Abstract
The application of genome-wide expression analysis to a large-scale, multicentered program in critically ill patients poses a number of theoretical and technical challenges. We describe here an analytical and organizational approach to a systematic evaluation of the variance associated with genome-wide expression analysis specifically tailored to study human disease. We analyzed sources of variance in genome-wide expression analyses performed with commercial oligonucleotide arrays. In addition, variance in gene expression in human blood leukocytes caused by repeated sampling in the same subject, among different healthy subjects, among different leukocyte subpopulations, and the effect of traumatic injury, were also explored. We report that analytical variance caused by sample processing was acceptably small. Blood leukocyte gene expression in the same individual over a 24-h period was remarkably constant. In contrast, genome-wide expression varied significantly among different subjects and leukocyte subpopulations. Expectedly, traumatic injury induced dramatic changes in apparent gene expression that were greater in magnitude than the analytical noise and interindividual variance. We demonstrate that the development of a nation-wide program for gene expression analysis with careful attention to analytical details can reduce the variance in the clinical setting to a level where patterns of gene expression are informative among different healthy human subjects, and can be studied with confidence in human disease.
Keywords: clinical studies, gene expression, inflammation, microarray, trauma
Our understanding of the biological basis for most complex human diseases remains incomplete. In an attempt to elucidate underlying pathophysiologies, the scientific and medical communities have often used reductionist approaches to recapitulate specific components of the biological process by employing model organisms like rodents or cell lines. Another approach has been to focus on an individual gene, signaling pathway, or mechanism in selected patient populations. Although these approaches have been very successful in the past, they often fail to provide critical information regarding complex interactions and networks during disease development. To improve our understanding of the integrated response to human disease, high-throughput genomics technologies have been recently developed, enabling the simultaneous determination of a large number of analytes from clinical samples (1, 2). For example, high-throughput technologies to survey the entire human transcriptome have been recently used to classify histologically similar tumors based on genome-wide expression patterns, as well as predicting clinical response to antineoplastic therapies (3-6).
Associated with these technologies are a number of theoretical and technical challenges that have delayed their widespread implementation in the clinical setting (1). These include (i) the requirement for a consortium of scientists and clinicians with diverse skill sets to develop an effective methodological strategy, (ii) the accumulation of sufficient technical expertise to generate high-quality, large-scale, biochemical, genetic, and physiological data, and, finally, (iii) the development of an effective mechanism and tools to properly store, disseminate, and analyze the data that will be generated from these large-scale scientific projects.
As a prelude to the National Institutes of Health “Road Map” for translational medicine (7), the National Institute of General Medical Sciences has supported a large-scale collaborative, clinical research program with the purpose of applying recently developed, high-throughput genomic and proteomic approaches to trauma and inflammation research. The overarching vision behind this program was to empower clinical scientists, biochemists, immunologists, and bioinformaticians to develop an organizational framework and appropriate infrastructure to introduce high-throughput, genome-wide expression technologies to a multicentered, hospital-based study. At the initiation of this program, considerable time and effort were spent establishing a network of communication among highly skilled individuals who had little experience at transdisciplinary, multicentered studies. The effective communication and exchange of expertise among all of the members of the program became the cornerstone for the successful implementation of these high-throughput genomic technologies in the clinical setting.
This report focuses on the programmatic effort to identify and develop strategies to minimize sources of variation in gene expression patterns from whole blood, leukocyte subpopulations, and skeletal muscle by using DNA oligonucleotide microarrays. The goal of these studies was to establish and implement standardized protocols that could be used in a clinical setting and to quantify their analytical variance. At the same time, the magnitude of analytical variance was determined in the context of interindividual variance in health and disease in an effort to ascertain the likelihood of obtaining meaningful expression data from investigations in hospitalized patients with severe trauma.
Materials and Methods
Organizational Structure. The experimental protocols were developed by a core of investigators comprising a total of eight participating institutions, and were approved by a programmatic Steering Committee. Studies in healthy subjects were conducted at four institutions: Washington University School of Medicine (St. Louis), University of Florida College of Medicine (Gainesville), University of Rochester School of Medicine (Rochester, NY), and the Robert Wood Johnson Medical School/University of Medicine and Dentistry of New Jersey (New Brunswick). Studies in severely traumatized patients were conducted at Harborview Medical Center (University of Washington, Seattle) and University of Rochester School of Medicine. Expression analyses were performed at the Stanford Genome Technology Center (Palo Alto, CA), University of Florida College of Medicine, and Washington University School of Medicine. Data were analyzed by an analytical core based at Massachusetts General Hospital (Cambridge), but also including Stanford Genome Technology Center and the University of Florida College of Medicine.
Patient Recruitment. Permission was obtained from healthy subjects and hospitalized patients to collect venous blood and/or waste skeletal muscle tissue in accordance with protocols approved by the Institutional Review Boards of the participating institutions. Obtaining informed consent early in the course of critical illness resulting from injury is a complex issue that was addressed first at the local level, then at the Program level. A complete description is available upon request. Blood or tissues were collected from a total of 23 healthy human subjects and 251 severely traumatized or burned patients (data from 34 of these subjects are reported herein). Universal Human Reference RNA (Stratagene) was used for the variance analyses of cRNA target synthesis and hybridization.
Blood and Tissue Processing. Blood and tissue samples were processed immediately at the clinical site and then frozen and shipped to the sample coordination site at the University of Florida. From there, depending on the experiment, frozen samples were either processed locally or sent to Stanford for processing of RNA and subsequent hybridization to either the U133A or U133 Plus GeneChip. Detailed descriptions for all of these protocols and specific laboratory methodologies can be obtained from published reports (8) and Supporting Text, which is published as supporting information on the PNAS web site; further details are available upon request.
Statistical Analyses. The statistical analyses are described in complete detail in Supporting Text. GeneChip expression signal normalization was performed with DNA Chip Analyzer (dChip v1.3, www.dchip.org) by using the perfect match algorithms. Probe sets whose apparent expression differed among groups were analyzed by Significance Analysis of Microarrays (SAM), using a false discovery rate of <0.001 based on 1,000 permutations of the data set (9).
Pearson's product moment correlation among all of the expression values for pairs of microarrays was used as a measure of variance within and between groups of microarrays. For selected groups of microarrays, the coefficient of variation for each probe set was computed as an additional measure of variance.
Results
Structural Organization of the Program. The complex and diverse nature of the program required the development of individual clinical, analytical, and administrative cores, which were comprised of clinicians, biochemists, immunologists, statisticians, and administrators. Each core had the direct responsibility to establish guidelines for the conduct of the clinical study, adherence to institutional responsibilities for clinical research, and development of analytical procedures. These issues included compliance with institutional and federal requirements for patient confidentiality, adequate training of the nursing and/or research staff in new technologies, sample transport, processing, and analysis of clinical materials at centralized analytical sites, and data transfer to a central data management site. Most importantly, decisions in each core were made by consensus, reduced to standard operating procedures, distributed among the participants, approved by a steering committee, and posted on the program's web site for reference (www.gluegrant.org). Communication among the participants was achieved through multiple approaches, consisting of weekly conference calls and, most importantly, quarterly face-to-face meetings. The latter provided a venue free from distractions and emphasized work product based on core-specific, quarterly deliverables.
Variance Caused by Microarray Platform and Target Generation. A significant limitation to the performance of genome-wide expression analysis in clinical studies is the quantity of blood or tissue available. Analytical methods to both amplify and label the RNA are required for hybridization to microarray platforms. We first determined the variance in apparent gene expression caused by the amplification, labeling and hybridization procedures required for the GeneChip platform (Standard Operating Procedure no. G007, see Supporting Text). A single sample of Universal Human Reference RNA underwent four simultaneous cRNA synthesis reactions using an initial 4 μg per reaction, and hybridization to separate U133A GeneChips. An additional single biotinylated cRNA target was also hybridized independently to four U133A GeneChips. As shown in Table 1, there was a high degree of correlation among replicates at both the level of target hybridization and generation of the cRNA target, with Pearson correlation coefficients of 0.997. When the concordance was examined by using the Universal Human Reference RNA as the starting material for the cRNA synthesis, the mean correlation coefficient was 0.996.
Table 1. Summary of concordance in gene expression.
| Pearson correlation coefficient | |
|---|---|
| From cRNA hybridization (n = 4) | 0.997 ± 0.0011 |
| From RNA starting material (n = 4) | 0.996 ± 0.0009 |
| Leukocyte gene expression from same healthy subject over 24h (n = 4 subjects, four to six time points per subject) | 0.991 ± 0.002 |
| Leukocyte gene expression from individual healthy subjects (n = 17) | 0.952 ± 0.0203 |
| Individual leukocyte populations in different healthy subjects (n = 5) | |
| T cells | 0.977 ± 0.0059 |
| Monocytes | 0.970 ± 0.0106 |
| Total WBCs | 0.968 ± 0.0122 |
| Comparing different cell types from same healthy subjects (n = 5) | |
| Monocytes vs. T cells | 0.879 ± 0.007 |
| T cells vs. total WBCs | 0.899 ± 0.011 |
| Monocytes vs. total WBCs | 0.942 ± 0.009 |
| Leukocyte gene expression from individual trauma patients (n = 14) | 0.919 ± 0.0349 |
WBC, white blood cell.
Variance Caused by Methods for Tissue Isolation. To evaluate the variance introduced by the method of isolating total cellular RNA from whole blood and a solid tissue in hospitalized patients, well accepted protocols were compared. In this case, blood was divided into three separate aliquots and processed according to the analytical techniques described in Materials and Methods. Not surprisingly, the concordance in apparent gene expression among subjects varied depending on the RNA isolation method (Table 2). Even more important was the lack of concordance in gene expression between PAXgene-derived samples and RNA samples obtained from the two leukocyte isolation protocols in the same subject.
Table 2. Concordance in gene expression due to sample preparation.
| Preparations | Pearson correlation coefficient |
|---|---|
| Human blood preparations | |
| Between subjects | |
| PAXgene | 0.934 ± 0.0242 |
| Lysis | 0.959 ± 0.0150 |
| Buffy coat | 0.906 ± 0.0965 |
| Between isolation methods | |
| PAXgene vs. lysis | 0.891 ± 0.041 |
| PAXgene vs. Buffy coat | 0.908 ± 0.046 |
| Buffy coat vs. lysis | 0.955 ± 0.061 |
| Human muscle preparations | |
| Between subjects (range) | |
| Snap frozen | 0.873 (0.824−0.942) |
| 70% ethanol | 0.878 (0.833−0.951) |
| RNAlater | 0.888 (0.855−0.948) |
| Between isolation methods | |
| RNAlater vs. snap frozen | 0.988 ± 0.005 |
| RNAlater vs. 70% ethanol | 0.991 ± 0.001 |
| Snap frozen vs. 70% ethanol | 0.982 ± 0.009 |
When different protocols were compared for immediate tissue preservation and isolation of RNA from human skeletal muscle, the results were very different from the blood isolation protocols. In this case, the concordance in gene expression from the same human muscle sample preserved by snap-freezing, immersion in ice-cold 70% ethanol, or immersion in RNAlater was markedly higher than the concordance in gene expression in muscle samples obtained from different burn subjects by using the same tissue preservation and RNA isolation protocol. This finding is not surprising because the samples were obtained from different subjects whose burn injuries and clinical course varied dramatically.
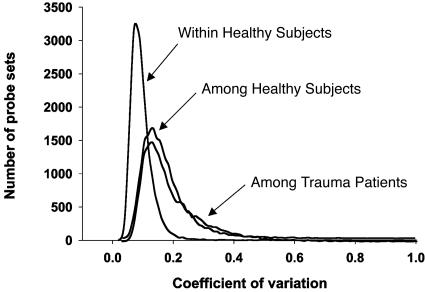
Variance Caused by Time, Cell Type, and Genotype in Healthy Subjects. Variance in apparent blood leukocyte gene expression was also examined in the same healthy subject over time, in different healthy subjects, and in different isolated enriched leukocyte populations from the same healthy subject. Four healthy subjects were admitted to the General Clinical Research Center at Robert Wood Johnson Medical School and, after an overnight fast, were placed in bed; blood was sampled six times over a 24 h period (0, 2, 4, 6, 9, and 24 h) (10). Most surprising was the high concordance in gene expression obtained from the same subject over the 24-h sampling period. As shown in Table 1 and Fig. 1, mean concordance at the probe set level was 0.991, very similar to the concordance seen in gene expression from a single Human Universal Reference RNA processed four times. This is best visualized in Table 3, where the mean coefficient of variation for RNA abundance across the 22,281 probe sets was 10%, and 90% of the probe sets had a coefficient of variation of <14%.
Fig. 1.
Variation in gene expression from healthy subjects and trauma patients. Blood leukocytes were obtained from four healthy subjects repeatedly over a 24-h study period, from 17 different healthy subjects, and from 14 patients after severe trauma. The coefficients of variation were determined at the probe set level and were plotted as a distribution curve.
Table 3. Inter- and intraindividual variance.
| Coefficient of variation | Within healthy subjects* (n = 4) | Among healthy subjects (n = 17) | Among trauma patients (n = 14) |
|---|---|---|---|
| Mean ± SD | 0.0965 ± 0.0482 | 0.1803 ± 0.1043 | 0.1998 ± 0.1267 |
| Median | 0.0881 | 0.1588 | 0.1621 |
| 90% | 0.1390 | 0.2720 | 0.3450 |
| 10% | 0.0586 | 0.1001 | 0.0972 |
The means and percentiles for the variation in gene expression among the three groups of subjects in Fig. 1.
Four to six replicates per subject.
In contrast, the variance in apparent gene expression among 17 different healthy subjects was considerably greater. These samples were obtained from individuals at three different institutions, and blood sampling was uncontrolled for time of day, physical activity, or prior nutritional intake. Mean concordance at the probe set level was 0.955, less than the 0.991 seen in the same subject over 24 h. In addition, in the 17 healthy subjects, the mean coefficient of variation in gene expression for each probe set was 18.0% (approximately twice that seen in the same subject over time), and 90% of the probe sets had a coefficient of variation of 27% or less.
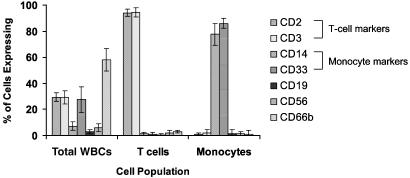
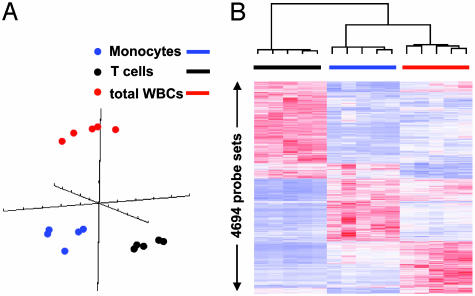
To examine the variance in apparent gene expression observed in leukocyte subpopulations, blood was obtained from a subset of five healthy subjects, and gene expression was determined in the total leukocyte population and enriched T cell and monocyte subpopulations. Fig. 2 summarizes the distribution of leukocyte subpopulations in the total leukocyte and in the enriched T cell and monocyte populations from these five healthy subjects. As shown in Table 1, the concordance in gene expression among enriched T cells and monocytes from the five healthy subjects was as good or better than the concordance in gene expression from the total leukocyte population (r = 0.977 and 0.970 vs. 0.968). However, as shown in Table 1, and more graphically in Fig. 3, the patterns in apparent gene expression among the total leukocyte population and enriched T cells and monocytes were dramatically different. The concordance between cell types varied from 0.879 to 0.942 among the three cell types. Principal component analysis (Fig. 3A) and hierarchical cluster analysis (Fig. 3B) revealed the considerable differences in apparent gene expression in the three leukocyte populations from the same healthy subjects. Table 4, which is published as supporting information on the PNAS web site, provides the lists of genes whose apparent expression discriminates among total leukocytes, monocytes, and T cells.
Fig. 2.
Leukocyte populations in total white blood cells (WBCs) and T cell- and monocyte-enriched populations. Whole blood obtained from five healthy subjects was subjected to either total WBC isolation or T cell or monocyte enrichment. The cell distribution was determined by flow cytometry using labeled antibodies to the cell surface markers identified as described (16). The total WBC preparation contained predominantly neutrophils (CD66b+), but also 32% T cells and ≈8% monocytes. T cell enrichment yielded ≈95% CD2+, CD3+ cells, and monocyte enrichment yielded ≈90% CD14+, CD33+ cells.
Fig. 3.
Principal component and hierarchical cluster analyses performed on leukocyte gene expression from buffy coat and T cell- and monocyte-enriched populations. Blood was obtained from five healthy subjects, and leukocyte populations were subjected to gene expression analysis with the U133A gene chip, as described in Materials and Methods. Principal component (A) and hierarchical cluster (B) analyses were performed on the hybridization signal intensities of probe sets significant with a false discovery rate of 0.001.
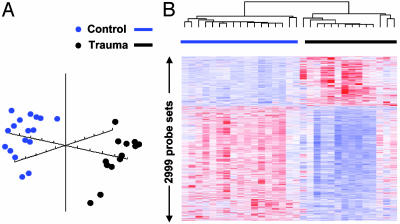
Variance Caused by Trauma. Gene expression profiles were also examined in 14 trauma subjects. Their clinical characteristics are provided in Table 5, which is published as supporting information on the PNAS web site. The variation in apparent gene expression in trauma patients was expectedly greater than the variation seen in healthy subjects. Concordance rates were lower at 0.919 vs. 0.952 (Table 1 and Fig. 4). However, concordance does not emphasize the very different patterns of apparent gene expression seen in the blood leukocytes from the trauma subjects. Comparing patterns between healthy and traumatized subjects reveals marked differences in apparent gene expression, as visualized by principal component and hierarchical cluster analyses (Fig. 4), allowing ready classification because of probe sets showing increased or decreased relative RNA abundance. Table 6, which is published as supporting information on the PNAS web site, includes those probe sets whose apparent expression was different between healthy subjects and traumatized patients. Further details are available in Table 7, which is published as supporting information on the PNAS web site.
Fig. 4.
Principal component and hierarchical cluster analyses performed on leukocyte gene expression from 14 trauma and 17 healthy subjects. Principal component (A) and hierarchical cluster (B) analyses were performed on the hybridization signal intensities of probe sets significant with a false discovery rate of 0.001.
Discussion
Conducting genome-wide expression analyses on blood and tissue samples obtained from hospitalized patients required the establishment of an infrastructure that not only supported the successful implementation of these analytical technologies, but also considered the constraints placed on clinical research in a critical care setting, which included the limited quantities and frequency of sample collection. Moreover, the dynamic nature of the host response to injury required logistics and a level of coordination not typical of previously reported multicentered clinical studies (e.g., the time element of cancer is less demanding). As a prerequisite for the application of genome-wide expression analysis to a multicenter clinical study, we recognized that the variation in apparent gene expression across the entire genome would need to be estimated, and protocols would have to be developed that were sufficiently sensitive, yet robust, when applied to a clinical setting. Success in developing the infrastructure for these multicenter studies was the direct result of frequent, open communications between the clinical personnel collecting and processing the samples, and the analytical personnel providing the genomic analyses.
Before the formal initiation of any experimental protocol, the clinical personnel were actively involved in acquiring the requisite skill sets and time commitment. Experimental protocols were subsequently developed and refined by a limited number of investigators with considerable expertise in clinical research, with the goal being to make the protocols as feasible as possible for a skilled but active clinical studies team. The protocols subsequently underwent testing and refinement at the clinical sites before their widespread implementation.
Choice of Platform. The GeneChip DNA microarray platform was chosen for these studies because of several advantages, including coverage of the entire human genome, access to probe sequences, probe redundancy (11 sequences per gene) to optimize fidelity of signal-to-noise ratio, ready commercial availability and quality control, available technical support, and relatively low cost per gene. Previous studies by Tan et al. (11) and Marshall (12) have emphasized the need to validate an analytical platform before its widespread implementation, and the difficulty in comparing expression data from different analytical platforms.
As shown in Table 1, the variance associated with the generation of the labeled, complementary nucleotide target from a standard human reference RNA, and its hybridization to the microarray, was minimal. Our results are not consistent with the earlier findings of Tan et al. (11), in that we attained near perfect unity in concordance with a reference RNA sample analyzed in replicate. Contrary to the conclusions of Tan et al. (11), and more recently Marshall (12), these findings suggest that the methodologies used to generate the labeled complementary nucleotides and the hybridization itself to the GeneChip platform introduced only minimal variance to the analyses.
Variance Associated with Tissue Isolation and Stabilization. One of the challenges associated with the use of genome-wide expression profiling in hospitalized patients has been the development of methodologies that could be used at diverse clinical sites to collect and store a high-quality RNA product (1, 13). We note that the concordance in apparent gene expression with samples using a whole blood RNA isolation protocol (PAXgene) were markedly different from the gene expression profiles seen with two leukocyte isolation protocols (Table 2). As we recently reported, this difference is likely due to the contribution of RNA from different cellular sources in the PAXgene preparations when compared to leukocyte isolation protocols (8).
Regarding the isolation of total RNA from a solid tissue specimen such as skeletal muscle, we compared three widely used techniques: snap-freezing or immersion of the tissue in ice-cold 70% ethanol or RNAlater. Concordance in gene expression in the same sample among the three techniques was much greater than the concordance among different burn subjects when a single analytical technique was used (Table 2). For solid tissues, snap-freezing in liquid nitrogen is cumbersome and poses potential health risks to staff collecting and transporting the tissues. Thus, collection of samples in a less problematic ice-cold 70% ethanol or RNAlater offers a number of practical advantages. Regardless, these studies emphasize the requirement to standardize protocols not only for the actual analysis of genome-wide expression, but also for the collection of tissues and isolation of cellular RNA.
Variation in Gene Expression Among Healthy Subjects. Before initiating the gene expression analyses in critically ill patients, we assessed the intra- and intersubject variance in gene expression among healthy subjects. Our studies showed a strong concordance in gene expression in the same subjects when measured repeatedly over a 24-h period. As shown in Fig. 1 and Tables 1 and 3, the level of concordance among the replicate analyses from the same subject exceeded 0.99, and the average coefficient of variation for >22,000 probe sets was only 9%. These results are highly encouraging because they suggest that, in healthy subjects committed to bed rest and a defined nutritional intake, variations in blood leukocyte gene expression over a 24-h period are remarkably small.
Variation in Gene Expression Among Leukocyte Subpopulations. One of the limitations associated with gene expression in whole blood or solid tissues is the heterogeneity of the cell population and the potential diversity of the cell-specific response. To explore the variance in apparent gene expression that is cell specific, we developed protocols (Standard Operating Protocol no. G029) to isolate enriched (>90%) T cell and monocyte populations based on negative selection techniques that are applicable to a clinical setting (available in Supporting Text). As shown in Figs. 2 and 3, the isolation of T cell and monocyte populations from healthy subjects did not appear to introduce any significant increase in the variance in gene expression. If anything, the variance in gene expression among different healthy subjects in the T cell and monocyte preparations was equivalent to or less than that seen in the total leukocyte preparations (Table 1). This finding would be expected if the greatest source of variation in gene expression was the differing proportion of leukocyte subpopulations in the total leukocyte preparation, as has been suggested by Whitney et al. (14). As shown in Fig. 3 and Table 4, the patterns of gene expression varied dramatically among T cell- and monocyte-enriched and total leukocyte populations. These data imply that distinct biological signals can be obtained not only from the total leukocyte population, but also from specific cell types, and isolation methodologies can be readily implemented in hospitalized patient populations. Moreover, the findings are consistent with the recent argument that a further understanding of the biological basis for changes in genome-wide expression analysis will require the identification of gene expression patterns from increasingly homogenous leukocyte subpopulations (2).
Variation in Gene Expression in Trauma. Based on the identified sources of variance, we applied these approaches to the first subset of samples obtained from patients after significant trauma, and asked whether the differences in gene expression in whole blood leukocytes produced by traumatic injury would be sufficiently great to distinguish injury specific patterns of gene expression independent of intersubject variability. As shown in Table 1, the intersubject variability in gene expression was greater in the trauma subjects than it was in healthy subjects. Such findings provide proof-of-principle in trauma patients, and are in agreement with the studies of others who also observed increased variance in gene expression in the blood of patients with cancer and infections (14).
The question that inevitably arises is whether this increased interindividual variability is sufficiently large in trauma patients to negate the detection of any injury-associated changes in gene expression. As shown in Fig. 4, the answer is unequivocally no. As demonstrated by both principal component and hierarchical cluster analysis, the patterns of RNA abundance from 14 severely traumatized patients differed dramatically from the 17 healthy subjects. These genes included those whose apparent expression is generally associated with blood neutrophil (MMP8, neutrophil elastase 2, bactericidal/permability increasing protein), monocyte (IL-1 receptor type II, TLR5, SOCS3), and T cell populations (CD28).
Conclusions. The findings suggest that a thorough exploration of the analytical and interindividual variations in genome-wide expression is required to successfully apply gene expression analyses to hospitalized patients. When these sources of variation are understood and controlled through the use of rigorously defined protocols, a detailed genome-wide picture of the gene expression response in human health and disease emerges. Although each specific gene expression platform or protocol has its advantages and limitations, gene expression profiling can provide meaningful data when conducted using methodologies that have been validated experimentally.
The major source of variance in apparent gene expression in the blood compartment is due to interindividual variance and not analytical noise. Our results reveal a notably high degree of reproducibility both with the analytical processes and in the same subject. The magnitude of the interindividual variance and the changes in gene expression produced by traumatic injury were somewhat greater than the variance associated with the sample processing and analysis in the same subject. Collectively, these findings and the infrastructure required to generate them provided the foundation necessary to explore differences in gene expression caused by systemic inflammation, severe trauma, and its complications by using the described platform and protocols in large-scale multicentered trials (15).
Supplementary Material
Acknowledgments
We thank the large number of nurse coordinators, technicians, and support personnel who contributed to the successful completion of these studies. In particular, we thank Julie Wilhelmy, Sandra MacMillan, Cynthia Tannahill, M. Cecilia Lopez, Amer Abouhamze, Derwin Hyde, and Dr. Philip Mason for their specific contributions. Special thanks are extended to Dr. Scott Somers for his critical review of the manuscript. This work was supported by National Institute of General Medical Sciences Injury and Host Response to Inflammation Large-Scale Collaborative Research Grant U54 GM-62119-03.
Author contributions: J.P.C., M.N.M., C.M.-G., S.E.C., H.V.B., W.X., B.H.B., D.N.H., S.F.L., D.H.S., L.L.M., R.W.D., and R.G.T. designed research; J.P.C., M.N.M., C.M.-G., S.E.C., H.V.B., K.L., W.X., B.H.B., D.N.H., R.V.M., and L.L.M. performed research; J.P.C., M.N.M., C.M.-G., S.E.C., H.V.B., K.L., W.X., B.H.B., C.M.E., D.L.H., D.N.H., S.F.L., R.V.M., D.H.S., L.L.M., R.W.D., and R.G.T. analyzed data; and J.P.C., M.N.M., S.E.C., H.V.B., W.X., C.M.E., D.L.H., S.F.L., R.V.M., L.L.M., R.W.D., and R.G.T. wrote the paper.
Data deposition: The array data reported in this paper have been deposited in the Gene Expression Omnibus database (accession nos. GSM42732-GSM42819; the entire series is accessible as GSE2328).
aDepartment of Molecular Genetics and Microbiology, University of Florida, Gainesville, FL 32606-0266; bDepartment of Surgery, University of Rochester Medical Center, Rochester, NY 14642; cDepartment of Surgery, University of Pittsburgh, Pittsburgh, PA 15213-2582; Departments of dGenetics and hSurgery, Washington University, St. Louis, MO 63110; eDepartment of Surgery, University of Medicine and Dentistry of New Jersey, New Brunswick, NJ 08903-0019; fPacific Northwest National Laboratory, Richland, WA 99352; gDepartment of Surgery, University of Alabama School of Medicine, Birmingham, AL 35294-0019; iDepartment of Biochemistry, Stanford University, Palo Alto, CA 94304; Departments of jBiostatistics, zMolecular Biology, and ccSurgery and ddBioMEMS Core Facility, Center for Engineering in Medicine, Massachusetts General Hospital, Boston, MA 02114; kDepartment of Surgery, Loyola University Stritch School of Medicine, Maywood, IL 60153; lDepartment of Surgery and yDivision of Trauma and General Surgery, University of Washington Harborview Medical Center, Seattle, WA 98104; mDepartment of Surgery, Northwestern University, Chicago, IL 60611; nBurn Services, University of Texas Medical Branch, Galveston, TX 77550; oDepartment of Surgery, pDivision of Burn, Trauma, and Critical Care, and xDepartment of Internal Medicine, Division of Infectious Diseases, University of Texas Southwestern Medical School, Dallas, TX 75390; qDepartment of Surgery, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115; rDepartment of Surgery, University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School, New Brunswick, NJ 08903; sTrauma Research Center and wDepartment of Surgery, University of Texas Medical School at Houston, Houston, TX 77030; tStanford Genome Technology Center, Stanford University Medical Center, Palo Alto, CA 94304-1103; uDepartment of Surgery, University of Florida College of Medicine, Gainesville, FL 32610-0286; vDepartment of Surgery, University of Colorado Health Sciences Center, Denver, CO 80262; aaDepartment of Medical Science, University of Michigan, Ann Arbor, MI 48109-0602; bbDepartment of Chemical Engineering and hhLaboratory of Human Nutrition, Massachusetts Institute of Technology, Cambridge, MA 02139-4307; eeDepartment of Medicine, Massachusetts General Hospital, Charlestown, MA 02129; ffDepartment of Surgery, University of Texas Shriners Burn Hospital, University of Texas Medical Branch, Galveston, TX 77550-2726; and ggStanford Genome Technology Center, Stanford University, Stanford, CA 94305-5440.
References
- 1.Steinmetz, L. M. & Davis, R. W. (2004) Nat. Rev. Genet. 5, 190-201. [DOI] [PubMed] [Google Scholar]
- 2.Hood, L., Heath, J. R., Phelps, M. E. & Lin, B. (2004) Science 306, 640-643. [DOI] [PubMed] [Google Scholar]
- 3.Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., Coller, H., Loh, M. L., Downing, J. R., Caligiuri, M. A., et al. (1999) Science 286, 531-537. [DOI] [PubMed] [Google Scholar]
- 4.Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503-511. [DOI] [PubMed] [Google Scholar]
- 5.Hedenfalk, I., Duggan, D., Chen, Y., Radmacher, M., Bittner, M., Simon, R., Meltzer, P., Gusterson, B., Esteller, M., Kallioniemi, O. P., et al.(2001) N. Engl. J. Med. 344, 539-548. [DOI] [PubMed] [Google Scholar]
- 6.Lapointe, J., Li, C., Higgins, J. P., van de Rijn, M., Bair, E., Montgomery, K., Ferrari, M., Egevad, L., Rayford, W., Bergerheim, U., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zerhouni, E. (2003) Science 302, 63-72. [DOI] [PubMed] [Google Scholar]
- 8.Feezor, R. J., Baker, H. V., Mindrinos, M., Hayden, D., Tannahill, C. L., Brownstein, B. H., Fay, A., Macmillan, S., Laramie, J., Xiao, W., et al. (2004) Physiol. Genomics 19, 247-254. [DOI] [PubMed] [Google Scholar]
- 9.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong, Y. M., Marano, M. A., Moldawer, L. L., Wei, H., Calvano, S. E., Kenney, J. S., Allison, A. C., Cerami, A., Shires, G. T. & Lowry, S. F. (1990) J. Clin. Invest. 85, 1896-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan, P. K., Downey, T. J., Spitznagel, E. L., Jr., Xu, P., Fu, D., Dimitrov, D. S., Lempicki, R. A., Raaka, B. M. & Cam, M. C. (2003) Nucleic Acids Res. 31, 5676-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall, E. (2004) Science 306, 630-631. [DOI] [PubMed] [Google Scholar]
- 13.Tumor Analysis Best Practices Working Group (2004) Nat. Rev. Genet. 5, 229-237. [DOI] [PubMed] [Google Scholar]
- 14.Whitney, A. R., Diehn, M., Popper, S. J., Alizadeh, A. A., Boldrick, J. C., Relman, D. A. & Brown, P. O. (2003) Proc. Natl. Acad. Sci. USA 100, 1896-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobb, J. P. & O'Keefe, G. E. (2004) Lancet 363, 2076-2083. [DOI] [PubMed] [Google Scholar]
- 16.De, A. K., Laudanski, K. & Miller-Graziano, C. L. (2003) J. Immunol. 170, 6355-6362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.