Abstract
Aberrant silencing of genes by DNA methylation contributes to cancer, yet how this process is initiated remains unclear. Using a murine model of inflammation-induced tumorigenesis, we tested the hypothesis that inflammation promotes recruitment of epigenetic proteins to chromatin, initiating methylation and gene silencing in tumors. Compared to normal epithelium and non-inflammation-induced tumors, inflammation-induced tumors gained DNA methylation at CpG islands, some of which are associated with putative tumor suppressor genes. Hypermethylated genes exhibited enrichment of repressive chromatin marks and reduced expression prior to tumorigenesis, at a time point coinciding with peak levels of inflammation-associated DNA damage. Loss of MutS homolog 2 (MSH2), a mismatch repair (MMR) protein, abrogated early inflammation-induced epigenetic alterations and DNA hypermethylation alterations observed in inflammation-induced tumors. These results indicate that early epigenetic alterations initiated by inflammation and MMR proteins lead to gene silencing during tumorigenesis, revealing a novel mechanism of epigenetic alterations in inflammation-driven cancer. Understanding such mechanisms will inform development of pharmacotherapies to reduce carcinogenesis.
Keywords: DNA methylation, mismatch repair, EZH2, inflammation, colon cancer
Introduction
Approximately 25% of all cancers are associated with chronic inflammation (1). Colorectal cancer (CRC) is a substantial contributor to morbidity and mortality worldwide (2). Although the vast majority of CRC cases are sporadic, 5–10% of cases have been attributed to hereditary conditions (3). In addition to family history, several risk factors associated with CRC have been identified including age, inflammatory bowel disease, bacterial infection, obesity, excessive alcohol consumption, and smoking (4). Most of these risk factors are characterized by chronic inflammation (5). Moreover, nonsteroidal anti-inflammatory drug use was reported to reduce the risk of CRC, reinforcing the notion that inflammation is a central contributor to CRC development (6).
In addition to inflammation, genetic mutations and epigenetic alterations play a key role in driving CRC (7, 8). Several genetic mutations that contribute to the initiation of CRC have been identified and well characterized (9). Epigenetic alterations also contribute to CRC initiation and progression. DNA methylation is the most extensively studied epigenetic alteration in cancer and cancer cells exhibit a global loss of DNA methylation that is thought to lead to genomic instability (8). Additionally, there is concurrent hypermethylation at distinct regions, often within promoter CpG islands of tumor suppressor genes (TSGs) (8). Such hypermethylation can lead to TSG silencing and contribute to cancer. While the fundamental importance of cancer-specific DNA methylation alterations is clear, how they are initiated is not well understood.
Our group previously demonstrated that treatment of cancer cells with the reactive oxygen species (ROS) hydrogen peroxide (H202) causes localization of an epigenetic silencing complex containing DNA methyltransferases DNMT1 and DNMT3B, and the chromatin silencers Sirtuin-1 (SIRT1) and Enhancer of Zeste-2 (EZH2) to sites of damaged chromatin. Notably, there was enrichment of these silencing proteins at the promoter CpG islands of TSGs, resulting in reduced gene expression (10). Based on these findings, we hypothesized that inflammation would induce similar events in vivo.
In the present study, we uncover a novel mechanism responsible for initiating cancer-relevant epigenetic alterations in a murine model of inflammation-induced tumorigenesis. Our results demonstrate that epigenetic alterations that occur during inflammation-driven tumorigenesis are distinct from those that occur during inflammation-independent tumorigenesis. To our knowledge, we are the first to demonstrate the involvement of the mismatch repair (MMR) pathway in initiating epigenetic alterations during inflammation-induced tumorigenesis. Since inflammation and epigenetic alterations are critical in the development of many cancer types and diseases, a deeper understanding of the interplay between these two factors, as gained from this study, is broadly relevant.
Materials and Methods
Animal model
C57BL/6J (Jackson labs) and MinApcΔ716+/− mice were handled and inoculated with ETBF as in Wu et al. (2009) (11). Msh2l/lVC are a result of crossing B6.Cg-MSH2tm2.1Rak/J and B6.Cg-Tg(Vil1-cre)997Gum/J mice (Jackson Labs) to create mice homozygous for MSH2tm2.1Rak and expressing the Vil1-cre transgene. Littermates not expressing Vil1-cre were used as WT controls. Msh2l/lVC/Min are the result of crossing Msh2l/lVC and MinApcΔ716+/− mice. For all experiments both males and females were used, mice were randomized between mock and ETBF groups, and mice of different genotypes were cohoused. Individual tumors were removed from dissected colons with the aid of a dissecting microscope and stored at −80°C until further analysis. Distal (0–2 cm measured from the rectum) and proximal (feathered portion adjacent to ceacum) epithelium was collected by scraping the mucosal surface of the dissected colon (after removal of any tumors), washed three times in PBS, and then subjected to the indicated protocol. Such scraping has been shown by others to be an effective method to obtain samples of intestinal epithelial cells (12). All mouse experiments were covered under protocol number 16-027, which was approved by the Indiana University Bloomington Animal Care and Use Committee in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International.
Methyl CpG binding domain (MBD)-seq
DNA was isolated from tumors or epithelium using the QIAamp DNA mini kit (Qiagen) following the manufacturer’s protocol. To identify differentially methylated regions (DMRs), MBD enrichment was performed from DNA from epithelium collected from different mice (n=5 WT/Min mock, n=3 WT/Min ETBF, n=3 Msh2l/lVC/Min mock) or individual tumors (n= 3 WT/Min mock, n=7 WT/Min ETBF, n=3 Msh2l/lVC/Min mock, n=3 Msh2l/lVC/Min ETBF) using Diagenode’s MethylCap kit. Libraries were prepared following Bioo Scientific’s Methyl Sequencing kit. Single-end 75 bp sequencing was performed using an Illumina Nextseq. Z-scores were calculated using a 500bp fixed sized bin spanning CpG islands based on the distribution of coverage from uniquely mapped reads. Z-ratios were derived from the comparison of z-scores for the different sample types for the 500bp regions. See Supplemental Methods for more detail.
Pyrosequencing and Quantitative Methylation Specific PCR (qMSP)
DNA was bisulfite treated (EZ DNA methylation-Gold kit, Zymo Research) and used for pyrosequencing. qMSP assays were first tested using a standard curve of bisulfite treated mixtures of unmethylated and methylated DNA (data not shown). Only methylated DNA assays with little to no amplification of unmethylated samples, close to 100% efficiency and R2 close to 1 were used further. See Table S1 for assays and primers used.
Gene expression
RNA was prepared from epithelium or tumors using Trizol followed by cleanup with a RNeasy kit (Qiagen). cDNA was prepared and qPCR was done using TaqMan assays (see Table S1 for assays used). Expression of candidate genes was normalized to expression of a housekeeping gene (PPIA or 18S).
Tight chromatin fractionation
Tight chromatin fractionation of washed epithelium was performed as described in Ding et al. (13). Blots presented are representative of three independent experiments.
Chromatin immunoprecipitation (ChIP)
ChIP was performed using anti-H3K27me3 (Diagenode) or anti-EZH2 (D2C9-Cell Signaling Technologies) and the iDeal CHIP-Seq kit for histones and transcription factors (Diagenode) according to the manufacturer’s instructions.
CoIP
coIPs were performed from nuclear extracts that were treated with oligoamines to release chromatin-bound proteins as described in Ding et al. (13). Antibodies used were rabbit IgG (Millipore) and anti-EZH2 (5246-Cell Signaling Technologies). Blots presented are representative of two independent experiments.
MSI
MSI analysis was performed as in Woerner et al. (14). MSI was determined by comparing DNA fragment analysis of mononucleotide markers in tumors relative to tail DNA from the same mouse.
16S microbiome sequencing
DNA was isolated from stool using the ZR Fecal DNA micro kit (Zymo Research). Libraries were made from 16S V1-V3 PCR amplicons from the stool DNA using the NEXTflex 16S V1-V3 kit (Bioo Scientific). 300 bp paired-end sequencing of pooled libraries was performed on a MiSeq. Read quality filtering was performed using mothur. AbundantOTU+ was used for clustering of sequences to Operational Taxonomic Units (OTUs) and further classification. MetagenomeSeq was used to determine differential abundance across samples using normalized OTU read counts. OTUs with significant (5% FDR) differential abundance between any of the sample groups were used to create the heatmap (65 OTUs).
Total CpG methylation
Total CpG methylation in DNA from epithelium and tumor samples was determined using an ELISA-based assay (MethylFlash Global DNA Methylation (5-mC) – Epigenetek).
Statistical Analysis
Expression data, densitometry, qMSP, and local ChIP are presented as the mean +/− standard error (SEM). These data are evaluated by one-tail t-test and considered statistically significant with a p-value < 0.05. Sample sizes are indicated in associated figure legends. MBD-seq statistical analysis is detailed in the Supplemental Methods.
Data availability
MBD and 16S sequencing data have been deposited at the Sequence Read Archive (SRA) data repository (project accession number SRP105286).
Results
Inflammation-induced tumors have a unique DNA hypermethylation signature
Enterotoxigenic Bacteroides fragilis (ETBF) is a clinically relevant strain of B. fragilis (15). When Multiple intestinal neoplasia (Min) mice, which are heterozygous for mutant adenomatous polyposis coli (ApcΔ716), are infected with ETBF, tumors rapidly form at the site of inflammation, in the distal colon (11). Studies have demonstrated that inflammation-induced tumors have unique DNA methylation alterations compared to uninflamed tissue therefore we evaluated DNA methylation in our model (16, 17). Min mice spontaneously develop rare colon tumors (18) allowing us to uniquely compare DNA methylation alterations of inflammation-induced ETBF tumors to methylation alterations of background Min (mock) tumors. Interestingly, relative to mock epithelium, the ETBF tumors compared to mock tumors had more hypermethylated DMRs (203 compared to 6) but fewer hypomethylated DMRs (194 compared to 700, Table 1, Figure 1A). Of the methylation gains that occurred in mock tumors, 5 overlap with the ETBF tumors (Table S2).
Table 1.
Number of 500 bp regions from the MBD-seq data that are statistically different in the indicated comparisons.
| Gains | Losses | Total | |
|---|---|---|---|
| Min mock tumor X Min mock epithelium | 6 | 700 | 706 |
| Min ETBF epithelium X Min mock epithelium | 40 | 265 | 305 |
| Min ETBF tumor X Min mock epithelium | 203 | 194 | 397 |
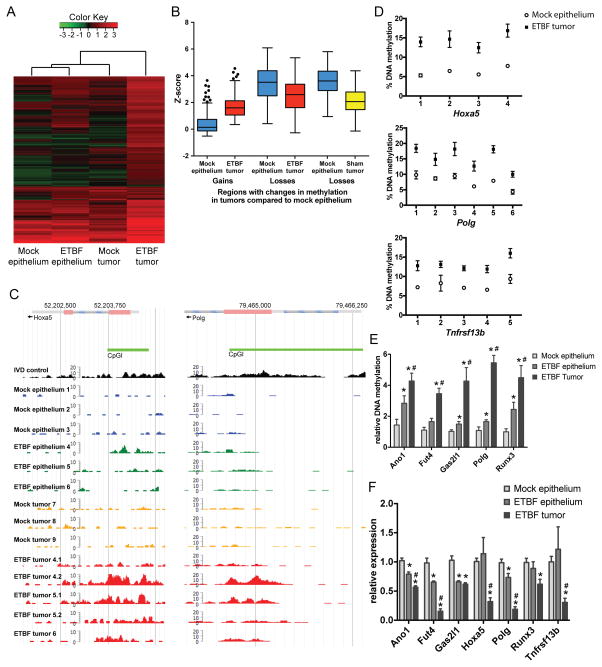
Figure 1. Inflammation-induced tumors have unique DNA hypermethylation alterations.
A) Unsupervised hierarchical clustering of MBD-seq z-scores of the 239 regions with DNA hypermethylation in one of the tumor groups relative to mock epithelium (rows). Each column corresponds to the indicated epithelium or tumor sample. The color of each cell reflects the degree of methylation. All tissue samples were collected 8 weeks after inoculation (N=3 mock and ETBF epithelium and mock tumors; N=5 ETBF tumors). B) Tukey box plots of z-scores of regions with gains or losses of methylation in ETBF or mock tumors relative to mock epithelium. C) MBD-seq data at representative regions for indicated epithelium and tumors. In vitro methylated DNA (IVD) serves as a positive control. D) Pyrosequencing of bisulfite treated DNA from indicated tissue in promoters of candidate genes. Mean +/− SEM. N≥6. *p<0.05. E) qMSP of samples as in D. *p<0.05 compared to mock epithelium. #p<0.05 compared to ETBF epithelium. F) Gene expression by qPCR of candidate genes relative to mock epithelium. Mean +/− SEM. N≥6. *, # as in E. See also Figure S1.
As Naumov et al. (19) demonstrated in human tumors, regions that gained methylation in ETBF tumors had low DNA methylation levels in mock epithelium (mean z-score of 0.4) whereas regions that lost methylation in ETBF or mock tumors had relatively high DNA methylation levels in mock epithelium (mean z-score 3.4 or 3.6, respectively) (Figure 1B). Both gains and losses occurred more often in CpG islands in exons or promoters of genes than introns, 3′ UTRs, or intragenic regions (Figure S1A). As expected, regions targeted for DNA hypermethylation were enriched for regions that are bivalent (containing both H3K4me3 and H3K27me3) in mouse embryonic stem cells (Figure S1B) (20–24).
Since chronic inflammation is known to induce DNA methylation changes in inflamed epithelium (16), we also assayed methylation changes in the distal colon epithelium of ETBF-infected mice (ETBF epithelium). ETBF epithelium had fewer hypermethylated DMRs in comparison to the ETBF tumors, but more than the mock epithelium and mock tumors (Table 1). Eleven of the 40 hypermethylated DMRs observed in the ETBF epithelium overlapped with those in the ETBF tumors (Table S2). When samples were clustered based on regions with DNA hypermethylation in tumors relative to mock epithelium, the ETBF epithelium clustered with the mock epithelium (Figure 1A). Interestingly, ETBF tumors clustered separately from the mock tumors and epithelium, suggesting that they have distinct methylation gains from the other samples.
Gene ontology (GO) analysis revealed that the genes associated with DMRs in the mock and ETBF tumors are involved in multiple biological processes many of which are associated with aspects of development, differentiation, and regulation of transcription (Table S3). Interestingly, several of the methylation gains observed in the ETBF tumors occurred within CpG islands of genes with known tumor suppressive function such as Hoxa5, Polg, Runx1, Runx3, CD37, Stx11, Tceb2, Lgr6, Cdx1, Fut4 (Figure 1C, Table S2) (25–34). CpG island hypermethylation at 18 out of 21 candidate genes in ETBF tumors versus mock epithelium were validated by pyrosequencing or qMSP (Figure 1D, 1E, S1C). No DNA methylation changes were detected in the promoter CpG island of Gapdh, a negative control (Figure S1C). 18 candidate genes are associated with human digestive organ tumors by Ingenuity Pathway Analysis, demonstrating their relevance to human disease (Figure S1D). Runx3, Gata2, and Hoxa5 are also known to undergo aberrant DNA hypermethylation in human CRC (35, 36). qMSP analysis at candidate genes demonstrated an intermediate level of methylation in the ETBF epithelium, between that observed in the mock epithelium and ETBF tumor (Figure 1E) consistent with the data in Figure 1A.
As hypothesized, many genes containing hypermethylated promoter CpG islands in ETBF tumors also had reduced mRNA expression (Figure 1F). Additionally, several candidate genes had lower expression in ETBF epithelium compared to ETBF tumors, but higher expression compared to mock epithelium (Figure 1F), consistent with the methylation results in Figure 1E. Genes that contained hypermethylated CpG islands in non-promoter exons had increased gene expression as has been demonstrated previously (Figure S1E – Cldn4, Spi1, Stx11) (37). Overall, these findings indicate that ETBF tumors contain distinct cancer-specific DNA hypermethylation alterations that distinguish them from mock and ETBF epithelium and mock tumors.
Early chromatin and transcriptional changes occur in genes that are DNA hypermethylated in inflammation-induced tumors
We hypothesized that ETBF-induced oxidative DNA damage may initiate epigenetic changes in the distal colon that ultimately result in the DNA hypermethylation observed in the inflammation-induced tumors. ETBF inoculation caused oxidative DNA damage (increased 8-oxoguanine) in the distal colon epithelium two days post-ETBF, which returned to background levels seven days post-ETBF (Figure 2A). We confirmed that the oxidative DNA damage was occurring in epithelial cells by assaying 8-oxoguanine in cells positive for the epithelial marker, EPCAM (Figure 2B).
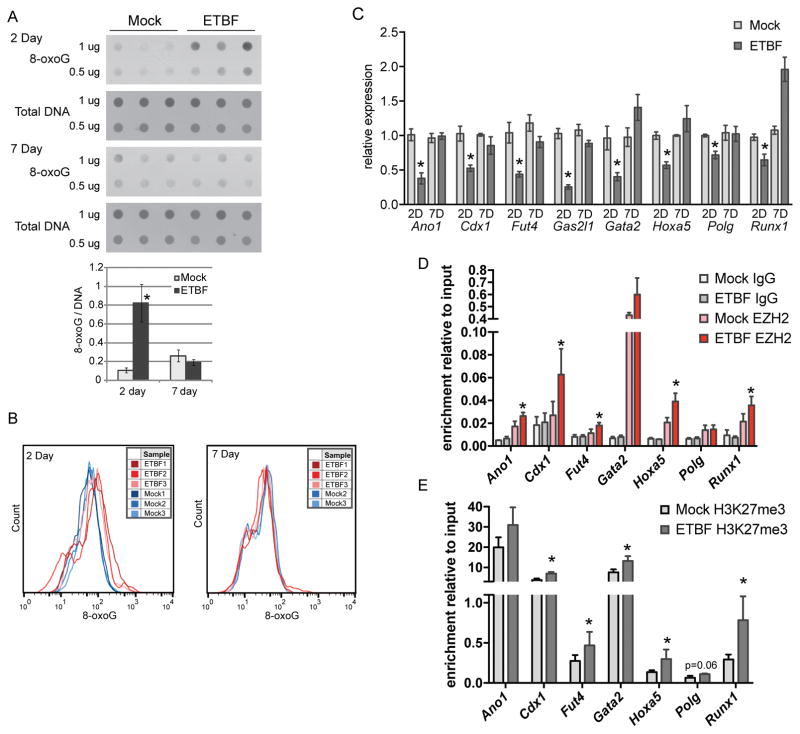
Figure 2. Genes that undergo methylation in tumors have reduced expression at the time point of highest oxidative damage.
A) 8-oxoG dot blot of DNA extracted from distal colon epithelium from mock or ETBF-inoculated mice, N=3. Bar graph of densitometry determined using ImageJ for dots. Mean +/− SEM. B) 8-oxoG staining by flow cytometry of Epcam-positive cells from mice treated as in A. C) Expression of candidate genes by qPCR in distal colon epithelium from mice at the indicated days post-mock or ETBF. Mean +/− SEM. N=5. *p<0.05. D) EZH2, IgG and E) H3K27me3 enrichment by ChIP relative to input at promoters of indicated genes in distal colon epithelium from mice 2 days post-mock or ETBF. Mean +/− SEM. N=3. *p<0.05. See also Figure S2.
The increased oxidative DNA damage observed in the distal colon epithelium two days post-ETBF corresponded to a decrease in the mRNA expression of several candidate genes that were found to be hypermethylated in the ETBF tumors with expression levels returning back to normal seven days post-ETBF (Figure 2C). To address whether these changes occurred specifically in epithelial cells, we assessed the purity of our colon epithelial samples. Our samples contained approximately 84% EPCAM-positive cells and fewer than 2% CD45-positive cells (Figure S2A), irrespective of ETBF treatment status. To probe this further we also cultured organoids from the distal colon epithelium following an established protocol (38), treated the epithelial organoids with H2O2 to mimic inflammation/oxidative damage in vitro, and examined expression of several candidate genes. Indeed, the candidate genes examined (Cdx1, Fut4, Hoxa5, Polg, Runx1) had reduced expression in response to oxidative stress (but not housekeeping genes Rpl0, Tbp), analogous to our in vivo results (Figure S2B). This result suggests that the changes observed in the colon epithelium in response to ETBF are likely occurring in epithelial cells and not an alternative cell type present in the mucosal surface.
Since EZH2 has been implicated in transcriptional repression at sites of oxidative DNA damage, we hypothesized that this protein would be enriched in the promoters of candidate genes that have reduced expression in the inflamed epithelium. Indeed, EZH2 was significantly enriched at several candidate genes in ETBF epithelium compared to mock epithelium two days post-ETBF (Figure 2D). These regions also had enrichment of the repressive chromatin mark catalyzed by EZH2, trimethyl H3K27 (H3K27me3) (Figure 2E). These findings indicate that ETBF initiates early epigenetic and transcriptional alterations in several candidate genes from the MBD-seq dataset, and these changes are temporally and spatially associated with the oxidative DNA damage induced by ETBF.
MSH2 interacts with EZH2 and plays a key role in the initiation of early ETBF-induced epigenetic alterations
The MSH2-MSH6 heterodimer facilitates repair of clustered oxidative DNA lesions in a PCNA-dependent and S-phase-independent manner (39). In accordance with this finding, our group observed in vitro that MSH2 and MSH6 become more tightly bound to chromatin in response to oxidative damage and are indispensable for the recruitment of epigenetic silencing proteins to damaged chromatin (13). Therefore, we hypothesized that MSH2 and MSH6 contribute to early epigenetic alterations that occur in response to ETBF.
First, we demonstrated that EZH2 co-immunoprecipitated with DNMT1, MSH2, MSH6, and PCNA in the distal colon epithelium two days after ETBF, suggesting an interaction between epigenetic proteins and MMR proteins in response to ETBF (Figure 3A). This interaction did not occur in mock epithelium. SUZ12 and EED are known EZH2 interacting partners and are positive controls for the EZH2 co-IP.
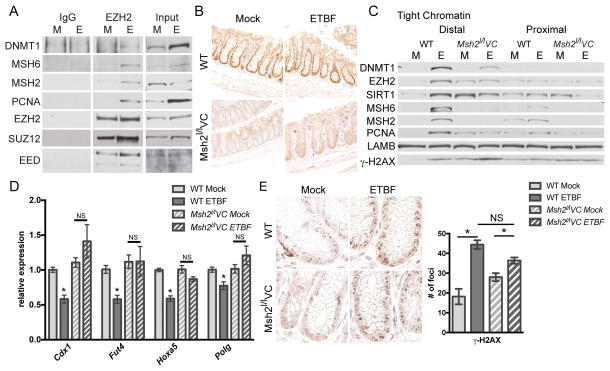
Figure 3. Acute inflammation-induced epigenetic changes are dependent on MSH2.
A) IgG or Ezh2 co-IP from distal colon epithelial cells 2 days post-mock (M) or ETBF (E) inoculation. B) Anti-MSH2 IHC of the distal colon of WT or Msh2l/lVC mice 2 days post-mock or ETBF. Images are representative of staining from 3 mice per group. C) Proteins that are tightly bound to chromatin (tight chromatin) from epithelium as in A. D) Expression of candidate genes by qPCR in distal colon epithelial cells from mice at 2 days post-mock or ETBF. Mean +/− SEM. N=5–7. *p<0.05. E) Anti-γ-H2AX IHC of the same tissue sections as in B with quantification of γ-H2AX foci per crypt. N≥45 crypts. Mean+/−SEM. *p<0.05 by Tukey’s multiple comparison test. See also Figure S3.
To evaluate whether the MMR pathway is involved in ETBF-mediated epigenetic alterations, we examined mice that lack expression of Msh2 in intestinal epithelial cells. In Msh2l/lVC mice, the Msh2 gene is flanked by LoxP sequences and Cre recombinase is driven by a Villin promoter leading to deletion of Msh2 in intestinal epithelial cells (40). We verified that MSH2 protein is present in colon crypt cells in mock and ETBF inoculated wild type (WT) mice, but not in crypts from Msh2l/lVC mice (Figure 3B). In distal colon epithelium from WT ETBF-infected mice, epigenetic silencing proteins (EZH2, DNMT1, and SIRT1) and MMR proteins (MSH2, MSH6 and PCNA) have a higher affinity for chromatin than in mock mice (Figure 3C). This enhanced affinity occurred in the distal colon and not in the proximal colon that lacks ETBF-mediated inflammation. Loss of Msh2 reduced the ETBF-mediated increase in binding of EZH2, DNMT1 and SIRT1 to chromatin (Figure 3C), highlighting the necessity of the MMR pathway in the response of epigenetic proteins to ETBF-induced damage. LAMB (Lamin-B), a loading control, was consistent across the samples. Levels of phosphorylated H2AX (γ-H2AX), a marker of DNA damage, were elevated in ETBF samples relative to mock samples. Interestingly, γ-H2AX levels were consistently higher in mock epithelium from Msh2l/lVC than WT mice, corresponding to a relative increase in binding of EZH2, SIRT1 and PCNA to chromatin. Moreover we examined the effect of loss of Msh2 on mRNA expression of several candidate genes and their expression was unaffected by ETBF in Msh2l/lVC mice (Figure 3D, S3A).
To rule out the possibility that loss of Msh2 alters the initial immune response to ETBF we examined the expression of several cytokines in response to ETBF two and seven days post-infection and found that their expression was unaltered by loss of Msh2 (Figure S3B, C). Cell proliferation measured by Ki-67 staining and phosphorylation of STAT-3 was also unaffected by loss of Msh2 (Figure S3D, E). ETBF-infected Msh2l/lVC mice also have similar numbers of γ-H2AX foci per crypt compared to ETBF-infected WT mice (Figure 3E). Collectively, these findings implicate a role for MSH2 in initiating early epigenetic and transcriptional alterations observed in the inflamed colon epithelium of mice two days post-ETBF.
Loss of Msh2 does not alter the background microbiota composition
It is possible that an alteration of the microbiota composition caused by loss of Msh2 could explain reduced ETBF-induced epigenetic alterations in Msh2l/lVC mice. Therefore, we examined whether loss of Msh2 alters the initial or ETBF-induced intestinal microbiota composition. 16S ribosomal RNA gene sequencing was performed on stool DNA and sequences were clustered into OTUs. Principal component analysis (PCA) and hierarchical clustering analysis based on the OTUs that had significantly differential abundance among the sample types revealed a high degree of similarity between the bacterial populations present in stool from mock-infected mice, regardless of genotype (Figure 4A and B). None of the OTUs were significantly different between the mock groups, suggesting that loss of Msh2 does not alter the background microbiota composition (Table S4). There was separation between samples from ETBF-infected and mock-infected mice, irrespective of genotype (Figure 4A and B, Table S4). PCA revealed a small degree of separation between ETBF-infected WT/Min versus ETBF-infected Msh2l/lVC/Min samples suggesting that loss of Msh2 might impact the ETBF-induced microbiota (Figure 4A and B, Table S4). However, loss of Msh2 correlates with a significant increased abundance of ETBF (Figure 4C) and colony formation units of ETBF per gram of stool (Figure S4), ruling out the possibility that loss of Msh2 reduces ETBF-mediated epigenetic alterations by reducing the abundance of ETBF. Altogether, loss of Msh2 did not alter the background microbiota of these mice, although it does alter the relative abundance of some bacterial populations upon ETBF-infection.
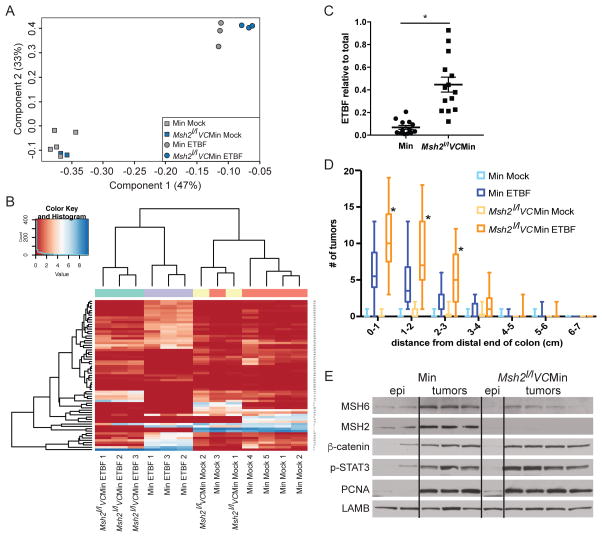
Figure 4. Inflammation-induced tumorigenesis is increased in the distal colon of mice with altered Msh2 expression.
A) PCA analysis of 16S microbiome sequencing of DNA from stool samples from WT/Min or Msh2l/lVC/Min mice 8 weeks post mock or ETBF. B) Heatmap representing the unsupervised hierarchical clustering of 65 OTUs found to be differentially abundant by one pair-wise comparison (rows) in individual stool samples (columns) from indicated mice treated as in A. C) ETBF abundance in stool relative to total bacterial DNA by qPCR. Symbols represent data from individual mice. Horizontal line is mean +/− SEM. N≥13. *p<0.05. D) Tukey box plots of tumor counts by cm in WT/Min or Msh2l/lVC/Min mice 8 weeks after mock or ETBF. N≥8. *p<0.05. E) Whole cell protein lysate from distal colon epithelium or tumors from mice of the indicated genotypes 8 weeks post-mock or ETBF inoculation, respectively, were blotted for the indicated proteins. See also Figure S4.
Loss of Msh2 in intestinal epithelial cells caused an increase in inflammation-induced tumors with microsatellite instability
Although loss of Msh2 reduced early ETBF-induced epigenetic alterations (Figure 3), sporadic and germline mutations in the MMR pathway are commonly implicated in CRC pathogenesis (4). Therefore, we examined the effect of Msh2 deletion from intestinal epithelial cells on tumorigenesis in our model. Mock Msh2l/lVC/Min mice were similar to uninfected WT/Min mice in terms of tumor burden (Figure 4D). Interestingly, ETBF-infected Msh2l/lVC/Min mice developed significantly more tumors in the distal colon than the WT/Min mice infected with ETBF, but the same regions had the highest number of tumors in both sets of mice (Figure 4D).
The Th17/STAT-3 immune response drives ETBF-mediated tumorigenesis in Min mice (11). Msh2 deletion did not alter ETBF-induced phosphorylation of STAT-3 in ETBF-induced tumors (Figure 4E). Furthermore, inactivation of APC leads to constitutive activation of the WNT/β-CATENIN signaling pathway in tumors in Min mice. Loss of Msh2 did not alter the magnitude of increase in levels of β-CATENIN or PCNA, a marker of proliferation, in ETBF-induced tumors (Figure 4E).
Mutations in MMR genes are known to cause genomic instability by altering the length of repetitive DNA sequences known as microsatellites (4). Interestingly, the mock and ETBF Msh2l/lVC/Min tumors tested had microsatellite instability (MSI) whereas most tumors from the WT/Min mice were microsatellite stable (MSS) (9/11) (Table 2). These results are consistent with the observation in humans that mutated MSH2 can lead to colon tumorigenesis (4) and suggest that ETBF-induced tumors from Msh2-deficient mice are more genetically unstable than ETBF-induced tumors from Msh2-sufficient mice.
Table 2.
Tumors from mice deficient in Msh2 are microsatellite instability (MSI) positive.
| Animal | Genotype | ETBF | U12235[A24] a | AC096777[A33] a | AA003063[A23] a | AC096777[T27] a | L24372[A27] a | MSI% b |
|---|---|---|---|---|---|---|---|---|
| 403 | Min | − | − | + | + | − | − | 40 |
| 410 | Min | − | − | − | + | − | − | 20 |
| 414 | Min | − | − | − | + | − | − | 20 |
| 7 | Min | + | − | − | − | − | − | 20 |
| 7 | Min | + | − | − | − | − | − | 20 |
| 408 | Min | + | − | − | − | − | − | 20 |
| 408 | Min | + | − | − | − | − | − | 20 |
| 408 | Min | + | − | − | − | − | − | 20 |
| 408 | Min | + | − | − | − | − | − | 20 |
| TR | Min | + | + | − | + | + | − | 60 |
| TR | Min | + | + | − | − | − | − | 20 |
| 331 | Msh2l/lVCMin | − | + | no data | + | + | + | 80 |
| 322 | Msh2l/lVCMin | − | + | + | − | + | + | 80 |
| 322 | Msh2l/lVCMin | − | + | + | − | + | + | 80 |
| 322 | Msh2l/lVCMin | − | + | − | − | + | + | 60 |
| 319 | Msh2l/lVCMin | + | + | + | − | + | + | 80 |
| 319 | Msh2l/lVCMin | + | − | + | − | + | + | 60 |
| 319 | Msh2l/lVCMin | + | + | + | − | + | + | 80 |
| 320 | Msh2l/lVCMin | + | − | + | − | + | + | 60 |
| 320 | Msh2l/lVCMin | + | − | + | − | + | + | 60 |
| 320 | Msh2l/lVCMin | + | − | + | − | + | + | 60 |
+ and − indicate positive or negative for instability, respectively, for the given assay.
tumors with greater than 20% of tested loci positive for instability are MSI+ (bold numbers).
Loss of Msh2 decreases CpG island hypermethylation and restores expression of candidate TSGs
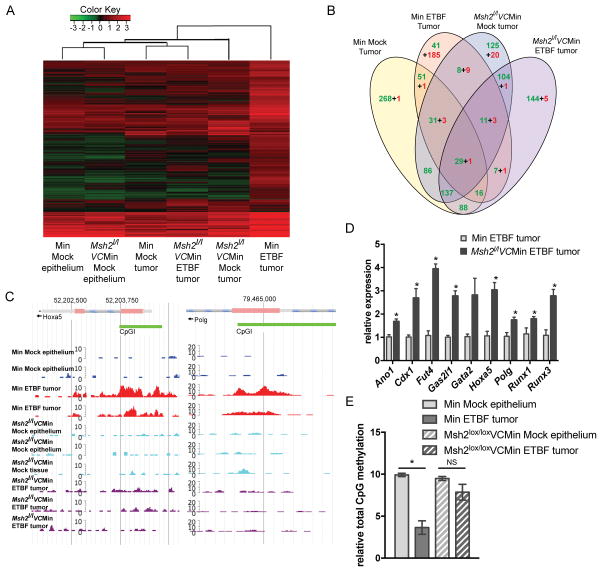
We initially demonstrated that Min ETBF tumors have a unique hypermethylation signature (Figure 1, Table 1). Since the Msh2l/lVC/Min mice still developed tumors in response to ETBF, this model provided us with a unique tool to answer the question: is MSH2 required for ETBF-induced tumor-specific DNA hypermethylation alterations during tumorigenesis? Therefore, we examined the effect of Msh2 deletion on methylation in ETBF tumors using MBD-seq. Importantly, loss of Msh2 dramatically reduced ETBF-mediated hypermethylation alterations with the Msh2l/lVC/Min ETBF tumors having only 11 DNA hypermethylated regions relative to Msh2l/lVC/Min mock epithelium (Figure 5A and B, Table S2). When samples are clustered using regions with DNA hypermethylation in tumors relative to the respective mock epithelium, the two mock epitheliums cluster together and the Msh2l/lVC/Min ETBF tumors cluster with the Min mock tumors. The WT/Min ETBF tumors fall on a distinct arm from the other samples suggesting that their DNA hypermethylation alterations are distinct from all other tumors types, including the ETBF-induced tumors from Msh2l/lVC/Min mice. Plots of the DNA methylation data for Hoxa5 and Polg demonstrate the reduction of promoter CpG island hypermethylation in Msh2l/lVC/Min ETBF tumors compared to WT/Min ETBF tumors (Figure 5C). This observation was validated at several candidate genes with tumor suppressive function including Hoxa5, Polg, Runx3, and Stx11 (Figure S5 A, B). Both the mock and ETBF Msh2l/lVC/Min tumors had many hypomethylated DMRs relative to Msh2l/lVC/Min mock epithelium (Figure 5B and Table S2), suggesting that the losses of methylation are general to the tumorigenesis process in these mice.
Figure 5. Msh2 deficiency abrogates inflammation-induced epigenetic changes in tumors.
A) Unsupervised hierarchical clustering of MBD-seq z-scores of regions with increased DMRs in one of the tumor groups relative to mock epithelium (rows). Each column corresponds to the indicated epithelium or tumor sample. The color of each cell reflects the degree of methylation. B) Numbers of DMRs for the indicated tumor group relative to the corresponding mock epithelium that overlap between comparisons. Green and red numbers are hypomethylated and hypermethylated regions, respectively. C) MBD-seq data at representative regions for indicated epithelium and tumors 8 weeks post-inoculation. D) Expression of candidate genes by qRTPCR relative to WT/Min ETBF tumors. Mean +/− SEM. N=5. *p<0.05. E) Total 5-mC content of DNA. Mean +/− SEM. N=3 mock, N=6 tumor. *p<0.05. See also Figure S5.
Furthermore, we examined the effect of loss of Msh2 on mRNA expression of several candidate genes in tumors that formed at sites of ETBF-mediated inflammation. Interestingly, loss of Msh2 partially restored expression of candidate genes in tumors that formed at sites of ETBF-mediated inflammation, including Cdx1, Fut4, Hoxa5, Polg, Runx1 and Runx3 (Figure 5D).
As mentioned, it has been widely reported that there is a global loss of DNA methylation in cancer. Therefore, we examined the effect of Msh2 deficiency on global CpG DNA methylation. Consistent with what is observed in humans, the WT/Min ETBF tumors exhibited a global loss of DNA CpG methylation. Interestingly, loss of Msh2 restored total CpG methylation back to mock epithelium levels (Figure 5D). Thus, not only did loss of Msh2 reduce regional hypermethylation alterations genome-wide, it also reduced ETBF-mediated global DNA hypomethylation. These results reinforce the notion that MSH2 is an essential player in both the recruitment of epigenetic proteins to chromatin during the early stages of inflammation and in the permanent silencing of several TSGs in tumors that form at sites of ETBF-mediated inflammation and oxidative DNA damage.
Discussion
Many studies have demonstrated a strong relationship between inflammation and epigenetic alterations (1). Although the mechanism connecting the two is not well understood, oxidative damage is a prominent feature associated with both. Based on previous work by our group, we hypothesized that sustained and/or repeated oxidative damage to chromatin, during inflammation, may result in epigenetic silencing (O’Hagan 2011). To test this hypothesis we used an established mouse model of inflammation-induced colon tumorigenesis. The bacterium ETBF has been shown to rapidly induce acute colitis and tumorigenesis in Min mice that resembles the pathology observed in humans with colorectal cancer (11, 41). Importantly, several studies revealed a strong association between ETBF colonization in the gut and colorectal cancer incidence in humans suggesting that this mouse model is highly relevant to human colon carcinogenesis and an appropriate model to use to investigate the mechanism underlying how inflammation initiates DNA methylation alterations (42, 43).
Whether ETBF-mediated inflammation can induce DNA methylation alterations in humans remains to be determined; however, colitis, which can be induced by ETBF, is strongly associated with aberrant DNA methylation alterations in humans (44). Furthermore, others have demonstrated an association between bacterial infection and DNA methylation alterations in both animals and humans. A well-characterized example is induction of aberrant DNA methylation alterations and gastric cancer in Mongolian gerbils due to exposure to the bacterium Helicobacter pylori (45). A human study reported that the gastric mucosa of H. pylori infected individuals had higher methylation levels at 8 marker CpG islands compared to uninfected individuals (46). The results presented in our study are highly consistent with these findings in animals and humans in that compared to mock colon epithelium, ETBF-induced tumors have altered DNA methylation, including hypermethylation of promoter CpG islands of several TSGs. Furthermore, the epithelium surrounding the tumors in ETBF-infected mice had more DNA hypermethylation alterations than the mock epithelium but still considerably fewer hypermethylation changes compared to ETBF tumors, confirming an inflammation-produced field effect that has previously been demonstrated (1). Interestingly, the DNA methylation pattern observed in ETBF-induced tumors was remarkably similar to what is observed in human cancers, including CRC, namely global hypomethylation concomitant with focal hypermethylation. As has been shown in human CRC, the DNA hypermethylation changes observed in our study occurred at regions that are bivalent in embryonic stem cells and genes associated with DMRs were enriched in processes associated with development and differentiation.
Min mice spontaneously develop occasional tumors in the colon. Conveniently, this allowed us to compare DNA methylation changes in inflammation-induced tumors to changes in non-inflammation-induced tumors. There were very few hypermethylation changes in the mock tumors compared to the ETBF tumors. A critical point here is that the vast majority of DNA hypermethylation alterations observed in our study in the ETBF-induced tumors are specifically driven by ETBF or ETBF-mediated inflammation and this is highly consistent with what has been reported in similar animal models and in humans.
Niwa et al. ruled out the possibility that H. Pylori itself is responsible for the induction of DNA methylation alterations in the gastric mucosa of Mongolian gerbils, and confirmed that innate immune-mediated inflammation resulting from H. pylori is the culprit (45). However, the molecular mechanism that underlies how inflammation initiates epigenetic alterations in cancer is not well understood. Our hypothesis in this study is that innate immune-mediated inflammation, caused by ETBF, induces oxidative DNA damage and thereby triggers early transient silencing of genes, rendering them susceptible to stable silencing through DNA methylation. Importantly, at the time point when ETBF-infection induces robust oxidative DNA damage, we see increased enrichment of EZH2 at promoters of genes that become methylated in tumors that form at sites of inflammation, coinciding with reduced expression. We demonstrate that Msh2 deletion in intestinal epithelial cells abrogates the ETBF-mediated recruitment of epigenetic proteins to chromatin and restores expression of candidate genes. Knowing that we now had a system where we could block the recruitment of epigenetic proteins to sites of oxidative DNA damage without affecting the inflammatory response or DNA damage levels, we could determine if DNA hypermethylation in the inflammation-induced tumors depends on this early epigenetic response to oxidative damage. Importantly, Msh2 deletion reduced ETBF-mediated genome-wide hypermethylation alterations and reversed ETBF-induced global hypomethylation in tumors that formed at sites of inflammation.
Intriguingly, loss of Msh2 led to increased tumorigenesis in response to ETBF, despite a lack of DNA hypermethylation and reexpression of TSGs. This result suggests that another mechanism, independent of aberrant epigenetic alterations, can drive tumorigenesis in the context of ETBF-infection. We speculate that increased genomic defects, due to MSI, underlie tumorigenesis in Msh2l/lVC/Min mice exposed to ETBF. To our advantage, the fact that Msh2l/lVC/Min mice develop tumors in response to ETBF allowed us to elucidate a mechanism to explain how epigenetic alterations occur during tumorigenesis, a task that could not have been achieved if tumors did not form in these mice. Whether or not DNA hypermethylation changes are necessary to drive inflammation-induced tumorigenesis in the context of a normal mutational burden needs to be studied further.
It remains to be determined precisely how the early ETBF-induced epigenetic alterations observed in our model are maintained and converted into permanent epigenetic alterations in tumors that form at sites of inflammation. We speculate that, in addition to the epithelial cells, these alterations occur and persist in the intestinal stem cells, which are known to eventually go on to transform into tumor cells (47). This hypothesis is supported by the observation that some epigenetic alterations that occur early in stem cells lead to permanent silencing of TSGs thereby contributing to predisposing the cell to malignant transformation (48).
The work described here provides a mechanism to explain how inflammation, in this case inflammation mediated by ETBF, a pathogen associated with CRC in humans, causes early epigenetic alterations, some of which persist in tumors that form at sites of inflammation. A challenge in elucidating the mechanism by which inflammation initiates epigenetic alterations, is that it is difficult to selectively modulate responses caused by inflammation without also modulating the initial inflammatory response altogether. We were able to prevent inflammation-induced hypermethylation in tumors without modulating the initial inflammatory response by using mice lacking Msh2. Our findings suggest a role for MSH2 in initiating early ETBF-mediated epigenetic alterations and maintaining DNA methylation alterations that occur in the ETBF-induced tumors. A recent study demonstrated that MSI tumors from patients with Lynch syndrome, a disease in which patients harbor genetic mutations in MMR genes and eventually develop CRC, have less DNA hypomethylation and fewer regional hypermethylation alterations (49), analogous to what we observed in our study.
While in sporadic CRC loss of expression of the MMR protein MLH1 is associated with CpG Island Methylator Phenotype (CIMP), we demonstrate here that loss of MSH2 or MSH6 plays an opposite role. This finding is supported by CRC TCGA data where, of the hypermutated tumors studied, those with MLH1 methylation were CIMP, but those with mutations in MSH2/6/3 and WT MLH1 expression were not CIMP (9).
The results presented herein fill in critical gaps in our understanding of the relationship between inflammation and epigenetic alterations by elucidating a role for inflammation-induced DNA damage and consequent MMR in initiating cancer-specific epigenetic alterations during tumorigenesis. Knowledge gained from this study has notably increased our understanding concerning how epigenetic alterations are initiated during tumorigenesis in the colon and this information can likely be extrapolated to other types of cancer. Interestingly, regions with DNA hypermethylation in the inflammation-induced tumors were surprisingly consistent between tumors from different mice and different experimental cohorts suggesting a strong selective pressure for these silencing events. We hypothesize that the mechanism of DNA hypermethylation is similar in other inflammatory-driven epithelial cancers, but the specific genes silenced may differ depending on the selective pressures of tumorigenesis in other tissues. Understanding the mechanism of initiation of DNA methylation changes will allow the scientific community to therapeutically target inflammation-induced epigenetic changes and potentially reduce the cancer risk of individuals with chronic inflammatory diseases.
Supplementary Material
Acknowledgments
We thank the Indiana University Center for Genomics and Bioinformatics and the Indiana Molecular Biology Institute for their assistance. We also thank Sue Childress for her assistance with tissue processing.
Footnotes
COI statement: The authors declare no potential conflicts of interest.
Additional Information:
Heather M. O’Hagan financial support: NIEHS (R01ES023183)
Cynthia L. Sears financial support: NCI (R01CA151325) and support from the Department of Medicine and the Bloomberg Kimmel Immunotherapy Institute.
References
- 1.Chiba T, Marusawa H, Ushijima T. Inflammation-Associated Cancer Development in Digestive Organs: Mechanisms and Roles for Genetic and Epigenetic Modulation. Gastroenterology. 2012;143:550–63. doi: 10.1053/j.gastro.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Jackson-Thompson J, Ahmed F, German RR, Lai SM, Friedman C. Descriptive epidemiology of colorectal cancer in the United States, 1998–2001. Cancer. 2006;107:1103–11. doi: 10.1002/cncr.22007. [DOI] [PubMed] [Google Scholar]
- 4.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 5.Carbonnel F, Jantchou P, Monnet E, Cosnes J. Environmental risk factors in Crohn’s disease and ulcerative colitis: an update. Gastroenterologie clinique et biologique. 2009;33(Suppl 3):S145–57. doi: 10.1016/S0399-8320(09)73150-1. [DOI] [PubMed] [Google Scholar]
- 6.Thun MJ, Namboodiri MM, Calle EE, Flanders WD, Heath CW., Jr Aspirin use and risk of fatal cancer. Cancer Res. 1993;53:1322–7. [PubMed] [Google Scholar]
- 7.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG islands. Cancer Cell. 2011;20:606–19. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–22. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ortega-Cava CF, Ishihara S, Rumi MA, Aziz MM, Kazumori H, Yuki T, et al. Epithelial toll-like receptor 5 is constitutively localized in the mouse cecum and exhibits distinctive down-regulation during experimental colitis. Clin Vaccine Immunol. 2006;13:132–8. doi: 10.1128/CVI.13.1.132-138.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding N, Bonham EM, Hannon BE, Amick TR, Baylin SB, O’Hagan HM. Mismatch repair proteins recruit DNA methyltransferase 1 to sites of oxidative DNA damage. Journal of molecular cell biology. 2016;8:244–54. doi: 10.1093/jmcb/mjv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woerner SM, Tosti E, Yuan YP, Kloor M, Bork P, Edelmann W, et al. Detection of coding microsatellite frameshift mutations in DNA mismatch repair-deficient mouse intestinal tumors. Mol Carcinog. 2015;54:1376–86. doi: 10.1002/mc.22213. [DOI] [PubMed] [Google Scholar]
- 15.Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22:349–69. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, et al. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–9. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abu-Remaileh M, Bender S, Raddatz G, Ansari I, Cohen D, Gutekunst J, et al. Chronic Inflammation Induces a Novel Epigenetic Program That Is Conserved in Intestinal Adenomas and in Colorectal Cancer. Cancer Research. 2015;75:2120–30. doi: 10.1158/0008-5472.CAN-14-3295. [DOI] [PubMed] [Google Scholar]
- 18.Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc Natl Acad Sci U S A. 1995;92:4482–6. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naumov VA, Generozov EV, Zaharjevskaya NB, Matushkina DS, Larin AK, Chernyshov SV, et al. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics : official journal of the DNA Methylation Society. 2013;8:921–34. doi: 10.4161/epi.25577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–42. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 22.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 23.Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 2012;22:271–82. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ordonez-Moran P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 Counteracts Stem Cell Traits by Inhibiting Wnt Signaling in Colorectal Cancer. Cancer Cell. 2015;28:815–29. doi: 10.1016/j.ccell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Singh KK, Ayyasamy V, Owens KM, Koul MS, Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet. 2009;54:516–24. doi: 10.1038/jhg.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voora D, Rao AK, Jalagadugula GS, Myers R, Harris E, Ortel TL, et al. Systems Pharmacogenomics Finds RUNX1 Is an Aspirin-Responsive Transcription Factor Linked to Cardiovascular Disease and Colon Cancer. EBioMedicine. 2016;11:157–64. doi: 10.1016/j.ebiom.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju X, Ishikawa TO, Naka K, Ito K, Ito Y, Oshima M. Context-dependent activation of Wnt signaling by tumor suppressor RUNX3 in gastric cancer cells. Cancer science. 2014;105:418–24. doi: 10.1111/cas.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Winde CM, Veenbergen S, Young KH, Xu-Monette ZY, Wang XX, Xia Y, et al. Tetraspanin CD37 protects against the development of B cell lymphoma. J Clin Invest. 2016;126:653–66. doi: 10.1172/JCI81041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida N, Tsuzuki S, Karube K, Takahara T, Suguro M, Miyoshi H, et al. STX11 functions as a novel tumor suppressor gene in peripheral T-cell lymphomas. Cancer science. 2015;106:1455–62. doi: 10.1111/cas.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rowbotham DA, Enfield KS, Martinez VD, Thu KL, Vucic EA, Stewart GL, et al. Multiple Components of the VHL Tumor Suppressor Complex Are Frequently Affected by DNA Copy Number Loss in Pheochromocytoma. International journal of endocrinology. 2014;2014:546347. doi: 10.1155/2014/546347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong X, Carmon KS, Lin Q, Thomas A, Yi J, Liu Q. LGR6 is a high affinity receptor of R-spondins and potentially functions as a tumor suppressor. PLoS One. 2012;7:e37137. doi: 10.1371/journal.pone.0037137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hryniuk A, Grainger S, Savory JG, Lohnes D. Cdx1 and Cdx2 function as tumor suppressors. J Biol Chem. 2014;289:33343–54. doi: 10.1074/jbc.M114.583823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moriwaki K, Miyoshi E. Fucosylation and gastrointestinal cancer. World journal of hepatology. 2010;2:151–61. doi: 10.4254/wjh.v2.i4.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim YH, Lee HC, Kim SY, Yeom YI, Ryu KJ, Min BH, et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Annals of surgical oncology. 2011;18:2338–47. doi: 10.1245/s10434-011-1573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–90. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 39.Zlatanou A, Despras E, Braz-Petta T, Boubakour-Azzouz I, Pouvelle C, Stewart GS, et al. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol Cell. 2011;43:649–62. doi: 10.1016/j.molcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 40.Kucherlapati MH, Lee K, Nguyen AA, Clark AB, Hou H, Jr, Rosulek A, et al. An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology. 2010;138:993–1002. e1. doi: 10.1053/j.gastro.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest. 2014;124:4166–72. doi: 10.1172/JCI72334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208–15. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–6. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 44.Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer research. 2001;61:3573–7. [PubMed] [Google Scholar]
- 45.Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–40. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 46.Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989–95. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- 47.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 48.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 49.Sahnane N, Magnoli F, Bernasconi B, Tibiletti MG, Romualdi C, Pedroni M, et al. Aberrant DNA methylation profiles of inherited and sporadic colorectal cancer. Clinical epigenetics. 2015;7:131. doi: 10.1186/s13148-015-0165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MBD and 16S sequencing data have been deposited at the Sequence Read Archive (SRA) data repository (project accession number SRP105286).