Abstract
Introduction: Fatty acid ethanolamides (FAEs) are a family of lipid mediators that participate in a host of biological functions. Procedures for the quantitative analysis of FAEs include organic solvent extraction from biological matrices (e.g., blood), followed by purification and subsequent quantitation by liquid chromatography–mass spectrometry (LC/MS) or gas chromatography–mass spectrometry. During the validation process of a new method for LC/MS analysis of FAEs in biological samples, we observed unusually high levels of the FAE, palmitoylethanolamide (PEA), in blank samples that did not contain any biological material.
Materials and Methods: We investigated a possible source of this PEA artifact via liquid chromatography coupled to tandem mass spectrometry, as well as accurate mass analysis.
Results: We found that high levels of a contaminant indistinguishable from PEA is present in new 5.75″ glass Pasteur pipettes, which are routinely used by laboratories to carry out lipid extractions. This artifact might account for discrepancies found in the literature regarding PEA levels in human blood serum and other tissues.
Conclusions: It is recommended to take into account this pitfall by analyzing potential contamination of the disposable glassware during the validation process of any method used for analysis of FAEs.
Keywords: : contamination, fatty acid ethanolamides, glass Pasteur pipettes, LC/MS, palmitoylethanolamide, sample preparation
Introduction
Fatty acid ethanolamides (FAEs) are a family of endogenous lipid mediators, whose chemical structures consist of a fatty acid moiety bound to ethanolamine by an amide linkage. These compounds are synthesized by cells throughout the body and control inflammation, appetite and food intake, learning and memory, and pain among other functions.1 Palmitoylethanolamide (PEA) and oleoylethanolamide (OEA) suppress inflammation by activating the ligand-operated transcription factor, peroxisome proliferator-activated receptor-α (PPAR-α).2 Anandamide (arachidonoylethanolamide [AEA]) acts as a partial agonist at cannabinoid receptor (CB) type 1 and 2 receptors and, therefore, belongs to the diverse family of lipid signaling molecules called endocannabinoids (ECBs).3,4
Due to their similar physicochemical properties, FAEs and other ECBs, such as 2-arachidonoyl-sn-glycerol (2AG), are usually coextracted from biological samples.5 The procedure for their analysis includes extraction with organic solvents (e.g., methanol and chloroform) followed by purification through solid-phase extraction (e.g., open-bed silica gel) and subsequent quantitation by liquid chromatography–mass spectrometry (LC/MS) or gas chromatography–mass spectrometry (GC/MS).
FAEs are present in blood serum or plasma in the pmol per mL scale and in biological tissues in concentration ranging from the pmol to nmol per gram scale. A review of the literature, however, reveals that data from different laboratories, reporting concentration of FAEs in human serum from healthy subjects, often do not corroborate one another. In particular, reported levels of PEA and OEA in serum or plasma of healthy human subjects differ by up to two orders of magnitude, from 5 to 30 pmol per mL6–17 up to 200 pmol per mL of serum or plasma.18,19
During the validation process of a new method for LC/MS analysis of FAEs and ECBs in human serum extracts, we observed unexpectedly high levels of PEA, as compared with data previously obtained in our laboratory.6 We suspected that these abnormal levels could be due to a recurrent contamination. We found that 5″¾ Pasteur pipettes of most, if not all, commercial brands, contain multiple contaminants detectable by LC/MS, including readily detectable quantities of a compound indistinguishable from PEA.
Materials and Methods
Materials
All solvents, including chloroform, methanol, and acetone, were of the highest purity commercially available, suggested for pesticide residue analysis, residue 1 mg/L max, from Burdick & Jackson (Honeywell International, Inc., Morris Plains, NJ). Water was high-performance liquid chromatography (HPLC) grade from Fisher Scientific (Thermo Fisher Scientific, Inc., Waltham, MA). Synthetic standards of FAEs, including deuterium-containing internal standards (ISTDs) for isotope dilution ([2H4]PEA, [2H4]OEA, [2H4]AEA and [2H5]2AG) were purchased from Cayman Chemical (Ann Arbor, MI). Ammonium acetate and acetic acid were Optima grade from Fisher Chemical (Thermo Fisher Scientific, Inc.). Disposable glass Pasteur pipettes (5″¾ and 9″ corresponding to 150 and 230 mm, respectively) were from the following vendors: Fisher Scientific (Thermo Fisher Scientific, Inc.), VWR (VWR International LLC, Radnor, PA), Corning (Corning, NY), and Wheaton (Wheaton Industries, Inc., Millville, NJ). Eight milliliter vials were from Thermo Scientific (Thermo Fisher Scientific, Inc.). LC amber vials with inserts were from Agilent Technologies (Santa Clara, CA). Beakers and cylinders were from Fisher Scientific (Thermo Fisher Scientific, Inc.). Glass syringes were from Hamilton (Reno, NV). Silica Gel was 60Å 230–400 Mesh ASTM from Whatman, Inc. (Sigma-Aldrich Corp., St. Louis, MO). Untreated glass wool was from Grace (Columbia, MD).
Extraction of FAEs from blood serum
Samples of human serum from healthy subjects were kindly provided by Dr. Hamid Moradi. The study protocol was approved by the Institutional Review Board of the University of California, Irvine and was completed with the assistance of the University of California, Irvine, General Clinical Research Center. Written informed consent was obtained from all subjects. Blood samples were collected in red top blood collection tubes (vacutainers, silicon-coated, and no additives), which were left sitting upright at room temperature for a minimum of 30 min to a maximum of 60 min to allow clotting. The blood sample was centrifuged immediately at the end of the clotting time in a horizontal rotor (swing-out head) for 20 min at 1100–1300 g at room temperature. The serum was transferred into labeled cryogenic vials, which were immediately stored upright in a rack at −80°C. Aliquot volume was 1 mL. This process was completed within 1 h of centrifugation. Aliquots were transported in dry ice, then slowly defrosted at 4°C and stirred for 30 sec prior use. A volume of serum of 0.6 mL was transferred in an 8-mL glass vial. Protein precipitation was performed by adding 1 mL of cold acetone containing deuterium-labeled ISTDs as follows: 1.2 pmol of [2H4]PEA, 1.2 pmol of [2H4]OEA, 0.6 pmol of [2H4]AEA, and 30 pmol of [2H5]2AG. Samples were stirred for 60 sec then left in a cold room (4°C) for 20 min, then centrifuged at 2000 rpm at 4°C for 20 min. The supernatants were transferred with a Pasteur pipette in another 8-mL glass vial before undergoing lipid extraction procedure. Lipid extraction was performed using a modified Folch procedure,20 as follows. The excess of acetone was evaporated under N2. Water was added up to 1 mL of total volume and the sample was vortexed for 60 sec. Then 1 mL of methanol was added and the sample vortexed for 60 sec. Finally, 2 mL of chloroform was added and the sample vortexed for 60 sec before centrifugation for 15 min at 3500 rpm at 4°C. The lower chloroform phase was collected with a Pasteur pipette and transferred in another glass vial, evaporated under N2 stream, and reconstituted in 2 mL of chloroform. Afterward, fractionation of the lipid extract through open-bed chromatography was performed as previously described.5 Briefly, the standardized extraction procedure employed in our laboratory is an open-bed silica-gel column chromatography (silica gel 60 230–400 mesh) that precedes LC/MS analyses (i.e., a custom-made solid-phase extraction procedure). Silica gel columns were prepared as follows: a 5″¾ glass Pasteur pipette was plugged with glass wool at the beginning of the thin tip; 1 mL of a slurry of silica gel (60Å 230–400 Mesh ASTM) in chloroform (1:1, v/v) was poured into the pipette held in a rack with a metal tray underneath; the column was washed with 2 mL of chloroform. The reconstituted 2 mL of lipid extract was loaded onto the column. FAEs were eluted with 2 mL of chloroform/methanol (9:1, v/v). The eluate was collected in another 8-mL glass vial, evaporated under N2, reconstituted in 60 μL of methanol, and transferred in a 250-μL insert of a 2-mL LC vial before LC/MS analysis. The final concentrations of ISTD were, therefore, the following: 20 nM [2H4]PEA and [2H4]OEA, 10 nM [2H4]AEA and 500 nM [2H5]2AG. The same concentrations were used for the calibration curves described in the LC/MS section below.
Pipette extraction
Disposable glass Pasteur pipettes were washed with 8 mL of chloroform. The chloroform was collected in an 8-mL vial, spiked with 20 moles of ISTD [2H4]PEA, evaporated under a stream of N2, reconstituted in 100 μL of methanol and transferred with a glass syringe in a 250-μL insert of a 2-mL LC vial before LC/MS analysis. The final concentration of [2H4]PEA ISTD was therefore 200 nM.
Foam extraction
Polyurethane foam is used by vendors to wrap the glass Pasteur pipettes to protect them from breakage. Small pieces of this plastic material were cut and weighed. Pieces of 1, 2.5, 5, and 10 mg were deposited in 8-mL glass vials that were previously washed with chloroform twice. Each vial was filled with 5 mL of chloroform containing 20 pmol of ISTD [2H4]PEA. Vials were stirred thoroughly. The chloroform was evaporated under N2 and the extracts were suspended in 100 μL of methanol and transferred with a glass syringe in a 250-μL insert of a 2-mL LC vial.
Transferring PEA from the foam to the glass
A PEA-free 9″ glass Pasteur pipette was further washed with chloroform. The pipette was warmed up at 60°C and wrapped in polyurethane foam used by vendors for packaging. Subsequently, a pipette extraction was performed as described above to check whether PEA could be transferred from the foam to the glass.
Liquid chromatography–mass spectrometry
LC/MS analyses of samples were performed using an Agilent 1200 LC system coupled to an Agilent G6410A triple quadrupole (QQQ) MS detector (Agilent Technologies) equipped with an electrospray ionization (ESI) interface. FAEs were separated using a XDB Eclipse C18 column (2.1×50 mm i.d., 1.8 μm), eluted with an isocratic method of methanol in water (A: 20% water+0.25% acetic acid+5 mM ammonium acetate and B: 80% methanol+0.25% acetic acid + 5 mM ammonium acetate in 8 min) at a flow rate of 0.4 mL/min. Column temperature was kept at 40°C. MS detection was in the positive mode, capillary voltage was set at 4 kV, fragmentor voltage varied from 120 to 140 V, and collision energy was 20 eV. Helium was used as collision gas, whereas nitrogen was used as a drying gas at a flow rate of 12 liters/min at 350°C. Nebulizer pressure was set at 50 PSI. In parallel, LC/MS and tandem mass spectrometry (MS/MS) data were also acquired on an ACQUITY UPLC I-Class system coupled to a Xevo TQ-S Micro Mass Spectrometer (Waters, Milford, MA) with accompanying ESI interface (data presented in Fig. 3B, Supplementary Figs. S5 and S7). On both set-ups we quantified FAEs with an isotope dilution method, monitoring protonated adducts of the molecular ions [M+H]+ in multiple reaction-monitoring (MRM) mode. The MRM transitions monitored for FAEs detection and quantitation were the following: PEA (m/z=300.3>62.1); OEA (m/z=326.3>62.1); AEA (m/z=348.3>62.1); 2AG (m/z=379.3>287.2); [2H4]PEA (m/z=304.3>66.1); [2H4]OEA (m/z=330.3>66.1); [2H4]AEA (m/z=352.3>66.1); and [2H5]2AG (m/z=384.3>287.2). Additional analytes were investigated on the Waters UPLC/MS/MS as possible contaminants in foam: docosahexaenoyl ethanolamide (DHEA, Supplementary Fig. S5G, H) (m/z=372.3>62.1), and docosahexaenoyl glycerol (DHG, Supplementary Fig. S5K, L) (m/z=403.4>311.2). We used two different sets of calibration curves, one for the serum analysis with low ISTD and one for pipettes and foam extracts with higher ISTD. In particular, we prepared standard calibration curves by adding a constant amount of deuterium-labeled standards to increasing amount of the corresponding unlabeled FAEs, followed by MS analysis as described above. The relative concentrations of unlabeled versus labeled ions were plotted against their relative response (i.e., peak area) and the calibration curves were constructed using linear regression. R2 was 0.998 for all analytes, indicating linear response. In the calibration curve for serum extracts analysis, the ISTDs were kept as follows: [2H4]PEA and [2H4]OEA at 20 nM, [2H4]AEA at 10 nM, and [2H5]2AG at 500 nM. In the calibration curve for foam and pipette extracts analysis, the ISTD [2H4]PEA was kept at 200 nM. In both cases the nondeuterated reference standard of PEA, OEA, AEA, and 2AG were at concentrations ranging from 1 nM to 2 μM, for a total of 11 points of calibration.
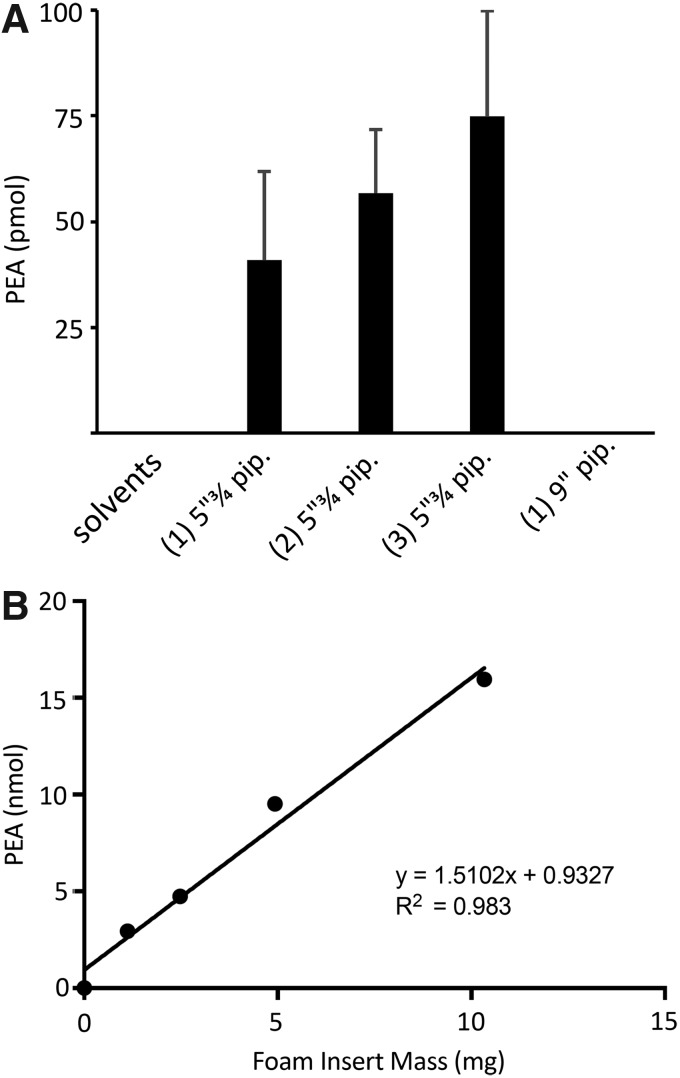
FIG. 3.
Quantitation of PEA in extracts of pipettes. (A) Each pipette was washed once with 8 mL of CHCl3. Number of pipettes washed in each experiment is indicated in brackets. Error bars represent SD, n=3. The Fisher Pasteur pipettes, 5″¾, were from lot n. 16168998 (Box 1). (B) Linear regression between mass of foam insert and quantified PEA values. SD, standard deviation.
Highly accurate mass data were obtained on a Shimadzu IT-TOF (ion trap–time-of-flight) coupled to Shimadzu Nexera HPLC system following a similar LC/MS method. A reverse-phased endcapped C18 column from Knauer was employed (Eurospher II 100 C18: 2×100 mm i.d., 3 μm, 100 A). The solvent system and oven temperature were the same as described above for the Agilent instrument. The LC method was 13 min long and proceeded as follows: from 85 to 98%B in 5 min, then kept at 98%B for 5 min, followed by a re-equilibration step at 85%B for 3 min. Source temperature was kept at 250°C, drying gas pressure was 103 kPa (10 L/min), detector voltage was 1.55 kV, and probe voltages were 4.5 and −3.5 kV. Data were acquired and analyzed with the Shimadzu software LCMS Solution (Acquisition and Analysis) and Formula Predictor. All LC/MS data showed in this article were obtained in the positive ion mode.
Results
Quantitation of FAEs and 2AG in human serum samples
We quantified PEA, OEA, AEA, and 2AG levels in serum samples from three healthy subjects. Table 1 contains a comparison of average FAEs and 2AG levels, measured in this work and by independent laboratories, in human serum/plasma from healthy subjects, by LC/MS analysis. For most compounds, concentrations measured in the present study were in close agreement with those previously reported by our group6 and others.7 PEA levels were similar to those reported by Bilgin et al.18 and by Sipe et al.,19 but 10 to 20 times higher than those reported by other groups7–17 and previously by us.6 This suggested that a contamination might be present.
Table 1.
Comparison of Human Serum/Plasma Concentrations of Fatty Acid Ethanolamides and 2-Arachidonoyl-sn-Glycerol Determined in This Study with Other Reports
| This work | Schreiber et al.6 | Gachet et al.7 | Balvers et al.8 | Balvers et al.9 | Lin et al. 201210 | Ozalp and Barroso11 | Ottria et al.12 | Lam et al.13 | Lam et al.13 | Jumpertz et al.14 | Caraceni et al.15 | Hill et al.16 | Schaefer et al.17 | Bilgin et al.18 | Sipe et al.19 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lipid species | S | S | P | P | P | P | P | P | P | S | P | P | S | S | P | P |
| AEA | 0.6 | 0.9 | 1.7 | 0.7 | 1.1 | 2.6 | 0.7 | 0.8 | 0.72 | 1.01 | 1.8 | 5.1 | 2.6 | 2.0 | 14.5 | 12.8 |
| OEA | 4.2 | 3.7 | 9.9 | 4.2 | 6.1 | 8.6 | 4.9 | 4.9 | 4.6 | 6.0 | 4.0 | 12.6 | 5.5 | 9.6 | 162.5 | 107.9 |
| PEA | 419.4* | 28.4 | 23 | 4.7 | 4.9 | 8.0 | 5.2 | 24.8 | 16.9 | 16.3 | – | 12.6 | 12.5 | 20.5 | 203.6 | 213.3 |
| 2AG | 12.1 | – | 16.5 | 18.8 | – | – | 18.8 | – | – | – | 32.8 | 1.7 | 39.0 | 10.0 | 11.8 | 5.6 |
Values are expressed in pmol per mL of serum. The PEA value obtained in this work is marked with an asterisk, indicating an artifact due to contamination. In the selected reports, when only plotted and not reported in the text, values were back calculated from digitized images.
2AG, 2-arachidonoyl-sn-glycerol; AEA, arachidonoylethanolamide, anandamide; OEA, oleoylethanolamide; P, plasma; PEA, palmitoylethanolamide; S, serum.
Identification of PEA in glass Pasteur pipettes and plastic foam
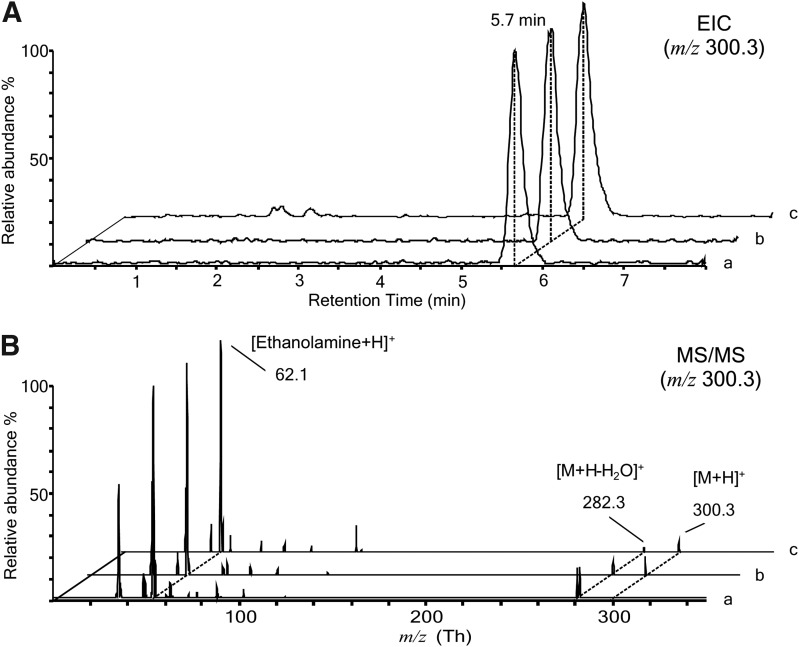
First, all solvents were carefully checked as a possible source of contamination. Skonberg et al.21 have found that chloroform from certain vendors contained small amounts of PEA. We could not confirm those findings: all solvents used during our procedure for extraction of FAEs and ECBs from serum were found to be FAEs free. This prompted us to carefully check all the glassware, disposable and not, used to carry out the extraction procedure. This included 2-mL LC vials, vial inserts, 8-mL disposable glass vials, glass beakers, glass solvents bottles, disposable glass Pasteur pipettes, and the polyurethane foam used to wrap them. LC-MS and MS/MS traces of authentic PEA standard and chloroform–methanol washout of glass pipettes and extracts of polyurethane foam were recorded in the positive ion mode. LC-MS data were acquired on two separate QQQ instruments (Agilent G6410A and Waters Xevo TQ-S Mass Spectrometer) and on a high accuracy IT-TOF mass spectrometer (Shimadzu IT-TOF) to confirm identification by accurate mass measurement. Figure 1 shows the comparative LC-MS (panel A) and LC-MS/MS (panel B) analysis of PEA standard (traces a), pipette washout (traces b), and foam extract (traces c) carried out on the Agilent QQQ. The MS1 analysis in panel A shows the extracted ion chromatograms (EIC) for the m/z 300.3 belonging to the PEA-positive pseudomolecular ion [M+H]+. These three chromatograms show coeluting peaks at retention time (RT) 5.7 min for the m/z 300.3. The MS2 analysis in Figure 1B shows the fragmentation pattern obtained by Collision-Induced Dissociation (Fragmentor 135 V, CID 20 eV) of the peaks at 300.3 in each chromatogram. The obtained MS/MS spectra are superimposable and show peaks of the positive pseudomolecular ion of PEA [M+H]+ at 300.3 Th, the fragment deriving from the water loss [M+H-H2O]+ at 282.3 Th, and the diagnostic fragment of ethanolamine [Ethanolamine+H]+ at 62.1 Th. The signal belonging to the ammonia loss (−17) at 283.3 Th is also visible except in trace a and b, but not c. Collectively, results in Figure 1 demonstrate that one of the contaminants, present in the pipette and in the foam extracts, gives rise to a signal detected by LC/MS analyses that is attributable to PEA, having the same RT and MS/MS fragmentation pattern of authentic standard.
FIG. 1.
Comparative LC/MS and LC-MS/MS analysis of PEA standard, pipette extract, and foam extract by triple quadrupole mass spectrometer. (A) Positive EIC chromatogram for the m/z 300.3 Th generated from a 5 μL injection of a 1 μM solution of PEA, 5 pmoles on column (trace a), of the pipette extract (trace b) and of the foam extract (trace c); (B) MS/MS analysis in the positive ion mode of the peaks in panel A at RT=5.7 min having m/z 300.3 Th. Panel A shows coeluting peaks at RT=5.7. In panel B the MS/MS spectra show peaks of the protonated PEA molecular ion [M+H]+ at m/z=300.3 Th and of its two fragment: the molecular ion arising from the water loss [M+H-H2O]+ at 282.3 Th and the ethanolamine-positive pseudomolecular ion [Ethanolamine+H]+ at 62.1 Th. EIC, extracted ion chromatograms; LC/MS, liquid chromatography–mass spectrometry; MS/MS, tandem mass spectrometry; PEA, palmitoylethanolamide; RT, retention time.
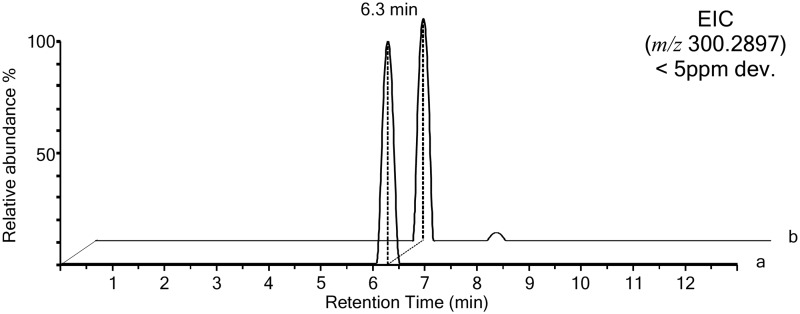
To confirm the presence of PEA as contaminant in the pipettes and foam extracts, we also analyzed the pure standard of PEA and the pipette extract on a different instrument capable of providing accurate mass measurement up to the fourth decimal and with a ppm deviation lower than 5 (Shimadzu IT-TOF). The EIC for the high accurate exact mass of the positive pseudomolecular ion of PEA [M+H]+ at 300.2897 Th is shown in Figure 2. The spectra show coeluting peaks at RT 6.3 min for the m/z 300.2897 with ppm deviation <5. The brute formula of this compound was predicted by the Formula Predictor software as C18H37NO2, both for the PEA standard and the contaminant in the pipette with a ppm deviation of 2.7 and 3.7, respectively (Supplementary Figs. S1 and S2). This experiment unambiguously demonstrates the presence of PEA as a contaminant in the glass Pasteur pipette extract.
FIG. 2.
Comparative LC/MS analysis by high-resolution accurate mass spectrometer. Positive EIC chromatogram for the m/z 300.2897 Th (ppm dev <5) generated from a 2 μL injection of a 1 μM solution of PEA, 2 pmoles on column (trace a) and of the pipette extract (trace b). The two chromatograms show coeluting peaks for the interrogated m/z.
Other possible contaminants
To determine whether PEA was the only contaminant present in glass pipettes, we analyzed the TIC of an MS1 scan of the pipette washout. We performed this analysis in the positive ion mode on the QQQ and in both positive and negative ion mode on the IT-TOF machine (Supplementary Figs. S3 and S4, respectively). The analysis in the negative ion mode (Supplementary Fig. S4, blue trace) clearly identified two peaks attributable to palmitic acid and stearic acid by RT and accurate exact mass (data not shown). Analysis in the positive ion mode showed a number of peaks on both instruments among which PEA is a contaminant. It is outside the scope of this article to provide a comprehensive identification of all contaminants, which would require extensive structure elucidation work by MS analysis. Importantly, however, analysis of other FAEs and monoacylglycerols commonly studied in the ECB field resulted in no evidence of their presence in foam (5 mg). These analytes included: PEA for reference (Supplementary Fig. S5A, 100 pmol; Supplementary Fig. S5B, foam), OEA (Supplementary Fig. S5C, 100 pmol; Supplementary Fig. S5D, foam), AEA (Supplementary Fig. S5E, 100 pmol; Supplementary Fig. S5F, foam), DHEA (Supplementary Fig. S5G, 100 pmol; Supplementary Fig. S5H, foam), 2AG (Supplementary Fig. S5I, 100 pmol; Supplementary Fig. S5J, foam), and DHG (Supplementary Fig. S5K, 100 pmol; Supplementary Fig. S5L, foam).
Quantitation of PEA in glass Pasteur pipettes and in plastic foam
We quantified the amount of PEA that can be extracted from a single glass pipette when washed with 8 mL of chloroform. Figure 3A shows proportional increase in concentrated extracts of 1, 2, and 3 pipettes. We also tested 9″ pipettes, where PEA was present in negligible trace amount or absent. To quantify the PEA content in each pipette we used an isotope dilution LC/MS method with acquisition in MRM mode as described in the experimental procedures. PEA was quantified in 100 μL of concentrated washout of each pipette and results are expressed as pmol of PEA in the reconstituted extract (Fig. 3A). We also quantified PEA content in pipettes from different vendors (Supplementary Fig. S5). Although the concentration of PEA increases with the number of pipettes used in the extraction procedure, the multiple experiments show a range of standard deviations (SDs) in triplicate experiments. This is observed also in pipettes produced by the same manufacturer and having the same lot number, but taken from different boxes (Fig. 1 as compared with groups 1–2 in Supplementary Fig. S6, showing independent experiments in triplicate). The average amount of PEA present in one pipette, calculated considering all nine experiments shown in Figure 3 and Supplementary Figure S6, is 33.4±4.02 pmol (mean±standard error of mean, n=9). Altogether, these results show that one wash with 8 mL of chloroform can extract an average amount of PEA of 33.4 pmol per pipette. Notably, the SD within all pipette extraction experiments ranges from 10 to 75% (average SD is around 30% in triplicate experiments). This considerable data scattering indicates that an extraction from a single pipette produced by the same manufacturer can give rise to PEA contaminations of very different extent. Finally, we employed our LC/MS method to quantify the amount of contaminant PEA present in the polyurethane foam used to wrap the glass pipettes. Figure 3B shows a strong linear relationship between mass of foam insert and calculated PEA values.
Transfer of PEA from the foam to the glass
A washed clean glass Pasteur pipette was incubated in contact with the foam as described in experimental procedures. Pipette extractions with subsequent LC/MS analysis were carried out before and after the washing step as well as after exposure to the foam. For reference, the intensity (ion counts) of 100 pmol PEA standard was 1.45e8 (Supplementary Fig. S7A). The PEA signal was present in the unwashed pipette at 2.19e7 (Supplementary Fig. S7B), absent in the washed pipette extract (Supplementary Fig. S7C) and present again in the pipette exposed to 5 mg foam at 1.56e6 (Supplementary Fig. S7D).
Discussion
A contaminant that is undistinguishable from PEA is present in glass Pasteur pipettes in amounts that are sufficient to interfere with analysis of biological samples. The contaminant was identified based on its LC retention time, accurate mass, and MS/MS fragmentation pattern, which were identical to those of authentic PEA. By contrast, only a negligible PEA contamination was found in 9″ Pasteur pipettes. Furthermore, we isolated the PEA contamination to the polyurethane foam used to package the pipettes, which is transferred to glass pipettes by contact. In line with this finding, Oddi et al.22 recently reported that FAEs can be absorbed by plastic materials during laboratory assays. It is therefore conceivable that FAEs incidentally absorbed by plastics during industrial processes can be released later in organic solvents. Lastly, no other commonly analyzed FAEs or monoacylglycerols were found to be present in the pipettes.
We published GC/MS23 and LC/MS5 analytical methods for the quantitation of ECBs and other related FAEs and monoacylglycerols in biological samples, including human serum.6 Prompted by the need for a novel quantitative LC/MS method to analyze ECBs in blood, we reviewed the literature and noticed discrepancies in the reported concentrations of FAEs and ECBs in human blood serum and plasma (Table 1). The EC50 for anandamide and 2AG vary depending upon assay and tissue; however, it is important to note that levels reported in Table 1 for both compounds in plasma/serum are below the apparent biologically active concentrations required to activate CB receptors (see for comprehensive review of specific assays used by a variety of research groups24).
Regarding relative levels of PEA and OEA, a number of studies reported very similar concentrations for both compounds,8–11,15 whereas others reported PEA approximately twice higher than OEA.6,7,12,13,16,17 Regarding absolute values, two separate laboratories reported levels of PEA and OEA in plasma that were excessively high,18,19 which reached or exceeded the concentrations needed by these ligands to engage PPAR-α as agonists. PEA and OEA are, in fact, considered high-potency ligands of PPAR-α; in heterologous expression systems, these FAEs engage the receptor with median effective concentration (EC50) values of 0.12 μM for OEA and 3 μM for PEA.25,26 In the abovementioned reports,18,19 although PEA levels did not exceed the EC50, levels of PEA were high relative to other reports.6–17 The steady-state concentrations of FAEs (and ECBs) in plasma/serum of healthy individuals possibly reflect an equilibrium of ECBs released by peripheral tissues and their enzymatic degradation in the blood stream. In animal tissues (e.g., brain and upper small intestine), levels of PEA and OEA are present in the same order of magnitude27,28; therefore, it was predictable to find a similar pattern in human serum or plasma, as also shown by the literature reports in Table 1. Surprisingly, in our preliminary experiments, the measured level of OEA was in agreement with most literature reports, whereas PEA was one order of magnitude higher than expected (Table 1). This finding prompted us to carefully screen all possible sources of contamination, including solvents, reagents, and glassware used for lipid extraction and quantitative analysis.
In this study, we identify glass Pasteur pipettes (5″¾) used to transfer solvents and lipid extracts as the source of PEA contamination. The contaminant was identified as PEA by its exact mass and RT in three similar but different chromatographic systems, as well as by its MS2 fragmentation pattern, which were identical to those of standard PEA. Furthermore, we show that PEA is present in the polyurethane foam that manufacturers use to wrap the pipettes before packing, from which it leaks onto the glass pipettes. Moreover, accurate exact mass measurements with ppm deviation lower than five unambiguously confirmed the identity of the contaminant as PEA. Quantitative assessment showed that the content of PEA is 33.4±4.02 pmol per pipette. Unfortunately, none of the various manufacturers, whose pipettes were tested, provides 5″¾ glass Pasteur pipette that are contaminant free (Supplementary Fig. S6). Only 9″ pipettes from one vendor were free of PEA traces (Fig. 3A), allowing the use of these consumables in the overall procedure.
The field of lipidomics is rapidly developing; however, reproducible standard procedures across laboratories are not established. Therefore, it is not uncommon for lipidomics data to differ among from independent laboratories.29 It is generally thought that these discrepancies are a result of the use of different instruments for lipid analysis, as well as differing extraction and separation protocols. In this study, however, all results were confirmed by two independent laboratories using different LC systems and QQQ mass spectrometers (Agilent 1200 LC system coupled to an Agilent G6410A QQQ Mass Spectrometer in the Piomelli laboratory, and the Waters ACQUITY UPLC I-Class system coupled to a Xevo TQ-S QQQ Mass Spectrometer in the DiPatrizio laboratory). Furthermore, accurate mass data were acquired on a third Shimadzu IT-TOF High-Resolution Mass Spectrometer for definitive confirmation that the contaminant was indeed PEA. Another well-known issue in lipidomics is that various sources of contamination can originate artifacts. Lipids, especially fatty acids, are common contaminants in detergents, mineral oils, greases, and plasticizers; hence, they are often present in laboratory equipment, including glassware and solvents. As shown in this study, assessment of FAEs, a group of lipids with diverse signaling properties, is not sheltered from this pitfall. We have shown that glass Pasteur pipettes, commonly used in lipidomic laboratories to transfer lipid extracts and organic solvents, can contain PEA as contaminant. This contamination gives rise to artifacts in the measurement of PEA in biological samples, especially when the procedure for sample preparation includes fractionation of the lipid extract, which concentrates the contaminant. The scope of this study is an alert to the ECB and FAE scientific community about possible PEA analytical artifacts and thus, great care is needed to exclude the possibility of contaminants when analyzing endogenous PEA levels in biological tissues.
Conclusion
In this study, we identified PEA as a quantitatively relevant contaminant present in glass pipettes as well as in the polyurethane foam used to wrap pipettes in the packaging. We strongly recommend examining glassware used to carry out the procedure by including in analysis a “blank” extraction, whereby one additional extract is subjected to the exact protocol of lipid extraction and processing, but with no biological sample present. In particular, we warn readers to be cautious with all plastic (foam) and glass materials that can release FAE-like compounds leading to an overestimation of concentrations in biological samples. To avoid these pitfalls it would be convenient to indicate vendors in the material section for all consumables, including glassware. High-quality certified disposable glassware for LC/MS analysis is already on the market, but limited to vials and inserts. The present article shows that there is an urgent need to expand the variety of high-quality disposable glassware, including glass pipettes, optimized for lipid extraction. In the meantime, we additionally recommend including a washing step of the glassware, with either chromosulfuric acid mixture or chloroform, in the laboratory workflow.
Supplementary Material
Abbreviations Used
- 2AG
2-arachidonoyl-sn-glycerol
- AEA
arachidonoylethanolamide, anandamide
- CB
cannabinoid receptor
- DHEA
docosahexaenoyl ethanolamide
- DHG
docosahexaenoyl glycerol
- ECBs
endocannabinoids
- EIC
extracted ion chromatograms
- ESI
electrospray ionization
- FAEs
fatty acid ethanolamides
- GC/MS
gas chromatography–mass spectrometry
- ISTDs
deuterium-containing internal standards
- IT-TOF
ion trap–time-of-flight
- LC/MS
liquid chromatography–mass spectrometry
- MRM
multiple reaction-monitoring
- MS/MS
tandem mass spectrometry
- OEA
oleoylethanolamide
- PEA
palmitoylethanolamide
- PPAR-α
peroxisome proliferator-activated receptor-α
- QQQ
triple quadrupole
- SD
standard deviation
Acknowledgments
The authors gratefully acknowledge funding support from the National Institutes of Health (NIH) grant DA034009 to N.V.D., and DA012413 to D.P. They thank Dr. Hamid Moradi for kindly providing human serum samples. They are grateful to Lucia D'Accolti for the availability of the ion trap–time-of-flight instrument and to Angela Corcelli for laboratory support. The human subjects project described was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, NIH, through grant UL1 TR000153. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Piomelli D. More surprises lying ahead: the endocannabinoids keep us guessing. Neuropharmacology. 2014;76:228–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pontis S, Ribeiro A, Sasso O, et al. Macrophage-derived lipid agonists of PPAR-alpha as intrinsic controllers of inflammation. Crit Rev Biochem Mol Biol. 2016;51:7–14 [DOI] [PubMed] [Google Scholar]
- 3.Devane WA, Hanus L, Breuer A, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949 [DOI] [PubMed] [Google Scholar]
- 4.Di Marzo V, Fontana A, Cadas H, et al. Formation and inactivation of endogenous cannabinoid anandamide in central neurons. Nature. 1994;372:686–691 [DOI] [PubMed] [Google Scholar]
- 5.Astarita G, Piomelli D. Lipidomic analysis of endocannabinoid metabolism in biological samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2755–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber D, Harlfinger S, Nolden BM, et al. Determination of anandamide and other fatty acyl ethanolamides in human serum by electrospray tandem mass spectrometry. Anal Biochem. 2007;361:162–168 [DOI] [PubMed] [Google Scholar]
- 7.Gachet MS, Rhyn P, Bosch OG, et al. A quantitiative LC-MS/MS method for the measurement of arachidonic acid, prostanoids, endocannabinoids, n-acylethanolamines and steroids in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 2015;976–977:6–18 [DOI] [PubMed] [Google Scholar]
- 8.Balvers MG, Verhoeckx KC, Witkamp RF. Development and validation of a quantitative method for the determination of 12 endocannabinoids and related compounds in human plasma using liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1583–1590 [DOI] [PubMed] [Google Scholar]
- 9.Balvers MG, Wortelboer HM, Witkamp RF, et al. Liquid chromatography-tandem mass spectrometry analysis of free and esterified fatty acid n-acyl ethanolamines in plasma and blood cells. Anal Biochem. 2013;434:275–283 [DOI] [PubMed] [Google Scholar]
- 10.Lin L, Yang H, Jones PJ. Quantitative analysis of multiple fatty acid ethanolamides using ultra-performance liquid chromatography-tandem mass spectrometry. Prostaglandins Leukot Essent Fatty Acids. 2012;87:189–195 [DOI] [PubMed] [Google Scholar]
- 11.Ozalp A, Barroso B. Simultaneous quantitative analysis of n-acylethanolamides in clinical samples. Anal Biochem. 2009;395:68–76 [DOI] [PubMed] [Google Scholar]
- 12.Ottria R, Ravelli A, Gigli F, et al. Simultaneous ultra-high performance liquid chromathograpy-electrospray ionization-quadrupole-time of flight mass spectrometry quantification of endogenous anandamide and related n-acylethanolamides in bio-matrices. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;958:83–89 [DOI] [PubMed] [Google Scholar]
- 13.Lam PM, Marczylo TH, Konje JC. Simultaneous measurement of three n-acylethanolamides in human bio-matrices using ultra performance liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2010;398:2089–2097 [DOI] [PubMed] [Google Scholar]
- 14.Jumpertz R, Wiesner T, Bluher M, et al. Circulating endocannabinoids and n-acyl-ethanolamides in patients with sleep apnea—specific role of oleoylethanolamide. Exp Clin Endocrinol Diabetes. 2010;118:591–595 [DOI] [PubMed] [Google Scholar]
- 15.Caraceni P, Viola A, Piscitelli F, et al. Circulating and hepatic endocannabinoids and endocannabinoid-related molecules in patients with cirrhosis. Liver Int. 2010;30:816–825 [DOI] [PubMed] [Google Scholar]
- 16.Hill MN, Miller GE, Carrier EJ, et al. Circulating endocannabinoids and n-acyl ethanolamines are differentially regulated in major depression and following exposure to social stress. Psychoneuroendocrinology. 2009;34:1257–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer C, Enning F, Mueller JK, et al. Fatty acid ethanolamide levels are altered in borderline personality and complex posttraumatic stress disorders. Eur Arch Psychiatry Clin Neurosci. 2014;264:459–463 [DOI] [PubMed] [Google Scholar]
- 18.Bilgin M, Bindila L, Graessler J, et al. Quantitative profiling of endocannabinoids in lipoproteins by LC-MS/MS. Anal Bioanal Chem. 2015;407:5125–5131 [DOI] [PubMed] [Google Scholar]
- 19.Sipe JC, Scott TM, Murray S, et al. Biomarkers of endocannabinoid system activation in severe obesity. PLoS One. 2010;5:e879–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biolog Chem. 1957;226:497–509 [PubMed] [Google Scholar]
- 21.Skonberg C, Artmann A, Cornett C, et al. Pitfalls in the sample preparation and analysis of n-acylethanolamines. J Lipid Res. 2010;51:3062–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oddi S, Fezza F, Catanzaro G, et al. Pitfalls and solutions in assaying anandamide transport in cells. J Lipid Res. 2010;51:2435–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giuffrida A, Piomelli D. Isotope dilution GC/MS determination of anandamide and other fatty acylethanolamides in rat blood plasma. FEBS Lett. 1998;422:373–376 [DOI] [PubMed] [Google Scholar]
- 24.Pertwee RG. Endocannabinoids and their pharmacological actions. Handb Exp Pharmacol. 2015;231:1–37 [DOI] [PubMed] [Google Scholar]
- 25.Fu J, Gaetani S, Oveisi F, et al. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-[alpha]. Nature. 2003;425:90–93 [DOI] [PubMed] [Google Scholar]
- 26.LoVerme J, La Rana G, Russo R, et al. The search for the palmitoylethanolamide receptor. Life Sci. 2005;77:1685–1698 [DOI] [PubMed] [Google Scholar]
- 27.Fu J, DiPatrizio NV, Guijarro A, et al. Sympathetic activity controls fat-induced oleoylethanolamide signaling in small intestine. J Neurosci. 2011;31:5730–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bardou I, DiPatrizio NV, Brothers HM, et al. Pharmacological manipulation of cannabinoid neurotransmission reduces neuroinflammation associated with normal aging. Health. 2012;4:679–684 [Google Scholar]
- 29.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat Rev Mol Cell Biol. 2010;11:593–598 [DOI] [PubMed] [Google Scholar]
References
Cite this article as: Angelini R, Argueta DA, Piomelli D, DiPatrizio NV (2017) Identification of a widespread palmitoylethanolamide contamination in standard laboratory glassware, Cannabis and Cannabinoid Research 2:1, 123–132, DOI: 10.1089/can.2017.0019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.