ABSTRACT
Thermococcus kodakarensis is a hyperthermophilic archaeon that harbors a complete set of genes for chitin degradation to fructose 6-phosphate. However, wild-type T. kodakarensis KOD1 does not display growth on chitin. In this study, we developed a T. kodakarensis strain that can grow on chitin via genetic and adaptive engineering. First, a chitinase overproduction strain (KC01) was constructed by replacing the chitinase gene promoter with a strong promoter from the cell surface glycoprotein gene, resulting in increased degradation of swollen chitin and accumulation of N-,N′-diacetylchitobiose in the medium. To enhance N-,N′-diacetylchitobiose assimilation in KC01, genes encoding diacetylchitobiose deacetylase, exo-β-d-glucosaminidase, and glucosamine-6-phosphate deaminase were also overexpressed to obtain strain KC04. To strengthen the glycolytic flux of KC04, the gene encoding Tgr (transcriptional repressor of glycolytic genes) was disrupted to obtain strain KC04Δt. In both KC04 and KC04Δt strains, degradation of swollen chitin was further enhanced. In the culture broth of these strains, the accumulation of glucosamine was observed. KC04Δt was repeatedly inoculated in a swollen-chitin-containing medium for 13 cultures. This adaptive engineering strategy resulted in the isolation of a strain (KC04ΔtM1) that showed almost complete degradation of 0.4% (wt/vol) swollen chitin after 90 h. The strain produced high levels of acetate and ammonium in the culture medium, and, moreover, molecular hydrogen was generated. This strongly suggests that strain KC04ΔtM1 has acquired the ability to convert chitin to fructose 6-phosphate via deacetylation and deamination and further convert fructose 6-phosphate to acetate via glycolysis coupled to hydrogen generation.
IMPORTANCE Chitin is a linear homopolymer of β-1,4-linked N-acetylglucosamine and is the second most abundant biomass next to cellulose. Compared to the wealth of research focused on the microbial degradation and conversion of cellulose, studies addressing microbial chitin utilization are still limited. In this study, using the hyperthermophilic archaeon Thermococcus kodakarensis as a host, we have constructed a strain that displays chitin-dependent hydrogen generation. The apparent hydrogen yield per unit of sugar consumed was slightly higher with swollen chitin than with starch. As gene manipulation in T. kodakarensis is relatively simple, the strain constructed in this study can also be used as a parent strain for the development and expansion of chitin-dependent biorefinery, in addition to its capacity to produce hydrogen.
KEYWORDS: chitin, chitinase, hyperthermophile, archaea, Thermococcus kodakarensis, Archaea, Thermococcus, chitinases, hyperthermophiles
INTRODUCTION
Chitin is a linear homopolymer of β-1,4-linked N-acetylglucosamine (GlcNAc) and is largely present in the exoskeleton of crustaceans and insects and in the cell walls of fungi. The annual steady-state amount of chitin formation has been estimated to be 1010 to 1011 tons (1), making it next in abundance to cellulose. However, the majority of chitin is unused by mankind and accumulates as part of the surplus biomass. Therefore, effective methods are desired to enhance the utilization of this unused biomass.
Microbial conversion of biomass into useful products provides an environmentally friendly alternative to the conventional manufacturing processes dependent on fossil fuel. In particular, the use of (hyper)thermophiles for this purpose has several advantages over use of their mesophilic counterparts as they are able to hydrolyze biomass at elevated temperatures, which promotes higher substrate solubility and catalytic rates, as well as decreasing the risk of contamination (2–6).
Thermococcus kodakarensis strain KOD1 is a hyperthermophilic archaeon, isolated near the coast of Kodakara Island, Kagoshima, Japan (7, 8). T. kodakarensis KOD1 can grow on a variety of carbon sources, such as amino acids, starch (maltodextrin), and pyruvate. The strain is also known to produce molecular hydrogen at a relatively high rate (9–12). The entire genome sequence of T. kodakarensis KOD1 has been determined (13), and gene manipulation systems based on homologous recombination and shuttle vectors have been developed (14–18). These have frequently been used to study gene function in vivo as well as to engineer T. kodakarensis for biotechnological purposes (10, 19).
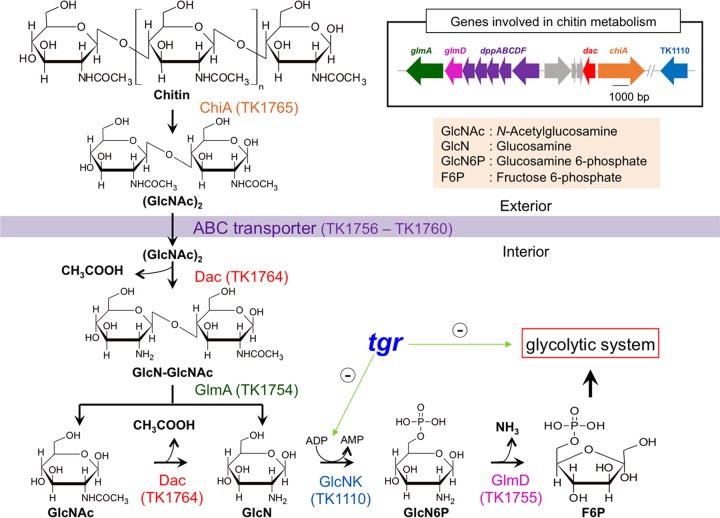
T. kodakarensis harbors a gene cluster for chitin degradation/assimilation on its genome (20). Our group has previously characterized the chitinase (ChiA; encoded by the TK1765 gene) (21–23), the diacetylchitobiose deacetylase (Dac; encoded by the TK1764 gene) (24), the exo-β-d-glucosaminidase (GlmA; encoded by the TK1754 gene) (24, 25), and the glucosamine-6-phosphate deaminase (GlmD; encoded by the TK1755 gene) (20). The chitin degradation pathway of T. kodakarensis is unique (Fig. 1). Chitin is first degraded into the disaccharide N-,N′-diacetylchitobiose [(GlcNAc)2] by ChiA, which possesses two catalytic domains, one exhibiting endo-type activity and the other exhibiting exo-type activity (21–23). The (GlcNAc)2 is then presumed to be taken up by an ABC transport system, whose genes (dppABCDF) are located adjacent to glmDA. The acetyl group of the nonreducing side of (GlcNAc)2 is first deacetylated by Dac, and the partially deacetylated disaccharide is hydrolyzed into glucosamine (GlcN) and GlcNAc by GlmA, followed by further deacetylation of GlcNAc to GlcN by Dac (24, 25). The gene cluster lacks a gene for the phosphorylation of GlcN to generate glucosamine 6-phosphate (GlcN6P). However, it is thought to be mediated by an ADP-dependent glucokinase (GK) encoded at a different locus (the TK1110 gene), as the orthologs from closely related archaea, Pyrococcus furiosus and Thermococcus litoralis, are capable of phosphorylating GlcN to GlcN6P in vitro (26). GlcN6P is further deaminated and converted to fructose 6-phosphate (F6P) by GlmD (20) flowing into the glycolytic pathway.
FIG 1.
Chitin metabolic pathway of T. kodakarensis. Chitin is converted into N-,N′-diacetylchitobiose, (GlcNAc)2, by extracellular chitinase (ChiA) encoded by the TK1765 gene. An ABC transporter (DppABCDF) encoded by the genes at locus TK1756 to TK1760 may allow (GlcNAc)2 to enter into the cell, where deacetylation of the nonreducing end occurs by diacetylchitobiose deacetylase (Dac) encoded by the TK1764 gene. Exo-β-d-glucosaminidase (GlmA) encoded by the TK1754 gene hydrolyzes the product (GlcN-GlcNAc) to glucosamine (GlcN) and N-acetylglucosamine (GlcNAc). Dac further converts GlcNAc to GlcN with release of one more acetate. In T. kodakarensis, there is no homolog for GlcN kinase (GlcNK) that produces glucosamine 6-phosphate (GlcN6P), but this reaction is thought to be performed by an ADP-dependent glucokinase encoded by the TK1110 gene. GlcN6P is further deaminated, leaving ammonium, by GlcN6P deaminase (GlmD) encoded by the TK1755 gene. The fructose 6-phosphate (F6P) formed in this reaction further enters into the glycolytic pathway. Tgr is a transcriptional repressor for glycolytic genes including TK1110 (green arrows with minus signs represent effects of Tgr on transcriptional repressions of glycolytic genes).
As described above, T. kodakarensis KOD1 harbors a complete set of genes for chitin degradation and assimilation. In addition, the transcription levels of the genes in the cluster are induced in the presence of (GlcNAc)2 (20, 24, 25). Intriguingly however, T. kodakarensis does not show robust growth on medium containing crystal/swollen chitin as the major carbon source. In the present study, we have engineered T. kodakarensis via repeated genetic manipulations followed by an adaptive engineering approach. As a result, a T. kodakarensis strain that can degrade and assimilate chitin was obtained that can efficiently produce molecular hydrogen dependent on the degradation of swollen chitin.
RESULTS
Construction of chiA overexpression strain KC01 and its characterization.
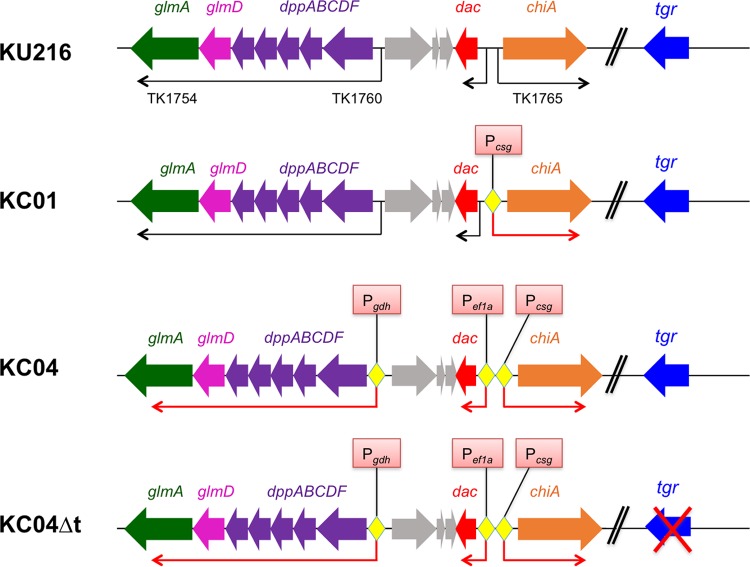
In order to enhance the chitin degradation capability of T. kodakarensis, a chitinase gene (chiA) overexpression strain (KC01) was constructed by replacing its original promoter with a strong promoter from the TK0895 gene (Fig. 2). TK0895 encodes a cell surface glycoprotein (Csg) that is strongly expressed in T. kodakarensis, and its promoter has previously been used to overexpress ChiAΔ4 (the C-terminal domain of T. kodakarensis ChiA) (19), endogenous pantoate kinase (27), and α-1,4-glucan phosphorylase from Sulfolobus solfataricus (28) in T. kodakarensis. The csg promoter was placed upstream of chiA (TK1765) in plasmid pUD3 (27) harboring pyrF used for transformant selection (see Fig. S1 in the supplemental material). The plasmid constructed (pUD3-Pcsg-chiA) (Fig. S2) was used to transform T. kodakarensis KU216 (host strain) to obtain strain KC01.
FIG 2.
Engineering of T. kodakarensis strains. KU216 is a T. kodakarensis host strain expressing GlmA, GlmD, ABC transporter, Dac, and ChiA with their native promoters. In strain KC01, the native promoter of ChiA was replaced with a strong promoter, Pcsg. KC01 was further modified to produce strain KC04 by replacing promoters of an operon (dppABCDF-glmDA) and the dac gene with strong promoters of Pgdh and of the EF-1α gene, respectively. Strain KC04 was further modified to produce strain KC04Δt by deleting tgr that is involved in transcriptional repression of glycolytic enzymes.
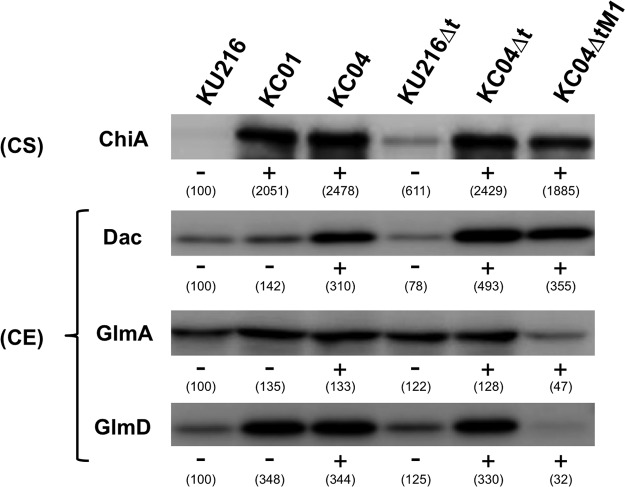
Protein expression levels of ChiA were analyzed using KU216 and KC01 cells cultivated in nutrient-rich medium containing chitin oligosaccharides (ASW-VMT-Mdx-CO medium, consisting of artificial salt water [ASW], vitamin mixture with trace minerals [VMT], maltodextrin [Mdx], and chitin oligomers [CO]) (Table 1). Culture supernatants and cell extracts were prepared and subjected to SDS-PAGE. Western blot analysis using anti-ChiA antibodies revealed that ChiA was overexpressed and secreted efficiently to the supernatant of KC01 cultures (Fig. 3). A chitinase assay of the culture supernatants using p-nitrophenyl-(GlcNAc)2 revealed the presence of chitinase activity only for KC01 (data not shown), indicating that ChiA secreted in the culture supernatant was active. In this strain, increased expression levels of GlmD, GlmA, and Dac were also observed. This is most likely caused by the generation of (GlcNAc)2, which is the product of the ChiA reaction, in the culture medium. The expression of GlmD, Dac, and GlmA has previously been shown to be induced in the presence of (GlcNAc)2 (20, 24, 25).
TABLE 1.
Compositions of culture media used in this study
| Medium type and name | Composition | Reference or source |
|---|---|---|
| Base media | ||
| ASW-YT | 0.8× ASW, 5.0 g/liter liter yeast extract, and 5.0 g liter−1 tryptone | 14 |
| ASW-AA | 0.8× ASW, a mixture of 20 amino acids, a vitamin mixture, and a modified Wolfe's trace minerals mixture | 14 |
| ASW-VMT | 0.8× ASW, 5.0 g liter−1 tryptone, a vitamin mixture, and a modified Wolfe's trace minerals mixture (including 3μM NiCl2 and 10μM Na2WO4) | This study |
| Culture media | ||
| ASW-YT-S0 | ASW-YT plus 2.0 g/liter sulfur powder | 27 |
| ASW-YT-Pyr | ASW-YT plus 5.0 g/liter sodium pyruvate | 27 |
| ASW-AA-S0 | ASW-AA plus 2.0 g/liter sulfur powder | 14 |
| ASW-VMT-SC | ASW-VMT plus 4.0 g/liter swollen chitin | This study |
| ASW-VMT-CO | ASW-VMT plus 1.0 g/liter chitin oligomers | This study |
| ASW-VMT-Mdx-CO | ASW-VMT plus 5.0 g/liter maltodextrin and 1.0 g/liter chitin oligomers | This study |
| ASW-VMT-Pyr | ASW-VMT plus 5.0 g/liter sodium pyruvate | This study |
FIG 3.
Expression levels of ChiA, Dac, GlmA, and GlmD in engineered T. kodakarensis strains. All strains were grown in ASW-VMT-Mdx-CO medium. Western blot analysis of ChiA was performed using culture supernatants (CS). Culture supernatant corresponding to a volume of 150 μl was loaded on each lane. Western blot analyses of Dac, GlmA, and GlmD were performed using cell extracts (CE). An equal amount (10 μg) of protein was loaded on each lane. A plus sign indicates overexpression of the enzyme performed by promoter replacement, and a minus sign indicates that there is no promoter replacement. Numbers in parentheses indicate band intensities (percent) relative to those of KU216 cultivated under the same medium condition (which is defined as 100%).
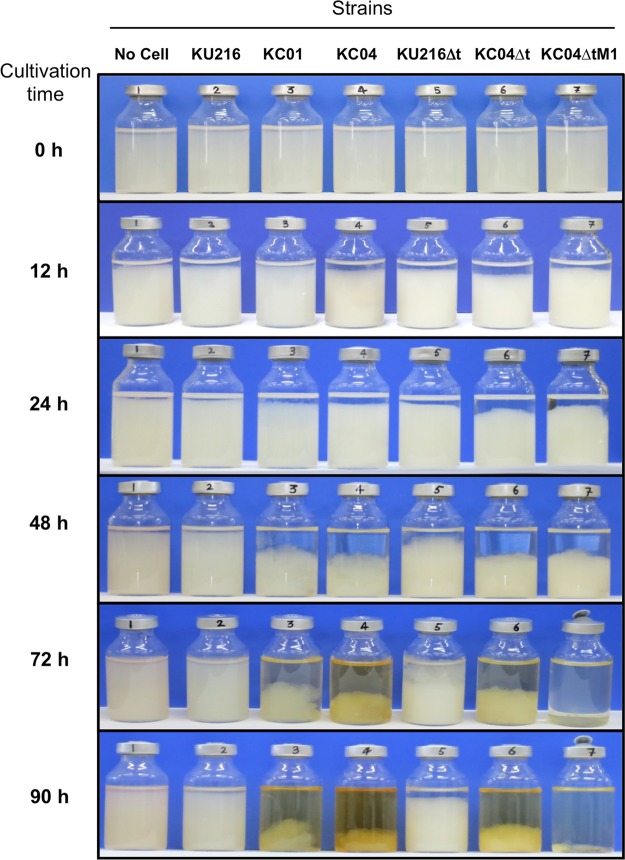
Chitin degradation abilities of KC01 were examined at 85°C using a medium containing 0.4% (wt/vol) swollen chitin (ASW-VMT-SC medium, where SC is swollen chitin) (Table 1). After 48 h of cultivation, a clear decrease in swollen chitin was detected for KC01, and almost half disappeared after 90 h, with a slight color change (yellowish) of the culture broth (Fig. 4). On the other hand, no sign of chitin degradation was observed for the host strain, KU216, even after 90 h.
FIG 4.
Degradation of swollen chitin by engineered strains of T. kodakarensis. Cultivations were performed in ASW-VMT-SC medium at 85°C, and degradation of swollen chitin was observed at different time points until 90 h. An increased capacity to degrade swollen chitin was observed in strain KC04ΔtM1.
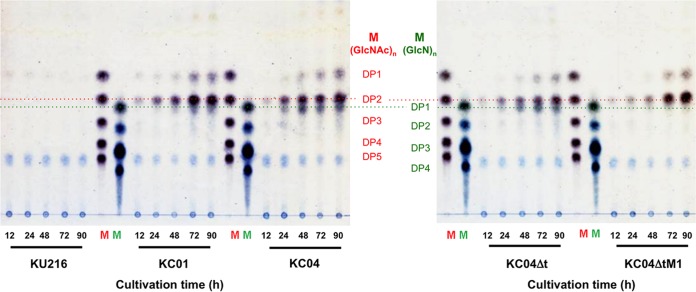
Degradation products of swollen chitin in the culture supernatants of KC01 were analyzed by thin-layer chromatography (TLC). Cells were cultivated at 85°C in ASW-VMT-SC medium, and culture supernatants were collected at different cultivation periods (until 90 h). In the culture supernatants of KC01, accumulation of (GlcNAc)2 as well as of GlcNAc was detected. The abundances of these products increased with cultivation time and reached their maxima at around 72 to 90 h (Fig. 5). On the other hand, no such degradation was detected with KU216 cells.
FIG 5.
TLC analysis of supernatants after cultivation of engineered T. kodakarensis strains in ASW-VMT-SC medium. Chitin degradation products in culture supernatant were analyzed at different time points until 90 h. Lane M, standard GlcNAc oligomers ranging from DP1 to DP5, or standard GlcN oligomers ranging from DP1 to DP4. DP indicates the degree of polymerization. Culture supernatants of KC04ΔtM1 cells displayed increased accumulation of (GlcNAc)2 with very little accumulation of GlcN.
Construction and characterization of strains KC04 and KC04Δt.
As we observed accumulation of (GlcNAc)2 in the culture supernatant of KC01, we next set out to enhance the production of enzymes involved in (GlcNAc)2 assimilation. Strain KC04 was constructed in which strong promoters, the csg promoter, the promoter of the EF-1α gene, and gdh promoter, were introduced upstream of chiA, dac (TK1764), and the dppABCDF-glmDA operon (TK1754-TK1760), respectively (Fig. 2).
Based on KC04, strain KC04Δt was further constructed by disrupting the TK1769 gene (Fig. 2). TK1769 encodes a transcriptional regulator, Tgr (TrmBL1), which controls the transcription levels of genes involved in glycolysis and gluconeogenesis in T. kodakarensis (29). In the case of glycolytic genes, Tgr binds to a sequence motif (TGM; Thermococcales glycolytic motif) that is located downstream of the BRE/TATA sequence (30). Binding of Tgr to the motif blocks transcription through the inhibition of RNA polymerase recruitment to the promoter. Binding of maltotrioses to Tgr triggers their release from the TGM sites, resulting in the derepression of glycolytic genes. Gene disruption of Tgr thus results in an increase in the transcript levels of the glycolytic genes even in the absence of α-glucans (29). If GK, encoded by the TK1110 gene, is also responsible for glucosamine kinase activity, the Tgr gene disruption should lead to an increase in glucokinase/glucosamine-kinase activity in the cells. Moreover, tgr disruption should also increase the total glycolytic flux of the cells, particularly from F6P to pyruvate. Therefore, we expected that KC04Δt would satisfy all the requirements for the breakdown and assimilation of chitin.
To examine the expression levels of ChiA, Dac, GlmD, and GlmA in T. kodakarensis KC04 and KC04Δt, Western blot analyses were performed. The two strains were grown in ASW-VMT-Mdx-CO medium. As a result, levels of ChiA were much higher in the culture supernatants of KC04 and KC04Δt than in the supernatant of KU216, and levels of Dac, GlmD, and GlmA were also higher in the cell extracts of KC04 and KC04Δt than in the cell extract of KU216 (Fig. 3).
Chitin degradation capabilities of strains KC04 and KC04Δt were examined using ASW-VMT-SC medium. In KC04, the degree of swollen chitin degradation was slightly higher than that in KC01, while the color of the culture broth looked darker (yellowish) after 90 h (Fig. 4). As for the KC04Δt strain, swollen chitin degradation was comparable to that of KC04, while the color of the culture broth was relatively light.
When degradation products of chitin in the culture supernatants were analyzed, we noticed the accumulation of GlcN in addition to (GlcNAc)2 in both strains (Fig. 5). This suggests that (GlcNAc)2 conversion to GlcN was enhanced, while a bottleneck reaction was present after GlcN generation.
The chitin degradation capacities of KC04 and KC04Δt were also estimated by examining acetate and ammonium concentrations in the culture supernatants after growth in ASW-VMT-SC medium for 72 h. Acetate levels in the cultures of KC04 and KC04Δt were similar to each other and were higher than those in the cultures of KC01 and KU216 (Fig. 6A). On the other hand, ammonium levels of KC04 and KC04Δt were almost equal to the level of KC01 (Fig. 6B).
FIG 6.
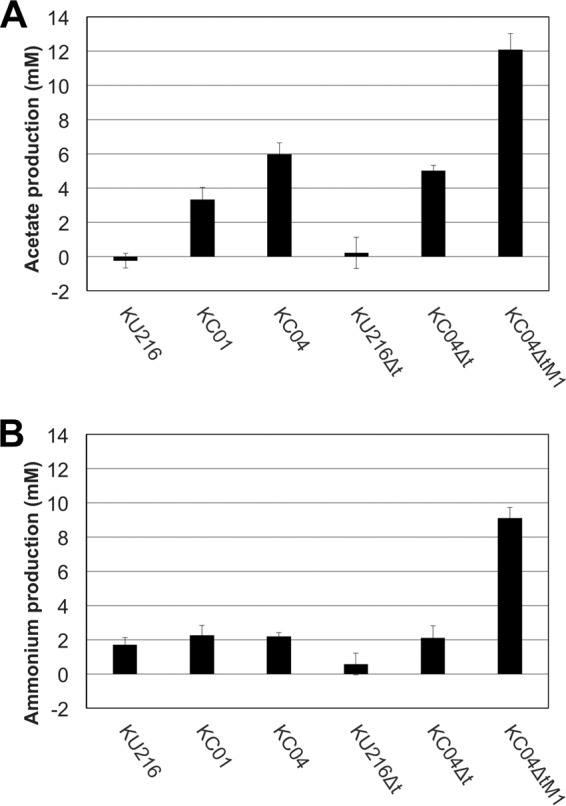
Quantification of acetate and ammonium concentrations in culture supernatants of engineered strains of T. kodakarensis growing on swollen chitin. Cultivations were performed in ASW-VMT-SC medium at 85 °C for 72 h, and culture supernatants prepared were analyzed. Results shown were determined by subtracting acetate or ammonium concentrations present in culture medium without inoculation. Error bars represent standard deviations of three independent measurements. KC04ΔtM1 showed increased production of both acetate and ammonium.
Overall, the studies on the metabolites indicated that the chitin degradation capacities of KC04 and KC04Δt were significantly increased by the genetic manipulations, but these strains still display GlcN accumulation in the culture broth. As GlcN contains an amino group and a reducing group, excess accumulation of GlcN at high temperatures should promote the Maillard reaction that leads to yellowish by-products, which agrees well with the yellowish appearance of the medium that we observed. The progression of the Maillard reaction should result in a substantial reduction in sugars that would otherwise be available and utilized for cell growth.
Adaptive engineering of T. kodakarensis on chitin.
The previous analyses suggested that minimizing GlcN accumulation might lead to a more efficient degradation and assimilation of chitin. As it seemed difficult to rationally optimize the balance between GlcN generation and utilization, we next employed an adaptive engineering approach. The KC04Δt strain was cultivated in ASW-VMT-SC medium up to eight serial cultivations. Although chitin degradation was observed for at least 90 h (Fig. 4), aliquots of the cultures were inoculated to the next medium after 24 h in order to enrich cells that displayed relatively faster growth on chitin. With the progression of the serial cultures, the time required to completely degrade the swollen chitin became shorter. After the eighth culture, an initial isolation was carried out on ASW-VMT-SC solid medium, and three isolates were subjected to a further five serial cultivations in ASW-VMT-SC medium. Isolation was carried out from the medium that displayed the most rapid degradation rate and resulted in the isolation of strain KC04ΔtM1. When cultures were performed with KC04ΔtM1 in ASW-VMT-SC medium, almost all of the medium became transparent by 72 h, with little color change (Fig. 4). Western blot analyses indicated that protein levels of GlmA and GlmD were unexpectedly much lower in KC04ΔtM1, while protein levels of ChiA and Dac were comparable to those in KC04Δt (Fig. 3). TLC analysis of chitin degradation products indicated that KC04ΔtM1 exhibited increased accumulation of (GlcNAc)2 but almost no accumulation of GlcN (Fig. 5). Moreover, accumulation of both acetate and ammonium was observed in the culture supernatant of KC04ΔtM1 (Fig. 6A and B), showing that both deacetylation and deamination processes were efficiently proceeding in this strain.
We next examined the possible cause of the lower expression levels of GlmD and GlmA in the KC04ΔtM1 strain. As genes for GlmD and GlmA are located in a single operon under the control of the gdh promoter, the promoter region was sequenced to examine whether it contains mutations. As a result, a 15-bp duplication (5′-ATCGAAAGGTTTATA-3′) was found that contains nearly a complete BRE/TATA sequence (Fig. S6). As the BRE/TATA sequence is involved in the process of transcriptional initiation, the duplication of BRE/TATA sequence may have caused a deleterious effect for transcription of this locus, resulting in decreased expression levels of both GlmA and GlmD.
Growth characteristics of the engineered T. kodakarensis strains.
We have described the construction of multiple strains of T. kodakarensis with the aim of developing a strain with enhanced chitin degradation capacity. This required higher expression of a relatively large number of genes related to chitin degradation as well as glycolysis. We examined if the increased expression of a high number of genes affected growth of these strains on a conventional carbon source for T. kodakarensis. We grew each strain in a typical nutrient-rich medium supplemented with pyruvate (ASW-YT-Pyr, composed of ASW, yeast extract and tryptone [YT], and pyruvate [Pyr]) (Table 1). Overall, we found that all strains displayed growth in ASW-YT-Pyr medium (Fig. S7). Growth of T. kodakarensis KOD1 exhibited two peaks in this medium, one at around 10 h and the other at around 18 h. When we compared specific growth rates among these strains, we observed a tendency that strains with a higher number of overexpressed genes exhibited lower specific growth rates (Table S1). KC04Δt, which overexpresses all of the genes related to chitin degradation and those involved in glycolysis, displayed a 25% decrease in specific growth rate compared to that of KU216. In terms of cell yield, we observed that the KC04Δt and KC04ΔtM1 strains displayed the lowest values among the strains (Fig. S7). The differences we observed can be presumed to be due to the investment of starting material and energy directed toward the overexpression of multiple proteins.
Chitin-dependent hydrogen production by T. kodakarensis KC04ΔtM1.
As T. kodakarensis is known to produce molecular hydrogen from maltooligosaccharides through primary metabolism (9, 12), we analyzed whether strain KC04ΔtM1 produced hydrogen from chitin. When T. kodakarensis KC04ΔtM1 and its host strain, KU216, were grown in ASW-VMT-SC medium, much higher levels (approximately 10-fold) of hydrogen were produced from KC04ΔtM1 than from KU216 (Fig. 7A). On the other hand, in a medium without swollen chitin (ASW-VMT medium) (Table 1), both strains produced only low levels of hydrogen (Fig. 7B), and this level was comparable to the hydrogen levels of KU216 grown in the presence of swollen chitin. This clearly indicates that most of the hydrogen produced from KC04ΔtM1 is dependent on the presence of chitin and the genetic modifications harbored in KC04ΔtM1.
FIG 7.
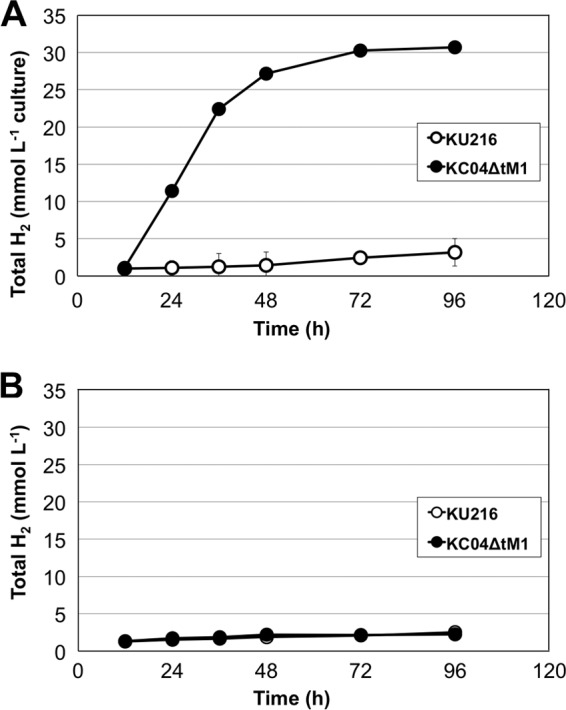
Comparison of chitin-dependent hydrogen (H2) production in KU216 and KC04ΔtM1 in ASW-VMT-SC medium (A) and in ASW-VMT medium (B). Total hydrogen represents the amount of molecular hydrogen present in the headspace of a culture bottle per milliliter of culture volume (medium volume was 15 ml). Error bars represent standard deviations of three independent cultivations. The results show that strain KC04ΔtM1 exhibits hydrogen production dependent on swollen chitin.
We estimated the amounts of chitin consumed and compared these levels with those of the production of acetate, ammonium, and hydrogen. Although we introduced 60 mg of chitin into each culture (15 ml), the results of the TLC analysis shown in Fig. 5 indicated that residual (GlcNAc)2 and GlcNAc were still present in the medium at the end of our cultures. By quantifying the residual sugars by high-performance liquid chromatography (HPLC), we found that 0.12 ± 0.01 mmol of chitin (expressed as GlcNAc units) was consumed during a 96-h culture. The concentration of hydrogen generated at this time was 30.1 ± 1.8 mmol liter−1 culture, corresponding to 0.46 ± 0.03 mmol hydrogen (Fig. 7A). We also examined the amounts of acetate and ammonium and found that 0.24 ± 0.01 mmol of acetate and 0.16 ± 0.00 mmol of ammonium were present in the medium. The maximum theoretical ratios of hydrogen/GlcNAc, acetate/GlcNAc, and ammonium/GlcNAc can be considered 4:1, 3:1, and 1:1, respectively. The results of our cultures with KC04ΔtM1 cells gave ratios of 3.79:1, 2.01:1, and 1.29:1, respectively. In order to exclude the amounts deriving from tryptone metabolism, we subtracted the values obtained in cultures with KU216 cells, resulting in ratios of 3.40:1 (hydrogen/GlcNAc), 2.01:1 (acetate/GlcNAc), and 0.99:1 (ammonium/GlcNAc).
Effect of tgr knockout on hydrogen production of KC04ΔtM1.
In order to analyze whether tgr disruption had effects on the hydrogen production of KC04ΔtM1, the tgr gene was reintroduced into KC04ΔtM1 using a pLC70-based plasmid vector (17). While introduction of pLC70M, a control vector without tgr, displayed virtually no effect on hydrogen production of KC04ΔtM1 in ASW-VMT-SC medium, reintroduction of the tgr gene resulted in a clear delay in the initiation of hydrogen generation (Fig. S8). However, hydrogen production was observed afterwards and reached levels comparable to those with KC04ΔtM1 and KC04ΔtM1/pLC70M cells. We confirmed that the reintroduction of the Tgr gene led to the production of Tgr protein in the cell (Fig. S9). These cells displayed levels of the glycolytic GK and phosphofructokinase (PFK) similar to those of strain KU216 when it was grown on pyruvate (ASW-VMT-Pyr medium), indicating that the Tgr protein in the reintroduced strain represses these two genes under gluconeogenic conditions, as expected (Fig. S10). However, when cells were grown in ASW-VMT-SC medium, we found that the effects of tgr reintroduction were less prominent, with the extents of GK and PFK repression much lower than those in cells grown on pyruvate (Fig. S9). Protein levels of Tgr were also lower than those in cells grown on pyruvate. This may be the reason why the tgr-complemented strain eventually displayed the ability to assimilate chitin and generate hydrogen.
DISCUSSION
In this study, we have constructed T. kodakarensis strains that exhibit improved abilities to degrade/assimilate chitin. In the case of strain KC04, chitin degradation was enhanced, but accumulation of GlcN in the culture supernatant as well as browning of the medium was observed, suggesting that the excess GlcN was subjected to the Maillard reaction (31, 32). Gene disruption of tgr (TK1769) was previously shown to increase activity levels of ADP-dependent GK (the TK1110 protein) in the cell extract of T. kodakarensis (29). If ADP-dependent GK also exhibits glucosamine kinase activity, activity levels of glucosamine kinase (as well as the activities of glycolytic enzymes) should also be higher in the KC04Δt strain. This might be the reason why the yellowish appearance of the medium was decreased in KC04Δt compared to that in KC04 (Fig. 4). However, the ammonium levels in the culture supernatants of KC04Δt and KC04 were similar, indicating that the flux of the following deamination reaction did not increase significantly and that chitin degradation was still incomplete (Fig. 6B).
KC04ΔtM1 exhibited the highest capacity of chitin degradation, and accumulation of GlcN was not observed (Fig. 4 and 5). Increases in both acetate and ammonium levels indicated that most of the chitin was converted through the chitin metabolic pathway (Fig. 6). As we observed only very little color change in the medium, GlcN accumulation and the progression of the Maillard reaction can be presumed to be minimal in this strain. Metabolites from chitin were efficiently assimilated and resulted in cell growth, as high levels of hydrogen production, which occur only in growing cells (12, 33), were observed. Western blot analyses revealed that expression levels of GlmD and GlmA were significantly decreased in KC04ΔtM1 compared to the level in the direct parent strain KC04Δt (Fig. 3). This was an unexpected result but can be interpreted as follows. The decrease in the levels of GlmA (exo-β-d-glucosaminidase), which generates GlcN, reduces its production rate. This can be expected to reduce the accumulation of GlcN, which is actually the case, as shown in Fig. 5. As GlcN accumulation readily causes progression of the Maillard reaction under high-temperature environments, a decrease in accumulation may prevent the loss of carbon and energy provided by GlcN. The prevention of the Maillard reaction can clearly be observed when the colors of the cultures of KC04Δt and KC04ΔtM1 at 90 h shown in Fig. 4 are compared. In addition to the loss of carbon and energy, the products of the Maillard reaction have a strong inhibitory effect on cell growth of the aerobic hyperthermophilic archaeon Aeropyrum pernix (34), and this may also be the case for T. kodakarensis.
To analyze the effect of tgr disruption on chitin assimilation of KC04ΔtM1, the tgr gene was reintroduced to the strain. This resulted in a delay in the initiation of hydrogen generation, but the strain still generated hydrogen in ASW-VMT-SC medium to a level similar to that observed with KC04ΔtM1 (see Fig. S8 in the supplemental material). The weak effect of tgr reintroduction in cells cultivated in ASW-VMT-SC medium agrees well with the fact that GK and PFK are only slightly repressed (∼30% decrease) (Fig. S9). By Western blotting, we observed that levels of Tgr protein in a tgr-complemented strain grown in ASW-VMT-SC medium was almost one-third of that grown in ASW-VMT-Pyr medium, in which levels of the repression of PFK and GK were much more significant (50 to 90% decrease) (Fig. S9). This is intriguing and may be due to the fact that the tgr gene is located on a plasmid and not on the genome, but further analysis will be necessary to elucidate the weakened repression by Tgr in chitin-grown cells.
We find it intriguing that T. kodakarensis, with a complete set of chitin-degrading genes, does not display robust growth on chitin. When we look for genes involved in chitin degradation in other hyperthermophilic archaea, we find that the distribution is very limited. Genomes that harbor homologs of the chitin-degrading enzymes of T. kodakarensis are found only in P. furiosus, Thermococcus chitonophagus, and Thermococcus nautili. In P. furiosus, two consecutive chitinase genes (Pf-chiA and Pf-chiB) are separated by a single base insertion, and the removal of this base leads to a fusion gene whose product is structurally similar to that of T. kodakarensis chiA (Tk-chiA) harboring two different catalytic domains (endo- and exo-type) (35, 36). The growth of P. furiosus on chitin seems to vary depending on the conditions of the medium or on how the cells are adapted to the presence of chitin (35–38). Recently, genetic engineering of P. furiosus was reported that conducted a deletion of the single base insertion located between Pf-chiA and Pf-chiB (39). The engineered strain with a single chitinase gene reached a maximum cell density of 1 × 108 ml−1 on colloidal chitin-containing medium, which is a 10-fold higher cell concentration than that of the wild-type strain. It is interesting that P. furiosus, that had lost its intact chitinase gene, could easily restore growth on chitin, whereas T. kodakarensis, with an intact chitinase gene, does not display growth on chitin.
T. chitonophagus is known to possess multiple chitinases (40–42). Recently, two groups of investigators have conducted genome analyses of this archaeon, and both predicted the presence of three chitinase genes (43, 44). Reclassification of the organism to the genus Pyrococcus has also been proposed (species name Pyrococcus chitonophagus) (43). Biochemical characterization of one of the T. chitonophagus chitinases, ChiD, has revealed that the enzyme is a structurally novel chitinase exhibiting exo-type activities and mainly releases (GlcNAc)2 units from the reducing ends of chitin chains (44). As T. kodakarensis ChiA contains an endo-chitinase domain and an exo-chitinase domain that recognizes only the nonreducing ends of chitin chains, introduction of the ChiD gene into T. kodakarensis KC04ΔtM1 may further improve its chitin degradation capabilities.
Using the engineered T. kodakarensis KC04ΔtM1 strain, a system for molecular hydrogen production from chitin has been established. According to our search, there is only one report on hydrogen production from chitin or chitin-containing materials. Evvyernie et al. has reported hydrogen production from a mesophilic bacterium, Clostridium paraputrificum M-21, using N-acetylglucosamine, chitin, and chitin-containing wastes (45, 46). From 1 g of raw chitinous wastes, the evolved hydrogen was reported as 5.2 to 7.6 mmol. T. kodakarensis KC04ΔtM1 exhibited a higher hydrogen conversion (18.7 ± 1.1 mmol of H2 per gram of swollen chitin [consumed]), but we should note that our study employed pure chitin.
The results of our cultures with KC04ΔtM1 cells gave ratios of 3.40:1 (hydrogen/GlcNAc), 2.01:1 (acetate/GlcNAc), and 0.99:1 (ammonium/GlcNAc). It has been shown that in the metabolism of T. kodakarensis, the pyruvate formed from GlcNAc can be directed to either acetate (with hydrogen production) or alanine (no hydrogen production) (9, 12). As the ratios of hydrogen/GlcNAc and acetate/GlcNAc, particularly the latter, are lower than the maximum values, we can expect that a portion of the pyruvate formed from GlcNAc is directed toward alanine formation. When soluble starch is used as the sugar source, the theoretical maximum ratios of hydrogen/glucose and acetate/glucose are 4:1 and 2:1, respectively. In a previous study, we observed hydrogen/glucose and acetate/glucose ratios of 3.33:1 and 1.12:1, respectively, indicating that the conversion of chitin to hydrogen is comparable to that of starch to hydrogen (9).
Our results indicate that hydrogen production ceased prior to the total consumption of (GlcNAc)2 in the medium. A possible reason for the cessation of hydrogen production and cell growth might be the accumulation of hydrogen itself. Another reason might be the drop in pH of the culture medium. We observed a decrease in pH from 6.4 to 5.4 after 72 h of cultivation in ASW-VMT-SC medium, which might be due to accumulation of acetate in the culture medium. Cultivation methods that reduce hydrogen and/or acetate accumulation (e.g., increase in the volume of the gas phase or cultivation using dialysis membrane) should contribute to further increasing the hydrogen conversion rate from chitin.
MATERIALS AND METHODS
Strains and media.
T. kodakarensis strains were cultivated with basically the same methods as described elsewhere (14, 15). T. kodakarensis KU216 (15) and its derivative strains were grown under anaerobic conditions at 85°C in either a nutrient-rich ASW-YT medium, a synthetic ASW-AA medium or a semisynthetic medium supplemented with tryptone (ASW-VMT medium). The compositions of all media used in this study are shown in Table 1. ASW-VMT-SC medium, which is ASW-VMT medium supplemented with 4.0 g liter−1 swollen chitin, was the medium used for adaptive engineering cultures and to evaluate chitin degradation of various T. kodakarensis strains. ASW-AA medium (where AA is amino acids) was used in transformation procedures. ASW-YT medium was mainly used for growth examinations and precultivation. ASW-YT, ASW-AA, and ASW-VMT media were supplemented with 2.0 g liter−1 of S0 (sulfur powder), sodium pyruvate (5.0 g liter−1), maltodextrin (5.0 g liter−1), or 1.0 g liter−1 chitin oligomers (NA-COS-Y; Yaizu Suisankagaku Industry, Yaizu, Japan) when necessary. The media used in individual experiments are indicated in the respective sections. For solid media, S0 was replaced with 2 ml of a polysulfide solution (10 g of Na2S·9H2O and 3 g of sulfur powder in 15 ml of H2O) per liter, and 10 g liter−1 of Gelrite was added to solidify the medium. When necessary, 4.0 g liter−1 swollen chitin was also added. All medium components, unless mentioned otherwise, were purchased from Wako Pure Chemical Industries (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan). Cell growth of T. kodakarensis strains grown in ASW-YT-Pyr medium at 85°C was continuously monitored at 660 nm in an incubator equipped with an optical density (OD) measurement apparatus (model OD-BR-43FH-VH; Taitec, Koshigaya, Japan) with water as a reference. Escherichia coli strain DH5α was cultivated in LB medium (47) at 37°C with 100 mg liter−1 ampicillin sodium salt. All strains and plasmids used in this study are listed in Table 2.
TABLE 2.
Strains and plasmids used in the study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Stratagene (La Jolla, CA) |
| T. kodakarensis | ||
| KOD1 | Wild type | 7, 8 |
| KU216 | KOD1 ΔpyrF | 20 |
| KC01 | KU216 Pcsg-chiA | This study |
| KC04 | KU216 Pcsg-chiA Pgdh-dppABCDF-glmDA EF-1α gene promoter-dac | This study |
| KU216Δt | KU216 Δtgr | This study |
| KC04Δt | KC04 Δtgr | This study |
| KC04ΔtM1 | KC04Δt, after adaptive engineering | This study |
| Plasmids | ||
| pUC118 | General cloning vector, Ampr | TaKaRa Bio (Otsu, Japan) |
| pUD3 | pUC118 derivative; pyrF marker cassette | 27 |
| pUD3(ΔNdeI) | pUD3 derivative; NdeI site was deleted | This study |
| pUD3-Pcsg-chiA | pUD3(ΔNdeI) derivative; Pcsg-chiA pyrF | This study |
| pUD3-dac-Pef-1α-Pcsg-chiA | pUD3(ΔNdeI) derivative; Pcsg-chiA EF-1α gene promoter-dac pyrF | This study |
| pUD3-Pgdh-dppA | pUD3(ΔNdeI) derivative; Pgdh-dppA pyrF | This study |
| pUD3_1769D | pUD3 derivative; for tgr disruption | This study |
| pLC70 | pLC64 derivative; hmg trpE | 17 |
| pLC70M | pLC70 derivative; hmg | This study |
| pLC-tgr | pLC70 derivative; hmg tgr | This study |
Preparation of swollen chitin.
Chitin from crab shells (2.5 g; Nacalai Tesque) was mixed with 125 ml of phosphoric acid (85%, wt/vol, in water) and stirred at 4°C for 24 h. The suspension was poured into 1.25 liters of deionized water and centrifuged (15,300 × g, 10 min, 4°C). The resulting precipitate was washed with deionized water several times until the pH of the suspension became neutral. The phosphoric acid treatment disrupts the hydrogen bond networks of chitin chains (48). The networks are only partially restored after washes with water, resulting in swollen chitin. A similar phosphate treatment procedure conducted at room temperature produced chitin samples with average degrees of polymerization (DP) varying from 1,300 to 110 (49). The swollen chitin was finally suspended with 100 ml of deionized water. The concentration of chitin in the suspension was determined by drying an aliquot and measuring the dry weight. TLC/HPLC analyses indicated that GlcN and GlcNAc oligomers were not present at detectable levels in our swollen-chitin preparation.
Plasmid construction.
Plasmids to construct overexpression strains for chiA (TK1765), dac (TK1764), and the dppABCDF-glmDA operon (TK1754-TK1760) were designed so that their native promoters were replaced with the following strong promoters: the cell surface glycoprotein gene (csg; TK0895) promoter (Pcsg), the promoter of the archaeal elongation factor 1α subunit gene (TK0308), and the glutamate dehydrogenase gene (gdh; TK1431) promoter (Pgdh), respectively. Gene replacements were performed via a single-crossover insertion/pop-out recombination technique (see Fig. S1 in the supplemental material) previously reported in T. kodakarensis (27, 50). Nucleotide sequences of all plasmids were examined to confirm the absence of any unintended mutations. Sequences of primers used in this study are listed in Table 3.
TABLE 3.
Primers used in the study
| Name | Sequence (5′–3′)a |
|---|---|
| ChiA_UP1_PstI | AAACTGCAGCCTTTCCTTTCTTATCACGG |
| PcsgR | GTGGTGGTGGTGGTGGTGCATATGACAACACCTCCTTGGGTT |
| OE-chiA-f-1 | AAAAGTCGACCATATGAAGAAGATTTGGACTTCA |
| OE-chiA-r-1 | AAAAGAATTCAGAGGATCAGCGTAGGTATC |
| dPchiA-inverse-f | AAAAAGATCTTATCGGCAAAAGGCGAATTATGTGT |
| dPchiA-inverse-r | ATGGTGTTTGAGGAGTTCAACAATT |
| PEF-1a-EcoRV-f | AAAAGATATCCAAACACCTCCATATTTTGGT |
| PEF-1a-r | AAAAAGATCTTGCGGGCTTTCTTCTTGTTCTCTC |
| ABCtransporter-f | AAAACTGCAGTATGTCGATGTAGTATGTACCGACG |
| ABCtransporter-r | AAAAGAATTCTGAGAAGGTTTTAACTGCCTCGTAG |
| ABCt-inverse-f | AAAAGGATCCCATATGAAGAAAGCTACCGCGGTTG |
| ABCt-inverse-r | AAAAGGATCCGGACACCACCACTTATAAGG |
| Pgdh-f | AAAAGTCGACCATATGTCATACCACCTCATTTCGGTAAT |
| Pgdh-r | AAAAGGATCCGCCCGTTGCCCGATGATTGGTTT |
| Δtk1769.1 | AAAAAAGTCGACGGTGGAAAACGCCGTCGAGTAC |
| Δtk1769.2 | TTTTTTGGATCCCGGTTATCACTTTCACGTTCTC |
| Δtk1769.3 | CCCATCATTTTTAATTTCTAAACTT |
| Δtk1769.4 | GGCTTAACCCCCAAAGACATTTAAG |
| TK1769compF1 | GGGAATTCGCGGCCGCCACTCTGCCGTGGATGAAGT |
| TK1769compR1 | GCATGGGCCCTAAAGCTATGCCCCAAAAATTCAGAAGAGAAATAGAAAAATGTAGAGGAATCACTCAAGGAGGATGAACT |
Restriction enzyme sites incorporated for genetic manipulation are underlined.
(i) Construction of a plasmid for chiA overexpression.
A chiA overexpression plasmid (pUD3-Pcsg-chiA) was constructed as follows (Fig. S2). A DNA fragment containing part of dac and the promoter region of chiA (PchiA) fused with Pcsg was amplified from pUD3-TK2141 (27) with the primer set ChiA_UP1_PstI/PcsgR and digested with PstI/NdeI (fragment A). A part of the chiA coding region (containing the start codon) was amplified with the primer set OE-chiA-f-1/OE-chiA-r-1, and the fragment was digested with NdeI/EcoRI (fragment B). Fragment A and fragment B were inserted into pUD3(ΔNdeI) digested with PstI/EcoRI, resulting in the chiA expression vector pUD3-Pcsg-chiA. pUD3(ΔNdeI) is derived from pUD3 (27) with a point mutation within the NdeI site of pUD3 (CATATG to CACATG).
(ii) Construction of a plasmid for overexpression of both chiA and dac.
The plasmid used to construct a T. kodakarensis strain that overexpresses both chiA and dac was constructed based on pUD3-Pcsg-chiA (Fig. S3). Inverse PCR was performed with the primer set dPchiA-inverse-f/dPchiA-inverse-r, followed by digestion with BglII. A 234-bp region of the EF-1α gene promoter was amplified with the primer set PEF-1a-EcoRV-f/PEF-1a-r, followed by digestion with EcoRV/BglII. These digested products were ligated to obtain the plasmid pUD3-dac-Pef-1α-Pcsg-chiA.
(iii) Construction of a plasmid for overexpression of an ABC transporter, glmD, and glmA.
For overexpression of the dppABCDF-glmDA operon, a fragment containing part of the dppA (TK1760) coding region and part of the neighboring β-glycosidase gene (TK1761), together with the promoter region located in between these genes, was amplified with the primer set ABCtransporter-f/ABCtransporter-r, followed by digestion with PstI/EcoRI (Fig. S4). The fragment was ligated with pUD3(ΔNdeI) digested with PstI/EcoRI. Inverse PCR of the plasmid was performed to separate dppA with its promoter region with the primer set ABCt-inverse-f/ABCt-inverse-r, and the amplified product was digested with NdeI/BamHI. The fragment was ligated with a 299-bp gdh promoter region that was amplified using genomic DNA of T. kodakarensis with the primer set Pgdh-f/Pgdh-r and digested with NdeI/BamHI. The plasmid was designated pUD3-Pgdh-dppA.
(iv) Construction of a tgr disruption vector.
The tgr gene (TK1769) encodes a transcriptional repressor of glycolytic enzymes (29). A disruption plasmid for the tgr gene (TK1769) (designated pUD3_1769D) was constructed as follows. The TK1769 gene together with its 5′- and 3′-flanking regions (∼1 kbp) was amplified from the T. kodakarensis genomic DNA using the primer set Δtk1769.1/Δtk1769.2. The amplified fragment was digested with SalI/BamHI and inserted into the respective sites of pUD3. To remove the tgr coding region from the resulting plasmid, inverse PCR was performed using the primer set Δtk1769.3/Δtk1769.4. The PCR product was self-ligated to construct pUD3_1769D.
(v) Construction of a tgr-containing complementation vector.
The tgr gene together with its promoter region was amplified from T. kodakarensis KOD1 genomic DNA using the primer set TK1769comp-F1/TK1769comp-R1. The primer TK1769comp-R1 contains the transcriptional terminator region (5′-TAAAGCTATGCCCCAAAAATTCAGAAGAGAAATAGAAAAATGTAGAGGAA-3′) of the P. furiosus glutamate dehydrogenase gene (PF1602). The amplified fragment contained the tgr gene flanked with its promoter region and the PF1602 terminator region and was digested with ApaI/NotI restriction enzymes. pLC70 is a shuttle vector which can autonomously replicate in both E. coli and T. kodakarensis (17). pLC70 was digested with ApaI/NotI and was ligated with the fragment containing the tgr gene, resulting in pLC-tgr (Fig. S5). A control plasmid was made to delete trpE by digestion of pLC70 using NotI/ApaI restriction enzymes, blunting, and self-ligation (pLC70M).
Transformation of T. kodakarensis.
T. kodakarensis KU216 strain (15) was the parent strain for constructing recombinant strains that can degrade and assimilate chitin. Transformation methods were performed basically as described previously (14, 15). Plasmid for transformation (3 μg) was added to the cell suspension, and the mixture was kept on ice for a further 1 h, followed by heat shock at 85°C for 45 s. The cells were transferred to ASW-AA medium supplemented with S0 (without uracil) and incubated at 85°C for 2 days. The cells were further cultivated for 2 days in the same medium to enrich the pyrF-containing transformants obtained via single-crossover recombination. Cells with pyrF deleted by pop-out recombination were selected in the presence of 5-fluoroorotic acid using ASW-AA or ASW-YT solid medium containing polysulfide. Genotypes of transformants were confirmed by PCR and direct sequencing. The shuttle plasmids pLC-tgr and pLC70M were introduced into T. kodakarensis KC04ΔtM1, and the transformants were selected by simvastatin resistance as described previously (16).
Western blot analysis of culture supernatants and cell extracts.
T. kodakarensis strains were cultivated at 85°C in ASW-VMT-Mdx-CO medium and ASW-VMT-Pyr medium for 12 h and in ASW-VMT-SC medium for 90 h. Culture supernatants were prepared by centrifugation (5,000 × g, 15 min, 4°C) of cell cultures twice. The resulting culture supernatants were concentrated 15-fold with a Vivaspin ultracentrifugation device (30-kDa-cutoff membrane; Sartorius Stedim Biotech, Göttingen, Germany). Cell pellets after the first centrifugation were resuspended in 50 mM Tris-HCl (pH 7.5) buffer and disrupted by vortexing for 5 min. The resulting suspension was centrifuged (20,400 × g, 15 min, 4°C), and the supernatant obtained was used as cell extract. The culture supernatants and cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis ([SDS-PAGE] 12.5% or 15% acrylamide concentration), followed by blotting to a polyvinylidene difluoride membrane (Hybond-P; GE Healthcare Biosciences, Chicago, IL). For detection of ChiA, Dac, GlmD, GlmA, GK (encoded by the TK1110 gene), phosphofructokinase (PFK) (encoded by the TK0376 gene), or Tgr (encoded by the TK1769 gene), rabbit anti-ChiA, anti-Dac, anti-GlmD, anti-GlmA, anti-GK, anti-PFK, or anti-Tgr antiserum was used, respectively. Horseradish peroxidase-conjugated recombinant protein G (Zymed Laboratories, San Francisco, CA) was used as the secondary antibody. For signal detection, ECL Select Western blotting detection reagent (GE Healthcare Biosciences) and ImageQuant LAS 500 (GE Healthcare Biosciences) were used.
Analysis of chitin degradation capabilities.
To examine the chitin degradation capabilities of recombinant strains, cells were precultivated in ASW-YT-S0 medium at 85°C for 12 h. Cells were inoculated into ASW-VMT-SC medium (total volume, 20 ml) to obtain a theoretical initial OD at 660 nm (OD660) of 0.0025 and cultivated at 85°C. The degrees of chitin degradation were examined at different time intervals (0, 12, 24, 48, 72, and 90 h).
Analysis of chitin metabolites in culture supernatants.
Culture supernatants were collected at different time intervals (12, 24, 48, 72, and 90 h) to analyze metabolites of chitin degradation and assimilation. Aliquots of broth were centrifuged (20,400 × g, 15 min, 4°C), and the supernatants were spotted onto a silica gel plate (DC Kieselgel 60; Merck Co., Berlin, Germany). Products were developed with 1-butanol–methanol–25% ammonia solution–water (5:4:2:1 [vol/vol/vol/vol]) and separated. Products were identified by spraying the plate with aniline-diphenylamine reagent (4 ml of aniline, 4 g of diphenylamine, 200 ml of acetone, and 30 ml of 85% phosphoric acid), followed by baking at 180°C for 3 min. The amounts of chitin oligomers in culture supernatants after 96 h of cultivation in ASW-VMT-SC medium were determined by HPLC. A 10-μl aliquot of culture supernatant was applied to an Asahipak NH2P-50 4E column (Showa Denko, Kanagawa, Japan). As a mobile phase, 70% (vol/vol) acetonitrile was used at a flow rate of 1.0 ml min−1 at 40°C, and detection was performed with a UV detector at a wavelength of 210 nm.
Analysis of acetate and ammonium levels in culture supernatants.
T. kodakarensis strains were cultivated in ASW-VMT-SC medium at 85°C, and culture supernatants were collected after 72 or 96 h. Concentrations of acetate and ammonium were determined enzymatically using F-kits for acetate and ammonium (Roche Diagnostics, Basel, Switzerland), respectively.
Analysis of hydrogen production.
Chitin-dependent production of molecular hydrogen (H2) was analyzed periodically (12, 24, 36, 48, 72, and 96 h) during cultivation in ASW-VMT-SC medium (total volume, 15 ml) at 85°C by a gas chromatograph (GC) equipped with thermal conductivity detectors (GC-17A, TCD-17 detector at 100°C, injector at 100°C [Shimadzu, Kyoto, Japan]). Hydrogen was measured by separation using a Sincarbon-T column (Shinwa Kako, Kyoto, Japan) at a temperature of 60°C using argon as the carrier gas with a flow rate of 30 ml min−1.
Supplementary Material
ACKNOWLEDGMENTS
We declare that there are no conflicts of interest.
This study was partially funded by the Core Research for Evolutional Science and Technology program of the Japan Science and Technology Agency to H.A. within the research area Creation of Basic Technology for Improved Bioenergy Production through Functional Analysis and Regulation of Algae and Other Aquatic Microorganisms. This work was also partially funded by JSPS KAKENHI grant number 26292038 (to T.K.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00280-17.
REFERENCES
- 1.Gooday GW. 1990. The ecology of chitin degradation. Adv Microb Ecol 11:387–430. doi: 10.1007/978-1-4684-7612-5_10. [DOI] [Google Scholar]
- 2.Niehaus F, Bertoldo C, Kähler M, Antranikian G. 1999. Extremophiles as a source of novel enzymes for industrial application. Appl Microbiol Biotechnol 51:711–729. doi: 10.1007/s002530051456. [DOI] [PubMed] [Google Scholar]
- 3.Ladenstein R, Antranikian G. 1998. Proteins from hyperthermophiles: stability and enzymatic catalysis close to the boiling point of water. Adv Biochem Eng. Biotechnol 61:37–85. doi: 10.1007/BFb0102289. [DOI] [PubMed] [Google Scholar]
- 4.Zeikus JG, Vieille C, Savchenko A. 1998. Thermozymes: biotechnology and structure-function relationships. Extremophiles 2:179–183. doi: 10.1007/s007920050058. [DOI] [PubMed] [Google Scholar]
- 5.Frock AD, Kelly RM. 2012. Extreme thermophiles: moving beyond single-enzyme biocatalysis. Curr Opin Chem Eng 1:363–372. doi: 10.1016/j.coche.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller M, Loder A, Basen M, Izquierdo J, Kelly RM, Adams MWW. 2015. Production of lignofuels and electrofuels by extremely thermophilic microbes. Biofuels 5:499–515. doi: 10.1080/17597269.2014.996729. [DOI] [Google Scholar]
- 7.Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl Environ Microbiol 60:4559–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263–267. doi: 10.1155/2004/204953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanai T, Imanaka H, Nakajima A, Uwamori K, Omori Y, Fukui T, Atomi H, Imanaka T. 2005. Continuous hydrogen production by the hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. J Biotechnol 116:271–282. doi: 10.1016/j.jbiotec.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Kanai T, Simons JR, Tsukamoto R, Nakajima A, Omori Y, Matsuoka R, Beppu H, Imanaka T, Atomi H. 2015. Overproduction of the membrane-bound [NiFe]-hydrogenase in Thermococcus kodakarensis and its effect on hydrogen production. Front Microbiol 6:847. doi: 10.3389/fmicb.2015.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanai T, Imanaka T, Atomi H. 2013. Hydrogen production by the hyperthermophilic archaeon Thermococcus kodakarensis. J Jpn Pet Inst 56:267–279. doi: 10.1627/jpi.56.267. [DOI] [Google Scholar]
- 12.Kanai T, Matsuoka R, Beppu H, Nakajima A, Okada Y, Atomi H, Imanaka T. 2011. Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol 193:3109–3116. doi: 10.1128/JB.01072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res 15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato T, Fukui T, Atomi H, Imanaka T. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol 185:210–220. doi: 10.1128/JB.185.1.210-220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato T, Fukui T, Atomi H, Imanaka T. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl Environ Microbiol 71:3889–3899. doi: 10.1128/AEM.71.7.3889-3899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumi R, Manabe K, Fukui T, Atomi H, Imanaka T. 2007. Disruption of a sugar transporter gene cluster in a hyperthermophilic archaeon using a host-marker system based on antibiotic resistance. J Bacteriol 189:2683–2691. doi: 10.1128/JB.01692-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santangelo TJ, Ĉuboňová L, Reeve JN. 2008. Shuttle vector expression in Thermococcus kodakaraensis: contributions of cis elements to protein synthesis in a hyperthermophilic archaeon. Appl Environ Microbiol 74:3099–3104. doi: 10.1128/AEM.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santangelo TJ, Ĉuboňová L, Reeve JN. 2010. Thermococcus kodakarensis genetics: TK1827-encoded β-glycosidase, new positive-selection protocol, and targeted and repetitive deletion technology. Appl Environ Microbiol 76:1044–1052. doi: 10.1128/AEM.02497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takemasa R, Yokooji Y, Yamatsu A, Atomi H, Imanaka T. 2011. Thermococcus kodakarensis as a host for gene expression and protein secretion. Appl Environ Microbiol 77:2392–2398. doi: 10.1128/AEM.01005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka T, Takahashi F, Fukui T, Fujiwara S, Atomi H, Imanaka T. 2005. Characterization of a novel glucosamine-6-phosphate deaminase from a hyperthermophilic archaeon. J Bacteriol 187:7038–7044. doi: 10.1128/JB.187.20.7038-7044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka T, Fujiwara S, Nishikori S, Fukui T, Takagi M, Imanaka T. 1999. A unique chitinase with dual active sites and triple substrate binding sites from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. Appl Environ Microbiol 65:5338–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka T, Fukui T, Imanaka T. 2001. Different cleavage specificities of the dual catalytic domains in chitinase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Biol Chem 276:35629–35635. doi: 10.1074/jbc.M105919200. [DOI] [PubMed] [Google Scholar]
- 23.Imanaka T, Fukui T, Fujiwara S. 2001. Chitinase from Thermococcus kodakaraensis KOD1. Methods Enzymol 330:319–329. doi: 10.1016/S0076-6879(01)30385-3. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka T, Fukui T, Fujiwara S, Atomi H, Imanaka T. 2004. Concerted action of diacetylchitobiose deacetylase and exo-β-d-glucosaminidase in a novel chitinolytic pathway in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Biol Chem 279:30021–30027. doi: 10.1074/jbc.M314187200. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T, Fukui T, Atomi H, Imanaka T. 2003. Characterization of an exo-β-d-glucosaminidase involved in a novel chitinolytic pathway from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J Bacteriol 185:5175–5181. doi: 10.1128/JB.185.17.5175-5181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koga S, Yoshioka I, Sakuraba H, Takahashi M, Sakasegawa S, Shimizu S, Ohshima T. 2000. Biochemical characterization, cloning, and sequencing of ADP-dependent (AMP-forming) glucokinase from two hyperthermophilic archaea, Pyrococcus furiosus and Thermococcus litoralis. J Biochem 128:1079–1085. doi: 10.1093/oxfordjournals.jbchem.a022836. [DOI] [PubMed] [Google Scholar]
- 27.Yokooji Y, Tomita H, Atomi H, Imanaka T. 2009. Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the Archaea. J Biol Chem 284:28137–28145. doi: 10.1074/jbc.M109.009696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller M, Takemasa R, Schwarz A, Atomi H, Nidetzky B. 2009. “Short-chain” α-1,4-glucan phosphorylase having a truncated N-terminal domain: functional expression and characterization of the enzyme from Sulfolobus solfataricus. Biochim Biophys Acta 1794:1709–1714. doi: 10.1016/j.bbapap.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Kanai T, Akerboom J, Takedomi S, van de Werken HJ, Blombach F, van der Oost J, Murakami T, Atomi H, Imanaka T. 2007. A global transcriptional regulator in Thermococcus kodakaraensis controls the expression levels of both glycolytic and gluconeogenic enzyme-encoding genes. J Biol Chem 282:33659–33670. doi: 10.1074/jbc.M703424200. [DOI] [PubMed] [Google Scholar]
- 30.van de Werken HJ, Verhees CH, Akerboom J, de Vos WM, van der Oost J. 2006. Identification of a glycolytic regulon in the archaea Pyrococcus and Thermococcus. FEMS Microbiol Lett 260:69–76. doi: 10.1111/j.1574-6968.2006.00292.x. [DOI] [PubMed] [Google Scholar]
- 31.Helou C, Marier D, Jacolot P, Abdennebi-Najar L, Niquet-Léridon C, Tessier FJ, Gadonna-Widehem P. 2014. Microorganisms and Maillard reaction products: a review of the literature and recent findings. Amino Acids 46:267–277. doi: 10.1007/s00726-013-1496-y. [DOI] [PubMed] [Google Scholar]
- 32.Hrynets Y, Ndagijimana M, Betti M. 2015. Studies on the formation of Maillard and caramelization products from glucosamine incubated at 37°C. J Agric Food Chem 63:6249–6261. doi: 10.1021/acs.jafc.5b02664. [DOI] [PubMed] [Google Scholar]
- 33.Sapra R, Bagramyan K, Adams MW. 2003. A simple energy-conserving system: proton reduction coupled to proton translocation. Proc Natl Acad Sci U S A 100:7545–7550. doi: 10.1073/pnas.1331436100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim KW, Lee SB. 2003. Inhibitory effect of Maillard reaction products on growth of the aerobic marine hyperthermophilic archaeon Aeropyrum pernix. Appl Environ Microbiol 69:4325–4328. doi: 10.1128/AEM.69.7.4325-4328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J, Bauer MW, Shockley KR, Pysz MA, Kelly RM. 2003. Growth of hyperthermophilic archaeon Pyrococcus furiosus on chitin involves two family 18 chitinases. Appl Environ Microbiol 69:3119–3128. doi: 10.1128/AEM.69.6.3119-3128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oku T, Ishikawa K. 2006. Analysis of the hyperthermophilic chitinase from Pyrococcus furiosus: activity toward crystalline chitin. Biosci Biotechnol Biochem 70:1696–1701. doi: 10.1271/bbb.60031. [DOI] [PubMed] [Google Scholar]
- 37.Driskill LE, Kusy K, Bauer MW, Kelly RM. 1999. Relationship between glycosyl hydrolase inventory and growth physiology of the hyperthermophile Pyrococcus furiosus on carbohydrate-based media. Appl Environ Microbiol 65:893–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huber R, Stöhr J, Hohenhaus S, Rachel R, Burggraf S, Jannasch HW, Stetter KO. 1995. Thermococcus chitonophagus sp. nov., a novel, chitin-degrading, hyperthermophilic archaeum from a deep-sea hydrothermal vent environment. Arch Microbiol 164:255–264. doi: 10.1007/BF02529959. [DOI] [Google Scholar]
- 39.Kreuzer M, Schmutzler K, Waege I, Thomm M, Hausner W. 2013. Genetic engineering of Pyrococcus furiosus to use chitin as a carbon source. BMC Biotechnol 13:9. doi: 10.1186/1472-6750-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andronopoulou E, Vorgias CE. 2003. Purification and characterization of a new hyperthermostable, allosamidin-insensitive and denaturation-resistant chitinase from the hyperthermophilic archaeon Thermococcus chitonophagus. Extremophiles 7:43–53. [DOI] [PubMed] [Google Scholar]
- 41.Andronopoulou E, Vorgias CE. 2004. Multiple components and induction mechanism of the chitinolytic system of the hyperthermophilic archaeon Thermococcus chitonophagus. Appl Microbiol Biotechnol 65:694–702. doi: 10.1007/s00253-004-1640-4. [DOI] [PubMed] [Google Scholar]
- 42.Andronopoulou E, Vorgias CE. 2004. Isolation, cloning, and overexpression of a chitinase gene fragment from the hyperthermophilic archaeon Thermococcus chitonophagus: semi-denaturing purification of the recombinant peptide and investigation of its relation with other chitinases. Protein Expr Purif 35:264–271. doi: 10.1016/j.pep.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Papadimitriou K, Baharidis PK, Georgoulis A, Engel M, Louka M, Karamolegkou G, Tsoka A, Blom J, Pot B, Malecki P, Rypniewski W, Huber H, Schloter M, Vorgias C. 2016. Analysis of the complete genome sequence of the archaeon Pyrococcus chitonophagus DSM 10152 (formerly Thermococcus chitonophagus). Extremophiles 20:351–361. doi: 10.1007/s00792-016-0826-x. [DOI] [PubMed] [Google Scholar]
- 44.Horiuchi A, Aslam M, Kanai T, Atomi H. 2016. A structurally novel chitinase from the chitin-degrading hyperthermophilic archaeon Thermococcus chitonophagus. Appl Environ Microbiol 82:3554–3562. doi: 10.1128/AEM.00319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evvyernie D, Yamazaki S, Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. 2000. Identification and characterization of Clostridium paraputrificum M-21, a chitinolytic, mesophilic and hydrogen-producing bacterium. J Biosci Bioeng 89:596–601. doi: 10.1016/S1389-1723(00)80063-8. [DOI] [PubMed] [Google Scholar]
- 46.Evvyernie D, Morimoto K, Karita S, Kimura T, Sakka K, Ohmiya K. 2001. Conversion of chitinous wastes to hydrogen gas by Clostridium paraputrificum M-21. J Biosci Bioeng 91:339–343. doi: 10.1016/S1389-1723(01)80148-1. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Wu T, Wang G, Gao C, Chen Z, Feng L, Wang P, Zeng X, Wu Z. 2016. Phosphoric acid-based preparing of chitin nanofibers and nanospheres. Cellulose 23:477–491. doi: 10.1007/s10570-015-0829-2. [DOI] [Google Scholar]
- 49.Vincendon M. 1997. Regenerated chitin from phosphoric acid solutions. Carbohydr Polym 32:233–237. doi: 10.1016/S0144-8617(97)00005-2. [DOI] [Google Scholar]
- 50.Hirata A, Kanai T, Santangelo TJ, Tajiri M, Manabe K, Reeve JN, Imanaka T, Murakami KS. 2008. Archaeal RNA polymerase subunits E and F are not required for transcription in vitro, but a Thermococcus kodakarensis mutant lacking subunit F is temperature-sensitive. Mol Microbiol 70:623–633. doi: 10.1111/j.1365-2958.2008.06430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.