Abstract
is one of the most widely used statistics in population genetics. Recent mathematical studies have identified constraints that challenge interpretations of as a measure with potential to range from 0 for genetically similar populations to 1 for divergent populations. We generalize results obtained for population pairs to arbitrarily many populations, characterizing the mathematical relationship between the frequency M of the more frequent allele at a polymorphic biallelic marker, and the number of subpopulations K. We show that for fixed K, has a peculiar constraint as a function of M, with a maximum of 1 only if for integers i with For fixed M, as K grows large, the range of becomes the closed or half-open unit interval. For fixed K, however, some always exists at which the upper bound on lies below We use coalescent simulations to show that under weak migration, depends strongly on M when K is small, but not when K is large. Finally, examining data on human genetic variation, we use our results to explain the generally smaller values between pairs of continents relative to global values. We discuss implications for the interpretation and use of
Keywords: allele frequency, FST, genetic differentiation, migration, population structure
GENETIC differentiation, in which individuals from the same subpopulation are more genetically similar than are individuals from different subpopulations, is a central concept in population genetics. It can arise from a large variety of processes, including from aspects of the physical environment such as geographic barriers, variable permeability to migrants, and spatially heterogeneous selection pressures, as well as from biotic phenomena such as assortative mating and self-fertilization. Genetic differentiation among populations is thus a pervasive feature of population-genetic variation.
To measure genetic differentiation, Wright (1951) introduced the fixation index defined as the “correlation between random gametes, drawn from the same subpopulation, relative to the total.” Many definitions of and related statistics have since been proposed (reviewed by Holsinger and Weir 2009). is often defined in terms of a ratio involving mean heterozygosity of a set of subpopulations, and “total heterozygosity” of a population formed by pooling the alleles of the subpopulations, (Nei 1973):
| (1) |
For a polymorphic biallelic marker whose more frequent allele has mean frequency M across K subpopulations, denoting by the frequency of the allele in subpopulation k, and
and related statistics have a wide range of applications. For example, is used as a descriptive statistic whose values are routinely reported in empirical population-genetic studies (Holsinger and Weir 2009). It is considered as a test statistic for spatially divergent selection, either acting on a locus (Lewontin and Krakauer 1973; Bonhomme et al. 2010) or, using comparisons to a corresponding phenotypic statistic on a trait (Leinonen et al. 2013). is also used as a summary statistic for demographic inference, to measure gene flow between subpopulations (Slatkin 1985); or via approximate Bayesian computation, to estimate demographic parameters (Cornuet et al. 2008).
Applications of generally assume that values near 0 indicate that there are almost no genetic differences among subpopulations, and that values near 1 indicate that subpopulations are genetically different (Hartl and Clark 1997; Frankham et al. 2002; Holsinger and Weir 2009). Mathematical studies, however, have challenged the simplicity of this interpretation, commenting that the range of values that can take is considerably restricted by the allele frequency distribution (Table 1). Such studies have highlighted a direct relationship between allele frequencies and constraints on the range of through functions of the allele frequency distribution such as the mean heterozygosity across subpopulations, The maximal has been shown to decrease as a function of both for an infinite (Hedrick 1999) and for a fixed finite number of subpopulations (Long and Kittles 2003; Hedrick 2005). Consequently, if subpopulations differ in their alleles but separately have high heterozygosity, then can be high and can be low; can be near 0 even if subpopulations are completely genetically different in the sense that no allele occurs in more than one subpopulation.
Table 1. Studies describing the mathematical constraints on FST.
| Reference | Number of alleles | Number of subpopulations | Variable in terms of which constraints are reporteda |
|---|---|---|---|
| Hedrick (1999) | unspecified value | ||
| Long and Kittles (2003) | unspecified value | fixed finite value | |
| Rosenberg et al. (2003) | 2 | 2 | δ |
| Hedrick (2005) | unspecified value | fixed finite value | |
| Maruki et al. (2012) | 2 | 2 | M |
| Jakobsson et al. (2013) | unspecified value | 2 | M |
| Edge and Rosenberg (2014) | fixed finite value | 2 | M |
| This article | 2 | fixed finite value | M |
and denote the within-subpopulation and total heterozygosities, respectively. δ denotes the absolute difference in the frequency of a specific allele between two subpopulations, and M denotes the frequency of the most frequent allele.
Instead of heterozygosities or some studies consider homozygosities or
Detailed mathematical results have clarified the relationship between allele frequencies and in the case of subpopulations. Considering a biallelic marker, Maruki et al. (2012) evaluated the constraint on by the frequency M of the most frequent allele: the maximal decreases monotonically from 1 to 0 with increasing M, Jakobsson et al. (2013) extended this result to multiallelic markers with an unspecified number of distinct alleles, showing that the maximal increases from 0 to 1 as a function of M when and decreases from 1 to 0 when in the manner reported by Maruki et al. (2012). Edge and Rosenberg (2014) generalized these results to the case of a fixed finite number of alleles, showing that the maximal differs slightly from the unspecified case when the fixed number of distinct alleles is odd.
In this study, we characterize the relationship between and the frequency M of the most frequent allele, for a biallelic marker and an arbitrary number of subpopulations K. We derive the mathematical upper bound on in terms of M, extending the biallelic two-subpopulation result to arbitrary K. To assist in interpreting the bound, we simulate the joint distribution of and M in the island migration model, describing its properties as a function of the number of subpopulations and the migration rate. The K-population upper bound on as a function of M facilitates an explanation of counterintuitive aspects of global human genetic differentiation. We discuss the importance of the results for applications of more generally.
Mathematical Constraints
Model
Our goal is to derive the range of values can take, the lower and upper bounds on as a function of the frequency M of the most frequent allele for a biallelic marker when the number of subpopulations K is a fixed finite value ≥2. We consider a polymorphic locus with two alleles, A and a, in a setting with K subpopulations contributing equally to the total population. We denote the frequency of allele A in subpopulation k by The frequency of allele a in subpopulation k is Each allele frequency lies in the interval
The mean frequency of allele A across the subpopulations is and the mean frequency of allele a is Without loss of generality, we assume that allele A is the more frequent allele in the total population, so that Because by assumption the locus is polymorphic,
We assume that the allele frequencies M and are parametric allele frequencies of the total population and subpopulations, and not estimated values computed from data.
FST as a function of M
Equation 1 expresses as a ratio involving within-subpopulation heterozygosity, and total heterozygosity, We substitute and in Equation 1 with their respective expressions in terms of allele frequencies:
| (2) |
Simplifying Equation 2 by noting that leads to:
| (3) |
For fixed M, we seek the vectors with and that minimize and maximize
We clarify here that in mathematical analysis of the relationship between and allele frequencies, we adopt an interpretation of as a “statistic” that describes a mathematical function of allele frequencies rather than as a “parameter” that describes coancestry of individuals in a population. Multiple interpretations of exist, giving rise to different expressions for computing it (e.g., Nei 1973; Nei and Chesser 1983; Weir and Cockerham 1984; Holsinger and Weir 2009). Our interest is in disentangling properties of the mathematical function by which true allele frequencies are used to compute from the population-genetic relationships among individuals and sampling phenomena that could be viewed as affecting the computation. As a result, it is natural to follow the statistic interpretation that has been used in earlier scenarios involving bounds in relation to allele frequencies viewed as parameters, rather than as estimates or outcomes of a stochastic process (Table 1), and in which such a disentanglement is possible. We return to this topic in the Discussion.
Lower bound
From Equation 3, for all setting in all subpopulations k yields The Cauchy–Schwarz inequality guarantees that with equality if and only if Hence, or by dividing both sides by to give requires Examining Equation 3, is thus the only vector that yields We can conclude that the lower bound on is equal to 0 irrespective of M, for any value of the number of subpopulations K.
Upper bound
To derive the upper bound on in terms of M, we must maximize in Equation 3, assuming that M and K are constant. Because all terms in Equation 3 depend only on M and K except the positive term in the numerator, maximizing corresponds to maximizing at fixed M and K.
Denote by the greatest integer less than or equal to x, and by the fractional part of x. Using a result from Rosenberg and Jakobsson (2008), Theorem 1 from Appendix A states that the maximum for satisfies
| (4) |
with equality if and only if allele A has frequency 1 in subpopulations, frequency in a single subpopulation, and frequency 0 in all other subpopulations. Substituting Equation 4 into Equation 3, we obtain the upper bound for
| (5) |
The upper bound on in terms of M has a piecewise structure, with changes in shape occurring when is an integer.
For define the interval by for in the case that K is odd and by for all other For has a constant value i. Writing so that for each interval the upper bound on is a smooth function:
| (6) |
where x lies in (or in for odd K and and i lies in
The conditions under which the upper bound is reached illuminate its interpretation. The maximum requires the most frequent allele to have frequency 1 or 0 in all except possibly one subpopulation, so that the locus is polymorphic in at most a single subpopulation. Thus, is maximal when fixation is achieved in as many subpopulations as possible.
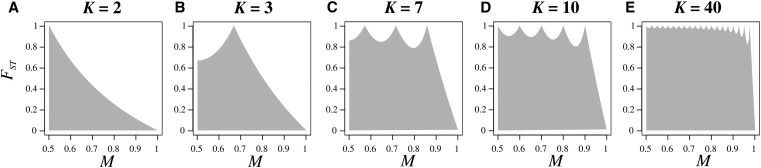
Figure 1 shows the upper bound on in terms of M for various values of K. It has peaks at values where it is possible for the allele to be fixed in all K subpopulations and for to reach a value of 1. Between and the function reaches a local minimum, eventually decreasing to 0 as M approaches 1. The upper bound is not differentiable at the peaks, and it is smooth and strictly <1 between the peaks. If K is even, then the upper bound begins from a local maximum at if K is odd, it begins from a local minimum at
Figure 1.
Bounds on as a function of the frequency of the most frequent allele, M, for different numbers of subpopulations K: (A) (B) (C) (D) and (E) The shaded region represents the space between the upper and lower bounds on The upper bound is computed from Equation 5; for each K, the lower bound is 0 for all values of M.
Properties of the upper bound
Local maxima:
We explore properties of the upper bound on as a function of M for fixed K by examining the local maxima and minima. The upper bound is equal to 1 on interval if and only if the numerator and denominator in Equation 6 are equal. Noting that this condition is equivalent to and hence, because Thus, on interval for M, the maximal is 1 if and only if is an integer.
has exactly integer values for Consequently, given K, there are exactly maxima at which can equal 1, at if K is odd and at if K is even.
This analysis finds that is only unconstrained within the unit interval for a finite set of values of the frequency M of the most frequent allele. The size of this set increases with the number of subpopulations K.
Local minima:
Equality of the upper bound at the right endpoint of each interval and the left endpoint of for each i from to demonstrates that the upper bound on is a continuous function of M. Consequently, local minima necessarily occur between the local maxima. If K is even, then the upper bound on has local minima, each inside an interval If K is odd, then the upper bound has local minima, the first in interval and each of the others in an interval with Note that because we restrict attention to we do not count the point at and as a local minimum.
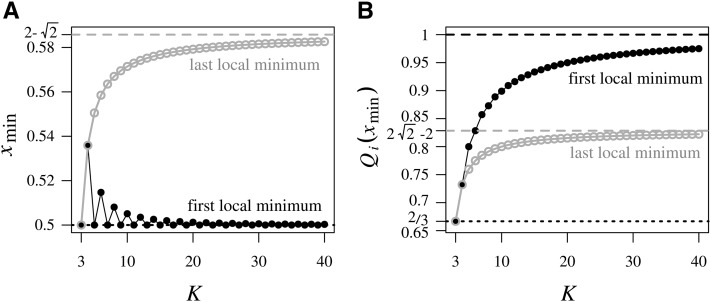
Theorem 2 from Appendix B describes the relative positions of the local minima within intervals as a function of the number of subpopulations K. From Proposition 1 of Appendix B, for fixed K, the relative position of the local minimum within interval increases with i; as a result, the leftmost dips in the upper bound (those near are less tilted toward the right endpoints of their associated intervals than are the subsequent dips (nearer The unique local minimum in interval lies either exactly at for the leftmost dip for odd K (Proposition 2), or slightly to the right of the midpoint of interval in other intervals, but no farther from the center than (Proposition 3; Figure B1).
The values of the upper bound on at the local minima as a function of i are computed in Appendix B (Equation B5) by substituting the positions M of the local minima into Equation 5. From Proposition 4 of Appendix B, for fixed K, the value of at the local minimum in interval decreases as i increases. The maximal among local minima increases as K increases (Proposition 5). The upper bound on at the local minimum closest to tends to 1 as (Proposition 5). The upper bound on at the local minimum closest to however, is always smaller than (Proposition 6).
In conclusion, although is constrained below 1 for all values of M in the interior of intervals the constraint is reduced as and in the limit it even completely disappears in the interval closest to Nevertheless, there always exists a value of for which the upper bound on is lower than
Mean range of possible FST values:
We now evaluate how strongly M constrains the range of as a function of K. Following similar computations for other settings where is considered in relation to a quantity that constrains it (Jakobsson et al. 2013; Edge and Rosenberg 2014), we compute the mean maximum across all possible M values. This quantity, denoted is the area between the lower and upper bounds on divided by the length of the domain for M, or near 0 indicates a strong constraint.
Because the lower bound on is 0 for all M between and 1, corresponds to the area under the upper bound on divided by or twice the integral of Equation 5 between and 1:
| (7) |
The integral is computed in Appendix C. We obtain
| (8) |
We also obtain an asymptotic approximation in Appendix C, where
| (9) |
Here, represents the Glaisher–Kinkelin constant.
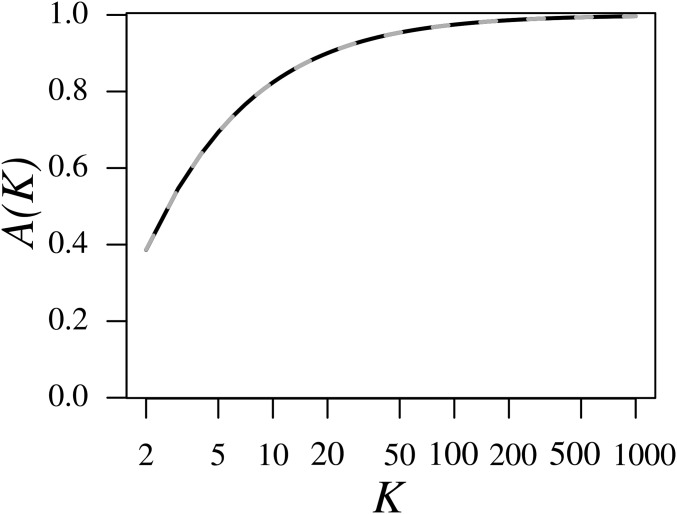
in accord with the case of Jakobsson et al. (2013). Interestingly, the constraint on the mean range of disappears as Indeed, from Equation 9, we immediately see that (Figure 2). As a mean of 1 indicates that ranges from 0 to 1 for all M (except possibly on a set of measure 0), for large K, the range of is approximately invariant with respect to M.
Figure 2.
The mean of the upper bound on over the interval as a function of the number of subpopulations K. is computed from Equation 8 (black line). The approximation is computed from Equation 9 (gray dashed line). A numerical computation of the relative error of the approximation as a function of K, finds that the maximal error for is 0.00174, achieved when The x-axis is plotted on a logarithmic scale.
The increase of with K is monotonic (Theorem 3 of Appendix C). By numerically evaluating Equation 8, we find that although for and for Nevertheless, although the mean of the upper bound on approaches 1, we have shown in Proposition 6 from Appendix B that for large K, values of M continue to exist at which the upper bound is constrained substantially below 1.
Evolutionary Processes and the Joint Distribution of M and for a Biallelic Marker and K Subpopulations
Thus far, we have described the mathematical constraint imposed on by M without respect to the frequency with which particular values of M arise in evolutionary scenarios. As an assessment of the bounds in evolutionary models can illuminate the settings in which they are most salient in population-genetic data analysis (Hedrick 2005; Whitlock 2011; Rousset 2013; Alcala et al. 2014; Wang 2015), we simulated the joint distribution of and M in three migration models, in each case relating the distribution to the mathematical bounds on This analysis considers allele frequency distributions, and hence values of M and generated by evolutionary models.
Simulations
We simulated independent SNPs under the coalescent, using the software MS (Hudson 2002). We considered a population of total size diploid individuals subdivided into K subpopulations of equal size N. At each generation, a proportion m of the individuals in a subpopulation originated from another subpopulation. Thus, the scaled migration rate is and it corresponds to twice the number of individuals in a subpopulation that originate elsewhere. We focus on the finite island model (Maruyama 1970; Wakeley 1998), in which migrants have the same probability to come from any specific other subpopulation. The finite rectangular and linear stepping-stone models generate similar results (Figures S1–S4 in File S1).
We examined three values of K (2, 7, 40) and three values of (0.1, 1, 10). Note that in MS, time is scaled in units of generations, so there is no need to specify the subpopulation sizes N. To obtain independent SNPs, we used the MS command “-s” to fix the number of segregating sites S to 1. For each parameter pair we performed 100,000 replicate simulations, sampling 100 sequences per subpopulation in each replicate. values were computed from the parametric allele frequencies.
Fixing and accepting all coalescent genealogies entails an implicit assumption that all genealogies have equal potential to produce exactly one segregating site. We therefore also considered a different approach to generating SNPs, assuming an infinitely-many-sites model with a specified scaled mutation rate θ and discarding simulations leading to We chose θ so that the expected number of segregating sites in a constant-sized population, or was 1. This approach produces similar results to the fixed-S simulation (Figure S5 in File S1).
Weak migration
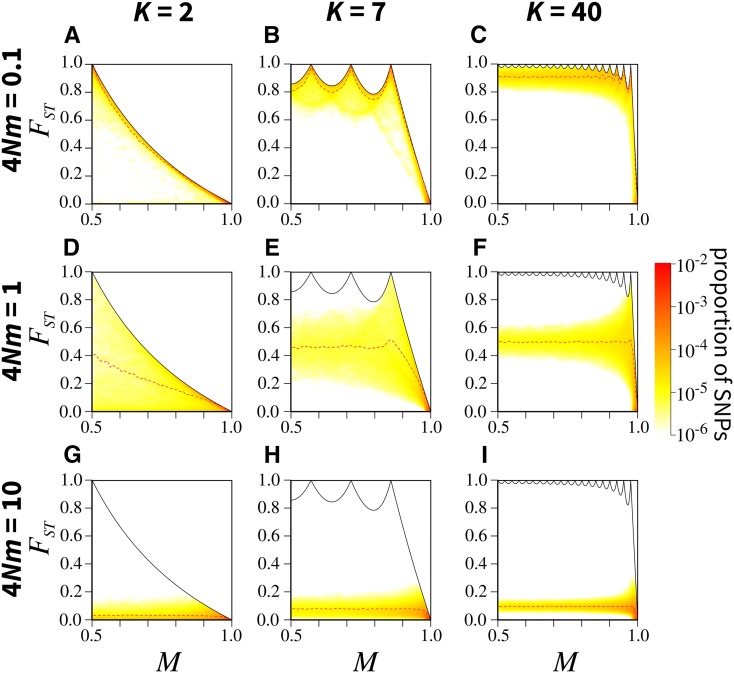
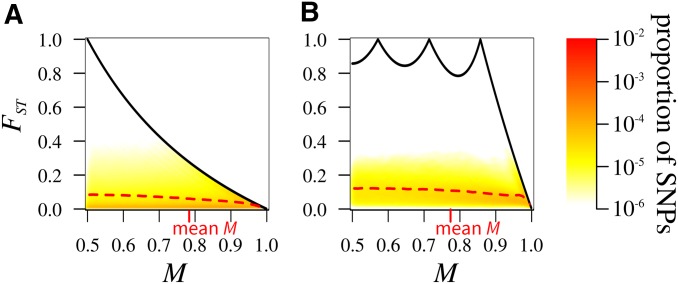
Under the island model with weak migration the joint distribution of M and is highest near the upper bound on in terms of M, for all K (Figure 3, A–C). For most SNPs have M near 0.5, representing fixation of the major allele in one subpopulation and absence in the other, and near 1 (Figure 3A). The mean in sliding windows for M closely follows the upper bound on For most SNPs have M near or representing fixation of the major allele in four, five, or six subpopulations and absence in the others, and (Figure 3B). The mean closely follows the upper bound. For most SNPs either have M near or and or and (Figure 3C). The mean follows the upper bound for For it lies below the upper bound and does not possess its characteristic peaks.
Figure 3.
Joint density of the frequency M of the most frequent allele and in the island migration model, for different numbers of subpopulations K and scaled migration rates (where N is the subpopulation size and m the migration rate): (A) (B) (C) (D) (E) (F) (G) (H) and (I) The black solid line represents the upper bound on in terms of M (Equation 5); the red dashed line represents the mean in sliding windows of M of size 0.02 (plotted from 0.51 to 0.99). Colors represent the density of SNPs, estimated using a Gaussian kernel density estimate with a bandwidth of 0.007, with density set to 0 outside of the bounds. SNPs are simulated using coalescent software MS, assuming an island model of migration and conditioning on one segregating site. See Figure S5 in File S1 for an alternative algorithm for simulating SNPs. Each panel considers 100,000 replicate simulations, with 100 lineages sampled per subpopulation. Figures S2 and S3 in File S1 present similar results under finite rectangular and linear stepping-stone migration models.
We can interpret these patterns using the model of Wakeley (1999), which showed that when migration is infrequent compared to coalescence, coalescence follows two phases. In the scattering phase, lineages coalesce in each subpopulation, leading to a state with a single lineage per subpopulation. In the collecting phase, lineages from different subpopulations coalesce. As a result, considering K subpopulations with equal sample size n, when , genealogies tend to have K long branches close to the root, each corresponding to a subpopulation and each leading to n shorter terminal branches. The long branches coalesce as pairs accumulate by migration in shared ancestral subpopulations. A random mutation on such a genealogy is likely to occur in one of two places. It can occur on a long branch during the collecting phase, in which case the derived allele will have frequency 1 in all subpopulations whose lineages descend from the branch, and 0 in the others. Alternatively, it can occur toward the terminal branches in the scattering phase, in which case the mutation will have frequency in one subpopulation and 0 in all others. These scenarios that are likely under weak migration, one allele fixed in some subpopulations or present only in one subpopulation, correspond closely to conditions under which the upper bound on is reached at fixed M. Thus, the properties of likely genealogies explain the proximity of to its upper bound.
Intermediate and strong migration
With intermediate migration for all K, the joint density of M and is highest at lower values of than with (Figure 3, D–F). For most SNPs have and the mean is almost equidistant from the upper and lower bounds on nearing the upper bound as M increases (Figure 3D). For most SNPs have as was seen for the mean is almost equidistant from the upper and lower bounds, moving toward the upper bound as M increases (Figure 3E). For the pattern is similar, most SNPs having (Figure 3F).
Under intermediate migration, migration is sufficient that more mutations than in the weak-migration case generate polymorphism in multiple subpopulations. A random mutation is likely to occur on a branch that leads to many terminal branches from the same subpopulation, but also to branches from other subpopulations. Thus, the allele is likely to have intermediate frequency in multiple subpopulations. This setting does not generate the conditions under which the upper bound on is reached, so that except at the largest M, intermediate migration leads to values farther from the upper bound than in the weak-migration case. For large M, the rarer allele is likely to be only in one subpopulation, so that is nearer to the upper bound.
With strong migration the joint density of M and nears the lower bound (Figure 3, G–I). For each K, most SNPs have and with the mean increasing somewhat as K increases. Under strong migration, because lineages can migrate between subpopulations quickly, they can also coalesce quickly, irrespective of their subpopulations of origin. As a result, a random mutation is likely to occur on a branch that leads to terminal branches in many subpopulations. The allele is expected to have comparable frequency in all subpopulations, so that is likely to be small. This scenario corresponds to the conditions under which the lower bound on is approached.
Proximity of the joint density of M and FST to the upper bound
To summarize features of the relationship of to the upper bound seen in Figure 3, we can quantify the proximity of the joint density of M and to the bounds on For a set of Z loci, denote by and the values of and M at locus z. The mean for the set, or is
| (10) |
Using Equation 5, a corresponding mean maximum given the observed denoted is
| (11) |
The ratio gives a sense of the proximity of the values to their upper bounds: it ranges from 0, when values at all SNPs equal their lower bounds, to 1, when values at all SNPs equal their upper bounds.
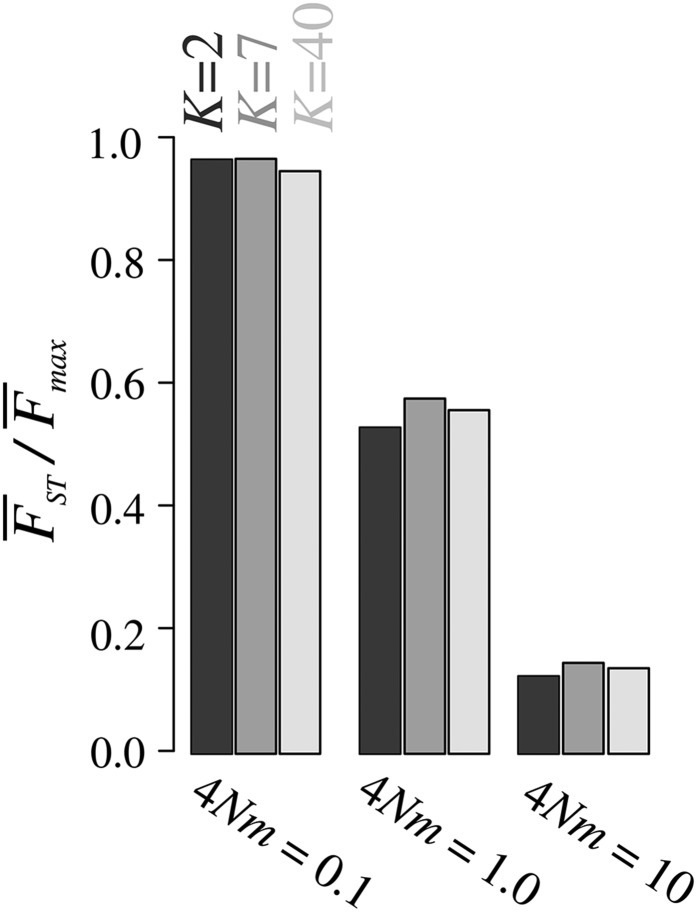
Figure 4 shows the ratio under the island model for different values of K and For each value of the number of subpopulations, decreases with This result summarizes the influence of the migration rate observed in Figure 3: values tend to be close to the upper bound under weak migration, and near the lower bound under strong migration. is only minimally influenced by the number of subpopulations K (Figure 4). Even though the upper bound on in terms of M is strongly affected by K, the proximity of to the upper bound is similar across K values.
Figure 4.
the ratio of the mean to the mean maximal given the observed frequency M of the most frequent allele, as a function of the number of subpopulations K and the scaled migration rate for the island migration model. Colors represent values of K. values are computed from coalescent simulations using MS for 10,000 independent SNPs and 100 lineages sampled per subpopulation. is computed from Equation 11. Figure S4 in File S1 presents similar results under rectangular and linear stepping-stone migration models.
Application to Human Genomic Data
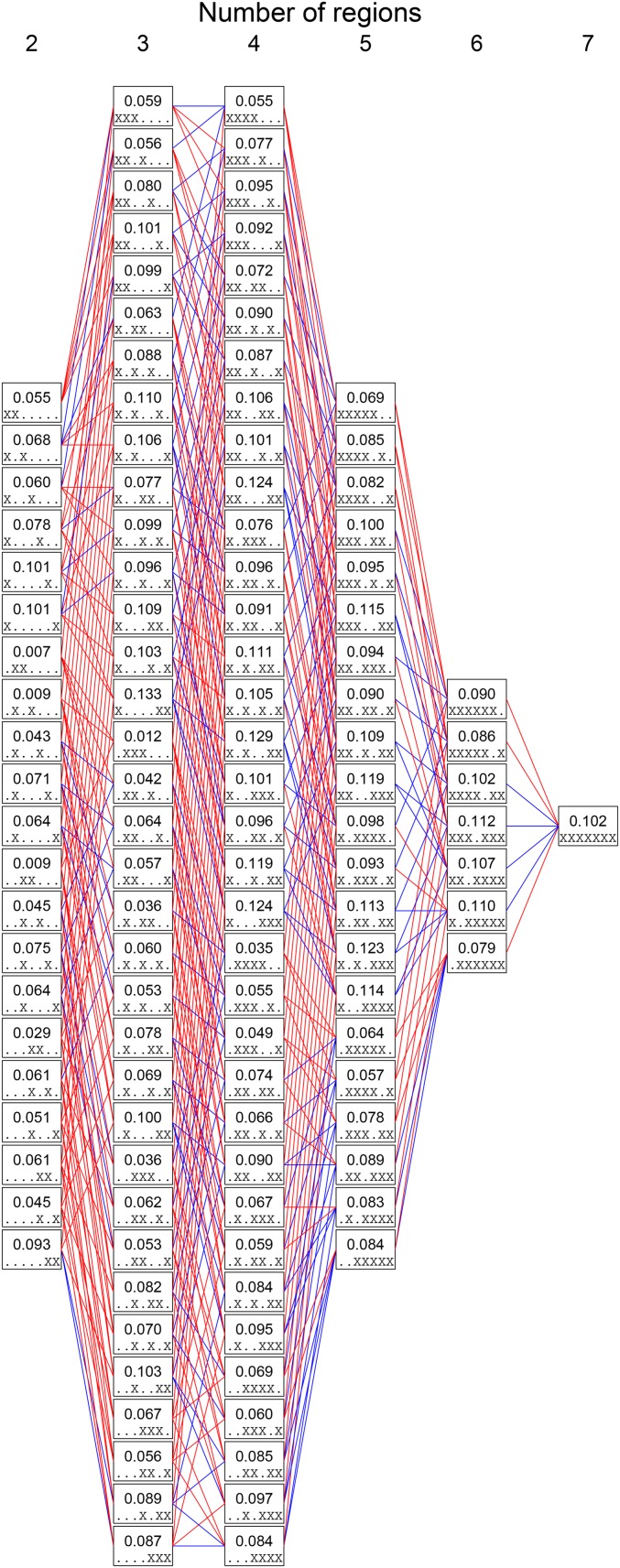
We now use our theoretical results to explain observed patterns of human genetic differentiation, and in particular, to explain the impact of the number of subpopulations. We examine data from Li et al. (2008) on 577,489 SNPs from 938 individuals of the Human Genome Diversity Panel (HGDP) (Cann et al. 2002), as compiled by Pemberton et al. (2012). We use the same division of the individuals into seven geographic regions that was examined by Li et al. (2008) (Africa, Middle East, Europe, Central and South Asia, East Asia, Oceania, and America). Previous studies of these individuals have used to compare differentiation in regions with different numbers of subpopulations sampled (Rosenberg et al. 2002; Ramachandran et al. 2004; Rosenberg 2011).
We computed the parametric allele frequencies for each region, averaging across regions to obtain the frequency M of the most frequent allele. We then computed for each SNP, averaging values across SNPs to obtain the overall for the full SNP set. To assess the impact of the number of subpopulations K on the relationship between M and we computed for all 120 sets of two or more geographic regions (Figure 5). The 21 pairwise values range from 0.007 (between Middle East and Europe) to 0.101 (Africa and America), with a mean of 0.057, SD of 0.027, and median of 0.061. is substantially larger for sets of three geographic regions. The smallest value is larger, 0.012 (Middle East, Europe, Central/South Asia); as is the largest value, 0.133 (Africa, Oceania, America); the mean of 0.076; and the median of 0.089. Among the ways of adding a third region to a pair of regions, 83 produce an increase in For 17 sets of three regions, the value of exceeds that of each of its three component pairs.
Figure 5.
Mean values across loci for sets of geographic regions. Each box represents a particular combination of two, three, four, five, six, or all seven geographic regions. Within a box, the numerical value shown is among the regions. The regions considered are indicated by the pattern of “.” and “X” symbols within the box, with X indicating inclusion and “.” indicating exclusion. From left to right, the regions are Africa, Middle East, Europe, Central/South Asia, East Asia, Oceania, and America. Thus, for example, X...X.. indicates the subset {Africa, East Asia}. Lines are drawn between boxes that represent nested subsets. A line is colored red if the larger subset has a higher value, and blue if it has a lower Computations rely on 577,489 SNPs from the HGDP.
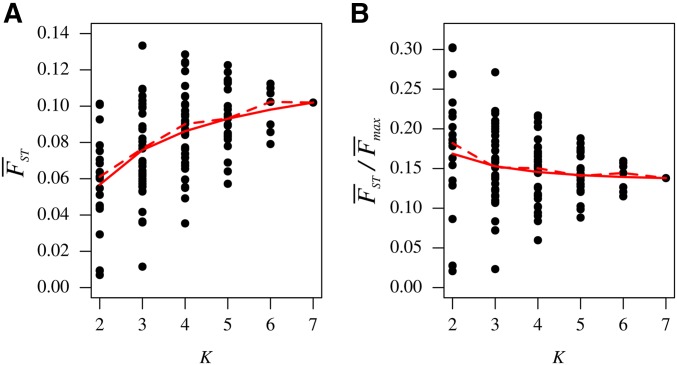
The pattern of increase of with the inclusion of additional subpopulations can be seen in Figure 6A, which plots the values from Figure 5 as a function of K. The magnitude of the increase is greatest from to decreasing with increasing K. From to 4, 82 of 140 additions of a region increase 54 of 105 produce an increase from to 5; 21 of 42 from to 6; and 3 of 7 from to 7. The seven-region of 0.102 exceeds all the pairwise values.
Figure 6.
values for sets of geographic regions as a function of K, the number of regions considered. (A) computed using Equation 10. (B) computed using Equation 11. For each subset of populations, the value of is taken from Figure 5. The mean across subsets for a fixed K appears as a solid red line, and the median as a dashed red line.
The larger values with increasing K can be explained by the difference in constraints on in terms of M (Figure 7). For fixed M, as we saw in the increase of with K (Figure 2), the permissible range of values is smaller on average for values computed among smaller sets of populations than among larger sets. For example, the maximal value at the mean M of 0.76 observed in pairwise comparisons is 0.33 for (black line in Figure 7A), while the maximal value at the mean M of 0.77 observed for the global comparison of seven regions is 0.86 for (Figure 7B). Given the stronger constraint in pairwise calculations, it is not unexpected that pairwise values would be smaller than the values computed with more regions, such as in the seven-region computation. Interestingly, the effect of K on is largely eliminated when values are normalized by their maxima (Figure 6B). The normalization, which takes both K and M into account, generates nearly constant means and medians of as functions of K, with higher values for
Figure 7.
Joint density of the frequency M of the most frequent allele and in human population-genetic data, considering 577,489 SNPs. (A) computed for pairs of geographic regions. The density is evaluated from the set of values for all 21 pairs of regions. (B) computed among geographic regions. The figure design follows Figure 3.
Data availability
MS commands for the coalescent simulations appear in File S2. See Li et al. (2008) for the human SNP data.
Discussion
We have evaluated the constraint imposed by the frequency M of the most frequent allele at a biallelic locus on the range of for arbitrarily many subpopulations. Although is unconstrained in the unit interval when for integers i satisfying it is constrained below 1 for all other M. We have found that the number of subpopulations K has considerable impact on the range of with a weaker constraint on as K increases. As shown by Jakobsson et al. (2013) for across possible values of M, is restricted to 38.63% of the possible space. For however, can occupy 97.47% of the space. Although the mean over M values of the permissible interval for approaches the full unit interval as for any K, an allele frequency exists for which the maximal is lower than
Multiple studies have highlighted the relationship between and M in two subpopulations for biallelic markers (Rosenberg et al. 2003; Maruki et al. 2012) and, more generally, for an unspecified (Jakobsson et al. 2013) or specified number of alleles (Edge and Rosenberg 2014). We have extended these results to the case of biallelic markers in a specified but arbitrary number of subpopulations, comprehensively describing the relationship between and M for the biallelic case. The study is part of an increasing body of work characterizing the mathematical relationship of population-genetic statistics with quantities that constrain them (Hedrick 1999, 2005; Rosenberg and Jakobsson 2008; Reddy and Rosenberg 2012). As we have seen, such relationships contribute to understanding the behavior of the statistics in evolutionary models and to interpreting counterintuitive results in human population genetics.
Properties of FST in evolutionary models
Our work extends classical results about the impact of evolutionary processes on values. Wright (1951) showed that in an equilibrium population, is expected to be near 1 if migration is weak, and near 0 if migration is strong. On the basis of our simulations, we can more precisely formulate this proposition: considering a SNP at frequency M in an equilibrium population, is expected to be near its upper bound in terms of M if migration is weak and near 0 if migration is strong. This formulation of Wright’s proposition makes it possible to explain why SNPs subject to the same migration process can display a variety of patterns; indeed, under weak migration, we expect values to mirror the considerable variation in the upper bound on in terms of M.
Lower FST values in pairwise comparisons than in comparisons of more subpopulations
values have often been compared across computations with different numbers of subpopulations. Such comparisons appear frequently, for example, in studies of domesticated animals such as horses, pigs, and sheep (Cañon et al. 2000; Kim et al. 2005; Lawson Handley et al. 2007). In human populations, table 1 of the microsatellite study of Rosenberg et al. (2002) presents comparisons of values for scenarios with K ranging from 2 to 52. Table 3 of Rosenberg et al. (2006) compares values for microsatellites and biallelic indels in population sets with K ranging from 2 to 18. Major SNP studies have also compared values for scenarios with and groups (Hinds et al. 2005; International HapMap Consortium 2005).
Our results suggest that such comparisons between values with different K can hide an effect of the number of subpopulations, especially when some of the comparisons involve the most strongly constrained case of For human data, we found that owing to a difference in the constraint for different K values, pairwise values between continental regions were consistently lower than values computed using three or more regions, and sets of three regions were identified for which the value exceeded the values for all three pairs of regions in the set. The effect of K might help illuminate why SNP-based pairwise human values (table S11 of 1000 Genomes Project Consortium 2012) are generally smaller than estimates that use all populations together (11.1% of genetic variance due to between-region or between-population differences; Li et al. 2008). We find that comparing values with different choices of K can generate as much difference—twofold—as comparing with different marker types (Holsinger and Weir 2009). This substantial impact of K on merits further attention.
Consequences for the use of FST as a test statistic
The effects of constraints on extend beyond the use of as a statistic for genetic differentiation. In FST-based genome scans for local adaptation, tracing to the work of Lewontin and Krakauer (1973), a hypothesis of spatially divergent selection at a candidate locus is evaluated by comparing at the locus with the distribution estimated from a set of putatively neutral loci. Under this test, values smaller or larger than expected by chance are interpreted as being under stabilizing or divergent selection, respectively. Modern versions of this approach compare values at single loci with the distribution across the genome (Beaumont and Nichols 1996; Akey et al. 2002; Foll and Gaggiotti 2008; Bonhomme et al. 2010; Günther and Coop 2013).
The constraints on in our work and the work of Jakobsson et al. (2013) and Edge and Rosenberg (2014) suggest that values strongly depend on the frequency of the most frequent allele. Consequently, we expect that outlier tests that do not explicitly take into account this constraint will result in a deficit of power at loci with high- and low-frequency alleles. Because pairwise and values in many populations have different constraints, we predict that the effect of the constraint on outlier tests relying on a single global (e.g., Beaumont and Nichols 1996; Foll and Gaggiotti 2008) will be smaller than in tests relying on pairwise (e.g., Günther and Coop 2013).
FST as a statistic or as a parameter
The perspective we used in obtaining bounds treats as a mathematical function of allele frequencies rather than as a population-genetic parameter. Thus, the starting point for our mathematical analysis (Equation 3) is that the allele frequencies are mathematical constants rather than random outcomes of an evolutionary process.
In the alternative perspective that is a parameter rather than a statistic, both the sample of alleles drawn from a set of subpopulations and the sample of subpopulations drawn from a larger collection of subpopulations are treated as random. An analysis of mathematical bounds analogous to our analysis of Equation 3 in terms of M would then investigate bounds on estimators of where the value of the estimator is bounded in terms of the largest sample allele frequency. In this perspective, the estimator of Weir and Cockerham (1984) for a biallelic locus Weir 1996, p. 173), under an assumption of equal sample sizes in the K subpopulations and either haploid data or a random union of gametes in diploids, is
| (12) |
Here, is the sample size per subpopulation (n diploid individuals or haploids), is the mean sample frequency of the most frequent allele across subpopulations, and is the empirical variance of the sample frequency of the most frequent allele across subpopulations.
Although Equation 12 has more terms than Equation 3, it can be shown that for fixed with and fixed and and hence are minimized and maximized under corresponding conditions to those that minimize and maximize Equation 3. In particular, Theorem 1 applies to with We then expect that corresponding mathematical results to those seen for as computed in Equation 3 will hold for from Equation 12. Such computations indicate that our “statistic” perspective on generates mathematical results of interest to a “parameter” interpretation of
Conclusions
Many recent articles have noted that often behaves counterintuitively (Whitlock 2011; Alcala et al. 2014; Wang 2015), for example, indicating low differentiation in cases in which populations do not share any alleles (Balloux et al. 2000; Jost 2008) or suggesting less divergence among populations than is visible in clustering analyses (Tishkoff et al. 2009; Algee-Hewitt et al. 2016). It has thus become clear that observed patterns often trace to peculiar mathematical properties of FST—in particular its relationship to other statistics such as homozygosity or allele frequency—instead of to biological phenomena of interest. Our work here, extending approaches of Jakobsson et al. (2013) and Edge and Rosenberg (2014), seeks to characterize those properties, so that the influence of mathematical constraints on can be disentangled from biological phenomena.
One response to the dependence of on M may be to compute only when allele frequencies lie in a specific class, such as Such choices can potentially avoid a misleading interpretation that a genetic differentiation measure is low in scenarios when minor alleles, though rare, are in fact private to single populations. We note, however, that the dependence of spans the full range of values of M, and exists for values of M both above and below a choice of cutoff. In addition, this dependence varies with the number of subpopulations K, so that use of the same cutoff could have a different effect on values in scenarios with different numbers of subpopulations.
In a potentially more informative approach, addressing the mathematical dependence of on the within-subpopulation mean heterozygosity Wang (2015) has proposed plotting the joint distribution of and to assess the correlation between the two statistics. Using the island model, Wang (2015) argued that when and are uncorrelated, is expected to be more revealing about the demographic history of a species than when they are strongly correlated and merely reflects the within-subpopulation diversity. Our results suggest a related framework: studies can compare plots of the joint distribution of M and with the bounds on in terms of M. This framework, which examines constraints on in terms of allele frequencies in the total population, complements that of Wang (2015), which considers constraints in terms of subpopulation allele frequencies. Such analyses, considering together with additional measures of allele frequencies, are desirable in diverse scenarios for explaining counterintuitive phenomena, for avoiding overinterpretation of values, and for making sense of comparisons across settings that have a substantial difference in the nature of one or more underlying parameters.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.199141/-/DC1.
Acknowledgments
We thank the editor M. Beaumont, K. Holsinger, and an anonymous reviewer for comments on an earlier version of the manuscript. Support was provided by National Science Foundation grants BCS-1515127 and DBI-1458059; National Institute of Justice grant 2014-DN-BX-K015; a postdoctoral fellowship from the Stanford Center for Computational, Evolutionary, and Human Genomics; and Swiss National Science Foundation Early Postdoc.Mobility fellowship P2LAP3_161869.
Appendix A: Demonstration of Equation 4
This appendix provides the derivation of the upper bound on as a function of K and M.
Theorem 1. Suppose and are specified, where K is an integer. Considering all sequences with and implies is maximal if and only if for and for and its maximum is
Proof. This theorem is a special case of lemma 3 from Rosenberg and Jakobsson (2008), which states (changing notation for some of the variables to avoid confusion): “Suppose and and that is denoted L. Considering all sequences with and implies is maximal if and only if for and for and its maximum is ”
In our special case, we apply the lemma with and We also restrict consideration to sequences of finite rather than infinite length; however, our condition for the number of terms in the sequence guarantees that the maximum in the case of infinite sequences, which requires nonzero terms, is attainable with sequences of the finite length we consider. For convenience in numerical computations, we state our result using the floor function rather than the ceiling function, requiring some bookkeeping to obtain our corollary.
If σ is not an integer, then in lemma 3 of Rosenberg and Jakobsson (2008), and the maximum occurs with and for equaling
If σ is an integer, then and the maximum occurs with and for equaling
In both cases, the maximum simplifies to noting that in the latter case.□
In our application of the theorem in the main text, the definition of M gives so that plays the role of σ. We thus obtain that the maximal value of for sequences with implies and is with equality if and only if for and for Considering all sequences with and not necessarily ordered such that implies the maximum is achieved when any terms equal 1, one term is and remaining terms are 0.
Appendix B: Local Minima of the Upper Bound on FST
This appendix derives the positions and values of the local minima in the upper bound on in terms of M (Equation 5).
Positions of the Local Minima
To derive the positions of the local minima of the upper bound on in terms of M, we study the function (Equation 6) on the interval for x, where and so that Recall that K and i are integers with and i in Note that ensures that in accord with our assumption of a polymorphic locus.
Theorem 2. Consider fixed integers and i in
has no local minimum on for or for
For and i in has a unique local minimum on the interval for x, with position denoted
For odd and
- For all other with and i in where
(B1)
Proof. We take the derivative of
| (B2) |
As the denominator in Equation B2 is nonzero, and is equivalent to a quadratic equation in x:
| (B3) |
If then the quadratic term in Equation B3 vanishes and Equation B3 becomes a linear equation in x, with solution That the solution is a local minimum follows from the continuity of on together with the fact that and for Consequently, if K is odd, then the local minimum for occurs at the lowest possible value of M. This establishes (iii).
Excluding for odd K, for all with Equation B3 has a unique solution in this solution has (Equation B1). The other root of Equation B3 exceeds 1. That represents a local minimum is again a consequence of the continuity of on together with and for This establishes (ii) and (iv).
For the case of Equation B3 has a double root at outside the permissible domain for x, on and approaches 0 as Consequently, has no local minimum for For is the only possible value of i, and has no local minimum. This establishes (i). □
Positions of the Local Minima for Fixed K as a Function of i
Having identified the locations of the local minima, we now explore how those locations change at fixed K with increasing i. For fixed we consider from Theorem 2 as a function of i on the interval It is convenient to define interval equaling for even K and for odd K.
Proposition 1. Consider a fixed integer
The function increases as i increases from to
Its minimum is if K is odd, and if K is even.
- Its maximum is for and for it is
(B4)
Proof. By Theorem 2, for fixed and is given by Equation B1. Treating i as continuous, we take the derivative:
where Because all other terms of are positive for i in has the same sign as
Rearranging terms, we have
Because for i in and Consequently, for all i in Thus, for all i in and increases with i in this interval.
For odd K and Equation B1 gives Thus, because by Theorem 2, is continuous at The function therefore increases with i in the closed interval This proves (i).
Because increases with i for all i in is minimal when i is minimal. For odd K, the minimal value of i is From Theorem 2, for all odd K. For even K, the minimal value of i is From Theorem 2, This proves (ii).
Similarly, is maximal when i is maximal. From Theorem 2, the maximal value of i for which there exists a minimum of is and the position of this local minimum is In particular, for so there is a unique local minimum at position This proves (iii). □
Positions of the First and Last Local Minima as Functions of K
We now fix i and examine the effect of K on the local minimum at fixed i. We first focus on the interval closest to the first local minimum of the upper bound on
Proposition 2. Consider integers
For odd K, the relative position of the first local minimum does not depend on K and is
For even K, the relative position of the first local minimum decreases as tends to and is bounded above by
Proof. For odd K, the interval closest to is In this interval, from Proposition 1ii, the minimum occurs at irrespective of K. This proves (i).
For even K, the interval for M closest to is In this interval, from Proposition 1ii, the minimum has position The derivative of this function is
which is negative for all Thus, decreases with K for In addition, as Because decreases with K, its maximum value is reached when K is minimal. The minimal even value of K is Thus, This proves (ii). □
By Proposition 2, if K is large and even, then the first local minimum lies near the center of the interval for M.
Proposition 3. For integers the relative position of the last local minimum increases as and tends to
Proof. From Theorem 2, for and there is a single local minimum. Hence, from Proposition 2, the position of the last local minimum is for and for The position of the last local minimum then increases from to
If from Proposition 1iii, the position of the last local minimum follows Equation B4. We take the derivative
For the denominator is positive and has the same sign as its numerator. Because for we have and a positive numerator. Then and increases for
From Equation B4, tends to as Thus, the last local minimum is not at the center of interval rather, it is nearer to the upper endpoint. Because increases with K, and the last local maximum has position bounded above by □
As we have shown in Proposition 1i that for fixed K, as i increases from to the relative position of the local minimum increases, this relative position is restricted in the interval Further, because from Proposition 2, for odd K and for even K and from Proposition 3, the relative positions of the local minima must be in the interval
Figure B1 illustrates as functions of K the relative positions of the first local minimum for odd K and for even K and the last local minimum The restriction of these positions to the interval is visible, with the first local minimum lying closer to the center of interval for x than the last local minimum. The decrease in the position of the first local minimum for even K alternating with values of for odd K (Proposition 2) and the increase in the position of the last local minimum (Proposition 3) are visible as well.
Values at the Local Minima
We obtain the value of the local minima of the upper bound on in each interval by substituting into Equation 6 the value of i for interval and its associated from Theorem 2. We obtain
| (B5) |
Note that for odd K, although is undefined at is continuous. Thus, is also defined and continuous for all We consider as a function of i on this interval.
Proposition 4. For fixed the local minima decrease as i increases from to
Proof. We take the derivative for fixed K and From Equations B5 and B1,
where and
For all the denominator of the derivative is positive, as are K, and Hence, has the same sign as
Because decreases for and We factor
where Because for all Thus, the sign of is given by the sign of
For
and hence,
| (B6) |
The term increases as a function of i for Hence, decreases with i. It is minimal at the largest value in the permissible domain for i, or with minimum The denominator of this quantity is positive and the numerator increases with K. It is thus minimal for at which This proves that for
We can then remove the absolute value in Equation B6 and rearrange to obtain Both sides of this inequality are positive for and we can take the square root to obtain Hence for for and the local minima decrease with i. □
Proposition 5. For the first local minimum has value for the first local minimum increases as a function of K and tends to 1 as
Proof. From Proposition 2i, for odd K, the first local minimum is reached for and and the upper bound on is Thus, for the first local minimum has value For even K, the first local minimum is reached if and with upper bound on equal to
| (B7) |
Denote by the difference between the first local minimum for even K and the first local minimum for odd and by the difference between the first local minimum for odd and the first local minimum for even K. To show that the first local minimum increases with K, we must show that for all even (i) and (ii)
For (i), subtracting from Equation B7, we have
Because all other terms are positive for has the same sign as Dividing by this quantity in turn has the same sign as This last quantity is positive for as when we multiply it by the positive the result reduces simply to the number 4. This proves (i).
For (ii), subtracting Equation B7 from we have
Because all other terms are positive for has the same sign as Dividing by this quantity has the same sign as which was shown to be positive in the proof of (i). This demonstrates (ii).
From (i) and (ii), the value of the upper bound on at the first local minimum increases with K for all To see that the limiting value is 1 as we note that the subsequence of values at odd K tends to 1 as As the sequence of values of the first local minimum with increasing K is monotonic and bounded above by 1, it is therefore convergent; as it has a subsequence converging to 1, the sequence converges to 1. □
Proposition 6. For the last local minimum has value for the last local minimum increases as a function of K and tends to as
Proof. From Proposition 5, for the single local minimum has value By Theorem 2, for the last local minimum is reached when and in which case from Equations B5 and B1 the upper bound on is
| (B8) |
where
We examine the derivative of with respect to K. For and
where For the derivative has the same sign as
We note that
with equality only at Because is positive for with equality only at Thus, and for and hence, the last local minimum increases with K.
For the limit as we take the limit of in Equation B8, obtaining □
Appendix C: Computing the Mean Range of FST
This appendix provides the computation of the integral (Equation 7) and its asymptotic approximation
Computing A(K)
To compute we break the integral into a sum over intervals. If K is even, then we consider intervals with If K is odd, then we use intervals and with
By construction of (Equation 6), in each interval the upper bound on is equal to with In the odd case, because is an integer, on interval has a constant value and and the upper bound is equal to Making the substitution we obtain and we can write Equation 7 in terms of
| (C1) |
We next use a partial fraction decomposition. For
For
For
Thus, when K is even,
When K is odd,
| (C2) |
The expressions for are equal for even and odd K. We can simplify further:
| (C3) |
Substituting the expression from Equation C3 into Equation C2 and simplifying, we obtain Equation 8.
Asymptotic Approximation for A(K) (Equation 9)
To asymptotically approximate we first need a large-K approximation of Because where is the hyperfactorial function, we can use classical results about the asymptotic behavior of
| (C4) |
where C is the Glaisher–Kinkelin constant. Because the logarithm function is continuous at C, has limit as Thus, if we write for two functions that satisfy then
| (C5) |
Substituting the expression from Equation C5 into Equation 8, we obtain function (Equation 9) and the relationship
Monotonicity of Equation 8 in K
Theorem 3. increases monotonically in K for
Proof. We must show that for all From the expression for in Equation 8, we have:
| (C6) |
To show that we find a lower bound for denoted and then show that
We first find a lower bound for From the Euler–Maclaurin summation formula, we have
For all positive integers
| (C7) |
This integral can be seen to be positive from the equivalence for of with This latter inequality follows from the inequality for from the Taylor expansion of noting that
Consequently, as the integral in Equation C7 is positive for each and
| (C8) |
As a result, the following function is a lower bound for
Dividing by and substituting for if and only if for It can be seen that this latter inequality holds by noting that and □
Figure B1.
The first and last local minima of as functions of the frequency M of the most frequent allele, for subpopulations. (A) Relative positions within the interval of the first and last local minima, as functions of K. The position of the local minimum in interval is computed from Equation B1. If K is odd, then this position is if K is even, then it is The position of the last local minimum is Dashed lines indicate the smallest value for of and the limiting largest value of (B) The value of the upper bound on at the first and last local minima, as functions of K. These values are computed from Equation 5, taking and with as in part (A). Dashed lines indicate the limiting values of 1 and for the first and last local minima, respectively.
Footnotes
Communicating editor: M. A. Beaumont
Literature Cited
- 1000 Genomes Project Consortium , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey J. M., Zhang G., Zhang K., Jin L., Shriver M. D., 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12: 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcala N., Goudet J., Vuilleumier S., 2014. On the transition of genetic differentiation from isolation to panmixia: what we can learn from GST and D. Theor. Popul. Biol. 93: 75–84. [DOI] [PubMed] [Google Scholar]
- Algee-Hewitt B. F. B., Edge M. D., Kim J., Li J. Z., Rosenberg N. A., 2016. Individual identifiability predicts population identifiability in forensic microsatellite markers. Curr. Biol. 26: 935–942. [DOI] [PubMed] [Google Scholar]
- Balloux F., Brünner H., Lugon-Moulin N., Hausser J., Goudet J., 2000. Microsatellites can be misleading: an empirical and simulation study. Evolution 54: 1414–1422. [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., Nichols R. A., 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. Ser. B Biol. Sci. 263: 1619–1626. [Google Scholar]
- Bonhomme M., Chevalet C., Servin B., Boitard S., Abdallah J., et al. , 2010. Detecting selection in population trees: the Lewontin and Krakauer test extended. Genetics 186: 241–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann H. M., de Toma C., Cazes L., Legrand M.-F., Morel V., et al. , 2002. A human genome diversity cell line panel. Science 296: 261–262. [DOI] [PubMed] [Google Scholar]
- Cañon J., Checa M. L., Carleos C., Vega-Pla J. L., Vallejo M., et al. , 2000. The genetic structure of Spanish Celtic horse breeds inferred from microsatellite data. Anim. Genet. 31: 39–48. [DOI] [PubMed] [Google Scholar]
- Cornuet J.-M., Santos F., Beaumont M. A., Robert C. P., Marin J.-M., et al. , 2008. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24: 2713–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge M. D., Rosenberg N. A., 2014. Upper bounds on FST in terms of the frequency of the most frequent allele and total homozygosity: the case of a specified number of alleles. Theor. Popul. Biol. 97: 20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M., Gaggiotti O., 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R., Ballou J. D., Briscoe D. A., 2002. Introduction to Conservation Genetics. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- Günther T., Coop G., 2013. Robust identification of local adaptation from allele frequencies. Genetics 195: 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., Clark A. G., 1997. Principles of Population Genetics. Sinauer, Sunderland, MA. [Google Scholar]
- Hedrick P. W., 1999. Highly variable loci and their interpretation in evolution and conservation. Evolution 53: 313–318. [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., 2005. A standardized genetic differentiation measure. Evolution 59: 1633–1638. [PubMed] [Google Scholar]
- Hinds D. A., Stuve L. L., Nilsen G. B., Halperin E., Eskin E., et al. , 2005. Whole-genome patterns of common DNA variation in three human populations. Science 307: 1072–1079. [DOI] [PubMed] [Google Scholar]
- Holsinger K. E., Weir B. S., 2009. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 10: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium , 2005. A haplotype map of the human genome. Nature 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M., Edge M. D., Rosenberg N. A., 2013. The relationship between FST and the frequency of the most frequent allele. Genetics 193: 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L., 2008. GST and its relatives do not measure differentiation. Mol. Ecol. 17: 4015–4026. [DOI] [PubMed] [Google Scholar]
- Kim T. H., Kim K. S., Choi B. H., Yoon D. H., Jang G. W., et al. , 2005. Genetic structure of pig breeds from Korea and China using microsatellite loci analysis. J. Anim. Sci. 83: 2255–2263. [DOI] [PubMed] [Google Scholar]
- Lawson Handley L. J., Byrne K., Santucci F., Townsend S., Taylor M., et al. , 2007. Genetic structure of European sheep breeds. Heredity 99: 620–631. [DOI] [PubMed] [Google Scholar]
- Leinonen T., McCairns R. J. S., O’Hara R. B., Merilä J., 2013. comparisons: evolutionary and ecological insights from genomic heterogeneity. Nat. Rev. Genet. 14: 179–190. [DOI] [PubMed] [Google Scholar]
- Lewontin R., Krakauer J., 1973. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics 74: 175–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Z., Absher D. M., Tang H., Southwick A. M., Casto A. M., et al. , 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104. [DOI] [PubMed] [Google Scholar]
- Long J. C., Kittles R. A., 2003. Human genetic diversity and the nonexistence of biological races. Hum. Biol. 75: 449–471. [DOI] [PubMed] [Google Scholar]
- Maruki T., Kumar S., Kim Y., 2012. Purifying selection modulates the estimates of population differentiation and confounds genome-wide comparisons across single-nucleotide polymorphisms. Mol. Biol. Evol. 29: 3617–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., 1970. Effective number of alleles in a subdivided population. Theor. Popul. Biol. 1: 273–306. [DOI] [PubMed] [Google Scholar]
- Nei M., 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Chesser R. K., 1983. Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 47: 253–259. [DOI] [PubMed] [Google Scholar]
- Pemberton T. J., Absher D., Feldman M. W., Myers R. M., Rosenberg N. A., et al. , 2012. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 91: 275–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Rosenberg N. A., Zhivotovsky L. A., Feldman M. W., 2004. Robustness of the inference of human population structure: a comparison of X-chromosomal and autosomal microsatellites. Hum. Genomics 1: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S. B., Rosenberg N. A., 2012. Refining the relationship between homozygosity and the frequency of the most frequent allele. J. Math. Biol. 64: 87–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., 2011. A population-genetic perspective on the similarities and differences among worldwide human populations. Hum. Biol. 83: 659–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Jakobsson M., 2008. The relationship between homozygosity and the frequency of the most frequent allele. Genetics 179: 2027–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Pritchard J. K., Weber J. L., Cann H. M., Kidd K. K., et al. , 2002. Genetic structure of human populations. Science 298: 2381–2385. [DOI] [PubMed] [Google Scholar]
- Rosenberg N. A., Li L. M., Ward R., Pritchard J. K., 2003. Informativeness of genetic markers for inference of ancestry. Am. J. Hum. Genet. 73: 1402–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Mahajan S., Gonzalez-Quevedo C., Blum M. G. B., Nino-Rosales L., et al. , 2006. Low levels of genetic divergence across geographically and linguistically diverse populations from India. PLoS Genet. 2: e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F., 2013. Exegeses on maximum genetic differentiation. Genetics 194: 557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., 1985. Rare alleles as indicators of gene flow. Evolution 39: 53–65. [DOI] [PubMed] [Google Scholar]
- Tishkoff S. A., Reed F. A., Friedlaender F. R., Ehret C., Ranciaro A., et al. , 2009. The genetic structure and history of Africans and African Americans. Science 324: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., 1998. Segregating sites in Wright’s island model. Theor. Popul. Biol. 53: 166–174. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 1999. Nonequilibrium migration in human history. Genetics 153: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., 2015. Does GST underestimate genetic differentiation from marker data? Mol. Ecol. 24: 3546–3558. [DOI] [PubMed] [Google Scholar]
- Weir B. S., 1996. Genetic Data Analysis II. Sinauer, Sunderland, MA. [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., 2011. and D do not replace Mol. Ecol. 20: 1083–1091. [DOI] [PubMed] [Google Scholar]
- Wright S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS commands for the coalescent simulations appear in File S2. See Li et al. (2008) for the human SNP data.