Abstract
The X-linked deubiquitinase USP9X has been implicated in multiple pathological disorders including malignancies and X-linked intellectual disability. However, its biological function and substrate repertoire remain to be investigated. In this study, we utilized the tandem mass tag labeling assay to identify USP9X-regulated proteins and revealed that the expression of multiple genes is altered in USP9X-deficient cells. Interestingly, we showed that USP9X promotes stabilization of centrosome proteins PCM1 and CEP55 through its catalytic activity. Remarkably, we demonstrated that USP9X is physically associated and spatially co-localized with PCM1 and CEP55 in the centrosome, and we revealed that either PCM1 or CEP55 loss resulted in impairment of USP9X centrosome localization. Moreover, we showed that USP9X is required for centrosome duplication, and this effect is dependent on its catalytic activity and its N-terminal module, which is responsible for physical association of USP9X with PCM1 and CEP55. Collectively, our experiments identified USP9X as an integral component of the centrosome where it functions to stabilize PCM1 and CEP55 and promote centrosome biogenesis.
Keywords: centrosome, deubiquitylation (deubiquitination), protein stability, protein-protein interaction, proteomics
Introduction
The centrosome is a microtubule organizing center, the primary site where microtubule nucleation and organization take place during interphase and mitosis in diploid cells (1, 2). Similar to the chromosomes, the centrosome is precisely reproduced, initiated around the time of S phase entry and completed by the end of G2 phase (3). During the prophase of mitosis, the centrosomes migrate to opposite poles of the cell, and the mitotic spindle forms between them. Upon division, each daughter cell receives one centrosome (4). The centrosome plays a crucial role in multiple cellular processes including cell division, polarity, and migration (1, 2). However, how this protein-based structure undergoes accurate duplication in a semiconservative manner, at least the molecular mechanism by which centrosome duplication is regulated, is still poorly understood.
As a non-membrane organelle in most mammalian cells, the centrosome is composed of two orthogonally arranged centrioles surrounded by an amorphous granular structure termed the pericentriolar material (PCM)4 where microtubules are nucleated from γ-tubulin ring complexes (5, 6). The formation of an organized microtubule array is in turn dependent on PCM proteins such as PCM1 (7–9), a scaffolding component in centrosome that acts to restrict PCM localization of centrosome-associated CEP family proteins like CEP90 (10, 11), CEP131 (10, 12), and CEP72 (13). These proteins are the active components of the centrosome and are believed to play critical roles in biogenesis and functionality of the centrosome (14–16). CEP55 is a member of the CEP family proteins (17), and it has been reported to be present at the centrosomes, mitotic spindle, spindle midzone, and midbody in somatic cells (18–21). CEP55 acts as a microtubule organizing center-associated protein regulating spindle organization and is important for the completion of cytokinesis (19, 21, 22).
Similarly to acetylation and methylation, protein ubiquitination is a reversible reaction and is constantly opposed by deubiquitinases (DUBs) with enzymatic activity that proteolytically removes polyubiquitin chains from substrates (23). The human genome encodes approximately 95 putative DUBs (23). Among these DUBs, the largest family is the ubiquitin-specific proteases (USPs). Loss of function of each USP is associated with functional consequences that are often severe (23, 24). One of the most studied USPs is the ubiquitin-specific peptidase 9 X-linked (USP9X; also known as FAM for Drosophila fat facets in mouse) (25). It is reported that USP9X acts in a substrate-specific manner and precisely regulates multiple cellular processes. In particular, USP9X has been reported to target dozens of proteins to regulate cellular processes that are fundamental to many aspects of development and disease, namely protein trafficking/endocytosis (26), apoptosis and death (27), polarity (28), autophagy (29, 30), cell growth and migration (31, 32), immune response (33, 34), and stem cell renewal and differentiation (35, 36). In responding to its diverse cellular functions, the intracellular localization of USP9X is versatile and dependent on cell type and status. Generally, USP9X predominantly resides in cytoplasmic and membrane-associated puncta (26, 28, 36); however, USP9X has also been detected at specific cellular compartments including Golgi apparatus, late endosomes, mitochondria, and nucleus (24). These observations point to an important cellular function of USP9X and highlight the need to interrogate USP9X substrates with less disruptive biochemical or genetic approaches.
In this study, we revealed that USP9X is physically associated with PCM1 and CEP55 and stabilizes these centrosome proteins through its deubiquitinase activity. We showed that USP9X is an integral component of the centrosome and required for proper centrosome duplication.
Results
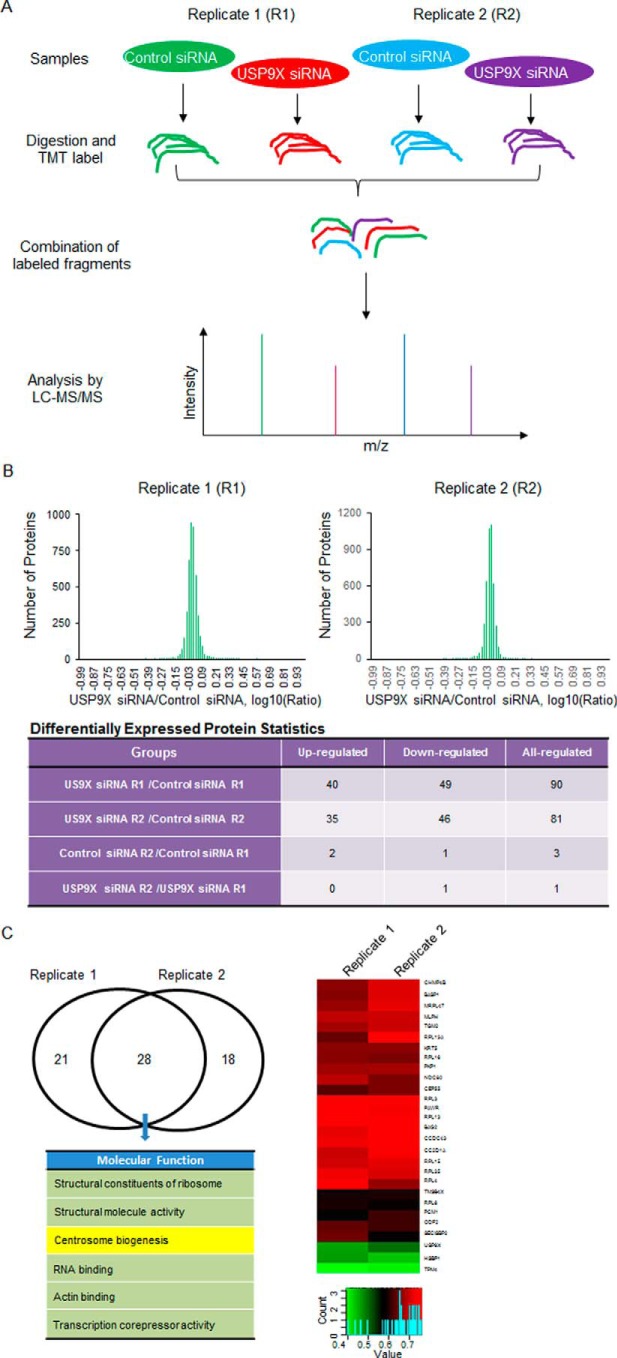
TMT labeling assay analysis of USP9X-regulated genes
To obtain unbiased information of USP9X-promoted protein stabilization and further characterize its biological functionality, a quantitative proteomics strategy based on the TMT labeling assay was performed to determine the down- and up-regulated proteins upon USP9X depletion in MCF-7 cells (Fig. 1A). Specifically, MCF-7 cells were transfected with control siRNA or USP9X siRNA, and the whole cellular proteins from biological duplicate samples were extracted and quantified. Following trypsin digestion of equal amounts of proteins, the resolved peptides were labeled with fourplex TMT kits, fractionated by HPLC, and analyzed by mass spectrometry. In total, 4452 proteins were identified and quantified in all of the four samples (supplemental Table 1). After modeling the normal distribution of the protein ratios, cutoff values of ≦0.770 and ≧1.3 with p values <0.05 were applied to identify proteins with significant expression changes upon USP9X depletion (Fig. 1B and supplemental Table 1). In summary, 27 proteins were up-regulated, and 28 proteins were down-regulated in replicate analyses (Fig. 1C and supplemental Table 1). Because the down-regulated proteins resulting from USP9X depletion could be candidate substrates of USP9X, we primarily focused on these proteins in this study. Functional enrichment-based clustering for overlapping hits from duplicate treatment indicated that USP9X deficiency is associated with alterations of multiple signaling pathways including centrosome biogenesis as manifested by down-regulated expression levels of centrosome components PCM1 and CEP55 upon USP9X loss of function (Fig. 1C and supplemental Table 1).
Figure 1.
TMT labeling assay analysis of USP9X-regulated genes. A, TMT labeling assay-based mass spectrometry analysis of USP9X-regulated protein expression. A flow chart describing the protocol to quantify USP9X depletion-associated protein expression changes from biological duplicate experiments is shown. B, summary of the TMT labeling assay results. By modeling the normal distribution of the protein ratios, a confidence interval of 95% and cutoff values of ≦0.770 and ≧1.3 were applied to identify the proteins with significant down- or up-regulation after USP9X depletion. C, functional analysis and clusterization of down-regulated proteins associated with USP9X depletion from overlapping hits in duplicate experiments. A color key and histogram of the relative expression level for 28 down-regulated proteins are shown.
USP9X promotes PCM1 and CEP55 stabilization through its enzymatic activity
To validate the TMT labeling assay data and test whether the abundance of PCM1 and CEP55 is truly controlled by USP9X, we examined the effect of USP9X on the expression of PCM1 and CEP55. Western blot analysis of the cellular lysates of MCF-7 cells transfected with two independent sets of siRNAs targeting different regions of USP9X revealed that although the expression of centrosome protein Pericentrin and loading control protein β-actin was essentially unchanged, the level of PCM1 and CEP55 was markedly decreased upon USP9X depletion (Fig. 2A and supplemental Fig. S1A). In addition, quantitative reverse transcription (qRT)-PCR measurements indicated that USP9X knockdown did not result in alterations in mRNA expression levels of PCM1 and CEP55 (Fig. 2A). Moreover, the reduction in PCM1 and CEP55 protein levels associated with USP9X depletion was probably a result of proteasome-mediated protein degradation as the effect could be effectively blocked by a proteasome-specific inhibitor, MG132 (Fig. 2B and supplemental Fig. S1B).
Figure 2.
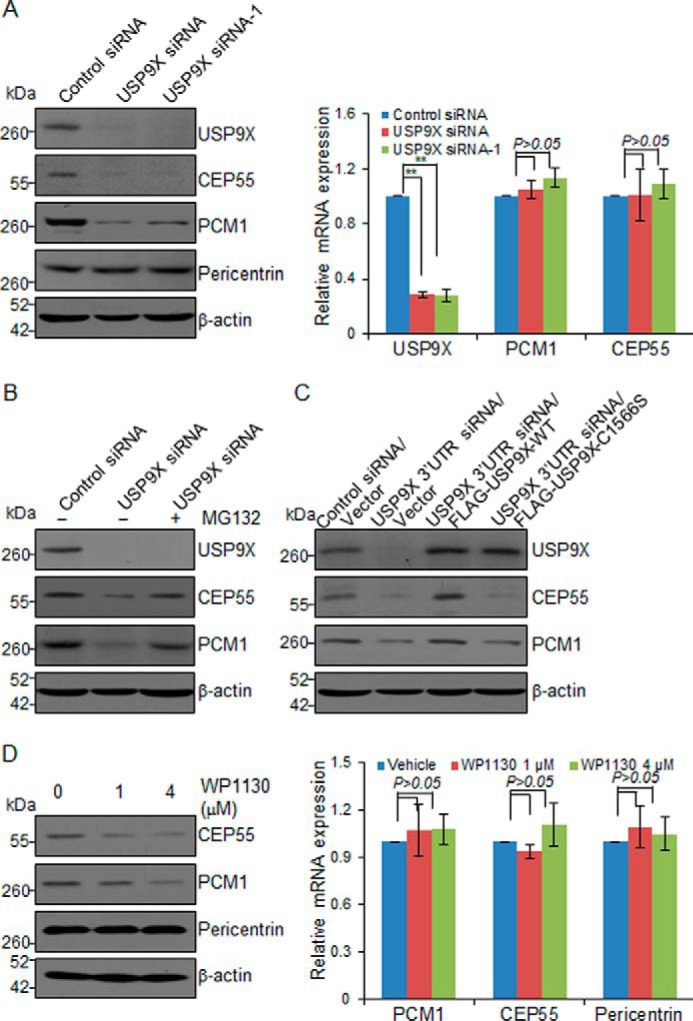
USP9X promotes PCM1 and CEP55 stabilization. A, MCF-7 cells were transfected with control siRNA or different sets of USP9X siRNAs. Cellular extracts or total RNA was prepared and analyzed by Western blotting (left panel) or qRT-PCR (right panel), respectively. Error bars represent S.D. for biological triplicate experiments. **, p < 0.01, one-way ANOVA. B, MCF-7 cells were transfected with control siRNA or USP9X siRNA followed by treatment with DMSO or proteasome inhibitor MG132 (10 μm). Cellular extracts were prepared and analyzed by Western blotting with antibodies as indicated. C, control U2OS cells or U2OS cells stably expressing USP9X-WT or catalytic mutant USP9X (USP9X-C1566S) were transfected with control siRNA or USP9X 3′-UTR siRNA for 96 h followed by Western blot analysis. D, MCF-7 cells were cultured in the absence or presence of WP1130 as indicated. Cellular extracts or total RNA was collected and analyzed by Western blotting (left panel) or qRT-PCR (right panel), respectively. Error bars represent S.D. for biological triplicate experiments. p values were determined by one-way ANOVA.
To gain molecular insights into the functional connection between USP9X and PCM1 or CEP55, we next investigated whether USP9X-promoted PCM1 and CEP55 stabilization is a consequence of protein deubiquitination. Control U2OS cells or U2OS cells stably expressing USP9X were transfected with siRNA targeting the 3′-UTR of USP9X mRNA, and Western blot analysis revealed that wild-type USP9X (USP9X-WT) was able to restore the expression of PCM1 and CEP55, whereas catalytic mutant USP9X (USP9X-C1566S) failed to do so (Fig. 2C and supplemental Fig. S1C). Furthermore, treatment of MCF-7 cells with WP1130, a deubiquitinase inhibitor reported to inhibit USP9X at low micromolar concentrations (37), resulted in a dose-dependent reduction in the levels of PCM1 and CEP55 protein but not that of Pericentrin, whereas it had no effect on the mRNA levels of these genes (Fig. 2D and supplemental Fig. S1D). Together, these results indicated that USP9X regulates the stability of PCM1 and CEP55 and that USP9X does so through its deubiquitinase activity.
USP9X is a centrosomal protein and regulates the abundance of PCM1 and CEP55 in centrosome
The functional association of deubiquitinase USP9X with centrosomal proteins PCM1 and CEP55 suggested that USP9X is a centrosome-associated protein. To test this hypothesis, immunofluorescent staining was performed to examine the subcellular localization of endogenous USP9X in U2OS cells. We found that USP9X was predominantly localized in centrosomes and co-localized with the centrosomal markers Centrin and γ-tubulin (Fig. 3A). In support of the finding that USP9X promotes stabilization of PCM1 and CEP55, immunofluorescence analysis indicated that USP9X was spatially co-localized with PCM1 (Fig. 3B) and CEP55 (Fig. 3C).
Figure 3.
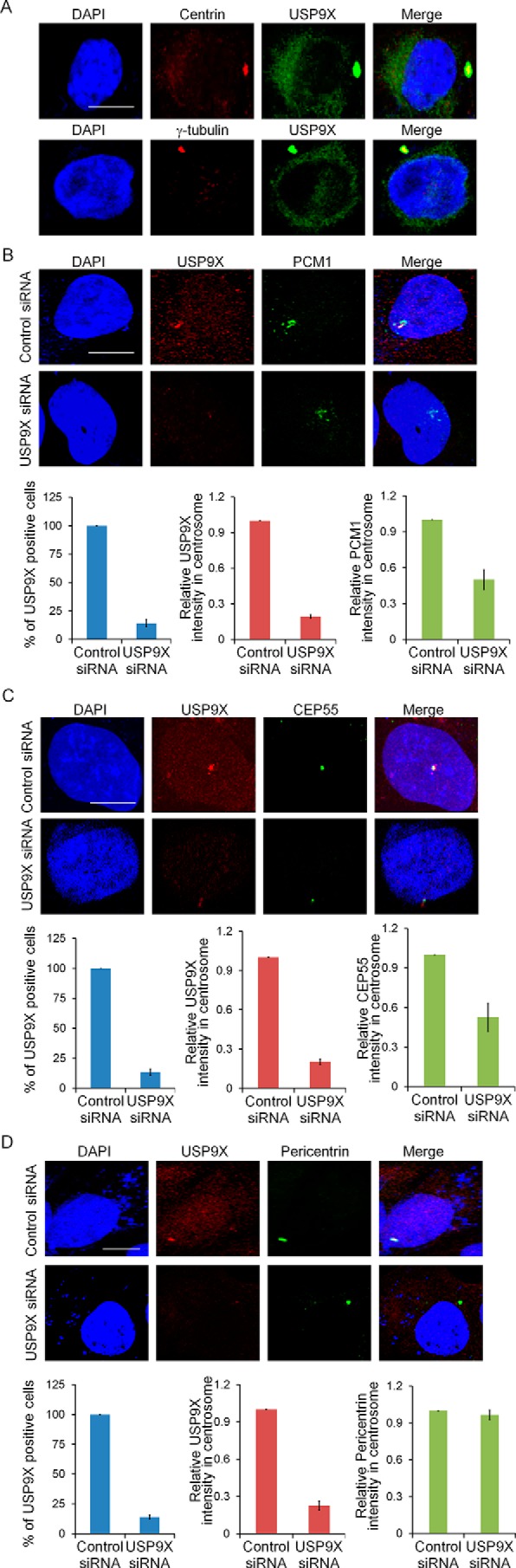
USP9X is a centrosomal protein and functions to regulate the abundance of PCM1 and CEP55 in centrosome. A, U2OS cells were fixed and stained with antibodies against USP9X and Centrin or γ-tubulin. Scale bar, 10 μm. B, U2OS cells transfected with control siRNA or USP9X siRNA were fixed and subjected to immunostaining with antibodies against PCM1 and USP9X. The percentage of cells with USP9X knockdown was counted, and the relative intensity of PCM1 or USP9X in the centrosome was analyzed by ImageJ software. Error bars represent S.D. for biological triplicate experiments. **, p < 0.01, Student's t test. Scale bar, 10 μm. C, U2OS cells transfected with control siRNA or USP9X siRNA were fixed and subjected to immunostaining with antibodies against CEP55 and USP9X. The percentage of cells with USP9X knockdown was counted, and the relative intensity of CEP55 or USP9X in the centrosome was analyzed by ImageJ software. Error bars represent S.D. for biological triplicate experiments. **, p < 0.01, Student's t test. Scale bar, 10 μm. D, U2OS cells transfected with control siRNA or USP9X siRNA were fixed and subjected to immunostaining with antibodies against Pericentrin and USP9X. The percentage of cells with USP9X knockdown was counted, and the relative intensity of Pericentrin or USP9X in the centrosome was analyzed by ImageJ software. Error bars represent S.D. for biological triplicate experiments. **, p < 0.01, Student's t test. Scale bar, 10 μm.
To further establish the functional link between USP9X and PCM1 or CEP55, we asked whether USP9X-promoted PCM1 and CEP55 stabilization has an effect on the localization or centrosome abundance of these proteins. To this end, immunofluorescent staining followed by confocal microscopy analysis was performed, and the results implied that depletion of USP9X resulted in lost staining of PCM1 (Fig. 3B) and CEP55 (Fig. 3C) in centrosomes, but the centrosomal localization of Pericentrin (Fig. 3D) was essentially unchanged. Together, these results suggested that USP9X is a centrosomal protein and further support the notion that USP9X controls the stabilization of PCM1 and CEP55 in the centrosome.
PCM1 and CEP55 regulate the centrosomal localization of USP9X
Because centrosome proteins exhibit a high degree of interdependence in their localizations, we next asked whether the centrosomal localization of USP9X is controlled by PCM1 or CEP55. To test this hypothesis, control siRNA, PCM1 siRNA, or CEP55 siRNA was transfected into U2OS cells. Immunofluorescent staining followed by confocal microscopy analysis indicated that knockdown of the expression of either PCM1 (Fig. 4A) or CEP55 (Fig. 4B) resulted in a diminished centrosomal localization of USP9X in U2OS cells, suggesting that the centrosomal localization of USP9X is dependent on PCM1 and CEP55.
Figure 4.
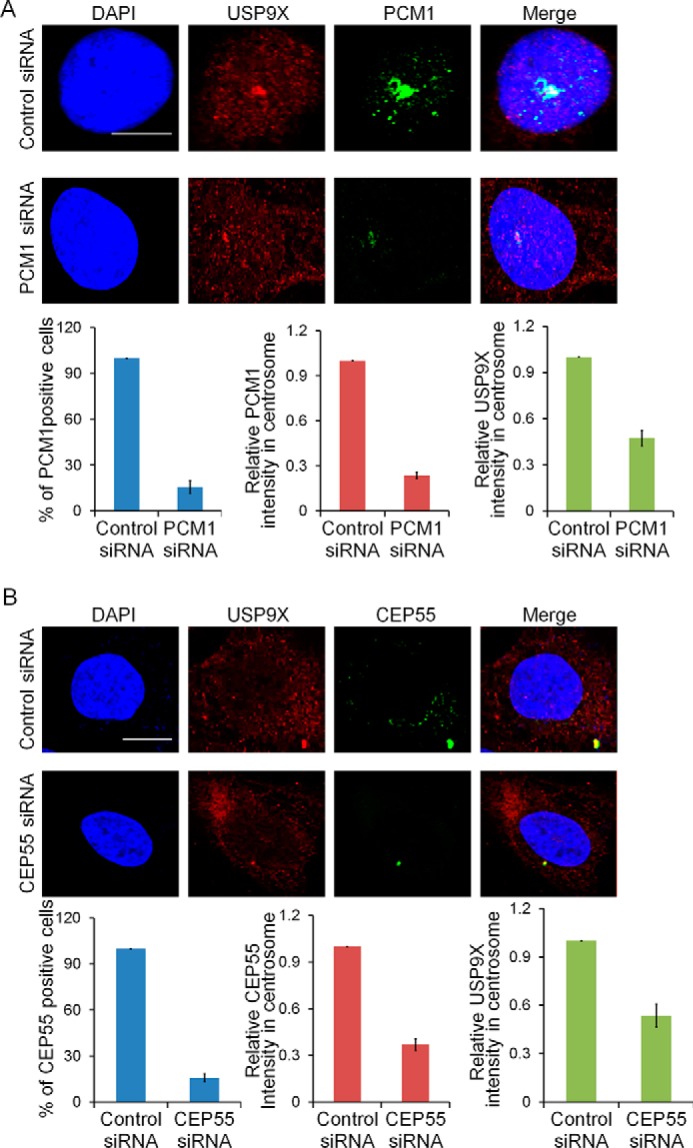
PCM1 and CEP55 are both required for centrosomal localization of USP9X. A, U2OS cells transfected with control siRNA or PCM1 siRNA were fixed and subjected to immunostaining with antibodies against PCM1 and USP9X. The percentage of cells with PCM1 knockdown was counted, and the relative intensity of PCM1 or USP9X in the centrosome was analyzed by ImageJ software. Error bars represent S.D. for biological triplicate experiments. **, p < 0.01, Student's t test. Scale bar, 10 μm. B, U2OS cells transfected with control siRNA or CEP55 siRNA were fixed and subjected to immunostaining with antibodies against CEP55 and USP9X. The percentage of cells with CEP55 knockdown was counted, and the relative intensity of CEP55 or USP9X in the centrosome was analyzed by ImageJ software. Error bars represent S.D. for biological triplicate experiments. **, p < 0.01, Student's t test. Scale bar, 10 μm.
USP9X is physically associated with PCM1 and CEP55
The functional link between USP9X and PCM1 or CEP55 prompted us to investigate whether USP9X is physically associated with PCM1 and CEP55. To test this hypothesis, co-immunoprecipitation experiments were performed with MCF-7 cell extracts. Immunoprecipitation (IP) with antibodies against PCM1 or CEP55 followed by immunoblotting (IB) with antibodies against USP9X demonstrated that USP9X was efficiently co-immunoprecipitated with PCM1 or CEP55 (Fig. 5A). Reciprocally, IP with antibodies against USP9X and IB with antibodies against PCM1 or CEP55 also revealed that either PCM1 or CEP55 was efficiently co-immunoprecipitated with USP9X (Fig. 5B). To further understand the molecular details of the association of USP9X with PCM1 and CEP55, we analyzed the domain structures of USP9X. In its N-terminal region, there is an α-α superhelix domain (Fig. 5C), which is supposed to mediate protein–protein interaction (38, 39). Indeed, we demonstrated that the N-terminal region of USP9X (USP9X-N) is necessary and sufficient for its association with PCM1 and CEP55 (Fig. 5C). In support of the above findings, fluorescence microscopy analysis revealed that USP9X-N is able to localize to centrosome, whereas lack of the N-terminal region impaired USP9X centrosome localization (Fig. 5D). These results indicated that USP9X and PCM1 or CEP55 co-exist in the same multiprotein-containing complex and are physically associated (could be indirectly or directly), which provides a meaningful explanation for the centrosomal localization of USP9X and its centrosome substrate preference.
Figure 5.
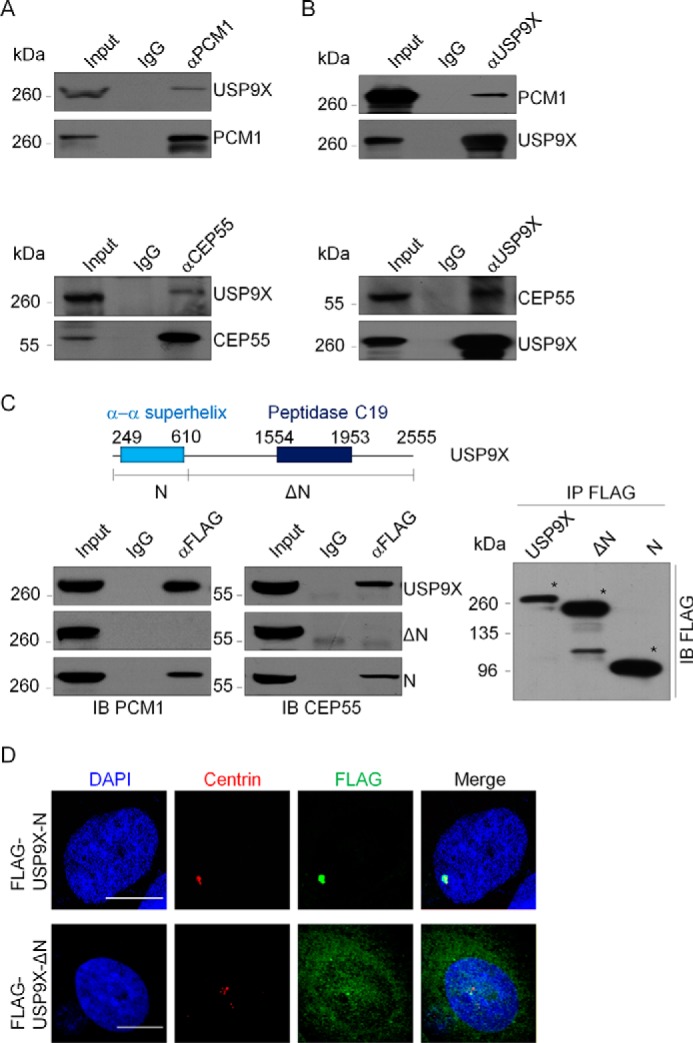
USP9X co-exist with PCM1 or CEP55. A, whole-cell lysates from MCF-7 cells were immunoprecipitated with antibody against PCM1 or CEP55 followed by IB with antibodies against the indicated proteins. B, whole-cell lysates from MCF-7 cells were immunoprecipitated with USP9X antibody followed by immunoblotting with antibodies against the indicated proteins. C, co-immunoprecipitation analysis of the molecular interface required for the association of USP9X with PCM1 and CEP55. FLAG-tagged USP9X or deletion mutants were transfected into HeLa cells. Whole-cell lysates were immunoprecipitated with FLAG antibody followed by immunoblotting with antibodies against the indicated proteins. The conserved domains of USP9X were determined by the Simple Modular Architecture Research Tool (SMART). The asterisk indicates proteins immunoprecipitated with FLAG antibody. D, FLAG-tagged USP9X-N or USP9X-ΔN was transfected into U2OS cells. The cells were then fixed and immunostained with antibodies against the indicated proteins. Scale bar, 10 μm.
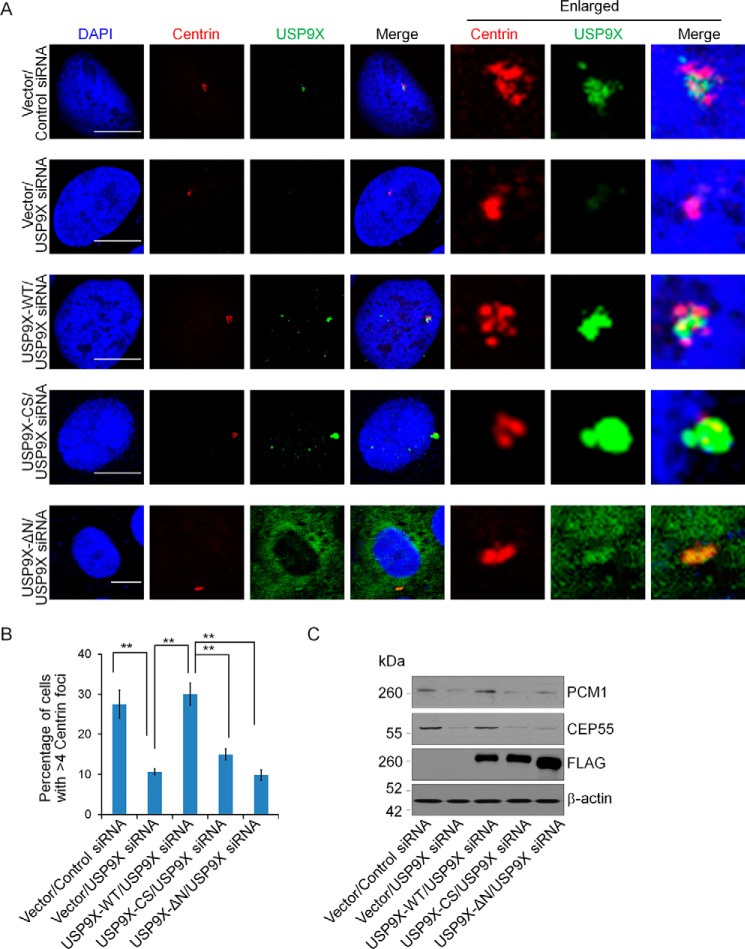
USP9X is required for centrosome biogenesis
To understand the biological significance of USP9X centrosomal localization, we next investigated the effect of USP9X on the biogenesis of centrosome. To this end, U2OS cells were transfected with control siRNA or siRNA targeting the 3′-UTR of USP9X mRNA followed by treatment with hydroxyurea to induce a prolonged S phase. Immunofluorescent staining of USP9X and Centrin in these cells showed that although control cells that were arrested in S phase underwent centrosome overduplication, manifested by the presence of more than four Centrin foci, USP9X depletion resulted in a significant reduction of centrosome amplification (Fig. 6, A and B). Importantly, the phenotype associated with USP9X deficiency could be rescued by overexpression of USP9X-WT but not its catalytic mutant USP9X-C1566S or USP9X lacking the N-terminal region (USP9X-ΔN) (Fig. 6, A and B). Consistent with the observation that USP9X-ΔN was largely diffused in the cytoplasm (Fig. 6A), we showed that USP9X-ΔN overexpression could not restore the expression of PCM1 and CEP55 in USP9X-deficient cells (Fig. 6C and supplemental Fig. S2). These results indicate that USP9X-promoted centrosome biogenesis is dependent on its catalytic activity and its N-terminal module that is responsible for physical association of USP9X with PCM1 and CEP55. Collectively, these results support the notion that USP9X is an important player in the regulation of centrosome duplication/biogenesis.
Figure 6.
USP9X promotes centrosome biogenesis. A, control U2OS cells or U2OS cells stably expressing USP9X-WT, catalytic mutant USP9X (USP9X-CS), or USP9X-ΔN were transfected with control siRNA or USP9X 3′-UTR siRNA for 96 h followed by hydroxyurea treatment. Then the cells were fixed and immunostained with antibodies against the indicated proteins. Scale bar, 10 μm. B, population of cells from A with the indicated numbers of Centrin foci was counted. Error bars represent S.D. for biological triplicate experiments. **, p < 0.01, one-way ANOVA. C, cellular extracts from A were prepared and analyzed by Western blotting with antibodies against the indicated proteins.
Discussion
In this study, we used proteomic screening and mass spectrometry analysis and found that depletion of the X-linked deubiquitinase USP9X led to expression alterations of multiple genes including centrosome proteins PCM1 and CEP55. Co-immunoprecipitation analysis demonstrated that USP9X is physically associated with PCM1 and CEP55. Fluorescence microscopy analysis revealed that USP9X is spatially restricted to the centrosome where it is co-localized with PCM1 and CEP55 and promotes the stabilization of these proteins, indicating that USP9X is an integral component of the centrosome.
Understanding the cellular distributions of USP9X is a prerequisite for characterization of its role in development and disease. Thus far, USP9X has been reported to be present at distinct cellular organelles including the mitochondria (40) and nucleus (41), although a significant portion is localized in the cytoplasm and membrane (26, 28). Our observation that USP9X is localized in the centrosome expands the spatial domains and thus the functionality of this deubiquitinase. Interestingly, disrupting protein trafficking or the Golgi in polarized epithelia results in relocation or accumulation of USP9X, implying that this protein circulates between organelles and vesicles with different resident times at each compartment (24). Although whether USP9X is able to shuttle between the centrosome and other organelles or vesicles remains to be investigated, the diversified cellular localization of USP9X reflects a dynamic nature of distribution of this protein in cells. Consistently, USP9X has been reported to interact with and deubiquitinate dozens of proteins in distinct cellular compartments (24).
Functionally, based on its targeted proteins and various signaling pathways, USP9X has been implicated in the regulation of multiple cellular activities (24). We showed that USP9X is spatially restricted to the centrosome by PCM1 and CEP55 and regulates centrosome duplication in its catalytic activity-dependent manner, although it remains to be explored whether and how PCM1 or CEP55 contributes to USP9X-promoted centrosome biogenesis in the future. In addition to our finding, several studies reported that the deubiquitination enzymes including BAP1, USP33, USP1, USP44, cylindromatosis (CYLD), and USP21 have been implicated in centrosome regulation (43–48), pointing to a role of deubiquitinases as key regulators in centrosome homeostasis. Possibly, PCM1 and/or CEP55 could be controlled by these enzymes. In support of our observation, a recent study reported that USP9X is capable of stabilizing centrosomal proteins CEP131 and PCM1 (49).
In parallel with the findings that USP9X dysregulation has been involved in multiple malignancies and considered as an oncogenic protein (24), CEP55 overexpression has been found to significantly correlate with tumor stage, aggressiveness, metastasis, and poor prognosis across multiple tumor types (19, 50, 51). Elucidation of whether CEP55 is a key substrate linking USP9X dysregulation with tumorigenesis will be helpful in understanding the biological function of USP9X and benefit tumor intervention.
Moreover, USP9X has been implicated in several pathological states including Turner syndrome (52), X-linked intellectual disability (31), seizures (53), and Parkinson disease (30). Interestingly, the centrosome is reported to be required for proper neurodevelopment (54–56), and recent genetic studies suggested that centrosomal dysfunction underlies risks for various neuropsychiatric disorders, namely schizophrenia and bipolar disorder (57–59). Because PCM1 is one of the reproducible linkage loci for schizophrenia and bipolar disorder (57) and we found that USP9X functions to stabilize PCM1, we postulate that the USP9X dysregulation associated with neurological disorders could possibly be attributed to its functionality and behavior in the centrosome. Therefore, it is important to explore the relationship between substrate diversity and cellular activities of USP9X and investigate the effect as well as the underlying mechanism of USP9X-promoted centrosome biogenesis in physiological or pathological states.
Materials and methods
Antibodies and reagents
The sources of antibodies against the following proteins were: PCM1 (sc-398365; for IF) from Santa Cruz Biotechnology; β-actin (A1978), CEP55 (SAB1409483), γ-tubulin (T6557), and USP9X (WH0008239M1; for IP) from Sigma; Pericentrin (ab28144; for IF) from Abcam; Centrin (04-1624) and Pericentrin (ABT59; for WB) from Millipore; USP9X (55054-1-AP; for IF, IP, and WB) and PCM1 (19856-1-AP; for WB and IP) from Proteintech. Hydroxyurea (H8627) and MG132 (SML1135) were purchased from Sigma. CHX (0970; working concentration, 50 μg/ml) was purchased from Tocris.
Plasmids
The human FLAG-tagged USP9X and USP9X-C1566S mutant carried by pLenti-Hygro vector were amplified from wild-type and catalytic mutant V5-tagged USP9X cDNAs kindly provided by Dr. Feng Cong (Novartis Institutes for BioMedical Research, Cambridge, MA), respectively. Truncations of FLAG-tagged USP9X were amplified from USP9X cDNA and subcloned into pLenti-Hygro vector.
Cell culture
MCF-7 and U2OS were obtained from the American Type Culture Collection (Manassas, VA) and cultured under the manufacturer's instructions. All of the cell lines have been tested for Mycoplasma contamination.
Western blotting
The experiments were performed according to standard procedures, and large scans of blots shown in the main figures are shown in supplemental Figs. S3 and S4.
TMT labeling assay
Protein extraction
Control MCF-7 cells and USP9X-deficient MCF-7 cells were sonicated three times on ice using a high-intensity ultrasonic processor (Scientz) in lysis buffer (8 m urea, 1% Triton X-100, 10 mm DTT, 50 mm Tris-HCl, and 1% protease inhibitor mixture). The remaining debris was removed by centrifugation at 20,000 × g at 4 °C for 10 min. Finally, the protein was precipitated with cold 15% TCA for 2 h at −20 °C. After centrifugation at 4 °C for 10 min, the supernatant was discarded. The remaining precipitate was washed with cold acetone three times. The protein was redissolved in buffer (8 m urea and 100 mm TEAB, pH 8.0), and the protein concentration was determined with a 2-D Quant kit (GE Healthcare) according to the manufacturer's instructions.
Trypsin digestion
For digestion, the protein solution was reduced with 10 mm DTT (Sigma-Aldrich) for 1 h at 37 °C and alkylated with 20 mm iodoacetamide (Sigma-Aldrich) for 45 min at room temperature in darkness. For trypsin digestion, the protein sample was diluted by adding 100 mm TEAB (Sigma-Aldrich) to urea with concentration less than 2 m. Finally, trypsin (Promega) was added at a 1:50 trypsin-to-protein mass ratio for the first digestion overnight and a 1:100 trypsin-to-protein mass ratio for a second 4-h digestion. Approximately 100 μg of protein for each sample was digested with trypsin for the following experiments.
TMT labeling
After trypsin digestion, peptide was desalted by Strata X C18 solid-phase extraction column (Phenomenex) and vacuum-dried. Peptide was reconstituted in 0.5 m TEAB and processed according to the manufacturer's protocol for the fourplex TMT kit (Thermo Fisher). Briefly, 1 unit of TMT reagent (defined as the amount of reagent required to label 100 μg of peptide) was thawed and reconstituted in 24 μl of acetonitrile (Fisher Chemical). The peptide mixtures were then incubated for 2 h at room temperature, pooled, desalted, and dried by vacuum centrifugation.
HPLC fractionation
The sample was then fractionated by high-pH reversed-phase HPLC using an Agilent 300Extend C18 column (5-μm particles, 4.6-mm inner diameter, 250-mm length). Briefly, peptides were first separated with a gradient of 2–60% acetonitrile in 10 mm ammonium bicarbonate, pH 10, over 80 min into 80 fractions. Then the peptides were combined into eight fractions using the pooling scheme as described in a study reported by Svinkina et al. (42). Pooled fractions were dried by vacuum centrifugation.
Quantitative proteomic analysis by LC-MS/MS
Peptides were dissolved in 0.1% formic acid, directly loaded onto a reversed-phase precolumn (Acclaim PepMap 100, Thermo Scientific). Peptide separation was performed using a reversed-phase analytical column (Acclaim PepMap RSLC, Thermo Scientific). The gradient comprised an increase from 6 to 10% solvent B (0.1% formic acid in 98% acetonitrile) over 3 min, 10–22% in 24 min, 22–35% in 8 min, climbing to 80% in 4 min, and then holding at 80% for the last 3 min, all at a constant flow rate of 300 nl/min on an EASY-nLC 1000 UPLC system, The resulting peptides were analyzed by a Q ExactiveTM hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). The peptides were subjected to the nanospray ionization source followed by tandem mass spectrometry (MS/MS) in a Q Exactive Plus (Thermo Fisher Scientific) coupled online to the UPLC system. Intact peptides were detected in the Orbitrap at a resolution of 70,000. Peptides were selected for MS/MS using a normal collision energy setting of 30; ion fragments were detected in the Orbitrap at a resolution of 17,500. A data-dependent procedure that alternated between one MS scan and 20 MS/MS scans was applied for the top 20 precursor ions above a threshold ion count of 2E4 in the MS survey scan with 30.0-s dynamic exclusion. The electrospray voltage applied was 2.0 kV. Automatic gain control was used to prevent overfilling of the ion trap; 5E4 ions were accumulated for generation of MS/MS spectra. For MS scans, the m/z scan range was 350–1800. Fixed first mass was set as 100 m/z.
Database search
The resulting MS/MS data were processed using the Mascot search engine (v.2.3.0). Tandem mass spectra were searched against the Swiss-Prot human database. Trypsin/P was specified as the cleavage enzyme allowing up to two missing cleavages. Mass error was set to 10 ppm for precursor ions and 0.02 Da for fragment ions. Carbamidomethyl on Cys, TMT fourplex on N terminus, and TMT fourplex on Lys were specified as fixed modification, and oxidation on Met was specified as a variable modification. The false discovery rate was adjusted to <1%, and peptide ion score was set ≥20. By modeling the normal distribution of the protein ratios, cutoff values of ≦0.770 and ≧1.3 and p < 0.05 examined by t test were applied to find the protein with significant down- or up-regulation after USP9X depletion.
Immunofluorescence
Cells on glass coverslips (BD Biosciences) were fixed with 2% paraformaldehyde and permeabilized with 0.2% Triton X-100 in PBS. Samples were then blocked in 5% donkey serum in the presence of 0.1% Triton X-100 and stained with the appropriate primary and secondary antibodies coupled to Alexa Fluor 488 or 594 (Invitrogen). To avoid bleed-through effects in double-staining experiments, each dye was scanned independently in a multitracking mode. Almost 100 cells in each treatment were scored in biological triplicate experiments.
Immunoprecipitation
Cell lysates were prepared by incubating the cells in NETN buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.2% Nonidet P-40, and 2 mm EDTA) in the presence of protease inhibitor mixtures (Roche Applied Science) for 20 min at 4 °C. This was followed by centrifugation at 14,000 × g for 15 min at 4 °C. For immunoprecipitation, about 500 μg of protein was incubated with control or specific antibodies (1–2 μg) for 12 h at 4 °C with constant rotation; 50 μl of 50% protein G magnetic beads (Invitrogen) was then added, and the incubation was continued for an additional 2 h. Beads were then washed five times using the lysis buffer. Between washes, the beads were collected by magnetic stand (Invitrogen) at 4 °C. The precipitated proteins were eluted from the beads by resuspending the beads in 2× SDS-PAGE loading buffer and boiling for 5 min. The boiled immune complexes were subjected to SDS-PAGE followed by immunoblotting with appropriate antibodies.
RNA interference
All siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer's recommendations. The final concentration of the siRNA molecules was 10 nm, and cells were harvested 72 or 96 h later according to the purposes of the experiments. Control siRNA (ON-TARGETplus Non-Targeting Pool, D-001810-10) and USP9X siRNA (ON-TARGETplus, L-006099-00-0005) were obtained from Dharmacon as a SMARTpool library, whereas the individual siRNAs against USP9X (USP9X siRNA-1/3′-UTR; GAGAGTTTATTCACTGTCTTA), CEP55 siRNA (GUGGGAAAGGAAAGCUGAC), and PCM1 siRNA (GGACAAAUUGCUUGGAGAA) were chemically synthesized by Sigma.
Real-time RT-PCR
Total cellular RNAs were isolated with TRIzol reagent (Invitrogen) and used for first-strand cDNA synthesis with a reverse transcription system (Roche Applied Science). Quantitation of all gene transcripts was done by quantitative PCR using Power SYBR Green PCR Master Mix (Roche Applied Science) and an ABI PRISM 7500 sequence detection system (Applied Biosystems) with the expression of GAPDH as the internal control. The primers used were: USP9X, AAGTGAAGCATGTCAGCGATT (forward) and GCCACACATAGCTCCACCA (reverse); PCM1, CGGAGTCGTCACCAGGAGT (forward) and GCTGTTGCTCTACCTTGGGAAT (reverse); CEP55, AGTAAGTGGGGATCGAAGCCT (forward) and CTCAAGGACTCGAATTTTCTCCA (reverse); Pericentrin, TTGTGGTCCCAGCTTGATTCT (forward) and CTCCAGTTCGGACTCATGCT (reverse); and GAPDH, GAAGGTGAAGGTCGGAGTC (forward) and GAAGATGGTGATGGGATTTC (reverse). The cycle threshold values (Ct values) were used to calculate the -fold differences by the ΔΔCt method.
Statistical analysis
Data from biological triplicate experiments are presented with error bars as S.D. Two-tailed unpaired Student's t test was used for comparing two groups of data. Analysis of variance (ANOVA) in conjugation with Bonferroni's correction was used to compare multiple groups of data except for the TMT labeling assay. Adjusted p values after Bonferroni's correction are shown. All of the statistical testing results except TMT labeling assay were determined by SPSS 19.0 software.
Author contributions
Q. W., Y. T., Y. Z., and W. G. designed the research studies. Q. W. and Y. T. conducted experiments. Q. W. and Y. T. acquired data. Q. W., Y. T., Y. X., Y. J., S. X., Q. D., Y. Z., and W. G. analyzed data. Q. W., Y. T., Y. Z., and W. G. wrote the manuscript.
Supplementary Material
This work was supported by National Natural Science Foundation of China Grants 81200763 and 81670963 (to W. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S4 and Table 1.
- PCM
- pericentriolar material
- TMT
- tandem mass tag
- CEP
- centrosomal protein
- DUB
- deubiquitinase
- USP
- ubiquitin-specific protease
- USP9X
- ubiquitin-specific peptidase 9 X-linked
- qRT
- quantitative reverse transcription
- IP
- immunoprecipitation
- IB
- immunoblotting
- USP9X-N
- N-terminal region of USP9X
- USP9X-ΔN
- USP9X lacking the N-terminal region
- IF
- immunofluorescence
- WB
- Western blotting
- TEAB
- tetraethylammonium bromide
- UPLC
- ultraperformance LC
- ANOVA
- analysis of variance.
References
- 1. Badano J. L., Teslovich T. M., and Katsanis N. (2005) The centrosome in human genetic disease. Nat. Rev. Genet. 6, 194–205 [DOI] [PubMed] [Google Scholar]
- 2. Conduit P. T., Wainman A., and Raff J. W. (2015) Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 16, 611–624 [DOI] [PubMed] [Google Scholar]
- 3. Baumann K. (2012) Cell cycle: maintaining centrosome copy number. Nat. Rev. Mol. Cell Biol. 13, 542. [DOI] [PubMed] [Google Scholar]
- 4. David R. (2011) Cell cycle: keeping centrosome numbers in check. Nat. Rev. Mol. Cell Biol. 12, 466. [DOI] [PubMed] [Google Scholar]
- 5. Lüders J. (2012) The amorphous pericentriolar cloud takes shape. Nat. Cell Biol. 14, 1126–1128 [DOI] [PubMed] [Google Scholar]
- 6. Loncarek J., Hergert P., Magidson V., and Khodjakov A. (2008) Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 10, 322–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hori A., and Toda T. (2017) Regulation of centriolar satellite integrity and its physiology. Cell. Mol. Life Sci. 74, 213–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Swaminathan S. (2004) Human disease: the centrosome connection. Nat. Cell Biol. 6, 383. [DOI] [PubMed] [Google Scholar]
- 9. Ou Y., Zhang M., and Rattner J. B. (2004) The centrosome: the centriole-PCM coalition. Cell Motil. Cytoskeleton 57, 1–7 [DOI] [PubMed] [Google Scholar]
- 10. Staples C. J., Myers K. N., Beveridge R. D., Patil A. A., Lee A. J., Swanton C., Howell M., Boulton S. J., and Collis S. J. (2012) The centriolar satellite protein Cep131 is important for genome stability. J. Cell Sci. 125, 4770–4779 [DOI] [PubMed] [Google Scholar]
- 11. Kim J., Krishnaswami S. R., and Gleeson J. G. (2008) CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 17, 3796–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Villumsen B. H., Danielsen J. R., Povlsen L., Sylvestersen K. B., Merdes A., Beli P., Yang Y. G., Choudhary C., Nielsen M. L., Mailand N., and Bekker-Jensen S. (2013) A new cellular stress response that triggers centriolar satellite reorganization and ciliogenesis. EMBO J. 32, 3029–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stowe T. R., Wilkinson C. J., Iqbal A., and Stearns T. (2012) The centriolar satellite proteins Cep72 and Cep290 interact and are required for recruitment of BBS proteins to the cilium. Mol. Biol. Cell 23, 3322–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bettencourt-Dias M., Hildebrandt F., Pellman D., Woods G., and Godinho S. A. (2011) Centrosomes and cilia in human disease. Trends Genet. 27, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dzhindzhev N. S., Yu Q. D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., and Glover D. M. (2010) Asterless is a scaffold for the onset of centriole assembly. Nature 467, 714–718 [DOI] [PubMed] [Google Scholar]
- 16. Wilkinson C. J., Carl M., and Harris W. A. (2009) Cep70 and Cep131 contribute to ciliogenesis in zebrafish embryos. BMC Cell Biol. 10, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fabbro M., Zhou B. B., Takahashi M., Sarcevic B., Lal P., Graham M. E., Gabrielli B. G., Robinson P. J., Nigg E. A., Ono Y., and Khanna K. K. (2005) Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell 9, 477–488 [DOI] [PubMed] [Google Scholar]
- 18. Kuo T. C., Chen C. T., Baron D., Onder T. T., Loewer S., Almeida S., Weismann C. M., Xu P., Houghton J. M., Gao F. B., Daley G. Q., and Doxsey S. (2011) Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 13, 1214–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeffery J., Sinha D., Srihari S., Kalimutho M., and Khanna K. K. (2016) Beyond cytokinesis: the emerging roles of CEP55 in tumorigenesis. Oncogene 35, 683–690 [DOI] [PubMed] [Google Scholar]
- 20. Martinez-Garay I., Rustom A., Gerdes H. H., and Kutsche K. (2006) The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics 87, 243–253 [DOI] [PubMed] [Google Scholar]
- 21. Doxsey S. J. (2005) Molecular links between centrosome and midbody. Mol. Cell 20, 170–172 [DOI] [PubMed] [Google Scholar]
- 22. van der Horst A., Simmons J., and Khanna K. K. (2009) Cep55 stabilization is required for normal execution of cytokinesis. Cell Cycle 8, 3742–3749 [DOI] [PubMed] [Google Scholar]
- 23. Komander D., Clague M. J., and Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 24. Murtaza M., Jolly L. A., Gecz J., and Wood S. A. (2015) La FAM fatale: USP9X in development and disease. Cell. Mol. Life Sci. 72, 2075–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wood S. A., Pascoe W. S., Ru K., Yamada T., Hirchenhain J., Kemler R., and Mattick J. S. (1997) Cloning and expression analysis of a novel mouse gene with sequence similarity to the Drosophila fat facets gene. Mech. Dev. 63, 29–38 [DOI] [PubMed] [Google Scholar]
- 26. Murray R. Z., Jolly L. A., and Wood S. A. (2004) The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and β-catenin. Mol. Biol. Cell 15, 1591–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nagai H., Noguchi T., Homma K., Katagiri K., Takeda K., Matsuzawa A., and Ichijo H. (2009) Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol. Cell 36, 805–818 [DOI] [PubMed] [Google Scholar]
- 28. Théard D., Labarrade F., Partisani M., Milanini J., Sakagami H., Fon E. A., Wood S. A., Franco M., and Luton F. (2010) USP9x-mediated deubiquitination of EFA6 regulates de novo tight junction assembly. EMBO J. 29, 1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grasso D., Ropolo A., Lo Ré A., Boggio V., Molejón M. I., Iovanna J. L., Gonzalez C. D., Urrutia R., and Vaccaro M. I. (2011) Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J. Biol. Chem. 286, 8308–8324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rott R., Szargel R., Haskin J., Bandopadhyay R., Lees A. J., Shani V., and Engelender S. (2011) α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc. Natl. Acad. Sci. U.S.A. 108, 18666–18671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Homan C. C., Kumar R., Nguyen L. S., Haan E., Raymond F. L., Abidi F., Raynaud M., Schwartz C. E., Wood S. A., Gecz J., and Jolly L. A. (2014) Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am. J. Hum. Genet. 94, 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M. J., Soligo S., Morsut L., Inui M., Moro S., Modena N., Argenton F., Newfeld S. J., et al. (2009) FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 [DOI] [PubMed] [Google Scholar]
- 33. Naik E., Webster J. D., DeVoss J., Liu J., Suriben R., and Dixit V. M. (2014) Regulation of proximal T cell receptor signaling and tolerance induction by deubiquitinase Usp9X. J. Exp. Med. 211, 1947–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park Y., Jin H. S., and Liu Y. C. (2013) Regulation of T cell function by the ubiquitin-specific protease USP9X via modulating the Carma1-Bcl10-Malt1 complex. Proc. Natl. Acad. Sci. U.S.A. 110, 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu J., Taya S., Kaibuchi K., and Arnold A. P. (2005) Spatially and temporally specific expression in mouse hippocampus of Usp9x, a ubiquitin-specific protease involved in synaptic development. J. Neurosci. Res. 80, 47–55 [DOI] [PubMed] [Google Scholar]
- 36. Jolly L. A., Taylor V., and Wood S. A. (2009) USP9X enhances the polarity and self-renewal of embryonic stem cell-derived neural progenitors. Mol. Biol. Cell 20, 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang S., Kollipara R. K., Srivastava N., Li R., Ravindranathan P., Hernandez E., Freeman E., Humphries C. G., Kapur P., Lotan Y., Fazli L., Gleave M. E., Plymate S. R., Raj G. V., Hsieh J. T., et al. (2014) Ablation of the oncogenic transcription factor ERG by deubiquitinase inhibition in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 111, 4251–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takayanagi H., Yuzawa S., and Sumimoto H. (2015) Structural basis for the recognition of the scaffold protein Frmpd4/Preso1 by the TPR domain of the adaptor protein LGN. Acta Crystallogr. F Struct. Biol. Commun. 71, 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taschner M., Kotsis F., Braeuer P., Kuehn E. W., and Lorentzen E. (2014) Crystal structures of IFT70/52 and IFT52/46 provide insight into intraflagellar transport B core complex assembly. J. Cell Biol. 207, 269–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwickart M., Huang X., Lill J. R., Liu J., Ferrando R., French D. M., Maecker H., O'Rourke K., Bazan F., Eastham-Anderson J., Yue P., Dornan D., Huang D. C., and Dixit V. M. (2010) Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463, 103–107 [DOI] [PubMed] [Google Scholar]
- 41. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., and Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Svinkina T., Gu H., Silva J. C., Mertins P., Qiao J., Fereshetian S., Jaffe J. D., Kuhn E., Udeshi N. D., and Carr S. A. (2015) Deep, quantitative coverage of the lysine acetylome using novel anti-acetyl-lysine antibodies and an optimized proteomic workflow. Mol. Cell. Proteomics 14, 2429–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zarrizi R., Menard J. A., Belting M., and Massoumi R. (2014) Deubiquitination of γ-tubulin by BAP1 prevents chromosome instability in breast cancer cells. Cancer Res. 74, 6499–6508 [DOI] [PubMed] [Google Scholar]
- 44. Li J., D'Angiolella V., Seeley E. S., Kim S., Kobayashi T., Fu W., Campos E. I., Pagano M., and Dynlacht B. D. (2013) USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature 495, 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jung J. K., Jang S. W., and Kim J. M. (2016) A novel role for the deubiquitinase USP1 in the control of centrosome duplication. Cell Cycle 15, 584–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., Foreman O., Wigle D. A., Kosari F., Vasmatzis G., Salisbury J. L., van Deursen J., and Galardy P. J. (2012) USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J. Clin. Investig. 122, 4362–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gomez-Ferreria M. A., Bashkurov M., Mullin M., Gingras A. C., and Pelletier L. (2012) CEP192 interacts physically and functionally with the K63-deubiquitinase CYLD to promote mitotic spindle assembly. Cell Cycle 11, 3555–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Urbé S., Liu H., Hayes S. D., Heride C., Rigden D. J., and Clague M. J. (2012) Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol. Biol. Cell 23, 1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li X., Song N., Liu L., Liu X., Ding X., Song X., Yang S., Shan L., Zhou X., Su D., Wang Y., Zhang Q., Cao C., Ma S., Yu N., et al. (2017) USP9X regulates centrosome duplication and promotes breast carcinogenesis. Nat. Commun. 8, 14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carter S. L., Eklund A. C., Kohane I. S., Harris L. N., and Szallasi Z. (2006) A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38, 1043–1048 [DOI] [PubMed] [Google Scholar]
- 51. Zhou W., Yang Y., Xia J., Wang H., Salama M. E., Xiong W., Xu H., Shetty S., Chen T., Zeng Z., Shi L., Zangari M., Miles R., Bearss D., Tricot G., and Zhan F. (2013) NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell 23, 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xu J., Taya S., Kaibuchi K., and Arnold A. P. (2005) Sexually dimorphic expression of Usp9x is related to sex chromosome complement in adult mouse brain. Eur. J. Neurosci. 21, 3017–3022 [DOI] [PubMed] [Google Scholar]
- 53. Paemka L., Mahajan V. B., Ehaideb S. N., Skeie J. M., Tan M. C., Wu S., Cox A. J., Sowers L. P., Gecz J., Jolly L., Ferguson P. J., Darbro B., Schneider A., Scheffer I. E., Carvill G. L., et al. (2015) Seizures are regulated by ubiquitin-specific peptidase 9 X-linked (USP9X), a de-ubiquitinase. PLoS Genet. 11, e1005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ayala R., Shu T., and Tsai L. H. (2007) Trekking across the brain: the journey of neuronal migration. Cell 128, 29–43 [DOI] [PubMed] [Google Scholar]
- 55. Tsai L. H., and Gleeson J. G. (2005) Nucleokinesis in neuronal migration. Neuron 46, 383–388 [DOI] [PubMed] [Google Scholar]
- 56. Gupta A., Tsai L. H., and Wynshaw-Boris A. (2002) Life is a journey: a genetic look at neocortical development. Nat. Rev. Genet. 3, 342–355 [DOI] [PubMed] [Google Scholar]
- 57. Gurling H. M., Critchley H., Datta S. R., McQuillin A., Blaveri E., Thirumalai S., Pimm J., Krasucki R., Kalsi G., Quested D., Lawrence J., Bass N., Choudhury K., Puri V., O'Daly O., et al. (2006) Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Arch. Gen. Psychiatry 63, 844–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hennah W., Tomppo L., Hiekkalinna T., Palo O. M., Kilpinen H., Ekelund J., Tuulio-Henriksson A., Silander K., Partonen T., Paunio T., Terwilliger J. D., Lönnqvist J., and Peltonen L. (2007) Families with the risk allele of DISC1 reveal a link between schizophrenia and another component of the same molecular pathway, NDE1. Hum. Mol. Genet. 16, 453–462 [DOI] [PubMed] [Google Scholar]
- 59. Ishizuka K., Paek M., Kamiya A., and Sawa A. (2006) A review of Disrupted-In-Schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions. Biol. Psychiatry 59, 1189–1197 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.