Abstract
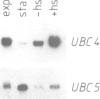
Ubiquitin-conjugating enzymes catalyse the covalent attachment of ubiquitin to target proteins. Members of this enzyme family are involved in strikingly diverse cellular functions: UBC2 (RAD6) is central to DNA repair, UBC3 (CDC34) is involved in cell cycle control. We have cloned the genes for two novel ubiquitin-conjugating enzymes, UBC4 and UBC5, from the yeast Saccharomyces cerevisiae. These enzymes mediate selective degradation of short-lived and abnormal proteins. UBC4 and UBC5 are closely related in sequence and complementing in function. Expression of UBC4 and UBC5 genes is heat inducible. UBC4 and UBC5 enzymes generate high mol. wt ubiquitin-protein conjugates in vivo consistent with previous studies which suggested that attachment of multiple ubiquitin molecules to proteolytic substrates is required for their selective degradation. UBC4 and UBC5 enzymes comprise a major part of total ubiquitin-conjugation activity in stressed cells. Turnover of short-lived proteins and canavanyl-peptides but not of long-lived proteins is markedly reduced in ubc4ubc5 mutants. Loss of UBC4 and UBC5 activity impairs cell growth, leads to inviability at elevated temperatures or in the presence of an amino acid analog, and induces the stress response.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ananthan J., Goldberg A. L., Voellmy R. Abnormal proteins serve as eukaryotic stress signals and trigger the activation of heat shock genes. Science. 1986 Apr 25;232(4749):522–524. doi: 10.1126/science.3083508. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Finley D., Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986 Oct 10;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bachmair A., Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989 Mar 24;56(6):1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- Ball E., Karlik C. C., Beall C. J., Saville D. L., Sparrow J. C., Bullard B., Fyrberg E. A. Arthrin, a myofibrillar protein of insect flight muscle, is an actin-ubiquitin conjugate. Cell. 1987 Oct 23;51(2):221–228. doi: 10.1016/0092-8674(87)90149-8. [DOI] [PubMed] [Google Scholar]
- Bienz M., Pelham H. R. Mechanisms of heat-shock gene activation in higher eukaryotes. Adv Genet. 1987;24:31–72. doi: 10.1016/s0065-2660(08)60006-1. [DOI] [PubMed] [Google Scholar]
- Bond U., Schlesinger M. J. Ubiquitin is a heat shock protein in chicken embryo fibroblasts. Mol Cell Biol. 1985 May;5(5):949–956. doi: 10.1128/mcb.5.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989 Mar 24;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Elias S., Heller H., Hershko A. "Covalent affinity" purification of ubiquitin-activating enzyme. J Biol Chem. 1982 Mar 10;257(5):2537–2542. [PubMed] [Google Scholar]
- Ciechanover A., Finley D., Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984 May;37(1):57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Finley D., Bartel B., Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989 Mar 30;338(6214):394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- Finley D., Ciechanover A., Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984 May;37(1):43–55. doi: 10.1016/0092-8674(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987 Mar 27;48(6):1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Goebl M. G., Yochem J., Jentsch S., McGrath J. P., Varshavsky A., Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988 Sep 9;241(4871):1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- Goff S. A., Goldberg A. L. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 1985 Jun;41(2):587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- Haas A. L., Bright P. M. The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J Biol Chem. 1985 Oct 15;260(23):12464–12473. [PubMed] [Google Scholar]
- Haas A. L., Bright P. M. The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J Biol Chem. 1988 Sep 15;263(26):13258–13267. [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Heller H., Haas A. L., Rose I. A. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Heller H., Eytan E., Reiss Y. The protein substrate binding site of the ubiquitin-protein ligase system. J Biol Chem. 1986 Sep 15;261(26):11992–11999. [PubMed] [Google Scholar]
- Hershko A., Heller H. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem Biophys Res Commun. 1985 May 16;128(3):1079–1086. doi: 10.1016/0006-291x(85)91050-2. [DOI] [PubMed] [Google Scholar]
- Hershko A., Leshinsky E., Ganoth D., Heller H. ATP-dependent degradation of ubiquitin-protein conjugates. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1619–1623. doi: 10.1073/pnas.81.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. Ubiquitin-mediated protein degradation. J Biol Chem. 1988 Oct 25;263(30):15237–15240. [PubMed] [Google Scholar]
- Hershko A. Ubiquitin: roles in protein modification and breakdown. Cell. 1983 Aug;34(1):11–12. doi: 10.1016/0092-8674(83)90131-9. [DOI] [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem. 1987 Jun 15;262(17):8303–8313. [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Leung D. W., Spencer S. A., Cachianes G., Hammonds R. G., Collins C., Henzel W. J., Barnard R., Waters M. J., Wood W. I. Growth hormone receptor and serum binding protein: purification, cloning and expression. Nature. 1987 Dec 10;330(6148):537–543. doi: 10.1038/330537a0. [DOI] [PubMed] [Google Scholar]
- Lindquist S., Craig E. A. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Matthews W., Driscoll J., Tanaka K., Ichihara A., Goldberg A. L. Involvement of the proteasome in various degradative processes in mammalian cells. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2597–2601. doi: 10.1073/pnas.86.8.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A. W., Solomon M. J., Kirschner M. W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989 May 25;339(6222):280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
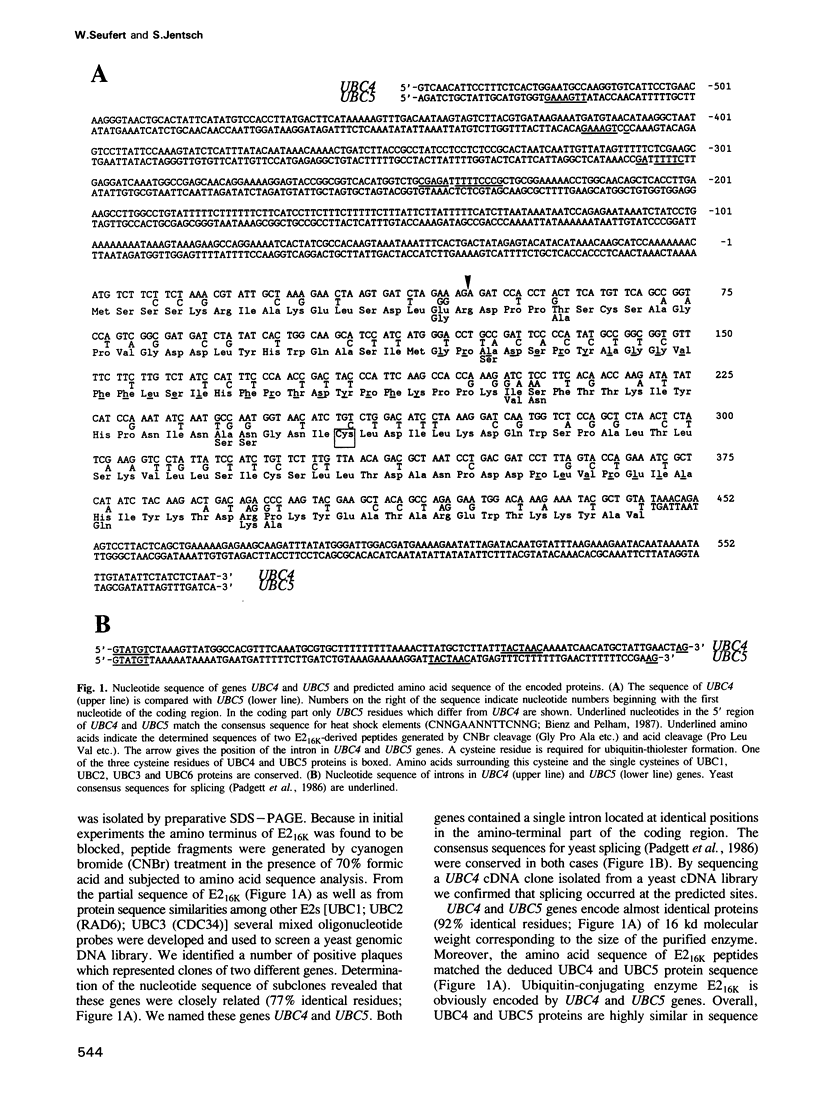
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parsell D. A., Sauer R. T. Induction of a heat shock-like response by unfolded protein in Escherichia coli: dependence on protein level not protein degradation. Genes Dev. 1989 Aug;3(8):1226–1232. doi: 10.1101/gad.3.8.1226. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell. 1986 Sep 26;46(7):959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- Pickart C. M., Rose I. A. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985 Feb 10;260(3):1573–1581. [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Shanklin J., Jabben M., Vierstra R. D. Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci U S A. 1987 Jan;84(2):359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegelman M., Bond M. W., Gallatin W. M., St John T., Smith H. T., Fried V. A., Weissman I. L. Cell surface molecule associated with lymphocyte homing is a ubiquitinated branched-chain glycoprotein. Science. 1986 Feb 21;231(4740):823–829. doi: 10.1126/science.3003913. [DOI] [PubMed] [Google Scholar]
- St John T., Gallatin W. M., Siegelman M., Smith H. T., Fried V. A., Weissman I. L. Expression cloning of a lymphocyte homing receptor cDNA: ubiquitin is the reactive species. Science. 1986 Feb 21;231(4740):845–850. doi: 10.1126/science.3003914. [DOI] [PubMed] [Google Scholar]
- Straus D. B., Walter W. A., Gross C. A. Escherichia coli heat shock gene mutants are defective in proteolysis. Genes Dev. 1988 Dec;2(12B):1851–1858. doi: 10.1101/gad.2.12b.1851. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman L., Fagan J. M., Goldberg A. L. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. J Biol Chem. 1987 Feb 25;262(6):2451–2457. [PubMed] [Google Scholar]
- Wilkinson K. D., Urban M. K., Haas A. L. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J Biol Chem. 1980 Aug 25;255(16):7529–7532. [PubMed] [Google Scholar]
- Wu R. S., Kohn K. W., Bonner W. M. Metabolism of ubiquitinated histones. J Biol Chem. 1981 Jun 10;256(11):5916–5920. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yarden Y., Escobedo J. A., Kuang W. J., Yang-Feng T. L., Daniel T. O., Tremble P. M., Chen E. Y., Ando M. E., Harkins R. N., Francke U. Structure of the receptor for platelet-derived growth factor helps define a family of closely related growth factor receptors. Nature. 1986 Sep 18;323(6085):226–232. doi: 10.1038/323226a0. [DOI] [PubMed] [Google Scholar]