Abstract
Several taxonomically distinct mammalian groups—certain microbats and cetaceans (e.g., dolphins)—share both morphological adaptations related to echolocation behavior and strong signatures of convergent evolution at the amino acid level across seven genes related to auditory processing. Aye-ayes (Daubentonia madagascariensis) are nocturnal lemurs with a specialized auditory processing system. Aye-ayes tap rapidly along the surfaces of trees, listening to reverberations to identify the mines of wood-boring insect larvae; this behavior has been hypothesized to functionally mimic echolocation. Here we investigated whether there are signals of convergence in auditory processing genes between aye-ayes and known mammalian echolocators. We developed a computational pipeline (Basic Exon Assembly Tool) that produces consensus sequences for regions of interest from shotgun genomic sequencing data for nonmodel organisms without requiring de novo genome assembly. We reconstructed complete coding region sequences for the seven convergent echolocating bat–dolphin genes for aye-ayes and another lemur. We compared sequences from these two lemurs in a phylogenetic framework with those of bat and dolphin echolocators and appropriate nonecholocating outgroups. Our analysis reaffirms the existence of amino acid convergence at these loci among echolocating bats and dolphins; some methods also detected signals of convergence between echolocating bats and both mice and elephants. However, we observed no significant signal of amino acid convergence between aye-ayes and echolocating bats and dolphins, suggesting that aye-aye tap-foraging auditory adaptations represent distinct evolutionary innovations. These results are also consistent with a developing consensus that convergent behavioral ecology does not reliably predict convergent molecular evolution.
Keywords: convergent evolution, comparative genomics, evolutionary ecology
Introduction
The aye-aye (Daubentonia madagascariensis) is an Endangered nocturnal lemur that is the only surviving member of the family Daubentoniidae. Among other unique adaptations, aye-ayes possess an elongated and highly flexible middle finger with which they tap rapidly along the surface of trees in the search for the internal mines of wood-boring insect larvae (Erickson 1991; Sterling and Povinelli 1999; Schwitzer et al. 2013; Thompson et al. 2016). The resulting differential soundings of a tree’s variable interior structures are received by aye-ayes’ large, alert, high-frequency attuned pinna (Coleman and Ross 2004; Ramsier and Dominy 2012) and processed by a relatively enlarged inferior colliculus in a brain that is also overall the largest relative to body size among all extant strepsirrhine primates (Smith and Jungers 1997; Kaufman et al. 2005). In addition to acoustic signals, tactile and olfactory cues are also hypothesized to play important roles in aye-aye foraging ecology (Erickson 1991; Bush and Allman 2004; Kaufman et al. 2005; Ramsier and Dominy 2012). Once a suitable location on the deadwood is detected, the aye-aye uses its large, continuously growing incisors to gnaw through the tree’s exterior and extract the larvae within using the flexible middle finger (Erickson 1991; Soligo 2005).
The tap-foraging adaptations of the aye-aye have led to analogies to woodpeckers, as a result of their similar extractive, insectivorous dietary niche (Sandwith 1859; Cartmill 1974). Woodpeckers investigate trees for potential food sources by systematically probing and excavating cavities within a copse for grubs before moving on to a new area (Lima 1983). To test whether aye-ayes conform to expectations of systematic and/or random cavity investigation, Erickson (1991) conducted a series of behavioral experiments with four wild-caught, captive aye-ayes at the Duke Lemur Center. The aye-ayes were significantly more likely to excavate cavities relative to solid areas of the block, regardless of whether the cavities contained larvae (Erickson 1991), indicating that aye-ayes can consistently distinguish true cavities from false indicators even in the absence of visual or olfactory cues by using the acoustic signals generated in tap-foraging to identify potential larval mines.
The neurological and genetic pathways involved in aye-aye tap-foraging behavior are currently unknown. Recent molecular analyses of ecologically and morphologically convergent, but phylogenetically distinct, taxa have shown that similar genetic changes may, in some cases underlie convergent biology (Stern 2013; Gallant et al. 2014). For example, RNASE1 gene duplications and subsequent amino acid substitutions occurred independently in folivorous colobine primates and ruminating cows, likely facilitating the digestion of large amounts of bacterial RNA in lower pH conditions associated with foregut fermentation (Zhang et al. 2002; Zhang 2003; Schienman et al. 2006; Zhou et al. 2014). The availability of aye-aye genomic sequence data (Perry, Reeves et al. 2012; Perry et al. 2013) facilitates cross-species analyses that may contribute to our understanding of the underlying biology.
Our present analysis is motivated by the observation of molecular convergence among echolocating bats (two phylogenetically divergent clades; suborders Yinpterochiroptera and Yangochiroptera) and between echolocating bats and the toothed whales (suborder Odontoceti). Bat and whale echolocators exhibit radical differences in their mechanisms of sound production, with the former relying on the standard mammalian laryngeal apparatus to emit vocalizations and the latter possessing a specialized nasal structure for sound production (Cranford et al. 1996; Au and Simmons 2007; Jakobsen et al. 2013). Despite using distinct organs for producing and propagating sounds, a striking pattern of convergent genetic evolution in seven genes involved in auditory functioning suggests similarities in at least some of the ways that the returning sounds are processed. Specifically, there is strong consensus evidence for seven auditory processing genes with convergent amino acid substitutions among the echolocating bats and dolphins (table 1), to such a degree that phylogenetic reconstructions of the predicted protein sequences of these genes produce monophyletic clades of all echolocators to the exclusion of their more closely related, nonecholocating sister taxa (Liu et al. 2010; Shen et al. 2012). Although recent suggestions of an even wider, cross-genome level of convergence between bats and dolphins (Parker et al. 2013) have not been supported by subsequent analyses (Thomas and Hahn 2015; Zou and Zhang 2015), the evidence for convergence at the seven genes listed in table 1 is robust.
Table 1.
Genes Previously Shown To Have Significantly Convergent Encoded Amino Acid Sequences among Echolocating Bats and Dolphins
| Gene Name (Symbol) | Gene Ontology | Sources |
|---|---|---|
| Cadherin-23 (CDH23) | Stereocilia organization and hair bundle formation | Shen et al. 2012 |
| Potassium channel, voltage gated KQT-like subfamily Q-4 (KCNQ4) | Cochlear neuronal excitability | Liu et al. 2011, 2012 |
| Otoferlin (OTOF) | Vesicle membrane fusion; mouse knockout causes deafness | Shen et al. 2012 |
| Protocadherin-15 (PCDH15) | Maintenance of normal retinal and cochlear function | Shen et al. 2012; Parker et al. 2013 |
| Deafness, autosomal recessive 59 (PJVK) | Required for proper function of auditory neurons | Davies et al. 2012 |
| Prestin (SLC26A5) | Cochlear hair cell motility and ion exchange | Li et al. 2008, 2010; Liu et al. 2010; Parker et al. 2013 |
| Transmembrane channel-like 1 (TMC1) | Required for normal functioning of cochlear hair cells | Davies et al. 2012; Parker et al. 2013 |
In this study, we tested whether the signals of convergent adaptation detected between the echolocating bat and dolphin lineages in these genes were shared with aye-ayes. Given that these patterns of convergence appear to be organized around a common reliance on interpreting complex auditory signals to detect prey in different media rather than mechanisms of sound production, we hypothesized that the relatively poorly understood neurological and mechanical pathways that aye-ayes use to process high-frequency acoustic signals might have similar genetic underpinnings. To test this hypothesis, we reconstructed all seven loci implicated in previous bat dolphin comparisons in aye-ayes and the diademed sifaka (Propithecus diadema), a sister lemur taxon, using both new and previously published genomic short read data (Perry, Reeves et al. 2012; Perry et al. 2013) and the Basic Exon Assembly Tool (BEAT), a newly developed pipeline that links multiple existing bioinformatics tools to rapidly extract consensus sequences for loci of interest from shotgun sequence data. We then evaluated the level of amino acid convergence among these lemurs and 12 additional species including echolocating bats and dolphins and their nonecholocating sister taxa.
Materials and Methods
Data Acquisition
For aye-aye sequences, sequence short-reads from Perry, Reeves et al. (2012) and Perry et al. (2013) were retrieved from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (www.ncbi.nlm.nih.gov/Traces/sra, accession numbers SRA066444 and SRA043766.1) and queried as compressed FASTQ files, comprising 32 lanes of data (3,842,334,284 reads). Tissue samples from two P. diadema individuals were provided by the Duke Lemur Center. Sifaka sequencing libraries were prepared using the method described in Meyer and Kircher (2010). Each individual was sequenced on one lane of the Illumina HiSeq 2500, for 150 cycles from each end (150×150 paired end sequencing; 547,279,426 reads in total for the two individuals). The diademed sifaka sequence read data generated for this study have been deposited in the Sequence Read Archive with the BioProject accession number PRJNA317769.
In addition to the aye-aye and sifaka data, our analysis included CDH23, KCNQ4, OTOF, PCDH15, PJVK, SLC26A5 (Prestin), and TMC1 (see table 1) gene sequence data that were provided by Parker et al. (2013) for 12 species, including six bats (echolocators: Pteronotus parnellii, Megaderma lyra, Rhinolophus ferrumequinum, and Myotis lucifugus; nonecholocators: Eidolon helvum and Pteropus vampyrus), bottlenose dolphins (Tursiops truncatus) and their nonecholocating relative domestic cattle (Bos taurus), and three nonecholocating mammals: House mouse (Mus musculus), African elephant (Loxodonta africana), and European rabbit (Oryctolagus cuniculus). TMC1 was not available for African elephants, so analyses involving this species are based on the six remaining genes. Other species from the original Parker et al. (2013) alignments with <75% sequence length coverage for any gene were excluded from our analyses. The original Parker et al. (2013) alignments were missing SLC26A5 for E. helvum; however, we were able to reconstruct this gene using short read data from reads accessioned by Parker et al. (2013) (NCBI accession number SRA091593) as the input and a Pteropus vampyrus ortholog. Gene nomenclatural differences between Parker et al.’s (2013) alignments and the NCBI’s nuccore database used for the assembly of D. madagascariensis and P. diadema sequences were standardized using the UCSC table browser (https://genome.ucsc.edu/cgi-bin/hgTables; last accessed July 10, 2017). Where more than one isoform of a gene has been reported, the homologous human sequences that D. madagascariensis and P. diadema short read sequences were mapped to with BEAT were chosen to match the isoforms used in the multi-species alignments from Parker et al. (2013).
Data Generation and Processing
BEAT (https://github.com/RBankoff/BEAT) is a set of perl shell scripts intended to quickly and reliably reconstruct a consensus sequence from a nonmodel organism for a genomic region of interest. The genomic region should be orthologous to a reasonably conserved reference sequence from the genome of a distantly related species. BEAT uses a set of raw Illumina paired-end short read data from the nonmodel species with sufficient depth of coverage to reliably call genotypes by alignment against the provided reference sequence. The capability to easily and accurately create assemblies for nonmodel species such as the aye-aye is critical to maximize data usability for endangered or otherwise difficult-to-study species, particularly for research groups not explicitly focused on bioinformatic analyses.
BEAT acts as a pipeline to coordinate the parallelization of multiple freely available programs (released under the GNU GPL3 License) in a UNIX/LINUX environment, as described below. BEAT does not introduce any novel techniques for assaying genomic information for quality or analyzing the data it generates; its purpose is to facilitate the efficient extraction of selected regions of interest from a large but otherwise unassembled set of raw reads in a single end-user step using parallelization of computationally intensive processes. BEAT can be run through the GNU parallel shell tool (Tange 2011) on a workstation or can take advantage of distributed cluster computing environments such as the TORQUE Process Manager to map multiple sets of paired-end short read sequence data from different flowcells and individuals sequenced on an Illumina platform to a high-quality reference genome in parallel. By linking directly to the Entrez database with the free EntrezDirect tool from NCBI (Kans 2011), BEAT can take user requests for specific loci and obtain reference coordinate and sequence information for orthologous sequences from a specified reference genome.
BEAT maps reads to either a reference genome (e.g., GRCh38) or other reasonably unique sequence(s) of interest by running the BWA-MEM algorithm (Li 2013) over each set of provided paired-end reads in parallel (see fig. 1). For example, a BEAT job that uses 20 paired-end short read files in 10 file pairs will be submitted to run as 10 simultaneous jobs. For larger short read data sets, BEAT will subdivide further, breaking them into small files (default 1 Gb) that can be further processed in parallel. The raw bam-formatted mapped output files are quality filtered to exclude unmapped reads with the samtools view -F4 flag (Li et al. 2009) before being merged into a master bam alignment file and passed through samtools rmdup to remove PCR duplicate reads that might otherwise bias consensus calls. Per-nucleotide genotype estimates are then generated using the samtools mpileup utility, which calls the read-specific genotype and read quality from the bam alignment file into a position-wise pileup output file. This master pileup file can subsequently be queried for the corresponding sequence call at any position along the length of its reference using the BEAT_consensus script. A novel component of BEAT, this script takes advantage of the read-by-read genotypes mapped to the ortholog in the mpileup output file to generate a consensus sequence based on the majority genotype across all reads mapping to a particular range of coordinates along the orthologs. The creation of consensus sequences is accompanied by the generation of a summary statistics file containing per subfeature (e.g., exon, promoter region, or other segment of interest) coverage, using the sum of position-wise read depth/length of subtarget. As we show below, consensus sequence output data produced by BEAT for a given region is more complete than that contained within a corresponding genome assembly from the same sequencing data, while requiring significantly lower computational investments.
Fig. 1.—
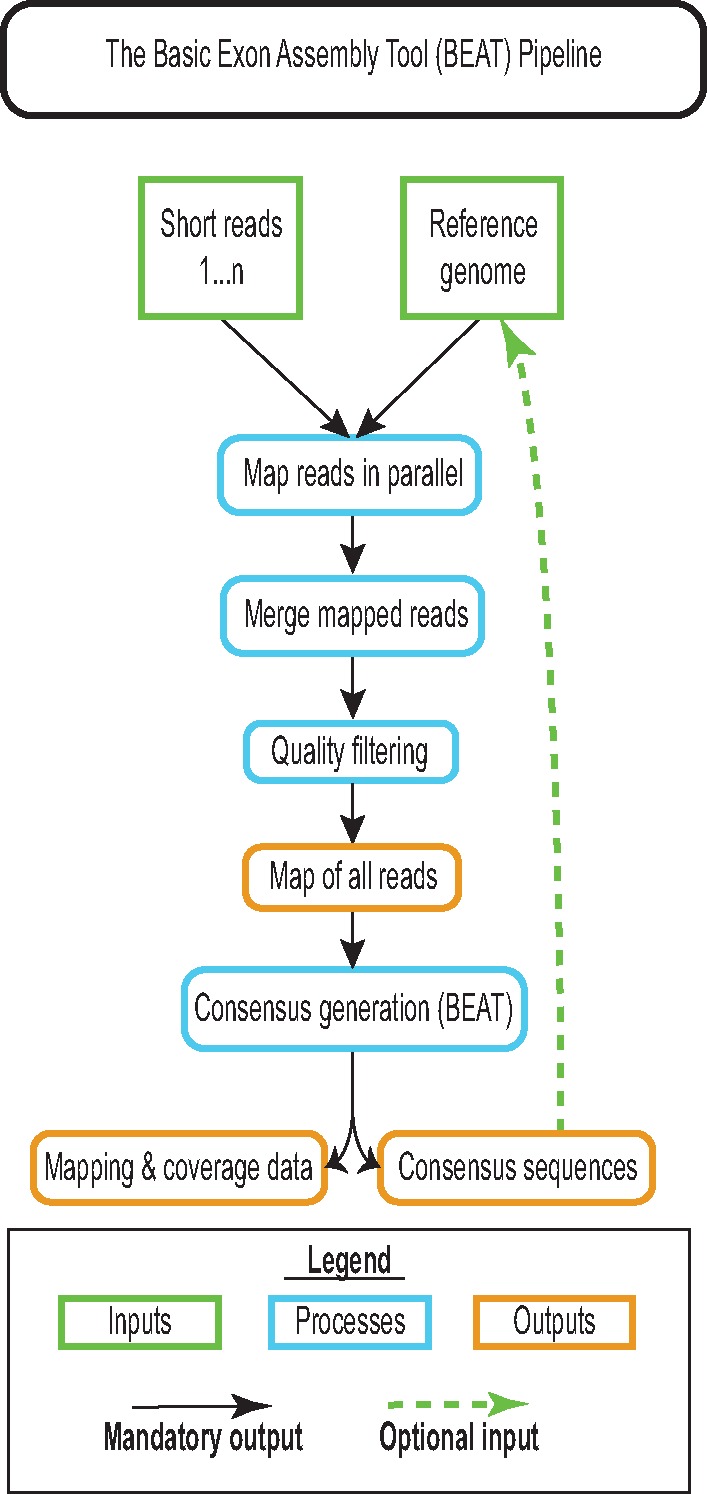
The basic workflow used by BEAT when assembling a single query sequence from a short read data set. BEAT maps all short-reads provided to a reference genome in parallel, removes low-quality and duplicate reads, then merges the mapped files to produce a map of all reads in the data set. Mapped reads are then scanned nucleotide-by-nucleotide against the reference, and a call for each position is generated by taking the quality-weighted median call of all mapping reads at positions with a depth of coverage >2. To avoid genotyping error when generating consensus sequences for loci of interest from a distantly related reference, BEAT should be run twice, with the second iteration using the consensus generated by the first as its reference.
The ideal parameters for running the BEAT pipeline include 16-Gb RAM and one computing core per set of ∼20 Gb combined compressed short read data files in FASTQ format. Lower-performance systems should still be able to run BEAT, but job runtime may differ significantly from those reported below, as parallelization is the key to BEAT’s efficiency. To limit potential consensus sequence inaccuracies that could theoretically result from high genetic distance between a reference genome and a potentially different species of interest, we recommend that all sequence reads be iteratively mapped to the first consensus generated by the process to generate a second, final consensus sequence for each locus. Another source of potential error for the BEAT consensus sequences could arise from the accidental inclusion of reads from paralogous or recently duplicated genes in addition to the orthologous gene of interest in the mapped file. We recommend generating a sample of consensus sequences for genes selected randomly from across the genome to obtain a distribution of sequence read depth, which can then compared with the distribution for the targeted genes of interest in order to mask any genes or gene regions with unusually high sequence coverage relative to the randomly sampled set of genes (e.g., >2× standard deviation). This process can be implemented using auxiliary scripts included in the BEAT package, with a user-selected parameter to determine the relative depth of coverage cutoff for masking.
For the evolutionary analyses performed in this study, consensus sequences for seven echolocation convergence candidate genes for both P. diadema and D. madagascariensis were generated with the BEAT pipeline, using human RefSeq mRNA sequences (assembly hg19/GRCh38) as the input reference orthologous sequences (table 1). Consensus calls for each nucleotide were calculated at each site with 2× unique mapped read coverage. Consensus sequences for each gene were generated on an exon-by-exon basis, aligned to the canonical mRNA transcript version of the human ortholog using Geneious version 8.0.5 (Kearse et al. 2012), and assembled into initial consensus sequences for the coding sequences of each gene and species. These consensus sequences were then used as the references for a second pass of BEAT, generating an iteratively mapped consensus sequence for each gene of interest for both aye-aye and sifaka. This step can be implemented to limit potential consensus-calling biases that could result from using a distant reference species for alignment. Additionally, 100 loci were selected randomly from a list of genes drawn from the UCSC Genome Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables; last accessed July 10, 2017) to serve as a baseline for expected sequence read depth per data set for both aye-aye and sifaka reads. After iterative mapping with BEAT and excluding 34 genes with consensus sequence recovery of <100-bp continuous sequence in either species, coverage depths for each species were calculated in 50-bp intervals with at least 3× coverage in both species for all sites included, using tunable scripts included in the BEAT package. Regions in the seven genes of interest with coverage exceeding twice the standard deviation of that from the randomly selected genes were considered potentially affected by paralogous mapping, and hence masked with “N”s and excluded from further analysis. Using this procedure, ∼1.74% of aye-aye sites for the genes of interest and ∼6.66% of sifaka sites were excluded (see supplementary table S3, Supplementary Material online).
The final consensus sequences for the coding sequence of each gene and species were aligned with the 14 species nucleotide alignments from Parker et al. (2013) in Geneious using the Geneious alignment algorithm, with a high gap open penalty of 25 but a low gap extension score of 2 to discourage fragmentation while allowing for indels. These alignments were manually assessed for quality, including removal of any codon frame-altering indels relative to the human reference sequence. Curated nucleotide alignments for each gene were translated into predicted amino acid sequences in Geneious. The multi-species consensus amino acid sequence for each gene was determined with a strict 50% cutoff (i.e., if the most common amino acid was observed in fewer than 50% of the species in the alignment, then no consensus was recorded for that position). All final multi-species amino acid and DNA alignments used in this study have been deposited in the Open Access Scholarsphere digital repository at https://scholarsphere.psu.edu/collections/rv042t11v. The methods used for our analyses were not dependent on Geneious; all of BEAT is freely available online, and the various processing steps for which we used Geneious in our analysis (data curation, alignment, depth assessment, translation, and visualization) can be fully replicated with various freely accessible bioinformatics tools (cf. Katoh and Standley 2013; Li 2013; Kumar et al. 2016).
Assessment of BEAT Performance
To test the targeted region assembly performance of BEAT compared with that of a de novo complete genome assembly generated using the same underlying sequence data, we assessed the percent coverage of consensus sequences generated by BEAT to that achieved by locating matching sequences using the BLASTn utility in BLAST+. We mapped D. madagascariensis short read data to the human orthologs of each of the seven genes of interest using BEAT and compared the percent coverage with that achieved by locating matching sequences using BLAST from the aye-aye genome assembly scaffolds (GenBank Accession AGTM000000000) published in Perry, Reeves et al. (2012). Although the consensus sequences generated by BEAT for the convergence analysis presented in the Results of this paper used additional short read data from Perry et al. (2013), for this assessment, consensus sequences were regenerated solely from the read data used to create the original scaffolds published in Perry, Reeves et al. (2012) to achieve full BEAT-scaffold comparability.
For the BEAT-generated data for this test, aye-aye short-reads from Perry, Reeves et al. (2012) were processed using the human orthologs used for mapping loci of interest in BEAT as BLAST baits. Short-reads that matched these orthologs were then mapped to the orthologous sequences using BEAT, and a consensus sequence was generated in BEAT_consensus for any nucleotides with more than two uniquely mapping reads that passed the minimum mapping quality threshold. After consensus sequence generation, we screened the consensus sequences for excessive underlying read depth relative to the read depth distribution from 66 randomly selected genes described above to cautiously mask regions that could potentially have been affected by paralogous read mapping.
For sequences from the Perry, Reeves et al. (2012) scaffolds, the same human orthologs for all seven genes were queried using BLASTn against the scaffolds with a minimum alignment score of 50, identity cutoff set at 80%, and a word length of 11. Sequences that BLAST matched to the orthologs were extracted from the scaffolds and aligned to the ortholog in Geneious. Of the 21,769 nucleotides mapping to genes of interest from the BLAST search, 2,948 were from regions where two or more nonidentical scaffolds aligned to the same location, indicating potential paralogous mapping or errors in the original assembly. Because there was no way to determine which of the scaffolds was correctly mapping to the reference, we compared the BEAT output with two consensus sequences for each gene: To the “Untrimmed” consensus derived from a map including both scaffolds, with positions where scaffolds disagreed excluded, and to the “Trimmed” consensus derived from a map using only identical and nonoverlapping scaffolds (see table 2 and supplementary table S4, Supplementary Material online).
Table 2.
Comparison of De Novo Genome Assembly and BEAT Methods for Reconstruction of Aye-Aye Gene Coding Regions Using the Same Shotgun Sequence Read Data for Both Methods
| Human Reference Coding Region Size (bp) | BEAT with Aye-Aye Sequence Readsa |
Untrimmed Aye-Aye De Novo Assemblyb |
Trimmed Aye-Aye De Novo Assemblyb |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | bp recovered | % coverage | % identity to reference | bp recovered | % coverage | % identity to reference | % identity to BEAT | bp recovered | % coverage | % identity to reference | % identity to BEAT | |
| CDH23 | 8976 | 8952 | 99.73 | 92.76 | 7863 | 87.60 | 92.81 | 98.99 | 7139 | 79.53 | 93.14 | 99.52 |
| KCNQ4 | 1713 | 1713 | 100 | 94.74 | 1013 | 59.14 | 90.97 | 95.63 | 815 | 47.58 | 91.78 | 96.93 |
| OTOF | 5634 | 5605 | 99.49 | 92.73 | 4190 | 74.37 | 91.63 | 98.56 | 3854 | 68.41 | 92.53 | 99.77 |
| PCDH15 | 4194 | 4189 | 99.88 | 94.84 | 4009 | 95.59 | 94.51 | 98.98 | 3581 | 85.38 | 94.86 | 99.75 |
| PJVK | 840 | 840 | 100 | 96.07 | 840 | 100 | 96.07 | 100 | 840 | 100 | — | — |
| SLC26A5 | 2016 | 2016 | 100 | 94.81 | 1968 | 97.62 | 94.70 | 98.93 | 1687 | 83.68 | 95.14 | 99.70 |
| TMC1 | 2116 | 1969 | 93.05 | 94.29 | 1886 | 89.13 | 92.21 | 96.04 | 905 | 42.77 | 94.59 | 99.11 |
Analysis based on BEAT reconstructions from the same shotgun sequencing read data used in Perry, Reeves et al. (2012), a subset of the sequence read data used in the full analysis of this paper.
Analysis based on BLAST reconstructions of scaffolds from aye-aye de novo assembly published in Perry, Reeves et al. (2012). Sequences assembled from the aye-aye de novo assembly were scanned for regions with more than one nonidentical mapping scaffold; these scaffolds were allowed to contribute to the consensus in the “Untrimmed” group, and removed from the “Trimmed” group.
Following BLAST hit extraction, BEAT mapping, and alignment to the ortholog in Geneious, per-gene percent coverage of scaffold- and BEAT-derived sequences was calculated as the percentage of nucleotides in the ortholog with a corresponding aligned (though not necessarily identical, given expected human and aye-aye sequence divergence) nucleotide from the test data sets. We then compared the percent coverage results for the seven BEAT-generated sequences with those for the scaffold-generated sequences for both Untrimmed and Trimmed sequences. We found that BEAT retrieves more sequence data on average than using BLASTn to query the de novo assembly genomic scaffolds from Perry, Reeves et al. (2012), with an overall mean 98.88% coverage for consensus sequences generated by BEAT versus 86.21% mean coverage for querying the genome assembly scaffolds with BLAST, and dropping to 72.48% mean coverage with potential paralogs trimmed (table 2). The proportion of total sites reconstructed in the final BEAT consensus sequences across these seven genes (0.989) was significantly greater than the proportion recovered from the BLAST analysis with the aye-aye de novo genome scaffolds, both for the maximum “untrimmed” BLAST data set (0.862; Fisher’s exact test; P < 2.2E-16) and the “trimmed” BLAST data set (0.725; Fisher’s exact test; P < 2.2E-16).
Of the 18,821 orthologous nucleotides reconstructed both by BEAT and the trimmed BLAST analysis, only 140 (0.746%) were different between the two methods. Although the observed proportion of differences is low, it is still higher than estimates of the proportion of heterozygous nucleotide sites and the average proportion of pairwise nucleotide sequence differences for aye-ayes, which range from 0.051% to 0.081% (Perry et al. 2007; Perry, Melsted et al. 2012; Perry, Reeves et al. 2012) which might otherwise explain differences between a genome assembly sequence and the BEAT consensus (i.e., if alternative SNP alleles were selected at some sites between the two methods). To investigate this larger-than-expected difference, we examined the alignments between the trimmed BLAST sequences and BEAT sequences. Most differences were localized to a few exons, rather than evenly spread, suggesting the inclusion of paralogous sequence in these regions in either the genome assembly scaffolds or the BEAT sequences. We thus examined all scaffolds containing more than one disagreement from the BEAT consensus sequence for each gene by comparing the number of sequence differences between both the genome assembly scaffolds and the BEAT consensus sequences with the putative human reference gene ortholog, and by mapping the aye-aye reads from Perry, Reeves et al. (2012) to the aye-aye genome assembly scaffolds using BWA-MEM. The read mapping analysis revealed that the differences between the assembly and the BEAT consensus are explained by either errors in the genome assembly or by paralogous rather than orthologous sequences represented in the genome assembly scaffolds (i.e., a distinct population of reads maps to the assembly scaffolds that is more dissimilar to the human reference sequence than the population of reads that forms the BEAT consensus; supplementary fig. S2, Supplementary Material online). BEAT-derived sequences were only produced for regions with >2 mapping reads that contained fewer than 2× the standard deviation of an empirically derived depth distribution from 66 genes randomly selected from across the genome. Using these criteria, 300 nucleotides were excluded from BEAT-derived Daubentonia sequences prior to subsequent analyses. However, none of the excluded regions overlapped with the two regions totaling 247 bp for which sequences from the trimmed BLAST data set included potentially paralogous scaffolds. In regions with likely BLAST paralogy, BLAST-derived scaffold sequence identity was 84.21% of the human reference, whereas BEAT-derived sequences’ shared identity with the human reference 93.52% of the time. As the average identity is 94.02% for BLAST-derived scaffolds and 94.32% for BEAT-derived sequences over all seven genes (see table 2 and supplementary fig. S2, Supplementary Material online) the discrepancy in average identities in these two regions demonstrates the robusticity of BEAT-derived sequences to paralogous reads relative to de novo assembly in regions with mapping paralogs. Thus, if a de novo genome assembly is incomplete, paralogous scaffolds can be mistakenly recruited by BLAST, whereas by directly assaying high-coverage shotgun sequence reads, BEAT can limit the introduction of errors that might confound the initial stages of consensus generation using whole-genome assemblies, rendering sequences produced by BEAT as good or better than the initial steps of whole genome-assembly.
Convergence Analyses
To evaluate the level of amino acid convergence across the seven candidate genes between D. madagascariensis and the echolocating bat and dolphin lineages, we devised a straightforward method to compare patterns of convergence between closely related species, illustrated with an example in figure 2. First, consider a set of three species from the alignment: For example, D. madagascariensis, P. diadema, and T. truncatus, where D. madagascariensis and P. diadema are more closely related to each other than either is to T. truncatus. In this example, D. madagascariensis and P. diadema form a species pair, with D. madagascariensis serving as the putative echolocator, P. diadema as the nonecholocating sister species, and T. truncatus as the test outgroup species against which the two lemur species are being evaluated for convergence. In the absence of convergent evolution between dolphins (T. truncatus) and one of the two lemur species (D. madagascariensis, P. diadema), T. truncatus is expected to share an approximately equal number of identical, aligned amino acids at each gene with D. madagascariensis but not P. diadema as it is with P. diadema but not D. madagascariensis. Only sites with shared amino acids between D. madagascariensis and T. truncatus that also differ from both the P. diadema sequence and the multi-species consensus sequence are considered convergent. In other words, we count the number of sites at which T. truncatus shares an amino acid with D. madagascariensis but not with P. diadema or the multi-species consensus sequence, relative to the total number of amino acids in the alignment. We then can statistically assess the probability that the observed number of convergent amino acids would occur by chance by comparing this result with the count of the number of amino acid shared by T. truncatus and P. diadema but not with D. madagascariensis or the multi-species consensus (again, relative to the total number of aligned amino acids). This comparison can be computed for any species pair-outgroup combination, can be computed for an individual gene or summed across multiple genes, and can be statistically evaluated with a Fisher’s exact test. The ratio between the number of convergent amino acids relative to the total number of amino acids in each species can be expressed as a “convergence ratio,” which simply describes the frequency of convergences in the test pair (in the above case, D. madagascariensis and T. truncatus) relative to the control pair (P. diadema and T. truncatus), but is not itself used for statistical tests. For the purposes of this analysis, we summed all convergent amino acids and divided by the sum of all amino acids across all seven genes for each species pair-outgroup combination.
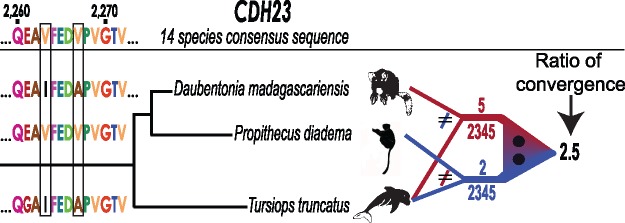
Fig. 2.—
Illustration of the pairwise convergent evolution analysis used in this study. The example shown is for a section of the CDH23 gene, here comparing two lemurs, Daubentonia and Propithecus diadema, with the echolocating dolphin Tursiops truncatus. For each lemur, we computed the number of positions out of the total aligned positions (2,345 for this gene) for which that species shared an amino acid with T. truncatus, but for which that amino acid was different than that of the other lemur species and the multi-species consensus sequence determined from the multiple-species alignment. A section of CDH23 is shown that contains two Daubentonia—T. truncatus convergent amino acids. For the whole gene, the ratio of convergent amino acids for Daubentonia and T. truncatus (5) and P. diadema and T. truncatus (2) = 2.5, providing a magnitude and directionality of relative convergence between aye-aye and dolphin, with a phylogenic correction based on the sister lemur species. These ratio values are depicted in figure 3 for all tested comparisons, summed across the seven genes analyzed in this study. The difference in the number of convergent amino acids is evaluated with a Fisher’s exact test computed based on the 2×2 contingency table, not the convergence ratio; for the illustrated CDH23 comparison, P = 0.4528, prior to Bonferroni multiple test correction.
To test the hypothesis of convergence between aye-ayes and the true echolocators at loci of interest with this method, we followed the above procedure separately for each echolocating species, that is, we used Fisher’s exact test to compare the proportion of D. madagascariensis and echolocator convergent amino acids (to the exclusion of P. diadema and the multi-species consensus) with the proportion of P. diadema and echolocator convergent amino acids (to the exclusion of Daubentonia and the multi-species consensus), separately for each echolocating species in the multi-species alignment. Similarly, we compared bottlenose dolphins (Tursiops truncatus) and a number of echolocating bat species with one another using domestic cattle (Bos taurus) and nonecholocating bats, respectively, as the nonecholocating sister species in the analysis. For each species pair—outgroup comparison, only sites without gaps or missing data in all three species’ sequences (and a called multi-species consensus amino acid) were counted toward the total number of amino acids used in calculating the comparison.
In addition to this comparison, we conducted a second version of the analysis in which the only sites scored as convergent were those at which 1) the test species shared an amino acid residue with the echolocating comparison species, 2) that residue was different than the multi-species consensus residue, and 3) the nonecholocating species in the pair had the same residue as the multi-species consensus sequence. This analysis was performed to ensure that the observed results could not be explained by the presence of nonecholocating lineages with high substitution rates leading to erroneous signatures of convergence between echolocators and test species.
We also analyzed these data with an alternative approach in which we normalized the number of observed convergent amino acids between a pair of taxa by their evolutionary distance. As in the above comparison, we compared the amino acid identities of a test species (e.g., Daubentonia) with each of a pair of species more closely related to each other, one of which can echolocate while the other cannot (e.g., the echolocating bat R. ferrumequinum and its nonecholocating relative E. helvum) and to the consensus of all species in the alignment. However, instead of comparing the number of convergent sites with the number of total comparable sites, we examined the number of convergent amino acids relative to divergent amino acids, with divergent amino acids defined as residues that differ between the test species (e.g., M. lucifugus) and the echolocating species (e.g., R. ferrumequinum) and for which neither of those residues are shared with the nonecholocating species (e.g., E. helvum) or the multi-species consensus. Convergence/divergence approaches such as this have been shown to be a conservative but reliable indicator of convergence while controlling for evolutionary rate (cf. Castoe et al. 2009; Thomas and Hahn 2015). To correct for the number of tests performed, we applied a Bonferroni correction by multiplying the individual Fisher’s exact test P values by the number of tests.
It must be noted our methods assume that there has not been a history of incomplete lineage sorting (ILS) among descendent species included in the analysis or of introgression between them. In general, ILS would not affect our primary results, given that the deep divergence times among the nonbat echolocator Tursiops, Daubentonia, and echolocating bats are far greater than those between echolocating bats and their nonecholocating relatives. In other words, we would not expect ILS or introgression to affect our analyses of convergent evolution between echolocating bats and cetaceans, bats and primates, or primates and cetaceans, all of which are separated from one another by deep phylogenetic gulfs (O’Leary et al. 2013; dos Reis et al. 2014). Within bats, gene trees of echolocation-related loci are discordant with the overall molecular phylogeny of the order, which could theoretically reflect convergent evolution, ILS, or hybridization and introgression, scenarios that are difficult to distinguish given the long branch lengths of these species (Hahn and Nakhleh 2016). Chiroptera’s two clades underwent successive swift radiation events in the early Paleocene (∼60 Ma for crown Chiroptera; Shi and Rabosky 2015; Lei and Dong 2016); if this rapid cladogenesis did result in ILS or other ancestral sorting of variation, signals of “convergence” between the different echolocating bat lineages as detected by our method may have been amplified by shared ancestry.
Results
Echolocation has likely evolved twice independently in bat species (Jones and Teeling 2006; Jones and Holderied 2007) and at least once in toothed whales (Geisler et al. 2014). Previous studies have shown that echolocating bats and dolphins share significantly more amino acids with each other at seven genes implicated in auditory processing than they do with phylogenetically closer nonecholocating taxa (Liu et al. 2011; Shen et al. 2012). These previous observations are strongly supported by the results from our echolocator versus nonecholocating sister group pairwise comparisons for these seven genes (fig. 3). For example, dolphins (T. truncatus) shared 67 convergent amino acids (out of 7,740 total aligned positions across the seven studied genes) with the echolocating bat Rhinolophus ferrumequinam to the exclusion of cattle (Bos taurus, more closely related to dolphins), whereas R. ferrumequinum and B. taurus shared only seven convergent amino acids to the exclusion of T. truncatus for a convergence ratio = 9.57, which is significantly greater than expected by chance (Fisher’s exact test, P = 1.941E-13; Bonferroni-corrected P = 2.329E-12). Similarly, the echolocating bat R. ferrumequinam and T. truncatus share 69 out of 7,669 comparable sites, whereas T. truncatus and E. helvum, a nonecholocating bat, share only 4 (convergence ratio = 16.3; P < 2.2E-16; Bonferroni-corrected P < 2.64E-15). Significant convergence was observed between all other echolocator–echolocator comparisons, but not between dolphins and nonecholocating bats (fig. 3 and supplementary table S1, Supplementary Material online). On a gene-by-gene basis, CDH23, OTOF, PCDH15, SLC26A5 (Prestin) display the strongest, most consistent signals of convergence in the echolocator–echolocator comparisons (see supplementary table S2, Supplementary Material online, for gene-by-gene comparison results).
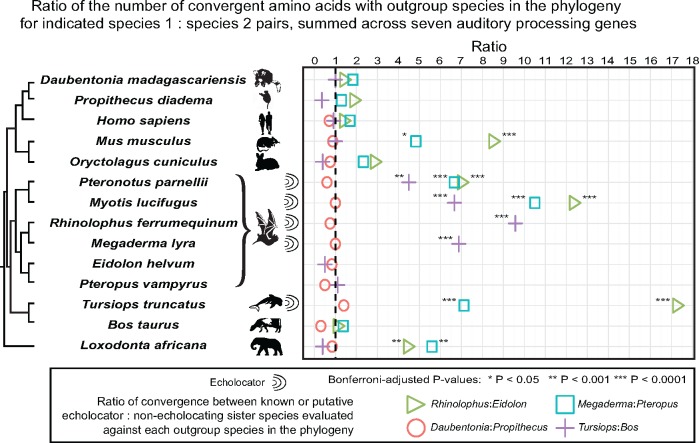
Fig. 3.—
Observed seven gene convergence ratios for three pairs of known echolocator: Nonecholocator sister taxa and for Daubentonia: Propithecus, each compared with other echolocating and nonecholocating mammalian taxa. Ratio of the number of convergent/total amino acids with outgroup species in the phylogeny for indicated species 1: Species 2 pairs, summed across seven auditory processing genes. Colored symbols represent the ratio between the number of convergent amino acids shared between the first species in the indicated pair and the species on the corresponding row of the phylogeny (the “test species”) relative to that for the second species in the indicated pair, after results were summed over the seven auditory genes considered in this analysis. All statistical tests were performed on a comparison of the sum of convergent/total amino acids for each species pair, not the ratio between the proportions thus formed. The dashed line indicates a ratio of 1, or no difference in the observed level of convergence with the outgroup test species between the two sister species. For the bat–bat comparisons, pairs were chosen at random from the set of four possible comparisons (Rhinolophus: Eidolon, Rhinolophus: Pteropus, Megaderma: Eidolon, Megaderma: Pteropus); similar results were obtained from the other possible pairs (see supplementary table S1, Supplementary Material online, for all comparisons). The indicated significance values have been corrected for multiple tests using the Bonferroni method, within each set of pairwise comparisons across the phylogeny.
In contrast to the significant results for the known echolocator versus known echolocator lineages, we did not observe a signature of convergent amino acid evolution between Daubentonia and the echolocating bats or dolphins relative to P. diadema (fig. 3 and supplementary table S1, Supplementary Material online). For example, in sum across the seven genes, D. madagascariensis and the echolocating bat M. lyra shared eight convergent amino acids (of 6,928 positions) to the exclusion of P. diadema, whereas P. diadema and M. lyra also shared eight convergent amino acids to the exclusion of D. madagascariensis (convergence ratio = 1; P = 1).
In addition to the above results from the planned comparisons that motivated our study, an unexpected finding also emerged from our multi-species analysis. Specifically, we observed significant convergent amino acid enrichment between the echolocating bats and both the African elephant (Loxodonta africana) and the mouse (Mus musculus) to the exclusion of the nonecholocating bats (fig. 3 and supplementary table S1, Supplementary Material online). For example, African elephants shared 31 out of 7,523 amino acids with R. ferrumequinum to the exclusion of E. helvum, compared with only 7 amino acids shared with E. helvum to the exclusion of R. ferrumequinum; the resulting convergence ratio is significantly greater than expected by chance (convergence ratio = 4.43; Fisher’s exact test, P = 1.14E-4, Bonferroni-corrected P = 1.37E-3). Likewise, mice shared 34 out of 8,214 amino acids with R. ferrumequinum to the exclusion of E. helvum yet only 4 with E. helvum to the exclusion of R. ferrumequinum (convergence ratio = 8.5; P = 5.89E-7; Bonferroni-corrected P = 7.07E-6). Adding the more stringent criterion that the nonecholocator in each pair must share an amino acid residue with the multi-species consensus sequence in order for the site to be considered convergent between the test species and the echolocating comparison species does not have a major effect on these results (supplementary fig. S3, Supplementary Material online). To confirm that the significant results observed in our analyses (fig. 3 and supplementary fig. S3, Supplementary Material online) reflect a signal of convergent evolution rather than an unknown systematic effect (that would then be expected to have similar effects on loci across the genome), we repeated the convergence analysis with the 66 randomly selected genes that were used to screen BEAT-generated for average expected read depth. The extreme ratios of convergent amino acids between echolocator–echolocator versus echolocator–nonecholocator species pairs observed in our analysis of seven auditory processing genes (fig. 3 and supplementary fig. S3, Supplementary Material online) were not observed in the analysis of the 66 randomly selected loci, and after correcting for multiple testing we observed no significant signal of convergence between any species pairs (supplementary fig. S4, Supplementary Material online).
To corroborate the results from the above analysis, we also assessed the number of convergent amino acids relative to the number of divergent amino acids between two lineages, to control for the higher convergence rate that might be expected for taxa with higher overall amino acid substitution rates. For this analysis, divergent amino acids are defined as residues that differ between the test species and the echolocating species, and for which neither of those residues are shared by the nonecholocating species or the multi-species consensus. Using this convergence/divergence approach, neither the proportions of convergent amino acids between mice and echolocating bats nor those between African elephants and echolocating bats were significantly different from those expected by chance; however, significant signals of convergence between all echolocating bats and dolphins were still observed (supplementary fig. S1, Supplementary Material online).
Discussion
We did not identify any signature of convergence between the aye-aye and true echolocators at the seven genes related to auditory processing that we examined. Although our analysis cannot account for the potential presence of nonidentical amino acid changes that are nonetheless convergent in biochemical function, the finding of strong convergent evolution for specific amino acid changes at these loci between echolocating dolphins and bats but not between aye-ayes and either of these lineages suggests that aye-aye tap-foraging has evolved from a distinct, as of yet unknown set of molecular adaptations compared with echolocating bats and dolphins. Dolphin–bat and most bat–bat convergences were supported under both the lenient and stringent versions of our main analysis (fig. 3 and supplementary fig. S3, Supplementary Material online) and under the convergence/divergence model intended to control for evolutionary rate (supplementary fig. S1, Supplementary Material online).
The lack of apparent convergence between aye-ayes and other echolocators is not wholly unexpected, given that echolocation is a complex behavior reliant on numerous anatomical and behavioral adaptations, and does not necessarily imply that the percussive foraging behavior of aye-ayes is wholly dissimilar to echolocation. In fact, we found evidence that convergence at these loci may not be solely associated with echolocation. Specifically, we observed unexpectedly significant enrichments for convergent amino acids between echolocating bats and mouse or elephant in four of the seven genes using both versions of the alternative approach shown in figure 3 (see also supplementary table S2 and fig. S3, Supplementary Material online). Intriguingly, mice have been observed to communicate in the ultrasound during courtship and mating (Musolf et al. 2010), with a frequency comparable with that used by many echolocating bats (∼70 kHz; White et al. 1998), and outside the range of aye-aye auditory ability (Ramsier and Dominy 2012). In contrast to mice, African elephants are known for their ability to hear low-frequency sounds in the infrasonic range (<20 Hz; Payne et al. 1986; Langbauer et al. 1991) rather than the high-frequency ultrasound (1–200 kHz) of echolocating bats (Au and Simmons 2007).
Accounting for these unanticipated results is challenging, especially considering the limited scope of the present investigation and the numerous loci presumably involved in complex processes of capturing and interpreting auditory signals. The significance of convergence at these loci in mouse and African elephants is supported by both the original analysis shown in figure 3 and the secondary analysis described in supplementary figure S3, Supplementary Material online. However, using the more conservative convergence/divergence approach shown in supplementary figure S1, Supplementary Material online, only bat–bat and bat–dolphin convergence is supported. Furthermore, these significant convergences do not appear to be shared between both mouse and elephant and cetaceans, who also have high-ultrasound processing demands via echolocation. The statistical and biological significance of mouse–bat and elephant–bat convergence observed under the first two methods is thus uncertain. If the convergent signals between these taxa do actually reflect as-of-yet unknown underlying biological similarities, then perhaps these shared amino acid changes play general roles in auditory signal conduction at frequency extremes, rather than only at high frequencies as part of echolocation behavior.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This research was supported by funds from the Penn State College of the Liberal Arts and the Huck Institutes of the Life Sciences (to G.H.P.), and the Pennsylvania State University Graduate Fellowship (to R.J.B.). The Duke Lemur Center kindly provided the diademed sifaka DNA samples (this is DLC publication # 1357), and Parker et al. (2013) provided access to the gene alignment data from their paper. We thank Craig Praul, Candace Price, and the Penn State Huck Institutes of the Life Sciences Genomics Core for generating the Illumina sequencing data, Stephen Johnson, Kate Thompson, David Villalta, Richard George, Alexis Sullivan, Arslan Zaidi, James Fisher, Stephen King, and Kristen Alldredge for discussions and feedback on the study and manuscript, and Emily Davenport for suggestions on the GitHub repository. This study was funded by a grant from the National Science Foundation (BCS-1554834 to G.H.P.) Computational resource instrumentation was funded by the National Science Foundation through grant OCI-0821527.
Literature Cited
- Au WWL, Simmons JA.. 2007. Echolocation in dolphins and bats. Phys Today [Internet] 60(9): 40–45. [Google Scholar]
- Bush EC, Allman JM.. 2004. The scaling of frontal cortex in primates and carnivores. Proc Natl Acad Sci U S A. 101(11): 3962–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartmill M. 1974. Daubentonia, Dactylopsila, woodpeckers and klinorhynchy In: Martin RD, Doyle GA, Walker AC, editors. Prosimian biology. London: Duckworth; p. 655–670. [Google Scholar]
- Castoe TA, et al. 2009. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci U S A. 106(22): 8986–8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MN, Ross CF.. 2004. Primate auditory diversity and its influence on hearing performance. Anat Rec A Discov Mol Cell Evol Biol. 281: 1123–1137. [DOI] [PubMed] [Google Scholar]
- Cranford TW, Amundin M, Norris KS.. 1996. Functional morphology and homology in the odontocete nasal complex: implications for sound generation. J Morphol. 228(3): 223–285. [DOI] [PubMed] [Google Scholar]
- Davies KTJ, Cotton JA, Kirwan JD, Teeling EC, Rossiter SJ.. 2012. Parallel signatures of sequence evolution among hearing genes in echolocating mammals: an emerging model of genetic convergence. Heredity 108(5): 480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Donoghue PCJ, Yang Z.. 2014. Neither phylogenomic nor palaeontological data support a Palaeogene origin of placental mammals. Biol Lett. 10(1): 20131003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson CJ. 1991. Percussive foraging in the aye-aye, Daubentonia madagascariensis. Anim Behav. 41(5): 793–801. [Google Scholar]
- Gallant JR, et al. 2014. Genomic basis for the convergent evolution of electric organs. Science 344(6191): 1522–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler JH, Colbert MW, Carew JL.. 2014. A new fossil species supports an early origin for toothed whale echolocation. Nature 508(7496): 383–386. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Nakhleh L.. 2016. Irrational exuberance for resolved species trees. Evolution 70(1): 7–17. [DOI] [PubMed] [Google Scholar]
- Jakobsen L, Brinkløv S, Surlykke A.. 2013. Intensity and directionality of bat echolocation signals. Front Physiol. 4: 89.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Holderied MW.. 2007. Bat echolocation calls: adaptation and convergent evolution. Proc Biol Sci. 274: 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Teeling EC.. 2006. The evolution of echolocation in bats. Trends Ecol Evol. 21(3): 149–156. [DOI] [PubMed] [Google Scholar]
- Kans J. 2011. Entrez direct: E-utilities on the UNIX command line. Curr Top Med Chem. 11: 2171–2179.21671877 [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4): 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman JA, Ahrens ET, Laidlaw DH, Zhang S, Allman JM.. 2005. Anatomical analysis of an aye-aye brain (Daubentonia madagascariensis, primates: Prosimii) combining histology, structural magnetic resonance imaging, and diffusion-tensor imaging. Anat Rec. 287A(1): 1026–1037. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12): 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbauer WR, Payne KB, Charif RA, Rapaport L, Osborn F.. 1991. African elephants respond to distant playbacks of low-frequency conspecific calls. J Exp Biol. 157: 35–46. [Google Scholar]
- Lei M, Dong D.. 2016. Phylogenomic analyses of bat subordinal relationships based on transcriptome data. Sci Rep. 6: 27726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, et al. 2008. The hearing gene Prestin reunites echolocating bats. Proc Natl Acad Sci U S A. 105(37): 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16): 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997. [Google Scholar]
- Li Y, Liu Z, Shi P, Zhang J.. 2010. The hearing gene Prestin unites echolocating bats and whales. Curr Biol. 20(2): R55–R56. [DOI] [PubMed] [Google Scholar]
- Lima SL. 1983. Downy woodpecker foraging behavior: foraging by expectation and energy intake rate. Oecologia 58(2): 232–237. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. 2010. Convergent sequence evolution between echolocating bats and dolphins. Curr Biol. 20(2): R53–R54. [DOI] [PubMed] [Google Scholar]
- Liu Z, et al. 2011. Parallel evolution of KCNQ4 in echolocating bats. PLoS One 6(10): e26618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Li GH, Huang JF, Murphy RW, Shi P.. 2012. Hearing aid for vertebrates via multiple episodic adaptive events on prestin genes. Mol Biol Evol. 29(9): 2187–2198. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M.. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010(6): pdb.prot5448.. [DOI] [PubMed] [Google Scholar]
- Musolf K, Hoffmann F, Penn DJ.. 2010. Ultrasonic courtship vocalizations in wild house mice, Mus musculus musculus. Anim Behav. 79(3): 757–764. [Google Scholar]
- O’Leary MA, et al. 2013. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339: 662–667. [DOI] [PubMed] [Google Scholar]
- Parker J, et al. 2013. Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502(7470): 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne KB, Langbauer WR Jr, Thomas EM.. 1986. Infrasonic calls of the Asian elephant (Elephas maximus). Behav Ecol Sociobiol. 18(4): 297–301. [Google Scholar]
- Perry GH, et al. 2013. Aye-aye population genomic analyses highlight an important center of endemism in northern Madagascar. Proc Natl Acad Sci U S A. 110(15): 5823–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Martin RD, Verrelli BC.. 2007. Signatures of functional constraint at aye-aye opsin genes: the potential of adaptive color vision in a nocturnal primate. Mol Biol Evol. 24(9): 1963–1970. [DOI] [PubMed] [Google Scholar]
- Perry GH, Melsted P, et al. 2012. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 22(4): 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Reeves D, et al. 2012. A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar. Genome Biol Evol. 4(2): 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsier MA, Dominy NJ.. 2012. Receiver bias and the acoustic ecology of aye-ayes (Daubentonia madagascariensis). Commun Integr Biol. 5(6): 637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandwith H. 1859. On the habits of the “aye‐aye” (Cheiromys madagascariensis, L., Cuv.). J Proc Linn Soc Lond Zool. 4(13): 28–30. [Google Scholar]
- Schienman JE, Holt RA, Auerbach MR, Stewart C-B.. 2006. Duplication and divergence of 2 distinct pancreatic ribonuclease genes in leaf-eating African and Asian colobine monkeys. Mol Biol Evol. 23(8): 1465–1479. [DOI] [PubMed] [Google Scholar]
- Schwitzer C, et al. 2013. Lemurs of Madagascar: a strategy for their conservation 2013–2016. IUCN SSC Primate Specialist Group. Arlington, VA. [Google Scholar]
- Shen Y-Y, Liang L, Li G-S, Murphy RW, Zhang Y-P.. 2012. Parallel evolution of auditory genes for echolocation in bats and toothed whales. PLoS Genet. 8(6): e1002788.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JJ, Rabosky DL.. 2015. Speciation dynamics during the global radiation of extant bats. Evolution 69(6): 1528–1545. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Jungers WL.. 1997. Body mass in comparative primatology. J Hum Evol. 32(6): 523–559. [DOI] [PubMed] [Google Scholar]
- Soligo C. 2005. Anatomy of the hand and arm in Daubentonia madagascariensis: a functional and phylogenetic outlook. Folia Primatol. 76(5): 262–300. [DOI] [PubMed] [Google Scholar]
- Sterling EJ, Povinelli DJ.. 1999. Tool use, aye-ayes, and sensorimotor intelligence. Folia Primatol. 70(1): 8–16. [DOI] [PubMed] [Google Scholar]
- Stern DL. 2013. The genetic causes of convergent evolution. Nat Rev Genet. 14(11): 751–764. [DOI] [PubMed] [Google Scholar]
- Tange O. 2011. Gnu parallel-the command-line power tool. The USENIX Magazine. 36(1): 42–47. [Google Scholar]
- Thomas GWC, Hahn MW.. 2015. Determining the null model for detecting adaptive convergence from genomic data: a case study using echolocating mammals. Mol Biol Evol. 32(5): 1232–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ET, Bankoff RJ, Louis EE, Perry GH.. 2016. Deadwood structural properties may influence aye-aye (Daubentonia madagascariensis) extractive foraging behavior. Int J Primatol. 37(2): 281–295. [Google Scholar]
- White NR, Prasad M, Barfield RJ, Nyby JG.. 1998. 40- and 70-kHz vocalizations of mice (Mus musculus) during copulation. Physiol Behav. 63(4): 467–473. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Y-P, Rosenberg HF.. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet. 30(4): 411–415. [DOI] [PubMed] [Google Scholar]
- Zhang J. 2003. Parallel functional changes in the digestive RNases of ruminants and colobines by divergent amino acid substitutions. Mol Biol Evol. 20(8): 1310–1317. [DOI] [PubMed] [Google Scholar]
- Zhou X, et al. 2014. Whole-genome sequencing of the snub-nosed monkey provides insights into folivory and evolutionary history. Nat Genet. 46(12): 1303–1310. [DOI] [PubMed] [Google Scholar]
- Zou Z, Zhang J.. 2015. No genome-wide protein sequence convergence for echolocation. Mol Biol Evol. 32(5): 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.