In this study, Zhang et al. investigated the mechanisms by which liver lipid metabolism is controlled by the timing of both the circadian clock and feeding. Their results show that liver-specific depletion of nuclear receptors RORα and RORγ, key components of the molecular circadian clock, up-regulate expression of lipogenic genes only under fed conditions, but not under fasting conditions, and establish ROR/Insig2/SREBP as a molecular pathway by which circadian clock components anticipatorily regulate lipogenic responses to feeding.
Keywords: ROR, liver, circadian, SREBP1c, metabolism
Abstract
Liver lipid metabolism is under intricate temporal control by both the circadian clock and feeding. The interplay between these two mechanisms is not clear. Here we show that liver-specific depletion of nuclear receptors RORα and RORγ, key components of the molecular circadian clock, up-regulate expression of lipogenic genes only under fed conditions at Zeitgeber time 22 (ZT22) but not under fasting conditions at ZT22 or ad libitum conditions at ZT10. RORα/γ controls circadian expression of Insig2, which keeps feeding-induced SREBP1c activation under check. Loss of RORα/γ causes overactivation of the SREBP-dependent lipogenic response to feeding, exacerbating diet-induced hepatic steatosis. These findings thus establish ROR/INSIG2/SREBP as a molecular pathway by which circadian clock components anticipatorily regulate lipogenic responses to feeding. This highlights the importance of time of day as a consideration in the treatment of liver metabolic disorders.
A molecular circadian clock related to the 24-h oscillation of internal and environmental factors, including light/dark cycles, time of food availability, temperature, and activity/rest cycles, exists in most cells of nearly every living organism (Bass and Takahashi 2010; Feng and Lazar 2012; Asher and Sassone-Corsi 2015). The master clock in mammals is located in the suprachiasmatic nucleus (SCN) of the hypothalamus, and many physiological processes are subject to circadian oscillations, including lipid and carbohydrate metabolism, hormone secretion, and feeding behaviors (Eckel-Mahan and Sassone-Corsi 2013; Liu et al. 2013; Adamovich et al. 2014). In the liver, the circadian clock is closely related to the rhythmicity of hepatic metabolism (Lin et al. 2008; Tahara and Shibata 2016). The autonomous rhythm of hepatocytes is entrained by not only the hormonal and neuronal signals from the master clock but also the nutrients and metabolites whose oscillation is in large part determined by feeding activity (Kornmann et al. 2007; Feng and Lazar 2012). Misalignment of peripheral clock and circadian behaviors, such as feeding behavior, in the case of shiftwork, repeated jet lag, and night eating conditions has been associated with increased incidence of metabolic disorders (Scheer et al. 2009; Dibner and Schibler 2015).
At a molecular level, the cellular circadian clock is maintained by a complex circuitry of transcriptional/translational regulatory loops (Papazyan et al. 2016b). In mammals, a positive limb is comprised of BMAL1 and CLOCK, which activate transcription of PER and CRY as well as the nuclear receptors Rev-erbα and Rev-erbβ (Preitner et al. 2002). The Rev-erbs directly bind to the Arntl (Bmal1) gene to repress its transcription (Preitner et al. 2002; Yin and Lazar 2005), and this is mitigated by ROR nuclear receptors that compete for genomic binding at sites containing a specific DNA sequence motif referred to as the RORE (Giguere et al. 1994; Harding and Lazar 1995). In addition to their roles in the core clock mechanism, Rev-erbs and RORs directly regulate metabolic genes in a cell type-specific manner, in some cases at nonoverlapping sites due to tethered recruitment by tissue-specific transcription factors (Zhang et al. 2015). The circadian expression of Rev-erbs and RORγ thus connects the clock to tissue-specific metabolic outputs (Everett and Lazar 2014; Cook et al. 2015).
ROR has three subtypes: RORα, RORβ, and RORγ. Only RORα and RORγ are highly expressed in the liver, while RORβ is expressed mainly in the central nervous system (Forman et al. 1994; Andre et al. 1998; Takeda et al. 2012). Studies of total body knockout mice reported that both RORα- and RORγ-deficient mice exhibited improved insulin sensitivity and glucose tolerance with decreased hepatic gluconeogenesis (Takeda et al. 2014a; Kadiri et al. 2015). RORα-deficient mice, which suffer from severe cerebellar defects and display a staggerer phenotype (Sidman et al. 1962; Dussault et al. 1998; Steinmayr et al. 1998), also exhibited lower total body fat and were resistant to age-induced white adipose tissue (WAT) and brown adipose tissue (BAT) hypertrophy (Kang et al. 2011). It has also been reported that RORs regulate rhythmic expression of several lipid metabolic genes, including the Elovl3 and Cyp8b1, by enhancing their expression around Zeitgeber time 20–4 (ZT20–ZT4) (Takeda et al. 2012, 2014b). However, studies of ROR regulation of hepatic triglyceride levels are contradictory, with some reporting that RORα- and RORγ-null mice had decreased hepatic triglycerides and lipogenic gene expression (Lau et al. 2008; Kang et al. 2011; Takeda et al. 2014b), while another reported that RORα-null mice had increased liver triglyceride accumulation and lipogenic gene expression (Wada et al. 2008).
The reasons for these inconsistencies are unclear but could pertain to experimental differences in the time of day and feeding conditions under which the mice were studied or the developmental effects of the gene deletions. Here, to elucidate the hepatic functions of the RORs, we conditionally knocked out RORα and RORγ singly or together in adult mouse livers. We found that the RORs redundantly control lipid metabolism by regulating lipogenic gene expression. These effects were strong at ZT22 (5:00 AM) in mice fed ad libidum but were not observed at ZT10 or under fasting conditions and were dependent on the induction of SREBP1c. Thus, the hepatic metabolic role of ROR depends on the time of day and nutritive status. These results demonstrate important considerations in the interpretation of metabolic studies and suggest that circadian rhythms should be taken into account when contemplating therapeutic intervention aimed at liver metabolism.
Results
RORα and RORγ redundantly regulate clock and metabolic genes
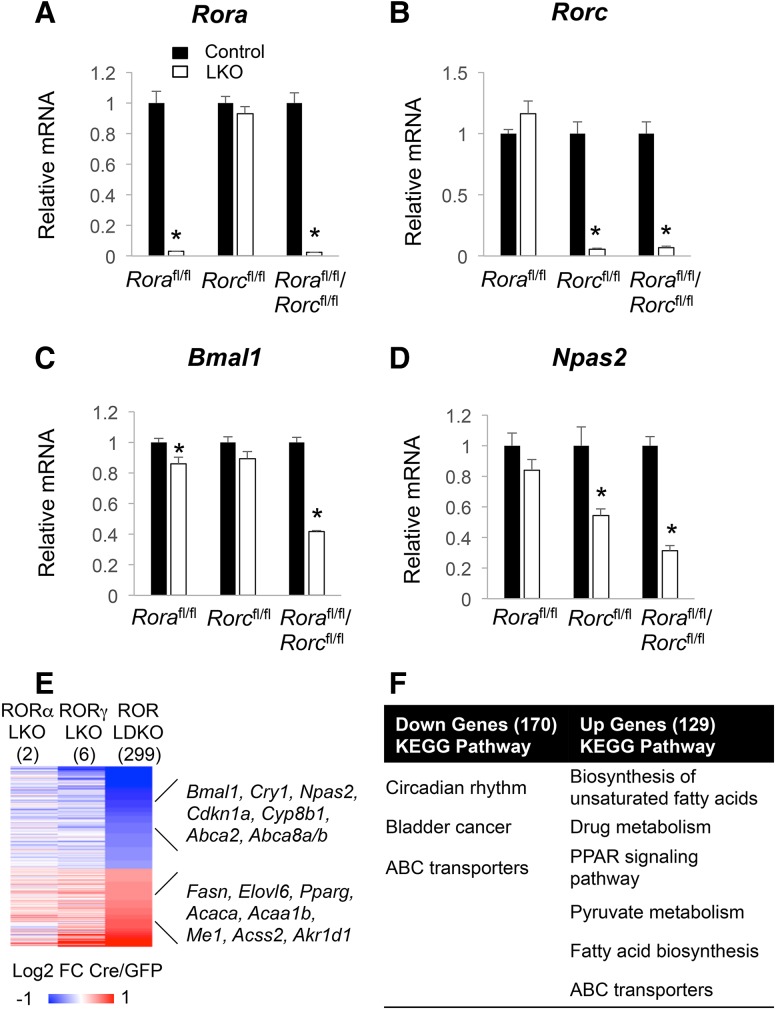
RORα and RORγ were knocked out singly as well as together in livers by injecting hepatocyte targeted AAV-TBG-Cre into Rorafl/fl, Rorcfl/fl or double-floxed (LDKO [liver-specific double-knockout]) mice (Zhang et al. 2015). The depletion of RORs was confirmed by quantitative RT–PCR (qRT–PCR) (Fig. 1A,B) and Western blot (Supplemental Fig. S1A,B) when compared with control floxed mice injected with AAV-TBG-GFP. Notably, while the expression of clock genes Bmal1 and Npas2 was weakly affected in single-knockout livers, it was down-regulated in ROR LDKO livers (Fig. 1C,D), suggesting redundant function of gene regulation by RORα and RORγ. To explore the functional roles of RORα and RORγ in the mouse liver, we compared the transcriptome of single-knockout and double-knockout mouse livers using microarray analysis on mice sacrificed at ZT22 (5:00 AM), which is the peak of RORγ expression (Takeda et al. 2012). Although very few genes were differentially expressed in single-knockout livers, 299 genes were altered in ROR LDKO livers, with 170 genes down-regulated and 129 genes up-regulated (Fig. 1E; Supplemental Table S2). The down-regulated genes are consistent with the canonical function of RORs as activators of gene expression and included known ROR target genes, such as Bmal1, Cry1, Npas2, and Cyp8b1, which were enriched for circadian rhythm pathways (Fig. 1F). The up-regulated genes, including Fasn, Elovl6, and Acaca, were enriched for lipid and fatty acid metabolism pathways (Fig. 1E,F). These results suggested that RORs might be involved in the regulation of hepatic lipid and fatty acid metabolism.
Figure 1.
RORα and RORγ redundantly regulate clock and metabolic genes. (A–D) Gene expression of Rora and Rorc (A,B) and clock genes Bmal1 and Npas2 (C,D) in ROR single-knockout or double-knockout livers at ZT22. Data are expressed as mean ± SEM (*) P < 0.05, Student's t-test. n = 4–5 per group. (E) Heat map of the ROR single-knockout and double-knockout liver microarray analysis at ZT22. The color bar indicates the log2 fold change ratio of Cre/GFP. The cutoff used here was fold change >1.3 with a 15% false discovery rate. (F) Gene ontology analysis of the down-regulated genes and up-regulated genes in the ROR LDKO livers at ZT22.
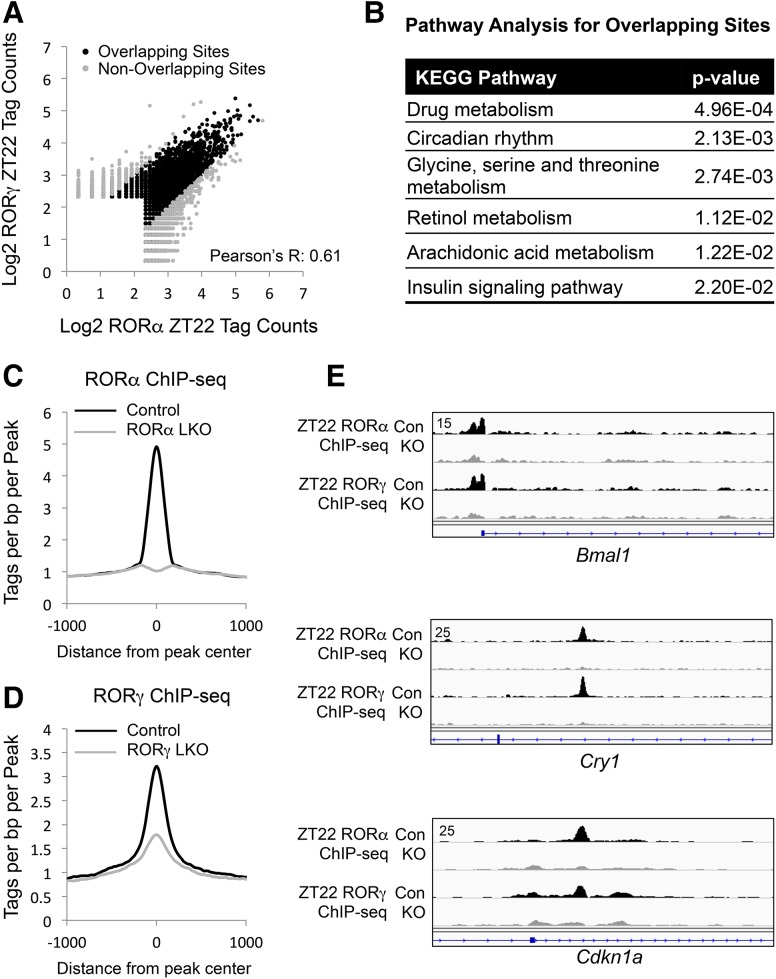
To further explore the functional redundancy of RORα and RORγ in the liver, we profiled and compared their cistrome by ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analysis. RORα and RORγ ChIP-seq peaks were highly overlapping (Fig. 2A), and the genes near these sites were enriched for drug metabolism and circadian rhythm pathways (Fig. 2B). The specificity of the ChIP as well as the efficiency of the knockout were confirmed by the marked reduction in ChIP signal in the RORα and RORγ knockout livers (Fig. 2C,D). Screenshots of RORα and RORγ binding to target genes, including Bmal1, Cry1, and Cdkn1a, and loss of this binding in the knockouts are shown in Figure 2E.
Figure 2.
Extensive correlation of liver RORα and RORγ cistromes. (A) Scatter plot of RORα and RORγ ChIP-seq signals at ZT22. All of the binding sites were filtered by the knockout sample (knockout/control > 3). (B) Gene ontology analysis of the genes closest to the overlapping binding sites. (C) Average profile of RORα ChIP-seq signal in the control mouse livers and their counterparts in the RORα LKO (liver-specific knockout) mouse livers. (D) Average profile of RORγ ChIP-seq signal in the control mouse livers and their counterparts in the RORγ LKO mouse livers. (E) Screenshot of the RORα- and RORγ-binding sites near the well-characterized target genes of RORs in the control samples and knockout samples, including Bmal1, Cry1, and Cdkn1a.
Hepatocyte-specific deletion of RORα and RORγ up-regulates lipogenic genes specifically around ZT22 to promote hepatosteatosis
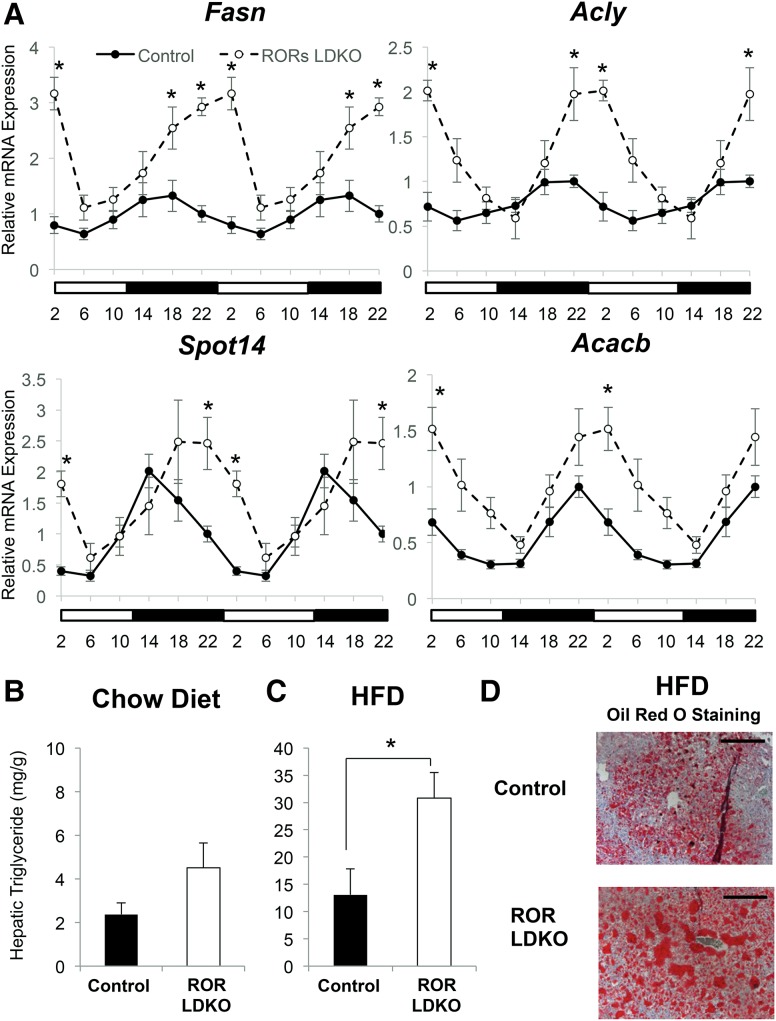
We next investigated the ROR regulation of lipid metabolic genes throughout the day by collecting mouse livers every 4 h for a period of 24 h. Remarkably, the up-regulation of metabolic genes in the ROR LDKO livers was restricted to ZT18–ZT2 (Fig. 3A), indicating that their regulation by RORs was limited to this time frame. This increase in amplitude of lipogenic gene expression was associated with a trend toward increased hepatic triglycerides at ZT22 in ROR LDKO mice on a chow diet (Fig. 3B), but, when challenged with a high-fat diet, the mice lacking liver RORs exhibited a marked increase in hepatic triglycerides at ZT22 (Fig. 3C), which was confirmed by Oil Red O staining (Fig. 3D). Thus, the heightened amplitude of circadian lipogenic gene expression in ROR LDKO livers predisposes the mice to hepatosteatosis.
Figure 3.
Hepatocyte-specific deletion of RORα and RORγ up-regulates lipogenic genes specifically around ZT22, favoring hepatosteatosis. (A) mRNA expression of Fasn, Acly, Spot14, and Acacb over a 24-h period in control and ROR LDKO mouse livers. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4–5 per group. Data are double-plotted for better visualization. (B,C) Hepatic triglyceride measurement of the livers at ZT22 from the control mice and ROR mutant mice fed a normal chow diet (B) and after 6 wk on a high-fat diet (C). (mg/g) Milligrams of triglycerides per gram of liver weight. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4–5 per group. (D) Oil Red O staining of livers from the control mice and ROR LDKO mice fed for 6 wk on a high-fat diet at a magnification of 200×. Bars, 100 µm.
Gene activation by RORα and RORγ is more commonly direct, while negative regulation tends to be indirect
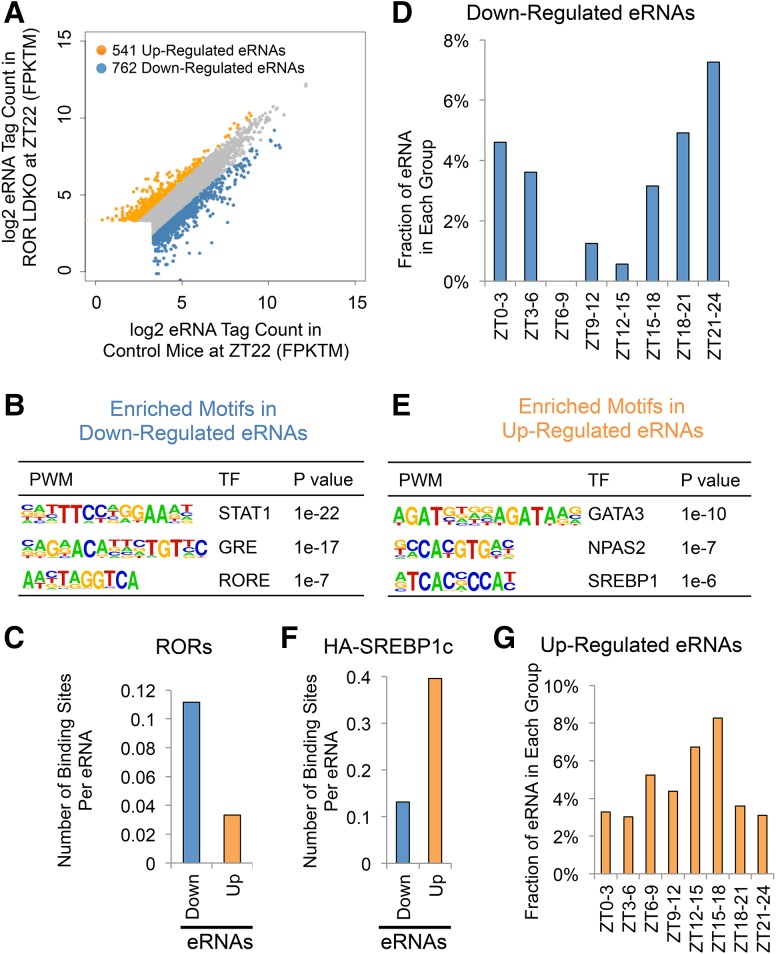
To better understand the mechanism of regulation of genes differentially expressed in ROR LDKO mice, we performed global run-on (GRO) followed by deep sequencing (GRO-seq) to measure nascent transcription in livers of control and ROR LDKO mice at ZT22. Measurement of divergent RNA transcription at enhancers provided a quantitative index of enhancer activity (Fang et al. 2014), and differential analysis revealed 762 down-regulated enhancer RNAs (eRNAs) and 541 up-regulated eRNAs in the ROR LDKO (Fig. 4A). The down-regulated eRNAs likely represent sites where RORs act in their classical activation mode. Indeed, the RORE motif was enriched at these sites (Fig. 4B), as were motifs for glucocorticoid receptor and STAT transcription factors, potentially due to collaborative interactions analogous to that between Rev-erbα and HNF6 (Ciofani et al. 2012; Zhang et al. 2015). Cross-comparison of the differentially expressed eRNAs with the RORα and RORγ overlapping cistrome showed that ROR binding is enriched near the down-regulated eRNAs relative to the up-regulated eRNAs, (Fig. 4C), consistent with the notion that these classically activated enhancers are more likely to be direct binding sites for ROR. Down-regulated eRNAs were also circadian, enriched around phase ZT21–ZT24 (Fig. 4D). The lower expression of the down-regulated eRNAs around phase ZT6–ZT12 is consistent with the peak of expression of Rev-erbα, which represses these RORE-containing enhancers, including the ones at clock genes such as Bmal1 and Npas2 (Fang et al. 2014).
Figure 4.
Gene activation by RORα and RORγ is more commonly direct, while negative regulation tends to be indirect. (A) Scatter plot of eRNA tag counts in control and ROR mutant mouse livers. Differentially expressed eRNAs are marked by different colors. (B) De novo motif analysis of the down-regulated eRNAs. (C) RORα and RORγ overlapping binding sites at the differentially expressed eRNAs. (D) The fraction of eRNAs that are up-regulated in ROR LDKO livers in each circadian phase in the wild-type mice as characterized previously (Fang et al. 2014). (E) De novo motif analysis of the up-regulated eRNAs. (F) HA-nSREBP1c-binding sites at the differentially expressed eRNAs. (G) The fraction of eRNAs that are down-regulated in ROR LDKO livers in each circadian phase in the wild-type mice as characterized previously (Fang et al. 2014).
We were particularly interested in the up-regulated enhancers, since lipogenic gene activation in the ROR LDKO was likely an indirect consequence of losing gene activation by RORs. The up-regulated eRNAs were enriched for E-boxes as well as the GATA motif (Fig. 4E), with the E-box being notable as the binding site of SREBP1, the master regulator of lipogenesis (Horton 2002; Jeon and Osborne 2012). Indeed, genomic binding of ectopically expressed HA-SREBP1c (Papazyan et al. 2016a) was highly enriched at the up-regulated eRNAs relative to the down-regulated eRNAs (Fig. 4F), consistent with the enrichment of the SREBP1 motif in the up-regulated eRNAs, which suggested a role of SREBP1 in the ROR LDKO-induced eRNAs. Notably, the up-regulated eRNAs were enriched around ZT15-ZT18, which is similar to the phase shown earlier for lipogenic gene induction in the ROR LDKO (Fig. 4G).
ROR LDKO activates SREBP1 through repression of Insig2 expression
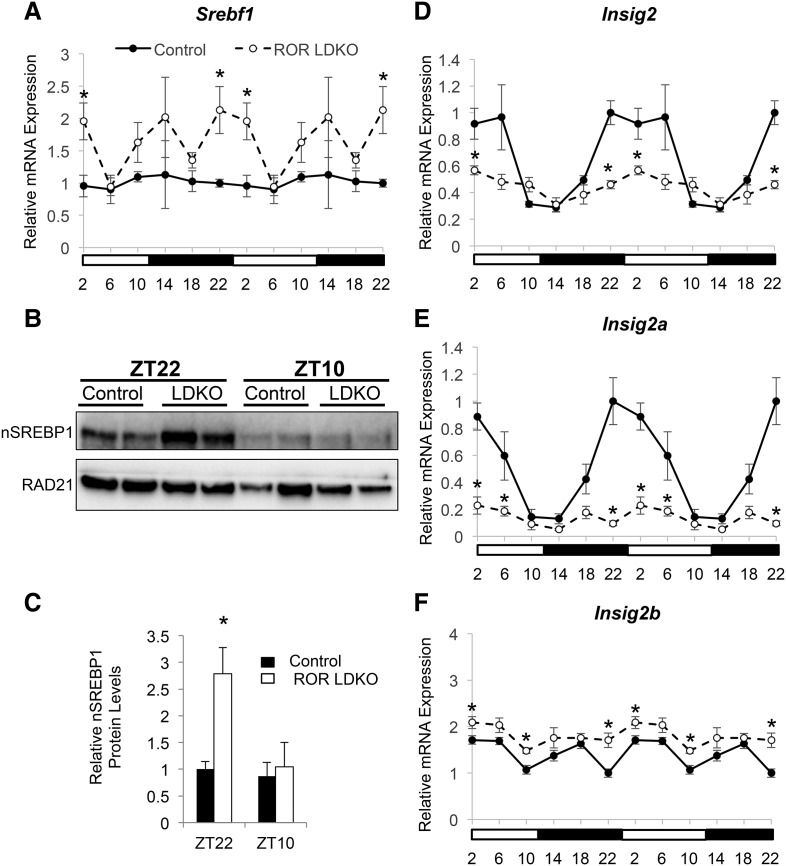
The enrichment of SREBP1 motifs in the up-regulated eRNAs in ROR LDKO livers prompted us to next investigate the activation status of lipogenic transcription factor SREBP1c. Indeed, SREBP1 gene expression was increased in the ROR LDKO around ZT22∼ZT2 (Fig. 5A). Moreover, the cleaved and active nuclear form of SREBP1c was induced at ZT22 in the ROR LDKO livers (Fig. 5B, quantitation of replicates in C). Increased nuclear SREBP1c was consistent with reduced expression of Insig2 (Fig. 5D), which is a negative regulator of SREBP post-translational cleavage and activation (Yabe et al. 2002). Indeed, Insig2 gene expression also displayed a circadian rhythm that peaked around ZT22–ZT6, in phase with the ROR activity and anti-phase with the peak expression of lipogenic genes, including Fasn, Acly, Spot14, and Acacb (Fig. 3A; Le Martelot et al. 2009). A further analysis of the different isoforms of Insig2 transcripts revealed that Insig2a, which is the predominant transcript in the liver and selectively down-regulated by insulin (Yabe et al. 2003), was significantly reduced around ZT22∼ZT6 in ROR LDKO livers, while Insig2b was slightly up-regulated (Fig. 5E,F). Together, these data suggested that RORs directly regulate Insig2 expression to influence SREBP1c activation around ZT22∼ZT6.
Figure 5.
ROR LDKO activates SREBP1 through repression of Insig2 expression. (A) mRNA expression of Srebf1 over a 24-h period in control and ROR LDKO mouse livers. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4 per group. Data are double-plotted for better visualization. (B) Western blot of the active form of nuclear SREBP1 and the loading control RAD21 in the livers of Rorafl/fl/Rorcfl/fl mice injected with GFP (control) and Cre (knockout) and harvested at ZT22 and ZT10. (C) Quantification of the protein level shown in the Western blot in B. SREBP1 protein level was normalized to the loading control RAD21. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 3–4 per group. (D) mRNA expression of Insig2 over a 24-h period in control and ROR LDKO mouse livers. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4 per group. Data are double-plotted for better visualization. (E) mRNA expression of Insig2a over a 24-h period in control and ROR LDKO mouse livers. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4 per group. Data are double-plotted for better visualization. (F) mRNA expression of Insig2b over a 24-h period in control and ROR LDKO mouse livers. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4 per group. Data are double-plotted for better visualization.
Increased lipogenic gene expression in ROR LDKO livers requires food intake
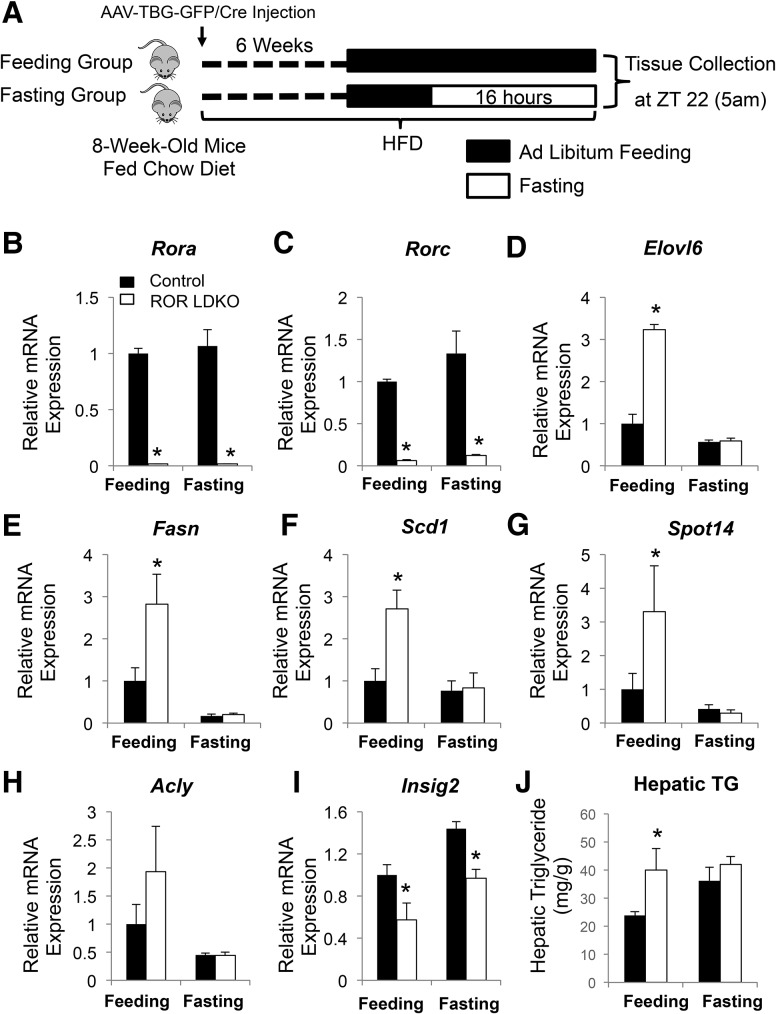
Feeding behavior is tightly regulated by the circadian clock and has a robust rhythm, with higher food intake during the active dark phase (ZT12–ZT24) as compared with intake levels during the inactive light phase (ZT0–ZT12) (Turek et al. 2005). In addition, feeding behavior also induced activation of SREBP as well as the lipogenic gene expression (Horton et al. 1998; Dentin et al. 2005). To investigate whether the time-specific regulation of lipid metabolic genes by RORs was related to feeding status, we compared the lipid metabolic phenotype of ROR LDKO mice in either ad libitum feeding or 16-h fasted conditions in mice fed a high-fat diet for 6 wk (Fig. 6A). Indeed, the marked activation of SREBP1c and induction of lipid metabolic genes in the ROR LDKO at ZT22 was almost completely abrogated under fasted conditions (Fig. 6B–H; Supplemental Fig. S2), although the regulation of Insig2 expression of ROR was irrespective of feeding/fasting status (Fig. 6I). Consistent with this, hepatic triglycerides were increased in the ROR LDKO livers under ad libitum feeding conditions, but the difference between control and ROR LDKO livers was abrogated in the fasting state (Fig. 6J).
Figure 6.
Increased expression of lipogenic genes in ROR LDKO livers occurs in the fed, but not fasted, state. (A) Study design and schedule for fasted and feeding study groups. (B–H) mRNA expression of RORs (B,C) and lipid metabolic genes, including Elovl6 (D), Fasn (E), Scd1 (F), Spot14 (G), and Acly (H), at ZT22, normalized to Arbp, as measured by RT-qPCR. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 3–5 per group. (I) mRNA expression levels of Insig2 in the control and ROR LDKO mouse livers under feeding or fasting condition. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 3–5 per group. (J) Hepatic triglyceride measurement of the livers from the fed and fasted control mice and ROR mutant mice fed a high-fat diet for 6 wk. (mg/g) Milligrams of triglycerides per gram of liver weight. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 3–5 per group.
Activation of lipid metabolic genes in the ROR LDKO requires SREBP1c
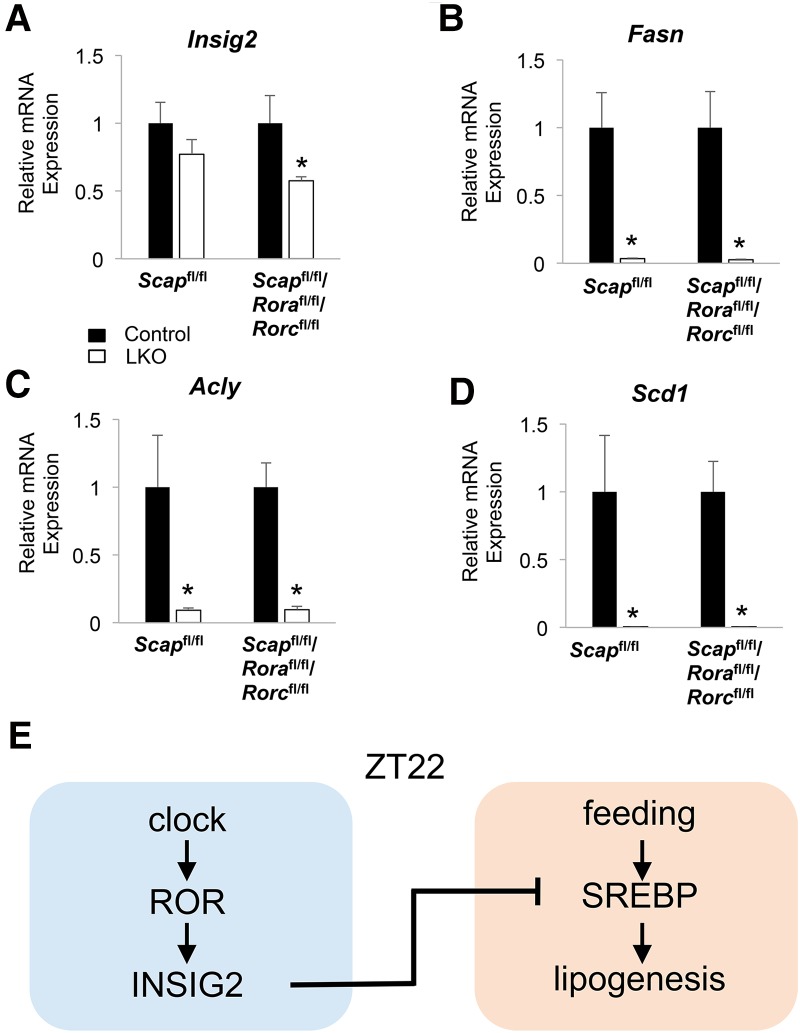
To test whether ROR LDKO-dependent induction of lipogenic genes was occurring entirely through SREBP activation, we generated mice lacking RORs and SREBP cleavage-activating protein (SCAP), which is a protein that stabilizes SREBPs and facilitates their cleavage and activation (Hua et al. 1996). Although, SCAP/ROR LTKO (liver-specific triple-knockout) livers had reduction of Insig2 mRNA levels similar to that in ROR LDKO (Fig. 7A), lipogenic genes such as Fasn, Acly, and Scd1 were no longer induced at ZT22 (Fig. 7B–D). Taken together, these results demonstrate that SREBP1c is required for the lipogenic effects of ROR LDKO, and ROR likely directly controls Insig2 expression to affect hepatic lipid metabolism (Fig. 7E).
Figure 7.
Activation of lipid metabolic genes in the ROR LDKO requires SREBP1c. (A) mRNA expression levels of Insig2 in SCAP LKO, SCAP/ROR LTKO, and control mouse livers at ZT22. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4–6 per group. (B–D) mRNA expression of Fasn (B), Scd1 (C), and Acly (D) in SCAP LKO, SCAP/ROR LTKO, and control mouse livers at ZT22. Data are expressed as mean ± SEM. (*) P < 0.05, Student's t-test. n = 4–6 per group. (E) Model depicting the hepatic circadian clock fine-tuning the lipogenic response through ROR regulation of SREBP1c and lipogenesis during feeding.
Discussion
Previous studies of RORα- or RORγ-null mice have suggested a role in the control of energy homeostasis and the regulation of lipid and glucose metabolism. However, since the regulation of energy and metabolism homeostasis is a complex process that involves multiple interrelated pathways in many organs involving the endocrine, immune, and nervous systems as well as the circadian clock and gut microbiome, it is difficult to determine whether the metabolic changes observed in ROR-deficient mice are directly controlled by RORs or are a result of systemic changes. Therefore, we used a liver-specific inducible ROR LDKO mouse model to elucidate the tissue-autonomous function of RORs in the adult mouse liver.
Previous reports on ROR regulation of liver metabolism using the total body ROR-deficient mice have generated contradicting results. Here, we showed that ROR regulation of lipid metabolism varies depending on the time of the day as well as the feeding conditions. Around ZT22, when the mice were at the end of their feeding phase in the dark, RORs repressed the lipid metabolic process through inhibition of the SREBP pathway. However, around ZT10 or during fasting even at ZT22, the effect of ROR LDKO on lipid metabolism was abolished. The rhythmicity of ROR repression of lipogenic genes such as Fasn and Acly fit well with the natural oscillation of their expression in the control mice, where Fasn and Acly mRNA levels peaked around ZT14–ZT18 and decreased rapidly at ZT22, at which time point RORs started to induce Insig2 expression. In the absence of RORs, the expression level of these lipogenic genes continued to rise until ZT6. It is interesting that both RORs account for these time-sensitive effects, yet only RORγ expression exhibits a high-amplitude circadian rhythm, whereas RORα expression is minimally circadian (Takeda et al. 2012; Zhang et al. 2015). The redundancy of both a circadian and noncircadian ROR subtype suggests that a threshold level of ROR activity is required, with the oscillation of RORγ exceeding the threshold at specific times of day. In addition, these findings highlight that, although RORs and Rev-erbs compete for binding at overlapping RORE-containing sites in clock genes, their binding and regulatory function at metabolic genes frequently differ due to differential recruitment requiring tissue-specific factors (Zhang et al. 2015, 2016). In this context, it is not surprising that lipid metabolism in the ROR knockout livers is not the phenotypic opposite of the lack of Rev-erbs.
Mechanistically, we found that loss of hepatic RORs led to a marked induction of SREBP1c protein, most likely induced by the reduction of Insig2 expression. A post-translational increase in cleaved SREBP1c is favored by loss of circadian INSIG2 expression in the ROR LDKO livers. Of note, ROR activation of Insig2 is restricted between ZT22 and ZT6, suggesting that Insig2 might be repressed by other factors outside this phase. Indeed, Rev-erbα and its associated nuclear corepressor (NCoR) and histone deacetylase 3 (HDAC3)-repressive complex have been reported to repress Insig2 transcription between ZT10 and ZT14 (Le Martelot et al. 2009; Bugge et al. 2012; Papazyan et al. 2016a), which might be the cause of the Insig2 expression trough in this phase. However, other lipid metabolic genes are induced in an SREBP1c-independent manner in the HDAC3 knockout livers, which are steatotic (Feng et al. 2011; Sun et al. 2012; Papazyan et al. 2016a). In addition, cholesterol and cholesterol derivatives have been cocrystallized with RORα and have been suggested to be ROR ligands that regulate ROR activities (Kojetin and Burris 2014; Santori et al. 2015). Interestingly, SREBP is also a major regulator of cholesterol synthesis, and its activity is inhibited by high levels of cholesterol (Horton et al. 2002). Therefore, its suppression by ROR might contribute to this negative feedback mechanism.
In summary, our study delineated a redundant role of RORs in regulating lipogenic metabolism at specific times of day and only when the mice are eating. These results highlight the importance of considering the time of day in interpreting the effects of environmental or genetic manipulations and suggest that therapies directed at ROR and other targets should be administered according to a schedule that corresponds to their biological rhythms in order to maximize the effectiveness and minimize the side effects of the therapy. This is a particularly relevant consideration in the treatment of nonalcoholic fatty liver disease and steatohepatitis, which are highly prevalent in association with the epidemics of obesity and diabetes and whose progression to cirrhosis is already the third most common indication for liver transplantation (Zezos and Renner 2014).
Materials and methods
Animals
Rorafl/fl mice were obtained from Mouse Clinical Institute/Institut Clinique de la Souris (MCI/ICS), and Rorcfl/fl mice were obtained from Jackson Laboratory. Scap floxed mice generated previously (Matsuda et al. 2001) were backcrossed to the C57BL/6J genetic background for at least seven to eight generations (T.F. Osborne). Mice were housed on a temperature-controlled specific pathogen-free facility with 12:12-h light–dark cycle (lights on at 07:00, lights off at 19:00). Experiments were carried out on 8- to 20-wk-old male mice. The Penn Vector Core generated the AAV vectors (AAV8-TBG-GFP for control and AAV8-TBG-Cre for knockout). We injected each AAV vector intravenously at 1.5 × 1011 genome copies per mouse and characterized the mice at 3–4 wk after AAV injection. All animal studies were performed with an approved protocol from the University of Pennsylvania Perelman School of Medicine Institutional Animal Care and Use Committee.
Western blot
For Western blot, nuclear fractionation was performed to determine nSREBP1, RORα, and RORγ levels in mouse livers as described previously (Wan et al. 2011). Lysates were resolved in 4%–12% Bis-Tris NUPAGE gradient gels with MOPS running buffer (Thermo Fisher Scientific). Proteins were transferred onto polyvinylidene difluoride membranes and blotted with anti-SREBP1 antibody from Abcam (ab3259), anti-RORα antibody from Santa Cruz Biotechnology (sc-28612), anti-RORγ antibody from Abcam (ab78007), and anti-Rad21 antibody from Cell Signaling Technology (4321S).
Gene expression analysis
Total RNA was extracted from liver tissue using Trizol reagent (Life Techologies) followed by RNeasy minikit (Qiagen). The RNA was reverse-transcribed using the High-Capacity cDNA reverse transcription kit (Applied Biosystems) and analyzed by qPCR. Gene expression was normalized to the mRNA levels of the housekeeping gene Arbp and the level of the gene of interest in the control samples.
qPCR
qPCR was performed with Power SYBR Green PCR Mastermix and the Prism 7500 instrument (Applied Biosystems), and analysis was performed by the standard curve method. Primers used in the qPCR are listed in Supplemental Table S1.
Microarray analysis
Microarray expression analyses of control versus ROR single LKO (liver-specific knockout) and LDKO livers at ZT22 were performed on livers from n = 4 mice per genotype. Total RNA was extracted from liver tissue using Trizol reagent (Life Techologies) followed by RNeasy minikit (Qiagen). RNA from each liver was individually processed with the Ambio WT expression kit and GeneChIP WT terminal labeling and control kit (Affymetrix) and hybridized to the Mouse Gene 2.0 ST arrays (Affymetrix). Array images were captured on a GCS3000 laser scanner (Affymetrix) and analyzed by the Penn Microarray Core using the Partek genomics suite. Subsequent data analysis was performed using the oligo package in R-BioConductor (Huber et al. 2015). Differentially regulated genes in the knockout were selected using a threshold of expression fold change >1.3 and false discovery rate of 15%. Microarray data are available in Gene Expression Omnibus (GSE101116).
GRO-seq
GRO-seq was performed using the control and ROR LDKO livers at ZT22 following the protocol described previously (Fang et al. 2014). Briefly, nuclei were extracted from muscles using hypotonic buffer. Nuclear run-on was performed in the presence of Br-UTP followed by enrichment with anti-Br-UTP antibodies, reverse transcription, and library preparation. Run-on reactions from four mice were pooled to make one sequencing library.
GRO-seq data processing
GRO-seq sequencing reads were aligned to the mm9 genome using Bowtie version 0.12.7. Uniquely mapped reads were extended to 150 base pairs (bp) in the 5′–3′ direction and used for downstream analysis. eRNA identification and quantification were performed according to previously established protocols (Fang et al. 2014). eRNAs with a more than twofold change in ROR LDKO livers than the control livers at ZT22 were considered differentially expressed. For calculating number of peaks per eRNA and number of eRNAs at each circadian time point, Bedtools intersect function was used. eRNAs were extended to 1000 bp from the center, and peaks/eRNAs overlapping at least 1 bp were considered overlapping.
ChIP
ChIP experiments were performed as described with minor changes (Zhang et al. 2015). Mouse livers were harvested, minced, and cross-linked in 1% formaldehyde for 20 min followed by quenching with 1/20 vol of 2.5 M glycine solution for 5 min and two washes with 1× PBS. Nuclear extracts were prepared by dounce homogenization in ChIP buffer (50 mM Tris-HCl at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% NaDOC). Chromatin fragmentation was performed by sonication in lysis buffer (50 mM Tris-HCL at pH 8.0, 0.1%SDS, 10 mM EDTA) using the Bioruptor (Diagenode). Proteins were immunoprecipitated in ChIP buffer using anti-RORα antibody from Santa Cruz Biotechnology (sc-28612) and RORγ antibody from Santa Cruz Biotechnology (sc-28559), cross-linking was reversed overnight at 65°C in SDS buffer (50 mM Tris-HCL, 10 mM EDTA, 1% SDS at pH 8), and DNA isolated using phenol/chloroform/isoamyl alcohol. Precipitated DNA was analyzed by qPCR or high-throughput sequencing.
ChIP-seq
ChIP experiments were performed independently on liver samples from individual mice harvested at the indicated times. DNA was amplified according to the ChIP-seq sample preparation guide provided by Illumina using adaptor oligo and primers from Illumina, enzymes from New England Biolabs, and PCR purification kit and MinElute kit from Qiagen. Deep sequencing was performed by the Functional Genomics Core of the Penn Diabetes Research Center using an Illumina HiSeq2000, and sequences were obtained using the Solexa Analysis Pipeline.
ChIP-seq data processing
Sequencing reads of biological replicates were pooled and aligned to the mm9 genome followed by peak calling as described previously (Zhang et al. 2015). Genome browser tracks of ChIP-seq data were generated using HOMER version 4.7 (Heinz et al. 2010) and visualized in Integrative Genomics Viewer (Robinson et al. 2011; Thorvaldsdottir et al. 2013). For the ROR cistrome analysis, peaks >0.5 read per million (RPM) and at least three times stronger than their counterparts in the LKO mouse livers were used. The binding sites that were commonly bound by RORα and RORγ with less than twofold difference were considered ROR overlapping binding sites. All ChIP-seq peaks were annotated by HOMER using the mapping within 50 kb of gene transcription start sites (TSSs). The pathway analysis of the ROR overlapping sites was performed using the TSSs nearest to the binding sites. Scatter plots and average profiles were created using HOMER and R package. ChIP-seq data are available in Gene Expression Omnibus (GSE101116).
Hepatic triglyceride assay
Liver samples were homogenized in the TissueLyser (Qiagen) with steel beads in tissue lysis buffer (140 mM NaCl, 50 mM Tris, 1% Triton-X at pH 8.0). Triglyceride concentration in the lysates was quantified using LiquiColor triglyceride procedure number 2100 (Stanbio).
Oil Red O staining
Five-micrometer frozen sections were prepared from snap-frozen liver tissues. The sections were stained with 0.5% Oil Red O in propylene glycerol overnight for lipid and then in hematoxylin for 5 sec. The procedures were performed by the Penn Digestive Disease Center Morphology Core.
Statistics
Microsoft Excel was used for graphing and statistical tests. Error bars represent the SEM, and statistical significance was determined by two-tailed type 2 t-test; a P-value of <0.05 was considered significant unless otherwise stated in the figure legends.
Accession number
ChIP-seq and microarray data have been deposited in the Gene Expression Omnibus (GSE101116).
Supplementary Material
Acknowledgments
We acknowledge the Functional Genomics Core and the Viral Vector Core of the Penn Diabetes Research Center (P30 DK19525) for next-generation sequencing and virus preparation, respectively. We thank the Penn Digestives Disease Center Morphology Core (P30 DK050306) for histology studies, and the Molecular Profiling Core for microarray analysis. This work was supported by National Institutes of Health grants R01 DK045586 (M.A.L.), F32 DK108555 (R.P.), and R01ES027544 (Z.S.), and the Cox Medical Research Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.302323.117.
References
- Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, Kraut-Cohen J, Wang M, Han X, Asher G. 2014. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab 19: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre E, Conquet F, Steinmayr M, Stratton SC, Porciatti V, Becker-Andre M. 1998. Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J 17: 3867–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Sassone-Corsi P. 2015. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell 161: 84–92. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. 2010. Circadian integration of metabolism and energetics. Science 330: 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. 2012. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev 26: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et al. 2012. A validated regulatory network for Th17 cell specification. Cell 151: 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DN, Kang HS, Jetten AM. 2015. Retinoic acid-related orphan receptors (RORs): regulatory functions in immunity, development, circadian rhythm, and metabolism. Nucl Receptor Res 2: 101185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentin R, Girard J, Postic C. 2005. Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie 87: 81–86. [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U. 2015. Circadian timing of metabolism in animal models and humans. J Int Med 277: 513–527. [DOI] [PubMed] [Google Scholar]
- Dussault I, Fawcett D, Matthyssen A, Bader JA, Giguere V. 1998. Orphan nuclear receptor RORα-deficient mice display the cerebellar defects of staggerer. Mech Dev 70: 147–153. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K, Sassone-Corsi P. 2013. Metabolism and the circadian clock converge. Physiol Rev 93: 107–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett LJ, Lazar MA. 2014. Nuclear receptor Rev-erbα: up, down, and all around. Trends Endocrinol Metab 25: 586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, Lazar MA. 2014. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159: 1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Lazar MA. 2012. Clocks, metabolism, and the epigenome. Mol Cell 47: 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. 2011. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Chen J, Blumberg B, Kliewer SA, Henshaw R, Ong ES, Evans RM. 1994. Cross-talk among RORα 1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol 8: 1253–1261. [DOI] [PubMed] [Google Scholar]
- Giguere V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. 1994. Isoform-specific amino-terminal domains dictate DNA-binding properties of RORα, a novel family of orphan hormone nuclear receptors. Genes Dev 8: 538–553. [DOI] [PubMed] [Google Scholar]
- Harding HP, Lazar MA. 1995. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol 15: 4791–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD. 2002. Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem Soc Transac 30: 1091–1095. [DOI] [PubMed] [Google Scholar]
- Horton JD, Bashmakov Y, Shimomura I, Shimano H. 1998. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc Natl Acad Sci 95: 5987–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Nohturfft A, Goldstein JL, Brown MS. 1996. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell 87: 415–426. [DOI] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon TI, Osborne TF. 2012. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab 23: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadiri S, Monnier C, Ganbold M, Ledent T, Capeau J, Antoine B. 2015. The nuclear retinoid-related orphan receptor-α regulates adipose tissue glyceroneogenesis in addition to hepatic gluconeogenesis. Am J Physiol Endocrinol Metab 309: E105–E114. [DOI] [PubMed] [Google Scholar]
- Kang HS, Okamoto K, Takeda Y, Beak JY, Gerrish K, Bortner CD, DeGraff LM, Wada T, Xie W, Jetten AM. 2011. Transcriptional profiling reveals a role for RORα in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol Genomics 43: 818–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojetin DJ, Burris TP. 2014. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov 13: 197–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. 2008. The orphan nuclear receptor, RORα, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem 283: 18411–18421. [DOI] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, Moschetta A, Schibler U. 2009. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol 7: e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JD, Liu C, Li S. 2008. Integration of energy metabolism and the mammalian clock. Cell Cycle 7: 453–457. [DOI] [PubMed] [Google Scholar]
- Liu S, Brown JD, Stanya KJ, Homan E, Leidl M, Inouye K, Bhargava P, Gangl MR, Dai L, Hatano B, et al. 2013. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature 502: 550–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Korn BS, Hammer RE, Moon YA, Komuro R, Horton JD, Goldstein JL, Brown MS, Shimomura I. 2001. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev 15: 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazyan R, Sun Z, Kim YH, Titchenell PM, Hill DA, Lu W, Damle M, Wan M, Zhang Y, Briggs ER, et al. 2016a. Physiological suppression of lipotoxic liver damage by complementary actions of HDAC3 and SCAP/SREBP. Cell Metab 24: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazyan R, Zhang Y, Lazar MA. 2016b. Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol 23: 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–260. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santori FR, Huang P, van de Pavert SA, Douglass EF Jr, Leaver DJ, Haubrich BA, Keber R, Lorbek G, Konijn T, Rosales BN, et al. 2015. Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab 21: 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, Shea SA. 2009. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci 106: 4453–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidman RL, Lane PW, Dickie MM. 1962. Staggerer, a new mutation in the mouse affecting the cerebellum. Science 137: 610–612. [DOI] [PubMed] [Google Scholar]
- Steinmayr M, Andre E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, Auclair N, Daniel H, Crepel F, Mariani J, Sotelo C, et al. 1998. staggerer phenotype in retinoid-related orphan receptor α-deficient mice. Proc Natl Acad Sci 95: 3960–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. 2012. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med 18: 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara Y, Shibata S. 2016. Circadian rhythms of liver physiology and disease: experimental and clinical evidence. Nat Rev Gastroenterol Hepatol 13: 217–226. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Jothi R, Birault V, Jetten AM. 2012. RORγ directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res 40: 8519–8535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Kang HS, Freudenberg J, DeGraff LM, Jothi R, Jetten AM. 2014a. Retinoic acid-related orphan receptor γ (RORγ): a novel participant in the diurnal regulation of hepatic gluconeogenesis and insulin sensitivity. PLoS Genet 10: e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Kang HS, Lih FB, Jiang H, Blaner WS, Jetten AM. 2014b. Retinoid acid-related orphan receptor γ, RORγ, participates in diurnal transcriptional regulation of lipid metabolic genes. Nucleic Acids Res 42: 10448–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. 2005. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308: 1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kang HS, Angers M, Gong H, Bhatia S, Khadem S, Ren S, Ellis E, Strom SC, Jetten AM, et al. 2008. Identification of oxysterol 7α-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor α (RORα) (NR1F1) target gene and a functional cross-talk between RORα and liver X receptor (NR1H3). Mol Pharmacol 73: 891–899. [DOI] [PubMed] [Google Scholar]
- Wan M, Leavens KF, Saleh D, Easton RM, Guertin DA, Peterson TR, Kaestner KH, Sabatini DM, Birnbaum MJ. 2011. Postprandial hepatic lipid metabolism requires signaling through Akt2 independent of the transcription factors FoxA2, FoxO1, and SREBP1c. Cell Metab 14: 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Brown MS, Goldstein JL. 2002. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci 99: 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. 2003. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc Natl Acad Sci 100: 3155–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Lazar MA. 2005. The orphan nuclear receptor Rev-erbα recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol 19: 1452–1459. [DOI] [PubMed] [Google Scholar]
- Zezos P, Renner EL. 2014. Liver transplantation and non-alcoholic fatty liver disease. World J Gastroenterol 20: 15532–15538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al. 2015. Gene regulation. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348: 1488–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fang B, Damle M, Guan D, Li Z, Kim YH, Gannon M, Lazar MA. 2016. HNF6 and Rev-erbα integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes Dev 30: 1636–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.