Abstract
Pelvic organ prolapse (POP) is a highly disabling condition common for a vast number of women worldwide. To contribute to existing knowledge in POP pathogenesis, we performed a systematic review of expression studies on both specific gene and whole-genome/proteome levels and an in silico analysis of publicly available datasets related to POP development. The most extensively investigated genes in individual studies were related to extracellular matrix (ECM) organization. Three premenopausal and two postmenopausal sets from two Gene Expression Omnibus (GEO) studies (GSE53868 and GSE12852) were analyzed; Gene Ontology (GO) terms related to tissue repair (locomotion, biological adhesion, immune processes and other) were enriched in all five datasets. Co-expression was higher in cases than in controls in three premenopausal sets. The shared between two or more datasets up-regulated genes were enriched with those related to inflammatory bowel disease (IBD) in the NHGRI GWAS Catalog. ECM-related genes were not over-represented among differently expressed genes. Up-regulation of genes related to tissue renewal probably reflects compensatory mechanisms aimed at repair of damaged tissue. Inefficiency of this process may have different origins including age-related deregulation of gene expression.
Introduction
Pelvic organ prolapse (POP), the dropping of the pelvic organs due to the loss of normal support of the vagina, is an age-related condition associated with enormous physical and emotional discomfort for a vast number of women worldwide. In total, POP affects 40–50% of women, being one of the most common reasons for gynecological surgery1–3. Despite a relatively large number of scientific papers on POP, mechanisms of its occurrence remain unclear2, whereas understanding of POP pathophysiology is necessary for its prevention and treatment.
Expression studies provide valuable information for deciphering molecular mechanisms of diseases. Several reviews on POP have included results of the investigations of expression changes in POP mainly focusing on the genes/proteins of collagen, elastin, matrix metalloproteinases and their tissue inhibitors4, 5. The comparison of the expression patterns of multiple genes in different studies may be useful for understanding of disease pathogenesis at the gene level. To the best of our knowledge, no research has been performed so far to analyze all available data from expression studies in POP, on both specific gene and whole-genome/proteome levels. In order to identify new genes and biological processes implicated in POP pathogenesis, we conducted a systematic review of the expression studies and an in silico analysis of publicly available data sets related to POP development.
Results
Overview of studies assessing individual gene expression profile in POP
From a total of 465 studies found on the theme of investigation by searching PubMed, Embase and Web of Knowledge resources (Supplementary Figure S1) 78 papers were selected for data analysis (Supplementary Table S1). One hundred twenty two gene or protein products were studied, 113 of them corresponded to specific genes (Supplementary Table S1). In general, papers on associations between studied genes or proteins and POP are characterized by high heterogeneity in experimental design and data presentation. The visualization of survey data for the genes/proteins investigated in five or more studies was conducted to display proportions of data for up-regulation, down-regulation or non-significant associations for all studied genes/proteins and POP (Fig. 1). Proportions are indicated for the number of studies and for the number of subjects in these studies. The least variable results with the largest number of studies being performed were found for MMP2/ MMP2 followed by MMP1/MMP1. Data for MMP1 were in agreement with results of the only meta-analysis in the field, namely with a higher expression level of MMP1 protein in POP cases in comparison with controls6. Decreased activity or non-significant results were mainly registered for TIMP Metallopeptidase Inhibitors, Collagen type I alpha 1, Lysyl oxidase, Fibulin 5 and Elastin. Other data appeared to be far more contradictory.
Figure 1.
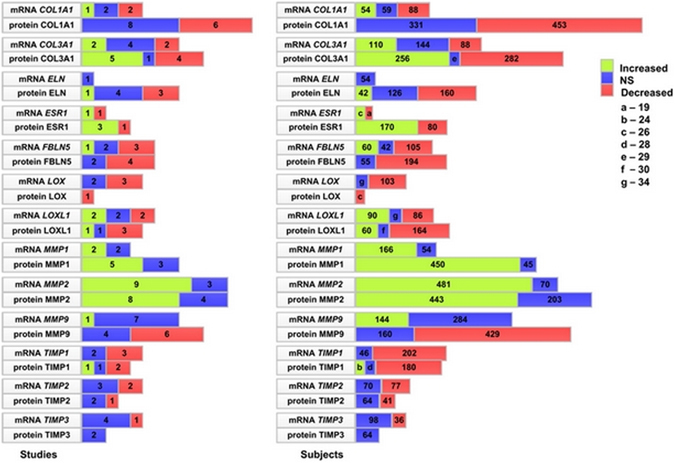
Summary of literature data on POP-related expression changes for selected genes. Numbers in boxes indicate the number of studies (Fig. 1a) and subjects (Fig. 1b) for each marker. Letter symbols (a–g) in the small boxes are decrypted in the right upper corner.
Next, we applied KOBAS 3.0 resource7 to perform GO (Gene Ontology)8 gene set enrichment analysis for the gene spectrum considered in the expression studies. REVIGO9 summary for the cluster GO representatives is provided in Fig. 2. As expected, the highest enrichment (the lowest P-value) was found for the term “extracellular matrix organization” which is the most specific relative to other displayed GO terms (has the lowest frequency in the underlying GO annotation database). From 13 genes presented in Fig. 1, 11 genes were related to extracellular matrix organization (ECM). These genes also contributed to the other biological processes which are indicated for the whole set of the studied genes (Fig. 2).
Figure 2.
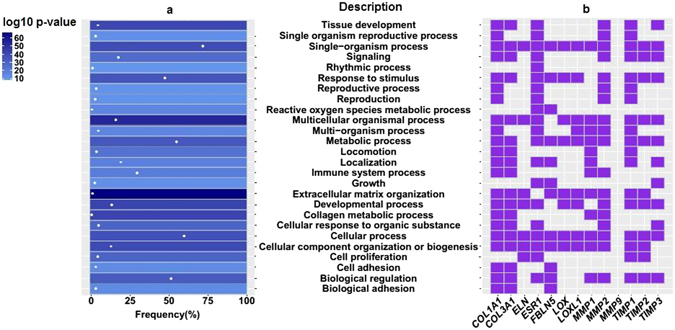
(a) Heat map for GO terms cluster representatives for genes considered in POP studies. (b) GO terms associated with the selected genes. Color indicates the user-supplied P- value; the term ‘frequency’ means frequency of the specific term in the underlying GO annotation database.
Overview of whole genome/proteome studies of POP
Papers assessing the whole genome/proteome expression profile in POP did not provide consistent results (Table 1). Two studies on pubococcygeus tissue in POP patients have shown both up- and down-regulation of cytoskeletal genes or proteins10, 11. In full-thickness vaginal wall biopsies, genes related to smooth muscle contraction, proteolysis, response to oxidative stress, transcriptional regulation, cytoskeletal organization, and lipid catabolism were over-expressed12. The analysis of 34 arrays (17 round ligaments (RLs) and 17 uterosacral ligaments (USLs)) has revealed that ‘immunity and defense’ genes were up-regulated in POP13. Expression changes of transcriptional response and signal transduction genes associated with estrogen were detected in the USLs of POP patients14. Genes relevant to cell cycle, proliferation and embryonic development as well as genes related to cell adhesion were down-regulated in USLs of females with uterine prolapse15. In USL samples, differently expressed genes (DEG) between POP patients and controls were significantly enriched with those related to canonical Wnt receptor signaling pathway (GO term) and neuroactive ligand-receptor interaction (pathway)16.
Table 1.
Characteristics of the whole genome/proteome studies of POP.
| Dataset | Number of samples (age/MP status) | Analysis | DEG (DEP) | Main findingsa | Confirmation of whole genome/proteome data | Reference |
|---|---|---|---|---|---|---|
| Pubococcygeus muscle in Caucasian POP patients and mixed ethnicity controls | 5 cases (58.9 ± 5.5); 5 controls (45.5 ± 11.4) | Microarray gene expression | Down-regulation in POP patients: FC > 2.0, 257 genes; FC > 5, 20 genes; FC > 10, 3 genes. Up-regulation in POP patients: FC > 2.0, 479 genes; FC > 5, 18 genes; FC > 10, 2 genes. | The genes MYBPH (FC 24.7), MYH3 (FC 17.4) and COMP (FC 6.0) were down-regulated, while smooth muscle myosin heavy chain (FC 11.8), MLCK (FC 5.77) and TNC (FC 5.1) genes were up-regulated in POP patients in comparison with controls. | No | 10 |
| Full-thickness vaginal wall biopsies from patients with POP and age-, parity -, and body mass index -matched controlsb | 5 PrM cases, 5 PrM controls | Microarray gene expression | Upregulated genes (n = 50) comprised those involved in smooth muscle contraction, proteolysis, response to oxidative stress, transcriptional regulation, cytoskeletal organization, and lipid catabolism. | The PPP1R12A and ADAMTS1 genes were up-regulated 2.4 and 4-fold in cases compared to controls. | mRNA analysis for 4 matched pairs of POP/control tissues | 12 |
| USLs and RLs from mixed ethnicity patients with uterine prolapse (POP-Q stage ≥ 2) and controls | 8 POP patients (5 PrM and 3 PM); 9 controls (6 PrM and 3 PM) | Microarray gene expression | In a combined analysis of expression changes in USLs and RLs, 1521 genes were up-regulated and 1193 genes were down-regulated with FC change 1.5. 249 up-regulated gene probes met FDR criteria ≤ 5.18. | After FDR correction for multiple testing, genes enriched for ‘immunity and defense’ were up-regulated in POP patients. | qRT-PCR Up-regulation in POP patients: IL-6 (FC 9.8), ATF3 (FC 2.6), THBS1 (FC 3.5) and PTGS2 (FC 2.4) | 13 |
| Pubocervical fascia from females with POP and SUI and from asymptomatic controlsb | 4 cases (PM); 3 controls (PM) | Proteomic analysis (two-dimensional electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry) | 7 proteins with an expression FC more than two (3.8 ─ 54.3-fold). | The expression levels of transgelin (FC 54.3), smooth muscle gamma-actin (FC 6.3), myosin light polypeptide 6 (FC 4.4), and alpha-1 antitrypsin precursor (FC 4.3) were higher in patients than in controls. | 2-DE western blot analysis for transgelin | 11 |
| USLs from Asian women with POP-Q stage 3-4 (cases) or 0-1 (controls) | 12 cases (PM), 5 controls (PM) | Microarray gene expression | 143 genes were up-regulated and 87 genes were down-regulated with FC > 1.5 (P < 0.05). | The expression levels of ESRRA (FC 0.434) decreased, while the expression levels of DAPK2 (FC 2.013), IL15 (FC 2.1) and STAP2 (FC 2.392) increased in POP patients compared with controls. | qRT-PCR for the genes ESRRA, DAPK2, IL15, STAP2 | 14 |
| USLs from patients with uterine prolapse and controlsb | 16 POP patients (15 patients PM); 9 controls (7 females PM) | Microarray gene expression | 21 up-regulated and 7 down-regulated genes with FC > 2.0 were found in patients with POP compared to the control group. | Down-regulation in POP patients compared with controls after FDR correction for multiple testing: NKX2-3 (FC 2.6) in the whole set; KIF11 (FC 1.3) in patients with ≥ 3 deliveries compared to patients with < 3 deliveries; UGT1A1 (FC 1.4), SCARB1 (FC 3.2) and NKX2-3 (FC 6.2) in PrM patients compared to PrM controls; UGT1A1 (FC 1.1) in PM patients compared to PM controls. | No | 15 |
| USL samples of patients with stage II to stage IV POP (mean age 61 years) and normal controls (mean age 55 years)b | No data | RNA-seq (a HiSeqTM 2500 platform (Illumina)) | After FDR-correction, a total of 81 genes had different expression patterns in POP patients and controls. Sixty-six DEGs did not differ between the POP samples with different stages | Canonical Wnt receptor signaling pathway was the most significantly enriched GO term (P- value = 3.33E-07), and neuroactive ligand-receptor interaction was the most significantly enriched pathway (P- value = 1.24E-03). | No | 16 |
aData are uncorrected for multiplicity, otherwise specified. bEthnicity is not specified.
Abbreviations: DEG, differently expressed genes; DEP, differently expressed proteins; FC, fold change; FDR, false discovery rate; PM, postmenopausal; PrM, premenopausal; RLs, round ligaments; SUI, stress urinary incontinence; USLs, uterosacral ligaments.
Overview of available GEO datasets for POP
Among published whole genome studies, only the study of Brizzolara13 has been represented in the GEO database repository17. An unpublished study assessing gene expression profile in the sites of prolapsed versus non-prolapsed vaginal tissues is also available in the resource (Table 2).
Table 2.
Characteristics of the POP datasets in the Gene Expression Omnibus (GEO) database repository.
| Dataset | Reference | Tissue | Number of samples | GEO accession no; Platform | Number of DEGa with FDR-corrected P - value < 0.05 | Number of DEGa with FC ≥ 2.0 and P - value < 0.05 (uncorrected for multiplicity) |
|---|---|---|---|---|---|---|
| PrM Caucasian women (cystocele POP-Q stage ≥ 2), n = 12 | Citation missing | AVW (POP site) versus precervical AVW (non POP site) | 12 women (24 samples) | GSE53868; GPL18142 Agilent-014850 WHGM 4 × 44K G4112F | 50 (↑ 31; ↓ 19) | 74 (↑ 58; ↓16) |
| PrM POP women, n = 5b | Brizzolara et al.13 | RLs | 5 | GSE12852; GPL2986 ABI Human Genome Survey Microarray Version 2 | PrM_RLs set 0 | PrM_RLs set 287 (↑ 246; ↓ 41) |
| USLs | 5 | |||||
| PrM controls, n = 6b | RLs | 6 | PrM_USLs set 0 | PrM_USLs set 972 (↑ 949; ↓23) | ||
| USL | 6 | |||||
| PM POP women, n = 3b | RL | 3 | PM_RLs set 0 | PM_RLs set 485 (↑ 345; ↓ 140) | ||
| USL | 3 | |||||
| PM controls, n = 3b | RL | 3 | PM_USLs set 2 (↑ 2; ↓ 0) | PM_USLs set 494(↑ 263; ↓ 231) | ||
| USL | 3 |
a↑ up-regulation; ↓ down-regulation; bMixed ethnicity.
Abbreviations: AVW, anterior vaginal wall; DEG, differently expressed genes; FC, fold change; FDR, false discovery rate; WHGM, whole human genome microarray; PM, postmenopausal; PrM, premenopausal; RLs, round ligaments; USLs, uterosacral ligaments.
Since the focus of our study was to find consistent patterns of gene expression profiles in different POP-related datasets, we subjected the whole dataset of Brizzolara13 stratified by the sort of ligaments (USLs or RLs) and the menopausal status (premenopausal (PrM) or postmenopausal (PM)) to the analysis. We were guided by the fact that the USLs, being the main supportive structures of the uterus and vagina18, have demonstrated higher tensile biomechanical properties (stiffness and maximum stress) than RLs19. These differences may be linked with distinctive features of gene expression profile. Additionally, in the study of Brizzolara13 USLs and RLs differed by smooth muscle cells composition; moreover, among the top 250 DEG genes only five genes coincided for USLs and RLs (the genes F5, NR4A3, CLC, SLC24A4 and GDF15 were up-regulated in both series). With regard to menopausal status, we have taken into account that expression patterns often differ between PrM and PM females20–25. Given that menopause is one of the main risk factors for POP, sample stratification based on menopausal status may provide better comparable results.
GO enrichment analysis for the GEO datasets
DEG were analyzed with the KOBAS 3.0 application. Data for the series of not less than five genes associated with GO terms in the studied sets are presented in Supplementary Table S2. The number of enriched terms was noticeably larger for up- than for down-regulated genes. Biological process gene ontology terms for the five sets of over-expressed genes were summarized with the REVIGO application (Fig. 3). This analysis showed massive enrichment for genes implicated in tissue renewal and regeneration.
Figure 3.
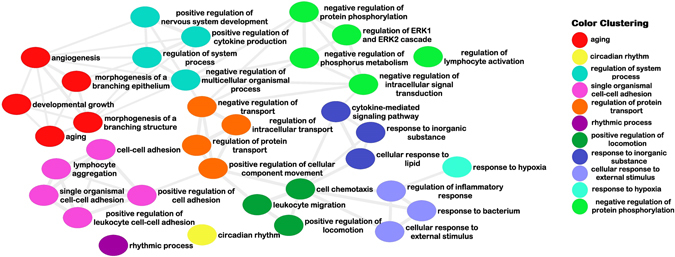
Graph for the results of enrichment analysis for five gene sets up-regulated in POP. More similar nodes are placed closer together. The line width indicates the degree of similarity between GO terms cluster representatives. Clustering by color into super clusters was obtained by using REVIGO TreeMap application.
Up-regulated genes were associated with a number of shared terms for all five gene sets. Interestingly, among the most specific GO categories (not more than 500 background genes in the GO database) terms that include the wording ‘positive regulation’ covered biological processes related to: adhesion (n = 3), locomotion (n = 4), activation (n = 3), transport (n = 2) as well as cytokine production, MAPK cascade and nervous system development. These mechanistic considerations also provide some evidence that genes that control tissue repair are over-represented among the up-regulated genes in all data sets under study.
A bird’s eye view on co-expression data for DEG
Co-expression analysis may give insights into altered regulatory mechanisms between disease and healthy controls, since co-regulated genes tend to exhibit similar expression patterns. Given that up-regulated genes in POP tissues were enriched with those relevant to tissue repair, we found it interesting to perform a comparative co-expression analysis. Pearson correlation coefficients were plotted in heat maps and density plots for pared genes (Fig. 4). Higher levels of co-expression in prolapsed versus healthy tissues were revealed among sets of up-regulated genes in all premenopausal groups (PrM_RLs, PrM_USLs, AVW) and down-regulated genes in the groups PrM_RLs and PrM_USLs. This observation may reflect the underlying activity of transcriptional networks, indirectly supporting the assumption on activation of regeneration processes in POP tissues; however, co-expression of down-regulated genes may reflect some intrinsic problems in realization of these processes. In postmenopausal sets (PM_RLs and PM_USLs), density plots for up- and down-regulated genes were somewhat similar, thus indicating relatively high levels of positive and negative correlations in both control and POP specimens. A possible explanation of this phenomenon is that in aging tissues disturbances in regulatory mechanisms can predominate and/or obscure the activity of the repair processes. The results may also be random due to the small samples size.
Figure 4.
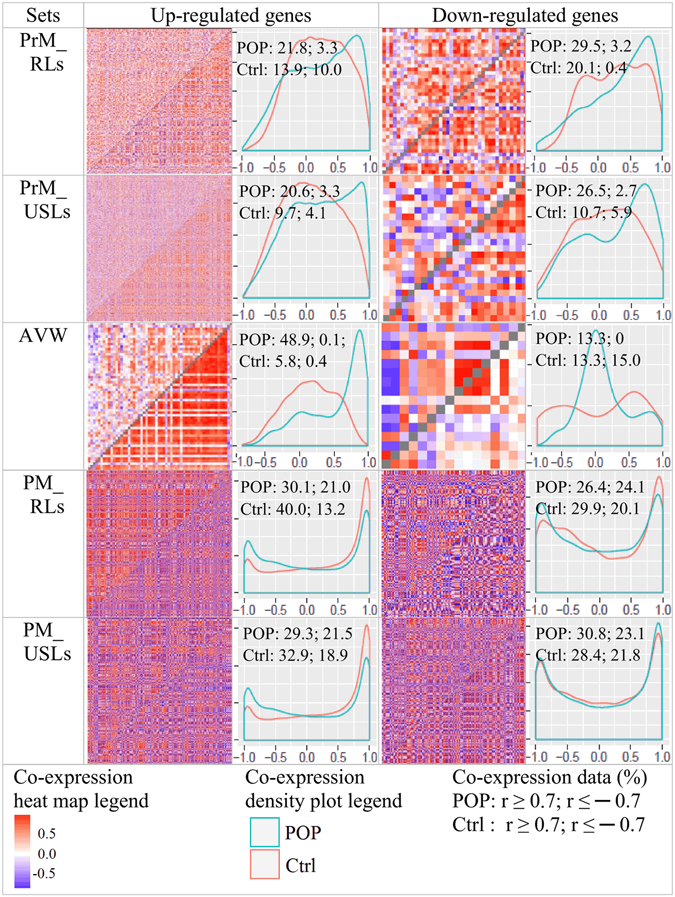
Co-expression heatmaps and density plots. Heatmaps display Pearson correlation coefficients for gene expression in controls (above the diagonal) and cases (below the diagonal). Density plots present correlation coefficients distribution for controls (blue) and cases (red) with addition of percentage of Pearson correlations (r-values) ≥ 0.7 and ≤ -0.7. Density plots x-axis: Pearson correlation coefficient (r), y-axis: density.
Enrichment analysis (GO, KEGG, GWAS Catalog) for the shared gene set
A total of 142 up-regulated and 12 down-regulated genes were shared between two or more datasets (Supplementary Table S3). All top DEG in the study of Brizzolara13 appeared to be in the list of shared genes.
The results of GO enrichment analysis for shared up-regulated genes correlated with those obtained for the individual sets (Fig. 5). For down-regulated genes, GO analysis did not yield significant terms.
Figure 5.
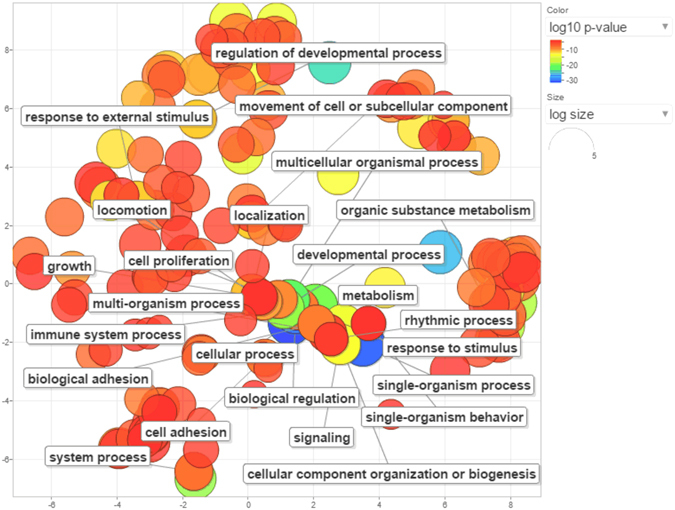
REVIGO scatterplot for GO terms cluster representatives for shared genes up-regulated in POP tissues. Bubble color indicates the user-supplied P - value; size shows the frequency of the GO term in the GO annotation database.
All shared up-regulated genes were tested for enrichment of metabolic pathways and association signals from GWAS data (Table 3). The enriched KEGG pathways were mainly linked with inflammatory and immune-mediated diseases. Of great interest were the results of the enrichment analysis of GWAS association data which indicated that genes associated with inflammatory bowel disease (IBD) and Crohn’s disease (one of the two main forms of IBD) were overrepresented among shared up-regulated genes. These genes (with the exception for the genes SLC22A4, BORCS5 and CNNM1) are involved in ‘immune system process’ (GO term).
Table 3.
The results of gene set enrichment analysis for shared genes up-regulated in POP tissues.
| Term | Input number | Background number | P - value | Corrected P - value | Genes |
|---|---|---|---|---|---|
| KEGG Pathway | |||||
| Malaria (hsa05144) | 7 | 49 | 1,25E-09 | 1,93E-07 | COMP, ICAM1, IL6, CXCL8, CCL2, THBS1, THBS4 |
| Influenza A (hsa05164) | 8 | 176 | 3,48E-07 | 2,69E-05 | NLRP3, ICAM1, IL6, CXCL8, NXF3, CCL2, SOCS3, RSAD2 |
| Rheumatoid arthritis (hsa05323) | 5 | 91 | 2,51E-05 | 1,19E-03 | CTSK, ICAM1, IL6, CXCL8, CCL2 |
| PI3K-Akt signaling pathway (hsa04151) | 8 | 342 | 3,91E-05 | 1,19E-03 | CDKN1A, COMP, GNGT1, IL6, MCL1, PPP2R2C, THBS1, THBS4 |
| AGE-RAGE signaling pathway in diabetic complications (hsa04933) | 5 | 101 | 4,04E-05 | 1,19E-03 | ICAM1, IL6, CXCL8, SERPINE1, CCL2 |
| Chagas disease (American trypanosomiasis) | 5 | 104 | 4,62E-05 | 1,19E-03 | IL6, CXCL8, SERPINE1, PPP2R2C, CCL2 |
| Hepatitis C (hsa05142) | 5 | 133 | 1,41E-04 | 2,43E-03 | CDKN1A, CXCL8, LDLR, PPP2R2C, SOCS3 |
| Hepatitis B (hsa05161) | 5 | 146 | 2,15E-04 | 3,32E-03 | CDKN1A, EGR2, EGR3, IL6, CXCL8 |
| Phagosome (hsa04145) | 5 | 155 | 2,80E-04 | 3,95E-03 | COMP, FCAR, THBS1, THBS4, COLEC11 |
| HTLV-I infection (hsa05166) | 6 | 259 | 3,90E-04 | 4,91E-03 | CDKN1A, EGR2, ICAM1, IL6, ATF3, IL1R2 |
| Transcriptional misregulation in cancer (hsa05202) | 5 | 180 | 5,44E-04 | 5,63E-03 | CDKN1A, IL6, CXCL8, IL1R2, NR4A3 |
| Herpes simplex infection (hsa05168) | 5 | 186 | 6,29E-04 | 6,09E-03 | IL6, NXF3, CCL2, TNFSF14, SOCS3 |
| Cytokine-cytokine receptor interaction (hsa04060) | 5 | 265 | 2,89E-03 | 2,03E-02 | IL6, CXCL8, CCL2, IL1R2, TNFSF14 |
| Pathways in cancer (hsa05200) | 6 | 397 | 3,31E-03 | 2,13E-02 | CDKN1A, CKS2, GNGT1, IL6, CXCL8, BDKRB1 |
| The NHGRI GWAS Catalog | |||||
| Inflammatory bowel disease | 9 | 271 | 8,03E-07 | 1,26E-04 | BORCS5, CCL2, CXCL8, ICAM1, IL1R2, NFIL3, PTPRC, SLC11A1, SLC22A4 |
| Crohn’s disease | 6 | 256 | 3,68E-04 | 1,15E-02 | SLC22A4, ICAM1, CCL2, TAGAP, PTPRC, CNNM1 |
Literature data supporting associations revealed for the shared genes
Several associations revealed in the set of shared genes were partially supported by literature data. Three different kinds of evidence were presented: (i) results on animal models, (ii) evidence from the studies of gene polymorphisms and (iii) expression data for any type of prolapse with the same direction of association. Usage of (iii) as an evidence was based on the rationale that heart valve, cartilage, tendon, and bone development share common regulatory pathways26. The genes ADAMTS1 12 , MYH3 27 and SERPINE1 28 were up-regulated in POP tissues. Polymorphic variants in the genes LIN28B 29 and AGT 30 were associated with POP and mitral valve prolapse (MVP) respectively. Nfil3-/- mice developed colitis with high prevalence of rectal prolapse31. High expression of Nlrp3 was found in Il10-/-mice with colitis combined with rectal prolapse32. The genes CTSK 33, MMP19 and THBS4 34 were over-expressed in MVP. Egr2-/- mice had features of human aortic valve disease, in particular excess of proteoglycan deposition and reduction of collagen fibres35. This information is given in more detail in Supplementary Table S4.
Discussion
This study highlighted some important biological processes and putative candidate genes most likely linked with POP development. We present below our vision of POP pathogenesis which is based on our findings and literature data.
The majority of individual gene expression studies in POP were focused on genes related to ECM. Summary data obtained on dozens and even hundreds of patients (Fig. 2) basically supported the common opinion on expression changes of these genes in POP. The results presented in the whole-genome/proteome studies as well as the results of the in silico analysis did not confirm these observations. The discrepancies may be linked with the small samples for the whole genome sets, with a highly variable design in terms of investigated tissue, menopausal status, ethnicity, experimental methods and other. Different designs were also used in the studies of candidate genes; however, in larger samples the differences are smoothed out resulting in more accurate parameter estimates, which, in turn, lead to a greater probability to find the desired results. Correction for multiplicity was rarely applied in the studies considering several individual genes; however, in the genome-based approach, any method of selecting the most significant genes was used always (FDR correction, top DEG, top gene sets) with a probability to miss less pronounced but biologically plausible correlations.
Our in silico analysis included three independent (AVW, PrM and PM) and two dependent (USL and RL from the same subjects) sets. The design was focused on the search of expression changes in one set and the validation of found differences in other sets. Genes related to ECM-structure were not represented among the sets of shared genes according to the GO results. The shared terms for up-regulated genes revealed the need for response to stimulus, locomotion, adhesion, immune, rhythmic, developmental and some other biological processes which should precede ECM synthesis. In this context, given the substantial histological differences between prolapsed versus non-prolapsed tissues36, 37, changes in expression for the most studied ECM genes may be found as a consequence of prolapse rather than an underlying cause38. This assumption is in line with the results of a few studies on the role of germline genetic variations in POP. Association studies on the whole-genome level have not revealed genes expected to be linked with connective tissue disorders29, 39. Meta-analyses in this field yielded unstable results40, 41. Contradictions in the studies of germline genetic variations may partially depend on the underestimation of important risk factors such as perineal trauma in childbirth in the majority of POP genetic association studies42, 43. The genetic component of prolapse is rather high44, 45 and, given the presence of different kinds of biological activities of proteins encoded by the studied ECM genes (Fig. 2), the question of whether they are only markers or to a certain extent causative genes for POP is still open.
Our findings that in POP tissues genes involved in tissue repair were up-regulated appeared to be unexpected but biologically plausible. Several levels of evidence supported this statement: (i) in all five gene sets under study as well as (ii) in the shared gene set, DEG were enriched with those involved in biological processes implicated in tissue regeneration; (iii) in three PrM sets there was a high co-expression (conceivably co-regulation) of DEG just in POP specimens. Compensatory mechanisms aimed at the recovery of damaged tissues require coordinated regulation of many biological processes with a crucial step being the recruitment of blood cells which mediate inflammatory and immune responses, promoting tissue repair. Inefficiency of these processes may be linked with many reasons, among which the most important are age-related changes. The proportion of women with pelvic floor disorders is dramatically increasing with age reaching up to 10% in women aged 20 to 39 years and up to 50% in women aged 80 years or older1.
ECM is constantly being remodeled by degrading and reassembling. In response to injury and other stimuli, remodeling rates increase significantly. ECM degradation products induce inflammation46, which is in turn associated with acceleration of proteolytic cascades leading to further destruction of ECM. Inflammation linked or non-linked with infection and other diseases increases with age and age-related clinical conditions. Balanced immune and anti-inflammatory response is crucial for successive regeneration of vaginal tissues. In youth and maturity clinically asymptomatic damage is compensated by repair processes and/or other components of the pelvic floor support mechanism. Aging and menopause are associated with oxidative stress and hormonal disturbances, both conditions strongly exacerbating ECM breakdown processes47–49. Tissue remodeling may be successful in youth and maturity but in old age many processes are deregulated and this factor may at least partially explain the delayed onset of the disease with major risk factors (parity and perineal trauma in childbirth) linked to youth.
An interesting finding in our work is that up-regulated genes in prolapsed tissues were enriched with those related to IBD in the GWAS Catalog. IBD includes Crohn’s disease and ulcerative colitis. Persons with constipation-predominant symptoms may suffer from pelvic floor muscular incoordination and failure of normal relaxation of pelvic floor muscles during attempted defecation. GWAS-implicated variants lie on genes that may be linked with immune-mediated disturbances in physiological homeostasis in both diseases. Other hypothesis interpreting shared susceptibility to both disorders might come from the results of animal studies: IBD frequently co-exists with rectal prolapse which is in turn associated with other types of genital prolapse since rectocele leads to overdistension of the perineal body50.
The study has some limitations with the main problem being in a small number of datasets and a small number of samples in these datasets. These data appeared to be insufficient for construction of co-expression networks. The results of the enrichment analysis for the overlapping up-regulated genes with GWAS association signals should be discussed as preliminary. These results raise a question rather than provide an answer on a possible shared genetic component for IBD and POP.
Finally, our analysis provided some in-depth data important for understanding POP pathogenesis. In terms of genetic overlap between IBD and POP, the work has translational impact. The study findings are biologically plausible; however, they require verification in independent studies.
Materials and Methods
Selection of studies assessing individual gene expression profile
The final search was performed on 11 January 2017 of the PubMed, EMBASE and Web of Science databases in compliance with PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analysis) guidelines51. The following keyword terms were used as criteriae for searching: pelvic organ prolapse, vaginal prolapse, genital prolapse, uterovaginal prolapse, uterine prolapse, prolapse of vaginal vault, pelvic floor dysfunction, pelvic floor disorder, cystocele, rectocele in combination with the terms: expression, production, secretion, gene and protein. Additional articles were identified by checking reference lists of relevant articles. We used the following inclusion criteria. The article had to be published in English and had to have evaluated POP-related expression changes in pelvic floor supportive tissues in a case-control study in vivo. For overlapping studies, we selected those with larger number of subjects.
Microarray data processing
Results of two studies submitted to the repository Gene Expression Omnibus (GEO) were processed with GEO2R – an interactive web tool which exploits Limma R packages from Bioconductor project17 for comparison of user-defined groups of samples under the same experimental conditions (http://www.ncbi.nlm.nih.gov/geo/geo2r/). The use of the ‘Value distribution’ option showed that the data have been normalized and therefore cross-comparable.
Gene set enrichment analysis
We generated gene sets of DEG from the GEO2R data by setting the cut-off P – value < 0.05 (without correction for multiplicity) and fold change ≥2.0. These gene sets were treated with KOBAS 3.0 resource7 for Gene Ontology (GO) enrichment analysis. The following settings were applied: the minimum number of genes per category was five, while Benjamini and Hochberg false discovery rate (FDR) corrected P - value threshold was 0.05. Web server REVIGO was utilized for summarizing GO terms, which was guided by the P-value9. We took into account the hierarchical structure among GO terms for data interpretation: when a gene is associated with a term, it is automatically associated with its parent terms8.
KOBAS 3.0 tool was additionally used for other types of enrichment analyses for shared genes between gene sets under study, namely metabolic pathways analysis (KEGG PATHWAY) and comparison with the NHGRI GWAS Catalog associations.
Statistical considerations
Our study was performed to identify common molecular features for POP phenotype. Many promising markers selected in a single data set appeared to be not so promising or even non-significant in independent sets (a winner’s curse problem). External/independent validation is recommended for large-scale (OMICS) studies52. Taking into account these recommendations, we searched for shared DEG with the same direction (down-regulation or up-regulation) of the association in independent sets.
Data analyses and visualization were conducted with the R statistical software53.
Compliance with ethical standards
As a secondary analysis of public data the study does not require IRB approval.
Electronic supplementary material
Acknowledgements
The study was supported by the Russian Foundation for Basic Research, Project 15-04-02378.
Author Contributions
L.E.S. conceived the idea. All authors contributed to the study design. M.B.K. and L.E.S. supervised the project. M.B.K., D.S.K. and L.E.S. analyzed data. M.B.K. and D.S.K. prepared figures. L.E.S. wrote the manuscript. D.S.K. revised the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-08185-6
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nygaard I, et al. Prevalence of symptomatic pelvic floor disorders in US women. JAMA. 2008;300:1311–1316. doi: 10.1001/jama.300.11.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fritel X, Varnoux N, Zins M, Breart G, Ringa V. Symptomatic pelvic organ prolapse at midlife, quality of life, and risk factors. Obstet Gynecol. 2009;113:609–616. doi: 10.1097/AOG.0b013e3181985312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerten KA, Markland AD, Lloyd LK, Richter HE. Prolapse and incontinence surgery in older women. J Urol. 2008;179:2111–2118. doi: 10.1016/j.juro.2008.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campeau L, Gorbachinsky I, Badlani GH, Andersson KE. Pelvic floor disorders: linking genetic risk factors to biochemical changes. BJU Int. 2011;108:1240–1247. doi: 10.1111/j.1464-410X.2011.10385.x. [DOI] [PubMed] [Google Scholar]
- 5.Lim VF, Khoo JK, Wong V, Moore KH. Recent studies of genetic dysfunction in pelvic organ prolapse: the role of collagen defects. Aust N Z J Obstet Gynaecol. 2014;54:198–205. doi: 10.1111/ajo.12169. [DOI] [PubMed] [Google Scholar]
- 6.Feng, Y., Wang, Y., Yan, B., Li, L. & Deng Y. Matrix metalloproteinase-1 expression in women with and without pelvic organ prolapse: a systematic review and meta-analysis. Clin Transl Sci. doi:10.1111/cts.12409 (2016). [DOI] [PMC free article] [PubMed]
- 7.Xie C, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.GO Consortium Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visco AG, Yuan L. Differential gene expression in pubococcygeus muscle from patients with pelvic organ prolapse. Am J Obstet Gynecol. 2003;189:102–112. doi: 10.1067/mob.2003.372. [DOI] [PubMed] [Google Scholar]
- 11.Athanasiou S, Lymberopoulos E, Kanellopoulou S, Rodolakis A, Vlachos G, Antsaklis A. Proteomic analysis of pubocervical fascia in women with and without pelvic organ prolapse and urodynamic stress incontinence. Int Urogynecol J. 2010;21:1377–1384. doi: 10.1007/s00192-010-1203-4. [DOI] [PubMed] [Google Scholar]
- 12.Stratford RR, Baumann SS, Jamroz RC, Kuehl TJ, Shull BL, Pierce LM. Poster 14: Histology and differential mRNA expression in vaginal connective tissue of women with pelvic relaxation. Journal of Pelvic Medicine & Surgery. 2005;11:S32. doi: 10.1097/01.spv.0000178878.12148.0d. [DOI] [Google Scholar]
- 13.Brizzolara SS, Killeen J, Urschitz J. Gene expression profile in pelvic organ prolapse. Mol Hum Reprod. 2009;15:59–67. doi: 10.1093/molehr/gan074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moon YJ, Bai SW, Jung CY, Kim CH. Estrogen-related genome-based expression profiling study of uterosacral ligaments in women with pelvic organ prolapse. Int Urogynecol J. 2013;24:1961–1967. doi: 10.1007/s00192-013-2124-9. [DOI] [PubMed] [Google Scholar]
- 15.Ak H, Zeybek B, Atay S, Askar N, Akdemir A, Aydin HH. Microarray gene expression analysis of uterosacral ligaments in uterine prolapse. Clin Biochem. 2016;49:1238–1242. doi: 10.1016/j.clinbiochem.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Xie R, Xu Y, Fan S, Song Y. Identification of differentially expressed genes in pelvic organ prolapse by RNA-Seq. Med Sci Monit. 2016;22:4218–4225. doi: 10.12659/MSM.900224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett T, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivaux G, Rubod C, Dedet B, Brieu M, Gabriel B, Cosson M. Comparative analysis of pelvic ligaments: a biomechanics study. Int Urogynecol J. 2013;24:135–139. doi: 10.1007/s00192-012-1861-5. [DOI] [PubMed] [Google Scholar]
- 19.Martins P, et al. Strength of round and uterosacral ligaments: a biomechanical study. Arch Gynecol Obstet. 2013;287:313–318. doi: 10.1007/s00404-012-2564-3. [DOI] [PubMed] [Google Scholar]
- 20.Kow N, Ridgeway B, Kuang M, Butler RS, Damaser MS. Vaginal expression of LOXL1 in premenopausal and postmenopausal women with pelvic organ prolapse. Female Pelvic Med Reconstr Surg. 2016;22:229–235. doi: 10.1097/SPV.0000000000000251. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, et al. Biomechanical properties and associated collagen composition in vaginal tissue of women with pelvic organ prolapse. J Urol. 2012;188:875–880. doi: 10.1016/j.juro.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Bortolini MA, et al. Expression of genes encoding smooth muscle contractile proteins in vaginal tissue of women with and without pelvic organ prolapse. Neurourol Urodyn. 2012;31:109–114. doi: 10.1002/nau.21175. [DOI] [PubMed] [Google Scholar]
- 23.Zbucka-Kretowska M, et al. Expression of estrogen receptors in the pelvic floor of pre- and post-menopausal women presenting pelvic organ prolapse. Folia Histochem Cytobiol. 2011;49:521–527. doi: 10.5603/FHC.2011.0073. [DOI] [PubMed] [Google Scholar]
- 24.Budatha M, et al. Extracellular matrix proteases contribute to progression of pelvic organ prolapse in mice and humans. J Clin Invest. 2011;121:2048–2059. doi: 10.1172/JCI45636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Söderberg MW, Byström B, Kalamajski S, Malmström A, Ekman-Ordeberg G. Gene expressions of small leucine-rich repeat proteoglycans and fibulin-5 are decreased in pelvic organ prolapse. Mol Hum Reprod. 2009;15:251–257. doi: 10.1093/molehr/gap011. [DOI] [PubMed] [Google Scholar]
- 26.Lincoln J, Lange AW, Yutzey KE. Hearts and bones: shared regulatory mechanisms in heart valve, cartilage, tendon, and bone development. Dev Biol. 2006;294:292–302. doi: 10.1016/j.ydbio.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 27.Hundley AF, Yuan L, Visco AG. Gene expression in the rectus abdominus muscle of patients with and without pelvic organ prolapse. Am J Obstet Gynecol. 2008;198(220):e1–7. doi: 10.1016/j.ajog.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 28.Budatha M, et al. Dysregulation of protease and protease inhibitors in a mouse model of human pelvic organ prolapse. PLoS One. 2013;8:e56376. doi: 10.1371/journal.pone.0056376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salnikova LE, Khadzhieva MB, Kolobkov DS. Biological findings from the PheWAS catalog: focus on connective tissue-related disorders (pelvic floor dysfunction, abdominal hernia, varicose veins and hemorrhoids) Hum Genet. 2016;135:779–795. doi: 10.1007/s00439-016-1672-8. [DOI] [PubMed] [Google Scholar]
- 30.Fatini C, et al. AGT and ACE genes influence classic mitral valve prolapse predisposition in Marfan patients. Int J Cardiol. 2008;123:293–297. doi: 10.1016/j.ijcard.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi T, et al. NFIL3-deficient mice develop microbiota-dependent, IL-12/23-driven spontaneous colitis. J Immunol. 2014;192:1918–1927. doi: 10.4049/jimmunol.1301819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L, et al. The pathogenic role of NLRP3 inflammasome activation in inflammatory bowel diseases of both mice and humans. J Crohns Colitis. 2016 doi: 10.1093/ecco-jcc/jjw219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabkin E, Aikawa M, Stone JR, Fukumoto Y, Libby P, Schoen FJ. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation. 2001;104:2525–2532. doi: 10.1161/hc4601.099489. [DOI] [PubMed] [Google Scholar]
- 34.Greenhouse DG, Murphy A, Mignatti P, Zavadil J, Galloway AC, Balsam LB. Mitral valve prolapse is associated with altered extracellular matrix gene expression patterns. Gene. 2016;586:56–61. doi: 10.1016/j.gene.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Odelin G, et al. Loss of Krox20 results in aortic valve regurgitation and impaired transcriptional activation of fibrillar collagen genes. Cardiovasc Res. 2014;104:443–455. doi: 10.1093/cvr/cvu233. [DOI] [PubMed] [Google Scholar]
- 36.De Landsheere L, et al. Histology of the vaginal wall in women with pelvic organ prolapse: a literature review. Int Urogynecol J. 2013;24:2011–2020. doi: 10.1007/s00192-013-2111-1. [DOI] [PubMed] [Google Scholar]
- 37.Vetuschi A, et al. Changes in muscularis propria of anterior vaginal wall in women with pelvic organ prolapse. Eur J Histochem. 2016;60:2604. doi: 10.4081/ejh.2016.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips CH, Anthony F, Benyon C, Monga AK. Collagen metabolism in the uterosacral ligaments and vaginal skin of women with uterine prolapse. BJOG. 2006;113:39–46. doi: 10.1111/j.1471-0528.2005.00773.x. [DOI] [PubMed] [Google Scholar]
- 39.Giri A, et al. Genetic determinants of pelvic organ prolapse among African American and Hispanic women in the Women’s Health Initiative. PLoS One. 2015;10:e0141647. doi: 10.1371/journal.pone.0141647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cartwright R, et al. Systematic review and metaanalysis of genetic association studies of urinary symptoms and prolapse in women. Am J Obstet Gynecol. 2015;212(199):e1–24. doi: 10.1016/j.ajog.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward RM, et al. Genetic epidemiology of pelvic organ prolapse: a systematic review. Am J Obstet Gynecol. 2014;211:326–335. doi: 10.1016/j.ajog.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dietz HP. Genetics of pelvic organ prolapse: comment. Int Urogynecol J. 2012;23:509–510. doi: 10.1007/s00192-011-1638-2. [DOI] [PubMed] [Google Scholar]
- 43.Khadzhieva MB, et al. Fibulin-5 (FBLN5) gene polymorphism is associated with pelvic organ prolapse. Maturitas. 2014;78:287–292. doi: 10.1016/j.maturitas.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 44.Buchsbaum G, Duecy E, Kerr L, Huang L–S, Perevich M, Guzick D. Pelvic organ prolapse in nulliparous women and their parous sisters. Obstet Gynecol. 2006;108:1388–1393. doi: 10.1097/01.AOG.0000245784.31082.ed. [DOI] [PubMed] [Google Scholar]
- 45.Altman D, Forsman M, Falconer C, Lichtenstein P. Genetic influence on stressurinary incontinence and pelvic organ prolapse. Eur Urol. 2008;54:918–922. doi: 10.1016/j.eururo.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 46.Antonicelli F, Bellon G, Debelle L, Hornebeck W. Elastin-elastases and inflamm-aging. Curr Top Dev Biol. 2007;79:99–155. doi: 10.1016/S0070-2153(06)79005-6. [DOI] [PubMed] [Google Scholar]
- 47.Bai SW, Chung DJ, Yoon JM, Shin JS, Kim SK, Park KH. Roles of estrogen receptor, progesterone receptor, p53 and p21 in pathogenesis of pelvic organ prolapse. Int Urogynecol J Pelvic Floor Dysfunct. 2005;16:492–496. doi: 10.1007/s00192-005-1310-9. [DOI] [PubMed] [Google Scholar]
- 48.Kim EJ, et al. Involvement of oxidative ress and mitochondrial apoptosis in the pathogenesis of pelvic organ prolapse. J Urol. 2013;189:588–594. doi: 10.1016/j.juro.2012.09.041. [DOI] [PubMed] [Google Scholar]
- 49.Liu C, et al. Collagen metabolic disorder induced by oxidative stress in human uterosacral ligament-derived fibroblasts: A possible pathophysiological mechanism in pelvic organ prolapse. Mol Med Rep. 2016;13:2999–3008. doi: 10.3892/mmr.2016.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefevre R, Davila GW. Functional disorders: rectocele. Clin Colon Rectal Surg. 2008;21:129–137. doi: 10.1055/s-2008-1075862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liberati A, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mischak H, et al. Recommendations for biomarker identification and qualification in clinical proteomics. Sci Transl Med. 2010;2:46ps42. doi: 10.1126/scitranslmed.3001249. [DOI] [PubMed] [Google Scholar]
- 53.Core Team R R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria http://www.R-project.org/ (2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


