Abstract
[FeFe]-hydrogenases are nature’s most prolific hydrogen catalysts, excelling at facilely interconverting H2 and protons. The catalytic core common to all [FeFe]-hydrogenases is a complex metallocofactor, referred to as the H-cluster, which is composed of a standard [4Fe-4S] cluster linked through a bridging thiolate to a 2Fe subcluster harboring dithiomethylamine, carbon monoxide, and cyanide ligands. This 2Fe subcluster is synthesized and inserted into [FeFe]-hydrogenase by three maturase enzymes denoted HydE, HydF, and HydG. HydE and HydG are radical S-adenosylmethionine enzymes and synthesize the nonprotein ligands of the H-cluster. HydF is a GTPase that functions as a scaffold or carrier for 2Fe subcluster production. Herein, we utilize UV–visible, circular dichroism, and electron paramagnetic resonance spectroscopic studies to establish the existence of redox active [4Fe-4S] and [2Fe-2S] clusters bound to HydF. We have used spectroelectrochemical titrations to assign iron-sulfur cluster midpoint potentials, have shown that HydF purifies with a reduced [2Fe-2S] cluster in the absence of exogenous reducing agents, and have tracked iron–sulfur cluster spectroscopic changes with quaternary structural perturbations. Our results provide an important foundation for understanding the maturation process by defining the iron-sulfur cluster content of HydF prior to its interaction with HydE and HydG. We speculate that the [2Fe-2S] cluster of HydF either acts as a placeholder for HydG-derived Fe(CO)2CN species or serves as a scaffold for 2Fe subcluster assembly.
Graphical abstract
Biological H2 metabolism is primarily accomplished by [FeFe]- and [NiFe]-hydrogenase, two evolutionarily unrelated enzyme families that have distinct active site metal clusters but similar iron-bound carbon monoxide (CO) and cyanide (CN−) ligands.1 [FeFe]-hydrogenases exhibit a lower tolerance for O2 than [NiFe]-hydrogenases, but the former generally catalyze the evolution of H2(g) more readily with turnover rates of 104 events per second.2–6 The complex metallocofactor at the active site of the [FeFe]-hydrogenases (HydA) is referred to as the H-cluster and is composed of a [4Fe-4S] cubane linked through a bridging cysteine thiolate to a diiron subcluster, with the remaining ligands to the latter subcluster comprising three CO, two CN−, and a bridging dithiomethylamine (DTMA) (Figure 1).7–9
Figure 1.
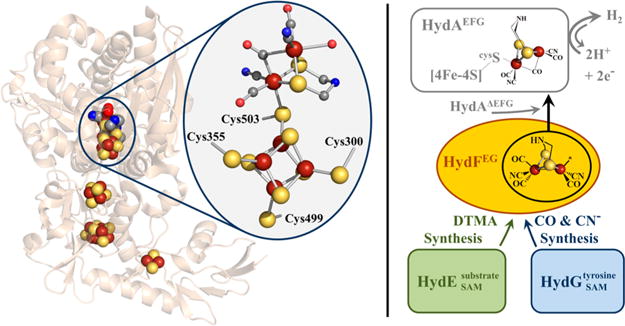
Left panel. X-ray crystal structure of [FeFe]-hydrogenase from Clostridium pasteurianum I (CpI) (PDB: 3C8Y). The H-cluster is highlighted within the oval. The H-cluster and accessory FeS clusters are depicted as spheres. Color scheme is as follows: iron, rust; sulfur, yellow; carbon, gray; oxygen, red; nitrogen, blue. Right panel. Hypothetical maturation scheme for 2Fe subcluster biosynthesis (see main text for additional details).
H-cluster biosynthesis is achieved by HydE, HydF, and HydG; the coexpression of all three proteins from Clostridium acetobutylicum (C.a.) in Escherichia coli (E. coli) was demonstrated to be an essential requirement to achieve the heterologous expression of active HydA.10–12 HydE and HydG belong to the radical S-adenosylmethionine (SAM) enzyme superfamily.10,13 While it was long presumed that HydE plays an essential role in H-cluster maturation,10,11 insight into its putative substrate and reaction mechanism has been provided only recently. We demonstrated that select compounds containing a thiol functional group exerted significant effects on the level of deuterium atom incorporation from D2O into 5′-deoxyadenosine.14 This observation led to the hypothesis that HydE catalyzes Cα–Cβ bond cleavage of two thiolcontaining molecules to generate two thioformaldehyde species that condense with ammonia to yield the DTMA bridge (Figure 1);14 this chemistry is analogous to in vitro DTMA synthesis in which formaldehyde and ammonia condense on an Fe2(SH)2(CO)6 scaffold.15 Biochemical characterization of HydG has offered direct insight into the role of this protein during H-cluster biosynthesis, as it utilizes tyrosine as substrate to form p-cresol16 along with the diatomic species CN−17 and CO (Figure 1).18 While the initial H atom abstraction from and cleavage of tyrosine to produce p-cresol occurs near the radical SAM [4Fe-4S] cluster in the N-terminal region of the protein, the subsequent chemical steps to produce CO and CN− appear to occur at or near a second iron-sulfur cluster in the C-terminal region of HydG, although the mechanistic aspects of this process require additional clarification.19–27
Sequence annotation of HydF shows the presence of Walker A P-loop and Walker B Mg2+ binding motifs associated with small RAS GTPases, as well as a putative FeS cluster-binding C-terminal CXHX46–53CXXC motif; both of these motifs were shown to be essential for achieving active HydA.11 The GTP hydrolysis functionality of HydF has been proposed to act in gating protein–protein interactions between the maturase proteins during H-cluster precursor assembly.21,28,29 A report on Thermotoga maritima (T.m.) HydF demonstrated coordination of a [4Fe-4S]+ cluster and GTP hydrolysis.30 Our studies have revealed that the heterologous coexpression of C.a. HydF with HydE and HydG (HydF) results in FTIR bands consistent with Fe—CO and Fe—CN− species bound to the purified protein,28 a result corroborated by similar studies on homologously overexpressed C.a. HydFEG.31 Significantly, our work has shown that HydFEG is capable of in vitro HydA activation when the latter is expressed in the absence of HydE, HydF, and HydG (HydAΔEFG), suggesting that HydF acts as a scaffold or carrier protein during H-cluster assembly (Figure 1).28,32,33 Clarification of HydF’s role as either a scaffold (wherein the 2Fe subcluster is synthesized on HydF) or carrier (wherein HydF accepts a preassembled 2Fe subcluster) requires additional experimentation. Supporting this scaffold/carrier hypothesis, purified HydF can be loaded with synthetic 2Fe subcluster analogues, which can then be transferred to HydAΔEFG.34,35
When C.a. HydF is expressed in the absence of HydE and HydG (HydFΔEG), binding of both [2Fe-2S] and [4Fe-4S] clusters has been suggested based on UV–visible absorbance32 and EPR spectra of the enzyme in as-isolated and reduced preparations.28 The [2Fe-2S] cluster (g = 2.00, 1.96) was suggested as a possible site for delivery and coordination of the nonprotein ligands synthesized by HydE and HydG during the assembly of the 2Fe subcluster of the H-cluster.28 The existence of a [2Fe-2S] cluster was challenged in a subsequent report on Shewanella oneidensis (S.o.) HydFΔEG, where a slow-relaxing g ≈ 2.0 species that disappeared upon reduction was instead attributed to a [3Fe-4S]+ cluster.36 In EPR studies performed with Thermotoga neapolitana (T.n.) and C.a. HydFΔEG, dithionite (DT) reduced enzyme preparations showed an axial [4Fe-4S]+ cluster signal with an overlapping g ≈ 2.0 feature; while the origin of the overlapping g ≈ 2.0 signal was not examined, it was presumed to arise from a protein-derived radical species resulting from treatment with DT.37,38
Here we report extensive spectroscopic (UV–vis, circular dichroism, EPR) analysis of freshly prepared HydF under both nonreducing and reducing conditions, and assign midpoint potentials to the FeS clusters. We also demonstrate the effects of sample handling (freeze/thaw, dialysis, concentration) on FeS cluster spectroscopic features and changes to the quaternary structure during these processes. The results have provided a more complete picture of the FeS cluster states associated with HydF prior to “loading” by HydE and HydG, and demonstrate the existence of a redox active [2Fe-2S] cluster bound to this protein. These results are significant given the likely mechanistic relevance of the iron–sulfur clusters bound to HydF and provide a foundation for understanding the pathway by which the loaded HydFEG is generated.
EXPERIMENTAL PROCEDURES
HydF Protein Expression and Purification
Constructs encoding Clostridium acetobutylicum hydF were transformed into E. coli BL21(DE3) (Stratagene) cells as previously described.32 Fresh plates were streaked and single colonies were chosen for small scale overnight culture growth in media supplemented with 30 μg/mL kanamycin. The following morning the seed cultures were used to inoculate 9 L of media which comprised 10 g/L tryptone, 5 g/L yeast extract, 5 g/L KCl, 5 g/L glucose, 50 mM potassium phosphate buffer pH 7.5, and 30 μg/mL kanamycin. Cell cultures were grown at 37 °C and 230 rpm shaking until OD600 values reached 0.5–0.6. The culture was then induced with IPTG (1 mM final concentration) and supplemented with ferrous ammonium sulfate (FAS, 0.16 mM final concentration). Cultures continued to shake at 230 rpm for 2.5 h at 37 °C and then were removed from the incubator and allowed to cool to room temperature. An additional FAS aliquot was added (0.16 mM final concentration), and the flasks were then transferred to a 4 °C refrigerator and sparged with N2(g) for ~15 h. Cells were harvested by centrifugation, and resulting cell pellets were immediately flash frozen in liquid N2. Composite cell pellet mass was recorded, and cells were stored at −80 °C until further use.
Cell lysis and protein purification were performed with minor modifications to previous methodologies.28 All techniques were accomplished under anaerobic conditions in a Coy anaerobic chamber (Grass Lake, MI) maintained with a 96% N2(g), 4% H2(g) atmosphere that was operated in a 4 °C walk-in refrigerator. Cell pellets were transferred to a Coy anaerobic chamber in a frozen state and were then thawed and resuspended in a lysis buffer containing 50 mM HEPES pH 7.4, 0.3 M KCl, 5% glycerol, and 10 mM imidazole (Buffer A). The cell lysis mixture was supplemented with 10 mM MgCl2, 1 mM PMSF, 0.8% Triton X-100, 0.07 mg DNase, and RNase per gram cell, and ~0.4 mg of lysozyme per gram cell. This mixture was then stirred for 70 min, loaded into gastight centrifuge bottles, and centrifuged at 18 000 rpm for 30 min. Then the supernatant was loaded onto a 5 mL HisTrap Ni2+-affinity column (GE Healthcare) using an ÄKTA Basic 100 FPLC (GE Healthcare). The column was pre-equilibrated in Buffer A, and following supernatant loading was washed with 15 column volumes of Buffer A or until baseline absorbance resumed. HydF elution was accomplished by using a step gradient of Buffer B (50 mM HEPES, pH 7.4, 0.3 M KCl, 5% glycerol, 500 mM imidazole). Absorbance intensities at both 280 and 415 nm were tracked, and the ratios revealed that substantially pure HydF eluted at both 20% and 50% Buffer B wash steps. In most cases, only the peak protein fraction (as determined by the fraction with the highest 415 to 280 nm ratio) eluting at 50% Buffer B was utilized for experiments. The protein eluting at 50% Buffer B with a buffer composition of 50 mM HEPES, pH 7.4, 0.3 M KCl, 5% glycerol, and 255 mM imidazole is defined as the “freshly purified” state. Purification fractions that were pooled together and either dialyzed or buffer exchanged into a 50 mM HEPES, pH 7.4, 0.3 M KCl, 5% glycerol buffer, prior to concentration with a Prochem BJP 10/40 (or EMD Millipore Minicon B15) static protein concentrator, will heretofore be referred to as the “as-isolated” state.
Freshly purified HydF was routinely spectroscopically analyzed via UV–vis and CD techniques immediately after elution from the HisTrap column following the procedures outlined in sections below. Aliquots from the parent fraction were taken, and while spectroscopic data collection was occurring, fractions would be simultaneously run over a gel filtration column. Immediately following spectral collection, samples were transferred into the MBraun chamber for preparation of EPR samples. Remnant aliquots of the freshly purified enzyme were flash frozen in liquid N2 and stored at −80 °C until further use.
Chemical reconstitution of HydFΔEG samples with Na2S and FeCl3 was carried out with minor modifications to our previously described protocol.28 Briefly, HydF samples (2.24 ± 0.49 Fe/dimer) were supplemented with 5 mM dithiothreitol (DTT), followed by a 6-fold excess of FeCl3 and Na2S·9H2O and allowed to incubate under stirring for 2.5 h. At this time, samples underwent centrifugation to remove bulk FeS particulates, and the clarified supernatant was then buffer exchanged using a Sephadex G25 resin column. The reconstitution process was monitored via UV–vis absorbance spectroscopy of HydF before and after treatment with iron and sulfide.
Protein concentration was calculated via Bradford assay using a bovine serum albumin standard solution (Thermo Scientific). All HydF protein concentration values reported herein are based on the dimeric content of samples; HydE and pyruvate formate lyase activating enzyme (PFL-AE) concentration values are based on the monomeric content of samples. Iron content was determined either through using the spectrophotometric method of Fish, wherein protein is digested with 4.5% (w/v) KMnO4 and 1.2 N HCl and soluble iron is then complexed with ferrozine, or via a Varian SpectrAA 220 FS flame atomic absorption spectrometer.
HydE and PFL-AE Protein Expression and Purification
HydE was purified and chemically reconstituted with Na2S and FeCl3 as previously published.14 HydE samples were made up in a 25 mM HEPES, pH 8.0, 0.5 M KCl, 5% glycerol buffer. PFL-AE was purified as previously described, and samples were prepared in 50 mM Tris, 200 mM NaCl, pH 7.5 buffer.40,41
UV-Visible Spectroscopic Analysis
Samples analyzed by UV–vis spectroscopy were loaded into a 1.4 mL anaerobic cuvette (Spectrocell Inc.) within either an MBraun glovebox or a Coy anaerobic chamber. A Cary 6000i UV/vis/near-IR spectrophotometer (Varian) was used to acquire spectra at ambient temperatures at a scan rate of 600 nm/min. All experimental spectra were graphed in OriginPro 8.5 (OriginLab Corp. Northampton, MA, USA).
Circular Dichroism Spectroscopy
AJasco-710 spectropolarimeter was utilized to collect CD spectra on HydF samples. Samples were loaded into a 650 μL, 1 cm path length anaerobic cuvette (Hellma Analytics). Spectra were recorded from 300–700 nm at a sensitivity of 100 millidegrees, data pitch of 0.1 nm, continuous scanning mode at a 100 nm/min scanning speed, and 10 spectral accumulations. Baseline spectra of the background buffer (50 mM HEPES, pH 7.4, 0.3 M KCl, 5% glycerol, and 255 mM imidazole) were collected and subtracted from the experimental spectra using OriginPro 8.5 software. All data were collected under ambient temperature and anaerobic conditions.
Gel Filtration
Freshly purified and as-isolated C.a. HydF samples were gel-filtered via Superose 12 (GE Healthcare) size-exclusion media packed in an HR 10/30 column (1 cm i.d., 30 cm length; GE Healthcare) housed within an anaerobic vinyl Coy chamber at room temperature. A mobile phase of 50 mM HEPES pH 7.4, 0.3 M KCl, and 5% glycerol was utilized at a flow rate of 0.2 mL/min using an ÄKTA Purifier FPLC (GE Healthcare); column wash steps comprised at least five column volumes of buffer equilibration and at least two column volumes of wash between samples. Freshly purified samples were injected directly onto the gel filtration column within minutes of elution from the HisTrap column. As-isolated samples were quickly thawed in the Coy chamber and then immediately loaded onto the column. Sample injections were typically performed in duplicate. A BioRad Standard (#151-1901) that contained thyroglobin (bovine), γ-globulin (bovine), ovalbumin (chicken), myoglobin (horse), and vitamin B12 was used to calibrate sample oligomeric content. HydF tetrameric (~189 kDa) and dimeric (~94.5 kDa) species eluted with retention volumes of ~9.5 mL and ~10.5 mL, respectively.
Electron Paramagnetic Resonance (EPR) Sample Preparation and Data Collection
EPR samples were prepared in an MBraun box at O2 levels ≤1 ppm. Freshly purified samples, in either an as-eluted or photoreduced state, were loaded into EPR tubes (Wilmad LabGlass, 4 mm OD, NJ, USA), capped with a rubber septum and flash frozen outside the box in either liquid N2 or liquid propane. Photoreduced samples were prepared by supplementing enzyme with 100 μM 5-deazariboflavin and 5 mM DTT in 50 mM Tris, pH 7.4 buffer. Samples were illuminated with a 300 W Xe lamp in an ice water bath for 1 h, then immediately removed from the MBraun box and flash frozen in the same manner as described above. All samples were stored in a liquid N2 dewar until data collection occurred.
Low temperature (≤70 K) EPR measurements were made using either a Bruker (Billerica, MA) EMX X-band (9.4 GHz) spectrometer equipped with a 4119HS resonator, ESR900 liquid helium cryostat, and temperature controller (ITC503) from Oxford instruments, or a Bruker Cold Edge (Sumitomo Cryogenics) 10 K waveguide cryogen free system with a Mercury iTC controller unit. Unless otherwise noted, typical EPR parameters were 9.38 GHz microwave frequency, 1 mW microwave power, 100 kHz modulation frequency, 10 G modulation amplitude, time constant settings that varied between 20.48 and 163.84 ms, and spectra were averaged over either four or eight scans. OriginPro 8.5 was utilized to baseline correct and plot all experimental spectra. Simulations of experimental data were carried out using the EasySpin software platform,42 and the resulting g-values are presented in the text and Supporting Information. For determination of spin concentration, double integrations were performed using a standard sample of PFL-AE (200 μM protein with 3.75 ± 0.08 Fe/protein) that was photoreduced for 1 h in the presence of 5-deazariboflavin to generate the [4Fe-4S]+ state.41 EPR spectra for standards were collected under identical temperature and spectrometer settings as HydF samples.
Temperature Relaxation and Power Saturation Profiles
For select samples of freshly purified, as-isolated, and photoreduced (both freshly purified and as-isolated) HydF, and as-isolated HydE and PFL-AE, spectra were collected under identical parameter settings at various temperatures. This allowed for direct comparison of Topt values (defined as the optimum temperature setting for maximum signal intensity) for [4Fe-4S]+, [3Fe-4S]+, and [2Fe-2S]+ cluster signals among the different samples. In order to determine the power for half saturation (P1/2) for [4Fe-4S]+, [3Fe-4S]+, and [2Fe-2S] + cluster signals, power dependence studies were performed on samples at constant temperatures by varying the microwave power attenuation. Data collected at temperatures ≤15 K allowed for P1/2 determination for all three cluster types ([4Fe-4S]+, [3Fe-4S]+, and [2Fe-2S]+).43 Data were also collected at 30 K because this is near Topt for [2Fe-2S]+ cluster signals; at this temperature [4Fe-4S]+ and [3Fe-4S]+ cluster signals exhibited substantial line broadening and thus did not measurably interfere with [2Fe-2S]+ cluster signals.43
Power saturation curves were constructed by plotting the log (S/√P) against log P, where P is the microwave power and S is the peak-to-peak derivative signal amplitude (peak height); the most intense features were chosen for measuring S. In the case of PFL-AE, the peak-to-peak height of the g ≈ 2.030, 2.009, 1.988 feature was chosen for the [3Fe-4S]+ signal (12 K), and the peak-to-peak height of the g ≈ 2.005 centered feature was chosen for the [2Fe-2S]+ signal (30 K) (Table S1). For HydE FeS cluster signals at 15 K, only the features at magnetic field values of ~3305 G ([3Fe-4S]+) and ~3332 G ([2Fe-2S]+) were plotted due to the overlap between these signals. The peak-to-peak height of the g ≈ 1.88 centered feature at 15 K (representing the [4Fe-4S]+ cluster signal) was plotted for DT reduced HydE.14 The peak-to-peak height of the g ≈ 2.006 centered feature was chosen for the [2Fe-2S]+ signal in HydE at 30 K (Table S1). For HydF FeS cluster signals at ≤15 K, only the features at magnetic field values of ~3315 G ([3Fe-4S]+) and ~3334 G ([2Fe-2S]+) were plotted due to the overlap between these signals. The peak-to-peak height of the g ≈ 1.89 centered feature at 13 K (representing the [4Fe-4S]+ cluster signal) was plotted for photoreduced HydF.28,44 The peak to peak height of the g ≈ 2.002 centered feature was chosen for the [2Fe-2S]+ signal in HydF at 30 K (Table S1). In plots of log (S/√P) versus log P, FeS signals that do not exhibit power saturation effects result in lines that are parallel to the abscissa, while FeS signals that exhibit power saturation effects have lines that slope toward the abscissa with increasing power.43 P1/2 values along with the inhomogeneity parameter (b) were both calculated from fits to lines using a form of the equation S = √P/(1+P/P1/2)0.5b.43 All curve fitting was performed in OriginPro 8.5.
Midpoint Potentiometric Titrations
The midpoint potentials of the FeS clusters associated with HydF were determined by titration experiments on freshly purified enzyme according to published protocols.45,46 Two titration experiments were performed using 121 μM protein at 2.14 ± 0.06 Fe/dimer and 132 μM protein at 2.28 ± 0.12 Fe/dimer, respectively. Mediated potentiometric titrations on the freshly purified HydF were carried out in an MBraun anaerobic chamber. The solution potential was continuously monitored using a Fisher Scientific Accumet Basic pH/mV meter coupled to a Ag/AgCl platinum ORP electrode (Orion, Thermo Scientific) filled with 4 M KCl; electrode calibration was checked against an Orion 967901 ORP standard solution. All Ag/AgCl values are adjusted to the standard hydrogen electrode scale by the addition of 200 mV. To facilitate the redox equilibrium, the following mediator cocktail was employed: indigo disulfonate (Em = −125 mV), phenosafranine (Em = −255 mV), benzyl viologen (Em = −361 mV), and methyl viologen (Em = −440 mV). The mediator cocktail was added to freshly purified HydF to a final concentration of 20–25 μM; the solution was buffered against a 50 mM HEPES, pH 7.4, 0.3 M KCl, 5% glycerol (w/v) solution and was kept under constant stirring for the duration of the titration experiment. Stock solutions of NaDT at 2, 4, 8, and 10 mM were prepared in 50 mM HEPES, pH 7.4, 0.3 M KCl, 5% glycerol (w/v) and were subsequently used to poise the solution potential. Following DT additions, the solution potential was allowed to reach equilibration and at appropriate intervals, 200 μL of the solution was removed and immediately added to an EPR tube, which was capped, removed from the MBraun, and flash frozen in a liquid propane bath in ≤30 s. EPR samples were subsequently stored in a liquid N2 dewar.
Peak-to-peak signal amplitudes for the g ≈ 2.0 ([2Fe-2S]+) and g ≈ 1.89 ([4Fe-4S]+) cluster signals were plotted versus solution potential. The [4Fe-4S] + midpoint potential was determined by fitting the data to the Nernst equation, wherein E is the system potential and Em is the midpoint reduction potential for a n = 1 reduced iron sulfur cluster.47 All data fitting was carried out in OriginPro 8.5.
Dithiothreitol versus Dithionite as Reducing Agent
In order to examine the effects of different reducing agents on the HydF FeS cluster signals, experiments were conducted wherein protein was either supplemented with (i) buffer alone, (ii) buffer containing DTT (5 mM final concentration), or (iii) buffer containing DT (5 mM final concentration). Final sample volumes were kept identical (250 μL) in order to directly compare signal intensities. Following mixing, samples were allowed to sit for a 10 min incubation period before being transferred to EPR tubes, capped, removed from the MBraun anaerobic chamber, and flash frozen in liquid N2.
RESULTS
HydF serves as a scaffold for assembly of the 2Fe subcluster of the H-cluster, as evidenced by the ability of HydFEG to activate HydAΔEFG containing a [4Fe-4S] cluster in its active site.12,32,48,49 Spectroscopic evidence for an assembled 2Fe subcluster on HydFEG includes the observation of FTIR bands associated with iron-bound CO and CN−, and XAS results pointing to a dinuclear iron unit, in addition to a [4Fe-4S] cluster.28,31,50 The nature of the iron–sulfur cluster species on HydF prior to its “loading” by HydE and HydG, however, is less well-defined. The presence of a [4Fe-4S] cluster in HydFΔEG is widely accepted and is supported by data on enzymes from various organisms (including C.a., T.m., S.o., and T.n.) reported by a number of different laboratories.28,30,31,36–38,44,51,52 Evidence for a [2Fe-2S] cluster on HydFΔEG has come from EPR and XAS spectroscopic studies;28,31,50 however, the EPR data have been disputed, with the suggestion that the g ≈ 2.0 EPR signal arose from a [3Fe-4S]+ cluster or a protein radical.36–38 The nature of the FeS cluster species bound to HydF prior to its interaction with HydE and HydG is directly relevant to the process of maturation, as it determines the identity of species that must be transferred from HydE and HydG, and the processes that must occur during assembly of the 2Fe precursor of the H-cluster on HydF. We therefore set out to provide a more definitive description of the clusters present on HydFΔEG.
Characterization of HydF Freshly Purified and After Further Handling
FeS cluster lability is likely an important aspect of the function of HydF in scaffolding the assembly of, and then delivering, the 2Fe precursor of the H-cluster; lability, however, makes characterization of the cluster states challenging. Over the course of many HydF purifications and experiments, we found that sample handling, such as concentration and freezing/thawing, affected FeS cluster spectroscopic properties. We therefore set out to define the cluster states of HydF in “freshly purified” protein, by which we mean HydF that has been eluted off the purification column without further handling of any kind, and to evaluate the cluster changes occurring upon further handling. Our results provide a description of HydF iron–sulfur clusters that unifies and provides context for the previously contradictory results from multiple laboratories.
HydF freshly eluted from the HisTrap column exhibits LMCT transitions in the visible region with a prominent λmax at 405 nm, and careful examination also shows bands in the 510–575 nm region, albeit at low intensity (Figures 2 and S1). Analysis of these samples by circular dichroism reveals positive features centered at 306, 387, and 449 nm, with negative bands centered at 359, 417, 474, and 512 nm (Figures 2 and S1). These UV–visible and CD spectral features change following either concentration or a single freeze/thaw event under anaerobic conditions (Figures 2, S1 and S2). Specifically, LMCT features in the 510–575 nm region in UV–vis spectra intensify upon successive freeze/thaw events, while CD analysis shows that positive bands at 306 and 387 nm and negative bands at 474 and 512 nm all lose intensity as features at 560 and 610 nm develop (Figures 2 and S1); these new spectroscopic features are consistent with an increase in [2Fe-2S]2+ cluster content.53
Figure 2.
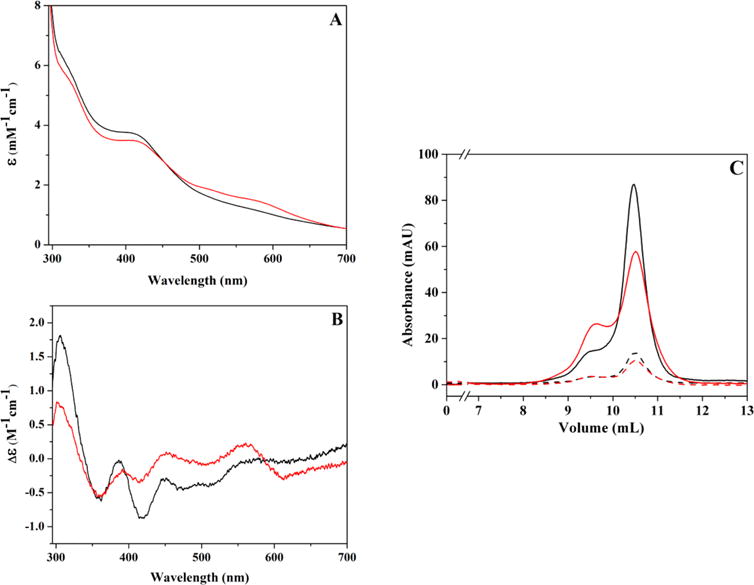
Changes in spectroscopic properties and oligomeric state accompanying a freeze/thaw event for a single sample of HydF. (A) UV–visible absorbance changes for freshly purified HydF before (black; 99 μM protein at 2.18 ± 0.08 Fe/dimer) and after a single freeze-thaw event (red). ε values are reported for the total Fe concentration in sample. (B) The corresponding circular dichroism spectra for the UV–vis spectra represented in panel A; Δε values shown are for total iron content in samples. (C) Gel filtration data for the samples shown in panels A and B. Solid lines represent the absorbance reading at 280 nm, while dashed lines represent absorbance reading at 426 nm. Color scheme is consistent in all panels.
Gel filtration analysis of freshly purified HydF shows that the protein is predominantly dimeric in nature with low amounts of tetramer species (Figures 2 and S2). Sample handling (concentration and/or freeze/thaw), however, results in an increase in tetramer/dimer ratio that occurs concomitantly with the FeS cluster spectroscopic changes described above. Isolation of low levels of pure dimer and pure tetramer HydF species by gel filtration was possible; however, over several hours these species re-equilibrated into a mixture of dimer and tetramer species (data not shown), demonstrating the presence of a dynamic equilibrium of protein quaternary states.
Low temperature (T ≈ 12 K) EPR spectra of freshly purified HydF samples reveal the existence of a nearly isotropic signal with g-values of 2.010, 2.002, and 1.963 (Figures S3, S4, and S5), along with a signal centered at g = 4.3 attributed to adventitiously bound high-spin Fe(III) species (data not shown). As the temperature is raised from 12 to 25 K, the g ≈ 2.00 signal intensifies before gradually diminishing with further temperature increase (Figure S4), behavior that is consistent with [2Fe-2S]+ clusters. EPR signals arising from [3Fe-4S]+ clusters have been observed in samples of as-isolated (following sample handling) HydF and are easily distinguished from [2Fe-2S]+ signals given their relaxation behavior; HydF [3Fe-4S]+ cluster signals exhibit fast temperature relaxation behavior and show substantial intensity loss between 12 and 20 K, consistent with the typical behavior of protein-bound [3Fe-4S]+ clusters (Table 1, Figure S4).40,54,55
Table 1.
EPR Spectroscopic Properties of the [2Fe-2S]+, [3Fe-4S]+, and [4Fe-4S]+ Clusters Associated with HydF, PFL-AE, and HydE
| enzyme | cluster type | TOPTb (K) | P1/2c (≤15 K) (mW) | bd (≤15 K) | P1/2 (30 K) (mW) | bd (30 K) |
|---|---|---|---|---|---|---|
| PFL-AE | ||||||
| as-isolated | [3Fe-4S]+ | <20 | 12.4 | 1.0 | N/Ac | N/A |
| as-isolated | [2Fe-2S]+ | ~30 | N/A | N/A | 8.90 | 1.4 |
| HydEa | ||||||
| DT reduced | [4Fe-4S]+ | <15 | 39.9 | 1.0 | N/A | N/A |
| as-isolated | [3Fe-4S]+ | <20 | ≥50.0 | N/A | N/A | N/A |
| as-isolated | [2Fe-2S]+ | ~30 | 0.720 | 1.1 | 13.0 | 1.0 |
| HydF | ||||||
| freshly purified | [2Fe-2S]+ | 25–30 | 0.270 | 1.0 | 11.5 | 1.1 |
| freshly purified, photoreducedf | [4Fe-4S]+ | <15 | ≥50.0 | N/A | N/A | N/A |
| freshly purified, photoreducedf | [2Fe-2S]+ | 25–30 | 0.100 | 1.3 | 12.5 | 1.4 |
| HydF | ||||||
| as-isolated | [3Fe-4S]+ | <20 | 14.0 | 1.0 | N/A | N/A |
| as-isolated | [2Fe-2S]+ | 25–30 | 0.190 | 1.4 | 10.4 | 1.2 |
| as-isolated, photoreducedf | [4Fe-4S]+ | <15 | ≥50.0 | N/A | N/A | N/A |
The [4Fe-4S]+ cluster signal properties reported here are for the reduced enzyme in the presence of exogenous SAM.
TOPT is defined as the temperature at which maximal peak to peak signal intensity is observed at 1 mW power.
Temperature values for reported P1/2 measurements range between 12–15 K.
The term “b” represents the inhomogeneity parameter, as described in ref 43.
N/A, not applicable.
Measurements made on samples following an anaerobic thaw and freeze event.
Freshly purified HydF photoreduced using 5-deazariboflavin, a catalytic source of low potential electrons,56 exhibits both a g ≈ 2 signal of low intensity that is comparable to that in the freshly purified enzyme and an overlapping axial [4Fe-4S]+ signal with g-values of 2.05, 1.89, and 1.86, similar to what we previously reported (Figure S3B).28,44 When the milder reducing agent DTT (E°′ = −0.33 V) is added to freshly purified enzyme, an intensification of the g ≈ 2 signal, but no overlapping [4Fe-4S]+ signal, is observed. In contrast, addition of the strong chemical reductant DT (E°′ = −0.66 V) causes significant diminution of the g ≈ 2 signal concomitant with the generation of the axial [4Fe-4S]+ signal (Figure S5). DT has been the reducing agent of choice for multiple HydF studies on enzyme from T.m.,30 T.n.,37 S.o.,36 and C.a.;37 our observation that DT significantly lessens the g ≈ 2 signal could well explain why many have failed to observe this signal.36,37 The existence of the g ≈ 2.00 signal in our anaerobic preparations of HydF, which do not include exogenous reducing agents, coupled with the intensification of this signal in the presence of DTT, is fully consistent with its assignment as arising from a [2Fe-2S] cluster, which would be expected to have a 2+/1+ potential in the range accessible by DTT.57 If the g ≈ 2 signal arose from either a [3Fe-4S]+ cluster or a protein radical as has been proposed by others,36,37 however, we would expect it to be lessened, not increased, by the addition of DTT. In chemically reconstituted samples of HydF, the [2Fe-2S]2+/+ cluster persists, as judged by the existence of LMCT features in UV–vis spectra in as-reconstituted samples (data not shown), as well as by the presence of a slow relaxing g ≈ 2.00 signal in DTT treated as-reconstituted protein (Figure 3). Table S2 reports the EPR spin quantitation of HydF [4Fe-4S]+ and [2Fe-2S]+ cluster signals in the various samples described above.
Figure 3.
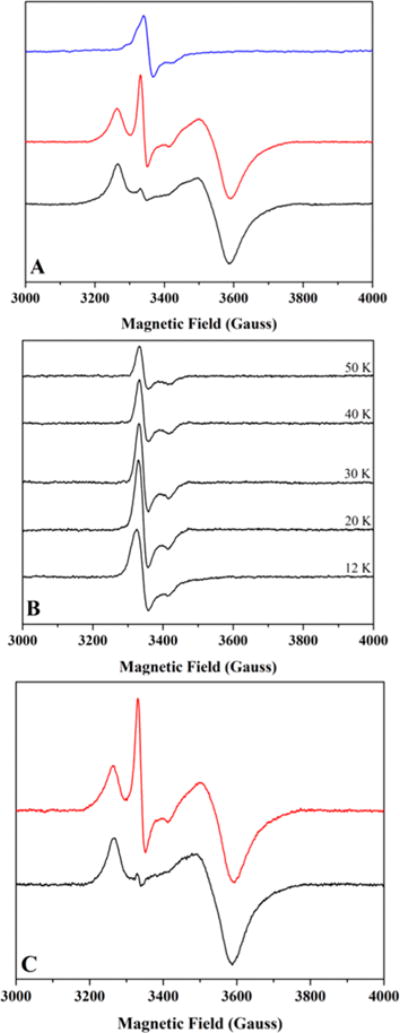
Low temperature CW X-band EPR spectroscopy of HydF samples. (A) As-isolated HydF (600 μM protein at 1.14 ± 0.08 Fe/dimer); spectra collected at 14 K. Data shown are for the same sample in its as-purified state (blue) and in its photoreduced state before (black) and after (red) a thaw/freeze event. (B) Temperature relaxation profile for as-reconstituted HydF (98 μM protein at 4.8 ± 0.8 Fe/dimer). (C) As-reconstituted, photoreduced HydF (110 μM protein at 4.8 ± 0.8 Fe/dimer) before (black) and after (red) a thaw/freeze event. Spectra recorded at 10.5 K.
Characterization of HydE and PFL-AE [2Fe-2S] Clusters
In order to provide a frame of reference for the [2Fe-2S]+ cluster signal in HydF, we analyzed [2Fe-2S]+ cluster signals in both PFL-AE and HydE by EPR. Our previous characterization of PFL-AE demonstrated that the as-isolated enzyme contained predominantly S = 1/2 [3Fe-4S]+, given the existence of a fast relaxing, nearly isotropic signal at 12 K.40 Some preparations of PFL-AE were noted to exhibit small contributions of a slow-relaxing axial signal with g-values of 2.01, 2.01, and 1.97 which was still observable at 100 K.58 Moreover, Mössbauer characterization showed the presence of small amounts of [2Fe-2S]2+ clusters in as-isolated enzyme.59 Taken together, these findings indicate that in some anaerobic PFL-AE preparations, partial reduction of [2Fe-2S]2+ clusters occurs during lysis, leading to the fortuitous existence of [2Fe-2S]+ clusters bound at the CX3CX2C radical SAM cluster site. Recent sample preparations reveal that a fast relaxing, nearly isotropic [3Fe-4S]+ signal with g-values of 2.030, 2.009, and 1.988 exists that accounts for the vast majority of signal intensity at 12 K; spectra obtained at higher temperatures (30–77 K) show the persistence of an axial [2Fe-2S]+ signal with g-values of 2.010, 2.005, and 1.964 (Figure S6, Tables S1 and S2). The existence of both cluster types in a single sample allowed for the definitive assignment of power saturation and temperature relaxation profiles for [3Fe-4S]+ and [2Fe-2S]+ clusters, which in turn provided a benchmark for HydF spectral assignments (Table 1).
HydE contains an accessory FeS cluster site comprising Cys311, Cys319, and Cys322 residues which are located ~20 Å from the CX3CX2C motif.60 An early X-ray structure of T.m. HydE treated with DTT revealed the presence of a [2Fe-2S] cluster in this accessory site; additional structures have been obtained where this site was either vacant or was occupied by a [4Fe-4S] cluster.60,61 We found that in our preparations of as-reconstituted C.a. HydE, this site was either empty or was occupied by a [2Fe-2S] cluster, depending on the iron number following the reconstitution process.14 The assignment as a [2Fe-2S]+ was based on temperature relaxation EPR profiles of different samples that consistently showed the presence of fast relaxing [3Fe-4S]+ cluster signals (presumably present in the radical SAM CX3CX2C motif) overlapping with variable amounts of slow relaxing [2Fe-2S]+ cluster signals (Figure S7). As was the case for PFL-AE, the existence of both cluster types in a single sample enabled the definitive assignment of power saturation and temperature relaxation profiles for [3Fe-4S]+ and [2Fe-2S]+ clusters (Table 1). Power saturation profiles for the various FeS clusters associated with PFL-AE and HydE are shown in Figure 4.
Figure 4.
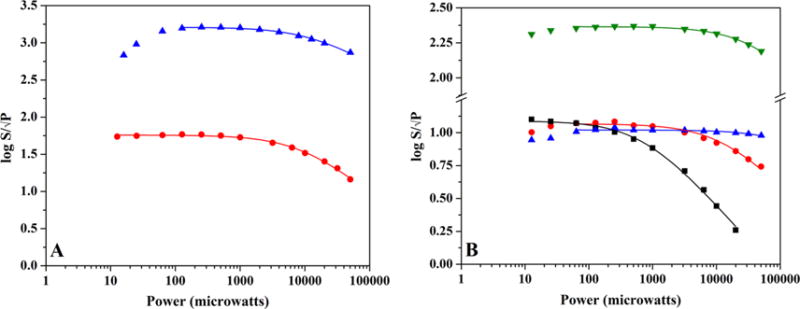
EPR power saturation curves for PFL-AE and HydE. (A) Power saturation behavior of as-isolated PFL-AE (1.68 mM protein with 2.70 ± 0.10 Fe/protein) FeS cluster signals ([3Fe-4S]+ cluster signal (blue), 12 K, gain setting of 1 × 102; [2Fe-2S]+ cluster signal (red), 30 K, gain setting of 1 × 103). (B) Power saturation behavior of FeS cluster signals in HydE. The [3Fe-4S]+ (blue) and [2Fe-2S]+ (black) cluster signals in as-reconstituted enzyme (344 μM protein at 7.64 ± 0.10 Fe/protein) are depicted for 15 K; the [2Fe-2S]+ cluster signal at 30 K (red) is also shown for comparative purposes. Also graphed is the [4Fe-4S]+ cluster signal at 15 K (green) from DT reduced enzyme with exogenous SAM added (275 μM protein at 7.64 ± 0.10 Fe/protein).
EPR Power Saturation Profiles Support Assignment of a [2Fe-2S]+ Cluster in HydF
In order to characterize the g ≈ 2 signal in HydF, we examined power saturation behavior in freshly purified, as-isolated, and photoreduced enzyme samples. As mentioned above, the g ≈ 2 signal in HydF exhibits Topt values (defined as the optimum temperature for signal intensity) between 25 and 30 K (Figures 3 and S4, Table 1). EPR power saturation curves were collected at different temperatures in order to better probe saturation effects on the g ≈ 2 signal. Figure 5 highlights the power saturation curves for freshly purified, as-isolated, and photoreduced HydF g ≈ 2 signals as a function of temperature. The spectral profiles reveal that the signals are more susceptible to increasing power at lower temperatures (≤15 K), while a noticeable shift occurs at 30 K where the signals are more resistant to increasing power levels (Figures 5 and S8). Similar temperature dependence of the power saturation behavior has been previously reported for [2Fe-2S]+-containing ferredoxin proteins from various sources43 and offers compelling experimental support that HydF coordinates a [2Fe-2S]+ cluster in freshly purified, as-isolated, and photoreduced enzyme states. Figure S9 shows power saturation curves for [2Fe-2S]+, [3Fe-4S]+, and [4Fe-4S]+ HydF cluster species.
Figure 5.
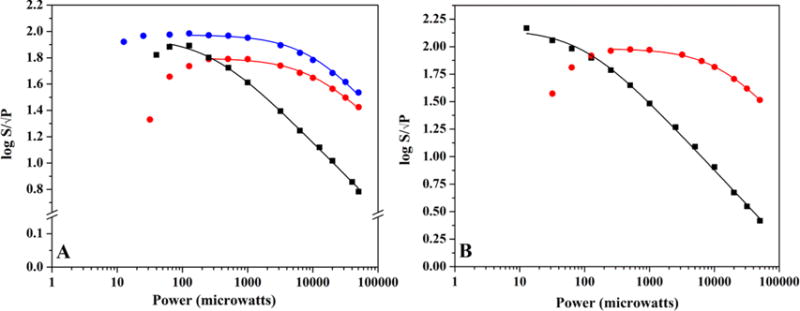
EPR power saturation curves of the g ≈ 2.0 signal in HydF. (A) The power saturation behavior of the g ≈ 2.0 signal in freshly purified HydF (111 μM protein at 2.50 ± 0.08 Fe/dimer) at 13 K (black) and 30 K (red). Also shown for comparative purposes is the power saturation behavior of the g ≈ 2.0 signal in as-isolated HydF (600 μM protein at 1.14 ± 0.08 Fe/dimer) at 30 K (blue). (B) The power saturation behavior of the g ≈ 2.0 signal in freshly purified photoreduced HydF (110 μM protein at 2.50 ± 0.08 Fe/dimer) that has undergone a thaw/freeze event (13 K, black; 30 K, red).
Compelling similarities exist between the power dependence of the g ≈ 2 signals of HydF, PFL-AE, and HydE: all three proteins have cluster signals that are more resistant to power saturation effects near TOPT values for the signals, and all exhibit similar P1/2 values at 30 K ranging between 9 and 13 mW (Table 1). Moreover, the shift in P1/2 for the HydE [2Fe-2S]+ cluster signal as a function of temperature mirrors the shift observed in HydF samples (Figures 4 and 5, Table 1); this observation is in line with previously published results with [2Fe-2S]+-containing ferredoxin proteins.43
Figure 6 shows the direct comparison of [2Fe-2S]+ cluster signals in as-isolated PFL-AE, as-reconstituted HydE, freshly purified HydF, freshly purified photoreduced HydF, as-isolated HydF, and chemically reconstituted HydF. The only remarkable distinction among these signals is the presence of the low field g = 2.035 feature most prominent in PFL-AE; this feature is assigned as arising from residual [3Fe-4S]+ component at 30 K, as it is absent at higher temperatures (Figure S10). In as-isolated HydF samples, a low intensity feature is observed with g-values of 2.045, 2.008, and 1.981; this signal does not appear to be residual [3Fe-4S]+ cluster given its persistence at temperatures above 30 K (Figure S4). Moreover, the power dependence of this feature appears to track with the main g ≈ 2.0 centered signal and is therefore more likely to be [2Fe-2S]+ in nature (Figure S8). Simulations demonstrate that the 30 K signals for freshly purified HydF and HydE are both adequately fit with a single [2Fe-2S]+ spin system, whereas the signals for PFL-AE and as-isolated HydF require two spin systems to fit the data ([3Fe-4S]+ and [2Fe-2S]+ in the case of PFL-AE and two [2Fe-2S]+ spin systems for HydF) (Figure S11). It is not currently clear why a second [2Fe-2S]+ cluster signal is apparent in the more concentrated HydF samples, but it could presumably arise from a low occupancy FeS cluster with a different ligation environment than the primary g = 2.010, 2.003, 1.961 [2Fe-2S]+ cluster (Table S1).
Figure 6.
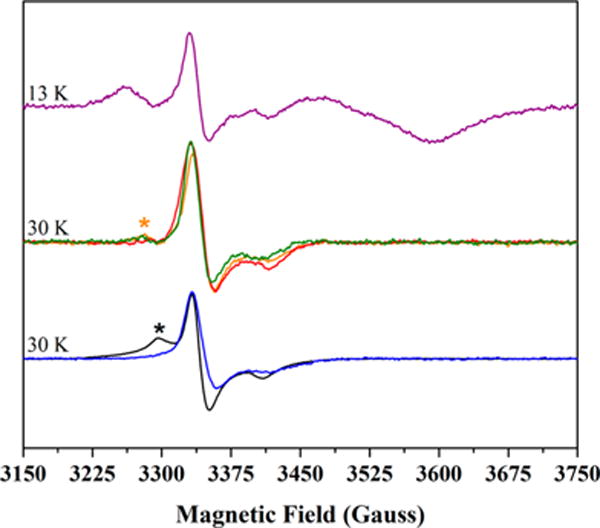
CW X-band EPR spectra of [2Fe-2S]+ cluster signals in HydF, PFL-AE, and HydE. The depicted spectra are as-isolated PFL-AE (1.68 mM protein with 2.70 ± 0.10 Fe/protein), black; as-reconstituted HydE (344 μM protein at 7.64 ± 0.10 Fe/protein), blue; freshly purified HydF (111 μM protein at 2.50 ± 0.08 Fe/dimer), green; as-isolated HydF (600 μM protein at 1.14 ± 0.08 Fe/dimer), orange; chemically reconstituted HydF (98 μM protein at 4.8 ± 0.8 Fe/dimer), red; photoreduced, freshly purified HydF (110 μM protein at 2.50 ± 0.08 Fe/dimer) following an anaerobic thaw/freeze event, purple. As this is a reduced sample, this latter spectrum also contains the axial [4Fe-4S]+ cluster signal, and this provides an internal frame of reference in the figure. Signal intensities for all data were arbitrarily normalized to scale spectra for direct comparison. Experimental temperature values for the various samples are provided in the figure; all spectra shown were collected at 1 mW power. The black asterisk denotes residual [3Fe-4S]+ content in the 30 K spectrum of PFL-AE (see Figures S6 and S10). The orange asterisk denotes a second [2Fe-2S]+ cluster signal that is most prevalent in the as-isolated HydF sample (refer to Figures S4, S8D, and S11D).
Gating [2Fe-2S]+ Cluster Signals in Reduced HydF Preparations
As illustrated in Figure 6, HydF exhibits an EPR signal we assign as arising from a [2Fe-2S]+ cluster and that strongly resembles the signal observed in PFL-AE and HydE. While we have demonstrated that HydF’s [2Fe-2S]+ cluster signal is dramatically intensified by addition of DTT to freshly purified enzyme (Figure S5), sample preparations in a reduced state (using either DT or photoreduction via 5-deazariboflavin) have a substantially diminished overlapping g ≈ 2.00 signal despite the prevalence of axial [4Fe-4S]+ cluster signals (Figures S3 and S5). Surprisingly, the g ≈ 2.00 signal intensity in photoreduced samples was affected by introducing a thaw/freeze event. When frozen photoreduced samples are thawed (under anaerobic conditions) and quickly refrozen, the spectra exhibit large [2Fe-2S]+ signal intensification (Figure 3). Importantly, these observations have been made using freshly purified (data not shown), as-isolated, and chemically reconstituted preparations of HydF (Figure 3). These data suggest that upon photoreduction, the [2Fe-2S] cluster is present as a diamagnetic species, perhaps in the diferrous [2Fe-2S]0 oxidation state, and that the thaw/freeze process results in partial oxidation to [2Fe-2S]+.
We have examined the effects of DT reduction on the appearance of paramagnetic signals following this treatment. As-isolated HydF that exhibited [2Fe-2S]+ cluster signals was treated with 5 mM DT, causing the loss of the [2Fe-2S]+ cluster signals with concomitant generation of a [4Fe-4S]+ cluster signal; treating this sample to a single anaerobic thaw/freeze event caused the appearance of a very weak overlapping [2Fe-2S]+ cluster signal (Figure S12). This result suggests that the appearance of the [2Fe-2S]+ signal in reduced preparations following anaerobic thaw/freeze treatment occurs substantially only in the absence of excess exogenous reducing agent (the reducing power of the photoreduced samples is lost in the absence of white light exposure); we conclude that the DT-treated sample, which retains its reducing power due to excess DT, keeps the [2Fe-2S] cluster in the diferrous state upon sample thawing.
Midpoint Potentials of the HydF [2Fe-2S]+ and [4Fe-4S]+ Clusters
The presence of a [2Fe-2S]+ cluster in freshly purified HydF (in the absence of exogenous reducing agents) prompted us to explore the midpoint potential of this cluster. EPR spectral analysis revealed the presence of the [2Fe-2S]+ cluster signal at the resting solution potential (prior to any addition of DT) (Figure 7). As the solution potential was gradually lowered with small additions of a concentrated DT solution, the [2Fe-2S]+ signal intensity substantially decreased in intensity until a point where the g ≈ 2.0 mediator radical signals began to appear concomitant with growth of the axial [4Fe-4S]+ signal at lower solution potentials. Two independent titration experiments yielded [4Fe-4S]2+/+ Em values of −380 ± 10 mV and −357 ± 3 mV (average Em = −368.5 ± 10.4 mV). From this data we can estimate the Em for the reduction of the [2Fe-2S]+ cluster (and by extension the [2Fe-2S]2+/+) to be ≥ −200 mV (Figure 7). This estimated potential for the [2Fe-2S] cluster would be consistent with the reduction from the 2+ to the 1+ state by DTT (Em = −330 mV vs NHE), as well as for the reduction from the 1+ to 0 state by DT (Em = −660 mV vs NHE).
Figure 7.
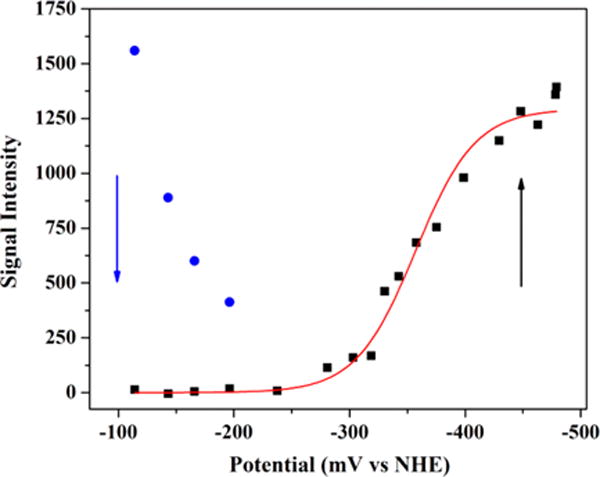
Potentiometric titration curve for freshly purified HydF. The plot shows the disappearance of the [2Fe-2S]+ cluster signal as a function of decreasing solution potential (blue), concomitant with the appearance of the [4Fe-4S]+ cluster signal (black). The [4Fe-4S]2+/+ midpoint potential was determined by fitting the increase in the signal intensity of the g = 1.89 feature as observed in 12.5 K, X-band EPR spectra to the Nernst equation for a 1 electron redox process (red line). HydF sample details are 132 μM protein at 2.28 ± 0.12 Fe/dimer.
DISCUSSION
HydF plays a central role in the H-cluster maturation process, and determining the FeS cluster state(s) of this protein is a pivotal aspect of defining the pathway by which HydE, HydF, and HydG assemble an active [FeFe]-hydrogenase. Moreover, clarifying the nature of the FeS cluster species bound to HydF prior to its interaction with HydE and HydG is directly relevant to this maturation process, as it informs on the species that must be transferred from HydE and HydG, and the processes that must occur during assembly of the 2Fe precursor of the H-cluster on HydF. Herein, we demonstrate that freshly purified, as-isolated, and chemically reconstituted preparations of HydFΔEG all coordinate a redox active [2Fe-2S] cluster that is amenable to spectroscopic and electrochemical characterization.
The results presented herein demonstrate that [2Fe-2S]+ cluster EPR signal intensity in HydF samples can be gated both by the use of reducing agent and by sample handling (Figures 3 and S5). While the addition of DTT to freshly purified HydF causes intensification of the [2Fe-2S]+ cluster signal, DT addition results in nearly complete abolition of [2Fe-2S]+ signal intensity; this signal loss suggests that the [2Fe-2S] cluster is susceptible to reduction to a diamagnetic state. Moreover, the ability to gate the appearance of the [2Fe-2S]+ cluster signal in reduced samples by introducing a thaw/freeze event (Figure 3) is intriguing, and our hypothesis is that this phenomenon is a consequence of quaternary structural changes to HydF that accompany sample handling (Figure 2). While we do not currently understand the exact mechanism for the reappearance of the [2Fe-2S]+ signal, as sample thawing should not inherently result in oxidation, it is clear that [2Fe-2S]+ signal gating is directly dependent upon the reducing power of the solution; samples reduced with DT do not exhibit [2Fe-2S]+ signal reappearance (Figure S12), whereas photoreduced samples that have lost their reducing power in the absence of exposure to light consistently show this behavior (Figure 3). It is possible that in the case of DT treatment, the excess strong reductant keeps the cluster in the diamagnetic diferrous state. There is precedence for protein bound all-ferrous [2Fe-2S] and [4Fe-4S] clusters, and these super-reduced states are stable and amenable to characterization,62–64 providing support for the notion that a diferrous [2Fe-2S] cluster in DT reduced samples of HydF could be stable to sample manipulation.
EPR spectral characterization of the [2Fe-2S]+ cluster signal in HydF shows that it exhibits similar TOPT and P1/2 values as the [2Fe-2S]+ cluster signals in PFL-AE and HydE (Table 1); additionally, the [2Fe-2S]+ signals also display similar shifts in the magnitude of P1/2 as a function of temperature, a result mirrored by [2Fe-2S] ferredoxin proteins.43 While we and others previously proposed that the as-isolated form of HydF exhibited [3Fe-4S]+ cluster signals based on the appearance of a g ≈ 2 component in low temperature EPR spectra in the absence of added reductant, 28,37,44 the analysis presented herein shows that this signal instead arises from a [2Fe-2S]+ cluster which is present in HydF preparations lacking exogenous reducing agents. The spectroscopic data of freshly purified HydF presented here demonstrate that the protein as-eluted with minimal sample handling contains [4Fe-4S]2+, [2Fe-2S]2+, and [2Fe-2S]+ clusters (Figures 2, 6, 7, S1, and S4). Subsequent handling of the protein induces intensification in [2Fe-2S]2+ LMCT features (Figures 2, S1, and S2) concomitant with changes to the quaternary structure as greater amounts of tetramer are observed (Figures 2 and S2). At this time we cannot determine if the dimer and tetramer forms of HydF bind distinct FeS cluster species, although this is an area of active research.
The estimated midpoint potentials for HydF’s [2Fe-2S]2+/+ (Em ≥ −200 mV) and [4Fe-4S]2+/+ (Em = −368.5 ± 10.4 mV) clusters fall near the positive end of the range typically measured for [2Fe-2S]2+/+ (−150 to −450 mV) and [4Fe-4S]2+/+ (−400 to −600 mV) clusters in biology.45,57,65–68 The observed [2Fe-2S] cluster potential is consistent with the freshly purified form of HydF containing some [2Fe-2S] + cluster despite the lack of any exogenous reducing agents in the purification process; intracellular reducing agents likely reduce HydF during the lysis procedure, and the employment of strictly anaerobic conditions during purification and sample handling enables the persistence of this [2Fe-2S]+ cluster. Moreover, this more positive [2Fe-2S] cluster potential is consistent with the ability to increase [2Fe-2S]+ cluster signal intensity upon treatment with DTT. The observation that sequential DT additions cause the [2Fe-2S]+ cluster signal intensity to decrease in a stepwise manner (Figure 7) suggests that the [2Fe-2S]+ state is being reduced to the [2Fe-2S]0 state or converted to another diamagnetic cluster form. This hypothesis is substantiated further by the lack of significant [2Fe-2S]+ cluster signal intensity in DT treated samples (Figures S5 and S12).
It is not clear at this time why the FeS clusters associated with HydF exhibit cluster midpoint potentials that are at the positive end of the range typically observed. The X-ray structure of HydF in a metal free state showed that the putative FeS cluster residues CXHX46–53HCXXC (see refs 10 and 11) from two monomeric subunits are located adjacent to one another in the tetrameric structure; FeS cluster coordination in the dimer state would presumably occur with residues from each monomeric subunit at the dimer interface.69 It is apparent that in both tetrameric and dimeric states, the FeS cluster(s) associated with HydF would be exposed and solvent accessible (although presumably to varying extents), and this is likely related to HydF’s in vivo function to transiently bind and transfer the 2Fe subcluster to HydAΔEFG.21 Furthermore, the FeS cluster binding ligands are found on loop regions within a cleft; this architecture would provide substantially more degrees of freedom for the rearrangement of the FeS cluster species.21 The accessibility of HydF’s [4Fe-4S] cluster has been corroborated by a recent hydrogen/deuterium exchange, 3p-ESEEM and HYSCORE spectroscopic study on T.n. HydFΔEG; this report shows that H2O molecules are bound in close proximity to the [4Fe-4S] cluster and that a protonated exogenous molecule (assigned as a hydroxyl group) acts as a ligand to the exchangeable, site-differentiated iron of the cluster (see Discussion below).38 Similar analysis of the [2Fe-2S] cluster environment on HydF is currently lacking, and it is therefore difficult to discern at this point in time if it is bound in a similar pocket as the [4Fe-4S] cluster. In other words, it is unknown if the [2Fe-2S] cluster also contains an exchangeable, site-differentiated iron, or if the [2Fe-2S] cluster site is as solvent exposed as the [4Fe-4S] cluster appears to be. If the dimer and tetramer states of HydF each coordinate distinct FeS cluster species, then these FeS clusters should be expected to reside in different environments despite the commonality in their protein-derived ligands. These putative differences may very well reflect distinct in vivo roles for the individual clusters during the maturation process.
The role of Cys304, Cys353, and Cys356 (numbering from C.a. sequence) in FeS cluster coordination has been examined by site-directed mutagenesis, and perturbations to these amino acids resulted in deleterious effects on either HydAΔEFG activation, iron quantitation, or FeS cluster binding, leading to the conclusion that these residues play a critical role in FeS cluster coordination.11,37,44 Remarkably, the presence of histidine coordination to the [4Fe-4S] cluster appears to be variable among HydF proteins from different sources.21 HYSCORE spectroscopy has demonstrated that in C.a. HydFΔEG, His352 is the fourth ligand to the [4Fe-4S] cluster, although a His352Ala variant was observed to bind a [4Fe-4S] cluster suggesting that on some level this amino acid is dispensable.31,37 It should be noted, however, that His306 and His352 residues in the C.a. protein play crucial roles, as expression of histidine variant HydFEG proteins does not afford HydAΔEFG activation.37,44
Interestingly, HYSCORE data with both T.m. and T.n. HydFΔEG proteins reveal that histidine coordination is absent; nitrogen coordination to the [4Fe-4S] cluster is, however, observed in the presence of exogenous imidazole, supporting the notion that one ligand site of the cluster is exchangeable and accessible to solvent.30,37,51 Along these lines, coordination to the [4Fe-4S] cluster in T.m. HydFΔEG by a histidine from the affinity tag present at the protein’s N-terminus has been observed.51 The studies described herein utilized wild type His-tagged C.a. HydFΔEG protein, and the enzyme analyzed in the freshly purified state contained exogenous imidazole. It is important to note that the [4Fe-4S]2+/+ and [2Fe-2S]2+/+ spectroscopic features described herein were observed in freshly purified, as-isolated, and chemically reconstituted samples of HydF; thus the FeS cluster spectroscopic signatures were retained when excess imidazole was removed either by dialysis or gel filtration. Moreover, we tested the effects of imidazole on freshly purified protein by immediately removing it from the protein via gel filtration after it was eluted from the HisTrap column; the sample was analyzed by UV–vis and CD spectroscopy concomitant with the preparation of EPR spectroscopic samples. The FeS cluster spectroscopic features of these samples were indistinguishable from those observed in freshly purified enzyme (Figure S13). Although we cannot unambiguously demonstrate that imidazole is not acting as a ligand to the FeS cluster(s) in our preparations of HydF, given that His352 is an FeS cluster ligand in the native Strep-tag II C.a. HydFΔEG protein37 and that the His-tagged C.a. HydFEG protein binds the 2Fe subcluster and activates HydAΔEFG,28,32,44 we expect that the spectroscopic features shown herein represent the native FeS cluster states associated with the protein.
It should be noted that the existence of an accessible site on the [4Fe-4S] cluster may implicate this location as being involved in the coordination (through a bridging ligand) of the 2Fe subcluster.21,34,50,51 It is intriguing to speculate that the [2Fe-2S] cluster observed herein may occupy this same site, either acting as a placeholder for HydG-derived Fe(CO)2CN units22 or serving as a scaffold for 2Fe subcluster assembly.28 A critical issue for delineating the pathway of 2Fe subcluster biosynthesis is to clarify the source of the two iron species. While it has been reported that HydG-derived 57Fe is transferred to HydA during the maturation process,22 careful examination of the experimental conditions underlying these results reveals that 57Fe in the HydG lysate used in activation could have accounted for the label ending up in the H-cluster; thus, it is difficult at this point to definitively rule out a [2Fe-2S] cluster precursor bound to HydFΔEG as the source of the iron species that are transformed into the 2Fe subcluster.28 Moreover, the apparent ability to cycle between oxidation states may support the notion that the [2Fe-2S] cluster functions in a scaffold-type role during biosynthesis, as the accessibility of lower cluster oxidation states may be an essential property of a cluster scaffold that sequentially accepts π-acid CO and CN− ligands. The experimental demonstration that the dimer and tetramer species of HydF are in dynamic equilibrium with one another raises the intriguing question as to which quaternary form interacts with HydE and HydG. We are currently working toward clarifying this issue, as well as demonstrating the FeS cluster states through Mössbauer spectroscopy. These future studies should help delineate the role that the redox active [2Fe-2S] cluster on HydFΔEG plays during maturation of the 2Fe subcluster.
CONCLUSIONS
EPR spectroscopic studies of HydFΔEG from different sources and prepared in multiple laboratories have provided consistent evidence for the presence of [4Fe-4S] clusters; however the evidence for [2Fe-2S] clusters in this protein has proved more controversial. EPR signals similar to what we originally assigned to a [2Fe-2S]+ cluster in reduced HydFΔEG28 have more recently been proposed to arise from a protein-derived radical species or a [3Fe-4S]+ cluster, despite temperature profiles that are inconsistent with either of these species.36–38 While a similar g ≈ 2 EPR signal was observed for S.o. HydF, the authors concluded that it could not arise from a [2Fe-2S]+ cluster because no exogenous reducing agents were used in sample preparations, and treatment with DT resulted in the disappearance of this signal.36 Here we have resolved these discrepancies by demonstrating that C.a. HydFΔEG purifies with a mixture of [2Fe-2S]2+/+ states in the absence of exogenous reducing agents, with the 1+ state giving rise to the g ≈ 2 EPR signal observed in HydFΔEG from multiple sources. Use of strong reducing agents such as DT can reduce this cluster to a diamagnetic state, thus explaining the absence of the [2Fe-2S]+ cluster signal, as well as its variable intensity, in many studies.30,36–38,51 The precise environment of, and role for, the [2Fe-2S] cluster of HydF has yet to be determined. We previously proposed that this cluster was the scaffold for assembly of the 2Fe subcluster precursor of the H-cluster upon delivery of the H-cluster ligands synthesized by HydE and HydG. It is also possible that the [2Fe-2S] cluster of HydF is a placeholder that is displaced upon delivery of “synthons” from HydG. Alternatively, the [2Fe-2S] cluster could play a role in electron transfer during assembly of the 2Fe subcluster on HydF. While our current study cannot differentiate between these possibilities, it provides the framework for a more complete understanding of the function of HydF. An important next step will be to determine whether the [2Fe-2S] cluster is near to, or even bridged to, the [4Fe-4S] cluster of HydF in a manner reminiscent of the H-cluster, as has been proposed by others.50
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Krista Shisler for providing the purified PFL-AE used for the EPR spectroscopic studies presented herein. The purification of PFL-AE was supported by the National Institutes of Health Grant R01 GM054608 (to J.B.B.), with all other work supported by U.S. Department of Energy Grant DE-FG02-10ER16194 (to J.B.B., J.W.P., and E.M.S.). J.N.B. would like to thank the United States Air Force Academy Faculty Pipeline Ph.D. Program for sponsoring him. We would also like to thank Dr. Robert Usselman for his guidance in setting up the midpoint potentiometric titration experiments, and Ms. Anna Scott for help in purifying HydE.
Funding
The work presented herein was supported by the U.S. Department of Energy Grant DE-FG02-10ER16194 (to J.B.B., J.W.P., and E.M.S.) and the National Institutes of Health (R01 GM54608 to J.B.B.).
ABBREVIATIONS
- C.a
Clostridium acetobutylicum
- C.p
Clostridium pasteurianum
- CpI
[FeFe]-hydrogenase I from Clostridium pasteurianum
- E. coli
Escherichia coli
- EPR
electron paramagnetic resonance spectroscopy
- FeS
iron sulfur
- FTIR
Fourier transform infrared spectroscopy
- GTP
guanosine triphosphate
- HydA
[FeFe]-hydrogenase
- HydAΔEFG
HydA expressed without HydE, HydF, and HydG maturases
- HydAEFG
HydA coexpressed with HydE, HydF, and HydG maturases
- HydFΔEG
maturase protein HydF expressed without HydE and HydG
- HydFEG
maturase protein HydF coexpressed with HydE and HydG
- HYSCORE
hyperfine sublevel correlation spectroscopy
- 3P-ESEEM
3-pulse electron spin echo envelope modulation
- P1/2
microwave power of half signal saturation
- PFL-AE
pyruvate formate lyase activating enzyme
- SAM
S-adenosyl-L-methionine
- S.o
Shewanella oneidensis
- T.m
Thermotoga maritima
- T.n
Thermotoga neopolitana
- WT
wild type
- XAS
X-ray absorption spectroscopy
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00528.
Additional UV–vis, CD, and gel filtration data of HydFΔEG samples. EPR data that include temperature relaxation profiles for HydF [2Fe-2S]+ cluster signals, the effects of exogenous reducing agents, microwave power saturation spectra and profiles, and [2Fe-2S]+ cluster EasySpin spectral simulations. Microwave power saturation spectra, temperature relaxation data, and EasySpin spectral simulations for PFL-AE and HydE [2Fe-2S]+ cluster signals. UV–vis, CD, and EPR spectral results for freshly purified HydF that was immediately gel filtered to remove imidazole. Table of simulated g-values for [2Fe-2S]+ and [3Fe-4S]+ cluster signals in HydF, PFL-AE, and HydE. Table of EPR spin integrations (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Peters JW, Schut GJ, Boyd ES, Mulder DW, Shepard EM, Broderick JB, King PW, Adams MW. [FeFe]-and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim Biophys Acta, Mol Cell Res. 2015;1853:1350–1369. doi: 10.1016/j.bbamcr.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 2.Adams MW. The structure and mechanism of iron-hydrogenases. Biochim Biophys Acta, Bioenerg. 1990;1020:115–145. doi: 10.1016/0005-2728(90)90044-5. [DOI] [PubMed] [Google Scholar]
- 3.Fritsch J, Scheerer P, Frielingsdorf S, Kroschinsky S, Friedrich B, Lenz O, Spahn CM. The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre. Nature. 2011;479:249–252. doi: 10.1038/nature10505. [DOI] [PubMed] [Google Scholar]
- 4.Goris T, Wait AF, Saggu M, Fritsch J, Heidary N, Stein M, Zebger I, Lendzian F, Armstrong FA, Friedrich B, Lenz O. A unique iron-sulfur cluster is crucial for oxygen tolerance of a [NiFe]-hydrogenase. Nat Chem Biol. 2011;7:310–318. doi: 10.1038/nchembio.555. [DOI] [PubMed] [Google Scholar]
- 5.Madden C, Vaughn MD, Diez-Perez I, Brown KA, King PW, Gust D, Moore AL, Moore TA. Catalytic turnover of [FeFe]-hydrogenase based on single-molecule imaging. J Am Chem Soc. 2012;134:1577–1582. doi: 10.1021/ja207461t. [DOI] [PubMed] [Google Scholar]
- 6.Swanson KD, Ratzloff MW, Mulder DW, Artz JH, Ghose S, Hoffman A, White S, Zadvornyy OA, Broderick JB, Bothner B, King PW, Peters JW. [FeFe]-hydrogenase oxygen inactivation is initiated at the H cluster 2Fe subcluster. J Am Chem Soc. 2015;137:1809–1816. doi: 10.1021/ja510169s. [DOI] [PubMed] [Google Scholar]
- 7.Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 8.Nicolet Y, Piras C, Legrand P, Hatchikian CE, Fontecilla-Camps JC. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure. 1999;7:13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Lemon BJ, Huang S, Swartz DJ, Peters JW, Bagley KA. Infrared studies of the CO-inhibited form of the Fe-only hydrogenase from Clostridium pasteurianum I: examination of its light sensitivity at cryogenic temperatures. Biochemistry. 2002;41:2036–2043. doi: 10.1021/bi011510o. [DOI] [PubMed] [Google Scholar]
- 10.Posewitz MC, King PW, Smolinski SL, Zhang L, Seibert M, Ghirardi ML. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem. 2004;279:25711–25720. doi: 10.1074/jbc.M403206200. [DOI] [PubMed] [Google Scholar]
- 11.King PW, Posewitz MC, Ghirardi ML, Seibert M. Functional studies of [FeFe] hydrogenase maturation in an Escherichia coli biosynthetic system. J Bacteriol. 2006;188:2163–2172. doi: 10.1128/JB.188.6.2163-2172.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGlynn SE, Ruebush SS, Naumov A, Nagy LE, Dubini A, King PW, Broderick JB, Posewitz MC, Peters JW. In vitro activation of [FeFe] hydrogenase: new insights into hydrogenase maturation. J Biol Inorg Chem. 2007;12:443–447. doi: 10.1007/s00775-007-0224-z. [DOI] [PubMed] [Google Scholar]
- 13.Rubach JK, Brazzolotto X, Gaillard J, Fontecave M. Biochemical characterization of the HydE and HydG iron-only hydrogenase maturation enzymes from Thermatoga maritima. FEBS Lett. 2005;579:5055–5060. doi: 10.1016/j.febslet.2005.07.092. [DOI] [PubMed] [Google Scholar]
- 14.Betz JN, Boswell NW, Fugate CJ, Holliday GL, Akiva E, Scott AG, Babbitt PC, Peters JW, Shepard EM, Broderick JB. [FeFe]-hydrogenase maturation: insights into the role HydE plays in dithiomethylamine biosynthesis. Biochemistry. 2015;54:1807–1818. doi: 10.1021/bi501205e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Rauchfuss TB. Iron carbonyl sulfides, formaldehyde, and amines condense to give the proposed azadithiolate cofactor of the Fe-only hydrogenases. J Am Chem Soc. 2002;124:726–727. doi: 10.1021/ja016964n. [DOI] [PubMed] [Google Scholar]
- 16.Pilet E, Nicolet Y, Mathevon C, Douki T, Fontecilla-Camps JC, Fontecave M. The role of the maturase HydG in [FeFe]-hydrogenase active site synthesis and assembly. FEBS Lett. 2009;583:506–511. doi: 10.1016/j.febslet.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Driesener RC, Challand MR, McGlynn SE, Shepard EM, Boyd ES, Broderick JB, Peters JW, Roach PL. [FeFe]-hydrogenase cyanide ligands derived from S-adenosylmethio-nine-dependent cleavage of tyrosine. Angew Chem, Int Ed. 2010;49:1687–1690. doi: 10.1002/anie.200907047. [DOI] [PubMed] [Google Scholar]
- 18.Shepard EM, Duffus BR, George SJ, McGlynn SE, Challand MR, Swanson KD, Roach PL, Cramer SP, Peters JW, Broderick JB. [FeFe]-hydrogenase maturation: HydG-catalyzed synthesis of carbon monoxide. J Am Chem Soc. 2010;132:9247–9249. doi: 10.1021/ja1012273. [DOI] [PubMed] [Google Scholar]
- 19.Driesener RC, Duffus BR, Shepard EM, Bruzas IR, Duschene KS, Coleman NJ, Marrison AP, Salvadori E, Kay CW, Peters JW, Broderick JB, Roach PL. Biochemical and Kinetic Characterization of Radical S-Adenosyl-l-methionine Enzyme HydG. Biochemistry. 2013;52:8696–8707. doi: 10.1021/bi401143s. [DOI] [PubMed] [Google Scholar]
- 20.Kuchenreuther JM, Myers WK, Stich TA, George SJ, Nejatyjahromy Y, Swartz JR, Britt RD. A radical intermediate in tyrosine scission to the CO and CN− ligands of FeFe hydrogenase. Science. 2013;342:472–475. doi: 10.1126/science.1241859. [DOI] [PubMed] [Google Scholar]
- 21.Shepard EM, Mus F, Betz JN, Byer AS, Duffus BR, Peters JW, Broderick JB. [FeFe]-Hydrogenase Maturation. Biochemistry. 2014;53:4090–4104. doi: 10.1021/bi500210x. [DOI] [PubMed] [Google Scholar]
- 22.Kuchenreuther JM, Myers WK, Suess DL, Stich TA, Pelmenschikov V, Shiigi SA, Cramer SP, Swartz JR, Britt RD, George SJ. The HydG enzyme generates an Fe(CO)2(CN) synthon in assembly of the FeFe hydrogenase H-cluster. Science. 2014;343:424–427. doi: 10.1126/science.1246572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duffus BR, Ghose S, Peters JW, Broderick JB. Reversible H atom abstraction catalyzed by the radical S-adenosylmethionine enzyme HydG. J Am Chem Soc. 2014;136:13086–13089. doi: 10.1021/ja504618y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolet Y, Pagnier A, Zeppieri L, Martin L, Amara P, Fontecilla-Camps JC. Crystal structure of HydG from Carboxydothermus hydrogenoformans: a trifunctional [FeFe]-hydrogenase maturase. ChemBioChem. 2015;16:397–402. doi: 10.1002/cbic.201402661. [DOI] [PubMed] [Google Scholar]
- 25.Dinis P, Suess DL, Fox SJ, Harmer JE, Driesener RC, De La Paz L, Swartz JR, Essex JW, Britt RD, Roach PL. X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly. Proc Natl Acad Sci U S A. 2015;112:1362–1367. doi: 10.1073/pnas.1417252112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suess DL, Burstel I, De La Paz L, Kuchenreuther JM, Pham CC, Cramer SP, Swartz JR, Britt RD. Cysteine as a ligand platform in the biosynthesis of the FeFe hydrogenase H cluster. Proc Natl Acad Sci U S A. 2015;112:11455–11460. doi: 10.1073/pnas.1508440112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagnier A, Martin L, Zeppieri L, Nicolet Y, Fontecilla-Camps JC. CO and CN− syntheses by [FeFe]-hydrogenase maturase HydG are catalytically differentiated events. Proc Natl Acad Sci U S A. 2016;113:104–109. doi: 10.1073/pnas.1515842113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shepard EM, McGlynn SE, Bueling AL, Grady-Smith CS, George SJ, Winslow MA, Cramer SP, Peters JW, Broderick JB. Synthesis of the 2Fe subcluster of the [FeFe]-hydrogenase H cluster on the HydF scaffold. Proc Natl Acad Sci U S A. 2010;107:10448–10453. doi: 10.1073/pnas.1001937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallese F, Berto P, Ruzzene M, Cendron L, Sarno S, De Rosa E, Giacometti GM, Costantini P. Biochemical analysis of the interactions between the proteins involved in the [FeFe]-hydrogenase maturation process. J Biol Chem. 2012;287:36544–36555. doi: 10.1074/jbc.M112.388900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brazzolotto X, Rubach JK, Gaillard J, Gambarelli S, Atta M, Fontecave M. The [Fe-Fe]-hydrogenase maturation protein HydF from Thermotoga maritima is a GTPase with an iron-sulfur cluster. J Biol Chem. 2006;281:769–774. doi: 10.1074/jbc.M510310200. [DOI] [PubMed] [Google Scholar]
- 31.Czech I, Silakov A, Lubitz W, Happe T. The [FeFe]-hydrogenase maturase HydF from Clostridium acetobutylicum contains a CO and CN− ligated iron cofactor. FEBS Lett. 2010;584:638–642. doi: 10.1016/j.febslet.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 32.McGlynn SE, Shepard EM, Winslow MA, Naumov AV, Duschene KS, Posewitz MC, Broderick WE, Broderick JB, Peters JW. HydF as a scaffold protein in [FeFe] hydrogenase H-cluster biosynthesis. FEBS Lett. 2008;582:2183–2187. doi: 10.1016/j.febslet.2008.04.063. [DOI] [PubMed] [Google Scholar]
- 33.Mulder DW, Shepard EM, Meuser JE, Joshi N, King PW, Posewitz MC, Broderick JB, Peters JW. Insights into [FeFe]-hydrogenase structure, mechanism, and maturation. Structure. 2011;19:1038–1052. doi: 10.1016/j.str.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Berggren G, Adamska A, Lambertz C, Simmons TR, Esselborn J, Atta M, Gambarelli S, Mouesca JM, Reijerse E, Lubitz W, Happe T, Artero V, Fontecave M. Biomimetic assembly and activation of [FeFe]-hydrogenases. Nature. 2013;499:66–69. doi: 10.1038/nature12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esselborn J, Lambertz C, Adamska-Venkatesh A, Simmons T, Berggren G, Noth J, Siebel J, Hemschemeier A, Artero V, Reijerse E, Fontecave M, Lubitz W, Happe T. Spontaneous activation of [FeFe]-hydrogenases by an inorganic [2Fe] active site mimic. Nat Chem Biol. 2013;9:607–609. doi: 10.1038/nchembio.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuchenreuther JM, Britt RD, Swartz JR. New insights into [FeFe] hydrogenase activation and maturase function. PLoS One. 2012;7:e45850. doi: 10.1371/journal.pone.0045850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berto P, Di Valentin M, Cendron L, Vallese F, Albertini M, Salvadori E, Giacometti GM, Carbonera D, Costantini P. The [4Fe-4S]-cluster coordination of [FeFe]-hydrogenase maturation protein HydF as revealed by EPR and HYSCORE spectroscopies. Biochim Biophys Acta, Bioenerg. 2012;1817:2149–2157. doi: 10.1016/j.bbabio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Albertini M, Berto P, Vallese F, Di Valentin M, Costantini P, Carbonera D. Probing the Solvent Accessibility of the [4Fe-4S] Cluster of the Hydrogenase Maturation Protein HydF from Thermotoga neapolitana by HYSCORE and 3p-ESEEM. J Phys Chem B. 2015;119:13680–13689. doi: 10.1021/acs.jpcb.5b03110. [DOI] [PubMed] [Google Scholar]
- 39.Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 40.Broderick JB, Henshaw TF, Cheek J, Wojtuszewski K, Smith SR, Trojan MR, McGhan RM, Kopf A, Kibbey M, Broderick WE. Pyruvate formate-lyase-activating enzyme: strictly anaerobic isolation yields active enzyme containing a [3Fe-4S](+) cluster. Biochem Biophys Res Commun. 2000;269:451–456. doi: 10.1006/bbrc.2000.2313. [DOI] [PubMed] [Google Scholar]
- 41.Henshaw TF, Broderick JB, Cheek J. The [4Fe-4S]1+ Cluster of Pyruvate Formate-Lyase Activating Enzyme Generates the Glycyl Radical on Pyruvate Formate-Lyase: EPR-Detected Single Turnover. J Am Chem Soc. 2000;122:8331–8332. [Google Scholar]
- 42.Stoll S, Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J Magn Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 43.Rupp H, Rao KK, Hall DO, Cammack R. Electron spin relaxation of iron-sulphur proteins studied by microwave power saturation. Biochim Biophys Acta, Protein Struct. 1978;537:255–269. doi: 10.1016/0005-2795(78)90509-3. [DOI] [PubMed] [Google Scholar]
- 44.Joshi N, Shepard EM, Byer AS, Swanson KD, Broderick JB, Peters JW. Iron-sulfur cluster coordination in the [FeFe]-hydrogenase H cluster biosynthetic factor HydF. FEBS Lett. 2012;586:3939–3943. doi: 10.1016/j.febslet.2012.09.028. [DOI] [PubMed] [Google Scholar]
- 45.Hinckley GT, Frey PA. Cofactor dependence of reduction potentials for [4Fe-4S]2+/1+ in lysine 2,3-aminomutase. Biochemistry. 2006;45:3219–3225. doi: 10.1021/bi0519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Usselman RJ, Fielding AJ, Frerman FE, Watmough NJ, Eaton GR, Eaton SS. Impact of mutations on the midpoint potential of the [4Fe-4S]+1,+2 cluster and on catalytic activity in electron transfer flavoprotein-ubiquinone oxidoreductase (ETF-QO) Biochemistry. 2008;47:92–100. doi: 10.1021/bi701859s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinckley GT, Frey PA. An adaptable spectroelectrochemical titrator: the midpoint reduction potential of the iron-sulfur center in lysine 2,3-aminomutase. Anal Biochem. 2006;349:103–111. doi: 10.1016/j.ab.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 48.Mulder DW, Ortillo DO, Gardenghi DJ, Naumov AV, Ruebush SS, Szilagyi RK, Huynh B, Broderick JB, Peters JW. Activation of HydA(DeltaEFG) requires a preformed [4Fe-4S] cluster. Biochemistry. 2009;48:6240–6248. doi: 10.1021/bi9000563. [DOI] [PubMed] [Google Scholar]
- 49.Mulder DW, Boyd ES, Sarma R, Lange RK, Endrizzi JA, Broderick JB, Peters JW. Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(DeltaEFG) Nature. 2010;465:248–251. doi: 10.1038/nature08993. [DOI] [PubMed] [Google Scholar]
- 50.Czech I, Stripp S, Sanganas O, Leidel N, Happe T, Haumann M. The [FeFe]-hydrogenase maturation protein HydF contains a H-cluster like [4Fe4S]-2Fe site. FEBS Lett. 2011;585:225–230. doi: 10.1016/j.febslet.2010.11.052. [DOI] [PubMed] [Google Scholar]
- 51.Berggren G, Garcia-Serres R, Brazzolotto X, Clemancey M, Gambarelli S, Atta M, Latour JM, Hernandez HL, Subramanian S, Johnson MK, Fontecave M. An EPR/HYSCORE, Mossbauer, and resonance Raman study of the hydrogenase maturation enzyme HydF: a model for N-coordination to [4Fe-4S] clusters. J Biol Inorg Chem. 2014;19:75–84. doi: 10.1007/s00775-013-1062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albertini M, Galazzo L, Maso L, Vallese F, Berto P, De Rosa E, Di Valentin M, Costantini P, Carbonera D. Characterization of the [FeFe]-Hydrogenase Maturation Protein HydF by EPR Techniques: Insights into the Catalytic Mechanism. Top Catal. 2015;58:708–718. [Google Scholar]
- 53.Stephens PJ, Thomson AJ, Dunn JB, Keiderling TA, Rawlings J, Rao KK, Hall DO. Circular dichroism and magnetic circular dichroism of iron-sulfur proteins. Biochemistry. 1978;17:4770–4778. doi: 10.1021/bi00615a026. [DOI] [PubMed] [Google Scholar]
- 54.Telser J, Lee HI, Hoffman BM. Investigation of exchange couplings in [Fe3S4]+ clusters by electron spin-lattice relaxation. J Biol Inorg Chem. 2000;5:369–380. doi: 10.1007/pl00010666. [DOI] [PubMed] [Google Scholar]
- 55.Shirakawa T, Takahashi Y, Wada K, Hirota J, Takao T, Ohmori D, Fukuyama K. Identification of variant molecules of Bacillus thermoproteolyticus ferredoxin: crystal structure reveals bound coenzyme A and an unexpected [3Fe-4S] cluster associated with a canonical [4Fe-4S] ligand motif. Biochemistry. 2005;44:12402–12410. doi: 10.1021/bi0508441. [DOI] [PubMed] [Google Scholar]
- 56.Massey V, Hemmerich P. Photoreduction of flavoproteins and other biological compounds catalyzed by deazaflavins. Biochemistry. 1978;17:9–16. doi: 10.1021/bi00594a002. [DOI] [PubMed] [Google Scholar]
- 57.Capozzi F, Ciurli S, Luchinat C. Coordination sphere versus protein environment as determinants of electronic and functional properties of iron-sulfur proteins. In: Hill HAO, Sadler PJ, Thomson AJ, editors. Metal Sites in Proteins and Models Redox Centres. Springer; Berlin: 2005. pp. 127–160. [Google Scholar]
- 58.Broderick JB, Duderstadt RE, Fernandez DC, Wojtuszewski K, Henshaw TF, Johnson MK. Pyruvate formate-lyase activating enzyme is an iron-sulfur protein. J Am Chem Soc. 1997;119:7396–7397. [Google Scholar]
- 59.Krebs C, Henshaw TF, Cheek J, Huynh BH, Broderick JB. Conversion of 3Fe-4S to 4Fe-4S clusters in native pyruvate formate-lyase activating enzyme: Mössbauer characterization and implications for mechanism. J Am Chem Soc. 2000;122:12497–12506. [Google Scholar]
- 60.Nicolet Y, Rubach JK, Posewitz MC, Amara P, Mathevon C, Atta M, Fontecave M, Fontecilla-Camps JC. X-ray structure of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima. J Biol Chem. 2008;283:18861–18872. doi: 10.1074/jbc.M801161200. [DOI] [PubMed] [Google Scholar]
- 61.Nicolet Y, Rohac R, Martin L, Fontecilla-Camps JC. X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. Proc Natl Acad Sci U S A. 2013;110:7188–7192. doi: 10.1073/pnas.1302388110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Im SC, Kohzuma T, McFarlane W, Gaillard J, Sykes AG. Formation, properties, and characterization of a fully reduced FeIIFeII form of spinach (and parsley) [2Fe-2S] ferredoxin with the macrocyclic complex [Cr(15-aneN(4))(H2O)(2)](2+) as reductant. Inorg Chem. 1997;36:1388–1396. doi: 10.1021/ic9608253. [DOI] [PubMed] [Google Scholar]
- 63.Yoo SJ, Meyer J, Munck E. Mössbauer evidence for a diferrous [2Fe-2S] cluster in a ferredoxin from Aquifex aeolicus. J Am Chem Soc. 1999;121:10450–10451. [Google Scholar]
- 64.Guo ML, Sulc F, Ribbe MW, Farmer PJ, Burgess BK. Direct assessment of the reduction potential of the [4Fe-4S](1+/0) couple of the Fe protein from Azotobacter vinelandii. J Am Chem Soc. 2002;124:12100–12101. doi: 10.1021/ja026478f. [DOI] [PubMed] [Google Scholar]
- 65.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. University Science Books; Mill Valley, CA: 1994. [Google Scholar]
- 66.Meyer J. Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J Biol Inorg Chem. 2008;13:157–170. doi: 10.1007/s00775-007-0318-7. [DOI] [PubMed] [Google Scholar]
- 67.Mapolelo DT, Zhang B, Naik SG, Huynh BH, Johnson MK. Spectroscopic and functional characterization of iron-sulfur cluster-bound forms of Azotobacter vinelandii (Nif)IscA. Biochemistry. 2012;51:8071–8084. doi: 10.1021/bi3006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi K, Fujikawa M, Kozawa T. Binding of promoter DNA to SoxR protein decreases the reduction potential of the [2Fe-2S] cluster. Biochemistry. 2015;54:334–339. doi: 10.1021/bi500931w. [DOI] [PubMed] [Google Scholar]
- 69.Cendron L, Berto P, D’Adamo S, Vallese F, Govoni C, Posewitz MC, Giacometti GM, Costantini P, Zanotti G. Crystal structure of HydF scaffold protein provides insights into [FeFe]-hydrogenase maturation. J Biol Chem. 2011;286:43944–43950. doi: 10.1074/jbc.M111.281956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


