Abstract
In recent years, the extent of our vulnerability to misinterpretation due to poorly characterized reagents has become an issue of great concern. Antibody reagents have been identified as a major source of error, contributing to the “reproducibility crisis.” In the current report, we define an additional dimension of the crisis; in particular, we define variation of the targets being analyzed. We report that natural variation in the immunoglobulin “constant” region alters the reactivity with commonly used subtype-specific anti-IgG reagents, resulting in cross-reactivity of polyclonal regents with inappropriate targets and blind spots of monoclonal reagents for desired targets. This raises the practical concern that numerous studies characterizing IgG subtypes in human disease may contain errors due to such previously unappreciated defects. These studies also focus attention on the broader concern that genetic variation may affect the performance of any laboratory or research test that uses antibodies for detection.
Keywords: Genetics, Immunology
Natural genetic variation in human immunoglobulin subclass alters sensitivity and specificity of commonly used detection reagents, affecting multiple studies in wide ranging fields.
Introduction
Antibodies represent an essentially ubiquitous tool in detection and characterization of gene product expression. In the past decade, concern about the “reproducibility crisis” has rapidly grown, and intrinsic inaccuracy in antibody reagents has recently been established to be a central part of the crisis, present at a higher level than previously appreciated. Prior to the advent of monoclonal antibody technologies, polyclonal antisera was the only option. Because individual animals will differ in their immune response and their background immunity, polyclonal antisera is an intrinsically variable reagent. Even within a given animal, polyclonal antisera will evolve over time, with changing titers, isotype and subtype composition, and evolution in affinities and specificity. The advent of monoclonal antibodies has allowed a stable source of consistent reagents. While focusing on a single recognized epitope typically increases specificity, due to removal of undesired antibodies against different targets, limiting reactivity to one target also decreases sensitivity. In addition, monoclonal antibodies can still have cross-reactivity due to their recognized epitope (or a chemically similar entity) being present on target molecules other than the intended entity. Poorly characterized antibody reagents (both monoclonal and polyclonal) — leading to failure of antibodies to recognize their ascribed targets and cross-reactivity with unanticipated targets — has been identified as a major problem in the generation of misinterpreted data (1). Indeed, Bradbury and Plückthun (along with 110 cosignatories) have called for a specific standardization of antibody reagents, in an effort to quantify (if not remedy) the problem (2).
Herein, we describe an additional problem, which is not included in the concerns already brought forth. In particular, natural genetic variation of target molecules themselves leads to a different performance of antibody reagents based upon genetic origin of the individual(s) from whom samples being analyzed are obtained. As a specific example of this problem, the performance of secondary antibodies to human Ig (e.g., anti-IgG) was investigated. Anti-IgG (commonly called Coombs reagent or anti–human-globulin [AHG]) is broadly used as the detection component in a wide variety of clinical and research assays to characterize human antibodies of a certain specificities. However, human IgG is not a single entity; rather, it is well known that there are 4 different subclasses of human IgG (IgG1–IgG4), each with different characteristics and specifics of effector function (3). As such, the relative quantitation of different IgG subtypes in a given serum has been a widely studied biology in multiple immune settings and plays an important role in diagnosis and research of human disease, including infectious disease serology (4–8), responses to vaccination, autoantibodies (9–12), alloimmunization to transfusion and transplantation antigens (13, 14), and maternal alloantibodies against fetal and paternal antigens (15–17). In addition, IgG subclass deficiency is among the more common immunodeficiencies in pediatric populations, resulting in increased rates of particular infections (18, 19).
Although the 4 different forms of IgG are widely known, less well appreciated is that, in addition to the canonical IgG1–4 subtypes, characterizations of allotypes and recent advances in human genomics have demonstrated at least 29 variants of IgG-constant regions in humans (hereafter called isoallotypes) (20), with 7, 4, 15, and 3 isoallotypes described for IgG1, IgG2, IgG3, and IgG4, respectively (20). Isoallotypes are not disease variants; they represent normal variation, now understood to exist throughout the genome of outbred members of a given species. Whereas each of the IgG subtype–specific AHG reagents are validated using canonical variants of human IgG1–IgG4, little is known as to how anti-IgG reagents react with the 29 different known IgG isoallotypes. Herein, we describe that commonly used monoclonal IgG subclass–specific reagents have problems of blind spots to some IgG isoallotypes within their subclass specificity. We also describe that polyclonal IgG subclass–specific reagents cross-react with IgG isoallotypes of different subtypes. Together, these findings demonstrate a previously unreported problem with the use of antibodies in detection of specific target molecules, both in clinical diagnostics and in research. In particular, the performance of even highly well-characterized reagents will vary as a function of genetic differences in the samples being analyzed. The presented case of IgG subtype determination raises a specific problem with regards to IgG serology, as well as the more general problem as it extends to antibody use in an outbred population in which natural variation in gene product sequence will inevitably exist.
Results
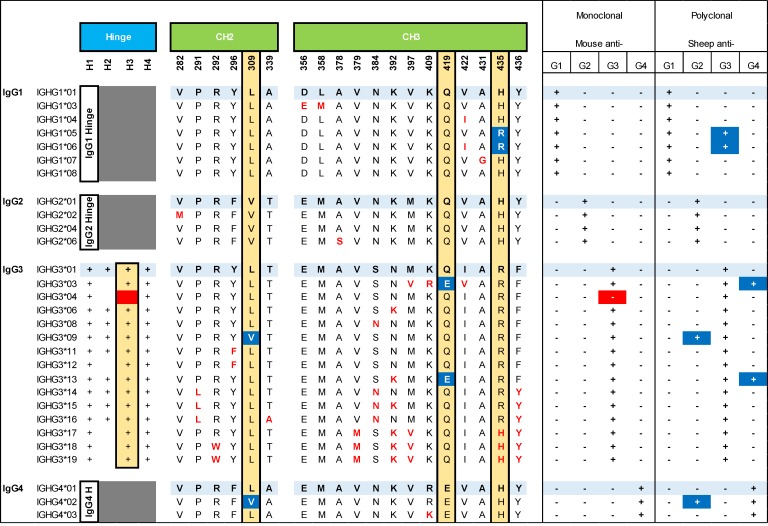
The heavy chain variable region of a monoclonal antibody against the human K1 RBC alloantigen was sequenced and cloned into expression vectors for IgG heavy chain switch variants of each of the known 29 IgG isoallotypes (21). The light chain variable region was cloned into a κ light chain vector and cotransfected with each heavy chain variant into CHO cells. In this way, each of the 29 known human IgG isoallotypes were isolated as an independent variable based upon 29 switch variants with identical antigen binding domains (21). A variety of available and commonly utilized IgG subclass–specific reagents (both monoclonal and polyclonal) were assessed with our panel of IgG variants, by flow cytometry using K1+ RBCs as a target. A synopsis of their reactivity is presented in Figure 1 and Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.94532DS1).
Figure 1. Human IgG isoallotype genetic variation and reactivities to anti-IgG subclass reagents.
Canonical sequences for each IgG subclass are shown in light blue, with all known intrasubclass variants within the hinge, CH2, and CH3 region shown below each canonical sequence. A summary of the reactivity with either monoclonal or polyclonal anti-IgG subclass–specific antibody is shown to the right, with blind spots (and their corrections) shown in red and cross-reactive antibodies (and their corrections) shown in blue. For clarity, variant amino acids that differ between IgG subclasses but that do not vary within a subclass (i.e., isoallotype) are not shown. For a more comprehensive view of all known amino acid differences between the 29 isoallotypes, please see ref. 21.
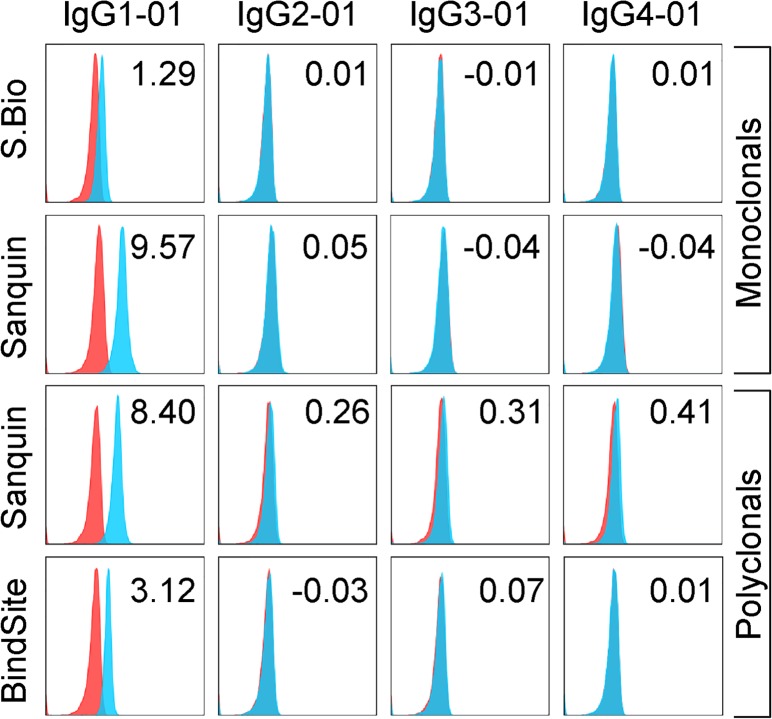
For anti-IgG1, each monoclonal and polyclonal preparation tested was appropriately reactive with, and specific for, all IgG1 isoallotypes currently known (Supplemental Table 1, Figure 1, and Figure 2). While only canonical subtype data is shown, no cross-reactivity was observed with any other isoallotype. Importantly, while there were no aberrant reactivities, there were distinct differences in sensitivity to the target antigen, as the monoclonal antibody from Southern Biotech bound much more weakly to all of the IgG1 subtypes than any of the other antibodies tested (Supplemental Table 1, ± symbol; Figure 2).
Figure 2. Reactivity of anti-IgG1 monoclonal and polyclonal reagents with canonical IgG isoallotypes.
Human RBCs with a phenotype of K+k+ (blue histograms) or K–k+ (red histograms; negative control) were stained with PUMA1 (specific for K+k+ cells) of the indicated human IgG subclass (labeled columns) and with the human IgG-specific reagents (labeled rows). Data shown are MFIs representative of 3 replicate experiments; stain indices are shown (upper right corner) for each histogram as indicator of staining intensity above background (see Methods). While only canonical isoallotypes are shown, all 29 isoallotypes were tested in a similar manner, and data are presented in Figure 1. S.Bio, Southern Biotech; BindSite, The Binding Site Group; Ortho, Ortho Clinical Diagnostics (see Methods for antibody details)
Antibodies specific for IgG2, IgG3, and IgG4 did not behave entirely as expected; each polyclonal preparation had unanticipated cross-reactivities with antibodies of the incorrect subclass, while certain monoclonal preparations did not react with an antibody from its cognate subclass, leading to a false negative. Details for each of these subclass-specific antibodies are discussed below.
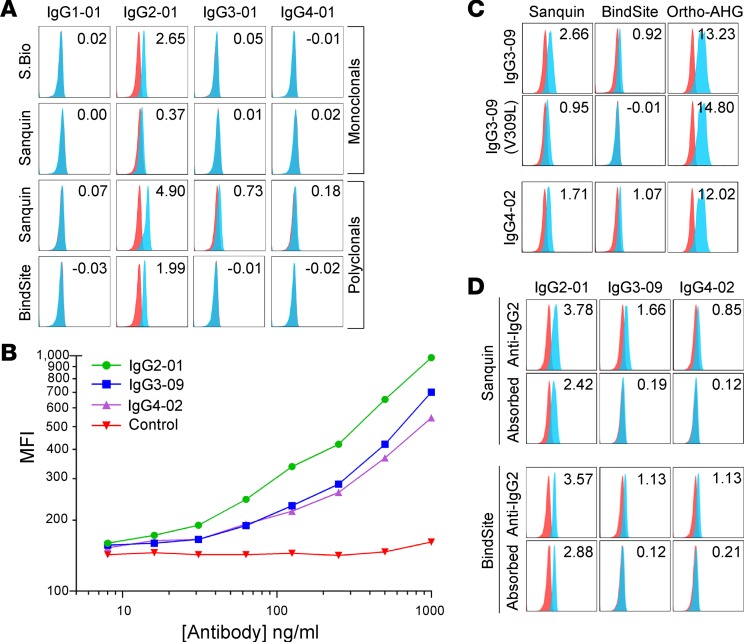
Each monoclonal and polyclonal anti-IgG2 preparation was appropriately reactive with and specific for IgG2 when looking at canonical IgG subtypes (Figure 3A). As seen with anti-IgG1, one of the monoclonal preparations had much weaker activity than the other antibodies examined (Supplemental Table 1, ± symbol; Figure 3A), even with concentrations as high as 100 μg/ml (data not shown). While the monoclonal anti-IgG2 antibodies maintained specificity when tested against all 29 isoallotypes, 2 different polyclonal preparations cross-reacted with antibodies of the incorrect isoallotype: IgG3-09 and IgG4-02 (Figure 3, B and C). Importantly, the cross-reactivities of IgG3-09 and IgG4-02 are weaker than the reaction with the correct IgG subclass. To test the possibility that lower concentrations of antisera would achieve IgG subtype specificity and eliminate cross-reactivity, without losing sensitivity, detailed titrations were carried out (Figure 3B). While the cross-reactive signal did decrease with titration, the correct reactivity also decreased in parallel, such that lack of cross-reactivity resulted in substantial decrease in sensitivity.
Figure 3. Reactivity of anti-IgG2 monoclonal and polyclonal reagents with the 29 IgG isoallotypes.
(A) Human RBCs with a phenotype of K+k+ (blue histograms) or K–k+ (red histograms; negative control) were stained with PUMA1 (specific for K+k+ cells) of the indicated human IgG subclass (labeled columns) and with the human IgG-specific reagents (labeled rows). While only canonical isoallotypes are shown, all 29 isoallotypes were tested in a similar manner, and data are presented in Figure 1. (B) Titration of the Sanquin anti-IgG2 polyclonal reagent with staining of canonical IgG2-01, as well as cross-reactive isoallotypes IgG3-09 and IgG4-02. (C) Cross-reactivity of IgG3-09 and IgG4-02 with polyclonal anti-IgG2 reagents is eliminated by mutating valine at amino acid position 309 in the CH2 region to leucine. Binding of the pan-IgG detection reagent Ortho-AHG is shown for each variant to indicate that loss of staining is not due to loss of antibody expression. (D) Adsorption of cross-reactive antibodies using IgG3-09 and IgG4-02 backbones fused to a variable domain that binds an epitope (HLA) not expressed on RBCs results in elimination of cross-reactive detection, while maintaining binding to canonical IgG2-01. Data shown are MFIs representative of 3 replicate experiments; stain indices are shown (upper right corner) for each histogram as indicators of staining intensity above background (see Methods). S.Bio, Southern Biotech; BindSite, The Binding Site Group; Ortho, Ortho Clinical Diagnostics (see Methods for antibody details)
All known IgG2 isoallotypes have a valine at amino acid position 309 of the constant heavy chain, and IgG3-09 and IgG4-02 are the only non-IgG2 isoallotypes that also have a valine at this position (all others have a leucine at position 309; Figure 1). To test the hypothesis that the V309L change was responsible for cross-reactivity, valine was mutated to leucine in IgG3-09, which either substantially reduced (monoclonal antibody from Sanquin) or completely eliminated (monoclonal antibody from The Binding Site) reactivity with anti-IgG2 (Figure 3C). This was not due to a lack of antibody expression or binding, as binding of the V309L IgG3-09 mutant still was detected by panreactive anti-IgG (Figure 3C, Ortho-AHG, right panel). Data for the V309L mutant of IgG4-02 is not shown here, as this sequence is the same as that represented by the IgG4-01 isoallotype, which did not cross-react. Based upon this epitope specificity of the polyclonal anti-IgG2 preparations for V309, we predicted that the specificity of polyclonal antisera could be improved by adsorbing the cross-reactive IgG variants. We cloned an antibody against an HLA epitope not expressed on RBCs and expressed it in IgG3-09 and IgG4-02 forms. Preincubating the anti-IgG2 detection reagent with these IgG3-09 and IgG4-02 antibodies, which are nonreactive in the RBC assay being used, eliminated the cross-reactivity without substantially decreasing signal intensity for detection of IgG2-01 (Figure 3D).
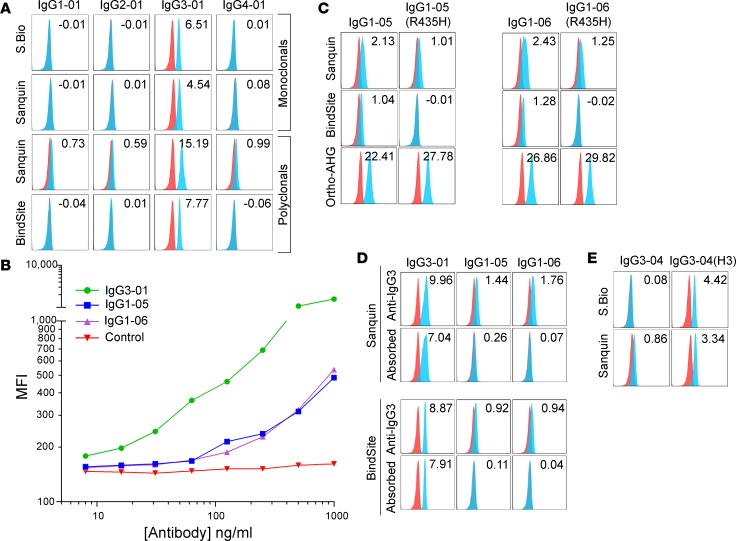
Similar to anti-IgG2, each monoclonal and polyclonal anti-IgG3 preparation was appropriately reactive with, and specific for, IgG3 when looking at canonical subtypes (Figure 4A). The Sanquin polyclonal antibody had a slight background signal for most of the 29 isoallotypes (as indicated by a slight peak shift and elevated stain index seen in Figure 4A); the polyclonal antibody from The Binding Site did not have this same issue with background. Regardless of the presence or absence of this background signal, both polyclonal anti-IgG3 preparations demonstrated inappropriate cross-reactivity to 2 isoallotypes of the IgG1 subclass: IgG1-05 and IgG1-06 (Figure 4, B and C). Similar to the cross-reactivity seen for anti-IgG2, antibody titration reduced not only binding to IgG1-05 and IgG1-06, but also reduced sensitivity for binding of the correct subtype IgG3-01 (Figure 4B).
Figure 4. Reactivity of anti-IgG3 monoclonal and polyclonal reagents with the 29 IgG isoallotypes.
(A) Human RBCs with a phenotype of K+k+ (blue histograms) or K–k+ (red histograms; negative control) were stained with PUMA1 (specific for K+k+ cells) of the indicated human IgG subclass (labeled columns) and with the human IgG-specific reagents (labeled rows). While only canonical isoallotypes are shown, all 29 isoallotypes were tested in a similar manner, and data are presented in Figure 1. (B) Titration of the Sanquin anti-IgG3 polyclonal reagent with staining of canonical IgG3-01, as well as cross-reactive isoallotypes IgG1-05 and IgG1-06. (C) Cross-reactivity of IgG1-05 and IgG1-06 with polyclonal anti-IgG3 reagents is eliminated by mutating argninine at amino acid position 435 in the CH3 region to histidine. Binding of the pan-IgG detection reagent Ortho-AHG is shown for each variant to indicate that loss of staining is not due to loss of antibody expression. (D) Adsorption of cross-reactive antibodies using IgG1-05 and IgG1-06 backbones fused to a variable domain that binds an epitope (HLA) not expressed on RBCs results in elimination of cross-reactive detection, while maintaining binding to canonical IgG3-01. (E) Monoclonal anti-IgG3 reagents do not detect the IgG3-04 isoallotype. This blind-spot in detection can be rescued by adding back one hinge repeat, indicating that this antibody is specific for the long IgG3 hinge. Data shown are MFIs representative of 3 replicate experiments; stain indices are shown (upper right corner) for each histogram as indicators of staining intensity above background (see Methods). S.Bio, Southern Biotech; BindSite, The Binding Site Group; Ortho, Ortho Clinical Diagnostics (see Methods for antibody details)
Sequence alignment of all 29 isoallotypes demonstrated that, similar to most of the IgG3 isoallotypes known, both IgG1-05 and IgG1-06 have an arginine instead of histidine at amino acid position 435 (Figure 1). An R435H mutation in IgG1-05 or IgG1-06 either substantially reduced (Sanquin monoclonal antibody) or completely eliminated (The Binding Site monoclonal antibody) reactivity with anti-IgG3 (Figure 4C). This was not due to a lack of antibody expression or binding, as binding of both the IgG1-05 and IgG1-06 R435H mutants was detected by panreactive anti-IgG (Figure 4C, Ortho-AHG, bottom panel). It should be noted that IgG3-17, IgG3-18, and IgG3-19 also each have a histidine at position 435; however, they also have multiple other differences from the IgG1 isoallotypes, some of which presumably compensate for the change to histidine at position 435. Based upon this epitope specificity of the polyclonal anti-IgG3 preparations for R435, we predicted that the specificity of polyclonal antisera could be improved by adsorbing the cross-reactive IgG variants. We cloned an antibody against an HLA epitope not expressed on RBCs and expressed it in IgG1-05 and IgG1-06 forms. Preincubating the anti-IgG3 detection reagent with these IgG1-05 and IgG1-06 antibodies, which are nonreactive in the RBC assay being used, eliminated the cross-reactivity without substantially decreasing signal intensity for detection of IgG3-01 (Figure 4D).
Unlike all of the other monoclonal antibodies tested, 2 separate monoclonal anti-IgG3 reagents both had a selective blind spot; neither reacted with IgG3-04 above background levels (Figure 4E). IgG3 is unique from the other antibody subtypes in that it has a hinge region of variable length repeats (3). IgG3-04 is the only IgG variant with fewer than 3 hinge region repeats (Figure 1). To test the hypothesis that this is the basis of the nonreactivity blind spot, a mutated IgG3-04 form was created in which one hinge repeat was added back (giving 3 hinge repeats total); the rest of the sequence was unaltered. The mutated IgG3-04(H3) gained reactivity with both monoclonal antibodies tested (Figure 4E), indicating that the failure of the anti-IgG3 to recognize authentic IgG3-04 was due to an insufficient hinge number, which suggests that the antibody recognizes a hinge-dependent epitope.
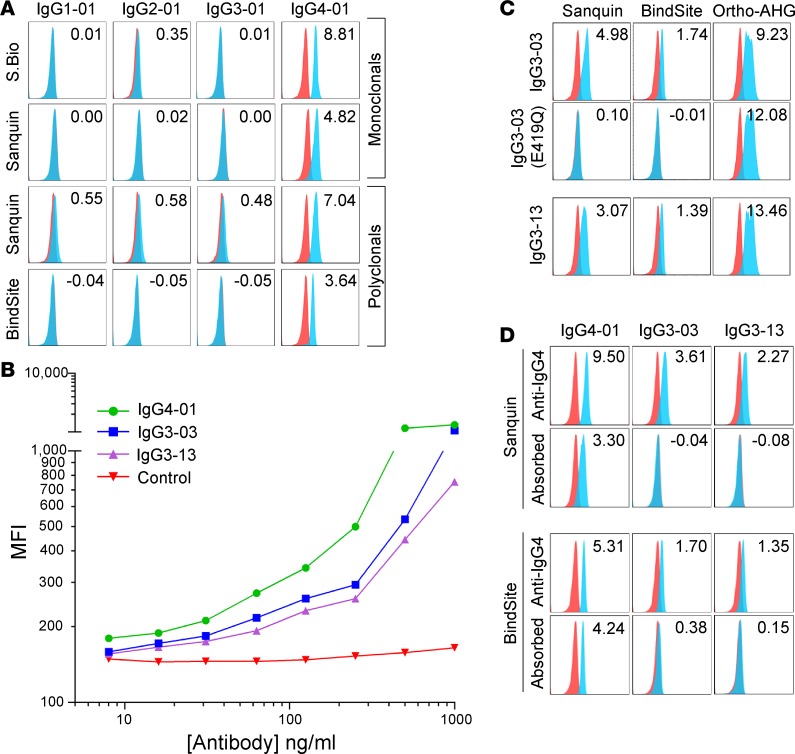
Each monoclonal and polyclonal anti-IgG4 preparation was appropriately reactive with, and specific for, canonical IgG subtypes, with the Sanquin polyclonal antibody once again showing low levels of background staining (Figure 5A). Two separate polyclonal anti-IgG4 reagents cross-reacted with IgG3-03 and IgG3-13 (Figure 5, B and C), which have a glutamic acid instead of glutamine at amino acid position 419 (Figure 1). Similar to the cross-reactivity seen for the other polyclonal antibodies examined, antibody titration reduced not only binding to IgG3-03 and IgG3-13, but also sensitivity for binding of the correct subtype IgG4-01 (Figure 5B). An E419Q mutation in IgG3-03 eliminated binding (Figure 5C). This was not due to a lack of antibody expression or binding, as binding of the IgG3-03 E419Q mutant was detected by panreactive anti-IgG (Figure 5C, Ortho AHG, right panels). No E419Q mutant of IgG3-13 was generated, as this sequence is already represented as IgG3-06. Based upon this epitope specificity of the polyclonal anti-IgG4 preparations for E419, we predicted that the specificity of polyclonal antisera could be improved by adsorbing the cross-reactive IgG variants. We cloned an antibody against an HLA epitope not expressed on RBCs and expressed it in IgG3-03 and IgG3-13 forms. Preincubating the anti-IgG4 detection reagent with these IgG3-03 and IgG3-13 antibodies, which are nonreactive in the RBC assay being used, eliminated the cross-reactivity without substantially decreasing signal intensity for detection of IgG4-01 (Figure 5D).
Figure 5. Reactivity of anti-IgG4 monoclonal and polyclonal reagents with the 29 IgG isoallotypes.
(A) Human RBCs with a phenotype of K+k+ (blue histograms) or K–k+ (red histograms; negative control) were stained with PUMA1 (specific for K+k+ cells) of the indicated human IgG subclass (labeled columns) and with the human IgG-specific reagents (labeled rows). While only canonical isoallotypes are shown, all 29 isoallotypes were tested in a similar manner, and data are presented in Figure 1. (B) Titration of the Sanquin anti-IgG4 polyclonal reagent with staining of canonical IgG4-01, as well as cross-reactive isoallotypes IgG3-03 and IgG3-13. (C) Cross-reactivity of IgG3-03 and IgG3-13 with polyclonal anti-IgG4 reagents is eliminated by mutating glutamic acid at amino acid position 419 in the CH3 region to glutamine. Binding of the pan-IgG detection reagent Ortho-AHG is shown for each variant to indicate that loss of staining is not due to loss of antibody expression. (D) Adsorption of cross-reactive antibodies using IgG3-03 and IgG3-13 backbones fused to a variable domain that binds an epitope (HLA) not expressed on RBCs results in elimination of cross-reactive detection while maintaining binding to canonical IgG4-01. Data shown are MFIs representative of 3 replicate experiments; stain indices are shown (upper right corner) for each histogram as an indicator of staining intensity above background (see Methods). S.Bio, Southern Biotech; BindSite, The Binding Site Group; Ortho, Ortho Clinical Diagnostics (see Methods for antibody details)
Discussion
Herein, we explore the details of a problem regarding the accuracy of diagnostic tests using antibody reagents for detection — in particular, that genetic variation of the person being tested can alter the binding specifics of the antibody. As a particular case, we demonstrate a widespread problem with available AHG reagents that have previously been considered specific for IgG subclass. This problem will not be evident by existing quality control data on reagents that have been validated against common genetic variants. Indeed, with regards to the classic canonical IgG forms, each of the tested reagents performed as predicted. However, when the naturally occurring human variants of IgG were tested, the reagents broke down at multiple levels. The cross-reactivity of polyclonal reagents was slightly weaker than the signal obtained on the intended targets (although anti-IgG2 reacted as strongly with IgG3-09 as authentic IgG2). However, the cross-reactivity was still substantial in all cases and would likely run the risk of detecting the incorrect IgG subtype in the serum of a patient or subject. Titration of the polyclonal antisera down to lower levels did not result in a condition where the intended target was detected while losing cross-reactivity, indicating that assay optimization is not a remedy (Figure 3B, Figure 4B, and Figure 5B). However, generation of refined polyclonal antibodies, either through modifying immunogens or by including the correct IgG variants in adsorptions, should remedy the problem, as indicated in our competition studies (Figure 3D, Figure 4D, and Figure 5D). In contrast, the problem with the monoclonal anti-IgG3 necessitates a new monoclonal antibody, as the blind spot observed in monoclonal anti-IgG3 is essentially complete, with no detectable binding. As an alternative, a blend of monoclonal antibodies may be of benefit, as it could produce a highly standardized virtual polyclonal reagent.
It is worth pointing out that all 4 known human IgG subtypes have a single known N-glycosylation site in their heavy chain constant region, at asparagine 297. However, none of the amino acid variants introduced in any of the isoallotypes expressed herein changed this Asparagine. Although it is possible that an allosteric change from a distant amino acid may alter glycosylation, it is unlikely, given that we have used mutagenesis to restore the binding of those natural variants that did not bind. In addition, CHO antibody–producing lines were not isolated; rather, bulk transient transfection was carried out for all antibody production, thus removing the possibility of isolating CHO lines with aberrant glycosylation during cloning. Therefore, while we cannot unequivocally rule out changes in glycosylation having an effect, it is not considered likely, given the above considerations. Future studies may explore the role of glycosylation (in particular) and posttranslational modification (in general).
The problem explored in this report is not restricted to IgG subtype–specific reagents; rather, we have recently reported a different blind spot in a common monoclonal AHG reagent, widely used in testing millions of patients for alloantibodies against RBC antigens to avoid incompatible transfusion (21). In addition, the issue identified herein will undoubtedly become more complicated as human genomics identifies additional IgG variants. Remedying the existing problem, paying careful attention to new IgG variants that may be problematic, and considering potential misinterpretation of the multitude of studies that analyze IgG subtype using the same (or similar) IgG subclass detecting reagents seem like appropriate responses.
In summary, these studies highlight what may be a broader concern about the role of antibody reagents in the reproducibility crisis. As advocated by many, it is fair to “blame it on the antibodies.” However, the blame is not only a case of reagents being poorly characterized and/or standardized against the known targets, as has been argued. Rather, as an additional problem, the targets themselves vary among different people. Variation among humans is considerable and is by no means restricted to IgG, as was studied in the current report. To the extent that all human antigens (of a protein nature) will vary among populations, both as a result of normal genetic variation and due to pathological genetic mutations, the prescribed remedy of better validating well-defined antibodies against defined antigens will not suffice as proposed, as a sole remedy (2, 22). Instead, as the defining antigens will not represent the panoply of variants likely to found across populations for essentially all gene products, validation must extend to consideration of variants of the target. This validation will be an ongoing process, with a need of frequent updating, as we gain better and better resolution of the variation among human genomes. Although the problem of genetic variation has been known for some time, the depth and breadth of the problem has likely not been fully appreciated, as is evidenced by the exploration of IgG subclass in this manuscript. This is the essence of personalized and precision medicine, which extends not only to personalizing treatment, but to general laboratory diagnosis, as well.
Methods
Generation and purification of antibody reagents.
The generation of the anti-K1 antibody and the 29 variants of human isoallotypes was performed as previously described (21). Mutations of IgG subtypes were carried out by in silico synthesis of the mutated region of each indicated heavy chain constant region, followed by standard restriction-based cloning methods to ligate to the K1 variable region (GeneWiz). All antibodies were synthesized by cotransfection of CHO cells with plasmids encoding heavy and light chains, as described (21).
Mice.
K transgenic mice (published as KEL1 mice) were generated and characterized as previously described (23) and were bred in the bloodworks vivarium. C57BL/6J (B6) mice were purchased from The Jackson Laboratory. All mice were maintained on standard rodent chow and water in a temperature-controlled and light-controlled environment. All murine studies were conducted under the approved BloodworksNW Research Institute IACUC protocol 105-01.
Isolation of anti-HLA antibody specific for HLA-A, -B, and -C.
Murine myeloma cell line HB-95 (W6/32) was purchased from ATCC and cultured according to instructions. RNA was isolated from the antibody-secreting hybridoma and was converted to 5′ RACE-ready cDNA using the SMARTer RACE 5′/3′ Kit (Clontech). Amplification of the heavy chain variable region was performed using primer CH1 (5′-GGCCAGTGGATAGACAGATGG-3′), while amplification of the light chain variable region was performed using primer Lk (5′-ACACTCATTCCTGTTGAAGCTCTT-3′) (primer sequences published previously; ref. 24). PCR products of the expected size (roughly 380 bp for the heavy chain and 650 bp for the light chain) were ligated into pGEM T-easy (Promega), and multiple isolates were sequenced. The predicted light chain variable region was synthesized de novo (GeneWiz) and cloned into pFUSE2-CLIg-hk (InvivoGen). The predicted heavy chain variable region was synthesized de novo and cloned into each of the following vector backbones: pFUSE-CHIg-hG1, pFUSE-CHIg-hG2, pFUSE-CHIg-hG3, and pFUSE-CHIg-hG4 (InvivoGen). Using the IgG1–4 backbones as substrates, derivative plasmids encoding each of the known 29 isoallotypes of IgG were synthesized by a commercial vendor (Genewiz).
Flow cytometry–based analysis of antibody binding.
Flow cytometry consisted of incubating test RBCs with PUMA1 variants, followed by the anti-IgG being evaluated and by a detection reagent. Test RBCs included RBCs from K mice and WT(B6) mice, as well as reagent RBCs from humans with the phenotype of (K+k+) or (K–k+). Test RBCs were resuspended in 50 μl of supernatants of PUMA1 isoallotypes. The samples were incubated at room temperature for 20 minutes and then washed 3 times with PBS.
Anti-IgG subclass antibodies.
Monoclonal mouse anti-IgG subclass antibodies conjugated to phycoerythrin were purchased from SouthernBiotech (catalog numbers 9056-09, 9070-09, 9210-09, and 9200-09) and were used at a dilution of 1/200. Sanquin Antibodies were purchased from Cell Science Inc.. Sanquin mouse monoclonal anti–human IgG1 (M1325), IgG2 (M1326), IgG3 (M1270), and IgG4 (M1271) were used at 1/300 dilution. Sanquin sheep anti–human IgG1, IgG2, IgG3, IgG4 subclass set of 4 (M1175) were used at 1/400 dilution. Sheep anti–human IgG1 (AF006), IgG2 (AF007), IgG3 (AF008), and IgG4 (AF009) — all fluorescein conjugated — were purchased from The Binding Site Group Ltd. and used at a dilution of 1/200. Ortho pan AHG, from Ortho Clinical Diagnostics Inc., was used undiluted. The RBCs were mixed with 50 μl of the indicated subclass antibody diluted in PBS. For the preincubation experiments, the test antibodies were diluted in the anti-HLA isoallotypes indicated in the text for 30 minutes prior to the addition to RBCs. All were incubated for 20 to 30 minutes at room temperature and then washed 3 times in PBS.
Detection reagents.
Goat anti–mouse Igs conjugated to allophycocyanin (550326) were purchased from BD Biosciences at a dilution of 1/100. Rabbit anti–sheep IgG FITC (6158-02) were purchased from SouthernBiotech at a dilution of 1/200. Donkey anti–rabbit IgG phycoerythrin (12-4739-81) were purchased from Affymetrix at a dilution of 1/200.
All antibodies were titrated to define working concentrations. Incubations were performed for 20 to 30 minutes at room temperature, followed by 3 washes with PBS. Flow cytometry was performed on an Accuri 4 color cytometer (BD Bioscience), and data were analyzed using FloJo version 10.
Statistics.
In order to numerically quantify the level of antibody staining above background, changes in antibody staining between different isoallotypes, and changes in staining following mutagenesis, we used the following equation to calculate a stain index for each antibody stain performed (25):
(meanpositive – meanbackground)/(2 × SD background)
This value is shown in the upper right corner of each histogram plot.
Study approval.
There were no human or animal subjects utilized in these studies. All processes and procedures were approved by the Institutional Biosafety Committee at BloodworksNW.
Author contributions
HLH and JCZ designed research studies. XW, LK, and JNL conducted experiments. HLH, XW, LK, JNL, LSE, and MD acquired and analyzed the data. HLH and JCZ wrote the manuscript.
Supplementary Material
Footnotes
Address correspondence to: James C. Zimring, BloodworksNW Research Institute, 1551 Eastlake Avenue E, Seattle, Washington 98102, USA. Phone: 206.568.2234; Email: jzimring@BloodworksNW.org.
Conflict of interest: BloodworksNW has filed intellectual properties around the PUMA1 reagents in this report (26904-30746/US and 26904-34310/US); HLH, XW, LK, JCZ, and MD are each employees of BloodworksNW. JCZ has a sponsored research agreement with Immucor (unrelated to the current work) and is on the scientific advisory board for Rubius Therapeutics.
Reference information: JCI Insight. 2017;2(13):e94532. https://doi.org/10.1172/jci.insight.94532.
Contributor Information
Meghan Delaney, Email: MeghanD@BloodWorksNW.org.
Xiaohong Wang, Email: XiaohongW@BloodWorksNW.org.
Lay See Er, Email: LaySeeE@BloodWorksNW.org.
Linda Kapp, Email: LindaK@BloodWorksNW.org.
References
- 1.Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521(7552):274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 2.Bradbury A, Plückthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015;518(7537):27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 3.Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao RY, et al. Human IgG subclasses against enterovirus Type 71: neutralization versus antibody dependent enhancement of infection. PLoS One. 2013;8(5):e64024. doi: 10.1371/journal.pone.0064024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavacini LA, Kuhrt D, Duval M, Mayer K, Posner MR. Binding and neutralization activity of human IgG1 and IgG3 from serum of HIV-infected individuals. AIDS Res Hum Retroviruses. 2003;19(9):785–792. doi: 10.1089/088922203769232584. [DOI] [PubMed] [Google Scholar]
- 6.Mathiesen T, Persson MA, Sundqvist VA, Wahren B. Neutralization capacity and antibody dependent cell-mediated cytotoxicity of separated IgG subclasses 1, 3 and 4 against herpes simplex virus. Clin Exp Immunol. 1988;72(2):211–215. [PMC free article] [PubMed] [Google Scholar]
- 7.Scharf O, et al. Immunoglobulin G3 from polyclonal human immunodeficiency virus (HIV) immune globulin is more potent than other subclasses in neutralizing HIV type 1. J Virol. 2001;75(14):6558–6565. doi: 10.1128/JVI.75.14.6558-6565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tay MZ, et al. Antibody-Mediated Internalization of Infectious HIV-1 Virions Differs among Antibody Isotypes and Subclasses. PLoS Pathog. 2016;12(8):e1005817. doi: 10.1371/journal.ppat.1005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapuy-Regaud S, Nogueira L, Clavel C, Sebbag M, Vincent C, Serre G. IgG subclass distribution of the rheumatoid arthritis-specific autoantibodies to citrullinated fibrin. Clin Exp Immunol. 2005;139(3):542–550. doi: 10.1111/j.1365-2249.2004.02708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huijbers MG, et al. The expanding field of IgG4-mediated neurological autoimmune disorders. Eur J Neurol. 2015;22(8):1151–1161. doi: 10.1111/ene.12758. [DOI] [PubMed] [Google Scholar]
- 11.Outschoorn IM, et al. Autoantibody heritability in thyroiditis: IgG subclass contributions. Autoimmunity. 2011;44(3):195–200. doi: 10.3109/08916934.2010.515275. [DOI] [PubMed] [Google Scholar]
- 12.Recke A, et al. Pathogenicity of IgG subclass autoantibodies to type VII collagen: induction of dermal-epidermal separation. J Autoimmun. 2010;34(4):435–444. doi: 10.1016/j.jaut.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Michaelsen TE, Kornstad L. IgG subclass distribution of anti-Rh, anti-Kell and anti-Duffy antibodies measured by sensitive haemagglutination assays. Clin Exp Immunol. 1987;67(3):637–645. [PMC free article] [PubMed] [Google Scholar]
- 14.Valenzuela NM, Hickey MJ, Reed EF. Antibody Subclass Repertoire and Graft Outcome Following Solid Organ Transplantation. Front Immunol. 2016;7:433. doi: 10.3389/fimmu.2016.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck LH. Lessons from a rare disease: IgG subclass and disease severity in alloimmune antenatal membranous nephropathy. Kidney Int. 2015;87(3):494–497. doi: 10.1038/ki.2014.367. [DOI] [PubMed] [Google Scholar]
- 16.Einarsdottir H, et al. H435-containing immunoglobulin G3 allotypes are transported efficiently across the human placenta: implications for alloantibody-mediated diseases of the newborn. Transfusion. 2014;54(3):665–671. doi: 10.1111/trf.12334. [DOI] [PubMed] [Google Scholar]
- 17.Einarsdottir HK, et al. On the perplexingly low rate of transport of IgG2 across the human placenta. PLoS ONE. 2014;9(9):e108319. doi: 10.1371/journal.pone.0108319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stiehm RE. The four most common pediatric immunodeficiencies. Adv Exp Med Biol. 2007;601:15–26. doi: 10.1007/978-0-387-72005-0_2. [DOI] [PubMed] [Google Scholar]
- 19. Zhao Y, Hammarstöm L. Selective IgG Subclass Deficiency. In: Lang F, ed. Encyclopedia of Molecular Mechanisms of Disease. Berlin: Springer Berlin Heidelberg; 2009:1913–1914. [Google Scholar]
- 20.Dugoujon JM, Hazout S, Loirat F, Mourrieras B, Crouau-Roy B, Sanchez-Mazas A. GM haplotype diversity of 82 populations over the world suggests a centrifugal model of human migrations. Am J Phys Anthropol. 2004;125(2):175–192. doi: 10.1002/ajpa.10405. [DOI] [PubMed] [Google Scholar]
- 21.Howie HL, et al. Serological blind spots for variants of human IgG3 and IgG4 by a commonly used anti-immunoglobulin reagent. Transfusion. 2016;56(12):2953–2962. doi: 10.1111/trf.13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bordeaux J, et al. Antibody validation. BioTechniques. 2010;48(3):197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibb DR, et al. Type I interferon is necessary and sufficient for inflammation-induced RBC alloimmunization in mice. Journal of Immunology. doi: 10.4049/jimmunol.1700401. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu TH, Yazdanbakhsh K, Oyen R, Smart E, Reid ME. Production and characterization of anti-kell monoclonal antibodies using transfected cells as the immunogen. Br J Haematol. 1999;106(3):817–823. doi: 10.1046/j.1365-2141.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 25.Maecker HT, Frey T, Nomura LE, Trotter J. Selecting fluorochrome conjugates for maximum sensitivity. Cytometry A. 2004;62(2):169–173. doi: 10.1002/cyto.a.20092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.