Abstract
At metaphase in mitotic cells, pulling forces at the kinetochore-microtubule interface create tension by stretching the centromeric chromatin between oppositely oriented sister kinetochores. This tension is important for stabilizing the end-on kinetochore microtubule attachment required for proper bi-orientation of sister chromosomes as well as for satisfaction of the Spindle Assembly Checkpoint and entry into anaphase. How force is coupled by proteins to kinetochore microtubules and resisted by centromere stretch is becoming better understood as many of the proteins involved have been identified. Recent application of genetically encoded fluorescent tension sensors within the mechanical linkage between the centromere and kinetochore microtubules are beginning to reveal – from live cell assays – protein specific contributions that are functionally important.
Keywords: centromeric chromatin, chromosome segregation, kinetochore, microtubules, mitosis, tension sensor
Introduction
Eukaryotic kinetochores have a highly conserved molecular architecture. A modified histone H3, CENP-A, within nucleosomes at the periphery of centromeric chromatin marks the site of kinetochore assembly. CENP-A recruits Constitutive Centromere Associated Protein Network (CCAN in human, COMA in yeast) to form the inner kinetochore and provide protein linkers to outer kinetochore proteins (budding yeast kinetochore Fig. 1A). Two important CCAN linkers are CENP-C (Mif2, Fig. 1A) and CENP-T (Cnn1) because their C-terminal ends are bound to chromatin and their N-terminal ends bind to the outer domain KMN network of proteins [1–5]. The KMN network includes the Knl1 complex (Knl1 and Zwint-1 human, Spc105 and Ydr532 yeast), the ~25 nm long Mis12/MIND complex (Mis12, Nsl1, Nnf1, and Dsn1, Fig. 1A) and the ~60 nm long Ndc80 complex (Ndc80, Nuf2, Spc24, Spc25) (Fig. 1B) (reviewed in [6]). A fraction of CENP-C binds the Mis12/MIND complex which is linked to the inner ends of the Knl1 and Ndc80 complexes, which is Spc24/Spc25 for the Ndc80 complex [3, 7]. In some cell types, CENP-T attaches the Ndc80 complex to kinetochores in two ways: one by directly binding Spc24/Spc25, and second by binding the inner end of the Mis12 complex [4–6, 8–11]. The Ndc80 complex (Ndc80c) has several critical functions. These include attachment to the plus ends of spindle microtubules to form kinetochore microtubules (kMTs) that mechanically link chromosomes to their spindle poles, force production coupled to kMT plus-end polymerization and depolymerization [12, 13], control of signaling by the spindle assembly checkpoint (SAC) through regulation of Knl1c activity [14, 15], and correction of errors in kMT attachment that prevent inaccurate chromosome segregation in anaphase [6] (Fig. 1A).
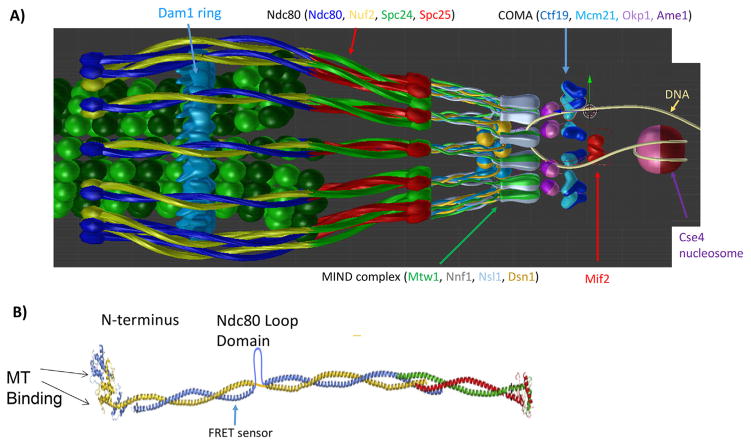
Figure 1.
Structural organization of the major kinetochore constituents at the microtubule plus-end. 3-D visualization of the metaphase budding yeast kinetochore-microtubule attachment as predicted by the protein localization data assuming a symmetric arrangement of kinetochore protein complexes around the cylindrical microtubule lattice. Ndc80c shown as blue/yellow and red/green coiled-coils in A and B with more detail. A: The MIND complex (Mtw1, Nnf1, Nsl1 and Dsn1) (gray/green ovoids and yellow/blue balls) provide the linkage from Ndc80c to COMA. COMA (Ctf19, Okp1, Mcm21, and Ame1) dk and lt blue triangles and purple spheres. Mif2 is depicted in red, the Cse4 containing nucleosome in shades of purple wrapped by a DNA double strand (yellow fiber). B: Structural view of Ndc80c, including the Ndc80 loop domain and the site of insertion of the FRET biosensor [53].
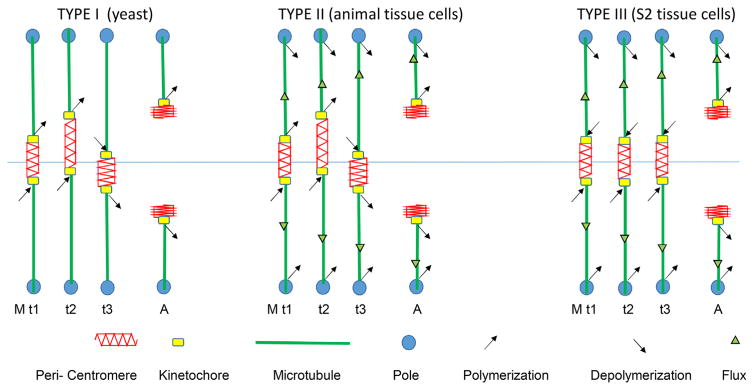
Pulling forces at kinetochores at metaphase are mainly resisted by the stretch of the chromatin between opposite facing kinetochores. Kinetochore pulling force depends on whether kMTs are in the polymerization or depolymerization phases of dynamic instability and whether spindle poleward microtubule flux exists [16, 17]. Kinetochore pulling force can also be “active” or “passive” [17–20]. Figure 2 shows examples of kinetochore, chromosome and microtubule dynamics in three different cell types at metaphase and shortly after anaphase chromosome separation. In Type I, kinetochores exhibit switching between persistent polymerization and depolymerization phases of their kMT dynamic instability that cause chromosome oscillation between the poles at meta-phase. During polymerization phases, kinetochores passively resist the tension of centromere stretch. There is no poleward flux. In Type II (typical of animal cells), at metaphase kinetochores exhibit directional instability as in Type I and they are also steadily pulled poleward by kMT poleward flux. At anaphase with loss of tension from centromere stretch, kMTs persist in depolymerization and their separated chromosomes are pulled poleward both by Pacman activity at kinetochores and poleward flux of their kMTs. In Type III, kinetochores at metaphase persist in polymerization driven by poleward microtubule flux. Pulling force for centromere stretch is produced by “passive” resistance to the sliding of the microtubule lattice through the kinetochore attachment site. In anaphase with the loss of centromere tension, kinetochores tend to switch to active force generation coupled to depolymerization (PacMan) [21], or remain in passive force generation, but with much slower polymerization [16, 22].
Figure 2.
Three different types of kinetochore microtubule and centromere stretch dynamics in metaphase (M) and anaphase A (A) exhibited by different kinds of mitotic or meiotic eukaryotic cells. Type I: No flux of kinetochore microtubules that extend between the kinetochore and the pole; kMTs lengthen and shorten at their kinetochore-attached plus ends (kinetochore directional instability), e.g. budding yeast. Type II: Flux of kinetochore microtubules and kinetochore directional instability, e.g. animal tissue cells. Type III: Flux of kinetochore microtubules coupled to polymerization at kinetochores until loss of tension in anaphase causes a switch to depolymerization or polymerization at kinetochores much slower than flux [42]); kinetochore oscillations are minimized, e.g. spermatocytes, oocytes, early embryos, and higher plants, Xenopus extract spindles, and Drosophila S2 tissue cells. The kinetochores (yellow box) are relatively stiff in metaphase.
Understanding force generation at kinetochores requires learning how active and passive force generation are coupled to deformation within the kinetochore itself, coupling to depolymerizing and polymerizing kMT ends respectively, and how chromatin between opposing kinetochores is made sufficiently stiff to produce the tension needed to drag kinetochores away from their poles and maintain the tension needed for control of kMT attachment and the spindle assembly checkpoint. The kinetochore contains minimally passive and active sites of force generation [18], and intra-kinetochore deformations can be observed through fluorescence co-localization with nanometer accuracy [14, 23–26]. At metaphase, after kMT formation and chromosome bi-orientation at the spindle equator, the protein linkage from the inner kinetochore to the MT lattice at the ends of kMTs becomes extended along the MT axis, and this linkage appears mechanically stiff in comparison to centromeric chromatin in between sister kinetochores [23, 26, 27]. However, changes in the deformation (intrakinetochore stretch) within the kinetochore protein linkage to the ends of kMTs on the order of 10–20 nm have been observed by measurements of fluorescence co-localization with nanometer accuracy between passive and active kinetochores [18], suppression of MT dynamics with taxol [27–29], and the loss of centromere tension at anaphase onset [23]. Whether these deformations represent changes in stretch of the linker molecules or their disorientation from the microtubule axis is currently controversial [25].
In contrast to the stiff kinetochore at metaphase, centromeric chromatin is compliant and heterogeneous in its mechanical properties. The tensile strength of centromeric chromatin is dictated in large part through SMC (structural maintenance of chromosome) proteins, cohesin and condensin. Cohesin and condensin are spatially segregated in the centromere and function to build a dense array of pericentromere chromatin loops in order to stiffen the linkage between sister kinetochores and provide resistance to the opposing force from kinetochore and spindle microtubules [30–32]. How the centromere spring is built, and is responsive to constant fluctuations in MT growth and shortening is starting to be understood.
In this essay we first review the pioneering kinetochore force measurements of Bruce Nicklas using calibrated microneedles in spermatocyte cells (Type III), then compare recent values for metaphase kinetochore force in a Type III tissue cell Drosophila S2, using fluorescent tension sensors integrated into the CENP-C linker to the Ndc80 complex. We then turn to measurements with a fluorescent tension sensor within the Ndc80 complex near its MT binding domains for mitotic budding yeast (Type I cell) that yield estimates for force at that site, but more importantly, provide data showing load sharing by other MT binding proteins that also bind Ndc80 inside of the tension sensor. Finally, we look at how fluorescent sensors in budding yeast are advancing knowledge of how the dynamic structure of centromeric chromatin generates stiffness and how crosslinkers, like cohesin and condensin make this chromatin very mechanically anisotropic.
Pioneering measurements of the magnitude of force at kinetochores
The founding experiments demonstrating the elastic properties of chromosomes were micromanipulation experiments of R.B. Nicklas on grasshopper spermatocytes [33, 34]. The maximum amount of force the spindle can generate at a kinetochore was determined by the amount of external force required to stop chromosome movement. Because the cell cortex offered no resistance in these spermatocytes, Nicklas was able to pull on chromosome arms and measure their extensions within dividing cells using microneedles from the outside. Chromosome extension was measured with a range of four force-calibrated needles. The elastic modulus of chromosome elasticity was derived from the equation EA = F l0/lΔ. The elastic modulus (Young’s modulus E) × cross-sectional area of chromosome = Force × (total length l0/change in length lΔ). Nicklas found that 700 pN was required to stall chromosome movement [35, 36]. There are 45 microtubules on average per kinetochore in these grasshopper cells. Nicklas estimated that only 1/3 of these extended from the kinetochore to the pole, and thus divided the total MT number by 3. We now know that microtubules can be cross-linked within the spindle and do not have to extend to the pole to generate force at the kinetochore [37, 38]. If we recalculate using 45 microtubules instead of the 15 he used, each kMT can exert ~15 pN of force, about a third of the 45 pN/kMT he calculated [35, 36]. How much force is actually required to segregate a chromosome in anaphase? To calculate this, one needs to estimate the drag force, fdrag = viscosity × shape × velocity (Stokes’ law). The force for chromosome segregation of a large Melanoplus chromosome is on the order of 0.2–0.5 pN [34], much smaller than the stall force [35, 36].
Estimate of metazoan kinetochore tension force at metaphase
To obtain the magnitude of tension at a multiple microtubule-attached kinetochore, Ye et al. [29] introduced a FRET sensor (TSMod) in the intrinsically disordered region of CENP-C in a Drosophila S2 cell line. In Drosophila, CENP-C currently appears to be the sole linkage between the inner kinetochore and the Ndc80 complex [3]. TSMod in vivo consists of genetically encoded fluorophores, mTurquoise2 and mVenus linked together by an unstructured amino acid chain that behaves like an entropic spring [39]. The average force per molecule was estimated to be 1–2 pN at bioriented kinetochores. In this and other studies with TSMod, the change in distance between fluorophores is converted to force based on in vitro calibration of a sensor with organic cyanine dyes, Cy3 and Cy5 [40, 41]. For the in vitro studies, Cy5 and Cy3 dyes were covalently attached to DNA that in turn was conjugated to the substrate and bead, respectively. The difference in linkages and dyes introduce uncertainties in how well the two force extension curves faithfully represent the correspondence between distance and force over a range of low to high extension. In addition the calibration using organic dyes does not consider the contribution of the 16 flexible amino acids as well as the physical size of the two fluorescent proteins and their relative orientation. To address this problem, Ye et al. [29] also measured force with a non-FRET fluorescent tension sensor inserted into CENP-C constructed of focal adhesion components talin and vinculin. Results from this assay also yielded 1–2 pN per CENP-C. The finding that both methods yielded comparable estimates of force is a strong validation of the approach and represents an important contribution in the field.
Ye et al. calculated a force of 12–62 pN per kMT and 144–764 pN/kinetochore based on their force estimates within CENP-C of 1–2 pN, 12–31 CENP-C (and Ndc80c) per kMT, and 12 kMTs per kinetochore in metaphase Drosophila S2 cells (33). These numbers are of the same order of magnitude as measured by Nicklas for kinetochores in meiotic spermatocytes as discussed previously, but their accuracy waits a direct measurement for the number of CENP-C per kinetochore.
Note, metaphase Drosophila S2 cells, like the spermatocyte cells have Type III spindles (Fig. 2) where kinetochore force on chromatin at metaphase is produced by resistance to poleward microtubule flux as their kMTs polymerize at the rate of flux [17, 42, 43]. We will consider next, kinetochores in budding yeast which like in mammalian cells (Type II, Fig. 2), exhibit directional instability and oscillations in the stretch of the centromere between sister kinetochores at metaphase, but unlike mammalian cells, have no poleward microtubule flux (Type I, Fig. 2) [44, 45].
Introduction of a live cell tension sensor in the Ndc80 microtubule binding complex
In budding yeast, the Ndc80 complex (Ndc80c, Fig. 1B) transmits force from an attached kMT to the inner centromere mainly through linkage of the C-terminal end of Spc24/Spc25 to the Mis12/MIND complex, which is bound to CCAN proteins Mif2 (CENP-C) and the COMA Complex (Ctf19, Okp1, Mcm21, Ame1, Chl4 (CENP-N), Iml3) [6]. At metaphase, each kinetochore binds one kMT and sister kinetochores become separated apart to opposite sides of the spindle equator where they exhibit directional instability as their kMT plus ends switch between persistent polymerization and depolymerization phases (Fig. 2, Type I). The number of kinetochore Ndc80 complexes per kMT has been estimated between 8 and 17 [46–49]. The Ndc80 complex has two MT binding domains [50–55]. One is the unstructured N-terminal tail of Ndc80 that binds the C-terminal tails of tubulin within MTs except when phosphorylated by Aurora B kinase to promote kMT detachment and to correct connections to the wrong pole [56, 57]. The second is the calponin homology (CH) domain in Ndc80 near the N-terminal tail, which is the primary Ndc80 MT binding domain.
Ndc80c also attaches to kMTs through two other microtubule binding proteins. Ndc80c recruits to kinetochores with attached kMTs, 1–2 Dam1 ring complexes and Stu2, a homolog of the TOG, XMAP215 family of MT binding proteins. Just where DAM1 ring(s) binds Ndc80c in vivo has been uncertain, but based on in vitro binding studies and in vivo protein mutation assays, there are multiple interaction sites that involve a domain just inside of the CH domains of Ndc80 and Nuf2, a domain near the loop domain in the middle of the Ndc80 complex and a domain further inside the loop domain (Fig. 1) [50–54]. Stu2 is proposed to bind the Ndc80 complex inside of the Ndc80 loop domain [55].
To understand how the N-terminal MT binding domains of Ndc80 contribute to kinetochore force, Suzuki et al. [58] inserted a FRET tension sensor (mYPet-28 unstructured amino acids-mCFP) into Ndc80 at amino acid 410, which is normally located within the alpha helical coiled-coil domain of the Ndc80/Nuf2 dimer, about 10 nm outside of the Ndc80 loop domain and toward the Ndc80 CH domain (Fig. 1B). Insertion of the FRET tension sensor at this site did not perturb the progression from prometaphase to anaphase in time-lapse imaging [58]. This site was the only one (of several tried) to accommodate the tension sensor and lies within a region found independently to be non-lethal to transposon mutagenesis [59]. The structural constraints of the coiled-coil heterodimer between Ndc80 and Nuf2 do not seem to be structurally inhibitory at this site. This feature of coiled-coils is not unprecedented, as the alpha-helical coiled coils of cytoplasmic dynein shift on the order of several nanometers during each power stroke [60].
Estimation of tension force at the Ndc80 MT binding domains of budding yeast at metaphase
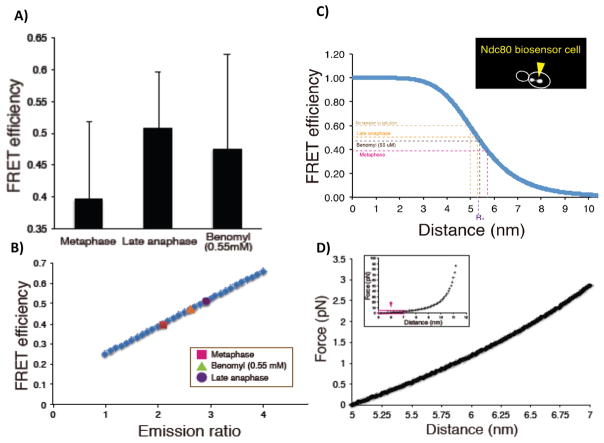
Suzuki et al. [58] calculated the absolute distance between the donor and acceptor fluorophores using mean values from FRET efficiency measurements (1 – mECFP channel emissionNdc80 biosensor/mECFP channel emissionNdc80 mECFP (410)). The FRET efficiency was 0.40 ± 0.12 (n = 69) in metaphase, 0.51 ± 0.09 (n = 54) in late anaphase, 0.45 (n = 118) in interphase, and 0.64 for the FRET probe attached to the C-terminus of Nuf2 (zero tension) (Fig. 3A and Table 1; adapted from [58]). These values for FRET efficiency were directly proportional to measured emission ratio values (FRET emission/mECFP emission) (Fig. 3B and C). This allowed estimates of FRET efficiency from measured emission ratios under conditions where FRET efficiency was not directly measured (Table 1). In solution, the Förster distance (R0, distance where energy transfer between donor and acceptor is 50%) for the FRET tension sensor was 5.3 nm for the mYPet-mECFP tension sensor [39]. Based on this value and FRET efficiency measurements, the mean separation between the donor and acceptor fluorophores in the FRET tension sensor was 5.7 nm in metaphase, 5.3 nm in late anaphase, 4.8 nm in Nuf2-FRET (zero tension), and 5.0 nm in solution (Table 1).
Figure 3.
Estimate mean force measured by FRET BioSensor. A: Measured values of FRET efficiency for control metaphase and late anaphase, and for cells treated with low-dose benomyl (55μM). B: FRET efficiency versus Emission Ratio shows linear dependency. C: FRET efficiency versus distance between fluorophores. Inset shows FRET in cell with the Ndc80 biosensor. The outline of the budded cell (white) and clusters of bi-oriented Ndc80 in metaphase are shown. The four dotted lines in the graph represent the position of No tension in solution (lt. brown), Late Anaphase (yellow), Benomyl 55μm (dk brown), and Metaphase (purple). D: Estimated force extension curve calculated using worm like chain (see WLC equation in text).
Table 1.
Summary of measured values for emission ratio and FRET efficiency to estimate force/kMT
| Emission ratio | FRET efficiency | K-K distance (nm) | Separation (nm) | Force/probe | Force/kMTa | ||
|---|---|---|---|---|---|---|---|
| Control | Metaphase | 2.12 ± 0.54 | 0.40 ± 0.12 | 891.1 ± 96.4 | 5.72 | ~0.43 pN | 3.44–7.44 pN |
| Late anaphase | 2.92 ± 0.73 | 0.51 ± 0.09 | – | 5.31 | ~0.23 pN | 1.84–4.01 pN | |
| Benomyl (55 μM) | Metaphase | 2.66 ± 0.62 | 0.47 ± 0.15 | 820.8 ± 126.3 | 5.44 | ~0.29 pN | 2.34–5.08 pN |
| Benomyl (165 μM) | Metaphase | 2.66 ± 0.62 | 0.47b | 610.4 ± 126.0 | 5.44 | ~0.29 pN | 2.34–5.08 pN |
| Benomyl (551 μM) | Metaphase | 2.61 ± 0.67 | 0.45b | ~0 | 5.47 | ~0.31 pN | 2.45–5.33 pN |
| Dam1-765 | Metaphase | 3.20 ± 0.88 | 0.53b | 988.4 ± 178.9 | 5.18 | ~0.17 pN | 1.36–2.95 pN |
| Nuf2 C-term FRET | Average of whole cell cycle | 3.99 ± 0.64 | 0.64b | – | 4.81 | ~0 pN | ~0 pN |
| FRET probe in solution | – | 0.59 | – | 5.00 | 0 pN | 0 pN |
Measurements of Ndc80 copy number varies from 8 to 17.4 molecules per kinetochore (kMT).
Estimated by Fig. 5C.
Estimates of the mean tension at the FRET sensor can be obtained using a force extension curve based on a worm-like chain model for the entropic tension produced by stretching the unstructured 28 amino acids interconnecting the two beta-barrels in the FRET biosensor [39, 61, 62]. Force, F, is calculated from [63]
where kT = 4.1 pN-nm, P, the persistence length = 1.09 nm, and L, the total contour length, = 12.5 nm. The total contour length is estimated from the 28 aa flexible segment (9.5 nm) plus 1.5 nm on each side to account for the distance from the outside of the fluorescent protein barrel to the fluorophore in the center giving a total contour length of 12.5 nm between donor and acceptor [39]. The x is separation distance minus the value in Nuf2-FRET (Control) under no tension = 4.8 nm. This value is very close to the value in solution (5.0 nm) measured in previous study using only the sensor probe, and fit with a worm-like-chain having a persistence length L = 1.1 nm [39]. Values from the worm-like chain equation indicated that the mean tension measured by the FRET biosensor per Ndc80 is ~0.45 pN in metaphase (Δx = 0.91), ~0.2 pN in late anaphase (Δx = 0.5), ~0.3 pN in interphase (Δx = 0.66) and ~0 pN when the FRET biosensor was bound to the C-terminus of Ndc80 (Δx = 0) and not under tension (Fig. 3D and Table 1). Using a value of ~13 Ndc80 complexes/kMT at metaphase based on the fluorescence of individual EGFP molecules in vitro or Cse4-EGFP in vivo [11, 47, 48], the total mean force at the FRET sensor per kMT is ~6 pN at metaphase, ~2.5 pN in late anaphase, ~4 pN in interphase cells (Fig. 3 and Table 1).
These forces remain rough estimates until in vitro calibrations with genetically encoded FRET sensors (mYPET, mECFP) can be performed. With current calibration standards, differences in the fluorophores (in vivo genetically encoded fluorescent proteins vs. in vitro organic dyes), possible compliant linkages within the beta barrels for mYPET and mECFP and effects from the crowded cellular environment remain sources of error for in vivo vs. in vitro force estimates. The ~5 pN per kMT at the Ndc80 MT binding domains in metaphase based on the WLC equation is comparable to in vitro measurements of 7–9 pN for the detachment force from MT ends of isolated yeast kinetochores as measured by Akiyoshi et al. [64]. It also is similar to the 7.5 pN value obtained from the stiffness measurements of stretched metaphase centromeres in budding yeast ([65], see below). Nevertheless, the actual in vivo mean kinetochore force maybe significantly higher than the above estimates because of load-bearing contributions of the Dam1 complex and Stu2 (see below). On the other hand, load-sharing by the Nuf2 coiled-coil domain within the Ndc80/Nuf2 dimer may be reducing the value measured by the FRET tension sensor. Finally, the moderate tension values at late anaphase and interphase 2.5–3 pN are an interesting puzzle. The nanoscale fluorescence co-localization measurements of Joglekar et al. [23] revealed that there was a shortening of the Ndc80 complex, perhaps through bending at its looped domain, indicative of a low tension state. However, the observed shortening could be a consequence of 2D versus 3D measurements that do not take into account geometrical changes in microtubule orientation in metaphase versus anaphase [66, 67].
The high tension values in anaphase and interphase are similar to the value measured when mitotic cells were treated with high (551 μM) doses of benomyl to inhibit MT polymerization and cause spindle collapse in metaphase (Table 1). In both interphase and high benomyl, kinetochores remain tethered to their poles by short, 50 nm kMTs [67]. Under both these conditions chromosomes are held close to the inside of the nuclear envelope where the poles are located. Perhaps kinetochore tension is produced by thermal repulsion of attached chromatin away from the inside of the nuclear envelope.
The FRET tension sensor within budding yeast Ndc80 protein reveals load sharing at metaphase between MT binding proteins at the kMT interface
It is important to note that the absolute value of force is not as significant as the changes in tension reported by the Ndc80 FRET sensor for discovering and testing mechanistic models of force coupling between kinetochores and kMT plus-ends. In budding yeast, the MT binding N-terminal tail of Ndc80 is not essential [68]. At metaphase, Suzuki et al. [58] found very low tension at metaphase through interphase in the Ndc80 tension sensor when the N-terminal tail was deleted, although centromeres were stretched to their normal mean length of ~800 nm at metaphase [58]. This showed that in vivo other MT binding proteins associated with the Ndc80 complex, like Dam1 and Stu2 [55, 69], have important roles in force coupling between kinetochores and MT ends.
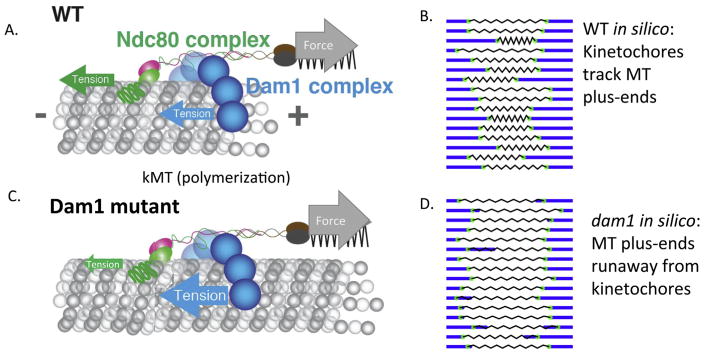
Suzuki et al. [58] also found that treatment of metaphase cells with low dose benomyl (55 μM) to suppress the extent of kMT dynamic instability results in a large drop in tension at the Ndc80 MT binding domains (Table 1), but no significant change in centromere stretch or spindle length [58]. These results suggested that the Ndc80 MT binding domains are under tension, being dragged along the kMT lattice away from the pole by centromere stretch during kMT polymerization, and under compression during the depolymerization phase of dynamic instability as the kinetochore moves poleward, stretching the centromere (Fig. 4).
Figure 4.
Models for the Ndc80 force coupler during polymerization in wild type and consequences of Dam1 mutants in which the kinetochore dislocates from the microtubule plus-end [58]. A: During polymerization, force from centromere stretch pulls the Ndc80 force coupler along kMTs with the microtubule binding domains of Dam1 and Ndc80 under tension. During depolymerization, forces from peeling protofilaments push the Dam1 and Ndc80 complexes along kMTs towards the pole to stretch the centromere; the MTBDs of both Dam1 and Ndc80 are under compression (not shown). C: Mutations in Dam1 that increase friction between Dam1 and the microtubule lattice may prevent kinetochores from keeping up with the ends of polymerizing microtubules, leading to dislocation of the kinetochore from the MT plus-end [70]. Ndc80 resistive tension (green arrow in C) from centromere stretch is reduced in this state with increased Dam1 friction (blue arrow in C). B and D: Snapshots of movies derived from 600 s duration simulations presented in [58], showing kinetics for all 16 sister kinetochore pairs and their centromere stretch. See Supporting Information to view the movies. B: WT, D: dam1 mutant.
To test this idea further, Suzuki et al. [58] used a Dam1 mutant (Dam1-765) isolated in a genetic screen for lethality when coupled with mutants that weakened the anchorage of microtubules in the spindle pole [70]. The rationale was that tight-binding kinetochore components would pull microtubules out of the pole, resulting in cell death. The key phenotypes of Dam1-765 were hyper-centromere stretch and kinetochores closer than normal to spindle poles at meta-phase with normal MT dynamics. Shimogawa et al. [70] proposed that the kinetochore was anchored to kMTs but uncoupled from the plus-end (Fig. 4A and C). Suzuki et al. [58] found lower Ndc80 tension than normal indicating the Ndc80 MT binding domains are under low tension in Dam1-765 cells as predicted by the enhanced affinity of the Dam1-765 mutant complex for the MT lattice and corresponding enhanced Dam1-765 drag force. The simulation in Suzuki et al. [58] indicates that depolymerizing ends push the Ndc80 force coupler poleward at normal rates because of the high forces available from curling protofilaments (Fig. 4D) [71]. During polymerization, the force from hyper centromere stretch pulls the kinetochore away from the pole at a much slower rate than the rate of kMT polymerization. The enhanced drag exerted by Dam1-765 results in compression of the Ndc80 complex, and thus the resistive force shown in Fig. 4C. This allows the kMT plus end to grow past the kinetochore until depolymerization re-engages the coupler (Fig. 4D). While the kinetochore is closely coupled to microtubule-plus ends in wild-type situation (Fig. 4B), the physical properties of the kinetochore can be tweaked in such a way to position the kinetochore along the lattice. This finding makes new testable predictions for the behavior of kinetochores dislocated from microtubule-plus-ends in metazoans.
The DAM1 ring complex is a well-established force coupler at depolymerizing MT ends in vitro, particularly when anchored by long alpha helical-coiled coiled filaments similar to the Ndc80 complex [71]. There is evidence that Stu2 also plays a critical role in force transduction in the kinetochore [69]. Stu2 is an essential protein recently shown to bind the Ndc80 complex [69]. It has roles in regulating both microtubule dynamics [72, 73] and attachment force. Upon depletion of the protein, microtubule dynamics (growth and shortening) are severely reduced [73]. Isolated kinetochores detach more frequently from assembling tips in the absence of Stu2 under load, and upon shortening kinetochores lacking Stu2 detach less frequently under a small load (0.5–2 pN) and more frequently if load exceeds 2.5 pN [69]. Thus Stu2 function is in some way dependent upon the state of MT dynamic instability and load on the kinetochore.
Finally the drop in force at the biosensor while maintaining metaphase centromere stretch with either tail deletion [58] or low dose benomyl (Table 1) indicates that load within the kinetochore can be transferred from one complex to another, depending on alterations within the complex, or constellation of MT-binding proteins at plus-ends. Thus the kinetochore is not a simple force transmitter, rather it is a more complex force coupler in which load is nonlinearly distributed among the protein sub-complexes that bind MTs, and sensitive to the dynamic state of the microtubule end. A role for Ndc80 binding proteins has been found in both fission yeast for the TOG homologs, Dis1 and Alp14, and in human cells for the Ndc80 loop binding protein Cdt1 and for the MT binding protein SKA, both of which, like the DAM1 complex in budding yeast, are only recruited to kinetochores at high concentrations after kMT formation [74–76]. How this recruitment occurs is an important issue to solve in future investigations.
The centromere spring
A major unsolved problem in mitosis is the organization of chromatin in the centromere and understanding how the centromere spring is constructed. Microtubules are very stiff polymers (persistence length, Lp = 6 mm), while DNA is five orders of magnitudes floppier (Lp = 50 nm). The spring constant for an entropic DNA spring is F = 3kBT/n(2Lp)2, where n = the number of Lp segments. For DNA length = 10 kb, the spring constant = 0.036 pN/μm, small indeed.
The prevailing model in the literature indicates that the structural maintenance of chromosome proteins (SMC 1 and 3), cohesin, is responsible for tethering sister chromatids and resisting outward microtubule pulling forces. While the mechanism of sister chromatid tethering is under debate (one cohesin complex/sister strands vs. two cohesin complexes/sister strands) there is ample evidence that loss of cohesin results in precocious sister chromatid separation [77, 78]. In the centromere, cohesin is 3X enriched but sister DNA strands are separated by several hundred nanometers [79–82]. Thus unlike in the chromosome arms, cohesin is not holding sister strands together in the pericentromere. Cohesin adopts a position radially displaced from the spindle axis, dictated by the size of the off-axis DNA loops [32, 83, 84].
Heterogeneity in the mechanical properties of the centromere
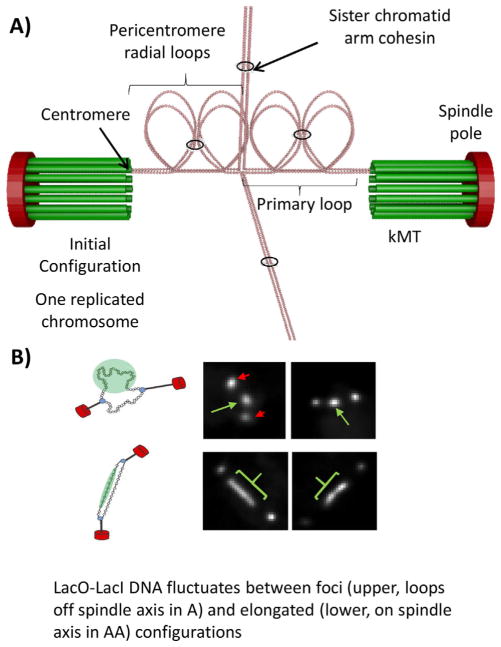
Metaphase sister kinetochore separation ranges from ~800 nm in budding yeast, to about 1,000 nm in human cells [85]. It has been proposed that conservation of this distance reflects the higher order organization of centromere DNA into loops (Fig. 5) [30, 83, 85]. Cohesin and condensin function to generate and/or increase the lifetime of chromatin loops throughout the genome and are enriched in the centromere region. Furthermore, these complexes are distributed in non-overlapping patterns within the centromere. Condensin resides proximal to the axis defined by the sister kinetochores (spindle axis) [32, 86], while cohesin is radially displaced from the spindle axis [32, 84, 86]. The data are consistent with a model wherein condensin binds to the base of centromere loops, while cohesin seeks regions of most disorder within the loops (Fig. 5A). This heterogeneous structure must be taken into account in studies used to estimate centromere spring force.
Figure 5.
Packing of pericentric chromatin loops between sister kinetochores in metaphase as a mechanism to increase stiffness. A: The pericentromeric chromatin from one replicated chromosome is shown. The DNA is depicted as beads on a string chromatin (pink circles). The centromeres (defined at the site of attachment to kMTs) from each sister chromatid are at the distal ends of the primary loop, in contact with a kMT (kMTs are in green). The 16 kMTs in the metaphase spindle emanate from the spindle pole body (red oval). The pericentric chromatin is organized as loops radiating from the primary loops (radial loops), depicted as four loops for each sister strand, a total of eight loops between the sister kinetochores. Cohesin (black rings) hold sister chromatid arms together, and pericentric cohesin links intra-strand loops. Chromosome arms (away from the pericentric region) extend north and south. B: Visualization of pericentric DNA in living cells. Left, schematic of spindle poles (red spheres) connected to a circular chromosomal DNA (small spheres) via kinetochore microtubules (black rods). The DNA will appear as a foci off the spindle axis (top) or as a linear array on the spindle axis (bottom). To the right of the schematic are shown images of live cells. The poles are indicated in red and the lacO DNA in green.
Force can be inferred from the thermal fluctuation of the DNA polymer [83, 87]. A segment of the centromere can be visualized through the insertion of lacO into the centromere bound by lacI-GFP. At metaphase in budding yeast, this fluorescently labeled DNA adopts several configurations ranging from foci to elongated filaments that are in dynamic equilibrium (Fig. 5B). The foci reside in the cohesin-rich region displaced from the spindle axis while the elongated fluorescence is aligned close to the spindle axis [83]. Using simplifying assumptions such as those to measure the spring constant of an optical trap (κ = kBT/variance), one can extract such a spring constant from the variance in foci spot fluctuation. Lawrimore et al. [83] have used this to estimate a metaphase value of 0.24 pN/μm for a budding yeast centromere linked DNA, 6.8 kb from the inner kinetochore and radially displaced from the spindle axis [83]. In the presence of azide, Lawrimore et al. [83] and Chacon et al. [65] find that the system stiffens considerably, reflecting inactivation of enzymatic processes and rigor binding of microtubule motor proteins. In the presence of azide, the metaphase value from spot measurements for the chromatin spring is 4 pN/μm. This value should not be taken as an estimate of centromere spring force between sister kinetochores due to the loop organization of centromere chromatin.
The DNA in loops is in thermal equilibrium with DNA along the spindle axis. When DNA approaches the spindle axis it does not adopt a random coil, rather the DNA becomes stretched between sister kinetochores, as evidenced by fluorescence of the LacI-GFP bound to elongated lacO DNA along the axis (see Fig. 5B). Using circular DNA of known size, Lawrimore et al. [83] has been able to estimate the degree of extension of a single molecule attached to a separated sister kinetochore pair at metaphase in budding yeast. The chromatin is stretched beyond the 7X compaction of nucleosomal DNA relative to B-form DNA, with approximately 30% of the nucleosomes evicted [83]. The force required to evict nucleosomes has been measured in a number of in vitro conditions. The consensus is that approximately 20 pN of force is required to “pop” histone octamers from chromatin in vitro in buffer [88]. However, these experiments are performed in buffer conditions lacking active nucleoplasm. In the presence of soluble protein extract from Xenopus eggs, the amount of force is on the order of 2 pN [89]. As the in vivo measurements are more comparable to the extract situation, we estimate that chromatin on the spindle axis experiences on the order of several piconewtons. This is within the range estimated from the metaphase force (3–7 pN) at the kinetochore (Table 1).
How cohesion and condensin strengthen and structure the pericentromeric chromatin spring
Using a combination of high resolution spatial mapping of labeled DNA (sub-diffraction limited) relative to the spindle axis, motion analysis of DNA foci vs. stretched chromatin, and coordinated motion and stretching of pericentromeric regions from different chromosomes, the pericentromere is best described as a cross-linked chromatin network [30, 31, 83, 90]. The network is in the form of a bottle brush, where chromatin loops (linked via cohesin) are radially displaced relative to the microtubule spindle axis. Condensin lies along the spindle axis, due in part to its interactions with tRNA genes and their regulatory factors [91]. Condensin and chromatin along the spindle axis, continuing the analogy to a bottle brush, define the primary load-bearing axis. The centromere is at the distal tip of the primary axis, projected onto the surface of the chromosome (Fig. 5).
Studies in polymer physics reveal that a bottle brush polymer, with a high number of sidechains (i.e. crowded), can generate extensional forces outward/poleward that under particular conditions are strong enough to break covalent bonds [92–94]. In a computational model of a bottle brush, experimental measurements including size, shape and uniformity of cohesin (SMC3-GFP) can be simulated in the pericentromere [30, 83].
The bottlebrush provides a mechanistic understanding for several outstanding problems. Firstly, the bottlebrush provides a physical basis for how a floppy DNA chain can be converted into a stiff (relative to an entropic chain) spring. A fluctuating chain in a thermal bath will find the most entropically favored state, that of a random coil. As a random coil the chain is its most disordered. The addition of side chains relative to the primary chain (or primary axis), limits the ability of the primary chain to adopt a random coil, through limiting the number of states the primary chain can adopt. Additional side chains further restrict the motion of the primary axis until a point where they generate tension along the axis. In this way, enthalpic energy put into making chains, results in entropic forces of the brush where tension in the chains amplifies tension along the primary axis. Thus chromatin loops significantly change the state of the centromere from a floppy chromatin polymer into a stiff chromatin network. Secondly, when chromosomes become bi-oriented, they continue to oscillate from pole to pole, accompanied by continual microtubule growth and shortening. Based on the FRET measurements, the drag forces (and tension) on Ndc80c and the Dam1 ring change direction depending on whether the microtubule is in a growth or shortening state [58]. Tension in Ndc80c will therefore fluctuate in concert with microtubule dynamics. Furthermore, whether in yeast where 16 kinetochores in each half spindle are clustered, or a metazoan kinetochore such as human, where there are multiple microtubule attachment sites per kinetochore, individual microtubules are fluctuating independent of one another. How the checkpoint copes with these stochastic fluctuations is not known. The bottlebrush network provides a solution if tension is integrated across the sister kinetochores, thus equalizing tension at the individual kinetochores.
Conclusions
The introduction of tension biosensors into a eukaryotic kinetochore provides a probe to query the relative contributions of the various sub-domains that in aggregate comprise the kinetochore. The N-terminal tail of Ndc80, while not essential for cellular life [68], makes a major contribution to force within the Ndc80 molecule [58]. Mutants in the Dam1 ring (Dam1-765) that maintain centromere stretch, alleviate tension within Ndc80. Tension could be transferred to the Dam1 ring in the Dam1-765 mutant, other kinetochore components, or reduced in the ensemble. In either of these cases, these studies indicate the complexity of force transduction through the kinetochore and the utility of tension biosensors and need to place them elsewhere in the kinetochore as has been recently accomplished for CENP-C in Drosophila [29]. There is still much to be learned about how the Ndc80 complex and the proteins that bind to it function at the kinetochore microtubule interface and control the spindle checkpoint through use of the Ndc80 tension sensor in mammalian kinetochores.
Centromere chromatin presents its own enigma. Molecular models have been built that provide a mechanistic basis for how megabases of floppy DNA can build tension. How the centromere builds and retains morphology (and cohesin its uniform barrel structure) in a dynamic system where individual chromatin chains are fluctuating and pulling microtubules lead to kinetochore deformation at the surface of the centromere [28] remains to be determined. Using polymer physics as our guide, slip link gels are known to retain their elastic and tensile moduli properties over several orders of magnitude [95, 96]. It has been known for almost half a century that chromosomes expand and contract upon removal and return of mono- and divalent ions [97], and depending on the treatment retain their original structure (e.g. such as their macroscopic banding pattern). The concentration of ring complexes such as cohesin and condensin in the pericentromere may be indicative of the slip-link property of centromeres.
Acknowledgments
We would like to thank Dr. Harold Erickson (Duke University), Dr. Aussie Suzuki (UNC-CH), Alyona Fulp (UNC-CH), and Joshua Lawrimore (UNC-CH) for their intellectual contributions and help with the manuscript and figures.
The authors have declared no conflicts of interest.
Abbreviation
- kMT
kinetochore microtubule
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- 1.Hori T, Amano M, Suzuki A, Backer CB, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–52. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Pekgoz Altunkaya G, Malvezzi F, Demianova Z, Zimniak T, et al. CCAN assembly configures composite binding interfaces to promote cross-linking of Ndc80 complexes at the kinetochore. Curr Biol. 2016;26:2370–8. doi: 10.1016/j.cub.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, et al. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Rago F, Gascoigne KE, Cheeseman IM. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol. 2015;25:671–7. doi: 10.1016/j.cub.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schleiffer A, Maier M, Litos G, Lampert F, et al. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol. 2012;14:604–13. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- 6.Musacchio A, Desai A. A molecular view of kinetochore assembly and function. Biology (Basel) 2017;6 doi: 10.3390/biology6010005. pii: E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen RL, Espelin CW, De Wulf P, Sorger PK, et al. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol Biol Cell. 2008;19:4480–91. doi: 10.1091/mbc.E08-03-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimitrova YN, Jenni S, Valverde R, Khin Y, et al. Structure of the MIND complex defines a regulatory focus for yeast kinetochore assembly. Cell. 2016;167:1014–27. e12. doi: 10.1016/j.cell.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huis In ’t Veld PJ, Jeganathan S, Petrovic A, Singh P, et al. Molecular basis of outer kinetochore assembly on CENP-T. Elife. 2016;5:e21007. doi: 10.7554/eLife.21007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Yu H. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J Cell Biol. 2015;208:181–96. doi: 10.1083/jcb.201407074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki A, Badger BL, Salmon ED. A quantitative description of Ndc80 complex linkage to human kinetochores. Nat Commun. 2015;6:8161. doi: 10.1038/ncomms9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 13.DeLuca JG, Gall WE, Ciferri C, Cimini D, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–82. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 14.Aravamudhan P, Goldfarb AA, Joglekar AP. The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat Cell Biol. 2015;17:868–79. doi: 10.1038/ncb3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joglekar AP. A cell biological perspective on past, present and future investigations of the spindle assembly checkpoint. Biology (Basel) 2016;5 doi: 10.3390/biology5040044. pii: E44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai A, Maddox PS, Mitchison TJ, Salmon ED. Anaphase A chromosome movement and poleward spindle microtubule flux occur At similar rates in Xenopus extract spindles. J Cell Biol. 1998;141:703–13. doi: 10.1083/jcb.141.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maddox P, Straight A, Coughlin P, Mitchison TJ, et al. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol. 2003;162:377–82. doi: 10.1083/jcb.200301088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumont S, Salmon ED, Mitchison TJ. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 2012;337:355–8. doi: 10.1126/science.1221886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khodjakov A, Rieder CL. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J Cell Biol. 1996;135:315–27. doi: 10.1083/jcb.135.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan X, Cimini D, Cameron LA, Salmon ED. The coupling between sister kinetochore directional instability and oscillations in centromere stretch in metaphase PtK1 cells. Mol Biol Cell. 2012;23:1035–46. doi: 10.1091/mbc.E11-09-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchison TJ, Salmon ED. Mitosis: a history of division. Nat Cell Biol. 2001;3:E17–21. doi: 10.1038/35050656. [DOI] [PubMed] [Google Scholar]
- 22.LaFountain JR, Jr, Cohan CS, Oldenbourg R. Functional states of kinetochores revealed by laser microsurgery and fluorescent speckle microscopy. Mol Biol Cell. 2011;22:4801–8. doi: 10.1091/mbc.E11-06-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–9. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magidson V, Paul R, Yang N, Ault JG, et al. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol. 2015;17:1134–44. doi: 10.1038/ncb3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CA, McAinsh AD, Burroughs NJ. Human kinetochores are swivel joints that mediate microtubule attachments. Elife. 2016;5:e16159. doi: 10.7554/eLife.16159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki A, Badger BL, Wan X, DeLuca JG, et al. The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev Cell. 2014;30:717–30. doi: 10.1016/j.devcel.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan X, O’Quinn RP, Pierce HL, Joglekar AP, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–84. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magidson V, He J, Ault JG, O’Connell CB, et al. Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells. J Cell Biol. 2016;212:307–19. doi: 10.1083/jcb.201412139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye AA, Cane S, Maresca TJ. Chromosome biorientation produces hundreds of piconewtons at a metazoan kinetochore. Nat Commun. 2016;7:13221. doi: 10.1038/ncomms13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrimore J, Aicher JK, Hahn P, Fulp A, et al. ChromoShake: a chromosome dynamics simulator reveals that chromatin loops stiffen centromeric chromatin. Mol Biol Cell. 2016;27:153–66. doi: 10.1091/mbc.E15-08-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens AD, Haggerty RA, Vasquez PA, Vicci L, et al. Pericentric chromatin loops function as a nonlinear spring in mitotic force balance. J Cell Biol. 2013;200:757–72. doi: 10.1083/jcb.201208163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stephens AD, Quammen CW, Chang B, Haase J, et al. The spatial segregation of pericentric cohesin and condensin in the mitotic spindle. Mol Biol Cell. 2013;24:3909–19. doi: 10.1091/mbc.E13-06-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicklas RB. A quantitative study of chromosomal elasticity and its influence on chromosome movement. Chromosoma. 1963;14:276–95. doi: 10.1007/BF00326816. [DOI] [PubMed] [Google Scholar]
- 34.Nicklas RB. Chromosome velocity during mitosis as a function of chromosome size and position. J Cell Biol. 1965;25:119–35. doi: 10.1083/jcb.25.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jannink G, Duplantier B, Sikorav JL. Forces on chromosomal DNA during anaphase. Biophys J. 1996;71:451–65. doi: 10.1016/S0006-3495(96)79247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicklas RB. Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol. 1983;97:542–8. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster PJ, Furthauer S, Shelley MJ, Needleman DJ. Active contraction of microtubule networks. Elife. 2015;4:e10837. doi: 10.7554/eLife.10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikirzhytski V, Magidson V, Steinman JB, He J, et al. Direct kinetochore-spindle pole connections are not required for chromosome segregation. J Cell Biol. 2014;206:231–43. doi: 10.1083/jcb.201401090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 2007;16:1429–38. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brenner MD, Zhou R, Conway DE, Lanzano L, et al. Spider silk peptide is a compact, linear nanospring ideal for intracellular tension sensing. Nano Lett. 2016;16:2096–102. doi: 10.1021/acs.nanolett.6b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grashoff C, Hoffman BD, Brenner MD, Zhou R, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–6. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaFountain JR, Jr, Cohan CS, Siegel AJ, LaFountain DJ. Direct visualization of microtubule flux during metaphase and anaphase in crane-fly spermatocytes. Mol Biol Cell. 2004;15:5724–32. doi: 10.1091/mbc.E04-08-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang D, Nicklas RB. The impact of chromosomes and centrosomes on spindle assembly as observed in living cells. J Cell Biol. 1995;129:1287–300. doi: 10.1083/jcb.129.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol. 1999;9:1423–6. doi: 10.1016/s0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- 46.Coffman VC, Wu P, Parthun MR, Wu JQ. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–72. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joglekar AP, Bouck DC, Molk JN, Bloom KS, et al. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–5. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–82. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCormick CD, Akamatsu MS, Ti SC, Pollard TD. Measuring affinities of fission yeast spindle pole body proteins in live cells across the cell cycle. Biophys J. 2013;105:1324–35. doi: 10.1016/j.bpj.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ciferri C, De Luca J, Monzani S, Ferrari KJ, et al. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–95. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 51.Ciferri C, Pasqualato S, Screpanti E, Varetti G, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–39. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–9. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 53.Wei RR, Schnell JR, Larsen NA, Sorger PK, et al. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–9. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–7. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aravamudhan P, Felzer-Kim I, Gurunathan K, Joglekar AP. Assembling the protein architecture of the budding yeast kinetochore-microtubule attachment using FRET. Curr Biol. 2014;24:1437–46. doi: 10.1016/j.cub.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics. 2009;183:1591–5. doi: 10.1534/genetics.109.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell. 2009;20:3818–27. doi: 10.1091/mbc.E09-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki A, Badger BL, Haase J, Ohashi T, et al. How the kinetochore couples microtubule force and centromere stretch to move chromosomes. Nat Cell Biol. 2016;18:382–92. doi: 10.1038/ncb3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tien JF, Fong KK, Umbreit NT, Payen C, et al. Coupling unbiased mutagenesis to high-throughput DNA sequencing uncovers functional domains in the Ndc80 kinetochore protein of Saccharomyces cerevisiae. Genetics. 2013;195:159–70. doi: 10.1534/genetics.113.152728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, et al. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–5. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rivetti C, Guthold M, Bustamante C. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J Mol Biol. 1996;264:919–32. doi: 10.1006/jmbi.1996.0687. [DOI] [PubMed] [Google Scholar]
- 62.Zhou HX. Polymer models of protein stability, folding, and interactions. Biochemistry. 2004;43:2141–54. doi: 10.1021/bi036269n. [DOI] [PubMed] [Google Scholar]
- 63.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–70. [Google Scholar]
- 64.Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–9. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chacon JM, Mukherjee S, Schuster BM, Clarke DJ, et al. Pericentromere tension is self-regulated by spindle structure in metaphase. J Cell Biol. 2014;205:313–24. doi: 10.1083/jcb.201312024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–31. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Demirel PB, Keyes BE, Chaterjee M, Remington CE, et al. A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces cerevisiae. Genetics. 2012;192:753–6. doi: 10.1534/genetics.112.143818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller MP, Asbury CL, Biggins S. A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell. 2016;165:1428–39. doi: 10.1016/j.cell.2016.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimogawa MM, Graczyk B, Gardner MK, Francis SE, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Volkov VA, Zaytsev AV, Gudimchuk N, Grissom PM, et al. Long tethers provide high-force coupling of the Dam1 ring to shortening microtubules. Proc Natl Acad Sci USA. 2013;110:7708–13. doi: 10.1073/pnas.1305821110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kosco KA, Pearson CG, Maddox PS, Wang PJ, et al. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol Biol Cell. 2001;12:2870–80. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pearson CG, Maddox PS, Zarzar TR, Salmon ED, et al. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol Biol Cell. 2003;14:4181–95. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grishchuk EL, Efremov AK, Volkov VA, Spiridonov IS, et al. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc Natl Acad Sci USA. 2008;105:15423–8. doi: 10.1073/pnas.0807859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, et al. Different assemblies of the DAM1 complex follow shortening micro-tubules by distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:6918–23. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, et al. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–90. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 77.Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 1995;9:587–99. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- 78.Strunnikov AV, Larionov VL, Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–48. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–59. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- 80.D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–27. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Megee PC, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–50. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- 82.Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–66. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lawrimore J, Vasquez PA, Falvo MR, Taylor RM, 2nd, et al. DNA loops generate intracentromere tension in mitosis. J Cell Biol. 2015;210:553–64. doi: 10.1083/jcb.201502046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yeh E, Haase J, Paliulis LV, Joglekar A, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bloom KS. Centromeric heterochromatin: the primordial segregation machine. Annu Rev Genet. 2014;48:457–84. doi: 10.1146/annurev-genet-120213-092033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stephens AD, Haase J, Vicci L, Taylor RM, 2nd, et al. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol. 2011;193:1167–80. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Verdaasdonk JS, Vasquez PA, Barry RM, Barry T, et al. Centromere tethering confines chromosome domains. Mol Cell. 2013;52:819–31. doi: 10.1016/j.molcel.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, et al. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci USA. 2002;99:1960–5. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan J, Maresca TJ, Skoko D, Adams CD, et al. Micromanipulation studies of chromatin fibers in Xenopus egg extracts reveal ATP-dependent chromatin assembly dynamics. Mol Biol Cell. 2007;18:464–74. doi: 10.1091/mbc.E06-09-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stephens AD, Snider CE, Haase J, Haggerty RA, et al. Individual pericentromeres display coordinated motion and stretching in the yeast spindle. J Cell Biol. 2013;203:407–16. doi: 10.1083/jcb.201307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Snider CE, Stephens AD, Kirkland JG, Hamdani O, et al. Dyskerin, tRNA genes, and condensin tether pericentric chromatin to the spindle axis in mitosis. J Cell Biol. 2014;207:189–99. doi: 10.1083/jcb.201405028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lebedeva NV, Nese A, Sun FC, Matyjaszewski K, et al. Anti-Arrhenius cleavage of covalent bonds in bottlebrush macromolecules on substrate. Proc Natl Acad Sci USA. 2012;109:9276–80. doi: 10.1073/pnas.1118517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Panyukov S, Zhulina EB, Sheiko SS, Randall GC, et al. Tension amplification in molecular brushes in solutions and on substrates (dagger) J Phys Chem B. 2009;113:3750–68. doi: 10.1021/jp807671b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Panyukov SV, Sheiko SS, Rubinstein M. Amplification of tension in branched macromolecules. Phys Rev Lett. 2009;102:148301. doi: 10.1103/PhysRevLett.102.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Granick S, Rubinstein M. Polymers: a multitude of macromolecules. Nat Mater. 2004;3:586–7. doi: 10.1038/nmat1199. [DOI] [PubMed] [Google Scholar]
- 96.Okumura Y, Ito K. The polyrotaxane gel: a topological gel by figure-of-eight cross-links. Adv Mater. 2001;13:485–7. [Google Scholar]
- 97.Lezzi M, Gilbert LI. Differential effects of K+ and Na+ on specific bands of isolated polytene chromosomes of Chironomus tentans. J Cell Sci. 1970;6:615–27. doi: 10.1242/jcs.6.3.615. [DOI] [PubMed] [Google Scholar]