Abstract
Background
Nano- and microscale copper oxide particles (CuO NP, CuO MP) are applied for manifold purposes, enhancing exposure and thus the potential risk of adverse health effects. Based on the pronounced in vitro cytotoxicity of CuO NP, systematic investigations on the mode of action are required. Therefore, the impact of CuO NP, CuO MP and CuCl2 on the DNA damage response on transcriptional level was investigated by quantitative gene expression profiling via high-throughput RT-qPCR. Cytotoxicity, copper uptake and the impact on the oxidative stress response, cell cycle regulation and apoptosis were further analysed on the functional level.
Results
Cytotoxicity of CuO NP was more pronounced when compared to CuO MP and CuCl2 in human bronchial epithelial BEAS-2B cells. Uptake studies revealed an intracellular copper overload in the soluble fractions of both cytoplasm and nucleus, reaching up to millimolar concentrations in case of CuO NP and considerably lower levels in case of CuO MP and CuCl2. Moreover, CuCl2 caused copper accumulation in the nucleus only at cytotoxic concentrations. Gene expression analysis in BEAS-2B and A549 cells revealed a strong induction of uptake-related metallothionein genes, oxidative stress-sensitive and pro-inflammatory genes, anti-oxidative defense-associated genes as well as those coding for the cell cycle inhibitor p21 and the pro-apoptotic Noxa and DR5. While DNA damage inducible genes were activated, genes coding for distinct DNA repair factors were down-regulated. Modulation of gene expression was most pronounced in case of CuO NP as compared to CuO MP and CuCl2 and more distinct in BEAS-2B cells. GSH depletion and activation of Nrf2 in HeLa S3 cells confirmed oxidative stress induction, mainly restricted to CuO NP. Also, cell cycle arrest and apoptosis induction were most distinct for CuO NP.
Conclusions
The high cytotoxicity and marked impact on gene expression by CuO NP can be ascribed to the strong intracellular copper ion release, with subsequent copper accumulation in the cytoplasm and the nucleus. Modulation of gene expression by CuO NP appeared to be primarily oxidative stress-related and was more pronounced in redox-sensitive BEAS-2B cells. Regarding CuCl2, relevant modulations of gene expression were restricted to cytotoxic concentrations provoking impaired copper homoeostasis.
Electronic supplementary material
The online version of this article (doi:10.1186/s12989-017-0209-1) contains supplementary material, which is available to authorized users.
Keywords: Copper oxide nanoparticles, Oxidative stress, Gene expression profiling, High-throughput RT-qPCR, DNA damage response, Copper uptake and intracellular distribution
Background
Due to desired material properties such as high temperature superconductivity, artificially synthesized CuO nanoparticles (NP) are increasingly applied in industry, for example as catalysts, additives in plastics and metal coatings, but also as antimicrobial agents (summarized in [1]). CuO microparticles (MP) are precursors during pigment production [2]. Furthermore, CuO fumes including ultrafine particles are generated in metal refining and processing [3–5].
In biological systems, copper represents the third common trace element next to zinc and iron. It serves as catalytic cofactor in more than 30 metalloenzymes like superoxide dismutase (SOD), cytochrome C oxidase (COX), lysyl oxidase, dopamine-β-hydroxylase and ceruloplasmin, thus playing critical roles within manifold biochemical pathways [6]. While the redox-activity is required for essential functions, copper ions may also be toxic due to the catalysis of Fenton-type reactions, which may lead to the formation of highly reactive hydroxyl radicals, provoking oxidative damage to lipids, proteins and DNA [7]. To ensure essential functions and to largely prevent toxic reactions, both systemic as well as cellular copper homoeostasis are tightly regulated by copper binding to proteins and/or to small chelating molecules, preventing the existence of “free” copper ions. On the systemic level, copper homeostasis is regulated via absorption in the small intestine and via storage, distribution and biliary excretion by the liver, avoiding overload or deficiency. Similarly, within cells, copper levels and protein binding are strictly controlled via uptake, storage, transfer and export proteins. Genetically determined disturbances in copper homeostasis lead to severe disease syndromes like Wilson’s or Menke’s disease [8]. Nevertheless, copper overload can also occur due to excessive uptake, for example via dietary supplements, or due to non-physiological exposure routes. While copper uptake occurs usually via food and drinking water by the oral route, which is tightly regulated, the situation may be different in case of inhalation of copper fumes or aerosols in exposed workers, leading potentially to cellular and/or systemic copper overload.
Different working groups reported an enhanced cytotoxic and genotoxic potential as well as a strong pro-inflammatory response of CuO NP in various, mainly lung-associated cell culture models when compared to other metal-based nanoparticles [9–11] as well as to bigger particles or water soluble copper compounds [12–15]. Thus, besides the elemental composition different physico-chemical characteristics appear to contribute to the pronounced toxicity. Important features may include size, morphology, solubility, surface area as well as the ability of the particles to associate with biomolecules from the environment to form a protein corona [16], with potential impact on interactions of the particles with cellular membranes, uptake and intracellular bioavailability, finally leading to elevated levels of ROS.
One explanation originally proposed by Limbach and co-authors in 2007 in case of Co3O4 nanoparticles and since then strengthened in several studies e.g. by Karlsson and co-workers for CuO NP has been termed the “Trojan horse type mechanism”. It proposes the endocytotic uptake of the particles and the processing of the resulting vesicles into acidic lysosomes, thus affecting the degree of metal release from the particles and provoking potential intracellular overload conditions [12–15, 17–19]. Supporting this theory, many NP have been reported to use endocytotic mechanisms as major import pathway into the cell [20–23], including nanosized CuO in A549 cells [19].
However, many aspects still need to be elucidated. Thus, while the high cytotoxicity of CuO NP has been repeatedly demonstrated, less attention has been given to the impact on genomic stability. Furthermore, also the degree and intracellular distribution of deliberated copper ions is still not known. With respect to the latter, copper uptake has frequently been determined in whole cells, baring the risk of irrelevant results due to remaining and barely removable particles at the outer membrane.
Within the present study, we used adenocarcinoma A549 and BEAS-2B lung cells since the lung is the major target organ of CuO NP toxicity upon inhalative exposure to assess the impact on the DNA damage response system and to link it to intracellular copper levels upon copper particle or water soluble copper exposure. In a previous study of our working group, a detailed investigation on the cyto- and genotoxicity as well as copper uptake of CuO NP as compared to CuO MP and CuCl2 was performed in A549 cells, demonstrating pronounced cytotoxicity in case of CuO NP and CuCl2, but no effect in case of CuO MP. Furthermore, apoptosis induction as well as the induction of DNA strand breaks analysed in the model cell line HeLa S3 were restricted to CuO NP. Also, strongest impairment of the DNA damage signalling reaction poly(ADP-ribosylation) mediated predominatly by poly(ADP-ribose) polymerase 1 was observed in case of CuO NP. Finally, highest intracellular copper levels, both in the cytoplasm and the nucleus, were found in case of CuO NP [14]. Following up on this investigation in A549 cells, the central aspect of the present study was to elucidate the impact of all three copper compounds on gene expression profiles by using a recently established high-throughput RT-qPCR system via the BioMark™ device. This procedure enables the parallel assessment of the impact of 96 samples on 95 genes crucial for maintaining genomic stability, consisting of selected genes coding for stress response, DNA damage response, specific DNA repair factors, cell cycle control, apoptosis and mitotic signalling [24]. The effects of CuO NP were compared with those evoked by CuO MP and water soluble CuCl2 in identical mass doses to differentiate between particle, nano-particle and copper-mediated effects. In addition to A549 cells we included BEAS-2B cells as a second lung cell model. Both cell lines display advantages and disadvantages; A549 cells are p53 proficient but as a tumor cell line they have lost some characteristics of the original epithelial type 2 cells and they display some persistent basal signalling deregulation. In contrast, BEAS-2B cells are non-tumorigenic, but functionally p53 deficient due to the immortalization [25]. Thus, the comparison of the two cell lines with different transformation status and the knowledge about their respective limitations may provide valuable hints for evaluating and interpreting the effects on gene expression profiles. Finally, different outcomes on the transcriptional response were followed up on the functional level, such as the impact on the oxidative stress response, cell cycle regulation and apoptosis.
Results
Particle characterization
The particulate copper compounds CuO NP and CuO MP were characterized in detail with respect to size, surface area and charge, chemical composition, crystallinity and morphology via dynamic light scattering (DLS), scanning electron microscopy (SEM), Brunauer-Emmet-Teller (BET) analysis, zeta potential (ZP), inductively coupled plasma mass spectrometry (ICP-MS), energy-dispersive X-ray spectroscopy (EDX) as well as X-ray diffraction (XRD) as previously described. Briefly, CuO NP as well as CuO MP were of high purity and of the same composition, free of endotoxins and did not alter the pH of cell culture media. CuO NP were approximately spherical, displayed a narrow size range (20–200 nm), showed a surface area of 17.23 m2/g with a calculated average diameter of 55 nm and were agglomerated in fluids. DLS measurements revealed a size distribution around 146 nm and a ZP of −13.1 mV in DMEM/FCS. CuO MP had a size distribution of 500 nm −10 μm, a surface area of 0.74 m2/g and a calculated average of 1289 nm. DLS analysis was not possible due to fast sedimentation of the particles [14].
Cytotoxicity
The impact on the cell number of BEAS-2B cells after 24 h incubation with CuO NP, CuO MP and CuCl2 was determined as a measure of cytotoxicity and to establish appropriate incubation conditions for subsequent analyses (Fig. 1). This approach was chosen since these cells do not form colonies and an interference by copper ions and copper-based particles with dye-based toxicity assays has been reported previously [26]. CuO NP provoked a strong concentration-dependent decrease of cell number. Thus, in case of 5 μg/mL a reduction to 79% compared to control was observed, reaching 30% and 10% in case of 20 μg/mL or 50 μg/mL, respectively. In contrast, incubation with CuO MP resulted in cell numbers above 80% up to 20 μg/mL and only in case of 50 μg/mL a pronounced reduction to 64% was observed. CuCl2 was only weakly cytotoxic with a maximum decrease of cell number to 81% throughout the analysed concentration range. In A549 cells the cytotoxicity of the same compounds has been investigated by colony forming ability and published previously; here, most pronounced cytotoxicity was also observed in case of CuO NP followed by CuCl2, whereas CuO MP were not cytotoxic throughout the analysed concentration range. As opposed to BEAS-2B cells, CuCl2 exhibited starting cytotoxicity at the lowest concentration tested, i.e. 63 μM corresponding to 5 μg/mL [14]. On the basis of these cytotoxicity results, subsequent investigations were performed mainly up to 20 μg/mL CuO and 252 μM CuCl2, respectively.
Fig. 1.

Cytotoxicity of CuO NP, CuO MP and CuCl2 . Cytotoxicity was determined via cell number after 24 h treatment with the different copper compounds in BEAS-2B cells. Shown are mean values of six determinations derived from three independent experiments ± SD. Statistically significant different from control: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (ANOVA-Dunnett’s T test)
Cellular uptake and intracellular distribution of copper
Cellular uptake and intracellular distribution of copper was analysed after 24 h incubation with different concentrations of CuO NP, CuO MP and CuCl2 via graphite furnace atomic absorption spectrometry (GF-AAS). When working with particles, one problem consists in the fact that particles attach and stick to the outer membrane, which may provide artefacts when determining the intracellular copper content. Thus, a special procedure was applied to quantify the copper content in the soluble fractions of the whole cell, the cytoplasm and the nucleus [14, 27].
As shown in Fig. 2a, the basal copper concentration in BEAS-2B cells was found to be 20 μM. Treatment with CuO NP resulted in a pronounced dose-dependent accumulation of intracellular copper. As low as 1 μg/mL provoked an 18-fold accumulation to 330 μM copper. After treatment with 5 μg/mL and higher intracellular concentrations up to 2 mM were observed. CuO MP also provoked a concentration-dependent but less pronounced increase in intracellular copper, resulting in a maximum of 330 μM. Incubation with CuCl2 was found to result in almost constant intracellular levels of 250–400 μM copper within the applied concentration range up to 252 μM.
Fig. 2.
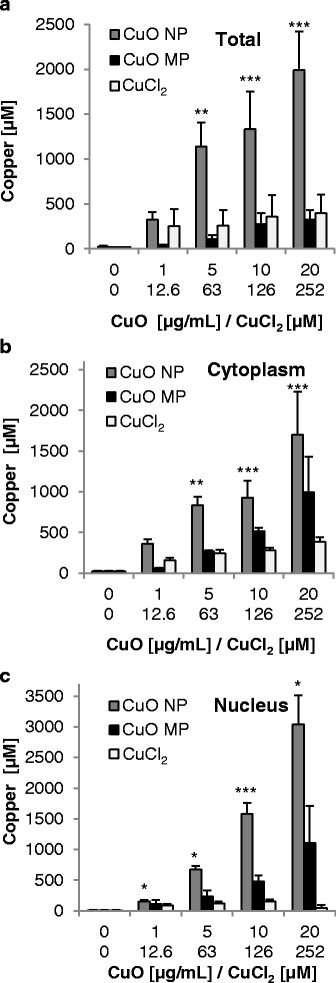
Cellular copper uptake and intracellular copper distribution after treatment with CuO NP, CuO MP and CuCl2. Copper content was determined in the soluble fraction of the total cell (a), the cytoplasm (b) and the nucleus (c) in BEAS-2B cells after 24 h treatment with the different copper compounds via GF-AAS. Shown are mean values of three independent experiments + SD ((a) and CuO NP (b,c)), mean values of six independent experiments + SD (10 μg/mL CuO NP (b,c)) or mean values of two independent experiments + R/2 (CuO MP and CuCl2 (b, c)). Statistically significant different from control: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 performed in case of (a) and CuO NP (b, c) (ANOVA-Dunnett’s T test). 20 μg/mL CuO are equal to 252 μM Cu2+. n.d.: not determined
The analysis of the cytoplasmic copper levels revealed a similar picture. Treatment with CuO NP resulted in copper levels of up to 1.7 mM in case of 20 μg/mL. Treatment with CuO MP displayed less pronounced dose-dependent increases in cytoplasmic copper as compared to CuO NP; at the highest dose, however, copper levels were higher than in the whole cells. This may be due to higher variations due to inhomogeneous particle uptake; thus, small differences in endocytosed particle numbers would be expected to result in pronounced differences in intracellular copper levels. Treatment with CuCl2 again revealed comparatively small increases up to 400 μM (Figure 2b). The basal copper concentration within the nuclear fraction of BEAS-2B cells was about 11 μM. Most strikingly, almost no increase in copper concentrations was observed in case of CuCl2, while CuO NP led to a markedly increased accumulation of nuclear copper up to 3 mM; this was even higher when compared to the cytoplasm. Intermediate levels were found in case of CuO MP. The cytoplasmic and nuclear copper levels add very well to the copper accumulation of the total cell. Since the volume of the nucleus accounts for only 10% of the cell volume, the cytoplasmic concentration should be close to the total cell volume. For example, in case of 20 μg/mL CuO NP respective values were 2000 μM (total), 1700 μM (cytoplasm), 3000 μM (nucleus); multiplied by the relative volumes results in an average concentration of 1830 μM, which fits quite reasonable to the copper content of the total cell. Similar agreements were found for the other concentrations applied.
Additionally, we quantified the copper content in whole cells including the outer membrane to get an impression of particles sticking to it, applying doses of 10 μg/mL CuO NP and CuO MP (Additional file 1). When not excluding the membrane, copper concentration was elevated more than 3.5-fold from 1350 μM up to 4800 μM in case of CuO NP and even more pronounced to more than 70-fold from 300 μM up to 22,000 μM in case of CuO MP. These findings demonstrate the importance of removing the membranes since otherwise the copper uptake would be considerably overestimated.
In A549 cells the copper uptake into cytoplasm and nucleus has been investigated previously; here, CuO NP and CuCl2 led to a concentration-dependent copper increase in the cytoplasm, even though less accumulation was seen when compared to BEAS-2B cells. In contrast, CuO MP led to higher, albeit highly variable cytoplasmic concentrations in A549 cells. With respect to the copper concentrations in the nucleus, effects of CuO NP and CuO MP were similar in A549 and BEAS-2B cells; in case of CuCl2, elevated copper levels were detected only in the former cell line, coincident with higher cytotoxicity [14].
Gene expression analyses
The impact of CuO NP, CuO MP and CuCl2 on gene expression profiles related to genomic stability was investigated in BEAS-2B and A549 cells after 24 h incubation via a high-throughput RT-qPCR technique [24]. The main observations obtained in both cell lines sorted in distinct signalling pathways and cellular processes are described in the following sections.
Impact on genes related to copper uptake and (oxidative) stress response
The three copper compounds induced a pronounced dose-dependent up-regulation of the metallothionein genes MT1X and MT2A (Fig. 3). Strongest induction was evident by CuO NP in BEAS-2B cells (Fig. 3a). MT1X was transcriptionally enhanced to 9-fold at 1 μg/mL and increasing to 29-fold at 20 μg/mL, whereas mRNA levels of MT2A were elevated up to 8-fold. The expression of these genes was also induced by CuO MP and CuCl2, albeit to a lesser extent, reaching 12-fold (MT1X) and 3-fold (MT2A) increases in case of CuO MP and CuCl2, respectively. In A549 cells (Fig. 3b) the increase of transcript levels was less pronounced as compared to BEAS-2B cells; both genes were induced dose-dependently in case of CuO NP up to 11- and 4-fold, respectively, while CuO MP showed no effect. CuCl2 enhanced transcription to a similar extent as CuO NP; however, induction was restricted to higher concentrations exerting considerable cytotoxicity in this cell line [14].
Fig. 3.
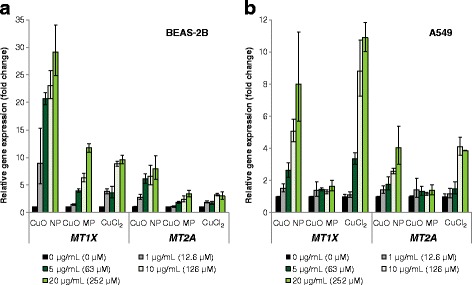
Impact of CuO NP, CuO MP and CuCl2 on gene expression of MT1X and MT2A related to copper uptake. BEAS-2B cells (a) or A549 cells (b) were treated with the different copper compounds for 24 h. Shown are linear fold changes of the relative gene expression from mean values of four determinations derived from two independent experiments ± SD. 20 μg/mL CuO are equal to 252 μM Cu2+. Please note the different scaling of the y-axis
An induction of the oxidative stress response system at the transcriptional level was again most pronounced after treatment with CuO NP, with strongest effects in BEAS-2B cells (Fig. 4). Thus, in BEAS-2B cells expression of the ROS-inducible gene HMOX1 was up-regulated concentration-dependently up to 53-fold by CuO NP and up to 10-fold by the other copper compounds. Transcription of the heat-shock sensitive HSPA1A was increased up to 28-fold or up to 8-fold by CuO NP or CuO MP, respectively, whereas no effects were observed after treatment with CuCl2 (Fig. 4a). In A549 cells HMOX1 and HSPA1A were almost equally enhanced by CuO NP on the transcriptional level to a maximum of 8-fold or 7-fold, respectively. CuCl2 and CuO MP induced no changes in gene expression, except for HMOX1 in case of CuCl2 at the highest concentration (Fig. 4b).
Fig. 4.

Impact of CuO NP, CuO MP and CuCl2 on gene expression of HMOX1 and HSPA1A related to oxidative stress response. BEAS-2B cells (a) or A549 cells (b) were treated with the different copper compounds for 24 h. Shown are linear fold changes of the relative gene expression from mean values of four determinations derived from two independent experiments ± SD. 20 μg/mL CuO are equal to 252 μM Cu2+. Please note the different scaling of the y-axis
Induction of inflammation marker IL8 was also far more pronounced in BEAS-2B cells as compared to A459 cells and most evident after treatment with CuO NP (Fig. 5). CuO NP caused a dose-dependent more than 100-fold increase in gene expression, while CuO MP and CuCl2 induced transcription only up to 16-fold or 4-fold, respectively. In A549 cells IL8 expression was activated 3-fold (CuO NP), 4-fold (CuCl2) or 2-fold (CuO MP); again CuCl2 displayed a slightly stronger effect than CuO NP, but restricted to cytotoxic concentrations.
Fig. 5.
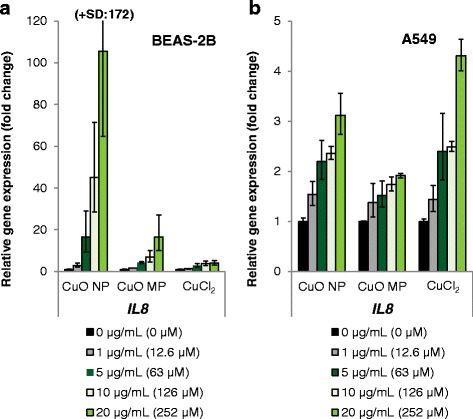
Impact of CuO NP, CuO MP and CuCl2 on gene expression of inflammation marker IL8. BEAS-2B cells (a) or A549 cells (b) were treated with the different copper compounds for 24 h. Shown are linear fold changes of the relative gene expression from mean values of four determinations derived from two independent experiments ± SD. 20 μg/mL CuO are equal to 252 μM Cu2+. Please note the different scaling of the y-axis
Impact on genes related to anti-oxidative defense, cell cycle regulation, apoptosis and DNA damage response and repair
The impact of CuO NP, CuO MP and CuCl2 on genes associated to the anti-oxidant defense system, cell cycle regulation and proliferation, apoptosis as well as DNA damage response and repair in both cell lines is summarized in a heat map view (Fig. 6). In contrast to the gene expression results shown above, changes in gene expression are presented as log2-fold changes with a value of 0 for the untreated control since a repression of transcription frequently observed within these signalling groups became clearer within this depiction.
Fig. 6.
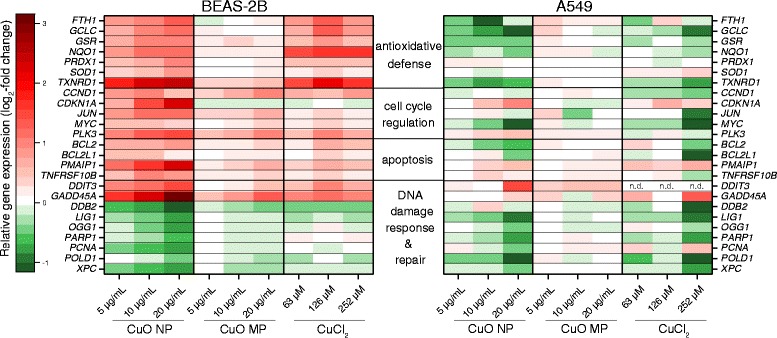
Impact of CuO NP, CuO MP and CuCl2 on gene expression related to anti-oxidative defense, cell cycle regulation, apoptosis and DNA damage response and repair. BEAS-2B cells or A549 cells were treated with the different copper compounds for 24 h. Shown are log2-fold changes of the relative gene expression in a heat map view from mean values of four determinations derived from two independent experiments (value of control = 0). 20 μg/mL CuO are equal to 252 μM Cu2+. n.d.: not determined
Most strikingly, strong modulations were observed in case of CuO NP already evident at the lowest concentration in BEAS-2B cells. Thus, a strictly dose-dependent induction of genes coding for anti-oxidative enzymes like FTH1, GCLC, GSR, NQO1, PRDX1, SOD1 and TXNRD1, as well as of the proliferation-associated JUN, MYC, PLK3 genes along with the induction of cell cycle inhibitor gene CDKN1A was observed in case of CuO NP in this cell line. Likewise, expression levels of the pro-apoptotic genes PMAIP1 and TNFRSF10B were induced together with enhanced transcription of the anti-apoptotic BCL2 and BCL2L1 genes, affecting both the extrinsic and the intrinsic pathway. Concerning the DNA damage response, transcription of the damage inducible genes DDIT3 and GADD45A was increased. However, the expression of specific DNA repair genes like the BER-associated LIG1, OGG1 and PARP1, the NER-associated DDB2 and XPC genes as well as PCNA was repressed by CuO NP in a concentration-dependent manner. With respect to CuO MP almost no modulation of the above mentioned genes was observed in BEAS-2B cells; by tendency altered transcript levels were at most restricted to the highest concentration. Intermediate effects occurred in case of CuCl2. Some anti-oxidative defense-associated as well as apoptosis related genes were induced together with a moderate induction of DDIT3 and GADD45A, while in case of the DNA repair genes only by tendency repressed transcript levels were observed. Most notably, no dose-dependency was obvious in case of CuCl2, since most relevant changes occurred in case of 126 μM and not 252 μM.
In A549 cells a completely different gene expression pattern was apparent. In case of CuO NP mainly repressed transcript levels were observed, especially with respect to the anti-oxidative defense-associated genes. Only genes coding for the cell cycle inhibitor p21 (CDKN1A), the pro-apoptotic Noxa (PMAIP1) and the DNA damage inducible DDIT3 (DDIT3) were transcriptionally slightly enhanced after treatment with CuO NP. With respect to pro-apoptotic signalling, effects were restricted to the intrinsic pathway (PMAIP1), along with slightly repressed mRNA levels of the anti-apoptotic genes BCL2 and BCL2L1. No effects at all were observed in case of CuO MP. CuCl2 displayed a gene expression modulation largely comparable to CuO NP; however, in contrast to CuO NP, pronounced effects were restricted to the highest, already cytotoxic concentration.
Intracellular GSH content
The induction of oxidative stress was investigated via modulations of the intracellular GSH levels after treatment with CuO NP, CuO MP and CuCl2 in BEAS-2B cells according to the method established by Tietze [28]. CuO NP led to slightly reduced intracellular GSH levels to 80% of control beginning at 10 μg/mL after 2 h (Fig. 7a ). In contrast, CuO MP or CuCl2 displayed no alterations at this time point. After a 24 h treatment period no changes were observed in case of the particulate copper compounds; however, CuCl2 displayed a pronounced increase up to 150–170% in the concentration range of 63–252 μM (Fig. 7b ).
Fig. 7.
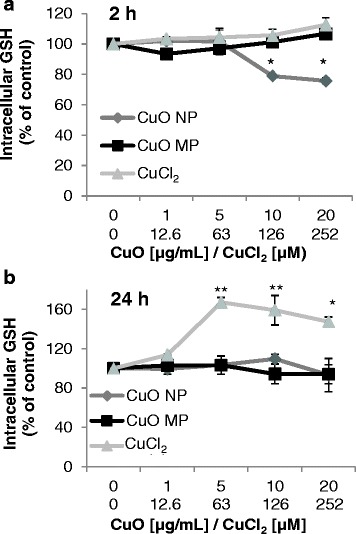
Impact of CuO NP, CuO MP and CuCl2 on the intracellular GSH level. BEAS-2B cells were treated for 2 h (a) or 24 h (b), respectively, with the different copper compounds. Shown are mean values of four determinations derived from two independent experiments ± SD. Statistically significant different from control: *p ≤ 0.05, **p ≤ 0.01 (ANOVA-Dunnett’s T test). 20 μg/mL CuO are equal to 252 μM Cu2+
Nrf2 activation
Furthermore, a potential activation of the redox-sensitive transcription factor Nrf2 was examined. Since nuclear Nrf2 levels were too low for quantification in BEAS-2B cells when using a reasonable cell number and A549 cells on the other hand exert a constitutively activated Nrf2 due to a dysfunction of its negative inhibitor Keap1 [29], these investigations were performed in HeLa S3 cells as a model. Experiments were accomplished by a TransAm Nrf2 ELISA assay, quantifying the binding capacity of nuclear located Nrf2 to the ARE consensus binding site (Fig. 8). Considering the time point of most pronounced activation of Nrf2 by the positive control sulforaphane (data not shown), cells were treated for 5 h with the respective copper compounds applying a concentration range from 5 to 40 μg/mL in case of CuO NP and CuO MP, or 63–504 μM in case of CuCl2. The concentrations were selected based on cytotoxicity of the copper compounds in this cell line as previously published [14]. While CuO MP were not cytotoxic within the entire concentration range applied, CuO NP and CuCl2 exerted strong cytotoxicity starting at 5 μg/mL or 126 μM, respectively. Furthermore, uptake studies with the copper compounds were performed in this cell line to assess the suitability as a model for Nrf2 activation (Additional file 2). Here, a concentration-dependent increase of intracellular copper levels reaching more than 2.5 mM was observed in case of CuO NP after 8 h and 24 h; copper accumulation was even more pronounced as compared to BEAS-2B cells. In contrast, copper levels were not altered by CuCl2 or CuO MP except for the high concentration of 50 μg/mL. Consequently, since cytotoxicity as well as copper uptake were generally comparable to BEAS-2B and A549 cells the use of HeLa S3 cells as a model was acceptable. Only CuO NP induced a strong activation of Nrf2 of 180–210% within the entire concentration range, whereas no activation of the transcription factor was observed in case of CuO MP; CuCl2-induced activation was restricted to the highest, cytotoxic concentration of 504 μM.
Fig. 8.
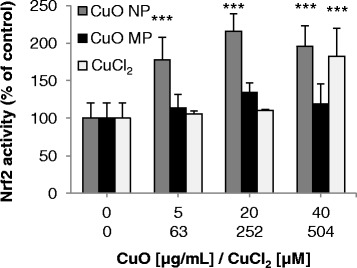
Impact of CuO NP, CuO MP and CuCl2 on Nrf2 activity. HeLa S3 cells were treated for 5 h with the different copper compounds. Shown are mean values of three independent experiments + SD. Statistically significant different from control: ***p ≤ 0.001 (ANOVA-Dunnett’s T test). 20 μg/mL CuO are equal to 252 μM Cu2+
Cell cycle distribution and apoptosis
The impact of CuO NP, CuO MP and CuCl2 on cell cycle distribution, apoptosis and necrosis at different times was investigated via flow cytometry in BEAS-2B cells. Cell cycle analyses were performed using the DNA intercalating fluorescent dye DAPI and the distinction between viable, apoptotic and necrotic cells took place using annexin V-FITC and propidium iodide (PI) staining.
The effect on cell cycle distribution was most evident after 8 h (Fig. 9). Thus, CuO NP led to a dose-dependent increase of cells in G0/G1 phase from 40% in the control up to more than 50% in case of 20 μg/mL. The amount of S phase cells did not alter, but cells in G2/M phase decreased accordingly. CuO MP and CuCl2 affected the cell cycle distribution in a similar manner. However, the modulation occurred to a lesser extent with relevant effects restricted to high concentrations starting at 20 μg/mL or 252 μM, respectively. After 24 h no alteration within cell cycle distribution was persistent (Additional file 3).
Fig. 9.
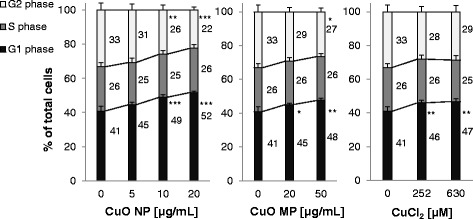
Impact of CuO NP, CuO MP and CuCl2 on cell cycle distribution. BEAS-2B cells were treated with the different copper compounds for 8 h. Shown are mean values of four determinations derived from two independent experiments + SD. Statistically significant different from control: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (ANOVA-Dunnett’s T test). The numbers right to each bar indicate the fraction of cells in the respective phase. Please note the different concentrations used for the respective copper compound. 20 μg/mL CuO are equal to 252 μM Cu2+
With respect to cell death induction, only weak effects were observed after 24 h (Additional file 4), whereas a distinct modulation was obvious after 48 h (Fig. 10). Thus, incubation with CuO NP concentration-dependently enhanced the amount of apoptotic cells to a maximum of nearly 10%, whereas CuO MP and CuCl2 displayed a maximum of 4% apoptotic cells. The amount of necrotic cells was close to 7% in case of CuO NP and 2% in case of CuO MP and CuCl2. Altogether, cell death was mediated about equally by apoptosis and necrosis.
Fig. 10.
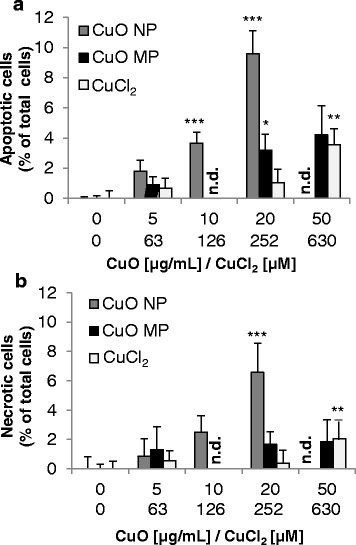
Induction of apoptosis (a) and necrosis (b) after treatment with CuO NP, CuO MP and CuCl2. BEAS-2B cells were treated with the different copper compounds for 48 h. Shown are mean values of four determinations derived from two independent experiments + SD. Statistically significant different from control: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (ANOVA-Dunnett’s T test). 20 μg/mL CuO are equal to 252 μM Cu2+. n.d.: not determined
Discussion
Within the present study, cytotoxicity, cellular uptake and intracellular copper accumulation after treatment with CuO NP as compared to CuO MP and water soluble CuCl2 were investigated in the immortalized human bronchial epithelial cell line BEAS-2B and compared to findings in the adenocarcinoma cell line A549 previously performed in our working group [14]. Moreover, the impact on cellular signalling pathways involved in the maintenance of genomic stability was analysed in both cell lines by systematic, concentration-dependent gene expression profiling via a high-throughput RT-qPCR approach. Some observations on mRNA level were pursued on the functional level by investigating induction of apoptosis, oxidative stress as well as modulation of cell cycle distribution in BEAS-2B cells. Main outcomes were a distinct accumulation of copper in the soluble fractions of the cytoplasm and the nucleus after treatment with CuO NP, leading to pronounced changes in gene expression related to metallothionein induction, oxidative stress response, DNA damage response, cell cycle regulation and apoptosis. Far less effects were observed in case of CuO MP. Interestingly, CuCl2 led to a copper accumulation in the cytoplasm, but not in the nucleus, and cellular effects were mainly restricted to cytotoxic concentrations with disturbed copper homeostasis.
The stronger cytotoxicity of CuO NP when compared to CuO MP or CuCl2 confirmed previous observations on the cytotoxicity of differently sized CuO particles and water-soluble copper compounds in lung-associated cell models [12, 13, 15, 30]. Additionally, CuO NP exerted more pronounced cytotoxic effects in BEAS-2B than in A549 cells, even though in the latter cell line the more sensitive colony forming ability was applied [14]. The higher sensitivity of BEAS-2B cells towards CuO NP as compared to A549 cells was also reported by Cronholm and colleagues [12]. This may be due to the higher GSH content of A549 compared to BEAS-2B cells [31–33]. However, interestingly, cytotoxicity of CuCl2 was higher in the former cell line.
As stated in the introduction, the major mechanism proposed for the high toxicity of CuO NP consists in the so-called “Trojan horse type mechanism” [17], comprising the endocytotic uptake of the particles, the solubilisation in lysosomes and the subsequent intracellular copper ion release. Endocytosis of particulate CuO has been shown in several studies [20–23]. While electron microscopy enables the detection of particles inside the cell and thus can demonstrate particle uptake, it does not allow an exact quantification of endocytosed particles. Considering the whole cell, a more efficient uptake of CuO NP in BEAS-2B cells as compared to A549 cells was already shown previously, determined by AAS [12]. However, since particles tend to bind to cell membranes and cannot be removed completely by cell washing, quantification of copper content of treated cells by AAS or ICP-MS does not allow to discriminate between copper within particles attached to the cells and deliberated copper ions inside the cell. Within the present study, therefore, copper was quantified by AAS, measuring total copper contents in cells and additionally by applying a specific two step lysis procedure. With respect to the latter, an approach has been chosen where the plasma membrane was lysed in the first step, yielding the soluble cytoplasmic fraction and cell nuclei. These were lysed in the second step, yielding the soluble fraction of the nuclei. Concerning the intracellular distribution, a strictly concentration-dependent copper accumulation up to millimolar concentrations both in the cytoplasm and even more pronounced in the nucleus was shown for the first time for CuO NP in BEAS-2B cells, exceeding again the values observed previously in A549 cells [14]. While the high copper contents in the cytoplasm could result both from remaining nanoparticles not yet completely solubilized in the lysosomes as well as from released copper ions, the even higher copper concentration in the nucleus strongly indicates the release of copper ions. These findings are in accordance with the far higher solubility of CuO NP in the acidic artificial lysosomal fluid (ALF) as compared to water or cell culture media [14]. CuO MP were taken up to a much lesser extent, but also in a concentration-dependent manner, yielding lower concentrations also in the cytoplasm and in the nucleus. Comparison to whole cell copper contents without removal of the outer membrane after particle treatment performed within this study demonstrated that overestimation of cellular copper levels would have been 3.5-fold in case of CuO NP and even 70-fold in case of CuO MP. In contrast to the particulate copper compounds, incubation with CuCl2 provoked comparatively slightly elevated copper levels in BEAS-2B cells. Especially in the nucleus almost no increase was observed, indicating a tight homeostatic control due to regulated uptake of copper ions. In contrast, higher nuclear copper levels have been observed in A549 cells [14], which may be explained by the more pronounced cytotoxicity in the latter cell line, leading to uncontrolled copper influx. Consequently, endocytotic uptake appears to circumvent copper homeostasis even at non-cytotoxic concentrations, while in case of water soluble copper compounds impaired homeostasis is restricted to cytotoxic concentrations. Altogether, the intracellular copper accumulation may be most decisive for the toxicity of CuO NP.
The systematic gene expression profiling via high-throughput RT-qPCR displayed a concentration-dependent impact of CuO NP on genes related to uptake and genomic stability. The extent of modulation of the uptake-related MT1X and MT2A genes, which are up-regulated in response to copper ions via the metal responsive transcription factor MTF-1 [34, 35], reflected the dose-dependent intracellular copper ion increase, exerting distinct differences among the cell lines as well as between the copper compounds. Thus, induction of MT1X and MT2A by CuO NP was very distinct in case of all investigated concentrations, higher than for CuO MP and CuCl2 as well as more pronounced in BEAS-2B cells as compared to A549 cells. Since metallothionein gene induction is mediated by copper ions and not the particulate compounds this is one clear indication of the marked intracellular release of copper ions by the particles.
Moreover, the up-regulation of oxidative and inflammatory response genes was more distinct in BEAS-2B than in A549 cells and again most pronounced in case of CuO NP; the concentration-dependent effects highly corresponded to the intracellular copper accumulation. The induction of the HMOX1 und HSPA1A genes is mediated via different redox-sensitive transcription factors like Nrf2, HSF-1 and AP-1 [36–38] and can be regarded as a model transcriptional response to the extent of ROS generation and activation of the respective transcription factors by the copper compounds. IL8 gene displayed highest induction of all investigated genes by CuO NP in BEAS-2B cells. This gene is known to be highly inducible by NF-ΚB and AP-1 in response to pro-inflammatory cytokines or cellular stress [39]. A pro-inflammatory response via IL-8 release coincident with an activation of AP-1 and NF-ΚB was further reported in case of CuO NP in A549 cells [40] and was also observed in A549 and HBEC cells using an in vitro air-liquid interface exposure system [41]. An induction of IL-8 was also described in response to TiO2 NP via NF-ΚB and AP-1 activation in different lung cell lines, indicating that oxidative stress may not be the only reason to induce respective signalling pathways [42].
The induction of genes of the anti-oxidant defense system, mainly regulated via Nrf2 [43], by CuO NP and to a lesser extent by CuCl2 was restricted to BEAS-2B cells, whereas the transcript levels were not altered or even repressed in A549 cells. These differences between the cell lines may be explained by lower levels of intracellular glutathione in BEAS-2B cells as compared to A549 cells [31–33, 44, 45] as well as by a persistent deregulation of the expression of those anti-oxidative genes due to constitutively active Nrf2 in A549 cells [29]. The proposed activation of Nrf2 was confirmed in case of CuO NP for all applied concentrations and in case of CuCl2 in high cytotoxic doses in HeLa S3 cells. These cells were chosen since they exert lower GSH levels as compared to A549 cells [46] and are thus in this respect comparable to BEAS-2B cells, and no dysfunctional regulation of Nrf2 has been reported. Since HeLa S3 cells are derived from cervical adenocarcinoma, results obtained in this cell line are not directly transferable to the lung cell models; however, the results show that Nrf2 is in principle activated by CuO NP and CuCl2. In support of using HeLa S3 cells as a model for this specific question, cytotoxicity as well as copper accumulation occurred to a comparable extent as in BEAS-2B and A549 cells. The induction of oxidative stress on the functional level was further analysed via changes in intracellular GSH levels in BEAS-2B cells. Accordingly, GSH depletion to 80% of control was limited to CuO NP after 2 h treatment; in case of CuCl2, after 24 h elevated intracellular GSH levels were observed, indicative of up-regulation of GSH synthesis and/or GSH reductase activity. These results confirm previous studies reporting oxidative stress induction, measured by enhanced ROS levels, changes in GSH and GSSG, lipid peroxidation or modulated activities of anti-oxidant enzymes by CuO NP at different time points [9, 47, 48].
With respect to genes associated with cell growth, cell cycle regulation and apoptosis, increased mRNA levels of the proto-oncogene JUN as well as the induction of cell cycle inhibitor gene CDKN1A were restricted to CuO NP in BEAS-2B cells. The positively auto-regulated JUN [49–51] together with the induced IL8 and HMOX1 genes described above strongly indicates an activation of AP-1 by CuO NP. In accordance with CDKN1A induction, a cell cycle arrest in G0/G1 phase was observed after 8 h predominantly in case of CuO NP on the functional level. The induction of the pro-apoptotic genes PMAIP1 und TNFRSF10B was restricted to CuO NP in BEAS-2B cells. Thus, genes coding for factors of both the intrinsic and the extrinsic cascade were activated in this cell line. However, the outcome concerning the intrinsic pathway was conflicting by the concurrent slight induction of genes of the anti-apoptotic Bcl-2 family. In contrast, in A549 cells only genes coding for the intrinsic pathway were modulated, indicating a weak, but clear pro-apoptotic signalling by a slight induction of PMAIP1, coincident with decreased anti-apoptotic BCL2 and BCL2L1 transcript levels. On the functional level, cell death mediated by CuO NP and to a lesser extent by CuO MP was necrotic and apoptotic, the latter slightly more pronounced. Consistent with the gene expression results the induction of apoptosis was restricted to CuO NP in BEAS-2B cells, first evident after 48 h treatment. In A549 cells previous analyses revealed the restriction of apoptosis to high concentrations of more than 20 μg/mL CuO NP [14].
Indicative for the induction of DNA damage, the DNA damage response inducible genes DDIT3 and GADD45A [52–54] were strongly up-regulated by CuO NP in BEAS-2B cells, while only slight effects were observed in case of CuO MP and CuCl2. Induction of these genes was also evident in A549 cells, albeit to a lesser extent and restricted to higher concentrations. The results are in agreement with previous observations, demonstrating enhanced expression of p38 and p53, indicative for DNA damage, and the induction of DNA damage inducible genes of the GADD family by CuO NP [19, 55]. The activation of the DNA damage response is consistent with the induction of DNA strand breaks by CuO NP measured via comet assay or alkaline unwinding [12, 14]. In addition to genotoxicity, copper ions appear to inactivate DNA repair processes. Thus, genes coding for specific proteins in DNA repair pathways such as BER and NER were down-regulated by CuO NP in BEAS-2B cells and by both CuO NP and CuCl2 in A549 cells. In addition, interactions of copper with distinct DNA repair proteins have been described on the functional level, e.g. XPA in NER and hOGG1 and PARP-1 in BER [56–58]. The inhibition of PARP-1 could further be confirmed by CuO NP and CuO MP [14]. Despite the DNA damage signalling, no activation of p53 appears to occur. Thus, no induction of its inhibitor MDM2, which is transcriptionally regulated by activated p53 [59], was observed (data not shown). In BEAS-2B cells p53 is inactivated as a consequence of the immortalization process of this cell line [25]. In A549 cells the missing activation was surprising since p53 is usually stabilized upon DNA damage, regulating cell cycle arrest and apoptosis. However, in accordance with our findings Hanagata and co-workers reported a minor role of p53 within gene expression induced by CuO NP [55].
As underlying mechanism of the observed oxidative and genotoxic stress response the increased generation of ROS due to elevated levels of redox-active copper ions can be assumed [12, 13, 15, 19, 48]. Further interactions leading for example to repressed transcript levels of DNA repair genes could include a decrease in mRNA stability, other post-translational mechanisms leading to diminished protein levels or the direct interaction with the respective transcription factors. Thus, due to the high thiol affinity and redox-activity of copper ions redox-sensitive cysteines inside or outside of zinc binding structures in transcription factors may display important targets of copper toxicity [60].
Conclusions
Within the present study, we confirmed the pronounced cytotoxicity of CuO NP as compared to microsize CuO particles or water soluble CuCl2 in human lung cell lines. A CuO NP-mediated copper overload in the cytoplasm and especially in the nucleus was shown to be of major importance for the cytotoxic and genotoxic potential as well as for dose-dependent transcriptional changes of genes related to copper overload, (oxidative) stress response, DNA damage response, cell growth and cell cycle regulation and apoptosis. The high-throughput RT-qPCR approach provided insight into the mode of action of CuO NP and revealed relevant differences not only between the copper compounds, but also between the different cell lines. Importantly, it allowed the identification of dose-dependency; thus, while CuO NP exerted effects already at low, non-cytotoxic concentrations, changes in gene expression mediated by water soluble CuCl2 were restricted to cytotoxic concentrations with disturbed copper homeostasis. One underlying mechanism appears to be increased intracellular amounts of “free” reactive copper ions, consistent with the “Trojan horse type mechanism”, which may lead to elevated levels of ROS, but also direct oxidation of redox-sensitive transcription factors such as critical cysteines in transcription factors. However, in addition to intracellular copper ion release, interactions of the CuO particles with the outer cell membrane cannot be completely excluded to contribute to the observed effects. Also, within this study, comparisons were made based on the copper content of the different compounds. Nevertheless, the impact of different surface areas of the nano- and microsized particles should be addressed. Finally, whether or not similar mechanisms account for the toxicity of other metal-based nanoparticles needs to be addressed on a case-to-case basis. These issues will be further investigated.
Methods
Materials
Chemicals, including agarose, salts, glycerol, leupeptine, phenylmethanesulfonyl-fluoride (PMSF), bovine serum albumin, acids, snap-on lid glasses and stirring bars were obtained from Carl Roth GmbH (Karlsruhe, Germany). Copper chloride was purchased in high purity (>99.9%) from Sigma-Aldrich Chemie GmbH (Steinheim, Germany). All PCR consumables including PCR tubes, strips and reaction tubes and tubules as well as cell culture dishes and flasks were obtained from Sarstedt (Nuembrecht, Germany). The primer pairs were synthesized by Eurofins (Ebersberg, Germany) or Fluidigm (San Francisco, USA). DMEM, trypsin, amphotericin B, trypsin inhibitor from Glycine max (soybean) (SBTI) and penicillin-streptomycin solutions are products of Sigma-Aldrich. Fetal calf serum (FCS) and LHC-9 media are products of Invitrogen GmbH (Darmstadt, Germany). Human fibronectin was obtained from Biopur (Reinach, Switzerland) and collagen from Roche (Mannheim, Germany). DNA suspension buffer, PCR certified water and TE buffer were obtained from Teknova (Hollister, USA). 2X Assay Loading Reagent and 20X DNA Binding Dye Sample Loading Reagent were purchased from Fluidigm (San Francisco, USA). Bio-Rad (Munich, Germany) provided the 2X SsoFast™ EvaGreen® Supermix with Low ROX and the 2X SYBR Green Supermix. The 2X TaqMan® PreAmp Master Mix was obtained from Applied Biosystems (Darmstadt, Germany) and the exonuclease I from New England Biolabs (Frankfurt am Main, Germany).
Particle characteristics
CuO NP (#544868, Lot #MKAA0633) and CuO MP (#208841, Lot #MKAA1788) were purchased from Sigma-Aldrich Chemie GmbH (Steinheim, Germany). A detailed particle characterization using DLS with respect to size, scanning electron microscopy (SEM) for size and morphology, BET for surface area, ZP for surface charge, ICP-MS, EDX and oxygen analysis for purity and composition as well as X-ray diffraction (XRD) for crystallinity was described previously. Additionally, the impact on pH in relevant media, endotoxin content and solubility in different fluid models like H2O, DMEM, DMEM/10% FCS, PBS, AAF and ALF was investigated [14].
Particle and CuCl2 incubation suspensions and dilutions
Fresh CuO NP, CuO MP suspensions and CuCl2 dilutions were prepared for each experiment. Particles, received as dry powder, were aliquoted by weighing into 1.5 mL polystyrene reaction tubes. Stock solutions of 1 mg/mL CuO were prepared into an endotoxin-free snap-on glass containing a stirring bar in DMEM containing 10% FCS. Dilutions in the range of 1–50 μg/mL were prepared by adding aliquots of the stirring stock solution into snap-on lid glasses filled with adequate volumes of fresh medium. Stirring took place at 900 rpm and room temperature on a multiphase stirrer (Variomag Poly, Carl Roth GmbH, Karlsruhe, Germany). CuCl2 was dissolved in bidistilled H2O (100 mM) and sterile-filtered. Dilutions in the range of 12.6–630 μM corresponding to the identical mass doses of copper per mL of the particulate CuO were prepared from the stock solution by dilution in adequate amounts of DMEM containing 10% FCS directly before incubation. 1 μg/mL CuO is equal to 0.2 μg/cm2 CuO and 12.6 μM Cu2+ in case of complete dissolution.
Cell culture and incubation
The adherent human adenocarcinoma cell line A549 (ATCC CCL-185) and human cervix adenocarcinoma cell line HeLa S3 (ATCC CCL-2.2) were obtained from ATCC and cultured as monolayer in DMEM containing 10% FCS, 100 U/mL penicillin and 100 mg/mL streptomycin. Human lung bronchial epithelial BEAS-2B cells (ATCC CRL-9609), immortalized with SV40 large T-antigen, were kindly provided by Dr. Carsten Weiss (Karlsruhe Institute of Technology, Karlsruhe, Germany). They were grown as monolayers in coated cell culture dishes (10 μg/mL human fibronectin, 30 μg/mL collagen and 10 μg/mL bovine serum albumin in PBS) in LHC-9 medium containing 2.5 μg/mL amphotericin B. Cells were incubated at 37 °C in a humidified atmosphere of 5% CO2 in air. For all experiments cells were seeded at a density of 16.600 cells/cm2. After 24 h of cultivation in case of A549 and HeLa S3 cells and 48 h in case of BEAS-2B cells the supernatant from the logarithmically growing cells was removed and replaced by the particle and CuCl2 incubation suspensions or dilutions (0.2 mL/cm2) in DMEM containing 10% FCS independent from the respective cell culture medium to ensure comparable incubation conditions for all applied cell lines.
Cell number
Logarithmically growing BEAS-2B cells were incubated for 24 h with 1–50 μg/mL CuO NP or CuO MP or 12.6–630 μM CuCl2, respectively, trypsinized and collected in DMEM containing 10% FCS. Cell number was determined via Casy® cell counter (OLS OMNI Life Science GmbH & Co. KG, Bremen, Germany).
Cellular uptake and intracellular distribution
Logarithmically growing BEAS-2B cells were treated with different concentrations of CuO NP, CuO MP and CuCl2 for 24 h in DMEM containing 10% FCS. The cells were trypsinized, collected in ice-cold PBS containing 10% FCS, washed twice with PBS and counted via Casy® cell counter for cell number and cell volume. For determination of copper in the soluble fraction of the whole cells, they were lysed in RIPA buffer (0.01 M Tris pH 7.6, 0.15 M NaCl, 0.001 M EDTA, 1% (v/v) Trition-X 100, 1% (v/v) DOC, 0.01% SDS, 0.001 M PMSF, 1 x protease-inhibitor mixture) for 30 min before centrifugation at 14.000 x g (1 h, 4 °C). The supernatant contained the soluble fraction and was used for graphite furnace atomic absorption spectrometry (GF-AAS). Soluble cytoplasmic and nuclear fractions were prepared using a method described previously [14, 27]. Briefly, cells were allowed to swell in cell lysis buffer (0.01 M HEPES pH 7.9, 0.01 M KCl, 0.0015 M MgCl2, 0.3 M saccharose, 0.0005 M dithiothreitol (DTT), 0.0006 M PMSF, 0.0065 mM Leupeptine) for 15 min before the addition of 25 μL 10% (v/v) IGEPAL CA 630 (Fluka) in H2O for cell lysis. The mixture was vortexed for 10 s and the nuclei pelleted at 1500 x g (15 min, 4 °C). The supernatant contained the soluble cytoplasmic fraction and was used for GF-AAS. The nuclei-containing pellet was washed twice with cell lysis buffer. Subsequently the volume and number of the nuclei was determined via Casy® cell counter and the soluble content was extracted by treatment with nuclear lysis buffer (0.01 M HEPES pH 7.9, 0.4 M KCl, 0.0015 M MgCl2, 15% (w/v) glycerol, 0.0005 M DTT, 0.0006 PMSF, 0.0065 mM Leupeptine) for 30 min on ice after mixing and subsequent centrifugation at 10.000 x g (30 min, 4 °C). The supernatant contained the soluble nuclear fraction and was used for GF-AAS. All cellular soluble fractions were evaporated at 95 °C, incubated with ashing mixture, 65% HNO3/30% H2O2 (1/1), evaporated at 95 °C again and resolved in water. Copper content was determined using GF-AAS (Perkin Elmer Atomic Absorption Spectrometer PinAAcle 900 T). Recovery rates were determined in the respective matrix using AAS elemental standard solutions (Carl Roth AG) and reached 101% ± 6% (external standard), 99% ± 4% (total fraction), 96% ± 6% (cytoplasmic fraction) and 99% ± 4% (nuclear fraction). Copper content was normalized to cell number and volume.
Gene expression analyses
Gene expression analyses via high-throughput RT-qPCR with Fluidigm dynamic arrays on the BioMark™ System were performed as described previously [24]. Briefly, 0.35–1 × 106 logarithmically growing A549 or BEAS-2B cells were treated with different concentrations of CuO NP, CuO MP and CuCl2 for 24 h in DMEM containing 10% FCS. After incubation, cells were trypsinized, resuspended in ice-cold PBS containing 10% FCS, collected by centrifugation, washed with ice-cold PBS and collected again by a second centrifugation step. Total RNA was isolated with MN NucleoSpin® RNA Plus KIT (Macherey-Nagel) according to the manufacturer’s instructions and quantified. 1 μg of total RNA was reverse transcribed in duplicate per sample into first-strand complementary DNA (cDNA) using qScript™ cDNA Synthesis Kit (Quanta) according to the manufacturer’s instructions. During high-throughput qPCR all pipetting steps were carried out in a separate room under decontaminated and sterile conditions in a PCR Workstation Pro (Peqlab, Erlangen, Germany). Before qPCR, specific target amplification (STA) and exonuclease I (E. coli) treatment had to be performed. A total of 5 μL STA mix was prepared containing 2.5 μL 2X TaqMan® PreAmp Master Mix, 0.5 μL of the 500 nM pooled primer mixture, 0.75 μL PCR certified water and 1.25 μL cDNA per reaction. A PCR certified water control (NTC-STA) and a non reverse-transcribed RNA control (NoRT) were included. STA was performed in a thermal cycler (T100, Bio-Rad Laboratories, Munich, Germany) using the following temperature program: 10 min at 95 °C followed by 12 cycles of 15 s at 95 °C and 4 min at 60 °C and a final holding temperature of 4 °C. Afterwards, 0.4 μL exonuclease I (Exo I) at 20 units/μL was diluted to 4 units/μL with 0.2 μL 10X Exonuclease I Reaction Buffer and 1.4 μL PCR certified water per reaction. 2 μL of the Exonuclease Reaction Mixture were added to the STA samples and digestion with Exo I at 4 units/μL was performed using the following temperature program: 40 min at 37 °C, 15 min at 80 °C and a final holding temperature at 4 °C. STA and Exo I-treated samples were diluted 5-fold with 18 μL TE buffer. For qPCR forward and reverse primers (100 μM) were diluted to 5 μM by adding 2.5 μL of each primer pair to 25 μL of 2X Assay Loading Reagent and 22.5 μL of DNA suspension buffer. The primer reaction mix was stored at −20 °C. For the sample mix, 2.25 μL of STA and Exo I-treated samples were mixed with 2.5 μL of 2X SsoFast™ EvaGreen® Supermix with Low ROX and 0.25 μL of 20X DNA Binding Dye Sample Loading Reagent. Preparation and loading of Fluidigm 96.96 Dynamic Array IFC (integrated fluidic circuit) was performed according to the manufacturer’s instructions. After priming, the chip was loaded with samples and the primer reaction mixes within 1 h to reduce the loss of the pressure within the chip. Thus, 5 μL of each primer reaction mix and each sample were pipetted into the respective inlets, avoiding the generation of air bubbles. Samples and primer reaction mixes were loaded into the chip by running the Load Mix (136×) script of the IFC Controller HX (Fluidigm, San Francisco, USA). After loading of the chip, potential dust particles were carefully removed from the surface of the chip using adhesive tape. The chip was transferred into the BioMark™ System (Fluidigm, San Francisco, USA) and qPCR and melting curve analysis were performed by running the following temperature program: 2400 s at 70 °C and 30 s at 60 °C, followed by a hot start for 60 s at 95 °C, 30 PCR cycles of 5 s at 96 °C for denaturation and 20 s at 60 °C for annealing and elongation. The melting curve analysis consisted of 3 s at 60 °C followed by heating up to 95 °C with a ramp rate of 1 °C/3 s. Data analysis and depiction was accomplished with the Fluidigm Real-Time PCR Analysis and with GenEx software. For normalization, five potential reference genes were available (ACTB, B2M, GAPDH, GUSB, and HPRT1). Finally, potential alterations of the transcript levels of the target genes under investigation were displayed as fold change compared to a control group by calculating relative quantities corresponding to the ΔΔCq method [61].
Quantification of intracellular glutathione
The quantification of intracellular glutathione levels was performed according to the method established by Tietze et al. [28]. Briefly, logarithmically growing BEAS-2B cells were incubated for 2 h or 24 h, respectively, with 1–20 μg/mL particulate CuO or 12.6–252 μM CuCl2, respectively, in DMEM containing 10% FCS. The cells were trypsinized, collected in ice-cold PBS containing 10% FCS, washed with PBS, counted via Casy® cell counter for cell number and cell volume and collected by centrifugation. 1 × 106 cells were collected in KP buffer (0.1 M KH2PO4, 0.1 M K2HPO4, 1 mM EDTA, pH 7.4). Cells were lyzed by 2 freeze-and-thawing cycles followed by sonification, followed by acidification with sulphosalicyclic acid and vortexed. Cells were centrifuged at 16.000 rpm for 20 min at 4 °C prior to measuring total GSH in the supernatant by reducing oxidized GSH content using GR enzyme (4 U/mL) and NADPH (0.3 mM). Thereby, GSH reacts with 5,5′-dithiobis-2-nitrobenzoic acid (DTNB) to form 2-nitro-5-thiobenzoate (TNB). The change in TNB absorbance was measured at 412 nm using a plate reader (Tecan). Data were compared to GSH standard calibration curves and normalized to cell volume.
Nrf2 activity
Logarithmically growing HeLa S3 cells were treated with different concentrations of CuO NP, CuO MP and CuCl2 for 5 h in DMEM containing 10% FCS. Nuclear extracts were prepared using a nuclear extract kit (Active Motif, Carlsbad, CA, USA) according to the manufacturer’s instructions. Thus, cells were washed with ice-cold PBS/phosphatase inhibitors, scraped into ice-cold PBS/phosphatase inhibitors buffer and collected by centrifugation. The cells were lyzed in hypotonic buffer and swelled on ice for 15 min. After addition of detergent and vortexing, cells were centrifuged at 14.000 x g for 30 s at 4 °C. The supernatant (containing the cytoplasmic fraction) was stored separately and the nuclear pellet was resuspended in Complete Lysis Buffer containing detergent, mixed and incubated for 30 min on a rocking platform at 150 rpm. Afterwards, the nuclear pellets were vortexed and centrifuged at 14.000 x g for 20 min at 4 °C. The supernatant containing the nuclear extract was used for the determination of Nrf2 activity after determining protein content according to the method established by Bradford [62]. Nrf2 activity was determined with the TransAM Nrf2 assay (Active Motif, Carlsbad, CA, USA) according to the manufacturer’s instructions. Nuclear extracts (5 μg) were incubated with ARE consensus site oligonucleotides (5′-GTCACAGTGACTCAGCAGAATCTG-3′) immobilized to 96-well plates. Bound protein levels were quantified by antibody specific to DNA-bound Nrf2 and visualized by colorimetric reaction catalysed by horseradish-peroxidase-conjugated secondary antibody. Resulting absorbance was measured at 405 nm with a reference wave length of 655 nm.
Cell cycle analysis and cell death induction
0.35–1 × 106 logarithmically growing BEAS-2B cells were treated with different concentrations of CuO NP, CuO MP and CuCl2 for 8, 24 or 48 h in DMEM containing 10% FCS. Cells were trypsinized, collected in fresh media, combined with the supernatant and centrifuged at 1300 rpm for 3 min at 4 °C. Respective parts of the cell suspension were used for cell death or cell cycle analysis. Concerning the latter, the pellet was resuspended in 1 mL ice-cold PBS before 3 mL ice-cold ethanol (96%) were added under vortexing, followed by fixation overnight at −20 °C. For flow cytometry analysis, the cells were collected by centrifugation, washed with 1 mL PBS and centrifuged again. The pellet was resuspended in 300 μL DAPI dye solution (CyStainDNA, Partec) and incubated for 30 min at 4 °C in the dark. For cell death analysis, the cell suspension was directly stained with a master mix (PI and annexin V-FITC in calcium buffer, pH 7.0) and incubated for 30 min at 4 °C in the dark. 10.000 or 30.000 cells per sample were analysed for cell death induction or DAPI intensity, respectively, via flow cytometry using a LSR Fortessa from Becton Dickinson. Excitation and emission detection of the applied fluorescence dyes were as follows: DAPI (405 nm - 450/50 nm), PI (488 nm – 695/40 nm) and annexin V-FITC (488 nm – 530/30 nm).
Statistics
Differences between control and treated samples were analysed by one-way analysis of variance (ANOVA) followed by Dunnett’s T posthoc test.
Additional files
Cellular copper content after 24 h treatment with CuO NP or CuO MP with and without removal of the outer membrane in BEAS-2B cells. (PPTX 66 kb)
Uptake studies on CuO NP, CuO MP and CuCl2 in HeLa S3 cells. (PPTX 71 kb)
Supporting information on cell cycle distribution after 24 h treatment with different copper compounds in BEAS-2B cells. (PPTX 98 kb)
Supporting information on cell death induction after 24 h treatment with different copper compounds in BEAS-2B cells. (PPTX 81 kb)
Acknowledgements
We thank Wera Hubele for her assistance in the copper uptake studies in HeLa S3 cells.
Availability of data and material
The relevant datasets supporting the conclusions of this article are included within the article and all datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported by the German Research Council DFG - Deutsche Forschungsgemeinschaft (Excellence Initiative KIT) and INST 121384/24–1 FUGG. BMS received a Scholarship Graduate Funding from the German States Programme (LGF) – Landesgraduiertenförderung.
Authors` contributions
AH supervised and coordinated the research project and finalized the manuscript. The study was performed by BMS and AH. BMS performed most of the experiments, analysed the data and drafted the manuscript. RKN performed some gene expression analyses and NLW performed a part of the uptake studies. All of the authors have read and approved the final manuscript.
Authors´ information
Not applicable.
Abbreviations
- AAF
Artificial alveolar fluid
- AIF
Apoptosis inducing factor
- ALF
Artificial lysosomal fluid
- AP-1
activator protein 1
- BER
Base excision repair
- BET
Brunauer-Emmett-Teller analysis
- Cq
Quantification cycle
- CuCl2
Copper chloride
- CuO MP
Copper oxide microparticles
- CuO NP
Copper oxide nanoparticles
- DLS
Dynamic light scattering
- EDX
Energy-dispersive X-ray spectroscopy
- GF-AAS
Graphite furnace atomic absorption spectrometry
- GSH
Glutathione
- GSSG
Glutathione disulfide
- hOGG1
8-oxoguanine DNA N-glycosylase 1
- ICP-MS
Inductively coupled plasma mass spectrometry
- Keap1
Kelch-Like ECH-Associated Protein 1
- MDM2
Mouse Double Minute 2 protein
- MTF-1
Metal-regulatory transcription factor 1
- NER
Nucleotide excision repair
- NF-ΚB
Nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells
- Nrf2
Nuclear factor (erythroid-derived 2)-like 2
- PARP-1
Poly(ADP-ribose)polymerase-1
- PI
Propidium iodide
- ROS
Reactive oxygen species
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- SEM
Scanning electron microscopy
- SOD
Superoxide dismutase
- TEM
Transmission electron microscopy
- XPA
Xeroderma Pigmentosum, Complementation Group A
- ZP
Zeta potential
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12989-017-0209-1) contains supplementary material, which is available to authorized users.
Contributor Information
Bettina Maria Strauch, Email: Bettina.Strauch@kit.edu.
Rebecca Katharina Niemand, Email: Rebecca.Niemand@kit.edu.
Nicola Lisa Winkelbeiner, Email: nicolawinkelbeiner@web.de.
Andrea Hartwig, Phone: +4972160847645, Email: Andrea.Hartwig@kit.edu.
References
- 1.Chang Y-N, Zhang M, Xia L, Zhang J, Xing G. The toxic effects and mechanisms of CuO and ZnO nanoparticles. Materials. 2012;5:2850–2871. doi: 10.3390/ma5122850. [DOI] [Google Scholar]
- 2.Shah ID, Ruzzi PL, Schluter RB. Decomposition of cupric oxide using a reducing scavenger. United States Pat 4,080,430. 1978.
- 3.Riffault V, Arndt J, Marris HLN, Mbengue S, Setyan A, Alleman LY, et al. Fine and ultrafine particles in the vicinity of industrial activities: a review. Crit Rev Environ Sci Technol. 2015;45:2305–2356. doi: 10.1080/10643389.2015.1025636. [DOI] [Google Scholar]
- 4.Charvet A, Bau S, Paez Coy NE, Bemer D, Thomas D. Characterizing the effective density and primary particle diameter of airborne nanoparticles produced by spark discharge using mobility and mass measurements (tandem DMA/APM) J Nanopart Res. 2014;16:1–11. doi: 10.1007/s11051-014-2418-y. [DOI] [Google Scholar]
- 5.EPA. Profile of the Nonferrous Metals Industry. Washington, DC: Environmental Protection Agency; 1995.
- 6.Arredondo M, Nunez MT. Iron and copper metabolism. Mol Asp Med. 2005;26:313–327. doi: 10.1016/j.mam.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Jomova K, Baros S, Valko M. Redox active metal-induced oxidative stress in biological systems. Transit Met Chem. 2012;37:127–134. doi: 10.1007/s11243-012-9583-6. [DOI] [Google Scholar]
- 8.Lopez de Romana D, Olivares M, Uauy R, Araya M. Risks and benefits of copper in light of new insights of copper homeostasis. J Trace Elem Med Biol. 2011;25:3–13. doi: 10.1016/j.jtemb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Karlsson HL, Cronholm P, Gustafsson J, Moller L. Copper oxide nanoparticles are highly toxic: a comparison between metal oxide nanoparticles and carbon nanotubes. Chem Res Toxicol. 2008;21(9):1726–1732. doi: 10.1021/tx800064j. [DOI] [PubMed] [Google Scholar]
- 10.Fahmy B, Cormier SA. Copper oxide nanoparticles induce oxidative stress and cytotoxicity in airway epithelial cells. Toxicol in Vitro. 2009;23:1365–1371. doi: 10.1016/j.tiv.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chusuei CC, Wu C-H, Mallavarapu S, Hou FYS, Hsu C-M, Winiarz JG, et al. Cytotoxicity in the age of nano: the role of fourth period transition metal oxide nanoparticle physicochemical properties. Chem Biol Interact. 2013;206:319–326. doi: 10.1016/j.cbi.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Cronholm P, Karlsson HL, Hedberg J, Lowe TA, Winnberg L, Elihn K, et al. Intracellular uptake and toxicity of Ag and CuO Nanoparticles: a comparison between Nanoparticles and their corresponding metal ions. Small. 2013;9:970–982. doi: 10.1002/smll.201201069. [DOI] [PubMed] [Google Scholar]
- 13.Midander K, Cronholm P, Karlsson HL, Elihn K, Moeller L, Leygraf C, et al. Surface characteristics, copper release, and toxicity of nano- and micrometer-sized copper and copper(II) oxide particles: a cross-disciplinary study. Small. 2009;5:389–399. doi: 10.1002/smll.200801220. [DOI] [PubMed] [Google Scholar]
- 14.Semisch A, Ohle J, Witt B, Hartwig A. Cytotoxicity and genotoxicity of nano - and microparticulate copper oxide: role of solubility and intracellular bioavailability. Part Fibre Toxicol. 2014;11:1–16. doi: 10.1186/1743-8977-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson HL, Gustafsson J, Cronholm P, Moller L. Size-dependent toxicity of metal oxide particles-a comparison between nano- and micrometer size. Toxicol Lett. 2009;188(2):112–118. doi: 10.1016/j.toxlet.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Elsaesser A, Howard CV. Toxicology of nanoparticles. Adv Drug Deliv Rev. 2012;64:129–137. doi: 10.1016/j.addr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Limbach LK, Wick P, Manser P, Grass RN, Bruinink A, Stark WJ. Exposure of engineered Nanoparticles to human lung epithelial cells: influence of chemical composition and catalytic activity on oxidative stress. Environ Sci Technol. 2007;41:4158–4163. doi: 10.1021/es062629t. [DOI] [PubMed] [Google Scholar]
- 18.Studer AM, Limbach LK, Van DL, Krumeich F, Athanassiou EK, Gerber LC, et al. Nanoparticle cytotoxicity depends on intracellular solubility: comparison of stabilized copper metal and degradable copper oxide nanoparticles. Toxicol Lett. 2010;197:169–174. doi: 10.1016/j.toxlet.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Wang ZY, Li N, Zhao J, White JC, Qu P, Xing BS. CuO Nanoparticle interaction with human epithelial cells: cellular uptake, location, export, and Genotoxicity. Chem Res Toxicol. 2012;25(7):1512–1521. doi: 10.1021/tx3002093. [DOI] [PubMed] [Google Scholar]
- 20.Canton I, Battaglia G. Endocytosis at the nanoscale. Chem Soc Rev. 2012;41(7):2718–2739. doi: 10.1039/c2cs15309b. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Size-dependent endocytosis of nanoparticles. Adv Mater. 2009;21:419–424. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunov O, Zablotskii V, Syrovets T, Roecker C, Tron K, Nienhaus GU, et al. Modeling receptor-mediated endocytosis of polymer-functionalized iron oxide nanoparticles by human macrophages. Biomaterials. 2010;32(2):547–555. doi: 10.1016/j.biomaterials.2010.08.111. [DOI] [PubMed] [Google Scholar]
- 23.Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer BM, Neumann D, Piberger AL, Risnes SF, Koeberle B, Hartwig A. Use of high-throughput RT-qPCR to assess modulations of gene expression profiles related to genomic stability and interactions by cadmium. Arch Toxicol. 2016;90:2745–2761. doi: 10.1007/s00204-015-1621-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine AJ. The common mechanisms of transformation by the small DNA tumor viruses: the inactivation of tumor suppressor gene products: p53. Virology. 2009;384:285–293. doi: 10.1016/j.virol.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 26.Semisch A, Hartwig A. Copper ions interfere with the reduction of the water-soluble Tetrazolium salt-8. Chem Res Toxicol. 2014;27:169–171. doi: 10.1021/tx400414c. [DOI] [PubMed] [Google Scholar]
- 27.Schwerdtle T, Hartwig A. Bioavailability and genotoxicity of soluble and particulate nickel compounds in cultured human lung cells. Mater Werkst. 2006;37:521–525. doi: 10.1002/mawe.200600030. [DOI] [Google Scholar]
- 28.Tietze F. Enzymic method for determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal Biochem. 1969;27:502–522. doi: 10.1016/0003-2697(69)90064-5. [DOI] [PubMed] [Google Scholar]
- 29.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:1865–1876. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berntsen P, Park CY, Rothen-Rutishauser B, Tsuda A, Sager TM, Molina RM, et al. Biomechanical effects of environmental and engineered particles on human airway smooth muscle cells. J R Soc Interface. 2010;7:331–340. doi: 10.1098/rsif.2010.0068.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lian F, Wang X-D. Enzymatic metabolites of lycopene induce Nrf2-mediated expression of phase II detoxifying/antioxidant enzymes in human bronchial epithelial cells. Int J Cancer. 2008;123:1262–1268. doi: 10.1002/ijc.23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietarinen-Runtti P, Raivio KO, Saksela M, Asikainen TM, Kinnula VL. Antioxidant enzyme regulation and resistance to oxidants of human bronchial epithelial cells cultured under hyperoxic conditions. Am J Respir Cell Mol Biol. 1998;19:286–292. doi: 10.1165/ajrcmb.19.2.2836. [DOI] [PubMed] [Google Scholar]
- 33.Carmichael J, Mitchell JB, Friedman N, Gazdar AF, Russo A. Glutathione and related enzyme activity in human lung cancer cell lines. Br J Cancer. 1988;58:437–440. doi: 10.1038/bjc.1988.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sims HI, Chirn G-W, Marr MT., II Single nucleotide in the MTF-1 binding site can determine metal-specific transcription activation. Proc Natl Acad Sci U S A. 2012;109:16516–16521. doi: 10.1073/pnas.1207737109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marr SK, Pennington KL, Marr MT. Efficient metal-specific transcription activation by drosophila MTF-1 requires conserved cysteine residues in the carboxy-terminal domain. Biochim Biophys Acta, Gene Regul Mech. 2012;1819:902–912. doi: 10.1016/j.bbagrm.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gozzelino R, Jeney V, Soares MP. Mechanisms of cell protection by heme oxygenase-1. Annu Rev Pharmacol Toxicol. 2010;50:323–354. doi: 10.1146/annurev.pharmtox.010909.105600. [DOI] [PubMed] [Google Scholar]
- 37.Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 38.Ryter SW, Choi AMK. Heme oxygenase-1: Redox regulation of a stress protein in lung and cell culture models. Antioxid Redox Signal. 2005;7:80–91. doi: 10.1089/ars.2005.7.80. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. J Leukoc Biol. 2002;72:847–855. [PubMed] [Google Scholar]
- 40.Cho W-S, Duffin R, Poland CA, Duschl A, Oostingh GJ, MacNee W, et al. Differential pro-inflammatory effects of metal oxide nanoparticles and their soluble ions in vitro and in vivo; zinc and copper nanoparticles, but not their ions, recruit eosinophils to the lungs. Nanotoxicology. 2012;6:22–35. doi: 10.3109/17435390.2011.552810. [DOI] [PubMed] [Google Scholar]
- 41.Jing X, Park JH, Peters TM, Thorne PS. Toxicity of copper oxide nanoparticles in lung epithelial cells exposed at the air-liquid interface compared with in vivo assessment. Toxicol in Vitro. 2015;29:502–511. doi: 10.1016/j.tiv.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ekstrand-Hammarström B, Akfur CM, Andersson PO, Lejon C, Österlund L, Bucht A. Human primary bronchial epithelial cells respond differently to titanium dioxide nanoparticles than the lung epithelial cell lines A549 and BEAS-2B. Nanotoxicology. 2012;6(6):623–634. doi: 10.3109/17435390.2011.598245. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi M, Yamamoto M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid Redox Signal. 2005;7:385–394. doi: 10.1089/ars.2005.7.385. [DOI] [PubMed] [Google Scholar]
- 44.Hatcher EL, Chen Y, Kang YJ. Cadmium resistance in A549 cells correlates with elevated glutathione content but not antioxidant enzymic activities. Free Radic Biol Med. 1995;19:805–812. doi: 10.1016/0891-5849(95)00099-J. [DOI] [PubMed] [Google Scholar]
- 45.Rahman I, Bel A, Mulier B, Lawson MF, Harrison DJ, MacNee W, et al. Transcriptional regulation of γ-glutamylcysteine synthetase-heavy subunit by oxidants in human alveolar epithelial cells. Biochem Biophys Res Commun. 1996;229:832–837. doi: 10.1006/bbrc.1996.1888. [DOI] [PubMed] [Google Scholar]
- 46.Chao CCK, Huang YT, Ma CM, Chou WY, Sue L. Overexpression of glutathione S-transferase and elevation of thiol pools in a multidrug-resistant human colon cancer cell line. Mol Pharmacol. 1992;41:69–75. [PubMed] [Google Scholar]
- 47.Seiffert JM, Baradez M-O, Nischwitz V, Lekishvili T, Goenaga-Infante H, Marshall D. Dynamic monitoring of metal oxide Nanoparticle toxicity by label free impedance sensing. Chem Res Toxicol. 2012;25:140–152. doi: 10.1021/tx200355m. [DOI] [PubMed] [Google Scholar]
- 48.Ahamed M, Siddiqui MA, Akhtar MJ, Ahmad I, Pant AB, Alhadlaq HA. Genotoxic potential of copper oxide nanoparticles in human lung epithelial cells. Biochem Biophys Res Commun. 2010;396:578–583. doi: 10.1016/j.bbrc.2010.04.156. [DOI] [PubMed] [Google Scholar]
- 49.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 50.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 51.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta, Rev Cancer. 1991;1072:129–157. doi: 10.1016/0304-419X(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 52.Fornace AJ, Jr, Nebert DW, Hollander MC, Luethy JD, Papathanasiou M, Fargnoli J, et al. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol Cell Biol. 1989;9:4196–4203. doi: 10.1128/MCB.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luethy JD, Holbrook NJ. Activation of the gadd153 promoter by genotoxic agents: a rapid and specific response to DNA damage. Cancer Res. 1992;52:5–10. [PubMed] [Google Scholar]
- 54.Zhan Q. Gadd45a, a p53- and BRCA1-regulated stress protein, in cellular response to DNA damage. Mutat Res Fundam Mol Mech Mutagen. 2005;569:133–143. doi: 10.1016/j.mrfmmm.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 55.Hanagata N, Zhuang F, Connolly S, Li J, Ogawa N, Xu MS. Molecular responses of human lung epithelial cells to the toxicity of copper oxide Nanoparticles inferred from whole genome expression analysis. ACS Nano. 2011;5(12):9326–9338. doi: 10.1021/nn202966t. [DOI] [PubMed] [Google Scholar]
- 56.Asmuss M, Mullenders LHF, Eker A, Hartwig A. Differential effects of toxic metal compounds on the activities of Fpg and XPA, two zinc finger proteins involved in DNA repair. Carcinogenesis. 2000;21:2097–2104. doi: 10.1093/carcin/21.11.2097. [DOI] [PubMed] [Google Scholar]
- 57.Schwerdtle T, Hamann I, Jahnke G, Walter I, Richter C, Parsons JL, et al. Impact of copper on the induction and repair of oxidative DNA damage, poly(ADP-ribosyl)ation and PARP-1 activity. Mol Nutr Food Res. 2007;51:201–210. doi: 10.1002/mnfr.200600107. [DOI] [PubMed] [Google Scholar]
- 58.Hamann I, Schwerdtle T, Hartwig A. Establishment of a non-radioactive cleavage assay to assess the DNA repair capacity towards oxidatively damaged DNA in subcellular and cellular systems and the impact of copper. Mutat Res Fundam Mol Mech Mutagen. 2009;669:122–130. doi: 10.1016/j.mrfmmm.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 59.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 60.Hartwig A. Metal interaction with redox regulation: an integrating concept in metal carcinogenesis? Free Radic Biol Med. 2013;55:63–72. doi: 10.1016/j.freeradbiomed.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 61.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cellular copper content after 24 h treatment with CuO NP or CuO MP with and without removal of the outer membrane in BEAS-2B cells. (PPTX 66 kb)
Uptake studies on CuO NP, CuO MP and CuCl2 in HeLa S3 cells. (PPTX 71 kb)
Supporting information on cell cycle distribution after 24 h treatment with different copper compounds in BEAS-2B cells. (PPTX 98 kb)
Supporting information on cell death induction after 24 h treatment with different copper compounds in BEAS-2B cells. (PPTX 81 kb)


