In vitro characterization of a thermostable Rubisco activase from Agave tequilana reveals a surface loop involved in the interaction between Rubisco activase and its substrate
Abstract
To maintain metabolic flux through the Calvin-Benson-Bassham cycle in higher plants, dead-end inhibited complexes of Rubisco must constantly be engaged and remodeled by the molecular chaperone Rubisco activase (Rca). In C3 plants, the thermolability of Rca is responsible for the deactivation of Rubisco and reduction of photosynthesis at moderately elevated temperatures. We reasoned that crassulacean acid metabolism (CAM) plants must possess thermostable Rca to support Calvin-Benson-Bassham cycle flux during the day when stomata are closed. A comparative biochemical characterization of rice (Oryza sativa) and Agave tequilana Rca isoforms demonstrated that the CAM Rca isoforms are approximately10°C more thermostable than the C3 isoforms. Agave Rca also possessed a much higher in vitro biochemical activity, even at low assay temperatures. Mixtures of rice and agave Rca form functional hetero-oligomers in vitro, but only the rice isoforms denature at nonpermissive temperatures. The high thermostability and activity of agave Rca mapped to the N-terminal 244 residues. A Glu-217-Gln amino acid substitution was found to confer high Rca activity to rice Rca. Further mutational analysis suggested that Glu-217 restricts the flexibility of the α4-β4 surface loop that interacts with Rubisco via Lys-216. CAM plants thus promise to be a source of highly functional, thermostable Rca candidates for thermal fortification of crop photosynthesis. Careful characterization of their properties will likely reveal further protein-protein interaction motifs to enrich our mechanistic model of Rca function.
Rubisco is responsible for all higher plant photosynthetic CO2 assimilation but exhibits slow and error-prone catalytic properties (Spreitzer and Salvucci, 2002; Tcherkez et al., 2006; Bracher et al., 2017). To support the enormous flux of carbon entering the biosphere, the enzyme is highly overexpressed in leaf tissue, at times comprising up to 50% of the total soluble protein (Parry et al., 2013). An additional unusual property of this enzyme concerns a high affinity for its substrate ribulose 1,5-bisphosphate and similar sugar phosphates, when the active site has not been primed by the cofactors CO2 and Mg2+ (Jordan and Chollet, 1983; Parry et al., 2008). In addition, the activated holoenzyme can also bind a range of molecules nonproductively (Andralojc et al., 2012). This situation leads to the formation of dead-end inhibited complexes that need to be remodeled by dedicated molecular chaperones known as Rubisco activases (Rcas; Portis, 2003; Mueller-Cajar et al., 2014; Carmo-Silva et al., 2015). Molecular motors exhibiting the Rca activity have evolved multiple times convergently in diverse autotrophic lineages to deal with the near universal property of Rubisco inhibition (Salvucci et al., 1985; Mueller-Cajar et al., 2011; Tsai et al., 2015; Mueller-Cajar, 2017).
In higher plants, the necessity of Rca to constantly clear blocked Rubisco active sites has been co-opted into a regulatory checkpoint, and the green-type Rcas are stimulated and inhibited by signals that correspond to high light (reducing equivalents) and low energy levels (ADP), respectively (Portis et al., 2008). It has been known for a long time that photosynthesis is inhibited at temperatures only slightly above the optimum (Berry and Bjorkman, 1980; Sage and Kubien, 2007; Yamori et al., 2014) and that this effect is correlated with a reduction in Rubisco activity (Weis, 1981; Kobza and Edwards, 1987). In the past two decades, numerous studies have linked an unusual thermolability of Rca to the reduction of photosynthesis at moderately elevated temperatures (Feller et al., 1998; Crafts-Brandner and Salvucci, 2000; Salvucci et al., 2001; Salvucci and Crafts-Brandner, 2004a, 2004b; Carmo-Silva and Salvucci, 2011). The notion that photosynthesis could be heat fortified by engineering Rca received significant experimental support, when Arabidopsis (Arabidopsis thaliana) plants expressing more thermostable Rca variants were shown to exhibit improved growth and biomass accumulation at supraoptimal temperatures (Kurek et al., 2007; Kumar et al., 2009). However, it should be noted that temperature stress does not solely affect Rca and photosynthesis, but also interferes with other processes in plants, such as anthesis and pollen tube growth (Wahid et al., 2007).
Rice (Oryza sativa) is the world’s most important food crop (Khush, 1997), and thus exploring the possibility of enhancing its thermotolerance is critical to ensure food security, especially when considering the anticipated effects of global climate change (Sage et al., 2008). Similar to other plants, photosynthesis in rice has also been shown to be Rubisco limited at temperatures over 33°C (Scafaro et al., 2012), and its Rca is thermolabile (Scafaro et al., 2016). Like most plants, rice possesses two isoforms of Rca, which are the product of alternative mRNA splicing (To et al., 1999) and yield a 46-kD Rcaα and a 43-kD Rcaβ gene product, which differ at the C termini. In rice, the two isoforms are not equally expressed; Rcaβ is more abundant than Rcaα in both O. sativa (Wang et al., 2010) and the wild rice Oryza meridionales (Scafaro et al., 2010). These studies also reported that in both species, heat stress resulted in a relative increased abundance of Rcaα; however, thermostability of the purified isoforms was found to be similar (Scafaro et al., 2016). In contrast to O. sativa, the wild rice Oryza australensis was shown to maintain photosynthetic activity at 45°C, which correlated with presence of Rca that, when measured in leaf extracts, maintained full activity up to 42°C compared to 36°C for O. sativa Rca (Scafaro et al., 2016). Interestingly, the recombinant O. australensis Rca presented with identical thermostability to the O. sativa Rca.
One hypothesis regarding the thermolability of Rubisco activase suggests that the enzyme may function as a fuse (Sharkey et al., 2001; Sharkey, 2005). As leaf temperatures rise, the deactivation of the activase will result in reduced Rubisco activity and thus a reduction in metabolic flux through the Calvin-Benson-Bassham cycle. In addition, plants from a high temperature environment may not require a particularly thermostable activase, since they often possess powerful leaf cooling mechanisms, especially transpiration (Wise et al., 2004; Carmo-Silva et al., 2012; Sage, 2013). We note that such a mechanism would not be available to plants using crassulacean acid metabolism (CAM) growing in a desert environment, where Calvin-Benson-Bassham cycle function will be taking place in the light, with the stomata closed to minimize water loss (Borland et al., 2014). Consequently, we reasoned that, like other enzymes required during daytime, CAM activases should be relatively thermostable (Brandon, 1967; Luttge, 2004). Any potential temperature fuse function should not be conserved in CAM Rca proteins.
While the lack of Rca thermostability and its physiological ramifications has received abundant attention, the molecular mechanism of Rca function was less amenable in the past, due to a lack of structural information. Only recently have atomic models of Rca become available, providing a structural framework to interpret earlier biochemical results (Henderson et al., 2011; Stotz et al., 2011; Hasse et al., 2015). Nevertheless, details of the green-type Rca mechanism remain mostly obscure (Wachter et al., 2013; Hauser et al., 2015). A current model based on biochemical results involves the disc-shaped Rca hexamer engaging inhibited Rubisco at the exposed βC-βD loop of the Rubisco large subunit N-terminal domain via the specificity helix H9 of the Rca’s α-helical subdomain (Larson et al., 1997; Ott et al., 2000; Li et al., 2005). In addition, the Rca N-terminal domain of approximately 60 residues and the pore loops of the hexamer have also been implicated in Rubisco remodeling (Esau et al., 1996; van de Loo and Salvucci, 1996; Stotz et al., 2011). It is evident that generating a more detailed mechanistic model requires the identification of additional residues involved in the Rca-Rubisco interaction.
Here we have utilized an in vitro biochemical approach to characterize the Rca isoforms from the CAM plant Agave tequilana and compared their properties to those from O. sativa. In addition to being highly thermostable, the CAM enzymes exhibit a high specific Rca activity due to enhanced affinity for rice Rubisco. Identification of a single residue change conferring this phenotype to rice Rca implicates a flexible loop in the Rca nucleotide binding domain as a new Rubisco interacting motif. Our results suggest that agave Rca is a suitable candidate to rigorously evaluate the effect of thermostable Rca function in rice, which may lead to enhanced productivity at moderately elevated temperatures.
RESULTS
Rubisco Activase from Agave Is Highly Functional at Activating Rice Rubisco
We aimed to explore our outlined hypothesis that CAM Rca proteins would need to be highly thermostable to maintain Rubisco function in high temperature environments in the absence of transpirational cooling. A sequence alignment of selected C3 and CAM Rca sequences did not yield systematic group-specific residue changes that could indicate parallel adaptive evolution as for instance seen in the phosphoenolpyruvate carboxylase of C4 plants (Supplemental Fig. S1; Christin et al., 2007). We therefore decided to test our hypothesis experimentally. We produced the agave (sequences mined from transcriptome data; Gross et al., 2013) and rice long (α) and short (β) Rca isoforms in Escherichia coli, followed by purification to homogeneity (Fig. 1A). The transcripts encoding the agave Rca isoforms result in amino acid sequences that are only 91% identical to each other and not consistent with alternative splicing of a single transcript. This indicates the existence of two separate Rca genes as described in cotton (Gossypium hirsutum; Salvucci et al., 2003). Both agave isoforms are 86% identical to the respective rice isoform.
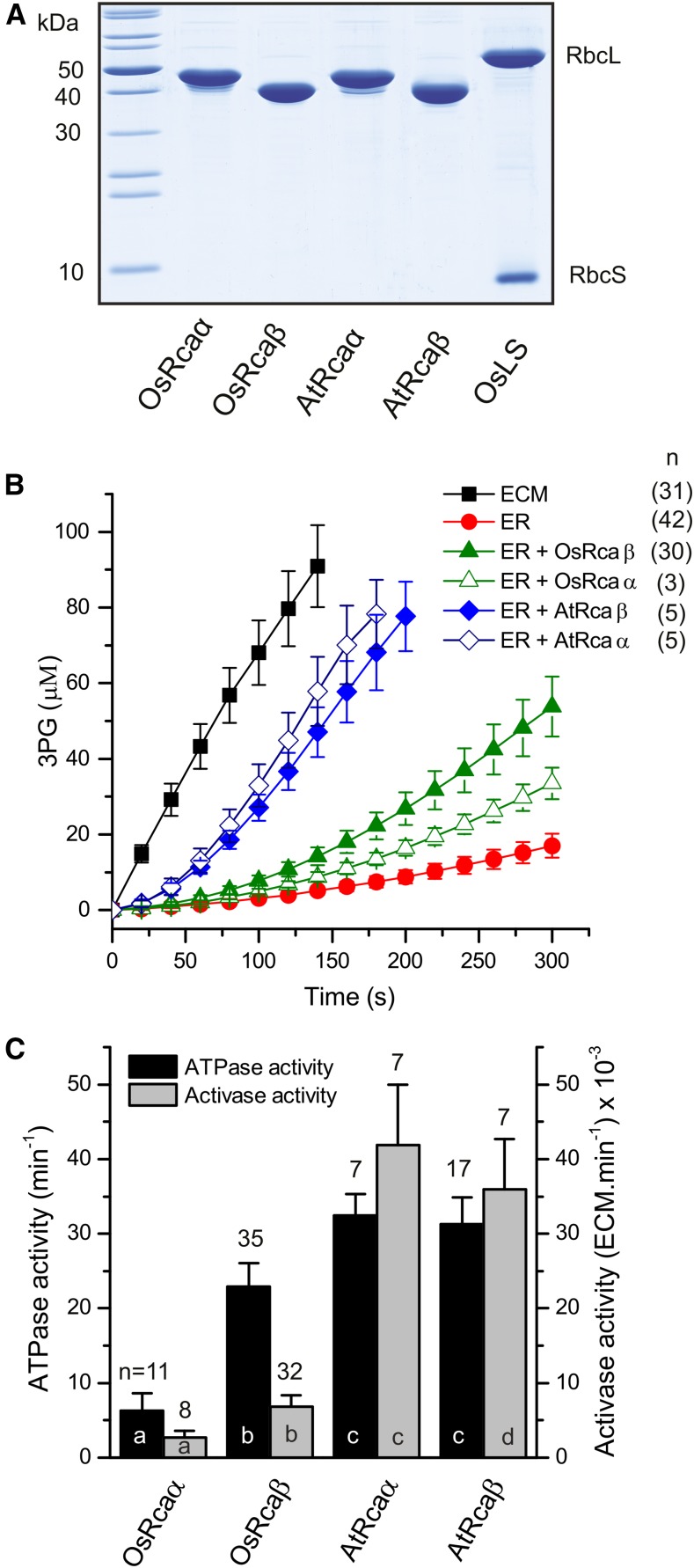
Figure 1.
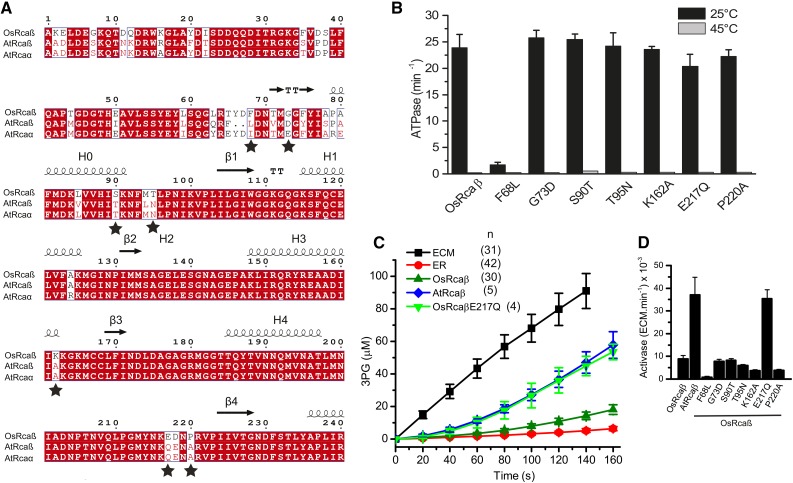
Biochemical properties of Rubisco activase isoforms from rice and agave. A, SDS-PAGE analysis of purified activases and rice Rubisco. A total of 4 µg of protein loaded per lane. B, Rubisco activation assays of RuBP-inhibited rice Rubisco (ER) performed at 25°C (0.5 µm Rubisco active sites, 5 µm activase protomer). C, ATPase activity assays (5 µm protomer, 25°C) and quantified Rubisco activase activity of the different activase isoforms. Error bars show mean and sd of independent experiments with n as indicated. Significant differences (P < 0.05) are denoted by different letters at the base of each bar.
Similar to that recently reported (Scafaro et al., 2016), the ATPase activity of the short rice Rubisco activase isoform (OsRcaβ) at 23 ± 3.2 min−1 protomer activase−1 was approximately3-fold higher than that observed for the long isoform (OsRcaα; Fig. 1C). In contrast both A. tequilana isoforms displayed a relatively high ATPase activity of approximately 32 min−1 (Fig. 1C). These numbers are very similar to those reported earlier for activases from other plant species (Hazra et al., 2015). All Rca isoforms presented with the concentration-dependent polydisperse oligomeric state typical for green-type Rca when analyzed by size exclusion chromatography (Supplemental Fig. S2; Blayney et al., 2011; Keown et al., 2013).
Plant activases tend to be broadly compatible, and so far incompatibility has only been demonstrated between activases and Rubiscos derived from Solanaceae and non-Solanaceae species (Wang et al., 1992). Consistent with this notion, the CAM activases were functional at catalyzing the conversion of inhibited rice Rubisco Enzyme-RuBP (ER) complexes to the holoenzyme (ECM). Strikingly, their comparative ability to do so was approximately 5-fold that of OsRcaβ under our assay conditions, with an activation rate of approximately 40 × 10−3 ECM.min−1 protomer activase−1 (Fig. 1, B and C). Supplemental Figure S3A illustrates an Rca activity quantification using our adaptation of the spectrophotometric Rubisco activase assay (Lan and Mott, 1991). The ability of Rca to function in vitro has previously been enhanced by inclusion of the macromolecular crowding agent polyethylene glycol (Salvucci, 1992), which increases the affinity between macromolecules due to excluded volume effects (Ellis, 2001). Inclusion of 5% v/v polyethylene glycol 3350 in the Rca assay almost eliminated the difference in Rca activity between OsRcaβ and AtRcaβ (Supplemental Fig. S3B). We therefore conclude that the enhanced Rca function exhibited by the agave isoforms are largely a consequence of an increased affinity between Rca and the carboxylase.
The Agave Activases Are Thermostable but Maintain High Activity at Lower Temperatures
Next we tested the thermostability of the different isoforms by incubating them at temperatures ranging from 25°C to 55°C for 10 mins, followed by an ATPase activity assay at 25°C. Consistent with our hypothesis, agave isoforms were around 10°C more thermostable than those from rice. AtRcaβ still maintained some functionality after incubation at 52°C, whereas OsRcaβ was completely inactive after incubation at 45°C (Fig. 2A). For both rice and agave, the long (α) isoforms were slightly less thermostable than the short (β) isoforms (Fig. 2A). In earlier work, it has been repeatedly found that bound ligands can increase the thermostability of Rca, for example (Salvucci et al., 2001; Henderson et al., 2013). In particular, the nucleotide analog ATPγS has an extraordinary effect on spinach (Spinacia oleracea) Rcaα, increasing the denaturation temperature from a low 30°C to 55°C (Crafts-Brandner et al., 1997; Keown and Pearce, 2014). An effect of this magnitude has not been described for any other Rca so far, and its physiological significance is unknown. For our isoforms, inclusion of Mg-ATPγS had only a very modest effect on thermostability (Supplemental Fig. S3C). Hence, the remainder of our thermostability characterizations were performed using the apo forms of our Rca proteins.
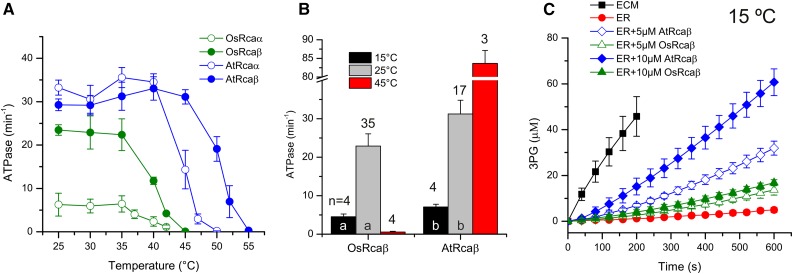
Figure 2.
Agave activases are highly thermostable. A, The activase isoforms were incubated for 10 min at the indicated temperature, and subsequently the remaining ATPase activity was assayed at 25°C (5 µm protomer). B, ATPase activity of OsRcaβ and AtRcaβ at various temperatures. Proteins were preincubated (10 min) and subsequently measured at the indicated temperatures under the conditions described in Figure 1C. Error bars show mean and sd of independent experiments with n indicated on top of the bars. Significant differences between activities at the same temperature (P < 0.05) are denoted by different letters at the base of each bar. C, AtRcaβ has higher Rubisco activase activity than OsRcaβ even at low temperature. Rubisco activation assays were performed as in Figure 1B at 15°C. A and C, error bars indicate mean and sd of independent experiments (n = 3–5).
It is commonly believed that increased thermostability of proteins in general trades off with functionality at lower temperatures (Zavodszky et al., 1998). To test whether this was the case for the CAM activases, we measured ATPase and activase activity at 15°C. As expected from physical principles, biochemical activity at the lower temperature was reduced, but in all cases the agave proteins retained their higher relative activities compared to their counterparts from rice (Fig. 2, B and C).
Agave Activases Form Functional Hetero-Oligomers with Rice Activases
Our results thus far indicated that agave Rca isoforms could be suitable candidates for expression in rice. In earlier work, Rcas from maize (Zea mays; of unknown thermostability) have been overexpressed in rice, which also contained the endogenous proteins (Yamori et al., 2012). Given the dynamic oligomeric state of Rubisco activase (Barta et al., 2010; Chakraborty et al., 2012), which includes rapid exchange of subunits (Salvucci and Klein, 1994; van de Loo and Salvucci, 1998; Stotz et al., 2011), it is clear that in many constellations a loss of function can be expected when subunits from different species are present and interact. Even if hetero-oligomerization does not impede biochemical function it is conceivable that heat-induced aggregation of the thermolabile isoform will cause the more thermostable isoforms to be recruited to the inclusion, in particular considering their presence in the same protein complex. To address these issues with regards to the rice-agave activase system, we performed a series of experiments. First, we characterized the biochemical properties of a 1:1 mixture of OsRcaβ and AtRcaβ. Both activase and ATPase activity of the subunits mixtures was intermediate to the homo-oligomers (Fig. 3, A and B). Although indicating that the activases do not impair their function, this result cannot distinguish between two distinct populations of homo-oligomers and the formation of functional hetero-oligomers. We therefore produced an Asp-173-Ala mutant of OsRcaβ. Asp-173 is the conserved Walker B acidic residue and when mutated, leads to loss of ATPase activity and nucleotide binding (van de Loo and Salvucci, 1998; Kuriata et al., 2014; Hasse et al., 2015; Fig. 3B). It is important to verify loss of functionality was not due to changes in oligomeric behavior. Therefore, all variants analyzed in this study (Supplemental Fig. S4A) were tested for the Rca-typical concentration-dependent oligomerization using analytical gel filtration (Supplemental Fig. S4B).
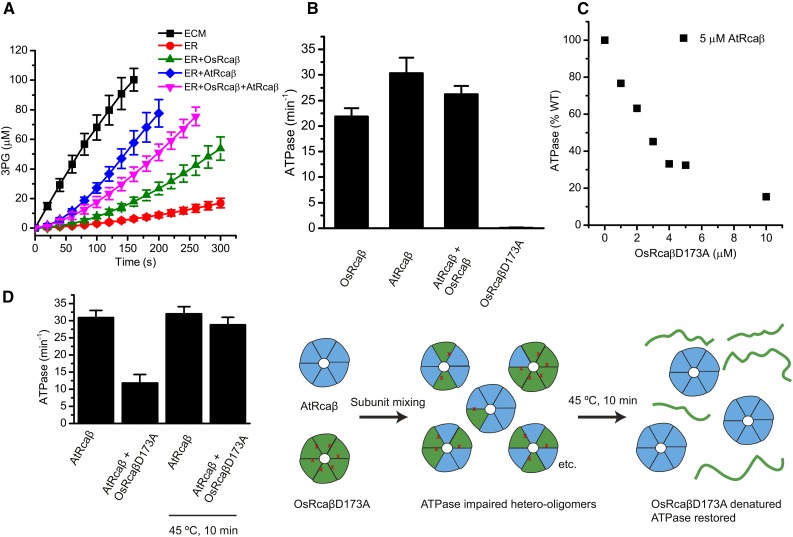
Figure 3.
Rice and agave activases form hetero-oligomers displaying intermediate functionality but subunit-specific thermostability. A and B, Rubisco activation assay (A) and ATPase assay (B) performed using 5 µm activase protomer (2.5 µm + 2.5 µm for AtRcaβ + OsRcaβ). ATPase is given per active site. C, Evidence for hetero-oligomerization. AtRcaβ was mixed with increasing amounts of ATPase inactive OsRcaβD173A, incubated for 10 min, and assayed for ATPase activity. D, Incubation at 45°C restores ATPase activity of impaired hetero-oligomers to levels of AtRcaβ wild type. C and D, ATPase activity was calculated as turnovers per wild-type active site. Error bars indicate mean and sd of independent experiments (A, n = 5–42; B and D, n = 4–7).
We then performed a mixing or poisoning experiment, where AtRcaβ concentration was kept constant and OsRcaβD173A concentration was titrated up, followed by measurement of the ATPase activity. Addition of the ATPase inactive rice subunits resulted in a strong decrease of the ATPase activity of the agave isoform (Fig. 3C) consistent with the cooperativity between subunits observed previously (van de Loo and Salvucci, 1998). Taken together, these results indicate that agave and rice activase form highly functional hetero-oligomers. We then decided to ask whether hetero-oligomerization would affect the thermostability of the agave subunits. The 1:1 mixture of AtRcaβ and OsRcaβD173A displayed a low ATPase activity of 11.8 ± 2.5 min−1 (per wild-type active site), consistent with the Walker B mutant poisoning the activity of the agave enzyme (Fig. 3D). Following a 10-min incubation of the mixture at 45°C, the agave activase regained full activity (28.8 ± 2.2 min−1). This result demonstrates that heat denaturation of the rice activase isoform, even when incorporated into hetero-oligomers, does not lead to reduced thermostability of the agave activase. Instead, denaturation of the rice subunits leads to formation of fully functional agave activase (Fig. 3D).
High Thermostability and Activity Maps to the N-Terminal Portion of Agave Rca
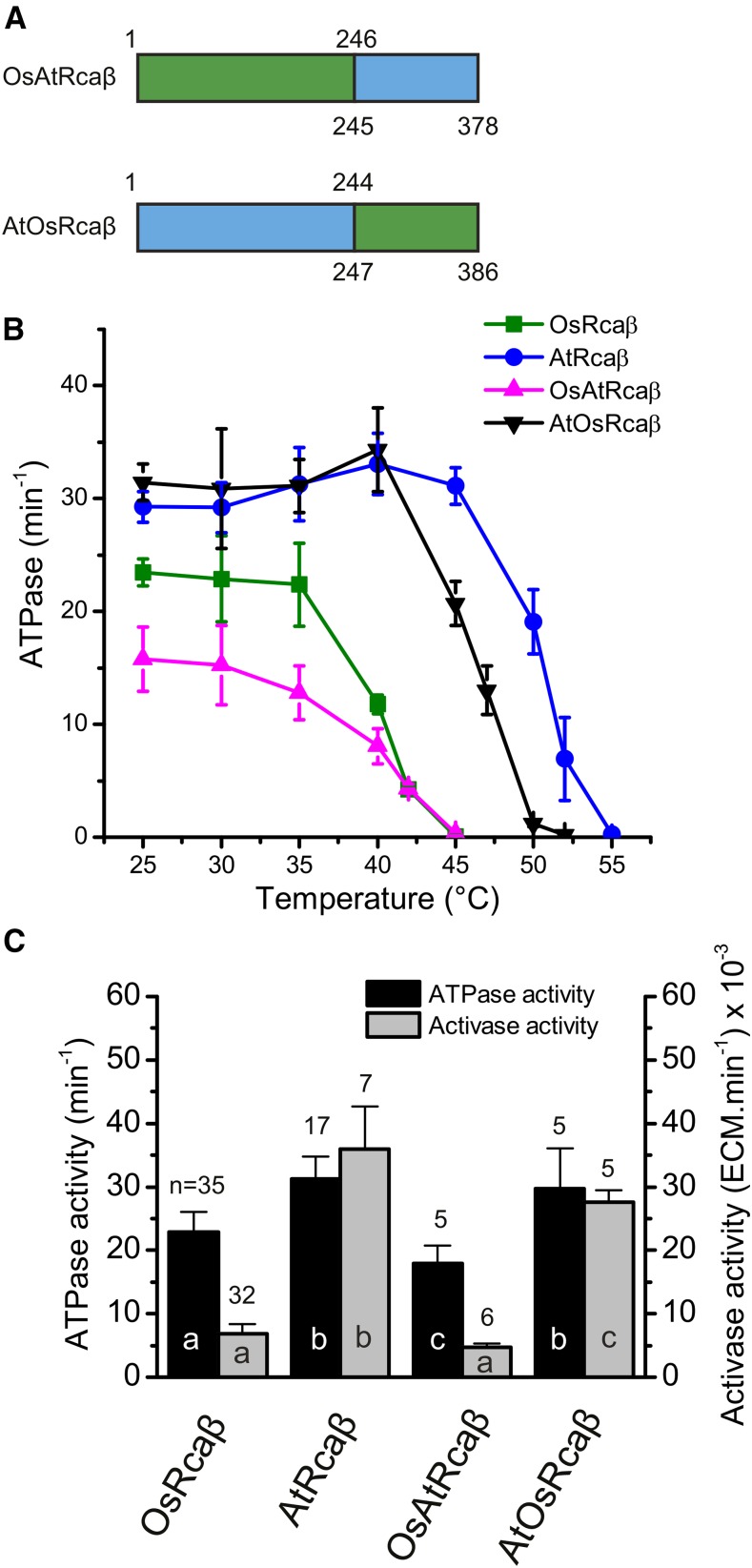
We next decided to map the determinants of agave Rca thermostability. As observed for activase homologs in previous studies (Esau et al., 1998; Kumar et al., 2009; Carmo-Silva and Salvucci, 2013), chimeric activases (Fig. 4A) could be purified and exhibited typical Rca oligomerization behavior (Supplemental Fig. S4, A and B). Characterization of domain-swapping chimeras indicated that both thermostability and higher activase activity of AtRcaβ mapped to its N-terminal 244 residues (crossover point indicated in Supplemental Fig. S1), which approximately encompasses the N-terminal domain and its nucleotide binding domain (Fig. 4, A–C). The chimeric activase AtOsRcaβ, comprising the N-terminal portion from agave and the C-terminal 140 residues from rice, maintained significant ATPase activity at 45°C but was inactive at 50°C and thus slightly less thermostable than AtRcaβ (Fig. 4B). It displayed a high activase activity equivalent to 80% of AtRcaβ (Fig. 4C). In contrast, the inverse construct OsAtRcaβ displayed very similar properties to OsRcaβ (Fig. 4, B and C). Thermostability and activase activity were identical between the two proteins, whereas the chimeric OsAtRcaβ possessed 75% of ATPase activity of OsRcaβ.
Figure 4.
Thermostability and high activase activity maps to residues 1 to 244 of AtRcaβ. A, Schematic representation of the chimeric activases. The crossover point corresponds approximately to the domain boundary of the nucleotide binding domain and the α-helical subdomain (residue 253 in OsRcaβ). Green, rice sequence; cyan, agave sequence. Numbering on top and bottom refers to residue numbers of the source sequence. B, The chimera containing the agave N-terminal portion (AtOsRcaβ) is thermostable. Thermostability assay performed as described in the legend to Figure 2A. Error bars indicate mean and sd of at least three independent experiments. C, The high activase activity of AtRcaβ maps to the N-terminal portion. Experimental conditions as in Figure 1C. Error bars show mean and sd of independent experiments with n = 3 to 4 (B) and as indicated on top of the bars (C). Significant differences (P < 0.05) are denoted by different letters at the base of each bar.
An inspection of a sequence alignment covering the identified N-terminal region of the rice and agave activase isoforms revealed only a small number of amino acids that differed systematically between the species (Fig. 5A). We therefore produced a series of seven OsRcaβ variants in an attempt to confer the agave isoforms’ properties to the rice enzyme. All seven variants were purified in soluble form (Supplemental Fig. S4A), and exhibited similar ATPase activity to the wild type, with the exception of F68L. This variant was severely impaired biochemically, as well as displaying atypical oligomerization (Fig. 5B; Supplemental Fig. S4B). Disappointingly, none of the variants was able to maintain functionality following a 10-min incubation at the nonpermissive temperature of 45°C (Fig. 5B). This result indicated that the relatively higher thermostability of AtRca compared to OsRca cannot be conferred by a single amino acid change. Instead it is determined by a combination of multiple residues that remain to be mapped. However, one of the variants, OsRcaβE217Q, possessed a similarly high activase activity as AtRcaβ (Fig. 5, C and D). Since the high activase activity of AtRcaβ is likely due to an increased affinity of the activase for its substrate Rubisco (Supplemental Fig. S3B), the phenotype of OsRcaβE217Q implicates this residue in protein-protein interactions.
Figure 5.
The Glu-217-Gln substitution confers high in vitro activase activity to OsRcaβ. A, Sequence alignment of the thermostability-conferring region of rice and agave activases. Residues indicated by stars were selected for mutagenesis. B, Thermostability assay of the Rca variants. ATPase assays were performed at 25°C and after a 10-min preincubation at 45°C. C and D, OsRcaβE217Q phenocopies the high in vitro activase activity of AtRcaβ. Error bars indicate mean and sd of independent experiments (B, n = 3–7 at 25°C, n = 1 at 45°C; C as shown; D, n = 3–5).
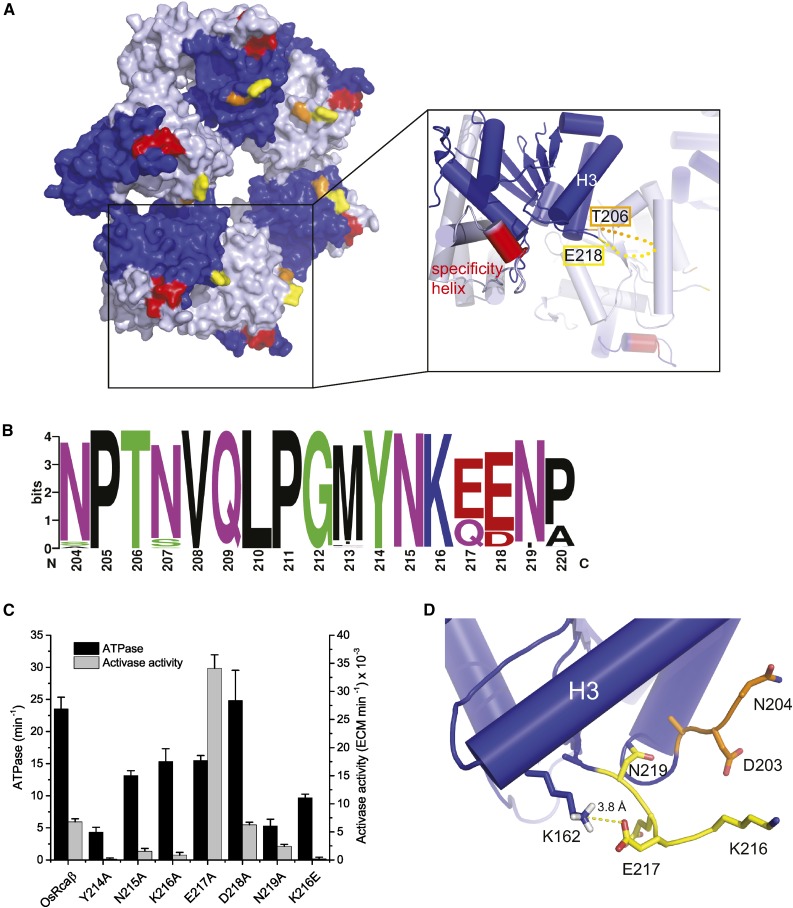
The α4-β4 Loop Is a New Rubisco-Rca Interacting Motif
Glu-217 is positioned in the loop connecting helix 4 to beta strand 4 in the nucleotide-binding domain (Supplemental Fig. S1). The two structural models of tobacco (Nicotiana tabacum) and Arabidopsis Rca are both missing 11 (albeit different) residues of the loop, suggesting that it is highly flexible (Stotz et al., 2011; Hasse et al., 2015). The loop is located on the top side of the Rca hexamer (Fig. 6A), the face exposing the nucleotide binding domains, which is commonly involved in substrate engagement of AAA+ proteins (Zhang and Wigley, 2008). This side is also predicted to be the surface utilized by the disc-shaped Rca hexamer to bind Rubisco (Stotz et al., 2011; Wachter et al., 2013) and exposes the Rubisco-interacting specificity helix H9. The increase in biochemical activity brought about by the E217Q substitution suggests that this variant possesses an enhanced affinity for rice Rubisco. Analysis of 220 plant Rca sequences shows that the loop is highly conserved (Fig. 6B), and we thus performed an Ala scan on the residues flanking E217. Substituting Y214 and N219 with Ala resulted in approximately 80% reductions in ATPase activity, suggesting that the loop can influence ATPase function but complicating analysis of the residues’ contribution to Rubisco remodeling (Fig. 6C). N219A, but not Y214A, exhibited an oligomerization defect (Supplemental Fig. S4B). E217A exhibited a similarly high activity phenotype to E217Q (Fig. 6C). This result suggests that residue 217 determines the conformation and Rubisco binding affinity of the loop rather than contributing to the Rca-Rubisco interaction directly. The Arabidopsis Rca structure includes information on E217 and K216, although the B-factors for the region are high (Hasse et al., 2015). The side chain carboxyl group of E217 is within salt bridge-forming distance of the amino group of K162, which is located at the C terminus of helix 3 (Fig. 6D). Hence, the E217A or Q substitution may disrupt this interaction, leading to enhanced flexibility and thus Rubisco affinity. The evidence for this interaction is strengthened by the observation that Rca sequences that encode E217 tend to possess a Lys at position 162. In contrast, sequences encoding Q217 present a variety of non-Lys residues at position 162 (indicated in Supplemental Fig. S1). However, it should be pointed out that the OsRcaβK162A variant, which cannot form the salt-bridge, did not exhibit the high Rca activity of AtRcaβ (Fig. 5D). This result indicates that mere disruption of the proposed salt-bridge is insufficient to confer the high activity phenotype to OsRca.
Figure 6.
Characterization of a new Rubisco interacting loop. A, Relative position of the Q217-containing loop in context of the Rca hexamer. A surface representation of the hexameric tobacco Rca model (PDB:3ZW6) is shown with alternate subunits colored in blue and light blue. The specificity helix H9 is colored red, and the last resolved residues flanking the flexible loop are shown in orange (T206) and yellow (E218). The close-up view shows the local secondary structure with helices represented by cylinders. The loop is indicated by a dotted line. B, Logo motif of the loop from an alignment of 220 plant Rca sequences generated using WebLogo (Crooks et al., 2004). C, Biochemical characterization of loop mutants was performed as in Figure 1C. Error bars indicate mean and sd of independent experiments with n = 4 to 9. D, The loop region in the Arabidopsis Rca structure (PDB:4W5W) suggests that the carboxyl group of E217 forms a salt-bridge with the amino group of K162. For simplicity, residue numbering in all panels of this figure corresponds to the OsRcaβ sequence. Structures were drawn using Pymol (www.pymol.org).
Both N215A and K216A presented with over 50% of wild-type ATPase activity, but exhibited strong reductions in Rca function. Supporting an important role for K216 in protein-protein interaction, the charge switch variant K216E retained 41% of ATPase activity, but was nonfunctional at activating Rubisco (Fig. 6C). Together these results indicate that the α4-β4 loop represents a new Rubisco interacting motif, where the side chains of N215 and K216 are likely to be directly involved in the protein-protein interaction. An E217-K162 salt-bridge may limit the flexibility of this loop and concomitantly reduce the affinity of OsRcaβ for its cognate Rubisco.
DISCUSSION
In this work, we aimed to test the hypothesis that plants utilizing the Calvin-Benson cycle in leaves experiencing high temperatures, such as those exhibiting the CAM photosynthetic syndrome, would encode a thermostable Rca protein to ensure Rubisco’s functionality. We made the unexpected discovery that the Rca isoforms from agave not only possessed the predicted properties, but also exhibited a much higher Rca activity against rice Rubisco than the cognate activase. In addition, the higher enzymatic activity of AtRcaβ was also demonstrated at lower temperatures, indicating that a flexibility-thermostability tradeoff does not apply in this instance. We hypothesize that there has been a selection pressure for this property to adapt agave photosynthesis to its desert environment. Following the night-time CO2 uptake Phase I, Phase II of CAM occurs during low morning temperatures, whereas Phase III corresponds to high day-time temperatures with closed stomata (Borland et al., 2014). Previous work has shown that Rubisco activation state increases from a low level during Phase II and is maintained during Phase III and IV in a number of CAM species (Maxwell et al., 1999; Griffiths et al., 2002, 2008; Davies and Griffiths, 2012), indicating Rca needs to be functional during these phases.
Earlier work reported relatively thermostable Rca from the C3 desert plant Larrea tridentata, which is found in a similar environment to agave (Salvucci and Crafts-Brandner, 2004b). However, it lost 50% of ATPase activity at 43°C (44°C in the presence of ATPγS) and thus is only slightly more thermostable than rice Rca. A very comprehensive analysis of cotton Rcaβ established conditions leading to high thermostability (apparent Tm of 50°C in presence of 8 mm ADP); however, the apo enzyme denatures below 40°C (Henderson et al., 2013). To our knowledge, agave Rca presents as the most thermostable green-type Rca in the absence of stabilizing ligands. Interestingly, the red-type Rca from the thermophilic red algae Cyanidioschyzon merolae tolerated incubation at 60°C (Loganathan et al., 2016); however, it is not compatible with plant Rubisco.
Our results imply that plants exhibiting the CAM syndrome growing in high temperature environments are in general likely to possess thermostable Rca proteins. Since CAM has evolved multiple times independently (Silvera et al., 2010), we can thus predict multiple unique solutions to thermostability to be available. Our inability to find a single amino acid substitution conferring thermostability to OsRca indicates multiple residue changes contributing subtle stabilizing effects (Matthews, 1993) will be necessary. In addition, it is possible that the observed high affinity of agave Rca for rice Rubisco is not due to use of a heterologous Rubisco. Instead it may be a consequence of the relatively low Rubisco content reported for CAM plants (4% of total soluble leaf protein in Agave victoriae; Galmes et al., 2014), which would necessitate a higher affinity (lower Km) of agave Rca for the Rubisco substrate assuming stromal Rubisco concentrations were also low. In contrast, rice possesses very high Rubisco levels of approximately 50% of total soluble leaf protein (Ishikawa et al., 2011), and Rca would thus require a lower affinity for Rubisco. To assess this point further, it will be necessary to measure the relative activation rate of agave Rubisco by AtRca. If AtRca also presents a high affinity for agave Rubisco, it follows that performing similar analyses of Rca proteins from plants containing low Rubisco levels will likely yield additional protein-protein interaction sites.
More generally, the ability to locate thermostable activases that also function robustly at low temperatures suggests that the thermolability of C3-plant Rca proteins could be under positive selection, which is relaxed in CAM plants. This implies a fitness defect may become apparent, should the hypothetical thermal fuse be overridden by thermostable Rca proteins (Sharkey, 2005). Nevertheless, organisms are prone to overreact to environmental stress, and under agricultural conditions our crops may well tolerate approaches that aim to “over-clock” metabolic pathways. Our work indicates that overexpression of AtRca in rice (for instance using a similar approach as described previously; Fukayama et al., 2012) should permit functional activase to persist at higher temperatures even in the presence of the endogenous Rca. This should expand the temperature range of photosynthesis of the transformants and possibly enhance growth at supraoptimal temperature similar to that seen in Arabidopsis (Kurek et al., 2007; Kumar et al., 2009). Experiments aiming to explore this possibility are underway. In addition, the rapid development of genome editing tools such as CRISPR-Cas9 (Belhaj et al., 2015) will permit more sophisticated modifications such as deletion of the endogenous OsRca. This would overcome problems encountered upon Rca overexpression earlier, such as reduced Rubisco levels (Fukayama et al., 2012).
The implication that the α4-β4 loop contributes to Rubisco activation helps in further constraining our mechanistic model of Rubisco activation (Wachter et al., 2013; Bhat et al., 2017) and firmly establishes the top surface of Rca as the Rubisco interacting face. Additional work will be needed to resolve the interacting residues on Rubisco’s surface. It will also be interesting to see whether the loop is a passive docking motif, or if loop movements are in fact coupled to rearrangement of the Rubisco active site.
By describing a highly functional, thermostable Rca that does not impair endogenous rice activase function, we hope to provide a tool that will enable a more detailed dissection of the role of Rca in the thermostability of photosynthesis. In addition, we intend to illustrate the utility of careful biochemical characterization of heterologous systems in the discovery of functional determinants, such as the described Rubisco interacting loop. We anticipate that the journey toward a more complete mechanistic understanding of plant Rca function will be informed by similar studies. Maintaining the function of the photosynthetic carboxylase is critical for our food and energy supplies, and increasing our appreciation of this process will outline feasible avenues toward maximizing CO2 fixation and enable their efficient implementation (Sharwood, 2017).
MATERIALS AND METHODS
Molecular Biology
Nucleotide sequences encoding Rcaα and Rcaβ from rice (Oryza sativa; uniprot: P93431-1, residues 48–466; P93431-2, residues 48–433) and agave (Agave tequilana; locus 3705, residues 57–474; locus 27298, residues 58–435; Gross et al., 2013) were synthesized (GenScript) and cloned into the SacII-HindIII site of pHue (Catanzariti et al., 2004). The QuikChange protocol (Stratagene) was used to introduce desired point mutations. An ApoI site was generated in the sequence encoding OsRcaβ using the silent G738A nucleotide substitution at the same position as found in AtRcaβ. This permitted construction of pHueAtOsRcaβ and pHueOsAtRcaβ encoding chimeric activases, by exchanging the ApoI-HindIII fragment of the two constructs. Plasmids and primers used in this study are listed in Supplemental Tables 1 and 2.
Protein Expression and Purification
Plasmids encoding the wild type and variant activases were transformed in Escherichia coli BL21 (DE3) cells, and proteins were produced and purified using immobilized metal affinity chromatography followed by precise His6-Ub cleavage and anion-exchange chromatography exactly as previously described for other His6-Ub tagged proteins (Tsai et al., 2015). The concentration of pure proteins was measured at 280 nm using extinction coefficients obtained using the Expasy protparam tool (web.expasy.org/protparam/). Rice Rubisco was purified from Oryza sativa L. ssp. japonica leaf tissue using a protocol similar to that described (Makino et al., 1983). Leaf powder was resuspended in buffer A (50 mm Tris-HCl, pH8.0, 50 mm NaCl, 1 mm Na2EDTA, 12.5% v/v glycerol) containing 1 mm phenylmethanesulfonyl fluoride, protease inhibitor cocktail (Pierce, ThermoScientific), 5 mm dithiothreitol (DTT), and 2% w/v polyvinylpolypyrrolidone). Following filtration by miracloth and removal of cell debris by centrifugation, 33% and 55% ammonium sulfate cuts were performed. The pellet obtained at 55% was resuspended in buffer A containing 5 mm DTT, followed by extensive dialysis against buffer B (20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 0.1 mm Na2EDTA, 1 mm DTT, 12.5% v/v glycerol). The dialyzed solution was applied to a MonoQ 10/100 column (GE healthcare) preequilibrated with buffer B and the protein eluted with a linear salt gradient to 500 mm NaCl. Fractions containing Rubisco were pooled and further purified by elution over a Superdex200 16/60 column equilibrated with buffer B.
Rca proteins were supplemented with 5% v/v glycerol prior to storage. All pure proteins were concentrated to >5 mg/mL, flash frozen in liquid nitrogen, and stored at −80°C.
Biochemical Assays
Rca activity was measured using an adaptation (Tsai et al., 2015) of the spectrophotometric Rubisco activase assay (Lan and Mott, 1991) and quantified as described (Loganathan et al., 2016; Supplemental Fig. S3A). Then 20 mm NaHCO3 and 1 mm RuBP were used as substrates. Ribose 5-phosphate was used to synthesize RuBP enzymatically (Horecker et al., 1958) and purified by anion-exchange chromatography (Kane et al., 1998). To generate ECM, 20 µm of Rubisco active sites were incubated (10 min, 25°C) with 20 mm NaHCO3 and 10 mm MgCl2 in buffer C (20 mm Tris-HCl, pH 8.0, 50 mm NaCl). ER was formed by incubating 20 µm of Rubisco active sites in buffer C containing 4 mm EDTA (10 min, 25°C), followed by addition of RuBP to 1 mm.
ATPase activity was assayed using a coupled spectrophotometric assay (Stotz et al., 2011) but did not contain DTT. Analytical gel filtration was performed by eluting the proteins in buffer C using a Superdex 200 PC3.2/30 Increase (GE healthcare) column.
To assess Rca thermostability, the purified enzymes (in concentrated form of approximately 10 mg/mL) were incubated for 10 min at the relevant temperature using a Biometra TB2 thermoblock before performing the ATPase assay at 25°C.
Statistical Analysis
Replicate measurements of biochemical assays were performed using the same protein preparations on different days. Statistical comparisons were performed using one-way ANOVA followed by the Tukey-Kramer method in OriginPro 8.5.1 (OriginLab). A value of P < 0.05 was considered significant.
Supplemental Materials
The following supplemental materials are available.
Supplemental Figure S1. Selected C3 and CAM Rca sequences were aligned using Clustal Omega.
Supplemental Figure S2. Rice and agave Rca proteins exhibit concentration-dependent polydispersity.
Supplemental Figure S3. Biochemical characterization of the Rca proteins.
Supplemental Figure S4. Quality control of Rca variants used in this study.
Supplemental Table S1. Plasmids used in this study.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank Prof. Prakash Kumar (National University of Singapore) for the gift of rice leaves. Maria Lapina, Lynette Liew, Xie Hua-Zhen, and Jediael Ng Zheng Ying are acknowledged for technical assistance.
Glossary
- CAM
crassulacean acid metabolism
- DTT
dithiothreitol
- ER
Rubisco Enzyme-RuBP
- MVB
Rca: chaperone Rubisco activase
Footnotes
This work was funded by a Nanyang Technological University startup grant and the Ministry of Education (MOE) of Singapore Tier 2 grant to O.M.-C. (MOE2013-T2-2-089).
Articles can be viewed without a subscription.
References
- Andralojc PJ, Madgwick PJ, Tao Y, Keys A, Ward JL, Beale MH, Loveland JE, Jackson PJ, Willis AC, Gutteridge S, et al. (2012) 2-Carboxy-D-arabinitol 1-phosphate (CA1P) phosphatase: evidence for a wider role in plant Rubisco regulation. Biochem J 442: 733–742 [DOI] [PubMed] [Google Scholar]
- Barta C, Dunkle AM, Wachter RM, Salvucci ME (2010) Structural changes associated with the acute thermal instability of Rubisco activase. Arch Biochem Biophys 499: 17–25 [DOI] [PubMed] [Google Scholar]
- Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V (2015) Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol 32: 76–84 [DOI] [PubMed] [Google Scholar]
- Berry J, Bjorkman O (1980) Photosynthetic response and adaptation to temperature in higher-plants. Annu Rev Plant Physiol Plant Mol Biol 31: 491–543 [Google Scholar]
- Bhat JY, Thieulin-Pardo G, Hartl FU, Hayer-Hartl M (2017) Rubisco activases: AAA+ chaperones adapted to enzyme repair. Front Mol Biosci 4: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blayney MJ, Whitney SM, Beck JL (2011) NanoESI mass spectrometry of Rubisco and Rubisco activase structures and their interactions with nucleotides and sugar phosphates. J Am Soc Mass Spectrom 22: 1588–1601 [DOI] [PubMed] [Google Scholar]
- Borland AM, Hartwell J, Weston DJ, Schlauch KA, Tschaplinski TJ, Tuskan GA, Yang X, Cushman JC (2014) Engineering crassulacean acid metabolism to improve water-use efficiency. Trends Plant Sci 19: 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher A, Whitney SM, Hartl FU, Hayer-Hartl M (2017) Biogenesis and metabolic maintenance of Rubisco. Annu Rev Plant Biol 68: 29–60 [DOI] [PubMed] [Google Scholar]
- Brandon PC. (1967) Temperature features of enzymes affecting crassulacean acid metabolism. Plant Physiol 42: 977–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Silva AE, Gore MA, Andrade-Sanchez P, French AN, Hunsaker DJ, Salvucci ME (2012) Decreased CO2 availability and inactivation of Rubisco limit photosynthesis in cotton plants under heat and drought stress in the field. Environ Exp Bot 83: 1–11 [Google Scholar]
- Carmo-Silva AE, Salvucci ME (2011) The activity of Rubisco’s molecular chaperone, Rubisco activase, in leaf extracts. Photosynth Res 108: 143–155 [DOI] [PubMed] [Google Scholar]
- Carmo-Silva AE, Salvucci ME (2013) The regulatory properties of Rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiol 161: 1645–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Silva E, Scales JC, Madgwick PJ, Parry MA (2015) Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ 38: 1817–1832 [DOI] [PubMed] [Google Scholar]
- Catanzariti A-M, Soboleva TA, Jans DA, Board PG, Baker RT (2004) An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci 13: 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Kuriata AM, Nathan Henderson J, Salvucci ME, Wachter RM, Levitus M (2012) Protein oligomerization monitored by fluorescence fluctuation spectroscopy: self-assembly of rubisco activase. Biophys J 103: 949–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P-A, Salamin N, Savolainen V, Duvall MR, Besnard G (2007) C4 Photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr Biol 17: 1241–1247 [DOI] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, Salvucci ME (2000) Rubisco activase constrains the photosynthetic potential of leaves at high temperature and CO2. Proc Natl Acad Sci USA 97: 13430–13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crafts-Brandner SJ, Van De Loo FJ, Salvucci ME (1997) The two forms of ribulose-1,5-bisphosphate carboxylase/oxygenase activase differ in sensitivity to elevated temperature. Plant Physiol 114: 439–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BN, Griffiths H (2012) Competing carboxylases: circadian and metabolic regulation of Rubisco in C3 and CAM Mesembryanthemum crystallinum L. Plant Cell Environ 35: 1211–1220 [DOI] [PubMed] [Google Scholar]
- Ellis RJ. (2001) Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci 26: 597–604 [DOI] [PubMed] [Google Scholar]
- Esau BD, Snyder GW, Portis AR Jr (1996) Differential effects of N- and C-terminal deletions on the two activities of rubisco activase. Arch Biochem Biophys 326: 100–105 [DOI] [PubMed] [Google Scholar]
- Esau BD, Snyder GW, Portis AR (1998) Activation of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) with chimeric activase proteins. Photosynth Res 58: 175–181 [Google Scholar]
- Feller U, Crafts-Brandner SJ, Salvucci ME (1998) Moderately high temperatures inhibit ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase-mediated activation of Rubisco. Plant Physiol 116: 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukayama H, Ueguchi C, Nishikawa K, Katoh N, Ishikawa C, Masumoto C, Hatanaka T, Misoo S (2012) Overexpression of rubisco activase decreases the photosynthetic CO2 assimilation rate by reducing rubisco content in rice leaves. Plant Cell Physiol 53: 976–986 [DOI] [PubMed] [Google Scholar]
- Galmés J, Kapralov MV, Andralojc PJ, Conesa MA, Keys AJ, Parry MA, Flexas J (2014) Expanding knowledge of the Rubisco kinetics variability in plant species: environmental and evolutionary trends. Plant Cell Environ 37: 1989–2001 [DOI] [PubMed] [Google Scholar]
- Griffiths H, Helliker B, Roberts A, Haslam RP, Girnus J, Robe WE, Borland AM, Maxwell K (2002) Regulation of Rubisco activity in crassulacean acid metabolism plants: better late than never. Funct Plant Biol 29: 689–696 [DOI] [PubMed] [Google Scholar]
- Griffiths H, Robe WE, Girnus J, Maxwell K (2008) Leaf succulence determines the interplay between carboxylase systems and light use during Crassulacean acid metabolism in Kalanchöe species. J Exp Bot 59: 1851–1861 [DOI] [PubMed] [Google Scholar]
- Gross SM, Martin JA, Simpson J, Abraham-Juarez MJ, Wang Z, Visel A (2013) De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC Genomics 14: 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasse D, Larsson AM, Andersson I (2015) Structure of Arabidopsis thaliana Rubisco activase. Acta Crystallogr D Biol Crystallogr 71: 800–808 [DOI] [PubMed] [Google Scholar]
- Hauser T, Popilka L, Hartl FU, Hayer-Hartl M (2015) Role of auxiliary proteins in Rubisco biogenesis and function. Nat Plants 1: 15065. [DOI] [PubMed] [Google Scholar]
- Hazra S, Henderson JN, Liles K, Hilton MT, Wachter RM (2015) Regulation of ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) activase: product inhibition, cooperativity, and magnesium activation. J Biol Chem 290: 24222–24236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JN, Hazra S, Dunkle AM, Salvucci ME, Wachter RM (2013) Biophysical characterization of higher plant Rubisco activase. Biochim Biophys Acta 1834: 87–97 [DOI] [PubMed] [Google Scholar]
- Henderson JN, Kuriata AM, Fromme R, Salvucci ME, Wachter RM (2011) Atomic resolution x-ray structure of the substrate recognition domain of higher plant ribulose-bisphosphate carboxylase/oxygenase (Rubisco) activase. J Biol Chem 286: 35683–35688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horecker BL, Hurwitz J, Weissbach A (1958) Ribulose diphosphate. Biochemical Preparations 6: 83–90 [Google Scholar]
- Ishikawa C, Hatanaka T, Misoo S, Miyake C, Fukayama H (2011) Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiol 156: 1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Chollet R (1983) Inhibition of ribulose bisphosphate carboxylase by substrate ribulose 1,5-bisphosphate. J Biol Chem 258: 13752–13758 [PubMed] [Google Scholar]
- Kane HJ, Wilkin JM, Portis AR, Andrews TJ (1998) Potent inhibition of ribulose-bisphosphate carboxylase by an oxidized impurity in ribulose-1,5-bisphosphate. Plant Physiol 117: 1059–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keown JR, Griffin MD, Mertens HD, Pearce FG (2013) Small oligomers of ribulose-bisphosphate carboxylase/oxygenase (Rubisco) activase are required for biological activity. J Biol Chem 288: 20607–20615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keown JR, Pearce FG (2014) Characterization of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase activase isoforms reveals hexameric assemblies with increased thermal stability. Biochem J 464: 413–423 [DOI] [PubMed] [Google Scholar]
- Khush GS. (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35: 25–34 [PubMed] [Google Scholar]
- Kobza J, Edwards GE (1987) Influences of leaf temperature on photosynthetic carbon metabolism in wheat. Plant Physiol 83: 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Li C, Portis AR Jr (2009) Arabidopsis thaliana expressing a thermostable chimeric Rubisco activase exhibits enhanced growth and higher rates of photosynthesis at moderately high temperatures. Photosynth Res 100: 143–153 [DOI] [PubMed] [Google Scholar]
- Kurek I, Chang TK, Bertain SM, Madrigal A, Liu L, Lassner MW, Zhu G (2007) Enhanced thermostability of Arabidopsis Rubisco activase improves photosynthesis and growth rates under moderate heat stress. Plant Cell 19: 3230–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriata AM, Chakraborty M, Henderson JN, Hazra S, Serban AJ, Pham TV, Levitus M, Wachter RM (2014) ATP and magnesium promote cotton short-form ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase hexamer formation at low micromolar concentrations. Biochemistry 53: 7232–7246 [DOI] [PubMed] [Google Scholar]
- Lan Y, Mott KA (1991) Determination of apparent K(m) values for ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) activase using the spectrophotometric assay of Rubisco activity. Plant Physiol 95: 604–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EM, O'Brien CM, Zhu G, Spreitzer RJ, Portis AR Jr (1997) Specificity for activase is changed by a Pro-89 to Arg substitution in the large subunit of Ribulose-1,5-bisphosphate carboxylase/oxygenase. J Biol Chem 272: 17033–17037 [DOI] [PubMed] [Google Scholar]
- Li C, Salvucci ME, Portis AR Jr (2005) Two residues of rubisco activase involved in recognition of the Rubisco substrate. J Biol Chem 280: 24864–24869 [DOI] [PubMed] [Google Scholar]
- Loganathan N, Tsai Y-CC, Mueller-Cajar O (2016) Characterization of the heterooligomeric red-type rubisco activase from red algae. Proc Natl Acad Sci USA 113: 14019–14024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüttge U. (2004) Ecophysiology of Crassulacean Acid Metabolism (CAM). Ann Bot (Lond) 93: 629–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Mae T, Ohira K (1983) Purification and storage of ribulose 1,5-bisphosphate carboxylase from rice leaves. Plant Cell Physiol 24: 1169–1173 [Google Scholar]
- Matthews BW. (1993) Structural and genetic analysis of protein stability. Annu Rev Biochem 62: 139–160 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Borland AM, Haslam RP, Helliker BR, Roberts A, Griffiths H (1999) Modulation of Rubisco activity during the diurnal phases of the Crassulacean acid metabolism plant Kalanchoe daigremontiana. Plant Physiol 121: 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O. (2017) The diverse AAA+ machines that repair inhibited Rubisco active sites. Front Mol Biosci 4: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O, Stotz M, Bracher A (2014) Maintaining photosynthetic CO2 fixation via protein remodelling: the Rubisco activases. Photosynth Res 119: 191–201 [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O, Stotz M, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M (2011) Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479: 194–199 [DOI] [PubMed] [Google Scholar]
- Ott CM, Smith BD, Portis AR Jr, Spreitzer RJ (2000) Activase region on chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase. Nonconservative substitution in the large subunit alters species specificity of protein interaction. J Biol Chem 275: 26241–26244 [DOI] [PubMed] [Google Scholar]
- Parry MA, Andralojc PJ, Scales JC, Salvucci ME, Carmo-Silva AE, Alonso H, Whitney SM (2013) Rubisco activity and regulation as targets for crop improvement. J Exp Bot 64: 717–730 [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Keys AJ, Madgwick PJ, Carmo-Silva AE, Andralojc PJ (2008) Rubisco regulation: a role for inhibitors. J Exp Bot 59: 1569–1580 [DOI] [PubMed] [Google Scholar]
- Portis AR., Jr (2003) Rubisco activase - Rubisco’s catalytic chaperone. Photosynth Res 75: 11–27 [DOI] [PubMed] [Google Scholar]
- Portis AR Jr, Li C, Wang D, Salvucci ME (2008) Regulation of Rubisco activase and its interaction with Rubisco. J Exp Bot 59: 1597–1604 [DOI] [PubMed] [Google Scholar]
- Sage RF. (2013) Photorespiratory compensation: a driver for biological diversity. Plant Biol (Stuttg) 15: 624–638 [DOI] [PubMed] [Google Scholar]
- Sage RF, Kubien DS (2007) The temperature response of C(3) and C(4) photosynthesis. Plant Cell Environ 30: 1086–1106 [DOI] [PubMed] [Google Scholar]
- Sage RF, Way DA, Kubien DS (2008) Rubisco, Rubisco activase, and global climate change. J Exp Bot 59: 1581–1595 [DOI] [PubMed] [Google Scholar]
- Salvucci ME. (1992) Subunit interactions of Rubisco activase: polyethylene glycol promotes self-association, stimulates ATPase and activation activities, and enhances interactions with Rubisco. Arch Biochem Biophys 298: 688–696 [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ (2004a) Mechanism for deactivation of Rubisco under moderate heat stress. Physiol Plant 122: 513–519 [Google Scholar]
- Salvucci ME, Crafts-Brandner SJ (2004b) Relationship between the heat tolerance of photosynthesis and the thermal stability of rubisco activase in plants from contrasting thermal environments. Plant Physiol 134: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME, Klein RR (1994) Site-directed mutagenesis of a reactive lysyl residue (Lys-247) of Rubisco activase. Arch Biochem Biophys 314: 178–185 [DOI] [PubMed] [Google Scholar]
- Salvucci ME, Osteryoung KW, Crafts-Brandner SJ, Vierling E (2001) Exceptional sensitivity of Rubisco activase to thermal denaturation in vitro and in vivo. Plant Physiol 127: 1053–1064 [PMC free article] [PubMed] [Google Scholar]
- Salvucci ME, Portis AR Jr, Ogren WL (1985) A soluble chloroplast protein catalyzes ribulosebisphosphate carboxylase/oxygenase activation in vivo. Photosynth Res 7: 193–201 [DOI] [PubMed] [Google Scholar]
- Salvucci ME, van de Loo FJ, Stecher D (2003) Two isoforms of Rubisco activase in cotton, the products of separate genes not alternative splicing. Planta 216: 736–744 [DOI] [PubMed] [Google Scholar]
- Scafaro AP, Gallé A, Van Rie J, Carmo-Silva E, Salvucci ME, Atwell BJ (2016) Heat tolerance in a wild Oryza species is attributed to maintenance of Rubisco activation by a thermally stable Rubisco activase ortholog. New Phytol 211: 899–911 [DOI] [PubMed] [Google Scholar]
- Scafaro AP, Haynes PA, Atwell BJ (2010) Physiological and molecular changes in Oryza meridionalis Ng., a heat-tolerant species of wild rice. J Exp Bot 61: 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafaro AP, Yamori W, Carmo-Silva AE, Salvucci ME, von Caemmerer S, Atwell BJ (2012) Rubisco activity is associated with photosynthetic thermotolerance in a wild rice (Oryza meridionalis). Physiol Plant 146: 99–109 [DOI] [PubMed] [Google Scholar]
- Sharkey TD. (2005) Effects of moderate heat stress on photosynthesis: importance of thylakoid reactions, rubisco deactivation, reactive oxygen species, and thermotolerance provided by isoprene. Plant Cell Environ 28: 269–277 [Google Scholar]
- Sharkey TD, Badger MR, von Caemmerer S, Andrews TJ (2001) Increased heat sensitivity of photosynthesis in tobacco plants with reduced Rubisco activase. Photosynth Res 67: 147–156 [DOI] [PubMed] [Google Scholar]
- Sharwood RE. (2017) Engineering chloroplasts to improve Rubisco catalysis: prospects for translating improvements into food and fiber crops. New Phytol 213: 494–510 [DOI] [PubMed] [Google Scholar]
- Silvera K, Neubig KM, Whitten WM, Williams NH, Winter K, Cushman JC (2010) Evolution along the crassulacean acid metabolism continuum. Funct Plant Biol 37: 995–1010 [Google Scholar]
- Spreitzer RJ, Salvucci ME (2002) Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol 53: 449–475 [DOI] [PubMed] [Google Scholar]
- Stotz M, Mueller-Cajar O, Ciniawsky S, Wendler P, Hartl FU, Bracher A, Hayer-Hartl M (2011) Structure of green-type Rubisco activase from tobacco. Nat Struct Mol Biol 18: 1366–1370 [DOI] [PubMed] [Google Scholar]
- Tcherkez GGB, Farquhar GD, Andrews TJ (2006) Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc Natl Acad Sci USA 103: 7246–7251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KY, Suen DF, Chen SCG (1999) Molecular characterization of ribulose-1,5-bisphosphate carboxylase/oxygenase activase in rice leaves. Planta 209: 66–76 [DOI] [PubMed] [Google Scholar]
- Tsai YC, Lapina MC, Bhushan S, Mueller-Cajar O (2015) Identification and characterization of multiple rubisco activases in chemoautotrophic bacteria. Nat Commun 6: 8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Loo FJ, Salvucci ME (1996) Activation of ribulose-1,5-biphosphate carboxylase/oxygenase (Rubisco) involves Rubisco activase Trp16. Biochemistry 35: 8143–8148 [DOI] [PubMed] [Google Scholar]
- van de Loo FJ, Salvucci ME (1998) Involvement of two aspartate residues of Rubisco activase in coordination of the ATP gamma-phosphate and subunit cooperativity. Biochemistry 37: 4621–4625 [DOI] [PubMed] [Google Scholar]
- Wachter RM, Salvucci ME, Carmo-Silva AE, Barta C, Genkov T, Spreitzer RJ (2013) Activation of interspecies-hybrid Rubisco enzymes to assess different models for the Rubisco-Rubisco activase interaction. Photosynth Res 117: 557–566 [DOI] [PubMed] [Google Scholar]
- Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61: 199–223 [Google Scholar]
- Wang D, Li XF, Zhou ZJ, Feng XP, Yang WJ, Jiang DA (2010) Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant. Physiol Plant 139: 55–67 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Snyder GW, Esau BD, Portis AR Jr, Ogren WL (1992) Species-dependent variation in the interaction of substrate-bound ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) and rubisco activase. Plant Physiol 100: 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis E. (1981) Reversible heat-inactivation of the calvin cycle: a possible mechanism of the temperature regulation of photosynthesis. Planta 151: 33–39 [DOI] [PubMed] [Google Scholar]
- Wise RR, Olson AJ, Schrader SM, Sharkey TD (2004) Electron transport is the functional limitation of photosynthesis in field-grown Pima cotton plants at high temperature. Plant Cell Environ 27: 717–724 [Google Scholar]
- Yamori W, Hikosaka K, Way DA (2014) Temperature response of photosynthesis in C3, C4, and CAM plants: temperature acclimation and temperature adaptation. Photosynth Res 119: 101–117 [DOI] [PubMed] [Google Scholar]
- Yamori W, Masumoto C, Fukayama H, Makino A (2012) Rubisco activase is a key regulator of non-steady-state photosynthesis at any leaf temperature and, to a lesser extent, of steady-state photosynthesis at high temperature. Plant J 71: 871–880 [DOI] [PubMed] [Google Scholar]
- Zavodszky P, Kardos J, Svingor A, Petsko GA (1998) Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc Natl Acad Sci U S A 95: 7406–7411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wigley DB (2008) The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nat Struct Mol Biol 15: 1223–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]