Abstract
Bisphenol-A is a ubiquitous environmental contaminant that is primarily metabolized by glucuronidation and associated with various human diseases including breast cancer. Here we identified UDP-glucuronosyltransferases (UGTs) and genetic polymorphisms responsible for interindividual variability in bisphenol-A glucuronidation in human liver and breast.
Hepatic UGTs showing the highest bisphenol-A glucuronidation activity included UGT2B15 and UGT1A9. Relative activity factor normalization indicated that UGT2B15 contributes >80% of activity at bisphenol-A concentrations under 5 µM, while UGT1A9 contributes up to 50% of activity at higher concentrations.
Bisphenol-A glucuronidation by liver microsomes (46 donors) ranged from 0.25 to 4.3 nmoles/min/mg protein. Two-fold higher glucuronidation (P = 0.018) was observed in UGT1A9 *22/*22 livers compared with *1/*1 and *1/*22 livers. However, no associations were observed for UGT2B15*2 or UGT1A1*28 genotypes.
Bisphenol-A glucuronidation by breast microsomes (15 donors) ranged from <0.2 to 56 fmoles/min/mg protein. Breast mRNA expression of UGTs capable of glucuronidating bisphenol-A was highest for UGT1A1, followed by UGT2B4, UGT1A9, UGT1A10, UGT2B7 and UGT2B15. Bisphenol-A glucuronidation was over 10-fold lower in breast tissues with the UGT1A1*28 allele compared with tissues without this allele (P = 0.006).
UGT2B15 and UGT1A9 contribute to glucuronidation variability in liver, while UGT1A1 is important in breast.
Keywords: Bisphenol-A, glucuronidation, liver, breast, breast cancer
INTRODUCTION
Bisphenol-A (BPA) is a by-product of producing certain plastics and epoxy resins that has been associated with various human diseases including breast cancer, prostate cancer, polycystic ovary disease, cardiovascular disease, diabetes, obesity, and thyroid dysfunction (Rezg et al. 2014; Rochester 2013; Schug et al. 2011). Differences in the susceptibility of people to the adverse effects of BPA may result from differences in the amount of BPA consumed and variability in the ability of individuals to effectively excrete BPA. After ingestion, BPA is taken up by the body and must be metabolized to enable efficient excretion. BPA is metabolized by two pathways, glucuronidation and sulfation (Kurebayashi et al. 2010). The main route of metabolism of BPA in humans is glucuronidation, which occurs primarily in the liver (Trdan Lusin et al. 2012) and is mediated by the UDP-glucuronosyltransferase (UGT) enzymes (Hanioka et al. 2008).
UGT2B15 has been proposed as the main catalyst of BPA glucuronidation in human liver since a recombinant enzyme screen showed highest activity for this isoform with a Km value (6 µM) that was similar to liver microsomes (9 µM) (Hanioka et al. 2008). However several other UGTs, including UGT1A1, UGT1A3, UGT1A9, UGT2B4, and UGT2B7, showed significant glucuronidation activities. On the other hand, UGT1A4, UGT1A6, and UGT2B17 showed no detectable BPA glucuronidation activity. A more recent study (Gramec Skledar et al. 2015) confirmed these results and also showed no detectable glucuronidation of BPA by UGT2A3 or UGT2B28. It is unknown whether the remaining UGTs known to be expressed in liver (UGT2B10 and UGT2B11) can glucuronidate BPA.
Significant glucuronidation of BPA could occur in extrahepatic tissues, such as intestines and kidney, which would limit systemic exposure after oral ingestion and enhance excretion (respectively). However, a recent report found only very low levels of BPA glucuronidation by microsomes from kidney (100 times lower) and intestines (400 times lower) compared with liver (Trdan Lusin et al. 2012) arguing against a significant role for human kidney and intestines in BPA glucuronidation. Regardless, several extrahepatic UGTs including UGT1A7, UGT1A8, UGT1A10, UGT2A1 and UGT2A2 have shown a capacity to glucuronidate BPA, although mostly with lower activity than UGT2B15 (Gramec Skledar et al. 2015). The exception was UGT2A1, which showed similar or higher activity than UGT2B15. However, the tissue expression of UGT2A1 is limited to nasal and airway epithelia, suggesting that this isoform may be important for inhaled BPA but not for ingested BPA (Gramec Skledar et al. 2015).
BPA glucuronidation could also occur in the breast and act to protect this hormone sensitive tissue from the effects of BPA circulating in the blood. Few studies have evaluated which UGTs are present in breast tissue. Expression of UGT2B10, UGT2B11, UGT2B15 and UGT2B17 (but not other UGTs) was detected in breast tissue RNA, but that study included tissue from only a single individual as part of a larger tissue panel (Court et al. 2012). Another study that included breast tissues from a large number of individuals showed expression of UGT1A10 and UGT2B7, but did not evaluate other UGTs (Starlard-Davenport et al. 2008). A third study showed UGT2B7, UGT2B10 and UGT2B11 expression by microarray in breast tissues from a range of individuals, but expression of other UGTs was not evaluated (Haakensen et al. 2010).
Polymorphisms of genes encoding UGTs that glucuronidate BPA may explain interindividual variability in risk for diseases associated with BPA exposure. An in vitro study showed that BPA glucuronidation was significantly lower for the expressed variant UGT2B15 85Y allozyme (UGT2B15*2) compared with the reference UGT2B15 85D allozyme (UGT2B15*1) (Hanioka et al. 2011). However, there are no reports of the association of the UGT2B15*2 polymorphism with human liver metabolism or with BPA pharmacokinetics in people. A common polymorphism in the promoter of the UGT1A1 gene (UGT1A1*28) that decreases UGT1A1 expression has been studied using genotyped human liver microsomes (Trdan Lusin et al. 2012). Although they found 80% lower BPA glucuronidation in liver microsomes with UGT1A1 *28/*28 genotype compared with *1/*1 and *1/*28 genotypes, the livers studied were from only one person with each genotype, and so the findings could have easily occurred by chance.
The purpose of this study was to use multiple approaches to identify UGTs that are responsible for BPA glucuronidation in the main organ metabolizing BPA (i.e. liver) and an important target organ for the endocrine effects of BPA (i.e. breast). In addition we evaluated effects of polymorphisms in the genes encoding these UGTs on BPA glucuronidation to provide perspective for prior and future epidemiologic studies of UGT polymorphism and BPA associated disease.
MATERIALS AND METHODS
Human tissues and recombinant enzymes
Liver samples from 46 European-American donors (35 men, 11 women) with no known liver diseases used for preparation of microsomes and extraction of DNA were obtained from the University of Chicago, the National Disease and Research Interchange, the International Institute for the Advancement of Medicine, and the University of Pittsburgh various sources with approval by the Investigation Review Board of Tufts University. Complete details regarding liver donor demographics are provided elsewhere (Court 2010). De-identified female breast tissue samples used to prepare microsomes, extraction of DNA and RNA were obtained through the Midwest Division of the Cooperative Human Tissue Network with approval by the Institutional Review Board of the University of Wisconsin-Madison as reported previously (Rhoads et al. 2011). Donors consisted of 4 Caucasian women, 3 African American women, and one Hispanic woman with a median age of 36 years and a range of 26–59 years. Race and age were not recorded for 7 donors.
Recombinant UGTs expressed in Sf9 insect cells were obtained from either BD Biosciences, Woburn, MA (UGTs 1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 2B4, 2B7, 2B10, 2B15, and 2B17), or were generated in Dr. Finel’s laboratory, Helsinki, Finland (UGTs 1A10, 2A1, 2A2, 2A3, 2B11) using techniques previously reported (Kurkela et al. 2003; Sneitz et al. 2009). Membranes prepared from HEK293 cells stably transfected with plasmids encoding the reference UGT2B15 p.85D and variant UGT2B15 p.85Y enzymes were kindly provided by Dr Chantal Guillemette, Laval University, Quebec as previously described (Court et al. 2004).
BPA glucuronidation assays
An assay was developed to measure BPA glucuronidation activities based on previously published methods (Court et al. 2002; Court et al. 2004). Unless otherwise indicated most incubation reagents were obtained from Sigma-Aldrich (St. Louis, MO). Briefly, reaction mixtures contained 1 to 1000 µM BPA, 5 mM UDP-glucuronic acid, 5 mM magnesium chloride, 5 µg alamethicin, and microsomes or recombinant UGTs (0.01 to 0.5 mg / mL), to a final volume of 100 µL with 50 mM phosphate buffer (pH 7.4). Incubations were conducted in a water bath at 37° C for 10 min. After adding 300 µL acetonitrile and 100 µL of 1 µM BPA sulfate in methanol as the internal standard the tubes were centrifuged at 15000g for 10 min, the supernatant was dried down in a centrifugal vacuum at 40° C. Once dry, 100 µL of potassium phosphate buffer (pH 7.4) was added and then transferred to vials for analysis by HPLC.
The HPLC apparatus (Agilent 1100) was run at a flow rate of 1 mL/min, an autoinjector, a 250 × 4.6 reverse phase C18 column (Phenomenex Synergi 4µM Hyro-RP 80A LC), and a UV absorbance detector set at 280 nm wavelength. The mobile phase was run at 1 mL/min and consisted of 20 mM potassium phosphate buffer (pH 4.5) mixed with varying proportions of acetonitrile (20 to 80% gradient over 14 minutes). Glucuronide peaks were quantified through use of standard curves of known quantities of BPA-glucuronide (Toronto Research Chemicals) and fixed amounts of internal standard (BPA-sulfate). Glucuronide formation rate was linear for up to 20 min incubation and 0.5 mg / mL protein concentration. Incubations lacking added BPA also showed no BPA-glucuronide formation, thereby indicating that there was no BPA contamination of the assay components. Initial studies also showed no significant effect of adding bovine serum albumin or saccharolactone (glucuronidase inhibitor) on glucuronidation activity and so this was not included in subsequent incubations. Data were expressed as nanomoles of glucuronide formed per minute per milligram microsomal protein.
BPA-glucuronide concentrations in breast microsomes incubations were much lower than for the liver incubations and so a more sensitive HPLC-mass spectrometry method was developed using an API 4000 mass spectrometer (Applied Biosystems, Framingham, MA). The mobile phase of 56% acetonitrile with 44% 20 mM ammonium formate (pH 4.75) was pumped at 0.35 mL per minute through a Synergi Hydro-RP 150 × 2-mm column (Phenomenex, Torrance, CA). Negative ion transitions monitored were m/z 403.3 → 227.1 for BPA-glucuronide, 307.2 → 227.1 for BPA-sulfate, and 227.1 → 133 for BPA.
Hepatic UGT probe activities for correlation and relative activity factor determination
Hepatic UGT selective activities including bilirubin, trifluoperazine, serotonin, propofol, zidovudine, and S-oxazepam glucuronidation were used for correlation analysis for UGT1A1, UGT1A4, UGT1A6, UGT1A9, UGT2B7, and UGT2B15 (respectively) as reported previously (Court 2005; Court 2010). Bilirubin, propofol, zidovudine, and S-oxazepam glucuronidation activities were also measured using recombinant UGT1A1, UGT1A9, UGT2B7, and UGT2B15 to calculate the relative contribution of these UGTs to hepatic BPA glucuronidation using a relative activity factor (RAF) approach as previously reported (Gibson et al. 2013). UGT1A4 and UGT1A6 RAF values were not determined since these isoforms did not glucuronidate BPA to any appreciable extent. RAF values (UGT probe activity divided by HLMs probe activity) were 0.93, 0.31, 1.79, and 0.19 for UGT1A1, UGT1A9, UGT2B7, and UGT2B15, respectively. RAF-adjusted recombinant enzyme BPA glucuronidation activities were then calculated by dividing by the unadjusted activity by the RAF value. The percent contribution of each UGT to total BPA glucuronidation was then calculated by dividing the RAF-adjusted UGT activity by the sum of all RAF-adjusted BPA glucuronidation activities and multiplying by 100.
UGT genotyping
DNA extracted from liver and breast tissues were genotyped for selected polymorphisms including UGT1A1*28 (rs34815109; n=40), UGT1A9*22 (rs45625337; n=40), and UGT2B15*2 (rs1902023; n=46). Methods included direct sequencing for UGT1A1*28 (Girard et al. 2005) and UGT1A9*22 (Girard et al. 2004) or Taqman allele discrimination assay for UGT2B15*2 (Assay C_27028164, Applied Biosystems, Foster City, CA).
mRNA concentrations
UGT1A, 2A and 2B subfamily and GAPDH mRNA concentrations in breast tissue total RNA were measured using a real-time polymerase chain reaction (PCR) assay on a Biorad CFX96 instrument as previously described in detail (Court et al. 2012). The lower limit of quantitation was defined as a threshold cycle number greater than 40 cycles. Data were normalized to GAPDH mRNA concentrations.
Enzyme kinetic and statistical analysis
Sigmaplot (version 12.0) was used to estimate Vmax and Km values by nonlinear regression curve fitting analysis. Enzyme kinetic models that were evaluated included simple Michaelis-Menten, sigmoidal, two enzymes, and substrate inhibition. The model that best fit the data was chosen based on evaluation of plots showing deviation of fitted from actual enzyme activities. Sigmaplot (version 12.0) was also used to evaluate differences in liver BPA glucuronidation activity related to genotype, gender, and alcohol use by one-way Kruskal-Wallis ANOVA on rank transformed data and Dunn’s multiple comparison method. The effect of donor gender and histories of alcohol use (>14 drinks per week) and smoking on each phenotype parameter was assessed using the Mann-Whitney rank sum test. A P value less than 0.05 was considered to be statistically significant.
RESULTS
Recombinant enzyme screen and hepatic relative activity factor normalization
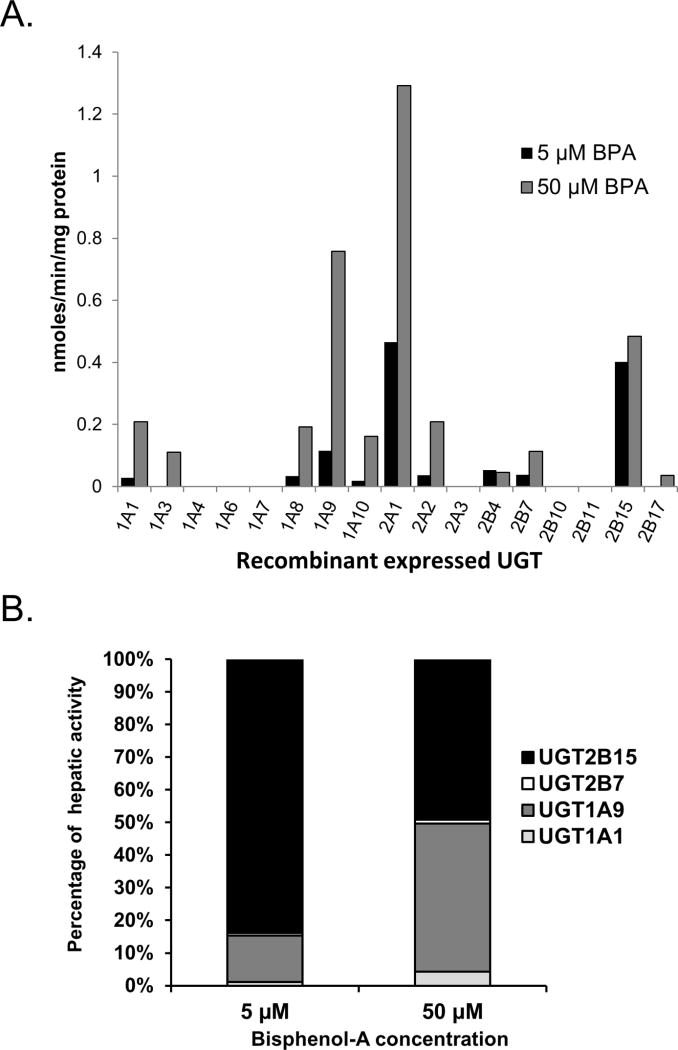
Recombinant UGTs were screened to identify those with the highest BPA glucuronidation activity at two BPA concentrations, 5 and 50 µM (Fig. 1A). These BPA concentrations were chosen to span the estimated Km value (~25 µM) for pooled HLMs determined in preliminary experiments (final estimates are given below). UGT2A1 showed highest activity out of all the UGTs screened at both 5 and 50 µM substrate concentrations. Other extrahepatic UGTs showing significant activity included UGT2A2, UGT1A8, and UGT1A10. Of those expressed in liver tissue, UGT2B15 and UGT1A9 showed the highest activity at both 5 and 50 µM BPA concentrations. Other hepatic UGTs including UGT1A1, UGT1A3, UGT2B4, UGT2B7 and UGT2B17 showed much lower activity, while UGTs 1A4, 1A6, 1A7, 2B10, and 2B11 showed no detectable BPA glucuronidation activity (LLOQ 20 pmoles/min/mg protein using HPLC with UV detection) at both of the substrate concentrations tested.
Figure 1.
(A) BPA glucuronidation activities of recombinant human UGTs. Each bar represents the average of duplicate determinations measured using either 5 µM (black bars) or 50 µM (gray bars) BPA concentration. (B) Predicted percent contribution of UGT2B15 (black bar), UGT2B7 (white bar), UGT1A9 (dark gray bar), and UGT1A1 (light gray bar) to BPA glucuronidation in liver at either 5 µM (left stacked bar) or 50 µM (right stacked bar) BPA concentration using a relative activity factor approach.
Since the enzyme activities of recombinant UGTs can under- or overestimate the activities of the native UGTs present measured in human tissues (when normalized to microsomal protein content), we used an RAF approach to adjust for these differences and thereby obtain an estimate of the relative contribution of different UGTs to hepatic BPA glucuronidation. We focused on the most active hepatic UGTs (UGT2B15 and UGT1A9), as well as UGTs with available isoform-selective probe activities that also showed significant capacity to glucuronidate BPA (UGT1A1 and UGT2B7). As shown in Fig. 1B, at 5 µM BPA concentration, the majority (84%) of BPA glucuronidation could be attributed to UGT2B15, with a smaller although substantial contribution (14%) from UGT1A9, but essentially no role of UGT1A1 or UGT2B7 (less than 2%). However at 50 µM BPA concentration, UGT2B15 (49%) and UGT1A9 (45%) contributed about equally to BPA glucuronidation, while again both UGT1A1 (4%) and UGT2B7 (1%) played only a minor role.
Enzyme kinetics of BPA glucuronidation by UGTs and HLMs
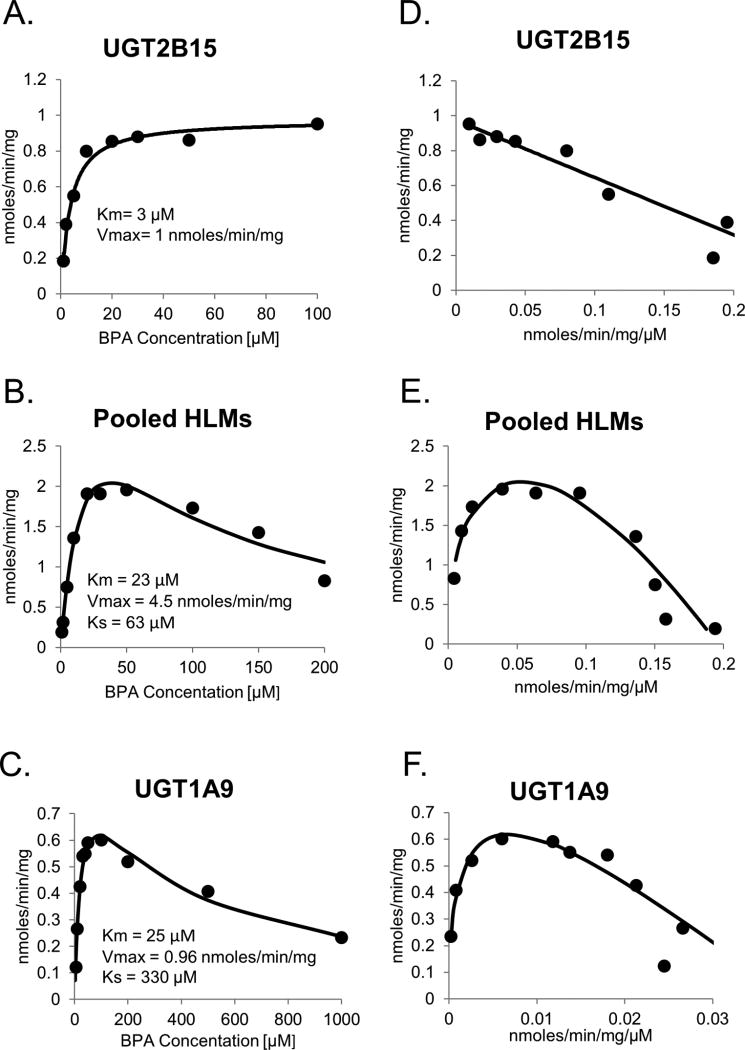
UGT2B15 and UGT1A9 were further studied to evaluate in more detail the effects of differing BPA substrate concentrations on activity in comparison with pooled HLMs. As shown in Figs. 2A and 2D, BPA glucuronidation activities for UGT2B15 was well described by a single enzyme Michaelis-Menten kinetic model over the substrate concentrations evaluated. On the other hand, both pooled HLMs (Figs. 2B and 2E) and recombinant UGT1A9 (Figs. 2C and 2F) showed significant substrate inhibition at BPA concentrations over 50 µM, and these data were better described using the uncompetitive substrate inhibition model. The Km values (mean +/− standard error of estimate) for pooled HLMs (23 +/− 8 µM) and UGT1A9 (25 +/− 5 µM) were quite similar and nearly 10 times higher than the Km for UGT2B15 (3.3 +/− 0.45 µM). The Vmax of UGT1A9 (1.0 +/− 0.1 nmoles / min / mg protein) was similar to UGT2B15 (0.97 +/− 0.03 nmoles / min / mg protein) but nearly 4 times lower than for pooled HLMs (4.5 +/− 1.0 nmoles / min / mg protein). The substrate inhibition constant (Ks) value for pooled HLMs (63 +/− 23 µM) was about 2 times higher than for UGT1A9 (30 +/− 65 µM).
Figure 2.
Enzyme kinetic plots of BPA glucuronidation by UGT2B15 (A), pooled HLMs (B), and UGT1A9 (C). Estimated enzyme kinetic parameters including Km and Vmax as well as the fitted curves using a one enzyme Michaelis-Menten model are shown on each plot. Eadie-Hofstee plots of the same data are shown in panels D, E, and F respectively.
BPA glucuronidation variability and correlation with UGT probe activities in HLMs
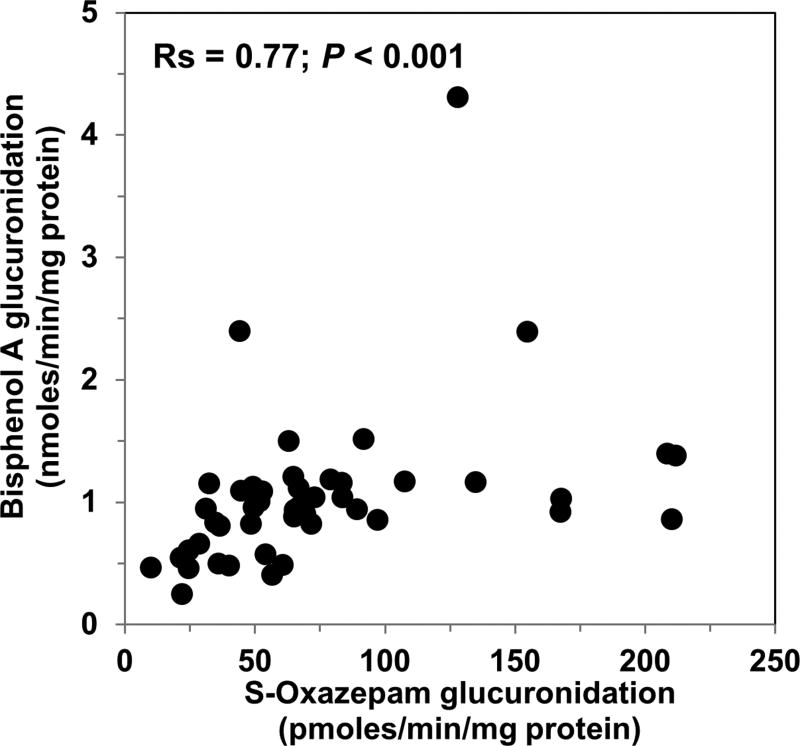
We then evaluated interindividual differences in hepatic BPA glucuronidation activities measured in a bank of HLMs (n=46) at a substrate concentration of 5 µM. Activities ranged from 0.25 nmoles/min/mg protein to 4.3 nmoles/min/mg (over 17-fold) with a mean (standard deviation, SD) activity of 1.1 (0.76) nmoles/min/mg, a median activity of 0.96 nmoles/min/mg protein, and a 90% confidence interval of 0.44 to 2.4 nmoles/min/mg protein. Correlation analysis was then performed to identify hepatic UGT isoforms that might account for this variability. Table 1 gives Spearman correlation coefficients comparing BPA glucuronidation with the 6 hepatic UGT probe activities evaluated. As shown in Fig. 3, the best correlation (Spearman correlation coefficient, Rs = 0.77; p<0.001) was seen with S-oxazepam glucuronidation activities, which is a probe for UGT2B15. Weaker correlations (Rs = 0.38 to 0.43; p<0.05) were observed with bilirubin (UGT1A1), propofol (UGT1A9), and zidovudine glucuronidation (UGT2B7), while there were no significant correlations with trifluoperazine (UGT1A4) or serotonin (UGT1A6) glucuronidation activities.
Table 1.
Correlation of BPA glucuronidation activities measured at 5 µM BPA concentration with UGT isoform selective glucuronidation activities measured in a bank of HLMs prepared from 46 different liver donors.
| Selective probe substrate |
UGT | Spearman correlation coefficient (Rs) |
P value |
|---|---|---|---|
| Bilirubin | 1A1 | 0.43 | 0.0034 |
| Trifluoperazine | 1A4 | 0.25 | 0.094 |
| Serotonin | 1A6 | 0.25 | 0.10 |
| Propofol | 1A9 | 0.40 | 0.012 |
| Zidovudine | 2B7 | 0.38 | 0.010 |
| S-Oxazepam | 2B15 | 0.77 | 0.0000002 |
Figure 3.
Correlation of BPA glucuronidation (measured at 5 µM BPA concentration) with S-oxazepam glucuronidation (a selective marker of UGT2B15 activity) measured in the same sample of 46 HLMs. Spearman correlation analysis indicated a moderately strong correlation of 0.77 (p<0.001).
Glucuronidation of BPA by UGT2B15-p.85D and UGT2B15-p.85Y variants
BPA glucuronidation activities were measured in HEK 293 cells stably transfected with plasmids encoding UGT2B15-p.85D and UGT2B15-p.85Y which correspond to the UGT2B15*1 and UGT2B15*2 alleles, respectively. At 5 µM substrate concentration, BPA glucuronide formation for UGT2B15-p.85D was 149 pmoles/min/mg protein, while at 50 µM substrate concentration, BPA glucuronide formation was 166 pmoles/min/mg protein. Under the same conditions, there was no detectable formation of BPA glucuronide by UGT2B15-p.85Y (LLOQ 20 pmoles/min/mg protein using HPLC with UV detection).
Correlation of BPA glucuronidation with liver donor demographics and UGT genotypes
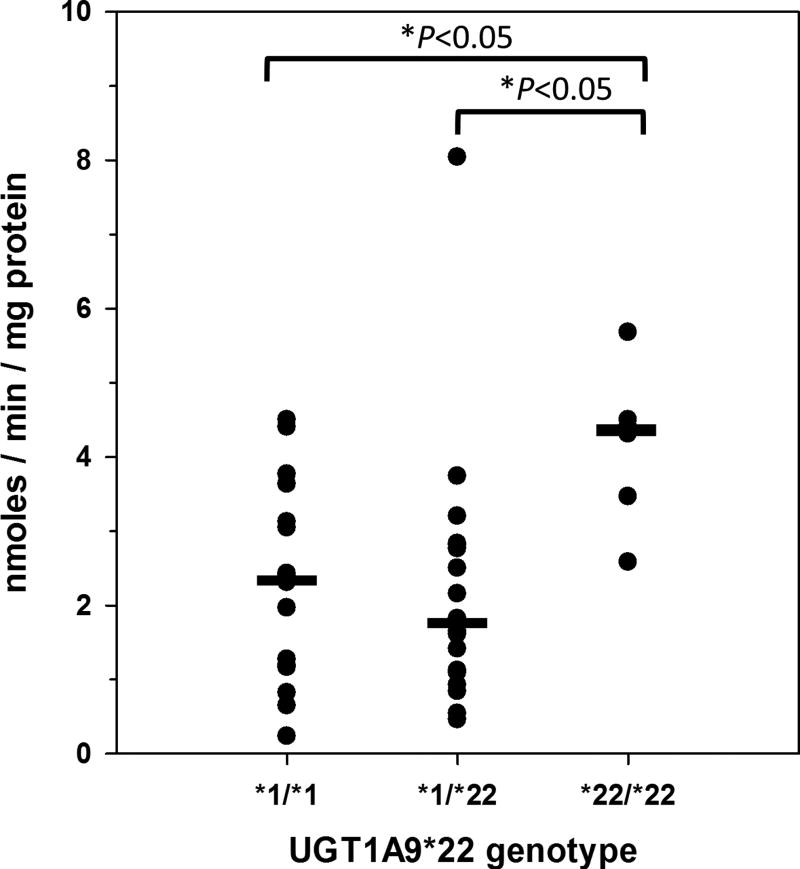
Table 2 shows mean (and SD) BPA glucuronidation activities measured using 5 µM BPA concentration in the human liver bank samples grouped by gender, alcohol consumption, smoking, and selected UGT genotypes, including UGT1A1*28, UGT1A9*22, and UGT2B15*2. Only UGT1A9*22 showed a significant association with BPA glucuronidation activity (P = 0.012; Kruskal-Wallis one way analysis of variance on ranks). As shown in Fig. 4, livers with the UGT1A9 *22/*22 genotype had significantly higher median (interquartile range; 25 – 75%) glucuronidation activity of 4.4 (3.2 – 4.8) nmoles/min/mg protein compared with 2.4 (1.2 – 3.5) nmoles/min/mg protein for livers with the *1/*1 genotype (P<0.05; Dunn’s test) and 1.7 (1.0 – 2.8) nmoles/min/mg protein for livers with the *1/*22 genotype (P<0.05; Dunn’s test). None of the other factors evaluated were associated with BPA glucuronidation. Multivariate analysis by multiple linear regression also failed to identify any factors (other than UGT1A9*22 genotype) that contributed to BPA glucuronidation variability (data not shown).
Table 2.
Analysis of differences in hepatic BPA glucuronidation activities associated with liver donor demographics and UGT genotypes.
| BPA glucuronidation rate (nmoles/min/mg protein) |
|||||
|---|---|---|---|---|---|
| Variable | Group | n | Median | Interquartile range | P value |
| Gender | Male | 35 | 2.4 | 1.2 – 3.8 | 0.11a |
| Female | 11 | 1.8 | 0.6 – 3.1 | ||
| Drinks (per week) | <14 | 27 | 1.8 | 0.8 – 3.6 | 0.46a |
| 14 or more | 12 | 2.3 | 1.7 – 2.8 | ||
| Smoking | Yes | 15 | 2.0 | 1.1 – 3.1 | 0.70a |
| No | 25 | 2.3 | 1.0 – 3.7 | ||
| UGT1A1*28 | *1/*1 | 21 | 2.6 | 1.5 – 4.0 | 0.51b |
| *1/*28 | 13 | 1.6 | 0.9 – 3.5 | ||
| *28/*28 | 6 | 2.4 | 1.2 – 3.5 | ||
| UGT1A9*22 | *1/*1 | 16 | 2.4 | 1.2 – 3.5 | 0.012b |
| *1/*22 | 18 | 1.7 | 1.0 – 2.8 | ||
| *22/*22 | 6 | 4.4 | 3.2 – 4.8 | ||
| UGT2B15*2 | *1/*1 | 8 | 3.1 | 2.4 – 5.2 | 0.23b |
| *1/*2 | 25 | 1.8 | 0.9 – 3.4 | ||
| *2/*2 | 13 | 2.0 | 1.2 – 3.0 | ||
Rank sum test
Kruskal-Wallis one way analysis of variance on ranks.
Figure 4.
Association of UGT1A9*22 genotype with BPA glucuronidation measured at 5 µM BPA concentration in 40 human liver microsomes samples. Shown are the glucuronidation activities determined for individual livers with UGT1A9 genotype *1/*1 (n=16), *1/*22 (n=18), and *2/*2 (n=6). Also shown are the median values (a horizontal bar) for each genotype group. Analysis of variance on ranks indicated a significant difference among groups (P = 0.012), and Dunn’s multiple comparison test showed that activities for livers with the UGT1A9 *2/*2 genotype were significantly higher than livers with either the *1/*1 or *1/*22 genotypes (*P < 0.05 for both comparisons).
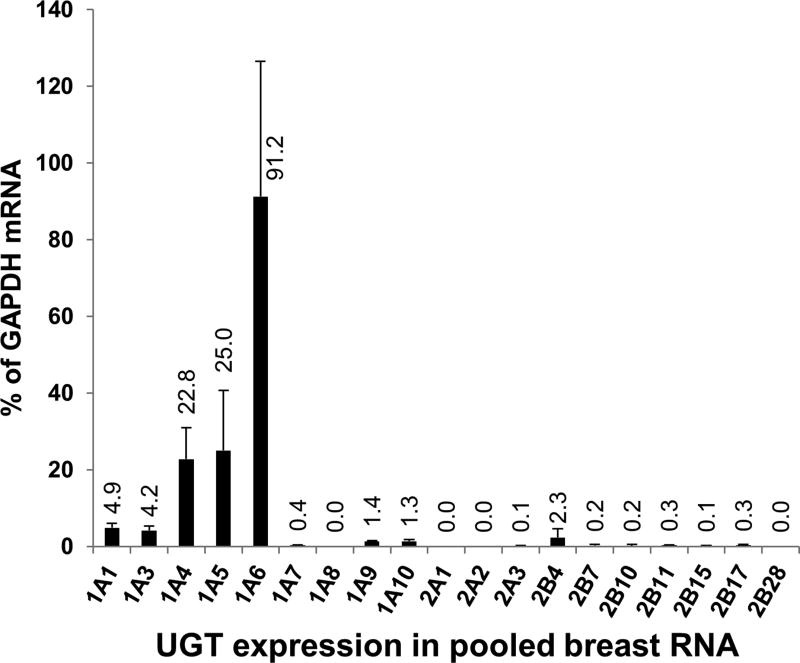
UGT mRNA expression in pooled breast RNA
Quantitative PCR of pooled (n = 15) breast RNA samples was used to determine whether breast tissue express mRNA encoding UGT enzymes capable of glucuronidating BPA. As shown in Fig. 5 relatively high expression was observed for UGT1A4, UGT1A5, and UGT1A6. Intermediate levels of expression were observed for UGT1A3, UGT1A1, UGT2B4, UGT1A10, and UGT1A9, while relatively low levels of expression were observed for most other UGT isoforms including UGT2B15. UGT1A8, UGT2A1, UGT2A2, and UGT2B28 were not detected. Of those UGTs that were found capable of glucuronidating BPA at 5 µM substrate concentration, the rank order of mRNA expression was UGT1A1 > UGT2B4 > UGT1A9 = UGT1A10 > UGT2B7 = UGT2B15.
Figure 5.
Expression of UGT mRNA in pooled human breast RNA samples measured by real-time quantitative PCR normalized to GAPDH mRNA content (mean and standard deviation of quadruplicate measurements).
BPA glucuronidation by breast microsomes and correlation with UGT genotype
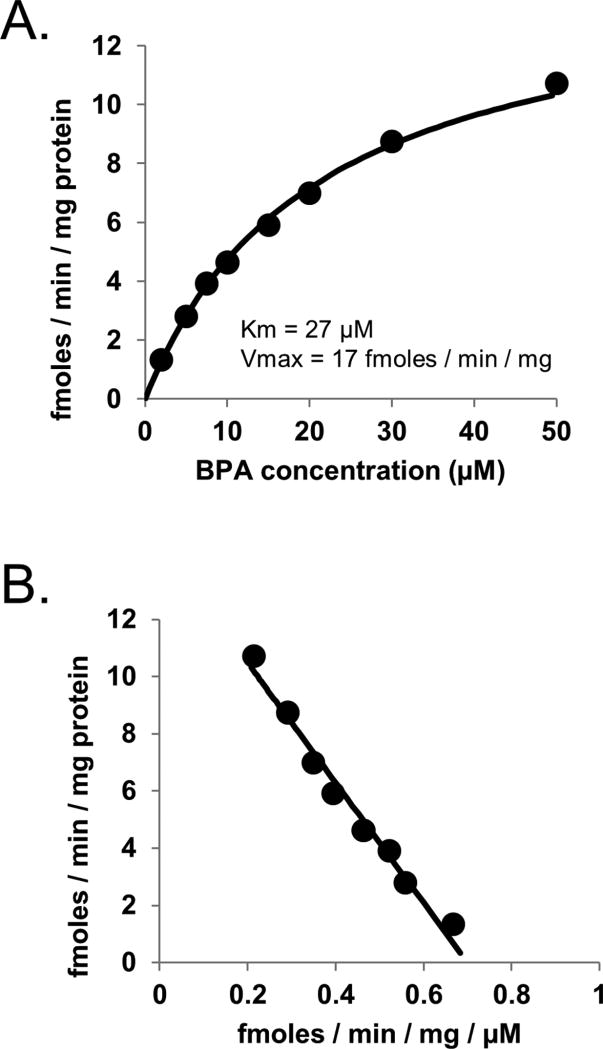
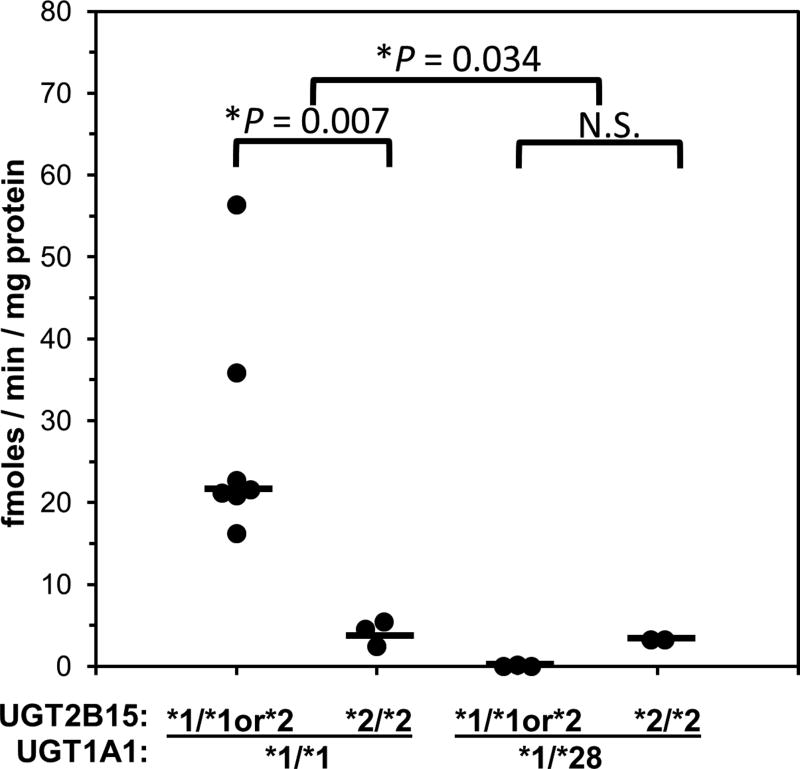
Enzyme kinetic analysis of pooled breast microsomes (Fig. 6) showed a hyperbolic profile with a Km value (27 µM) that was similar to pooled HLMs and a Vmax value (17 fmoles/min/mg protein) that was much lower than pooled HLMs. BPA glucuronidation activities (using 50 µM BPA concentration to enhance sensitivity) and UGT1A1*28, UGT1A9*22 and UGT2B15*2 polymorphism genotypes for individual breast samples are given in Table 3. Glucuronidation activities ranged from 56 fmoles/min/mg protein to 0.2 fmoles/min/mg protein for 13 of the 15 samples evaluated. Two of the samples (donors 17 and 29) showed no detectable BPA glucuronidation activity (LLOQ 0.2 fmoles/min/mg protein using HPLC with mass spectrometry detection). Correlation of these activities with UGT genotypes (data given in Table 4) showed significantly lower (by over 10 times) activities in breast samples with the UGT1A1 *1/*28 genotype compared with samples with the *1/*1 genotype (P = 0.006; rank sum test). No breast samples had the UGT1A1 *28/*28 genotype. None of the other genotypes evaluated were independently associated with BPA glucuronidation activity (P > 0.05; Kruskal-Wallis one way analysis of variance on ranks). However, as shown in Fig. 7, two-way analysis of variance (P = 0.034 for UGT1A1*28; P = 0.11 for UGT2B15*2; P = 0.044 for interaction) indicated that the UGT2B15 *2/*2 genotype identified a subgroup of breast samples (donors 159, 169 and 172) within samples with the UGT1A1 *1/*1 genotype that had much lower median (interquartile range) BPA glucuronidation activities of 4.5 (2.5 – 5.4) fmoles/min/mg protein compared with breast samples with UGT1A1 *1/*1 genotype and either UGT2B15 *1/*1 or *1/*2 genotypes with 28 (4.8 – 51) fmoles/min/mg protein (P = 0.007; Dunn’s test).
Figure 6.
Enzyme kinetic plots (Michael-Menten plot, panel A; and Eadie-Hofstee plot of the same data, panel B) of BPA glucuronidation by pooled breast microsomes (n=15). Also shown are the estimated enzyme kinetic parameters (Km and Vmax) as well as the fitted curves using a one enzyme Michaelis-Menten model.
Table 3.
BPA glucuronidation activities and UGT genotypes and determined for 15 breast tissue samples. Samples are ordered in the table from highest to lowest BPA glucuronidation activity.
| BPA glucuronidation rate | Polymorphism genotype
|
|||
|---|---|---|---|---|
| Sample ID | (fmoles/min/mg protein) | UGT1A1*28 | UGT1A9*22 | UGT2B15*2 |
| 165 | 56 | *1/*1 | *1/*22 | *1/*2 |
| 47 | 36 | *1/*1 | *1/*22 | *1/*2 |
| 160 | 23 | *1/*1 | *22/*22 | *1/*1 |
| 171 | 22 | *1/*1 | *22/*22 | *1/*1 |
| 156 | 21 | *1/*1 | *1/*22 | *1/*2 |
| 168 | 21 | *1/*1 | *1/*22 | *1/*1 |
| 164 | 16 | *1/*1 | ? | *1/*1 |
| 172 | 5.4 | *1/*1 | *1/*22 | *2/*2 |
| 159 | 4.5 | *1/*1 | *1/*22 | *2/*2 |
| 167 | 3.3 | *1/*28 | *1/*1 | *2/*2 |
| 20 | 3.3 | *1/*28 | *1/*22 | *2/*2 |
| 169 | 2.5 | *1/*1 | *1/*1 | *2/*2 |
| 158 | 0.2 | *1/*28 | *22/*22 | *1/*2 |
| 29 | 0a | *1/*28 | *1/*1 | *1/*2 |
| 17 | 0a | *1/*28 | *1/*22 | *1/*1 |
Less than the lower limit of quantitation (LLOQ, 0.2 fmoles/min/mg protein)
unable to determine genotype
Table 4.
Analysis of differences in breast tissue BPA glucuronidation activities associated with donor UGT genotypes.
| BPA glucuronidation rate (fmoles/min/mg protein) |
|||||
|---|---|---|---|---|---|
| UGT | Genotype | n | Median | Interquartile range | P value |
| UGT1A1*28 | *1/*1 | 10 | 21 | 5.2 – 26 | 0.006a |
| *1/*28 | 5 | 0.2 | 0.1 – 3.3 | ||
| UGT1A9*22 | *1/*1 | 3 | 2.5 | 0.1 – 3.3 | 0.25b |
| *1/*22 | 8 | 13 | 3.6 – 32 | ||
| *22/*22 | 3 | 22 | 0.2 – 23 | ||
| UGT2B15*2 | *1/*1 | 5 | 21 | 8.2 – 22 | 0.47b |
| *1/*2 | 5 | 21 | 0.15 – 46 | ||
| *2/*2 | 5 | 3.3 | 2.9 – 5.0 | ||
Rank sum test
Kruskal-Wallis one way analysis of variance on ranks
Figure 7.
Association of UGT1A1*28 and UGT2B15*2 genotype with BPA glucuronidation measured at 50 µM BPA concentration in 15 human breast microsomes samples. Shown are the glucuronidation activities determined for individual breast samples grouped by UGT1A1*28 genotype (*1/*1 versus *1/*28) and by UGT2B15*2 genotype (*1/*1 or *1/*2 versus *2/*2). Also shown are the median values (a horizontal bar) for each genotype group. Two-way ANOVA showed significantly lower activities in UGT1A1 *1/*28 versus *1/*1 breast samples (*P = 0.034), as well as an interaction (P = 0.04) between genotypes such that lower activities were observed for UGT2B15 *2/*2 samples compared with *1/*1 or *1/*2 samples (*P = 0.007, Dunn’s test) only in those samples that also had the UGT1A1*1/*1 genotype.
DISCUSSION
The results of our study confirm the importance of UGT2B15 in the glucuronidation of BPA in human liver, but also indicate a role for UGT1A9 particularly at higher BPA concentrations. Out of eleven hepatic UGTs screened, recombinant UGT2B15 and UGT1A9 showed the highest and second highest BPA glucuronidation activities, respectively. Adjustment for hepatic UGT abundance using a relative activity factor approach indicated that >80% of activity results from UGT2B15 at low substrate concentrations but nearly 50% of activity may result from UGT1A9 at higher concentrations. Comparative enzyme kinetic analysis showed similar Km values for HLMs and recombinant UGT1A9, although higher than for UGT2B15. Furthermore, both HLMs and UGT1A9 demonstrated significant inhibition of activity at BPA concentrations over 50 µM, while this was not observed for UGT2B15. Although we found the highest correlation between S-oxazepam glucuronidation (UGT2B15 marker activity) and BPA glucuronidation in HLMs, the strength of the correlation was only moderate (Rs = 0.77) and we did observe statistically significant although weaker (Rs = 0.4) correlation with propofol glucuronidation (UGT1A9 marker). On the other hand, we did not find lower BPA glucuronidation activity in HLMs with the UGT2B15 *2/*2 genotype as we have reported for several other UGT2B15 substrates including oxazepam and lorazepam (Court 2005; Court et al. 2004). This was despite showing a marked reduction in BPA glucuronidation by the recombinant UGT2B15*2 variant allozyme compared with the UGT2B15*1 reference allozyme. However, we did observe significantly higher BPA-glucuronidation in HLMs with the UGT1A9 *22/*22 genotype, which is consistent with previous reports describing the effect of this polymorphism on UGT1A9 gene expression (Yamanaka et al. 2004). Taken together, these findings indicate that UGT1A9 as well as UGT2B15 are responsible for BPA glucuronidation in human liver.
In support of our work, the results of a recent study (Fay et al. 2015) using a novel mouse gene knockout model suggest that the Ugt1a subfamily enzymes may also be important for BPA metabolism and clearance in other (non-human) species. Specifically it was shown that complete deletion of the murine Ugt2 gene locus (including all genes encoding the Ugt2a and Ugt2b subfamily enzymes) had no effect on BPA glucuronidation activities in liver microsomes or on the rate of excretion of BPA glucuronide into bile when compared with wild-type mice. However, as yet it is unknown which murine Ugt1a enzyme is capable of glucuronidating BPA.
Another major novel finding of our study is that human breast tissue is clearly capable of glucuronidating BPA, although with glucuronidation activities that are much lower (by more than 100,000-fold) compared with liver, the main metabolic organ. The low activity is probably a reflection of the heterogenous but focused distribution of the UGTs within breast tissue. In support of this it has been shown by immunohistochemistry that several UGTs, including UGT2B7 (Gestl et al. 2002) and UGT1A9 (Thibaudeau et al. 2006) are highly expressed in the ductal cells but not in other cell types in breast tissue. It was speculated that this focused distribution has physiological (and pathophysiological) relevance since the enzymes are concentrated within the hormone-sensitive cell types (Thibaudeau et al. 2006). Although the cellular distribution of other UGTs has not been reported (to our knowledge), our quantitative mRNA data indicate that both UGT1A1 and UGT2B15 are expressed in breast tissue (in addition to UGT1A9), and that UGT1A1 is the most highly expressed out of all of the isoforms capable of glucuronidating BPA we have identified. Consequently we proceeded to evaluate the association of breast microsome BPA glucuronidation activities with UGT1A1*28, UGT1A9*22, and UGT2B15*2 genotypes.
Our results suggest that lower BPA glucuronidation activity in breast tissue is associated with UGT1A1*28 genotype and possibly UGT2B15*2 genotype (at least after controlling for UGT1A1*28 effects). However, it should be pointed out that the number of samples we studied was relatively small (n=15 donors) and so this result will need to be confirmed through study of a larger sample size. Regardless, our data suggest that UGT1A1 and UGT2B15 could contribute to BPA metabolism in breast, and that individuals with low activity alleles for these enzymes could have low BPA glucuronidation activity, resulting in increased exposure of breast tissue to BPA, and an increased risk of developing breast cancer.
Nine different studies have evaluated the association of UGT1A1*28 with breast cancer. Four studies have shown an elevated risk for breast cancer with the UGT1A1*28 allele in African American (Guillemette et al. 2000), Chinese (Adegoke et al. 2004), Russian (Shatalova et al. 2006) and German (Marie-Genica 2010) women. However, four other studies have shown no association of UGT1A1*28 with breast cancer risk in Taiwanese (Cheng et al. 2005), Greek (Tsezou et al. 2007), or European American (Guillemette et al. 2001) women, or in a mixed population of American and Swedish women (Clendenen et al. 2013). Furthermore, one study showed a decreased risk of UGT1A1*28 for breast cancer in Nigerian women (Huo et al. 2008). A meta-analysis study published in 2010 (Yao et al. 2010), which aggregated 5,746 cases and 8,365 controls from most of these studies, concluded that there was evidence for enhanced risk for breast cancer in Caucasian women with the UGT1A1*28 allele. Consequently our finding of lower BPA glucuronidation in breast tissue with the UGT1A1*28 may explain in part these observations of enhanced breast cancer risk. However, it should also be noted that UGT1A1 also glucuronidates estrogens, carcinogenic catechol estrogens, and other carcinogens that may also contribute to breast cancer risk (Cheng et al. 1998; Girard et al. 2005).
Only one study could be identified that has reported on the association of UGT2B15*2 with breast cancer risk (Sun et al. 2012). They did not find any association with the UGT2B15*2 allele in women of African ancestry from the United States, Barbados, and Nigeria. However the results of our study suggest that the influence of the UGT2B15*2 allele may only be evident in individuals lacking the UGT1A1*28 allele.
There are some limitations to this study that need to be addressed. Serum concentrations of unconjugated BPA reported in people with exposure by environmental contamination ranges from 0.001 µM to 0.10 µM, which is somewhat lower than the BPA substrate concentration range we evaluated in this study (Vandenberg et al. 2007). However, the enzyme kinetic values we have determined for BPA glucuronidation should allow for extrapolation of activities to lower substrate concentrations. Although we did not measure the free (unbound) substrate concentration in this study, published work indicates that the unbound fraction of BPA for liver microsomes is quite high (0.94; (Kuester and Sipes 2007)). Consequently, significant binding of substrate to microsomes is unlikely to have affected the accuracy of our Km estimates. Finally, while glucuronidation is the primary metabolic pathway of BPA in humans, 10–20% of BPA metabolism can also occur via sulfation and so this pathway should also be studied in future work to assess variation in BPA metabolism and effects (Kurebayashi et al. 2010).
CONCLUSIONS
Our results indicate that both UGT2B15 and UGT1A9 are important determinants of variable BPA glucuronidation in human liver. However, UGT1A1 (and possibly UGT2B15) may determine interindividual variability in BPA glucuronidation in breast tissue.
Acknowledgments
This work was supported by the US National Institutes of Health grant GM102130 (M.H.C.), the William R. Jones endowment to Washington State University College of Veterinary Medicine (M.H.C.), the Auvil Research Fellowship (C.M.S.), and the Sigrid Juselius Foundation (M.F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding organizations. The authors thank Dr Chantal Guillemette (Laval University, Quebec) who provided the UGT2B15 allozymes and Dr Lauren Trepanier (University of Wisconsin, Madison, Wisconsin) who provided the breast tissue microsomes, DNA and RNA samples.
Footnotes
DECLARATION OF INTEREST
The authors report no declarations of interest.
References
- Adegoke OJ, Shu XO, Gao YT, Cai Q, Breyer J, Smith J, Zheng W. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 (UGT1A1) and risk of breast cancer. Breast Cancer Res Treat. 2004;85:239–45. doi: 10.1023/B:BREA.0000025419.26423.b8. [DOI] [PubMed] [Google Scholar]
- Cheng TC, Chen ST, Huang CS, Fu YP, Yu JC, Cheng CW, Wu PE, Shen CY. Breast cancer risk associated with genotype polymorphism of the catechol estrogen-metabolizing genes: a multigenic study on cancer susceptibility. Int J Cancer. 2005;113:345–53. doi: 10.1002/ijc.20630. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Rios GR, King CD, Coffman BL, Green MD, Mojarrabi B, Mackenzie PI, Tephly TR. Glucuronidation of catechol estrogens by expressed human UDP-glucuronosyltransferases (UGTs) 1A1, 1A3, and 2B7. Toxicol Sci. 1998;45:52–7. doi: 10.1006/toxs.1998.2494. [DOI] [PubMed] [Google Scholar]
- Clendenen T, Zeleniuch-Jacquotte A, Wirgin I, Koenig KL, Afanasyeva Y, Lundin E, Arslan AA, Axelsson T, Forsti A, Hallmans G, et al. Genetic variants in hormone-related genes and risk of breast cancer. PLoS One. 2013;8:e69367. doi: 10.1371/journal.pone.0069367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH. Isoform-selective probe substrates for in vitro studies of human UDP-glucuronosyltransferases. Methods Enzymol. 2005;400:104–16. doi: 10.1016/S0076-6879(05)00007-8. [DOI] [PubMed] [Google Scholar]
- Court MH. Interindividual variability in hepatic drug glucuronidation: studies into the role of age, sex, enzyme inducers, and genetic polymorphism using the human liver bank as a model system. Drug Metab Rev. 2010;42:209–24. doi: 10.3109/03602530903209288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court MH, Duan SX, Guillemette C, Journault K, Krishnaswamy S, Von Moltke LL, Greenblatt DJ. Stereoselective conjugation of oxazepam by human UDP-glucuronosyltransferases (UGTs): S-oxazepam is glucuronidated by UGT2B15, while R-oxazepam is glucuronidated by UGT2B7 and UGT1A9. Drug Metab Dispos. 2002;30:1257–65. doi: 10.1124/dmd.30.11.1257. [DOI] [PubMed] [Google Scholar]
- Court MH, Hao Q, Krishnaswamy S, Bekaii-Saab T, Al-Rohaimi A, von Moltke LL, Greenblatt DJ. UDP-glucuronosyltransferase (UGT) 2B15 pharmacogenetics: UGT2B15 D85Y genotype and gender are major determinants of oxazepam glucuronidation by human liver. J Pharmacol Exp Ther. 2004;310:656–65. doi: 10.1124/jpet.104.067660. [DOI] [PubMed] [Google Scholar]
- Court MH, Zhang X, Ding X, Yee KK, Hesse LM, Finel M. Quantitative distribution of mRNAs encoding the 19 human UDP-glucuronosyltransferase enzymes in 26 adult and 3 fetal tissues. Xenobiotica. 2012;42:266–77. doi: 10.3109/00498254.2011.618954. [DOI] [PubMed] [Google Scholar]
- Fay MJ, Nguyen MT, Snouwaert JN, Dye R, Grant DJ, Bodnar WM, Koller BH. Xenobiotic Metabolism in Mice Lacking the UDP-Glucuronosyltransferase 2 Family. Drug Metab Dispos. 2015 doi: 10.1124/dmd.115.065482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestl SA, Green MD, Shearer DA, Frauenhoffer E, Tephly TR, Weisz J. Expression of UGT2B7, a UDP-glucuronosyltransferase implicated in the metabolism of 4-hydroxyestrone and all-trans retinoic acid, in normal human breast parenchyma and in invasive and in situ breast cancers. Am J Pathol. 2002;160:1467–79. doi: 10.1016/S0002-9440(10)62572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CR, Lu P, Maciolek C, Wudarski C, Barter Z, Rowland-Yeo K, Stroh M, Lai E, Nicoll-Griffith DA. Using human recombinant UDP-glucuronosyltransferase isoforms and a relative activity factor approach to model total body clearance of laropiprant (MK-0524) in humans. Xenobiotica. 2013;43:1027–36. doi: 10.3109/00498254.2013.791761. [DOI] [PubMed] [Google Scholar]
- Girard H, Court MH, Bernard O, Fortier LC, Villeneuve L, Hao Q, Greenblatt DJ, von Moltke LL, Perussed L, Guillemette C. Identification of common polymorphisms in the promoter of the UGT1A9 gene: evidence that UGT1A9 protein and activity levels are strongly genetically controlled in the liver. Pharmacogenetics. 2004;14:501–15. doi: 10.1097/01.fpc.0000114754.08559.27. [DOI] [PubMed] [Google Scholar]
- Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, Hao Q, von Moltke LL, Greenblatt DJ, Guillemette C. UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology. 2005;42:448–57. doi: 10.1002/hep.20770. [DOI] [PubMed] [Google Scholar]
- Gramec Skledar D, Troberg J, Lavdas J, Peterlin Masic L, Finel M. Differences in the glucuronidation of bisphenols F and S between two homologous human UGT enzymes, 1A9 and 1A10. Xenobiotica. 2015;45:511–9. doi: 10.3109/00498254.2014.999140. [DOI] [PubMed] [Google Scholar]
- Guillemette C, De Vivo I, Hankinson SE, Haiman CA, Spiegelman D, Housman DE, Hunter DJ. Association of genetic polymorphisms in UGT1A1 with breast cancer and plasma hormone levels. Cancer Epidemiol Biomarkers Prev. 2001;10:711–4. [PubMed] [Google Scholar]
- Guillemette C, Millikan RC, Newman B, Housman DE. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and association with breast cancer among African Americans. Cancer Res. 2000;60:950–6. [PubMed] [Google Scholar]
- Haakensen VD, Biong M, Lingjaerde OC, Holmen MM, Frantzen JO, Chen Y, Navjord D, Romundstad L, Luders T, Bukholm IK, et al. Expression levels of uridine 5’-diphospho-glucuronosyltransferase genes in breast tissue from healthy women are associated with mammographic density. Breast Cancer Res. 2010;12:R65. doi: 10.1186/bcr2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanioka N, Naito T, Narimatsu S. Human UDP-glucuronosyltransferase isoforms involved in bisphenol A glucuronidation. Chemosphere. 2008;74:33–6. doi: 10.1016/j.chemosphere.2008.09.053. [DOI] [PubMed] [Google Scholar]
- Hanioka N, Oka H, Nagaoka K, Ikushiro S, Narimatsu S. Effect of UDP-glucuronosyltransferase 2B15 polymorphism on bisphenol A glucuronidation. Arch Toxicol. 2011;85:1373–81. doi: 10.1007/s00204-011-0690-5. [DOI] [PubMed] [Google Scholar]
- Huo D, Kim HJ, Adebamowo CA, Ogundiran TO, Akang EE, Campbell O, Adenipekun A, Niu Q, Sveen L, Fackenthal JD, et al. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and breast cancer risk in Africans. Breast Cancer Res Treat. 2008;110:367–76. doi: 10.1007/s10549-007-9720-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuester RK, Sipes IG. Prediction of metabolic clearance of bisphenol A (4,4 ‘-dihydroxy-2,2-diphenylpropane) using cryopreserved human hepatocytes. Drug Metab Dispos. 2007;35:1910–5. doi: 10.1124/dmd.107.014787. [DOI] [PubMed] [Google Scholar]
- Kurebayashi H, Okudaira K, Ohno Y. Species difference of metabolic clearance of bisphenol A using cryopreserved hepatocytes from rats, monkeys and humans. Toxicol Lett. 2010;198:210–5. doi: 10.1016/j.toxlet.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Kurkela M, Garcia-Horsman JA, Luukkanen L, Morsky S, Taskinen J, Baumann M, Kostiainen R, Hirvonen J, Finel M. Expression and characterization of recombinant human UDP-glucuronosyltransferases (UGTs). UGT1A9 is more resistant to detergent inhibition than other UGTs and was purified as an active dimeric enzyme. J Biol Chem. 2003;278:3536–44. doi: 10.1074/jbc.M206136200. [DOI] [PubMed] [Google Scholar]
- Marie-Genica Genetic polymorphisms in phase I and phase II enzymes and breast cancer risk associated with menopausal hormone therapy in postmenopausal women. Breast Cancer Res Treat. 2010;119:463–74. doi: 10.1007/s10549-009-0407-0. [DOI] [PubMed] [Google Scholar]
- Rezg R, El-Fazaa S, Gharbi N, Mornagui B. Bisphenol A and human chronic diseases: current evidences, possible mechanisms, and future perspectives. Environ Int. 2014;64:83–90. doi: 10.1016/j.envint.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Rhoads K, Sacco JC, Drescher N, Wong A, Trepanier LA. Individual variability in the detoxification of carcinogenic arylhydroxylamines in human breast. Toxicol Sci. 2011;121:245–56. doi: 10.1093/toxsci/kfr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester JR. Bisphenol A and human health: a review of the literature. Reprod Toxicol. 2013;42:132–55. doi: 10.1016/j.reprotox.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Schug TT, Janesick A, Blumberg B, Heindel JJ. Endocrine disrupting chemicals and disease susceptibility. J Steroid Biochem Mol Biol. 2011;127:204–15. doi: 10.1016/j.jsbmb.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatalova EG, Loginov VI, Braga EA, Kazubskaia TP, Sudomoina MA, Blanchard RL, Favorova OO. [Association of polymorphisms in SULT1A1 and UGT1A1 Genes with breast cancer risk and phenotypes in Russian women] Mol Biol (Mosk) 2006;40:263–70. [PubMed] [Google Scholar]
- Sneitz N, Court MH, Zhang X, Laajanen K, Yee KK, Dalton P, Ding X, Finel M. Human UDP-glucuronosyltransferase UGT2A2: cDNA construction, expression, and functional characterization in comparison with UGT2A1 and UGT2A3. Pharmacogenet Genomics. 2009;19:923–34. doi: 10.1097/FPC.0b013e3283330767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starlard-Davenport A, Lyn-Cook B, Radominska-Pandya A. Identification of UDP-glucuronosyltransferase 1A10 in non-malignant and malignant human breast tissues. Steroids. 2008;73:611–20. doi: 10.1016/j.steroids.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Huo D, Nemesure B, Hennis A, Leske MC, Wu SY, Niu Q, Olopade OI, Di Rienzo A. Lack of association between common UGT2B nonsynonymous single-nucleotide polymorphisms and breast cancer in populations of African ancestry. Int J Cancer. 2012;130:2740–2. doi: 10.1002/ijc.26300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaudeau J, Lepine J, Tojcic J, Duguay Y, Pelletier G, Plante M, Brisson J, Tetu B, Jacob S, Perusse L, et al. Characterization of common UGT1A8, UGT1A9, and UGT2B7 variants with different capacities to inactivate mutagenic 4-hydroxylated metabolites of estradiol and estrone. Cancer Res. 2006;66:125–33. doi: 10.1158/0008-5472.CAN-05-2857. [DOI] [PubMed] [Google Scholar]
- Trdan Lusin T, Roskar R, Mrhar A. Evaluation of bisphenol A glucuronidation according to UGT1A1*28 polymorphism by a new LC-MS/MS assay. Toxicology. 2012;292:33–41. doi: 10.1016/j.tox.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Tsezou A, Tzetis M, Giannatou E, Gennatas C, Pampanos A, Kanavakis E, Kitsiou-Tzeli S. Genetic polymorphisms in the UGT1A1 gene and breast cancer risk in Greek women. Genet Test. 2007;11:303–6. doi: 10.1089/gte.2007.0020. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Hauser R, Marcus M, Olea N, Welshons WV. Human exposure to bisphenol A (BPA) Reprod Toxicol. 2007;24:139–77. doi: 10.1016/j.reprotox.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Yamanaka H, Nakajima M, Katoh M, Hara Y, Tachibana O, Yamashita J, McLeod HL, Yokoi T. A novel polymorphism in the promoter region of human UGT1A9 gene (UGT1A9*22) and its effects on the transcriptional activity. Pharmacogenetics. 2004;14:329–32. doi: 10.1097/00008571-200405000-00008. [DOI] [PubMed] [Google Scholar]
- Yao L, Qiu LX, Yu L, Yang Z, Yu XJ, Zhong Y, Yu L. The association between TA-repeat polymorphism in the promoter region of UGT1A1 and breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;122:879–82. doi: 10.1007/s10549-010-0742-1. [DOI] [PubMed] [Google Scholar]