Abstract
Neuronal Calcium Sensor 1 (NCS1) is a multi-functional Ca2+-binding protein that affects a range of cellular processes beyond those related to neurons. Functional characterization of NCS-1 in neuronal model systems suggests that NCS-1 may influence oncogenic processes. To this end, the biological role of NCS-1 was investigated by altering its endogenous expression in MCF-7 and MB-231 breast cancer (BCa) cells. Overexpression of NCS-1 resulted in a more aggressive tumor phenotype demonstrated by a marked increase in invasion and motility, and a decrease in cell-matrix adhesion to collagen IV. Overexpression of NCS-1 was also shown to increase the efficacy of paclitaxel-induced cell death in a manner that was independent of cellular proliferation. To determine the association between NCS-1 and clinical outcome, NCS-1 expression was measured in two independent BCa cohorts by the Automated Quantitative Analysis (AQUA) method of quantitative immunofluorescence. Elevated levels of NCS-1 were significantly correlated with shorter survival rates. Furthermore, multivariate analysis demonstrated that NCS-1 status was prognostic, independent of Estrogen Receptor (ER), Progesterone Receptor (PR), Human Epidermal Growth Factor 2 Receptor (HER2), and lymph node status. These findings indicate that NCS-1 plays a role in the aggressive behavior of a subset of breast cancers and has therapeutic or biomarker potential.
Keywords: Neuronal Calcium Sensor-1, breast cancer, biomarkers, survival, prognosis, drug response, and quantitative immunofluorescence
INTRODUCTION
Calcium (Ca2+), a ubiquitous second messenger molecule, is involved in numerous physiological processes associated with tumor progression (e.g. proliferation, adhesion, migration, and invasion (1–4)). The dysregulation of Ca2+ homeostasis has been implicated as an important driver in the development of metastatic phenotypes (5). Numerous studies demonstrate that Ca2+-binding proteins (e.g., S100 proteins (6–9), calmodulin (10, 11), calcineurin (12, 13)) have the potential to promote tumor progression.
Neuronal Calcium Sensor-1 (NCS-1) is a Ca2+-binding protein that is expressed at high concentrations in neurons (14). The characterization of NCS-1 in neuronal models demonstrates its involvement in Ca2+-signaling transduction (15–18), exocytosis (19–21), membrane trafficking (22, 23), cell survival (24), and functional regulation of a diverse group of proteins (Supplemental Figure 1) (25–29). NCS-1 has also been identified as a novel binding partner of the chemotherapeutic drug, paclitaxel (30), and this binding interaction was shown to alter the biological activity of NCS-1 in vitro and in vivo (31–33).
In addition to neurons, NCS-1 (14) has been identified in non-neuronal tissues (34–36). The widespread expression of this protein suggests that it may have other functions in addition to its characterized role in neuronal models. For example, the functional properties of NCS-1 indicate that it may influence oncogenic processes. Recently, NCS-1 has been shown to act as a survival factor through the indirect activation of the PI3K/AKT signaling pathway (24), a pathway known to be perturbed in many forms of cancer (37). Based upon its potential to influence oncogenic processes and its established relationship with taxanes (30, 38), we hypothesize that increased expression of NCS-1 may be associated with tumor aggressiveness, taxane response, and poor clinical outcome in breast cancer patients. In this study, we assess the role of NCS-1 in breast cancer biology and drug response by altering the endogenous expression of this protein in MCF-7 and MB-231 cell lines. Additionally, we examined the clinical association between NCS-1 expression and survival in breast cancer patients.
MATERIALS AND METHODS
Validation and Reproducibility Assessment of the NCS-1 Antibody
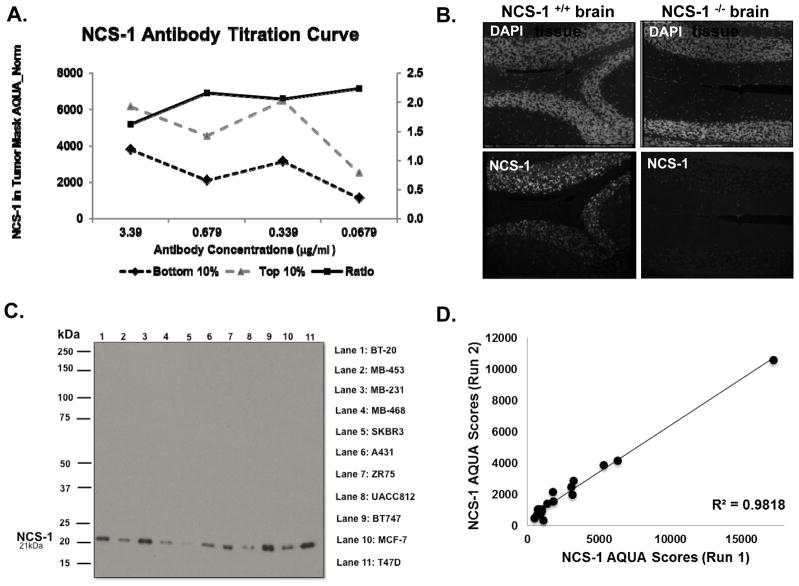
A rabbit monoclonal antibody targeting NCS-1 (Abcam, Cambridge, MA, USA, catalog number: ab129166) was validated for specificity, selectivity, and reproducibility using in-house techniques, previously described (39). The optimized signal-to-noise ratio for the NCS-1 antibody was 0.339 μg/ml (Figure 1A). Antibody specificity, was assessed by staining for NCS-1 expression in formalin-fixed, paraffin-embedded (FFPE)-brain tissues from NCS-1 knockout mice and wild-type controls (Figure 1B). Specificity was further evaluated by examining the expression of NCS-1 in a panel of cancer cell lines (Figure 1C). Reproducibility was assessed by comparing NCS-1 staining from serial sections run on different days. The regression of these control Tissue Microarrays (TMAs) showed a high linear regression coefficient (R2= 0.98, Figure 1D).
Figure 1. The NCS-1 Antibody is Specific and Reproducible.
A. Antibody titration curve of commercially available NCS-1 antibody reveals optimal antibody concentration of 0.339 μg/ml. B. Photomicrograph of cerebellum tissue from wild type and NCS-1 knockout mice stained for NCS-1 and DAPI reveals specificity of NCS-1 antibody. C. Immunoblot analysis of NCS-1 in multiple cancer cell lines, show NCS-1 at predicted 21 kDa. D. Linear regression of control TMAs stained on two different days. Pearson’s coefficient, R2 values > 0.90 denotes assay reproducibility.
Breast Cancer Cell Line Culture Maintenance
Breast cancer cell lines (MCF-7, ATCC® HTB-22™ and MB-231, ATCC® HTB-26™) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). ATCC validates all cell lines by Short Tandem Repeat Analysis. Upon receipt, cell lines were used within 6 months. MCF-7 cell lines were maintained at 37°C, 5% CO2 in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 1% MEM nonessential amino acids, 1% sodium pyruvate, and 0.01 mg/ml human recombinant insulin. MB-231 cell lines were maintained at 37°C, 0% CO2 in Leibovitz’s L-15 Medium (L-15) supplemented with 10% FBS and 1% penicillin/streptomycin.
Lentiviral Plasmid, Particles, and Production
A Human NCS-1 (hNCS1) lentiviral plasmid was purchased from Applied Biological Materials (Richmond, BC, Canada, Catalog no.: LV800661). Control (Catalog no.: sc-108080) and NCS-1 shRNA lentiviral particles (Catalog no.: sc-36019-V) were purchased from Santa Cruz Biotechnology Inc. (Paso Robles, CA, USA). For the hNCS-1 plasmid, lentiviral particles were produced through the transfection of HEK-293T cells. Briefly, HEK-293T cells were seeded and transfected with a mixture of hNCS1 lentiviral plasmid and second-generation packaging plasmids using Lipofectamine 2000 per manufacturers’ instructions. After 18–24 h, transfection media was removed and replaced with fresh media. Cells were then incubated for additional 24 h. After this period, lentiviral-containing supernatant was collected, centrifuged, and filtered through a 0.45-μm filter. The virus was stored at −80°C until the day of lentiviral transduction.
Lentiviral Transduction
Cells were seeded in a 6-well plate at a density of 5 x 105 cells/well 24 h prior to transduction. The following day, media was replaced with fresh media supplemented with polybrene at a concentration of 8μg/ml. For hNCS-1 lentiviral particles, 0.5 ml of virus was directly added to each well. For the more concentrated control and NCS-1 shRNA lentiviral particles 10μl of virus was added to each well. Following transduction, cells were incubated at 37°C. After 24 h media was replaced with fresh media. After 48 h puromycin was added at a pre-determined optimal concentration. After two weeks, puromycin concentrations were reduced to a maintenance concentration of 0.5 μg/ml.
Immunofluorescence
Cells were seeded on sterile glass coverslips and grown to 80% confluency. Fixation was performed for 15 min at room temperature (RT) with a 4% paraformaldehyde (PFA) solution. Following several washes in 1X Phosphate-buffered saline (PBS) supplemented with 0.1% Tween-20 (PBST), cells were permeabilized in 0.1% Triton-X 100 for 5 min. After permeabilization, cells were washed and then blocked for 1 h at RT in a blocking solution that consisted of 1X PBST supplemented with 10% normal goat serum (NGS; Cell Signaling Technology Inc., Danvers, MA, USA). Following block, cells were incubated with a rabbit anti-NCS-1 monoclonal antibody diluted in blocking solution (Abcam, Cambridge, MA, USA; diluted 1:1000) overnight (O/N) at 4°C. After extensive washing in PBST, cells were incubated with an AlexaFluor-546 goat anti-rabbit secondary antibody (Invitrogen, Carlsbad, CA, USA; diluted 1:1000) for 1 h at RT in the dark. Cells were then washed in PBST before being mounted on glass slides with anti-fade medium ProLong Gold with DAPI (Invitrogen, Carlsbad, CA, USA). Slides were cured O/N before images were captured with a confocal microscope (Zeiss LSM 710 Duo).
Quantitative Immunoblotting
Cells were lysed in ice cold M-PER mammalian Protein Extraction Reagent (Thermo Scientific, Rockford, IL, USA) supplemented with protease inhibitors. To determine protein concentration a Bradford assay was conducted using the Bio-Rad protein assay reagent (Bio-Rad, San Diego, CA, USA). Proteins (30 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane (GE Healthcare, Arlington Heights, IL, USA). The resulting blots were blocked for 1 h at RT in 5% skimmed dry milk diluted in 1X Tris-buffered saline supplemented with Tween-20 (TBST). Blots were incubated at 4°C O/N in primary antibodies specific for NCS-1 (Abcam, Cambridge, MA, USA; diluted 1:1000) and β-tubulin (Cell Signaling, Boston, MA, USA; diluted 1:500). Following incubation, blots were washed with 5% milk/TBST before incubation with a horseradish peroxidase, labeled goat anti-rabbit IgG (Santa Cruz Biotechnology Inc., Paso Robles, CA, USA; diluted 1:1000) at RT for 1 h. Blots were washed with 5% milk/TBST and bands were visualized using electrochemiluminescence detection reagents (Thermo Scientific, Rockford, IL, USA). Immunoblots were then quantified by scanning densitometry using the Un-Scan-It software (Silk Scientific, Orem, UT, USA).
Scratch Motility Assay
Scratch motility assays were performed by growing cells to confluency in a 6-well plate. Cells were starved in serum-free media (SFM) supplemented with mitomycin C two hours prior to the start of the assay. In each well three T-shaped wounds were induced with a p200 tip. Images were captured at baseline (0 h) and at the endpoints indicated: 48 h (MCF-7) and 36 h (MB-231). The percent open image area for each T-shaped wound was measured using TScratch Software (40). This assay was repeated in triplicate.
Cell Invasion and Chemotaxis Assay
Invasion assays were performed in 8μm-pore, Matrigel-coated Transwell chambers (Corning, Oneonta, NY, USA). Cells (1 X 104) were seeded in the upper chamber, which contained SFM. Cells were allowed to migrate towards the chemoattractant (media supplemented with 10% FBS) in the lower chamber. At the endpoints indicated, 72 h (MCF-7) or 24 h (MB-231), cells were removed from the top surface of the chamber. Cells that remained on the lower surface of the chamber were fixed in 4% PFA and stained with a 0.5% crystal violet solution. A pictograph was taken on EVOS FL cell imaging system (Thermo Scientific, Rockford, IL, USA). Cells were counted from the digitized images. For MCF-7 cells, the total cell number in each chamber was quantified at 4X magnification. For MB-231 cells, the average cell number from three random fields of view in each chamber was quantified at 10X magnification. All assays were performed in triplicate and a companion control, an uncoated 8μm-pore Transwell chamber, was used in parallel to assess the rate of chemotaxis.
Adhesion Assay
Adhesion assays were performed using a colorimetric-based assay (CytoSelect 48-Well Cell Adhesion Assay; Cell Biolabs Inc., San Diego, CA, USA) according to the manufacturer’s instructions. Briefly, cells were serum starved for 24 h prior to seeding on Collagen IV coated adhesion plate at a concentration of 1.0 x 106 cells/ml in SFM. Cells were incubated for 180 min (MCF-7) or 90 min (MB-231). Non-adherent cells were gently removed with several washes of 1X PBS. Cells were then fixed and stained. An extraction solution was then added and a portion of this solution was transferred to a well of a 96-well plate. The absorbance of this solution was read at 560 nm in a microplate reader.
Cell Viability Assay
Cell viability assays were performed by serum starving cells for 24 h prior to seeding in a 96-well plate at a the indicated concentrations: 1 × 104 cells/well (MCF-7) or 5 × 103 cells/well (MB-231). Cells were allowed to recover O/N. Fresh culture medium containing various concentrations of paclitaxel (0.1pM to 10,000nM) was then added. After 72 h of treatment, luminescent measurements were made using the CellTiter-Glo reagent (Promega, Madison, WI, USA) per manufacturer’s instructions. Briefly, plates were removed from the incubator and allowed to equilibrate at RT for 0.5 h. Equal volumes of CellTiter-Glo reagent were added directly to each well. Cell lysis was induced by gently shaking plates on orbital shaker for 2 min. Luminescent signal (RLU; relative light units) was stabilized for 10 min at RT before measurements were recorded. The percentage of cell death was calculated using the equation below. For the cell viability assay the positive control was media supplemented with 20% DMSO and the negative control was media supplemented with 0.05% DMSO.
Cell Proliferation Assay
Cell proliferation was quantified by the metabolic reduction of WST-8 (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) (CCK-8 cell proliferation assay, Dojindo Molecular Technologies, Rockville, MD, USA) to generate a growth curve over six days. Cell lines were serum starved for 24 h prior to seeding in a 96-well plate at the indicated concentrations: 2.5 x 103 cells/well (MCF-7) and 1 x 103 cells/well (MB-231). Cells recovered for 24 h before a baseline absorbance recording was determined by WST-8. Briefly, 10 μl of WST-8 was added to 100 μl media in each well. Plates were incubated at 37 °C for 4 h before absorbance was read at 450 nm. Each subsequent day, cell number was determined using WST-8. This absorbance value was then normalized to the baseline recording.
Human Subject Use
This study was carried out in accordance with the recommendations in the U.S. National Institutes of Health Guide for the Care and Use of Human Subjects. All cases obtained had signed consent for tissue use, or waiver of consent, under the approved Yale University human investigation committee protocol #9505008219.
Patient Cohorts
The retrospective studies were conducted using two Yale Tissue Microarrays (YTMAs) consisting of a cohort of archival breast cancer cases; YTMA 49 contained 619 breast cancer patients diagnosed from 1962 to 1982. YTMA 201 contained 405 breast cancer patients diagnosed from 1976 to 2005. Detailed information regarding these cohorts has previously been published (41). The clinico-pathological features of these cohorts are summarized in Table 1.
Table 1.
Clinicopathological Characteristics of YTMA 49 and YTMA 201
| YTMA 49 N (total, 448) |
% | YTMA 201 N (total, 287) |
% | |
|---|---|---|---|---|
|
Characteristics Age at Diagnosis |
||||
|
| ||||
| <50 | 122 | 27.2 | 95 | 33.1 |
| ≥50 | 326 | 72.8 | 192 | 66.9 |
|
| ||||
| Tumor Size | ||||
|
| ||||
| ≤2 cm | 132 | 29.5 | 167 | 58.2 |
| 2–5 cm | 227 | 50.7 | 100 | 34.8 |
| ≥5 cm | 51 | 11.4 | 1 | 0.3 |
| Unknown | 38 | 8.5 | 19 | 6.6 |
|
| ||||
| Nuclear Grade | ||||
|
| ||||
| Grade 1 | 65 | 14.5 | --- | --- |
| Grade 2 | 232 | 51.8 | --- | --- |
| Grade 3 | 121 | 27.0 | --- | --- |
| Unknown | 30 | 6.7 | --- | --- |
|
| ||||
| Nodal Status | ||||
|
| ||||
| Positive | 248 | 55.4 | 63 | 21.9 |
| Negative | 200 | 44.6 | 158 | 55.1 |
| Unknown | --- | --- | 66 | 23 |
|
| ||||
| ER Status | ||||
|
| ||||
| Positive | 230 | 51.3 | 164 | 57.1 |
| Negative | 206 | 45.9 | 92 | 32.1 |
| Unknown | 12 | 2.7 | 31 | 10.8 |
|
| ||||
| PR Status | ||||
|
| ||||
| Positive | 213 | 47.5 | 34 | 11.8 |
| Negative | 210 | 46.9 | 218 | 75.9 |
| Unknown | 25 | 5.6 | 35 | 12.2 |
|
| ||||
| HER2 Status | ||||
|
| ||||
| Positive | 75 | 16.7 | 30 | 10.5 |
| Negative | 354 | 79.0 | 209 | 72.8 |
| Unknown | 19 | 4.5 | 48 | 16.7 |
ER: Estrogen Receptor, PR: Progesterone Receptor, HER2: Human Epidermal Growth Factor Receptor 2, NCS-1: Neuronal Calcium Sensor-1
Immunohistochemical (IHC) Staining
For IHC staining, YTMAs were deparaffinized in an incubator for 0.5 h at 60°C, followed by two xylene washes. After deparaffinization, slides were rehydrated in decreasing concentrations of ethanol washes. Antigen retrieval was then performed by heating the slides in a LabVision PT module (LabVision, Fremont, CA) containing 7.5 mM sodium citrate (pH 6.0) supplemented with Tween-20 at 97°C for 20 min. To block endogenous peroxides and permeabilize the tissues, slides were incubated in a 2.5% H2O2/ methanol solution for 0.5 h at RT. Following this step, slides were washed in distilled water before incubation with a 0.3%BSA/ TBST solution for 1 h at RT to reduce any non-specific background staining. Slides were then incubated at 4°C O/N with BSA/TBST solution containing the following primary antibodies: a mouse anti-cytokeratin monoclonal antibody (clone AE1/AE3, DAKO, Carpinteria, CA; diluted 1:100) and a rabbit anti-NCS-1 monoclonal antibody (Abcam, Cambridge, MA, USA; diluted 1:1000). After O/N incubation, slides were washed in a series of Tris-buffered saline (TBS)/TBST baths before incubation with the indicated secondary antibodies for 1 h at RT: AlexaFluor goat anti-mouse 546 (Invitrogen, Carlsbad, CA, USA; diluted 1:100) diluted in anti-rabbit poly HRP antibody Envision (DAKO, Carpinteria, CA, USA). Following incubation, slides were again washed in a series of TBS/TBST baths before incubation with Cy5 tyramide at RT for 10 min. Slides were washed before being mounted onto glass coverslips with anti-fade medium Prolong Gold with DAPI (Invitrogen, Carlsbad, CA, USA)
Automated Quantitative Analysis (AQUA)
Automated quantitative analysis (AQUA®) was used for image acquisition and data analysis as previously described (42). Briefly, high-resolution images of the YTMA histospots were captured using a PM-2000 image workstation (Genoptix, Carlsbad, CA, USA). For each histospot, three images were obtained that corresponded to DAPI, AlexaFluor 546 (cytokeratin), and CY5 fluorophores (NCS-1). These images were analyzed by AQUA software and a ‘tumor mask’ was generated from anti-cytokeratin signal to differentiate tumor areas from stromal compartments. Nuclear and NCS-1 compartments were, respectively, created from DAPI and Cy5 signals. An AQUA score was calculated for each histospot by dividing the signal intensity by the total area under the tumor mask. The scores were normalized for exposure time, bit depth, and lamp hours. To reduce bias, data collection was blinded to the clinical outcome for each patient.
SurvExpress: A Biomarker Validation Tool, Used in the Assessment of the Prognostic Utility of NCS-1 mRNA from Available Breast Cancer Datasets
The prognostic utility of NCS-1 mRNA was assessed using 31 publicly available breast cancer datasets from SurvExpress, a biomarker validation tool (43). Results were obtained using the average score from available probe sets and using the original quantile-normalized format. Statistical analyses and graphical output were set using available endpoints (e.g. survival, recurrence-free survival (RFS), or metastasis) to obtain 2 maximized risk groups.
Statistical Analysis
Values are expressed as mean ± standard error of the mean (S.E.M). For the in vitro assays, significant differences between groups were assessed using two-sided student’s t test. For quantitative immunoblotting, significant differences between groups were assessed by ANOVA. Analyses were performed using GraphPad Prism 4 software (GraphPad Software Inc., La Jolla, CA, USA) and statistical significance was set at a p-value < 0.05. Survival analysis was performed using the StatView software platform (SAS Institute, Cary, NC, USA), Kaplan-Meier analyses were performed on each cohort (disease-specific survival [DSS]) and statistical significance was determined by using the log-rank test. Fisher’s exact test and Chi-square testing were also used to determine the significance of clinico-pathological factors (grade, hormone receptor status, HER2 status, and treatment) in predicting survival.
RESULTS
Generation of Stable Breast Cancer Cell Lines with Altered Expression of NCS-1
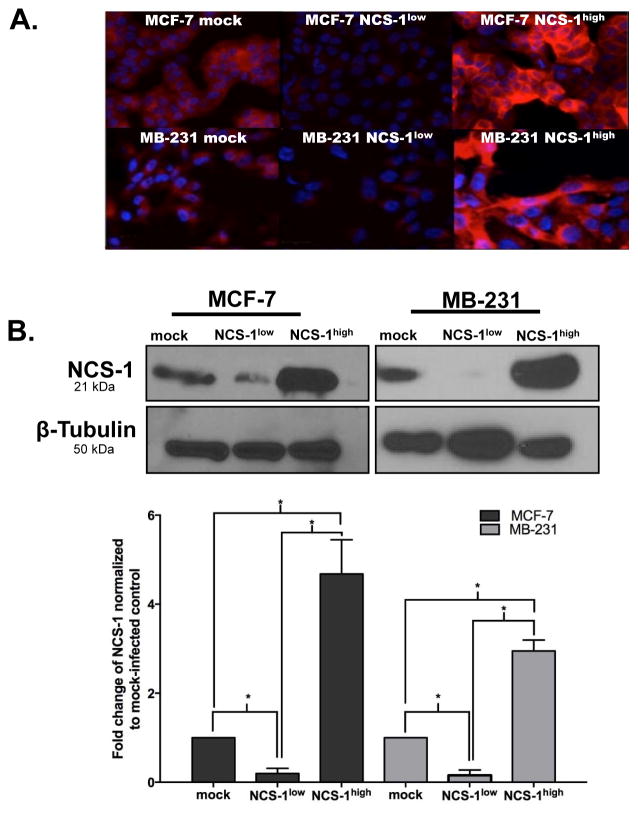
To determine if the expression of NCS-1 could have a role in oncogenesis or metastasis, we altered the endogenous expression of this protein in MCF-7 and MB-231 cell lines. Using lentiviral particles, cells were genetically modified to overexpress (MCF-7 NCS-1high or MB-231 NCS-1high) or knockdown (MCF-7 NCS-1low or MB-231 NCS-1low) the NCS-1 protein. Immunofluorescent (IF) staining qualitatively revealed that expression of NCS-1 was up-regulated in MCF-7 NCS-1high and MB-231 NCS-1high cell lines, compared to their mock-infected controls. IF staining also qualitatively revealed a reduction of NCS-1 in the MCF-7 NCS-1low and MB-231 NCS-1low cell lines (Figure 2A). No visible differences in the morphologic phenotype of either cell lines were observed by altering the expression of NCS-1. Quantitative immunoblotting revealed that NCS-1 expression in the MCF-7 NCS-1high cell line was increased by 4.7-fold (p=0.01) compared to mock-infected control (Figure 2B). In the MCF-7 NCS-1low was NCS-1 expression was reduced by a fold change of 0.20, (p=0.01). Similar results were observed in the MB-231 NCS-1high (fold change: 3.0, p=0.04) and MB-231 NCS-1low cell lines (fold change: 0.16, p=0.01).
Figure 2. Validation of Stable Breast Cancer Cell Lines.
A. Representative immunofluorescent images of MCF-7 and MB-231 cell lines that were generated with commercially available lentiviral particles to overexpress or knockdown the NCS-1 protein. Cells were fixed and stained for NCS-1 (red) and DAPI (blue). B. Representative immunoblots show the relative amounts of NCS-1 and β-tubulin in the stable breast cancer cell lines; (below) band densitometry was analyzed using UN-Scan IT, bar graphs, illustrates NCS-1 expression normalized to β-tubulin and expressed as a fold-change relative to mock-infected controls. Results shown are the average of three biological replicates.
NCS-1 Promotes Tumor Aggressiveness by Altering Invasion, Chemotaxis, and Adhesion
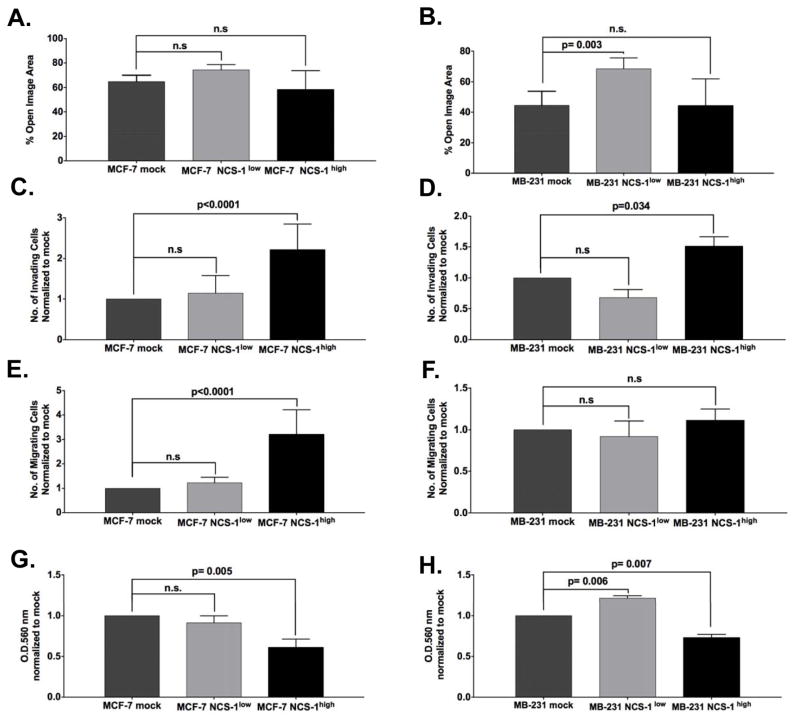
To examine the effects of NCS-1 on migration, we performed wound closure assays. In the MCF-7 cell line, no significant change was observed between the percent open image area for mock, MCF-7 NCS-1low, or MCF-7 NCS-1high: 65 ± 3 %, 74 % ± 3%, 58 ± 9%, respectively (Figure 3A). However, knockdown of NCS-1 significantly reduced migration in the highly motile MB-231 cell line (Figure 3B, p=0.003). The percent open image area after 36 h for MB-231 mock and MB-231 NCS-1low were 45% ± 5.% and 68% ± 4%, respectively. No significant change in percent open image area was observed between mock and MB-231 NCS-1high cell lines (p= 0.98, 44% ± 10%).
Figure 3. NCS-1 Promotes Tumor Aggressiveness by Altering Invasion, Chemotaxis, and Adhesion.
Motility was assessed in A. MCF-7 and B. MB-231 cell lines through scratch motility assays. A monolayer of cells was pre-treated with mitomycin C before a T-shaped scratch was mechanically induced with a sterile 200-μl pipette tip. The % open image areas were calculated using T-scratch software. Invasion and chemotaxis were assessed in C–E. MCF-7 and D–F. MB-231 cell lines by Transwell invasion assays. Media supplemented with 10% FBS was used as the chemoattractant in the lower chamber. Cell-matrix adhesion to Collagen IV was assessed in G. MCF-7 and H. MB-231 using Cell BioLabs adhesion kit, according to manufacturer’s instruction. The O.D. at 560 nm was normalized to mock. For all in vitro experiments, assays were performed in triplicate and the results shown are depicted as the mean ± S.E.M.
A Transwell Matrigel invasion assay, using media supplemented with 10% FBS as the chemoattractant in the lower chamber, revealed a significant effect of NCS-1 overexpression on cell invasion compared to mock-infected controls (p <0.0001 for MCF-7 and p=0.034 for MB-231 in student’s t-test, Figure 3C and D, respectively). Additionally, an increase in cell chemotaxis was observed in the MCF-7 NCS-1high cell lines (p< 0.0001, Figure 3E). Notably, no significant difference in chemotaxis was observed in MCF-7 NCS-1low, MB-231 NCS-1low, and MB-231 NCS-1high cell lines compared to mock-infected controls. (Figure 3E and 3F). To examine the effects of NCS-1 on cell-matrix adhesion we compared the ability of cells to bind to a Collagen IV matrix. Significant reductions in cell adherence to Collagen IV were observed in NCS-1high mutants compared to controls for both MCF-7 (fold change: 0.6 ± 0.06, p= 0.005, Figure 3G) and MB-231 cell lines (fold change: 0.7 ± 0.04 p= 0.007 Figure 3H). Conversely, an increase in adherence was observed in the MB-231 NCS-1low cell line (fold change: 1.2 ±0.03 p= 0.006 Figure 3H), whereas no significant change was observed in the MCF-7 NCS-1low line (Figure 3G). Results from in vitro assays are summarized in Supplementary Table 1 and representative images from each assay are highlighted in Supplementary Figures 2–4.
NCS-1 Increases the Efficacy of Paclitaxel without Altering Rates of Proliferation
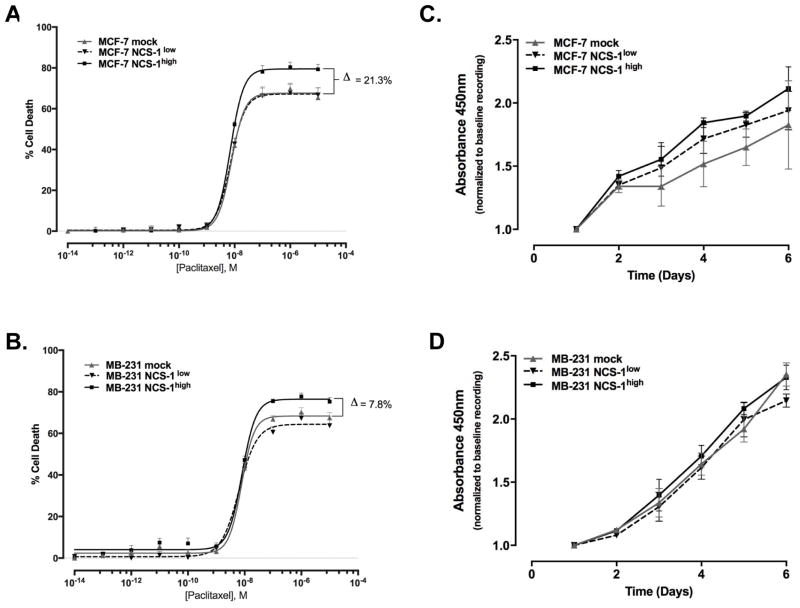
NCS-1 has recently been identified as a novel binding partner of the taxane, paclitaxel (30, 38). As paclitaxel is a standard part of most chemotherapeutic treatment strategies for breast cancer patients, it was of great interest to determine if NCS-1 could alter cellular response to paclitaxel in vitro. To test this we conducted dose response assays with paclitaxel. No significant change was observed in the LogIC50 of paclitaxel (nM) for either cell line (LogIC50 values for MCF-7 mock: −8.1 ± 0.04, MCF-7 NCS-1low: −8.1 ± 0.02, MCF-7 NCS-1high: −8.2 ± 0.01 Figure 4A and IC50 values for MB-231 mock: −8.1 ± 0.05, MB-231 NCS-1low: −8.2 ± 0.04, MB-231 NCS-1high: −8.1 ± 0.05 4B). Although the LogIC50 values did not differ, we did observe a difference in the maximal response to paclitaxel. In the MCF-7 NCS-1high cell line the maximal response, in terms of cell death, was 21% greater compared to mock and MCF-7 NCS-1low cell lines. The same trend was demonstrated in the MB-231 NCS-1high cell line, which had an 8% increase in maximum response compared to mock. Paclitaxel is reported to induce cell death through inhibition of cellular proliferation. Given the results of the dose response assay, the effect of NCS-1 expression on cellular proliferation was examined. Compared to mock-infected controls, no significant difference in proliferation rates were observed in either cell line (Figure 4C and 4D).
Figure 4. NCS-1 may Promote Paclitaxel-induced Cell Death.
Dose response curves were generated by CellTiterGlo assay after treatment with concentrations of paclitaxel (0.1 pM to 10,000 μM) for 72 hours for A. MCF-7 and B. MB-231 cell lines. The percentage of cell death was calculated using the equation previously described. Cell proliferation was determined by CCK-8 assay to generate a growth curve over six days for C. MCF-7 and D. MB-231 lines. The average absorbance values were calculated, normalized to baseline, and compared in a two-sided student’s t-test.
NCS-1 Predicts Survival, Recurrence, and Metastasis in Breast Cancer Patients
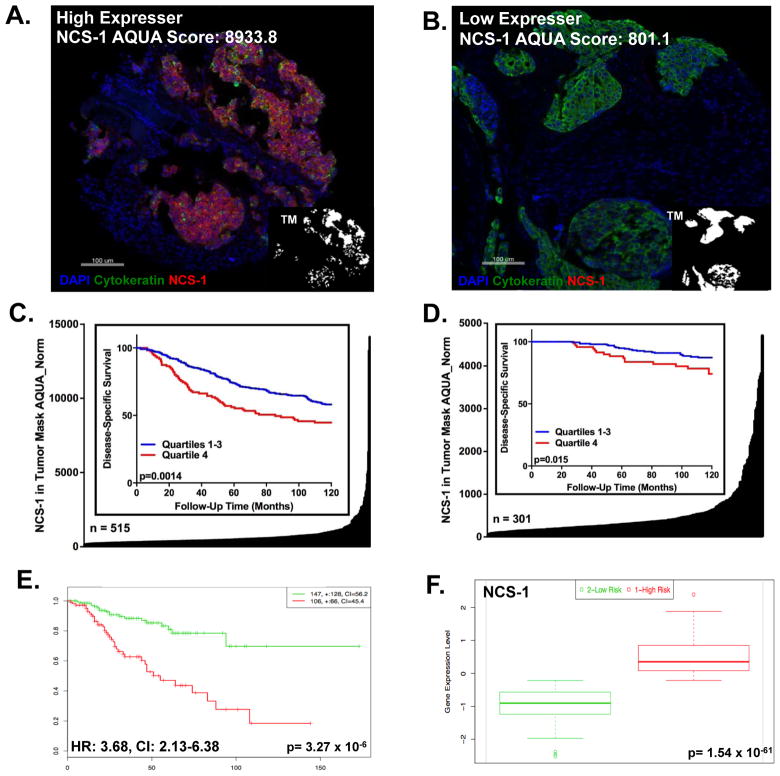
Given the association between NCS-1 expression and aggressive behavior in breast cancer cell lines, we assessed the expression of this protein among breast cancer patients. First, NCS-1 expression was measured in YTMA 49, a cohort consisting of 515 breast cancer cases serially collected from the 1962–1982. NCS-1 was then measured in YTMA 201, a cohort consisting of 301 breast cancer cases serially collected from 1976–2005. The expression of NCS-1 was measured by the AQUA method of quantitative immunofluorescence (QIF). Results from QIF revealed a small proportion of cases with very high expression of NCS-1 among many lower expressing cases (Figure 5A and B). The distribution of NCS-1 suggested that division of the cohort into quartiles, where patients in Quartiles 1–3 (Q1–3) have lower expression of NCS-1 and patients in Quartile 4 (Q4) have the highest expression of NCS-1, would provide the optimal cut-point. For YTMA 49, Kaplan Meier analysis revealed that patients belonging to Q4 (n= 112) had shorter disease-specific survival rates than patients with lower NCS-1 expression levels (Q1–3) (Figure 5C). This result was upheld in YTMA 201 (Figure 5D). Additionally, elevated NCS-1 was significantly associated with both local and distant disease recurrence (log rank p=0.009, data not shown). Using multivariable Cox proportional hazards regression model, risk estimates related to age, lymph node status, tumor size, ER, PR, HER-2 and NCS-1 status were estimated. In both cohorts, the hazard ratios (HR) for NCS-1 were statistically significant and indicative of increased odds of death by disease (Table 2).
Figure 5. High Expression of NCS-1 Predicts Poor Prognosis in Breast Cancer Patients.
Representative NCS-1 staining from a breast cancer patient who is a A. high NCS-1 expresser compared to a breast cancer patient who is a B. low NCS-1 expresser C. YTMA 49 (log rank p<0.001) and D. YTMA 201 (log rank p=0.015) Distribution plots showing AQUA scores for NCS-1 in tumor mask in the y-axis, for each patient within the cohort, x-axis. Insets: Kaplan Meier Curve showing AQUA score for NCS-1 that was stratified into quartiles: Q1–Q3, lower expressing and Q4, highest expressing. E. Kaplan Meier Analysis from online biomarker validation tool, SurvExpress, shows recurrence free survival among the low-risk (green) and high-risk (red) groups of the Prat-Perou GSE18229 cohort (HR: 3.68, CI: [2.13–6.38], p=3.27 x 10−6). F. Box Plot from SurvExpress showing the expression of NCS-1 among each risk group, low-risk (green) and high-risk (red).
Table 2.
Multivariate and Univariate Analysis for YTMA 49 and YTMA 201
| Multivariate | Univariate | Multivariate | Univariate | |||||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |
|
Characteristics Age at Diagnosis |
YTMA 49 | YTMA 201 | ||||||
|
| ||||||||
| <50 | 1 | 0.100 | 1 | 0.5013 | 1 | 0.407 | 1 | 0.609 |
| ≥50 | 1.373 (0.940–2.004) | 1.129(0.792–1.610) | 1.458(0.592–3.552) | 0.820(0.384–1.752) | ||||
|
| ||||||||
| Tumor Size | ||||||||
|
| ||||||||
| ≤2 cm | 1 | <0.0001 | 1 | <0.0001 | 1 | 0.030 | 1 | 0.005 |
| 2–5 cm | 3.680 (2.265–5.977) | 4.198(2.610–6.753) | 2.791 (1.103–7.061) | 3.022(1.401–6.522) | ||||
|
| ||||||||
| Nuclear Grade | ||||||||
|
| ||||||||
| Grade 1 | 1 | 0.267 | 1 | 0.0089 | ------ | ------ | ||
| Grade 2 | 1.213(0.863–1.705) | 0.642(0.461–0.895) | ||||||
|
| ||||||||
| Nodal Status | ||||||||
|
| ||||||||
| Negative | 1 | 0.0003 | 1 | <0.0001 | 1 | 0.006 | 1 | 0.002 |
| Positive | 1.967 (1.361–2.843) | 2.490 (1.764–35.16) | 3.456 (1.420–8.414) | 3.815(1.646–8.840) | ||||
|
| ||||||||
| ER Status | ||||||||
|
| ||||||||
| Negative | 1 | 0.075 | 1 | <0.0001 | 1 | 0.532 | 1 | 0.061 |
| Positive | 0.719 (0.501–1.033) | 0.536 (0.393–0.732) | 0.749 (0.303–1.852) | 0.492 (0.231–1.003) | ||||
|
| ||||||||
| PR Status | ||||||||
|
| ||||||||
| Negative | 1 | 0.220 | 1 | 0.0004 | 1 | 0.370 | 1 | 0.147 |
| Positive | 0.813 (0.584–1.132) | 0.565 (0.412–0.773) | 0.388 (0.049–3.083) | 0.228 (0.031–1.680) | ||||
|
| ||||||||
| HER2 Status | ||||||||
|
| ||||||||
| Negative | 1 | 0.104 | 1 | 0.1882 | 1 | 0.363 | 1 | 0.263 |
| Positive | 1.406 (0.932–2.121) | 1.297 (0.881–1.910) | 1.597 (0.582–4.379) | 1.737 (0.660–4.573) | ||||
|
| ||||||||
| NCS-1 Expression (Q1–3 vs. Q4) | ||||||||
|
| ||||||||
| Q1–3 | 1 | 0.020 | 1 | <0.0001 | 1 | 0.004 | 1 | 0.004 |
| Q4 | 1.532 (1.070–2.193) | 2.036 (1.472–2.815) | 3.677 (1.509–8.956) | 3.008 (1.400–6.327) | ||||
HR: Hazard Ratio, CI: Confidence Interval, ER: Estrogen Receptor, PR: Progesterone Receptor, HER2: Human Epidermal Growth Factor Receptor 2, NCS-1: Neuronal Calcium Sensor-1
To confirm the clinical association observed between NCS-1 protein expression and clinical outcome, we assessed the clinical utility of NCS-1 mRNA expression using 26 cohorts from the SurvExpress Bioinformatic database. Of these 26 cohorts, seven had survival annotations, 18 had recurrence-free survival annotations, and one had metastasis annotations. In 81% of these cohorts (21/26) we found that increased NCS-1 mRNA expression was significantly elevated in the higher-risk group. Additionally, high NCS-1 expression was predictive of poor clinical outcome in terms of survival, recurrence, and metastasis (Prat-Perou Breast Cohort, GSE18229, Figure 5E and F, additional Kaplan Meier Plots shown in Supplementary Figures 5A–Y).
DISCUSSION
Overall, our data is consistent with the hypothesis that NCS-1 expression is associated with tumor aggressiveness, taxane response, and poor clinical outcome in breast cancer patients. This is supported by cell biological and retrospective correlative studies. In vitro we found that overexpression of NCS-1 led to a marked increase in the number of invading and chemotactic cells, and a decrease in cell adhesion to Collagen IV. Invasion, chemotaxis, and adhesion are parameters associated with metastasis (44, 45). Alterations in these tumor phenotypes provides a rational explanation for the clinical association between NCS-1 and poor clinical outcome, which we observed in this study. Specifically, we found that high expression of NCS-1 was associated with shorter survival rates independent of ER, PR, HER-2, and lymph node status. Additionally, multivariate analysis showed the hazard ratios and log rank p-values of NCS-1 outperformed routinely used breast cancer biomarkers (ER, PR, and HER-2).
A provocative and paradoxical finding of this study was the observation that NCS-1 overexpression increased the efficacy of paclitaxel, without altering the IC50 of this drug. The primary mechanism of action of paclitaxel is reported to occur through direct binding to tubulin. This inhibits proliferation by preventing the disassembly of microtubules, thereby inducing cell death (46). However, no significant differences in proliferation rates were observed when NCS-1 expression was altered. Studies suggest that there are alternative and complementary mechanisms of action of paclitaxel (47–50). The recent identification of NCS-1 as a novel binding partner of the taxane, paclitaxel (30) coupled with its demonstrated impact on paclitaxel efficacy in vitro, suggests that NCS-1 may promote paclitaxel-induced cell death independent of microtubule stabilization. In sum, although patients who overexpress NCS-1 may have more aggressive tumors, they may also be more likely to respond to taxane-based therapies.
This study provides strong evidence in support of an association between NCS-1 expression and tumor aggressiveness; however, there are a number of limitations that should be considered when interpreting the findings presented. The first and most significant limitation is that tumor cell analyses were limited to two standard breast cancer cell lines. We believe that the results shown in these two cell lines are representative. However, future studies may expand upon these findings through the use of additional cell lines or in vivo mouse models. The second limitation of this study was that the patient cohorts used represent single institutional retrospective collections that were not uniformly treated. Thus, the predictive value of NCS-1 in breast cancer patients with respect to taxane therapy could not be assessed. Therefore, conclusions related to the value of NCS-1 as a predictive breast cancer biomarker of taxane response are limited, and require further validation.
Ultimately, our study shows that NCS-1 plays a role in the aggressive behavior of a subset of breast cancers and has therapeutic and biomarker potential. These findings are preliminary, but provocative, and suggest that NCS-1 could be used in a clinical setting as a prognostic biomarker to identify patients who have a greater risk of death by disease. Our study also indicates that NCS-1 may be a predictive biomarker of taxane response. Currently, there are no predictive biomarkers for taxanes. Given that taxane treatment can cause substantial morbidity and life-threatening toxicities (e.g. taxane-induced peripheral neuropathy (TIPN), neutropenia, and cardiac arrhythmia), the in vitro association we found between NCS-1 and taxanes is of great clinical interest. In conclusion, validating these observations and elucidating a molecular mechanism behind these findings may ultimately allow for the individualization of patient treatment by identifying patients who have a greater risk of death by disease, who may also benefit from taxane-based therapies.
Supplementary Material
Implications.
NCS-1, a calcium binding protein, is associated with clinicopathological features of aggressiveness in breast cancer cells and worse outcome in two breast cancer patient cohorts.
Acknowledgments
Special thanks to Lori Charette and Yale Tissue Pathology Services for providing the TMAs used within this study. This research was supported in part by an NRSA Pre-doctoral Fellowship (F31 CA183571-01), the Breast Cancer Research Foundation, and NIH training grant at Yale University (2T32GM007324-36A1).
Footnotes
DISCLOSURE/DUALITY OF INTEREST
Dr. David L. Rimm has served as a consultant to Biocept, BMS, Cernostics, Clearsight, FivePrime, Genoptix/Novartis, Cell Signaling Technology, Ultivue, and Perkin Elmer. Cepheid, Genoptix, Gilead Sciences, Pierre Fabre and OncoplexDx fund research in Dr. Rimm’s lab. The other authors have no conflicts of interest to disclose.
References
- 1.Lehnert M. Chemotherapy resistance in breast cancer. Anticancer Res. 1998;18(3C):2225–6. [PubMed] [Google Scholar]
- 2.Berridge MJ. Calcium signalling and cell proliferation. Bioessays. 1995;17(6):491–500. doi: 10.1002/bies.950170605. [DOI] [PubMed] [Google Scholar]
- 3.Prevarskaya N, Skryma R, Shuba Y. Calcium in tumour metastasis: new roles for known actors. Nat Rev Cancer. 2011;11(8):609–18. doi: 10.1038/nrc3105. [DOI] [PubMed] [Google Scholar]
- 4.Minton K. Cell migration: Coordinating calcium signalling. Nat Rev Mol Cell Biol. 2014;15(3):152. doi: 10.1038/nrm3761. [DOI] [PubMed] [Google Scholar]
- 5.Chen YF, et al. Remodeling of calcium signaling in tumor progression. J Biomed Sci. 2013;20:23. doi: 10.1186/1423-0127-20-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, et al. S100A6 as a potential serum prognostic biomarker and therapeutic target in gastric cancer. Dig Dis Sci. 2014;59(9):2136–44. doi: 10.1007/s10620-014-3137-z. [DOI] [PubMed] [Google Scholar]
- 7.Duan L, et al. S100A6 stimulates proliferation and migration of colorectal carcinoma cells through activation of the MAPK pathways. Int J Oncol. 2014;44(3):781–90. doi: 10.3892/ijo.2013.2231. [DOI] [PubMed] [Google Scholar]
- 8.Leclerc E, Heizmann CW. The importance of Ca2+/Zn2+ signaling S100 proteins and RAGE in translational medicine. Front Biosci (Schol Ed) 2011;3:1232–62. doi: 10.2741/223. [DOI] [PubMed] [Google Scholar]
- 9.Donato R, et al. Functions of S100 proteins. Curr Mol Med. 2013;13(1):24–57. [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmussen CD, Means AR. Calmodulin is involved in regulation of cell proliferation. EMBO J. 1987;6(13):3961–8. doi: 10.1002/j.1460-2075.1987.tb02738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coticchia CM, et al. Calmodulin modulates Akt activity in human breast cancer cell lines. Breast Cancer Res Treat. 2009;115(3):545–60. doi: 10.1007/s10549-008-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quang CT, et al. The calcineurin/NFAT pathway is activated in diagnostic breast cancer cases and is essential to survival and metastasis of mammary cancer cells. Cell Death Dis. 2015;6:e1658. doi: 10.1038/cddis.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, et al. Calcineurin promotes proliferation, migration, and invasion of small cell lung cancer. Tumour Biol. 2010;31(3):199–207. doi: 10.1007/s13277-010-0031-y. [DOI] [PubMed] [Google Scholar]
- 14.Schaad NC, et al. Direct modulation of calmodulin targets by the neuronal calcium sensor NCS-1. Proc Natl Acad Sci U S A. 1996;93(17):9253–8. doi: 10.1073/pnas.93.17.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss JL, Hui H, Burgoyne RD. Neuronal calcium sensor-1 regulation of calcium channels, secretion, and neuronal outgrowth. Cell Mol Neurobiol. 2010;30(8):1283–92. doi: 10.1007/s10571-010-9588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan J, et al. Modulation of CaV2.1 channels by neuronal calcium sensor-1 induces short-term synaptic facilitation. Mol Cell Neurosci. 2014;63:124–31. doi: 10.1016/j.mcn.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dragicevic E, et al. Cav1.3 channels control D2-autoreceptor responses via NCS-1 in substantia nigra dopamine neurons. Brain. 2014;137(Pt 8):2287–302. doi: 10.1093/brain/awu131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakao S, Wakabayashi S, Nakamura TY. Stimulus-dependent regulation of nuclear Ca2+ signaling in cardiomyocytes: a role of neuronal calcium sensor-1. PLoS One. 2015;10(4):e0125050. doi: 10.1371/journal.pone.0125050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koizumi S, et al. Mechanisms underlying the neuronal calcium sensor-1-evoked enhancement of exocytosis in PC12 cells. J Biol Chem. 2002;277(33):30315–24. doi: 10.1074/jbc.M201132200. [DOI] [PubMed] [Google Scholar]
- 20.Kapp-Barnea Y, et al. Neuronal calcium sensor-1 and phosphatidylinositol 4-kinase beta regulate IgE receptor-triggered exocytosis in cultured mast cells. J Immunol. 2003;171(10):5320–7. doi: 10.4049/jimmunol.171.10.5320. [DOI] [PubMed] [Google Scholar]
- 21.Zheng Q, et al. Neuronal calcium sensor-1 facilitates neuronal exocytosis through phosphatidylinositol 4-kinase. J Neurochem. 2005;92(3):442–51. doi: 10.1111/j.1471-4159.2004.02897.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X, et al. Interaction of neuronal calcium sensor-1 (NCS-1) with phosphatidylinositol 4-kinase beta stimulates lipid kinase activity and affects membrane trafficking in COS-7 cells. J Biol Chem. 2001;276(43):40183–9. doi: 10.1074/jbc.M104048200. [DOI] [PubMed] [Google Scholar]
- 23.Haynes LP, Thomas GM, Burgoyne RD. Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase beta and trans-Golgi network-plasma membrane traffic. J Biol Chem. 2005;280(7):6047–54. doi: 10.1074/jbc.M413090200. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura TY, et al. Novel role of neuronal Ca2+ sensor-1 as a survival factor up-regulated in injured neurons. J Cell Biol. 2006;172(7):1081–91. doi: 10.1083/jcb.200508156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haynes LP, et al. Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin delta. Proteomics. 2006;6(6):1822–32. doi: 10.1002/pmic.200500489. [DOI] [PubMed] [Google Scholar]
- 26.Pandalaneni S, et al. Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-protein-coupled Receptor Kinase 1 (GRK1) Peptides Using Different Modes of Interactions. J Biol Chem. 2015;290(30):18744–56. doi: 10.1074/jbc.M114.627059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlecker C, et al. Neuronal calcium sensor-1 enhancement of InsP3 receptor activity is inhibited by therapeutic levels of lithium. J Clin Invest. 2006;116(6):1668–74. doi: 10.1172/JCI22466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haynes LP, et al. Specificity, promiscuity and localization of ARF protein interactions with NCS-1 and phosphatidylinositol-4 kinase-III beta. Traffic. 2007;8(8):1080–92. doi: 10.1111/j.1600-0854.2007.00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taverna E, et al. Neuronal calcium sensor 1 and phosphatidylinositol 4-OH kinase beta interact in neuronal cells and are translocated to membranes during nucleotide-evoked exocytosis. J Cell Sci. 2002;115(Pt 20):3909–22. doi: 10.1242/jcs.00072. [DOI] [PubMed] [Google Scholar]
- 30.Boehmerle W, et al. Paclitaxel induces calcium oscillations via an inositol 1,4,5-trisphosphate receptor and neuronal calcium sensor 1-dependent mechanism. Proc Natl Acad Sci U S A. 2006;103(48):18356–61. doi: 10.1073/pnas.0607240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boehmerle W, et al. Chronic exposure to paclitaxel diminishes phosphoinositide signaling by calpain-mediated neuronal calcium sensor-1 degradation. Proc Natl Acad Sci U S A. 2007;104(26):11103–8. doi: 10.1073/pnas.0701546104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K, et al. Paclitaxel accelerates spontaneous calcium oscillations in cardiomyocytes by interacting with NCS-1 and the InsP3R. J Mol Cell Cardiol. 2010;49(5):829–35. doi: 10.1016/j.yjmcc.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo M, et al. Prevention of paclitaxel-induced peripheral neuropathy by lithium pretreatment. FASEB J. 2012;26(11):4696–709. doi: 10.1096/fj.12-214643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss JL, Archer DA, Burgoyne RD. Neuronal Ca2+ sensor-1/frequenin functions in an autocrine pathway regulating Ca2+ channels in bovine adrenal chromaffin cells. J Biol Chem. 2000;275(51):40082–7. doi: 10.1074/jbc.M008603200. [DOI] [PubMed] [Google Scholar]
- 35.Blasiole B, et al. Neuronal calcium sensor-1 gene ncs-1a is essential for semicircular canal formation in zebrafish inner ear. J Neurobiol. 2005;64(3):285–97. doi: 10.1002/neu.20138. [DOI] [PubMed] [Google Scholar]
- 36.Gromada J, et al. Neuronal calcium sensor-1 potentiates glucose-dependent exocytosis in pancreatic beta cells through activation of phosphatidylinositol 4-kinase beta. Proc Natl Acad Sci U S A. 2005;102(29):10303–8. doi: 10.1073/pnas.0504487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fresno Vara JA, et al. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30(2):193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Benbow JH, DeGray B, Ehrlich BE. Protection of neuronal calcium sensor 1 protein in cells treated with paclitaxel. J Biol Chem. 2011;286(40):34575–82. doi: 10.1074/jbc.M111.265751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bordeaux J, et al. Antibody validation. Biotechniques. 2010;48(3):197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geback T, et al. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques. 2009;46(4):265–74. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- 41.Welsh AW, et al. Quantitative analysis of estrogen receptor expression shows SP1 antibody is more sensitive than 1D5. Appl Immunohistochem Mol Morphol. 2013;21(2):139–47. doi: 10.1097/PAI.0b013e31825d73b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–7. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 43.Aguirre-Gamboa R, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8(9):e74250. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hazan RB, et al. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol. 2000;148(4):779–90. doi: 10.1083/jcb.148.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 46.Horwitz SB. Taxol (paclitaxel): mechanisms of action. Ann Oncol. 1994;5(Suppl 6):S3–6. [PubMed] [Google Scholar]
- 47.Huang Y, et al. The possible correlation between activation of NF-kappaB/IkappaB pathway and the susceptibility of tumor cells to paclitaxel-induced apoptosis. Oncol Res. 2002;13(2):113–22. [PubMed] [Google Scholar]
- 48.Dziadyk JM, et al. Paclitaxel-induced apoptosis may occur without a prior G2/M-phase arrest. Anticancer Res. 2004;24(1):27–36. [PubMed] [Google Scholar]
- 49.Chen TS, et al. Taxol induces caspase-independent cytoplasmic vacuolization and cell death through endoplasmic reticulum (ER) swelling in ASTC-a-1 cells. Cancer Lett. 2008;270(1):164–72. doi: 10.1016/j.canlet.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 50.Liao PC, Lieu CH. Cell cycle specific induction of apoptosis and necrosis by paclitaxel in the leukemic U937 cells. Life Sci. 2005;76(14):1623–39. doi: 10.1016/j.lfs.2004.09.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.