Abstract
Aneuploidy and polyploidy are a form of Genomic Instability (GIN) known as Chromosomal Instability (CIN) characterized by sporadic abnormalities in chromosome copy numbers. Aneuploidy is commonly linked to pathological states. It is a hallmark of spontaneous abortions and birth defects and it is observed virtually in every human tumor, therefore being generally regarded as detrimental for the development or the maturation of tissues under physiological conditions. Polyploidy however, occurs as part of normal physiological processes during maturation and differentiation of some mammalian cell types. Surprisingly, high levels of aneuploidy are present in the brain, and their frequency increases with age suggesting that the brain is able to maintain its functionality in the presence of high levels of mosaic aneuploidy. Because somatic aneuploidy with age can reach exceptionally high levels, it is likely to have long-term adverse effects in this organ. We describe the mechanisms accountable for an abnormal DNA content with a particular emphasis on the CNS where cell division is limited. Next, we briefly summarize the types of GIN known to date and discuss how they interconnect with CIN. Lastly we highlight how several forms of CIN may contribute to genetic variation, tissue degeneration and disease in the CNS.
Keywords: Genomic instability, Aneuploidy, DNA damage, Tissue degeneration, Brain, Polyploidy, Whole chromosome instability (W-CIN)
1. Introduction
Accurate replication of DNA and its equal distribution into daughter cells are essential to safeguard the balance of genetic content in eukaryotic cells (Masai et al., 2010). Failure or deficiency in these processes results in genome alterations in daughter cells relative to parental DNA (Shen, 2011). Genomic instability (GIN) is defined as the predisposition of genomes to undergo alterations under physiological conditions or when pathways responsible for genome maintenance become impaired. Accumulation of genomic damage deregulates cell division by prompting irreversible mitotic arrest (senescence), cell death, or uncontrolled proliferation and cancer (Maslovand Vijg, 2009; Shen, 2011; Vijg and Suh, 2013). Not surprisingly, GIN has been associated with the etiology of a plethora of human diseases, and it is believed to be a driver of tumorigenesis (Russo et al., 2015). Accumulation of DNA damage throughout life is a common denominator of the aging process, therefore GIN is also recognized as a hallmark of aging (Lopez-Otin et al., 2013). Unrepaired DNA damage is deemed a contributor to GIN and the aging process, which might be especially relevant in the central nervous system (CNS) due to some evidence for a low repair capacity of terminally differentiated neurons (Nouspikel and Hanawalt, 2000). One form of GIN is Chromosome Instability (CIN), a cellular state with a higher propensity for chromosome mis-segregation.
Abnormal ploidy is often the result of failures in segregating chromosomes during mitosis or completion of cytokinesis. Whole Chromosome Instability (W-CIN) can have a dramatic effect on cellular physiology due to the simultaneous gene dosage alteration of the many genes mapping to the unbalanced chromosomes. The term aneuploidy (an - not + eu-well + ploid-fold) was first proposed in 1922 by Gunnar Täckholm to describe a chromosome number that is not an exact multiple of the haploid content (Täckholm, 1922). While aneuploidy has been historically linked to disease, recent evidence suggests a possible role also in general tissue degeneration during aging. In fact aneuploidy has been proposed as a feature of aging because of the premature aging phenotypes associated with some CIN mouse models (Baker et al, 2004; Baker et al., 2006; Matsuyama et al., 2013; Ricke and van Deursen, 2013; Tanaka et al., 2015). A large body of literature now makes it clear that mosaic somatic aneuploidy is more common than previously anticipated (Jacobs et al., 2012; Torres et al., 2008). Indeed, aneuploidy cells have been shown to accumulate with age in mammalian tissues, particularly the brain (Faggioli et al., 2012), the liver (Duncan et al., 2012b; Faggioli et al., 2011a), blood lymphocytes (Jacobs et al., 1961) and oocytes (Jones, 2008).
Polyploidy, a cellular state in which two or more complete sets of chromosomes are present, is part of the normal differentiation process of some mammalian cell types like developing neurons in the cerebral cortex (see below), hepatocytes, osteoclasts (Davoli and de Lange, 2011), keratinocytes (Zanet et al., 2010), and vascular smooth muscle cells (VSMCs) (Jones and Ravid, 2004). Polyploidy has been observed under physiological conditions in lactating mammary gland, urothelium, mesothelium, and Purkinje neurons (Davoli and de Lange, 2011), but it is also associated with stress, aging and disease. While a clear reason for the presence of a polyploid genome remains obscure, it is likely to provide advantages, such as gene redundancy and protection against loss of heterozygosity (LOH) (Comai, 2005; Ganem et al., 2007).
2. GIN at the chromosomal level: routes to aneuploidy and polyploidy
Aneuploidy and polyploidy can be generated by defects in several genome maintenance mechanisms and their origins can be related. Here we review known routes to aneuploidy and polyploidy (Fig. 1).
Fig. 1.
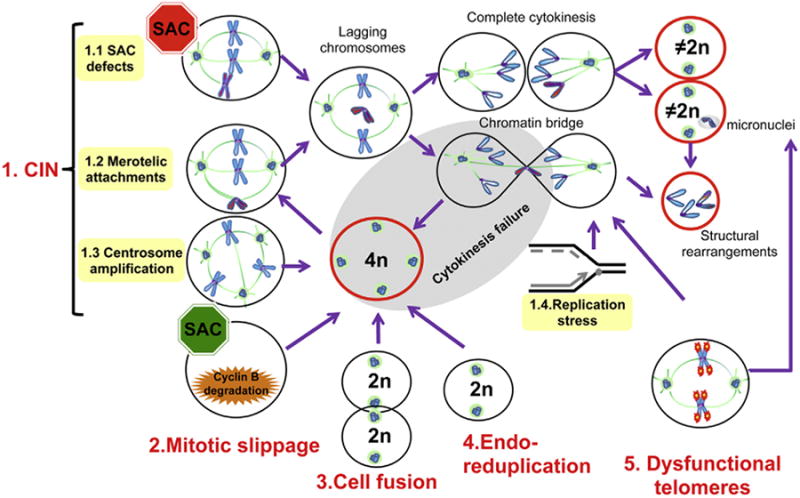
Routes to aneuploidy and tetraploidy. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) In a CIN state (1, top left) deficiency in SAC signaling (1.1) and merotelic attachments (1.2) result in lagging chromosomes that can generate mononucleated aneuploid cells, aneuploid cells containing a micronucleus, ortetraploid cells (cells in red) due to chromatin bridging and cytokinesis failure (grey area). Tetraploid cells arising from different mechanisms present centrosome amplification (1.3), which in turn can lead to multipolar mitotic spindles and lagging chromosomes. Finally, replication stress (1.4) is a trait of CIN cells that can lead to chromatin bridging and thus, cytokinesis failure or structural rearrangements. Mitotic slippage (2) occurs when the SAC is activated for an extended period of time due to unattached kinetochores and slow Cyclin B degradation. This can result in mitotic exit without cytokinesis and thus tetraploidy. Cell fusion (3) of diploid cells can form atetraploid/polyploid cell. Endoreduplication (4) occurs when DNAis duplicated but mitosis fails to occur, generating a tetraploid and/or polyploid cell. Dysfunctional telomeres (5) can undergo BFB cycles that can lead to chromatin bridging, structural rearrangements and/or cytokinesis failure. Chromosomes with eroded telomeres undergo frequent mis-segregation, and are also a source of aneuploidy. Because micronuclei have been linked to s-CIN and chromothripsis, all defects leading to their formation are hazardous and enable further GIN.
CIN is caused by defects in genes involved in SAC signaling, chromosome condensation, sister-chromatid cohesion, kinetochore assembly, spindle formation, cytokinesis and other mitotic events that facilitate segregation of chromosomes or rearrangements (Holland and Cleveland, 2012b; Ricke et al., 2008).
SAC defects arise due to depletion or malfunction of SAC components. The SAC is a surveillance mechanism that regulates chromosome segregation by delaying anaphase progression until all kinetochores are stably attached to spindle microtubules, which is essential for proper segregation of chromosomes. The Anaphase Promoting Complex/Cyclosome (APC/C) is a multi-subunit E3 ubiq-uitin ligase that degrades Securin and Cyclin B1, allowing sister chromatid separation and mitosis exit, respectively. APC/C requires a specificity factor (Cell-Division-Cycle 20 homologue - CDC20) to be able to recognize and interact with its substrates. Unattached chromosomes activate the SAC and recruit its components (BUB1, BubR1, BUB3, Mad1, Mad2) in a step-wise fashion to the outer kinetochore surface, inducing the formation of a complex betweenBubR1, active Mad2, BUB3 and CDC20. This tight association between SAC proteins and CDC20 prevents the activity of APC/C and thus prevents anaphase progression, allowing time for all spindle-kinetochores attachments to occur. Once all chromosomes are properly attached, CDC20 dissociates from the SAC components and binds APC/C, triggering mitosis exit (Kops et al., 2005; Lara-Gonzalez and Taylor, 2012; Suijkerbuijk and Kops, 2008). Exactly how the SAC module is assembled remains unknown, but it seems established that aneuploidy is a consequence of a deficient SAC (Gjoerup et al, 2007; Lentini et al., 2011; Liet al, 2010; Musio et al., 2003).
Merotelic attachments occur when a single kinetochore is bound to microtubules originating from opposite spindle poles, which often results in chromosomes lagging behind the segregating DNA during anaphase. It can be caused by defects in chromosome condensation, cohesion, kinetochore structure, and spindle assembly (Gregan et al., 2011). Lagging chromosomes have multiple fates: miss-segregation and generation of aneuploid daughter cells, failure to be incorporated in the main nucleus and subsequent formation of micronuclei, or chromatin bridges that inhibit the closure of the contractile ring, resulting in cytokinesis failure and tetraploidy. Of note, these erroneous chromosome attachments are not sensed by the SAC because the kinetochores are attached to both poles (Davoli and de Lange, 2011; Holland and Cleveland, 2012b; Russo et al., 2015; Zasadil et al., 2013).
Centrosome dysfunction arises when centrosomes undergo structural and/or numerical alterations, the latter generally more frequent and best documented (Montagna et al., 2002; Pihan, 2013). Centrosome amplification occurs when cells have more than two centrosomes due to over - duplication, cytokinesis failure or cell fusion (Vitre and Cleveland, 2012). Supernumerary centrosomes are observed in polyploid normal hepatocytes (Duncan et al., 2012b; Faggioli et al., 2011a), but are a common feature of cancer cells. Extra centrosomes can lead to catastrophic mitosis due to formation of multipolar spindles, resulting in chaotic chromosome segregation into two or more daughter cells and aneuploidy. Cancer cells and hepatocytes were both shown to be able to cluster supernumerary centrosomes into a bipolar spindle to overcome this problem. However, merotelic attachments are enriched by the multipolarity before clustering, leading to mis-segregation (Ganem et al., 2009, 2007).
Replication stress arises in CIN cells due to impaired replication fork progression prompted by unrepaired DNA damage, topological stress or limiting nucleotides (Burrell et al., 2013; Ichijimaet al., 2010; Meena et al., 2015). If sustained for long periods it can lead to incompletely duplicated chromosomes that could undergo fragmentation or generate chromatin bridges, which in turn sustain chromosome rearrangements or tetraploidy (Burrell et al., 2013; Donley and Thayer, 2013; Russo et al., 2015).
Mitotic slippage is the consequence of the slow degradation of Cyclin B when prolonged arrest mediated by the SAC occurs. It results in mitosis exit without anaphase or cytokinesis completion, leading to the formation of tetraploid cells (Brito and Rieder, 2006; Ganem et al., 2007).
Cell fusion can be physiologically generated during the normal development of specific tissues such as the liver (Faggioli et al., 2008), resulting in terminally differentiated polyploid cells. It can also be induced by viral infections, radiation exposure, inflammation, and chemotherapeutic drugs, all of which potentially increase the risk for transformation (Davoli and de Lange, 2011; Ganem et al., 2007; Holland and Cleveland, 2012b). This phenomenon is also observed in the brain, where bone marrow-derived cells (BMDCs) are shown to fuse with Purkinje cells with a frequency increasing with age (Lapham, 1968; Nern et al., 2009). Fusion is believed to rescue injured neuronal cells that cannot undergo cell division bycontributing genetic material from BM to restore tissue homeostasis (Kemp et al., 2014) (see also Section 7).
Endoreduplication is the replication of DNA without subsequent cell division and is associated with the terminal differentiation of non-dividing polyploid cells, such as megakaryocytes (Davoli and de Lange, 2011; Fox and Duronio, 2013; Ganem et al., 2007) and keratinocytes (Zanet et al., 2010). This process has been shown to generate tetraploid neurons in the normal vertebrate retina (Frade, 2010). It can also be induced by persistent DNA damage signaling, thus also leading to polyploidy under non-physiological conditions (Davoli and de Lange, 2011).
Dysfunctional telomeres result from sustained cell proliferation in the absence of telomerase activity and are the natural consequence of the inability of the cell replication machinery to duplicate the DNA all the way to the end of a chromosome (Olovnikov, 1996). Critically eroded telomeres can undergo BFB cycles that generate chromatin bridges, enabling chromosome structural rearrangements and cytokinesis failure (Gisselsson et al., 2001; Holland and Cleveland, 2012b; Rode et al., 2015). Chromosomes with short telomeres are more frequently involved in miss-segregation events, through both non-disjunction and anaphase lagging of dicentric chromatids, yielding whole chromosome aneuploidy (Pampalona et al., 2010b).
Chromatin bridges in the cleavage furrow is considered the main cause of cytokinesis failure and tetraploidy. DNA trapped in the furrow can be a consequence of lagging chromosomes, replication stress or dysfunctional telomeres, thus converging defects from multiple pathways into a similar outcome, that is, the formation of a tetraploid cell (Davoli and de Lange, 2011; Ganem et al., 2007; Holland and Cleveland, 2012b; Russo et al., 2015).
Mechanisms that prevent GIN are extensive and evolved to safeguard the integrity of the DNA sequence and genome content. However, under some conditions (i.e., aging or disease) the genome defensive system becomes vulnerable due to loss of function, reduced expression level or misregulation of proteins important for genome maintenance. For example, decreased transcripts of SAC components Bub1 and Mad2 and reduced levels of cohesin proteins were observed during normative aging in oocytes (Schwarzer et al., 2014; Steuerwald et al., 2001; Tsutsumi et al., 2014), which might be related to the increased aneuploidy with age in these cells. Gene expression analysis of murine tissues previously shown to undergo age-related aneuploidization (Baker et al., 2013; Faggioli et al., 2012) revealed age-associated down-regulation of proteins important for chromosome segregation, such as components of the SAC and centromere proteins (Zahn et al., 2007)(Fig. 2). Furthermore, components of the nuclear pore complexes within the Nuclear Envelope (NE) were shown to be damaged by oxidative post-translational modifications (OPTMs) and this phenomenon was associated with loss of nuclear integrity during aging, which in turn can result in GIN (D'Angelo et al., 2009).
Fig. 2.
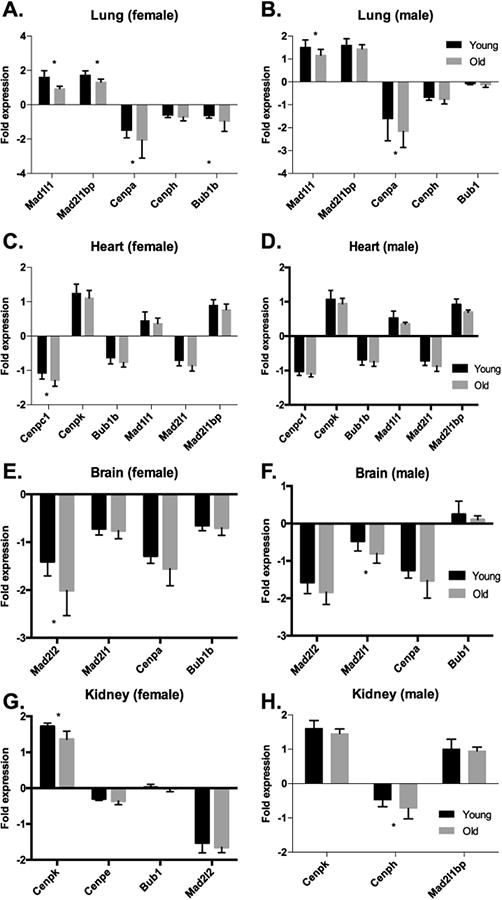
Down-regulation of components of the SAC and centromere proteins is observed during aging. A trend for reduced expression levels of ploidy-related genes occurs in old mice (16or24-months-old) relative to young (1 month-old) in tissues known to accumulate ploidy changes with age. Female mice (A) lung, (C) heart, (E) brain and (G) kidney. Male mice(B) lung, (D) heart, (F) brain and (H) kidney. Asterisks represent statistically significant differences (p<0.05). Data was plotted from (Zahn et al., 2007).
Aging is accompanied by an increase in oxidative stress (OS) (Kregel and Zhang, 2007; Romano et al., 2010) and long standing evidence suggests an involvement of ROS in the generation of ploidy changes and CIN, yet the exact mechanisms remain unclear (Dephoure et al., 2014; Estrada et al., 2013; Gentric et al., 2015; Limoli and Giedzinski, 2003; Nicotera et al., 1985; Samper et al., 2003; Tarin, 1995; Tormos et al., 2015). Among the proposed causes for OS-induced ploidy alterations are abrogation of SAC function, energy deficiency, centrosome amplification and defects in microtubules-kinetochore dynamics (D'Angiolella et al., 2007; Ikawa-Yoshida et al., 2013; Wang et al., 2013). More recently, it was proposed that OPTMs on proteins important for mitosis and cytoskeleton dynamics could result in cell cycle perturbations and interfere with cytokinesis (Mailankot et al., 2008). This hypothesis is supported by observations suggesting that some structural proteins (Vimentin, Actin and Tubulin) are consistently modified by ROS in different organs systems, including brain, and during in vitro senescence (Baraibar and Friguet, 2012). OPTMs promote disruption of both actin and the microtubule cytoskeleton (Banan et al., 2000a,b), which could result in spindle formation defects, chromosome miss-segregation and cytokinesis failure. Proteomic profiling of HeLa cells exposed to mild OS conditions revealed enrichment of oxidized proteins involved in kinetochore/spindle machinery and centrosome organization (Bollineni et al., 2014). In view of these findings, it is tempting to speculate that one of the mechanisms driving ploidy changes upon OS or during aging is the oxidative injury of components of the mitotic machinery and cytoskeleton components. This hypothesis is particularly relevant for the brain, a tissue that requires high levels of oxygen for normal functioning and has been demonstrated to accumulate aneuploidy with age (Faggioli et al., 2012; Uttara et al., 2009) (Fig. 3).
Fig. 3.
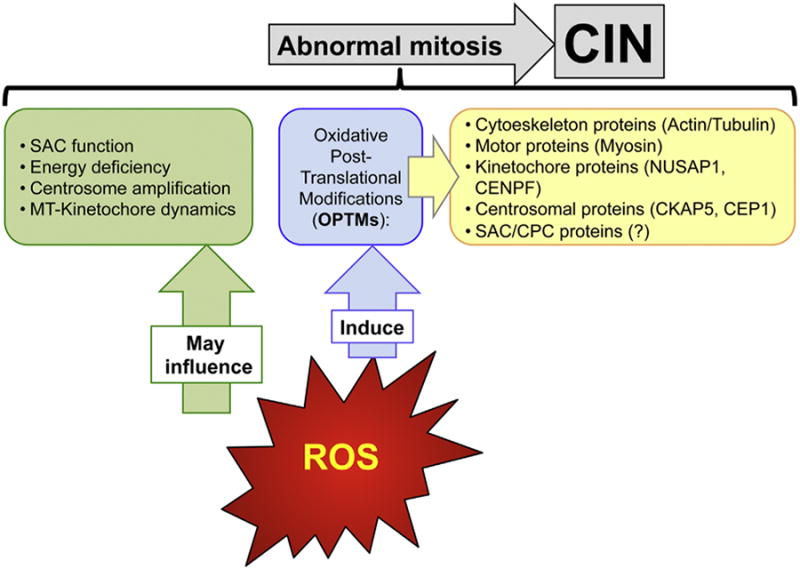
ROS are associated with the generation of CIN. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) ROS can interfere with multiple aspects of the mitotic pathway leading to changes in ploidy (green box). ROS can also induce oxidative post-translational modifications (OPTMs) of proteins important for mitosis and/or cytokinesis, which can result in protein unfolding and malfunction. Thus, oxidative damage to mitotic components could affect their proper activity, leading to abnormal mitosis and CIN. Proteins of the cytoskeleton, molecular motors, kinetochore and centrosomal proteins are found oxidized in a context of oxidative stress (yellow box). It remains to be determined if components of the SAC signaling or the chromosome passenger complex (CPC) are also prone to OPTMs.
3. CIN can generate DNA damage
Abnormal mitosis is associated with the acquisition of DNA damage, either directly during the mitotic process itself or indirectly upon miss-segregation of chromosomes. For instance, prolonged mitotic arrest due to drug exposure or genetic manipulation can generate γ-H2AX foci - indicative for DNA double-strand breaks - that gradually accumulate and persist after mitotic slippage (Ganem and Pellman, 2012; Hayashi and Karlseder, 2013). In this section we will review the evidence supporting the hypothesis that DNA damage can be induced as a consequence of chromosome miss-segregation.
DNA damage as an outcome of CIN is well documented (Ganem and Pellman, 2012; Hayashi and Karlseder, 2013; Janssen et al.,2011; Meena et al., 2015; Ohashi et al., 2015) and can occur through multiple non-exclusive mechanisms. Because aneuploidy is a state of unbalanced karyotype, levels of proteins essential for DNA replication, condensation, repair or cell division are also expected to be unbalanced, which in turn could promote the induction of DNA damage and replication defects (Holland and Cleveland, 2012b; Meena et al, 2015; Nicholson and Cimini, 2015). Yeast cells, aneuploid for one single chromosome, show higher rates of point mutations and mitotic recombinations (Sheltzer et al., 2011). Telomerase expression in aneuploid mammalian cells abrogates aneuploidy-induced telomere replication stress and also decreases aneuploidy-induced DNA damage accumulation (Meena et al., 2015).
As previously described, lagging of chromosomes produced by a variety of mechanisms can induce chromatin bridges between daughter cells or lead to the formation of micronuclei, both of which can promote further damage to the DNA. It remains unclear if microtubule-generated pulling forces are strong enough to cause breakage of intact lagging chromosomes, even though this was shown to be possible in a scenario where a centrosome-localized protein (Dido gene product) was absent (Ganem and Pellman, 2012). Chromosomes that miss-segregate are often damaged during cytokinesis by cleavage furrow-generated forces resulting in daughter cells staining positive for DSB foci. Broken chromosomes are susceptible to error-prone repair by NHEJ, and the resulting unbalanced translocations can lead to additional structural alterations (Janssen et al., 2011). DNA trapped in the cleavage furrow can lead to failure of cytokinesis, resulting in tetraploidy. Tetraploid cells enable further CIN due to their supernumerary centrosomes and consequent higher rates of chromosomemiss-segregation, multipolar spindle formation, and increased tolerance to chromosome alterations (Davoli and de Lange, 2011; Holland and Cleveland, 2012b; Russo et al., 2015). Moreover, because these cells contain twice the genomic content of diploid cells, the amount of DNA damage generated during S phase is also doubled (Ganem et al., 2007).
Micronuclei are created mainly when lagging chromosomes fail to be incorporated in the main nucleus and frequently contain high levels of DNA damage as assessed by γ-H2AX staining (Hayashi and Karlseder, 2013; Russo et al, 2015; Santaguida and Amon, 2015). Even though they can persist over generations as a distinct organelle, they eventually fuse with the main nucleus, which poses a threat for genome integrity. The processes of DNA replication and repair are defective within micronuclei, allowing mutations, chromosome breaks and rearrangements to accumulate.
Lastly, aneuploidy leads to metabolic alterations, and CIN has been associated with persistent OS (Kumari et al, 2014; Limoli et al., 2003; Oromendia and Amon, 2014). Accordingly, oxidative damage to the DNA (e.g., 8-hydroxy-2′-deoxyguanosine: 8-oxo-dG adducts) was shown to be a consequence of aneuploidy in the absence of DSB (Liet al, 2010). CIN can also result in RNA oxidative damage (Andriani et al, submitted), likely because this molecule is highly susceptible to attack by ROS and mechanisms for its repair or removal are so far unknown (Kong and Lin, 2010).
4. GIN-CIN complexity: a two sided coin
While GIN and CIN have been historically defined by two different terms, it is apparent that they are interconnected (Yurov et al., 2009b). GIN is a consequence of malfunction in DNA duplication, repair, genome distribution into daughter cells, defects in cell cycle progression, and checkpoint control (Shen, 2011). CIN is a cellular state with a higher propensity for chromosome mis-segregation. Just like CIN can cause DNA damage that is regarded as a main cause of GIN, it is often difficult to dissect causes and consequences of various forms of GIN. Accumulation of GIN, both at the nucleotide (Maynard et al., 2015) and chromosome level (Ricke and van Deursen, 2013; Yurov et al., 2014), is associated with neurodegenerative diseases (ND). Below we summarize succinctly the main types of GIN, how they are generated, their known correlation with CIN, and how they relate to aging and ND.
4.1. Nuclear and mtDNA mutations
Mutations, affecting single or few nucleotide bases, often result from DNA replication errors. Mutations can also occur byinaccurate repair of damaged bases resulting in irreversible modifications, which are known to accumulate with age (Dexheimer, 2013; Maslov and Vijg, 2009; Vijg and Suh, 2013). Many oxidative base lesions are mutagenic (Cooke et al., 2003), which is important in tissues sensitive to oxidative damage, such as the brain (Martin, 2008; Uttara et al., 2009).
Similar to nuclear DNA, discussed above in details, the mitochondrial genome can accumulate point mutations and/or large deletions. The mutation frequency of mitochondrial DNA (mtDNA) is much higher relative to nuclear DNA, likely due to its proximity to ROS and the limited availability of DNA repair enzymes (Khrapko and Vijg, 2009). Mitochondrial dysfunction caused by damage to mtDNA or constituent mitochondrial proteins has been associated with the aging process and ND (Table 1) (Greaves et al., 2012; Reeve et al., 2008). Experimental evidence supports a strong correlation between aneuploidy and mtDNA damage. Detailed studies have been performed in oocytes because of the well-known association between increased aneuploidy with maternal age. Significantly elevated levels of total mtDNA have been reported in aneuploid embryos when compared to control diploid embryos (Fragouli et al., 2015). Mitochondrial activity is essential for the assembly of the mitotic spindle and for chromosome segregation, and dysfunctional mitochondria have been implicated in higher error rates of chromosome miss-segregation (Coskun and Busciglio, 2012). Therefore, it is conceivable that mtDNA mutations may impair mitochrondrial function and, therefore, ultimately also affect chromosome segregation. Mutations in mtDNA play important causal roles in neurological disorders because of the high-energy metabolism of neurons (Carelli and Chan, 2014); thus we anticipate that like in oocytes, a direct association between mtDNA mutations and aneuploidy in the brain may be a contributor to neuronal dysfunction and neurodegeneration (ND).
Table 1.
Types of GIN and their association with aging, CIN and neurodegeneration.
| Type of GIN | Association with aging | Association with CIN | Association with ND |
|---|---|---|---|
| DNA mutations | Mutations accumulate with age, likely through reduction in efficiency of NER, BERand NHEJ repair pathways (Coppede and Migliore, 2010; Maslov and Vijg, 2009; Maynard et al., 2015). Age-related accumulation of DNA oxidation occurs in vitro and in vivo. Mutations in the NER pathway lead to premature aging disorders XP and CS (Cooke et al., 2003; Coppede and Migliore, 2010; Maynard et al., 2015). | Prolonged mitotic arrest generates γ-H2AX foci (Ganem and Pellman, 2012; Hayashi and Karlseder, 2013). Aneuploidy triggers unbalanced levels of proteins for genome maintenance (Holland and Cleveland, 2012b; Meena et al., 2015; Nicholson and Cimini, 2015). Cleavage furrow damage (Ganem and Pellman, 2012; Janssen et al., 2011). |
In vivo models of ND show DNA damage (mainly oxidative) as a common precursor of motor neuron death (Martin, 2008; Uttara et al., 2009). Mutations in genes involved in DNA repair or DNA-damage response are associated with neurodegenerative diseases, including XP, CS, TTD and AT (Coppede and Migliore, 2010; Martin, 2008). Increased DNA and RNA oxidation occurs in AD and PD brains, and high DNA oxidation in found in ALS samples (Coppede and Migliore, 2015). |
| mtDNA damage | mtDNA alterations accumulate during aging in some tissues. Deletions are associated with respiratory chain defects and they increase with age in the CNS (Reeve et al., 2008). mtDNA mutagenesis alone can cause degenerative age-related symptoms (Greaves et al., 2012). mtDNA mutations are associated with nuclear signaling pathways and influences the process of aging (Cha et al., 2015). |
Aneuploid cells have increased levels of mtDNA (Fragouli et al., 2015). Dysfunctional mitochondria have been associated with higher chromosome segregation errors (Coskun and Busciglio, 2012). | High levels of mtDNA deletions in neurons have been found in ND (Reeve et al., 2008). Mitochondrial dysfuntion induced by mtDNA mutations cause degeneration of dopaminergic neurons. Huntingtin-expressing and HD cells have higher levels of mtDNA damage (Cha et al., 2015). mtDNA damage is present in PD (Coppede and Migliore, 2015; Greaves et al., 2012; Reeve et al., 2008), AD (Cha et al., 2015; Coppede and Migliore, 2015), and ALS patients (Coppede and Migliore, 2015). |
| CNVs | Deletions are associated with mortality at old age (Kuningas et al., 2011; Nygaard et al., 2015). Deletion in Contactin-Associated-Protein-Like 4 negatively correlates with female survival at the age of 80 (Iakoubovet al., 2013). Negative association between CNV in Contactin-Associated-Protein-Like 2 is known healthy aging males (Iakoubovet al., 2015). |
CNVs target genes that control the mitotic cell cycle progression (Iourov et al., 2015). | Large deletions are associated with cognitive impairment in AD (Guffanti et al., 2013). CNV of glucose transporter SLC2A3 delays the onset of HD (Vittori et al., 2014). Triplication of the α-Synuclein locus is the cause of autosomal dominat familial PD, and CNVs in the Parkin gene are associated with recessive inheritance of PD (Singleton et al., 2003; Toft and Ross, 2010). |
| DMs | Limited reports. Increased frequency in murine aged kidney (Martin et al., 1985). No increase observed in in vitro aging of fibroblasts (Reis et al., 1985). |
Unclear. | Linked to therapy resistance: high frequency in neuroblastomas (Fan et al., 2011). Myc amplification in medulloblastoma, EGFR and other genes in glioma (Gebhart, 2005). 50% of high-grade astrocytomas and glioblastomas have DMs (Giollant et al., 1996). |
| Micronuclei | Age-related increase in micronuclei frequency has been associated with loss of sex chromosomes (Bolognesi et al., 1999; Bukvic et al.,2001; Guttenbach et al., 1994; Hando et al., 1994; Nath et al., 1995). Micronuclei are frequent in HGPS cells, premature aging disorders, and incidence increase with age (Bridger and Kill, 2004; Migliore et al., 2011). |
Can arise from mitotic spindle dysfunction (lagging chromosomes or from acentric chromosomal fragments) and can cause aneuloidy (Balmus et al., 2015; Cimini, 2008). They function as a mode to eliminate structurally abnormal or supernumerary gene copies (Ambros et al., 1997; Casati et al., 1995). | Peripheral lymphocytes from AD and PD patients and skin fibroblasts from AD and HD patients have increased frequency of micronuclei (Coppede and Migliore, 2015; Migliore et al., 2011). Neuroblastoma cell lines can actively elimininate extra copies of MYCN via micronuclei (Ambros et al., 1997). |
| Chromotripsis | Not defined association with aging or longevity. | Micronuclei have been proposed as an intermediate step leading to chromothripsis through the defective DNA repair mechanism and replication fork collapse present in micronuclei (Crastaet al., 2012; Rausch et al., 2012). | Occurs in 18% of high-stage neuroblastoma (Molenaaret al., 2012). Associated with gene amplification of receptor tyrosine kinases and modulators o TP53 and RB1 pathways in glioblastoma (Furgason et al., 2015). |
| Dysfunctional telomeres | Most human tissues but the bain, show significant telomere shortening during aging. Some age-related diseases are associated with telomere shortening (Jiang et al., 2007). DC patients carry mutation in telomerase (TERT and TERC) and present accelerated aging (Sahin and Depinho, 2010). Centenarians and their offspring maintain longer telomeres (Atzmon et al., 2010). | Bridge chromatids resulting from telomere fusion may be involved in missegregation events resulring in aneuploidy (Pampalona et al., 2010a). Aneuploidy may confer the ability to survive telomerase insufficiency (Millet et al., 2015). | No consistent relationship between telomere length and AD or PD but their shortening is associated with dementia and cognitive decline (Eitan et al., 2014). Short telomeres due to accelerated erosion have been reported in FA patients' leukocytes, and cerebellar tissues (Anjomani Virmouni et al., 2015). Telomerase reactivation reverses neurodegeneration phenotypes in aged telomerase-deficient mice (Jaskelioff et al., 2011). |
| Abnormal nuclear architecture | Premature aging disorders like HGPS are caused by mutations in LMNA or pre-lamins. Progerin, a truncated form of A-type lamin, accumulates with age (Gonzalo, 2014; Kudlow et al., 2007; Worman et al., 2009). | In vitro downregulation of lamin A/C cause aneuploidy (Capo-chichi et al., 2011a,b). | Accumulation of Huntingtin aggregates causes focal distortion of the nuclear envelope (Chapple et al., 2008). PD-associated mutation in LAMNA leads to abnormal nuclear architecture (Liu et al., 2012). Disruption of lamin A/C normal architecture is found in the nucleus in FXTAS (Arocena et al., 2005). |
4.2. Copy number variations (CNVs)
CNVs are alterations in genomic segments of 10-300 kb, often containing repeat elements with a high degree of identity to enable crossover and rearrangements through non-allelic homologous recombination. Their presence is linked to duplications, deletions, and/or translocations resulting in gene dosage imbalances, gene fusion and/or interruption. CNVs have been linked to replication fork stalling, retrotransposition, formation of non-B-DNA structures such as G-quadruplexes and CpG islands (Colnaghi et al., 2011; Zhang et al., 2009). CNVs are widespread in the human genome and represent a major source of genomic variation (Iafrate et al., 2004, Sebat et al., 2004). This holds true also for the brainwhere as many as 41% of neurons have been found exhibiting at least one megabase of de novo CNVs (McConnell et al., 2013) with studies suggesting that some CNVs may be shared by multiple neurons in both normal and pathological conditions (Cai et al., 2014). CNVs have been implicated in the pathogenesis of Neurodegenerative Diseases (NDs) with alterations being linked to Parkinson's Disease (PD) (Polymeropoulos et al., 1996), Alzheimer's Disease (AD) (Goate et al, 1991), Mental Retardation (MR), Autism, and Schizophrenia (Shi et al, 2009; Guffanti et al, 2013; Toft and Ross, 2010; Vittori et al., 2014) (Table 1).
A link between CNVs and Somatic Copy Number Alterations/Aberrations (SCNAs), which are changes in copy number that have arisen in somatic tissue, has emerged from studies of Iourov and colleagues who examined the frequency of CNVs mapping to the cell cycle pathway within a cohort of patients with intellectual disability, autism, and/or epilepsy (Iourov et al., 2015). The rationale for this study was based on the knowledge that somatic mosaicism is likely the consequence of alterations in cell division and genome maintenance pathways (as described in Section 2). CNVs assessed in 124 genes that control the mitotic cell cycle progression suggests that 71% of the 225 samples analyzed harbored CNVs in these pathways (mostly one copy per individual but with some cases revealing up to four CNVs per locus with some subjects including CNVs mapping to more than one locus). These findings suggest that CNVs act as a contributor to chromosome instability and somatic mosaicism. It is tempting to speculate that CNVs affecting genes involved in the control of the cell cycle may be responsible, at least in part, for the generation of somatic aneuploidies.
4.3. Double-minute (DM) chromosomes, homogeneously staining regions (HSRs), micronuclei, and chromothripsis
At the chromosome level, structural modifications, such as translocations, deletions, insertions, inversions, breaks, sister chromatid exchanges, chromothripsis and shortening of telomeres are considered forms of GIN. Alterations in the numbers of whole chromosomes (aneuploidy and polyploidy, with tetraploidy a common form of polyploidy) can result in the formation of micronuclei, which is also a form of GIN (Langie et al., 2015; Russo et al, 2015; Vijg and Suh, 2013). CIN defines cellular states in which cells are prone to miss-segregation of chromosomes or structural chromosome rearrangements (Ricke et al., 2008).
DMs are small autonomous circular chromatin bodies of a few mega-base pairs in size that replicate and segregate in the absence of centromeres or telomeres. HSRs are highly repetitive heterochromatic chromosomal regions that stain uniformly in G-banding. DMs and HSR are frequently observed in solid tumors with variable numbers per cell. They generally contain hundreds of copies of oncogenes and genes involved in drug resistance (Barker, 1982; Cowell, 1982; Hahn, 1993). For instance, MYC and MYCN amplification in the form of DMs are frequently found in medulloblastoma and neuroblastoma, while DM amplification of EGFR is associated with gliomas (Gebhart, 2005) (Table 1). The molecular mechanisms for their origin are not fully understood, but it is hypothesized that DNA DSBs that form in close proximity of amplified regions become miss-repaired, resulting in Breakage-Fusion-Bridge (BFB) cycles that eventually lead to DMs. Premature chromosome condensation has also been suggested as a driver of DMs (Gebhart, 2005; Hahn, 1993).
Micronuclei, extranuclear segments of chromatin, can arise from unrepaired DNA breaks but also from mitotic spindle dysfunction and are a recognized form of GIN (Balmus et al, 2015). They can be generated from lagging chromosomes or from acentric chromosomal fragments and are known to cause aneuploidy (Cimini, 2008). Micronuclei frequency increases with age and its occurrence was shown to be associated with loss of sex chromosomes in human lymphocytes, resulting in aneuploidy (Bolognesi et al, 1999; Bukvic et al., 2001; Guttenbach et al., 1994; Hando et al., 1994; Nath et al, 1995). Micronuclei was also demonstrated as a mode to eliminate chromosomes with structural rearrangements (Casati et al., 1995), or removal of supernumerary gene copies (like DMs) (Ambros et al, 1997). Because replication within the micronucleus is asynchronous with the nucleus, enclosed chromosomes tend to shatter during condensation and the resulting fragments are repaired by NHEJ. Disassembly of the micronuclear envelope in the subsequent cell cycle allows incorporation of a heavily rearranged chromosome into the nuclear genome. Higher incidence of micronuclei correlates with ND (Table 1) (Holland and Cleveland, 2012a).
Chromothripsis is a form of GIN that comprises tens to thousands of chromosomal rearrangements that localize to a limited number of genomic regions on single or few chromosomes (Stephens et al, 2011). Chromosomes are shattered into pieces and as a consequence, inaccurate repair of DSBs by NHEJ can lead to highly derivative chromosomes. This phenomenon can be caused by IR, BFB cycles, telomere dysfunction, and premature chromosome condensation within micronuclei (Crasta et al., 2012; Leibowitz et al., 2015; Rode et al., 2015; Zhang et al, 2015). In the CNS, high rates of chromotripsis have been found in neuroblastoma (Molenaar et al., 2012), glioblastoma (Furgason et al, 2015) and medulloblastoma (Rausch et al, 2012) (Table 1). While NHEJ and microhomology-mediated break-induced replication (MMBIR) have been proposed as the mechanisms leading to chromothripsis (Forment et al, 2012), it has recently been suggested that micronuclei could be an important step leading to chromothripsis through the defective DNA repair mechanism and replication fork collapse present in micronuclei (Crasta et al., 2012; Rausch et al., 2012). Therefore, based on these reports, one could reason that defects in chromosome segregation may trigger chromothripsis via GIN induced by micronuclei.
4.4. Telomere dysfunction
Dysfunctional telomeres can lead to dicentric chromosomes that undergo BFB cycles (Artandi et al, 2000). These highly-unstable chromosomes containing two centromeres are pulled to opposite spindle poles leading to chromatin bridge formation. During cytokinesis, mitotic spindle forces and the cytokinetic ring closure can cause breakage of dicentric chromosomes at multiple sites, and subsequent fusion created by NHEJ reinitiates the BFB cycle leading to continuous accumulation of rearrangements (Holland and Cleveland, 2012b; Rode et al, 2015; Russo et al., 2015). Telomere shortening is associated with the functional decline that accompanies aging (Atzmon et al., 2010; Jiang et al., 2007; Sahin and Depinho, 2010) and some studies suggests their involvement inND (Table 1) (Anjomani Virmouni et al., 2015; Eitan et al., 2014; Jaskelioff et al., 2011).
Telomere erosion and loss of telomere capping function triggers BFB cycles (McClintock, 1941), but experimental evidence suggests a broader link between GIN and telomere dysfunction. In fact, in human breast cancer, telomere-based BFB events concur in concomitance with a burst of chromosomal instability that is associated with the transition from benign to malignant cancers (Chin et al, 2004). Evidence generated using in vitro models supports the hypothesis that the origin of cytogenetic alterations may be concurrent with telomere crisis (Chin et al., 2004). DNA fragmentation may not be the only outcome of BFB cycles. In vitro studies suggest that bridge chromatids resulting from telomere fusion may be more frequently involved in mis-segregation events than chromosomes of normal telomere length (Pampalona et al., 2010b). This may be possible assuming that the forces generated by pulling anaphasebridges toward opposite poles are sufficient to detach a chromosome from the microtubules of one or both spindle poles. Therefore, telomere-driven instability can promote not only the appearance of chromosomal rearrangements but also W-CIN. Interestingly, newly reported evidence suggests that, at least in yeast cells, aneuploidy may confer, through a stress adaptation mechanism, the ability to survive telomerase insufficiency (Millet et al, 2015).
4.5. Nuclear envelope (NE) defects
The nuclear envelope (NE) is a highly regulated membrane barrier that separates the nucleus from the cytoplasm in eukaryotic cells. Its main components, A-type and B-type lamins and lamin-associated proteins, function as genome caretakers. A variety of human diseases have been associated with mutations in lamins and lamin-interacting proteins (termed laminopathies) and they have in common an abnormal nuclear morphology and increased GIN (Scaffidi and Misteli, 2008; Worman et al, 2009). Hutchinson-Gilford Progeria Syndrome (HGPS), caused by mutations in LMNA that generate an abnormal protein called progerin, which is also found accumulated in old healthy individuals (Coppede and Migliore, 2010; Pollex and Hegele, 2004). Besides structural scaffolding functions, A-type lamins have also been implicated in regulation of DNA replication and transcription, DNAdamage repair (NHEJ and HR), telomere homeostasis and dynamics. Alterations in A-type lamins interfere with the DNA damage response and specific repair mechanisms such as impaired recruitment or transcription regulation of downstream effector proteins (Gonzalo, 2014). Loss of lamin A/C expression is often found in cancer cells (Ho and Lammerding, 2012), where a direct link between nuclear envelope structural defects and W-CIN has been established. In vitro down-regulation of lamin A/C in non-cancerous primary ovarian or breast epithelial cells using shRNA resulted in increased nuclear size, ane-uploidy and polyploidy (Capo-chichi et al., 2011a,b). Abnormalities in the nuclear envelope have been associated with ND. However, to our knowledge nothing is known as yet of nuclear envelope defects and their possible correlation with aneuploidy (Table 1) (Arocena et al., 2005; Chapple et al., 2008; Liu et al., 2012).
5. Polyploidy in the brain
Chromosome segregation defects have been proposed as an underlying mechanism leading to aneuploidy in the developing brain (Westra et al., 2008; Yang et al., 2003). However, neurogenesis in the adult brain is limited and restricted, under normal physiological conditions, to the subgranular zone (SGZ) and the subventricular zone (SVZ) (Gage, 2000; Paton and Nottebohm, 1984). Therefore, the age-related accumulation of aneuploidy in the brain cannot be simply explained by mitotic defects. As described in Section 2, cell fusion is a physiological process in mammalian cells (Davoli and de Lange, 2011; Faggioli et al., 2008; Ganem et al., 2007; Holland and Cleveland, 2012b). Experimental evidence suggeststhat cell fusion occurs in the brain, which could contribute to the accumulation of aneuploidy in post-mitotic tissues. Evidence for this phenomenon stems from observations dating to the mid 1960s, suggesting the presence of hyperdiploid Purkinje cells (Lapham, 1968; Nern et al., 2009). It was later demonstrated that Purkinje cells have the ability to fuse with BMDCs and generate bi-nucleated Purkinje-like cells (Alvarez-Dolado et al., 2003; Weimann et al., 2003). Using parabiosis experiments connecting a wild-type recipient and a transgenic ubiquitously expressing GFP donor, Johansson and colleagues showed that transplanted BMDCs fuse with Purkinje cells with a surprisingly high frequency (∼30 folds) in a model of autoimmune encephalitis. Interestingly, the genetic programming of Purkinje neurons seems to be predominant in the bi-nucleated Purkinje/BMDCs heterokaryons, as suggested by the evidence that fused cells repress the transcription of genes of the haematopoietic lineage (Johansson et al., 2008; Weimann et al., 2003).
Bi-nucleated neurons have been described in a variety of human CNS pathologies, including AD, neuro-Behcet's disease, multiple sclerosis, Kuru, and spino olivo-ponto-cerebello-nigral atrophy (Kemp et al., 2014). In the case of AD it has been proposed that neuronal death is the consequence of an abortive cell cycle, possibly with intermediate tetraploid cells. However, experimental findings to support this hypothesis are controversial (Iourov et al., 2011). While cell cycle-related nuclear proteins (Ki-67) have been reported with increased frequency in the dentate gyrus of the hippocampus of AD patients versus controls (Nagy et al., 1997), interphase FISH studies suggest that the frequency of tetraploid nuclei in AD and control brains is similar (Westra et al., 2009). In addition, tetraploid nuclei were exclusively non-neuronal, contrasting with an absence of tetraploid neurons in AD patients.
A large body of evidence now suggests that ploidy changes play an important role not only during physiological development and disease, but also as a driver of tissue degeneration. While this field is expanding, more work is required to elucidate how mechanisms that control genome stability become altered and lead to CIN in the CNS. Moreover, the functional consequences of CIN at the single cell level and in the context of whole tissues and organisms are only now beginning to be understood.
6. Whole chromosome aneuploidies in the brain during aging and age-associated diseases
In the next three sections of this review we will describe the work associating aneuploidy with tissue degeneration and diseases with a particular focus on the brain (see also Table 2).
Table 2.
Types of CIN and their association with aging and neurodegeneration.
| Type of GIN | Association with aging | Association with ND |
|---|---|---|
| CIN, W-CIN, s-CIN | Age-related accumulation of ploidy changes have been reported inseveral mammalian tissues: lymphocytes (Jacobs et al., 1963), liver(Faggioli et al., 2011a), brain (Faggioli et al., 2011b; Faggioli et al., 2012), vascularsmooth muscle cells (Jones and Ravid, 2004), oocytes (Jones, 2008) andkeratinocytes (Zanet et al., 2010). Fibroblasts from premature aging syndromes have increasedaneuploidy (Mukherjee and Costello, 1998). Some CIN mouse models show premature aging phenotypes (Bakeret al., 2004; Bakeret al., 2006; Matsuyama et al., 2013; Tanaka et al., 2015), andprotection against aneuploidy by overexpression of BubR1 extendslifespan (Baker et al., 2013). |
Increased CIN can result in ND, as stochastic aneuploidy is present in the cerebellum and the cortex of AT patients (Coppede and Migliore, 2015; Iourov et al., 2009a,b). Increased aneuploidy in AD samples (Mainly chromosomes 18, 21, and X) (Geller and Potter, 1999; Iourov et al., 2009b; Yurovet al., 2014). Some CIN syndromes are associated with ND or neuropathology: AT, NBS, XP, TTD, CS, FAN, WS, BS and RTS (Mathur et al., 2000; Rolig and McKinnon, 2000). |
Studies aimed to better understand mechanisms underlying the generation of neuronal variability and complexity have led to the discovery that the brain is essentially a mosaic tissue composed of euploid and aneuploid neural cells (Faggioli et al., 2011b; Yurov et al., 2007). Structural genomic variation of the neuronal genome has long been proposed as a pivotal mechanism underling neuronal diversity. The first evidence for the presence of aneuploidycells in developing neurons came from studies by Rehen et al., who found by using a combination of FISH and SKY approaches that in the mouse ∼33% of neuroblasts analyzed between E11 and E17 underwent defective mitosis and were aneuploid for one or more chromosomes, the far majority of which were hypodiploid (Rehen et al., 2001). Building on these initial observations several other important discoveries in the field of brain aneuploidy were generated, suggesting that a surprisingly high number of neural cells may be aneuploid (38-47%) (Pack et al, 2005). First, it was observed that neural progenitor cells (NPCs) developing along the ventricular zone (VZ) of E12 and E14 embryos had supernumerary centrosomes, and their increased presence correlated with aneuploidy (∼33% of the analyzed cells) (Yang et al, 2003). Next, in the same year, Yourov's and Rehen's groups, using FISH and/or whole chromosome paint probes targeting several human chromosomes, demonstrated that aneuploid neurons are not a unique characteristic of murine tissues, but are also found in the developing and adult brain of humans at various levels (Yurov et al., 2005). Rehen and colleagues generated a map of mosaic aneuploidy across different brain regions: frontal cortex, occipital cortex and hippocampus, reporting varying levels of aneuploidy, ranging from 3.2 to 5.2% (Rehen et al., 2005). This study also provided a first clue that accumulation of mosaic aneuploidy may be age dependent (4.8% and 5.2% of cells aneuploid for chromosome 21 were observed in hippocampal cells of a 77 and 86 year old versus 3.2% observed in 2 y.o.). Understanding the mechanisms underlying the generation of neuronal variability and complexity remains a fundamental question in neuroscience. Therefore, the discovery of exceptionally high levels of mosaic aneuploidy in the developing nervous system prompts the question as to whether aneuploid neurons may be functionally active and integrated into brain circuitry. Kingsbury and colleagues used retrograde neuronal tracer techniques combined with paint for chromosome X and Y to demonstrate that aneuploid neurons express markers of functionally active cells (Egr-1 and c-Fos), therefore suggesting that neurons aneuploid for these two chromosomes are integrated in the brain circuit (Kingsbury et al., 2005).
The notion that aneuploidy is not merely a feature of the developing brain but that it accumulates during normative aging has now been suggested by several studies, with similar observations also in other tissues (Faggioli et al, 2012; Jacobs et al., 1961; Jones, 2008; Jones and Ravid, 2004; Yurov etal., 2009a; Zanetet al, 2010). In fact, a genome-wide study on blood and buccal swabs of cancer-free control individuals revealed a surprisingly high frequency of aneuploidy or copy-neutral LOH increasing with age (from 0.23% under 50 years up to 1.91% between 75 and 79 y.o.) (Jacobs et al., 2012). Aneuploidization is generally considered a deleterious process; indeed, aneuploid cells show defects in cell cycle progression and have altered metabolic properties (Sheltzer and Amon, 2011; Williams et al., 2008). There are, however, exceptions. Aneuploidy is well known to occur in disease-free tissues like the liver, where hepatocytes have a very complex ploidy status with a high frequency of cells deviating from the normal ploidy, also against a background of a polyploid genome (Duncan et al., 2012a, 2010; Faggioli et al., 2008, 2011a). It has been suggested that this high deviation from diploidy in hepatocytes may grant the tissue a selective advantage against a variety of assaults and stresses (Duncan et al, 2012a). Aneuploidy is therefore emerging as a phenomenon more common and complex than originally thought and potentially occurring in dividing as well as non-dividing cells (Mosch et al., 2007).
Brain aneuploidy is associated with disease and numerous studies suggest its involvement in the etiology of NDs (Coppede and Migliore, 2015; Kormann-Bortolotto et al., 1993; Potter, 1991; Rolig and McKinnon, 2000). Chromosome 21 has been reported as a prevalent aneuploidy in the brain and in peripheral tissues of ADpatients (Iourov et al., Migliore et al., 2006). This is concordant with the knowledge that nearly all Down's syndrome (DS) individuals older than 50 years of age contain plaques and tangles within their brains that are similar in form, number, and distribution to those seen in AD patients (Mann and Esiri, 1988). Of the other autosomes that have been tested so far in AD, chromosome 18 and X, in addition to 21, appear to have slightly higher aneuploidy frequency in AD patients compared to controls (Geller and Potter, 1999; Yurov et al., 2014). The Amyloid Beta (A4) Precursor Protein (APP) maps to chromosome 21, making mosaic aneuploidies resulting in gain of this specific chromosome a likely cause of increased synthesis of this protein, and therefore increased production of Aβ42 peptide that possibly responsible for pathological changes in the AD brain (Oyama et al, 1994).
Aneuploidy has also been found associated with CIN in Ataxia Telangiectasia (AT). Iourov and colleagues, using a combination of mFISH and chromosome paint probes, reported that the mean level of aneuploidy in the AT cerebral cortex was three-fold higher when compared to age- and sex-matched controls. In sum, these data suggest that 2-50% of cells in the cerebral cortex of AT patients may be aneuploid for one or more chromosomes (Iourov et al, 2009b; Yurov et al., 2009a).
Although aneuploidy seems to be linked to NDs, it is not clear yet how ploidy changes would occur in post-mitotic cells, such as neurons. One possibility is that neurons degenerate because they re-enter a lethal cell cycle. This is supported by the presence of FISH signals indicative of large genomic regions suggestive of ongoing replication in hippocampal pyramidal and basal fore-brain neurons of AD patients relative to age matched controls (Yang et al., 2001). Another possibility is the presence of genomic variation in genes that control, for example, chromosome segregation. This is suggested by the observation that young (<35 years of age) mothers of DS children have a statistically significant increase of micronucleated lymphocytes, a phenomenon that is indicative of the predisposition of these women to chromosomal nondisjunction leading to mosaic trisomy 21 (Migliore et al., 2006).
7. Structural genomic variation in the disease-free adult brain
Studies aimed to analyze mosaic GIN requires analysis at the single cell level because of the heterogenic nature of the alterations, which are often unique to one or few cells within a tissue ororgan. Karyotyping techniques are not well suited for the study of non-proliferating cells (Faggioli et al, 2014). Interphase FISH approaches enable the analysis of genomic variation at the single cell level in large number of cells in a relatively short period of time, and have been instrumental to the groundbreaking discoveries that opened the field of mosaic aneuploidies in the brain (Fig. 4). However, they have a limitation given that they allow the analysis of only few pre-selected loci. This feature limits the opportunity to study CNAs across all chromosomes at once and also to analyze segmental (i.e., across a portion of the chromosome) copy number alterations. The most suitable approach to overcome these limitations is whole genome single cell DNA sequencing (Gundry et al., 2012). Over the past few years ultra-low whole genome sequencing (∼0.1-0.5× coverage) has become a reliable method for the measurement of GIN at the level of whole chromosome aneuploidies or for the detection of large regions of copy number imbalance (Xie et al, 2013). Due to the development of techniques for single cell DNA amplification, these approaches are now emerging as effective tools for the analysis of CNVs. This, combined with a reduced cost of genome sequencing (Caulfield et al, 2013), enables the analysis of a large number of cells representative of the genomic heterogeneity that may be present in tissues. McConnell and colleagues performed aneuploidy analysis using Next Generation Sequencing (NGS) approaches to study neuron-derived human-induced pluripotent stem cells and human postmortem frontal cortex neurons (McConnell et al., 2013). Somatic sub-chromosomal deletionsand amplifications were mostly unique to each neuron, suggesting that in these cells CNVs were not clonal. Of the 116 neurons analyzed, 41% had frequent somatic CNVs up to 75 Mb, with three cases presenting large copy number alterations affecting more than 50% of the chromosome. The ability to analyze CNVs in the genome at the single cell level allows the future understanding of the biological implications of aneuploidy. For example, CNVs were found more frequently at telomeric regions but seemed not to affect other genomic features important for genome stability (such as transposons, segmental duplications or fragile sites). Losses were twice as common than chromosomal gains. Interestingly, it appears that a small percentage of cells (∼15%) contain a high frequency of CNVs (73% of all the detected alterations), suggesting that mosaic aneuploidy, at least in neurons, targets a relatively low number of cells but at high frequency (McConnell et al., 2013) (Fig. 5).
Fig. 4.
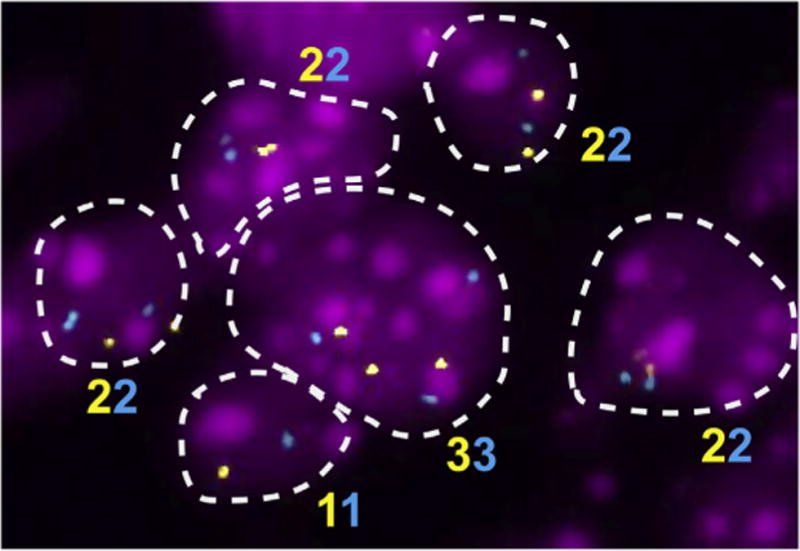
Aneuploidy assessed by four-color interphase FISH in E13.5 embryonic cerebral cortex of WT animals. Interphase FISH is a quantitative and sensitive approach for the analysis of ploidy at the single-cell level, irrespective of the proliferating status of the tested tissue. Examples of diploid and aneuploid cells are highlighted by the dotted line. Ploidy is shown as assigned by the colors of the fluorophores used to label FISH probes.
Fig. 5.
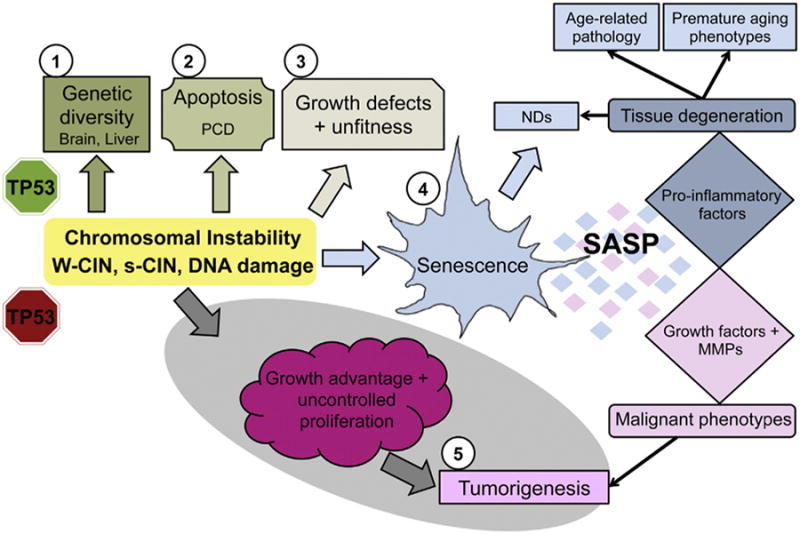
General model of functional consequences of CIN. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) When TP53 is present and functional (top portion of scheme), CIN and CIN-associated DNA damage triggers different types of responses depending on the cell type and on the severity of abnormalities. (1) Aneuploidy provides genetic diversity during neuronal maturation and also selective advantage against stresses in hepatocytes. (2) High CIN has been associated with increased apoptotic frequency and PCD during neural development likely to eliminate cells with abnormal DNA content. (3) Aneuploidy slows proliferation rate and it is detrimental to cell and organismal fitness. (4) CIN generation in vitro and in vivo has also been linked to the induction of senescence. Senescent cells have the ability to change their microenvironment through the secretion of SASP, which comprises several signaling molecules, pro-inflammatory and growth factors. The inflammation generated upon SASP expression has been implicated in the pathology of age-related diseases, early onset of premature aging phenotypes and also in NDs such as AD and PD. Cellular senescence is also thought to contribute directly to NDs because it causes neural stem cell depletion in the adult (blue arrow). In circumstances where TP53 is not functional (bottom portion of scheme), CIN can be a driver of tumorigenesis due to gene dosage imbalances or due to the higher propensity to generate GIN in these cells. In both cases the outcome is growth advantages relative to diploid counterparts. Cells prone to transformation can acquire malignant phenotypes upon interaction with the SASP (i.e., epithelial to mesenchymal transition, increased invasiveness and motility). Induction of senescence in the brain by elevated CIN provides a possible unifying mechanism to explain age-related increase in inflammation and neurodegeneration, as well as the higher incidence of brain tumors that is associated with age (5).
The power of NGS has recently enabled a better characterization of somatic retrotransposition in the brain, a mechanism long proposed as a source of genotypic variation among neurons (Coufal et al, 2009). Upton and colleagues, using single-cell retrotransposon capture sequencing (RC-seq) for the analysis of hippocampal neurons, glia, and cortical neurons (Upton et al, 2015), found that hippocampal neurons have a high frequency of L1 mosaicism and reported an average of 13.7% somatic L1 insertions per hippocampal neuron. This frequency is surprisingly high and agrees with a previously reported estimate of 80 insertions per brain cell (Coufal et al., 2009). However other studies have estimated somatic L1 insertions to be <0.6 per neuron (Evrony et al, 2012). A clear explanation for such a significant discrepancy is lacking and underscores the need for additional studies. Regardless of frequency and extent of L1 retro transposition activity, the ability to map L1 insertion sites with accuracy in somatic cells will soon shed light on mechanisms of neuronal function in the adult brain. In particular, the observation that L1 sequences may preferentially insert near protein-coding regions and highly transcribed genes suggests that they have the potential, like structural CNVs, to alter proper function of the adult brain (Upton et al., 2015).
8. Considerations on functional consequences of genomic instability
Murine models of aneuploidy support the hypothesis that ploidy changes contribute to tissue degeneration and premature aging phenotypes (Baker et al., 2004; Baker et al., 2006; Matsuyama et al., 2013; Tanakaet al., 2015). Indeed, mice expressing ∼10% of normal levels of the BubR1 protein (BubR1 hypomorphic), progressively accumulate aneuploidy in the spleen, develop a series of age-related pathologies and have reduced lifespan (Baker et al., 2004). Surprisingly, we have recently reported that the adult brain of these segmental progeroid mice contains similar levels of aneuploidy as age-matched controls (∼1%), suggesting that not all aspects of normative aging are recapitulated or that high levels of aneuploidy may be present during development and are eliminated later in life. Indeed, during embryonic development (E13.5), the cerebral cortex of BubR1 hypomorphic mice contained significantly higher levels of aneuploid and TUNEL positive nuclei than WT, suggesting that these mice generate high levels of CIN cells that possibly trigger extensive cell death or activate an immune response to prompt their elimination from the tissue (Andriani et al., 2016). These findings are in agreement with the hypothesis that one function of the programmed cell death (PCD) observed in the mammalian developing CNS is the protection against GIN (Bushman and Chun, 2013; Yurov et al., 2007). Interestingly, inhibition of PCD results in hyperplasia and higher aneuploidy (Peterson et al., 2012). Thus, removal of cells deviating from diploidy might be an important mechanism to maintain tissue homeostasis and genomic stability, because CIN can generate further DNA damage that possibly induce apoptosis, senescence or transformation. Accordingly, aneuploidy is compatible with life, yet at the cost of fitness, as clearly demonstrated by the survival of embryos with specific germline trisomies (14,18 and 21 in humans) and from individual carriers of the mosaic variegated aneuploid syndrome (MVA), a rare autosomal recessive disorder characterized by mosaic aneuploidy, predisposition to cancer and progeroid features (Jacquemont et al., 2002). MVA patients have mutations in the SAC components BUBR1 or CEP57, resulting in chromosome miss-segregation and thus aneuploidy.
That aneuploidy plays a role in aging is a relatively new discovery in the field, and the extent of this phenomenon, the complexity of aneuploidy and the chromosomes affected remains largely unknown. Aging is accompanied by a gradual impairment of cognitive functions, such as attention and memory (Glisky, 2007), and it remains to be determined if aneuploidy contributes to these outcomes. Most aneuploidy syndromes are associated with brain phenotypes that affect learning, language and behavior (Dierssen et al., 2009). Likewise, DS's patients, harboring an extra copy of chromosome 21, undergo memory loss and neurodegeneration with age (Lockrow et al., 2012). Interestingly, mice trisomic for genomic regions associated with DS exhibit DS-related neurological defects and impaired cognitive behaviors, likely due to gene dosage-imbalances that deregulate multiple pathways (Pereira et al., 2009; Yu et al., 2010). In addition, buccal cells isolated from individuals diagnosed with mild cognitive impairment or AD were found containing significantly higher DNA content than controls (Francois et al., 2014). Collectively these findings suggest a causative link between aneuploidy and age-related changes in memory and cognition, but more studies are necessary to establish a definitive link and to shed light on the mechanisms involved in this process.
The presence of somatic mosaicism relative to ploidy changes has been known for several decades. It was primarily reported in cases of mental retardation and affecting sex chromosomes (Ford et al., 1959), and later confirmed by genome-wide studies (Jacobs et al., 2012; Machiela et al., 2015). However, because a detailed characterization of ploidy changes and other forms of GIN in somatic tissue requires a single cell approach, this field of investigation remains in its infancy. Some studies are emerging suggesting widespread ploidy changes in the brain (McConnell et al., 2013), but others are contradictory (Knouse et al., 2014), suggesting that aneuploidy in somatic tissues may be present at lower levels than previously anticipated. This represents an emerging field associated with technical challenges. In fact, detection of CNVs in single nuclei is inherently challenging, since all methods for analysis of single cells require DNA amplification, which can be notoriously error-prone.
These results provoke a discussion about the severity of aneuploidy levels and functional consequences. On one hand, aneuploidy is associated with impaired fitness (Thorburn et al., 2013; Torres et al., 2008; Williams et al., 2008), as well as with early onset of senescence and premature aging-phenotypes (Baker et al., 2004; Baker et al., 2006). On the other hand, aneuploidy is a feature of most tumors and has been proposed as a driver of tumorigenesis (Boveri, 1902; Holland and Cleveland, 2012b; Ricke et al., 2008). It seems, therefore, that these two aspects contradict each other. In vitro transformation of aneuploid cells is associated with loss of function of the TP53 pathway (Soussi, 2010), which is also the most common altered pathway in tumors (2013) and its inactivation believed to be permissive for aneuploidy and GIN. Recent evidence also suggests a direct role of TP53 in regulating aneuploidy by prompting centrosome separation and, thus, when down-regulated, resulting in the formation of anaphase bridges and lagging chromosomes (Nam and van Deursen, 2014). Yet, aneuploidy in a murine model of induced aneuploidy by inactivation of a component of the chromosome segregation machinery (centromere protein E) suggests that while aneuploidy can promote tumorigenesis, high levels in certain tissues have tumor protective functions (Weaver et al., 2007). Solid tumors are characterized by specific patterns of chromosome gain and loss and, therefore, specific chromosomes may be of particular relevance for transformation in certain tissues, cell types or specific developmental stages (Forozan et al., 1997; Ried et al., 1995). In fact, in vitro experiments support this hypothesis (Santaguida and Amon, 2015; Williams et al., 2008).
Nevertheless, aneuploidy is not necessarily prejudicial to the organism. Studies in mature hepatocytes revealed that specific loss of chromosome 16 may function as an adaptive mechanism against chronic liver injury, suggesting that aneuploidy can be advantageous in pathological conditions (Duncan et al., 2012a). In the brain, aneuploid cells were shown to be capable of differentiating into neuronal and glial lineages and to constitute active neural circuitry, suggesting that they provide cellular diversity to the brain without impairing its functionality (Kaushal et al., 2003; Rehen et al., 2001). Therefore, it may be difficult to dissect the balance between a beneficial effect of aneuploidy and a detrimental outcome, such as tissue degeneration and cancer. Perhaps the presence of functional TP53 is the first determinant of cell fate upon aneuploidy/polyploidy generation, which can result in apoptosis, growth defects and senescence arrest. When TP53 is absent or mutated, non-diploid cells can proliferate without checkpoint activation and thrive, which could facilitate cancer initiation (Li et al.,2010; Thompson and Compton, 2010). However, most senescent cells activate a Senescence-Associated Secretory Phenotype (SASP) that affects neighboring cells by creating a pro-inflammatory tissue milieu, which contributes to aging-related pathologies and promote malignant phenotypes and tumor growth in vivo (Coppe et al., 2010). Recent evidence suggests that cellular senescence is a potential contributor to the age-related inflammation observed in the brain through the secretion of SASP factors and/or by abrogating neurogenesis during adulthood. Microglia undergo telomere shortening with aging and microglia-mediated up-regulation of SASP components is associated with normal brain aging and age-related NDs (Chinta et al., 2013; Chinta et al., 2015; Tan et al, 2014). Senescent-like phenotypes have also been observed in Purkinje, cortical and hippocampal neurons in response to DNA damage (Tan et al., 2014). Expression of both p16 and SASP metallopeptidase MMP3 were increased in astrocytes from aged and AD autopsied human brain tissues relative to young healthy controls, and higher amounts of p16 and γ-H2AX positive cells were also observed in PD samples. These findings suggest that senescence can occur in the mammalian brain and in a manner that makes it more prominent at old age and in ND (Chinta et al., 2015). In this view, senescence induction in the brain, through accumulation of GIN could be a unifying mechanism that may explain age-related inflammation and neurodegeneration, as well as the age-related increase in brain tumor incidence (Flowers, 2000).
Apart from the functional consequences one must also consider the mechanisms that lead to accumulation of somatic ploidy changes during aging. One hypothesis is that aging may be associated with a relaxation of the control of components of the chromosome segregation machinery, as demonstrated for oocytes (Leland et al. 2009), or with the accumulation of oxidative damage as suggested above. This would lead to the stochastic establishment of aneuploidy with a random pattern of chromosome gains and losses that could undergo selective pressure similar to what is observed in cancer cells. Indeed, our studies support this hypothesis suggesting that, at least in the cerebral cortex, aneuploidy may be chromosome-specific with higher frequencies observed for chromosomes 7,18 and Y (Faggioli et al., 2012).
9. Summary and future prospects
A large body of work demonstrates that mosaic aneuploidy resulting in tissues containing mixed diploid, aneuploid and euploid cells throughout an organism is not merely a feature of disease. While now a generally accepted phenomenon, this discovery is relatively recent and therefore represents a growing field of investigation. Several important questions still remain unanswered, while other are only beginnings to be explored. First, the extent in term of frequency of non-diploid cells, the number of chromosomes affected, the tissues and the cell subtypes affected within an organism remains largely unknown. While these studies would be descriptive, this knowledge would be important to understand the extent of this occurrence and vital to suggest possible links to functional consequences. In this regards technological advances are instrumental to tackle this problem. The establishment of improved murine lines, especially in a background of lineage tracing model, make it now possible to access enriched subpopulation of cells from genetically engineered mice in a variety of models of GIN and W-CIN. Because mosai c aneuploidy studies require the analysis at the single cell level, genome wide approaches like ultra low-coverage sequencing will be at the forefront to advance this field. Next a better understanding of the mechanisms responsible for mosaic aneuploidies, especially in the CNS, will be valuable to design new models to study the functional consequences at the cell, tissue and whole organism levels.In this respect the ultimate question as why mosaic aneuploidy is more prominent in some tissues or under certain conditions, why some tissue or cell subtypes tolerate aneuploidy better than other, still remains largely unexplored.
Acknowledgments
Funding: CM. and J.V. work is partially supported by grants from the National Institutes of Health ‘AG017242 to J.V’ and from the Albert Einstein Cancer Center Support Grant of the National Institutes of Health ‘P30CA013330’.
Abbreviations
- AD
Alzheimer's disease
- ALS
amyotrophic lateral sclerosis
- AT
ataxia telangiectasia
- BER
base excision repair
- BFB
breakage-fusion-bridge
- BMDCs
bone marrow-derived cells
- BS
Bloom syndrome
- CIN
chromosome instability
- CNS
central nervous system
- CPC
chromosome passenger complex
- CRC
colorectal cancer
- CS
Cockayne's syndrome
- DC
dyskeratosis congenita
- DS
Down's syndrome
- DSB
double-strand break
- DM
double-minute chromosome
- DNMT
DNA methyltransferases
- FA
Friedreich's ataxia
- FAN
Fanconi anemia
- FISH
fluorescence in situ hybridization
- FXTAS
fragile X-associated tremor/ataxia syndrome
- GIN
genomic instability
- HAT
histone acetyltransferase
- HD
Huntington's disease
- HDAC
histone deacetylases
- HGPS
Hutchinson-Gilford progeria syndrome
- HR
homologous recombination
- HSR
homogeneously staining region
- IR
ionizing radiation
- MMR
mismatch repair
- MR
mental retardation
- mtDNA
mitochondrial DNA
- MVA
mosaic variegated aneuploidy syndrome
- NBS
Nijmegen breakage syndrome
- ND
neurodegenerative diseases
- NER
nucleotide excision repair
- NGS
next generation sequencing
- NHEJ
non-homologous end joining
- NPC
neural progenitor cells
- OPTMs
oxidative post-translational modifications
- PCD
programmed cell death
- PD
Parkinson's disease
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RTS
Rothmund-Thomson syndrome
- SAC
spindle assembly checkpoint
- s-CIN
structural instability
- SGZ
subgranular zone
- SKY
spectral karyotype
- SNCA
alpha synuclein
- SCNAs
somatic copy number alteration/aberrations
- SVZ
subventricular zone
- TTD
trichothiodystrophy
- UV
ultraviolet
- VSMC
vascular smooth muscle cell
- VZ
ventricular zone
- W-CIN
whole chromosome instability
- WRN
Werner syndrome, recQ helicase-like
- WS
Werner syndrome
- XP
xeroderma pigmentosum
References
- Alvarez-Dolado M, Pardal R, Garcia-Verdugo JM, Fike JR, Lee HO, Pfeffer K, Lois C, Morrison SJ, Alvarez-Buylla A. Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature. 2003;425:968–973. doi: 10.1038/nature02069. [DOI] [PubMed] [Google Scholar]
- Ambros M, Rumpler S, Luegmayr A, Hattinger CM, Strehl S, Kovar H, Gadner H, Ambros PF. Neuroblastoma cells can actively eliminate supernumerary MYCN gene copies by micronucleus formation—sign of tumourcell revertance? Eur J Cancer. 1997;33(12):2043–2049. doi: 10.1016/s0959-8049(97)00204-9. [DOI] [PubMed] [Google Scholar]
- Andriani GA, Faggioli F, Baker D, Dolle ME, Sellers RS, Hebert JM, van Steeg H, Hoeijmakers J, Vijg J, Montagna C. Whole chromosome aneuploidy in the brain of Bub1bH/H and Ercc1-/Delta7 mice. Hum Mol Genet. 2016;25(4):755–765. doi: 10.1093/hmg/ddv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjomani Virmouni S, Al-Mahdawi S, Sandi C, Yasaei H, Giunti P, Slijepcevic P, Pook MA. Identification of telomere dysfunction in Friedreich ataxia. Mol Neurodegener. 2015;10:22. doi: 10.1186/s13024-015-0019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arocena DG, Iwahashi CK, Won N, Beilina A, Ludwig AL, Tassone F, Schwartz PH, Hagerman PJ. Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum Mol Genet. 2005;14:3661–3671. doi: 10.1093/hmg/ddi394. [DOI] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Atzmon G, Cho M, Cawthon RM, Budagov T, Katz M, Yang X, Siegel G, Bergman A, Huffman DM, Schechter CB, Wright WE, Shay JW, Barzilai N, Govindaraju DR, Suh Y. Evolution in health and medicine Sackler colloquium: genetic variation in human telomerase is associated with telomere length in Ashkenazi centenarians. Proc Natl Acad Sci USA. 2010;107(Suppl. 1):1710–1717. doi: 10.1073/pnas.0906191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, van Deursen JM. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Dawlaty MM, Wijshake T, Jeganathan KB, Malureanu L, van Ree JH, Crespo-Diaz R, Reyes S, Seaburg L, Shapiro V, Behfar A, Terzic A, vande Sluis B, van Deursen JM. Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan. Nat Cell Biol. 2013;15:96–102. doi: 10.1038/ncb2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmus G, Karp NA, Ng BL, Jackson SP, Adams DJ, McIntyre RE. Ahigh-throughput in vivo micronucleus assay for genome instability screening in mice. Nat Protoc. 2015;10:205–215. doi: 10.1038/nprot.2015.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banan A, Fields JZ, Decker H, Zhang Y, Keshavarzian A. Nitric oxide and its metabolites mediate ethanol-induced microtubule disruption and intestinal barrier dysfunction. J Pharmacol Exp Ther. 2000a;294:997–1008. [PubMed] [Google Scholar]
- Banan A, Zhang Y, Losurdo J, Keshavarzian A. Carbonylation and disassembly of the F-actin cytoskeleton in oxidant induced barrier dysfunction and its prevention by epidermal growth factor and transforming growth factor alpha in a human colonic cell line. Gut. 2000b;46:830–837. doi: 10.1136/gut.46.6.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraibar MA, Friguet B. Changes of the proteasomal system during the aging process. Prog Mol Biol Transl Sci. 2012;109:249–275. doi: 10.1016/B978-0-12-397863-9.00007-9. [DOI] [PubMed] [Google Scholar]
- Barker PE. Double minutes in human tumor cells. Cancer Genet Cytogenet. 1982;5:81–94. doi: 10.1016/0165-4608(82)90043-7. [DOI] [PubMed] [Google Scholar]
- Bollineni RC, Hoffmann R, Fedorova M. Proteome-wide profiling of carbonylated proteins and carbonylation sites in HeLa cells under mildoxidative stress conditions. Free Radic Biol Med. 2014;68:186–195. doi: 10.1016/j.freeradbiomed.2013.11.030. [DOI] [PubMed] [Google Scholar]
- Bolognesi C, Lando C, Forni A, Landini E, Scarpato R, Migliore L, Bonassi S. Chromosomal damage and ageing: effect on micronuclei frequency in peripheral blood lymphocytes. Age Ageing. 1999;28:393–397. doi: 10.1093/ageing/28.4.393. [DOI] [PubMed] [Google Scholar]
- Boveri T. Ueber mehrpolige Mitosen als Mittel zur Analyse des Zellkerns. Verhandlungen der physicalisch-medizinischen Gesselschaft zu Würzburg. 1902;35:67–90. [Google Scholar]
- Bridger JM, Kill IR. Aging of Hutchinson-Gilford progeria syndrome fibroblasts is characterised by hyperproliferation and increased apoptosis. Exp Gerontol. 2004;39:717–724. doi: 10.1016/j.exger.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs viacyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukvic N, Gentile M, Susca F, Fanelli M, Serio G, Buonadonna L, Capurso A, Guanti G. Sex chromosome loss, micronuclei, sister chromatid exchange and aging: a study including 16 centenarians. Mutat Res. 2001;498:159–167. doi: 10.1016/s1383-5718(01)00279-0. [DOI] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, Chew SK, Rowan AJ, Schenk A, Sheffer M, Howell M, Kschischo M, Behrens A, Helleday T, Bartek J, Tomlinson IP, Swanton C. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman DM, Chun J. The genomically mosaic brain: aneuploidy and more in neural diversity and disease. Semin Cell Dev Biol. 2013;24:357–369. doi: 10.1016/j.semcdb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Evrony GiladD, Lehmann HillelS, Elhosary PrincessC, Mehta BhavenK, Annapurna Poduri Walsh, Christopher A. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the humanbrain. Cell Rep. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-chichi CD, Cai KQ, Simpkins F, Ganjei-Azar P, Godwin AK, Xu XX. Nuclear envelope structural defects cause chromosomal numerical instability and aneuploidy in ovarian cancer. BMC Med. 2011a;9:28. doi: 10.1186/1741-7015-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-chichi CD, Cai KQ, Smedberg J, Ganjei-Azar P, Godwin AK, Xu XX. Loss of A-type lamin expression compromises nuclear envelope integrity in breast cancer. Chin J Cancer. 2011b;30:415–425. doi: 10.5732/cjc.010.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli, Chan DC. Mitochondrial DNA: impacting central and peripheral nervous systems. Neuron. 2014;84:1126–1142. doi: 10.1016/j.neuron.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati A, Riboni R, Caprioli J, Nuzzo F, Mondello C. Condensation anomalies and exclusion in micronuclei of rearranged chromosomes in human fibroblasts cultured in vitro. Chromosoma. 1995;104:137–142. doi: 10.1007/BF00347696. [DOI] [PubMed] [Google Scholar]
- Caulfield T, Evans J, McGuire A, McCabe C, Bubela T, Cook-Deegan R, Fishman J, Hogarth S, Miller FA, Ravitsky V, Biesecker B, Borry P, Cho MK, Carroll JC, Etchegary H, Joly Y, Kato K, Lee SS, Rothenberg K, Sankar P, Szego MJ, Ossorio P, Pullman D, Rousseau F, Ungar WJ, Wilson B. Reflections on the cost of low-cost whole genome sequencing:framing the health policy debate. PLoS Biol. 2013;11:e1001699. doi: 10.1371/journal.pbio.1001699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha MY, Kim DK, Mook-Jung I. The role of mitochondrial DNA mutation on neurodegenerative diseases. Exp Mol Med. 2015;47:e150. doi: 10.1038/emm.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple JP, Bros-Facer V, Butler R, Gallo JM. Focal distortion of the nuclear envelope by huntingtin aggregates revealed by lamin immunostaining. Neurosci Lett. 2008;447:172–174. doi: 10.1016/j.neulet.2008.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo WL, Ljung BM, Chew K, Myambo K, Miranda M, Krig S, Garbe J, Stampfer M, Yaswen P, Gray JW, Lockett SJ. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- Chinta SJ, Lieu CA, Demaria M, Laberge RM, Campisi J, Andersen JK. Environmental stress, ageing and glial cell senescence: a novel mechanistic link to Parkinson's disease? J Intern Med. 2013;273:429–436. doi: 10.1111/joim.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinta SJ, Woods G, Rane A, Demaria M, Campisi J, Andersen JK. Cellular senescence and the aging brain. Exp Gerontol. 2015;68:3–7. doi: 10.1016/j.exger.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Colnaghi R, Carpenter G, Volker M, O'Driscoll M. The consequences of structural genomic alterations in humans: genomic disorders, genomic instability and cancer. Semin Cell Dev Biol. 2011;22:875–885. doi: 10.1016/j.semcdb.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppede F, Migliore L. DNA repair in premature aging disorders and neuro degeneration. Curr Aging Sci. 2010;3:3–19. doi: 10.2174/1874609811003010003. [DOI] [PubMed] [Google Scholar]
- Coppede F, Migliore L. DNA damage in neurodegenerative diseases. Mutat Res. 2015;776:84–97. doi: 10.1016/j.mrfmmm.2014.11.010. [DOI] [PubMed] [Google Scholar]
- Coskun PE, Busciglio J. Oxidative stress and mitochondrial dysfunction in Down's syndrome: relevance to aging and dementia. Curr Gerontol Geriatr Res. 2012;2012:383170. doi: 10.1155/2012/383170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell JK. Double minutes and homogeneously staining regions: gene amplification in mammalian cells. Annu Rev Genet. 1982;16:21–59. doi: 10.1146/annurev.ge.16.120182.000321. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V, Santarpia C, Grieco D. Oxidative stress overrides the spindle checkpoint. Cell Cycle. 2007;6:576–579. doi: 10.4161/cc.6.5.3934. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O'Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. Quantitative proteomic analysis reveals post translational responses to aneuploidy in yeast. eLife. 2014;3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexheimer TS. DNA repair pathways and mechanisms. In: Mathews LA, Cabarcas SM, Hurt EM, editors. DNA Repair of Cancer Stem Cells. Springer; Netherlands: 2013. pp. 19–32. [Google Scholar]
- Dierssen M, Herault Y, Estivill X. Aneuploidy: from a physiological mechanism of variance to Down syndrome. Physiol Rev. 2009;89:887–920. doi: 10.1152/physrev.00032.2007. [DOI] [PubMed] [Google Scholar]
- Donley N, Thayer MJ. DNA replication timing, genome stability and cancer: late and/or delayed DNA replication timing is associated with increased genomic instability. Semin Cancer Biol. 2013;23:80–89. doi: 10.1016/j.semcancer.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, Grompe M. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest. 2012a;122:3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Hanlon Newell AE, Smith L, Wilson EM, Olson SB, Thayer MJ, Strom SC, Grompe M. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012b;142:25–28. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitan E, Hutchison ER, Mattson MP. Telomere shortening in neurological disorders: an abundance of unanswered questions. Trends Neurosci. 2014;37:256–263. doi: 10.1016/j.tins.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada JC, Torres Y, Benguria A, Dopazo A, Roche E, Carrera-Quintanar L, Perez RA, Enriquez JA, Torres R, Ramirez JC, Samper E, Bernad A. Human mesenchymal stem cell-replicative senescence and oxidative stress are closely linked to aneuploidy. Cell Death Dis. 2013;4:e691. doi: 10.1038/cddis.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, Park PJ, Walsh CA. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggioli F, Sacco MG, Susani L, Montagna C, Vezzoni P. Cell fusion is a physiological process in mouse liver. Hepatology. 2008;48:1655–1664. doi: 10.1002/hep.22488. [DOI] [PubMed] [Google Scholar]
- Faggioli F, Vezzoni P, Montagna C. Single-cell analysis of ploidy and centrosomes underscores the peculiarity of normal hepatocytes. PLoS One. 2011a;6:e26080. doi: 10.1371/journal.pone.0026080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggioli F, Vijg J, Montagna C. Chromosomal aneuploidy in the aging brain. Mech Ageing Dev. 2011b;132:429–436. doi: 10.1016/j.mad.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggioli F, Wang T, Vijg J, Montagna C. Chromosome-specific accumulation of aneuploidy in the aging mouse brain. Hum Mol Genet. 2012;21:5246–5253. doi: 10.1093/hmg/dds375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggioli F, Vijg J, Montagna C. Four-color FISH for the detection of low-level aneuploidy in interphase cells. Methods Mol Biol. 2014;1136:291–305. doi: 10.1007/978-1-4939-0329-0_14. [DOI] [PubMed] [Google Scholar]
- Fan Y, Mao R, Lv H, Xu J, Yan L, Liu Y, Shi M, Ji G, Yu Y, Bai J, Jin Y, Fu S. Frequency of double minute chromosomes and combined cytogenetic abnormalities and their characteristics. J Appl Genet. 2011;52:53–59. doi: 10.1007/s13353-010-0007-z. [DOI] [PubMed] [Google Scholar]
- Flowers A. Brain tumors in the older person. Cancer Control. 2000;7:523–538. doi: 10.1177/107327480000700604. [DOI] [PubMed] [Google Scholar]
- Ford CE, Jones KW, Miller OJ, Mittwoch U, Penrose LS, Ridler M, Shapiro A. The chromosomes in a patient showing both mongolism and the Klinefelter syndrome. Lancet. 1959;1:709–710. doi: 10.1016/s0140-6736(59)91891-4. [DOI] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP. Genome screening by comparative genomic hybridization. Trends Genet. 1997;13:405–409. doi: 10.1016/s0168-9525(97)01244-4. [DOI] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade JM. Somatic tetraploidy invertebrate neurons: implications in physiology and pathology. Commun Integr Biol. 2010;3:201–203. doi: 10.4161/cib.3.2.11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, Kokocinski F, Cohen J, Munne S, Wells D. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet. 2015;11:e1005241. doi: 10.1371/journal.pgen.1005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois M, Leifert W, Hecker J, Faunt J, Martins R, Thomas P, Fenech M. Altered cytological parameters in buccal cells from individuals with mildcognitive impairment and Alzheimer's disease. Cytometry A. 2014;85:698–708. doi: 10.1002/cyto.a.22453. [DOI] [PubMed] [Google Scholar]
- Furgason JM, Koncar RF, Michelhaugh SK, Sarkar FH, Mittal S, Sloan AE, Barnholtz-Sloan JS, Bahassi el M. Whole genome sequence analysis links chromothripsis to EGFR, MDM2, MDM4, and CDK4 amplification in glioblastoma. Oncoscience. 2015;2:618–628. doi: 10.18632/oncoscience.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. Linking abnormal mitosis to the acquisition of DNA damage. J Cell Biol. 2012;199:871–881. doi: 10.1083/jcb.201210040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy: aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhart E. Double minutes cytogenetic equivalents of gene amplification, in human neoplasia—a review. Clin Transl Oncol. 2005;7:477–485. doi: 10.1007/BF02717000. [DOI] [PubMed] [Google Scholar]
- Geller LN, Potter H. Chromosome missegregation and trisomy 21 mosaicism in Alzheimer's disease. Neurobiol Dis. 1999;6:167–179. doi: 10.1006/nbdi.1999.0236. [DOI] [PubMed] [Google Scholar]
- Gentric G, Maillet V, Paradis V, Couton D, L'Hermitte A, Panasyuk G, Fromenty B, Celton-Morizur S, Desdouets C. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Investig. 2015;125:981–992. doi: 10.1172/JCI73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giollant M, Bertrand S, Verrelle P, Tchirkov A, du Manoir S, Ried T, Mornex F, Dore JF, Cremer T, Malet P. Characterization of double minute chromosomes' DNA content in a human high grade astrocytoma cell line by using comparative genomic hybridization and fluorescence in situ hybridization. Hum Genet. 1996;98:265–270. doi: 10.1007/s004390050205. [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P, Hoglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjoerup OV, Wu J, Chandler-Militello D, Williams GL, Zhao J, Schaffhausen B, Jat PS, Roberts TM. Surveillance mechanism linking Bub1 loss to the p53 pathway. Proc Natl Acad Sci USA. 2007;104:8334–8339. doi: 10.1073/pnas.0703164104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisky EL. Changes in cognitive function in human aging. In: Riddle DR, editor. Brain Aging: Models, Methods, and Mechanisms. CRC Press/Taylor & Francis; Boca Raton (FL): 2007. [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Gonzalo S. DNA damage and lamins. Adv Exp Med Biol. 2014;773:377–399. doi: 10.1007/978-1-4899-8032-8_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves LC, Reeve AK, Taylor RW, Turnbull DM. Mitochondrial DNA and disease. The Journal of pathology. 2012;226:274–286. doi: 10.1002/path.3028. [DOI] [PubMed] [Google Scholar]
- Gregan J, Polakova S, Zhang L, Tolic-Norrelykke IM, Cimini D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 2011;21:374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti G, Torri F, Rasmussen J, Clark AP, Lakatos A, Turner JA, Fallon JH, Saykin AJ, Weiner M, ADNI the Alzheimer's Disease Neuroimaging Initiative. Vawter MP, Knowles JA, Potkin SG, Macciardi F. Increased CNV-region deletions in mild cognitive impairment (MCI) and Alzheimer's disease (AD) subjects in the ADNI sample. Genomics. 2013;102:112–122. doi: 10.1016/j.ygeno.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundry M, Li W, Maqbool SB, Vijg J. Direct, genome-wide assessment of DNA mutations in single cells. Nucleic Acids Res. 2012;40:2032–2040. doi: 10.1093/nar/gkr949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenbach M, Schakowski R, Schmid M. Aneuploidy and ageing: sex chromosome exclusion into micronuclei. Hum Genet. 1994;94:295–298. doi: 10.1007/BF00208287. [DOI] [PubMed] [Google Scholar]
- Hahn PJ. Molecular biology of double-minute chromosomes. Bioessays. 1993;15:477–484. doi: 10.1002/bies.950150707. [DOI] [PubMed] [Google Scholar]
- Hando JC, Nath J, Tucker JD. Sex chromosomes, micronuclei and aging in women. Chromosoma. 1994;103:186–192. doi: 10.1007/BF00368011. [DOI] [PubMed] [Google Scholar]
- Hayashi MT, Karlseder J. DNA damage associated with mitosis and cytokinesis failure. Oncogene. 2013;32:4593–4601. doi: 10.1038/onc.2012.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Lammerding J. Lamins at a glance. J Cell Sci. 2012;125:2087–2093. doi: 10.1242/jcs.087288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med. 2012a;18:1630–1638. doi: 10.1038/nm.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012b;13:501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Iakoubov L, Mossakowska M, Szwed M, Duan Z, Sesti F, Puzianowska-Kuznicka M. A common copy number variation (CNV) polymorphism in the CNTNAP4 gene: association with aging in females. PLoS One. 2013;8:e79790. doi: 10.1371/journal.pone.0079790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoubov L, Mossakowska M, Szwed M, Puzianowska-Kuznicka M. A common copy number variation polymorphism in the CNTNAP2gene: sexual dimorphism in association with healthy aging and disease. Gerontology. 2015;61:24–31. doi: 10.1159/000363320. [DOI] [PubMed] [Google Scholar]
- Ichijima Y, Yoshioka K, Yoshioka Y, Shinohe K, Fujimori H, Unno J, Takagi M, Goto H, Inagaki M, Mizutani S, Teraoka H. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS One. 2010;5:e8821. doi: 10.1371/journal.pone.0008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa-Yoshida A, Ando K, Oki E, Saeki H, Kumashiro R, Taketani K, Ida S, Tokunaga E, Kitao H, Morita M, Maehara Y. Contribution of BubR1 to oxidative stress-induced aneuploidy in p53-deficient cells. Cancer Med. 2013;2:447–456. doi: 10.1002/cam4.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iourov Y, Vorsanova SG, Liehr T, Kolotii AD, Yurov YB. Increased chromosome instability dramatically disrupts neural genome integrity and mediates cerebellar degeneration in the ataxia-telangiectasia brain. Hum Mol Genet. 2009a;18:2656–2669. doi: 10.1093/hmg/ddp207. [DOI] [PubMed] [Google Scholar]
- Iourov Y, Vorsanova SG, Liehr T, Yurov YB. Aneuploidy in the normal, Alzheimer's disease and ataxia-telangiectasia brain: differential expression and pathological meaning. Neurobiol Dis. 2009b;34:212–220. doi: 10.1016/j.nbd.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Iourov Y, Vorsanova SG, Yurov YB. Genomic landscape of the Alzheimer's disease brain: chromosome instability-aneuploidy, but not tetraploidy-mediates neurodegeneration. Neurodegenerative diseases. 2011;8:35–37. doi: 10.1159/000315398. discussion 38-40) [DOI] [PubMed] [Google Scholar]
- Iourov Y, Vorsanova SG, Zelenova MA, Korostelev SA, Yurov YB. Genomic copy number variation affecting genes involved in the cell cycle pathway: implications for somatic mosaicism. Int J Genom. 2015;2015:757680. doi: 10.1155/2015/757680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs PA, Court Brown WM, Doll R. Distribution of human chromosome counts in relation to age. Nature. 1961;191:1178–1180. doi: 10.1038/1911178a0. [DOI] [PubMed] [Google Scholar]
- Jacobs PA, Brunton M, Court Brown WM, Doll R, Goldstein H. Change of human chromosome count distribution with age: evidence for a sex differences. Nature. 1963;197:1080–1081. doi: 10.1038/1971080a0. [DOI] [PubMed] [Google Scholar]
- Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, Liu C, Horner MJ, Cullen M, Epstein CG, Burdett L, Dean MC, Chatterjee N, Sampson J, Chung CC, Kovaks J, Gapstur SM, Stevens VL, Teras LT, Gaudet MM, Albanes D, Weinstein SJ, Virtamo J, Taylor PR, Freedman ND, Abnet CC, Goldstein AM, Hu N, Yu K, Yuan JM, Liao L, Ding T, Qiao YL, Gao YT, Koh WP, Xiang YB, Tang ZZ, Fan JH, Aldrich MC, Amos C, Blot WJ, Bock CH, Gillanders EM, Harris CC, Haiman CA, Henderson BE, Kolonel LN, Le Marchand L, McNeill LH, Rybicki BA, Schwartz AG, Signorello LB, Spitz MR, Wiencke JK, Wrensch M, Wu X, Zanetti KA, Ziegler RG, Figueroa JD, Garcia-Closas M, Malats N, Marenne G, Prokunina-Olsson L, Baris D, Schwenn M, Johnson A, Landi MT, Goldin L, Consonni D, Bertazzi PA, Rotunno M, Rajaraman P, Andersson U, Beane Freeman LE, Berg CD, Buring JE, Butler MA, Carreon T, Feychting M, Ahlbom A, Gaziano JM, Giles GG, Hallmans G, Hankinson SE, Hartge P, Henriksson R, Inskip PD, Johansen C, Landgren A, McKean-Cowdin R, Michaud DS, Melin BS, Peters U, Ruder AM, Sesso HD, Severi G, Shu XO, Visvanathan K, White A, Wolk Zeleniuch-Jacquotte AW, Zheng DT, Silverman M, Kogevinas JR, Gonzalez O, Villa D, Li EJ, Duell HA, Risch SH, Olson C, Kooperberg BM, Wolpin L, Jiao M, Hassan W, Wheeler AA, Arslan Bueno-de-Mesquita HBCS, Fuchs S, Gallinger MD, Gross EA, Holly AP, Klein A, LaCroix MT, Mandelson G, Petersen Boutron-Ruault MCPM, Bracci F, Canzian K, Chang M, Cotterchio EL, Giovannucci M, Goggins JA, Hoffman Bolton M, Jenab KT, Khaw V, Krogh RC, Kurtz RR, McWilliams JB, Mendelsohn KG, Rabe E, Riboli A, Tjonneland GS, Tobias D, Trichopoulos JW, Elena H, Yu L, Amundadottir RZ, Stolzenberg-Solomon P, Kraft F, Schumacher D, Stram SA, Savage L, Mirabello IL, Andrulis JS, Wunder Patino Garcia AL, Sierrasesumaga DA, Barkauskas RG, Gorlick M, Purdue WH, Chow LE, Moore KL, Schwartz FG, Davis AW, Hsing SI, Berndt A, Black N, Wentzensen LA, Brinton J, Lissowska B, Peplonska KA, McGlynn MB, Cook BI, Graubard CP, Kratz MH, Greene RL, Erickson DJ, Hunter G, Thomas RN, Hoover FX, Real JF, Fraumeni NE, Jr, Caporaso M, Tucker N, Perez-Jurado RothmanLA, Chanock SJ. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Boceno M, Rival JM, Mechinaud F, David A. High risk of malignancy in mosaic variegated aneuploidy syndrome. Am J Med Genet. 2002;109:17–21. doi: 10.1002/ajmg.10281. (discussion 16) [DOI] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, Cadinanos J, Horner JW, Maratos-Flier E, Depinho RA. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2011;469:102–106. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Ju Z, Rudolph KL. Telomere shortening and ageing. Zeitschrift fur Gerontologie Geriatrie. 2007;40:314–324. doi: 10.1007/s00391-007-0480-0. [DOI] [PubMed] [Google Scholar]
- Johansson CB, Youssef S, Koleckar K, Holbrook C, Doyonnas R, Corbel SY, Steinman L, Rossi FM, Blau HM. Extensive fusion of haematopoietic cells with Purkinje neurons in response to chronic inflammation. Nat Cell Biol. 2008;10:575–583. doi: 10.1038/ncb1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Ravid K. Vascular smooth muscle polyploidization as a biomarker foraging and its impact on differential gene expression. J Biol Chem. 2004;279:5306–5313. doi: 10.1074/jbc.M308406200. [DOI] [PubMed] [Google Scholar]
- Jones KT. Meiosis in oocytes: predisposition to aneuploidy and its increased incidence with age. Hum Reprod Update. 2008;14:143–158. doi: 10.1093/humupd/dmm043. [DOI] [PubMed] [Google Scholar]
- Kaushal D, Contos JJ, Treuner K, Yang AH, Kingsbury MA, Rehen SK, McConnell MJ, Okabe M, Barlow C, Chun J. Alteration of gene expression by chromosome loss in the postnatal mouse brain. J Neurosci. 2003;23:5599–5606. doi: 10.1523/JNEUROSCI.23-13-05599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp K, Wilkins A, Scolding N. Cell fusion in the brain: two cells forward, one cell back. Acta Neuropathol. 2014;128:629–638. doi: 10.1007/s00401-014-1303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapko K, Vijg J. Mitochondrial DNA mutations and aging: devils in the details? Trends Genet. 2009;25:91–98. doi: 10.1016/j.tig.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, Kaushal D, Chun J. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc Natl Acad Sci USA. 2005;102:6143–6147. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Whittaker CA, Amon A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci USA. 2014;111:13409–13414. doi: 10.1073/pnas.1415287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Lin CL. Oxidative damage to RNA: mechanisms consequences, and diseases. Cell Mol Life Sci. 2010;67:1817–1829. doi: 10.1007/s00018-010-0277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Kim Y, Weaver BA, Mao Y, McLeod I, Yates JR, 3rd, Tagaya M, Cleveland DW. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormann-Bortolotto MH, de Arruda Cardoso Smith M, Toniolo Neto J. Alzheimer's disease and ageing: a chromosomal approach. Gerontology. 1993;39:1–6. doi: 10.1159/000213508. [DOI] [PubMed] [Google Scholar]
- Kregel KC, Zhang HJ. An integrated view of oxidative stress in aging: basic mechanisms, functional effects, and pathological considerations. Am J Physiol Regul Integr Comp Physiol. 2007;292:R18–R36. doi: 10.1152/ajpregu.00327.2006. [DOI] [PubMed] [Google Scholar]
- Kudlow BA, Kennedy BK, Monnat RJ., Jr Werner and Hutchinson-Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nat Rev Mol Cell Biol. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- Kumari G, Ulrich T, Krause M, Finkernagel F, Gaubatz S. Induction of p21CIP1 protein and cell cycle arrest after inhibition of Aurora B kinase is attributed to aneuploidy and reactive oxygen species. J Biol Chem. 2014;289:16072–16084. doi: 10.1074/jbc.M114.555060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuningas M, Estrada K, Hsu YH, Nandakumar K, Uitterlinden AG, Lunetta KL, van Duijn CM, Karasik D, Hofman A, Murabito J, Rivadeneira F, Kiel DP, Tiemeier H. Large common deletions associate with mortality at old age. Hum Mol Genet. 2011;20:4290–4296. doi: 10.1093/hmg/ddr340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langie SA, Koppen G, Desaulniers D, Al-Mulla F, Al-Temaimi R, Amedei A, Azqueta A, Bisson WH, Brown DG, Brunborg G, Charles AK, Chen T, Colacci A, Darroudi F, Forte S, Gonzalez L, Hamid RA, Knudsen LE, Leyns L, Lopez de Cerain Salsamendi A, Memeo L, Mondello C, Mothersill C, Olsen AK, Pavanello S, Raju J, Rojas E, Roy R, Ryan EP, Ostrosky-Wegman P, Salem HK, Scovassi AI, Singh N, Vaccari M, Van Schooten FJ, Valverde M, Woodrick J, Zhang L, van Larebeke N, Kirsch-Volders M, Collins AR. Causes of genome instability: the effect of low Dose chemical exposures in modern society. Carcinogenesis. 2015;36(Suppl. 1):S61–88. doi: 10.1093/carcin/bgv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapham LW. Tetraploid DNA content of Purkinje neurons of human cerebellar cortex. Science. 1968;159:310–312. doi: 10.1126/science.159.3812.310. [DOI] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Taylor SS. Cohesion fatigue explains why pharmacological inhibition of the APC/C induces a spindle checkpoint-dependent mitotic arrest. PLoS One. 2012;7:e49041. doi: 10.1371/journal.pone.0049041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz ML, Zhang CZ, Pellman D. Chromothripsis: a new mechanism for rapid karyotype evolution. Annu Rev Genet. 2015;49:183–211. doi: 10.1146/annurev-genet-120213-092228. [DOI] [PubMed] [Google Scholar]
- Leland S, Nagarajan P, Polyzos A, Thomas S, Samaan G, Donnell R, Marchetti F, Venkatachalam S. Heterozygosity for a Bub1 mutation causes female-specific germ cell aneuploidy in mice. Proc Natl Acad Sci USA. 2009;106:12776–12781. doi: 10.1073/pnas.0903075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini L, Barra V, Schillaci T, Di Leonardo A. MAD2 depletion triggers premature cellular senescence in human primary fibroblasts by activating a P53 pathway preventing aneuploid cells propagation. J Cell Physiol. 2011;227(9):3324–3332. doi: 10.1002/jcp.24030. [DOI] [PubMed] [Google Scholar]
- Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, Han S, van Deursen JM, Zhang P. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci USA. 2010;107:14188–14193. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli CL, Giedzinski E. Induction of chromosomal instability by chronic oxidative stress. Neoplasia. 2003;5:339–346. doi: 10.1016/S1476-5586(03)80027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli CL, Giedzinski E, Morgan WF, Swarts SG, Jones GD, Hyun W. Persistent oxidative stress in chromosomally unstable cells. Cancer Res. 2003;63:3107–3111. [PubMed] [Google Scholar]
- Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, Wagner U, Kim A, Ren B, Li Y, Goebl A, Kim J, Soligalla RD, Dubova I, Thompson J, Yates J, 3rd, Esteban CR, Sancho-Martinez I, Izpisua Belmonte JC. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012;491:603–607. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockrow JP, Fortress AM, Granholm AC. Age-related neurodegeneration and memory loss in down syndrome. Curr Gerontol Geriatr Res. 2012:463909. doi: 10.1155/2012/463909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Zhou W, Sampson JN, Dean MC, Jacobs KB, Black A, Brinton LA, Chang IS, Chen C, Chen C, Chen K, Cook LS, Crous Bou M, De Vivo I, Doherty J, Friedenreich CM, Gaudet MM, Haiman CA, Hankinson SE, Hartge P, Henderson BE, Hong YC, Hosgood HD, 3rd, Hsiung CA, Hu W, Hunter DJ, Jessop L, Kim HN, Kim YH, Kim YT, Klein R, Kraft P, Lan Q, Lin D, Liu J, Le Marchand L, Liang X, Lissowska J, Lu L, Magliocco AM, Matsuo K, Olson SH, Orlow I, Park JY, Pooler L, Prescott J, Rastogi R, Risch HA, Schumacher F, Seow A, Setiawan VW, Shen H, Sheng X, Shin MH, Shu XO, VanDen Berg D, Wang JC, Wentzensen N, Wong MP, Wu C, Wu T, Wu YL, Xia L, Yang HP, Yang PC, Zheng W, Zhou B, Abnet CC, Albanes D, Aldrich MC, Amos C, Amundadottir LT, Berndt SI, Blot WJ, Bock CH, Bracci PM, Burdett L, Buring JE, Butler MA, Carreon T, Chatterjee N, Chung CC, Cook MB, Cullen M, Davis FG, Ding T, Duell EJ, Epstein CG, Fan JH, Figueroa JD, Fraumeni JF, Jr, Freedman ND, Fuchs CS, Gao YT, Gapstur SM, Patino-Garcia A, Garcia-Closas M, Gaziano JM, Giles GG, Gillanders EM, Giovannucci EL, Goldin L, Goldstein AM, Greene G, Harris CC, Henriksson R, Holly EA, Hoover RN, Hu N, Hutchinson A, Jenab M, Johansen C, Khaw KT, Koh WP, Kolonel LN, Kooperberg C, Krogh V, Kurtz RC, LaCroix A, Landgren A, Landi MT, Li D, Liao LM, Malats N, McGlynn KA, McNeill LH, McWilliams RR, Melin BS, Mirabello L, Peplonska B, Peters U, Petersen GM, Prokunina-Olsson L, Purdue M, Qiao YL, Rabe KG, Rajaraman P, Real FX, Riboli E, Rodriguez-Santiago B, Rothman N, Ruder AM, Savage SA, Schwartz AG, Schwartz KL, Sesso HD, Severi G, Silverman DT, Spitz MR, Stevens VL, Stolzenberg-Solomon R, Stram D, Tang ZZ, Taylor PR, Teras LR, Tobias GS, Viswanathan K, Wacholder S, Wang Z, Weinstein SJ, Wheeler W, White E, Wiencke JK, Wolpin BM, Wu X, Wunder JS, Yu K, Zanetti KA, Zeleniuch-Jacquotte A, Ziegler RG, de Andrade M, Barnes KC, Beaty TH, Bierut LJ, Desch KC, Doheny KF, Feenstra B, Ginsburg D, Heit JA, Kang JH, Laurie CA, Li JZ, Lowe WL, Marazita ML, Melbye M, Mirel DB, Murray JC, Nelson SC, Pasquale LR, Rice K, Wiggs JL, Wise A, Tucker M, Perez-Jurado LA, Laurie CC, Caporaso NE, Yeager M, Chanock SJ. Characterization of large structural genetic mosaicism in human autosomes. Am J Hum Genet. 2015;96:487–497. doi: 10.1016/j.ajhg.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailankot M, Smith D, Howell S, Wang B, Jacobberger JW, Stefan T, Nagaraj RH. Cell cycle arrest by kynurenine in lens epithelial cells. Investig Ophthalmol Vis Sci. 2008;49:5466–5475. doi: 10.1167/iovs.08-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DM, Esiri MM. The site of the earliest lesions of Alzheimer's disease. N Engl J Med. 1988;318:789–790. doi: 10.1056/NEJM198803243181218. [DOI] [PubMed] [Google Scholar]
- Martin GM, Smith AC, Ketterer DJ, Ogburn CE, Disteche CM. Increased chromosomal aberrations in first metaphases of cells isolated from the kidneys of aged mice. Isr J Med Sci. 1985;21:296–301. [PubMed] [Google Scholar]
- Martin LJ. DNA damage and repair: relevance to mechanisms of neurodegeneration. J Neuropathol Exp Neurol. 2008;67:377–387. doi: 10.1097/NEN.0b013e31816ff780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim Biophys Acta. 2009;1790:963–969. doi: 10.1016/j.bbagen.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Chowdhury MR, Singh G. Recent advances in chromosome breakage syndromes and their diagnosis. Indian Pediatr. 2000;37:615–625. [PubMed] [Google Scholar]
- Matsuyama M, Tanaka H, Inoko A, Goto H, Yonemura S, Kobori K, Hayashi Y, Kondo E, Itohara S, Izawa I, Inagaki M. Defect of mitotic vimentin phosphorylation causes microophthalmia and cataract via aneuploidy and senescence in lens epithelial cells. J Biol Chem. 2013;288:35626–35635. doi: 10.1074/jbc.M113.514737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard S, Fang EF, Scheibye-Knudsen M, Croteau DL, Bohr VA. DNA damage, DNA, repair, aging, and neurodegeneration. Cold Spring Harb Perspect Med. 2015;5:1–18. doi: 10.1101/cshperspect.a025130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zeamays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, Gage FH. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena JK, Cerutti A, Beichler C, Morita Y, Bruhn C, Kumar M, Kraus JM, Speicher MR, Wang ZQ, Kestler HA, d'Adda di Fagagna F, Gunes C, Rudolph KL. Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion. EMBO J. 2015;34:1371–1384. doi: 10.15252/embj.201490070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliore L, Boni G, Bernardini R, Trippi F, Colognato R, Fontana I, Coppede F, Sbrana I. Susceptibility to chromosome malsegregation in lymphocytes of women who had a Down syndrome child in young age. Neurobiol Aging. 2006;27:710–716. doi: 10.1016/j.neurobiolaging.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Migliore L, Coppede F, Fenech M, Thomas P. Association of micronucleus frequency with neurodegenerative diseases. Mutagenesis. 2011;26:85–92. doi: 10.1093/mutage/geq067. [DOI] [PubMed] [Google Scholar]
- Millet C, Ausiannikava D, Le Bihan T, Granneman S, Makovets S. Cell populations can use aneuploidy to survive telomerase insufficiency. Nat Commun. 2015;6:8664. doi: 10.1038/ncomms9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, Ebus ME, Haneveld F, Lakeman A, Schild L, Molenaar P, Stroeken P, van Noesel MM, Ora I, Santo EE, Caron HN, Westerhout EM, Versteeg R. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- Montagna C, Andrechek ER, Padilla-Nash H, Muller WJ, Ried T. Centrosome abnormalities, recurring deletions of chromosome 4, and genomic amplification of HER2/neu define mouse mammary gland adenocarcinomas induced by mutant HER2/neu. Oncogene. 2002;21:890–898. doi: 10.1038/sj.onc.1205146. [DOI] [PubMed] [Google Scholar]
- Mosch B, Morawski M, Mittag A, Lenz D, Tarnok A, Arendt T. Aneuploidy and DNA replication in the normal human brain and Alzheimer's disease. J Neurosci. 2007;27:6859–6867. doi: 10.1523/JNEUROSCI.0379-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee AB, Costello C. Aneuploidy analysis in fibroblasts of human premature aging syndromes by FISH during in vitro cellular aging. Mech Ageing Dev. 1998;103:209–222. doi: 10.1016/s0047-6374(98)00041-4. [DOI] [PubMed] [Google Scholar]
- Musio A, Montagna C, Zambroni D, Indino E, Barbieri O, Citti L, Villa A, Ried T, Vezzoni P. Inhibition of BUB1 results in genomic instability and anchorage-independent growth of normal human fibroblasts. Cancer Res. 2003;63:2855–2863. [PubMed] [Google Scholar]
- Nagy Z, Esiri MM, Smith AD. Expression of cell division markers in the hippocampus in Alzheimer's disease and other neurodegenerative conditions. Acta Neuropathol. 1997;93:294–300. doi: 10.1007/s004010050617. [DOI] [PubMed] [Google Scholar]
- Nam HJ, van Deursen JM. Cyclin B2 and p53 control proper timing of centrosome separation. Nat Cell Biol. 2014;16:538–549. doi: 10.1038/ncb2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath J, Tucker JD, Hando JC. Y chromosome aneuploidy, micronuclei, kinetochores and aging in men. Chromosoma. 1995;103:725–731. doi: 10.1007/BF00344234. [DOI] [PubMed] [Google Scholar]
- Nern C, Wolff I, Macas J, von Randow J, Scharenberg C, Priller J, Momma S. Fusion of hematopoietic cells with Purkinje neurons does not lead to stable heterokaryon formation under noninvasive conditions. J Neurosci. 2009;29:3799–3807. doi: 10.1523/JNEUROSCI.5848-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JM, Cimini D. Link between aneuploidy and chromosome instability. Int Rev Cell Mol Biol. 2015;315:299–317. doi: 10.1016/bs.ircmb.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Nicotera TM, Block AW, Gibas Z, Sandberg AA. Induction of superoxide dismutase, chromosomal aberrations and sister-chromatid exchanges by paraquat in Chinese hamster fibroblasts. Mutat Res. 1985;151:263–268. doi: 10.1016/0027-5107(85)90078-8. [DOI] [PubMed] [Google Scholar]
- Nouspikel T, Hanawalt PC. Terminally differentiated human neurons repair transcribed genes but display attenuated global DNA repair and modulation of repair gene expression. Mol Cell Biol. 2000;20:1562–1570. doi: 10.1128/mcb.20.5.1562-1570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard M, Debrabant B, Tan Q, Deelen J, Andersen-Ranberg K, de Craen AJ, Beekman M, Jeune B, Slagboom PE, Christensen K, Christiansen L. Copy number variation associates with mortality in long-lived individuals: a genome-wide assessment. Aging Cell. 2015:49–55. doi: 10.1111/acel.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi A, Ohori M, Iwai K, Nakayama Y, Nambu T, Morishita D, Kawamoto T, Miyamoto M, Hirayama T, Okaniwa M, Banno H, Ishikawa T, Kandori H, Iwata K. Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated postmitotic apoptosis in SAC-impaired cells. Nat Commun. 2015;6:7668. doi: 10.1038/ncomms8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov AM. Telomeres, telomerase, and aging: origin of the theory. Exp Gerontol. 1996;31:443–448. doi: 10.1016/0531-5565(96)00005-8. [DOI] [PubMed] [Google Scholar]
- Oromendia AB, Amon A. Aneuploidy: implications for protein homeostasis and disease. Dis Models Mech. 2014;7:15–20. doi: 10.1242/dmm.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama F, Cairns NJ, Shimada H, Oyama R, Titani K, Ihara Y. Down's syndrome: up-regulation of beta-amyloid protein precursor and tau mRNAs and their defective coordination. J Neurochem. 1994;62:1062–1066. doi: 10.1046/j.1471-4159.1994.62031062.x. [DOI] [PubMed] [Google Scholar]
- Pack SD, Weil RJ, Vortmeyer AO, Zeng W, Li J, Okamoto H, Furuta M, Pak E, Lubensky IA, Oldfield EH, Zhuang Z. Individual adult human neurons display aneuploidy: detection by fluorescence in situ hybridization and single neuron PCR. Cell Cycle. 2005;4:1758–1760. doi: 10.4161/cc.4.12.2153. [DOI] [PubMed] [Google Scholar]
- Pampalona J, Soler D, Genesca A, Tusell L. Telomere dysfunction and chromosome structure modulate the contribution of individual chromosomes in abnormal nuclear morphologies. Mutat Res. 2010a;683:16–22. doi: 10.1016/j.mrfmmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Pampalona J, Soler D, Genesca A, Tusell L. Whole chromosome loss is promoted by telomere dysfunction in primary cells. Genes Chromosomes Cancer. 2010b;49:368–378. doi: 10.1002/gcc.20749. [DOI] [PubMed] [Google Scholar]
- Paton JA, Nottebohm FN. Neurons generated in the adult brain are recruited into functional circuits. Science. 1984;225:1046–1048. doi: 10.1126/science.6474166. [DOI] [PubMed] [Google Scholar]
- Pereira PL, Magnol L, Sahun I, Brault V, Duchon A, Prandini P, Gruart A, Bizot JC, Chadefaux-Vekemans B, Deutsch S, Trovero F, Delgado-Garcia JM, Antonarakis SE, Dierssen M, Herault Y. A new mouse model for the trisomy of the Abcg1-U2af1 region reveals the complexity of the combinatorial genetic code of down syndrome. Hum Mol Genet. 2009;18:4756–4769. doi: 10.1093/hmg/ddp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Yang AH, Bushman DM, Westra JW, Yung YC, Barral S, Mutoh T, Rehen SK, Chun J. Aneuploid cells are differentially susceptible to caspase-mediated death during embryonic cerebral cortical development. J Neurosci. 2012;32:16213–16222. doi: 10.1523/JNEUROSCI.3706-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihan GA. Centrosome dysfunction contributes to chromosome instability, chromoanagenesis, and genome reprograming in cancer. Front Oncol. 2013;3:277. doi: 10.3389/fonc.2013.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollex RL, Hegele RA. Hutchinson-Gilford progeria syndrome. Clin Genet. 2004;66:375–381. doi: 10.1111/j.1399-0004.2004.00315.x. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos MH, Higgins JJ, Golbe LI, Johnson WG, Ide SE, Di Iorio G, Sanges G, Stenroos ES, Pho LT, Schaffer AA, Lazzarini AM, Nussbaum RL, Duvoisin RC. Mapping of a gene for Parkinson's disease to chromosome 4q21-q23. Science. 1996;274:1197–1199. doi: 10.1126/science.274.5290.1197. [DOI] [PubMed] [Google Scholar]
- Potter H. Review and hypothesis: Alzheimer disease and Down syndrome-chromosome 21 nondisjunction may underlie both disorders. Am J Hum Genet. 1991;48:1192–1200. [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, Pfaff E, Tica J, Wang Q, Massimi L, Witt H, Bender S, Pleier S, Cin H, Hawkins C, Beck C, von Deimling A, Hans V, Brors B, Eils R, Scheurlen W, Blake J, Benes V, Kulozik AE, Witt O, Martin D, Zhang C, Porat R, Merino DM, Wasserman J, Jabado N, Fontebasso A, Bullinger L, Rucker FG, Dohner K, Dohner H, Koster J, Molenaar JJ, Versteeg R, Kool M, Tabori U, Malkin D, Korshunov A, Taylor MD, Lichter P, Pfister SM, Korbel JO. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve AK, Krishnan KJ, Turnbull D. Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann N Y Acad Sci. 2008;1147:21–29. doi: 10.1196/annals.1427.016. [DOI] [PubMed] [Google Scholar]
- Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH, Chun J. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci USA. 2001;98:13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, Fontanoz M, Chun J. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis RS, Srivastava A, Goldstein S. Macromolecular correlates of cellular senescence and cancer. In: Pullman B, Ts'o PP, Schneider E, editors. Interrelationship Among Aging, Cancer and Differentiation. Springer; Netherlands: 1985. pp. 121–132. [Google Scholar]
- Ricke RM, van Deursen JM. Aneuploidy in health, disease, and aging. J Cell Biol. 2013;201:11–21. doi: 10.1083/jcb.201301061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried T, Just KE, Holtgreve-Grez H, du Manoir S, Speicher MR, Schrock E, Latham C, Blegen H, Zetterberg A, Cremer T, et al. Comparative genomic hybridization of formalin-fixed, paraffin-embedded breast tumors reveals different patterns of chromosomal gains and losses in fibroadenomas and diploid and aneuploid carcinomas. Cancer Res. 1995;55:5415–5423. [PubMed] [Google Scholar]
- Rode A, Maass KK, Willmund KV, Lichter P, Ernst A. Chromothripsis in cancer cells: an update. Int J Cancer. 2015:2322–2333. doi: 10.1002/ijc.29888. [DOI] [PubMed] [Google Scholar]
- Rolig RL, McKinnon PJ. Linking DNA damage and neurodegeneration. Trends Neurosci. 2000;23:417–424. doi: 10.1016/s0166-2236(00)01625-8. [DOI] [PubMed] [Google Scholar]
- Romano AD, Serviddio G, de Matthaeis A, Bellanti F, Vendemiale G. Oxidative stress and aging. J Nephrol. 2010;23(Suppl. 15):S29–36. [PubMed] [Google Scholar]
- Russo A, Pacchierotti F, Cimini D, Ganem NJ, Genesca A, Natarajan AT, Pavanello S, Valle G, Degrassi F. Genomic instability: crossing pathways at the origin of structural and numerical chromosome changes. Environ Mol Mutagen. 2015;56:563–580. doi: 10.1002/em.21945. [DOI] [PubMed] [Google Scholar]
- Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samper E, Nicholls DG, Melov S. Mitochondrial oxidative stress causes chromosomal instability of mouse embryonic fibroblasts. Aging Cell. 2003;2:277–285. doi: 10.1046/j.1474-9728.2003.00062.x. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16:473–485. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer C, Siatkowski M, Pfeiffer MJ, Baeumer N, Drexler HC, Wang B, Fuellen G, Boiani M. Maternal age effect on mouse oocytes: new biological insight from proteomic analysis. Reproduction. 2014;148:55–72. doi: 10.1530/REP-14-0126. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z. Genomic instability and cancer: an introduction. J Mol Cell Biol. 2011;3:1–3. doi: 10.1093/jmcb/mjq057. [DOI] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- Soussi T. The history of p53: a perfect example of the drawbacks of scientific paradigms. EMBO Rep. 2010;11:822–826. doi: 10.1038/embor.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA, Campbell PJ. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuerwald N, Cohen J, Herrera RJ, Sandalinas M, Brenner CA. Association between spindle assembly checkpoint expression and maternal age in human oocytes. Mol Hum Reprod. 2001;7:49–55. doi: 10.1093/molehr/7.1.49. [DOI] [PubMed] [Google Scholar]
- Suijkerbuijk SJ, Kops GJ. Preventing aneuploidy: the contribution of mitotic checkpoint proteins. Biochim Biophys Acta. 2008;1786:24–31. doi: 10.1016/j.bbcan.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Täckholm G. Zytologische Studien überdie Gattung Rosa. Acta horti Bergiani. 1922;7:97–381. [Google Scholar]
- Tan FC, Hutchison ER, Eitan E, Mattson MP. Are there roles for brain cell senescence in aging and neurodegenerative disorders? Biogerontology. 2014;15:643–660. doi: 10.1007/s10522-014-9532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Goto H, Inoko A, Makihara H, Enomoto A, Horimoto K, Matsuyama M, Kurita K, Izawa I, Inagaki M. Cytokinetic failure-induced tetraploidy develops into aneuploidy, triggering skin aging in phosphovimentin-deficient mice. J Biol Chem. 2015;290:12984–12998. doi: 10.1074/jbc.M114.633891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarin JJ. Aetiology of age-associated aneuploidy: a mechanism based on the ‘free radical theory of ageing’; Hum Reprod. 1995;10:1563–1565. doi: 10.1093/humrep/10.6.1563. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, Amon A. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol Biol Cell. 2013;24:1274–1289. doi: 10.1091/mbc.E12-07-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft M, Ross OA. Copy number variation in Parkinson's disease. Genome Med. 2010;2:62. doi: 10.1186/gm183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormos AM, Talens-Visconti R, Bonora-Centelles A, Perez S, Sastre J. Oxidative stress triggers cytokinesis failure in hepatocytes upon isolation. Free Radic Res. 2015;49:927–934. doi: 10.3109/10715762.2015.1016019. [DOI] [PubMed] [Google Scholar]
- Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi M, Fujiwara R, Nishizawa H, Ito M, Kogo H, Inagaki H, Ohye T, Kato T, Fujii T, Kurahashi H. Age-related decrease of meiotic cohesins in human oocytes. PLoS One. 2014;9:e96710. doi: 10.1371/journal.pone.0096710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, Vanderver A, Faulkner GJ. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–239. doi: 10.1016/j.cell.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttara B, Singh AV, Zamboni P, Mahajan RT. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Current Neuropharmacol. 2009;7:65–74. doi: 10.2174/157015909787602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Suh Y. Genome instability and aging. Annu Rev Physiol. 2013;75:645–668. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- Vitre BD, Cleveland DW. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr Opin Cell Biol. 2012;24:809–815. doi: 10.1016/j.ceb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittori A, Breda C, Repici M, Orth M, Roos RA, Outeiro TF, Giorgini F, Hollox EJ REGISTRY investigators of the European Huntington's Disease Network. Copy-number variation of the neuronal glucose transporter gene SLC2A3 and age of onset in Huntington's disease. Hum Mol Genet. 2014;23:3129–3137. doi: 10.1093/hmg/ddu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Liu LN, Zhao ZB. The role of ROS toxicity in spontaneous aneuploidy in cultured cells. Tissue Cell. 2013;45:47–53. doi: 10.1016/j.tice.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Weimann JM, Johansson CB, Trejo A, Blau HM. Stable reprogrammed heterokaryons form spontaneously in Purkinje neurons after bone marrow transplant. Nat Cell Biol. 2003;5:959–966. doi: 10.1038/ncb1053. [DOI] [PubMed] [Google Scholar]
- Westra JW, Peterson SE, Yung YC, Mutoh T, Barral S, Chun J. Aneuploid mosaicism in the developing and adult cerebellar cortex. J Comp Neurol. 2008;507:1944–1951. doi: 10.1002/cne.21648. [DOI] [PubMed] [Google Scholar]
- Westra JW, Barral S, Chun J. A reevaluation of tetraploidy in the Alzheimer's disease brain. Neuro Degener Dis. 2009;6:221–229. doi: 10.1159/000236901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Investig. 2009;119:1825–1836. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Tan Y, Li X, Lin G, Jiang H, Chen F, Zhang C, Gong F, Pan X, Chen S, Lu G, Wang W, Zhang X. Rapid detection of aneuploidies on a benchtop sequencing platform. Prenat Diagn. 2013;33:232–237. doi: 10.1002/pd.4049. [DOI] [PubMed] [Google Scholar]
- Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer's disease. J Neurosci. 2001;21:2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AH, Kaushal D, Rehen SK, Kriedt K, Kingsbury MA, McConnell MJ, Chun J. Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. J Neurosci. 2003;23:10454–10462. doi: 10.1523/JNEUROSCI.23-32-10454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Li Z, Jia Z, Clapcote SJ, Liu C, Li S, Asrar S, Pao A, Chen R, Fan N, Carattini-Rivera S, Bechard AR, Spring S, Henkelman RM, Stoica G, Matsui S, Nowak NJ, Roder JC, Chen C, Bradley A, Yu YE. A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum Mol Genet. 2010;19:2780–2791. doi: 10.1093/hmg/ddq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Monakhov VV, Soloviev IV, Vostrikov VM, Vorsanova SG. The variation of aneuploidy frequency in the developing and adult human brain revealed by an interphase FISH study. J Histochem Cytochem. 2005;53:385–390. doi: 10.1369/jhc.4A6430.2005. [DOI] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Kutsev SI, Pellestor F, Beresheva AK, Demidova IA, Kravets VS, Monakhov VV, Soloviev IV. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS One. 2007;2:e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Vorsanova SG. Neurodegeneration mediated by chromosome instability suggests changes in strategy for therapy development in ataxia-telangiectasia. Med Hypotheses. 2009a;73:1075–1076. doi: 10.1016/j.mehy.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Yurov YB, Vorsanova SG, Iourov IY. GIN'n'CIN hypothesis of brain aging: deciphering the role of somatic genetic instabilities and neural aneuploidy during ontogeny. Mol Cytogenet. 2009b;2:23. doi: 10.1186/1755-8166-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurov YB, Vorsanova SG, Liehr T, Kolotii AD, Iourov IY. X chromosome aneuploidy in the Alzheimer's disease brain. Mol Cytogenet. 2014;7:20. doi: 10.1186/1755-8166-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-Mamczarz K, Lakatta EG, Boheler KR, Xu X, Mattson MP, Falco G, Ko MS, Schlessinger D, Firman J, Kummerfeld SK, Wood AB, 3rd, Kim SK, Becker KG. AGEMAP: a gene expression database foraging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J, Freije A, Ruiz M, Coulon V, Sanz JR, Chiesa J, Gandarillas A. A mitosis block links active cell cycle with human epidermal differentiation and results in endoreplication. PLoS One. 2010;5:e15701. doi: 10.1371/journal.pone.0015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadil LM, Britigan EM, Weaver BA. 2n or not 2n: aneuploidy, polyploidy and chromosomal instability in primary and tumor cells Semin. Cell Dev Biol. 2013;24:370–379. doi: 10.1016/j.semcdb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genom Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]


