Abstract
Analysis of EST and cDNA collections from a number of metazoan species has identified genes encoding long polyadenylated transcripts that do not contain ORFs of lengths typical for protein-encoding mRNAs. Noncoding functions of such polyadenylated transcripts have been elucidated in only a few examples. The corresponding genes neither contain hallmark sequence motifs nor appear to have been conserved across phyla. Thus, it is impossible to systematically identify new members of this class of gene by using sequence homology and traditional gene-finding algorithms that depend on protein-coding potential. Consequently, even their approximate number has not been established for any metazoan genome. We curated polyadenylated transcripts with limited protein-coding capacity from intergenic regions of the Drosophila melanogaster genome. We used RT-PCR assays, hybridization to RNA blots and whole-mount embryos, and computational analyses to characterize candidate transcripts. We verify the structures and expression of 17 distinct, likely non-protein-coding polyadenylated transcripts. We show that the expression of many of these transcripts is conserved in other Drosophila species, indicating that they have important biological functions.
Keywords: genome, noncoding RNA, noncoding RNA, Drosophila pseudoobscura, Drosophila virilis
The number of described noncoding RNA (ncRNA) genes has undergone recent and dramatic expansion as novel untranslated RNA molecules have been discovered in many metazoan genomes. Many of these are short species of RNA: small-interfering RNAs that direct mRNA degradation, microRNAs that are implicated in other posttranscriptional regulation, and small nuclear RNAs and small nucleolar RNAs involved in splicing and ribosomal RNA modification. Another class of nuclear untranslated RNA molecules is composed of spliced, polyadenylated transcripts whose lengths are typical of protein-coding mRNAs. However, these RNAs contain ORFs whose extents represent unusually small fractions of the full transcripts. Because of their structural similarities to protein-coding transcripts, these longer ncRNAs are sometimes referred to as “mRNA-like ncRNA” (1).
Although long ncRNAs in Drosophila melanogaster were first described more than two decades ago (2), their functions generally remain elusive (reviewed in ref. 3). The ncRNAs that have been best studied to date in D. melanogaster appear to function as part of RNA–protein complexes, but they do not share obvious sequence motifs or secondary structures. The RNA on the X (roX1 and roX2) transcripts function in localizing the chromatin remodeling activity of the male-specific lethal complex by binding specifically to male-specific lethal proteins (reviewed in ref. 4). Transcripts from the fly Heat-shock RNA-ω (Hsr-ω) locus, produced in response to cellular heat stress, have been proposed to aid in organizing heterogeneous nuclear RNA-binding proteins (5). In addition, ncRNA produced by the polar granule component (pgc) gene and found in the pole plasm of embryos (6) recently has been shown to mediate transcriptional repression, apparently as a result of transcription factor sequestration (7). However, the interaction motifs used by these ncRNAs are not recognizable in other genes and cannot yet be used to define classes of ncRNAs. Additionally, systematic identification and annotation of long noncoding transcripts has not been possible because traditional gene-finding algorithms rely on coding potential and sequence homology.
Another approach to new ncRNA gene discovery is positional curation, searching for evidence of specific transcription in segments of the genome that lack other annotations. Microarray data from several metazoan species indicate that much more of the genome is transcribed than can be accounted by annotated gene transcripts (8, 9). But how much of this transcriptional activity produces specific and discrete RNAs remains unclear. Positional curation has been used to identify novel candidate ncRNA sequences in the Arabidopsis and mouse genomes (10, 11). The reannotated fruitfly genome (12) and sequences from the Drosophila Gene Collection (13), a repository of high-quality full-insert cDNA sequences, offer a rich resource for discovery of novel transcripts with potentially noncoding functions.
Here, we present the results of a positional curation effort that identifies 17 previously undescribed, long putative ncRNA genes by employing RT-PCR assays, Northern analysis, and in situ hybridization to characterize 72 candidate sequences. Comparative genome analyses indicate that the very small ORFs contained in these transcripts are unlikely to encode proteins. In several cases, we have obtained direct experimental evidence for expression of orthologous transcripts in diverged Drosophila species, Drosophila pseudoobscura and Drosophila virilis, indicating that the expression of those genes as specific transcripts has been conserved during evolution.
Materials and Methods
We began our curation with 7,972 individual cDNA sequences from the Berkeley Drosophila Genome Project cDNA data set. These sequences were the basis for the Release 3.1 annotation and consisted of clones sequenced to high quality, with an error rate of <1 in 50,000 (13). Additionally, we aligned sequences in our final curation set to the Release 3.1 genomic sequence (sequencing error rate <1 in 100,000) (14) and upon inspection found two sequences with polymorphisms or sequencing errors causing frameshifts; neither the genomic nor cDNA sequence predicted a substantial ORF, and these were corrected to match the genomic sequence in final curation. To isolate candidate ncRNA genes, we screened for cDNAs that did not intersect existing annotations (12, 15). Additional analyses (transcript length, ORF length and composition, initiating codon, polyadenylation length and consensus sites, genomic extent of transcription unit, and splice site prediction) were accomplished by using perl scripts and the wu-blast 2.0 (http://blast.wustl.edu) and sim4 (16) algorithms. Pairwise Ka/Ks analysis between possible translations of D. melanogaster ncRNA transcripts and either the D. pseudoobscura (17) or D. virilis genome (www.genome.gov/11008080) used paml 3.12 (18); alignments for this comparison were derived from tblastn analysis by using the parameters E = 0.0001, wordmask = seg, W = 5, T = 25, Y = 140,000,000. Comparison between models employing a fixed Ka/Ks value of 1 and those with unconstrained Ka/Ks results were used to calculate significance by using the χ2 test. Results with P < 0.05 were retained. qrna (19, 20) analysis also used the D. pseudoobscura genome. To identify D. pseudoobscura and D. virilis sequences orthologous to ncRNA candidate transcripts, we concatenated syntenic and/or overlapping blastn highest scoring pair results to nonrepetitive sequence with expectation values <1e-05. primer3 (21) was used for primer design.
For descriptions of molecular biology methods, oligonucleotide sequences, and experimental observations, see Supporting Materials and Methods and Data Sets 1–7, which are published as supporting information on the PNAS web site.
Results
Curation of 72 Candidate ncRNAs. Our approach to curation of novel ncRNA genes in D. melanogaster relied on two public resources: the Release 3.1 set of annotated genes (12) and the extensive collection of full-insert cDNA sequences that comprise the Drosophila Gene Collection (13). Human curators annotated the fruitfly genome by using extensive EST and cDNA data sets (12); by using sequences corresponding to Drosophila Gene Collection cDNAs, we searched 7,972 sequences for evidence of transcription not overlapping existing annotations. This primary computational screen returned 193 sequences: 134 transcripts that did not intersect existing protein-coding annotations on either strand and 59 transcripts that partially overlapped an annotation on the opposite strand. Transcripts were evenly distributed among the four Drosophila chromosomes, with one transcript located in heterochromatic sequence. We calculated ORF lengths for the longest Met-initiated ORF in each transcript and eliminated sequences with ORFs of >100 codons, leaving 120 transcripts.
Because generation of cDNA clones by internal oligo(dT) priming of transcripts is a common artifact of cDNA library-construction, we disqualified 48 of 120 sequences whose 3′ poly(A)-RNA tracts appeared encoded in genomic sequence, leaving a final 72 candidate cDNAs (Table 1; see also Table 3, which is published as supporting information on the PNAS web site, for additional details).
Table 1. Summary of expression analyses for 72 candidate noncoding transcripts.
| Results of expression analyses | No. of candidates | Positive embryos in situ | Positive RT-PCR |
|---|---|---|---|
| cDNA is full-length* | |||
| Spliced | 13 | 4 | 13 |
| Unspliced | 4 | 1 | Not tested |
| Nuclear RNA detected in situ† | |||
| Spliced | 1 | 1 | 1 |
| Unspliced | 2 | 2 | Not tested |
| cDNA is not full-length‡ | |||
| Spliced | 7 | 3 | 6§ |
| Unspliced | 21 | 10 | Not tested |
| Detected only by RT-PCR | |||
| Spliced | 6 | 0 | 6 |
| Transcript not detected¶ | |||
| Spliced | 4 | 0 | 0 |
| Unspliced | 14 | 0 | Not tested |
Candidates are categorized into nonoverlapping classes of experimental observation. Data on individual transcripts are presented in Tables 3 and 4.
Length is approximate, as determined by Northern blot. Additional, longer transcript species also were detected in two cases.
Transcript not detected by Northern.
Indicated by Northern analysis. Average transcript length according to Northern analyses was ≈6 kb; observed range in transcript length was ≈600 bases to > 10 kb.
See Table 4, GM01206.
Not detected by Northern, in situ, or RT-PCR.
We examined transcripts for evidence of splicing. Alignment of the 72 candidate cDNA sequences to the Drosophila genome predicted that 39 sequences represented spliced transcripts and contained an average of 1.6 introns each (Table 3). To evaluate whether computationally predicted splice sites were likely to reflect transcript processing in vivo, we subsequently analyzed each of these 39 transcripts for the presence of consensus 5′ donor-site and 3′ acceptor-site sequences and for the presence of a branch-point sequence and a polypyrimidine tract. This analysis predicted that 31 candidates were likely to represent spliced transcripts; predictions for the other 8 candidates represented either nonconsensus splice sites or misalignments to the genome (Table 3). In addition, the directionality of splice junctions allowed us to confirm the 5′ to 3′ orientation of the cDNAs predicted by the positions of poly(A)-RNA tails.
To facilitate evolutionary analyses, we searched for sequences that were homologous to our transcripts in two other Drosophila species by using blastn. Retaining for analysis the highest scoring pairs with expectation values of 1e-5 or less, we identified considerable homology between D. melanogaster candidate ncRNA transcripts and regions of the D. pseudoobscura genome, which diverged from D. melanogaster ≈25 million to 30 million years ago (22): 63 (86%) candidate ncRNA transcripts had conserved regions, with most transcripts showing homology along their entire lengths. Additional analysis indicated that regions reflecting noncoding transcript orthology were usually syntenic with neighboring gene annotations. We observed similarly strong conservation of nucleotide sequence when we repeated the analysis by using an assembly of the D. virilis genome for comparison. D. virilis is more distantly related to D. melanogaster than is D. pseudoobscura (22); accordingly, we found 44 transcripts (60%) that contained homologous regions. These comparative findings support the hypothesis that the candidate transcripts represent conserved genes. On average, the candidate D. melanogaster transcripts shared 60% sequence identity with D. pseudoobscura sequence and shared 61% with D. virilis, similar to the 61% identity reported in a comparison of representative D. melanogaster and D. pseudoobscura protein-coding sequences (23).
In the more distantly related honey bee, Apis mellifera, (www.hgsc.bcm.tmc.edu/projects/honeybee) and mosquito, Anopheles gambiae (24), genomes we found sequence conservation for 32 (44%) and 23 (32%) of candidate cDNAs, respectively. Even in these evolutionarily distant genomes, homologous regions encompassed most of the transcript lengths.
Likelihood of Translation. Because these 72 candidate ncRNA transcripts are structurally similar to protein-coding mRNAs, it was important to assess their potential for encoding polypeptides. To make this assessment, we first considered ORF lengths and initiating codons. Next, we measured whether the sequence of each ORF was more conserved compared with untranslated sequences from the same transcript. Finally, we used two interspecies comparative analysis methods, Ka/Ks and qrna, to ascertain whether codon structure in each transcript was significantly conserved.
Having calculated the longest possible Met-initiated translation for each transcript (Table 3), we next randomized each transcript sequence and again calculated the longest ORF. After six randomization trials, we found that for 37% of the transcripts the average longest ORF length of randomized sequence was the same or longer than that of the native, nonrandomized sequence (Table 3). For transcripts with native translations longer than those found in the average of randomized trials, the native ORFs were longer by an average of only 23 codons. We conclude that the short ORFs that occur in our candidate ncRNAs are similar in length to ORFs that occur by chance in random sequence.
We examined each transcript for the longest non-Met-initiated ORF to address two issues: first, the rare occurrence of non-Met-initiated translation, and second, the possibility that a cDNA may not represent the true 5′ end of a transcript and thus may not include a used start codon. Within our data set, the non-Met-initiated ORFs are rarely significantly longer than the 100-codon curation criterion we applied for Met-initiated reading frames (Table 3). In addition, we ultimately excluded from our analysis any candidate cDNA that, based on Northern experiments (see below), did not appear to correspond to a full-length transcript; thus, we are unlikely to have missed a significant portion of any ORF due to a truncation of the transcript.
Although the ORFs encoded by our candidate ncRNAs are indeed short and their translated sequences have no similarity to known proteins (blastx analysis of these sequences was conducted as part of the D. melanogaster 3.1 annotation pipeline) (12, 15), the possibility remains that some of these sequences encode novel small peptides. To assess this possibility, we asked whether sequence conservation in the D. pseudoobscura genome is greater within the ORF than in the remainder of the transcript. We compared blastn results for the non-ORF regions in each transcript with results for its ORF. Of the 63 transcripts conserved in D. pseudoobscura, only 14 displayed greater sequence conservation in their longest ORF than in the remainder of their sequence (Table 3).
We next examined the ORFs of our transcript data set for evolutionary conservation of codon structure. We calculated the Ka/Ks ratio for the longest Met-initiated translation for each sequence in our transcript data set, comparing nonsynonymous (Ka) to synonymous (Ks) substitutions in codon structure (25). Although it has been shown that nearly 95% of D. melanogaster protein coding exons have conserved coding information as determined by Ka/Ks (23), we were able to find only seven transcripts (10%) in our data set with statistically significant conservation of any codon structure within a transcript ORF (Table 3).
Even though Ka/Ks analysis indicated that the majority of our transcript data set does not have any conserved codon structure, we used an additional method to evaluate the coding potential of the ncRNA candidate transcripts. The qrna algorithm relies upon stochastic pair grammars to evaluate substitutions between aligned homologous sequences (19, 20). Based on the pattern of mutations between conserved regions, the software assigns each nucleotide to one of three states: protein-coding, structural RNA, and other (potentially novel ncRNA). qrna analysis indicated only seven candidate transcripts that contain any conserved sequence that approximates codon structure (Table 3). Three of the seven qrna transcripts that show possible conservation of potential protein-coding sequence also have Ka/Ks results indicating conservation of codon structure (Table 3). Of these three transcripts, only one has an ORF that is more conserved than its flanking untranslated sequence.
In the absence of functional evidence, the very short ORFs found in these long transcripts, coupled with a lack of consistent support for conserved reading frames by independent methods of comparative analysis, sustain the current classification of these transcripts as likely non-protein-coding.
RT-PCR and Microarray Analyses of Candidate Noncoding Transcripts. To verify that candidate cDNAs represent expressed and processed transcripts and to validate the predicted splice junctions, we tested each of 31 putatively spliced transcripts with a RT-PCR assay, applying primers designed to amplify across predicted exon boundaries to RNA pooled from a broad range of Drosophila stages (see Table 4, which is published as supporting information on the PNAS web site). For 26 of the 31 putatively spliced transcripts, the sequences of amplification products primed by oligonucleotide pairs flanking predicted exon boundaries verify at least one splice junction (Table 4; for example, see Fig. 1 A and B); in one case a candidate transcript could be detected by RT-PCR but represented unspliced RNA. The four remaining putatively spliced candidates were not detected under our RT-PCR conditions. For 26 spliced transcripts detectable in this assay, single RT-PCRs were primed from the 5′- and 3′-most exons of those transcripts. The entire structure of the candidate cDNA was validated in this manner for 21 transcripts (Table 4). The Affymetrix Drosophila Genome 2.0 Gene Chip expression array contains all of the candidate noncoding candidate transcripts described in this study; 92% of candidate transcripts are detected in at least one RNA sample by hybridization to this array (Table 4).
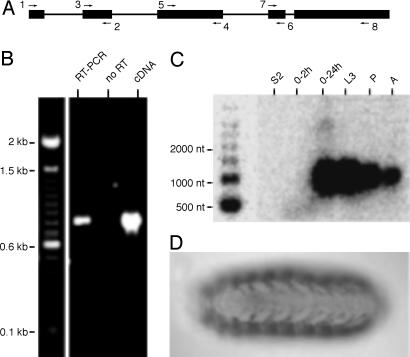
Fig. 1.
Experimental verification of candidate noncoding transcript RE28911, pncr003:2L.(A) Schematic diagram of predicted gene structure and PCR primer positions. The products of RT-PCR with primer pairs 1 and 2, 3 and 4, 5 and 6, and 7 and 8, respectively, were sequenced to confirm the gene structure. (B) PCR product generated by primers 1 and 8 in an reverse transcriptase-dependent RT-PCR is the same length as the product of a PCR by using the RE28911 cDNA as template; its identity and structure of product were confirmed by DNA sequencing. (C) Northern blot analysis of pncr010:2L; probe detects a transcript whose length corresponds to the RE28911 cDNA. Poly(A)+ RNA samples were isolated from Schneider S2 cultured cells, 0-to-2-h and 0-to-24-h embryos, late third-instar larvae (L3), pupae (P), and adults (A). (D) Antisense RNA in situ hybridization to whole-mount, late-stage embryo shows staining of muscle system, dorsal view, anterior to the left.
In Situ Hybridization Analysis of Candidate Noncoding Transcripts. To further confirm transcript expression and to determine developmental dynamics of transcript accumulation for candidate noncoding transcripts, antisense RNA in situ hybridization experiments were performed in whole-mount embryos for each of the 72 candidate cDNAs. Specific hybridization was detected for 21 candidates, or 29% of the data set (Table 4; for example, see Fig. 1D), approximately half the rate seen for protein-coding transcripts (26). Hybridization patterns ranged from that of simple maternal transcript deposition to a variety of complex zygotic restrictions in cell types derived from all germ layers. These data can be found in the Berkeley Drosophila Genome Project in situ expression database (www.fruitfly.org/cgi-bin/ex/insitu.pl).
Northern Blot Analysis of Candidate Noncoding Transcripts. Northern blot analysis was performed for each of the 72 candidates to determine which correspond to discrete RNA species and whether the cDNA clone representing each of those transcripts was full length (Table 4). Radiolabeled probes corresponding to 45 candidates detect transcripts on Northern blots of poly(A)+ RNA samples representing a broad range of developmental stages and a cultured Drosophila cell line (Fig. 1C). In 17 cases, the transcript length corresponded to that of the curated cDNA (for example, see Fig. 1C); two of these probes also detected transcripts whose molecular weights are higher (Table 4).
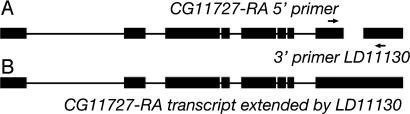
In the remaining 28 cases where a transcript was detected by Northern blotting, the RNA appears significantly longer than the corresponding cDNA, indicating that the predominant transcript represented by that clone is longer than the original cDNA. We designed RT-PCR experiments to amplify hypothetical RNAs that would bridge candidate ncRNA transcripts and proximal exons of protein-coding genes or nearby ESTs (for example, see Fig. 2), testing a single hypothetical gene structure for most of these 28 candidates. In five cases, we observed that the candidate transcript derived from an adjacent protein-coding annotation (data not shown), emphasizing the value of Northern analysis of transcripts first identified within EST collections.
Fig. 2.
RT-PCR links curated ncRNA candidate and proximal protein-coding annotation. (A) Schematic diagram of candidate ncRNA LD11130 and neighboring annotation CG11727. (B) Schematic diagram indicating revised annotation of CG11727 based on RT-PCR verification of transcript structure.
Assessment of Experimental Results and Curation of pncr Genes. Our criteria for experimental validation of a candidate ncRNA transcript required that the transcript be detected as a discrete species by Northern blot analysis and that its length agree with the curated cDNA clone; in this circumstance, the cDNA sequence can be analyzed with confidence that it represents a complete or nearly full-length transcript. We note that in principle, candidate transcripts not meeting these strict experimental validation requirements may still represent ncRNAs; however, they will require additional investigation. Seventeen of the 72 candidates considered in this study fulfilled our criteria (Tables 1 and 4), constituting a set of putative ncRNA genes. With evolutionary analyses arguing strongly against conserved protein-coding functions for these 17 genes, we have named them putative noncoding RNA (pncr) (Table 2). Overall, spliced and unspliced candidates were nearly equally likely to be detected in Northern and in situ assays. However, the 31 spliced candidates were categorized as previously unrecognized pncr genes at greater than four times the rate of the unspliced ones (Table 1). Six of the 17 pncr transcripts were detected by embryo in situ hybridization.
Table 2. Interspecies comparative analyses of pncr genes.
|
Dp
|
Dv
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene name | BDGP cDNA ID | cDNA length, bp | Spliced | Dm ORF len. | Dp Ka/Ks* | Dv Ka/Ks† | Dp QRNA‡ | Dm embryo in situ | BLASTN score (%)§ | Northern¶ | Embryo in situ¶ | BLASTN expect (%)§ | Northern |
| pncr001:3R | LD11162 | 1,549 | - | 49 | 0.1156 | 0.1393 | O1040 | Zyg | 5.0e-39 (98)∥ | Pos. | Zyg** | 8.0e-38 (92) | Pos. |
| pncr002:3R | LP03188 | 553 | + | 59 | No TB | No TB | O475 | ND | 7.4e-40 (90)∥†† | Pos. | Zyg | 1.1e-08 (76) | Pos. |
| pncr003:2L | RE28911 | 1,005 | + | 44 | No TB | No TB | O856 | Zyg | 7.3e-67 (100) | Pos. | Zyg** | 18.3e-12 (74) | Pos. |
| pncr004:X | RE54004 | 562 | + | 26 | No TB | No TB | O406 | ND | 1.2e-12 (94) | Pos. | ND | No Dm ortho‡‡ | Pos. |
| pncr005:2R | RE63504 | 331 | + | 38 | No TB | No TB | O96 | ND | 2.4e-18 (56)†† | Pos. | Mat/Zyg | 2.0e-15 (47) | Pos. |
| pncr006:X | RH45340 | 407 | + | 48 | 0.0488 | No TB | P16O129 | Mat/Zyg | 1.9e-15 (31) | Pos. | Mat/Zyg** | 6.1e-08 (64) | Pos. |
| pncr007:3R | RH63361 | 424 | - | 62 | Fχ2 | Fχ2 | Q72 | ND | 4.6e-19 (84) | Pos. | Mat | 1.0e-10 (99) | Pos. |
| pncr008:3L | RH62830 | 326 | + | 40 | 0.1831 | 0.0527 | O259 | ND | 1.3e-11 (79) | Pos. | ND | 1.2e-08 (64) | Neg. |
| pncr009:3L | RH57193 | 981 | + | 33 | 0.2853 | No TB | O684 | Mat/Zyg | 2.5e-08 (39) | Neg. RT-PCR | Neg. RT-PCR | 2.9e-10 (73) | Pos. |
| pncr010:3L | AT24650 | 578 | + | 77 | No TB | No TB | No NT A | ND | No ortho. | — | — | No ortho | — |
| pncr011:3L | GH03576 | 931 | + | 38 | No TB | No TB | O326 | ND | 2.6e-09 (76)∥†† | Neg. RT-PCR | Neg. RT-PCR | No ortho | — |
| pncr012:2L | GH14469 | 695§§ | + | 40 | No TB | No TB | O518 | ND | 1.5e-06 (90)∥ | Neg. RT-PCR | Neg. RT-PCR | No ortho | — |
| pncr013:4 | GM01028 | 1,157§§ | + | 70 | 0.1056 | No TB | No NT A | Mat | No ortho∥ | — | — | No ortho | — |
| pncr014:3L | LD13184 | 2,222 | - | 88 | No TB | No TB | O992 | ND | 3.8e-10 (36) | Neg. RT-PCR | Neg. RT-PCR | No ortho | — |
| pncr015:3L | LP12023 | 313 | - | 59 | No TB | No TB | No NT A | ND | No ortho. | — | — | No ortho | — |
| pncr016:2R | RH09485 | 518 | + | 40 | No TB | No TB | No NT A | ND | No ortho. | — | — | No ortho | — |
| pncr017:3R | SD10988 | 1,415 | + | 80 | No TB | No TB | O1182 | ND | 1.4e-29 (91) | —¶¶ | —¶¶ | No ortho | — |
Analyses of the 17 putative noncoding RNA (pncr) transcripts that appeared on a Northern blot as discrete species, matching the lengths of the corresponding sequenced cDNAs. D. melanogaster (Dm) transcript length, splicing (confirmed by RT-PCR), longest Met-initiated ORF, Ka/Ks, and QRNA result, and embryo in situ hybridization results are shown, as well as sequence similarity of D. melanogaster transcripts to the genomes of D. pseudoobscura (Dp) and D. virilis (Dv) and experimental results in these two species. BDGP, Berkeley Drosophila Genome Project; Zyg, zygotic; Pos., positive; Neg., negative; No TB, no TBLASTN; ND, not detected; Mat, maternal; F χ2, failed χ2 test; No NT A, no NT ALIGN; —, not tested.
Values <0.5 (italics) signify conservation of condon structure; sequences that fail χ2 data fitness test or fail to produce a protein alignment are noted.
As in *, with D. virilis used as the comparative genome.
P, protein coding (italics); Q, RNA secondary structure; O, other. The number after the letter indicates the number of nucleotides assigned by QRNA to this category; sequences that fail to produce a nucleotide alignment are noted (no NT ALIGN).
The fraction of the D. melanogaster sequence (with a BLASTN expectation < 1e-05) aligned as a percentage of transcript length is given in parentheses; transcripts lacking nonrepetitive homology above this cutoff are labeled “no ortholog” (No ortho).
RT-dependent RT-PCR products were used as probes for Northern and in situ analyses. Cases where an orthologous transcript was not detected by RT-PCR are designated “negative RT-PCR” (Neg.), and Northern and in situ experiments were not performed. In cases where no putative ortholog was observed by sequence analysis or where homology was to repetitive regions, RT-PCR was not attempted.
D. melanogaster transcript sequence conserved in A. mellifera.
Patterns of expression for D. pseudoobscura transcripts are similar to those observed in D. melanogaster.
D. melanogaster transcript sequence conserved in A. gambiae.
This D. virilis ortholog could not be identified by homology to the D. melanogaster cDNA; instead, the D. pseudoobscura orthologous sequence was used for this purpose.
Additional longer RNA species are detected by Northern analysis.
D. melanogaster SD10988, pncr017:3R was only detected in Schneider S2 cells; no RT-PCR experiment was performed for this putative D. pseudoobscura ortholog.
An additional three candidates, falling into a second class of experimental observation, seem worthy of interest with respect to potential noncoding functions (Table 1). These transcripts are not detected in any sample in our Northern assay; however, in situ analyses reveal specific, punctate hybridization to subnuclear foci that may represent the nascent transcripts (Tables 1 and 4). Such observations may be consistent with a noncoding function for a subfragment of the transcript represented by the curated cDNA. In this regard, it is worth noting that whereas microRNAs are derived from long polyadenylated precursors (27), none of our 72 candidates correspond to the 43 genomic locations known to encode microRNAs (28).
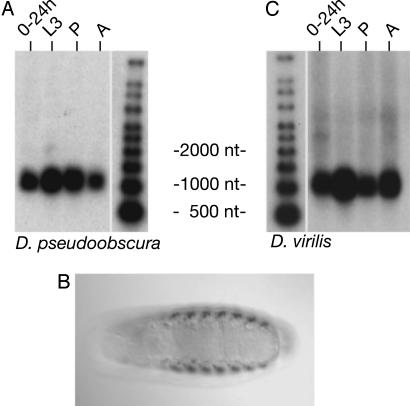
pncr Orthologs in D. pseudoobscura and D. virilis. Of the 17 D. melanogaster pncr genes, we were able to identify orthologous sequences in 13 cases in D. pseudoobscura and 9 cases in D. virilis (Table 2; for alignment of one example, see Fig. 4, which is published as supporting information on the PNAS web site). We used syntenic highest-scoring pairs to construct models of putative orthologous transcripts for both of these species. We next investigated whether these conserved sequences were expressed in D. pseudoobscura and D. virilis as discrete transcripts. By applying primers based on gene models to RNA isolated from D. pseudoobscura and D. virilis, RT-PCR was used to generate cDNAs whose identities were verified by sequencing. We were able to detect a putative ortholog in this way for 9 of the 13 D. melanogaster pncr transcripts for which genomic sequence conservation was observed. We used these RT-PCR products as probes for Northern blots in both related Drosophila species and in each case detected an orthologous transcript expressed in at least one of these two related Drosophila species; for seven pncr genes, an orthologous transcript was detected in both related species. The D. pseudoobscura and D. virilis orthologs exhibited lengths that roughly correspond to their cognate D. melanogaster transcripts (Data Sets 4 and 5). Stage-specific expression of transcripts was often observed to mirror D. melanogaster patterns (for example, see Fig. 3; other data not shown). In situ hybridizations also were carried out in D. pseudoobscura embryos (Table 2); expression patterns of three orthologs were nearly identical to those of respective D. melanogaster transcripts (for example, see Fig. 3).
Fig. 3.
Experimental verification of orthologs of candidate noncoding transcript RE28911, pncr003:2L in D. pseudoobscura and D. virilis. (A) D. pseudoobscura ortholog of pncr010:2L is detected on a Northern blot by using poly(A)+ RNA isolated from 0-to-24-h embryos, late third-instar larvae (L3), pupae (P), and adults (A). (B) Antisense RNA in situ hybridization to whole-mount, late-stage D. pseudoobscura embryo shows staining of muscle system, dorsal view, anterior to the left. (C) D. virilis ortholog of pncr010:2L is detected by Northern analysis; RNA sample sources are as in A. For alignment of orthologous sequences, see Fig. 4.
Discussion
Research over the past two decades in Drosophila has identified eight mRNA-like ncRNAs (roX1 and roX2, Hsr, pgc, bxd, αγ-element, iab-4, and bft) (2, 4–7, 29). In this work, starting from a set of 72 computationally curated candidates, we have identified 17 additional mRNA-like ncRNAs that produce distinct transcripts, tripling the number of described mRNA-like ncRNAs in Drosophila. No systematic approach to gene-finding for long, noncoding mRNA-like RNA genes in fruitfly has been previously reported. We describe previously unrecognized candidate ncRNAs that are both spliced and unspliced and, in some cases, conserved in other Dipteran genomes. We examined the expression and structures of these genes by multiple experimental approaches and demonstrated the expression of discrete orthologous transcripts of a subset of these genes in two other Drosophila species. Further investigation of the pncr set of Drosophila genes classified by this study may reveal novel RNA-mediated functions among their transcripts.
With the aim of evaluating independent curations of new ncRNAs, we applied our experimental approach to a small sampling of purported ncRNAs in mouse from the RIKEN fantom2 collection (http://fantom2.gsc.riken.go.jp) of recently identified cDNA transcripts (11). Information on these efforts can be found in Supporting Appendix, which is published as supporting information on the PNAS web site, and Data Sets 6 and 7.
Determination of protein-coding status is the most challenging task in noncoding gene curation. Pseudogenes, truncated clones, or errors in sequence determination could give rise to transcript sequences with reduced coding capacity. Additionally, a short ORF might still be translated to produce a small peptide. To minimize cDNA artifacts, we started with high-quality sequences and screened out truncated and reversed clones by using various computational and experimental methods. We then used homology searches to eliminate pseudogenes and extensive comparative studies to assess for conservation of protein-coding potential from the short ORFs. It is important to point out, however, that the argument that the pncr transcripts identified in this study are noncoding relies solely on the lack of positive evidence supporting the alternative hypothesis that these transcripts encode proteins. Demonstration of any RNA-mediated functions awaits further investigation.
Our efforts to curate long, mRNA-like ncRNA genes were nonsaturating because of our reliance on existing cDNA resources. EST and cDNA sequences have proven to be extremely valuable for the identification of protein-coding genes and their alternatively spliced transcripts (12, 30, 31). As demonstrated by this work and by ncRNA curation efforts in plant and mouse (10, 11), these resources are even more important for the identification and characterization of ncRNA genes. Although genomic hybridization technologies have begun to provide extensive evidence for transcription not accounted for by annotated protein-coding genes (9, 32), production of high-quality EST and cDNA sequence data, and experimental data such as Northern analyses, is essential for distinguishing which of these transcribed sequences encode discrete RNAs.
Because we have used EST and cDNA sequences as a source for candidate ncRNA curation, any estimate that we make of the total number of ncRNAs encoded in the Drosophila genome will depend on the extent of EST representation in our data set. In this regard, it is worth noting that the D. melanogaster control genes roX1, roX2, Hsr-w, and pgc (the most well characterized fruitfly mRNA-like ncRNAs) each have multiple ESTs (Table 3). However, >20% of annotated protein-coding genes, as well as most of the other described mRNA-like ncRNA genes, did not have corresponding ESTs at the time of the Release 3.1 reannotation (12).
Analysis of fruitfly 5′ ESTs that are not associated with annotations reveals almost 500 distinct clusters containing one or more spliced EST reads (J. Carlson, personal communication). We expect that analyses of these sequences by using the methods we have used here to characterize our 193 initial candidates will identify additional mRNA-like ncRNAs similar in properties to the 17 pncr transcripts we describe. Taking all these factors into account, our guess is that there may be a total of 50–100 long, mRNA-like ncRNA genes encoding discrete transcripts in the D. melanogaster genome. Based on the candidates we have examined in this work, many of these genes will be evolutionarily conserved, suggesting that they have important biological functions.
Supplementary Material
Acknowledgments
We thank Yuji Kageyama and colleagues for communication of unpublished results. We thank Pavel Tomancak, Amy Beaton, Elaine Kwan, and Richard Weiszmann for embryo in situ analysis; Garson Tsang for performing microarray experiments; and Ken Wan for DNA sequencing support. Chris Mungall, Mark Yandell, Joe Carlson, Chris Smith, George Hartzell, Josh Kaminker, Simon Prochnik, Mark Stapleton, and Shen-Qiang Shu contributed software and database resources and advice on their use. The manuscript was improved as a result of the comments of Sean Eddy. This work was supported by National Institutes of Health Grants HG002673 (to S.E.C.) and HG007500 (to G.M.R.) and by the Howard Hughes Medical Institute.
Author contributions: J.L.T., A.M.B., C.W.S., S.E.C., and G.M.R. designed research; J.L.T., A.M.B., G.D., M.E.-H., and C.W.S. performed research; J.L.T., A.M.B., G.D., M.E.-H., C.W.S., S.M., and G.M.R. analyzed data; J.L.T., A.M.B., and G.M.R. wrote the paper; and A.M.B., S.E.C., and G.M.R. supervised research.
Abbreviation: ncRNA, noncoding RNA.
References
- 1.Erdmann, V. A., Szymanski, M., Hochberg, A, Groot, N. & Barciszewski, J. (2000) Nucleic Acids Res. 28, 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipshitz, H. D., Peattie, D. A. & Hogness, D. S. (1987) Genes Dev. 1987, 1, 307–322. [DOI] [PubMed] [Google Scholar]
- 3.Celniker, S. E. & Rubin, G. M. (2003) Annu. Rev. Genomics Hum. Genet. 4, 89–117. [DOI] [PubMed] [Google Scholar]
- 4.Kelley, R. L. (2004) Dev. Biol. 269, 18–25. [DOI] [PubMed] [Google Scholar]
- 5.Prasanth, K. V., Rajendra, T. K., Lal, A. K. & Lakhotia, S. C. (2000) J. Cell Sci. 19, 3485–3497. [DOI] [PubMed] [Google Scholar]
- 6.Nakamura, A., Amikura, R., Mukai, M., Kobayashi, S. & Lasko, P. F. (1996) Science 274, 2075–2079. [DOI] [PubMed] [Google Scholar]
- 7.Martinho, R. G., Kunwar, P. S., Casanova, J. & Lehmann, R. (2004) Curr. Biol. 14, 159–165. [DOI] [PubMed] [Google Scholar]
- 8.Kapranov, P., Cawley, S. E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. & Gingeras, T. R. (2002) Science 296, 916–919. [DOI] [PubMed] [Google Scholar]
- 9.Stolc, V., Gauhar, Z., Mason, C., Halasz, G., van Batenburg, M. F., Rifkin, S. A., Hua, S., Herreman, T., Tongprasit, W., Barbano, P. E., et al. (2004) Science 306, 655–660. [DOI] [PubMed] [Google Scholar]
- 10.MacIntosh, G. C., Wilkerson, C. & Green, P. J. (2001) Plant Physiol. 127, 765–776. [PMC free article] [PubMed] [Google Scholar]
- 11.Numata, K., Kanai, A., Saito, R., Kondo, S., Adachi, J., Wilming, L. G., Hume, D. A., Hayashizaki, Y., Tomita, M., the RIKEN Genome Exploration Research Group & Genome Science Laboratory members (2003) Genome Res. 13, 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misra, S., Crosby, M. A., Mungall, C. J., Matthews, B. B., Campbell, K. S., Hradecky, P., Huang, Y., Kaminker, J. S., Millburn, G. H., Prochnik, S. E., et al. (December 31, 2002) Genome Biol. 3, research0083.1–research0083.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stapleton, M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, K., et al. (2002) Genome Biol. 3, research80.1–research80.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celniker, S. E., Wheeler, D. A., Kronmiller, B., Carlson J. W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S. P., Frise, E., et al. (2002) Genome Biol. 3, research79.1–research79.14. [Google Scholar]
- 15.Mungall, C. J., Misra, S., Berman, B. P., Carlson, J., Frise, E., Harris, N., Marshall, B., Shu, S., Kaminker, J. S., Prochnik, S. E., et al. (2002) Genome Biol. 3, research81.1–research81.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Florea, L., Hartzell, G., Zhang, Z., Rubin, G. M. & Miller, W. (1998) Genome Res. 8, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards, S., Liu, Y., Bettencourt, B. R., Hradecky, P., Letovsky, S., Nielsen, R., Thornton, K., Hubisz, M. J., Chen, R., Meisel, R. P., et al. (2005) Genome Res. 15, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang, Z. (2000) Phylogenetic Analysis by Maximum Likelihood (paml) 3.0. (University College, London).
- 19.Rivas, E. & Eddy, S. R. (2001) BMC Bioinformatics 2, 8.1–8.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivas, E., Klein, R. J., Jones, T. A. & Eddy, S. R. (2001) Curr. Biol. 11, 1369–1373. [DOI] [PubMed] [Google Scholar]
- 21.Rozen, S. & Skaletsky, H. (2000) in Bioinformatics Methods and Protocols: Methods in Molecular Biology, eds. Krawetz, S., Misener, S. & Totowa, N. J. (Humana, Clifton, NJ), pp. 365–386.
- 22.Piccin, A., Couchman, M., Claytona, J. D., Chalmers, D., Costab, R. & Kyriacoua, C. P. (2000) Genetics 154, 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergman, C. M., Pfeiffer, B. D., Rincon-Limas, D. E., Hoskins, R. A., Gnirke, A., Mungall, C. J., Wang, A. M., Kronmiller, B., Pacleb, J., Park, S., et al. (2002) Genome Biol. 3, research86.1–research86.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., Wincker, P., Clark, A. G., Ribeiro, J. M., Wides, R., et al. (2002) Science 298, 129–149.12364791 [Google Scholar]
- 25.Nekrutenko, A., Makova, K. D. & Li, W. H. (2002) Genome Res. 12, 198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomancak, P., Beaton, A., Weiszmann, R., Kwan, E., Shu, S., Lewis, S. E., Richards, S., Ashburner, M., Hartenstein, V., Celniker, S. E., et al. (2002) Genome Biol. 3, research88.1–research88.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H. & Kim, V. N. (2004) EMBO J. 23, 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai, E. C., Tomancak, P., Williams, R. W. & Rubin, G. M. (2003) Genome Biol. 4, research42.1–research42.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.FlyBase Consortium (2002) Nucleic Acids Res. 30, 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas, B. J., Volfovsky, N., Town, C. D., Troukhan, M., Alexandrov, N., Feldmann, K. A., Flavell, R. B., White, O. & Salzberg, S. L. (2002) Genome Biol. 3, research29.1–research29.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imanishi, T., Itoh, T., Suzuki, Y., O'Donovan, C., Fukuchi, S., Koyanagi, K. O., Barrero, R. A., Tamura, T., Yamaguchi-Kabata, Y., Tanino, M., et al. (2004) PLoS Biol. 2, 856–875. [Google Scholar]
- 32.Ishkanian, A. S., Malloff, C. A., Watson, S. K., DeLeeuw, R. J., Chi, B., Coe, B. P., Snijders, A., Albertson, D. G., Pinkel, D., Marra, M. A., et al. (2004) Nat. Genet. 36, 299–303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.