ABSTRACT
Periodontitis is a microbial infection that destroys the structures that support the teeth. Although it is typically a chronic condition, rapidly progressing, aggressive forms are associated with the oral pathogen Aggregatibacter actinomycetemcomitans. One of this bacterium's key virulence traits is its ability to attach to surfaces and form robust biofilms that resist killing by the host and antibiotics. Though much has been learned about A. actinomycetemcomitans since its initial discovery, we lack insight into a fundamental aspect of its basic biology, as we do not know the full set of genes that it requires for viability (the essential genome). Furthermore, research on A. actinomycetemcomitans is hampered by the field's lack of a mutant collection. To address these gaps, we used rapid transposon mutant sequencing (Tn-seq) to define the essential genomes of two strains of A. actinomycetemcomitans, revealing a core set of 319 genes. We then generated an arrayed mutant library comprising >1,500 unique insertions and used a sequencing-based approach to define each mutant's position (well and plate) in the library. To demonstrate its utility, we screened the library for mutants with weakened resistance to subinhibitory erythromycin, revealing the multidrug efflux pump AcrAB as a critical resistance factor. During the screen, we discovered that erythromycin induces A. actinomycetemcomitans to form biofilms. We therefore devised a novel Tn-seq-based screen to identify specific factors that mediate this phenotype and in follow-up experiments confirmed 4 mutants. Together, these studies present new insights and resources for investigating the basic biology and disease mechanisms of a human pathogen.
IMPORTANCE Millions suffer from gum disease, which often is caused by Aggregatibacter actinomycetemcomitans, a bacterium that forms antibiotic-resistant biofilms. To fully understand any organism, we should be able to answer: what genes does it require for life? Here, we address this question for A. actinomycetemcomitans by determining the genes in its genome that cannot be mutated. As for the genes that can be mutated, we archived these mutants into a library, which we used to find genes that contribute to antibiotic resistance, leading us to discover that antibiotics cause A. actinomycetemcomitans to form biofilms. We then devised an approach to find genes that mediate this process and confirmed 4 genes. These results illuminate new fundamental traits of a human pathogen.
KEYWORDS: Aggregatibacter, Tn-seq, antibiotics, biofilms, essential genome
INTRODUCTION
One of the longest-studied human microbiomes is the “animalcules” (bacteria), first observed by van Leeuwenhoek (1), that inhabit the oral cavity. Oral bacteria readily attach to tooth surfaces and develop into spatially organized, multispecies biofilms (2). These communities also persist beneath the gum line in the subgingival pocket, where they are kept in check by host immune cells (3). Though normally benign, subgingival communities can accumulate opportunistic oral pathogens that hyperactivate the immune response (3). Over time, this inflamed state, known as periodontitis, slowly destroys the gums and tooth socket, deepening the subgingival pocket and ultimately leading to tooth loss (3). As >400 bacterial species can colonize the subgingival pocket (4), the cause of periodontitis long eluded microbiologists, but now, specific pathogens and microbial consortia (5) are firmly linked to the disease. Although periodontitis can often be alleviated by eradicating these pathogens, it is estimated to affect 11% of the global population (743 million people in 2010) (6).
Periodontitis is generally a chronic disease that manifests only in adults; however, aggressive periodontitis (AgP) can progress 3 to 4 times faster (7) and afflict adolescents. Although AgP is rare, occurring at <1% on a population-wide level, it targets subpopulations of African descent (8, 9). For example, 8% of adolescents have AgP in Morocco (8), and 15 times more black (2.05%) than white (0.14%) adolescents have AgP in the United States (9). AgP is distinct clinically because it attacks only the incisors and first molars and features only a sparse biofilm that does not correspond with the observed tissue damage (10). The putative cause of AgP is Aggregatibacter actinomycetemcomitans, a Gram-negative, facultatively anaerobic, nonmotile, rod-shaped bacterium (11). A. actinomycetemcomitans colonizes <20% of healthy children but is harbored by >90% of juvenile AgP patients (12, 13). In addition to periodontitis, A. actinomycetemcomitans can spread from the oral cavity and establish infections throughout the body, including endocarditis (14), rheumatoid arthritis (15), and brain abscesses (16). In fact, A. actinomycetemcomitans was first described in 1912 after it was isolated from mixed-species lesions with Actinomyces (11). In its many infections, A. actinomycetemcomitans uses an arsenal of virulence factors to colonize (17), proliferate (18), and resist the host immune system (19). Most notably, these include a potent leukotoxin that can lyse white and red blood cells (20) and that is overexpressed in a highly virulent strain endemic to northern Africa (21).
A less well recognized A. actinomycetemcomitans virulence factor is antibiotic resistance. Many A. actinomycetemcomitans infections are refractory (22) and can be cured only by coupling periodontal surgery with antibiotic therapy since neither procedure alone can fully treat the infection (23). One of the most commonly used antibiotics to treat periodontal infections is macrolides (24), such as erythromycin (25). These antibiotics inhibit protein synthesis by targeting the 50S subunit of the ribosome (26). However, many strains of A. actinomycetemcomitans are intrinsically resistant to erythromycin (27), questioning its viability as a treatment option. Despite the urgency of this problem, we are only beginning to understand how A. actinomycetemcomitans resists antibiotics. These mechanisms include modifying the antibiotic's target (28) and, as seen in many other bacteria (29), finding shelter in highly resistant biofilms (30, 31).
Recently, our overall knowledge of A. actinomycetemcomitans has improved through genomic approaches such as transcriptome sequencing (RNA-seq) (32), transposon mutant sequencing (Tn-seq) (33), proteomics (34), and whole-genome sequencing (WGS). WGS, for instance, unveiled that strains of A. actinomycetemcomitans are genetically diverse. In addition to 2,034 core genes, the A. actinomycetemcomitans pan-genome features 1,267 flexible genes (35), often on horizontally acquired genomic islands (36), which are present in some but not all strains. Phylogenetically, strains of A. actinomycetemcomitans cluster by serotype (36). Among the 7 serotypes currently recognized (37), serotypes a, b, and c are the most common (38, 39), and reflecting their phylogeny, these serotypes differ in key phenotypes, such as toxin production (21) and natural competence (40).
One fundamental gap in our knowledge of A. actinomycetemcomitans is its essential genome, or the set of genes that it requires for life. Essential genomes have been defined for many model pathogens (41, 42) and can provide new avenues for discovering drug targets (43, 44). Furthermore, our knowledge of A. actinomycetemcomitans would benefit from an arrayed mutant collection for conducting screens and quickly obtaining mutants of interest. Here, we used transposon mutant sequencing (Tn-seq) (45) to define the essential genome in two strains of A. actinomycetemcomitans, and Cartesian pooling-coordinate sequencing (CP-CSeq) (46) to construct an ordered transposon mutant library. While screening the library for genes that mediate antibiotic resistance, we discovered that subinhibitory erythromycin induces A. actinomycetemcomitans to form biofilms, leading us to devise a novel Tn-seq-based screen to identify and confirm 4 genes that are involved in this bacterial defense behavior.
RESULTS
Generating saturating mutant pools in A. actinomycetemcomitans.
A. actinomycetemcomitans is a genetically diverse species, with the genome content between any two strains varying by as much as 16% (>300 genes) (36). Because of this, defining the essential genome of any single strain may not be representative of the entire species. Therefore, we set out to make dense transposon mutant pools in two divergent strains of A. actinomycetemcomitans, VT1169 (47) and 624. VT1169 is a serotype b strain with a 2.1-Mb genome, whereas 624 is a serotype a strain with a 2.4-Mb genome (Table 1 shows additional strain characteristics). For the two strains, we used the same transposon, mariner, since it has low site specificity, requiring only a TA dinucleotide for insertion. We also used the same mutagenesis procedure, conjugation with Escherichia coli on rich, undefined medium (tryptic soy agar supplemented with yeast extract [TSAYE]). Of note, we performed conjugations under both aerobic and anaerobic conditions, reasoning that this would increase mutant diversity since many genes in A. actinomycetemcomitans are required specifically under high or low oxygen levels (33). After generating the mutant pools, we defined the location and abundance of the insertions via Tn-seq. We sequenced two aliquots of each mutant pool and found the replicates to be highly correlated (correlation coefficient, ≥0.94) (see Fig. S1 in the supplemental material). In total, we identified 32,344 high-confidence insertions in VT1169 (coverage of ∼1 insert per 66 bp) and 26,530 high-confidence insertions in 624 (coverage of ∼1 insert per 89 bp) (see Table S1 in the supplemental material).
TABLE 1.
A. actinomycetemcomitans strains used in this study
| Name | Genome size (bp) | Serotype | Natural competence | Passaging | Antibiotic resistance | Fimbriation |
|---|---|---|---|---|---|---|
| VT1169 | 2,129,092 | b | Not competent | High | Nalidixic acid, rifampin | Smooth |
| 624 | 2,367,908 | a | Competent | Low | Rough |
Defining the essential genome in multiple strains of A. actinomycetemcomitans.
We next used a Monte Carlo-based approach (41) to define the essential genomes of VT1169 and 624 with statistical rigor. In this approach, the actual mariner insertions identified in the genome (observed data) are compared to “expected” simulated data, where each gene is assumed to be fitness neutral (equally capable of being disrupted by a transposon). To generate the expected data, the actual insertions are randomly reassigned to any of the possible TA sites in the genome. This process is then repeated numerous times, with the same insertion coverage as the observed data in each simulation. The advantage of this repeated simulation (Monte Carlo) approach is that, for any given coverage, it accurately estimates how often each gene should be disrupted when neutral for fitness. Genes in the observed data that have fewer insertions can therefore be considered required for fitness and part of the essential genome.
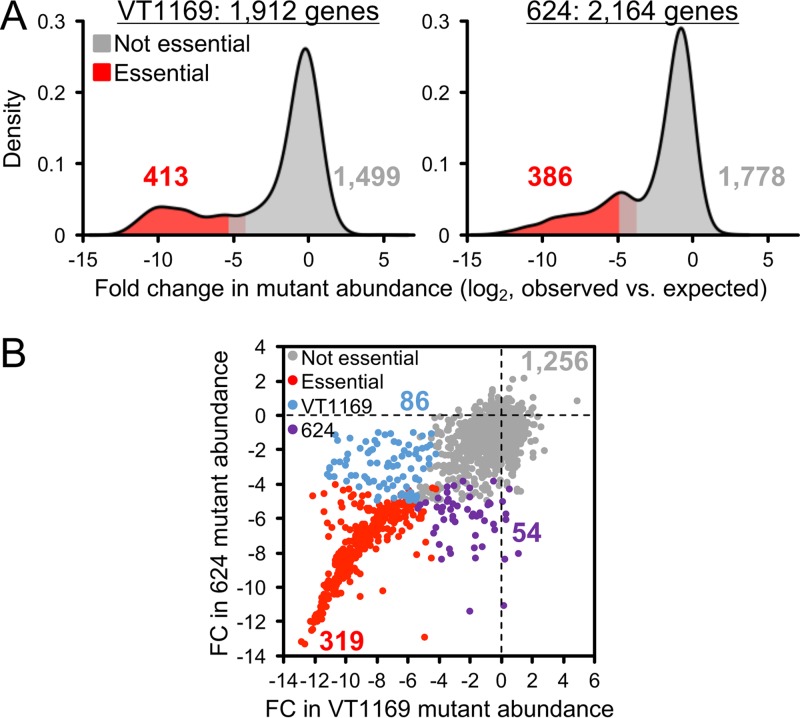
To apply this approach to A. actinomycetemcomitans, we generated expected data consisting of 100 simulations per strain. We then compared the expected to the observed data and observed that the fold changes in mutant abundance per gene follow typical (41, 48) bimodal distributions (Fig. 1A). The lower mode in these distributions corresponds to genes that cannot sustain insertions. As described previously (41), we determined a gene as essential if (i) it contained significantly fewer insertions than expected and (ii) clustered significantly with the lower of the two modes. Using these criteria, we identified 413 (22% of total) coding genes as essential in VT1169 and 386 (18% of total) coding genes as essential in strain 624 (Fig. 1A). These numbers are similar to how many essential genes are found in other bacterial species (for example, 336 in Pseudomonas aeruginosa [41] and 358 in Haemophilus influenzae [42]).
FIG 1.
The A. actinomycetemcomitans essential genome. (A) Density plots of the fold change in mutant abundance compared to expected value in VT1169 (left) and 624 (right). Essential genes (red) have significantly fewer mutants than expected (gray). (B) Scatter plot of the log2 fold change (FC) in mutant abundance compared to expected in VT1169 (x axis) versus 624 (y axis). Each point corresponds to an ortholog. Gray, not essential in either strain; red, essential in both strains; blue, essential in VT1169 only; purple, essential in 624 only.
The core A. actinomycetemcomitans essential genome.
As our ultimate goal was to define a core essential genome not specific to either strain, we next directly compared the essentiality of each gene in VT1169 and 624. To do so, we determined the set of orthologous genes that are shared between the two strains. In all, we identified 1,715 orthologs, and of these, 319 (19% of total) orthologs were essential in both strains and therefore could be assigned to the core essential genome (Fig. 1B). These orthologs represented most of the total essential genes in both strains, but as we anticipated, several orthologs were essential in only one of the two strains (see Table S3 in the supplemental material). This observation justifies our decision to define the essential genome of more than one strain. For example, we found that threonine biosynthesis is essential in strain 624 but not VT1169 (Fig. S2). As a result, we successfully excluded this metabolic pathway from the core A. actinomycetemcomitans essential genome. Interestingly, a small number of accessory (nonorthologous) genes were also essential in each strain. These genes warrant further study since most (15 of 21) encode hypothetical proteins with unknown function.
Functional capacity of the A. actinomycetemcomitans essential genome.
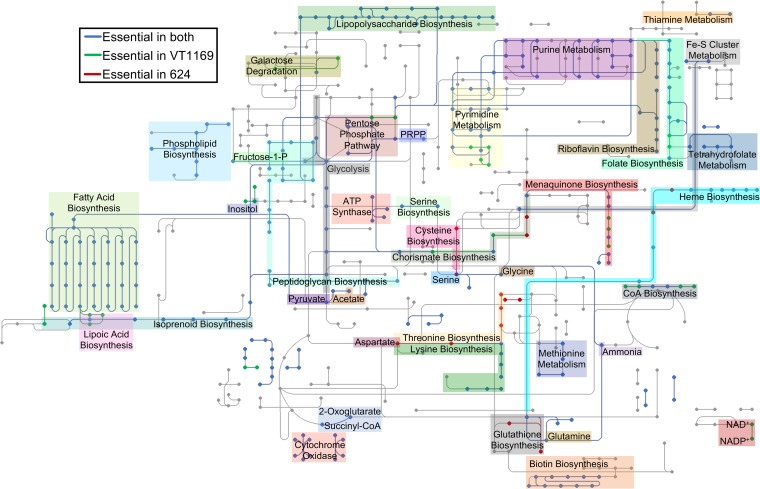
To gain an overview of its main functions, we next analyzed the A. actinomycetemcomitans essential genome for enriched Clusters of Orthologous Groups (COG) functional categories (49). As expected, VT1169 and 624 were enriched for functions known to be vital for bacterial life (50), such as lipid metabolism and cell wall biogenesis (COGs I and M) (Fig. S3). In contrast, some functions were depleted, such as carbohydrate (COG G) and inorganic ion metabolism (COG P) (Fig. S3), likely as a result of functional redundancy and the complex medium on which the mutant pools were generated. To examine the essential genome in more detail, we projected it as a KEGG (51)-style metabolic network to highlight specific essential pathways (Fig. 2). Not surprisingly, many of these pathways were functionally related to the same COGs that were enriched, such as fatty acid and peptidoglycan biosynthesis (related to COGs I and M, respectively). Other essential pathways were not apparent from our COG analysis, such as the biosynthesis of several amino acids (e.g., glutamine, glycine, and lysine) and vitamins (e.g., riboflavin, folate, and heme). As predicted by our ortholog analysis (Fig. 1B), some essential pathways were strain specific (e.g., lipoate biosynthesis in VT1169 and zinc transport in 624). In total, we identified over 40 metabolic pathways and functions that are essential in A. actinomycetemcomitans (Table S4 in the supplemental material).
FIG 2.
The A. actinomycetemcomitans essential metabolic network. Dots and lines represent compounds and reactions, respectively. Blue, pathways essential in both strains; green, pathways essential in VT1169 only; red, pathways essential in 624 only. The shaded regions indicate specific metabolites or metabolic pathways. PRPP, phosphoribosyl pyrophosphate; CoA, coenzyme A.
Conservation of the core A. actinomycetemcomitans essential genome.
Next, we determined whether the essential orthologs shared between VT1169 and 624 are conserved in the genomes of other strains of A. actinomycetemcomitans. We reasoned that if these genes do in fact represent the core essential genome, they should be highly conserved. To perform this analysis, we used a collection of 15 strains, with representatives from the 7 currently known A. actinomycetemcomitans serotypes (Table 2). As anticipated, each of the 319 genes in the core essential genome contained an ortholog in at least 12 of the 15 tested strains, with 311 (97.5%) containing an ortholog in at least 14 strains (Fig. S4A). On average, 99% of the core essential genome was conserved in each strain (Fig. S4B). These results suggest that the essential genome that we defined using only 2 strains is a close approximation of the essential genome among all A. actinomycetemcomitans strains.
TABLE 2.
A. actinomycetemcomitans strains used for testing conservation of the essential genome
| Isolation source and serotype | Strain | No. of coding genes in strain | No. of orthologsa |
|---|---|---|---|
| Human | |||
| a | D7S-1 | 2,041 | 317 |
| H5P1 | 1,978 | 311 | |
| b | HK1651 | 2,327 | 318 |
| I23C | 1,894 | 315 | |
| SCC1398 | 1,847 | 317 | |
| c | D11S-1 | 2,041 | 317 |
| SCC2302 | 1,829 | 315 | |
| d | I63B | 2,010 | 314 |
| SA508 | 1,991 | 314 | |
| e | SC1083 | 2,161 | 315 |
| SCC393 | 1,998 | 307 | |
| f | D18P-1 | 2,020 | 318 |
| SC29R | 2,166 | 317 | |
| g | NUM4039 | 2,364 | 318 |
| Rhesus macaque, serotype b | RhAA1 | 2,150 | 318 |
Number of orthologs to the 319 VT1169/624 core essential genes present in the indicated strain.
We next explored whether the essential genome of A. actinomycetemcomitans is conserved among the essential genomes of other bacterial species. To accomplish this, we used the Database of Essential Genes (DEG) (52). This database comprises >50,000 genes experimentally determined as essential in 38 bacterial strains and species. We found that nearly all of the core essential genes in both VT1169 (313 of 319) and strain 624 (314 of 319) contain at least one ortholog in DEG. However, a few genes lacked an ortholog, suggesting that they encode functions that are essential specifically to A. actinomycetemcomitans (Table S5 in the supplemental material). Many of these genes were related to metabolism, including transporters for glutamate and thiamine and a regulator of fatty acid metabolism. Next, we examined conservation with individual strains/species, finding that A. actinomycetemcomitans shares the most (243 of 319) essential orthologs with Salmonella enterica serovar Typhi and the fewest (74 of 319) with Bacillus thuringiensis (Fig. S5). We then grouped the species in DEG by phylogenetic class and examined each class's conservation with the A. actinomycetemcomitans essential genome (Fig. S5). We predicted that the classes would rank by their relatedness to A. actinomycetemcomitans, and as we predicted, the phylogenetic class to which A. actinomycetemcomitans belongs, the Gammaproteobacteria, ranked the highest, with less closely related classes such as the Bacilli ranking lower (Fig. S5). Together, these results show that though some functions may be required specifically in A. actinomycetemcomitans, most of this bacterium's required features are shared with other Gammaproteobacteria.
Constructing an A. actinomycetemcomitans ordered library.
Although many tools for genetic manipulation have been developed for A. actinomycetemcomitans, making targeted mutations can still be a time-consuming process. As shown above, Tn-seq can be valuable for defining the essential genome and rapidly conducting other large-scale screens. However, because the hallmark of Tn-seq is using a pooled mutant mixture, retrieving mutants of interest to follow up a screen is not possible. For this purpose, the standard genetic resource is an ordered mutant library in which individual mutants are arrayed into microtiter (e.g., 96-well) plates. The challenge of ordered libraries, however, is identifying the mutant in each of the (potentially thousands of) microtiter wells in the library. Because of this, constructing an ordered library generally represents a herculean though achievable task.
Previously, a simple method, Cartesian pooling-coordinate sequencing (CP-CSeq) (46), was developed for efficiently constructing ordered libraries. To understand how CP-CSeq works, it is best to imagine the ordered library as a stack of 96-well plates. The goal of CP-CSeq is to find the position (Cartesian coordinate) of each mutant in the stack. Each mutant's X and Y coordinates correspond to its row and column, and each mutant's Z coordinate corresponds to its plate within the stack. In CP-CSeq, these coordinates are found by subpooling the mutants from each of the stack's rows (8 pools), columns (12 pools), and plates (number of pools depends on library). More specifically, the row and column pools are generated by collapsing the stack into a single master plate, whereas the plate pools are generated by collecting each plate into an individual well of a second master plate (Fig. S6). These master plates are then profiled by Tn-seq, ultimately providing data for finding each mutant's coordinates. For example, the mutant in row A4 of plate 39 should be uniquely detected by Tn-seq in the row A pool, the column 4 pool, and the plate 39 pool (Fig. S6).
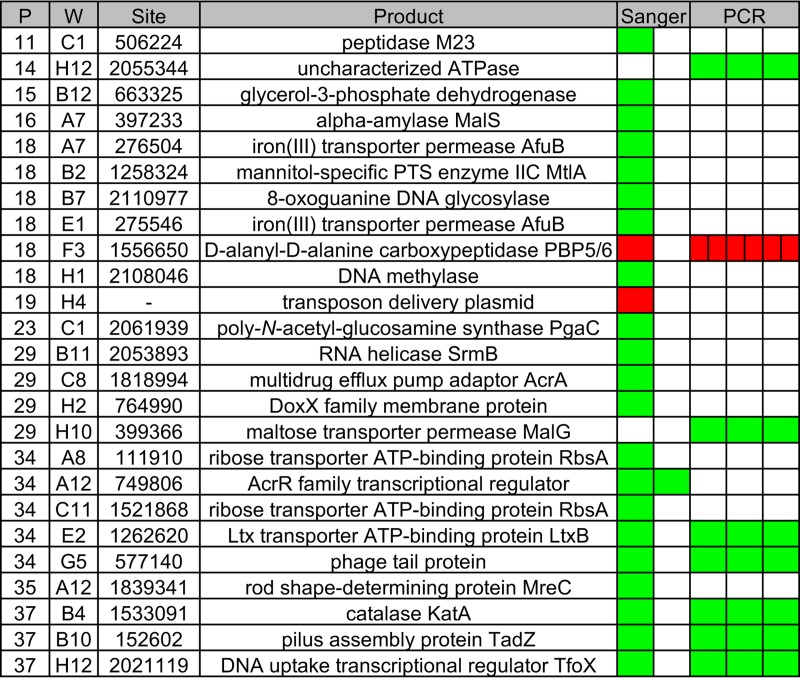
We used CP-CSeq to construct an ordered library in A. actinomycetemcomitans VT1169. We chose VT1169 because it is not as clumpy as clinical isolates like 624 (17) and therefore is more likely to produce clonal colonies. First, we serially diluted aliquots of the VT1169 mutant pool that we used to define the essential genome, and then, we hand-picked individual colonies into 96-well plates. In total, we picked 3,744 colonies into 39 plates. Using CP-CSeq, this number of plates required 33 total Tn-seq reactions to deduce the coordinates of each mutant (Table S6 in the supplemental material). After analyzing the CP-CSeq data, we were pleased to find that most wells in the library (1,971) contain only 1 to 2 mutants, and overall, we found that the library comprises 1,531 unique mutants. Of these, we found that 990 map internally to 626 total coding genes, indicating that the library encompasses mutants for 33% of the coding genes in the VT1169 genome (Table S7 in the supplemental material). To validate the library, we isolated individual colonies from 25 wells that spanned 11 plates in the library. We then used Sanger sequencing and/or PCR to confirm that each mutant was the same as predicted by CP-CSeq. Of the 25 wells, we successfully validated 23 (Fig. 3). Together, these results show that though the library is not comprehensive, it is a useful tool for quickly obtaining individual mutants of interest. As the CP-CSeq method is easily scalable, efforts are under way in our laboratory to expand the library's coverage.
FIG 3.
Validation of the A. actinomycetemcomitans ordered library. P, plate; W, well; Site, transposon insertion site in the VT1169 genome; Product, gene product nearest the transposon insertion; PTS, phosphotransferase; Ltx, leukotoxin. Each box under the Sanger and PCR headings indicates a replicate. Green, positive Sanger/PCR result; red, negative Sanger/PCR result.
Screening the ordered library for factors that contribute to antibiotic resistance.
As little is known about mechanisms of antibiotic resistance in A. actinomycetemcomitans, we next decided to use the ordered library to screen for genes involved in erythromycin resistance. Macrolides, such as erythromycin, inhibit formation of the 50S ribosomal subunit (26) and represent one of the most commonly used classes of antibiotics for treating periodontal disease (24). To perform this screen, we first determined that the MIC of erythromycin for A. actinomycetemcomitans is 2 μg/ml, which falls within previously reported ranges (27). Next, we revived the ordered library and grew each well individually with erythromycin at half-MIC (1 μg/ml). We chose half-MIC erythromycin since it would permit fully resistant strains to grow but inhibit mutants that are more susceptible to erythromycin. We then visually examined each well for lack of growth and identified two hypersensitive mutants (one of which is pictured in Fig. S7A). These mutants were disrupted for the genes encoding AcrA and AcrB, subunits of a multidrug efflux system shown in many bacteria to mediate antibiotic resistance (53, 54).
Screening the library for factors that contribute to antibiotic-induced attachment.
During the erythromycin screen, we were struck by an interesting phenomenon. We noticed that erythromycin causes A. actinomycetemcomitans to attach more to surfaces. Biofilm formation is a common stress response in many bacteria (55), and we hypothesized that a similar response was occurring in A. actinomycetemcomitans when exposed to erythromycin. A few factors that mediate antibiotic-induced attachment have been characterized (56, 57). However, to our knowledge such factors have not been explored systematically. Therefore, we designed a simple Tn-seq-based screen to identify mutants that fail to attach in response to antibiotic. We reasoned that in the presence of antibiotic, nondefective mutants would attach to surfaces while attachment-defective mutants would remain in the liquid phase (Fig. S7B). Therefore, the screen is conducted by growing the mutant pool in the absence and presence of antibiotic and then simply comparing the liquid phases of each culture. Mutants identified as enriched in the antibiotic-treated liquid phase are potentially defective for attachment. With this screen, we also anticipated identifying mutants that are susceptible to erythromycin, not only acrA and acrB but (due to Tn-seq's sensitivity) less severely attenuated mutants.
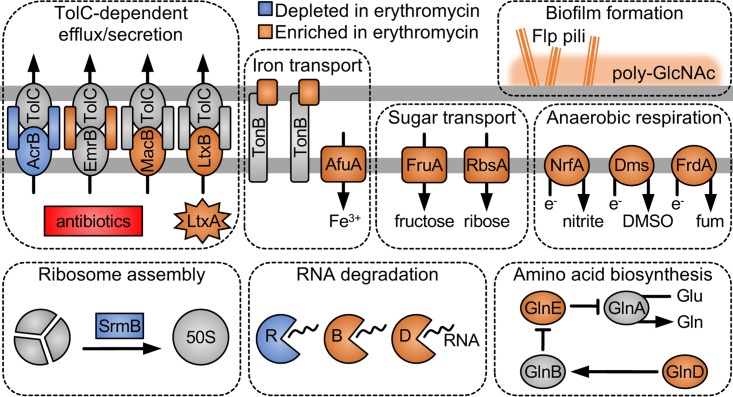
To perform the screen, we decided to use the ordered library since it would allow us to easily retrieve mutants for follow-up experiments. First, we collected the ordered library into a single pool, and then, we grew it both with and without half-MIC erythromycin. After comparing the two treatments via Tn-seq, we identified mutants in 9 genes that decreased in abundance in the presence of erythromycin compared to the control (Table S8 in the supplemental material). As expected, acrA and acrB were among the mutants that decreased in abundance (Fig. 4), confirming the results from our original screen. Other mutants that decreased were disrupted for srmB, an RNA helicase involved in 50S ribosome assembly (58), and rnr, an RNase involved in degrading ribosome-stalling mRNA (59) (Fig. 4). As these genes both interact with the ribosome, they likely mediate intrinsic resistance to erythromycin, a ribosome-targeting antibiotic.
FIG 4.
A. actinomycetemcomitans processes affected by subinhibitory erythromycin. The horizontal gray lines represent the outer (top line) and inner (bottom line) membranes. LtxA, leukotoxin; e−, electron; DMSO, dimethyl sulfoxide; fum, fumarate; Gln, glutamine; Glu, glutamate; poly-GlcNAc, poly-N-acetylglucosamine. R, B, and D represent RNases.
Surprisingly, mutants in 10 times as many (90) genes increased in abundance in the presence of erythromycin compared to the control (Table S8 in the supplemental material). As we anticipated, a number of these mutants were disrupted for genes associated with A. actinomycetemcomitans biofilm formation, including 3 genes in the tight adherence (tad) locus, involved in assembling Flp pili (17), and 1 gene in the pga locus, involved in synthesizing the biofilm polysaccharide poly-N-acetylglucosamine (60) (Fig. 4). However, it was unclear what, if any, role in biofilm formation was played by the many other genes in which mutants increased in abundance in the presence of erythromycin. Alternatively, disruption of these genes could somehow enhance fitness (for example, by increasing growth rates) when erythromycin is present, especially since many of these genes were metabolic in nature (Fig. 4; also see Discussion).
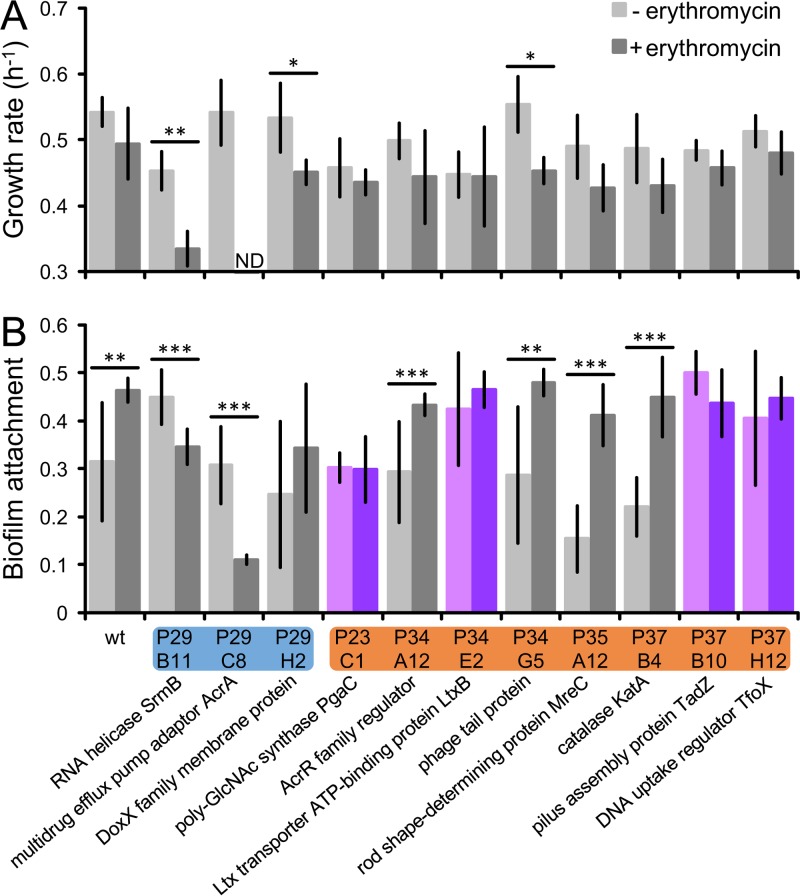
To explore these possibilities, we measured growth rates and attachment levels for 11 mutants (Fig. 5). We included both mutants that increased and mutants that decreased in abundance in the presence of erythromycin. As in the screen, we used subinhibitory (half-MIC) erythromycin and observed that in the presence of antibiotic, the wild type does not grow significantly slower. In contrast, mutants that decreased in abundance in the presence of erythromycin did grow slower, and in fact, the acrA mutant showed no signs of growth. Among the mutants that increased in abundance in the presence of erythromycin, we observed none that grew faster (Fig. 5A), ruling out enhanced fitness as an explanation for why these mutants were more abundant in the presence of erythromycin.
FIG 5.
A. actinomycetemcomitans factors that mediate antibiotic-induced attachment. Growth rates (A) and attachment levels (B) were measured for the wild type (wt) and mutants in half-MIC (+) erythromycin or none (−). y axis in panel A, doublings per hour; y axis in panel B, absorbance (A620) of crystal violet bound to biofilm; blue, mutants that decreased in abundance after erythromycin exposure; orange, mutants that increased in abundance after erythromycin exposure; purple, defective for antibiotic-induced attachment; ND, not detected. Data labels underneath indicate each mutant's disrupted gene product and coordinates in the ordered library. poly-GlcNAc, poly-N-acetylglucosamine; Ltx, leukotoxin. Error bars represent standard deviations (n = 3 to 4 for growth rate; n = 11 to 16 for attachment). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed Student's t test).
We next used a standard crystal violet assay (61) to examine biofilm formation. In this assay, attached cells are stained with crystal violet, which is then solubilized and measured using absorbance. A higher absorbance reading therefore indicates higher attachment. As we had observed qualitatively, the quantitative crystal violet assay showed that the wild type attaches at higher levels in response to erythromycin (Fig. 5B; see also Fig. S8). In contrast, the erythromycin-sensitive mutants attached at lower levels, likely because of growth inhibition. Similarly to the wild type, some nonsensitive mutants also increased in attachment. However, we identified 4 mutants that did not show this response (Fig. 5B), indicating that the corresponding genes mediate antibiotic-induced attachment. These results confirm that our screen successfully discovered factors that contribute to antibiotic-induced attachment.
DISCUSSION
Severe gum disease, or periodontitis, is one of the most common infectious diseases worldwide (6). It occurs when opportunistic oral pathogens overrun the attached biofilm communities (dental plaque) that form on tooth surfaces. Though periodontitis is treatable by eliminating plaque physically, many cases of periodontitis are highly refractory and necessitate chemical treatment with antibiotics (22, 23). However, antibiotic resistance among periodontal pathogens is widespread (24), motivating a search for alternative therapies and a better understanding of bacterial resistance mechanisms.
Essential genomes are the full set of genes required for the viability of an organism, and they can be leveraged to discover novel drug targets (43, 44). Here, we used a Monte Carlo approach (41) to define the essential genome in two strains of the periodontal pathogen A. actinomycetemcomitans, VT1169 and 624. To maximize mutant diversity, we generated the mutant pools in both strains on a rich, undefined medium. A defined medium would have undoubtedly restricted what genes could be disrupted. For example, on a medium with a defined carbon source, genes involved in utilizing that carbon source would not have been disrupted, giving the false impression that these genes are obligately essential. In total, we discovered 319 genes that are essential in both strains (Fig. 1); however, 161 genes were essential in only VT1169 or 624 (see Table S3 in the supplemental material). As VT1169 is undergoing genome reduction compared to 624 (36, 40), these uniquely essential genes may reflect changes in how the strains are genetically wired (62).
The primary advantage of using two strains was that it enabled us to establish a core set of metabolic functions that are widely conserved across strains (Fig. 2; see also Fig. S4)—information crucial for developing new strain-independent therapies. These conserved functions included the transport of thiamine and cystine (Table S4), both of which were previously shown to strongly stimulate the growth of A. actinomycetemcomitans in culture media (63). Metabolic targets such as these could therefore be exploited in future drug development, as exemplified by cancer researchers who have engineered enzymes, such as cyst(e)inases, to degrade amino acids that support auxotrophic cancers (64). Cystine is only conditionally essential in humans (65), but thiamine requires dietary uptake, making its depletion potentially toxic. However, as periodontitis is confined to the oral cavity, toxicity could be avoided by administering metabolic therapies directly to dental plaque.
In this report, we also present an ordered transposon mutant library for strain VT1169. We constructed the library by applying an efficient genomic method, CP-CSeq (46), originally developed for Mycobacterium bovis. Validation experiments proved the library to be highly accurate, with >90% of mutants (23 out of 25 wells) matching the predicted result (Fig. 3). Though it is not comprehensive, the library comprises >1,500 unique mutants, which are publicly available, and so we hope this resource will accelerate periodontitis research.
Because macrolide resistance is widespread among periodontal pathogens (24), we demonstrated the library's utility by conducting a screen for genes that confer resistance to erythromycin. We discovered that the main defense of A. actinomycetemcomitans against erythromycin is the multidrug efflux pump AcrAB (Fig. S7A). AcrAB can export not only antibiotics but also host substrates, such as bile and steroids, and so in many pathogens AcrAB acts as a virulence factor (54). Although AcrAB is not essential for A. actinomycetemcomitans survival in abscesses (33), it may play an important role in the oral cavity.
During the screen, we noticed that subinhibitory erythromycin induces A. actinomycetemcomitans biofilm formation. This clinically relevant behavior could prime A. actinomycetemcomitans for higher, more lethal antibiotic concentrations during infection. In other bacteria, similar phenomena are mediated by cyclic di-GMP signaling (56) and extracellular DNA release (57). To discover factors that mediate this process in A. actinomycetemcomitans, we developed a novel Tn-seq-based screen (Fig. S7B) and confirmed through biofilm assays that up to 50% of the screen's hits may mediate erythromycin-induced attachment (Fig. 5B). The remaining genes identified by the screen likely have other functions that, when disrupted, enhance fitness in the presence of erythromycin. Though these mutants did not grow faster (Fig. 5A), we suspect that their fitness advantage would be more discernible in competition experiments with the wild type.
While examining these genes further, we found that they are functionally enriched for inorganic ion transport (e.g., transferrin and ferric iron transporters) and defense mechanisms (see Table S9 in the supplemental material for summary of COG enrichment analysis). Other prominent cellular functions included multiple RNases, sugar transport, amino acid (glutamine, lysine, proline, and tryptophan) biosynthesis, and anaerobic respiration (Fig. 4). To our surprise, the enriched defense mechanisms were primarily other, non-Acr multidrug efflux systems (Fig. 4). Like the Acr system, the Emr system nonspecifically effluxes multiple substrates (66), whereas the Mac system specifically effluxes macrolides (67). All 3 systems, however, require the outer membrane channel TolC to function (66, 67). Since of these systems Acr has the lowest affinity for TolC (68), we hypothesize that disrupting Emr or Mac enhances Acr's efflux potential by increasing its access to TolC. In support of this, other TolC-dependent systems were also enriched in the presence of erythromycin, including the Rax (69) and leukotoxin (70) type I secretion systems (Fig. 4).
As expected, 2 of the 4 genes found to mediate antibiotic-induced attachment (Fig. 5B) encoded products known to assist in A. actinomycetemcomitans biofilm formation—Flp pili (tadZ [17]) and poly-GlcNAc (pgaC [60]) (Fig. 4). The remaining 2 genes, however, encoded less well recognized biofilm determinants—leukotoxin (ltxB [19]) and the DNA uptake transcriptional regulator TfoX (71). Leukotoxin is widely regarded as the primary virulence factor of A. actinomycetemcomitans, as it lyses both white and red blood cells (20), but recently it was also shown to control adherence. Specifically, disrupting ltxA diminishes tadA and pgaC expression and, as a result, binding to hydroxyapatite (calcium-based tooth mineral) (72). Therefore, leukotoxin appears to indirectly mediate antibiotic-induced biofilm formation.
Perhaps the screen's most surprising result was TfoX (71), since it is unclear how natural transformation is connected to biofilm formation. However, previously it was shown that in biofilms, A. actinomycetemcomitans expresses higher levels of tfoX when exposed to calcium, resulting in higher transformation frequencies (73). Furthermore, this calcium-induced transformation is dependent on the pga gene cluster (73). These findings therefore provide a functional link between natural transformation and biofilm formation. In another report, transcriptome analysis of a pga mutant revealed substantial differential gene expression compared to the wild type (74). As the pga regulon does not encompass tfoX (74), this suggests that tfoX may potentially regulate, or at least act upstream of, pga. Unfortunately, a tfoX consensus binding site is not available (75), and so we were unable to determine if tfoX binds to the pga promoter.
A final remaining question is how A. actinomycetemcomitans senses and transduces erythromycin as a signal to ultimately enhance biofilm formation. In Vibrio cholerae, natural transformation is induced by chitin, which is recognized by a membrane sensor that activates tfoX (76). We hypothesize that in A. actinomycetemcomitans, a similar sensor recognizes membrane stress (77, 78) caused by calcium, erythromycin, and potentially other antibiotics. Experiments are under way in our laboratory to test this hypothesis.
In summary, we provide in this report the essential genome, an ordered mutant library, and a new screen for attachment factors in A. actinomycetemcomitans. These resources should help to advance research on this devastating human pathogen.
MATERIALS AND METHODS
Strains and growth conditions.
The A. actinomycetemcomitans strains used were VT1169 (79) and 624. VT1169 is a serotype b laboratory strain; 624 is a serotype a clinical isolate. Additional strain characteristics are provided in Table 1. A. actinomycetemcomitans was routinely grown in filter-sterilized (80) tryptic soy broth plus 0.5% (wt/vol) yeast extract (TSBYE medium) with shaking at 250 rpm or on autoclaved tryptic soy agar plus 0.5% (wt/vol) yeast extract (TSAYE medium). Under oxic conditions, A. actinomycetemcomitans was incubated in a 5% CO2 atmosphere at 37°C. Under anoxic conditions, A. actinomycetemcomitans was incubated in a vinyl anaerobic chamber (Coy Laboratory Products) with an 85% N2, 10% CO2, and 5% H2 atmosphere at 37°C.
A. actinomycetemcomitans mutant pools.
A. actinomycetemcomitans was mutagenized by conjugation largely as described previously (81). The E. coli donor strain was MFDpir (82), a diaminopimelate (DAP) auxotroph. MFDpir harbored pMR361-K, a mariner minitransposon delivery plasmid. Conjugations were performed on TSAYE plus 0.3 mM DAP plus 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 to 8 h and counterselected on TSAYE plus 40 μg/ml kanamycin for 3 to 5 days. Conjugations and counterselections were done under both oxic and anoxic conditions. Mutants from independent conjugations were pooled, amplified under oxic and anoxic conditions, and then aliquoted to generate the final mutant pools. Because of contamination, the VT1169 pool was additionally amplified on TSAYE plus 40 μg/ml kanamycin, 50 μg/ml nalidixic acid, and 100 μg/ml rifampin. Aliquots of the 624 and VT1169 mutant pools before and after amplification were used for Tn-seq and treated as replicates for essential genome analyses.
Tn-seq libraries (essential genome).
DNA was extracted from each mutant pool largely as described previously (83). An aliquot (1-ml glycerol stock) was resuspended in 1 ml 1× buffer A (84) plus 0.1% SDS and 0.1% sodium deoxycholate, transferred to a Lysing Matrix B 2-ml tube (MP Biomedicals), homogenized in a Mini-Beadbeater (BioSpec), and digested overnight at 55°C with 1 mg/ml proteinase K. DNA was then extracted with 1 ml 25:24:1 phenol-chloroform-isoamyl alcohol (pH 8.0), purified from the aqueous phase by isopropanol precipitation, washed with 75% ethanol, and dissolved in 500 μl distilled water (diH2O). NaCl was then added to 50 mM, and DNA was treated with 0.1 mg/ml RNase A for 1 h at 37°C. DNA was then reextracted with 500 μl phenol-chloroform-isoamyl alcohol and purified by ethanol precipitation.
Tn-seq libraries were prepared as described previously (41) (Table 3 lists primer sequences). Libraries were sequenced at The University of Texas at Austin Genome Sequencing and Analysis Facility on an Illumina NextSeq 500 1-by-75 single-end run.
TABLE 3.
Primer sequences used in this study
| Primer | Function | Reaction or coordinatesb (plate/well) | Sequence (5′–3′) |
|---|---|---|---|
| olj376 | Preparing Tn-seq libraries (essential genome) | PCR-1 | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGGGGGGGGGGGGGGGG |
| olj510_mariner (mariner-1) | PCR-1 | ACTCACTATAGGAGGGCGGGAATCATTTGAAGGTTGGTAC | |
| BC_a | PCR-2 | CAAGCAGAAGACGGCATACGAGATxxxxxxGTGACTGGAGTTCAGACGTGTG | |
| olj511_mariner (mariner-2) | PCR-2 | Biotin-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNGTGTCAGACCGGGGACTTATCAG | |
| VT1169_0386-F | Validating ordered library | 29/H10 | GACGCTTCCTGATGAACAAAAGTGC |
| VT1169_0386-R | 29/H10 | CCACCATCATCTTCTTGCTCGTAGG | |
| VT1169_1989-F | 14/H12 | GTAAAGTCTGCGTTAGAATAGCGAGAAATCC | |
| VT1169_1989-R | 14/H12 | GTCACCCCATACAACGGCAC | |
| VT1169_0152-F | 37/B10 | GGTCCATAACCCTCAATCTCATCAGC | |
| VT1169_0562-R | 34/G5 | CGGACATAAACTGCCAAATCCAGC | |
| VT1169_1175-R | 34/E2 | ACGCAAATTCGTTAGCAACTACTGC | |
| VT1169_1449-R | 37/B4 | GTTTTGTGCGAATGGCGTTTGC | |
| VT1169_1462-R | 18/F3 | GACGCTAACGGTGAGCTTTCC | |
| VT1169_1954-F | 37/H12 | GACCCCGCCTTCAATATCATTTGG | |
| mariner-2-check | GTGTCAGACCGGGGACTTATCAG |
The primer is named after the Illumina barcode that is used (e.g., the x's in the sequence for BC01 are replaced with Illumina barcode 1).
Coordinates in the ordered library from which individual colonies were isolated.
Tn-seq analysis.
The location and frequency of transposon insertions were determined largely as described previously (33, 83). See Tables S1 and S2 for details and a summary of the analysis. Software/language versions were fqgrep v0.4.3 (https://github.com/indraniel/fqgrep), cutadapt v1.11 (85), python v2.7.9, bowtie2 v2.2.5 (86), and R v3.3.1.
Essential genome analysis.
The essential genome of each strain of A. actinomycetemcomitans was determined largely as described previously (41, 48) using the following two replicates per strain: (i) Tn-seq on an aliquot of the mutant pool before amplification and (ii) Tn-seq on an aliquot of the mutant pool after amplification (see “A. actinomycetemcomitans mutant pools”). After correcting for polymerase slippage as described previously (33, 87), the countif function in Microsoft Excel was used to filter for insertions present in both replicates (see Table S1). Local smoothing (LOESS) was then used as described previously (41, 48) to correct for how multifork replication can inflate the abundance of insertions close to the origin of replication (see Table S2).
An “expected” data set was then generated using a previously described (41) Monte Carlo method, where for each simulation the location of each insertion and its associated read counts were randomly reassigned to any of the possible TA sites in the genome. The locations of TA sites were identified using the Find Motif tool in the Integrative Genomics Viewer v2.3.80 (88). In total, the expected data set comprised 100 simulations: 50 simulations used the read counts from the first Tn-seq replicate, and 50 simulations used the read counts from the second Tn-seq replicate.
Next, the counts per coding DNA sequence (CDS) were tallied using the intersectBed function in BEDTools v2.24.0 (89), the sumif function in Microsoft Excel, and a genome annotation with the 3′ 10% of each gene truncated (to avoid analyzing insertions that may not disrupt gene function). Insertions intersecting 0 or 2 CDSs were excluded from the tallying step, and tallies were increased by 1 to avoid later potentially dividing by 0.
DESeq2 v1.12.4 (90) was then used with default parameters (except for turning off Cook's distance cutoff for outlier removal) to calculate differential mutant abundance between the observed (n = 2 Tn-seq replicates) and expected (n = 100 simulations) data sets, and mclust v5.2 (91) was used to categorize each CDS as either reduced or unchanged compared to the expected data set.
Essentiality was determined according to the following parameters: for VT1169, (i) observed versus expected DESeq2 log2 fold change of <0, (ii) DESeq2 adjusted P value of <0.001, (iii) mclust mode of 1, and (iv) mclust P value of <0.001; for 624, (i) observed versus expected DESeq2 log2 fold change of <0, (ii) DESeq2 adjusted P value of <0.01, (iii) mclust mode of 1, and (iv) mclust P value of <0.01. Stricter cutoffs were used for VT1169 because it underwent an additional passaging step to remove contamination (see “A. actinomycetemcomitans mutant pools”).
Furthermore, to avoid miscategorizing genes with high sequence similarity as essential, the Tn-seq analysis for each strain was also run for reads with MAPQ (uniqueness) scores below the cutoff (with MAPQ score of ≤39), and the low and high MAPQ data sets were directly compared using DESeq2. Genes were considered “duplicated,” and therefore not capable of being essential, according to the following parameters: (i) low-MAPQ versus high-MAPQ DESeq2 log2 fold change of >10 and (ii) DESeq2 adjusted P value of <0.001. See Data Set S1 for full results.
Ortholog analysis.
Orthologs between A. actinomycetemcomitans VT1169 and 624 were determined using InParanoid v4.1 (92) with BLAST v2.2.16 (93). The InParanoid bootstrapping option and the BLAST substitution matrix BLOSUM45 were used. Input protein sequences for InParanoid were generated as follows. Coding DNA sequences (CDSs) were extracted from A. actinomycetemcomitans genomes using the getfasta function in BEDTools v2.24.0 (89), and translated to protein sequences using EMBOSS Transeq (94). InParanoid paralogs and “duplicated” genes (see “Essential genome analysis”) were excluded from the final ortholog list.
Density plots.
Density plots in Fig. 1A were generated using the general R function density() with default parameters.
Functional enrichment analysis.
Genes were assigned to Clusters of Orthologous Groups (COGs) as follows. A. actinomycetemcomitans VT1169 and 624 were first annotated with K numbers as described previously (33) using the KEGG Automatic Annotation Server (KAAS) (95). K numbers were then converted to COGs (49) using the binary relationship file provided on the KEGG website (http://www.genome.jp/kegg/files/ko2cog.xl). Assigned COGs were then grouped into functional single-letter categories using the table provided on NCBI's COG website (ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/COG0303/cogs.csv). Finally, COGs were tallied for each strain's full and essential genomes using functions in Microsoft Excel. The significance of each COG's enrichment (or depletion) was determined as described previously (32) using a Fisher exact test add-in written for Microsoft Excel (http://www.obertfamily.com/software/fisherexact.html).
Metabolic network.
The metabolic network in Fig. 2 was generated using KEGG (51) as follows. The KEGG application programming interface (API) first was used to find (i) the list of R numbers associated with each K number (http://rest.kegg.jp/link/rn/ko) and (ii) the list of C numbers associated with each R number (http://rest.kegg.jp/link/cpd/rn) in the KEGG database. These lists were then used with the countif function in Microsoft Excel to find the list of C numbers (via the R numbers) associated with each K number in the A. actinomycetemcomitans VT1169 and 624 genomes (obtained using KAAS [95]). The KEGG Mapper Search and Color Pathway tool was then used to generate a metabolic map for the combined list of A. actinomycetemcomitans K and C numbers, both of which were colored black. This map was exported as a PNG file and copied into Microsoft PowerPoint, where gray background was removed by setting the brightness to 20%. Next, the KEGG Mapper Reconstruct Pathway tool was used to generate a metabolic map for the subset of A. actinomycetemcomitans K numbers that are essential. A. actinomycetemcomitans VT1169 was set to organism 1 to color its K numbers green, and A. actinomycetemcomitans 624 was set to organism 2 to color its K numbers red. K numbers essential in both genomes were colored blue. This map was also exported as a PNG file, copied into Microsoft PowerPoint, and adjusted to remove gray background. The transparency of the Reconstruct Pathway map was then set to 40%, and it was aligned over the Search and Color Pathway map. The final map was made by manually adding green, red, and blue dots (since compounds cannot be included in the Reconstruct Pathway tool), whiting out dots not connected to lines, and labeling specific pathways. See Table S4 for a list of metabolic pathways that are essential in A. actinomycetemcomitans.
Essential gene conservation analysis.
Orthologs to the core essential genes in A. actinomycetemcomitans VT1169 were identified in other strains of A. actinomycetemcomitans (Table 2 lists the strains) using InParanoid v4.1 (92) (as described in “Ortholog analysis”). Protein sequences from the other A. actinomycetemcomitans strains were downloaded (in February 2017) from GenBank (ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/Aggregatibacter_actinomycetemcomitans/latest_assembly_versions/). See Data Set S1 for full results. Orthologs to the core essential genes in A. actinomycetemcomitans VT1169 and 624 were identified in the essential genomes of other bacterial strains/species using the Database of Essential Genes (DEG) v14.7 (52) with default blastp parameters. In cases where DEG provided multiple entries for the same strain, the entry containing the most orthologs was used.
Ordered library.
The ordered library for A. actinomycetemcomitans VT1169 was generated as follows. Aliquots of the A. actinomycetemcomitans VT1169 mutant pool were thawed, diluted into <1 ml TSBYE, and incubated at 37°C for <5 h. Revived cells were then serially diluted, plated onto TSAYE (either plain or, to help prevent contamination, with 10 to 50 μg/ml nalidixic acid and/or 10 to 40 μg/ml kanamycin), and incubated under oxic conditions. Once visible, individual colonies were hand-picked using pipette tips into 96-well plates filled with TSBYE. Picked 96-well plates were grown overnight under oxic conditions, mixed 1:1 with 50% glycerol, and stored at −80°C. This procedure was repeated for a total of 39 plates.
CP-CSeq.
The 39 plates were revived by pin replication, first onto TSAYE and then into TSBYE in 96-well plates. We found that first reviving A. actinomycetemcomitans onto TSAYE helped increase the number of wells per 96-well plate that could be revived. Once revived, the 39 plates were pooled into 2 master plates as described for the Cartesian pooling-coordinate sequencing (CP-CSeq) method (46). Briefly, an aliquot of each well of each plate was added to the corresponding well in master plate 1, whereas all 96 wells per plate were first pooled and then each added to a single well in master plate 2 (see Fig. S6). Afterward, each row and column of each master plate were collected into separate pools, resulting in a total of 33 pools (20 [8 rows + 12 columns] from master plate 1 + 13 [5 rows + 8 columns] from master plate 2) (see Fig. S6).
Tn-seq libraries (ordered library).
Tn-seq libraries were made for each of the 33 ordered library pools as described above [see “Tn-seq libraries (essential genome”)] with minor modifications. DNA was extracted from each pool using the DNeasy blood and tissue kit (Qiagen) according to the manufacturer's protocol for pretreatment of Gram-negative bacteria. Terminal deoxynucleotidyltransferase reactions were scaled to 20 μl and used 1 μg sheared DNA as input. DNA was purified using the MinElute reaction cleanup kit (Qiagen) and eluted into 20 to 50 μl diH2O. Libraries were sequenced at The University of Texas at Austin Genome Sequencing and Analysis Facility on an Illumina HiSeq 4000 1-by-50 single-end run. The pool made from row E of master plate 2 was sequenced on an Illumina NextSeq 500 1-by-75 single-end run.
CP-CSeq analysis.
The 33 CP-CSeq Tn-seq libraries were analyzed as described above (see “Tn-seq analysis”) with one major exception. Because the libraries were sequenced on a 1-by-50 run instead of a 1-by-75 run, the reads were not long enough to be mapped to the A. actinomycetemcomitans genome. This was because our Tn-seq primers are designed such that reads off a 1-by-50 run extend only 12 bp past the transposon into the genome. Therefore, we were forced to initially analyze these 12-bp “tags,” instead of numeric insertion sites, when determining the coordinates of each mutant in the library. (The Tn-seq library for row E of master plate 2 was sequenced on a 1-by-75 run, so to make it equivalent to the other libraries, it was trimmed to 1 by 50 using fastx_trimmer [http://hannonlab.cshl.edu/fastx_toolkit/index.html].)
We found that each of the 33 pools contained >10,000 tags, even after increasing the stringency of our Tn-seq analysis (see Table S6). Since this was vastly more than expected, we decided to limit our analysis to only the most abundant tags in each pool. Along these lines, we generated lists of the most abundant tags in each pool that corresponded to how many tags would be expected if each well in the library contained a single mutant (see Table S6).
Next, we used the Unix command ‘grep −Fx −f list1 list2′ to find the tags that were in common (i) between each row and column of master plate 1 (providing lists of the tags that are in each well of the library, e.g., a single list for all tags that are in all wells A1, a single list for all tags that are in all wells A2, etc.), (ii) between each row and column of master plate 2 (providing lists of the tags that are in each plate of the library, e.g., a list for all tags in plate 1, a list for all tags in plate 2, etc.), and then (iii) between each list of well tags and list of plate tags (generated in the previous two steps) to finally identify the tag(s) that is in each well of each plate in the library.
To associate each of these tags with a numeric transposon insertion site in the A. actinomycetemcomitans genome, we collected the entire ordered library into a single pool, made a Tn-seq library, and sequenced it on a 1-by-75 run (see “Erythromycin screen” below). We then used the results to make a binary relationship file for associating each tag with an insertion site. In making this file (from the bowtie2 SAM output file), we were careful to extract the first 12 bases from sequences mapped to the positive strand and the last 12 bases from sequences mapped to the negative strand.
We then used the binary relationship file along with a custom shell script, mimicking the vlookup function in Microsoft Excel (see script 1), to associate the tag(s) in each well of the library with an insertion site. As some tags are associated with multiple insertion sites (i.e., the same 12-bp sequence is next to different insertion sites in the genome), we associated each tag with the more abundant insertion site by sorting the binary relationship file in order of lowest to highest insertion site (because of how the vlookup shell script works). Finally, we used the BEDTools v2.24.0 (89) closestBed function to find the closest coding gene to each insertion site.
A spreadsheet is provided in Data Set S1 that lists for each mutant (i) its library coordinate, (ii) tag, (iii) how many unique sites are associated with that tag, (iv) insertion site, (v) locus tag of closest gene, (vi) start site and (vii) stop site of the closest gene, and (viii) distance from the insertion site to the closest gene.
Ordered library validation.
Each well chosen to validate the library (Fig. 3) was struck out onto TSAYE, and isolated colonies were confirmed—either by Sanger sequencing or by colony PCR—as the mutant predicted by CP-CSeq.
Sanger sequencing.
Isolated colonies were cultured in TSBYE and then used to make Tn-seq libraries as described above [see “Tn-seq libraries (ordered library)”]. Libraries were sequenced using the primer mariner-2 (Table 3) on an Applied Biosystems 3730 DNA analyzer at The University of Texas at Austin DNA Sequencing Facility. Sanger sequencing data were aligned to the VT1169 chromosome using bowtie2 (86).
Colony PCR.
Isolated colonies were picked into diH2O, incubated at 95°C for 5 min in a thermocycler, and then added as the template to PCR mixtures using mariner-2-check and the indicated primer (Table 3). Genomic DNA purified from VT1169 was used as a control. PCRs using primers targeting both sides of the gene were confirmed if the PCR product was larger than the genomic DNA control. PCRs using a primer targeting the transposon (mariner-2-check) and a primer targeting only one side of the gene were confirmed if a PCR product was amplified for the mutant but not for the genomic DNA control.
Erythromycin MIC.
An overnight TSBYE culture of A. actinomycetemcomitans VT1169 was diluted to an optical density (OD) of 0.1 into a 2-fold serial dilution of TSBYE plus erythromycin in a 96-well plate. After the plate was grown overnight (up to 24 h), the MIC was determined as the lowest antibiotic concentration that did not permit growth. This procedure was repeated twice.
Erythromycin screen.
The ordered library was screened for altered erythromycin susceptibility both as individual mutants and as a pool.
Individual mutant screen.
The ordered library was revived by pin replicating batches of 4 to 9 plates onto TSAYE. After 2 to 3 days, colonies were pin replicated into TSBYE in 96-well plates and grown overnight. From these plates, 5 μl from each well was diluted into 95 μl of TSBYE plus half-MIC erythromycin in 96-well plates. After these plates were grown overnight (up to 24 h), individual wells were examined for lack of growth.
Pooled mutant screen.
Per batch of revived plates, 2 to 10 μl of each well was combined and thoroughly mixed. In the antibiotic treatment, 5 μl of the mixture was diluted into 95 μl of TSBYE plus half-MIC erythromycin in each of 24 wells of a 96-well plate. In the control treatment, 5 μl of the mixture was diluted into 95 μl of TSBYE lacking antibiotic in each of 24 wells of the same 96-well plate. The 96-well plate was grown overnight, and the 2 sets of wells were separately collected and stored. After screening of the full library, the antibiotic and control treatments from each batch were collected into separate pools. Tn-seq libraries were made from each pool as described above [see “Tn-seq libraries (ordered library)”]. Libraries were sequenced at The University of Texas at Austin Genome Sequencing and Analysis Facility on an Illumina NextSeq 1-by-75 single-end run.
Tn-seq analysis (erythromycin screen).
The location and frequency of transposon insertions were determined largely as described previously (33, 83). See Tables S1 and S2 for details and a summary of the analysis. Software/language versions were fqgrep v0.4.3 (https://github.com/indraniel/fqgrep), cutadapt v1.12 (85), python v2.7.9, bowtie2 v2.2.5 (86), and R v3.3.1. Differential mutant abundance was tested at the site and gene level as described previously (33, 83) with minor modifications. Read counts for the control and antibiotic treatments were merged by insertion site using the full_join() dplyr function in R. Read counts for sites present in one sample but not the other were set to 1. When tallying the site counts per gene, the 3′ 10% of genes was excluded to prevent analysis of insertions that may not disrupt gene function. DESeq2 v1.12.4 (90) was used with default parameters. Sites/genes were considered differentially abundant according to the following parameters: log2 fold change of >1 and P of <0.05. See Data Set S1 for full results and Table S8 for a summary.
Growth curves.
Overnight cultures were diluted to an OD at 600 nm (OD600) of 0.1 in 5 ml TSBYE with or without half-MIC erythromycin in test tubes and then grown under oxic conditions for 8 h, during which OD600 measurements were made at 2-h intervals. Growth curves were performed for the A. actinomycetemcomitans VT1169 wild type and mutants (Fig. 5) on at least 3 different days. For each growth curve, the generation time was calculated in Microsoft Excel as the average of every possible generation time that could be calculated from at least 3 data points with an R2 (square of Pearson product moment correlation coefficient) of >0.99.
Attachment assays.
Overnight cultures were diluted to an OD600 of 0.1 in 100 μl TSBYE with or without erythromycin (0.001, 0.01, 0.1, 0.5, or 1× MIC) in 96-well plates and then grown overnight (21 to 22 h) under oxic conditions. Afterward, TSBYE and loosely attached cells were removed by pipetting, and 100 μl 0.1% (wt/vol) crystal violet was added to each well. After incubation of the plates for 10 min, unbound crystal violet was removed by submerging the plates three to four times in diH2O. The plates were then placed in a flowing fume hood, and once they were dried, bound crystal violet was solubilized for 10 min with 200 μl 95% ethanol. Afterward, 100 μl was transferred to a new plate, and the absorbance (A620) was measured on a Synergy Mx microplate reader (BioTek). Attachment assays were performed for the A. actinomycetemcomitans VT1169 wild type and mutants (Fig. 5) in at least triplicate on at least 3 different days.
Computational analyses.
Computational analyses were performed both locally and on the Stampede system of the Texas Advanced Computing Center.
Accession number(s).
Raw sequencing data were deposited into the National Center for Biotechnology Information Sequence Read Archive under the accession number SRP099146. The sequence for the mariner minitransposon delivery plasmid, pMR361-K, was deposited into GenBank under the accession number KY767032.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by National Institutes of Health grants awarded to M.W. (R01DE023193) and A.S. (F31DE024931). M.W. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
A.M.N. performed experiments. M.M.R. constructed pMR361-K. A.S. generated the strain 624 mutant pool, designed experiments, performed analyses, and wrote the manuscript. M.W. designed experiments and edited the manuscript.
We thank Patricia Barnabie and Kelly Michie for generating the VT1169 mutant pool, Daniel Cornforth for thoughtful discussions, and Gina Lewin for critically reading the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00797-17.
REFERENCES
- 1.Gest H. 2004. The discovery of microorganisms by Robert Hooke and Antoni Van Leeuwenhoek, fellows of the Royal Society. Notes Rec R Soc Lond 58:187–201. doi: 10.1098/rsnr.2004.0055. [DOI] [PubMed] [Google Scholar]
- 2.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. 2016. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A 113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajishengallis G. 2014. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol 35:3–11. doi: 10.1016/j.it.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J Bacteriol 183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fine DH, Markowitz K, Fairlie K, Tischio-Bereski D, Ferrendiz J, Furgang D, Paster BJ, Dewhirst FE. 2013. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol 51:2850–2861. doi: 10.1128/JCM.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kassebaum NJ, Bernabe E, Dahiya M, Bhandari B, Murray CJ, Marcenes W. 2014. Global burden of severe periodontitis in 1990–2010: a systematic review and meta-regression. J Dent Res 93:1045–1053. doi: 10.1177/0022034514552491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baer PN. 1971. The case for periodontosis as a clinical entity. J Periodontol 42:516–520. doi: 10.1902/jop.1971.42.8.516. [DOI] [PubMed] [Google Scholar]
- 8.Haubek D, Ennibi OK, Poulsen K, Poulsen S, Benzarti N, Kilian M. 2001. Early-onset periodontitis in Morocco is associated with the highly leukotoxic clone of Actinobacillus actinomycetemcomitans. J Dent Res 80:1580–1583. doi: 10.1177/00220345010800062001. [DOI] [PubMed] [Google Scholar]
- 9.Loe H, Brown LJ. 1991. Early onset periodontitis in the United States of America. J Periodontol 62:608–616. doi: 10.1902/jop.1991.62.10.608. [DOI] [PubMed] [Google Scholar]
- 10.Armitage GC, Cullinan MP. 2010. Comparison of the clinical features of chronic and aggressive periodontitis. Periodontol 2000 53:12–27. doi: 10.1111/j.1600-0757.2010.00353.x. [DOI] [PubMed] [Google Scholar]
- 11.Fine DH, Kaplan JB, Kachlany SC, Schreiner HC. 2006. How we got attached to Actinobacillus actinomycetemcomitans: a model for infectious diseases. Periodontol 2000 42:114–157. doi: 10.1111/j.1600-0757.2006.00189.x. [DOI] [PubMed] [Google Scholar]
- 12.Slots J, Reynolds HS, Genco RJ. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun 29:1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zambon JJ, Christersson LA, Slots J. 1983. Actinobacillus actinomycetemcomitans in human periodontal disease. Prevalence in patient groups and distribution of biotypes and serotypes within families. J Periodontol 54:707–711. [DOI] [PubMed] [Google Scholar]
- 14.Yew HS, Chambers ST, Roberts SA, Holland DJ, Julian KA, Raymond NJ, Beardsley J, Read KM, Murdoch DR. 2014. Association between HACEK bacteraemia and endocarditis. J Med Microbiol 63:892–895. doi: 10.1099/jmm.0.070060-0. [DOI] [PubMed] [Google Scholar]
- 15.Konig MF, Abusleme L, Reinholdt J, Palmer RJ, Teles RP, Sampson K, Rosen A, Nigrovic PA, Sokolove J, Giles JT, Moutsopoulos NM, Andrade F. 2016. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci Transl Med 8:369ra176. doi: 10.1126/scitranslmed.aaj1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahamat-Langendoen JC, van Vonderen MG, Engstrom LJ, Manson WL, van Winkelhoff AJ, Mooi-Kokenberg EA. 2011. Brain abscess associated with Aggregatibacter actinomycetemcomitans: case report and review of literature. J Clin Periodontol 38:702–706. doi: 10.1111/j.1600-051X.2011.01737.x. [DOI] [PubMed] [Google Scholar]
- 17.Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol 9:429–437. doi: 10.1016/S0966-842X(01)02161-8. [DOI] [PubMed] [Google Scholar]
- 18.Ramsey MM, Rumbaugh KP, Whiteley M. 2011. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog 7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kachlany SC. 2010. Aggregatibacter actinomycetemcomitans leukotoxin: from threat to therapy. J Dent Res 89:561–570. doi: 10.1177/0022034510363682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balashova NV, Crosby JA, Al Ghofaily L, Kachlany SC. 2006. Leukotoxin confers beta-hemolytic activity to Actinobacillus actinomycetemcomitans. Infect Immun 74:2015–2021. doi: 10.1128/IAI.74.4.2015-2021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haubek D, Poulsen K, Kilian M. 2007. Microevolution and patterns of dissemination of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect Immun 75:3080–3088. doi: 10.1128/IAI.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renvert S, Wikstrom M, Dahlen G, Slots J, Egelberg J. 1990. On the inability of root debridement and periodontal surgery to eliminate Actinobacillus actinomycetemcomitans from periodontal pockets. J Clin Periodontol 17:351–355. doi: 10.1111/j.1600-051X.1990.tb00030.x. [DOI] [PubMed] [Google Scholar]
- 23.Mandell RL, Tripodi LS, Savitt E, Goodson JM, Socransky SS. 1986. The effect of treatment on Actinobacillus actinomycetemcomitans in localized juvenile periodontitis. J Periodontol 57:94–99. doi: 10.1902/jop.1986.57.2.94. [DOI] [PubMed] [Google Scholar]
- 24.Soares GM, Figueiredo LC, Faveri M, Cortelli SC, Duarte PM, Feres M. 2012. Mechanisms of action of systemic antibiotics used in periodontal treatment and mechanisms of bacterial resistance to these drugs. J Appl Oral Sci 20:295–309. doi: 10.1590/S1678-77572012000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helovuo H, Paunio K. 1989. Effects of penicillin and erythromycin on the clinical parameters of the periodontium. J Periodontol 60:467–472. doi: 10.1902/jop.1989.60.8.467. [DOI] [PubMed] [Google Scholar]
- 26.Usary J, Champney WS. 2001. Erythromycin inhibition of 50S ribosomal subunit formation in Escherichia coli cells. Mol Microbiol 40:951–962. doi: 10.1046/j.1365-2958.2001.02438.x. [DOI] [PubMed] [Google Scholar]
- 27.Slots J, Evans RT, Lobbins PM, Genco RJ. 1980. In vitro antimicrobial susceptibility of Actinobacillus actinomycetemcomitans. Antimicrob Agents Chemother 18:9–12. doi: 10.1128/AAC.18.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roe DE, Weinberg A, Roberts MC. 1996. Mobile rRNA methylase genes coding for erythromycin resistance in Actinobacillus actinomycetemcomitans. J Antimicrob Chemother 37:457–464. doi: 10.1093/jac/37.3.457. [DOI] [PubMed] [Google Scholar]
- 29.de la Fuente-Nunez C, Reffuveille F, Fernandez L, Hancock RE. 2013. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol 16:580–589. doi: 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi N, Ishihara K, Kato T, Okuda K. 2007. Susceptibility of Actinobacillus actinomycetemcomitans to six antibiotics decreases as biofilm matures. J Antimicrob Chemother 59:59–65. doi: 10.1093/jac/dkl452. [DOI] [PubMed] [Google Scholar]
- 31.Fine DH, Furgang D, Barnett ML. 2001. Comparative antimicrobial activities of antiseptic mouthrinses against isogenic planktonic and biofilm forms of Actinobacillus actinomycetemcomitans. J Clin Periodontol 28:697–700. doi: 10.1034/j.1600-051x.2001.028007697.x. [DOI] [PubMed] [Google Scholar]
- 32.Jorth P, Trivedi U, Rumbaugh K, Whiteley M. 2013. Probing bacterial metabolism during infection using high-resolution transcriptomics. J Bacteriol 195:4991–4998. doi: 10.1128/JB.00875-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stacy A, Fleming D, Lamont RJ, Rumbaugh KP, Whiteley M. 2016. A commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. mBio 7:e00782-16. doi: 10.1128/mBio.00782-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith KP, Fields JG, Voogt RD, Deng B, Lam YW, Mintz KP. 2015. The cell envelope proteome of Aggregatibacter actinomycetemcomitans. Mol Oral Microbiol 30:97–110. doi: 10.1111/omi.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kittichotirat W, Bumgarner RE, Asikainen S, Chen C. 2011. Identification of the pangenome and its components in 14 distinct Aggregatibacter actinomycetemcomitans strains by comparative genomic analysis. PLoS One 6:e22420. doi: 10.1371/journal.pone.0022420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kittichotirat W, Bumgarner RE, Chen C. 2016. Evolutionary divergence of Aggregatibacter actinomycetemcomitans. J Dent Res 95:94–101. doi: 10.1177/0022034515608163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takada K, Saito M, Tsuzukibashi O, Kawashima Y, Ishida S, Hirasawa M. 2010. Characterization of a new serotype g isolate of Aggregatibacter actinomycetemcomitans. Mol Oral Microbiol 25:200–206. doi: 10.1111/j.2041-1014.2010.00572.x. [DOI] [PubMed] [Google Scholar]
- 38.Zambon JJ, Slots J, Genco RJ. 1983. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun 41:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C, Wang T, Chen W. 2010. Occurrence of Aggregatibacter actinomycetemcomitans serotypes in subgingival plaque from United States subjects. Mol Oral Microbiol 25:207–214. doi: 10.1111/j.2041-1014.2010.00567.x. [DOI] [PubMed] [Google Scholar]
- 40.Jorth P, Whiteley M. 2012. An evolutionary link between natural transformation and CRISPR adaptive immunity. mBio 3:e00309-12. doi: 10.1128/mBio.00309-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A 112:4110–4115. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A 106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mobegi FM, van Hijum SA, Burghout P, Bootsma HJ, de Vries SP, van der Gaast-de Jongh CE, Simonetti E, Langereis JD, Hermans PW, de Jonge MI, Zomer A. 2014. From microbial gene essentiality to novel antimicrobial drug targets. BMC Genomics 15:958. doi: 10.1186/1471-2164-15-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moule MG, Hemsley CM, Seet Q, Guerra-Assuncao JA, Lim J, Sarkar-Tyson M, Clark TG, Tan PB, Titball RW, Cuccui J, Wren BW. 2014. Genome-wide saturation mutagenesis of Burkholderia pseudomallei K96243 predicts essential genes and novel targets for antimicrobial development. mBio 5:e00926-13. doi: 10.1128/mBio.00926-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chao MC, Abel S, Davis BM, Waldor MK. 2016. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol 14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandewalle K, Festjens N, Plets E, Vuylsteke M, Saeys Y, Callewaert N. 2015. Characterization of genome-wide ordered sequence-tagged Mycobacterium mutant libraries by Cartesian pooling-coordinate sequencing. Nat Commun 6:7106. doi: 10.1038/ncomms8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mintz KP, Fives-Taylor PM. 2000. impA, a gene coding for an inner membrane protein, influences colonial morphology of Actinobacillus actinomycetemcomitans. Infect Immun 68:6580–6586. doi: 10.1128/IAI.68.12.6580-6586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zomer A, Burghout P, Bootsma HJ, Hermans PW, van Hijum SA. 2012. ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PLoS One 7:e43012. doi: 10.1371/journal.pone.0043012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gil R, Silva FJ, Pereto J, Moya A. 2004. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev 68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao F, Luo H, Zhang CT, Zhang R. 2015. Gene essentiality analysis based on DEG 10, an updated database of essential genes. Methods Mol Biol 1279:219–233. doi: 10.1007/978-1-4939-2398-4_14. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez L, Pan W, Vinas M, Nikaido H. 1997. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J Bacteriol 179:6855–6857. doi: 10.1128/jb.179.21.6855-6857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piddock LJ. 2006. Multidrug-resistance efflux pumps—not just for resistance. Nat Rev Microbiol 4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 55.Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 57.Kaplan JB, Izano EA, Gopal P, Karwacki MT, Kim S, Bose JL, Bayles KW, Horswill AR. 2012. Low levels of beta-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 3:e00198-12. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol 48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 59.Richards J, Mehta P, Karzai AW. 2006. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol Microbiol 62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- 60.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, Knobloch JK, Ramasubbu N. 2004. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol 186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Toole GA. 2011. Microtiter dish biofilm formation assay. J Vis Exp doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Opijnen T, Dedrick S, Bento J. 2016. Strain dependent genetic networks for antibiotic-sensitivity in a bacterial pathogen with a large pan-genome. PLoS Pathog 12:e1005869. doi: 10.1371/journal.ppat.1005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sreenivasan PK, Meyer DH, Fives-Taylor PM. 1993. Factors influencing the growth and viability of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol 8:361–369. doi: 10.1111/j.1399-302X.1993.tb00612.x. [DOI] [PubMed] [Google Scholar]
- 64.Cantor JR, Panayiotou V, Agnello G, Georgiou G, Stone EM. 2012. Engineering reduced-immunogenicity enzymes for amino acid depletion therapy in cancer. Methods Enzymol 502:291–319. doi: 10.1016/B978-0-12-416039-2.00015-X. [DOI] [PubMed] [Google Scholar]
- 65.Sturman JA, Gaull G, Raiha NC. 1970. Absence of cystathionase in human fetal liver: is cystine essential? Science 169:74–76. doi: 10.1126/science.169.3940.74. [DOI] [PubMed] [Google Scholar]
- 66.Nikaido H. 1996. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol 178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yum S, Xu Y, Piao S, Sim SH, Kim HM, Jo WS, Kim KJ, Kweon HS, Jeong MH, Jeon H, Lee K, Ha NC. 2009. Crystal structure of the periplasmic component of a tripartite macrolide-specific efflux pump. J Mol Biol 387:1286–1297. doi: 10.1016/j.jmb.2009.02.048. [DOI] [PubMed] [Google Scholar]
- 68.Tikhonova EB, Dastidar V, Rybenkov VV, Zgurskaya HI. 2009. Kinetic control of TolC recruitment by multidrug efflux complexes. Proc Natl Acad Sci U S A 106:16416–16421. doi: 10.1073/pnas.0906601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.da Silva FG, Shen Y, Dardick C, Burdman S, Yadav RC, de Leon AL, Ronald PC. 2004. Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol Plant Microbe Interact 17:593–601. doi: 10.1094/MPMI.2004.17.6.593. [DOI] [PubMed] [Google Scholar]
- 70.Crosby JA, Kachlany SC. 2007. TdeA, a TolC-like protein required for toxin and drug export in Aggregatibacter (Actinobacillus) actinomycetemcomitans. Gene 388:83–92. doi: 10.1016/j.gene.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhattacharjee MK, Fine DH, Figurski DH. 2007. tfoX (sxy)-dependent transformation of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Gene 399:53–64. doi: 10.1016/j.gene.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Velusamy SK, Sampathkumar V, Godboley D, Fine DH. 2016. Profound effects of Aggregatibacter actinomycetemcomitans leukotoxin mutation on adherence properties are clarified in in vitro experiments. PLoS One 11:e0151361. doi: 10.1371/journal.pone.0151361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hisano K, Fujise O, Miura M, Hamachi T, Matsuzaki E, Nishimura F. 2014. The pga gene cluster in Aggregatibacter actinomycetemcomitans is necessary for the development of natural competence in Ca(2+)-promoted biofilms. Mol Oral Microbiol 29:79–89. doi: 10.1111/omi.12046. [DOI] [PubMed] [Google Scholar]
- 74.Shanmugam M, El Abbar F, Ramasubbu N. 2015. Transcriptome profiling of wild-type and pga-knockout mutant strains reveal the role of exopolysaccharide in Aggregatibacter actinomycetemcomitans. PLoS One 10:e0134285. doi: 10.1371/journal.pone.0134285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Macfadyen LP, Dorocicz IR, Reizer J, Saier MH Jr, Redfield RJ. 1996. Regulation of competence development and sugar utilization in Haemophilus influenzae Rd by a phosphoenolpyruvate:fructose phosphotransferase system. Mol Microbiol 21:941–952. doi: 10.1046/j.1365-2958.1996.441420.x. [DOI] [PubMed] [Google Scholar]
- 76.Dalia AB, Lazinski DW, Camilli A. 2014. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. mBio 5:e01028-13. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee S, Hinz A, Bauerle E, Angermeyer A, Juhaszova K, Kaneko Y, Singh PK, Manoil C. 2009. Targeting a bacterial stress response to enhance antibiotic action. Proc Natl Acad Sci U S A 106:14570–14575. doi: 10.1073/pnas.0903619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bosse JT, Sinha S, Li MS, O'Dwyer CA, Nash JH, Rycroft AN, Kroll JS, Langford PR. 2010. Regulation of pga operon expression and biofilm formation in Actinobacillus pleuropneumoniae by sigmaE and H-NS. J Bacteriol 192:2414–2423. doi: 10.1128/JB.01513-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mintz KP, Brissette C, Fives-Taylor PM. 2002. A recombinase A-deficient strain of Actinobacillus actinomycetemcomitans constructed by insertional mutagenesis using a mobilizable plasmid. FEMS Microbiol Lett 206:87–92. doi: 10.1111/j.1574-6968.2002.tb10991.x. [DOI] [PubMed] [Google Scholar]
- 80.Bhattacharjee MK, Sugawara K, Ayandeji OT. 2009. Microwave sterilization of growth medium alleviates inhibition of Aggregatibacter actinomycetemcomitans by Maillard reaction products. J Microbiol Methods 78:227–230. doi: 10.1016/j.mimet.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 81.Mintz KP. 2004. Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen. Microbiology 150:2677–2688. doi: 10.1099/mic.0.27110-0. [DOI] [PubMed] [Google Scholar]
- 82.Ferrieres L, Hemery G, Nham T, Guerout AM, Mazel D, Beloin C, Ghigo JM. 2010. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol 192:6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet 10:e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goodman AL, Wu M, Gordon JI. 2011. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Protoc 6:1969–1980. doi: 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12. [Google Scholar]
- 86.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Quinlan AR. 2014. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics 47:11.12.1–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scrucca L, Fop M, Murphy TB, Raftery AE. 2016. mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J 8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 92.Remm M, Storm CE, Sonnhammer EL. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 93.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 94.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 95.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.