Abstract
Scope
β,β-carotene-9’,10’-dioxygenase 2 (BCO2) is a carotenoid cleavage enzyme localized to the inner mitochondrial membrane in mammals. This study was aimed to assess the impact of genetic ablation of BCO2 on hepatic oxidative stress through mitochondrial function in mice.
Methods and Results
Liver samples from 6 week old male BCO2−/− knockout (KO) and isogenic wild-type (WT) mice were subjected to proteomics and functional activity assays. Compared to the WT, KO mice consumed more food (by 18 %) yet displayed significantly lower body weight (by 12 %). Mitochondrial proteomic results demonstrated that loss of BCO2 was associated with quantitative changes of the mitochondrial proteome mainly shown by suppressed expression of enzymes and/or proteins involved in fatty acid β–oxidation, the tricarboxylic acid cycle, and the electron transport chain (ETC). The mitochondrial basal respiratory rate, proton leak, and ETC complex II capacity were significantly elevated in the livers of KO compared to WT mice. Moreover, elevated reactive oxygen species and increased mitochondrial protein carbonylation were also demonstrated in liver of KO mice.
Conclusions
Loss of BCO2 induces mitochondrial hyperactivation, mitochondrial stress and changes of the mitochondrial proteome, leading to mitochondrial energy insufficiency. BCO2 appears to be critical for proper hepatic mitochondrial function.
Keywords: β, β-carotene -9’, 10’-oxygenase 2 (BCO2); electron transport chain; liver; mitochondrial dysfunction; mitochondrial proteome; oxidative stress
Introduction
The imbalance between the systemic production of reactive oxygen species (ROS) and the systemic capacity to quench and/or detoxify ROS production results in oxidative stress (1, 2). ROS can damage many cellular components, such as lipids, proteins, and nucleotides (3). Mitochondria are the power “engine” of the cell involved in numerous cellular processes including redox homeostasis, cellular signaling, development, growth, differentiation and even cell death (4–6). Leakage of protons and/or electrons from the mitochondrial electron transport chain (ETC) results in enhanced generation of ROS and subsequent mitochondrial oxidative stress and decreased efficiency of ATP production, cellular oxidative stress (7) and subsequent inflammation (8). Mitochondrial oxidative stress has been well established as a causal factor in the development of chronic diseases, such as diabetes (9–11) and obesity (12). In diabetes, oxidative stress is associated with tissue-specific changes of the mitochondrial proteomes (13), impaired insulin function and disrupted blood glucose control (14, 15). Additionally, systemic oxidative stress is closely associated with chronic diseases, such as obesity in humans (16). Thus, properly functioning of mitochondria is crucial for the maintenance of cellular homeostasis in mammals.
β, β-carotene-9’, 10’-oxygenase 2 (BCO2), a carotenoid cleavage enzyme that is conserved in primates, catalyzes the asymmetrical cleavage of carotenoids at 9’, 10’ and 9, 10 double bond to form various apocarotenoid cleavage products (17,18,19). Interestingly, in humans the BCO2 gene locus is closely linked to the proinflammatory mediator interleukin-18 (IL-18) (20). Variants at the BCO2 locus are associated with the IL-18 level, but not carotenoids in either plasma or macula (20). Studies have revealed that mutations in human BCO2 have been linked to macular degeneration (21), insulin resistance and obesity (22), and prostate cancer (23). Further, the loss of BCO2 protein expression is associated with the development of anemia in zebrafish, though the anemia results not from impaired erythropoiesis, but instead from increased apoptosis of erythrocyte precursors (24). BCO2 expression is also significantly reduced in obese and diabetic mice (25, 26) . Accordingly, increased intake of a high-fat diet rich in carotenoids increase the expression of BCO2 protein and was associated with increased insulin sensitivity and attenuation of fatty liver in obese C57BL/6J mice (25). Collectively, these findings indicate that BCO2 likely plays roles other than just as a carotenoid cleavage enzyme in mammals.
BCO2 is widely expressed in many types of tissues and is localized to in the inner mitochondrial membrane (18, 27, 28). Exogenous H2O2 induces BCO2 protein expression in HepG2 cells though the function of this protein under oxidative stress is not clear (29). Lutein and/or zeaxanthin-enriched diets cause hepatic mitochondrial dysfunction in BCO2 KO mice due to oxidative stress induced by accumulation of carotenoids (29). The role of BCO2 in retinal carotenoid metabolism is still controversial (17, 19). It remains unknown whether BCO2 functions as a structural protein in the mitochondrial homeostasis. In this study, the liver tissues of wild type (WT) and BCO2−/− knockout (KO) mice fed a standard chow diet were analyzed by mitochondrial proteome profiling and functional activity assays, production of reactive oxygen species (ROS), and mitochondrial protein carbonylation. We sought to determine whether BCO2 protein was critical to mitochondrial function in liver, and to investigate how ablation of BCO2 caused hepatic mitochondrial dysfunction and triggers cellular oxidative stress.
Materials and methods
Animals, animal care and fasting blood glucose test
The original BCO2 knockout mouse (29) was backcrossed to the wild-type 129S6 (WT) for seven generations to obtain a BCO2−/− knockout (KO) strain in a 129S6 genetic background (Taconic, New York). The breeding colonies of both WT and KO strains were maintained in the Animal Resources Facility at Oklahoma State University. After weaning at 21 days, mice had free access to water and food (e.g., ad libitum) throughout the study. All animal experiments and procedures were performed in accordance with our protocol approved by the Oklahoma State University Institutional Animal Care and Use Committee (OSU IACUC protocol # HS-14-4). Male mice were group-housed (3 mice/cage) in a controlled environment with a 12-h light/dark cycle (7:00 -19:00 light/19:00-7:00 dark) and fed a standard chow diet for 3 weeks (CD, 10% kcal from fat, catalog # D12450B, Research Diets, Inc., New Brunswick, NJ). The macronutrient composition of the CD is 10% kcal from fat, 20% kcal from protein, and 70% kcal from carbohydrate. Food intake and body weight were monitored weekly at 8:30-9:30 AM starting at weaning (at 3 weeks of age) to 6 weeks of age (n=12). The mean of weekly food intake between 5 and 6 weeks and the body weight at 6 weeks of age were presented. WT and KO mice at 6 weeks of age were used for tissue collection and laboratory analysis. Animals were sacrificed by CO2 at 8:30-9:30 AM for experiments after being fasted for 5-6 hr. Before necropsy, the fasting blood glucose was tested by tail snip using the Precision Xtra blood glucose monitoring system (Alameda, CA, USA).
Measurement of plasma free fatty acids (FFA)
Plasma FFA was analyzed using a BioLis 24i automated analyzer (Carolina Chemistry, NC, USA). The FFA level is expressed as milliequivalents per liter (mEq/L).
Cellular ROS level
The hepatic ROS level was measured using dichlorodihydrofluorescein (DCF) as previously described (26). Briefly, 25 µL carboxy-DCF and H2-DCF dyes at 125 µmol/L were added into freshly prepared liver homogenates (100 µg protein/100 µL probes) in a 96-well plate. Changes in fluorescence intensity were recorded by the Bio-Tek Synergy HT fluorescence microplate reader (Winooski, VT) at excitation 485 nm/emission 530 nm. The ROS level was expressed as increased absorbance values (relative fluorescent unit OD 485/530, OD at 45 min - OD at 0 min).
Hepatic mitochondrial isolation
After euthanization, liver tissue were immediately used to assess mitochondrial function. Approximately 60 mg liver tissue was minced in 0.5 mL mitochondrial isolation buffer at pH 7.2 (210 mmol/L mannitol, 70 mmol/L sucrose, 5 mmol/L HEPES, 1 mmol/L EGTA and 0.5% (w/v) fatty acid-free bovine serum albumin) and homogenized (Qiagen tissueruptor, Cat. No. 9001271, Limburg, CA) for 10 sec at the lowest speed on ice. The homogenate was centrifuged at 600×g for 5 min at 4°C and the supernatant was transferred to new microfuges tubes centrifuged at 5000×g for 5 min at 4°C. After discarding the supernatant, the mitochondrial pellet was resuspended in 1 mL mitochondrial assay solution (1X-MAS, pH7.2: 220 mmol/L mannitol, 70 mmol/L sucrose, 5 mmol/L MgCl2, 2 mmol/L HEPES, 10 mmol/L KH2PO4, 1 mmol/L EGTA, 0.2% (w/v) fatty acid-free BSA). Mitochondrial fractions were isolated and the total protein concentrations were determined as previously (26). Non-mitochondrial contaminants from lysosomes and peroxisomes (30) in the mitochondrial pellets were confirmed by immunoblotting using antibodies against lysosome-associated membrane protein 1 (LAMP-1) and catalase, respectively, while ATP synthase α subunit 1 (ATP5A1) and citrate synthase were used as mitochondrial markers.
Mitochondrial respiration assay
Freshly isolated mitochondria were then used to assess cellular respiration using a coupled assay to determine mitochondrial ETC efficiency and proton leak (30). All processing steps were conducted on ice unless otherwise indicated. Briefly, diluted isolated mitochondria (4 µg protein/90 µL 1X- MAS) were loaded into individual wells of a Seahorse XFe 96 cell culture microplate. The pre-warmed (37°C) coupling substrates solution composed of 1X-MAS supplemented with 10 mmol/L pyruvate, 10 mmol/L malate and 10 mmol/L succinate was added into each cell. The oxygen consumption rate (OCR) of isolated hepatic mitochondria was measured using the Extracellular Flux XFe 96 Analyzer according to the manufacturer’s instructions (Seahorse Bioscience Inc., North Billerica, MA). Injections were as follows: Port A, 20 µL of 40 mmol/L ADP (4 mmol/L final concentration); Port B, 22 µL of 15 µmol/L oligomycin (OLIGO, 1.5 µmol/L final concentration); Port C, 24 µL of 40 µmol/L carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP, 4 µmol/L final concentration); Port D, 26 µL of 40 µmol/L antimycin A (AA, 4 µmol/L final concentration). The basal reading of OCR determined the basal respiration rate, while the difference between the OCR reading after OLIGO and AA injections were used to calculate the proton leak. The non-mitochondrial respiration OCR reading was eliminated from the coupling OCR readings (for additional details, see Supplemental Fig. S1A, the original Seahorse OCR graph enclosed as supplemental Fig. S1C, biomarkers for the lysosomes and peroxisomes shown in supplemental Fig. 1E). To measure electron flow through the electron transport chain, pre-warmed (37°C) electron flow substrate solution containing 1X-MAS supplemented with 10 mmol/L pyruvate, 10 mmol/L malate and 4 µmol/L FCCP. The injection protocol for electron flow was as follows: Port A, 20 µL of 20 µmol/L rotenone (2 µmol/L final concentration); Port B, 22 µL of 100 mmol/L succinate (1.5 µmol/L final concentration); Port C, 24 µL of 40 µmol/L AA (4 µmol/L final concentration); Port D, 26 µL of 100 mmol/L ascorbate/1 mmol/L 5,10,15,20-tetrakis(2,4,6-trimethylphenyl)porphyrin (TMP) (final concentrations of 10 mmol/L and 100 µmol/L, respectively). The OCR difference before and after rotenone injection was used to determine the capacity of complex I capacity. The OCR difference before and after AA administration reflected the complex II capacity, and the capacity of complex IV was calculated by subtracting OCR post-AA injection from the OCR post-ascorbate/TMP. The non-mitochondrial respiration OCR reading was substrated from the electron flow OCR readings (for additional details, see Supplemental Fig. S1B, the original Seahorse OCR graph enclosed as supplemental Fig. S1D. The instrumental and injection protocols for both mitochondrial functional assays is described in the Supplemental Table S1.
Detection of mitochondrial protein carbonylation
Mitochondrial protein carbonylation was detected as described (31) using an OxyBlot protein oxidation detection kit (catalog # S7150, EMD Millipore, Temecula, CA, USA) according to the manufacturer’s instructions. Protein samples were reacted with 2,4-dinitrophenylhydrazine (DNP-hydrazine, DNPH) to derivatize the carbonyl groups to 2,4-dinitrophenylhydrazone (DNP-hydrazone). Samples without DNPH treatment were used as a negative control (background). The DNPH-treated and non-treated samples were separated electrophoretically on SDS-PAGE followed by Western blotting using anti-DNP antibody. Total protein loading was normalized using succinate dehydrogenase subunit α (SDHA) as a loading control.
Mitochondrial proteomics and bioinformatics analysis
Hepatic mitochondria from fresh tissues were isolated by use of a Qproteome mitochondrial isolation kit (Qiagen, Limburg, CA) and used for the mitochondrial proteomics and aconitase activity assay. Equal amounts of mitochondrial samples (25.0 µg/sample) were digested with trypsin using standard methodologies, and the resulting peptides were analyzed by LC-MS/MS on an LTQ-OrbitrapXL mass spectrometer as previously described (32) using 40 cm analytical columns. Extracts from each mouse (n=3/group) were analyzed at four technical replicates. The resulting data were used for Mascot searching against mouse sequences in the Uniprot protein sequence database (> 46,000 sequences). Search results were validated using the Peptide Prophet and Protein Prophet algorithms in Scaffold (Proteome Software, Inc, Portland, OR), using a cut-off threshold of < 1% protein and peptide false discoveries. The resulting proteins were quantitated by spectrum counting. Proteins that were differentially expressed at a significant level were selected with the P value smaller than 0.05 using a two-tailed student’s t-test.
The DAVID 6.7 Bioinformatics tool (http://david.abcc.ncifcrf.gov/) was used for gene functional categorization. The proteins from mitochondrial proteomic data showing significant changes in expression were loaded into DAVID and analyzed to determine protein functional annotation to identify GO (gene-gene ontology) categories of protein functions that were enriched in the set of differentially expressed proteins based on these selected proteins (33). The selected protein functional analysis was also conducted by Ingenuity Pathway Analysis (IPA) which provided canonical pathways, upstream regulator analysis, top Toxicity (Tox) analysis and downstream effects analysis based on these selected proteins (34, 35).
Mitochondrial aconitase activity assay
Hepatic mitochondrial samples from the preparation above using a Qproteome mitochondrial isolation kit (Qiagen, Limburg, CA) were also subjected to the mitochondrial aconitase activity assay according to the manufacturer’s instruction (Biovision Inc., catalog # K716-100, http://www.biovision.com/aconitase-activity-colorimetric-assay-kit-2947.html, Milpitas, CA, USA). The activity was expressed as mU/mg protein.
Immunoblot Analysis
Proteins were extracted from liver tissues and/or isolated mitochondria with the lysis buffer containing 20 mmol/L Tris-HCl (pH 7.5), 0.5 mmol/L EDTA, 0.5 mmol/L EGTA, 0.5% (v/v) Triton X-100, and 0.1% (v/v) protease and phosphatase inhibitor cocktails, respectively (Sigma-Aldrich). Immunoblot analysis was conducted as previously described (24, 25). Total pixel intensity of each protein band was normalized to a loading control for graphing and statistical analysis. Antibodies against fatty acid synthase (FASN) (catalog # 10624-2-Ap), ATP synthase α subunit 1 (ATP5A1) (catalog # 14676-1-AP), citrate synthase (catalog # 16131-1-AP), hydroxyacyl-coenzyme A dehydrogenase (HADH) (catalog # 19828-1-AP), succinate dehydrogenase α (SDHA) (catalog # , 14865-1-AP), and nicotinamide nucleotide transhydrogenase (NNT) (catalog # 13442-2-AP) were purchased from ProteinTech Group (Chicago, IL, USA); Antibodies against lysosome-associated membrane protein 1 (LAMP1) (catalog # sc-20011), catalase (catalog # sc-50508), NNT (catalog # sc-163154), glutathione reductase (GSR) (catalog # sc-133245) were purchased from Santa Cruz Biotech (Dallas, TX, USA); peroxisome proliferator-activated receptor α (PPARα) antibody (catalog # 101710) was purchased from Cayman (Ann Arbor, MI, USA); Antibodies against β-actin (catalog # 4967), phosphor-Thr172-AMP-activated protein kinase α (pT172-AMPKα) (catalog # 2535), and AMPKα (catalog # 2603) were purchased from Cell Signaling Technology (Danvers, MA, USA); voltage-dependent anion-selective channel protein 1 (VDAC1) antibody (catalog # PA1780) was purchased from Boster Biosciences (Pleasanton, CA, USA).
Statistical analysis
The results from the mitochondrial respiration assay were analyzed by the XFe wave software (Seahorse Bioscience Inc., MA), and displayed as OCR (pmol/µg protein/min). Experimental results were analyzed by student’s t-test. Data were presented as mean ± SD, n=6, unless otherwise stated in the text. Significance was set at P < 0.05. p values were corrected for multiple testing.
Results
Overall nutrient metabolism is changed in BCO2 KO mice
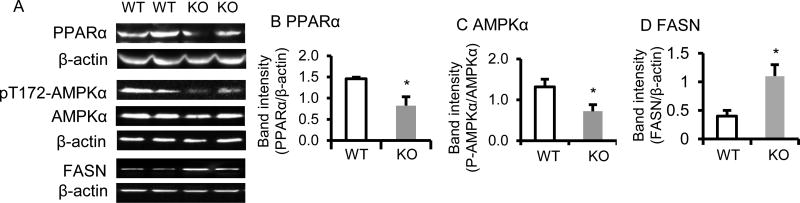
β–carotene 15, 15’-oxygenase 1 (BCO1) protein level was not altered in BCO2 KO compared to WT mice (data not shown). The daily food intake was measured from postnatal week 3 to week 6, and the result showed that BCO2 KO mice consumed more chow diet (by 15–18 %) compared to the WT, for instance, the daily food intake between 5 and 6 weeks of age was significantly higher in BCO2 KO (2.6 ± 0.1 g/mouse/day) than WT mice (2.2 ± 0.1 g/mouse/day) (n=12 mice/group, p <0.001). Interestingly, despite a significantly higher food intake, the body weight of BCO2 KO mice at 6 weeks of age (18.6 ± 1.5 g) was significantly lower (by approximately 12 %), compared to age-matched WT mice ( 21.2 ± 1.0 g) (p < 0.01, n=12 mice/group).The higher fasting blood glucose (70.2 ± 13.15 mg/dL in WT vs 111.67 ± 10.50 mg/dL in KO, p<0.001, n=12 mice/group) and FFA levels (0.70 ± 0.11 mEq/L in WT vs 0.95 ± 0.06 mEq/L in KO, p<0.01, n=6 mice/group) were observed in male BCO2 KO mice at 6 weeks of age, compared to the age and gender matched WT. Further, PPARαand AMPK, two key proteins in the energy metabolism pathway (36, 37) were analyzed by immunoblotting. The protein level of hepatic PPARα was significantly decreased in the BCO2 KO compared to the WT mice (Fig. 1 A and B). Additionally, the activation of AMPK, as indicated by phosphorylation of Thr172 in the AMPKα catalytic domain was significantly lower in the liver of BCO2 KO compared to WT mice (Fig. 1 A and C). Moreover, the elevated expression of fatty acid synthase protein (FASN, Fig. 1 A and D) was found in liver of BCO2 KO mice, compared to WT.
Figure 1. Expression of energy metabolism-related proteins is altered in BCO2 KO mice.
The expression of hepatic PPARα protein (A and B), AMPKα and pT172-AMPKα (A and C), and FASN (A and D) were determined by immunoblotting on the left and band quantification was shown on the right. The activation of AMPKα (pT172-AMPKα /AMPKα) was reduced in the BCO2 KO mice compared to WT mice. Values are means ± SD, n=6 mice/group. * Different from WT, p < 0.05. KO, knockout; WT wild type. PPARα, PPAR, peroxisome proliferator-activated receptor α; AMPKα, AMP-activated protein kinase α; FASN, fatty acid synthase
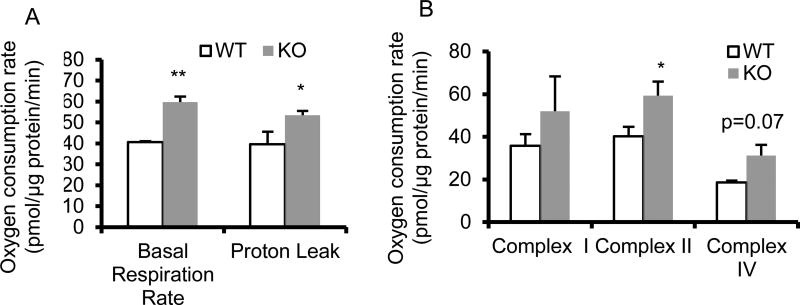
Mitochondrial proton leak, basal respiration rate, and capacity of complex II are altered in BCO2 KO mice
Mitochondrial function was measured via mitochondrial coupling and electron flow assays. These analyses revealed significant increases in the basal respiration rate, proton leak (Fig. 2A), and complex II capacity (Fig. 2B) in BCO2 KO compared to WT mice.
Figure 2. Functional analysis of isolated hepatic mitochondria from 6-week-old male BCO2 KO and WT mice.
(A) The basal respiration rate and proton leak were increased significantly in BCO2 KO mice. (B) No change was observed in the capacities of complex I and complex IV while complex II capacity was significantly increased in BCO2 KO, compared to WT mice. Values are means ± SD, n=6 mice/group, with 6 technical replicates/mouse. * Different from WT, p < 0.05. ** Different from WT, p < 0.01. KO, knockout; WT wild type.
The mitochondrial proteome is changed in BCO2 KO mice
In order to determine potential mechanisms by which mitochondrial functional changes were associated with mitochondrial protein expression in BCO2 KO mice, a comparison of the WT and BCO2 KO hepatic mitochondrial proteomes was performed (n=3 mice/group with 4 technical replicates/mouse). Thirty four mitochondrial proteins were identified to have significant changes with 13 proteins being up-regulated and 21 proteins being down-regulated in the hepatic mitochondrial fractions of BCO2 KO compared to WT mice (p < 0.05) (Table 1). Bioinformatic analysis using DAVID functional annotation tools revealed that fatty acid oxidation, amino acid metabolism, the tricarboxylic acid (TCA) cycle, and the electron transport chain (ETC)/oxidative phosphorylation (OXPHOS) were affected most in BCO2 KO mice, which are all related to energy metabolism. The detailed results were summarized in Table 1. The raw MS/MS data were uploaded as the online supplemental materials (Supplemental Table S2). Differences revealed by proteomic analysis were confirmed by immunoblot analysis for the mitochondrial proteins, hydroxyacyl-coenzyme A dehydrogenase (HADH), voltage-dependent anion-selective channel protein 1 (VDAC1)(Supplemental Fig. S2A and B), and nicotinamide nucleotide transhydrogenase (NNT) (Fig. 3C).
Table 1.
Functional categorization of proteins differentially expressed in hepatic mitochondria of BCO2 knockout (KO) vs. 129S6 wild type (WT) mice identified by spectrum counting. Values are means, n=3 mice/group with 4 technical replicates/mouse.
| Gene symbol | Protein name | Ratio(KO/WT) | p-values |
|---|---|---|---|
|
Fatty Acid Metabolism
| |||
| ATP11C | Probable phospholipid-transporting ATPase 11C | 0.73 | 0.04 |
| HADH | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 0.87 | 0.02 |
| ECI1 | Enoyl-CoA delta isomerase 1, mitochondrial | 1.59 | 0.01 |
|
| |||
|
Amino Acid Metabolism
| |||
| GLDC | Glycine dehydrogenase (decarboxylating), mitochondrial | 0.45 | 0.03 |
| MSRA | Isoform 2 of Mitochondrial peptide methionine sulfoxide reductase | 0.51 | 0.02 |
| GLS2 | Glutaminase liver isoform, mitochondrial | 0.72 | 0.03 |
| SARDH | sarcosine dehydrogenase, mitochondrial | 0.83 | 0.02 |
| GLYATL | Glycine N-acyltransferase-like protein | 0.85 | 0.02 |
| AMT | Aminomethyltransferase, mitochondrial | 1.43 | 0.04 |
| OAT | Ornithine aminotransferase, mitochondrial | 1.69 | 0.03 |
|
| |||
|
TCA Cycle
| |||
| SUCLG2 | Succinyl-CoA ligase [GDP-forming] subunit beta, mitochondrial | 0.81 | 0.04 |
| CS | Citrate synthase | 1.24 | 0.01 |
| DHTKD1 | Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial | 2.87 | 0.002 |
|
| |||
|
ETC
| |||
| NDUFA10 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | 0.84 | 0.05 |
| SDHA | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 0.87 | 0.03 |
| SUOX | Sulfite oxidase, mitochondrial | 1.30 | 0.003 |
| NNT | NAD(P) transhydrogenase, mitochondrial | 87.67/0 | 0.001 |
|
| |||
|
Mitochondria Protein Translation
| |||
| AK4 | Adenylate kinase 4, mitochondrial | 0.50 | 0.03 |
| RPL7 | ribosomal protein L7 | 0.72 | 0.004 |
|
| |||
|
Vitamin A and Drug Metabolism
| |||
| UGT1A5 | MCG14318, isoform CRAh | 0.71 | 0.02 |
| CYP1A2 | CYP1A2 cytochrome P450, family 1, subfamily A, polypeptide 2 | 1.15 | 0.03 |
| CYP2D9 | cytochrome P450, family 2, subfamily d, polypeptide 9 | 1.18 | 0.01 |
| CYP2C40 | Cytochrome P450 2C40 | 3.03 | 0.04 |
| CYP7B1 | 25-hydroxycholesterol 7-alpha-hydroxylase | 12.63 | 0.04 |
|
| |||
|
Protease
| |||
| CLPX | ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial | 0.74 | 0.02 |
|
| |||
|
Transport Protein
| |||
| TIMM50 | Mitochondrial import inner membrane translocase subunit TIM50 | 0.60 | 0.03 |
| VDAC3 | Voltage-dependent anion-selective channel protein 3 | 0.79 | 0.002 |
| VDAC1 | Isoform Mt-VDAC1 of Voltage-dependent anion-selective channel protein 1 | 0.91 | 0.01 |
|
| |||
|
Misc
| |||
| SOD2 | Superoxide dismutase [Mn], mitochondrial | 0.83 | 0.04 |
| HYOU1 | Hypoxia up-regulated protein 1 protein folding HSP70 family | 0.84 | 0.05 |
| CES2A | Pyrethroid hydrolase Ces2a | 0.92 | 0.04 |
| CPS1 | Carbamoyl Phosphate Synthetase I | 0.94 | 0.003 |
| ALDH1B1 | Aldehyde dehydrogenase X, mitochondrial | 1.11 | 0.02 |
| EPHX1 | Epoxide hydrolase 1 | 1.13 | 0.05 |
| ETHE1 | Persulfide dioxygenase ETHE1, mitochondrial | 1.28 | 0.002 |
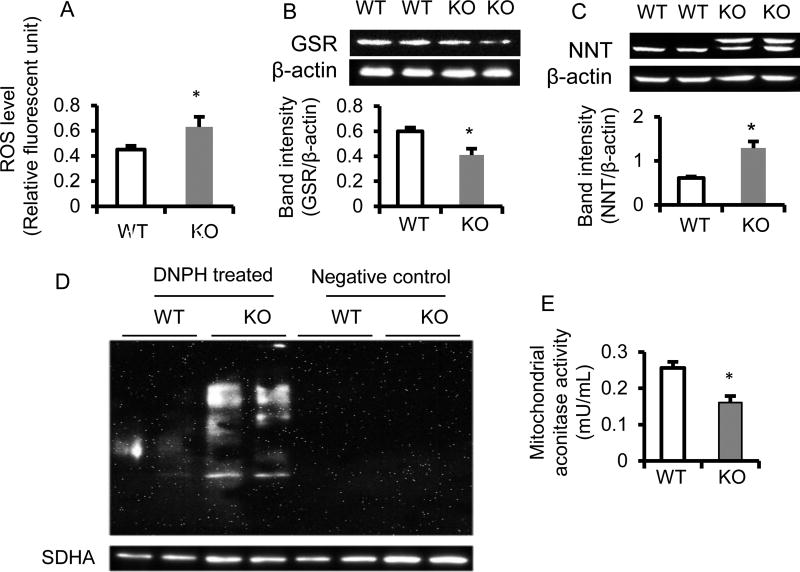
Figure 3. Cellular and mitochondrial oxidative stress are elevated in the liver of BCO2 KO compared to WT mice.
The level of hepatic cellular reactive oxygen species (ROS) (A), hepatic protein expression of glutathione reductase (GSR) (B), nicotinamide nucleotide transhydrogenase (NNT) (C) by immunoblotting. Mitochondrial protein carbonylation was assessed by the OxyBlot protein oxidation detection kit (catalog # S7150, EMD Millipore, Temecula, CA, USA) according to the manufacturer’s protocol (D). Succinate dehydrogenase α (SDHA) as a loading control. Hepatic mitochondrial aconitase activity was measured using an aconitase activity colormetric assay kit (Biovision Inc) (E). The results are presented as mean ±SD (n=6 mice/group, * Different from WT, p<0.05). KO, knockout; WT, wild type. DNPH, 2,4-dinitrophenylhydrazine
The mitochondrial proteome data were also subjected to the IPA Top Tox analysis indicating that mitochondrial function and oxidative phosphorylation were mostly affected in BCO2 KO liver (Proteomic raw data enclosed as the online Supplemental Table S2, Supplemental Table S3). In ETC/OXPHOS, the expression of proteins of mitochondrial complex I, complex II, complex IV and complex V was diminished in BCO2 KO mice compared to WT mice, whereas cytochrome c was increased in the BCO2 KO compared to the WT mice (Supplemental Fig. S3A).
Elevated cellular oxidative stress in the liver of BCO2 KO mice
The results in Fig. 3A showed a significant elevation of the ROS level in hepatic homogenates from the BCO2 KO (0.63 ± 0.08 OD 485/530 (relative fluorescent unit)), as compared to the WT (0.45 ± 0.03 OD 485/530 (relative fluorescent unit)) (n=6 mice/group, p < 0.05). In parallel, the mitochondrial proteomic data also showed elevation of oxidative stress in the liver of KO mice. For instance, the antioxidant enzymes, manganese superoxide dismutase (SOD2) (Table 1) and glutathione-disulfide reductase (glutathione reductase, GSR) (Fig. 3B) were both repressed. In contrast, the levels of peroxiredoxin 3 (PRX3) and PRX5 (38, 39) (Supplemental Fig. S3B), and nicotinamide nucleotide transhydrogenase (NNT) (Table 1, and Fig. 3C), were increased significantly in BCO2 KO compared to WT mice. Furthermore, the mitochondrial protein carbonylation levels were much higher in BCO2 KO than those in WT mice (Fig. 3D). Mitochondrial aconitase is an iron-sulfur protein containing a [4Fe-4S]2+ cluster in its active site that catalyzes the stereospecific dehydration–rehydration in the TCA cycle. It has been considered as a measure of oxidative damage (40). The result revealed that the mitochondrial aconitase activity was also declined in BCO2 KO mice (Fig. 3E). Collectively, the results indicated hepatic mitochondrial dysfunction and cellular oxidative stress in BCO2 KO mice.
4. Discussion
Mitochondrial dysfunction has been considered one of the major causal factors in the development of chronic diseases such as obesity, type 2 diabetes, and other metabolic disorders (40, 41). BCO2 resides in the inner mitochondrial membrane. In this study, the impact of BCO2 on mitochondrial function was investigated using WT and BCO2 KO mice. Importantly, there was no changes in either BCO1 protein level, hepatic retinol and retinal contents (data not shown), or accumulation of carotenoids (including β–carotene, α-carotene, β-cryptoxanthin, lutein, zeaxanthin, and lycopene) observed in the liver of BCO2 KO mice fed a standard chow diet (data not shown), suggesting that the loss of BCO2 did not affect hepatic vitamin A and carotenoid metabolism at 6 weeks of age, if there was any trace carotenoids in the diet. Whether this observation indicates the presence of a compensatory pathway or that it takes longer than 6 weeks to accumulate carotenoids remains unclear. Despite no apparent changes in carotenoid metabolism, the loss of BCO2 triggered alterations in various metabolic pathways including, but not limited to fatty acid oxidation, TCA cycle, and ETC/OXPHOS. Alterations in these pathways likely contributed to increased oxidative stress and elevation of fasting blood glucose and FFA levels that were observed in BCO2 KO mice and indicated an unknown role of BCO2 that was independent of its enzymatic carotenoid cleavage function.
Food intake is dynamic and regulated largely by whole-body energy metabolism and circadian rhyme. In this present study, the loss of BCO2 was hypothesized to result in mitochondrial dysfunction and contribute to the disturbance of energy metabolism in mice. Based on the proteomics analysis, the expression of ETC/OXPHOS complex I, II, and IV proteins was reduced in the livers of BCO2 KO mice, though the expression of cytochrome c was increased in the BCO2 KO, compared to WT mice. These observations may suggest that a change of mitochondrial proteome occurred in BCO2 KO mice (42–49). Decreased protein expression of the mitochondrial proteome has been linked to hyperactivation of hepatic mitochondria in diabetes and obesity (13, 46). The low aconitase activity occurs in obese mice with higher mitochondrial respiration activities (44). Ischemic hearts have a lower mitochondrial complex OCR activity though the mitochondrial complex I protein contents are higher (42). Mitochondria are far more complex in form and function than anticipated and are controlled by multiple factors such as circadian, nutrient availability (e.g., fasting and fed), and protein expressions (47, 49). Despite this potential compensatory mechanism of mitochondrial changes in BCO2 KO animals, the increased proton leak from the mitochondrial inner membrane space into the matrix observed in this study indicated the possibility that energy efficiency was impaired in BCO2 KO mice (50, 51).
Hyperactivation of mitochondrial respiration links to oxidative stress and metabolic disorders. Deficiency of BCO2 caused changes of the mitochondrial proteome and hyperactivation of the mitochondrial function in BCO2 KO mice. The results further demonstrated that hepatic cells experienced mitochondrial and cellular oxidative stress as shown in Fig. 3. Increased respiratory capacity of complex II leads to overproduction of ROS (52), which could also be supported by the result on the mitochondrial protein carbonylation (Fig. 3D). Oxidative stress leads to modification of native amino acid residues in proteins to carbonyl derivatives known as protein carbonylation (31). Indeed, the elevation of ROS production could further impair mitochondrial function (53).
Elevated ROS can be eliminated by a variety of enzymes and/or antioxidants, such as SOD, catalase, glutathione peroxidase (GPx) and glutathione (GSH). GSR catalyzes the reduction of glutathione disulfide (GSSG) to regenerate the sulfhydryl for GSH. NNT is a key mitochondrial antioxidant enzyme, utilizing the mitochondrial proton gradient to generate NADPH from NADH and NADP+ (54, 55). In this study, enhanced expression of NNT protein (two bands in BCO2 KO mice shown by immunoblotting) might suggest that BCO2 depletion might cause the altered expression of NNT variants under oxidative stress (56). It is clear that a mild increase in mitochondrial ROS production induces activation of long-lasting cytoprotective pathways, for instance activation of antioxidant enzymes (57), while pro-longed and/or severe oxidative stress results in suppression of a large variety of gene expression, including antioxidant enzymes, such as mitochondrial SOD2 (57, 58). Genetic depletion of BCO2 triggered mitochondrial and subsequent cellular oxidative stress which might account for decreased expression of SOD2, GSR, and other antioxidants. The reduction of GPx level would in turn conserve GSH in favor of forming GSSG. The further study is warranted to confirm the hypothesis above. Elevation of blood glucose and free fatty acids could also be considered as additional markers of disruption of nutrient metabolism and oxidative stress in BCO2 KO mice.
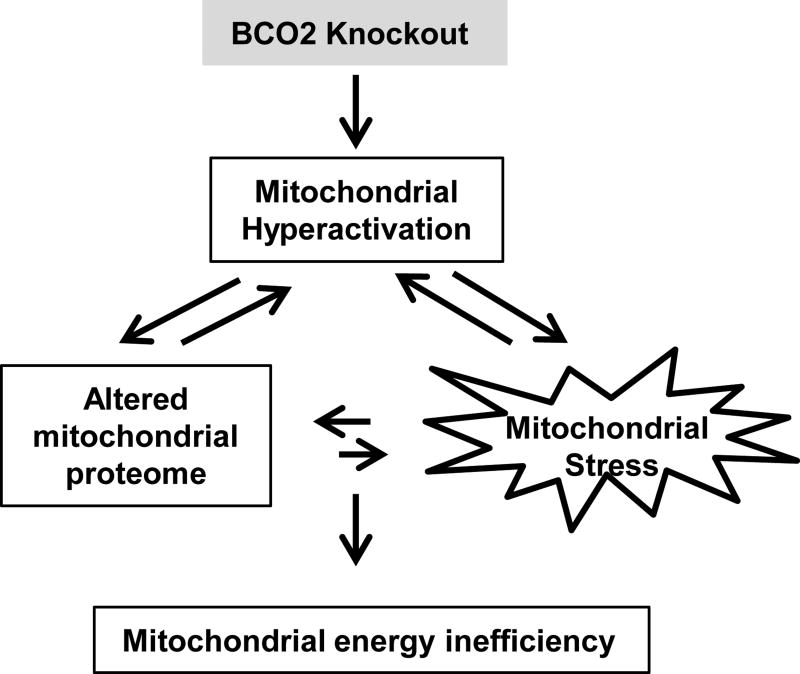
In conclusion, the results presented herein indicated that the loss of BCO2 was associated with changes of the hepatic mitochondrial proteome and function shown a decrease in the ETC efficiency and increases in proton leak, basal respiration activity, and complex II capacity– all of which could contribute to mitochondrial hyperactivation and subsequently mitochondrial and cellular oxidative stress. That in turn led to mitochondrial energy insufficiency and disruption of nutrient metabolism in BCO2 KO, compared to WT mice (Fig. 4). The findings suggested that BCO2 may play a structural role in the inner membrane of mitochondria through regulation of the ETC complexes activity, and/or metabolize unknown substrates that subsequently altered mitochondrial function. Further studies are warranted to determine if BCO2 represents a potential therapeutic target to reduce the risk of or impact chronic oxidative stress associated with diseases such as diabetes, obesity, and cardiovascular disease.
Figure 4. Schematic diagram showing the potential mechanism of BCO2 in mitochondrial function and oxidative stress.
The loss of BCO2 was associated with changes of the hepatic mitochondrial proteome and function shown as a decrease in the ETC efficiency and increases in proton leak, basal respiration activity, and complex II capacity– all of which could contribute to mitochondrial hyperactivation and subsequent mitochondrial and cellular oxidative stress, which in turn led to mitochondrial energy insufficiency and disruption of nutrient metabolism in BCO2 KO, compared to WT mice
Supplementary Material
Acknowledgments
Acknowledgment/funding
Mass spectrometry analyses were performed in the DNA/Protein Resource Facility at Oklahoma State University, using resources supported by the NSF MRI and EPSCoR programs (award DBI/0722494). This project was made possible in part by support from the USDA National Institute of Food and Agriculture 2015-67018-23176, USDA NIFA Hatch Project OKL02992, National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM103648, and Oklahoma State University faculty start-up project (to DL)
Abbreviations
- AMPK
AMP-activated protein kinase
- ATP5A1
ATP synthase α subunit 1
- BCO1
β–carotene-15, 15’-oxygenase 1
- BCO2
β, β-carotene-9’, 10’-dioxygenase 2
- BHMT
betaine homocysteine methyltransferase
- DMG
dimethylglycine
- ETC
electron transport chain
- FASN
fatty acid synthase
- GS
glutathione synthetase
- GSH
glutathione
- GSR
glutathione disulfide reductase
- GSSG
glutathione disulfide
- HADH
hydroxyacyl-coenzyme A dehydrogenase
- HSP60
heat shock protein 60
- IL-18
interleukin 18
- IPA
ingenuity pathway analysis
- KO
knockout
- LAMP1
lysosome-associated membrane protein 1
- MAS
mitochondrial assay solution
- NNT
nicotinamide nucleotide transhydrogenase
- OCR
oxygen consumption rate
- OXPHOS
oxidative phosphorylation
- PPAR
peroxisome proliferator-activated receptor
- PRX
peroxiredoxin
- ROS
reactive oxygen species
- SDHA
succinate dehydrogenase α
- SOD
superoxide dismutase
- SOD2
manganese superoxide dismutase, mitochondria
- TCA
tricarboxylic acid cycle
- VDAC1
voltage-dependent anion-selective channel protein 1
- WT
wild type
Footnotes
Author contributions
DMM, WW, SLC, EL, BJS, JL, and DL designed the study; LW, XG, SH, KP, AD, and HH conducted the research; LW, SH, KP analyzed the data; LW, DMM, WW, SLC, EL, BJS, JL, and DL wrote and revised the manuscript; and LW and DL had primary responsibility for final content. All authors read and approved the final manuscript.
Conflict of interest
All authors have no conflicts of interest
References
- 1.Hybertson BM, Gao B, Bose SK, McCord JM. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol. Aspects Med. 2011;32:234–246. doi: 10.1016/j.mam.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Borges CG, Canani CR, Fernandes CG, Zanatta A, Seminotti B, Ribeiro CA, Leipnitz G, Vargas CR, Wajner M. Reactive nitrogen species mediate oxidative stress and astrogliosis provoked by in vivo administration of phytanic acid in cerebellum of adolescent rats: A potential contributing pathomechanism of cerebellar injury in peroxisomal disorders. Neurosci. 2015;304:122–132. doi: 10.1016/j.neuroscience.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 3.Negi R, Pande D, Karki K, Kumar A, Khanna RS, Khanna HD. Association of oxidative DNA damage, protein oxidation and antioxidant function with oxidative stress induced cellular injury in pre-eclamptic/eclamptic mothers during fetal circulation. Chem-biol. Interactions. 2014;208:77–83. doi: 10.1016/j.cbi.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell. Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 5.Starkov AA. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. New York Acad. Sci. 2008;1147:37–52. doi: 10.1196/annals.1427.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams GS, Boyman L, Chikando AC, Khairallah RJ, Lederer WJ. Mitochondrial calcium uptake. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10479–10486. doi: 10.1073/pnas.1300410110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoss SE, Bahr GM, Echtay KS. Lopimune-induced mitochondrial toxicity is attenuated by increased uncoupling protein-2 level in treated mouse hepatocytes. Biochem. J. 2015;468:401–407. doi: 10.1042/BJ20150195. [DOI] [PubMed] [Google Scholar]
- 8.Bae J, Ricciardi CJ, Esposito D, Komarnytsky S, Hu P, Curry BJ, Brown PL, Gao Z, Biggerstaff JP, Chen J, et al. Activation of pattern recognition receptors in brown adipocytes induces inflammation and suppresses uncoupling protein 1 expression and mitochondrial respiration. Am. J. Physiol. Cell Physiol. 2014;306:C918–30. doi: 10.1152/ajpcell.00249.2013. [DOI] [PubMed] [Google Scholar]
- 9.Thrush AB, Zhang R, Chen W, Seifert EL, Quizi JK, McPherson R, Dent R, Harper ME. Lower mitochondrial proton leak and decreased glutathione redox in primary muscle cells of obese diet-resistant versus diet-sensitive humans. J. Clin. Endocrinol. Metab. 2014;99:4223–4230. doi: 10.1210/jc.2014-1726. [DOI] [PubMed] [Google Scholar]
- 10.Crescenzo R, Bianco F, Mazzoli A, Giacco A, Liverini G, Iossa S. Alterations in proton leak, oxidative status and uncoupling protein 3 content in skeletal muscle subsarcolemmal and intermyofibrillar mitochondria in old rats. BMC Geriatrics. 2014;14:79. doi: 10.1186/1471-2318-14-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- 12.Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, Nakayama O, Makishima M, Matsuda M, Shimomura I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 2004;114:1752–1761. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugger H, Chen D, Riehie C, Soto J, Theobald H, H X, et al. Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic Akita mice. Diebetes. 2009;58:1986–1997. doi: 10.2337/db09-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudich A, Tirosh A, Potashnik R, Hemi R, Kanety H, Bashan N. Prolonged oxidative stress impairs insulin-induced GLUT4 translocation in 3T3-L1 adipocytes. Diabetes. 1998;47:1562–1569. doi: 10.2337/diabetes.47.10.1562. [DOI] [PubMed] [Google Scholar]
- 15.Maddux BA, See W, Lawrence JC, Jr, Goldfine AL, Goldfine ID, Evans JL. Protection against oxidative stress-induced insulin resistance in rat L6 muscle cells by mircomolar concentrations of alpha-lipoic acid. Diabetes. 2001;50:404–410. doi: 10.2337/diabetes.50.2.404. [DOI] [PubMed] [Google Scholar]
- 16.Keaney JF, Jr, Larson MG, Vasan RS, Wilson PW, Lipinska I, Corey D, Massaro JM, Sutherland P, Vita JA, Benjamin EJ, et al. Obesity and systemic oxidative stress: clinical correlates of oxidative stress in the Framingham Study. Arterioscler. Thromb. Vasc. Biol. 2003;23:434–439. doi: 10.1161/01.ATV.0000058402.34138.11. [DOI] [PubMed] [Google Scholar]
- 17.Babino D, Palczewski G, Widjaja-Adhi MA, Kiser PD, Golczak M, von Lintig J. Characterization of the role of β-carotene 9,10-dioxygenase in macular pigment metabolism. J. Biol. Chem. 2015;290:24844–24857. doi: 10.1074/jbc.M115.668822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palczewski G, Amengual J, Hoppel CL, von Lintig J. Evidence for compartmentalization of mammalian carotenoid metabolism. FASEB J. 2014;28:4457–4469. doi: 10.1096/fj.14-252411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B, Vachali PP, Gorusupudi A, Shen Z, Sharifzadeh H, Besch BM, Nelson K, Horvath MM, Frederick JM, Baehr W, et al. Inactivity of human beta, beta-carotene-9',10'-dioxygenase (BCO2) underlies retinal accumulation of the human macular carotenoid pigment. Proc. Natl. Acad. U. S. A. 2014;111:10173–10178. doi: 10.1073/pnas.1402526111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He M, Cornelis MC, Kraft P, van Dam RM, Sun Q, Laurie CC, Mirel DB, Chasman DI, Ridker PM, Hunter DJ, et al. Genome-wide association study identifies variants at the IL18-BCO2 locus associated with interleukin-18 levels. Arterioscler. Thromb. Vasc. Biol. 2010;30:885–890. doi: 10.1161/ATVBAHA.109.199422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers KJ, Mares JA, Igo RP, Jr, Truitt B, Liu Z, Millen AE, Klein M, Johnson EJ, Engelman CD, Karki CK, et al. Genetic evidence for role of carotenoids in age-related macular degeneration in the Carotenoids in Age-Related Eye Disease Study (CAREDS) Invest. Ophthalmol. Vis. Sci. 2014;55:587–599. doi: 10.1167/iovs.13-13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brocato J, Hernandez M, Laulicht F, Sun H, Shamy M, Alghamdi MA, et al. In vivo exposures to particulate matter collected from Saudi Arabia or nickel chloride display similar dysregulation of metabolic syndrome genes. J. Toxicol. Environ Health A. 2015;78:1421–1436. doi: 10.1080/15287394.2015.1095689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong X, Marisiddaiah R, Zaripheh S, Wiener D, Rubin LP. Mitochondrial beta-carotene 9, 10 oxygenase modulates prostate cancer growth via NF-kappa B inhibition: a lycopene-independent function. Mol. Cancer Res. 2016 Jul 12; doi: 10.1158/1541-7786.MCR-16-0075. pii: molcanres.0075.2016. [DOI] [PubMed] [Google Scholar]
- 24.Lobo GP, Isken A, Hoff S, Babino D, von Lintig J. BCDO2 acts as a carotenoid scavenger and gatekeeper for the mitochondrial apoptotic pathway. Develop. 2012;139:2966–2977. doi: 10.1242/dev.079632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, Wark L, Ji H, Willard L, Jaing Y, Han J, He H, Ortiz E, Zhang Y, Medeiros DM, et al. Dietary wolfberry upregulates carotenoid metabolic genes and enhances mitochondrial biogenesis in the retina of db/db diabetic mice. Mol. Nutr. Food Res. 2013;57:1158–1169. doi: 10.1002/mnfr.201200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin D, He H, Ji H, Willis J, Willard L, Jiang Y, Medeiros DM, Wark L, Han J, Liu Y, et al. Wolfberries potentiate mitophagy and enhance mitochondrial biogenesis leading to prevention of hepatic steatosis in obese mice: the role of AMP-activated protein kinase alpha2 subunit. Mol. Nutr. Food Res. 2014;58:1005–1015. doi: 10.1002/mnfr.201300186. [DOI] [PubMed] [Google Scholar]
- 27.Kiefer C, Hessel S, Lampert JM, Vogt K, Lederer MO, Breithaupt DE, von Lintig J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 2001;276:14110–14116. doi: 10.1074/jbc.M011510200. [DOI] [PubMed] [Google Scholar]
- 28.Lindqvist A, He YG, Andersson S. Cell type-specific expression of beta-carotene 9',10'-monooxygenase in human tissues. J. Histochem. Cytochem. 2005;53:1403–1412. doi: 10.1369/jhc.5A6705.2005. [DOI] [PubMed] [Google Scholar]
- 29.Amengual J, Lobo GP, Golczak M, Li HN, Klimova T, Hoppel CL, Wyss A, Palczewski K, von Lintig J. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 2011;25:948–959. doi: 10.1096/fj.10-173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Darshi M, Ma Y, Perkins GA, Shen Z, Haushalter KJ, et al. Quantitative proteomic and functional analysis of liver mitochondria from high fat diet (HFD) diabetes mice. Mol. Cell. Proteomics. 2013;12:3744–3758. doi: 10.1074/mcp.M113.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalle-Donne I, Rossi R, Giustarini D, Milzani A, Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta. 2003;329:23–38. doi: 10.1016/s0009-8981(03)00003-2. [DOI] [PubMed] [Google Scholar]
- 32.Voruganti S, Lacroix JC, Rogers CN, Rogers J, Matts RL, Hartson SD. The anticancer drug AUY922 generates a proteomics fingerprint that is highly conserved among structurally diverse Hsp90 inhibitors. J. Proteome Res. 2013;12:3697–3706. doi: 10.1021/pr400321x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Yu F, Shen XY, Fan L, Yu ZC. Genome-wide analysis of genetic variations assisted by Ingenuity Pathway Analysis to comprehensively investigate potential genetic targets associated with the progression of hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2014;18:2102–2108. [PubMed] [Google Scholar]
- 35.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prosdocimo DA, John JE, Zhang L, Efraim ES, Zhang R, Liao X, Jain MK. KLF15 and PPARalpha cooperate to regulate cardiomyocyte lipid gene expression and oxidation. PPAR. Res. 2015;2015:201625. doi: 10.1155/2015/201625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fentz J, Kjobsted R, Birk JB, Jordy AB, Jeppesen J, Thorsen K, Schjerling P, Kiens B, Jessen N, Viollet B, et al. AMPKalpha is critical for enhancing skeletal muscle fatty acid utilization during in vivo exercise in mice. FASEB J. 2015;29:1725–1738. doi: 10.1096/fj.14-266650. [DOI] [PubMed] [Google Scholar]
- 38.Oka S, Hirai J, Yasukawa T, Nakahara Y, Inoue YH. A correlation of reactive oxygen species accumulation by depletion of superoxide dismutases with age-dependent impairment in the nervous system and muscles of Drosophila adults. Biogerontol. 2015;16:485–501. doi: 10.1007/s10522-015-9570-3. [DOI] [PubMed] [Google Scholar]
- 39.Freeman H, Shimomura K, Cox RD, Ashcroft FM. Nicotinamide nucleotide transhydrogenase: a link between insulin secretion, glucose metabolism and oxidative stress. Biochem. Soc. Trans. 2006;34:806–810. doi: 10.1042/BST0340806. [DOI] [PubMed] [Google Scholar]
- 40.Yan L-J, Levine RL, Sohai RS. Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. U.S.A. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hojlund K, Mogensen M, Sahlin K, Beck-Nielsen H. Mitochondrial dysfunction in type 2 diabetes and obesity. Endocrinol. Metab. Clin. North Am. 2008;37:713–731. doi: 10.1016/j.ecl.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Liu T, Chen L, Kim E, Tran D, Phinney BS, Knowlton AA. Mitochondrial proteome remodeling in ischemic heart failure. Life Sci. 2014;101:27–36. doi: 10.1016/j.lfs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valdecantos MP, Perez-Matute P, Gonzalez-Muniesea P, Prieto-Hontoria PL, Moreno-Aliaga MJ, Martinez JA. Lipoic acid administration prevents nonalcoholic steatosis linked to long-term high-fat feeding by modulating mitochondrial function. J. Nutr. Biochem. 2012;23:1676–1684. doi: 10.1016/j.jnutbio.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Crescenzo R, Bianco F, Coppola P, Mazzoli A, Cigliano L, Liverini G, Iossa S. Increased skeletal muscle mitochondrial efficiency in rats with fructose-induced alteration in glucose tolerance. Br. J. Nutr. 2013;110:1996–2003. doi: 10.1017/S0007114513001566. [DOI] [PubMed] [Google Scholar]
- 45.Segref A, Kevei É, Pokrzywa W, Schmeisser K, Mansfeld J, Livnat-Levanon N, et al. Pathogenesis of human mitochondrial diseases is modulated by reduced activity of the ubiquitin/proteasome system. Cell Metab. 2014;19:642–652. doi: 10.1016/j.cmet.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Koliaki C, Szendroedi J, Kaul K, Jelenik T, Nowotny P, Jankowiak F, et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab. 2015;21:739–746. doi: 10.1016/j.cmet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Neufeld-Cohen A, Robles MS, Aviram R, Manella G, Adamovich Y, Ladeuix B, et al. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1673–1682. doi: 10.1073/pnas.1519650113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akude E, Zherebitskaya E, Chowdhury SK, Smith DR, Dobrowsky RT, Fernyhough P. Diminished superoxide generation is associated with respiratory chain dysfunction and changes in the mitochondrial proteome of sensory neurons from diabetic rats. Diabetes. 2011;60:288–297. doi: 10.2337/db10-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pagliarini DJ1, Rutter J. Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 2013;27:2615–2627. doi: 10.1101/gad.229724.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Busiello RA, Savarese S, Lombardi A. Mitochondrial uncoupling proteins and energy metabolism. Front. Physiol. 2015;6:36. doi: 10.3389/fphys.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porter RK. Mitochondrial proton leak: a role for uncoupling proteins 2 and 3? Biochim. Biophys. Acta. 2001;1504:120–127. doi: 10.1016/s0005-2728(00)00246-2. [DOI] [PubMed] [Google Scholar]
- 52.Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, Brand MD. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J. Biol. Chem. 2012;287:27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cui H, Kong Y, Zhang H. Oxidative stress, mitochondrial dysfunction, and aging. J. Signal Transduction. 2012 doi: 10.1155/2012/646354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopert P, Patel M. Nicotinamide nucleotide transhydrogenase (Nnt) links the substrate requirement in brain mitochondria for hydrogen peroxide removal to the thioredoxin/peroxiredoxin (Trx/Prx) system. J. Biol. Chem. 2014;289:15611–15620. doi: 10.1074/jbc.M113.533653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nickel AG, von Hardenberg A, Hohl M, Loffler JR, Kohlhaas M, Becker J, Reil JC, Kazakov A, Bonnekoh J, Stadelmaier M, et al. Reversal of mitochondrial transhydrogenase causes oxidative stress in heart failure. Cell Metab. 2015;22:472–484. doi: 10.1016/j.cmet.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 56.Wong N, Blair AR, Morahan G. Andrikopoulos, The deletion variant of nicotinamide nucleotide transhydrogenease (NNT) does not affect insulin secretion or glucose tolerance. Endocrinol. 2010;151:96–102. doi: 10.1210/en.2009-0887. [DOI] [PubMed] [Google Scholar]
- 57.Willems PHGM, Rossignol R, Dieteren CEJ, Purphy MP, Kopman WJH. Redox homeostasis and mitochondrial dynamics. Cell Metab. 2015;22:207–217. doi: 10.1016/j.cmet.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 58.Giu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.