Abstract
Histone acetylation is a reversible posttranslational modification that plays a fundamental role in regulating eukaryotic gene expression and chromatin structure/function. Key enzymes for removing acetyl groups from histones are metal (zinc)-dependent and NAD+-dependent histone deacetylases (HDACs). The molecular function of HDACs have been extensively characterized by various approaches including chemical, molecular, and structural biology, which demonstrated that HDACs regulate cell proliferation, differentiation, and metabolic homeostasis, and that their alterations are deeply involved in various human disorders including cancer. Notably, drug discovery efforts have achieved success in developing HDAC-targeting therapeutics for treatment of several cancers. However, recent advancements in proteomics technology have revealed much broader aspects of HDACs beyond gene expression control. Not only histones but also a large number of cellular proteins are subject to acetylation by histone acetyltransferases (HATs) and deacetylation by HDACs. Furthermore, some of their structures can flexibly accept and hydrolyze other acyl groups on protein lysine residues. This review mainly focuses on structural aspects of HDAC enzymatic activity regulated by interaction with substrates, co-factors, small molecule inhibitors, and activators.
Keywords: acetylation, acylation, cancer, chromatin, epigenetics, sirtuin
Introduction
Covalent modifications of core histones play an important role in the modulation of chromatin structure and function. It has been proposed that site-specific histone modifications play a distinct role in diverse cellular functions and that their combinations constitute histone codes.1,2) Histone acetylation is a well-characterized modification regulated by opposing activities of histone acetyltrasferases (HATs) and histone deacetylases (HDACs), and serves as one of the major components in epigenetic regulation of gene expression. In the years since Vincent Allfrey (1921–2002) et al. discovered that histone acetylation levels correlate with gene activity in 1964,3) core histones have become the best-established protein target for reversible acetylation. Acetylation of histones occurs on lysine residues present in the N-terminal tails of core histones and is shown to associate with transcriptionally active chromatin.4) HDACs are enzymes that remove the acetyl functional group from histones, and are divided into two major families, zinc-dependent and NAD+-dependent families.5) HDACs are evolutionarily conserved from bacteria to humans. Although bacteria do not contain histones, they also have HAT- and HDAC-like proteins, which may function as enzymes regulating acetylation of non-histone proteins.6) As it has been demonstrated that mammalian HDACs can accept not only histones but also a variety of non-histone cellular proteins as their substrates, they are collectively called protein-lysine (K) deacetylases (KDACs).7) Furthermore, recent studies have shown that lysine residues are targeted by other acyl group modifications and that some HDAC enzymes are responsible for their deacylation.8–10) Thus, the catalytic reactions and biological functions of HDACs have become broader than originally thought.
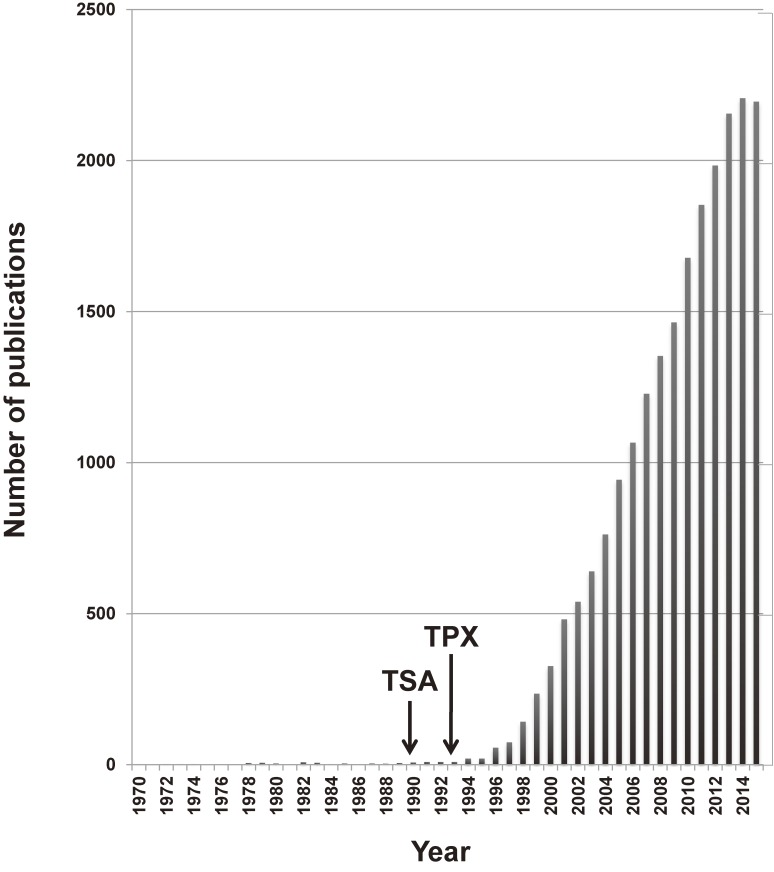
Importantly, identification of specific HDAC inhibitors trichostatin A (TSA) and trapoxin (TPX) A and B in the early 1990s triggered and accelerated the boom in HDAC research (Fig. 1).11) These HDAC inhibitors induced dramatic changes in gene expression and cellular phenotypes.12) Because aberrant acetylation of histones has been associated with many diseases including cancer, inflammation, and neuronal degeneration, HDACs attract broad attention as promising targets for therapy.13–15) Indeed, four distinct HDAC inhibitors have been approved by US Food and Drug Administration (FDA) and used clinically for chemotherapy against T-cell lymphoma or multiple myeloma. Structural biology of HDACs revealed the molecular mechanisms as to how the catalytic reaction occurs and is inhibited by these compounds.16) Thus, HDACs have become recognized as important enzymes for not only controlling cellular functions but also diseases in humans. Currently, the number of scientific papers containing the keyword “HDAC(s)” or “histone deacetylase(s)” exceeds 2,000 per year (Fig. 1).
Figure 1.
Number of publications regarding histone deacetylases per year. Data were obtained from the PubMed (https://www.ncbi.nlm.nih.gov/pubmed) based on the keywords of “histone deacetylase”, “histone deacetylases”, “HDAC”, and “HDACs”. The years of identification of TSA and TPX as HDAC inhibitors are indicated.
Discovery of deacetylases and their inhibitors
The first report on HDAC activity appeared in 1969 from Japan.17) Since then, great efforts have been made to characterize their enzymatic activities in vitro, however attempts to purify these enzymes to homogeneity were unsuccessful for more than 25 years. Furthermore, the basic physiological roles of HDACs in gene expression in cells and tissues remained elusive until the 1990s because there were no tools available to specifically control their cellular activity. The major breakthrough came from the chemical biological study of HDACs. During biochemical analysis of HDAC activity in nuclei in the 1970s, it was found that millimolar concentrations of n-butyrate (Fig. 2A) induced accumulation of acetylated histones in cells.18) Soon after this finding, it was reported that n-butyrate inhibits HDAC activity both in vitro and in vivo.19,20) Unfortunately, it was unclear whether the phenotypic consequences observed, such as cell cycle inhibition, were caused by n-butyrate-induced histone hyperacetylation because of its non-specific action on other enzymes and membranes.21) In 1990, potent HDAC inhibition by the natural product TSA (Fig. 2A) was discovered by our group.22) TSA, which had been isolated from a Streptomyces strain as an antifungal antibiotic23) was rediscovered as a powerful inducer of murine erythroleukemia cell differentiation.24,25) TSA inhibited the activity of partially purified HDACs with a low nanomolar inhibition constant. TSA has a hydroxamic acid group, which can chelate a metal ion, suggesting that HDACs are metal-containing enzymes. Importantly, TSA-resistant mutant cells possessing a TSA-resistant HDAC enzyme were found, providing genetic evidence for the causal relationship between HDAC inhibition and TSA-induced cell cycle arrest.22) Thus, TSA has become a key chemical probe widely used for analyzing the role of histone acetylation in various biological systems.11) TPX A and B (Fig. 2A), fungal cyclic peptides that had been identified as inducers of morphological change in transformed cells26) were identified as another type of strong HDAC inhibitors.27) Unlike TSA, TPX A and B irreversibly inhibit HDAC activity depending on their epoxyketone moiety. Discovery of these inhibitors from microbial origins opened a new avenue of research on histone acetylation, as researchers could investigate phenotypic changes induced by histone hyperacetylation in a variety of biological systems (Fig. 3).11) Histone acetylation was tightly associated with transcriptional control and treatment of cells with HDAC inhibitors induced a global change in gene expression. The potent ability of TPX to bind to HDACs was used for isolating the first HDAC molecule (HDAC1) by means of a TPX B-affinity matrix.28) HDAC1 was found to be a homolog of RPD3, a transcriptional regulator in the budding yeast Saccharomyces cerevisiae. Since the initial cloning of HDAC1, more than 20,000 papers have been published on HDACs to date.
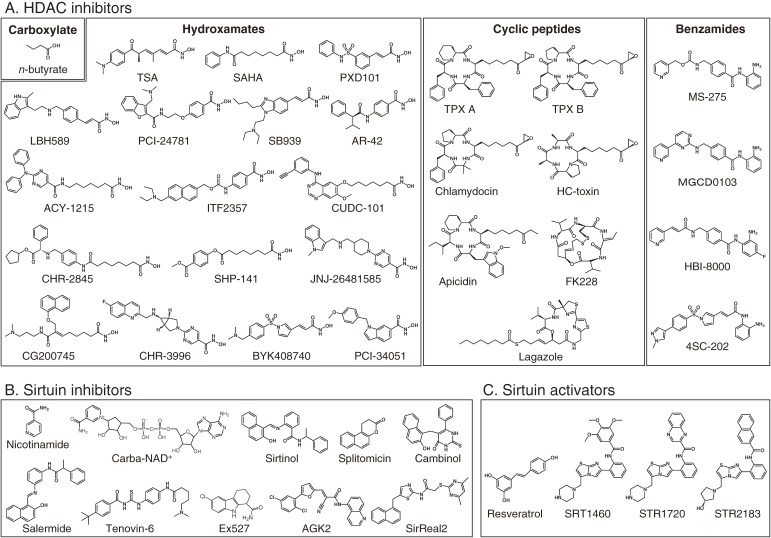
Figure 2.
Structures of small molecule modulators of lysine deacetylases. (A) Structures of HDAC inhibitors. HDAC inhibitors are classified according to the structural signatures. (B) Structures of sirtuin inhibitors. (C) Structures of sirtuin activators. Structures of SIRT1460, STR1720 and STR2183 are shown as representatives of STACs.
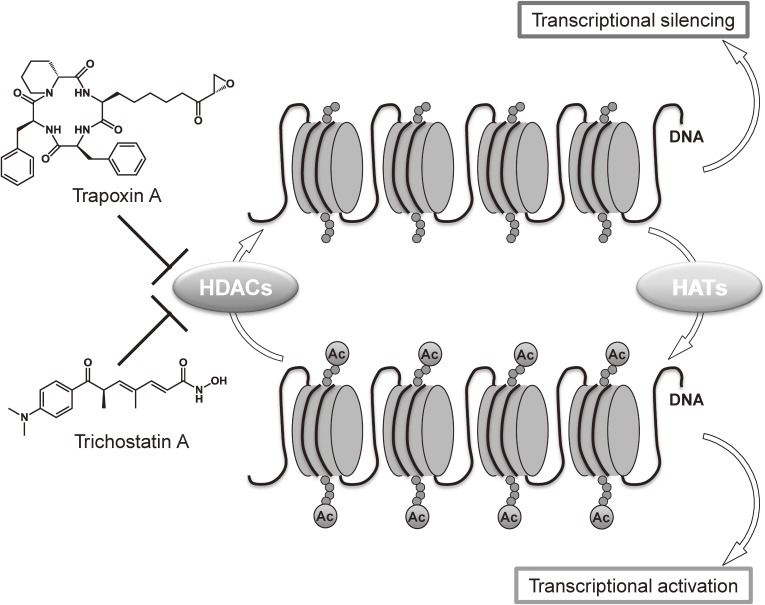
Figure 3.
Chromatin status regulated by histone acetylation. Histone acetyltransferases (HATs) are recruited by transcriptional regulators such as transcription factors and co-activators to the particular genetic loci, which form potentially active chromatin. On the other hand, histone deacetylases (HDACs) are recruited by transcriptional repressors or co-repressors and are associated with transcriptionally inactive chromatin. Specific HDAC inhibitors such as TSA and TPX A inhibit HDACs, leading to histone hyperacetylation in vivo.
Based on the homology to HDAC1, eleven orthologs in this family were identified in humans. These enzymes are divided into three classes: RPD3-like class I (HDAC1, HDAC2, HDAC3, and HDAC8); HDA1-like class II (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10); and Hos3-like class IV (HDAC11).29) The class II enzymes are further divided into two subclasses, class IIa (HDAC4, HDAC5, HDAC7, and HDAC9) and class IIb (HDAC6 and HDAC10). All of these HDACs are zinc-dependent enzymes that can be inhibited by TSA. Importantly, however, class IIb enzymes are relatively resistant to some classes of HDAC inhibitors such as TPX A and B.
The cellular- and tissue-level functions of zinc-dependent HDACs have been extensively studied by biochemical and genetic approaches.29) Knockout mouse lines have been developed for all of the HDAC genes and their phenotypes were characterized (Table 1).30–39) Class I HDACs are ubiquitously expressed in all tissues and localized mainly in the nucleus. Deletion of the HDAC1 gene resulted in embryonic lethality in mice and reduced proliferation of embryonic stem (ES) cells.30) Cell cycle inhibition by HDAC inhibitors or HDAC1 disruption appears to be largely due to the increased expression of the cyclin-dependent kinase (CDK) inhibitor protein p21, as deletion of the p21 gene partially rescued the growth inhibitory phenotype of both cancer and ES cells.40,41) HDAC3 knockout mice were also embryonic lethal with abnormal heart development.32) On the other hand, HDAC2 deletion did not cause embryonic lethality, but the mice died soon after birth due to cardiac defects.31) HDAC8-deficient mice also died in the perinatal period because of cranial defects.37) Class IIa HDACs are expressed in a tissue-specific manner. HDAC4 knockout mice showed abnormal hyperosteogeny, while HDAC5 and HDAC9 knockouts exhibited severe cardiac defects.33,34) HDAC7 disruption caused embryonic lethality due to vascular system defects.36) In contrast, class IIb HDACs appear dispensable for animal growth. HDAC6, a unique enzyme containing two active catalytic domains, was nonessential for mouse development and its knockout mice developed normally without any visible phenotypes35) although the enzyme is ubiquitously expressed.
Table 1.
Apparent phenotypes of gene knockouts
| Genes | Phenotypes in knockout mice | Reference |
|---|---|---|
| HDAC1 | Embryonic lethality due to severe proliferation defects | 14 |
| HDAC2 | Perinatal lethality due to severe cardiac defects | 15 |
| HDAC3 | Embryonic lethality | 16 |
| HDAC4 | Death by postnatal day 10. Premature ossification of developing bones | 17 |
| HDAC5 | Cardiac deficiency | 18 |
| HDAC6 | Viable and no obvious phenotype | 19 |
| HDAC7 | Embryonic lethality due to dilatation and rupture of blood vessels | 20 |
| HDAC8 | Perinatal lethality due to skull instability | 21 |
| HDAC9 | Cardiac deficiency | 18 |
| HDAC10 | Viable | 22 |
| HDAC11 | Viable | 23 |
| SIRT1 | Mostly early postnatal lethality | 30 |
| SIRT2 | Viable | 34 |
| SIRT3 | Viable and no obvious phenotype | 35 |
| SIRT4 | Viable and no obvious phenotype | 36 |
| SIRT5 | Viable and no obvious phenotype | 35 |
| SIRT6 | Death at about 4 weeks by severe metabolic defects | 31 |
| SIRT7 | Shorter life span | 32, 33 |
In 2000, S. cerevisiae Sir2, which had been originally identified in a genetic screen for genes involved in controlling expression of silent mating type loci, was found to deacetylate histones in a distinct mechanism dependent on nicotinamide adenine dinucleotide (NAD+).42,43) Yeast Sir2 is required for transcriptional silencing in the chromosomal regions around heterochromatin. Yeast has four other Sir2-like proteins (sirtuins), all of which function as deacetylases. Sirtuins are categorized as the third class of HDACs (class III), which require NAD+ as a cofactor for enzymatic activity. Sirtuins are also conserved from bacteria to mammals and there are seven homologues (SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7) in humans. Based on the phylogenic relationship of sirtuins having a conserved core domain with several sequence motifs, they are further categorized into several classes and subclasses. Of the seven mammalian sirtuins (SIRT1–SIRT7), SIRT1, 2, and 3 have robust deacetylase activity on a wide variety of non-histone substrates, whereas SIRT7 is a highly specific deacetylase for histone H3K18Ac.44) Nicotinamide (Fig. 2B), a byproduct of the sirtuin enzymatic reaction, is a physiological inhibitor that decreases gene silencing, increases rDNA recombination, and accelerates aging in yeast. Therefore, nicotinamide is widely used as a global inhibitor of sirtuins, although millimolar concentrations are required for inhibition of their activity in cell culture.
Mammalian sirtuin members exhibit different subcellular localizations, substrate specificities and are involved in diverse biological processes such as metabolism, inflammation, oxidative stress and senescence. SIRT1, SIRT6, and SIRT7 are localized in the nucleus,45–47) while SIRT2 is essentially cytoplasmic,48) although it can translocate into the nucleus during a particular cell cycle phase.49) On the other hand, SIRT3, SIRT4, and SIRT5 are located in mitochondria.50) Knockout mice of each sirtuin isoform have been developed and their phenotypes have been examined (Table 1). Deletion of any of the nuclear sirtuin genes (i.e., SIRT1, SIRT6, and SIRT7) in mice resulted in severe growth defects or shorter lifespans. SIRT1-deficient mice exhibit early postnatal lethality and developmental defects of the retina and heart.51) SIRT6-deficient mice are born at the Mendelian ratio without obvious abnormalities, but exhibit age-related degeneration, severe metabolic defects, and eventually die four weeks after birth.52) SIRT6-deficient cells show increased sensitivity to DNA damage and genomic instability. SIRT7 knockout mice exhibit partial embryonic lethality and a shorter lifespan with a progeroid-like phenotype.53,54) Depletion of SIRT7 in cultured cells consistently resulted in increased replication stress and impaired DNA repair largely due to reduced efficiency of non-homologous end joining.54) In contrast to depletion of the nuclear sirtuin genes in mice, mice lacking SIRT2 or the mitochondrial sirtuins including SIRT3, SIRT4, and SIRT5, were viable and did not display an obvious abnormality at the steady state.55–57)
Some sirtuins have been shown to possess another type of enzymatic activity, mono-ADP-ribosyltransferase.45,58) Interestingly, mono-ADP-ribosyltransferase activity was found to be the main enzymatic activity of SIRT4. Mutational analysis demonstrated that a conserved histidine residue was essential not only for NAD+-dependent deacetylase activity but also for mono-ADP-ribosyltransferase activity.59) Recently, it has been shown that sirtuins have a broad spectrum of de-acylation activity on proteins with acyl-lysine residues (see below).
Structural and catalytic mechanism of deacetylases
HDLP.
Successful determination of the X-ray crystal structure of histone deacetylase-like protein (HDLP) from the hyperthermophilic bacterium Aquifex aeolicus60) provided the first insight into the molecular mechanism underlying the TSA-sensitive, zinc-dependent enzymatic reaction of HDACs (classes I, II, and IV) (Fig. 4A). Despite its bacterial origin, HDLP showed more than 35% identity with human HDAC1 and TSA-sensitive histone deacetylase activity. The overall structure of HDLP showed topology similar to arginase, with an α/β fold and an 8-stranded parallel β-sheet (Fig. 4B).61) Arginase is a Mn2+-containing enzyme that hydrolyzes arginine to produce urea and ornithine, which is required for the urea cycle, suggesting that HDACs evolved from a common metalloprotein ancestor.
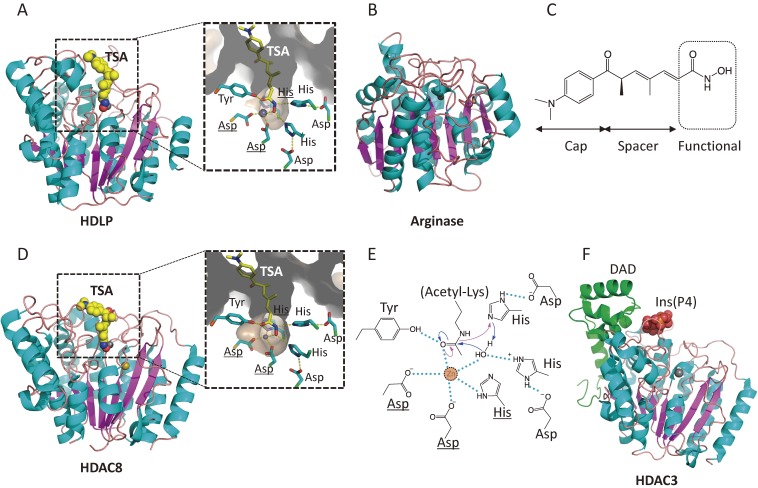
Figure 4.
Crystal structures and catalytic mechanisms of HDAC proteins. (A) HDLP (PDB id: 1C3R). TSA and zinc ion are represented as space-filled spheres, in which C, N, O, and Zn atoms are colored in yellow, blue, red, and grey, respectively. Close-up view of the active site is shown in the right panel. Two asparagine residues and histidine (underlined) coordinate the active site Zn2+ ion. (B) Arginase (1RLA). (C) The chemical structure of TSA, consisting of a cap, a spacer, and a zinc-binding functional group. (D) HDAC8 (1T64). K+ ion (Na+ ion in this structure due to the crystallization conditions) is colored in khaki. (E) Catalytic mechanism of class I HDACs. The catalytic histidine residue facilitates the nucleophilic attack at the substrate carbonyl by activating a water molecule (blue arrow). Hydrogen bond interactions are drawn in dotted lines. (F) HDAC3 in complex with inositol-tetraphosphate and DAD (4A69). The inositol phosphate is represented as space-filled spheres, in which C, O, and P atoms are depicted in yellow, red, and orange, respectively. The DAD is colored in green.
The HDLP catalytic core consists of a tubular pocket, a zinc-binding site, and active-site residues of a tyrosine and two histidines that make hydrogen bonds with two aspartic acids (Fig. 4A). Based on the structure, it was proposed that one of the two His-Asp charge relay systems facilitates serine protease-like nucleophilic attack at the substrate carbonyl by activating a water molecule coordinated with the zinc ion. Co-crystallization of HDLP with TSA demonstrated that TSA binds by inserting its long aliphatic chain into the tube-like pocket in HDLP and inhibits enzymatic activity by interacting with the zinc and active-site residues through its hydroxamic acid at one end of the aliphatic chain (Fig. 4A).60) Thus, TSA acts as a substrate mimetic. As a functional group of the inhibitor, the hydroxamic acid residue coordinates the zinc through its carbonyl and hydroxyl groups, resulting in the formation of a penta-coordinated zinc. Indeed, suberoylanilide hydroxamic acid (SAHA) (Fig. 2A), the first HDAC inhibitor that was approved as an anticancer drug by FDA, was also shown to coordinate the zinc with its hydroxamic acid residue.60) On the other hand, the aromatic group at the other end of TSA or SAHA has contact with the hydrophobic residues surrounding the entrance of the active site pocket, acting as a cap.60) Thus, the chemical structure consisting of a cap, spacer and a zinc-binding functional group, is the shared architecture of potent HDAC inhibitors (Fig. 4C).
Class I HDACs.
Subsequent studies by X-ray crystallography of mammalian HDAC8 showed essentially the same catalytic domain structure as HDLP (Fig. 4D).62,63) The residues constituting the active site pocket are highly conserved among the zinc-dependent HDAC enzymes. However, further crystallographic analysis of active site residue mutants of HDAC8 as well as molecular dynamic simulations, prompted a revised model for the catalytic mechanism anticipated by the HDLP structure.64,65) In the revised model, H143 serves as the catalytic base in the nucleophilic attack, which is a similar role for the catalytic histidine residue in conventional metalloproteases (Fig. 4E). The role of metal ions for HDAC8 was also structurally and biochemically analyzed, and not only zinc but also iron is suggested to act as a catalytic metal in both in vitro and in vivo reactions.66,67) Quantum mechanics/Molecular mechanics (QM/MM) molecular dynamics simulations suggest that the K+ ion, which was found about 7 Å away from the catalytic zinc ion in HDAC8, has a role in substrate binding and the basicity of the catalytic H143 residue.65)
Class I HDACs have been shown to form large complexes in cells. HDAC1 and HDAC2 exist together and constitute at least three distinct complexes called mammalian Sin3 complex (mSin3), nucleosome remodeling and deacetylating complex (NuRD) and co-repressor for element-1-silencing transcription factor (CoREST) (reviewed by Ayer, Trends Cell Biol, 199968)). HDAC3 is a catalytic subunit of silencing mediator for retinoid or thyroid-hormone receptors (SMRT) and nuclear receptor co-repressor (N-CoR) complexes, which were identified as nuclear hormone receptor co-repressors. These multiprotein complexes recruit class I HDACs to specific gene loci for transcriptional repression. In addition, complex formation enhances their enzymatic activity.29)
The mechanism underlying catalytic activation by co-repressor binding was investigated by crystal structure analysis of HDAC3 complexed with the deacetylase activation domain (DAD) from the human SMRT co-repressor. Surprisingly, there was inositol-tetraphosphate (Ins(1,4,5,6)P4) in the interface between the two proteins, acting as a molecular glue (Fig. 4F).69) Ins(1,4,5,6)P4 was also detected in the complex of HDAC1 with Metastasis-associated protein 1 (MTA1) from the NuRD complex, which may function to recruit nucleosomes to the active site of the enzyme.70) Further biochemical and structural studies on inositol-phosphates suggest that there is allosteric communication between the inositol-binding site and the active site, and that the role of inositol-phosphate in complex formation as a bona fide regulator of class I HDACs provides novel therapeutic opportunities to control gene expression.71)
Class II HDACs.
Class II HDACs are subdivided into class IIa and IIb depending on sequence similarity. The enzymatic activity of the catalytic domain of class IIa HDACs is weak, and its overall activity depends on SMRT or N-CoR multiprotein co-repressor complex containing HDAC3.72) The first structural information on the class II HDAC family was provided from the crystal structure of a bacterial homologue, FB188 histone deacetylase-like amidohydrolase (FB188 HDAH).73) FB188 HDAH has significant similarity to HDAC6, exhibiting 30% identity with the first catalytic domain of HDAC6 (CD1) and 35% identity with the second (CD2), suggesting that FB188 HDAH is a cognate enzyme with HDAC6. Following FB188 HDAH, structures of human class IIa enzymes such as HDAC7 and HDAC4 were solved.74,75) Although the active site structures of zinc-dependent HDACs are generally well conserved, the active site tyrosine residue in class IIa enzymes is replaced by a histidine residue. This tyrosine residue appears to be important for stabilizing the tetrahedral intermediate with the zinc ion. Therefore, the most likely reason for the weak catalytic activity of class IIa HDACs is the lack of the catalytic tyrosine residue. Indeed, the catalytic activity of HDAC4 was enhanced by the single His to Tyr substitution, reaching levels comparable to that of class I enzymes.76) The physiological role of this weak catalytic activity remains unknown. Furthermore, a novel zinc-binding domain unique to class IIa enzymes was found near the active site in the crystal structures, which results in modified catalytic properties, active site pocket enlargement, and association with HDAC3 and co-repressor complexes such as SMRT and N-CoR. It is likely that the activity of class IIa enzymes is regulated through the formation of multi-subunit complexes with other proteins, but further investigation is needed for elucidating the detailed mechanism.
Class III HDACs (Sirtuins).
Sirtuins are categorized into the deoxyhypusine synthase (DHS)-like NAD/FAD-binding domain superfamily of proteins. Structural studies of Sir2 homologues have shown the mechanism of NAD+-dependent catalysis. Sirtuin proteins consist of two domains, a large domain having a Rossmann-fold and a small zinc-binding domain.77–79) The conserved catalytic pocket resides in a groove between the two domains, forming a protein-tunnel in which the substrate interacts with NAD+ (Fig. 5A–C). The reaction mechanism for deacetylation includes nicotinamide cleavage and ADP-ribose transfer.80,81) After nucleophilic addition of the acetamide oxygen to the C1′ position of the nicotinamide ribose to form a C1′-O-alkylamidate intermediate and free nicotinamide, the 1′,2′-cyclic intermediate formation occurs, which is then attacked by an activated water molecule, resulting in the formation of deacetylated lysine and 2′-O-acetyl-ADP-ribose (Fig. 5D). 2′-O-acetyl-ADP-ribose can be readily converted to 3′-O-acetyl-ADP-ribose in aqueous solution by non-enzymatic intramolecular transesterification. Thus, nicotinamide, the deacetylated peptide and a mixture of 2′- and 3′-O-acetyl-ADP-ribose are the final reaction products.
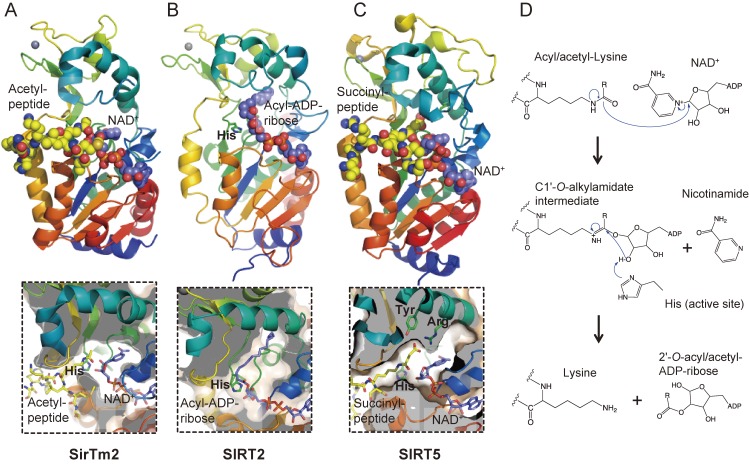
Figure 5.
Crystal structures and catalytic mechanism of sirtuin family proteins. (A) SirTm2 in complex with acetyl lysine peptide and NAD+ (PDB id: 2H4F), (B) SIRT2 in complex with acyl-ADP-ribose, an intermediate of the reaction between an acyl-peptide and NAD+ (4Y6Q), and (C) SIRT5 in complex with N-succinyl lysine peptide (3RIY). Active site histidine and substrates are represented as stick models, in which yellow, blue, red, and orange represent C, N, O, and P atoms, respectively. C atoms of NAD+ and acyl-ADP-ribose are in purple for clarity. Zn2+ ions are represented as space-filled spheres in grey. Close-up views of the active sites are shown in lower panels. (D) Catalytic mechanism of sirtuin family proteins. Acyl/acetyl-lysine residue is deacyl/deacetylated by nucleophilic addition of the acetamide oxygen to the C1′ position of the nicotinamide ribose. Nicotinamide and a deacylated/deacetylated peptide and a 2′-O-acyl/acetyl-ADP-ribose are final reaction products.
SIRT1 (also known as Sir2α) is the closest mammalian homologue of yeast Sir2. In human SIRT1, which is composed of 747 residues, the catalytic domain is located in the central region while the N- and C-terminal regions regulate catalytic activity.82) In particular, 25 residues at the C-terminal region (aa 631-655) are required for catalytic activity83) and compete with Deleted in bladder cancer protein 1 (DBC1), a negative regulator of SIRT1.84,85) Recently, the crystal structure of human SIRT1 in complex with the C-terminal region was determined.86) Without NAD+ and the substrate, SIRT1 adopts an open form, while in the presence of NAD+ it adopts a closed conformation with rotation of the small domain. The C-terminal region binds to the lower edge of the large domain to complement the Rossman fold, and functions allosterically to stabilize the catalytic core.
Small molecule inhibitors/activators
HDAC inhibitors.
n-Butyrate was the first chemical tool to inhibit cellular HDAC activity, but required a high concentration for HDAC inhibition.19,20) Subsequent discovery of natural products that specifically inhibit HDAC activity has greatly contributed to the elucidation of structures and functions of HDACs and their role in gene expression and diseases. TSA and TPX are structurally unrelated microbial metabolites that were originally isolated as antifungal antibiotics, suggesting that HDACs are potential targets for antibiotics. Indeed, chlamydocin (Fig. 2A), HC-toxin (Fig. 2A), and a number of other cyclic tetrapeptides with an epoxyketone have been identified as cytotoxic compounds from natural sources and were later shown to have anti-HDAC activity.11,87) Apiciidin (Fig. 2A), which had been identified as a potent, broad-spectrum antiprotozoal agent containing an ethyl ketone, was also shown to be an HDAC inhibitor.88) We hypothesized that the hydroxamic acid of TSA and the electrophilic ketone group of TPX-related cyclic tetrapeptides are common features that inhibit HDAC activity, and designed a series of novel cyclic tetrapeptides containing the hydroxamic acid moiety instead of epoxy ketones. These synthetic hybrid compounds turned out to be very potent inhibitors with increased stability.87,89) The potential of HDACs as molecular targets for cancer therapeutics was widely studied using variety of natural and synthetic HDAC inhibitors and genetic mutations, and class I HDACs might be promising targets since it was shown that class I HDACs act as repressors of cyclin-dependent kinase (CDK) inhibitors, differentiation factors, and proapoptotic factors.90)
In 1998, HDAC inhibition by two clinically important compounds, SAHA (vorinostat) (Fig. 2A) and FK228 (romidepsin) (Fig. 2A), was reported.91,92) SAHA was originally synthesized starting from hybrid polar compounds such as hexamethylenebisacetamide and reported as a potent inducer of murine erythroleukemia cell differentiation.93) Richon et al. found the HDAC inhibitory activity of SAHA according to its structural and biological similarity to TSA. FK228, an antitumor cyclic depsipeptide isolated from Chromobacterium violaceum, was also shown to inhibit HDACs by our group.92) Phase I clinical trials conducted by Bates et al. in the National Cancer Institute revealed that FK228 is effective against cutaneous T-cell lymphoma (CTCL).94) Because no effective drug for CTCL therapy was available, the development of HDAC inhibitors as therapeutic drugs was accelerated. SAHA was the first HDAC inhibitor approved for cancer chemotherapy in 2006.95) Following SAHA, FK228 was also approved in 2009. FK228 has an intramolecular disulfide bond, which is reduced in cells to form a thiol group that can interact with the active-site zinc ion.96) Thus, FK228 acts as a prodrug, which can be activated in cancer cells (Fig. 6). Largazole (Fig. 2A), another natural product that contains a thioester bond, is also activated by hydrolysis in cells.97) Another class of clinically important HDAC inhibitors is benzamides, including entinostat/SNDX-275/MS-275 (Fig. 2A). MS-275 is the first synthetic benzamide compound with HDAC inhibitory activity.98) Molecular interaction between the zinc ion in the active site and the functional groups of inhibitors including not only hydroxamic acids, but also benzamides and thiols, was confirmed by X-ray co-crystallography.99,100) Thus, a large number of structurally diverse HDAC inhibitors have been discovered from natural sources or synthetically developed, many of which are being studied clinically.101) Recently, belinostat/PXD101 (Fig. 2A) and panobinostat/LBH589 (Fig. 2A) were also approved for treatment of T-cell lymphoma and multiple myeloma, respectively. In the case of panobinostat/LBH589, treatment of multiple myeloma in combination with bortezomib (velcade), a proteasome inhibitor, is recommended because of the synergistic cytotoxicity to multiple myeloma due to the simultaneous inhibition of HDAC6-dependent aggresome formation followed by autophagy-mediated degradation and proteasomal degradation of misfolded proteins.102) More than 20 compounds are currently undergoing clinical trials: mocetinostat/MGCD0103, abexinostat/PCI-24781, entinostat/SNDX-275/MS-275, SB939, CS055/HBI-8000, resminostat/4SC-201/BYK408740, givinostat/ITF2357, quisinostat/JNJ-26481585, CHR-2845, CHR-3996, CUDC-101, AR-42, DAC-060, EVP-0334, MGCD-290, CXD-101/AZD-9468, CG200745, arginine butyrate, 4SC-202, rocilinostat/ACY-1215 and SHP-141 (Fig. 2A).103,104)
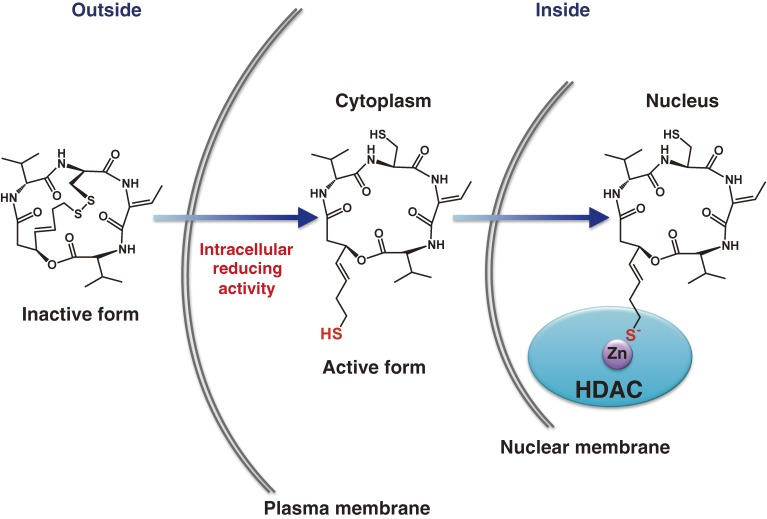
Figure 6.
A unique mode of action of FK228. FK228 is activated by cellular reducing activity in cells, forming thiol groups from its intramolecular disulfide bond. One the thiols may act as a zinc-binding ligand to inhibit HDAC.
The mode of action of some of these inhibitors has been elucidated by structural analysis of co-crystals. The first generation of HDAC inhibitors such as TSA and SAHA inhibit essentially all zinc-dependent HDACs including HDAC6. In contrast, a subset of HDAC inhibitors such as n-butyrate, TPX, FK228 and MS-275 cannot inhibit HDAC6, likely due to the difference in the structure of the active site pocket. Consistent with this, a bacterial HDAC6 homologue, FB188 HDAH, with significant difference in the area around the active site entrance, showed an inhibitor sensitivity spectrum similar to HDAC6. Tubacin, an HDAC6 specific inhibitor, strongly inhibited the enzymatic activity of FB188, while chlamydocin and HC-toxin, cyclic tetrapeptide inhibitors related to TPX, failed to inhibit FB188.105) Based on the flexible structure of the surface around the active site, the HDAC8-specific inhibitor, PCI-34051 (Fig. 2A), was synthesized and induced apoptosis in T-cell lymphoma without increasing histone and α-tubulin acetylation.106) Using structural information about differences between the shapes of the HDAC1 and HDAC3 internal cavities, novel biaryl derivatives of benzamide inhibitors were designed, which showed selective HDAC1/HDAC2 inhibitory activity.107)
Despite a large number of reports on anticancer activity of HDAC inhibitors, the precise mechanism by which HDAC inhibitors exert therapeutic effects still remains ambiguous. Aberrant histone acetylation is associated with dysregulation of gene expression in cancer. Indeed, loss-of-function mutations or translocations of CBP, p300, and MOZ genes encoding HAT enzymes have been found in some cancers. Individuals with the Rubinstein-Taybi syndrome, who carry a loss-of-function mutation in one of the alleles of CBP, have a high risk of cancer.108) In some cases of acute myeloid leukemia, fusion of the MOZ and p300 genes has been found.109) Altered expression of HDAC genes have also been reported in various types of cancers, but does not always correlate with malignancy. Mutations in HDAC genes are rarely found in cancer.110) In the case of HDACs, aberrant activities of HDACs are more likely to link to oncogenic events rather than mutations or aberrant gene expression. It is reported that aberrant transcriptional repression through the recruitment of HDACs by oncogenic translocation products such as PML-RARα is associated with specific forms of leukemia and lymphoma.111) HDAC inhibitor treatment induces hyperacetylation of histones, resulting in altered expression of specific genes, including tumor suppressors, which were epigenetically repressed in cancer cells. As a result, multiple events are induced in cancer cells, including cell cycle arrest, apoptosis, senescence, differentiation, immunogenicity, and inhibition of angiogenesis.112) HDAC inhibitors arrest the cell cycle of normal cells in G1 and G2 phases without severe cytotoxicity113) by increasing the expression of p21 cell cycle inhibitor protein.114,115) In contrast, apoptotic cell death was induced in cancer cells by HDAC inhibitors through the increased expression of proapoptotic factors and the reduced expression of antiapoptotic factors.116) In addition to histones, HDAC inhibitors appear to exhibit antitumor activities through hyperacetylation of non-histone proteins including transcriptional factors and chaperon proteins.117) Increased acetylation of transcriptional factors with tumor suppressor function such as p53 by HDAC inhibitors enhances their transcriptional activity, which alters target gene expression leading to tumor suppression. HDAC inhibitors also induce hyperacetylation of HSP90, which inhibits its chaperon activity, causing destabilization of a number of client proteins including oncoproteins.118)
Sirtuin inhibitors.
The deacetylase activity of sirtuins is insensitive to TSA, due to difference in their catalytic mechanisms and structures. Sirtuin reactions require hydrolysis of NAD+, and its byproduct nicotinamide is an intrinsic inhibitor. Carba-NAD+ (Fig. 2B), a non-hydrolyzable NAD+ analogue, is a competitive inhibitor of sirtuins. Several acetyl-lysine analogues have also been shown to inhibit sirtuins as mechanism-based inhibitors. For instance, thioacetyl-peptide acts as a tight-binding inhibitor that blocks reaction of the ADP-ribose-peptide enzyme intermediate by forming 1′-S-alkylamidate.119) Inhibitors that form complexes with sirtuins have been useful for understanding the mechanisms of enzymatic reactions.
Involvement of sirtuins in various human diseases such as leukemia and Parkinson’s disease has been reported and prompted many groups to discover pharmacological inhibitors of sirtuins. A class of small molecule Sir2 inhibitors was discovered by a high throughput, phenotypic screen in yeast cells. Sirtinol (Fig. 2B) and splitomicin (Fig. 2B) were identified through a screen based on telomere silencing in yeast (e.g., Bedalov et al., 2001120)). Following these, a number of compounds including cambinol, salermide, tenovin, EX-527, AGK2, etc. (Fig. 2B) have been reported as sirtuin inhibitors. Sirtuin inhibitors may be roughly divided into two groups, those interacting with the NAD+ (nicotinamide and ADP-ribose) binding site and those interacting with the acetyl-lysine binding site. These inhibitors have been applied not only to investigate the role of sirtuins in diverse biological processes but also to validate sirtuins as therapeutic targets. Using a selective inhibitor for instance, SIRT2 emerged as a potential therapeutic target for Parkinson’s disease, since AGK2 rescues neuronal cell death in models of Parkinson’s disease.121) Recently, Sirtuin-rearranging ligand 2 (SirReal2) (Fig. 2B) was reported as a specific small molecule inhibitor of SIRT2. Binding of SirReal2 elicited a drastic conformational rearrangement to generate a large hydrophobic cavity to accept the inhibitor.122)
Sirtuin activators.
It has been established that caloric restriction can induce lifespan extension in a wide range of organisms from yeasts to primates. Caloric restriction upregulates cellular NAD+ levels, suggesting that sirtuins are activated under calorie restriction conditions. Following the first observation that Sir2 and NAD+ are required for lifespan extension in the budding yeast Saccharomyces cerevisiae in 2000.123) SIR2 has been one of the candidate genes involved in the regulation of aging through energy metabolism in metazoans.124) SIRT1 is the mammalian Sir2 homologue with the highest sequence similarity, and is reported to regulate aging in mice. However, the role of sirtuins in the lifespan extension has been controversial. The longevity of Caenorhabditis elegans and Drosophila melanogaster induced by Sir2 overexpression in early observations was re-investigated and found to be independent of Sir2.125) In contrast, overexpression of SIRT6 but not SIRT1 caused a significantly longer lifespan than wild-type in male but not female mice.126)
In 2003, resveratrol (Fig. 2C) and related polyphenols were identified as sirtuin activators, which mimic caloric restriction and delay the aging process.127) Since resveratrol and related compounds in grapes have been postulated to reduce the risk of ischemic cardiac disease, sirtuins became regarded as one of the key regulators for human health span. Furthermore, synthetic small molecule activators of SIRT1 (STACs) (Fig. 2C), chemically unrelated to resveratrol, showed pharmacological activity to protect mice from metabolic disorders such as diet-induced obesity, type 2 diabetes, or nonalcoholic fatty liver disease.128) However, the validity of resveratrol and STACs in directly activating SIRT1 has been widely debated because these small molecules increased enzymatic activity only when a fluorescently labeled peptide substrate was used.129,130) On the other hand, another group demonstrated STAC-mediated allosteric activation of SIRT1 towards a subset of specific substrates. Indeed, STACs significantly upregulated SIRT1 enzyme activity to deacetylate specific substrates containing hydrophobic residue(s) at subsite +1 or +6, such as in PGC-1α and FOXO3a.131) Furthermore, a single point mutation at E230 was identified to attenuate STAC-mediated activation of SIRT1, suggesting a direct effect on SIRT1.132) Recently, an X-ray crystallographic structure of mini-hSIRT1, a minimal structure enabling STAC-mediated activation, was analyzed in complex with STACs. The STAC-binding domain was located in the N-terminal region apart from the active site, but could intramolecularly interact with the active site by electrostatic interaction between E230 and R446, supporting the hypothesis of a direct activation mechanism by small molecules.133)
However, it is still possible that resveratrol action depends on other cellular pathways, as resveratrol shows pleiotropic actions including antioxidant, AMP kinase activation and phosphodiesterase 4 (PDE4) inhibition.134,135) Recent studies demonstrated that another PDE4 inhibitor reproduced most of the resveratrol actions including prevention of diet-induced obesity and an increase in mitochondrial function, physical stamina and glucose tolerance in mice. Furthermore, resveratrol also strongly binds to the active site of human tyrosyl tRNA synthetase (TyrRS), which in turn activates poly(ADP-ribose) polymerase 1 (PARP1) by stimulating auto-ADP-ribosylation, thereby activating stress tolerance.136)
Non-histone substrates
Although acetylation of some non-histone proteins including ribosomal proteins and α-tubulin had been reported more than 30 years ago,137,138) their functional relevance was unknown. In 1997, p53, a tumor suppressor protein, was reported to be acetylated: p300 catalyzes acetylation, which enhanced the transcriptional activity of p53 by conformational change.139) PCAF was also reported to acetylate p53,140) whereas HDAC1, HDAC2, HDAC3, and SIRT1 were involved in deacetylation of p53.141,142) It was shown that acetylation promotes the DNA binding activity of p53. In vivo sites of acetylation by p300 and PCAF are targets for DNA damage-induced acetylation, indicating that acetylation of p53 mediates response to cellular DNA damage. Importantly, some of these acetylation sites overlap with those of ubiquitination sites, suggesting that acetylation regulates both the activity and stability of p53.143) Immediately after the discovery of p53 acetylation, acetylation of many other proteins (mostly nuclear proteins such as transcription factors) was reported including Ku70, nuclear factor κB (NF-κB) subunit RelA, and protooncogene protein c-Myc, most of which have been determined to be substrates of class I and III enzymes.117) SIRT1 was also reported to deacetylate p53,142) NF-κB,144) FOXO145) and PPARγ,146) in addition to core histones. These non-histone protein substrates of SIRT1 are important transcriptional regulators involved in aging and metabolism, and their activity is regulated by SIRT1.
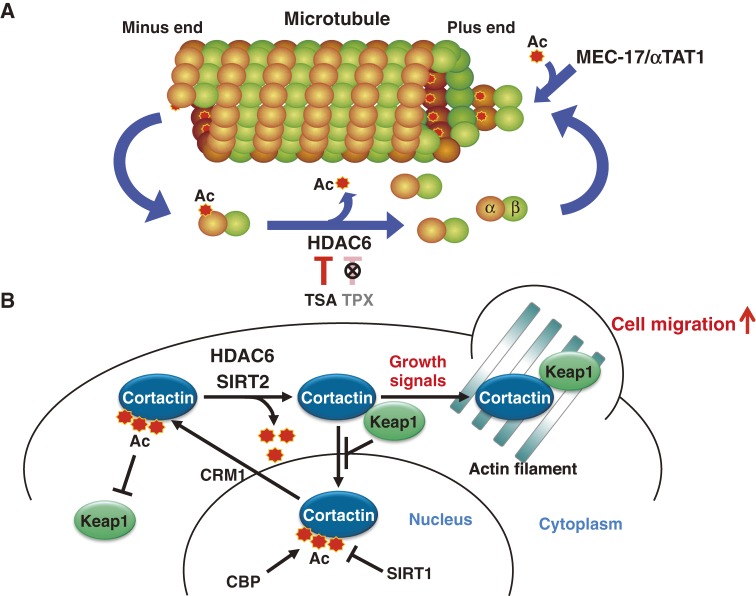
In addition to nuclear acetylated proteins regulated by conventional HDACs, a number of cytoplasmic proteins are also targets of acetylation. HDAC6 is a unique enzyme that is specifically localized in the cytoplasm.147) We found that TPX A and B cannot inhibit HDAC6, whereas TSA inhibits all HDACs including HDAC6.87) This differential target enzyme specificity provided an experimental setting for determining the substrate of HDAC6. As a result of screening for proteins whose acetylation is increased by TSA but not TPX B, we identified α-tubulin as a specific substrate for HDAC6 (Fig. 7A).148,149) Acetylation of the ε-amino group of K40 on α-tubulin has long been known, but the enzymes responsible for acetylation and deacetylation remained elusive. Thus, HDAC6 was the long-awaited deacetylase for α-tubulin. In addition, SIRT2 was found to co-localize with HDAC6 and microtubules, and was shown to be another deacetylase of α-tubulin.150) Hyperacetylation of α-tubulin by HDAC inhibitors or HDAC6 knockdown increased the stability of the dynamic pool of microtubules. It was also reported that HDAC6 is involved in recruiting ubiquitinated unfolded proteins to dynein on microtubules through its ubiquitin-binding domain, thereby facilitating aggresome formation and subsequent clearance by autophagic degradation.151,152) In 2010, MEC-17 (also known as αTAT1), a protein related to the Gcn5 histone acetyltransferases and required for the function of touch receptor neurons in Caenorhabditis elegans, was identified as an α-tubulin acetyltransferase (Fig. 7A).153,154)
Figure 7.
Regulation of cytoplasmic proteins by reversible acetylation. (A) Identification of HDAC6 as a tubulin deacetylase. By using the difference in target enzyme specificity between TSA and TPX, HDAC6 was identified as a tubulin deacetylase. TSA treatment increases α-tubulin acetylation, while TPX, which cannot inhibit HDAC6, fails to increase the α-tubulin acetylation. (B) Regulation of cell motility by cortactin acetylation and interaction with Keap1. Keap1, known as a negative regulator of the transcription factor Nrf2, was identified as a novel partner of cortactin, which regulates nuclear-cytoplasmic transport of cortactin. In addition, cortactin interaction with Keap1 is required for cortical localization of cortactin. Acetylation of cortactin inhibits the interaction with Keap1, thereby controlling cell migration. Cortactin is acetylated by acetyltransferase activity of the nuclear CBP, which is also by regulated by deacetylase activity of SIRT1 in the nucleus. Upon growth stimulation, cortactin is deacetylated by two cytoplasmic deacetylases, HDAC6 and SIRT2, which promotes cortical translocation and cell migration.
HDAC6 was also shown to deacetylate several cytoplasmic proteins such as Hsp90 and cortactin.118,155) Hyperacetylation of Hsp90 by the loss of HDAC6 activity resulted in the inhibition of its chaperone activity, thereby suppressing Hsp90-mediated protein stability and growth signaling pathways.118) Cortactin (cortical actin binding protein) is an actin-binding protein promoting polymerization and rearrangement of the actin cytoskeleton, which has emerged as a key factor in aggressive cancers.156) We found that cortactin shuttles between the cytoplasm and the nucleus in a manner dependent on chromosome region maintenance 1 (CRM1), a general nuclear export factor. We also identified Kelch-like ECH-associated protein 1 (Keap1) as a novel binding partner of cortactin, responsible for tethering cortactin in the cytoplasm, promoting cortical localization of cortactin and subsequent actin remodeling in response to growth signaling (Fig. 7B). Acetylation occurs on conserved lysine residues in the repeat domain required for the interaction with Keap1. Therefore, cortactin acetylation inhibits binding to Keap1, leading to inhibition of cortical translocation of cortactin necessary for cancer cell motility and invasion (Fig. 7B).155,157) Importantly, both HDAC6 and SIRT2 were involved in deacetylation of cortactin, as they are with α-tubulin, suggesting that these two enzymes share part of a physiological function and form a common complex. Inhibition of HDAC6 by genetic or pharmacological means reduces cell migration,148,158) suggesting that HDAC6 and SIRT2 are attractive targets for anti-metastasis therapy.
The functional consequences of protein acetylation are diverse and include changes in activity, subcellular localization, and stability. These changes are attributed at least in part, to inhibition of nuclear import or export, regulation of protein-protein interactions and competition with other protein lysine post-translational modifications such as ubiquitination. Recent technological advancements in high-resolution mass spectrometry have allowed for comprehensive analysis of cellular acetylated proteins, called the “acetylome”. Using stable isotope labeling by amino acids in cell culture (SILAC), more than 3,600 acetylation sites of 1,750 proteins were determined from cells treated with HDAC inhibitors.159) Gene ontology analysis demonstrates that proteins regulated by acetylation are widespread, e.g., many proteins that control the cell cycle, RNA splicing, and ribosome are acetylated, in addition to histones and factors involved in chromatin remodeling. For instance, acetylation of SMC3 in S-phase, one of the core proteins of the cohesin complex, is important for sister chromatid cohesion.160) HDAC8 was identified as the enzyme required for deacetylation of SMC3, and loss of HDAC8 activity by genetic mutation increases SMC3 acetylation and causes Cornelia de Lange syndrome.161)
In mitochondria, which are postulated to be derived form symbiotic bacteria, many proteins are also subject to acetylation. SIRT3 knockout mice for instance, exhibited striking mitochondrial protein hyperacetylation, indicating that SIRT3 is a major mitochondrial deacetylase.162) Like other sirtuins, SIRT3 requires NAD+ for its enzymatic activity, suggesting a role of SIRT3 as a metabolic sensor in mitochondria. The first identified substrate of SIRT3 was acetyl-CoA synthase 2 (AceCS2), which is involved in utilization of acetate under fasting conditions.163,164) The enzymatic activity of AceCS2 was completely inhibited by acetylation, while it was reactivated by SIRT3-mediated deacetylation. Long-chain acyl-CoA dehydrogenase (LCAD) is also regulated by reversible acetylation. Deacetylation of LCAD by SIRT3 activates its catalytic activity, facilitating mitochondrial fatty acid oxidation in response to fasting.162) Advanced acetylome analysis also revealed a large number of acetylated proteins in bacteria that do not have histones.165)
Defatty-acylation: Novel functions of HDACs
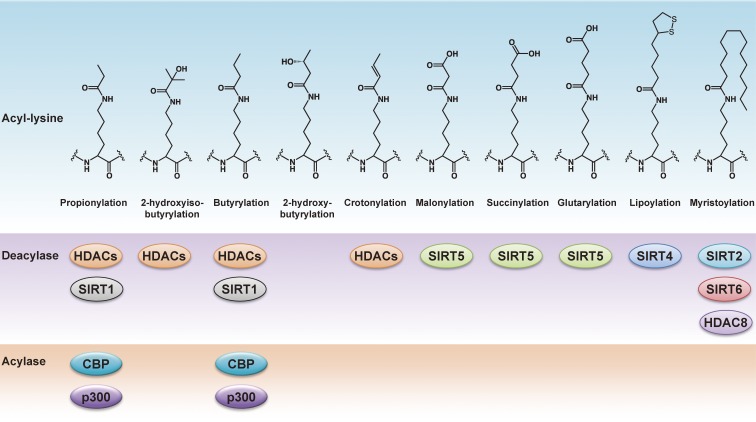
Protein lysine residues are subject to a variety of posttranslational modifications. Recent advancements in mass spectrometry have enabled the finding of new acyl-modifications on lysine residues such as: propionylation, butyrylation, succinylation, malonylation, crotonylation and glutarylation (Fig. 8). In addition to acetylation, HDACs are responsible for removal of these short-chain acyl modifications. In mammalian histones, propionylation, butyrylation, succinylation, malonylation, crotonylation and 2-hydroxyisobutyrylation are also recognized lysine acyl-modifications.8,166–168) Multiple acyl-modifications often target the same positions, suggesting the presence of differential regulation. It is suggested by mutational analysis in budding yeast that succinylation of histone H4K77 has a role in negatively regulating gene silencing.167) Histone crotonylation is enriched in active promoters and predicted enhancer regions, and is suggested to function as a specific mark of active sex chromosome-linked genes that escape sex chromosome inactivation after completion of meiosis during spermatogenesis.166) Histone 2-hydroxyisobutyrylation shows distinct genomic distributions from acetylation or crotonylation during male germ cell differentiation. 2-hydroxyisobutyrylation of histone H4K8 is associated with active gene transcription in meiotic and post-meiotic cells.168) On the other hand, the roles of histone propionylation, butyrylation, and malonylation in epigenetic gene regulation remain to be elucidated. HDAC1, 2, and 3 are suggested to be responsible for decrotonylation.166) Cyclic-AMP-response-element-binding protein (CREB) binding protein (CBP) and p300 were shown to be responsible for propionylation and butyrylation in addition to acetylation.8)
Figure 8.
Summary of lysine N-acyl modifications and related enzymes.
Emerging evidence demonstrates that sirtuins have general functions not only as deacetylases but also deacylases capable of acting on several acylated substrates. It is reported that the bacterial sirtuin CobB, which is the only known deacetylase in E. coli, has both deacetylase and desuccinylase activities.169) Recent in vitro studies have further expanded the known enzymatic activities of mammalian sirtuins (Fig. 8). SIRT1 was identified as the first enzyme to depropionylate and debutyrylate.9) The catalytic mechanism for depropionylation was investigated with Sir2Tm from Thermotoga maritimea in complex with a propionylated p53 peptide. The crystal structure of Sir2Tm showed that the binding of propionylated peptide induces structural rearrangements to accommodate the bulkier acyl group in the active site compared to the acetylated peptide-bound structure.170) In addition, SIRT1 and SIRT2 act as efficient histone decrotonylases,171) and SIRT1, SIRT2, and SIRT3 can remove several long-chain fatty acyl groups from lysine.172) We have analyzed the X-ray crystal structures of SIRT2 co-crystalized with myristoylated peptides and found that the myristoylated lysine residue was accommodated in a hydrophobic cavity, which was formed upon substrate binding. Interestingly, simultaneous incubation with NAD+ resulted in the detection of O-myristoylated ADP-ribose, an intermediate in the transition state, in the crystals (Fig. 5B).173) The mitochondrial SIRT5 displays very weak deacetylase activity, but has potent desuccinylase and demalonylase activities, and was also recently found to have deglutarylase activity.174–177) Some lysine sites for acetylation, succinylation, malonylation, and glutarylation are overlapped.177) A crystal structure in complex with a succinate peptide showed that SIRT5 has a unique acyl pocket in which the carboxylate of succinate interacts with the tyrosine and arginine residues (Fig. 5C).175) SIRT5, together with the major mitochondrial deacetylase SIRT3, modulates key metabolic enzymes and functions as a metabolic sensor in response to nutrient availability or the energy status of the cell. SIRT6 efficiently removes long-chain fatty acyl modifications, such as myristoyl, from lysine residues. SIRT6 promotes the secretion of tumor necrosis factor-α (TNF-α) by removing the fatty acyl group on K19 and K20 of TNF-α. The crystal structure of SIRT6 revealed a large hydrophobic pocket that can accommodate long-chain fatty acyl groups.178) SIRT6 also undergoes auto-ribosylation, which might contribute to the self-regulation of catalytic functions.45) Though the mitochondrial SIRT4 has no HDAC activity but weak ADP-ribosyltransferase activity, the loss of SIRT4 affects glutamate dehydrogenase activity and insulin secretion, suggesting its physiological significance.57,179) Recently, SIRT4 was shown to exhibit lipoamidase activity, which removes the lipoyl group from the pyruvate dehydrogenase complex responsible for generation of acetyl-CoA in mitochondria.180) Most recently, it was reported that HDAC8, a zinc-dependent class I HDAC, has defatty-acylation activity from octanoyl, dodecanoyl, and myristoyl lysine residues.181) These observations suggest that both NAD+- and zinc-dependent HDACs have an evolutionary role in removing a variety of acyl groups ranging from acetyl to long-chain fatty acyl groups on protein lysine residues that often occur in the presence of acyl-CoA.
In contrast to the discovery of enzymes responsible for the removal of a variety of acyl groups from protein lysine substrates, apparently no enzyme for acyl transfer to lysine residues has been reported, except histone acetyltransferase CBP/p300, which is also responsible for histone propionylation and butyrylation.8) In bacteria and mitochondria, non-enzymatic protein acylation may occur by using reactive thioester or acyl phosphate compounds such as acetyl-CoA, succinyl-CoA and acetyl phosphate.182,183) The microenvironment of the mitochondrial matrix with an alkaline pH and abundant acyl-CoA may also facilitate non-enzymatic protein acylation.182) Such uncontrolled protein acylation could interfere with protein function and thus is proposed to be a form of “carbon stress”.184) In this context, lysine deacylases may function to maintain protein quality by removing acyl groups. It is also hypothesized that bacterial or mitochondrial acylation acts as fat storage reflecting metabolic memory.
Conclusion
Over four decades, the fundamental roles of histone acetylation in gene expression and epigenetics have been elucidated thanks to extensive studies at the molecular, cellular, and animal levels. In the first half of this history, however, there were no chemical and molecular tools, which made it difficult to find convincing evidence for the causal relationship between histone acetylation and gene expression. The key discovery leading to identification of deacetylase enzymes and development of novel therapeutic opportunities was achieved from microbial screening aimed at anticancer drug discovery in the 1990s. The identified natural products including TSA, TPX, and FK228 paved the way to molecular studies on HDACs by serving as definitive chemical tools for biochemistry, molecular cloning, and cancer therapy, respectively. This is a typical example indicative of the power of chemical biology for unraveling biologically complex phenomena. Recent structural, chemical, and molecular biological studies of HDACs revealed new roles of this enzyme family including regulation of metabolism, aging, and homeostasis, leading to the next phase of HDAC research. Indeed, discovery of the enzymatic activity of HDACs to remove a variety of acyl groups from protein lysine residues shed new light on the functional origin and evolution of the enzyme family. Identification of endogenous regulatory molecules and development of isoform specific small molecule inhibitors will further accelerate the progress of HDAC research for understanding the bona fide functions conserved from bacteria to humans.
Acknowledgments
We thank many collaborators for their tremendous works on the HDAC inhibitors described in this review. We also thank Elliot Bradshaw and Mayumi Arata for critical reading of the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (M.Y.).
Profile
Minoru Yoshida was born in Tokyo in 1957, and brought up in Aichi Prefecture. He received his Ph.D. degree in 1986 from the University of Tokyo, where he started mode of action studies of natural products. He identified histone deacetylase and CRM1 exportin 1 as the specific targets of trichostatin A and leptomycin B, respectively, which greatly contributed to the field of epigenetics and nuclear transport. After he was promoted to Associate Professor in 1995 in the Department of Biotechnology at the University of Tokyo, he moved to RIKEN and started the Chemical Genetics Laboratory as Chief Scientist in 2002. In 2013, he was also appointed as Group Director, Chemical Genomics Research Group, RIKEN Center for Sustainable Resource Science. Currently, he has joint appointments in the Department of Biotechnology at the University of Tokyo and Graduate School of Science and Engineering at Saitama University. At RIKEN, he identified small molecules with unique targets, including spliceostatin, an inhibitor of pre-mRNA splicing, and theonellamide, which binds sterols to cause phase separation of lipid membranes. His major interest is the interface between chemistry and biology. He received many awards and commendations including The Sumiki-Umezawa Memorial Award, Prizes for Science and Technology, the Commendation for Science and Technology by the Minister of MEXT, Japan, The Japan Society for Bioscience, Biotechnology, and Agrochemistry Award, Charles E. Dohme Memorial Lectureship Award, the Japan Academy Prize, and the Princess Takamatsu Cancer Research Fund Prize.
References
- 1).Turner B.M. (2000) Histone acetylation and an epigenetic code. BioEssays 22, 836–845. [DOI] [PubMed] [Google Scholar]
- 2).Jenuwein T., Allis C.D. (2001) Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 3).Allfrey V.G., Faulkner R., Mirsky A.E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Wade P.A., Pruss D., Wolffe A.P. (1997) Histone acetylation: chromatin in action. Trends Biochem. Sci. 22, 128–132. [DOI] [PubMed] [Google Scholar]
- 5).Khochbin S., Verdel A., Lemercier C., Seigneurin-Berny D. (2001) Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 11, 162–166. [DOI] [PubMed] [Google Scholar]
- 6).Jones J.D., O’Connor C.D. (2011) Protein acetylation in prokaryotes. Proteomics 11, 3012–3022. [DOI] [PubMed] [Google Scholar]
- 7).Allis C.D., Berger S.L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., Shilatifard A., Workman J., Zhang Y. (2007) New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636. [DOI] [PubMed] [Google Scholar]
- 8).Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S.C., Falck J.R., Peng J., Gu W., Zhao Y. (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Cheng Z., Tang Y., Chen Y., Kim S., Liu H., Li S.S., Gu W., Zhao Y. (2009) Molecular characterization of propionyllysines in non-histone proteins. Mol. Cell. Proteomics 8, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Albaugh B.N., Arnold K.M., Denu J.M. (2011) KAT(ching) metabolism by the tail: insight into the links between lysine acetyltransferases and metabolism. ChemBioChem 12, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Yoshida M., Horinouchi S., Beppu T. (1995) Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays 17, 423–430. [DOI] [PubMed] [Google Scholar]
- 12).Yoshida M., Matsuyama A., Komatsu Y., Nishino N. (2003) From discovery to the coming generation of histone deacetylase inhibitors. Curr. Med. Chem. 10, 2351–2358. [DOI] [PubMed] [Google Scholar]
- 13).Yoshida M., Horinouchi S. (1999) Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export. Ann. N. Y. Acad. Sci. 886, 23–36. [DOI] [PubMed] [Google Scholar]
- 14).Marks P.A., Richon V.M., Rifkind R.A. (2000) Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J. Natl. Cancer Inst. 92, 1210–1216. [DOI] [PubMed] [Google Scholar]
- 15).Kazantsev A.G., Thompson L.M. (2008) Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug Discov. 7, 854–868. [DOI] [PubMed] [Google Scholar]
- 16).Marmorstein R. (2001) Structure of histone deacetylases: insights into substrate recognition and catalysis. Structure 9, 1127–1133. [DOI] [PubMed] [Google Scholar]
- 17).Inoue A., Fujimoto D. (1969) Enzymatic deacetylation of histone. Biochem. Biophys. Res. Commun. 36, 146–150. [DOI] [PubMed] [Google Scholar]
- 18).Riggs M.G., Whittaker R.G., Neumann J.R., Ingram V.M. (1977) n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 268, 462–464. [DOI] [PubMed] [Google Scholar]
- 19).Candido E.P., Reeves R., Davie J.R. (1978) Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 14, 105–113. [DOI] [PubMed] [Google Scholar]
- 20).Vidali G., Boffa L.C., Bradbury E.M., Allfrey V.G. (1978) Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 75, 2239–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Rubenstein P., Sealy L., Marshall S., Chalkley R. (1979) Cellular protein synthesis and inhibition of cell division are independent of butyrate-induced histone hyperacetylation. Nature 280, 692–693. [DOI] [PubMed] [Google Scholar]
- 22).Yoshida M., Kijima M., Akita M., Beppu T. (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17174–17179. [PubMed] [Google Scholar]
- 23).Tsuji N., Kobayashi M., Nagashima K., Wakisaka Y., Koizumi K. (1976) A new antifungal antibiotic, trichostatin. J. Antibiot. 29, 1–6. [DOI] [PubMed] [Google Scholar]
- 24).Yoshida M., Iwamoto Y., Uozumi T., Beppu T. (1985) Trichostatin C, a new inducer of differentiation of Friend leukemic cells. Agric. Biol. Chem. 49, 563–565. [Google Scholar]
- 25).Yoshida M., Nomura S., Beppu T. (1987) Effects of trichostatins on differentiation of murine erythroleukemia cells. Cancer Res. 47, 3688–3691. [PubMed] [Google Scholar]
- 26).Itazaki H., Nagashima K., Sugita K., Yoshida H., Kawamura Y., Yasuda Y., Matsumoto K., Ishii K., Uotani N., Nakai H., Terui A., Yoshimatsu S., Ikenishi Y., Nakagawa Y. (1990) Isolation and structural elucidation of new cyclotetrapeptides, trapoxins A and B, having detransformation activities as antitumor agents. J. Antibiot. 43, 1524–1532. [DOI] [PubMed] [Google Scholar]
- 27).Kijima M., Yoshida M., Sugita K., Horinouchi S., Beppu T. (1993) Trapoxin, an antitumor cyclic tetrapeptide, is an irreversible inhibitor of mammalian histone deacetylase. J. Biol. Chem. 268, 22429–22435. [PubMed] [Google Scholar]
- 28).Taunton J., Hassig C.A., Schreiber S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411. [DOI] [PubMed] [Google Scholar]
- 29).Seto E., Yoshida M. (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6, a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Lagger G., O’Carroll D., Rembold M., Khier H., Tischler J., Weitzer G., Schuettengruber B., Hauser C., Brunmeir R., Jenuwein T., Seiser C. (2002) Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 21, 2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Montgomery R.L., Davis C.A., Potthoff M.J., Haberland M., Fielitz J., Qi X., Hill J.A., Richardson J.A., Olson E.N. (2007) Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 21, 1790–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Montgomery R.L., Potthoff M.J., Haberland M., Qi X., Matsuzaki S., Humphries K.M., Richardson J.A., Bassel-Duby R., Olson E.N. (2008) Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J. Clin. Invest. 118, 3588–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Vega R.B., Matsuda K., Oh J., Barbosa A.C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J.M., Richardson J.A., Karsenty G., Olson E.N. (2004) Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119, 555–566. [DOI] [PubMed] [Google Scholar]
- 34).Chang S., McKinsey T.A., Zhang C.L., Richardson J.A., Hill J.A., Olson E.N. (2004) Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24, 8467–8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Zhang Y., Kwon S., Yamaguchi T., Cubizolles F., Rousseaux S., Kneissel M., Cao C., Li N., Cheng H.L., Chua K., Lombard D., Mizeracki A., Matthias G., Alt F.W., Khochbin S., Matthias P. (2008) Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 28, 1688–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Chang S., Young B.D., Li S., Qi X., Richardson J.A., Olson E.N. (2006) Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 126, 321–334. [DOI] [PubMed] [Google Scholar]
- 37).Haberland M., Mokalled M.H., Montgomery R.L., Olson E.N. (2009) Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 23, 1625–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Stammler D., Eigenbrod T., Menz S., Frick J.S., Sweet M.J., Shakespear M.R., Jantsch J., Siegert I., Wolfle S., Langer J.D., Oehme I., Schaefer L., Fischer A., Knievel J., Heeg K., Dalpke A.H., Bode K.A. (2015) Inhibition of histone deacetylases permits lipopolysaccharide-mediated secretion of bioactive IL-1β via a caspase-1-independent mechanism. J. Immunol. 195, 5421–5431. [DOI] [PubMed] [Google Scholar]
- 39).Sahakian E., Powers J.J., Chen J., Deng S.L., Cheng F., Distler A., Woods D.M., Rock-Klotz J., Sodre A.L., Youn J.I., Woan K.V., Villagra A., Gabrilovich D., Sotomayor E.M., Pinilla-Ibarz J. (2015) Histone deacetylase 11: A novel epigenetic regulator of myeloid derived suppressor cell expansion and function. Mol. Immunol. 63, 579–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Kim Y.B., Ki S.W., Yoshida M., Horinouchi S. (2000) Mechanism of cell cycle arrest caused by histone deacetylase inhibitors in human carcinoma cells. J. Antibiot. 53, 1191–1200. [DOI] [PubMed] [Google Scholar]
- 41).Zupkovitz G., Grausenburger R., Brunmeir R., Senese S., Tischler J., Jurkin J., Rembold M., Meunier D., Egger G., Lagger S., Chiocca S., Propst F., Weitzer G., Seiser C. (2010) The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol. Cell. Biol. 30, 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).Imai S., Armstrong C.M., Kaeberlein M., Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800. [DOI] [PubMed] [Google Scholar]
- 43).Landry J., Sutton A., Tafrov S.T., Heller R.C., Stebbins J., Pillus L., Sternglanz R. (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U.S.A. 97, 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Barber M.F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., Tennen R.I., Paredes S., Young N.L., Chen K., Struhl K., Garcia B.A., Gozani O., Li W., Chua K.F. (2012) SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487, 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Liszt G., Ford E., Kurtev M., Guarente L. (2005) Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 280, 21313–21320. [DOI] [PubMed] [Google Scholar]
- 46).Tanno M., Sakamoto J., Miura T., Shimamoto K., Horio Y. (2007) Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 282, 6823–6832. [DOI] [PubMed] [Google Scholar]
- 47).Kiran S., Chatterjee N., Singh S., Kaul S.C., Wadhwa R., Ramakrishna G. (2013) Intracellular distribution of human SIRT7 and mapping of the nuclear/nucleolar localization signal. FEBS J. 280, 3451–3466. [DOI] [PubMed] [Google Scholar]
- 48).Vaquero A., Scher M.B., Lee D.H., Sutton A., Cheng H.L., Alt F.W., Serrano L., Sternglanz R., Reinberg D. (2006) SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 20, 1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).North B.J., Verdin E. (2007) Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One 2, e784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Huang J.Y., Hirschey M.D., Shimazu T., Ho L., Verdin E. (2010) Mitochondrial sirtuins. Biochim. Biophys. Acta 1804, 1645–1651. [DOI] [PubMed] [Google Scholar]
- 51).Cheng H.L., Mostoslavsky R., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F. (2003) Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 100, 10794–10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M., Mills K.D., Patel P., Hsu J.T., Hong A.L., Ford E., Cheng H.L., Kennedy C., Nunez N., Bronson R., Frendewey D., Auerbach W., Valenzuela D., Karow M., Hottiger M.O., Hursting S., Barrett J.C., Guarente L., Mulligan R., Demple B., Yancopoulos G.D., Alt F.W. (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329. [DOI] [PubMed] [Google Scholar]
- 53).Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T., Braun T., Bober E. (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102, 703–710. [DOI] [PubMed] [Google Scholar]
- 54).Vazquez B.N., Thackray J.K., Simonet N.G., Kane-Goldsmith N., Martinez-Redondo P., Nguyen T., Bunting S., Vaquero A., Tischfield J.A., Serrano L. (2016) SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 35, 1488–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Lo Sasso G., Menzies K.J., Mottis A., Piersigilli A., Perino A., Yamamoto H., Schoonjans K., Auwerx J. (2014) SIRT2 deficiency modulates macrophage polarization and susceptibility to experimental colitis. PLoS One 9, e103573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Lombard D.B., Alt F.W., Cheng H.L., Bunkenborg J., Streeper R.S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A., Yang Y., Chen Y., Hirschey M.D., Bronson R.T., Haigis M., Guarente L.P., Farese R.V., Jr., Weissman S., Verdin E., Schwer B. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 27, 8807–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Haigis M.C., Mostoslavsky R., Haigis K.M., Fahie K., Christodoulou D.C., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., Karow M., Blander G., Wolberger C., Prolla T.A., Weindruch R., Alt F.W., Guarente L. (2006) SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 126, 941–954. [DOI] [PubMed] [Google Scholar]
- 58).Frye R.A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260, 273–279. [DOI] [PubMed] [Google Scholar]
- 59).Tanny J.C., Dowd G.J., Huang J., Hilz H., Moazed D. (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99, 735–745. [DOI] [PubMed] [Google Scholar]
- 60).Finnin M.S., Donigian J.R., Cohen A., Richon V.M., Rifkind R.A., Marks P.A., Breslow R., Pavletich N.P. (1999) Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401, 188–193. [DOI] [PubMed] [Google Scholar]
- 61).Kanyo Z.F., Scolnick L.R., Ash D.E., Christianson D.W. (1996) Structure of a unique binuclear manganese cluster in arginase. Nature 383, 554–557. [DOI] [PubMed] [Google Scholar]
- 62).Vannini A., Volpari C., Filocamo G., Casavola E.C., Brunetti M., Renzoni D., Chakravarty P., Paolini C., De Francesco R., Gallinari P., Steinkuhler C., Di Marco S. (2004) Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. U.S.A. 101, 15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63).Somoza J.R., Skene R.J., Katz B.A., Mol C., Ho J.D., Jennings A.J., Luong C., Arvai A., Buggy J.J., Chi E., Tang J., Sang B.C., Verner E., Wynands R., Leahy E.M., Dougan D.R., Snell G., Navre M., Knuth M.W., Swanson R.V., McRee D.E., Tari L.W. (2004) Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12, 1325–1334. [DOI] [PubMed] [Google Scholar]
- 64).Gantt S.L., Joseph C.G., Fierke C.A. (2010) Activation and inhibition of histone deacetylase 8 by monovalent cations. J. Biol. Chem. 285, 6036–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65).Wu R., Wang S., Zhou N., Cao Z., Zhang Y. (2010) A proton-shuttle reaction mechanism for histone deacetylase 8 and the catalytic role of metal ions. J. Am. Chem. Soc. 132, 9471–9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66).Gantt S.L., Gattis S.G., Fierke C.A. (2006) Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion. Biochemistry 45, 6170–6178. [DOI] [PubMed] [Google Scholar]
- 67).Dowling D.P., Gattis S.G., Fierke C.A., Christianson D.W. (2010) Structures of metal-substituted human histone deacetylase 8 provide mechanistic inferences on biological function. Biochemistry 49, 5048–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Ayer D.E. (1999) Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9, 193–198. [DOI] [PubMed] [Google Scholar]
- 69).Watson P.J., Fairall L., Santos G.M., Schwabe J.W. (2012) Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Millard C.J., Watson P.J., Celardo I., Gordiyenko Y., Cowley S.M., Robinson C.V., Fairall L., Schwabe J.W. (2013) Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol. Cell 51, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71).Watson P.J., Millard C.J., Riley A.M., Robertson N.S., Wright L.C., Godage H.Y., Cowley S.M., Jamieson A.G., Potter B.V., Schwabe J.W. (2016) Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nat. Commun. 7, 11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Fischle W., Dequiedt F., Hendzel M.J., Guenther M.G., Lazar M.A., Voelter W., Verdin E. (2002) Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45–57. [DOI] [PubMed] [Google Scholar]
- 73).Nielsen T.K., Hildmann C., Dickmanns A., Schwienhorst A., Ficner R. (2005) Crystal structure of a bacterial class 2 histone deacetylase homologue. J. Mol. Biol. 354, 107–120. [DOI] [PubMed] [Google Scholar]
- 74).Schuetz A., Min J., Allali-Hassani A., Schapira M., Shuen M., Loppnau P., Mazitschek R., Kwiatkowski N.P., Lewis T.A., Maglathin R.L., McLean T.H., Bochkarev A., Plotnikov A.N., Vedadi M., Arrowsmith C.H. (2008) Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J. Biol. Chem. 283, 11355–11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75).Bottomley M.J., Lo Surdo P., Di Giovine P., Cirillo A., Scarpelli R., Ferrigno F., Jones P., Neddermann P., De Francesco R., Steinkuhler C., Gallinari P., Carfi A. (2008) Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J. Biol. Chem. 283, 26694–26704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76).Lahm A., Paolini C., Pallaoro M., Nardi M.C., Jones P., Neddermann P., Sambucini S., Bottomley M.J., Lo Surdo P., Carfi A., Koch U., De Francesco R., Steinkuhler C., Gallinari P. (2007) Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 104, 17335–17340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77).Finnin M.S., Donigian J.R., Pavletich N.P. (2001) Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 8, 621–625. [DOI] [PubMed] [Google Scholar]
- 78).Min J., Landry J., Sternglanz R., Xu R.M. (2001) Crystal structure of a SIR2 homolog-NAD complex. Cell 105, 269–279. [DOI] [PubMed] [Google Scholar]
- 79).Zhao K., Chai X., Clements A., Marmorstein R. (2003) Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 10, 864–871. [DOI] [PubMed] [Google Scholar]
- 80).Avalos J.L., Boeke J.D., Wolberger C. (2004) Structural basis for the mechanism and regulation of Sir2 enzymes. Mol. Cell 13, 639–648. [DOI] [PubMed] [Google Scholar]
- 81).Zhao K., Harshaw R., Chai X., Marmorstein R. (2004) Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD+-dependent Sir2 histone/protein deacetylases. Proc. Natl. Acad. Sci. U.S.A. 101, 8563–8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82).Pan M., Yuan H., Brent M., Ding E.C., Marmorstein R. (2012) SIRT1 contains N- and C-terminal regions that potentiate deacetylase activity. J. Biol. Chem. 287, 2468–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Kang H., Suh J.Y., Jung Y.S., Jung J.W., Kim M.K., Chung J.H. (2011) Peptide switch is essential for Sirt1 deacetylase activity. Mol. Cell 44, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84).Kim J.E., Chen J., Lou Z. (2008) DBC1 is a negative regulator of SIRT1. Nature 451, 583–586. [DOI] [PubMed] [Google Scholar]
- 85).Zhao W., Kruse J.P., Tang Y., Jung S.Y., Qin J., Gu W. (2008) Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86).Davenport A.M., Huber F.M., Hoelz A. (2014) Structural and functional analysis of human SIRT1. J. Mol. Biol. 426, 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Furumai R., Komatsu Y., Nishino N., Khochbin S., Yoshida M., Horinouchi S. (2001) Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc. Natl. Acad. Sci. U.S.A. 98, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88).Darkin-Rattray S.J., Gurnett A.M., Myers R.W., Dulski P.M., Crumley T.M., Allocco J.J., Cannova C., Meinke P.T., Colletti S.L., Bednarek M.A., Singh S.B., Goetz M.A., Dombrowski A.W., Polishook J.D., Schmatz D.M. (1996) Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. U.S.A. 93, 13143–13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89).Komatsu Y., Tomizaki K.Y., Tsukamoto M., Kato T., Nishino N., Sato S., Yamori T., Tsuruo T., Furumai R., Yoshida M., Horinouchi S., Hayashi H. (2001) Cyclic hydroxamic-acid-containing peptide 31, a potent synthetic histone deacetylase inhibitor with antitumor activity. Cancer Res. 61, 4459–4466. [PubMed] [Google Scholar]
- 90).Glozak M.A., Seto E. (2007) Histone deacetylases and cancer. Oncogene 26, 5420–5432. [DOI] [PubMed] [Google Scholar]
- 91).Richon V.M., Emiliani S., Verdin E., Webb Y., Breslow R., Rifkind R.A., Marks P.A. (1998) A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 95, 3003–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92).Nakajima H., Kim Y.B., Terano H., Yoshida M., Horinouchi S. (1998) FR901228, a potent antitumor antibiotic, is a novel histone deacetylase inhibitor. Exp. Cell Res. 241, 126–133. [DOI] [PubMed] [Google Scholar]
- 93).Richon V.M., Webb Y., Merger R., Sheppard T., Jursic B., Ngo L., Civoli F., Breslow R., Rifkind R.A., Marks P.A. (1996) Second generation hybrid polar compounds are potent inducers of transformed cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 93, 5705–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94).Piekarz R.L., Robey R., Sandor V., Bakke S., Wilson W.H., Dahmoush L., Kingma D.M., Turner M.L., Altemus R., Bates S.E. (2001) Inhibitor of histone deacetylation, depsipeptide (FR901228), in the treatment of peripheral and cutaneous T-cell lymphoma: a case report. Blood 98, 2865–2868. [DOI] [PubMed] [Google Scholar]
- 95).Bolden J.E., Peart M.J., Johnstone R.W. (2006) Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 5, 769–784. [DOI] [PubMed] [Google Scholar]
- 96).Furumai R., Matsuyama A., Kobashi N., Lee K.H., Nishiyama M., Nakajima H., Tanaka A., Komatsu Y., Nishino N., Yoshida M., Horinouchi S. (2002) FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 62, 4916–4921. [PubMed] [Google Scholar]
- 97).Ying Y., Taori K., Kim H., Hong J., Luesch H. (2008) Total synthesis and molecular target of largazole, a histone deacetylase inhibitor. J. Am. Chem. Soc. 130, 8455–8459. [DOI] [PubMed] [Google Scholar]
- 98).Saito A., Yamashita T., Mariko Y., Nosaka Y., Tsuchiya K., Ando T., Suzuki T., Tsuruo T., Nakanishi O. (1999) A synthetic inhibitor of histone deacetylase, MS-27-275, with marked in vivo antitumor activity against human tumors. Proc. Natl. Acad. Sci. U.S.A. 96, 4592–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99).Bressi J.C., Jennings A.J., Skene R., Wu Y., Melkus R., De Jong R., O’Connell S., Grimshaw C.E., Navre M., Gangloff A.R. (2010) Exploration of the HDAC2 foot pocket: Synthesis and SAR of substituted N-(2-aminophenyl)benzamides. Bioorg. Med. Chem. Lett. 20, 3142–3145. [DOI] [PubMed] [Google Scholar]
- 100).Cole K.E., Dowling D.P., Boone M.A., Phillips A.J., Christianson D.W. (2011) Structural basis of the antiproliferative activity of largazole, a depsipeptide inhibitor of the histone deacetylases. J. Am. Chem. Soc. 133, 12474–12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101).Khan O., La Thangue N.B. (2012) HDAC inhibitors in cancer biology: emerging mechanisms and clinical applications. Immunol. Cell Biol. 90, 85–94. [DOI] [PubMed] [Google Scholar]
- 102).Catley L., Weisberg E., Kiziltepe T., Tai Y.T., Hideshima T., Neri P., Tassone P., Atadja P., Chauhan D., Munshi N.C., Anderson K.C. (2006) Aggresome induction by proteasome inhibitor bortezomib and α-tubulin hyperacetylation by tubulin deacetylase (TDAC) inhibitor LBH589 are synergistic in myeloma cells. Blood 108, 3441–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103).Mottamal M., Zheng S., Huang T.L., Wang G. (2015) Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules 20, 3898–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104).Murugan K., Sangeetha S., Ranjitha S., Vimala A., Al-Sohaibani S., Rameshkumar G. (2015) HDACiDB: a database for histone deacetylase inhibitors. Drug Des. Devel. Ther. 9, 2257–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105).Hildmann C., Wegener D., Riester D., Hempel R., Schober A., Merana J., Giurato L., Guccione S., Nielsen T.K., Ficner R., Schwienhorst A. (2006) Substrate and inhibitor specificity of class 1 and class 2 histone deacetylases. J. Biotechnol. 124, 258–270. [DOI] [PubMed] [Google Scholar]
- 106).Balasubramanian S., Ramos J., Luo W., Sirisawad M., Verner E., Buggy J.J. (2008) A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 22, 1026–1034. [DOI] [PubMed] [Google Scholar]
- 107).Methot J.L., Chakravarty P.K., Chenard M., Close J., Cruz J.C., Dahlberg W.K., Fleming J., Hamblett C.L., Hamill J.E., Harrington P., Harsch A., Heidebrecht R., Hughes B., Jung J., Kenific C.M., Kral A.M., Meinke P.T., Middleton R.E., Ozerova N., Sloman D.L., Stanton M.G., Szewczak A.A., Tyagarajan S., Witter D.J., Secrist J.P., Miller T.A. (2008) Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2). Bioorg. Med. Chem. Lett. 18, 973–978. [DOI] [PubMed] [Google Scholar]
- 108).Murata T., Kurokawa R., Krones A., Tatsumi K., Ishii M., Taki T., Masuno M., Ohashi H., Yanagisawa M., Rosenfeld M.G., Glass C.K., Hayashi Y. (2001) Defect of histone acetyltransferase activity of the nuclear transcriptional coactivator CBP in Rubinstein-Taybi syndrome. Hum. Mol. Genet. 10, 1071–1076. [DOI] [PubMed] [Google Scholar]
- 109).Kitabayashi I., Aikawa Y., Yokoyama A., Hosoda F., Nagai M., Kakazu N., Abe T., Ohki M. (2001) Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia 15, 89–94. [DOI] [PubMed] [Google Scholar]
- 110).Lafon-Hughes L., Di Tomaso M.V., Mendez-Acuna L., Martinez-Lopez W. (2008) Chromatin-remodelling mechanisms in cancer. Mutat. Res. 658, 191–214. [DOI] [PubMed] [Google Scholar]
- 111).Pandolfi P.P. (2001) Oncogenes and tumor suppressors in the molecular pathogenesis of acute promyelocytic leukemia. Hum. Mol. Genet. 10, 769–775. [DOI] [PubMed] [Google Scholar]
- 112).West A.C., Johnstone R.W. (2014) New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 124, 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113).Yoshida M., Beppu T. (1988) Reversible arrest of proliferation of rat 3Y1 fibroblasts in both the G1 and G2 phases by trichostatin A. Exp. Cell Res. 177, 122–131. [DOI] [PubMed] [Google Scholar]
- 114).Archer S.Y., Meng S., Shei A., Hodin R.A. (1998) p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc. Natl. Acad. Sci. U.S.A. 95, 6791–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115).Kim Y.B., Lee K.H., Sugita K., Yoshida M., Horinouchi S. (1999) Oxamflatin is a novel antitumor compound that inhibits mammalian histone deacetylase. Oncogene 18, 2461–2470. [DOI] [PubMed] [Google Scholar]
- 116).Marks P., Rifkind R.A., Richon V.M., Breslow R., Miller T., Kelly W.K. (2001) Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer 1, 194–202. [DOI] [PubMed] [Google Scholar]
- 117).Glozak M.A., Sengupta N., Zhang X., Seto E. (2005) Acetylation and deacetylation of non-histone proteins. Gene 363, 15–23. [DOI] [PubMed] [Google Scholar]
- 118).Kovacs J.J., Murphy P.J., Gaillard S., Zhao X., Wu J.T., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.P. (2005) HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell 18, 601–607. [DOI] [PubMed] [Google Scholar]
- 119).Smith B.C., Denu J.M. (2007) Mechanism-based inhibition of Sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry 46, 14478–14486. [DOI] [PubMed] [Google Scholar]
- 120).Bedalov A., Gatbonton T., Irvine W.P., Gottschling D.E., Simon J.A. (2001) Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. U.S.A. 98, 15113–15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121).Outeiro T.F., Kontopoulos E., Altmann S.M., Kufareva I., Strathearn K.E., Amore A.M., Volk C.B., Maxwell M.M., Rochet J.C., McLean P.J., Young A.B., Abagyan R., Feany M.B., Hyman B.T., Kazantsev A.G. (2007) Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson’s disease. Science 317, 516–519. [DOI] [PubMed] [Google Scholar]
- 122).Rumpf T., Schiedel M., Karaman B., Roessler C., North B.J., Lehotzky A., Olah J., Ladwein K.I., Schmidtkunz K., Gajer M., Pannek M., Steegborn C., Sinclair D.A., Gerhardt S., Ovadi J., Schutkowski M., Sippl W., Einsle O., Jung M. (2015) Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat. Commun. 6, 6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123).Lin S.J., Defossez P.A., Guarente L. (2000) Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128. [DOI] [PubMed] [Google Scholar]
- 124).Tissenbaum H.A., Guarente L. (2001) Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230. [DOI] [PubMed] [Google Scholar]
- 125).Burnett C., Valentini S., Cabreiro F., Goss M., Somogyvari M., Piper M.D., Hoddinott M., Sutphin G.L., Leko V., McElwee J.J., Vazquez-Manrique R.P., Orfila A.M., Ackerman D., Au C., Vinti G., Riesen M., Howard K., Neri C., Bedalov A., Kaeberlein M., Soti C., Partridge L., Gems D. (2011) Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126).Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., Bar-Joseph Z., Cohen H.Y. (2012) The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221. [DOI] [PubMed] [Google Scholar]
- 127).Howitz K.T., Bitterman K.J., Cohen H.Y., Lamming D.W., Lavu S., Wood J.G., Zipkin R.E., Chung P., Kisielewski A., Zhang L.L., Scherer B., Sinclair D.A. (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196. [DOI] [PubMed] [Google Scholar]
- 128).Milne J.C., Lambert P.D., Schenk S., Carney D.P., Smith J.J., Gagne D.J., Jin L., Boss O., Perni R.B., Vu C.B., Bemis J.E., Xie R., Disch J.S., Ng P.Y., Nunes J.J., Lynch A.V., Yang H., Galonek H., Israelian K., Choy W., Iffland A., Lavu S., Medvedik O., Sinclair D.A., Olefsky J.M., Jirousek M.R., Elliott P.J., Westphal C.H. (2007) Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129).Kaeberlein M., McDonagh T., Heltweg B., Hixon J., Westman E.A., Caldwell S.D., Napper A., Curtis R., DiStefano P.S., Fields S., Bedalov A., Kennedy B.K. (2005) Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 280, 17038–17045. [DOI] [PubMed] [Google Scholar]
- 130).Pacholec M., Bleasdale J.E., Chrunyk B., Cunningham D., Flynn D., Garofalo R.S., Griffith D., Griffor M., Loulakis P., Pabst B., Qiu X., Stockman B., Thanabal V., Varghese A., Ward J., Withka J., Ahn K. (2010) SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 285, 8340–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131).Dai H., Kustigian L., Carney D., Case A., Considine T., Hubbard B.P., Perni R.B., Riera T.V., Szczepankiewicz B., Vlasuk G.P., Stein R.L. (2010) SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J. Biol. Chem. 285, 32695–32703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132).Hubbard B.P., Gomes A.P., Dai H., Li J., Case A.W., Considine T., Riera T.V., Lee J.E., E S.Y., Lamming D.W., Pentelute B.L., Schuman E.R., Stevens L.A., Ling A.J., Armour S.M., Michan S., Zhao H., Jiang Y., Sweitzer S.M., Blum C.A., Disch J.S., Ng P.Y., Howitz K.T., Rolo A.P., Hamuro Y., Moss J., Perni R.B., Ellis J.L., Vlasuk G.P., Sinclair D.A. (2013) Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science 339, 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133).Dai H., Case A.W., Riera T.V., Considine T., Lee J.E., Hamuro Y., Zhao H., Jiang Y., Sweitzer S.M., Pietrak B., Schwartz B., Blum C.A., Disch J.S., Caldwell R., Szczepankiewicz B., Oalmann C., Yee Ng P., White B.H., Casaubon R., Narayan R., Koppetsch K., Bourbonais F., Wu B., Wang J., Qian D., Jiang F., Mao C., Wang M., Hu E., Wu J.C., Perni R.B., Vlasuk G.P., Ellis J.L. (2015) Crystallographic structure of a small molecule SIRT1 activator-enzyme complex. Nat. Commun. 6, 7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134).Dasgupta B., Milbrandt J. (2007) Resveratrol stimulates AMP kinase activity in neurons. Proc. Natl. Acad. Sci. U.S.A. 104, 7217–7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135).Park S.J., Ahmad F., Philp A., Baar K., Williams T., Luo H., Ke H., Rehmann H., Taussig R., Brown A.L., Kim M.K., Beaven M.A., Burgin A.B., Manganiello V., Chung J.H. (2012) Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 148, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136).Sajish M., Schimmel P. (2015) A human tRNA synthetase is a potent PARP1-activating effector target for resveratrol. Nature 519, 370–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137).Liew C.C., Gornall A.G. (1973) Acetylation of ribosomal proteins. I. Characterization and properties of rat liver ribosomal proteins. J. Biol. Chem. 248, 977–983. [PubMed] [Google Scholar]
- 138).L’Hernault S.W., Rosenbaum J.L. (1985) Chlamydomonas α-tubulin is posttranslationally modified by acetylation on the ε-amino group of a lysine. Biochemistry 24, 473–478. [DOI] [PubMed] [Google Scholar]
- 139).Gu W., Roeder R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 140).Sakaguchi K., Herrera J.E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C.W., Appella E. (1998) DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141).Juan L.J., Shia W.J., Chen M.H., Yang W.M., Seto E., Lin Y.S., Wu C.W. (2000) Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275, 20436–20443. [DOI] [PubMed] [Google Scholar]
- 142).Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159. [DOI] [PubMed] [Google Scholar]
- 143).Ito A., Kawaguchi Y., Lai C.H., Kovacs J.J., Higashimoto Y., Appella E., Yao T.P. (2002) MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21, 6236–6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144).Yeung F., Hoberg J.E., Ramsey C.S., Keller M.D., Jones D.R., Frye R.A., Mayo M.W. (2004) Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145).Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Hu L.S., Cheng H.L., Jedrychowski M.P., Gygi S.P., Sinclair D.A., Alt F.W., Greenberg M.E. (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 146).Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M.W., Guarente L. (2004) Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147).Verdel A., Curtet S., Brocard M.P., Rousseaux S., Lemercier C., Yoshida M., Khochbin S. (2000) Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr. Biol. 10, 747–749. [DOI] [PubMed] [Google Scholar]
- 148).Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., Yoshida M., Wang X.F., Yao T.P. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458. [DOI] [PubMed] [Google Scholar]
- 149).Matsuyama A., Shimazu T., Sumida Y., Saito A., Yoshimatsu Y., Seigneurin-Berny D., Osada H., Komatsu Y., Nishino N., Khochbin S., Horinouchi S., Yoshida M. (2002) In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 21, 6820–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150).North B.J., Marshall B.L., Borra M.T., Denu J.M., Verdin E. (2003) The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444. [DOI] [PubMed] [Google Scholar]
- 151).Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., Yao T.P. (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738. [DOI] [PubMed] [Google Scholar]
- 152).Iwata A., Riley B.E., Johnston J.A., Kopito R.R. (2005) HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280, 40282–40292. [DOI] [PubMed] [Google Scholar]
- 153).Akella J.S., Wloga D., Kim J., Starostina N.G., Lyons-Abbott S., Morrissette N.S., Dougan S.T., Kipreos E.T., Gaertig J. (2010) MEC-17 is an α-tubulin acetyltransferase. Nature 467, 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154).Shida T., Cueva J.G., Xu Z., Goodman M.B., Nachury M.V. (2010) The major α-tubulin K40 acetyltransferase αTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. U.S.A. 107, 21517–21522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155).Zhang X., Yuan Z., Zhang Y., Yong S., Salas-Burgos A., Koomen J., Olashaw N., Parsons J.T., Yang X.J., Dent S.R., Yao T.P., Lane W.S., Seto E. (2007) HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell 27, 197–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156).Ammer A.G., Weed S.A. (2008) Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil. Cytoskeleton 65, 687–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157).Ito A., Shimazu T., Maeda S., Shah A.A., Tsunoda T., Iemura S., Natsume T., Suzuki T., Motohashi H., Yamamoto M., Yoshida M. (2015) The subcellular localization and activity of cortactin is regulated by acetylation and interaction with Keap1. Sci. Signal. 8, ra120. [DOI] [PubMed] [Google Scholar]
- 158).Haggarty S.J., Koeller K.M., Wong J.C., Grozinger C.M., Schreiber S.L. (2003) Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. U.S.A. 100, 4389–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159).Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840. [DOI] [PubMed] [Google Scholar]
- 160).Zhang J., Shi X., Li Y., Kim B.J., Jia J., Huang Z., Yang T., Fu X., Jung S.Y., Wang Y., Zhang P., Kim S.T., Pan X., Qin J. (2008) Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31, 143–151. [DOI] [PubMed] [Google Scholar]
- 161).Deardorff M.A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D., Cole K.E., De Baere E., Decroos C., Di Donato N., Ernst S., Francey L.J., Gyftodimou Y., Hirashima K., Hullings M., Ishikawa Y., Jaulin C., Kaur M., Kiyono T., Lombardi P.M., Magnaghi-Jaulin L., Mortier G.R., Nozaki N., Petersen M.B., Seimiya H., Siu V.M., Suzuki Y., Takagaki K., Wilde J.J., Willems P.J., Prigent C., Gillessen-Kaesbach G., Christianson D.W., Kaiser F.J., Jackson L.G., Hirota T., Krantz I.D., Shirahige K. (2012) HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489, 313–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162).Hirschey M.D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D.B., Grueter C.A., Harris C., Biddinger S., Ilkayeva O.R., Stevens R.D., Li Y., Saha A.K., Ruderman N.B., Bain J.R., Newgard C.B., Farese R.V., Jr., Alt F.W., Kahn C.R., Verdin E. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163).Schwer B., Bunkenborg J., Verdin R.O., Andersen J.S., Verdin E. (2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164).Hallows W.C., Lee S., Denu J.M. (2006) Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165).Hentchel K.L., Escalante-Semerena J.C. (2015) Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol. Mol. Biol. Rev. 79, 321–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166).Tan M., Luo H., Lee S., Jin F., Yang J.S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167).Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y., Boeke J.D., Zhao Y. (2012) Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168).Dai L., Peng C., Montellier E., Lu Z., Chen Y., Ishii H., Debernardi A., Buchou T., Rousseaux S., Jin F., Sabari B.R., Deng Z., Allis C.D., Ren B., Khochbin S., Zhao Y. (2014) Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 10, 365–370. [DOI] [PubMed] [Google Scholar]
- 169).Colak G., Xie Z., Zhu A.Y., Dai L., Lu Z., Zhang Y., Wan X., Chen Y., Cha Y.H., Lin H., Zhao Y., Tan M. (2013) Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol. Cell. Proteomics 12, 3509–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170).Bheda P., Wang J.T., Escalante-Semerena J.C., Wolberger C. (2011) Structure of Sir2Tm bound to a propionylated peptide. Protein Sci. 20, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171).Bao X., Wang Y., Li X., Li X.M., Liu Z., Yang T., Wong C.F., Zhang J., Hao Q., Li X.D. (2014) Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. eLife 3, e02999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172).Feldman J.L., Baeza J., Denu J.M. (2013) Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288, 31350–31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173).Feldman J.L., Dittenhafer-Reed K.E., Kudo N., Thelen J.N., Ito A., Yoshida M., Denu J.M. (2015) Kinetic and structural basis for acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry 54, 3037–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174).Nakagawa T., Lomb D.J., Haigis M.C., Guarente L. (2009) SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175).Du J., Zhou Y., Su X., Yu J.J., Khan S., Jiang H., Kim J., Woo J., Kim J.H., Choi B.H., He B., Chen W., Zhang S., Cerione R.A., Auwerx J., Hao Q., Lin H. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176).Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M., Luo H., Zhang Y., He W., Yang K., Zwaans B.M., Tishkoff D., Ho L., Lombard D., He T.C., Dai J., Verdin E., Ye Y., Zhao Y. (2011) The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteomics 10, M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177).Tan M., Peng C., Anderson K.A., Chhoy P., Xie Z., Dai L., Park J., Chen Y., Huang H., Zhang Y., Ro J., Wagner G.R., Green M.F., Madsen A.S., Schmiesing J., Peterson B.S., Xu G., Ilkayeva O.R., Muehlbauer M.J., Braulke T., Muhlhausen C., Backos D.S., Olsen C.A., McGuire P.J., Pletcher S.D., Lombard D.B., Hirschey M.D., Zhao Y. (2014) Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178).Jiang H., Khan S., Wang Y., Charron G., He B., Sebastian C., Du J., Kim R., Ge E., Mostoslavsky R., Hang H.C., Hao Q., Lin H. (2013) SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179).Argmann C., Auwerx J. (2006) Insulin secretion: SIRT4 gets in on the act. Cell 126, 837–839. [DOI] [PubMed] [Google Scholar]
- 180).Mathias R.A., Greco T.M., Oberstein A., Budayeva H.G., Chakrabarti R., Rowland E.A., Kang Y., Shenk T., Cristea I.M. (2014) Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell 159, 1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181).Aramsangtienchai P., Spiegelman N.A., He B., Miller S.P., Dai L., Zhao Y., Lin H. (2016) HDAC8 catalyzes the hydrolysis of long chain fatty acyl lysine. ACS Chem. Biol. 11, 2685–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182).Wagner G.R., Payne R.M. (2013) Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288, 29036–29045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183).Weinert B.T., Iesmantavicius V., Wagner S.A., Scholz C., Gummesson B., Beli P., Nystrom T., Choudhary C. (2013) Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51, 265–272. [DOI] [PubMed] [Google Scholar]
- 184).Wagner G.R., Hirschey M.D. (2014) Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 54, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]