Abstract
Blastocladiella emersonii is an aquatic fungus of the chytridiomycete class which diverged early from the fungal lineage and is notable for the morphogenetic processes which occur during its life cycle. Its particular taxonomic position makes this fungus an interesting system to be considered when investigating phylogenetic relationships and studying the biology of lower fungi. To contribute to the understanding of the complexity of the B. emersonii genome, we present here a survey of expressed sequence tags (ESTs) from various stages of the fungal development. Nearly 20,000 cDNA clones from 10 different libraries were partially sequenced from their 5′ end, yielding 16,984 high-quality ESTs. These ESTs were assembled into 4,873 putative transcripts, of which 48% presented no matches with existing sequences in public databases. As a result of Gene Ontology (GO) project annotation, 1,680 ESTs (35%) were classified into biological processes of the GO structure, with transcription and RNA processing, protein biosynthesis, and transport as prevalent processes. We also report full-length sequences, useful for construction of molecular phylogenies, and several ESTs that showed high similarity with known proteins, some of which were not previously described in fungi. Furthermore, we analyzed the expression profile (digital Northern analysis) of each transcript throughout the life cycle of the fungus using Bayesian statistics. The in silico approach was validated by Northern blot analysis with good agreement between the two methodologies.
Blastocladiella emersonii is a saprobic aquatic fungus, belonging to the class Chytridiomycetes (35), whose life cycle suffers dramatic biochemical and morphological changes during the following two stages of cell differentiation: germination and sporulation. The life cycle begins with the zoospore, a motile uninucleated nongrowing cell, which germinates rapidly and synchronously upon exposure to nutrient medium or an inorganic salt solution containing certain monovalent cations (43), cyclic AMP (12), or other inducers (13). During the first 20 min of germination at 27°C, the zoospore retracts its flagellum and forms a cell wall of chitin. The resulting round cell converts into a germling cell, with the formation of a germ tube that elongates and begins to branch at approximately 60 min, giving rise to a rhizoidal system through which nutrients are absorbed (29). During vegetative growth, B. emersonii cells go through intense nuclear division without cytokinesis, generating single-celled coenocytes. Nutrient starvation at any time during growth induces the other transitional stage, the sporulation, that after 3.5 to 4 h at 27°C culminates with the intracellular formation of the zoospores, which are then released to the medium through an opening in the cell wall denominated discharge papilla (29).
B. emersonii is a primitive fungus which has diverged early in the fungal lineage (17, 49). Based on rRNA data, it seems clear which groups form the fungal monophyletic clade; however, the phylogenetic relationships among the various fungal taxa remain doubtful (19, 36, 49). Similarly, the relationships among the various crown taxa remain not well resolved. In the same way that molecular phylogenies based on rRNA have alternatively placed either plants or fungi as more closely related to animals, different works, mainly based on protein sequences (elongation factors 1α and 2, actin, and tubulins), have supported Cavalier-Smith's proposal that animals and fungi are sister groups (2, 48).
Despite the particular taxonomic position and the significance as an important ecological group that involves saprobes as well as plant, animal, and fungal pathogens (35), the chytrids remain poorly characterized. Although B. emersonii has become one member of the group that has been extensively studied at different levels, present knowledge about its expressed genes is limited to the rRNA genes and eight protein coding sequences (4, 5, 9, 10, 31, 37, 38, 42, 45, 49).
An efficient way to obtain information about gene expression and coding sequences of uncharacterized genomes is to sequence a large number of expressed sequence tags (ESTs). If obtained from nonnormalized libraries, EST sequencing analysis (also known as digital Northern analysis) can represent the expression profile, including complexity and abundance levels of transcripts from different tissues, cell types, and developmental stages (8).
We report here a high-throughput cDNA sequencing program which is the first approach to the understanding of gene complexity in B. emersonii. We have produced 16,984 high-quality sequences corresponding to the 5′ ends of cDNAs from 10 different libraries constructed using mRNA isolated from synchronized cells, collected at different stages along the B. emersonii life cycle. The set includes 4,873 putative unique sequences, among which 2,306 were annotated in at least one of the three Gene Ontology (GO) project terms: biological process, molecular function and cellular component. A total of 1,680 ESTs were classified in different biological processes. We also tested previously selected proteins to reconstruct the eukaryotic phylogeny based on the neighbor-joining method. At the same time, we conducted an in silico analysis to evaluate differential gene expression throughout B. emersonii life cycle, which was validated by Northern blot for eight selected genes. This first large-scale sequencing project of a chytridiomycete transcriptome represents an important set of expressed sequences for studies of phylogeny as well as growth and differentiation in lower fungi.
MATERIALS AND METHODS
Culture conditions.
Cultures of B. emersonii were maintained on plates containing 0.13% peptone, 0.13% yeast extract, 0.3% glucose, and 1% agar. For RNA extraction, zoospores were inoculated (3 × 105 cells per ml) in defined DM3 medium (30) and then grown for 16 h at 18°C with agitation. Vegetative cells were then starved by filtering them through a Nitex cloth, rinsing and resuspending the cells in sporulation solution (1 mM Tris-maleate buffer [pH 6.8], 1 mM CaCl2) at a density of 5 × 105 cells per ml. After incubation for 3.5 to 4 h at 27°C with agitation, the zoospores were released from the cells and separated from the empty zoosporangia by filtering through Nitex cloth. Zoospore germination was initiated by inoculation of the cells at a density of 106 cells per ml in DM3 medium and incubation with agitation at 27°C. The progress and synchrony of both sporulation and germination were monitored by taking samples and examining cell types under a light microscope (29, 44).
Construction of cDNA libraries and DNA sequencing.
Total RNA was isolated using Trizol LS (Life Technologies), and mRNA was purified by employing the Oligotex-dT mRNA mini kit (QIAGEN). RNA samples were isolated from zoospores, germinating cells 30, 60, 90, and 120 min after induction of germination at 27°C, vegetative cells after 16 h of growth at 18°C, and sporulating cells 30, 60, 90, and 120 min after induction of sporulation at 27°C, totalling 10 different samples along the fungal life cycle. Libraries were denominated as follows: ZSP (zoospores), G30-G120 (germlings from 30 to 120 min of germination), NSV (vegetative cells), and E30-E120 (zoosporangia from 30 to 120 min of sporulation). DNA was synthesized using 1 to 5 μg of poly(A)+ RNA and size-fractionated in Sephacryl S-500 HR (Life Technologies) columns, and cDNA fractions containing fragments larger than 500 bp were pooled. We constructed nonnormalized cDNA libraries in the vector pSPORT I using the SuperScript plasmid system (Life Technologies), resulting in directional cloning. We also prepared a nondirected cDNA library from vegetative cells in pGEMT-Easy (Promega) using a PCR-select cDNA subtraction kit (Clontech) without the subtraction step. We carried out the link between the adapter sequences provided for subtraction and the cDNA sample, cloned the resulting fragments into the plasmid, and amplified the inserts by PCR, using the adapter sequences as primers. Sequencing reactions were performed with 100 to 200 ng of plasmid DNA prepared in a 96-well format at all stages, from bacterial growth through sequencing reaction purification. Sequencing reactions were carried out using the ABI Prism BigDye Terminator sequencing kit (Applied Biosystems) and analyzed on ABI377 or ABI3100 automatic sequencer instruments.
EST processing pipeline, annotation, and database construction.
The EST sequence chromatograms were stored, processed, and trimmed through a web-based service (34; http://bioinfo.iq.usp.br/estweb) including base calling and quality control by PHRED (6, 7) and trimming (which included the removal of poly(A) tails, low-quality sequences, and vector and adapter regions) by Cross Match (14; http://www.phrap.org/phrap_documentation.html). The accepted sequences were chosen to contain a minimum of 150 bases, with at least 75 bases within a window of 100 bases with a PHRED quality value of ≥15. The sequences were filtered using a local BLASTN analysis (1) to eliminate sequences that matched ribosomal and mitochondrial sequences or that matched bacterial sequences. Assembly of ESTs into contigs was carried out using the CAP3 program (18; http://genome.cs.mtu.edu/cap/cap3.html) set with default parameters. The redundancy was estimated as the number of assembled ESTs minus the number of contigs divided by the total number of ESTs and transformed to a percentage. We annotated putative protein products for assembling results based on BLASTX best hits against a nonredundant database at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/BLAST/), and we automatically assigned Gene Ontology terms (http://www.geneontology.org) to the resulting contigs and singlets using BLASTX against curated databases (Swiss-Prot and TrEMBL) available at the Swiss Institute of Bioinformatics (http://www.expasy.org). We used a bit score cutoff of 55 for the NCBI annotation (this score corresponds roughly to an E-value ≤ 10−6 considering the nonredundant database size) and an E value of ≤10−6 as the BLASTX cutoff for the Swiss-Prot and TrEMBL annotation. We included both annotation browsers in our online integrated database available at the project website (http://blasto.iq.usp.br), as well as a dynamic searching tool. Annotation was also manually checked. The unigenes showing matches with any of the first 10 amino acids (aa) of a protein present in the SwissProt database (BLASTX hit with an E value of <10−6), and with a minimum of 30 nucleotides (nt) 5′ to the matching amino acids, if not the initiator methionine, were assumed to be full-length transcripts.
Alignments and construction of trees.
We aligned protein sequences using Clustal W version 1.82, and alignments were manually checked for final evaluation (47; http://www.ebi.ac.uk/clustalw). We constructed neighbor-joining trees (39) based on a gamma model using the software package MEGA version 2.1 (25; http://www.megasoftware.net) and performed bootstrap analysis on 3,000 samples to estimate the tree support.
Analysis of gene expression profiles.
To evaluate changes in EST abundance among the sequenced ESTs from different libraries, we used the so-called digital Northern analysis or in silico transcriptional profiling, which can be performed by counting the number of sequenced ESTs for a given gene within the whole sequenced EST population (normalized counts), obtained from nonnormalized libraries. From a mathematical viewpoint, this in silico approach is a small-scale equivalent of the SAGE (serial analysis of gene expression) technique. As a counting sampling process, digital Northern analysis is easily modeled using basic Bayesian statistics. This model was used to build credibility intervals (error bars) for abundance results, according to Vêncio et al. (50). We chose a noninformative a priori probability density function and reported 68% credibility intervals. To group gene expression level profiles, we used the well-known hierarchical clustering algorithm with the unweighted-pair group tree-building method with arithmetic means and a correlation metric (16) using Spotfire for Functional Genomics software.
Differentially expressed transcripts were evaluated by considering their error bars. To validate the results obtained in the digital Northern analysis, classical Northern blots were carried out for eight genes selected by considering their different expression profiles and using biological replicates. Total RNA was extracted from synchronized cells at different stages of the B. emersonii developmental cycle, using the Trizol method (Life Technologies), and Northern blot analysis was performed as previously described (42). We blotted the RNA to Hybond N+ membranes (Amersham) and used as probes EcoRI/NotI fragments from sequenced library clones corresponding to the selected transcripts. We normalized the hybridization signals obtained using the bands corresponding to the 28S and 18S rRNAs, visualized in the agarose gels by ethidium bromide staining.
Nucleotide sequence accession numbers.
All sequences described in this study have been submitted to the GenBank EST section and the accession numbers assigned to them were CO961503 to CO978552 and dbEST-Id 25065829 to 25082878.
RESULTS
Generation and assembly of ESTs.
We have generated 19,928 sequences from 10 different cDNA libraries that were constructed with mRNA extracted from synchronized cells isolated at different times during the B. emersonii developmental cycle. After trimming low-quality sequences and poly(A) tail, vector, and adapter sequences and removing contaminant bacterial, ribosomal, and mitochondrial DNA, we obtained a total of 16,984 accepted high-quality sequences (Table 1), which represent 85% of usable ESTs. The number of ESTs sequenced from each library ranged from 1,020 to 2,260, depending on the redundancy of each particular library. The estimated redundancy of the whole project was 71%, a relatively high value even for nonnormalized libraries. We preferred to continue sequencing, even observing a relatively high redundancy, as long as the percentage of new ESTs in the 96 clone sets was not lower than 10%. A particular case was the library NSV, which had a PCR step. The NSV library redundancy was 63%, and the estimated redundancy due to amplification was 12%. Despite these percentages, this library contributed 91 singlets, about 3% of the total for the project.
TABLE 1.
B. emersonii EST summary
| Parameter | Value |
|---|---|
| Total no. of sequences | 19,928 |
| Total no. of high-quality sequencesa | 16,985 |
| Average insert sizeb (bp) | 1,000 |
| Average sequence sizec (nt) | 573 |
| No. of unigenesd | 4,873 |
| No. of contigs | 1,975 |
| No. of singlets | 2,898 |
| Estimated no. of full-length cDNAs | 982 |
| No. (%) of ESTs matching GenBank database sequences | 2,535 (52) |
| No. (%) of ESTs without match | 2,338 (48) |
Sequences with a minimum of 150 nt and PHRED quality value of ≥15.
Assessed by PCR assay of 1,500 clones from different cDNA libraries.
Average sequence size after trimming.
Unigenes represent putative unique sequences.
The range of insert sizes (200 to 2,500 bp) suggested that the libraries contained a fraction of full-length cDNAs. We have confirmed this expectation for the best hits obtained during EST alignments with proteins with highly conserved amino-terminal regions. We have also estimated the number of complete cDNAs covered by the overlapped assembled sequences, which added up to 982, about 20% of the unigenes. The average sequence size (573 nt) indicated that the majority of the reads extended over part of the coding sequence (Table 1), considering the size of the 5′untranslated region of seven previously characterized B. emersonii genes, determined to vary between 65 and 362 nt. We have identified 4,873 putative unique transcripts, which amount to nearly half of the number of predicted protein-coding genes in the Neurospora crassa genome (11), and about as many genes as in Schizosaccharomyces pombe (51).
Roughly 52% of the assembled sequences (2,535) represent novel orthologs from B. emersonii (ESTs with matches in GenBank), whereas 48% (2,338) have no putative identification. The high percentage of sequences with no match reflects the early stage of fungal genome exploration, in particular in the chytridiomycete class. Previously known protein-coding genes from B. emersonii are represented in our database, and their contribution amounts to about 0.16%. In addition, we have found one new paralog corresponding to the catalytic subunit of the cyclic AMP-dependent protein kinase and four paralogs corresponding to the heat shock protein HSP70.
Biological survey of B. emersonii ESTs.
As the result of GO annotation, a total of 2,306 assembled sequences (47%) were classified in 4,861 nodes distributed among the three ontologies of the GO structure: molecular function, biological process, and cellular component (46). Hypothetical proteins have no designation within this hierarchy because they do not represent a biological entity. However, the number of hypothetical proteins (a total of 989) identified in the NCBI database dropped to nearly half (468) after they were classified in one or more ontologies. Pure hypothetical proteins, i.e., open reading frames without any evidence of being transcribed and without significant similarity with any other well-characterized expressed sequence, represented the other half, which were not considered in the GO classification. Despite the difficulty in establishing a direct comparison between the two annotations, this number probably represents the majority of the observed differences between both assignments.
About 35% (1,680) of the annotated unigenes corresponded to biological process ontology. Among the main represented nodes, 18% corresponded to nucleic acid metabolism, 21% corresponded to protein metabolism, and 20% corresponded to cell growth and/or maintenance. Transcription and RNA processing (8%), protein biosynthesis (9%), and transport (11%) were the most prevalent processes related to main nodes, respectively (see Fig. S1 in the supplemental material). Among the 20 most abundant ESTs, 15 are involved in the protein biosynthesis process (GO:0006412), 14 of which encode ribosomal proteins, including 5 misidentified according to the first best match, suggesting the importance of this overrepresentation for B. emersonii protein synthesis (Table 2).
TABLE 2.
The 20 most abundant ESTs from B. emersonii
| No. of ESTs (frequency %) | Best matcha | Swiss-Prot/TrEMBL code | GO code for biological process |
|---|---|---|---|
| 291 (1.82) | No match | ||
| 165 (1.03) | Hypothetical protein (Neurospora crassa) | Q7RWD9 | GO:0006412 |
| 157 (0.98) | GTP-binding protein SAS1 (Dictyostelium discoideum) | P20790 | GO:0007264 |
| 155 (0.97) | Elongation factor 1 α long form (Monosiga brevicollis) | Q95UT9 | GO:0006412 |
| 137 (0.86) | 40S ribosomal protein S9 (Podospora anserina) | P52810 | GO:0006412 |
| 128 (0.80) | Ribosomal protein S4 Y isoform (Monodelphis domestica) | O62739 | GO:0006412 |
| 127 (0.79) | 40S ribosomal protein S18 (Branchiostoma belcheri) | Q81SP0 | GO:0006412 |
| 123 (0.77) | Polyubiquitin (Schizosaccharomyces pombe) | O74819 | |
| 122 (0.76) | 60S ribosomal protein L1-B (Schizosaccharomyces pombe) | O74836 | GO:0006412 |
| 119 (0.74) | Ubiquitin (Ciona savignyi) | Q8MY15 | GO:0006412 |
| 114 (0.71) | AgCP12166 (Anopheles gambiae strain PEST) | Q7PXS7 | GO:0006412 |
| 109 (0.68) | Ribosomal protein 22 of the small subunit (Phaffia rhodozyma) | Q9UUP9 | GO:0006412 |
| 108 (0.67) | AgCP1729 (Fragment) (Anopheles gambiae strain PEST) | Q7QC42 | GO:0006412 |
| 104 (0.65) | 40S ribosomal protein S3a (Rattus norvegicus) | P49242 | GO:0006412 |
| 100 (0.62) | No match | ||
| 100 (0.62) | ENSANGP00000023750 (Anopheles gambiae strain PEST) | Q7PSW8 | GO:0006412 |
| 97 (0.61) | Guanine nucleotide-binding protein beta subunit-like (RACK1) (Homo sapiens) | P25388 | GO:0007205 GO:0007165 |
| 91 (0.57) | 40S ribosomal protein L25B (Arabidopsis thaliana) | Q9FJX2 | GO:0006412 |
| 90 (0.56) | Ribosomal protein S12 (Branchiostoma belcheri) | Q81SP4 | GO:0006412 |
| 85 (0.53) | Hypothetical protein (Neurospora crassa) | Q7SD62 | GO:0006412 |
The BLASTX tool against the Swiss-Prot and TrEMBL databases considered a BLASTX cutoff of 10−6.
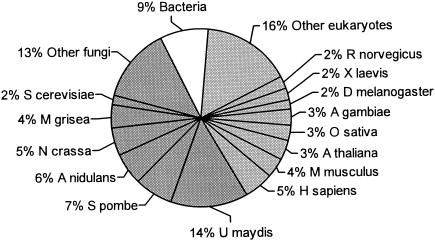
Among the alignments reported by BLASTX output against the nonredundant database at GenBank, we recorded the best hits obtained with fungi and other nonfungal organisms (Fig. 1). From the 2,535 assembled groups compared, 51% represented best hits with fungal predicted proteins, whereas 49% corresponded to other organisms, distributed among different nonfungal eukaryotes (40%) and bacteria (9%). The percentage of fungal best hits, representing 27% of the total B. emersonii unigenes, is smaller than the 33% found for N. crassa putative proteins (11). According to these first results, B. emersonii could have nearly as many homologues (orthologs and paralogs) in fungal as in nonfungal organisms, but our analysis could be biased since we produced a partial transcriptome of B. emersonii, and the number of available sequences in public databases from the majority of the organisms is still limited. However, these results could indicate a closer proximity between B. emersonii and early-diverging nonfungal eukaryotes than between B. emersonii and fungi so distant as ascomycetes (highly represented in the GenBank database).
FIG. 1.
Summary of BLASTX analysis of B. emersonii predicted proteins. The proportions of B. emersonii predicted proteins with BLASTX best hits (E values of ≤10−6) to fungal and nonfungal eukaryotes and bacteria are indicated. The fungal and nonfungal eukaryotes with the highest percentages of BLASTX best hits to B. emersonii predicted proteins are shown. Abbreviations: M. grisea, Magnaporthe grisea; A. nidulans, Aspergillus nidulans; R. norvegicus, Rattus norvegicus; X. laevis, Xenopus laevis; D. melanogaster, Drosophila melanogaster; A. gambiae, Anopheles gambiae; A. thaliana, Arabidopsis thaliana; M. musculus, Mus musculus; H. sapiens, Homo sapiens.
First a chytrid, then a fungus: some features of B. emersonii predicted proteins.
The highest percentage (14%) of best matches of B. emersonii assembled ESTs was with predicted proteins of Ustilago maydis (Fig. 1), one of the few basidiomycetes with an important representation among the fungal hits. This observation probably reflects again the scarce genomic information in public databases from chytridiomycetes and zygomycetes, which are phylogenetically closer to B. emersonii. On the other hand, the better representation of U. maydis among the best hits than that of different ascomycetes agrees with the closer proximity of basidiomycetes to chytridiomycetes based on rRNA and protein molecular phylogenies (36, 49).
According to rRNA sequence analysis, B. emersonii has diverged early in the fungal lineage (49). Thus, we wondered whether our results could be offering a new perspective to construct phylogenetic trees based on different protein sequences. Towards this end, we decided to look for previously selected proteins used in phylogenetic studies among our ESTs, and tested them in the construction of neighbor-joining trees (39). We selected three proteins used or indicated in the literature for which complete cDNAs were present in our libraries: elongation factor 1α, enolase, and β-tubulin (2, 36).
Animals and fungi all share a 12-aa insertion in EF-1α and a 2-aa deletion located approximately 53 residues N-terminal to the 12-aa insertion, relative to other eukaryotes (2, 36). Our amino acid sequence alignments indicated that B. emersonii EF-1α also presents the conserved 12-aa insertion (Fig. 2A) but with an additional amino acid (13-aa insertion). Furthermore, B. emersonii EF-1α does not share the 2-aa deletion but has three different amino acid insertions of 8, 4, and 5 amino acids not found in fungi. These differences are also shared by the choanoflagellate Monosiga brevicollis EF-1α long-form sequence, the best match obtained against the nonredundant GenBank database after the alignment done with the BLASTX tool (Fig. 2A). Additionally, B. emersonii and M. brevicollis EF-1α sequences have C-terminal regions 18 to 20 amino acids shorter than those of the other sequences.
FIG. 2.
Fragments of alignments between EF-1α (A) and enolase (B) sequences from B. emersonii and those of selected organisms. (A) A 12-aa insertion shared by animals and fungi (B. emersonii has one extra amino acid) and three insertions (8, 5, and 4 aa) shared only by B. emersonii and M. brevicollis. (B) Four gaps (1-aa deletion, 2-aa insertion, 1-aa deletion, and 5-aa deletion) shared by animals and fungi. B. emersonii has two extra amino acids in the 2-aa insertion. Alignments were made with the Clustal W program and manually revised. The numbers above the alignments indicate B. emersonii sequence positions. Abbreviations: *, residues identical in all sequences; :, conserved substitutions; ., semiconserved substitutions; Ch, choanoflagellates; Fu, fungi; Me, metazoa; Vp, viridiplantae; Eu, euglenozoa; Mb, M. brevicollis (gi 16554295); Be, B. emersonii (gi number pending); Um, U. maydis (gi 49068500); Sc, S. cerevisiae (gi 171454); Hs, Homo sapiens (gi 15421129, gi 338694); Dm, Drosophila melanogaster (gi 7915, gi 158738); Le, Lycopersicon esculentum (gi 295810, gi 19280); Zm, Zea mays (gi 2282584, gi:22272); Eg, Euglena gracilis (gi 119148, gi 18438).
It was not possible to construct an informative tree including B. emersonii and M. brevicollis sequences. In this sense, a very recent report has presented evidence that the M. brevicollis EF-1α long form is in fact an elongation factor-like protein (EFL), which is a class of proteins very similar to but distinct from canonical EF-1α that can replace it in diverse organisms (23). Thus, it is most probable that the full-length sequence presented here is an EFL sequence. In agreement with this hypothesis, a neighbor-joining tree constructed with B. emersonii EF-1α and the sequences used by Keeling and Inagaki (23) resulted in a group containing EFL sequences which included B. emersonii EF-1α (see Fig. S2A in the supplemental material).
Previous analysis of enolase sequences has revealed three small gaps (two deletions and one insertion) that join together fungi and animals, leaving plants out (2). Clustal W alignments, manually checked, indicated the previously reported conservation of the 1-aa deletion and the 2-aa insertion, except in the case of B. emersonii that showed a 4-aa insertion (Fig. 2B). We also observed two conserved 1- and 5-aa deletions, approximately 15 and 23 residues C-terminal to this insertion, which had not been reported before (Fig. 2B). Furthermore, we noticed that the last 1-aa deletion reported by Baldauf and Palmer (2) was not found in four fungal sequences (Alternaria alternata, Aspergillus oryzae, N. crassa, and Tuber borchii), whereas another 2-aa deletion (between G314 and I315 from Saccharomyces cerevisiae) was observed, except for three fungi: B. emersonii, Neocallimastix frontalis and Candida albicans (data not shown). Protein neighbor-joining phylogeny based on 21 sequences clustered plants and fungi, including B. emersonii and another chytrid, N. frontalis, with 53 and 59% of bootstrap support, respectively. Animals were resolved in a group that excluded nematodes, which branched in a second group (with 71 and 100% of bootstrap support, respectively), but all clustered closer to fungi than plants (with 62% of bootstrap support) (see Fig. S2B in the supplemental material).
We also evaluated 31 β-tubulin sequences, among which we included the partial sequence of Allomyces macrogynus, a member of the order Blastocladiales, which is closer to B. emersonii than the other chytrids analyzed above, and the complete sequence of M. brevicollis. The distance tree was informative, resolving plant, animal, and fungal groups (95, 81, and 50% of bootstrap support, respectively) with animals and fungi in the same clade (with bootstrap confidence of 91%) (see Fig. S2C in the supplemental material).
Putative higher eukaryote receptor proteins in B. emersonii.
Several B. emersonii ESTs presented best matches with different receptor proteins. Interestingly, two best hits were found with plant hormone receptors: the ethylene receptor CS-ETR2 of Cucumis sativus (gi 6136818) and the putative phytosulfokine receptor precursor of Oryza sativa (japonica cultivar group; gi 48717048). The corresponding cDNAs were sequenced from both ends and were found to be incomplete. Nevertheless, we identified the GAF domain characteristic of the ethylene receptor using PfamB and also identified the three leucine-rich repeats as well as the cysteine pair that flanks the leucine-rich repeat domain (33) characteristic of the phytosulfokine receptor using PfamA. These results constitute the first description of such putative receptors in an organism other than a plant.
Another receptor best hit was found to the mannose-6-phosphate/insulin-like growth factor II receptor (MPG/IGF2R) from Didelphis virginiana (gi 16506700), a North American opossum. The MGP/IGF2R has homologues identified in reptiles, amphibians, fish, birds, and mammals. However, no DNA sequence information concerning homologues in other animals or microorganisms exists in public databases. In B. emersonii, two incomplete cDNAs with similarity to MPG/IGF2R were found, and Pfam analysis has confirmed the presence of a cation-independent mannose-6-phosphate receptor domain.
Looking for genes with different expression profiles.
We have considered nine cDNA libraries to evaluate differential gene expression, excluding the NSV library which had a PCR amplification step and whose EST normalized counts thus could not represent the real transcript abundance. Therefore, for digital Northern analysis purposes, the EST cluster assignment was carried out again without the NSV library. This new cDNA library-assembling process produced 1,897 clusters and 2,857 singlets, with a total of 4,754 putative unigenes.
Initially, we analyzed the global patterns of EST diversity by comparing the sporulation and germination stages using the distribution of clusters among different levels of abundance. We included the zoospore at the end of the sporulation stage for this analysis, since zoospore-expressed messages represent the last part of the sporulation-transcribed EST population (29). The two stages presented quite different distributions, as shown in Fig. S3 in the supplemental material, and were validated by a P value of 0.00 in the two-sample Kolmogorov-Smirnov test. The normalized histograms indicate that sporulating cells express higher EST diversity than germinating cells, as reflected by the presence of a larger number of singlets and low-abundance contigs in the libraries from sporulation and a larger number of high-abundance contigs in the libraries from germination.
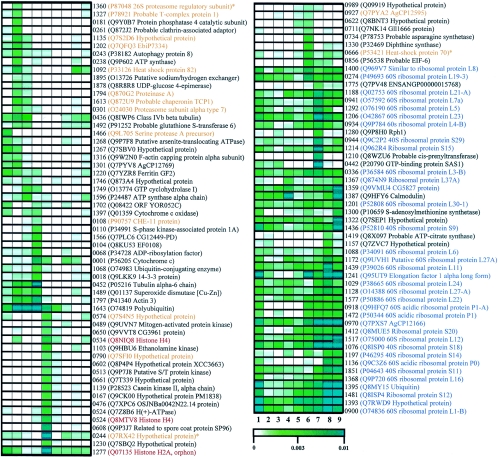
As a second approach, we analyzed the expression profile of each transcript throughout the life cycle of the fungus in a time series framework. We used a Bayesian statistical model to account for sampling error in transcript abundance and produced qualitative classes (representing different reliability classes) constructing credibility intervals (error bars) for transcript abundance. Afterwards, we performed a profile hierarchical clustering by using a correlation metric to identify coexpressed transcripts and selected 109 contigs, based on previously adopted error bars, which were hierarchically reclustered (Fig. 3). A similar reassembling was carried out with the complete set of contigs, which can be assessed using the project web site (http://blasto.iq.usp.br). After this final step, we were prepared to look for putative developmentally regulated genes, i.e., assembled sequences presenting different abundance levels during specific stages of the fungal life cycle.
FIG. 3.
Abundance profiles of 109 different transcripts during the B. emersonii life cycle. Distinct profiles were selected from digital Northern analysis data and subjected to hierarchical clustering. The abundance (from 0 to 0.01) was estimated by considering the number of ESTs sequenced for a given transcript divided by the total number of ESTs sequenced in each library as follows: 1, E30; 2, E60; 3, E90; 4, E120; 5, ZSP; 6, G30; 7, G60; 8, G90; 9, G120. The library names are as given in Materials and Methods. The first column shows the identification number of selected contigs from digital Northern analysis. The descriptions in parentheses are the best hits for each contig using BLASTX against the Swiss-Prot database. Transcripts highlighted in orange have the GO classifications GO:0006508 and GO:0006457, which correspond to the biological processes proteolysis or peptidolysis and protein folding, respectively; in blue are transcripts classified as GO:0006412, which corresponds to protein biosynthesis; in red are transcripts classified as GO:0006334, which corresponds to chromosome structure. *, genes manually annotated to GO:0006508 and GO:0006457.
Considering that, at least for some cell processes, functionally related genes might present significant similarities in their expression patterns, we looked for clusters with similar profiles and associated with a specific GO biological process. Several contigs classified in the same biological process were predominantly clustered together. For instance, contigs with putative function related to proteolysis or peptidolysis and to protein folding processes were more abundant during the beginning of the sporulation stage. Transcripts related to chromosome structure presented higher levels in the zoospore. Furthermore, contigs related to protein biosynthesis showed high abundance in the beginning of sporulation and during germination.
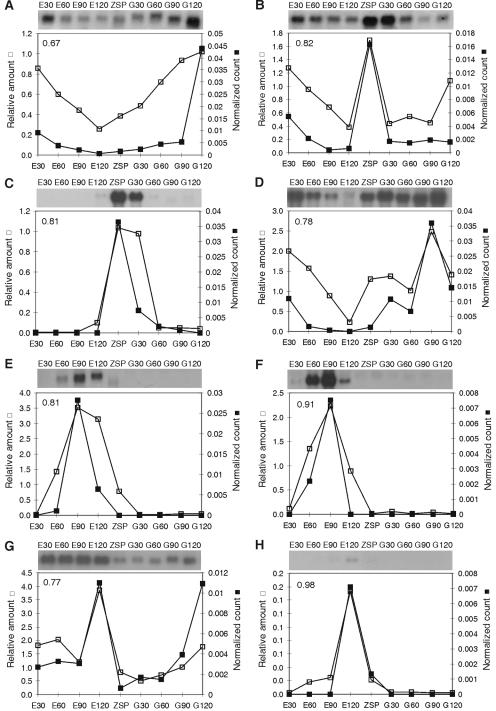
In addition, we confirmed the in silico profiles for eight genes by Northern blot analysis, as depicted in Fig. 4. A good correlation was observed between the in silico profiles and normalized Northern blot hybridization signals.
FIG. 4.
Expression patterns of selected transcripts differentially expressed during the B. emersonii life cycle. The top portions of each panel shows autoradiograms of Northern blot experiments using total RNA isolated from synchronized cells at the indicated stages of the life cycle. The bottom portions show line plots comparing data obtained from the Northern blot (□) with data obtained from the digital Northern analysis (▪). The number in the left top corner of each plot corresponds to the Pearson correlation. Putative proteins encoded by each transcript are as follows: 40S ribosomal protein S9 (A), histone H2A (B), unknown (C), elongation factor 1α long form (D), unknown (E), unknown (F), calmodulin (G), and unknown (H). The results obtained by densitometric analysis of Northern blot hybridization signals confirm the in silico expression profiles.
DISCUSSION
We have conducted a large-scale EST sequencing program that represents the first approach to understand the genome complexity of the chytridiomycete B. emersonii, which constitutes an exclusive model among the chytrids because of previous biochemical and physicochemical studies. Approximately 52% of the putative unique transcripts characterized present similarity with other publicly available sequences, but 48% (2,338) have no putative homologue assigned (Table 1). This high percentage of sequences with no matches agrees with reports from other eukaryotic transcriptome studies. These results reflect the still-partial knowledge that we have about genomes in general and also remind us of the limits of using sequencing data to identify some distant protein homologues, which are better related by structure than amino acid sequence.
Multiple levels of protein synthesis regulation, involving both transcriptional and translational controls, seem to occur during the sporulation and germination stages of the B. emersonii life cycle (40, 41). We were able to associate the assembled expressed sequences reported in the present work with different biological processes using the GO system and found that transcription and RNA processing (8%) and protein biosynthesis (9%) were among the most prevalent biological processes. Furthermore, the most frequent ESTs corresponded to sequences encoding putative proteins directly related to the protein biosynthesis machinery (15 of the first 20). Among them, except for elongation factor 1α, all were ribosomal protein transcripts. The level of expression of EF-lα correlates with the rate of cell growth and proliferation (24). Indeed, the noteworthy expression profile of EF-1α mRNA, similar to that of ribosomal proteins, indicated its developmental regulation with increasing levels during the germination stage. Furthermore, EF-1α is always present in vast molar excess compared to the other essential components of translation, raising the possibility that EF-1α may have additional functions in the cell (27). In B. emersonii, one such additional function could be the reallocation of zoospore-stored mRNAs for their regulated translation during germination.
The relationships among fungal lineages as well as among the crown taxa are still not well resolved. Multiple molecular data have suggested that fungi and animals are sister groups forming a unique clade together with choanoflagellates (2, 36, 48). We have tested three commonly used protein sequences for the reconstruction of phylogenetic trees, EF-1α, enolase, and β-tubulin, which were found to have been completely sequenced in our study, with the goal of doing a first reevaluation of B. emersonii branching position and a first evaluation of our data source.
The B. emersonii EF-1α amino acid sequence presented several insertions shared with the choanoflagellate M. brevicollis but not found in EF-1α from other organisms. The recently described class of eukaryotic GTPase proteins named EFL, which are similar to canonical EF-1α but clearly distinct from it and which included the M. brevicollis protein, shed light upon our observation (23). EFL and EF-1α seem to have similar functions and expression levels and tend to be mutually exclusive in distribution. Organisms that possess EFL are typically closely related to other organisms that possess EF-1α and lack EFL. In this sense, B. emersonii EF-1α reported here could be an EFL sequence and thus not useful for tree reconstruction. Furthermore, a canonical EF-1α sequence is probably absent from the fungus genome.
The trees we constructed based on enolase and β-tubulin sequences better resolved the phylogenies in agreement with several studies, indicating animals as a sister group of fungi (2, 36, 48) and including M. brevicollis within the clade, in the case of the β-tubulin tree. Larger data sets, many gene sequences considered together, and other methods of phylogenetic analysis would contribute to clarify these initial results.
The novel information about B. emersonii led us to confirm how much more we need to learn about this chytrid. We generated several ESTs that show similarity to receptor proteins, some of them never described for fungi. Features like that were previously reported for B. emersonii, as is the case of the P-type ATPases related to animal Na+/K+-ATPase characterized in our laboratory (5, 9). Another interesting finding from our study is an MGP/IGF2R best hit that was described for animals but not previously identified in fungi. The primary functions of MGP/IGF2R are intracellular traficking of lysosomal enzymes, and the internalization of IGF2 and other extracellular ligands to the lysosomes for degradation (20).
Other findings have revealed features of B. emersonii that are closer to those of plants than of animals. For instance, previous data indicating the association of the α and β subunits of B. emersonii mitochondrial processing peptidase with the mitochondrial inner membrane (38), as described for plants, instead of their localization in the mitochondrial matrix, as verified in S. cerevisiae and mammals. In this context, two ESTs found in our libraries showed best matches with receptors described only in plants: the putative phytosulfokine receptor precursor and the ethylene receptor CS-ETR2. Phytosulfokine is an intercellular signal that plays a key role in cellular dedifferentiation and proliferation in plants (32), whereas ethylene is a plant hormone that regulates many aspects of plant growth and development (15).
The mRNAs of a typical somatic cell are distributed in three abundance classes: superprevalent, intermediate, and complex (3). Considering that the proportions among these three classes are presumably represented in nonnormalized cDNA libraries, we conducted an evaluation of the expression profiles of the sequenced ESTs, with the goal of determining some biologically significant expressed genes in B. emersonii. The normalized histogram of gene expression levels can give an overall idea of the organism transcriptome (26). In the case of B. emersonii, the normalized histograms of EST counts comparing the sporulation and germination stages indicated a greater diversity of transcripts during sporulation, which is in agreement with the higher mRNA turnover rate observed during this stage (21, 40).
Furthermore, we were able to identify various highly abundant clusters, which are more represented in the germination libraries, by looking at the most abundant cloned ESTs and the expression profiles corresponding to these messages and concluding that mRNAs related to the protein biosynthesis process are in part responsible for the different distribution. This is more significant when taking into account two Northern blot experiments (for a ribosomal protein and for EF-1α) that confirmed the in silico expression profiles. These patterns indicate the importance of transcriptional regulation for the protein biosynthesis machinery when the growth rate is increasing.
Conversely, we observed contigs associated with the proteolysis or peptidolysis process with correlated expression profiles during the sporulation, which is in agreement with a previous report (28). The probable biological significance for this correlation could be the necessity to provide amino acids for new protein synthesis and to remove proteins which could impair the progress of cell differentiation (28).
In addition to the expression profiles related to protein synthesis and degradation, we reported other expressive correlated patterns. For instance, histone gene expression (with a peak in zoospores) could be a marker of a particular moment during the cell cycle, as shown for histone H3 in alfalfa cells (22). Furthermore, at least 1% of all contigs considered in the digital Northern analysis with no putative identity showed relatively narrow credibility intervals and expression profiles which were hierarchically clustered using a correlation metric. The statistical identification of significant similarities between expression patterns of known and unknown genes could help in ascribing functions to these anonymous sequences.
In conclusion, the unique collection of ESTs presented here represents a valuable resource not only for studies in lower fungal biology and phylogeny but also for future microarray construction and subsequent high-throughput transcriptional studies in diverse biological contexts.
Supplementary Material
Acknowledgments
This work was supported by a grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). S.L.G. was partially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
We thank Sergio Verjovski-Almeida, Abimael A. Machado, and Milton Y. Nishiyama, Jr., for their help with data processing, Apuã C. M. Paquola for his advice with the ESTWeb package, J. Adriano A. Freitas for his assistance with the database, Tie Koide for her advice with the Spotfire software, and Rosangela Campanhã and Mara Silvestri for their help in the beginning of this work. K.F.R. and S.M.S.-I. are doctoral fellows of Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). R.C.G. and R.Z.N.V. are doctoral fellows of FAPESP.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldauf, S. L., and J. D. Palmer. 1993. Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. USA 24:11558-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, J. O., J. G. Morton, M. Rosbash, and M. Richardson. 1974. Three abundance classes in HeLa cell messenger RNA. Nature 250:199-204. [DOI] [PubMed] [Google Scholar]
- 4.de Oliveira, J. C., A. C. Borges, M. V. Marques, and S. L. Gomes. 1994. Cloning and characterization of the gene for the catalytic subunit of cAMP-dependent protein kinase in the aquatic fungus Blastocladiella emersonii. Eur. J. Biochem. 219:555-562. [DOI] [PubMed] [Google Scholar]
- 5.de Souza, F. S., and S. L. Gomes. 1998. A P-type ATPase from the aquatic fungus Blastocladiella emersonii similar to animal Na, K-ATPases. Biochim. Biophys. Acta 1383:183-187. [DOI] [PubMed] [Google Scholar]
- 6.Ewing, B., and P. Green. 1998. Base-calling of automatic sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 7.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automatic sequencer traces using phred. I. Accuracy assesment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 8.Ewing, R. M., A. B. Kahla, O. Poirot, F. Lopez, S. Audic, and J. M. Claverie. 1999. Large-scale statistical analyses of rice ESTs reveal correlated patterns of gene expression. Genome Res. 9:950-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fietto, L. G., L. Pugliese, and S. L. Gomes. 2002. Characterization and expression of two genes encoding isoforms of a putative Na, K-ATPase in the chytridiomycete Blastocladiella emersonii. Biochim. Biophys. Acta 1576:59-69. [DOI] [PubMed] [Google Scholar]
- 10.Forster, H., M. D. Coffey, H. Elwood, and M. L. Sogin. 1990. Sequence analysis of the small subunit ribosomal RNAs of three zoosporic fungi and implications for fungal evolution. Mycologia 82:306-312. [Google Scholar]
- 11.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, V. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 12.Gomes, S. L., L. Mennucci, and J. C. C. Maia. 1980. Calcium efflux during germination of Blastocladiella emersonii. Dev. Biol. 77:157-166. [DOI] [PubMed] [Google Scholar]
- 13.Gottschalk, W. K., and D. R. Sonneborn. 1982. Phenotypic dissections of the Blastocladiella emersonii zoospore's developmental choice. Dev. Biol. 93:165-180. [DOI] [PubMed] [Google Scholar]
- 14.Green, P. 1996. PHRAP documentation. University of Washington, Seattle, USA http://bozeman.mbt.washington.edu/phrap.docs/phrap.html.
- 15.Guo, H., and J. Ecker. 2004. The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7:40-49. [DOI] [PubMed] [Google Scholar]
- 16.Hartigan, J. A. 1975. Clustering algorithms. John Wiley & Sons, New York, N.Y.
- 17.Heckman, D. S., D. M. Geiser, B. R. Eidell, R. L. Stauffer, N. L. Kardos, and S. B. Hedges. 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293:1129-1133. [DOI] [PubMed] [Google Scholar]
- 18.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James, T. Y., D. Porter, C. A. Leander, R. Vilgalys, and J. E. Longcore. 2000. Molecular phylogenetics of the Chytridiomycota supports the utility of ultrastructural data in chytrid systematics. Can. J. Bot. 78:336-350. [Google Scholar]
- 20.Jirtle, R. L. 1999. Mannose-6-phosphate receptors, p. 1441-1447. In G. Collins (ed.), Encyclopedia of molecular biology. Wiley-Liss, Inc., New York, N.Y.
- 21.Johnson, S. A., and J. S. Lovett. 1984. Gene expression during development of Blastocladiella emersonii. Exp. Mycol. 8:132-145. [Google Scholar]
- 22.Kapros, T., L. Bögre, K. Németh, L. Bakó, J. Györgyey, S. C. Wu, and D. Dudits. 1992. Differential expression of histone H3 gene variants during cell cycle and somatic embryogenesis in alfalfa. Plant Physiol. (Rockville) 98:621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeling, P. J., and Y. Inagaki. 2004. A class of eukaryotic GTPase with a punctate distribution suggesting multiple functional replacements of translation elongation factor 1alpha. Proc. Natl. Acad. Sci. USA 101:15380-15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieg, P. A., S. M. Varnum, W. M. Wormington, and D. A. Melton. 1989. The mRNA encoding elongation factor 1-alpha (EF-1 alpha) is a major transcript at the midblastula transition in Xenopus. Dev. Biol. 133:93-100. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA 2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 26.Kuznetsov, V. A. 2002. Statistics of the numbers of transcripts and protein sequences encoded in the genome, p. 334-543. In W. Zhang and I. Shmulevich (ed.), Computational and statistical approaches to genomics. Kluwer Academic Publishers, Boston, Mass.
- 27.Liu, G., B. T. Edmonds, and J. Condeelis. 1996. pH, EF-la and the cytoskeleton. Trends Cell Biol. 6:168-171. [DOI] [PubMed] [Google Scholar]
- 28.Lodi, W. R., and D. R. Sonneborn. 1974. Protein degradation and protease activity during the late cycle of Blastocladiella emersonii. J. Bacteriol. 117:1035-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovett, J. S. 1975. Growth and differentiation of the water mold Blastocladiella emersonii: cytodifferentiation and the role of ribonucleic acid and protein synthesis. Bacteriol. Rev. 39:345-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maia, J. C., and E. P. Camargo. 1974. cAMP phosphodiesterase activity during growth and differentiation in Blastocladiella emersonii. Cell Differ. 3:147-155. [DOI] [PubMed] [Google Scholar]
- 31.Marques, M. V., and S. L. Gomes. 1992. Cloning and structural analysis of the gene for the regulatory subunit of cAMP-dependent protein kinase in Blastocladiella emersonii. J. Biol. Chem. 267:17201-17207. [PubMed] [Google Scholar]
- 32.Matsubayashi, Y., M. Ogawa, A. Morita, and Y. Sakagami. 2002. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296:1470-1472. [DOI] [PubMed] [Google Scholar]
- 33.Napier, R. 2004. Plant hormone binding sites. Ann. Bot. (London) 93:227-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paquola, A., M. Y. Nishiyama, Jr., E. M. Reis, A. M. da Silva, and S. Verjovski-Almeida. 2003. ESTWeb: bioinformatics services for EST sequencing projects. Bioinformatics 19:1587-1588. [DOI] [PubMed] [Google Scholar]
- 35.Powell, M. J. 1993. Looking at mycology with a Janus face: a glimpse at chytridiomycetes active in the environment. Mycologia 85:1-20. [Google Scholar]
- 36.Ragan, M. A., C. A. Murphy, and T. G. Rand. 2003. Are Ichthyosporea animals or fungi? Bayesian phylogenetic analysis of elongation factor 1 alpha of Ichthyophonus irregularis. Mol. Phylogenet. Evol. 29:550-562. [DOI] [PubMed] [Google Scholar]
- 37.Rocha, C. R. C., and S. L. Gomes. 1998. Isolation, characterization, and expression of the gene encoding the beta subunit of the mitochondrial processing peptidase from Blastocladiella emersonii. J. Bacteriol. 15:3967-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rocha, C. R. C., and S. L. Gomes. 1999. Characterization and submitochondrial localization of the alpha subunit of the mitochondrial processing peptidase from the aquatic fungus Blastocladiella emersonii. J. Bacteriol. 181:4257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 40.Silva, A. M., J. C. C. Maia, and M. H. Juliani. 1986. Developmental changes in translatable RNA species and protein synthesis during sporulation in the aquatic fungus Blastocladiella emersonii. Cell Differ. 18:263-274. [DOI] [PubMed] [Google Scholar]
- 41.Silva, A. M., J. C. C. Maia, and M. H. Juliani. 1987. Changes in the pattern of protein synthesis during zoospore germination in Blastocladiella emersonii. J. Bacteriol. 169:2069-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simão, R. C., and S. L. Gomes. 2001. Structure, expression, and functional analysis of the gene coding for calmodulin in the chytridiomycete Blastocladiella emersonii. J. Bacteriol. 183:2280-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soll, D. R., and D. R. Sonneborn. 1972. Zoospore germination in Blastocladiella emersonii. IV. Ion control over cell differentiation. J. Cell Sci. 10:315-333. [DOI] [PubMed] [Google Scholar]
- 44.Soll, D. R., R. Bromberg, and D. R. Sonneborn. 1969. Zoospore germination in the water mold Blastocladiella emersonii. I. Measurement of germination and sequence of subcellular morphological changes. Dev. Biol. 20:183-217. [DOI] [PubMed] [Google Scholar]
- 45.Stefani, R. M., and S. L. Gomes. 1995. A unique intron-containing hsp70 gene induced by heat shock and during sporulation in the aquatic fungus Blastocladiella emersonii. Gene 152:19-26. [DOI] [PubMed] [Google Scholar]
- 46.The Gene Ontology Consortium. 2001. Creating the gene ontology resource: design and implementation. Genome Res. 11:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van de Peer, Y., S. L. Baldauf, W. F. Doolittle, and A. Meyer. 2000. An updated and comprehensive rRNA phylogeny of (crown) eukaryotes based on rate-calibrated evolutionary distances. J. Mol. Evol. 51:565-576. [DOI] [PubMed] [Google Scholar]
- 49.Van der Auwera, G., and R. De Wachter. 1996. Large-subunit rRNA sequence of the chytridiomycete Blastocladiella emersonii, and implications for the evolution of zoosporic fungi. J. Mol. Evol. 43:476-483. [DOI] [PubMed] [Google Scholar]
- 50.Vêncio, R. Z. N., H. Brentani, and C. A. B. Pereira. 2003. Using credibility intervals instead of hypothesis tests in SAGE analyses. Bioinformatics 19:2461-2464. [DOI] [PubMed] [Google Scholar]
- 51.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, D. Basham, S. Bowman, K. Brooks, D. Brown, S. Brown, T. Chillingworth, C. Churcher, M. Collins, R. Connor, A. Cronin, P. Davis, T. Feltwell, A. Fraser, S. Gentles, A. Goble, N. Hamlin, D. Harris, J. Hidalgo, G. Hodgson, S. Holroyd, T. Hornsby, S. Howarth, E. J. Huckle, S. Hunt, K. Jagels, K. James, L. Jones, M. Jones, S. Leather, S. McDonald, J. McLean, P. Mooney, S. Moule, K. Mungall, L. Murphy, D. Niblett, C. Odell, K. Oliver, S. O'Neil, D. Pearson, M. A. Quail, E. Rabbinowitsch, K. Rutherford, S. Rutter, D. Saunders, K. Seeger, S. Sharp, J. Skelton, M. Simmonds, R. Squares, S. Squares, K. Stevens, K. Taylor, R. G. Taylor, A. Tivey, S. Walsh, T. Warren, S. Whitehead, J. Woodward, G. Volckaert, R. Aert, J. Robben, B. Grymonprez, I. Weltjens, E. Vanstreels, M. Rieger, M. Schafer, S. Muller-Auer, C. Gabel, M. Fuchs, A. Dusterhoft, C. Fritzc, E. Holzer, D. Moestl, H. Hilbert, K. Borzym, I. Langer, A. Beck, H. Lehrach, R. Reinhardt, T. M. Pohl, P. Eger, W. Zimmermann, H. Wedler, R. Wambutt, B. Purnelle, A. Goffeau, E. Cadieu, S. Dreano, S. Gloux, V. Lelaure, S. Mottier, F. Galibert, S. J. Aves, Z. Xiang, C. Hunt, K. Moore, S. M. Hurst, M. Lucas, M. Rochet, C. Gaillardin, V. A. Tallada, A. Garzon, G. Thode, R. R. Daga, L. Cruzado, J. Jimenez, M. Sanchez, F. del Rey, J. Benito, A. Dominguez, J. L. Revuelta, S. Moreno, J. Armstrong, S. L. Forsburg, L. Cerutti, T. Lowe, W. R. McCombie, I. Paulsen, J. Potashkin, G. V. Shpakovski, D. Ussery, B. G. Barrell, P. Nurse, and L. Cerrutti. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 21:871-880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.