Abstract
Rhodiola crenulata, a well-known medicinal Tibetan herb, is mainly grown in high-altitude regions of the Tibet, Yunnan, and Sichuan provinces in China. In the past few years, increasing numbers of studies have been published on the potential pharmacological activities of R. crenulata, strengthening our understanding into its putitive active ingredient composition, pharmacological activity, and mechanism of action. These findings also provide strong evidence supporting the important medicinal and economical value of R. crenulata. Consequently, some Rhodiola species are becoming endangered because of overexploitation and environmental destruction. However, little is known about the genetic and genomic information of any Rhodiola species. Here we report the first draft assembly ofthe R. crenulata genome, which was 344.5 Mb (25.7 Mb Ns), accounting for 82% of the estimated genome size, with a scaffold N50 length of 144.7 kb and a contig N50 length of 25.4 kb. The R. crenulata genome is not only highly heterozygous but also highly repetitive, with ratios of 1.12% and 66.15%, respectively, based on the k-mer analysis. Furthermore, 226.6 Mb of transposable elements were detected, of which 77.03% were long terminal repeats. In total, 31 517 protein-coding genes were identified, capturing 86.72% of expected plant genes in BUSCO. Additionally, 79.73% of protein-coding genes were functionally annotated. R. crenulata is an important medicinal plant and also a potentially interesting model species for studying the adaptability of Rhodiola species to extreme environments. The genomic sequences of R. crenulata will be useful for understanding the evolutionary mechanism of the stress resistance gene and the biosynthesis pathways of the different medicinal ingredients, for example, salidroside in R. crenulata.
Keywords: Rhodiola crenulata, genomics, genome assembly, annotation
Data Description
Background information
Genus Rhodiola, in the family Crassulaceae, is a perennial herbaceous flowering plant and is mainly grown in the cool climate of subarctic areas, such as North America, Northern and Central Europe, and mountainous regions of southwest and northwest China. In general, Rhodiola species have similar morphology, causing difficulty and confusion in their taxonomic identification and classification [1]. Although many Rhodiola species have been used as traditional medicines for a long time, some being widely used for therapies of cardiovascular disease, hypobaric hypoxia, microbial infection, tumour and muscular weakness, the precise pharmacological mechanisms of actions are still unclear [1–6]. In China, in comparison with other Rhodiola species, R. crenulata is the most popular and in demand, but the supply of R. crenulata is limited due to its stringent growing requirement. The high selling price of R. crenulata causes serious problems of R. crenulata adulteration in the market. In order to improve the understanding of Rhodiola species, we have sequenced the whole genome of R. crenulata, and have subsequently completed the genomic assembly and annotation.
Sample collection and sequencing
According to protocol 1 (Additional File 2), genomic DNA was isolated from the leaf tissue of a single male R. crenulata (NCBI taxonomy ID: 242839)(Fig. 1), which was collected from Shangri-La, located in the northwest of Yunnan province, China. Three paired-end libraries with insert sizes of 250 bp, 500 bp, and 800 bp and three mate-pair libraries (5 kb, 10 kb, and 20 kb) were subsequently constructed with the standard protocol provided by Illumina (San Diego, CA, USA) and sequenced on an Illumina HiSeq 2000/4000 platform using a whole genome shotgun sequencing (WGS) strategy. A total of 162.08 Gb (∼380X) of raw sequence reads were generated (Additional File 1: Table S1). To reduce the effect of sequencing errors to the assembly, SOAPfilter (v. 2.2), a package from SOAPdenovo2 (SOAPdenovo2, RRID:SCR_014986) [7], was used to filter reads with adapters, low quality, undersize insert size, and PCR duplication. Finally, 123.47 Gb (∼290X)of clean data were obtained (Additional File 1: Table S1).
Figure 1:

Example of R. crenulata (image from Shifeng Li).
RNA was extracted from the root, stem, and leaf tissues, respectively, of a single male R. crenulata, which was collected from the Jade Dragon Snow Mountain, located in the northwest of Yunnan province, China, according to the protocol 2 (Additional File 2). Single-end libraries were constructed subsequently using standard protocol provided by BGI (BGI-Shenzhen) and then sequenced on the BGISEQ-500 platform [8,9]. In total, 13.54 Gb of raw data was obtained, and after filtering by SOAPnuke (v. 1.5.6; https://github.com/BGI-flexlab/SOAPnuke), we finally produced 13.23 Gb of high-quality clean data (Additional File 1: Table S2). In this study, different sequencing platforms were used, taking into consideration the efficiency of data generation and also allowing the consistency of data for analysis.
Assembly
First, the genome size, 420.2 Mb, was estimated based on the 17-mer analysis [10] using 34.4 Gbof clean data from 250 bp insert library, as well as the repetitive and heterozygous ratio with 66.15% and 1.12%, respectively (Additional File 1: Table S3; Fig. S1). We also found that our estimated genome size of R. crenulata was relatively close to the median genome size of species inthe family Crassulaceae based on existing data in the C-values database [11], which range from 142 Mb to 8.9 Gb (Additional File 1: Table S4). Given the high heterozygosity, Platanus (v. 1.2.4) [12],which is efficient for the assembly of highly heterozygous genomes, was used to assemble the genome by performing “assemble, scaffold, gap_close” modes orderly with “k = 35.” As a result, 345.1 Mb (containing 65.9 Mb Ns) of draft assembly with a contig N50 length of 6.3 kb and a scaffold N50 length of 145.1 kb was generated (Additional File 1: Table S5). To further improve the quality of our assembly genome, GapCloser (v. 1.10) [7] was implemented with all six libraries of data. Finally, we obtained the 344.5 Mb (containing 25.7 Mb Ns) of assembly genome, representing 82% of the estimated genome size, with contig and scaffold N50 lengths of 25.4 kb and 144.7 kb, respectively (Table 1). Meanwhile, we also ran other prevalent de novo assemblers, such as SOAPdenovo2 [7]and ABySS (v. 1.9.0; ABySS, RRID:SCR_010709) [13], with various modifications of parameters. But the results based on these assemblers were not better (Additional File 1: Table S5). More methodological information is available in protocol 3 (Additional File 2).
Table 1:
Statistics of the final assembly using Platanus and Gapcloser.
| Type | Scaffold | Contig |
|---|---|---|
| Total number | 150 003 | 161 878 |
| Total length (bp) | 344 513 827 | 318 807 120 |
| N50 length (bp) | 144 749 | 25 360 |
| N90 length (bp) | 1003 | 877 |
| Max length (bp) | 1 309 315 | 300 573 |
| GC content (%) | 39.68 | 39.68 |
Repeat annotation and gene prediction
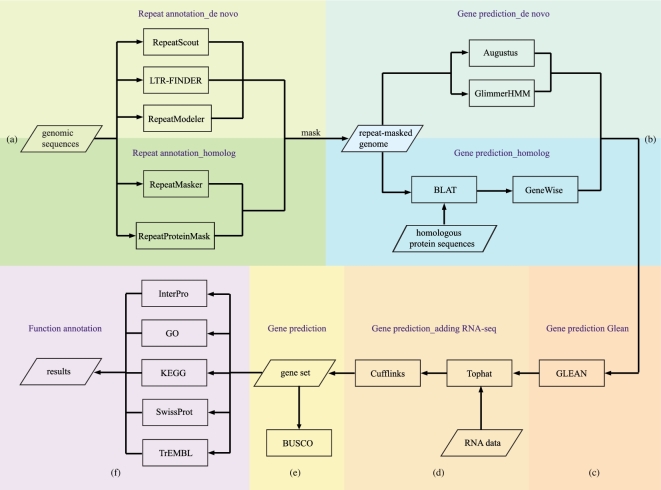
De novo and homolog-based methods were conducted in combination to identify the transposable elements (TEs) and predict the protein-coding genes inthe R. crenulata genome according to protocol 3 (Additional File 2), which is also illustrated in Fig. 2.
Figure 2:
An overview of the annotation workflow. The workflow begins with assembled genomic sequences, and it produces results of the repeat annotation, protein-coding gene prediction, and functional annotation. (a) Repeat annotation: repeats in the genome are detected in two different methods: de novo and homolog based. In the de novo method, RepeatScout, LTR-FINDER, and RepeatModeler are used to build de novo repeat libraries and further classified by RepeatMasker. In the homolog-based method, RepeatMasker and RepeatProteinMask are performed to search TEs by aligning sequences against existing libraries. (b) Gene prediction: before the gene prediction, TEs are totally masked. Augustus and GlimmerHMM are used to perform de novo prediction; BLAT and GeneWise are executed to predict gene models based on homologous protein sequences. (c) GLEAN is performed to obtain a consensus gene set. (d) In combination with the clean RNA sequenced reads, a more comprehensive gene set is integrated finally. (e) Estimation of the completeness of the gene set using BUSCO. (f) Functional annotation.
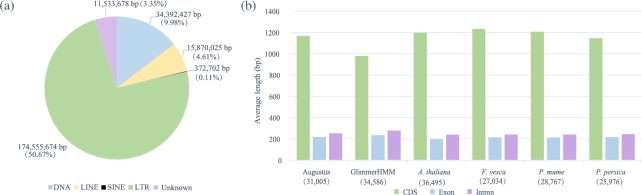
Briefly, in terms of the repeat detection, first, RepeatScout (v. 1.0.5; RepeatScout, RRID:SCR_014653) [14], LTR-FINDER (v. 1.0.5) [15], and RepeatModeler (v. 1.0.5) [16] were used to builda de novo library on the basis of our genome sequences, and then, by using the library as database, RepeatMasker (v. 3.3.0; RepeatMasker, RRID:SCR_012954) [16] was utilized to classify the types of repetitive sequences (Additional File 1: Table S6). On the other hand, TEs in DNA and protein levels were identified by aligning genome sequences against the Repbase TE library (v. 17.01) [17, 18] and TE protein database with RepeatMasker and RepeatProteinMask (v. 3.3.0) (Additional File 1: Table S7)[16]. Overall, 226.6 Mb of TEs (65.77% of the assembly) were detected, containing 174.6 Mb (50.67% of the assembly) of LTR (Fig. 3a; Additional File 1: Table S7).
Figure 3:
Summary statistics of the repeats and gene models. (a) The lengths of different types of TEs and proportions in the genome. LTR is the most predominant element. (b) The numbers of predicted genes and average lengths of CDS, exon, and intron predicted in different methods. The green, blue, and purple bars represent the CDS, exon, and intron, respectively. The gene numbers in each de novo or homolog-based method are listed in parentheses.
Before gene prediction, TEs observed above were masked to reduce the interference. Regarding the de novo gene prediction, Augustus (v. 2.5.5; Augustus: Gene Prediction, RRID:SCR_008417) [19, 20] and GlimmerHMM (v. 3.0.1; GlimmerHMM, RRID:SCR_002654) [21] were conducted with the Arabidopsis training set, and 31 005 and 34 586 protein-coding genes were predicted, respectively (Fig. 3b; Additional File 1: Table S8). With respect to the homolog-based methods, because of the lack of accessible genome sequences in the family Crassulaceae, we downloaded the protein sequences of model organism Arabidopsis thaliana (https://www.ncbi.nlm.nih.gov/genome/?term=Arabidopsis+thaliana) and the relatively closely related species Fragaria vesca (https://www.ncbi.nlm.nih.gov/genome/3314?genome_assembly_id=34435), Prunus mume (https://www.ncbi.nlm.nih.gov/genome/13911?genome_assembly_id=44389), and Prunus persica (https://www.ncbi.nlm.nih.gov/genome/388?gen-ome_assembly_id=28754) in rosids, and then aligned these against the repeat-masked genome using BLAT [22]. GeneWise (v. 2.2.0) [23], whose algorithm was derived from a principled combination of hidden Markov models, was subsequently used to merge these mapping results and predict gene structures, resulting in 36 495, 27 034, 28 767, and 25 976 protein-coding genes, respectively. In addition, all average lengths of CDS, exon, and intron predicted in different methods were similar (Fig. 3b; Additional File 1: Table S8). We then performed GLEAN [24] to integratethe genes predicted above and got a non-redundant gene set containing 28 981 protein-coding genes. Also, we discarded those genes with an overlapping ratio of less than 0.8 when comparing with homolog-based evidence. A total of 27 107 genes remained. Additionally, to further improve credibility, sequenced transcriptomes data from three R. crenulata tissues were mapped to the consensus gene set by TopHat (v. 2.1.0; TopHat, RRID:SCR_013035) [25], and then Cufflinks (v. 2.2.1; Cufflinks, RRID:SCR_014597) [26] were executed to assemble and merge transcripts based on the mapping results. Finally, a gene set with 31 517 protein-coding genes was generated, of which 79.73% of genes could be functionally annotated with SWISS-PROT [27], TrEMBL [27], and KEGG (KEGG, RRID:SCR_012773) [28, 29] databases, and using InterProScan (v. 4.7; InterProScan, RRID:SCR_005829) (Additional File 1: Table S9)[30, 31].
Completeness of the gene set and assembly
To evaluate the completeness of the gene set and assembly, BUSCO (BUSCO, RRID:SCR_015008) [32] was performed with “-OGS” and “-genome” modes, respectively. The results showed that 86.72% of reference genes were captured as complete single-copy BUSCOs when searching our gene set; meanwhile, regarding the assembly, 91.63% of the 956 expected plant genes were detected as complete (Table 2). Additionally, RNA sequence reads were mapped to our genome assembly by TopHat (v. 2.1.0) [25], and the average mapping ratio was almost 81.5% (Additional File 1: Table S10).
Table 2:
Statistics of the BUSCO assessment.
| Gene Set | Assembly | |||
|---|---|---|---|---|
| Types of BUSCOs | Number | Percentage | Number | Percentage |
| Complete single-copy BUSCOs | 829 | 86.72 | 876 | 91.63 |
| Fragmented BUSCOs | 37 | 3.87 | 35 | 3.66 |
| Missing BUSCOs | 90 | 9.41 | 45 | 4.71 |
| Total BUSCO groups searched | 956 | 100 | 956 | 100 |
In summary, the R. crenulata genome that we have sequenced, assembled, and annotated here was the first published genome in the genus Rhodiola and family Crassulaceae. The R. crenulata genome should serve as an important resource for comparative genomic studies, for further investigations of the adaptability of Rhodiola species in an extreme environment, and for the elucidation of the biosynthesis pathways of pharmacologically active metabolites in Rhodiola species.
Additional files
Additional File 1: Supplementary Tables and Figures.docx
Additional File 2: Protocols.io.xls
Abbreviations
bp: base pair; CDS: coding sequence; Gb: giga base; kb: kilo base; Mb: mega base; SRA: Sequence Read Archive; TE: transposable elements; WGS: whole genome shotgun sequencing.
Funding
This work was supported by the National High Technology Research and Development Program of China (NO.2014AA10A602-4) and Basic Research Program Support by the Shenzhen Municipal Government (No. JCYJ20150831201123287) and Key Research & Development Program of Jiangsu Province (BE2016002-3).
Availability of supporting data
The DNA sequencing data have been deposited into the NCBI Sequence Read Archive (SRA) under the ID SRA538315. The RNA sequencing data are under ID SRA539059. Supporting data are also available from the GigaScience database (GigaDB) [33]. DNA/RNA extraction and assembly and annotation protocols presented here are also archived in protocols.io [34].
Conflicts of interest
The authors declare that they have no competing interests.
Authors’ contributions
S.M.Y.L., X.L., X.S., and X.X. designed the project. Y.F., L.L., S.H., R.G., G.F., H.W., W.C., and H.Z. analyzed the data. Y.F., S.M.Y.L., X.L., G.F., and C.S. wrote the manuscript. G.L., J.W., L.M., J.Y., X.N., and Z.Y. prepared the samples and conducted the experiments.
Supplementary Material
Supplementary Tables and Figures.docx
Protocols.io.xls
References
- 1. Recio MC, Giner RM, Manez S. Immunmodulatory and antiproliferative properties of Rhodiola species. Planta Med 2016;82(11–2):952–60. [DOI] [PubMed] [Google Scholar]
- 2. Zhu C, Guan F, Wang C et al. The protective effects of Rhodiola crenulata extracts on Drosophila melanogaster gut immunity induced by bacteria and SDS toxicity. Phytother Res 2014;28(12):1861–6. [DOI] [PubMed] [Google Scholar]
- 3. Bassa LM, Jacobs C, Gregory K et al. Rhodiola crenulata induces an early estrogenic response and reduces proliferation and tumorsphere formation over time in MCF7 breast cancer cells. Phytomedicine 2016;23(1):87–94. [DOI] [PubMed] [Google Scholar]
- 4. Dudek MC, Wong KE, Bassa LM et al. Antineoplastic effects of Rhodiola crenulata treatment on B16-F10 melanoma. Tumour Biol 2015;36(12):9795–805. [DOI] [PubMed] [Google Scholar]
- 5. Cai Z, Li W, Wang H et al. Antitumor effects of a purified polysaccharide from Rhodiola rosea and its action mechanism. Carbohydr Polym 2012;90(1):296–300. [DOI] [PubMed] [Google Scholar]
- 6. Panossian A, Wikman G, Sarris J. Rosenroot (Rhodiola rosea): traditional use, chemical composition, pharmacology and clinical efficacy. Phytomedicine 2010;17(7):481–93. [DOI] [PubMed] [Google Scholar]
- 7. Luo R, Liu B, Xie Y et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 2012;1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016;17(6):333–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuzuki D. BGISEQ-500 debuts at the International Congress of Genomics 10. Next Generation Technologist, 24 October2015. [Google Scholar]
- 10. Li R, Fan W, Tian G et al. The sequence and de novo assembly of the giant panda genome. Nature 2010;463(7279):311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bennett M, Leitch I. Plant DNA C-values database (release 6.0, December 2012). http://data kew org/cvalues/ (14 October 2014, date last accessed). [Google Scholar]
- 12. Kajitani R, Toshimoto K, Noguchi H et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res 2014;24(8):1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simpson JT, Wong K, Jackman SD et al. ABySS: a parallel assembler for short read sequence data. Genome Res 2009;19(6):1117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics 2005;21(suppl 1):i351–8. [DOI] [PubMed] [Google Scholar]
- 15. Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 2007;35(web server issue):W265–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 2009; chapter 4: unit 4 10. doi:10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- 17. Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA 2015;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jurka J, Kapitonov VV, Pavlicek A et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 2005;110(1–4):462–7. [DOI] [PubMed] [Google Scholar]
- 19. Stanke M, Keller O, Gunduz I et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 2006;34(web server issue):W435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keller O, Kollmar M, Stanke M et al. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 2011;27(6):757–63. [DOI] [PubMed] [Google Scholar]
- 21. Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 2004;20(16):2878–9. [DOI] [PubMed] [Google Scholar]
- 22. Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res 2002;12(4):656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res 2004;14(5):988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Elsik CG, Mackey AJ, Reese JT et al. Creating a honey bee consensus gene set. Genome Biol 2007;8(1):R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25(9):1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trapnell C, Roberts A, Goff L et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7(3):562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res 2000;28(1):45–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M, Goto S, Sato Y et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014;42(database issue):D199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jones P, Binns D, Chang HY et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 2014;30(9):1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001;17(9):847–8. [DOI] [PubMed] [Google Scholar]
- 32. Simao FA, Waterhouse RM, Ioannidis P et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015;31(19):3210–2. [DOI] [PubMed] [Google Scholar]
- 33. Fu Y, Li L, Hao S et al. Supporting data for “Draft genome sequence of the Tibetan medicinal herb, Rhodiola crenulata.” Gigascience Database 2017; http://dx.doi.org/10.5524/100301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fu Y, Li L, Hao S et al. Protocols for “Draft genome of the Tibetan medicinal herb, Rhodiola crenulata”. 2017, protocols.io. http://dx.doi.org/10.17504/protocols.io.hrkb54w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables and Figures.docx
Protocols.io.xls