Abstract
RNA folding has been studied extensively in vitro, typically under dilute solution conditions and abiologically high salt concentrations of 1 M Na+ or 10 mM Mg2+. The cellular environment is very different, with 20–40% crowding and only 10–40 mM Na+, 140 mM K+ and 0.5–2.0 mM Mg2+. As such, RNA structures and functions can be radically altered under cellular conditions. We previously reported that tRNAphe secondary and tertiary structures unfold together in a cooperative two-state fashion under crowded in vivo-like ionic conditions, but in a non-cooperative multi-state fashion under dilute in vitro ionic conditions unless in non-physiologically high concentrations of Mg2+. The mechanistic basis behind these effects remains unclear, however. To address the mechanism that drives RNA folding cooperativity, we probe effects of cellular conditions on structures and stabilities of individual secondary structure fragments comprising the full-length RNA. We elucidate effects of a diverse set of crowders on tRNA secondary structural fragments and full-length tRNA at three levels: at the nucleotide level by temperature-dependent in-line probing (ILP), at the tertiary structure level by small angle X-ray scattering (SAXS), and at the global level by thermal denaturation. We conclude that cooperative RNA folding is induced by two overlapping mechanisms: increased stability and compaction of tertiary structure through effects of Mg2+, and decreased stability of certain secondary structure elements through effects of molecular crowders. These findings reveal that despite having very different chemical makeups RNA and protein can both have weak secondary structures in vivo leading to cooperative folding.
Keywords: RNA folding, cellular conditions, cooperativity
Introduction
RNA structure serves many roles in biology including catalysis, small molecule recognition, and gene regulation.1 High concentrations of monovalent (1 M Na+)2,3 and divalent salts (up to 50 mM Mg2+)4,5 have typically been used to study RNA folding in vitro. Rationales for these conditions are to force simple RNA duplexes to fold in a two-state-like fashion, which simplifies thermodynamic interpretation of data, or to induce catalytic RNAs to fully adopt a tertiary structure. However, the cellular environment is very different. In cells, the predominant monovalent ion is K+ and its concentration is only ~140 mM;6 Mg2+ concentrations are low at just 0.5–1.0 mM in eukaryotic cells and 1.5–2.0 mM in prokaryotic cells,7–11 and there is an estimated 10–40 mM Na+.12,13 There are also membranous and non-membranous compartments that concentrate RNA and an estimated 20% – 40% molecular crowding in the cytoplasm14,15.
There are manifold reasons to study RNA folding under cellular-like conditions. First, genome-wide studies that probe RNA structure in cells have shown that certain classes of RNAs adopt very different structures in vivo than predicted in silico or measured in vitro16 and there appears to be less extensive RNA structure in vivo than in vitro.17 Nonetheless, despite being less stable in vivo, short hairpin structures were recently found to form in vivo downstream of the poly(A) signal, which aids in the cleavage and polyadenylation of human mRNAs.18 Based on their free energy, these hairpin structures are predicted to be no more stable than random sequence indicating that even weakly folding hairpins can fold in vivo in certain regions of the mRNA. Likewise, studies in model cytoplasms indicate that in solutions that mimic the cellular environment, certain RNAs can form more native-like structures, have improved functions, and fold through different pathways.19–22 For instance, the adenine riboswitch adopts a more stable and compact structure under cellular conditions;19 a small group I intron with lower flexibility has a larger free energy gap for folding in Mg2+;20 the CPEB3 ribozyme cleaves with a faster rate and is stable, even under semi-denaturing conditions, in cellular conditions;21 and tRNA folds in a two-state, cooperative manner when in the background of cellular mimics. 22
Functional RNAs typically fold in a hierarchical manner, with secondary structures forming first and tertiary structures forming later.23 This order is necessitated by the global architecture of RNA, wherein secondary structures provide a structural framework, and tertiary structures typically assemble out of that framework. Free energy parameters for RNA folding in the presence of 1 M NaCl have been measured for many model duplexes and are exceptionally favorable. For instance, 4–5 base pair RNA helices can have folding free energies of ≤ –10 kcal/mol. Such strong secondary structures have been suggested to drive RNA folding,24 which led to the notion that RNAs fold in a non-cooperative manner wherein weak tertiary structures unfold at lower temperatures and strong secondary structures at higher temperatures.
Secondary structures of RNA may not be as stable in vivo as they are in vitro. Indeed, RNA and DNA secondary structures have been reported to be destabilized in the presence of high concentrations of crowders and small osmolytes similar to small molecules that can be found in cells. For example, in 20% crowding agents and cosolutes, both DNA oligomer duplexes and hammerhead ribozyme secondary structures have a decrease in melting temperature.25,26 In the case of the hammerhead ribozyme, this decrease is accompanied by an increase in the rate of self-cleavage.26 In addition, diverse small osmolytes have been reported to destabilize nearly all RNA secondary structures while having mixed effects on the thermostability of tertiary structures.27 The above experiments, while leading to keen insight into RNA stability in crowded solutions, were performed in non-physiological concentrations of divalent, monovalent, or small osmolyte concentrations; moreover, these studies did not focus on RNA folding cooperativity.
In the protein-folding community, folding cooperativity is often defined as the coupling of secondary and tertiary structure unfolding, and we make an effort to understand RNA folding cooperativity in these terms herein. We differentiate this definition of cooperativity from the important but separate viewpoint of folding cooperativity amongst various RNA tertiary structural elements.28,29 Woodson and co-workers showed that RNA tertiary structure can couple with secondary structure and guide proper formation of secondary structure. While pioneering, these studies were conducted in the absence of physiological concentrations of monovalent ions, crowders, or cosolutes.30 Recent studies from our lab showed that under in vivo-like conditions that mimic cellular crowding and ionic conditions, functional RNAs unfold in a cooperative all-or-none-like fashion in which tertiary and secondary structures unfold together.22 However, the contributions of secondary structure destabilization and tertiary structure stabilization to the cooperative unfolding in RNA under in vivo-like conditions remain unclear.
Although we are strong proponents of studying RNA folding directly in vivo,16,31 it is very difficult to perform thermodynamic studies in living cells. To complement our direct in vivo studies, we have adopted a detailed thermodynamic and structural study of RNA folding in model cytoplasms that mimic the crowding and ionic conditions of the cellular environment.32 We report here studies on full-length yeast tRNAphe and its secondary structural fragments under in vivo-like conditions, which reveal that tRNA folds in a cooperative manner owing to a combination of destabilizing secondary structures and stabilizing tertiary structure.
Materials and Methods
Chemicals
PEG200, PEG4000, PEG8000, and PEG20000, MgCl2, HEPES, and sodium cacodylate were purchased from Sigma. KCl was purchased from J. T. Baker. Calf intestinal phosphatase and polynucleotide kinase were purchased from NEB.
RNA Constructs and Preparation
Wild type (WT) tRNAphe was purchased from Sigma and purified on a 10% denaturing PAGE gel and recovered by a crush and soak/ethanol precipitation procedure.
T7 full length (FL) tRNA was transcribed from a hemi-duplex DNA template from IDT (Coralville, IA) that was used without further purification. The T7 promoter binding site is underlined in the DNA template. The T7 promoter and DNA template were annealed at 95°C for 3 min in 100 mM NaCl and cooled at room temperature for 10 min. The tRNAphe was transcribed using T7 polymerase in 40 mM Tris (pH 7.5), 25 mM MgCl2, 2 mM DTT, 1 mM spermidine, and 3 mM NTPs, with incubation at 37°C for 4 h. The RNA was purified by 10% denaturing PAGE gel and recovered by a crush and soak/ethanol precipitation procedure.
-
T7 promoter:
5′d(TAATACGACTCACTATA)
-
FL tRNAphe DNA Template:
5′d(TGGTGCGAATTCTGTGGATCGAACACAGGACCTCCAGATCTTCAGTCTGGCGCTCTCCCAACTGAGCT AAATCCGCTATAGTGAGTCGTATTA)
-
FL tRNAphe:
5′GCGGAUUUAGCUCAGUUGGGAGAGCGCCAGACUGAAGAUCUGGAGGUCCUGUGUUCGAUCCACAGAAUUCGCACCA
The four secondary structure RNA fragments derived from the arms of FL tRNA were synthesized and HPLC-purified by GE Dharmacon (Lafayette, CO), deblocked upon receiving, and dialyzed into 10 mM sodium cacodylate (pH 7.0) and 140 mM KCl using a microdialysis system (Gibco-BRL Life Technologies).
Acceptor stem RNA: 5′ GCGGAUUUUUUUUUUAAUUCGC
D stem-loop RNA: 5′ GCUCAGUUGGGAGAGC
Anticodon (AC) stem-loop RNA: 5′ CCAGACUGAAGAUCUGG
TΨC stem-loop RNA: 5′ CUGUGUUCGAUCCACAG
Thermal Denaturation and Data Analysis
RNA was renatured by incubating at 95° C for 3 min and cooled at room temperature over 10 min in the presence of KCl and sodium cacodylate (pH 7.0). Polyethylene glycol (PEG), the crowding agent, and/or MgCl2 was added to the RNA solution, and the sample was incubated at 55 °C for 3 min and allowed to cool at room temperature for 10 min. Prior to thermal denaturation experiments, the sample was spun down at 14,000 rpm for 5 min at 4 °C to remove air bubbles and particulates.
Thermal denaturation experiments were performed on a Gilford Response II spectrophotometer, with a data point collected every 0.5 °C and a heating rate of ~0.6 °C/min with absorbance detection at 260 nm. This method is referred to as ‘optical melting’. Single-transition melt data were fit to a two-state model using sloping baselines. Data were smoothed with an 11-point window prior to taking the derivative. Samples were 1.0 μM RNA in 10 mM sodium cacodylate (pH 7.0), 140 mM KCl, 0–2.0 mM MgCl2, and 0–40% (w/v) crowder. To analyze the unfolding of the four secondary structures as a whole and compare them to FL tRNA, raw absorbance data for the melting curves of each secondary structure were summed (eq 1), smoothed with 11-point smoothing, and then the derivative was taken.
| (eq 1) |
These processed data are referred to as the “sum of the secondary structure fragments” or “SSS”, and the TMs reported for these data are apparent TMs. The melting temperature of each RNA transition acquired by optical melting in each condition was found using a non-linear Marquardt algorithm in Kaleidagraph33 using equation 2,
| (eq. 2) |
where mu and mf are the slopes of the upper (unfolded) and lower (folded) baselines, bu and bf are the y-intercepts of the upper and lower baselines, ΔH is the enthalpy of folding in kcal mol-1, and TM is the melting temperature in degrees Celsius. R is the gas constant of 0.001987 kcalK−1mol−1.
Temperature-Dependent In-line Probing and Data Analysis
The triphosphate on the 5′ end of FL tRNA was removed by incubation with calf intenstinal phosphatase (CIP) (NEB) at 37° C for 20 min, and the RNA was recovered by phenol/chloroform extraction and ethanol precipitation. This RNA was labeled on its 5′ end using [γ-32P]ATP and polynucleotide kinase (NEB), with incubation at 37°C for 30 min. Labeled RNA was purified by 10% PAGE followed by a crush and soak and ethanol precipitation procedure.
In-line probing (ILP) experiments on FL tRNA were performed at 12 temperatures between 35 and 75° C using a Biometra Tgradient thermocycler as an incubator. Before beginning ILP experiments, 1 μL of 500,000 cpm/μL 32P-labeled RNA was renatured with Tris buffer (pH 8.3) and KCl for 1 min at 95°C and cooled at room temperature for 10 min. Subsequently, MgCl2 and/or crowder were added, and the sample was heated at 55°C for 1 min and cooled at room temperature for 10 min. Final sample conditions were 20 mM Tris (pH 8.3), 140 mM KCl, 0.5 or 2.0 mM MgCl2, and 0 or 20% crowder; the slightly elevated pH aids RNA self-cleavage without alkaline denaturing the sample. The RNA was then incubated in the thermocycler at the appropriate temperature and aliquots were removed at a specified time. To achieve relatively even degradation across different temperatures, shorter time points were used for higher temperatures. At 35.0°C, 38.6°C, and 42.2°C, the 36 h time point was analyzed; at 45.8°C, 49.5°C, and 53.2°C, the 24 h time point was analyzed; at 56.8°C and 60.4°C, the 5 h time point was analyzed; at 64.0°C and 67.7°C, the 3 h time point was analyzed; and at 71.4°C and 75.0°C, the 1 h time point was analyzed. Time points were quenched with 2X formamide loading dye containing 50 mM Tris (pH 7.0) and 20 mM EDTA, which lowered the pH and sequestered Mg2+ ions. RNA was fractionated on 10% PAGE gels, which were visualized using a PhosphorImager.
Gel data were evaluated by semi-automated footprinting analysis (SAFA) software34 to provide reactivities of individual bands. The ILP reactivities output from SAFA were analyzed for percent reacted and corrected for loading differences by normalizing the raw data to nucleotides 34–36 in the anticodon loop, which were single stranded at all temperatures tested. Normalized ILP data were fit to the two-state melting model Marquardt algorithm for non-linear curve fitting (eq. 2)33 but were fit in IgorPro to allow the option of global fitting. When non-cooperative folding of FL tRNA was observed by ILP (e.g. in buffer with 0.5 mM Mg2+), non-global fitting of the nucleotide traces of each secondary structure was performed for separate TM’s. However, when cooperative folding of FL tRNA was observed by ILP (e.g. in solutions of 20% PEG200 with 0.5 and 2.0 mM Mg2+ and buffer with 2.0 mM Mg2+), global fitting of the base-paired nucleotides was performed to obtain a single TM for either a given helix or for FL tRNA unfolding, while slopes, y-intercepts and ΔH were still fit separately for each nucleotide.
SAXS Data Collection
FL tRNA was purified and precipitated as described above. RNA was buffer exchanged into 1X SAXS buffer (25 mM HEPES (pH 7.5) and 140 mM KCl) using an Amicon ultracentrifugal filter (3 kDa molecular weight cutoff). Stock solutions of PEG8000 and MgCl2 were prepared in the same 1X SAXS buffer to ensure buffer matching. Prior to data collection, the RNA was renatured in the presence of HEPES (pH 7.5) by heating at 95°C for 3 min and cooling at room temperature. After cooling, Mg2+ and PEG8000 were added to the solution, which was heated at 55°C for 3 min and cooled at room temperature for 10 min. Samples were centrifuged at 14k rpm for 10 min to minimize aggregation and remove dust particles. SAXS data were collected on G1 station at MacCHESS35,36—the solution scattering beamline at the Cornell High Energy Synchrotron Source (CHESS). The detector for data recording was a dual 100K-S SAXS/WAXS detector (Pilatus). The sample capillary-to-detector distance setup allowed for simultaneous collection of small- and wide-angle scattering data, covering a broad momentum-transfer range (q range) of 0.0075 – 0.8 Å−1 (q = 4πsin(θ)/λ, where 2θ is the scattering angle). The energy of the X-ray beam was 9.8528 keV (1.2548 Å) and the synchrotron X-ray beam diameter was 250 μm × 250 μm.
Data were collected for SAXS either by in-line size-exclusion (SEC) or hand mixing. SEC helps determine if aggregates are present at SAXS concentrations. The SEC SAXS data were collected on FL tRNA in buffer with 0.5 mM Mg2+ at 4° C. Samples were injected into a Shodex KW420.5-4F size-exclusion column using a GE HPLC (AKTApurifier) that routed sample through the column and into the BioSAXS flow cell where scattering images were collected. Sample flowed at a rate of 0.15 mL/min, and each frame was collected with 2 sec exposures. RNA elution was monitored by UV-vis detection in line with scattering detection. SEC-SAXS was not performed with PEG8000 because the X-ray beam causes the polymer to aggregate on the capillary tube walls and the viscous solution would result in high column pressure.20 No aggregation was observed at RNA concentrations injected at 0.2 mg/mL, which is diluted by ~10-fold during the SEC run.
For non-SEC samples, plugs of ~40–45 μL were delivered by hand to the quartz capillary tube, as the MacCHESS robot had trouble pipetting viscous samples with PEG. To test for sample aggregation in hand-loaded experiments, samples was prepared at 0.1, 0.15, 0.2, and 0.4 mg/mL RNA and 0, 0.5, or 2.0 mM Mg2+ with and without 20% PEG8000, and a buffer scattering curve was measured before and after each sample set. To ensure absence of PEG aggregation on the sample cell, buffer scans before and after sample acquisition were checked for matching. As with the SEC SAXS samples, only RNA concentrations of 0.2 mg/mL or lower are reported herein due to RNA aggregation.
To reduce radiation damage and known X-ray induced aggregation of PEG, a computer-controlled syringe pump was used to keep the hand-loaded sample oscillating in the X-ray beam. Ten scattering images were collected per sample. In the absence of PEG, we collected 1 sec exposures, while in the presence of PEG the exposure time was reduced to 0.5 sec and the oscillating rate was increased. In addition, after each PEG-containing sample was exposed to the X-ray beam, the sample cell was washed with several mLs of Hellmanax III solution, rinsed with several mLs of water and ethanol, and dried with forced air.
SAXS Data Analysis
Scattering curves were analyzed with BioXTAS RAW software.37 The collected scattering images were examined for signs of X-ray damage, which manifested itself as increases in signal at low q-range, and images containing non-damaged samples were averaged. The scattering curves of the buffer were subtracted from the scattering curves of the RNA. The linear region of the ln(I) vs q2 plot, wherein qmaxRg<1.3, was identified for each sample scattering curve using the Guinier analysis, and the radius of gyration (Rg) and molecular weight of the sample were determined. The pairwise distribution function in GNOM,38 using the ATSAS software package,39 was used to determine Rg using the Porod approximation and the maximum particle dimension (Dmax).
Bead models were created by putting the GNOM output file into the online server for DAMMIF.40 Twenty individual DAMMIF bead models were created per reaction condition and DAMCLUST39 was used to cluster individual DAMMIF models, which resulted in 2–3 groups of similar models that were used to assess the ambiguity of the reconstructions. For all experimental conditions, the DAMCLUST bead models were very similar to each other, so a consensus model was calculated by averaging all the DAMMIF models using DAMAVER.41 The output bead model from DAMAVER was aligned with the crystal structure of tRNAphe (PDB ID: 1ehz) using SUPCOMB39 and compared in PyMOL.42 As an additional check on our experimental bead models, we generated a theoretical bead model directly from a tRNA crystal structure (PDB ID: 1ehz), which was done by first generating a theoretical scattering curve from the crystal structure using FoXS software and then converting that scattering curve into a bead model. We then compared this theoretical bead model and its Rg and Dmax values with our experimental data.
Results and Discussion
We previously examined the folding of FL tRNA under solution conditions similar to typical in vitro environments, as well as solution conditions that emulate the cellular crowding and ionic conditions of the cell.22 In the presence of physiological Mg2+ concentrations, the RNA folded in a simple two-state manner with molecular crowding, but in a complex multi-state manner in dilute solution. Folding cooperativity, defined here as the simultaneous unfolding of tertiary and all secondary RNA structure, could arise from stabilization of tertiary structure, destabilization of secondary structures, or both. We test these models by mechanistic investigations of the helical fragments (HFs) that comprise the secondary structure of tRNA.
Effects of Mg2+ and Crowder are Similar for WT and FL tRNA
The folding properties of wild-type (WT), which has the natural modifications, and T7 transcribed full-length (FL) tRNAphe (Figure S1) were compared in different concentrations of Mg2+ and crowder in the background of physiological 140 mM K+ and 10 mM Na+. We used optical melting to compare the unfolding of WT and FL tRNA. Notably, WT and FL tRNA have the same tertiary structure and rate of aminoacylation.43
WT and FL tRNA have similar folding properties in buffer and crowded conditions at physiological Mg2+ (Figure S2). In buffer as the concentration of Mg2+ is increased from 0 to 2.0 mM, both RNAs fold more cooperatively. This is seen as sharpened melting transitions that occur at higher temperature, albeit with higher amplitudes for WT tRNA. In 20% PEG8000, similar behavior of WT and FL tRNA is observed, as indicated by the amplitudes and TMs of the transitions. Given the modest effect of nucleotide modifications on tRNA folding and function, we conducted studies on unmodified tRNA and its fragments.
Cooperative tRNA Folding Can be Induced by Mg2+-Driven Tertiary Structure Stabilization
To separate out the effects of physiological conditions on RNA secondary and tertiary structures, the FL tRNA and four secondary structure HFs that comprise tRNAphe—the acceptor stem, D SL, AC SL, and TΨC SL—were prepared as separate model RNA hairpins (Figure 1). Each HF is designed from the parent tRNA, with the exception of the acceptor stem, where a stretch of 8 Us was inserted into the loop to create a hairpin structure. In this and the following section, these HFs were melted in the presence of 140 mM K+ and 10 mM Na+ with either 0, 0.5, or 2.0 mM Mg2+, with or without various crowding agents, and absorbance curves were summed together as the “sum of the secondary structure fragments” or “SSS” (SSS) (see Materials and Methods), and the TMs reported for these species are apparent TMs. These are compared to the melting of FL tRNA under identical solution conditions. Melting temperatures are compiled in Tables S1–S3.
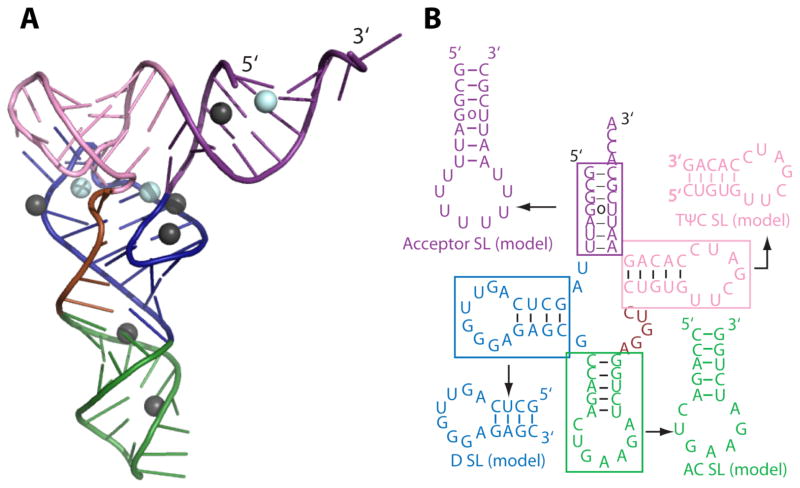
Figure 1.
Structures of FL yeast tRNAphe and its model helical fragments (HF) with their predicted folds. (A) Three-dimensional folded structure of yeast FL tRNAphe (PDB: 1ehz) and (B) FL tRNA secondary structure and the model HF derived from each stem. In panel (A) Mg2+ (cyan) and Mn2+ (black) ions associated with tRNA are shown as spheres. The colors of each structural element are the same in both panels. FL tRNA tertiary contacts are provided in Figure S1. The sequences of the model HF are the same as in FL tRNAphe and are predicted to form the shown hairpins by the Mfold server for each model sequence.
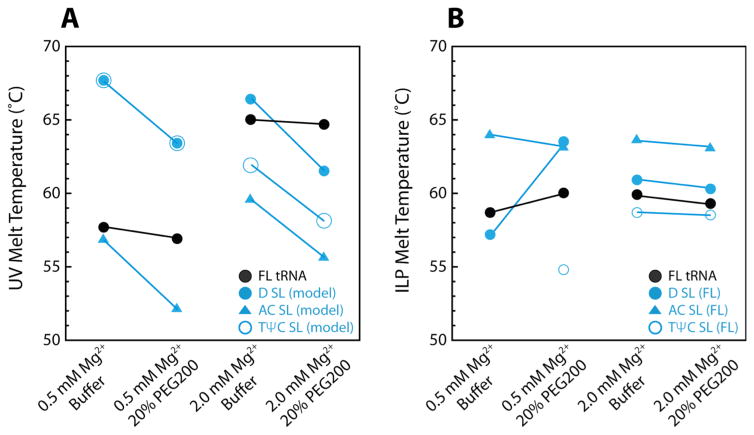
In dilute buffer-only conditions as the concentration of Mg2+ increased, FL tRNA folded cooperatively and at a higher TM, while secondary structure stability remained largely unchanged (Figure 2A). For instance, in buffer as the concentration of Mg2+ is increased from 0 to 2.0 mM, FL tRNA is stabilized by ~12 °C, yet SSS is stabilized by only ~4 °C (Table S1–S3). Because the cooperative transition encompasses both secondary and tertiary structure unfolding and because secondary structure is largely unchanged in stability, tertiary structure must be stabilized. In other words, Mg2+ is primarily stabilizing tertiary structure rather than secondary structure in buffer. The stabilization of RNA structures by Mg2+ in buffer is a well-studied phenomenon.44
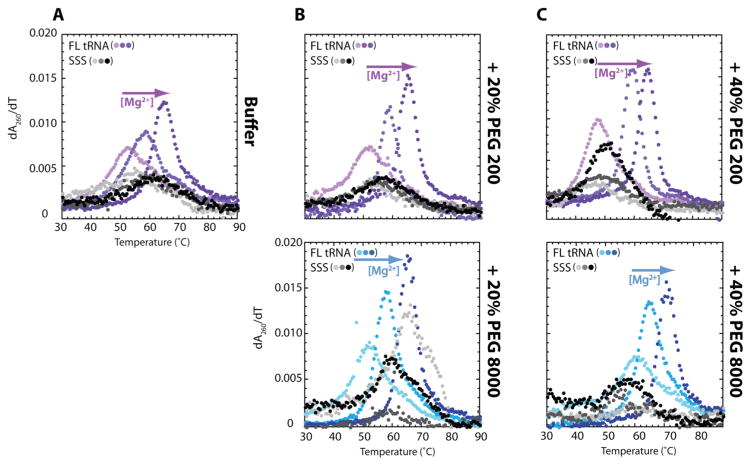
Figure 2.
First derivative curves of thermal denaturation experiments of FL tRNA and SSS under physiological ionic and crowded conditions. First derivative curves of thermal denaturation experiments in (A) 0% PEG200, (B) 20% PEG200 or PEG8000, and (C) 40% PEG200 or PEG8000 with increasing concentrations of Mg2+ from 0 to 0.5 to 2.0 mM (light to dark colors). The TM’s are provided in Tables S1, S2 and S3. Solutions contain a background of 10 mM sodium cacdoylate (pH 7.0) and 140 mM KCl.
This same phenomenon of Mg2+-driven tertiary structure stabilization holds in the background of 20 or 40% of crowder (Figure 2B, C). In particular, in 20% PEG200, the melting transition of SSS remained broad and the TM increased by only 4 °C, as the concentration of Mg2+ is increased from 0 to 2.0 mM (Figure 2B). Under the same crowded conditions, FL tRNA has a sharp transition in the derivative curve and an increase in thermostability of 12 °C upon raising the Mg2+ concentration. The large stabilization of FL tRNA compared to the secondary structures in cellular conditions indicates that Mg2+ stabilizes tertiary structure even in the presence of crowder. In 40% PEG200, similar effects are observed (Figure 2C). In sum, low millimolar concentrations of free Mg2+, like those found in vivo, strongly stabilize RNA tertiary structure under in vivo-like monovalent and crowding conditions.
To test if the effects are peculiar to PEG200, additional crowders were tested (Figure 2, 3, Tables S1–S3). In general, at 0 mM Mg2+, lower molecular weight crowders, such as PEG200 and PEG4000, tend to destabilize, or have little effect on, secondary and tertiary structures, and larger crowders, such as PEG8000 and PEG20000 tend to stabilize those structures (Figure 3A). These observations are unique to 0 mM Mg2+, which is non-physiological. We thus examined physiological Mg2+ and crowding effects. Similar to PEG200, cooperativity of FL tRNA is induced by the addition of Mg2+ in 20 and 40% PEG8000, while SSS stability is largely unaffected (Figure 2B, 2C). Furthermore, at 0.5 and 2.0 mM Mg2+ almost all crowding agents (PEG200, PEG4000, PEG8000, and PEG20000) increase the stability of FL tRNA, resulting in a higher melting temperature than in buffer and more cooperative folding (Figure 3B, 3C) indicating that the effect is not peculiar to one crowding agent.
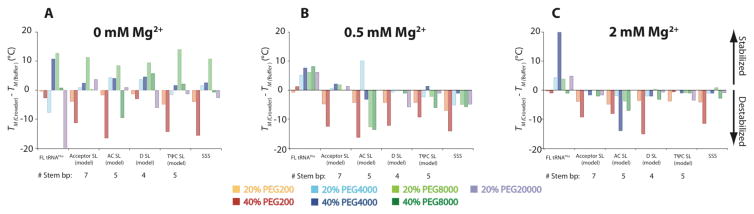
Figure 3.
Difference in TM of FL tRNA, SSS, and each of the four model HF in crowder compared to buffer. TM in crowder minus TM in buffer in (A) 0, (B) 0.5, and (C) 2.0 mM Mg2+. In all panels TM differences in 20% PEG200, 40% PEG200, 20% PEG4000, 40% PEG4000, 20% PEG8000, 40% PEG8000, and 20% PEG20000 are in orange, red, light blue, dark blue, light green, dark green, and purple, respectively. The TM’s are shown on the plot and in Tables S1, S2 and S3. Solutions contain a background of 10 mM sodium cacdoylate (pH 7.0) and 140 mM KCl.
Cooperative tRNA Folding Can be Induced by Crowder-Driven Secondary Structure Destabilization
The previous section found that Mg2+ can stabilize RNA tertiary structure in buffer conditions, a phenomenon that is well known,44 as well as in crowded conditions. Increasing Mg2+ up to physiological concentrations led to cooperative folding because the stability of the underlying secondary structural framework was largely unaffected by Mg2+. Another way to induce cooperative folding of RNA, at least in principle, is by weakening secondary structure. Ken Dill defines protein folding cooperativity as destabilization of folding intermediates.45 We therefore measured effects of various crowders on the folding of FL tRNA and the SSS in different background Mg2+ concentrations.
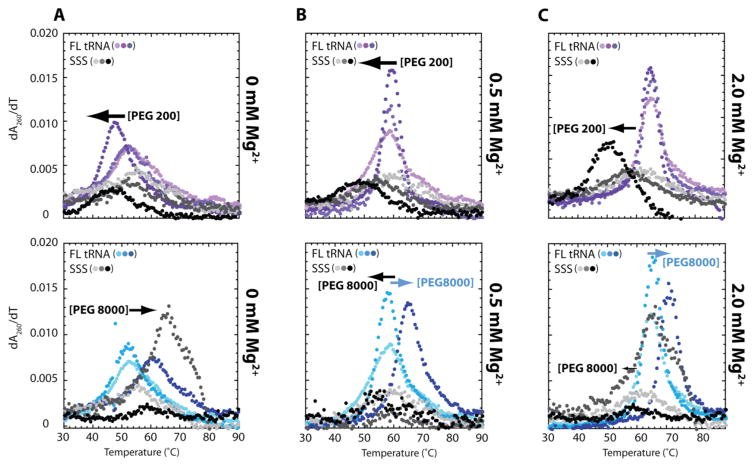
Increasing the amount of crowding agent at constant physiological Mg2+ induced more cooperative folding of FL tRNA (Figure 4A–C). Remarkably, the observed increase in FL tRNA cooperativity is achieved by decreasing the stability of the secondary structures (Figures 3, 4, Tables S1–S3). When the concentration of PEG200 is increased from 0 to 40%, the SSS TM decreased by almost 14 °C in 0.5 mM Mg2+ (Figure 4B) and 12 °C in 2 mM Mg2+ (Figure 4C), yet the overall structure stability is largely unchanged. Because the cooperative transition encompasses both secondary and tertiary structure unfolding and because secondary structure is destabilized, tertiary structure must be stabilized. Similar effects are observed when the concentration of PEG8000 is increased from 0 to 40% in 0.5 mM Mg2+ (Figure 4B) and 2.0 mM Mg2+ (Figure 4C), although destabilization of secondary structure is less dramatic. In 0.5 mM Mg2+, the addition of 20% PEG8000 decreased SSS TM by 5 °C, and addition of 40% PEG8000 decreased SSS TM by 6 °C. The weakening of SSS in PEG8000 is accompanied by an increase in overall structure stability and cooperative folding of FL tRNA. In 2.0 mM Mg2+, secondary structure is still destabilized by the addition of PEG8000, albeit less than that observed in 0.5 mM Mg2+, and tertiary structure is stabilized. PEG4000 and PEG20000 show similar effects (Figure 3). These findings suggest that the mechanism for cooperative folding changes with the molecular weight of the crowder. Crowders with low MW induce cooperativity mainly through secondary structure destabilization, while higher MW crowders induce cooperativity through both secondary structure destabilization and tertiary structure stabilization as depicted in Figure 4. A general destabilization of RNA secondary structure under in vivo conditions is consistent with genome-wide data comparing in vivo and in vitro data sets.17
Figure 4.
Cooperative folding can be induced through crowder-driven secondary structure destabilization. First-derivative curves of thermal denaturation experiments of FL tRNA and SSS with increasing concentrations of (upper row) PEG200 and (lower row) PEG8000 in (A) 0, (B) 0.5 and (C) 2.0 mM Mg2+, with increasing concentrations of PEG200 or PEG8000 from 0% to 20% to 40% (w/v). The TM’s are provided in Tables S1, S2 and S3. Solutions contain a background of 10 mM sodium cacdoylate (pH 7.0) and 140 mM KCl.
We were curious as to how crowding agents could act on specific secondary structures. We found that crowding agents have differing effects on stability of model HF depending on base-pair lengths. For instance, in 0.5 mM Mg2+ the short HF models of the anticodon (AC), D, and TΨC SL models are largely destabilized by the addition of 20 or 40% PEG200, PEG4000, PEG8000, and PEG20000 (Figure 3B). These SL models contain only 4 or 5 base pairs. On the contrary, the addition of these crowders, with the exception of PEG200, marginally stabilize the acceptor SL model, which is a longer hairpin comprised of 7 base pairs. Similarly, in 2.0 mM Mg2+, the addition of the above concentration of crowders destabilizes all HF (Figure 3C) and shorter SL structure are generally more affected, specifically the AC SL and D SL. Similar trends have been made using DNA and RNA model helices,25,26 in which cellular-like conditions have bigger effects for helices with fewer base pairs.
Cooperative and Non-Cooperative Folding are Observed on the Nucleotide Level
The previous two sections showed that cooperative folding of FL tRNA arises both from tertiary structure stabilization and secondary structure destabilization. According to these observations, in a cooperatively folding environment all the nucleotides involved in secondary structure and tertiary structure interactions should be unfolding over approximately the same temperature range. To determine if FL tRNA is folding in a two-state manner at the nucleotide level, we turned to a technique with nucleotide resolution. Temperature-dependent in-line probing (ILP) was carried out on 5′-end labeled FL tRNA in both buffer and model cytoplasms. FL tRNA was incubated at 12 temperatures between 35 and 75 °C, and the length of incubation at each temperature was chosen to achieve even RNA degradation across temperatures (see Materials and Methods).
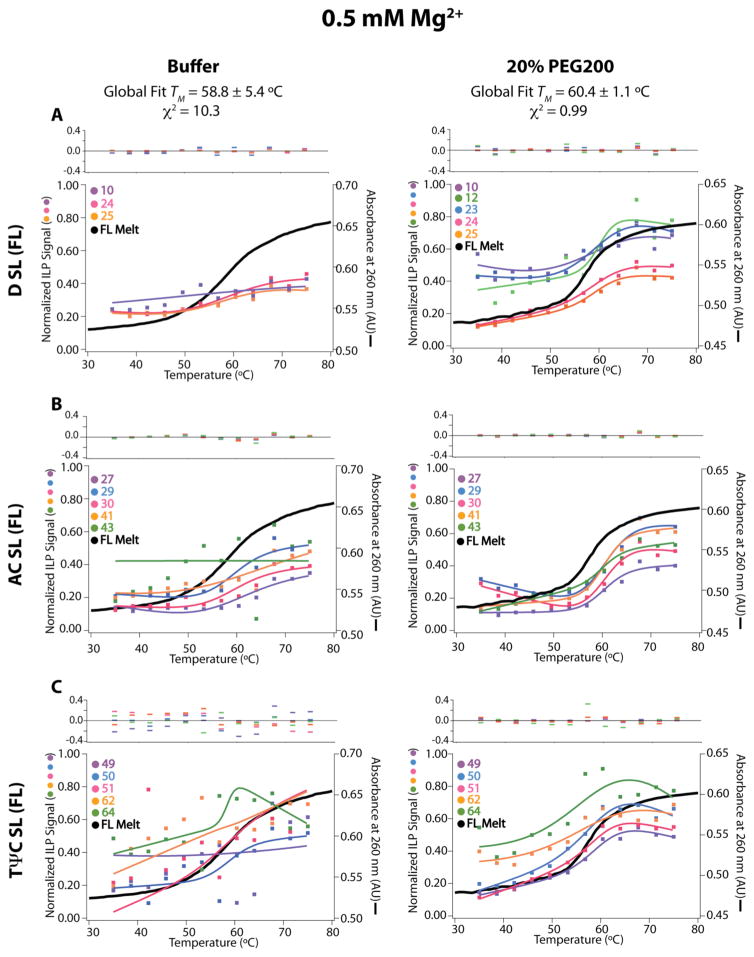
When fractionated on a sequencing gel, the normalized ILP reactivities patterned according to the cloverleaf structure of tRNA, with higher reactivity observed at single-stranded than double-stranded regions (Figure S3, S4). Normalized ILP reactivity for FL tRNA was plotted versus temperature and fit to a two-state unfolding model for individual nucleotides, helices (global fit), or FL tRNA (global fit) as appropriate at both 0.5 (Figures 5, 6) and 2.0 mM Mg2+ (Figures S5, S6).
Figure 5.
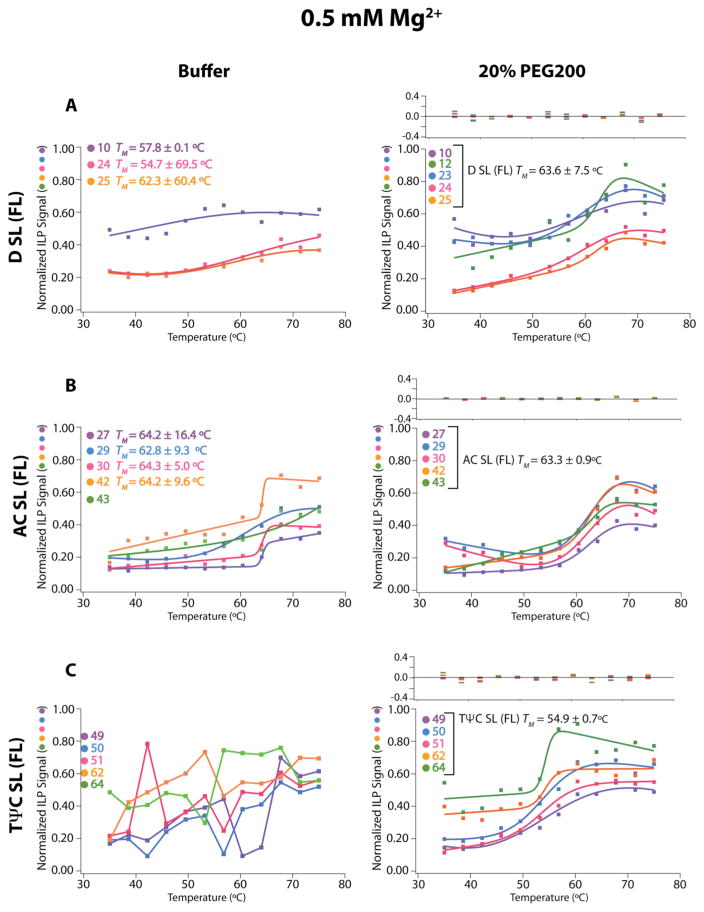
Nucleotide and helical stem fitting of temperature-dependent ILP data in buffer and 20% PEG200 with 0.5 mM Mg2+. Nucleotide fits (non-global fitting) and helical fits (globally fit for nucleotides shown with a bracket) were performed on buffer and 20% PEG200 samples, respectively, to obtain a TM for unfolding of each nucleotide or each stem in (A) D SL, (B) AC SL, and (C) TΨC SL. The TM values and residuals from the fits are provided in each figure and in Table 1. Fits could not be obtained in panel C in buffer; points are connected by lines to guide the eye.
Figure 6.
Global fitting of temperature-dependent ILP data in buffer and 20% PEG200 with 0.5 mM Mg2+. The same ILP data Figure (5) were fit globally across each stem of the (A) D SL, (B) AC SL, and (C) TΨC SL for a single TM of the RNA and to look for two-state behavior. The TM values and residuals from the fits are provided in each figure and in Table 1.
For FL tRNA in the background of 0.5 mM Mg2+ and uncrowded conditions, non-cooperative folding is observed at the nucleotide level (Figure 5). Some of the base-paired nucleotides could not be fit to standard melting equations. For those base-paired nucleotides exhibiting two-state unfolding behavior, we fit them to obtain a TM. Varying stability is observed within and between each HF at 0.5 mM Mg2+. For example, the D SL (FL), representing the D SL in the context of full length tRNA, unfolds between 55 °C and 62 °C (Figure 5A, LH; Table 1) and the AC SL (FL) unfolds between 63 °C and 64°C, while the TΨC SL is unstable. As might be expected, ILP data of different HF were fit poorly globally (Figure 6, LH), as indicated by a high error on the fit TM, a large χ2 of 10.3, and large residuals for the fit. Both the varying stability of the nucleotide fits and the large error on the global fit in 0.5 mM Mg2+ without crowder are indicative of non-cooperative folding.
Table 1.
Melting temperatures of FL tRNA and Secondary Structure Fragments as Determined by Optical Melting and ILP in Buffer and 20% PEG200 with 0.5 and 2.0 mM Mg2+.
| Structure Element |
TM (°C) Optical Melting |
TM (°C) ILP |
|---|---|---|
| Buffer with 0.5 mM Mg2+ | ||
|
| ||
| FL tRNA | 57.8 | 58.8 ± 5.4a |
| D Stem-Loop | (1)60.5, (2) 71.0 | 54.7–62.3b |
| AC Stem-Loop | 63.0 | 62.8–64.2b |
| TΨC Stem-Loop | 67.8 | -- |
|
| ||
| 20% PEG200 with 0.5 mM Mg2+ | ||
|
| ||
| FL tRNA | 57.0 | 60.4 ± 1.1a |
| D Stem-Loop | 63.5 | 63.6 ± 7.5c |
| AC Stem-Loop | 52.3 | 63.3 ± 0.9c |
| TΨC Stem-Loop | 63.5 | 54.9 ± 0.7c |
|
| ||
| Buffer with 2.0 mM Mg2+ | ||
|
| ||
| FL tRNA | 65.1 | 60.0 ± 0.6a |
| D Stem-Loop | 66.5 | 61.0 ± 0.9c |
| AC Stem-Loop | 59.7 | 63.8 ± 1.0c |
| TΨC Stem-Loop | 62.0 | 58.8 ± 1.0c |
|
| ||
| 20% PEG200 with 2.0 mM Mg2+ | ||
|
| ||
| FL tRNA | 64.8 | 59.4 ± 0.4a |
| D Stem-Loop | 61.6 | 60.4 ± 1.0c |
| AC Stem-Loop | 55.8 | 63.2 ± 0.7c |
| TΨC Stem-Loop | 58.2 | 58.6 ± 0.4c |
Solutions contain 10 mM sodium cacodylate (pH 7.0) and 140 mM KCl. There is an additional 0.5 or 2.0 mM MgCl2, and 0% or 20% PEG200 as indicated in the table.
Normalized ILP reactivities of all base-paired nucleotides were fit globally to find a single TM for the entire RNA (see Figs. 6 and S6).
Individual nucleotide TMs were obtained by fitting each base-paired nucleotide in each stem to obtain a single TM for each nucleotide (see Fig. 5, LH).
The parameters and conclusions made from optical melting of each HF and of each helix from ILP of FL in the background of 0.5 mM Mg2+ and uncrowded conditions largely agree with on another (Table 1). We consider the D SL, AC SL, and TΨC SL HFs in turn. In buffer at 0.5 mM Mg2+, two distinct transitions are observed using optical melting for the D SL (model), a low temperature transition at 60.5°C and a high temperature transition at 71.0 °C (Table 1); in agreement, two transitions were also observed for the D SL (FL) in the context of FL tRNA using ILP with nucleotide fitting: a low temperature transition between around 55 °C and a high temperature transition around 62 °C (Table 1, TFigure 5A). The AC SL (model) has an optical M of 63.0 °C and a similar ILP TM between 62.8 – 64.2 °C (Table 1, Figure 5A). From optical melting in buffer at 0.5 mM Mg2+ the TΨC SL (model) has a TM of 67.8 °C, while in the context of FL tRNA using ILP the entire RNA melts out below this transition (Figure 5C, LH). The wide range of optical melt TM’s of the model HFs and the poor global fit of the ILP data are supportive of non-cooperative folding in dilute and low Mg2+ conditions.
For FL tRNA in the same background of 0.5 mM Mg2+ but with 20% PEG200, cooperative folding is now observed at the nucleotide level. Global fitting of ILP data within each helical stem reveals that all HFs unfold with a single transition with TM’s clustered between 55 and 64 °C depending on HF (Figure 5, RH), which is similar to the single transition in optical melting transition for FL tRNA of 57.0 °C (Table 1). Furthermore, global fitting of ILP data across all stems reveals a single transition of 60.4 °C (Figure 6, RH). The good quality of this global fit of the data is evidenced by the low error of the fit, low residuals, and low χ2 of 0.99. Agreement in the TM between the helical level fits of stems and global fits, as well as their agreement with the optical melting TM of FL tRNA, strongly supports cooperative unfolding.
Next we consider 2.0 mM Mg2+ data. Optical melting studies on FL tRNA, showed that cooperative folding of FL is induced in buffer containing 2.0 mM Mg2+ (Figure 2A) and that the addition of a crowding agent increases the cooperative folding nature (Figure 2B, C, Figure 4). The same trend is observed with temperature-dependent ILP in these conditions. In both uncrowded and 20% PEG200 conditions in the background of 2.0 mM Mg2+, helical fitting of ILP data within each stem shows that each HF unfolds in a single transition with TM’s tightly clustered between 58.8 and 63.8 °C in the absence of PEG200 and between 58.6 and 63.2 °C in 20% PEG200 (Figure S5). The range of TM’s both with and without crowder is just ~5 °C showing that all secondary structures melt with similar TM’s and are very close to the optical melting temperature of FL tRNA in 2.0 mM Mg2+ without (65.1°C) and with 20% PEG200 (64.8°C) (Table 1). Global fitting of ILP data across all stems (for same TM) reveals that all stems can be well described with a single transition of 60.0 °C and 59.4 °C in buffer and 20% PEG200, respectively (Figure S6, Table 1). The residuals for the global fits in 2.0 mM Mg2+ in both uncrowded and crowded are low, indicating that the global fits represent the data well. These fits are also validated by the low error, the low residuals, and the low χ2 values of 0.33 and 0.63 for the global fits in 2.0 mM Mg2+ without and with PEG200.
The data in this section and the previous section are summarized in Figure 7. The stabilization of tertiary structure upon the addition of Mg2+ and the destabilization of secondary structure upon the addition of crowding agent is clear when the TMs obtained by UV or ILP melting are plotted versus solution conditions. As the concentration of PEG200 increases in the background of 0.5 or 2.0 mM Mg2+, the UV- or ILP-detected TM of FL tRNA is relatively unchanged, suggesting crowding is not affecting overall stability (Figure 7). Yet, when the model HFs are probed with optical melting dramatic destabilization of TM is observed upon the addition of crowding agent in 0.5 or 2.0 mM Mg2+, indicating that crowding destabilizes secondary structure (Figure 7A). In contrast, when ILP TM s are plotted, as conditions become more cooperative, the secondary structures have shifts in their TM closer to that of FL tRNA, indicative of cooperativity (Figure 7B). In other words, the various helices are stabilized by the presence of FL tRNA.
Figure 7.
Melting temperatures of FL tRNA and HF obtained by optical melting and ILP in buffer and crowded conditions. TM values were obtained by (A) optical melting on FL tRNA or the model HF, or by (B) ILP on FL tRNA, data from which were globally fit for a TM of either FL tRNA or the HF in FL tRNA. Blue closed and open circles overlapped in panel A left-hand side.
tRNAphe Adopts a More Compact Structure Under In Vivo-Like Conditions
The above sections provided evidence that both crowding agents and physiological concentrations of Mg2+ can induce cooperative folding of FL tRNA, which is observed on both the global and nucleotide levels. To assess tertiary structure directly, we used small angle X-ray scattering (SAXS). In-line size-exclusion chromatography (SEC) SAXS was collected on FL tRNA in buffer with 0.5 mM Mg2+ to determine if the RNA forms aggregates. (SEC SAXS could not be done with PEG; see Materials and Methods.) At FL tRNA concentrations of 0.4 and 0.6 mg/mL, two peaks were observed in both the absorbance- and scattering-detected SEC traces, indicating population of two distinct tRNA species (Figure S7). Molecular weight analysis in BioXTAS RAW indicated that the two species are a dimer (MW: 50 kDa) and a monomer (MW: 27 kDa; expected monomer MW of 25 kDa). At 0.2 mg/mL tRNA, the dimer peak disappeared in the scattering traces and is barely detectable in the absorbance traces, indicating that concentrations of ≤0.2 mg/mL are adequate concentrations to collect data in complex solution conditions.
Compaction of FL tRNA was observed upon an increase of Mg2+ or crowder in solution. This is evidenced by decreasing radius of gyration (Rg), Dmax, and excluded volume values as the concentration of Mg2+ or PEG increases (Figure S8, Table 2). As Mg2+ concentration increases from 0 to 0.5 to 2.0 mM, the Rg decreases from 31.9 to 25.0 to 24.2 Å respectively (Table 2), and the Dmax decreases from 112 to 87 to 82 Å, all indicating that the addition of Mg2+ compacts tRNA. A similar compaction is observed in the presence of a crowding agent. In 0.5 mM Mg2+ when 20% PEG8000 is added, Dmax decreases from 87 to 83 Å, and in 2.0 mM Mg2+ when 20% PEG8000 is added, both Rg decreases from 25.1 to 23.9 Å and Dmax decreases from 82 to 74 Å (Table 2). A theoretical scattering curve of the tRNA crystal structure was generated and found to have Rg and Dmax values of 23.1 and 82 Å, respectively, which are very similar to the values collected experimentally (Table 2). Using FoXS the theoretical and experimental scattering curves were overlaid (Figure S9A–C), and for all experimental conditions a low χ2 value was obtained indicating that the data fit well to the model.
Table 2.
Structural parameters obtained for FL tRNAphe using SAXS.
| Solution Condition | MWa (kDa) | Rga (Å) | Rgb (Å | Dmaxb (Å) | Excluded Volumec (Å3) |
|---|---|---|---|---|---|
| 0 mM MgCl2 | 32.2 | 31.9 | 33.8 | 112 | 63,700 |
| 0.5 mM MgCl2 | 28.9 | 25.0 | 26.1 | 87 | 55,400 |
| 2.0 mM MgCl2 | 28.5 | 24.2 | 25.1 | 82 | 47,700 |
| 0.5 mM MgCl2 + 20% PEG | 25.7 | 25.9 | 25.7 | 83 | 67,900 |
| 2.0 mM MgCl2 +20% PEG | 41.7 | 21.2 | 23.9 | 74 | 39,600 |
| 1ehz cyrstal structured | 25.9 | 23.1 | 25.2 | 82 | 47,000 |
Solutions contain 25 mM HEPES (pH 7.5), 140 mM KCl, and 0, 0.5, or 2.0 mM MgCl2. Samples containing 20% PEG8000 are indicated.
These parameters were obtained by analysis of the experimental scattering curves using BioXTAS RAW software. These values are reported in the text.
These parameters were obtained by analysis of the experimental scattering cruves using the pairwise distribution function in GNOM, using the ATSAS software package.
The excluded volume was found using the alignment program SUPCOMB when the DAMAVER bead models were aligned to the tRNA crystal structure (PDB ID: 1ehz).
The scattering curve for the tRNA crystal structure (PDB ID: 1ehz) was calculated using FoXS and processed similar to experimentally collected scattering data.
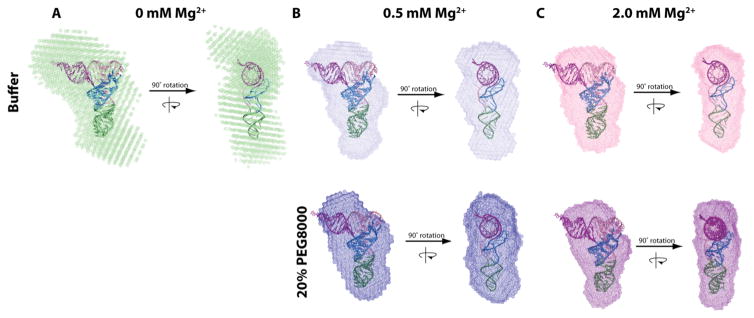
Next we generated bead models from SAXS data and compared them to the crystal structure of tRNA (Figure 8). All of the bead models align well to the crystal structure, as assessed by the RMSD and visual inspection of the alignment, with the exception of the bead model for tRNA in buffer with 0 and 0.5 mM Mg2+ in which tRNA is more extended than the crystal structure (Figure 8; Table S4). The bead models, as well as the above-discussed Rg and Dmax values, show a compaction of tRNA upon the addition of Mg2+ or crowder (Figure 8). A bead model was also generated with the theoretical scattering curve of the tRNA crystal structure (Figure S9D), and this bead model has a similar shape to the experimental bead models, which benchmarks our bead model approach.
Figure 8.
Physiological crowded and ionic conditions favor a compact folded state of FL tRNA. The tRNAphe crystal structure (PDB ID: 1ehz) was aligned with FL tRNA SAXS bead models in (A) 0, (B) 0.5, and (C) 2.0 mM Mg2+, without (top) and with (bottom) 20% PEG8000. The bead models were made using DAMMIF and DAMAVER and overlayed with the crystal structure using SUPCOMB.
The compaction of tRNA under in vivo-like conditions coincides with an increase in folding cooperativity under the same in vivo-like conditions. For instance, increasing the concentration of Mg2+ in the absence of crowder induces thermostability and more cooperative folding behavior of FL tRNA according to optical melts (Figure 2). These same changes in conditions also induce compaction of the RNA. For example, scattering experiments show that the addition of Mg2+ in the absence of crowder induces compaction of FL tRNA (Table 2, Figure 8). Similar agreement between optical melting and scattering experiments is found in the presence of 20% PEG200 (Figures 2 and 8). Similarities between folding cooperativity and compaction suggest that the excluded volume effect of the molecular crowders on FL tRNA help induce two-state folding to the more stabilized folded structure.
Conclusion
Our data indicate that there are two distinct mechanisms for inducing tRNAphe folding cooperativity. The first is a Mg2+-induced increase in tertiary structure stability. The second is a crowder-induced decrease in secondary structure stability. Under in vivo-like conditions, RNA folds cooperatively by a combination of these mechanisms. This study was performed on a single tRNA construct and thus additional studies will be needed to test the generality of these findings.
It is of interest to consider the chemical and biological importance, as well as the ultimate origin, of such two-state-like folding of functional RNA. Strong tertiary structure can hold together intrinsically unstable secondary structures (Figure 7). This has chemical importance because it can assemble the secondary structural framework in a way that creates binding pockets for ligands and active sites for catalysts. Weakening of secondary structures, observed in the presence of crowders, may assist the search for a native tertiary structure by smoothing the RNA folding funnel. For proper function, RNAs need to fold into the correct structures on a biologically relevant time scale. Several studies in vitro under high ionic conditions used to fold RNA, have shown that misfolded states can last from minutes to hours.46,47 These misfolds can have free energy values that are very similar to the native state and structural changes to reach the native state can be very large involving breaking of bonds.48 Weakened secondary structures, like those that occur under in vivo-like conditions, should help smooth out the folding landscape and lead to faster adoption of the native state.
There is possible biological relevance of the helical fragments and conditions studied herein. Fragments of fully processed tRNAs regulate protein coding genes, RNA metabolism, and RNA interference in prokaryotes and eukaryotes during stress response49 suggesting that there may be direct biological relevance of understanding the folding of tRNA fragments. In addition, some organisms, such as plants, can increase the concentrations of monovalent ions and compatible cosolutes dramatically during abiotic stress, which could aid RNA folding.50,51
RNA and proteins have starkly different chemical makeups and intermolecular forces. This has led to the belief that they fold differently. Indeed, unfolding of small functional RNAs has been widely regarded as non-cooperative, with weak tertiary structure unfolding before strong secondary structure,24 while proteins fold in a two-state manner and have weak secondary structure and strong tertiary structure.45 However, our work suggests that RNA and proteins have similar folding properties under cellular conditions. We show that, like their protein counterparts, small RNAs unfold cooperatively under cellular conditions and that this is driven by weakened secondary structural elements and strengthened tertiary structure. These findings suggest that evolutionary forces may drive biopolymers, regardless of their chemical composition, to fold in a cooperative fashion.
Supplementary Material
Acknowledgments
We thank Dr. Richard Gillian and Dr. Jesse Hopkins for help with small angle X-ray scattering experiments. This work was supported by U. S. National Institutes of Health Grant R01-GM110237 (P. C. B). This work is based on research conducted at the Cornell High Energy Synchrotron Sourse (CHESS), which is supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences under NSF award DMR-0936384, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by GM-103485 from the National Institutes of Health, through its National Institute of General Medical Sciences. We thank Erica Frankel for her help with global fitting of ILP data using IgorPro. We thank Elizabeth Whitman for collecting preliminary data on the RNA hairpins.
Footnotes
The Supporting Information is available free of charge on the ACS Publications website at DOI: XXX. Supplemental tables are available on melting temperatures of tRNA, individual helical fragments, and sum of secondary structures, as well as SAXS parameters. Supplemental figures are available showing tRNA modifications, folding of WT modified tRNA, and in-line probing of FL tRNAphe, as well as 2 mM Mg2+ analysis of ILP data, and supporting SAXS figures.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 4.Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophil ribozyme. 1 Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 5.Tanner MA, Cech TR. Activity and thermostability of the small self-splicing group I intron in the pre-tRNAIle of the purple bacterium Azoarcus. RNA. 1996;2:74–83. [PMC free article] [PubMed] [Google Scholar]
- 6.Feig AL, Uhlenbeck OC. The role of metal ions in RNA biochemistry. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. pp. 287–320. [Google Scholar]
- 7.Lusk JE, Williams RJ, Kennedy EP. Magnesium and the growth of Escherichia Coli. J Biol Chem. 1968;243:2618–2624. [PubMed] [Google Scholar]
- 8.Truong DM, Sidote DJ, Russell R, Lambowitz AM. Enhanced group II intron retrohoming in magnesium-deficient Escherichia coli via selection of mutations in the ribozyme core. Proc Natl Acad Sci USA. 2013;110:E3800–E3809. doi: 10.1073/pnas.1315742110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alberts B, Bray D, Lewis J, Roberts K, Watson JD. Molecular biology of the cell. 3 1994. [Google Scholar]
- 10.London RE. Methods for measurement of intracellular magnesium: NMR and fluorescence. Annu Rev Physiol. 1991;53:241–258. doi: 10.1146/annurev.ph.53.030191.001325. [DOI] [PubMed] [Google Scholar]
- 11.Romani AM. Magnesium homeostasis in mammalian cells. Front Biosci. 2007;12:308–331. doi: 10.2741/2066. [DOI] [PubMed] [Google Scholar]
- 12.Nagata S, Adachi K, Shirai K, Sano H. 23Na NMR spectroscopy of free Na+ in the halotolerant bacterium Brevibacterium sp. and Escherichia coli. Microbiology (Reading, England) 1995;141(Pt 3):729–736. doi: 10.1099/13500872-141-3-729. [DOI] [PubMed] [Google Scholar]
- 13.Hirota N, Imae Y. Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J Biol Chem. 1983;258:10577–10581. [PubMed] [Google Scholar]
- 14.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strulson CA, Molden RC, Keating CD, Bevilacqua PC. RNA catalysis through compartmentalization. Nat Chem. 2012;4:941–946. doi: 10.1038/nchem.1466. [DOI] [PubMed] [Google Scholar]
- 16.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. 35. [DOI] [PubMed] [Google Scholar]
- 17.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X, Bartel DP. Widespread Influence of 3′-End Structures on Mammalian mRNA Processing and Stability. Cell. 2017;169:905–917. doi: 10.1016/j.cell.2017.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular envirnment stabilized adenine riboswitch RNA structure. Biochemistry. 2013;52:8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilburn D, Roh JH, Behrouzi R, Briber RM, Woodson SA. Crowders perturb the entropy of RNA energy landscapes to favor folding. J Am Chem Soc. 2013;135:10055–10063. doi: 10.1021/ja4030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strulson CA, Yennawar NH, Rambo RP, Bevilacqua PC. Molecular crowding favors reactivity of a human ribozyme under physiological ionic conditions. Biochemistry. 2013;52:8187–8197. doi: 10.1021/bi400816s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strulson CA, Boyer JA, Whitman EE, Bevilacqua PC. Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions. RNA. 2014;20:331–347. doi: 10.1261/rna.042747.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 24.Tinoco IJ, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 25.Nakano S-i, Karimata HT, Ohmichi T, Kawakami J, Sugimoto N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J Am Chem Soc. 2004;126:14330–14331. doi: 10.1021/ja0463029. [DOI] [PubMed] [Google Scholar]
- 26.Nakano S-i, Karimata HT, Kitagawa Y, Sugimoto N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J Am Chem Soc. 2009;131:16881–16888. doi: 10.1021/ja9066628. [DOI] [PubMed] [Google Scholar]
- 27.Lambert D, Draper DE. Effects of osmolytes on RNA secondary and tertiary structure stabilities and RNA-Mg2+ ion interactions. J Mol Biol. 2007;370:993–1005. doi: 10.1016/j.jmb.2007.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. J Am Chem Soc. 2008;130:6085–6087. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behrouzi R, Roh JH, Kilburn D, Briber RM, Woodson SA. Cooperative tertiary interaction network guides RNA folding. Cell. 2012;149:348–357. doi: 10.1016/j.cell.2012.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J Am Chem Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. Determination of in vivo RNA structure in low-abundance transcripts. Nat Commun. 2013;4:3971–3983. doi: 10.1038/ncomms3971. [DOI] [PubMed] [Google Scholar]
- 32.Leamy KA, Assmann SM, Mathews DH, Bevilacqua PC. Bridging the gap between in vitro and in vivo RNA folding. Q Rev Biophys. 2016;49:e10–e36. doi: 10.1017/S003358351600007X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegfried NA, Bevilacqua PC. Thinking inside the box: designing, implementing, and interpreting thermodynamic cycles to dissect cooperativity in RNA and DNA folding. Methods Enzymol. 2009;455:365–393. doi: 10.1016/S0076-6879(08)04213-4. 36. [DOI] [PubMed] [Google Scholar]
- 34.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acerbo AS, Cook MJ, Gillilan RE. Upgrade of MacCHESS facility for X-ray scattering of biological macromolecules in solution. J Synchrotron Radiat. 2015;22:180–186. doi: 10.1107/S1600577514020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skou S, Gillilan RE, Ando N. Synchrotron-based small-angle X-ray scattering (SAXS) of proteins in solution. Nat Protoc. 2014;9:1727–1739. doi: 10.1038/nprot.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen SS, Toft KN, Snakenborg D, Jeppesen MG, Jacobsen JK, Vestergaard B, Kutter JP, Arleth L. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J Appl Crystallogr. 2009;42:959–964. [Google Scholar]
- 38.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 39.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeLano WL. The PyMOL Molecular Graphics System 2002 [Google Scholar]
- 43.Hall KB, Sampson JR, Uhlenbeck OC, Redfield AG. Structure of an unmodified tRNA molecule. Biochemistry. 1989;28:5794–5801. doi: 10.1021/bi00440a014. [DOI] [PubMed] [Google Scholar]
- 44.Stein A, Crothers DM. Conformational changes of transfer RNA. The role of magnesium(II) Biochemistry. 1976;15:160–168. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- 45.Dill KA, Bromberg S, Yue K, Fiebig KM, Yee DP, Thomas PD, Chan HS. Principles of protein folding--a perspective from simple exact models. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Banerjee AR, Turner DH. The time dependence of chemical modification reveals slow steps in the folding of a Group I ribozyme. Biochemistry. 1995;34:6504–6512. doi: 10.1021/bi00019a031. [DOI] [PubMed] [Google Scholar]
- 47.Chadalavada DM, Senchak SE, Bevilacqua PC. The folding pathway of the genomic hepatitis delta virus ribozyme is dominated by slow folding of the pseudoknots. J Mol Biol. 2002;317:559–575. doi: 10.1006/jmbi.2002.5434. [DOI] [PubMed] [Google Scholar]
- 48.Mitchell DI, Jarmoskaite I, Seval N, Seifert S, Russell R. The long-range P3 helix of the Tetrahymena ribozyme is disrupted during folding between the native and misfolded conformations. J Mol Biol. 2013;425:2670–2686. doi: 10.1016/j.jmb.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–1806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greenway H, Munns R. Mechanisms of salt tolerance in nonhalophytes. Annu Rev Plant Physiol. 1980;31:149–190. [Google Scholar]
- 51.Handa S, Bressan RA, Handa AK, Carpita NC, Hasegawa PM. Solutes contributing to osmotic adjustment in cultured plant cells adapted to water stress. Plant Physiol. 1983;73:834–843. doi: 10.1104/pp.73.3.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.