Abstract
We show that the microRNA miR-155 can be processed from sequences present in BIC RNA, a spliced and polyadenylated but non-protein-coding RNA that accumulates in lymphoma cells. The precursor of miR-155 is likely a transient spliced or unspliced nuclear BIC transcript rather than accumulated BIC RNA, which is primarily cytoplasmic. By using a sensitive and quantitative assay, we find that clinical isolates of several types of B cell lymphomas, including diffuse large B cell lymphoma (DLBCL), have 10- to 30-fold higher copy numbers of miR-155 than do normal circulating B cells. Similarly, the quantities of BIC RNA are elevated in lymphoma cells, but ratios of the amounts of the two RNAs are not constant, suggesting that the level of miR-155 is controlled by transcription and processing. Significantly higher levels of miR-155 are present in DLBCLs with an activated B cell phenotype than with the germinal center phenotype. Because patients with activated B cell-type DLBCL have a poorer clinical prognosis, quantification of this microRNA may be diagnostically useful.
Keywords: diffuse large B cell lymphoma, Invader assays, microRNAs
Inappropriate expression of a protooncogene or inactivation of a tumor suppressor gene can contribute to cancer. An example is the BIC gene, which was originally identified as a common site for insertion of proviral DNA in avian leukosis virus-induced lymphomas (1, 2). Activation of the BIC gene can accelerate the pathogenesis of lymphomas and leukemias that are associated with the up-regulation of c-MYC, showing that BIC functions in the etiology of these diseases (3). Expression of BIC RNA is low in normal lymphoid tissues but elevated in Hodgkin and children's Burkitt lymphoma and in in-vitro-activated B and T cells (4–6).
The BIC RNAs of avian, mouse, and human cells are spliced and polyadenylated ≈1.7-kb [counting poly(A)] RNAs, presumably generated by RNA polymerase II. However, they lack long ORFs, and their short, putative ORFs are not conserved, leading to the proposal that BIC RNA functions as a non-protein-coding RNA (7). Recently, a mouse microRNA (miRNA), miR-155 (8), was found to be encoded within the only phylogenetically conserved region of BIC RNA (7). miRNAs are ≈22-nt molecules that function in posttranscriptional down-regulation of gene expression in plants, vertebrates, and invertebrates (9–11). Thus, miR-155 could be responsible for the oncogenic activity attributed to BIC RNA.
In animal cells, endogenous miRNAs are produced from primary RNA polymerase II transcripts by sequential processing in the nucleus and cytoplasm (reviewed in ref. 12). Nuclear precursor RNAs are cleaved by the endonuclease Drosha in a “microprocessor complex,” releasing pre-miRNAs, which are short, 60- to 70-nt imperfect hairpin structures. After transport to the cytoplasm by exportin-5, pre-miRNAs are processed by the endonuclease DICER, generating ≈22-nt duplexes, one strand of which is the mature miRNA. The conserved region of the BIC transcript encoding miR-155 can form an imperfect hairpin structure (7) (Fig. 6, which is published as supporting information on the PNAS web site), suggesting that miR-155 is generated by this pathway.
Changes in the levels of miRNAs may alter the control of growth or apoptosis in some cancers (13, 14). Reductions in the levels of miR-15a and miR-16, let-7a, or miR-143 plus miR-145 have been reported in chronic lymphocytic leukemia (CLL) (15), lung cancer (16), and colon carcinoma (17), respectively. Although BIC RNA is up-regulated in some human lymphomas (5, 6), very little is known about the levels of miR-155 in these cancers.
Diffuse large B cell lymphoma (DLBCL), an aggressive B cell neoplasm accounting for 30–40% of all lymphoma cases (18), can be categorized immunohistochemically into groups with significantly different clinical outcomes (19). The prognosis is poorer for patients having DLBCL with an activated B cell (ABC) than a nonactivated germinal center (GC) phenotype. So far, a relationship has not been examined between miR-155/BIC RNA levels and the phenotypes of this most frequent of all lymphoid neoplasms.
Here, we report on the processing, localization, and quantification of BIC RNA and miR-155 in several established tumor cell lines and in clinical isolates of B cell lymphomas. Our quantitative assays reveal that, relative to the levels in control B cells, the copy numbers of miR-155 and BIC RNA are greatly increased (although to different extents) in clinical lymphoma isolates. Furthermore, primary DLBCL cells with the ABC phenotype have 2- to 3-fold higher levels of miR-155 than do primary DLBCL cells with a GC phenotype, suggesting that quantification of this miRNA could be used diagnostically.
Methods
Lymphoid Cell Lines and Cultures. The cell lines Ramos, JY25, CB33, U266, Jurkat, K562, and HL60 and Hodgkin lymphoma (HL) cell lines HDLM2, L428, KMH2, L591, and L1236 were cultured in RPMI medium 1640 with 10% heat-inactivated FCS (Invitrogen), and human embryonic kidney (HEK)-293T cells were maintained in DMEM with 10% heat-inactivated calf serum. Daudi cells or OCI-Ly1 and OCI-Ly8 cells and OCI-Ly3 cells were grown in Iscove's modified Dulbecco medium supplemented with 10% heat-inactivated serum, 20% FBS, or 10% human serum (NABI Biopharmaceuticals, Boca Raton, FL), respectively.
Human Tissues and Peripheral Blood B cells. Tissue samples of DLBCL, CLL, and marginal zone were from the Department of Pathology, Weill Medical College, Cornell University. Specimens from all patients were obtained according to the protocol approved by the Institutional Review Board. All lymphoma cases were reviewed and classified according to World Health Organization classification. For DLBCLs, only de novo cases were selected. Human B cell controls were purified from the lymphocyte fraction of whole blood (buffy coat) obtained from human donors by positive selection with CD19+ beads (MACS, Miltenyi Biotec, Auburn, CA) and cultured in RPMI medium 1640 with 10% FCS.
Plasmids. To generate pcDNA3.BIC, BIC exon 3 sequences (Fig. 6) were amplified by PCR with 5′ and 3′ primers with flanking NheI and XbaI sites, respectively. The PCR-generated fragment was digested with NheI, Klenow-filled, digested with XbaI, and subcloned into EcoRI/blunt and XbaI sites of pcDNA3 (Invitrogen). The final construct was confirmed by sequencing.
RNA Isolation. Total RNA from cultured cells or cryostat tissue sections was isolated by TRIzol (Invitrogen), and RNA integrity was monitored by electrophoresis in 8% denaturing polyacrylamide gels. Nuclear and cytoplasmic RNA were isolated according to standard protocols (20). For extraction of nuclear RNA, TRIzol was added directly to the nuclear pellet (21).
Northern Blot Analysis of miRNAs. Northern blot analysis was performed as described in ref. 8 by using 20 μg of total RNA. As a loading control, tRNAs were detected by ethidium bromide staining of the gels before transfer. The 5′ end-labeled Northern probe, 5′-CCCCTATCACGATTAGCATTAA, is antisense to human miR-155.
Semiquantitative RT-PCR. We used 2 μg of DNaseI-treated total RNA for reverse transcription in a total volume of 20 μl with the SuperScript First-Strand Synthesis system (Invitrogen). We used 1/10th of the RT reaction mix for PCR amplification with primers indicated in Fig. 6. Primers for amplification of β-actin sequences were 5′-CTGTGCTATCCCTGTACGCCTC and 5′-CATGATGGAGTTGAAGGTAGTTTCGT. All PCR reactions were performed with an annealing temperature of 58°C and verified to be in the linear range by analyses (in 2% agarose gels) of the reaction products at different cycle numbers.
Immunohistochemistry. Immunohistochemical analyses were performed on representative sections of formalin-fixed paraffin-embedded tissues from DLBCL cases by staining with antibodies against phenotypic markers BCL-6, CD10, multiple myeloma 1/IFN regulatory factor 4, and CD138, by using the avidin–biotin–peroxidase technique with antigen epitope enhancement by pressure cooker heating (19). Each marker was considered positive if >20% of the neoplastic lymphocytes were stained positive. Criteria for classification as GC or ABC phenotype are listed in the legend to Table 1, which is published as supporting information on the PNAS web site.
Statistical Analysis. Student's t test for comparison of miR-155 expression levels between the two categories of DLBCL and the regression plots of miR-155 vs. BIC RNA were done by using the statistical package statview (SAS Institute, Cary, NC).
Invader Assays. Invader mRNA assays specific for spliced (splice junction 2–3) and spliced/unspliced (exon 3) BIC RNAs and miRNA assays specific for miR-155, miR-15a, miR-16, and let-7a were performed as described in refs. 22 and 23. Invader probe sets are depicted in Fig. 6; detailed reaction conditions are provided in Supporting Methods, which is published as supporting information on the PNAS web site; and oligonucleotides are listed in Table 1. Cleavase enzyme was from Third Wave Technologies (Madison, WI). All Invader reactions were performed by using 20–40 ng of total cell RNA, and fluorescence signal was converted to copy number per cell by using a standard curve of known amounts of synthetic target RNA and assuming 20 pg of total RNA per cell (rather than U6 RNA levels, which varied between cell types; Fig. 7, which is published as supporting information on the PNAS web site). Expression of BIC RNA and miR-155 in control U266 myeloma cells was undetectable (less than ≈10 copies per cell of BIC RNA and ≈50 copies per cell of miR-155).
Results
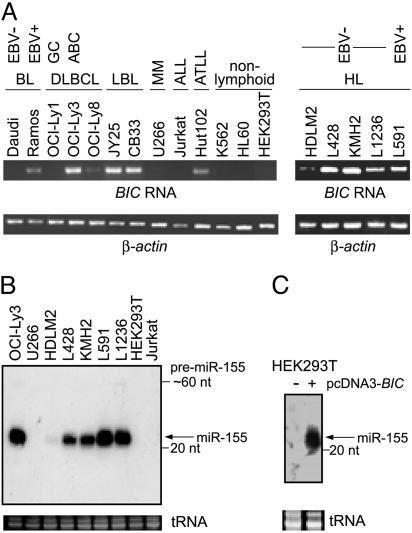
Elevated Levels of BIC RNA and miR-155 in Activated Lymphoid Cells. Initially, we used semiquantitative RT-PCR to probe for BIC RNA and Northern blotting to probe for miR-155 in RNAs of several human lymphoid and nonlymphoid cell lines. Cells of the GC-related DLBCL line OCI-Ly1 contained low levels of BIC RNA (Fig. 1A), consistent with a previous report (6). In contrast, cells of non-GC DLBCL lines had intermediate (OCI-Ly8) or very high (OCI-Ly3) levels of BIC RNA, which may be related to their more active ABC phenotype. Six other Epstein–Barr virus (EBV)-immortalized lymphoblastoid lines that resemble in vitro ABCs also had high levels of BIC RNA (e.g., Fig. 1, JY25 and CB33, and data not shown). Higher BIC RNA levels are independent of EBV infection, given that they were also observed in EBV-negative HL lines L428, KMH2, and L1236, which had BIC RNA levels comparable with the EBV-positive HL cell line L591 (Fig. 1 A) (6). Elevated BIC RNA was also detected in human T cell leukemia virus type-1 transformed T cells (e.g., HUT102) but not in hematopoietic cell lines, such as a myeloma (U266), preT (Jurkat), chronic myelogenous leukemia (K562), and acute promyelocytic leukemia (HL60), or of nonhematopoietic cell lines, such as HEK-293T. These results agree well with previous observations indicating that BIC RNA accumulates during activation of B and T cells (4–6).
Fig. 1.
BIC RNA and miR-155 in cell lines. (A) Total RNAs of different cell lines were analyzed by semiquantitative RT-PCR for BIC RNA and β-actin (as a normalization control) by using the primer pair A/C (Fig. 6) and the primers described in Methods, respectively. PCR products obtained after 30 cycles (BIC) and 24 cycles (β-actin) of reaction were analyzed by gel electrophoresis. BL, Burkitt lymphoma; LBL, EBV-immortalized lymphoblastoid cell line; MM, multiple myeloma; ALL, acute lymphoblastic leukemia; ATLL, adult T cell leukemia/lymphoma (human T cell leukemia virus type-1-positive). (B) Northern blot of total RNA isolated from different cell lines was probed with a [32P]5′ end-labeled oligonucleotide with sequence complementary to the sequence of human miR-155. tRNAs served as a loading control. The 22-nt miR-155 is indicated. The predicted ≈60-nt pre-miR-155 could be detected as a weak band in some samples. (C) Northern analysis of miR-155 in HEK-293T cells and HEK-293T transfected with the vector pcDNA3.BIC expressing BIC exon 3 sequences (see Fig. 6).
To determine whether elevated levels of BIC RNA were accompanied by increased accumulation of miR-155, we probed for the presence of the mature ≈22-nt miRNA by Northern blot analysis (Fig. 1B). miR-155 was detected only in cells that expressed BIC RNA. As with BIC RNA, the levels of accumulated miR-155 differed between cell lines, and changes in the relative levels of miR-155 and BIC RNA resembled each other qualitatively.
Processing of BIC Transcripts into miR-155. To test directly whether a BIC transcript could be processed into miR-155, we transfected HEK-293T cells with pcDNA3.BIC, which harbors a CMV promoter driving synthesis of a 417-nt RNA starting ≈90 nt to the 5′ side of the miR-155 coding sequence (Fig. 6). Cells transfected with this plasmid produced readily detectable amounts of mature miR-155 (Figs. 1C and 7), supporting the idea that a nonspliced, partial copy of BIC RNA can be processed into miR-155, which is consistent with recent studies of other similarly truncated human miRNA genes (24).
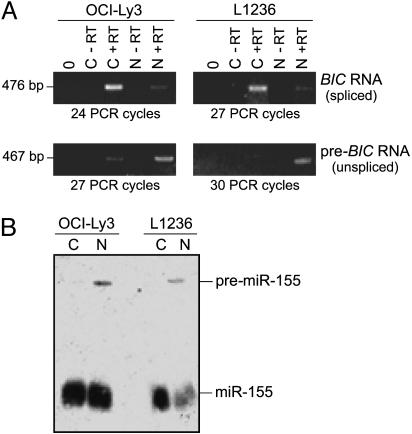
Intracellular Localization of BIC RNA and miR-155. Intron-free BIC RNA can be processed into miR-155, but it is unclear whether normal substrates for Drosha are the primary ≈12-kb transcript or the accumulated spliced ≈1.7-kb BIC RNA detected by Northern blot and/or RT-PCR analyses (4–7) (Fig. 1 A). Being both spliced and polyadenylated, BIC RNA resembles mRNA (although it lacks a long ORF) and may be rapidly exported to the cytoplasm by means of an mRNA export pathway (25). To determine the intracellular localization of BIC RNA, total RNAs from the cytoplasmic or nuclear fractions of OCI-Ly3 and L1236 cells were analyzed by RT-PCR, with primers specific for unspliced or spliced BIC RNAs (Fig. 6). As predicted, the intron-containing transcript was predominantly nuclear (Fig. 2A). In contrast, the much more abundant spliced BIC RNA was primarily cytoplasmic and, hence, no longer accessible to the nuclear Drosha microprocessor complex. Thus, most of the detectable BIC RNA cannot be processed into miR-155 by any currently known miRNA-processing pathway (12).
Fig. 2.
Intracellular localization of BIC RNA and miR-155. (A) Nuclear (N) and cytoplasmic (C) RNAs isolated from OCI-Ly3 and L1236 cells were reverse-transcribed and amplified with primers specific for spliced or unspliced BIC RNA (primer pairs A/C and B/C, producing 476- and 467-bp products, respectively) (see Fig. 6). The reactions were performed with and without reverse transcriptase (+RT and -RT) and in the absence of template (0). PCR was performed for the indicated number of cycles. (B) Northern analysis was performed on total RNA isolated from the nuclear and cytoplasmic fractions of OCI-Ly3 and L1236. The 22-nt miR-155 and the ≈60-nt predicted precursor are marked.
Pre-miR-155 (predicted to be ≈62 nt long) (Fig. 6) was detected only in the nuclear fractions (Fig. 2B), indicating that it is processed rapidly after export to the cytoplasm. The presence of mature miR-155 in nuclear and cytoplasmic fractions resembles distributions reported for other miRNAs (26, 27) and may be due, in part, to contamination of nuclei by perinuclear cytoplasm, which is rich in miRNAs (E.L. and J.E.D., unpublished results).
Quantification of BIC RNA and miR-155. Most of the detectable BIC RNA is cytoplasmic (Fig. 2) and unlikely to serve as a precursor of miR-155, so BIC RNA levels may not be valid predictors of miR-155. To test the proportionality of these RNAs, we measured their levels by using Invader mRNA and miRNA assays, which accurately quantify mRNAs and miRNAs in as little as 0.1–20 ng of total cell RNA (depending on the type of RNA being measured and its expression level) (22, 23). The Invader probes used for detection of BIC RNA were specific for the spliced RNA (Fig. 6) and did not detect unspliced precursor. Other probes that measured the spliced plus unspliced BIC RNA transcripts yielded comparable copy numbers, confirming that the precursor of BIC RNA was present in very low amounts (data not shown). The miRNA Invader assay was specific for the mature miR-155 (22) (Fig. 8, which is published as supporting information on the PNAS web site).
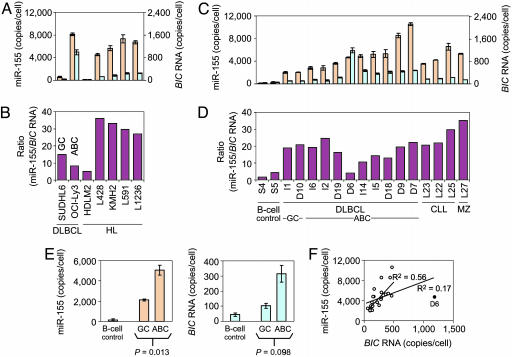
The copy numbers of BIC RNA and miR-155 per cell of some of the established lines ranged from ≈20 to ≈1,000 molecules of BIC RNA and from ≈100 to ≈8,000 molecules of miR-155 (Fig. 3A and Table 2, which is published as supporting information on the PNAS web site). Consistently, the levels of BIC and miR-155 RNAs, as quantified by the Invader assays, agreed well with the results of the semiquantitative RT-PCR and Northern blot analyses (Fig. 1). Notably, the molar ratios of miR-155 to BIC RNA, which varied from ≈4 to ≈35 (Fig. 3B), did not correlate with absolute copy numbers of either molecule and thus are likely to be governed by a combination of the rates of synthesis, processing, nuclear export, and turnover of the RNAs.
Fig. 3.
Numbers of molecules (copies per cell) of BIC RNA and miR-155 in lymphoma cells. Quantification was by miRNA and mRNA Invader assays designed to detect miR-155 and BIC RNA, respectively (see Methods). Samples were tested in triplicate. (A, C, and E) Orange bars and aqua bars are miR-155 and BIC RNA levels, respectively. (A–D) Samples tested in A and C are indicated on the x axes of B and D, respectively, and abbreviations are as described for Fig. 1. (A) miR-155 and BIC RNA levels in several cultured lymphoma cell lines. Note the expanded scale for BIC RNA (right axis). Error bars represent one standard deviation. (B) Ratio of copy number per cell of miR-155 and BIC RNA in cultured lymphoma cells shown in A. (C) miR-155 and BIC RNA levels in cells of clinical isolates of lymphomas. Scales and error bars are as described for A. Samples S4 and S5, normal circulating B cells, are taken as normal controls. (D) Ratio of copy numbers per cell of miR-155 and BIC RNA in the samples shown in C. (E) Comparison of the copy numbers per cell in DLBCL cells exhibiting the GC vs. ABC phenotypes for miR-155 and BIC RNA. A total of 23 cases were tested (11 shown in C and D and reported in Table 2 plus 12 reported in Table 3). The indicated P values are calculated from a t test between the GC phenotype (n = 4) and the ABC phenotype (n = 19). Error bars represent one standard error. (F) Lack of correlation between amounts of miR-155 and BIC RNA in clinical isolates of lymphomas. The graph represents the 23 cases shown in E. Linear regression (R2 values are indicated) was performed with and without sample D6 (filled circle).
Previous in situ hybridization studies indicated that the amount of BIC RNA in DLBCL was very low or undetectable (6). However, the Invader assays showed that detectable amounts of BIC RNA and miR-155 were present in SUDHL6 cells, a prototypic GC-type DLBCL line, and that their levels were ≈25- and ≈15-fold higher, respectively, in OCI-Ly3 cells, a prototypic ABC-type DLBCL line (Fig. 3A). Thus, BIC RNA and miR-155 are detectable in both types of DLBCLs, but their amounts are particularly elevated in cells with the ABC phenotype.
Increased Accumulation of BIC RNA and miR-155 in Clinical B Cell Lymphomas. To determine whether clinically isolated DLBCL cells also had elevated levels of BIC RNA and miR-155, we quantified both RNAs in 23 clinically isolated DLBCL samples (Fig. 3C, Table 2, and Table 3, which is published as supporting information on the PNAS web site). As controls, we used normal, circulating CD19+ B cells (Fig. 3, samples S4 and S5) in which BIC RNA expression was shown in other experiments to increase upon mitogen stimulation (WT, data not shown).
Relative to the control B cells, BIC RNA levels were elevated from 2- to 10-fold in DLBCL cells, with one sample (Fig. 3C, sample D6) showing an increase of >20-fold. miR-155 levels were increased to even greater extents, generally ≈12- to ≈30-fold but ranged as high as 50- to 60-fold for samples I9, D7, and D9 (≈8,500–10,000 copies per cell) (Fig. 3C and Table 3). Significantly, the levels of both miR-155 and BIC RNA were, on average, 2- to 3-fold higher in DLBCL cells with the ABC phenotype than with the GC phenotype (Fig. 3E). Thus, the levels of miR-155 (and BIC RNA) appear to correspond with clinically significant subtypes of DLBCLs, and quantification of miR-155 levels may be a useful prognostic indicator.
For comparison, we also analyzed three cases (L22, L23, and L25) of CLL and one case (L27) of marginal zone B cell lymphoma. In all cases, miR-155 and BIC RNA levels were comparable with those observed for DLBCL cells with the ABC phenotype, indicating that increased accumulation of miR-155 and BIC RNA is likely to be a common feature of B cell lymphomas.
The levels of BIC RNA sequences in clinical samples have been assumed by others to be an indirect measure of miR-155 levels (5). To examine the predictive value of BIC RNA levels, we calculated the molar ratios of the two RNAs and found that they ranged greatly, from ≈4 to ≈35 (Fig. 3D). A regression plot (Fig. 3F) showed only a weak correlation (R2 = 0.17) between the levels of miR-155 and BIC RNA, even when sample D6 was excluded (R2 = 0.56). These data indicate that the amount of BIC RNA in a cell should not be taken as an accurate measure of the amount of miR-155, which is likely to be the active gene product.
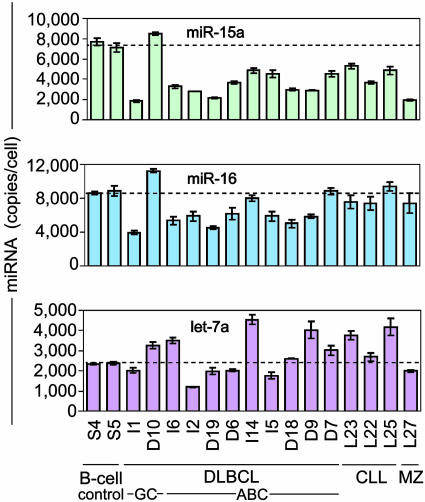
Quantification of Other miRNAs. Recently, the levels of miR-15a and miR-16 were reported to be reduced in clinical isolates of CLL cells (15), and lower levels of let-7a RNA were detected in lung cancer biopsies that had poor clinical outcome (16). To determine whether lymphoma cells exhibited similar changes in the levels of these miRNAs, we quantified their levels by using Invader miRNA assays (Fig. 4 and Table 2). In most of the DLBCL isolates, miR-15a levels were reduced by ≈30–70% compared with circulating B cells (Fig 4 Top), and miR-16 levels were reduced ≈25% on average (Fig. 4 Middle), but let-7a levels showed no consistent pattern of change (Fig. 4 Bottom). In no case did the levels of these miRNAs exhibit the ≈30-fold increase that we observed for miR-155.
Fig. 4.
Quantification of miR-15a, miR-16, and let-7a miRNAs in the normal B cells and clinical isolates of lymphoma cells analyzed in Fig. 3C by using Invader miRNA assays. For a summary of the quantification of miRNAs in cultured lymphoma cell lines analyzed in Fig. 3A, see Fig. 10, which is published as supporting information on the PNAS web site. Samples were measured in triplicate (see Table 2 for values), and error bars represent one standard deviation.
Discussion
Activation of BIC transcription by retroviral provirus insertion contributes to the development of lymphomas in chickens (1), indicating that BIC RNA accumulation is likely to promote, rather than result from, cell transformation. In certain human B cell lymphomas and leukemias, the spliced and polyadenylated BIC RNA accumulates, as does its encoded miRNA, miR-155 (Fig. 1B), which is likely to be the functional oncogenic product of the non-protein-encoding BIC gene.
We were able to accurately quantify the levels of miR-155 in clinical DLBCL samples because, unlike other types of miRNA measurements (28), Invader miRNA assays require very small amounts of total RNA (20–80 ng) and directly measure the ≈22-nt miRNA but not hairpin precursors or primary transcripts (Fig. 8). In contrast to a previous report indicating that expression of human BIC RNA accumulation was limited to HLs (6), we detected elevated levels of BIC RNA and miR-155 in several types of B cell lymphomas. Thus, the Invader miRNA assay may find utility as a diagnostic test, for example, to detect elevated levels of miR-155 in other B cell lineages or to help differentiate between ABC and GC phenotypes in DLBCL cases.
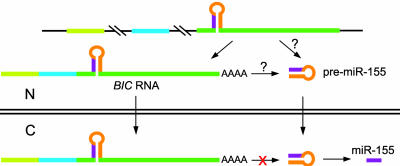
miR-155 and BIC RNA are processed from the same primary transcript, so it is not surprising that the amounts of both RNAs increase when BIC transcription is up-regulated in lymphomas (refs. 1 and 2 and W.T. and L.S., unpublished observations). Nevertheless, ratios of the amounts of the two RNAs ranged from ≈4:1 to ≈35:1 (Figs. 3 B and D). Variations in the relative amounts of miR-155 and BIC RNA between samples may result from alterations in RNA processing, turnover, or export. Generation of the ≈62-nt pre-miR-155 presumably occurs within the nucleus (12) by processing of the primary (unspliced) BIC transcript, the spliced BIC RNA (before it is exported), or both. Export of BIC RNA may diminish the pool of potential precursors of pre-miR-155 (Fig. 5), perhaps explaining why BIC RNA levels are not reliable indicators of miR-155 levels.
Fig. 5.
Pathways for generation of pre-miR-155. Pre-miR-155 may be generated by the nuclear Drosha-containing microprocessor complex acting on the primary BIC gene transcript or on spliced and polyadenylated BIC RNA that has not yet been exported from the nucleus. Cytoplasmic BIC RNA does not have access to this complex and, thus, cannot be used to make pre-miR-155.
Most accumulated BIC RNA is unlikely to have any known function because, being cytoplasmic (Fig. 2A), it is physically separated from the nuclear microprocessor complex. The short ORFs in BIC RNAs are not phylogenetically conserved (7), so they probably do not encode functional polypeptides. Thus BIC RNA that accumulates in cells may be the dead-end product of a system that controls the efficiency of pre-miR-155 production in the nucleus by export of this potential precursor.
Generation of several other pre-miRNAs or miRNAs is controlled. For example, human embryonic stem cells contain measurable amounts of the primary transcript encoding let-7a-1 but lack mature let-7a RNA (29). Like BIC RNA, many miRNA precursors resemble mRNAs or pre-mRNAs (30, 31), so their processing may be controlled by splicing (32), modification (33), and, as proposed here, nuclear export.
Our analyses of lymphoma samples and cell lines show that an elevation in the amount of miR-155 occurs in a wide range of lymphomas derived from B cells of different developmental stages. Increased miR-155 levels (≈2,000–10,000 copies per cell vs. ≈150 in normal circulating B cells) were observed in aggressive (DLBCL) and more indolent (CLL and MZ) lymphomas, and in non-HLs and HLs. Thus, miR-155 may play a role in the pathogenesis of B cell lymphomas in general. In clinical isolates of DLBCL and established cell lines, significantly higher levels of miR-155 were present in cells with the ABC phenotype than in cells with the GC phenotype (Fig. 3E and Tables 2 and 3). Thus, miR-155 levels may be useful diagnostically because ABC-type lymphomas have worse prognoses (34–35).
Croce and colleagues (15) reported reduced levels of miR-15a and/or miR-16 in most CLL isolates. We found that the levels of miR-15a were similarly reduced in DLBCL isolates, which shows that reduction in the level of this miRNA is not specific to CLL. In both DLBCL and the other samples studied here, the levels of miR-16 and let-7a RNA showed no consistent pattern of increase or decrease relative to the control B-cells, indicating that these latter two miRNAs do not change appreciably in lymphomas.
To our knowledge, miR-155 is the first miRNA shown to increase in cancer. Because miRNAs act as posttranscriptional down-regulators of gene expression, an elevated level of miR-155 might directly or indirectly reduce the synthesis of a protein with tumor suppressor or proapoptotic function. Recently, the mRNA of transcription factor PU.1, which is required for late differentiation of B cells (36), was identified as a possible target for miR-155 (37). We find similar miR-155 target sequences in the 3′ UTRs of PU.1 mRNAs of other mammals and chickens (Fig. 9, which is published as supporting information on the PNAS web site) and propose that the absence of PU.1 protein from Reed–Sternberg cells of HL (38, 39) is, at least in part, a consequence of the elevated levels of miR-155 in these cells. Also, the mRNA of another transcription factor that is controlled during B cell development, C/EBPβ (40), has a potential target site for miR-155 in its 3′ UTR (Fig. 9). It will be interesting to learn the extent to which changes in miRNA levels observed in this and other neoplasias contribute to the etiology of the cancers.
Supplementary Material
Acknowledgments
We thank Paul Bertics for reading the manuscript and Victor Lyamichev and Hatim Allawi (Third Wave Technologies) for discussions and generous gifts of some reagents used in the miR-15a, miR-16, and let-7a assays. This work was supported by National Institutes of Health Grant GM-30220 (to J.E.D.) and the Department of Pathology and Laboratory Medicine of Cornell University (to W.T.).
Author contributions: P.S.E., W.T., E.L., and J.E.D. designed research; P.S.E., W.T., L.S., A.C., Z.L., M.F.G., and E.L. performed research; P.S.E., W.T., and E.L. contributed new reagents/analytic tools; P.S.E., W.T., E.L., and J.E.D. analyzed data; and P.S.E., W.T., E.L., and J.E.D. wrote the paper.
Abbreviations: miRNA, microRNA; DLBCL, diffuse large B cell lymphoma; CLL, chronic lymphocytic leukemia; GC, germinal center; ABC, activated B cell; EBV, Epstein–Barr virus; HEK, human embryonic kidney.
References
- 1.Tam, W., Ben-Yehuda, D. & Hayward, W. S. (1997) Mol. Cell. Biol. 17, 1490-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clurman, B. E. & Hayward, W. S. (1989) Mol. Cell. Biol. 9, 2657-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam, W., Hughes, S. H., Hayward, W. S. & Besmer, P. (2002) J. Virol. 76, 4275-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haasch, D., Chen, Y. W., Reilly, R. M., Chiou, X. G., Koterski, S., Smith, M. L., Kroeger, P., McWeeny, K., Halbert, D. N., Mollison, K. W., et al. (2002) Cell. Immunol. 217, 78-86. [DOI] [PubMed] [Google Scholar]
- 5.Metzler, M., Wilda, M., Busch, K., Viehmann, S. & Borkhardt, A. (2004) Genes Chromosomes Cancer 39, 167-169. [DOI] [PubMed] [Google Scholar]
- 6.van den Berg, A., Kroesen, B. J., Kooistra, K., de Jong, D., Briggs, J., Blokzijl, T., Jacobs, S., Kluiver, J., Diepstra, A., Maggio, E. & Poppema, S. (2003) Genes Chromosomes Cancer 37, 20-28. [DOI] [PubMed] [Google Scholar]
- 7.Tam, W. (2001) Gene 274, 157-167. [DOI] [PubMed] [Google Scholar]
- 8.Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W. & Tuschl, T. (2002) Curr. Biol. 12, 735-739. [DOI] [PubMed] [Google Scholar]
- 9.Bartel, D. P. (2004) Cell 116, 281-297. [DOI] [PubMed] [Google Scholar]
- 10.He, L. & Hannon, G. J. (2004) Nat. Rev. Genet. 5, 522-531. [DOI] [PubMed] [Google Scholar]
- 11.Pasquinelli, A. E. (2002) Trends Genet. 18, 171-173. [DOI] [PubMed] [Google Scholar]
- 12.Cullen, B. R. (2004) Mol. Cell 16, 861-865. [DOI] [PubMed] [Google Scholar]
- 13.McManus, M. T. (2003) Semin. Cancer Biol. 13, 253-258. [DOI] [PubMed] [Google Scholar]
- 14.Xu, P., Guo, M. & Hay, B. A. (2004) Trends Genet. 20, 617-624. [DOI] [PubMed] [Google Scholar]
- 15.Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., Aldler, H., Rattan, S., Keating, M., Rai, K., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takamizawa, J., Konishi, H., Yanagisawa, K., Tomida, S., Osada, H., Endoh, H., Harano, T., Yatabe, Y., Nagino, M., Nimura, Y., et al. (2004) Cancer Res. 64, 3753-3756. [DOI] [PubMed] [Google Scholar]
- 17.Michael, M. Z., O'Conner, S. M., van Holst Pellekaan, N. G., Young, G. P. & James, R. J. (2003) Mol. Cancer Res. 1, 882-891. [PubMed] [Google Scholar]
- 18.Anonymous (1997) Blood 89, 3909-3918.9166827 [Google Scholar]
- 19.Chang, C. C., McClintock, S., Cleveland, R. P., Trzpuc, T., Vesole, D. H., Logan, B., Kajdacsy-Balla, A. & Perkins, S. L. (2004) Am. J. Surg. Pathol. 28, 464-470. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 21.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- 22.Allawi, H. T., Dahlberg, J. E., Olson, S., Lund, E., Olson, M., Ma, W. P., Takova, T., Neri, B. P. & Lyamichev, V. I. (2004) RNA 10, 1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eis, P. S., Olson, M. C., Takova, T., Curtis, M. L., Olson, S. M., Vener, T. I., Ip, H. S., Vedvik, K. L., Bartholomay, C. T., Allawi, H. T., et al. (2001) Nat. Biotechnol. 19, 673-676. [DOI] [PubMed] [Google Scholar]
- 24.Chen, C. Z., Li, L., Lodish, H. F. & Bartel, D. P. (2004) Science 303, 83-86. [DOI] [PubMed] [Google Scholar]
- 25.Erkmann, J. A. & Kutay, U. (2004) Exp. Cell Res. 296, 12-20. [DOI] [PubMed] [Google Scholar]
- 26.Zeng, Y. & Cullen, B. R. (2003) RNA 9, 112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G. & Tuschl, T. (2004) Mol. Cell 15, 185-197. [DOI] [PubMed] [Google Scholar]
- 28.Calin, G. A., Liu, C. G., Sevignani, C., Ferracin, M., Felli, N., Dumitru, C. D., Shimizu, M., Cimmino, A., Zupo, S., Dono, M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 11755-11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suh, M. R., Lee, Y., Kim, J. Y., Kim, S. K., Moon, S. H., Lee, J. Y., Cha, K. Y., Chung, H. M., Yoon, H. S., Moon, S. Y., et al. (2004) Dev. Biol. 270, 488-498. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez, A., Griffiths-Jones, S., Ashurst, J. L. & Bradley, A. (2004) Genome Res. 14, 1902-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smalheiser, N. R. (2003) Genome Biol. 4, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bracht, J., Hunter, S., Eachus, R., Weeks, P. & Pasquinelli, A. E. (2004) RNA 10, 1586-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luciano, D. J., Mirsky, H., Vendetti, N. J. & Maas, S. (2004) RNA 10, 1174-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gascoyne, R. D. (2004) Curr. Opin. Oncol. 16, 436-441. [DOI] [PubMed] [Google Scholar]
- 35.Lossos, I. S., Czerwinski, D. K., Alizadeh, A. A., Wechser, M. A., Tibshirani, R., Botstein, D. & Levy, R. (2004) N. Engl. J. Med. 350, 1828-1837. [DOI] [PubMed] [Google Scholar]
- 36.Loddenkemper, C., Anagnostopoulos, I., Hummel, M., Johrens-Leder, K., Foss, H. D., Jundt, F., Wirth, T., Dorken, B. & Stein, H. (2004) J. Pathol. 202, 60-69. [DOI] [PubMed] [Google Scholar]
- 37.John, B., Enright, A. J., Aravin, A., Tuschl, T., Sander, C. & Marks, D. S. (2004) PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hertel, C. B., Zhou, X. G., Hamilton-Dutoit, S. J. & Junker, S. (2002) Oncogene 21, 4908-4920. [DOI] [PubMed] [Google Scholar]
- 39.Jundt, F., Kley, K., Anagnostopoulos, I., Schulze Probsting, K., Greiner, A., Mathas, S., Scheidereit, C., Wirth, T., Stein, H. & Dorken, B. (2002) Blood 99, 3060-3062. [DOI] [PubMed] [Google Scholar]
- 40.Xie, H., Ye, M., Feng, R. & Graf, T. (2004) Cell 117, 663-676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.