Abstract
Toxicity and its detection in the dinoflagellate fish predators Pfiesteria piscicida and Pfiesteria shumwayae depend on the strain and the use of reliable assays. Two assays, standardized fish bioassays (SFBs) with juvenile fish and fish microassays (FMAs) with larval fish, were compared for their utility to detect toxic Pfiesteria. The comparison included strains with confirmed toxicity, negative controls (noninducible Pfiesteria strains and a related nontoxic cryptoperidiniopsoid dinoflagellate), and P. shumwayae strain CCMP2089, which previously had been reported as nontoxic. SFBs, standardized by using toxic Pfiesteria (coupled with tests confirming Pfiesteria toxin) and conditions conducive to toxicity expression, reliably detected actively toxic Pfiesteria, but FMAs did not. Pfiesteria toxin was found in fish- and algae-fed clonal Pfiesteria cultures, including CCMP2089, but not in controls. In contrast, noninducible Pfiesteria and cryptoperidiniopsoids caused no juvenile fish mortality in SFBs even at high densities, and low larval fish mortality by physical attack in FMAs. Filtrate from toxic strains of Pfiesteria spp. in bacteria-free media was cytotoxic. Toxicity was enhanced by bacteria and other prey, especially live fish. Purified Pfiesteria toxin extract adversely affected mammalian cells as well as fish, and it caused fish death at environmentally relevant cell densities. These data show the importance of testing multiple strains when assessing the potential for toxicity at the genus or species level, using appropriate culturing techniques and assays.
Keywords: toxigenic dinoflagellates, Pfiesteria piscicida, Pfiesteria shumwayae
Outbreaks of toxic Pfiesteria piscicida and Pfiesteria shumwayae, dinoflagellates that prey upon fish and other organisms, in the two largest estuaries on the U.S. mainland during the 1990s provide a compelling example of linkages between fish kills/disease and impacts on human health (1–3). Previous research has established that toxicity is highly variable among strains within a given toxic algal species (4), including Pfiesteria spp., ranging from strains with negligible toxicity to highly toxic strains (1, 5–7). Pfiesteria strains can cause larval fish death by physical attack (1). Their toxicity status was operationally defined by a multiagency/academic consensus group (8). Under conditions conducive to toxicity expression (9–11), actively toxic (TOX-A) strains, grown with live finfish, are capable of killing fish with toxin involvement at low to moderate cell densities (≥4 × 102 to 103 cells per ml, minutes to several hours). Impacts are exacerbated by Pfiesteria physical attack (6, 7, 12). The same strains separated from live fish for days or longer can sometimes produce sufficient exotoxin to cause death of sensitive larval stages without physical contact (7). Noninducible (NON-IND) strains apparently are incapable of killing fish with toxin (1, 6, 8).
A hydrophilic Pfiesteria toxin (PfTx), isolated in 1997 (13) and consisting of a metallated organic complex (see the supporting information, which is published on the PNAS web site), has been shown to affect fish and mammals (1, 13–15). After 7 years, PfTx has been purified and a pharmacological mode of action has been described (16). The time course for analysis is within the range for other dinoflagellate toxins, limited by the quantity of available toxic culture: e.g., ciguatoxin and maitotoxin required 23 years (17, 18) and 17 years (19, 20), respectively, from isolation to purification and structural elucidation. Additional PfTx is being produced (chemical structure being published elsewhere) so that standards can be developed for routine use. In the interim, various fish assays have been used in attempts to detect toxic Pfiesteria strains (5, 6, 10–12, 21–25). Although some techniques have provided evidence of toxicity to fish (5, 10, 14, 24), researchers using other methods have concluded that Pfiesteria is not toxigenic at the species [P. shumwayae (21, 22)] or genus (23, 25) levels. A comparative technique analysis was warranted to resolve these divergent conclusions, which have important policy implications in coastal resource management (3, 26).
Here we compare the two techniques most frequently used to detect toxic Pfiesteria, standardized fish bioassays (SFBs) with juvenile fish (10) and fish microassays (FMAs) with larval finfish (1, 21) or shellfish (7), for utility in detecting actively toxic Pfiesteria strains. SFBs, often maintained for weeks, originally were calibrated by using Pfiesteria spp. strains capable of rapid fish-killing at low to moderate cell densities (4 × 102 to 103 cells per ml) and confirmed to produce hydrophilic PfTx (10, 14). SFBs were not developed to distinguish between toxicity and physical attack as factors in fish death; rather, they were designed to detect toxic Pfiesteria strains, known to routinely prey upon fish (1, 10). Small-scale assays were also used to test for toxicity of culture filtrate from SFBs. FMAs, maintained for hours to several days, have been proposed for use in ruling out the presence of toxic Pfiesteria (21, 23) based on lack of fish death in culture filtrate. The following hypotheses were tested: (i) Pfiesteria spp. strains toxic to fish vary in PfTx release and can produce toxin in bacteria-free media; and (ii) toxicity is significantly enhanced by bacteria and other prey, especially live fish.
Materials and Methods
Dinoflagellate Cultures and SFBs with Juvenile Fish. Pfiesteria strains [P. piscicida CAAE2200 isolated in 1996 from Beaufort Point, NC, cloned and tested in 1999–2001 by A.S.G. and H.G.M.; P. shumwayae CAAE1024C, referred to as 101272 in ref. 11, isolated in 2000 from Marshall Creek, Chincoteague Bay, MD, and cloned (1); “CAAE” indicates the collection of the Center for Applied Aquatic Ecology] were grown with benign algal prey (Cryptomonas sp.) (ref. 1 and supporting information). Algae-fed Pfiesteria cultures were added to separate sets of SFBs (7 liters, n = 3–10, depending on the experiment, ≈70 flagellated cells per ml) in a biohazard level III containment system, following ref. 10. They were maintained for ≤14 weeks (2000–2004; toxicity to fish and PfTx verified as in refs. 10 and 14) with tilapia (Oreochromis mossambica or Oreochromis nilauticus, 3–10 juveniles per container depending on the experiment, total length 4–5 cm). P. shumwayae strain CCMP2089 [isolated from the Pamlico Estuary, NC, in 1999 (21); “CCMP” indicates the collection of the Culture Center for Marine Phytoplankton, Bigelow Laboratory, West Boothbay Harbor, ME], previously used by other researchers to negate toxigenicity in this species (21) and genus (23), was cultured and assayed as above (techniques in refs. 10 and 11).
To compare Pfiesteria toxicity when SFBs were inoculated with algae-versus fish-fed cultures, one of the SFBs that exhibited fish death (≥4 h as in ref. 11) involving toxic Pfiesteria was randomly selected and Pfiesteria isolates were inoculated from this SFB into a second set of SFBs (n = 5). SFBs were monitored for fish mortality (excluding occasional cannibalism), water quality, Pfiesteria abundance, and other microbes as in ref. 10. Rapid removal of dead fish maintained good water quality as in ref. 10 [i.e., total ammonia <1 mg/liter; pH 7.6–8.2 (supporting information)]. After repeated rapid fish death occurred, Pfiesteria strains were recloned and cultured with cryptomonads or with a fish cell line (27) in bacteria-free conditions to assess whether Pfiesteria can show toxicity in the absence of bacteria.
Negative controls included SFBs of juvenile tilapia treated identically but without dinoflagellates. Feeding controls (attempt to account for dinoflagellate physical attack of fish) included SFBs with cryptoperidiniopsoids [CAAE543A-AC1 (CCMP2302), a nontoxic Pfiesteria-like organism (1)], and SFBs with NON-IND Pfiesteria [P. piscicida clone CAAE1036C; clone from isolate CAAE2200, tested in 2003; and CCMP1832 (supporting information)]. Cryptoperidiniopsoids and NON-IND as well as toxic Pfiesteria strains prey upon fish by raptorial feeding (1, 28). Feeding control isolates were inoculated into SFBs as above. Dinoflagellates from fish- and algae-fed cultures were identified by plate tabulation as in ref. 1 and sequence-specific 18S ribosomal DNA-based PCR molecular probes (29, 30).
Filtrate from positive SFBs [fish death in ≤4 h with ≥3.0 × 102 to 9.28 × 103 cells per ml of P. piscicida (CAAE2200, in 2001) or P. shumwayae (CAAE1024C and CCMP2089, in 2001–2003)] and negative controls were tested for fish-killing activity by A.S.G. and H.G.M. within a biohazard level III containment system [10–20 liters gently filtered (0.2-μm porosity) and pooled; one juvenile tilapia per 100-ml container, total length ≈2 cm; gentle aeration; n = 10]. Whole cultures were tested for comparison. Toxic activity was inferred from mortality in ≤24 h. Fish were also examined for epidermal integrity (n = 32; supporting information).
Microbial Communities. PCR amplification of rDNA,§§ followed by denaturing gradient gel electrophoresis (DGGE) (ref. 31 and supporting information), was used to evaluate microbial composition in the water column. Prokaryotes (≥0.2-5.0-μm and ≥5-μm size fractions) in algae-fed cultures plus bacteria, control (minus dinoflagellates, and plus feeding controls), and positive SFBs were compared.
FMAs with Larval Fish. P. shumwayae strains CAAE1024C and CCMP2089 (algae- or fish-fed) were tested in FMAs (one fish per 10-ml well, n = 6, in triplicate; supporting information) by using Cyprinodon variegatus (age <25 days). Two sets of FMAs were used, with (7, 12, 21) versus without (1, 12) a filter partition (0.4-μm porosity, Costar 3450) to prevent physical contact of dinoflagellates and fish. A cell density gradient was imposed by adding filtered media (0.22-μm porosity, 15 practical salinity units) to effect initial Pfiesteria densities of 0 (controls), 0.25 × 102, 0.5 × 102, 0.8 × 102, 1 × 103, 2 × 103, 3 × 103, and 5 × 103 cells per ml. Values for the 50% lethal concentration (LC50) were determined at 24, 48, and 72 h. NON-IND Pfiesteria and cryptoperidiniopsoids were also tested (density gradient up to 4 × 104 cells per ml).
Tests for Pfiesteria Culture Purity. A standard mix of antibiotics (Sigma P4083) was inoculated into one culture each of algae-fed P. shumwayae and P. piscicida [final concentrations 100 units of penicillin per ml, 0.10 mg of streptomycin per ml, 0.20 mg of neomycin per ml (ref. 32 and supporting information)]. After 48 h, the culture was aseptically transferred to sterile nutrient-enriched broth (1 ml, 23°C) and agar media (0.1 ml, 37°C), incubated in darkness, and checked daily (8 d) for bacterial and fungal growth. Bacteria-free P. shumwayae fed fish cells was prepared as described in ref. 27. Uninoculated broth tubes or agar plates served as controls. Cultures were also examined under light microscopy (phase-contrast, ×750) after 7–10 d (32). Growth was not detected; thus, cultures were considered bacteria- and fungus-free, defined as lacking demonstrable unwanted prokaryotes and eukaryotes (32–34), and culture filtrates were tested for cytotoxicity.
Toxicity, Toxin Production, and Environmental Relevance. Filtrate (0.45-μm porosity) from SFBs, algae-fed cultures, and negative controls were encoded and sent to P.D.M. and J.S.R. for blind analysis of PfTx according to ref. 14. The toxin(s) from Pfiesteria spp. cultures have yielded no diagnostic UV chromophore at concentrations typically present in culture extracts. High-performance liquid chromatography analysis was carried out to test for the presence/absence of PfTx by using bioassay-guided fractionation, i.e., retention time and associated cytotoxicity and/or fish assays. 13C NMR spectra of the PfTx were also obtained (supporting information). Additional controls were equivalent or higher volumes (≥750 ml) of filtered seawater (0.22-μm porosity; 15 practical salinity units with deionized water), and algal culture (≈1.500 × 104 cells per ml). Samples were processed for toxin extraction, purification, cytotoxicity, and fish toxicity. For cytotoxicity assays, the rat pituitary GH4C1 cell line (ref. 13 and supporting information) and the colorimetric assay of ref. 14 were used. Also, filtrate from SFBs and algae-plus bacteria-fed Pfiesteria cultures (3-μm filter porosity, to remove Pfiesteria and cryptomonads) was refiltered (0.22-μm porosity), and retained bacteria were resuspended in fresh medium and tested for cytotoxic activity. An elutropic solvent scheme of increasing polarity was used to partition the soluble extract derived from Pfiesteria culture (supporting information). Data for PfTx concentrations are reported on a per-cell basis as dry residue weight taken up in standard volumes (dry weight of PfTx residue obtained from the total number of cells in a given sample, added to a known volume), to enable comparison of Pfiesteria isolates.
For fish assays with PfTx, 50 μlof PfTx extract in 100% methanol carrier was added to 2-ml FMAs (15 practical salinity units) with larval C. variegatus [age 7 d, one fish per FMA, 24 h, n = 6, two sets (supporting information)]. Extract from control cultures was also tested. We normalized fish mortality on a picograms of toxic extract per Pfiesteria cell basis, and calculated the cell density required for sufficient PfTx to kill test fish. The data were compared with Pfiesteria field densities based on semiquantitative PCR of water samples from two estuarine fish kills, and also compared with published data for other algal toxins (supporting information). For the standard curve in semiquantitative PCR, cell numbers of a serially diluted, mixed clonal P. piscicida culture were quantified in light microscopy, and the DNA was extracted and quantified. P. piscicida cells were also quantified in a set of internal spikes [linear relationship between cell numbers and DNA, R2 = 0.9518 (supporting information)].
Results
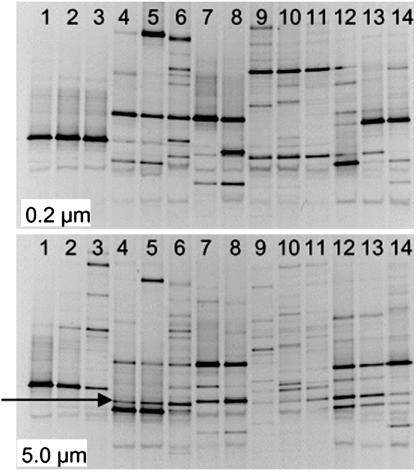
Environmental Conditions and Microbial Communities. Water quality in control and test SFBs with TOX-A P. shumwayae was comparable and followed recommended conditions for toxicity expression in Pfiesteria spp. (ref. 10 and supporting information). Presumptive Vibrio spp. generally were higher in negative control SFBs than in test SFBs plus toxic Pfiesteria strains (Student's t test, P < 0.1; supporting information). PCR/DGGE analyses indicated no significant pattern of specific bacterial growth in control versus test SFBs (Fig. 1) (supporting information). No bands were consistently, exclusively present in all lanes of the toxic Pfiesteria samples or in negative controls except for band 10, which was also present in several toxic samples.
Fig. 1.
DGGE images of PCR products, amplified for prokaryotic 16S rDNA by using eubacterial primers (31). DNA template was isolated from size-fractionated media from SFBs [0.2- to 5-μm fraction (Upper) and >5-μm fraction (Lower)]. Lanes 1–6, toxic P. shumwayae (two cultures evaluated, n = 3: CAAE1024C in lanes 1–3, CCMP2089 in lanes 4–6); lane 7, cryptoperidiniopsoid CAAE543A-AC1; lane 8, P. piscicida CAAE1332T-AC2 (CCMP2361); lanes 9–14, negative controls (media from fish cultures minus dinoflagellates). The presence of band 10 (arrow) in lanes 4–6 (toxic P. shumwayae CCMP2089) and lanes 9–14 (negative controls) suggests that this bacterium does not play a role in Pfiesteria toxicity.
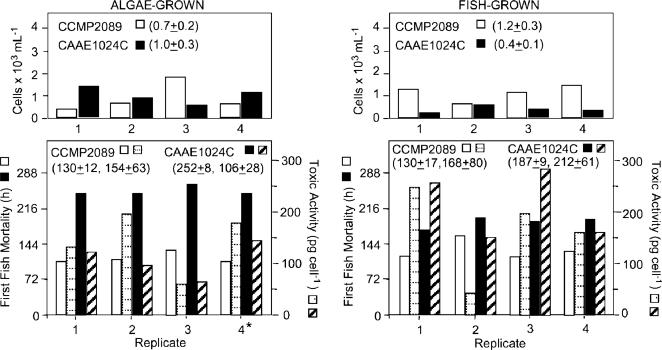
SFBs with Juvenile Fish. There was 100% fish survival and fish appeared active and healthy in all controls (fish minus dinoflagellates, fish plus cryptoperidiniopsoids, and fish plus NON-IND Pfiesteria). Feeding controls did not cause juvenile fish death even at high densities (up to 2.00 × 104 cells per ml; supporting information). In contrast, at low to moderate densities (≥4 × 102 to 103 cells per ml), there was 100% fish mortality in SFBs with Pfiesteria spp. strains verified as actively toxic (CAAE2200, CAAE1024C, and CCMP2089) (Table 1 and Figs. 2, 3, and 4). Fish death was usually preceded by lethargy alternating with hyperactivity as in refs. 1 and 12. Thus, when these strains were grown under appropriate conditions in SFBs [pH >7, total ammonia <1 ppm, etc. (10)], they expressed toxicity to fish. There was no significant correlation between initial or final Pfiesteria cell densities and time to first fish death in SFBs (P > 0.1). Fish-fed CAAE1024C (P < 0.00005), but not CCMP2089, killed additional fish more rapidly and had higher toxicity per cell than algae-fed subclones (P < 0.01) (Fig. 2). Whereas CCMP2089 killed fish more rapidly in SFBs (P < 0.0008), fish-acclimated CAAE1024C killed at lower cell densities than fish-acclimated CCMP2089 (P < 0.02) (Fig. 3).
Table 1. Toxic activity of purified PfTx extract from filtrate of Pfiesteria isolates in FMAs with larval C. variegatus, and cytotoxicity to mammalian cells.
| Culture
|
|||||
|---|---|---|---|---|---|
| Culture and year tested | Volume, liters | Cells per ml | Toxic activity,* pg of toxin extract per cell | 50 μl of PfTx extract yields time to fish death (cell equivalents)† | Cytotoxicity (GH4C1 rat pituitary cells) |
| Cultures from positive SFBs | |||||
| P. shumwayae CAAE1024C | |||||
| Trial 1 (algae-fed, then SFBs, 2003) | 1 | 1.5 ± 0.3 × 103 | 58–202 | <30 min (41 ce) | + |
| Trial 2 (fish-fed, SFBs, 2003) | 1 | 0.8 ± 0.2 × 102 | 43–251 | NA | + |
| Trial 3 (fish-fed, SFBs, 2004) | 6.4 | 11.63×104 | 47 | <28 min (817 ce) | + |
| Trial 4 (fish cell-fed – bacteria, 2004) | 0.7 | 3.5×105 | — | NA | + |
| P. shumwayae CCCMP2089 | |||||
| Trial 1 (algae-fed, then SFBs, 2003) | 1 | 1.2 ± 0.2 × 103 | 67–142 | <28 min (64 ce) | + |
| Trial 2 (fish-fed, SFBs, 2003) | 1 | 1.4 ± 1.1 × 103 | 150–284 | NA | + |
| Trial 3 (fish-fed, SFBs, 2004) | 6.4 | 1.1×104 | 0.5 | <24 h (555 ce) | + |
| Trial 4 (algae-fed, 2002) | 90 | 3.00×104 | 5 | <40 min (1,500 ce) | + |
| Controls (no fish death, SFBs) | |||||
| P. piscicida (fish-fed, SFBs, CCMP1832, NON-IND, 2003) | 300 | 2.50×104 | 6 | <90 min (1,250 ce) | + |
| P. piscicida (fish-fed, SFBs, CAAE2200, NON-IND, 2003) | 1 | 1.3×103 | ND | No mortality | — |
| Cryptoperidiniopsoid (fish-fed, SFBs, CAAE543-AC1, 2003) | 1 | 3×102 | — | No mortality | — |
| Fish – dinoflagellates (2003) | 7 | — | No mortality | — | |
All control fish remained active with no signs of stress. NA, not available; ND, not detectable.
Toxicity activity, based on mass of PfTx fraction purified from HW40F size-exclusion chromatography
Cell equivalents (ce) = μl of purified PfTx extract (derived from size-exclusion chromatography) × the cell density used to produce that purified extract
Fig. 2.
Pfiesteria cell densities and cytotoxic activity in SFBs. (Left) SFBs of four subclones of algae-fed P. shumwayae CCMP2089 and CAAE1024C, showing water-column cell densities at the time of first fish mortality (Upper) and time of first fish mortality (left ordinate) and estimated cytotoxic activity per cell at time of second fish mortality (right ordinate) (Lower). (Right) Data from SFBs of four subclones previously fed fish (from subclone 4 at left).
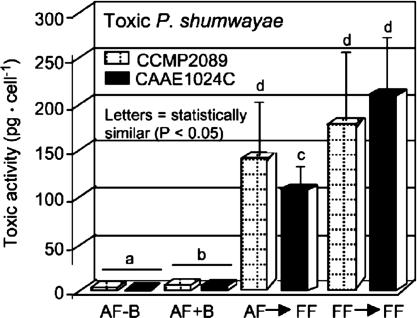
Fig. 3.
Estimated cytotoxic activity per flagellated cell (examples, P. shumwayae CAAE1024C and CCMP2089), from testing purified PfTx extract residues with GH4C1 rat pituitary cells: AF-B, algae-fed, bacteria-free culture; AF+B, algae-fed culture plus bacteria (for both clones significantly higher than in bacteria-free culture; P < 0.025); AF→FF, previously algae-fed subclones taken into SFBs; and FF→FF, fish-fed subclones inoculated into additional SFBs.
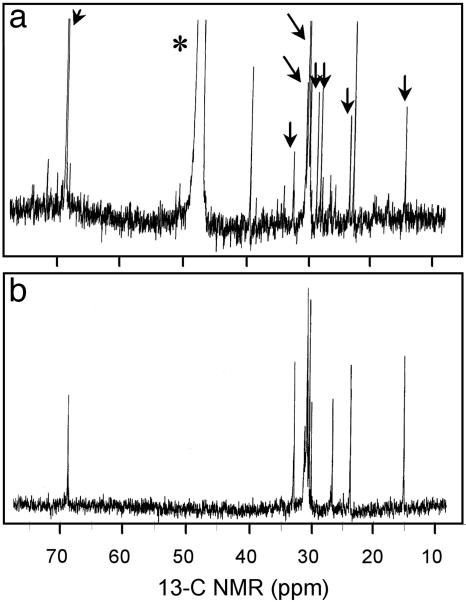
Fig. 4.
13C NMR spectra of PfTx from Pfiesteria. (a) PfTx from P. shumwayae (CAAE1024C) after size exclusion (HW40F column) and subsequent passage through three sequential C18 columns using d4 methanol (*) as a solvent (supporting information. This material, from a fish-fed culture, shows multiple molecular species (congeners) compared with b; arrows indicate peaks in common with b. (b) PfTx from algae-fed P. piscicida (CCMP1832) after the same chromatographic steps used to generate a, followed by passage through a bidentate column using 100% HPLC-grade water as a solvent (supporting information).
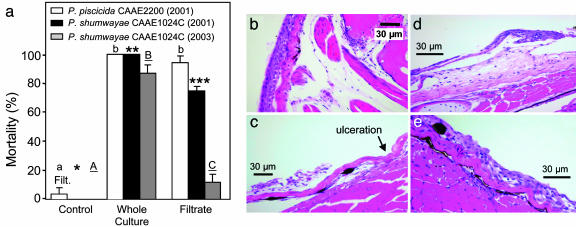
Fish death occurred in ≤24 h in filtrates, and in supernatant media after centrifugation, from SFBs conducted in 2000–2001 with P. piscicida CAAE2200 and P. shumwayae CAAE1024C (e.g., Fig. 5a). However, mortality was markedly lower than in unfiltered water plus Pfiesteria cells. Toxic activity of filtrates from positive SFBs of these strains (fish death in ≥4 h) was similar. CCMP2089 was less toxic, indicated by no fish mortality or epidermal lesions (as in ref. 21) in culture filtrate. However, additional tests yielded low fish mortality in filtrate from SFBs with CCMP2089 (6%, n = 13 trials, 8 fish per trial; significantly different from controls with 0% fish death, P < 0.05) (24). Beginning at 3 h of exposure, fish exposed to CAAE1024C culture and filtrate had necrosis and loss of large areas of epidermis (Fig. 5 c and d). Mild to moderate erosions predominated, but scattered areas had a focal breach of the basement membrane (ulceration). Edges of eroded or ulcerated areas showed moderate to severe swelling of Malpighian epithelial cells, with epithelial lifting/intraepidermal clefting, and occasional apoptotic cells. The underlying dermis was expanded multifocally by edema. After 4–12 h of exposure, fish had more severe skin erosions, but no time-dependent increase in severity was noted after 4 h. No remarkable gross lesions were discerned, and no remarkable microscopic abnormalities were found in controls. CAAE1024C had lost most toxicity by 2003 [14 ± 7% fish death in filtrate from SFBs (24)], compared with its toxicity in 2001 (80 ± 9% fish death in filtrate from SFBs with CAAE1024C) (Table 1 and Fig. 5a).
Fig. 5.
Experiments with fish in whole cultures of Pfiesteria versus in culture filtrate. (a) Juvenile tilapia killed (percent) by whole cultures and filtrate of P. shumwayae CAAE1024C and P. piscicida CAAE2200 from positive SFBs (means ± 1 SE; n = 10 fish per treatment; repeated 6 time for whole cultures, 10 times for filtrates; letters and asterisks indicate significant differences (P < 0.01 to 0.05]; modified from ref. 11). P. piscicida and P. shumwayae 2000–2002 tests were compared with retests in 2003. The P. piscicida strain had lost toxicity (no fish death in whole-culture SFBs or filtrate; data not shown). The P. shumwayae strain had lower toxicity [caused some death in whole-culture SFBs and filtrate (24)]. (b) Skin and underlying musculature from an unexposed control fish (pectoral area, epithelial cells four to six cell layers thick). At the dermal–epidermal interface there was mild artifactual separation, accentuating the stratum spongiosum. Deep to the skin were normal subcutis, skeletal muscle bundles, and bone. (c–e) Eroded and ulcerated skin from fish exposed for 8 h to cell-free filtrate from toxic P. shumwayae CAAE1024C, showing necrosis and fragmentation of eroded epidermis (lifted from the basement membrane) (c); dermal–epidermal cleft (blister-like) with underlying area of severe dermal edema that expanded the stratum spongiosum (d and e). Necrosis of Malpighian epithelial cells in the epidermis and dermal edema suggest that these changes were not artifacts of handling, immersion fixation, or processing. (e) After 4 h, increased edema between Malpighian epithelial cells and spongiosis. Occasional apoptotic cells and intracellular edema are consistent with early epithelial injury.
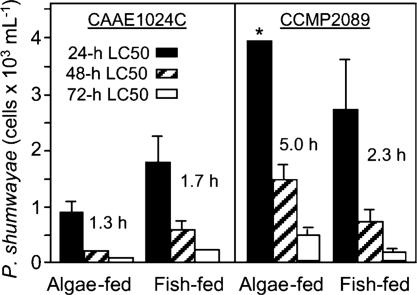
FMAs with Larval Fish. There was no larval fish death in negative controls, and only one feeding control strain tested (CAAE1036C) caused larval fish death in FMAs when allowed physical contact. At 72 h there was 83 ± 10% mortality at the highest cell density of this strain (3.881 × 104 cells per ml), 33 ± 10% mortality at 1.538 × 104 cells per ml, 22 ± 15% mortality at 8.74 × 103 cells per ml, 1 fish death (of 18 in total) at 3.64 × 103 cells per ml, and no fish mortality at lower cell densities. FMAs with toxic strains that were allowed physical contact with fish yielded variable data (Fig. 6). For P. shumwayae strain CAAE1024C, time to first fish death was more rapid at lower cell densities when subpopulations had been acclimated to algal prey before the FMAs; for CCMP2089, the reverse was observed. For both strains, higher cell densities were required for 24-h LC50s than for 48- to 72-h LC50s, and inoculum cell density was directly related to mortality (linear regression, R2 = 0.72–0.98). Although each strain had been capable of killing juvenile tilapia in SFBs in ≤4 h (with PfTx presence and toxicity to fish confirmed), and also killed some juvenile fish in filtrate from SFBs (this study and refs. 11 and 24), there was no larval fish death in FMAs unless these strains were allowed physical access (as in ref. 21).
Fig. 6.
LC50 values (24, 48, and 72 h) in FMAs of algae- and (juvenile) fish-fed P. shumwayae allowed physical contact with larval fish, comparing two toxic clones (means + 95% confidence intervals; h indicates time to first fish death; *, 24-h LC50 was greater than the highest density evaluated).
Cytotoxicity. Cytotoxicity assays with GH4C1 cells (14), coupled with sequential chromatographic analyses, showed that strains of Pfiesteria spp. that tested positive for fish mortality and toxin in SFBs also were toxic in algae-fed cultures with or without extracellular bacteria, although bacteria and prey enhanced toxicity (e.g., Fig. 3). Cytotoxic activity per cell was significantly lower from algae-fed than fish-fed toxic Pfiesteria (Fig. 3; P < 0.0001). No cytotoxicity or fish toxicity was found from lipophilic fractions, or from media with or without bacteria without Pfiesteria (supporting information).
Fish Response to Purified PfTx Extract. PfTx extracts yielded similar 13C NMR spectra for P. piscicida and P. shumwayae, and they consistently caused larval fish mortality, whereas there was no mortality of control fish exposed to carrier solvent alone (Table 1 and Fig. 4). Controls did not contain PfTx, except for feeding control NON-IND Pfiesteria CCMP1832 (Fig. 4). Although that strain did not kill fish in SFBs or FMAs, PfTx extracted from a large culture volume rapidly killed larval fish (Table 1). Thus, some NON-IND Pfiesteria strains produce this ichthyotoxin at very low levels, whereas others produce undetectable PfTx. The picograms of toxic extract per cell data for Pfiesteria were comparable to published data for other toxic algae (supporting information). Densities of Pfiesteria/Pfiesteria-like cells [generally ≥3 × 102 to 1.4 × 103 cells per ml (1)] or of Pfiesteria cells [estimated by semiquantitative PCR at ≈300–500 P. piscicida cells per ml (supporting information)] in fish kills involving toxic Pfiesteria were more than cell densities required to produce sufficient PfTx to kill fish in these assays (Table 1).
Discussion
This study demonstrates several points about Pfiesteria and, more generally, toxic dinoflagellates. Most importantly, it underscores the high variability in toxicity expression among strains within a given toxic algal species (reviewed in ref. 6). Also, Pfiesteria toxicity and its detection depended on the assay conditions and prey type. SFBs, originally standardized with actively toxic Pfiesteria strains (5, 10, 14), were reliable in detecting toxic Pfiesteria, based on confirmation of PfTx, but cytotoxicity assays and toxin assays were more sensitive for detecting PfTx than fish death in SFBs or FMAs. Because of the variability in toxin release shown by toxic strains of Pfiesteria spp. under different conditions, use of fish assays to form conclusions about presence versus absence of toxin production by a given strain should be accompanied by appropriate PfTx detection assays. Other important findings are confirmation of toxin production as indicated by cytotoxic activity by filtrate from Pfiesteria toxic strains in bacteria-free media, and of strain-dependent stimulation of Pfiesteria toxicity by live fish. In support of previous research (1, 11, 15), PfTx was also shown to be lethal to fish and toxic to mammals, and fish mortality from Pfiesteria was exacerbated by physical attack during predation (6). Physical attack by Pfiesteria was earlier hypothesized to facilitate PfTx entry into fish tissues and to generally weaken fish prey (6).
CCMP2089, like various other Pfiesteria spp. strains (1, 11, 14, 16), was toxic to fish and to mammalian cells, and at environmentally relevant cell densities. We suggest that previous conclusions that this strain did not produce exotoxin (21–23) were based on large-scale fish assay conditions that were not conducive to Pfiesteria toxicity expression [e.g., pH as low as 5.6, ammonia as high as 30 mg/liter (refs. 21 and 23 and methods ref. 35, versus methods for SFBs in ref. 10)], and/or tests for toxin/toxicity that were insufficient (21–23). Moreover, algae-fed CCMP2089, used as a culture source for FMAs in refs. 21 and 22, and an older strain used in ref. 25 (≈7 years in culture), would have had very low PfTx activity, based on the data from this study. The present findings indicate that lack of fish death in filtrate of FMAs (used in refs. 21 and 22) and SFBs (used in ref. 25) is not a sufficiently sensitive indicator of toxin presence/absence. Here, when filtrates from CCMP2089 were analyzed for PfTx by using cytotoxicity and ichthyotoxin assays, PfTx was confirmed. Thus, in FMAs, P. shumwayae strain CCMP2089 produced exotoxin that would have contributed to larval fish stress, but it did not produce sufficient exotoxin to kill fish as a toxin effect alone (21–23) or, in some tests, only occasionally did so (24).
The assumption previously had also been made (23), without precedent (4), that toxicity in all dinoflagellates is inexorably linked to polyketide synthase (PKS) genes. An insensitive molecular assay was used (23), given the high heterogeneity among known PKS genes (36), and when a PKS gene in strain CCMP2089 was not detected, it was concluded that Pfiesteria spp. are not toxigenic (23). Yet, when a degenerate nonribosomal peptide synthase primer set (37) was used in the same study, a PKS-encoding gene accidentally was amplified in CCMP2089 (23). A related study (21) misstated that fish lesions had been attributed only to Pfiesteria in Pfiesteria-related fish kills (in refs. 1 and 38) and concluded (21), solely on the basis of strain CCMP2089, that Pfiesteria cannot cause fish lesions as a toxic effect. Here, however, filtrate from a toxic strain did cause epidermal lesions in fish with toxin involvement, without physical attack by Pfiesteria cells. It was also previously asserted (23) that the only bioactive substance found in Pfiesteria cultures was a lipophilic plasticizer artifact from synthetic salts (supporting information). Yet, the reference cited in support of that assertion (14) had described isolation of hydrophilic PfTx. As shown by comparison of this study with refs. 21–23 and 25, recognition of variability in toxicity expression and other traits among strains (6), use of culture conditions conducive to toxicity expression (5, 10, 11), and appropriate assays for toxin will remain critical in forming sound insights about toxicity of harmful algal species and genera.
Many questions remain about controls on toxicity expression in Pfiesteria, such as the role of bacteria. Pfiesteria is capable of producing toxin in bacteria-free media, yet Pfiesteria toxin production was much higher in the presence of extracellular bacteria plus algal or fish prey. Similar results have been found for other toxic algae (e.g., ref. 39). Work with Pfiesteria in other SFBs has indicated differences in the bacteria flora (K.J.C. and S.C.C., unpublished data). Dynamic interactions between Pfiesteria and bacteria, perhaps including endosymbiotic bacteria, may play a role in stimulating Pfiesteria toxin production and/or release. For example, bacterial metabolism may convert exudates from Pfiesteria cells into toxic compounds, or enzymes released from bacteria may facilitate release of toxin from Pfiesteria cells. Bacteria may also play a role in loss of toxicity in Pfiesteria toxic strains over time in culture, as observed for other toxic algae (reviewed in ref. 1). Determination of molecular and environmental controls on its toxin production and release versus retention will further advance scientific understanding about toxic Pfiesteria.
Supplementary Material
Acknowledgments
G. Scott (National Ocean Service, Charleston, SC) counseled in assessment of environmental relevance of PfTx. Funding was provided by the National Science Foundation (Grants OCE-9912088 and OCE-9912089 to J.M.B.), the Environmental Protection Agency (Grant CR827831-01-0 to J.M.B.); the Centers for Disease Control and Prevention through the North Carolina Department of Health and Human Services (Grant 01501-04 to J.M.B.); the Virginia Department of Health and Department of Environmental Quality; and the National Oceanic and Atmospheric Administration (Grants NA-860P0510 and NA-96OP0335 to J.M.B.; and Grant NA-160P2805 to A.S.G. and J.M.B.).
Author contributions: J.M.B., A.S.G., P.D.M., J.M.L., K.J.C., A.J.L., H.G.M., N.J.D., S.C.C., J.W.K., and P.A.R. designed research; A.S.G., P.D.M., J.M.L., K.J.C., N.J.D., J.W.K., and S.L.M. performed research; J.W.K. contributed new reagents/analytic tools; J.M.B., A.S.G., P.D.M., J.M.L., K.J.C., A.J.L., J.S.R., H.G.M., N.J.D., S.C.C., J.W.K., and P.A.R. analyzed data; and J.M.B., A.S.G., P.D.M., J.M.L., K.J.C., A.J.L., and H.G.M. wrote the paper.
Abbreviations: CAAE, Center for Applied Aquatic Ecology; CCMP, Culture Center for Marine Phytoplankton; DGGE, denaturing gradient gel electrophoresis; FMA, fish microassay; LC50, 50% lethal concentration; NON-IND, noninducible strain, unable to kill fish with toxin; PfTx, hydrophilic Pfiesteria toxin; SFB, standardized fish bioassay; TOX-A, actively toxic.
Footnotes
References
- 1.Burkholder, J. M., Glasgow, H. B. & Deamer-Melia, N. J. (2001) Phycologia 40, 186-204. [Google Scholar]
- 2.Grattan, L. M., Oldach, D., Perl, T. M., Lowitt, M. H., Matuszak, D. L., Dickson, C., Parrott, C., Shoemaker, R. C., Kauffman, C. L., Wasserman, M. P., et al. (1998) Lancet 352, 532-539. [DOI] [PubMed] [Google Scholar]
- 3.Samet, J., Bignami, G. S., Feldman, R., Hawkins, W., Neff, J. & Smayda, T. (2001) Environ. Health Perspect. 109, 639-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plumley, F. G. (1997) Limnol. Oceanogr. 42, 252-264. [Google Scholar]
- 5.Marshall, H. G., Gordon, A. S., Seaborn, D. W., Dyer, B., Dunstan, W. M. & Seaborn, A. (2000) J. Exp. Mar. Biol. Ecol. 255, 65-74. [DOI] [PubMed] [Google Scholar]
- 6.Burkholder, J. M., Glasgow, H. B., Deamer-Melia, N. J., Springer, J., Parrow, M. W., Zhang, C. & Cancellieri, P. (2001) Environ. Health Perspect. 109, 667-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Springer, J., Shumway, S. E., Burkholder, J. M. & Glasgow, H. B. (2002) Mar. Ecol. Progr. Ser. 245, 1-10. [Google Scholar]
- 8.Turgeon, D. D., Higgins, J. L., Luttenberg, D., White, H., Orencia, M., Craig, N. & Bushaw-Newton, K. (2001) Protocols for Monitoring Pfiesteria and Related Health and Environmental Conditions in U.S. Coastal Waters (National Oceanic and Atmospheric Administration–National Ocean Service, Silver Spring, MD).
- 9.Burkholder, J. M. & Glasgow, H. B. (2002) J. Phycol. 38, 1261-1267. [Google Scholar]
- 10.Burkholder, J. M., Marshall, H. G., Glasgow, H. B., Seaborn, D. W. & Deamer-Melia, N. J. (2001) Environ. Health Perspect. 109, 745-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, A. S., Dyer, B. J., Seaborn, D. & Marshall, H. G. (2002) Harmful Algae 1, 85-94. [Google Scholar]
- 12.Burkholder, J. M. & Glasgow, H. B. (1997) Limnol. Oceanogr. 42, 1052-1075. [Google Scholar]
- 13.Fairey, E. R., Edmunds, J. S., Deamer-Melia, N. J., Glasgow, H. B., Johnson, F. M., Moeller, P. D. R., Burkholder, J. M. & Ramsdell, J. S. (1999) Environ. Health Perspect. 107, 711-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moeller, P. D. R., Morton, S. L., Mitchell, B. A., Sivertsen, S. K., Fairey, E. R., Mikulski, T. M., Glasgow, H., Deamer-Melia, N. J., Burkholder, J. M. & Ramsdell, J. S. (2001) Environ. Health Perspect. 109, 739-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin, E. D., Blackwelder, W. P., Glasgow, H. B., Burkholder, J. M., Moeller, P. D. R. & Ramsdell, J. S. (2003) Neurotoxicol. Teratol. 25, 419-426. [DOI] [PubMed] [Google Scholar]
- 16.Melo, A. C., Moeller, P. D. R., Glasgow, H. B., Burkholder, J. M. & Ramsdell, J. S. (2001) Environ. Health Perspect. 109, 731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheuer, P. J., Takahashi, W., Tsutsumi, J. & Yoshida, T. (1967) Science 155, 1267-1268. [DOI] [PubMed] [Google Scholar]
- 18.Murata, M., Legrand, A. M., Ishibashi, Y., Fukui, M. & Yasumoto, T. (1990) J. Am. Chem. Soc. 112, 4380-4386. [Google Scholar]
- 19.Yasumoto, T., Bagnis, R. & Vernoux, J. P. (1976) Bull. Jpn. Sci. Fish. 42, 359-365. [Google Scholar]
- 20.Murata, M., Naoki, H., Iwashita, T., Matsunaga, S., Sasaki, M., Yokoyama, A. & Yasumoto, T. (1993) J. Am. Chem. Soc. 115, 2060-2062. [Google Scholar]
- 21.Vogelbein, W. K., Lovko, V. J., Shields, J. D., Reece, K. S., Mason, P. L., Haas, L. A. & Walker, C. C. (2002) Nature 418, 967-970. [DOI] [PubMed] [Google Scholar]
- 22.Lovko, V. J., Vogelbein, W. K., Shields, J. D., Haas, L. W. & Reece, K. S. (2003) J. Phycol. 39, 600-609. [Google Scholar]
- 23.Berry, J. P., Reece, K. S., Rein, K. S., Baden, D. G., Haas, L. W., Ribeiro, W. L., Shields, J. D., Snyder, R. V., Vogelbein, W. K. & Gawley, R. E. (2002) Proc. Natl. Acad. Sci. USA 99, 10970-10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon, A. & Dyer, B. (2005) Harmful Algae 4, 423-431. [Google Scholar]
- 25.Drgon, T., Saito, K., Gillevet, P. M., Sikaroodi, M., Whitaker, B., Krupatkina, D. N., Argemi, F. & Vasta, G. R. (2005) Appl. Environ. Microbiol. 79, 519-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magnien, R. E. (2001) Bioscience 51, 843-852. [Google Scholar]
- 27.Parrow, M. W., Burkholder, J. M., Deamer, N. J. & Ramsdell, J. S. (2005) Aquat. Microb. Ecol., in press.
- 28.Parrow, M. W. & Burkholder, J. M. (2003) J. Phycol. 39, 678-696. [Google Scholar]
- 29.Oldach, D. W., Delwiche, C. F., Jakobsen, K. S., Tengs, T., Brown, E. G., Kempton, J. W., Schaefer, E. F., Bowers, H. A., Glasgow, H. B., Jr., Burkholder, J. M., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coyne, K. J., Hutchins, D. A., Hare, C. E. & Cary, S. C. (2001) Aquat. Microb. Ecol. 24, 275-285. [Google Scholar]
- 31.Muyzer, G. de Waal, E. C. & Uitterlinden, A. G. (1993) Appl. Environ. Microbiol. 59, 695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guillard, R. R. L. & Keller, M. D. (1984) in Dinoflagellates, ed. Spector, D. L. (Academic, London), pp. 391-442.
- 33.Hoshaw, R. W. & Rosowski, J. R. (1973) in Handbook of Phycological Methods, ed. Stein, J. (Cambridge Univ. Press, London), pp. 53-68.
- 34.Guillard, R. R. L. (1975) in Culture of Marine Invertebrate Animals, eds. Smith, W. L. & Chanley, M. H. (Plenum, New York), pp. 29-60.
- 35.Vogelbein, W. K., Shields, J. D., Haas, L. W., Reece, K. S. & Zwerner, D. E. (2001) Environ. Health Perspect. 109, 687-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bentley, R. & Bennett, J. W. (1999) Annu. Rev. Microbiol. 53, 411-446. [DOI] [PubMed] [Google Scholar]
- 37.Marahiel, M. A., Stachelhaus, T. & Mootz, H. D. (1997) Chem. Rev. 97, 2651-2674. [DOI] [PubMed] [Google Scholar]
- 38.Glasgow, H. B., Burkholder, J. M., Mallin, M. A., Deamer-Melia, N. J. & Reed, R. E. (2001) Environ. Health Perspect. 109, 715-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bates, S. S., Douglas, D. J., Doucette, G. J. & Leger, C. (1995) Nat. Toxins 3, 428-435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.