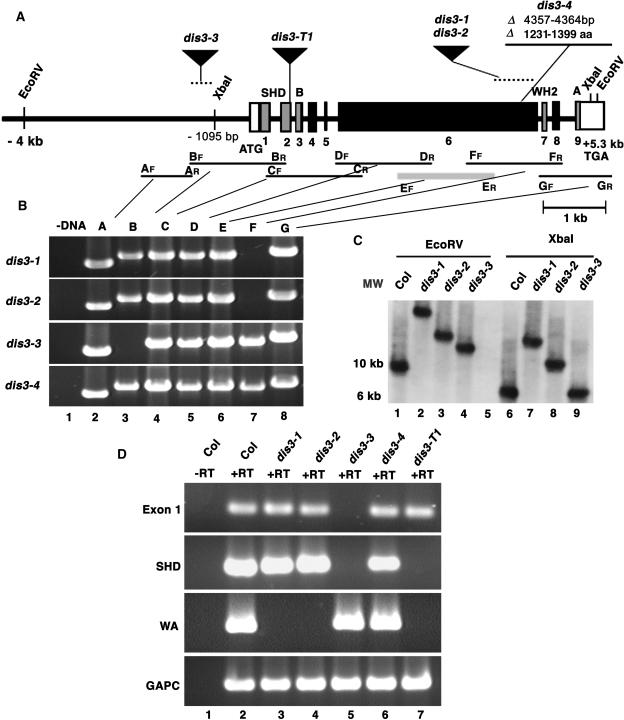
Complementation tests and mapping data showed that dis3-1 did not correspond to any of the eight known distorted group loci (Schwab et al., 2003). The dis3-1 allele was closely linked to a T-DNA insertion in AT2G34680. The failure to detect DNA sequence polymorphisms in AT2G34680 in a subset of dis3 alleles and the lack of a trichome phenotype in AT2G34680 knockout lines led us to test the idea that DIS3 corresponded to either of the linked genes AT2G34150 (SCAR1) or AT2G38440 (SCAR2). These genes were candidates because both encoded proteins with a putative N-terminal SCAR homology domain and a C-terminal WA domain. The SCAR homology and acidic regions of SCAR/WAVE proteins mediate WAVE complex assembly (Echarri et al., 2004; Innocenti et al., 2004) and Arp2/3 activation (Machesky and Insall, 1998; Machesky et al., 1999), respectively. In the three dis3 backgrounds tested, SCAR1 was not affected (data not shown), but SCAR2 coding information was disrupted in each case. The dis3-4 allele deleted eight nucleotides of AT2G38440 spanning +4357 to +4364 (relative to the A+1TG start codon; Figure 3A). The dis3-4 deletion caused a reading frame shift after amino acid 1230, the coding of eight amino acids of non-sense, and then a stop codon. The dis3-1, dis3-2, and dis3-3 alleles were isolated as untagged T-DNA alleles. Based on PCR and DNA gel blot analyses, dis3-3 contained an insertion of ∼3.5 kb located in the interval from −1209 to −1095 (Figures 3B and 3C). The dis3-3 transcript lacked coding sequences for the SCAR homology domain but did encode an intact WA domain (Figure 3D). Perhaps the upstream insertion in dis3-3 unmasks or generates a cryptic transcription start site downstream from the SCAR homology domain. The weaker phenotype of dis3-3 compared with other dis3 alleles suggested that a partially functional protein accumulates. The dis3-1 and dis3-2 alleles contained a large DNA insertion of ∼10 and 5 kb, respectively (Figures 3B and 3C). Both insertions resided in exon 6, caused premature termination of transcription, and eliminated the WA coding region (Figure 3D). To isolate a likely null dis3 allele, we analyzed the T-DNA insertion line SALK_057481 (Alonso et al., 2003), which contained a T-DNA insertion in the second exon of AT2G38440 (Figure 3A) and caused premature transcriptional termination. The SCAR homology domain was truncated, and the downstream WA coding region was removed (Figure 3D). Lines that were homozygous for SALK_057481 had distorted trichomes, shoot epidermal cell adhesion defects, and reduced hypocotyl elongation phenotypes that were indistinguishable from the other dis3 alleles. The failure of homozygous SALK_057481 lines to complement dis3-1 in crosses proved that it was an allele of DIS3 and subsequently was renamed dis3-T1. The molecular characterization of five independent dis3 alleles provided strong proof that DIS3 corresponded to AT2G38440. Therefore, we renamed DIS3 to DIS3/SCAR2.
Figure 3.
Physical Map and Molecular Characterization of DIS3/SCAR2 Mutant Alleles.
(A) The physical structure of the DIS3/SCAR2 gene. The transcribed region of DIS3/SCAR2 is labeled with boxes (exons) and lines (introns). The DIS3/SCAR2 exons that encode conserved domains or motifs are gray and are labeled with text: SHD, SCAR homology domain; B, basic region; WH2, G-actin binding region; A, acidic region. The location and nature of dis3-1, dis3-2, dis3-3, dis3-4, and dis3-T1 alleles are defined. Nucleotides are numbered relative to the first nucleotide of the start codon defined as +1. aa, amino acids.
(B) The dis3-1 through dis3-4 alleles disrupt the DIS3/SCAR2 gene. Genomic DNA was isolated from dis3-1, dis3-2, dis3-3, and dis3-4 alleles, and overlapping pairs of PCR primers that spanned the DIS3/SCAR2 locus were used to locate the affected regions of the gene. The pairs of PCR primers are named according to their order along the gene and their direction, either forward (F) or reverse (R). Lane 1 includes a representative no template DNA control PCR reaction, and lanes 2 to 8 contain the PCR products obtained using PCR primer pairs A through G, respectively. The location of each primer pair relative to the DIS3/SCAR2 gene is indicated in (A).
(C) The dis3-1, dis3-2, and dis3-3 alleles contain DNA insertions. DNA gel blot analysis was conducted on genomic DNA that was isolated from Col, dis3-1, dis3-2, and dis3-3 seedlings. Genomic DNA was digested with EcoRV (lanes 1 to 4) and XbaI (lanes 6 to 9). The probe was constructed using PCR product E shown in (A). MW, DNA size standards.
(D) The dis3 alleles that harbor DNA insertions affect the DIS3/SCAR2 transcript. RNA was isolated from Col and dis3 mutant seedlings and subjected to RT-PCR analyses using primer pairs located within different regions of the DIS3/SCAR2 gene, including exon 1, primers that flank the encoded SCAR homology domain, and primers that flank the WA domain. Glyceraldehyde-3-phosphate dehydrogenase C subunit (GAPC) was used as a positive control. Lane 1, a representative no reverse transcriptase (−RT) control; lanes 2 to 7, plus reverse transcriptase (+RT) for Col, dis3-1, dis3-2, dis3-3, dis3-4, and dis3-T1, respectively.
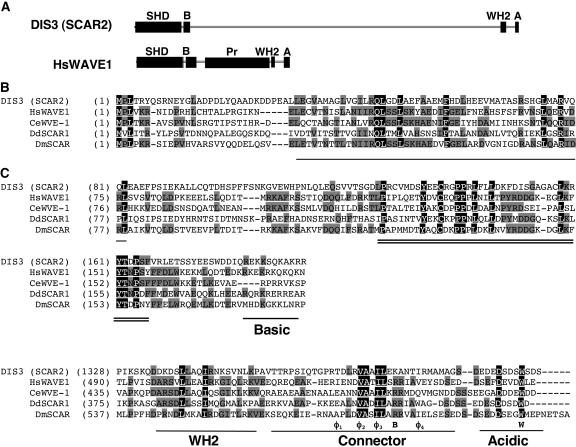
We sequenced a full-length DIS3/SCAR2 cDNA clone, Kazusa clone RZL03H01, to define the encoded polypeptide (GenBank accession number AY817016). The deduced amino acid sequence of DIS3/SCAR2 agreed perfectly with the predicted gene model and encoded a 1399–amino acid polypeptide with a pI of 4.5. DIS3/SCAR2 is one of five Arabidopsis proteins that share amino acid identity with the N-terminal SCAR homology domain of the WAVE/SCAR family of Arp2/3 activators (Brembu et al., 2004; Deeks et al., 2004). The level of amino acid identity between the SCAR homology domain of DIS3/SCAR2 and non-plant Scar/WAVEs was highest toward the N- and C-terminal ends (Figure 4B). The N-terminal subregion of the SCAR homology domain of Scar/WAVE1 is necessary and sufficient for an interaction with the WAVE complex subunit Abi-1 (Echarri et al., 2004) and may also bind to the WAVE complex subunit HSPC300 (Innocenti et al., 2004). DIS3/SCAR2, like non-plant homologs, contained a short stretch of basic amino acids adjacent to the SCAR homology domain (Figure 4A).
Figure 4.
DIS3/SCAR2 Is a SCAR Homology and WA Domain–Containing Protein.
(A) The domain organization and relative sizes of the DIS3/SCAR2 and human WAVE1. The abbreviations of the domains are as in Figure 3. Pr, Pro-rich region of human WAVE.
(B) The SCAR homology domains of DIS3/SCAR2 contain blocks of highly conserved amino acid sequences. Amino acid sequences of the SCAR homology domain of DIS3/SCAR2 and other WAVE/SCAR family proteins were aligned using ClustalW. The bold black line and the double lines mark two highly conserved blocks toward the N- and C-terminal ends of the SCAR homology domains, respectively. A region enriched in basic amino acids present in all SCAR/WAVE proteins is underlined and labeled.
(C) DIS3/SCAR2A contains a C-terminal WA domain. Amino acid sequences of DIS3/SCAR2 and other WAVE/SCAR family proteins were aligned using ClustalW. According to published alignments and functional studies of the WA domain (Panchal et al., 2003), the alignments were anchored using a conserved Arg (marked B below the alignment) and Trp (marked W in the alignment) residues, and conserved hydrophobic amino acids (marked Φ) in the connector region are labeled (Panchal et al., 2003). The WH2 G-actin binding region and acidic regions are labeled. The GenBank accession numbers used for alignments are AY817016 (DIS3/SCAR2), AAD33052 (human WAVE1), NP_493028 (C. elegans WVE-1), AAD29083 (D. discoideum Scar1), and AAF74194 (D. melanogaster SCAR).
All known Scar/WAVE proteins contain a WA domain. SCAR1, DIS3/SCAR2, SCAR3, and SCAR4 have a WA domain, and the WA domain of SCAR1 appears to bind G-actin (Deeks et al., 2004). Although the WA domain of DIS3/SCAR2 encodes key conserved acidic and hydrophobic residues that are present in all WASP/Scar/WAVE family members, there is some variation in the length and composition of the connector region (Figure 4C). DIS3/SCAR2 does not encode an extended Pro-rich region that is characteristic of non-plant WASP/Scar/WAVE family members. Instead, the analogous region of DIS3/SCAR2 contained a large central region of no known function (Figure 4A).
DIS3/SCAR2 Functions within a WAVE-ARP2/3 Pathway
The similar cell shape and actin cytoskeleton defects of dis3 and ARP2/3 and WAVE complex subunit mutants strongly suggested that DIS3/SCAR2 functions within the same pathway. We constructed the dis3-1 dis2-1 double mutant and assayed its trichome phenotypes. The double mutant displayed a strong distorted trichome phenotype that was indistinguishable from dis2-1 (Table 1). However, the mean branch length of dis3-1 dis2-1 trichomes was reduced significantly when compared with dis3-1 alone.
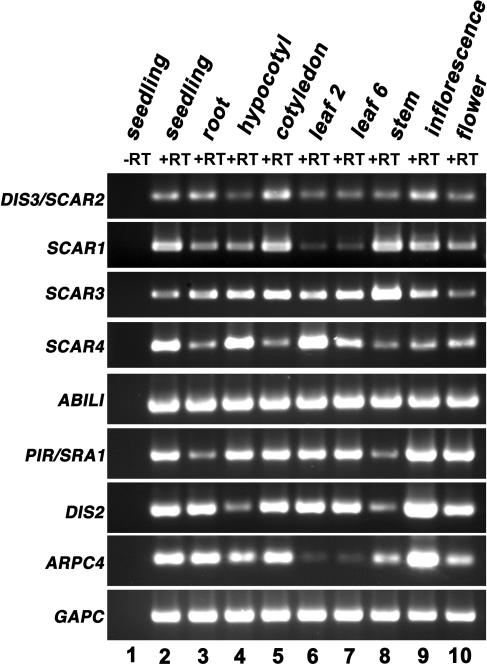
We wanted to determine if the gene expression pattern of DIS3/SCAR2 and other SCAR genes overlapped with each other and with known WAVE and ARP2/3 subunit genes. All of the SCAR family members assayed, like PIR (SRA-1) and DIS2 (ARPC2), were widely expressed in all of the major organs examined (Figure 5). There were modest differences in the relative levels of SCAR1 through SCAR4 expression in different organs, but each of the SCAR genes were expressed in all of the major organs that were analyzed. These data are consistent with the idea that the SCAR genes function in the same organs, but the cell- and tissue-specific gene expression patterns of these genes need to be examined in detail. DIS3/SCAR2 expression levels did not correlate with the growth rates of leaves. Expression levels in actively growing (Figure 5, lane 7) versus fully expanded leaves (Figure 5, lane 6) were similar.
Figure 5.
SCAR Genes Are Expressed throughout Plant Development.
Total RNA isolated from the organs listed at the top of the figure was analyzed by RT-PCR using gene-specific primers for SCAR1, SCAR2, SCAR3, and SCAR4. PIR (SRA-1) expression was analyzed to provide an example for a WAVE complex gene. DIS2 (ARPC2) and ARPC4 were used as examples of ARP2/3 subunit genes. For each primer pair the PCR cycle number was optimized to reveal relative differences in expression between organs and were SCAR1 (35 cycles), SCAR2 (35 cycles), SCAR3 (35 cycles), SCAR4 (35 cycles), ABIL1 (25 cycles), PIR (SRA-1) (22 cycles), DIS2 (ARPC2) (20 cycles), ARPC4 (21 cycles), and GAPC (glyceraldehyde-3-phosphate dehydrogenase C subunit) (20 cycles). Lane 1, no reverse transcriptase (−RT) control; lanes 2 to 10, various RNA samples subjected to RT treatment. Control experiments that lacked RT were conducted on all RNA samples, but only the seedling experiment is shown.
WAVE and ARP2/3 subunit mutants fail to organize a population of loosely aligned actin bundles at the earliest stages of branch elongation (Basu et al., 2004; El-Assal et al., 2004b). Therefore, we assayed the actin-based phenotypes of dis3 cells at the onset of the cell swelling phenotype during early stage 4. Representative actin localization results are shown using the dis3-1 allele, and similar actin defects were observed in dis3-T1 and dis3-4 trichomes (data not shown). The actin cytoskeleton of WAVE and ARP2/3 subunit mutants have been analyzed using different probes to label actin, with different results. Using green fluorescent protein (GFP):TALIN as a probe, several groups have described the actin cytoskeleton in ARP2/3 and WAVE complex mutant trichomes as excessively bundled (Li et al., 2003; Mathur et al., 2003a, 2003b; Brembu et al., 2004). When the actin cytoskeleton is visualized in WAVE and ARP2/3 mutants using antibodies or phalloidin, or in living cells probed with GFP:ABD2, a disorganization of actin bundles is revealed at early stages as well as a reduction in the levels of actin filaments at later stages (Le et al., 2003; Basu et al., 2004; Deeks et al., 2004; El-Assal et al., 2004a, 2004b).
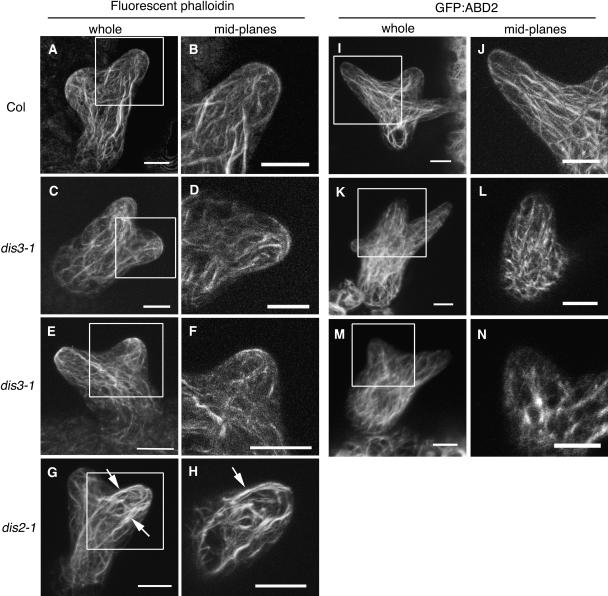
In our hands, the use of phalloidin-labeled fixed cells and GFP:ABD2 (Sheahan et al., 2004) yielded the most consistent actin labeling results. Wild-type stage 3/4 trichomes contain poorly developed branch buds and branches with a blunt tip morphology. At this stage, the branches are densely cytoplasmic and lack an enlarged central vacuole. As shown previously, fixed wild-type stage 3/4 trichomes labeled with phalloidin contained a population of actin filaments and bundles that reside entirely within the core branch cytoplasm and were loosely aligned with the long axis of the branch (eight of nine branches, 10 to 25 μm in length) (Figures 6A and 6B). As shown previously (Basu et al., 2004; El-Assal et al., 2004a), and confirmed in this study, very few similarly staged dis2-1 trichomes (3 of 13 branches, 10 to 25 μm in length) have organized bundles in the core cytoplasm, and many of the core bundles were continuous with cortical bundles (Figures 6G and 6H). The actin defects of stage 3/4 dis3-1 trichomes were subtle and resembled those of pir (sra-1) and grl (nap1) (Basu et al., 2004; Deeks et al., 2004; El-Assal et al., 2004b). More than half of the phalloidin-labeled dis3-1 stage 3/4 branches (10 of 16, 10- to 25-μm branches) contained a relatively normal looking array of polarized cytoplasmic bundles (Figures 6C and 6D). The remaining dis3 stage3/4 branches of this size class contained randomly oriented actin filaments or bundles (Figures 6E and 6F).
Figure 6.
A Subpopulation of dis3 Trichomes Fails to Generate and Maintain a Normal Population of Cytoplasmic Actin Bundles.
The actin cytoskeletons of whole-mounted fixed cells and living stage 3/4 trichomes were detected using Alexa-488 phalloidin ([A] to [H]) and GFP:ABD2 ([I] to [N]) and visualized using confocal microscopy. For each actin probe, the panels on the left are maximum projections of an entire cell, and those on the right include the midplanes of longitudinal optical sections through the branch that is boxed in the maximum projection. Arrows in (G) and (H) indicate actin bundles that have a full or partial cortical localization. Bar =10 μm in all panels.
(A) and (B) F-actin organization in a wild-type stage 3/4 trichome visualized using fluorescent phalloidin.
(C) and (D) A dis3-1 stage 3/4 trichome displaying a wild type–like parallel actin bundle organization.
(E) and (F) A dis3-1 trichome displaying disorganized actin filaments and/or bundles.
(G) and (H) A dis2-1 (arpc2) trichome displaying mislocalized cytoplasmic actin filaments.
(I) and (J) Actin organization of a wild-type stage 3/4 trichome visualized using GFP:ABD2.
(K) and (L) A dis3-1 GFP:ABD2-expressing plant containing a stage 3/4 trichome displaying a wild type–like parallel actin bundle organization.
(M) and (N) A dis3-1 GFP:ABD2-expressing plant—a representative example of a stage 3/4 trichome lacking a population of loosely aligned cytoplasmic bundles.
When crossed into the wild-type (Figures 6I and 6J) and dis3-1 (Figures 6K to 6N) backgrounds and imaged in living cells, GFP:ABD2 yielded similar results. In general, compared with phalloidin-labeled cells, the actin cytoskeleton visualized with GFP:ABD2 appeared slightly less bundled and diminished in the extreme apical regions of developing branches. We have previously documented the slightly increased bundling of phalloidin-labeled filaments compared with antibody-labeled filaments in fixed cells (Le et al., 2003). Approximately half of the dis3-1 trichomes had a dense population of bundles that resembled the wild type (Figures 6K and 6L). Another population of dis3-1 trichomes contained cytoplasmic bundles but failed to generate an organized array (Figures 6M and 6N). Therefore, using two different probes to detect actin, we detected clear actin bundle disorganization in the smallest size class of branches.
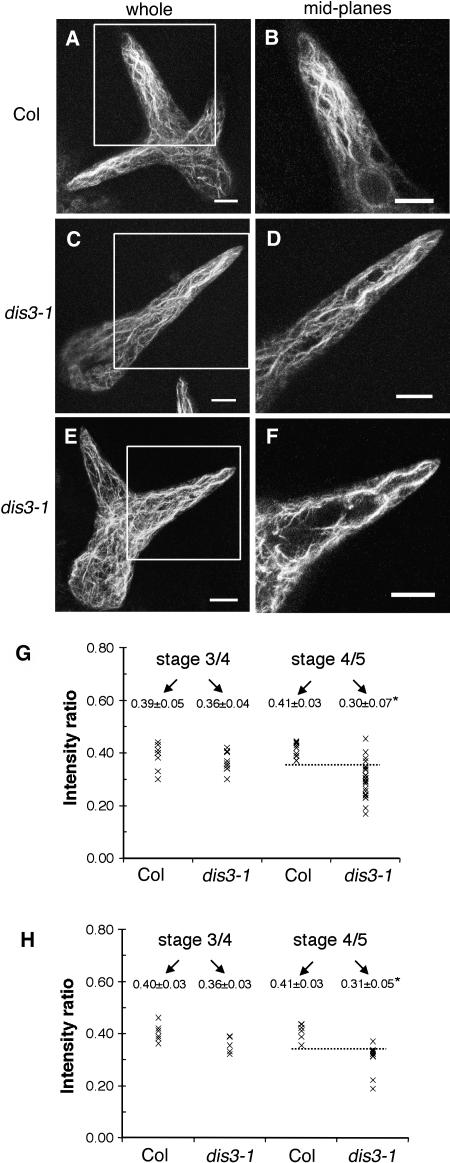
At later stages of branch development (branches ranging from 26 to 75 μm), two classes of actin organization were observed in dis3-1 trichomes. Ten of 18 branches contained a population of aligned core bundles in the apical region of the branch (Figures 7C and 7D). The remaining dis3 branches in this size class had a core cytoplasm that was dominated by the vacuole with no obvious actin bundle organization in the branch apex (Figures 7E and 7F). All of the wild-type trichomes in this size class maintained a dense population of cytoplasmic bundles in the branch apex (10 of 10 cells; Figures 7A and 7B). A quantitative measurement of the relative amount of cytoplasmic actin filaments in elongated dis3 branches was consistent with the localization data. In both phalloidin (Figure 7G) and GFP:ABD2 (Figure 7H) labeling experiments, a subset of stage 4/5 dis3 trichomes had reduced relative actin filament levels in the core cytoplasm.
Figure 7.
Elongating Stage 5 dis3-1 Trichomes Have Defects in Actin Filament Organization and Vacuole Positioning.
Whole-mounted fixed trichomes belonging to this stage were identified based on a pointed tip morphology, the absence of papillae, a cell height of at least 40 μm, and a branch length between 30 and 75 μm. Left panels are maximum projections of the entire branch, and those on the right include longitudinal optical sections through the middle of the branch within the boxed region.
(A) and (B) Actin organization in a wild-type trichome.
(C) and (D) A dis3-1 stage 5 trichome displaying aligned core actin filaments.
(E) and (F) A dis3-1 trichome in which the core cytoplasm lacks actin bundles and is occupied by the central vacuole. Bar = 10 μm in (A) to (F).
(G) Quantitation of the relative amounts of cortical and core cytoplasmic actin in Alexa 488 phalloidin–labeled samples. Total integrated intensity of core: total actin signal was measured in several image planes centered on the branch midpoint. Intensity ratios for Col and dis3 branches were measured in stage 3/4 and stage 4/5 trichomes. Mean value of each measured branch is plotted and the grand mean ± sd for each genotype is listed in the plot.
(H) Same as in (G) but calculated using GFP:ABD as a probe. Asterisks indicate significant difference compared with the wild type according to a Student's t test (P value < 0.05).
It has been reported that ARP2/3 subunit genes affect the microtubule cytoskeleton (Schwab et al., 2003; Saedler et al., 2004). To determine if microtubules were affected at the onset of the cell swelling phenotype in dis3, we assayed for transverse cortical microtubules in stage 3/4 branches that were 10 to 25 μm in length (Figure 8A). In the wild type, 88% (n = 16) of the 10- to 25-μm branches had a clear transverse orientation of microtubules relative to the long axis. Signs of a reorientation of microtubules from a transverse to a helical or longitudinal arrangement were first detected in branches that were ∼30 μm in length, and 50% (n = 10) of the wild-type branches between 26 and 75 μm in length retained a transverse alignment. As wild-type branches elongated to lengths >50 μm, the frequency of cells having a helical or longitudinal alignment increased (Figure 8B). When dis3-1 trichomes at similar developmental stages were assayed for transverse microtubules, the proportion of cells having a transverse alignment was nearly identical: 90% (n = 10) transverse in 10- to 25-μm branches and 60% transverse (n = 10) in 26- to 75-μm branches. The timing of the transition from transverse to helical was similar in dis3-1 cells because, like the wild type, the transition from transverse to helical was first detected in 30-μm-long branches. An example of a transverse cortical array in normal looking dis3 stage 4 trichomes is illustrated in Figure 8C. In swollen dis3-1 stage 4 cells, branches with an obviously reduced length had a transverse microtubule array (Figure 8D, arrow). In both stage 4 (Figure 8D, arrowhead) and stage 5 trichomes (Figure 8E, arrowhead), the extent of random microtubule organization correlated with the extent of cell swelling. Normally shaped stage 5 dis3 trichomes displayed a highly organized microtubule cytoskeleton (Figure 8E, arrow).
Figure 8.
Microtubule Organization in Wild-Type and dis3 Trichomes at Different Stages.
Microtubules were visualized in living cells using the GFP:MBD probe. All images are maximum projections of confocal images. Bar = 10 μm in all panels.
(A) Transversely aligned cortical microtubules in a stage 4 wild-type trichome.
(B) Oblique alignment of microtubules in a wild-type stage 5 trichome.
(C) Microtubule organization in a dis3-1 stage 4 trichome with a normal branch morphology.
(D) Microtubule organization in a dis3-1 stage 4 trichome that displays a reduced branch length (arrow) and branch swelling (arrowhead).
(E) Microtubule arrangements in stage 5 dis3-1 trichomes. Projections of two cells on the same leaf that have either no obvious microtubule defect (arrow) or a swollen morphology and randomly organized microtubules (arrowhead).
DIS3/SCAR2 May Assemble into a WAVE Complex
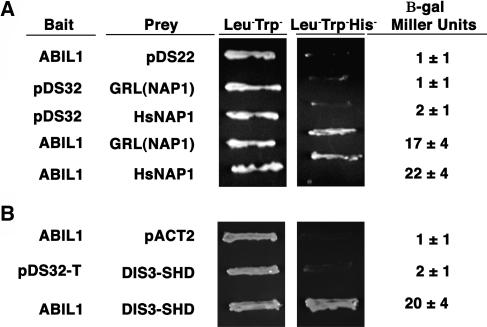
The presence of a SCAR homology domain and a WA domain in DIS3/SCAR2 suggested that the protein assembles into a WAVE complex and positively regulates ARP2/3. We have shown that PIR (SRA-1) and GRL (NAP1) encode WAVE complex subunits that interact physically (Basu et al., 2004; El-Assal et al., 2004b). If a five-subunit WAVE complex assembles in plants, an Abi-1–like function may be required to bind both NAP1 and SCAR/WAVE-like subunits (Gautreau et al., 2004; Innocenti et al., 2004). Arabidopsis encodes at least four proteins that share limited amino acid sequence identity with Abi-1 (Figure 9). One of them, Abi-1–like 1 (ABIL1 [AT2G46225]), was expressed throughout the plant (Figure 5) and is a reasonable candidate as a WAVE complex subunit. We tested the ability of ABIL1 to interact with GRL (NAP1) and the SCAR homology domain of DIS3/SCAR2 in the yeast two-hybrid assay. Because of the interchangeable functions of human and Arabidopsis WAVE complex subunits (Basu et al., 2004; El-Assal et al., 2004b), human WAVE subunits serve as a useful control. In the yeast two-hybrid assay, full-length ABIL1 interacted with both GRL (NAP1) and human NAP1, as judged by the ability of yeast strains to grow on Leu−Trp−His− media and the activation of the β-galactosidase (β-Gal) reporter gene more than 15-fold relative to controls (Figure 10A). We next tested the ability of ABIL1 to interact with the conserved SCAR homology domain of DIS3/SCAR2. Yeast strains that harbored SCAR homology domain of DIS3/SCAR2 and ABIL1 fusion proteins grew on Leu−Trp−His− media and activated the β-Gal reporter gene more than 10-fold relative to controls (Figure 10B). A set of control experiments showed that none of the fusion proteins yielded growth on His− media or activated the β-Gal reporter on their own. The ABIL1–SCAR homology domain interaction was specific because ABIL1 failed to interact with human WAVE1 (SCAR), the C terminus of DIS3/SCAR2, or ROP2 in the same assay (data not shown). These data indicated that ABIL1 has the ability to recruit DIS3/SCAR2 to a WAVE complex.
Figure 9.
Amino Acid Sequence Alignment of the Four Arabidopsis Proteins that Share Limited Amino Acid Identity with the N Terminus of Human Abi-1.
The N-terminal region of conserved amino acid identity between the Arabidopsis proteins and human Abi-1 is underlined. The SH3 domain of Abi-1 is labeled with a dashed line. GenBank accession numbers are listed in Methods.
Figure 10.
An Arabidopsis Abi-1–Like Protein May Mediate the Assembly of DIS3/SCAR2 into a WAVE Complex.
(A) ABIL1 interacts with GRL (NAP1) and human NAP1 in the yeast two-hybrid assay. The bait and prey plasmids that were cotransformed into yeast are defined to the left of the corresponding yeast patches, and β-galactosidase assay results are shown to the right.
(B) The SHD domain of DIS3/SCAR2 interacts with ABIL1 in the yeast two-hybrid assay. The bait and prey plasmids that were cotransformed into yeast are defined to the left of the corresponding yeast patches, and β-galactosidase assay results are shown to the right.
DIS3/SCAR2 Binds to ARPC3 and Activates Arp2/3
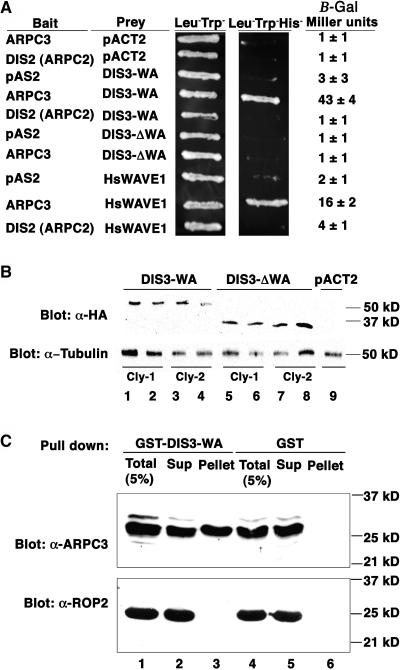
We wanted to test directly the ability of DIS3/SCAR2 to bind to and activate ARP2/3. The identity of WASP and Scar as Arp2/3 activators was initially based on the ability of these proteins to bind to the ARPC3 subunit of Arp2/3 complex in a yeast two-hybrid assay (Machesky and Insall, 1998). A strong yeast two-hybrid interaction was detected between a C-terminal fragment of DIS3/SCAR2 spanning amino acids 1032 to 1399 that contained the WA domain (DIS3-WA) and ARPC3 (Figure 11A). Neither DIS3-WA nor human WAVE1 (SCAR) interacted with DIS2 (ARPC2). We constructed a DIS3/SCAR2 2-hybrid construct that deleted amino acids 1239 to 1399 and mimicked the dis3-4 allele (DIS3-ΔWA). The DIS3-ΔWA fusion protein did not interact with ARPC3, even though DIS3-ΔWA fusion protein levels in yeast extracts were on average twofold higher than the WA-containing construct (Figure 11B). To confirm that the interaction between DIS3-WA and ARPC3 was direct, binding between recombinant glutathione S-transferase (GST)-DIS3-WA and His-tagged ARPC3 was assayed in GST pull-down assays. GST-DIS3-WA (Figure 11C, top panel, lane 3), but not GST alone (Figure 11C, top panel, lane 6), was able to sediment ARPC3. The interaction was specific, because GST-DIS3-WA did not physically interact with the similarly sized His-tagged ROP2 protein (Figure 11C, bottom panel, lane 3).
Figure 11.
The WA Domain of DIS3/SCAR2 Is Required for Direct Binding to ARPC3.
(A) DIS3/SCAR2 and human WAVE1 interact with ARPC3 in a yeast two-hybrid assay. The bait and prey plasmids that were cotransformed into yeast are defined to the left of the corresponding yeast patches and β-galactosidase assay results to the right. DIS3-WA contains amino acids 1032 to 1399; DIS3-ΔWA contains amino acids 1032 to 1238; ARPC3, DIS2 (ARPC2), and HSWAVE1 are full-length proteins.
(B) The DIS3-ΔWA and DIS3-WA two-hybrid constructs are expressed at comparable levels in yeast. Top, lanes 1 to 9, duplicate yeast protein soluble fractions probed with anti-HA antibody. Bottom, same blot probed with control anti-tubulin antibody. Lanes 1-4, duplicate protein samples extracted from two individual colonies (cly-1 and cly-2) of the yeast strain expressing HA-tagged DIS3-WA; lanes 5 to 8, same as 1 to 4, but expressing HA-tagged DIS3-ΔWA; lane 9, yeast extracts from the strain harboring the empty vector pACT2. All the strains used also contained the ARPC3 bait construct.
(C) Direct interaction of DIS3/SCAR2 and ARPC3 in a GST pull-down assay. Top, lanes 1 to 6, fractions probed with an anti-ARPC3 antibody; bottom, fractions probed with an anti-ROP2 antibody. Lane 1, 5% of total binding reaction; lane 2, unbound; lane 3, GST-DIS3-WA–associated fraction; lane 4, 5% of total binding reaction with GST alone control beads; lane 5, unbound; lane 6, GST bead-associated fraction.
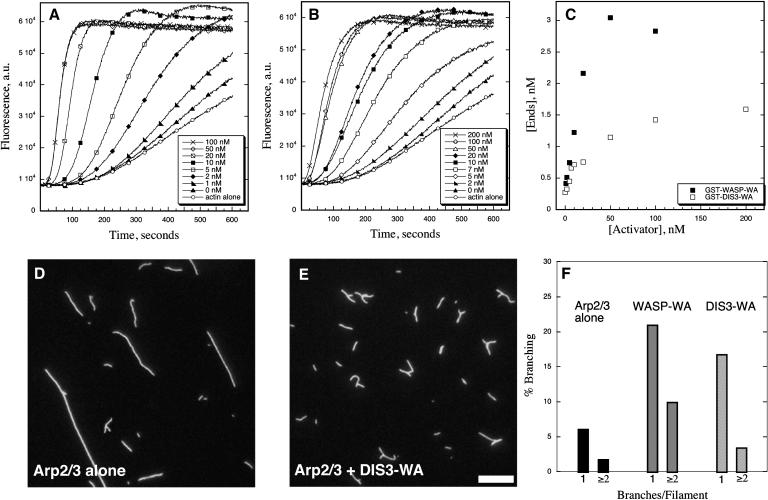
If DIS3/SCAR2 is a bona fide ARP2/3 activator, it should increase the actin filament nucleation and branching activities of the complex. In an actin polymerization assay, bovine Arp2/3 alone does not affect the kinetics of actin filament assembly (Figures 12A and 12B). Known Arp2/3 activators, such as the WA domain of WASP, decreased the lag phase of actin polymerization and enhanced the polymerization rate in a dose-dependent manner (Figure 12A). In identical assays, a DIS3-WA protein fragment efficiently activated Arp2/3 at nanomolar concentrations (Figure 12B). One method to standardize the ability of different proteins to activate Arp2/3 is to plot the concentration of the activator versus the number of barbed ends produced at a defined point in the polymerization reaction. At saturating levels of activator (Figure 12C), the amount of ends produced by DIS3-WA was comparable to values that have been reported for human Scar-1 under similar experimental conditions (Blanchoin et al., 2000). The efficiency of DIS3-WA–dependent nucleation was less than that of WASP-WA. However, this result is expected because human SCAR/WAVE proteins are also less efficient Arp2/3 activators when compared with WASP (Zalevsky et al., 2001). We also assayed the ability of DIS3-WA to increase the actin filament branching activity of Arp2/3. Direct observation of the actin filaments generated in the presence of Arp2/3 alone yielded a very low fraction of branched actin filaments (Figures 12D and 12F). In the presence of DIS3-WA, the number of filaments that form branched structures was increased approximately threefold greater (Figures 12E and 12F).
Figure 12.
The WA Domain of DIS3/SCAR2 Efficiently Activates Vertebrate Arp2/3.
(A) to (C) Actin (4 μM) (5% pyrene labeled) with or without 50 nM Arp2/3 and various amounts of activators was allowed to polymerize and the kinetics of assembly monitored by fluorimetry. Pyrene fluorescence (a.u., arbitrary units), which increases when actin assembles into filaments, is plotted versus time after addition of G-actin to initiate polymerization. Nucleation activity is evident as a decrease in the lag period required before actin assembly ensues and an increase in the initial rate of polymerization.
(A) Polymerization of actin in the presence of 50 nM Arp2/3 and a GST fusion protein with the WA domain of human N-WASP (GST-WASP-WA) at various concentrations: 0, 1, 2, 5, 10, 20, 50, and 100 nM (see legend).
(B) Similar experiments performed in the presence of the WA domain from DIS3 (GST-DIS3-WA) at various concentrations: 0, 2, 5, 7, 10, 20, 50, 100, and 200 nM (see legend). DIS3-WA was comprised of amino acids 1084 to 1399 of DIS3/SCAR2.
(C) Dependence of the concentration of apparent ends ([Ends]) on the concentration of WASP-WA (closed squares) and DIS3-WA (open squares) calculated from the rate of polymerization at the time where 50% (2.0 μM) of the actin was polymerized. These values were calculated from the data shown in (A) and (B) using the equation described in Methods.
(D) to (F) Activators of Arp2/3 were also analyzed by fluorescence microscopy for the ability to stimulate branch formation. Conditions: 4 μM G-actin was polymerized in the presence of 50 nM Arp2/3 with or without activators. Equimolar rhodamine-phalloidin was added to the reactions to label filaments, and reactions were diluted into fluorescence buffer after 30 min.
(D) Arp2/3 alone (50 nM), no activator
(E) Arp2/3 (50 nM) + 100 nM GST-DIS3-WA. Bar = 2.5 μm.
(F) The bar graph shows quantification of branch frequency counted from at least five different micrographs and >300 filaments for each treatment. Percentage of branching was binned into three categories: no branches, one branch, and two or more branches. Given on the graph are the latter two subcategories.
DISCUSSION
The distorted group of trichome mutants defines a pathway of WAVE and ARP2/3 subunit genes that translate morphogenetic signals into well orchestrated cytoskeletal responses. DIS3/SCAR2 is a Scar/WAVE homolog that directly activates Arp2/3. Assembly of DIS3/SCAR2 into a WAVE complex may occur through a physical interaction with an Abi-1–like intermediate, ABIL1. DIS3/SCAR2 is required for the normal morphogenesis of several cell types in the shoot and functions within an ARP2/3 pathway. The viability of Arabidopsis WAVE and ARP2/3 subunit mutants and their distinct trichome phenotypes provide an ideal experimental system to develop and test hypotheses for WAVE and ARP2/3 functions in cells, tissues, and organs.
DIS3/SCAR2 Functions in a WAVE-ARP2/3 Pathway in Arabidopsis
The WAVE complex is unique to multicellular organisms and defines an important mechanism for actin filament nucleation and morphogenesis. Strong genetic and cytological data place DIS3/SCAR2 within a WAVE-ARP2/3 pathway. The phenotypes of dis3 plants are identical to ARP2/3 and WAVE subunit mutants. All of the above mutants display stage-specific trichome distortion, defective epidermal cell–cell adhesion, and reduced hypocotyl elongation in the dark. Our double mutant analyses in this report indicate that dis3 dis2 (arpc2) plants have a phenotype that is indistinguishable from dis2 (arpc2) and provide good evidence that both genes function in a common pathway. The mild disorganized actin bundle phenotypes of dis3 trichome branches were very similar to those of the WAVE subunit mutants pir (sra-1) (Basu et al., 2004) and grl (nap1) (Deeks et al., 2004; El-Assal et al., 2004b). One explanation for these results is that PIR (SRA-1), GRL (NAP1), and DIS3/SCAR2 positively regulate ARP2/3 during epidermal development. In vertebrates, the RAC1-WAVE2-Arp2/3 signaling pathway is a major and essential route for actin filament nucleation (Yan et al., 2003). WAVE-dependent positive regulation of Arp2/3 is thought to occur through stabilization of the Scar/WAVE subunit (Blagg et al., 2003; Kunda et al., 2003; Rogers et al., 2003) and through RAC1-dependent relocalization of a fully assembled WAVE complex to subcellular locations of high ARP2/3 activity (Innocenti et al., 2004). Convincing genetic (Blagg et al., 2003) and biochemical data (Eden et al., 2002) suggest an alternative mechanism by which WAVE complex subunits negatively regulate SCAR/WAVE activity. In this model SCAR/WAVE is negatively regulated in trans by association with the WAVE complex. Activating signals dissociate Scar/WAVE from the WAVE complex and unmask its ability to activate Arp2/3 (Eden et al., 2002). If Arabidopsis WAVE complex positively regulates the activity of ARP2/3, the presence of a SCAR/WAVE-like subunit is expected.
DIS3/SCAR2 Encodes Conserved SCAR Homology and WA Domains That Are Required for in Vivo Function
The identity of DIS3 as a Scar-like protein is based on strong genetic evidence. We isolated four independent dis3 alleles (dis3-1 through dis3-4) in our mutant screen. Each dis3 allele altered the coding information of the DIS3/SCAR2 transcript by truncating either the conserved WA (dis3-1, dis3-2, and dis3-4) or SCAR homology (dis3-3) domain. As further proof of gene identity, we demonstrated that the SALK_057481 line (dis3-T1) was a likely null allele of DIS3/SCAR2, caused trichome distortion in homozygous lines, and was an allele of dis3. Taken together, these data prove that DIS3 and SCAR2 are the same gene.
The domain organization of the predicted DIS3/SCAR2 protein is similar to Scar/WAVE family members. First, the N terminus of DIS3/SCAR2 shares ∼20% identity with its human homologs. The SCAR homology domain of human WAVE (SCAR) can bind to both Abi-1 and HSPC300/Brk1 subunits (Echarri et al., 2004; Innocenti et al., 2004). There are two subregions within the SCAR homology domain of plant and animal SCAR proteins that show a higher level of amino acid sequence conservation (Figure 4B). The N-terminal conserved block corresponds to the region of human WAVE1 (SCAR) that is necessary and sufficient for Abi-1 binding (Echarri et al., 2004). Perhaps this region corresponds to a subdomain of plant and animal SCAR proteins that binds directly to Abi-1–like proteins. An intact SHD appears to be essential for DIS3/SCAR2 function. The dis3-3 allele removes the SCAR homology domain but encodes an intact WA domain (Figure 3D). Most aspects of the dis3-3 phenotype are equivalent to the null alleles. This suggests that interaction of DIS3/SCAR2 with the WAVE complex is an important aspect of its regulation. Association of DIS3/SCAR2 with a WAVE complex could stabilize the protein or regulate its subcellular localization in response to upstream signals. It is also possible that binding of DIS3/SCAR2 to WAVE complex subunits stimulates its ability to activate ARP2/3, as has been shown for human SCAR/WAVE (Innocenti et al., 2004).
The WH2 and A regions of WASP/WAVE proteins bind to G-actin and Arp2/3, respectively, and both activities are required for activation (Zalevsky et al., 2001). An intact WA domain appears to be essential for DIS3/SCAR2 function. Several dis3 alleles remove the WA domain coding information (Figure 3D). The dis3-4 allele is the best characterized allele, deleting the WA domain, but leaving the remainder of the protein intact. If dis3-4 protein accumulates, this would provide strong evidence that the WA domain is essential. The DIS3-WA protein fragment has the expected biochemical properties. DIS3-WA interacted physically with the ARPC3 subunit (Figure 11). The WA region was essential for the interaction with ARPC3 because a deletion construct that mimicked the dis3-4 allele failed to bind ARPC3. Importantly, we found that the C-terminal region of DIS3/SCAR2 efficiently activated the nucleation activity of bovine Arp2/3 in actin polymerization assays. As expected, DIS3/SCAR2 also stimulated the actin filament branching activity of Arp2/3. Although DIS3/SCAR2 clearly activates vertebrate Arp2/3, it will be important to test the efficiency with which it activates the Arabidopsis complex.
A Conserved Abi-1 Binding Domain May Recruit DIS3/SCAR2 into a WAVE Complex
DIS3/SCAR2 is likely to function as an ARP2/3 activator in the context of a WAVE complex. Human SCAR/WAVE is recruited to the WAVE complex via a direct interaction with Abi-1 (Gautreau et al., 2004; Innocenti et al., 2004). A highly diverged Arabidopsis Abi1-like protein, ABIL1, appears to mediate similar interactions. ABIL1 shares 20% amino acid identity with the N-terminal region of human Abi-1 that mediates NAP1 and SCAR/WAVE binding but lacks the C-terminal SH3 domain that is present in Abi-1 (Figure 9). Despite the low level of amino acid identity, ABIL1 interacted with both human NAP1 and GRL (NAP1) in the yeast two-hybrid assay. In addition, full-length ABIL1 interacted with the SCAR homology domain of DIS3/SCAR2. Although these experiments do not define completely the composition of WAVE complexes in vivo, the data strongly suggest that Arabidopsis WAVE subunits have retained the domains needed for complex assembly. BRK1 is coexpressed with WAVE and ARP2/3 family members (El-Assal et al., 2004b) and is expected to bind to the SCAR homology domain of SCAR proteins. In maize (Zea mays), mutation of Brick1 causes epidermal shape defects (Frank and Smith, 2002); however, the involvement of BRK1 in Arabidopsis WAVE complex function is not known.
Multiple Pathways to ARP2/3 Activation
Based on our phenotypic measurements, we rank the distorted mutants in the following order of decreasing phenotypic severity: dis1 (arp3) ≈ dis2 (arpc2) > pir (sra-1) ≈ grl (nap1) > dis3/scar2. The moderate trichome branch length reduction and actin phenotypes of null pir (sra-1) and grl (nap1) compared with ARP2/3 subunit mutants suggest that WAVE complex-independent pathways for ARP2/3 activation exist (Basu et al., 2004; El-Assal et al., 2004b). The existence of multiple activating pathways converging on Arp2/3 is a common theme in eukaryotic cells. For example, in Drosophila, WASP and SCAR/WAVE activators control dramatically different developmental processes (Ben-Yaacov et al., 2001; Zallen et al., 2002). In an insect cell RNA interference assay, SCAR is essential for normal lamellipodia morphology, whereas WASP has no clear effect (Kunda et al., 2003; Rogers et al., 2003). Budding yeast uses three different Arp2/3 activators, Pan1p, Las17p (WASP-like), and Abp1p, in a common endocytic pathway (Kaksonen et al., 2003). However, the timing of activator assembly and disassembly varies with respect to the different stages of plasma membrane internalization.
Multiple SCAR genes are expected to activate Arabidopsis ARP2/3. The null dis3 phenotype is weak compared with those of pir (sra-1) and grl (nap1) plants. Given the presence of multiple genes that encode both SCAR and WA homology domains, it seems likely that additional SCAR proteins function as WAVE complex subunits during morphogenesis. This appears to be the case in vertebrates. Within the WAVE subfamily, WAVE isoform mutants have distinct phenotypes (Dahl et al., 2003; Yan et al., 2003). SCAR proteins may have distinct functions within the cell. Multiple WAVE isoforms present in the same cell type have distinct localization patterns (Nozumi et al., 2003). It will be important to determine the extent to which Arabidopsis SCAR proteins function interchangeably.
The Actin Cytoskeleton and Potential Functions of DIS3/SCAR2
Although much recent progress has been made in the cloning of distorted group genes, uncertainty remains concerning the in vivo roles for WAVE and ARP2/3. This stems in part from the lack of knowledge concerning the localization of WAVE and ARP2/3 complexes in cells. However, a few simple conclusions about the roles WAVE and ARP2/3 play during actin-dependent growth can be made. The following genetic arguments hold only if null alleles of individual WAVE and ARP2/3 subunit mutants reflect the complete loss of function phenotype for the complexes. However, in many cases, loss of a single WAVE or Arp2/3 subunit is lethal (Winter et al., 1999; Hudson and Cooley, 2002; Stevenson et al., 2002; Zallen et al., 2002; Sawa et al., 2003; Yan et al., 2003). The first general observation is that the WAVE-ARP2/3 pathway is not the major pathway for actin filament nucleation in Arabidopsis. WAVE and ARP2/3 subunit mutants have an extensive actin cytoskeleton with subtle defects in its organization. We have mentioned previously the possibility that FORMINs are the major actin filament nucleators in Arabidopsis (Le et al., 2003; El-Assal et al., 2004a). Second, the WAVE-ARP2/3 pathway is not the primary means by which plant cells control cell shape or growth. The reduced fresh weight and hypocotyl length of WAVE and ARP2/3 subunit mutants suggest a general role for these genes during plant growth, but the mutants have a normal overall architecture. In addition tip growing cells, such as pollen tubes and root hairs, are not clearly affected in ARP2/3 and WAVE mutants (although subtle root hair defects have been reported; Mathur et al., 2003b). Similarly, the effects of ARP2/3 and WAVE subunit mutations on pavement cell lobing are subtle and may vary depending on growth conditions. Trichomes, perhaps because of their intricate shape and constrained requirements for a properly organized actin cytoskeleton, may be the only cell type that has strict requirements for intact WAVE and ARP2/3 complexes during morphogenesis.
In non-plant cells, Arp2/3-dependent actin polymerization is physically linked to the distortion of membranes, whether it be dendritic actin filament networks (Wang, 1985; Svitkina and Borisy, 1999) or actin bundles (Svitkina et al., 2003) that push on the plasma membrane or actin comet tails that drive organelle motility (Taunton et al., 2000) or membrane internalization (Kaksonen et al., 2003). Although the force-generating potential of an actin filament network is high, the energy of turgor pressure is believed to be the primary driving force for plant cell expansion (Cosgrove, 1987). Perhaps the energy of ARP2/3-dependent actin polymerization and turgor pressure have complementary functions during plant cell expansion. Current ideas of WAVE and ARP2/3 function rely heavily on the observed actin phenotypes of WAVE and ARP2/3 mutants, with no direct information about which of the numerous actin arrays in the cell are ARP2/3 dependent. For example, groups that use GFP:TALIN to visualize actin filaments report excessive actin filament bundling in WAVE and ARP2/3 subunit mutants. The hypothesized primary function of the wild-type genes is to spatially restrict ARP2/3-dependent polymerization and subsequent vesicle delivery to specific cortical domains that are growing actively (Li et al., 2003; Mathur et al., 2003a, 2003b; Brembu et al., 2004). Groups that have employed antibody and phalloidin labeling of fixed cells and GFP:ABD2 in live cell imaging experiments do not detect excessive bundling (Basu et al., 2004; Deeks et al., 2004; El-Assal et al., 2004b); rather, WAVE and ARP2/3 subunit mutants have disorganized core cytoplasmic bundles at the onset of the mutant phenotype.
Although the disorganization of cytoplasmic actin bundles correlates with the onset of the cell swelling, the defects may be because of an indirect effect: an upstream actin-dependent process, such as altered membrane trafficking or organelle biogenesis, could have pleiotropic effects on the cell. We have speculated that ARP2/3-dependent polymerization is linked directly to core cytoplasmic bundle formation (Le et al., 2003; El-Assal et al., 2004a). The aligned bundles in developing branches may reflect or generate a membrane trafficking pathway to the vacuole—one in which ARP2/3 has multiple functions. The internalization step of endocytosis, the transport and fusion of compartments in an endosomal pathway, and the dynamics and/or positioning of the tonoplast may involve ARP2/3. Trichomes elongate at the greatest rate in the apical region of the branch (Schwab et al., 2003). Perhaps highly efficient membrane trafficking in the apical region of the branch is required to maintain polarized growth, and in its absence, unpredictable defects in cell shape control ensue. Colocalization of WAVE and ARP2/3 complexes with markers for different endomembrane compartments will help to define functional relationships.
METHODS
Plant Strains and Growth Conditions
The dis3-2, dis3-3, and dis3-4 alleles are in the Col-0 background and were isolated from T-DNA insertion pools obtained from the ABRC stock center. The dis3-1 allele was a gift from David Marks and is in the Col-0 background. zwi-3 was a gift from David Oppenheimer and is in the Col-0 background. The dis3-T1 allele (Salk_057481) was also obtained from the ABRC stock center. All alleles were backcrossed at least twice before use. Soil-grown plants were grown in Scotts Redi-Earth (Hope, AR) that was overlaid on an equal volume of coarse vermiculite under continuous illumination (110 μmol photons m−2 s−1) at 25°C. Tissue culture–grown plants were grown on half-strength MS plates with 1% sucrose at 24°C under continuous illumination (110 μmol photons m−2 s−1). The data reported for hypocotyl growth and actin localization were obtained using tissue culture–grown plants. Cell–cell adhesion defects were assayed using both soil- and tissue culture–grown plants. Trichome morphometry used soil-grown plants.
Plasmid Construction and Protein Expression in Escherichia coli
The DIS3-SHD (amino acids 1 to 283) and DIS3-ΔWA (amino acids 1032 to 1238) clones were made using gene-specific primers and Pfu DNA polymerase (Statagene, La Jolla, CA) to amplify the coding regions from cDNA. The name and sequence of all oligonucleotide primers used in this article are given in Table 4. The oligonucleotide primers used to amplify DIS3-SHD fragment had introduced EcoRI and XhoI sites, and those used to amplify DIS3-ΔWA had introduced NcoI and EcoRI sites. PCR products were cloned into pCRII-TOPO (Invitrogen, Carlsbad, CA) to generate pTOPO-DIS3-SHD and pTOPO- DIS3-ΔWA. The two-hybrid DIS3-WA construct (amino acids 1032 to 1399) was amplified similarly with oligonucleotide primers with introduced NcoI and EcoRI sites and was cloned as above to generate pTOPO-DIS3-WA. The DIS3-WA fragment used for recombinant protein expression in E. coli included amino acids 1084 to 1399 and was amplified from EST 171I23 (GenBank accession number R65505). BamHI and XhoI sites were introduced into the oligonucleotide primers and were used to clone the PCR product into the Gateway vector pENTR/D/TOPO (pEN) (Invitrogen) to generate pEN-1084WA. To create a full-length human WAVE1 clone, 1680 bp of the coding region was amplified from full-length cDNA (GenBank accession number D87459), with oligonucleotide primers having introduced NcoI and EcoRI sites, and cloned into pCRII-TOPO to generate pTOPO-HsWAVE1. Full-length ARPC3 and ABIL1 coding regions were amplified from cDNA and cloned into Gateway vector pENTR/D/TOPO (pEN) (Invitrogen) to generate pEN-ARPC3 and pEN-ABIL1. For ARPC3 amplification, NcoI and EcoRI sites, and for ABIL1 amplification, EcoRI and BamHI were introduced in the oligonucleotide primers. All PCR-generated clones were sequenced on both strands before use, and the introduction of restriction sites added two amino acids to the N termini of the fusion proteins. pTOPO-DIS3-SHD, pTOPO-DIS3-WA, pTOPO-DIS3-ΔWA, and pTOPO-HSWAVE1 were subcloned into the yeast two-hybrid GAL4-activation vector (prey) pACT2 using the appropriate restriction enzymes to generate pAT-DIS3-SHD, pAT- DIS3-WA, pAT- DIS3-ΔWA, and pAT-HSWAVE1, respectively. pEN-DIS2 (El-Assal et al., 2004a) and pEN-ARPC3 were subcloned into the yeast two-hybrid GAL4-DNA binding vector (bait) pAS2 using the appropriate restriction enzymes to generate pAS-DIS2 and pAS-ARPC3. pEN-ABIL1 was recombined into Gateway destination yeast two-hybrid bait (GAL4 DNA binding domain) vectors pDEST32 and pDEST32-Trp to generate pDS32-ABIL1 and pDS32-ABIL1-T, respectively. pDS22-ATNAP and pDS22-HSNAP constructs are as previously described (Basu et al., 2004).
Table 4.
The Names and Sequences of the Oligonucleotide Primers That Were Used in This Article
| Name | Sequence (5′−3′) |
|---|---|
| Primers used for DIS3 allele sequencing: | |
| DIS3-F1 | CTCATATGCCAACACAAGATCCTCCA |
| DIS3-F2 | TCCGCCGTCTTGAATCCAC |
| DIS3-F3 | GTGATGAAGGTTTGCTGCTGAG |
| DIS3-F4 | GTTTGAAACGCTACACCGTACC |
| DIS3-F5 | GGAGGAGCATTTAGAGGCTCAT |
| DIS3-F6 | GACAATTATGTGGACGCACCAG |
| DIS3-F7 | GCATCAGAAGTCGTTGAAACTTCT |
| DIS3-F8 | CTCTGGATGATGAAACCGATTGT |
| DIS3-F9 | CTGATGCTGGAGAGATTTTCCAT |
| DIS3-F10 | CCTGAGACTATTCCGCCCAAT |
| DIS3-F11 | GTCGATGCTGTTGCAGCTCATGA |
| DIS3-F12 | CCCAGATGGTCCTGTGAACAGA |
| DIS3-F13 | AATCGATGATCGTTGGCTTTTC |
| DIS3-R1 | GGTGGTGCCAGCCGAGAA |
| DIS3-R2 | CCAATGACTGTATGTATTTAATGTTATATGT |
| DIS3-R3 | CGTGTTGTTACTGCCGGCTTTAG |
| DIS3-R4 | GATCATCGCTTGAACTTTGAGATT |
| DIS3-R5 | CCTGCTGATTGTGGTTCAAACTCTAA |
| DIS3-R6 | CAACTGTATCATGGAAAATCTCTCC |
| DIS3-R7 | CACAATCGGTTTCATCATCCAG |
| DIS3-R8 | GGAGAAGTTTCAACGACTTCTGAT |
| DIS3-R9 | CTGTTCCTGTTTCCGACTCCATG |
| DIS3-R10 | TGATGAGCCTCTAAATGCCCTC |
| DIS3-R11 | AGCAATCTAACCTTTGTTAGAGAA |
| DIS3-R12 | CATCTCAGCAGCAAACCTTCAT |
| DIS3-R13 | CAAGACGGCGGATATAGGTAGG |
| Primers used to make different plasmid constructs: | |
| pTOPO-DIS3-WA/F | CACCATGGAGCCTAAGCAAGAGTTGAATCTG |
| pTOPO-DIS3-WA/R | GAATTCTCAAGAATCATCCAACTATCTG |
| pTOPO-DIS3-ΔWA/F | CACCATGGAGCCTAAGCAAGAGTTGAATCTG |
| pTOPO-DIS3-ΔWA/R | GAATTCTCACTTTGTTGATTGAGTTGC |
| pTOPO-HSWAVE1/F | CACCATGGACATGCCGCTAGTGAAAAGAAACATCG |
| pTOPO-HSWAVE1/R | GGATCCTTACTCCAACCAATCTACTTCATC |
| pTOPO-DIS3-SHD/F | CACCAGAATTCACATGCCGTTGACGAGGTACCAATC |
| pTOPO-DIS3-SHD/R | CTCGAGCCATTCCTCCACTGAGATGCTCTCC |
| pEN-ATARPC3/F | CACCATGGTTTATCACTCGAGTTTTGT |
| pEN-ATARPC3/R | GAATTCTCATAGGACGACCACGTTCA |
| pEN-ATABIL1/F | CACCGAATTCATGGAAACGGAGATAT |
| pEN-ATABIL1/R | GGATCCTCACGAGACATAGCCAGCTT |
| pEN-1084WA/F | CACCGGATCCGTCGAAACCAGTAGTTCTGCTTTATCAGC |
| pEN-1084WA/R | CCATCTCGAGTCAAGAATCACTCCAACTATCTGAATCTTC |
| Primers used for gene expression study (RT-PCR): | |
| ATSCAR1-Nterm/F | CACCAGAATTCACATGTATGACGAATTGCCACCG |
| ATSCAR1-Nterm/R | CTCGAGGTTTAGCCACAGAGGCAAGTCTTG |
| DIS3-Nterm/F (An) | CACCAGAATTCACATGCCGTTGACGAGGTACCAATC |
| DIS3-Nterm/R | CTCGAGCCATTCCTCCACTGAGATGCTCTCC |
| ATSCAR3-Nterm/F | CACCATGGAATGCCACGGAATGTATACGG |
| ATSCAR3-Nterm/R | GAATTCGGCCAAACGTGACATATCTCAAC |
| ATSCAR4-Nterm/F | CACCGAATTCATGGCATTGACGAGATACCAG |
| ATSCAR4-Nterm/R | CTCGAGCGTCTCTTTAATAGTTGTGTGAG |
| ATSCAR5-Nterm/F | CACCATGGACATGCCGTTGGTGAGGTTCAAG |
| ATSCAR5-Nterm/R | GGATCCCACTTTGTTCTTGCGCACCTGAC |
| ATPIR (SRA-1)/F | CGCCTTCTGACTCTTCCAGC |
| ATPIR (SRA-1)/R | CCATTGTCCAAGGCAAAGAGAA |
| pEN-ATABIL1/F | CACCGAATTCATGGAAACGGAGATAT |
| pEN-ATABIL1/R | GGATCCTCACGAGACATAGCCAGCTT |
| ATARPC2/F | ATGATACTATTGCAGTCACATTCA |
| ATARPC2/R | AACTTGACGTGATAACTAACATAAG |
| ATARPC4/F | TAGGACATGGCAAACTCATTAGGG |
| ATARPC4/R | ACTGGCTTCCTCCTCAGTACTTGAAA |
| GAPC/F | CACTTGAAGGGTGGTGCCAAG |
| GAPC/R | CCTGTTGTCGCCAACGAAGTC |
For protein expression in E. coli, pEN-1084WA was subcloned into the GST expression vector pGEX6P-1 (GE Healthcare, Piscataway, NJ) as a BamHI and XhoI fragment to generate pGEX6P-1084WA. pEN-ARPC3 was subcloned into the N-terminal HIS expression vector pET30a (Novagen, Madison, WI) as a NcoI and EcoRI fragment to generate pET30a-ARPC3. To produce GST-DIS3-WA and ARPC3, the plasmids were freshly transformed into the E. coli strain Rossetta (Novagen), grown to an OD600 of 0.6 to 0.7, and induced with 200 μM isopropylthio-β-galactoside at 37°C for 3.5 h. GST-DIS3-WA and His-ARPC3 were purified with glutathione (Sigma-Aldrich, St. Louis, MO) or Ni2+ (Novagen) agarose beads, respectively, according to standard protocols.
Yeast Two-Hybrid and GST Pull-Down Assays
For yeast two-hybrid assays, pairs of bait and prey plasmids defined in the figures were cotransformed into the Saccharomyces cerevisiae strain Y190 using the lithium acetate transformation method. The transformants were selected on Leu−Trp− medium. A two-hybrid interaction was determined by colony formation on Leu−Trp−His− medium containing 40 mM 3-amino-1,2,4-triazole. For each two-hybrid plasmid pair, liquid β-galactosidase assays were done in triplicate from three independent colonies using standard protocols (Ausubel et al., 1994). To quantify the expression levels of DIS3-WA and DIS3-ΔWA in yeast two-hybrid strains, the soluble protein fraction of Y190 strains containing pAS-ARPC3 and pAT-DIS3-WA, pAT-DIS3-ΔWA, or pACT2 were extracted from 10 mL of overnight culture using a glass-bead extraction procedure, and protein gel blots were probed with anti-HA (BabCo, Richmond, CA) or anti-budding yeast α-Tubulin (Developmental Studies Hybridoma Bank, Iowa City, IA) antibodies. Based on densitometry of the protein gel blot, the mean ratio of total integrated signal from HA-tagged fusion proteins and tubulin was calculated for each fusion protein.
In pull-down assays, bead-bound GST-DIS3-WA (1 μM) was mixed with 1 μM of His-ARPC3 in binding buffer containing 50 mM Hepes/KOH, pH 7.6, 20 mM KCl, and 5 mM MgCl2. Binding reactions were incubated at 4°C for 2 h. Total, bead-bound, and unbound fractions were separated by SDS-PAGE and transferred to nitrocellulose membranes. His-ARPC3 was detected with polyclonal anti-ARPC3 antibody (1:1000), and blots were quantified using densitometry. Anti-ARPC3 peptide antibodies were made in rabbits using amino acids 2 to 15 (CVYHSSFVDEEGVTK) and 25 to 41 (CKSHIKGPAPVSEQDKTD) of ARPC3 as antigens. As a control for nonspecific binding of GST-DIS3-WA to His-tagged proteins, 1 μM GST-DIS3-WA was mixed with 1 μM HIS-ROP2 and analyzed as above. HIS-ROP2 was detected with a rabbit polyclonal antibody. Protein concentrations were determined using the Bradford assay.
Amino Acid Sequence Comparisons
Protein amino acid sequences were aligned using ClustalW. In ClustalW, the gap-opening and gap-extension penalties were set at 0.20 and 10, respectively, and the alignments were refined manually.
Plant Morphometry
Trichome branch lengths were measured using scanning electron microscopy images and images captured using a Nikon Coolpix 4500 digital camera (Tokyo, Japan) mounted on a Leica SMZ 12.5 stereo microscope (Wetzlar, Germany) fitted with a 1.6× 1.2 NA objective. Measurements of trichome shape parameters were taken from at least 10 different trichomes using ImageJ software version 1.3 (http://rsb.info.nih.gov/ij). For branch length measurement, trichomes with three visible branches that were approximately parallel to the image plane were selected. The epidermal cells of distorted group mutants are desiccation sensitive. We therefore used cryo-fixed samples and scanning electron microscopy to visualize gaps between adjacent cells. Pavement cell gaps were also visualized using reflected light from epi-illumunated seedlings that were mounted on a Nikon E800 Eclipse microscope and visualized using a Nikon 50X LU 0.55 NA metallurgical lens. For this method, shoots were removed, and the hypocotyl was immediately mounted in a blob of pure Vaseline on a glass slide. Images were captured on a SPOT RT CCD camera (Diagnotics, Sterling Heights, MI). To measure the cell–cell gap frequency in 12-DAG cotyledons, 540 × 390-μm boxes were overlaid onto three nonoverlapping regions of a scanning electron microscopy image taken from the apical third of the cotyledon. The mean gaps/mm2 within the three boxes was calculated for each cotyledon, and the reported gap frequency for each gentoype is the mean gaps/mm2 of three different measurements. For the etiolated hypocotyl elongation assay, wild-type and mutant seeds were sown onto the surface of half-strength MS in square plates. After 2 to 3 d of cold treatment and 12 h of light exposure, plates were wrapped with double layers of aluminum foil and placed vertically in a growth chamber at 22°C. Total hypocotyl length was measured after 7 d. The length of the cells in the middle region of the hypocotyls was measured from differential interference contrast images. Fresh weight measurements were taken from wild-type and mutant plants in segregating populations. Seedlings were selected based on synchronous germination, but independent of phenotype at 3 DAG, and were harvested for fresh weight determination at 16 DAG.
DIS3 Allele Characterization and Sequencing
PCR products were generated from genomic DNA isolated from the mutant plants. PCR primer pairs included the following: DIS3-F1/DIS3-R11, DIS3-F3/DIS3-R9, DIS3-F5/DIS3-R7, DIS3-F7/DIS3-R5, DIS3-F9/DIS3-R3, and DIS3-F11/DIS3-R1. The PCR products were sequenced using 13 forward primers (DIS3-F1 to DIS3-F13) and 13 reverse primers (DIS3-R1 to DIS3-R13; see Table 4). DNA sequencing was done using Bigdye version 3.1 (PE Biosystems, Foster City, CA), and unincorporated nucleotides were removed with a Centriflex TM gel filtration cartridge (Edge Biosystems, Gaithersburg, MD). Sequencing reactions were analyzed at the Purdue Genomics Center. For DNA gel blots, DNA extractions were performed using the CTAB (N-cetyl trimethyl ammonium bromide) method. Six micrograms of each sample were separated on a 0.9% agarose gel and blotted onto a positively charged nylon membrane (Nytron transfer membranes; Midwest Scientific, Valley Park, MO). The DNA probe was labeled using DIG High Prime DNA labeling and detection starter kit II (Roche, Penzberg, Germany). Hybridization was performed overnight at 42°C in a hybridization incubator (Robbins Scientific model 400; Sunnyvale, CA). Washes were performed twice at 65°C in 2× SSC (0.3 M NaCl and 0.03 M sodium citrate) with 0.1% SDS for 15 min, followed by two washes in 0.2× SSC with 0.1% SDS for 15 min. Signal was visualized using chemiluminescence detection.
RT-PCR
Total RNA for RT-PCR was isolated using the RNeasy plant mini kit from Qiagen (Chatsworth, CA). All RNA extractions and RT-PCR analyses were repeated at least two times. Two micrograms of total RNA was treated with five units of RQ1 RNase-free DNase (Promega, Madison, WI). For first-strand cDNA synthesis reactions, 25 μL of total RNA was annealed with random hexanucleotides (20 ng/μL) (New England Biolabs, Beverly, MA) and then reverse transcribed with 200 units of M-MLV RT (Invitrogen). Thereafter, 1/25 volume of the cDNA was used as a template for PCR. To analyze the expression of different DIS3 alleles, we isolated RNA from 15-DAG seedlings.
Fluorescence Microscopy
Actin filaments polymerized in vitro were observed by epi-fluorescence illumination under a Nikon Microphot SA microscope equipped with a 60×, 1.4 NA Planapo objective and digital images were collected with a Hamamatsu ORCA-ER 12-bit CCD camera (Osaka, Japan). Confocal images were collected using an MRC Bio-Rad 2100 laser scanning confocal microscope (Hercules, CA) mounted on a Nikon Eclipse E800 microscope with a Nikon 60× 1.2 NA Planapo water immersion objective. Images were processed and analyzed using MetaMorph 6.0 (Universal Imaging, Downingtown, PA) or ImageJ (http://rsb.info.nih.gov/ij) software.
Cytoskeleton Localization and Quantification
Tissues were fixed in 2% formaldehyde in 100 mM Pipes-KOH, 5 mM EGTA, and 4 mM MgCl2, pH 6.9 (PEM), for 30 min. After two washes with PEM, the tissues were incubated in PEM containing 1% glycerol and 0.198 μM Alexa Fluor 488 phalloidin (Molecular Probes, Eugene, OR) overnight at 4°C. For live cell imaging, intact GFP:ABD2 (Sheahan et al., 2004) or GFP:MBD (Marc et al., 1998) expressing 10- to 14-DAG seedlings were transferred to 2- to 3-mm deep chambered slides and submerged in water. GFP was excited using a 488-nm argon-ion laser line and ∼300 μW of laser power at the sample and a 515 ± 30-nm band-pass filter. Images were collected at 500 lines/s with two or three iterations of Kahlman averaging. Seedlings were scanned for appropriately staged cells for up to 30 min before they were discarded and a new sample was made. The methods for the quantification of the relative amounts of core cytoplasmic actin filaments in phallodin labeled cells and GFP:ABD2 lines were as described previously (Le et al., 2003; El-Assal et al., 2004a). Briefly, the relative amounts of core cytoplasmic actin filaments to total actin filaments were measured at the midpoint ± 1.3 μm of stage 3/4 and stage 4/5 branches. The cortical domain of the branch was defined as a 2.5-μm-wide band that included the plasma membrane at the outer edge. Core cytoplasm was the excluded inner cytoplasmic volume. Total is defined as cortical + core. Stage 3/4 branches were defined as those that had lengths between 10 and 15 μm. Stage 4/5 branches were defined as those that had branches between 16 and 50 μm in length. To ensure that late stage dis3 cells with a reduced branch length were not included in the analyses, no cells with a total vertical height of more than 40 μm were included in the measurements. For microtubule quantitation, a branch having a majority of microtubules at an approximate angle of 90° relative to the long axis was defined as transverse. A branch having a majority of microtubules with angles <45° from the long axis of the branch was defined as oblique or longitudinal.
Actin Polymerization
Actin was purified from rabbit skeletal muscle. Oligomers and minor actin binding protein contaminants were removed by gel filtration chromatography on Sephacryl S-300 (MacLean-Fletcher and Pollard, 1980) and the resulting monomeric actin stored in Buffer G (1× is 5 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT, and 0.1 mM NaN3) at 4°C. Pyrene derivatization of G-actin on Cys374 was performed as modified by Pollard (1984). Assembly of actin (5% pyrene labeled) was monitored with a PTI Quantamaster spectrofluorimeter (QM-2000-SE; Photon Technology International, South Brunswick, NJ) with excitation and emission wavelengths of 365 and 407 nm, respectively.
Actin nucleation assays were performed essentially as described by Higgs et al. (1999). Arp2/3 was purchased from Cytoskeleton (Denver, CO). Two activators of Arp2/3 were compared. The WA domains of human N-WASP (GST-WASP-WA) and DIS3/SCAR2 (GST-DIS3-WA) were purified by affinity chromatography on glutathione agarose (the original GST-N-WASP-VCA expression plasmid [Egile et al., 1999] was kindly provided by Marie-France Carlier). To test the ability of WASP-WA and DIS3-WA to activate vertebrate Arp2/3 complex, we measured their effect on the initial phase of actin polymerization. Monomeric actin was converted to MgATP-G-actin by addition of 0.2 mM EGTA and 50 μM MgCl2 and stored on ice. Reactions (150 μL) in KMEI (1× contains 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole-HCl, pH 7.0) included 50 nM Arp2/3 complex and varying amounts of GST-WASP-WA or GST-DIS3-WA, as described in the figure legend. Polymerization was initiated by the addition of G-actin to a final concentration of 4 μM (5% pyrene labeled) and fluorescence monitored for at least 600 s.
The concentration of barbed ends [N] generated during the nucleation reaction was calculated according to the equation [N] = elongation rate/(k+[A]), where k+ is the association rate constant for the barbed end at pH 7.0 (11.6 μM−1 s−1), [N] is the concentration of barbed ends, and [A] is the concentration of actin monomers (Higgs et al., 1999). The elongation rate was taken from the slope of the assembly curves at half maximal polymerization.
Actin Filament Branching Assay
Individual actin filaments labeled with fluorescent phalloidin were imaged by fluorescence microscopy according to Blanchoin et al. (2000). Actin at 4 μM alone, or with 50 nM Arp2/3 and/or 100 nM activator (GST-WASP-WA or DIS3-WA), was polymerized in 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 0.2 mM CaCl2, 0.5 mM DTT, 3 mM NaN3, and 10 mM imidazole, pH 7.0, at 25°C for 30 min and labeled with an equimolar amount of rhodamine-phalloidin (Sigma-Aldrich) during polymerization. The polymerized F-actin was diluted to 10 nM in fluorescence buffer containing 10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 100 mM DTT, 100 μg/mL of glucose oxidase, 15 mg/mL of glucose, 20 μg/mL of catalase, and 0.5% methylcellulose. A dilute sample of 3 μL was applied to a 22 × 22-mm cover slip coated with poly-l-Lys (0.01%).
The amino acid sequences of SHD, basic, and WA domains from Arabidopsis thaliana have been deposited with the EMBL/GenBank data libraries under the following accession numbers: DIS3/SCAR2 (GenBank accession number AY817016), ABIL1 (AT2G46225, GenBank accession number AY17012), ABIL2 (AT3G49290, GenBank accession number AY17013), ABIL3 (AT5G24310, GenBank accession number AY17014), ABIL4 (AT5G42030, GenBank accession number AY17015), and Hs-Abi-1 (GenBank accession number AAN28379).
Acknowledgments
We would like to thank David McCurdy for seeds from GFP:ABD2-expressing lines. We also thank David Marks for the dis3-1 allele and Laurent Blanchoin for good advice on the Arp2/3 nucleation and branching assays. Phillip SanMiguel and Purdue Genomics Center provided excellent DNA sequencing services. Debbie Sherman acquired the scanning electron microscopy images at the Purdue Life Sciences Microscopy Center. Work in the lab of D.B.S. was supported by a grant from the Department of Energy–Energy Biosciences Division (DE-FG02-02ER15357) and a Purdue Agricultural Research Program fellowship. Work in the lab of C.J.S. was supported by grants from the USDA–National Research Initiative (2002-35304-12412) and the Department of Energy–Energy Biosciences Division (DE-FG02-04ER15526).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Daniel B. Szymanski (dszyman@purdue.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.027987.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current protocols in molecular biology. (New York: John Wiley & Sons).
- Basu, D., El-Assal, S.E., Le, J., Mallery, E.L., and Szymanski, D.B. (2004). Interchangeable functions of Arabidopsis PIROGI and the human WAVE complex subunit SRA-1 during leaf epidermal morphogenesis. Development 131, 4345–4355. [DOI] [PubMed] [Google Scholar]
- Ben-Yaacov, S., Le Borgne, R., Abramson, I., Schweisguth, F., and Schejter, E. (2001). Wasp, the Drosophila Wiskott-Aldrich syndrome gene homologue, is required for cell fate decisions mediated by notch signaling. J. Cell Biol. 152, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagg, S.L., and Insall, R.H. (2004). Solving the WAVE function. Nat. Cell Biol. 6, 279–281. [DOI] [PubMed] [Google Scholar]
- Blagg, S.L., Stewart, M., Sambles, C., and Insall, R.H. (2003). PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 13, 1480–1487. [DOI] [PubMed] [Google Scholar]
- Blanchoin, L., Amann, K.J., Higgs, H.N., Marchand, J.B., Kaiser, D.A., and Pollard, T.D. (2000). Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404, 1007–1011. [DOI] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Bogdan, S., Grewe, O., Strunk, M., Mertens, A., and Klambt, C. (2004). Sra-1 interacts with Kette and Wasp and is required for normal neuronal and bristle development. Development 16, 3981–3989. [DOI] [PubMed] [Google Scholar]
- Bogdan, S., and Klambt, C. (2003). Kette regulates actin dynamics and genetically interacts with Wave and Wasp. Development 130, 4427–4437. [DOI] [PubMed] [Google Scholar]
- Brembu, T., Winge, P., Seem, M., and Bones, A.M. (2004). NAPP and PIRP encode subunits of a putative Wave regulatory protein complex involved in plant cell morphogenesis. Plant Cell 16, 2335–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings, D.A., Harper, J.D., and Vaughn, K.C. (2003). The association of peroxisomes with the developing cell plate in dividing onion root cells depends on actin microfilaments and myosin. Planta 218, 204–216. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1987). Wall relaxation and the driving forces for cell expansive growth. Plant Physiol. 84, 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, J.P., Wang-Dunlop, J., Gonzales, C., Goad, M.E., Mark, R.J., and Kwak, S.P. (2003). Characterization of the WAVE1 knock-out mouse: Implications for CNS development. J. Neurosci. 23, 3343–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks, M.J., Kaloriti, D., Davies, B., Malho, R., and Hussey, P.J. (2004). Arabidopsis NAP1 is essential for ARP2/3-dependent trichome morphogenesis. Curr. Biol. 14, 1410–1414. [DOI] [PubMed] [Google Scholar]
- Echarri, A., Lai, M.J., Robinson, M.R., and Pendergast, A.M. (2004). Abl interactor 1 (Abi-1) Wave-binding and SNARE domains regulate its nucleoplasmic shuttling, lamellipodium localization and Wave-1 levels. Mol. Cell. Biol. 24, 4979–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden, S., Rohatgi, R., Podtelejnikov, A.V., Mann, M., and Kirschner, M.W. (2002). Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793. [DOI] [PubMed] [Google Scholar]
- Egile, C., Loisel, T.P., Laurent, V., Li, R., Pantaloni, D., Sansonetti, P.J., and Carlier, M.-F. (1999). Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 146, 1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assal, S.E., Le, J., Basu, D., Mallery, E.L., and Szymanski, D.B. (2004. a). DISTORTED2 encodes an ARPC2 subunit of the putative Arabidopsis ARP2/3 complex. Plant J. 38, 526–538. [DOI] [PubMed] [Google Scholar]
- El-Assal, S.E., Le, J., Basu, D., Mallery, E.L., and Szymanski, D.B. (2004. b). Arabidopsis GNARLED encodes a NAP125 homologue that positively regulates ARP2/3. Curr. Biol. 14, 1405–1409. [DOI] [PubMed] [Google Scholar]
- Frank, M.J., and Smith, L.G. (2002). A small, novel protein highly conserved in plants and animals promotes the polarized growth and division of maize leaf epidermal cells. Curr. Biol. 12, 849–853. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Wu, G., and Yang, Z. (2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152, 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau, A., Ho, H.Y., Steen, H., Gygi, S.P., and Kirschner, M.W. (2004). Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA 101, 4379–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N., Friml, J., Stierhof, Y.D., Jurgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428. [DOI] [PubMed] [Google Scholar]
- Gibbon, B.C., Kovar, D.R., and Staiger, C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11, 2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gournier, J.H., Goley, E.D., Niederstrasser, H., Trinh, T., and Welch, M.D. (2001). Reconstitution of human Arp 2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol. Cell 8, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Higgs, H.N., Blanchoin, L., and Pollard, T.D. (1999). Influence of the C terminus of Wiscott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 38, 15212–15222. [DOI] [PubMed] [Google Scholar]
- Hudson, A.M., and Cooley, L. (2002). A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J. Cell Biol. 156, 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti, M., Zucconi, A., Disanza, A., Frittoli, E., Areces, L., Steffen, A., Stradal, T.E.B., Di Fiore, P.P., Carlier, M., and Scita, G. (2004). Abi1 is essential for the formation and activation of a WAVE2 signaling complex mediating Rac-dependent actin remodeling. Nat. Cell Biol. 6, 319–327. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Sun, Y., and Drubin, D.G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475–487. [DOI] [PubMed] [Google Scholar]
- Kunda, P., Craig, G., Dominguez, V., and Baum, B. (2003). Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13, 1867–1875. [DOI] [PubMed] [Google Scholar]
- Le, J., El-Assal, S.E., Basu, D., Saad, M.E., and Szymanksi, D.B. (2003). Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr. Biol. 13, 1341–1347. [DOI] [PubMed] [Google Scholar]
- Li, S., Blanchoin, L., Yang, Z., and Lord, E.M. (2003). The putative Arabidopsis Arp2/3 complex controls leaf cell morphogenesis. Plant Physiol. 132, 2034–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L., and Insall, R. (1998). Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp 2/3 complex. Curr. Biol. 8, 1347–1356. [DOI] [PubMed] [Google Scholar]
- Machesky, L., Mullins, D., Higgs, H., Kaiser, D., Blanchoin, L., Hall, M., and Pollard, T. (1999). Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp 2/3 complex. Proc. Natl. Acad. Sci. USA 96, 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky, L.M., Atkinson, S.J., Ampe, C., Vandekerckhove, J., and Pollard, T.D. (1994). Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J. Cell Biol. 127, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean-Fletcher, S., and Pollard, T.D. (1980). Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem. Biophys. Res. Commun. 96, 18–27. [DOI] [PubMed] [Google Scholar]
- Marc, J., Granger, C.L., Brincat, J., Fisher, D.D., Kao, T., McCubbin, A.G., and Cyr, R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10, 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kernebeck, B., and Hulskamp, M. (2003. a). Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15, 1632–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kirik, V., Kernebeck, B., Srinivas, B.P., and Hulskamp, M. (2003. b). Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130, 3137–3146. [DOI] [PubMed] [Google Scholar]
- Nebenfuhr, A., Gallagher, L.A., Dunahay, T.G., Frohlick, J.A., Mazurkiewicz, A.M., Meehl, J.B., and Staehelin, L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121, 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozumi, M., Nakagawa, H., Miki, H., Takenawa, T., and Miyamoto, S. (2003). Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J. Cell Sci. 116, 239–246. [DOI] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Pollock, M.A., Vacik, J., Szymanski, D.B., Ericson, B., Feldmann, K., and Marks, M.D. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94, 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal, S.C., Kaiser, D.A., Torres, E., Pollard, T.D., and Rosen, M.K. (2003). A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat. Struct. Biol. 10, 591–598. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D. (1984). Polymerization of ADP-actin. J. Cell Biol. 99, 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., and Borisy, G.G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- Qiu, J.L., Jilk, R., Marks, M.D., and Szymanski, D.B. (2002). The Arabidopsis SPIKE1 gene is required for normal cell shape control and tissue development. Plant Cell 14, 101–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.L., Wiedemann, U., Stuurman, N., and Vale, R.D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi, R., Ho, H.H., and Kirschner, M.W. (2000). Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150, 1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler, R., Mathur, N., Srinivas, B.P., Kernebeck, B., Hulskamp, M., and Mathur, J. (2004). Actin control over microtubules suggested by DISTORTED2 encoding the Arabidopsis ARPC2 subunit homolog. Plant Cell Physiol. 45, 813–822. [DOI] [PubMed] [Google Scholar]
- Sawa, M., Suetsugu, S., Sugimoto, A., Miki, H., Yamamoto, K., and Takenawa, T. (2003). Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J. Cell Sci. 116, 1505–1518. [DOI] [PubMed] [Google Scholar]
- Schwab, B., Mathur, J., Saedler, R., Schwarz, H., Frey, B., Scheidegger, C., and Hulskamp, M. (2003). Regulation of cell expansion by the DISTORTED genes in Arabidopsis thaliana: Actin controls the spatial organization of microtubules. Mol. Genet. Genomics 269, 350–360. [DOI] [PubMed] [Google Scholar]
- Sheahan, M.B., Rose, R.J., and McCurdy, D.W. (2004). Organelle inheritance in plant cell division: The actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 37, 379–390. [DOI] [PubMed] [Google Scholar]
- Staiger, C.J., and Hussey, P.J. (2004). Actin and actin-modulating proteins. In The Plant Cytoskeleton in Cell Differentiation and Development, P. Hussey, ed (Oxford: Blackwell), pp. 32–80.
- Stevenson, V., Hudson, A., Cooley, L., and Theurkauf, W. (2002). Arp 2/3-dependent psuedocleavage furrow assembly in syncytial Drosophila embryos. Curr. Biol. 12, 705–711. [DOI] [PubMed] [Google Scholar]
- Stradal, T.E., Rottner, K., Disanza, A., Confalonieri, S., Innocenti, M., and Scita, G. (2004). Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 14, 303–311. [DOI] [PubMed] [Google Scholar]
- Svitkina, T., and Borisy, G. (1999). Arp2/3 and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in Lamellipodia. J. Cell Biol. 145, 1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina, T.M., Bulanova, E.A., Chaga, O.Y., Vignjevic, D.M., Kojima, S., Vasiliev, J.M., and Borisy, G.G. (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, D.B., Jilk, R.A., Pollock, S.M., and Marks, M.D. (1998). Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Marks, M.D., and Wick, S.M. (1999). Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11, 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton, J., Rowning, B.A., Coughlin, M.L., Wu, M., Moon, R.T., Mitchison, T.J., and Larabell, C.A. (2000). Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol. 148, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.L. (1985). Exchange of actin subunits at the leading edge of living fibroblasts: Possible role of treadmilling. J. Cell Biol. 101, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys, G.O., and Galway, M.E. (2003). Remodeling the cytoskeleton for growth and form: An overview with some new views. Annu. Rev. Plant Biol. 54, 691–722. [DOI] [PubMed] [Google Scholar]
- Welch, M.D., and Mullins, R.D. (2002). Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18, 247–288. [DOI] [PubMed] [Google Scholar]
- Winter, D.C., Choe, E.Y., and Li, R. (1999). Genetic dissection of the budding yeast Arp 2/3 complex: A comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. USA 96, 7288–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C., et al. (2003). WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 22, 3602–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky, J., Lempert, L., Kranitz, H., and Mullins, R.D. (2001). Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr. Biol. 11, 1903–1913. [DOI] [PubMed] [Google Scholar]
- Zallen, J.A., Cohen, Y., Hudson, A.M., Cooley, L., Wieschaus, E., and Schejter, E.D. (2002). SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 156, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- While this work was under review, Frank et al. (2004) reported that plant SCAR proteins bind to BRK1 (HSPC300) and can activate vertebrate Arp2/3.
- Frank, M., Egile, C., Dyachok, J., Djakovic, S., Nolasco, M., Li, R., and Smith, L.G. (2004). Activation of Arp2/3 complex-dependent actin polymerization by plant proteins distantly related to Scar/WAVE. Proc. Natl. Acad. Sci. USA 101, 16379–16384. [DOI] [PMC free article] [PubMed] [Google Scholar]