Abstract
Nonsense-mediated RNA decay (NMD) is a crucial post-transcriptional regulatory mechanism that recognizes and eliminates aberrantly processed transcripts, and mediates the expression of normal gene transcripts. In this study, we report that in the filamentous fungus Neurospora crassa, the NMD factors play a conserved role in regulating the surveillance of NMD targets including premature termination codon (PTC)-containing transcripts and normal transcripts. The circadian rhythms in all of the knockout strains of upf1-3 genes, which encode the Up-frameshift proteins, were aberrant. The upf1 knockout strain displays a shortened circadian period, which can be restored by constantly expressing exogenous Up-frameshift protein 1 (UPF1). UPF1 regulates the circadian clock by modulating the splicing of the core clock gene frequency (frq) through spliceosome and spliceosome-related arginine/serine-rich splicing factors, which partly account for the short periods in the upf1 knockout strain. We also demonstrated that the clock genes including White Collar (WC)-1, WC-2, and FRQ are involved in controlling the diurnal growth rhythm, and UPF1 may affect the growth rhythms by mediating the FRQ protein levels in the daytime. These findings suggest that the NMD factors play important roles in regulating the circadian clock and diurnal growth rhythms in Neurospora.
Keywords: NMD, Neurospora crassa, circadian clock, frq, alternative splicing, diurnal rhythm
NONSENSE-MEDIATED RNA decay (NMD) governs the surveillance of aberrantly processed transcripts and controls the expression of normal transcripts (Kervestin and Jacobson 2012; Schweingruber et al. 2013; Fatscher et al. 2015). UPF1–3 in mammals and Saccharomyces cerevisiae, and the homologs SMG (suppressor with morphogenetic effect on genitalia) -2, SMG-3, and SMG-4 in Caenorhabditis elegans, are the core NMD components. Some other factors including SMG proteins and eukaryotic release factors (eRFs) factors, which vary in different species, are also critical components or regulators for NMD. Among NMD factors, UPF1 possesses RNA helicase activity and it is the central regulator in NMD. A number of SMG proteins (SMG-1, SMG5, SMG6, and SMG7) mediate the phosphorylation and dephosphorylation of UPF1 (Rehwinkel et al. 2006; Isken and Maquat 2008). During NMD, the target transcripts containing premature termination codons (PTCs) are first recognized by UPF1 after the transition from the nucleus to the cytoplasm. In mammals, during the pioneer round of translation, the exon junction complex (EJC) located downstream of the termination codon retains its association with the PTC-containing mRNA and recruits UPF1 and other NMD factors. UPF1 then recruits the machineries responsible for mRNA decay to eliminate the aberrant mRNAs. The pioneer round of translation mechanism involves the cap-binding complex (CBC), which binds to the 5′-m7GpppN cap structure of newly synthesized mRNAs. CBC interacts with UPF1 and enhances NMD. In contrast, in S. cerevisiae, NMD appears to occur during any round of translation (Chang et al. 2007; Chamieh et al. 2008; Maquat et al. 2010). Recently, in the filamentous fungus Neurospora crassa, it has been demonstrated that NMD could be elicited by transcripts bearing upstream open reading frames (uORFs) or 3′-UTR introns. During RNA surveillance, NMD factors are sufficient to eliminate transcripts with uORFs, while additional factors including EJC and CBC factors are required to eliminate transcripts harboring 3′-UTR introns (Zhang and Sachs 2015).
Circadian clocks control a broad spectrum of physiological and behavioral processes including development and growth, in which circadian clocks have been identified in most of the interrogated organisms across different kingdoms (Bellpedersen et al. 2005; Baker et al. 2012). In Neurospora, White Collar 1 (WC-1) and WC-2 are the two positive components of the circadian clock, and FREQUENCY (FRQ) is the negative element (Baker et al. 2012). The positive and negative elements constitute a transcription–translational negative feedback loop to drive circadian rhythms at the molecular level (Guo et al. 2010; Hunt et al. 2010; Baker et al. 2012; Hurley et al. 2013; Lauinger et al. 2014).
In recent years, increasing data have shown that post-transcriptional regulation plays multiple roles in mediating the expression of clock genes and maintaining the proper rhythmicity. Post-transcriptional regulation occurs after or along with the process of transcription at the RNA level, which includes splicing, 5′ and 3′ processing, transporting, localization, surveillance, and turnover. Almost all these processes have been linked to the regulation of the circadian clock in various species (Guo et al. 2009; Kojima et al. 2011; Lim and Allada 2013; Nolte and Staiger 2015; Zhang et al. 2015).
It has been reported that the NMD is involved in the regulation of the circadian clock in various species (Morgan and Feldman 1997, 2001; Schoning et al. 2007; Weischenfeldt et al. 2012). In Neurospora, the prd-6 mutant bears a mutation in the locus of upf1, which exhibits a short circadian period and abnormality in the temperature compensation of its circadian rhythms (Morgan and Feldman 1997, 2001; Adhvaryu et al. 2016). The involvement of UPF1/PRD-6 in FRQ-less oscillator(s) (FLOs) has also been investigated. On mediums supplemented with choline or geraniol, lack of PRD-6 led to shorter FRQ-less conidiation periods in a frq-null background (Lombardi et al. 2007; Adhvaryu et al. 2016). Whereas, when grown on low choline medium or treated with heat pulse, no remarkable effects on FRQ-less rhythms were observed in the strain lacking prd-6 (Lombardi et al. 2007; Adhvaryu et al. 2016).
In Arabidopsis, Atgrp7 or Atgrp8, both of which encode glycine-rich RNA-binding proteins, are two clock-controlled genes. These two genes harbor PTCs and their expression is under NMD control (Heintzen et al. 1997; Staiger et al. 2003; Schoning et al. 2007). A transcriptomic analysis revealed a widespread occurrence of PTCs in Arabidopsis clock genes that is well-conserved across different plants (Filichkin and Mockler 2012). For instance, CCA1, which encodes a negative element of the Arabidopsis clock, comprises a PTC and retention of the PTC-containing intron leads to the translation of nonfunctional products (Filichkin et al. 2010), and the retention of the PTC-containing intron of CCA1 transcripts is subject to change under some specific environmental stress conditions (Filichkin and Mockler 2012). Additionally, the 3′-UTR regions of transcripts of the clock genes LHY, LCL1, and REV8/LCL5 contain introns, indicating that they are potential NMD substrates (Filichkin and Mockler 2012). These findings suggest that the NMD factors may play an important role in regulating the expression and function of both clock and clock-controlled genes.
Although certain NMD-targeted clock genes have been identified, the role and mechanism of NMD factors in controlling circadian rhythms remain largely unclear. In this work, we demonstrate that the NMD factors play conserved roles in regulating the elimination of PTC-containing transcripts, and we also show that NMD factors, especially UPF1, are also critical for the regulation of the circadian clock in a complex fashion.
Materials and Methods
Strains, constructs, and race tube assay
301-5 (bd, a) was used as the wild-type (WT) strain in this study. The ku70RIP strain was used as the host strain to generate different Hygromycin B (hph) knockout transformants (Zhao et al. 2009). Microconidia purification to obtain homokaryon progeny was conducted with 0.5-μm filter membranes (Ebbole and Sachs 1990). For the strains that require the addition of quinic acid (qa) in the mentioned experiments (Geever and Giles 1983), the concentration of qa is 0.01 M. The Neurospora crassa unit (NCU) numbers of the Neurospora upf1, upf2, and upf3 genes are NCU04242, NCU05267 and NCU03435.
To generate the upf1KO, bar-qa-upf1 strain, which constantly expresses upf1 in the background of a upf1-null strain, a fragment bearing the qa-2 promoter and upf1 ORF was inserted into the BamHI–NotI sites of pBARKS1 (Morgan et al. 2003). The derived qa-upf1-bar construct was transformed into the upf1KO strains and transformants (upf1KO, bar-qa-upf1) were selected on the media supplemented with basta/ignite (200 μg/ml). The resistance against basta/ignite was conferred by the bar gene (Larrondo et al. 2012). The transformants were further validated by a series of biochemical analyses described in context below.
The promoter region of the vivid (vvd) gene was used to drive the light-induced expression of either FRQ protein (Hurley et al. 2012). For these purposes, the vvd promoter was fused with the frequency (frq) ORF and cloned into pRMP62. The derived constructs of Pvvd-frq was transformed into frq10 (bd, his-3) at the his-3 locus, to generate the frq10, Pvvd-frq strain (Aronson et al. 1994).
Race tubes are long glass tubes containing a layer of solid media. In a race tube assay, the Neurospora conidia is inoculated on the surface of the solid media at one end of the race tube and it grows toward the other end. During growth, Neurospora yields conidiation bands, which are controlled by the circadian clock. In this way, the race tube assay allows for the calculation of the circadian parameters including periods, amplitudes, and phases (Baker et al. 2012). For the study of growth rhythms in light/dark (LD)12:12 cycles (light 12 hr vs. dark 12 hr, repeatedly), the growth fronts of Neurospora hyphae were labeled at the time points of LD transition points, twice a day.
All the mentioned strains are listed in Supplemental Material, Table S1 in File S1.
DNA, RNA, and protein analysis
DNA was extracted with CTAB solution (Rehman et al. 2008). RNA extraction and northern blot analyses were performed as described previously (Aronson et al. 1994). Primers spanning the PTC-containing introns were synthesized to amplify the spliced and unspliced transcripts that contain PTCs in the unspliced transcripts. In measuring the mRNA stability of frq, thiolutin (Sigma [Sigma Chemical], St. Louis, MO) was used to inhibit transcription and the concentration was 12 μg/ml (Guo et al. 2009).
Protein extraction, quantification, western blot analysis, and co-immunoprecipitation (Co-IP) assays were performed as described previously (Görl et al. 2001). To inhibit translation, cycloheximide (CHX; Sigma) was used to treat samples for 3 hr and the final concentration was 200 μg/ml (Linde et al. 2007).
RNA-seq and analysis
The upf1KO and WT strains were grown and the RNA samples were from independent triplicate pools, respectively. The pooled RNA samples were processed to prepare the mRNA-seq library using the standard Illumina protocol. The sequencing was performed on an Illumina HiSeq 2000 at Shanghai Biotechnology Corporation, China. The read lengths were > 90 nt. The parameters of number of clean reads, clean reads ratio, and mapping ratio were 3.12G, 84.1%, and 94.7% for WT and 6.24G, 85.8%, and 95.6% for the upf1KO strain, respectively. Tophat was used as the aligner to map the reads to the reference genome [N. crassa OR74A (NC12)]. Cufflinks was used to reconstruct the transcripts and estimate the gene expression levels (Trapnell et al. 2010).
Generation and validation of UPF1 and UPF2 antibodies
The entire ORFs of the Neurospora upf1 and upf2 genes were cloned into pET-28a (+) and expressed in the BL21 (DE3) Escherichia coli strain. Purified His-tagged proteins were used to immunize rabbits. The obtained antibodies were validated by western blot analysis with the knockout strains.
Sucrose fractionation analysis
Sucrose density gradients (10–30%) were prepared, and 4 mg total protein samples were loaded for each analysis. The gradients were centrifuged at 175,000 × g (SW-40 Rotor; Beckman) for 18 hr at 4°. Twelve equal fractions were collected, and 450 μl of each fraction was subjected to an RNA analysis. The samples were treated with DNase I prior to RT-PCR to determine the U5 levels (Fury and Zieve 1996; Zhang et al. 2015).
Luciferase reporter assay
The luciferase reporter construct (frq-luc-bar) (from Yi Liu, and originally from Jay Dunlap) was transformed into the corresponding sites of pBARKS1 of the described strains. The transformants were screened using basta/ignite (200 μg/ml) resistance conferred by the bar gene (Zhou et al. 2013). The luciferase signal was recorded with LumiCycle (Actimetrics) and analyzed as previously described (Gooch et al. 2008). The luciferase signals were normalized and analyzed as previously described (Zhou et al. 2013).
Statistics
Significant differences were determined by applying the Student’s t-tests and two-way ANOVA analysis. Data are mean ± SD or mean ± SE as indicated. n ≥ 3. * represents the P-value of the statistical tests being less than a significance level of 0.05 (P < 0.05); ** P < 0.01, and # P < 0.001. n.s. denotes no significance.
Data availability
Strains used are available upon request. The RNA-seq data sets are available at the GEO database (GSE97157).
Results
Roles of UPF genes in the NMD pathway
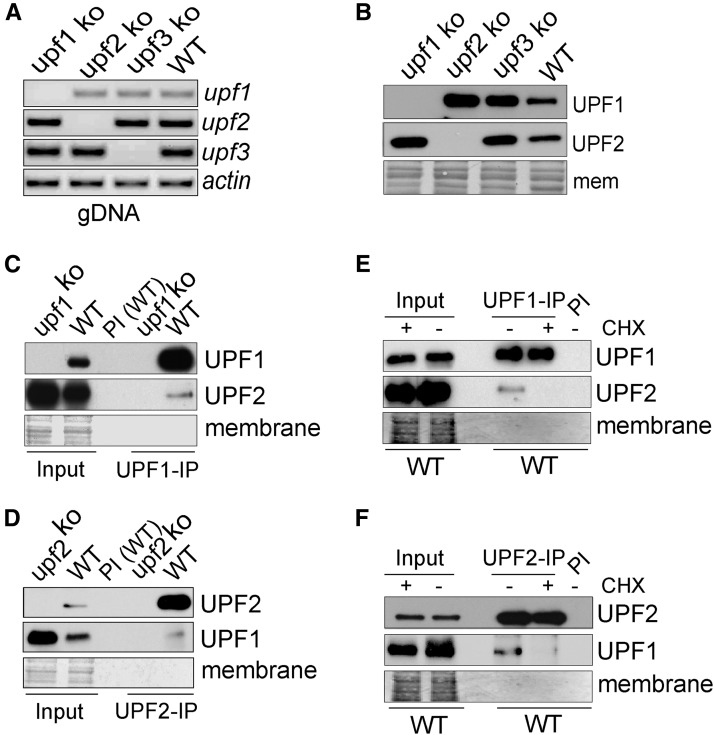
We first validated the involvement of UPF genes in the Neurospora NMD pathway. To this end, the knockout strains of upf1–3 were constructed by a homologous recombination strategy and referred to as upf1KO, upf2KO and upf3KO. The obtained knockout strains were further purified by microconidia filtration to obtain homeokaryons (Ebbole and Sachs 1990). PCR amplification of the genomic DNA showed that upf1–3 genes were deleted, respectively (Figure 1A), and RT-PCR amplification of cDNAs from the knockout strains showed no detectable expression of these genes in corresponding knockout strains (Figure S1 in File S1). Western blot analyses were further conducted to measure the expression of UPF1 and UPF2 proteins with the antibodies generated in this work, and the results consistently showed no detectable expression (Figure 1B). The Neurospora strains with individual deletion of upf1, upf2 and upf3 were viable, although they grew slowly. Similarly, UPF1 is not required for the viability of S. cerevisiae and C. elegans, but a lack of UPF1/SMG-2 led to defects in fermentation in S. cerevisiae and reproduction abnormality in C. elegans (Culbertson et al. 1980; Hodgkin et al. 1989).
Figure 1.
Characterization of UPF1 and UPF2 interaction. (A) Results of PCR amplification of gDNA samples from the indicated ko strains. Actin served as control. (B) Western blot results of UPF1 and UPF2 in upf1–3 knockout strains and WT. Membrane stained with amido black served as control. (C and D) Co-IP results demonstrating that UPF1 is in complex with UPF2. The results were IP with UPF1 antibody (C) and UPF2 antibody (D). IP with the preimmune (PI) serum was conducted as a control. (E and F) CHX treatment inhibits the association between UPF1 and UPF2. The results were IP with UPF1 antibody (E) and UPF2 antibody (F) IP with the PI serum was conducted as a control. Amido black staining served as a loading control. CHX, cycloheximide; Co-IP, co-immunoprecipitation; gDNA, genomic DNA; IP, immunoprecipitation; ko, knockout; PI, preimmune; WT, wild-type.
The association between NMD factors is required for the processing of PTC-containing transcripts (He et al. 1997; Kurosaki et al. 2014; Fatscher et al. 2015). We used the UPF1 and UPF2 antibodies to test the association between UPF1 and UPF2, and the Co-IP results showed that UPF1 and UPF2 can be reciprocally pulled down (Figure 1, C and D and Figure S2 in File S1). The first run of translation is essential for NMD, and the inhibition of translation, e.g., by CHX, causes the repression of NMD (Ishigaki et al. 2001; Zhang and Sachs 2015). We treated the cultured strains with CHX and then extracted proteins and conducted Co-IP, and the results showed that the association between UPF1 and UPF2 was repressed (Figure 1, E and F). These data suggest that Neurospora UPF1 and UPF2 are in a complex to exert their function.
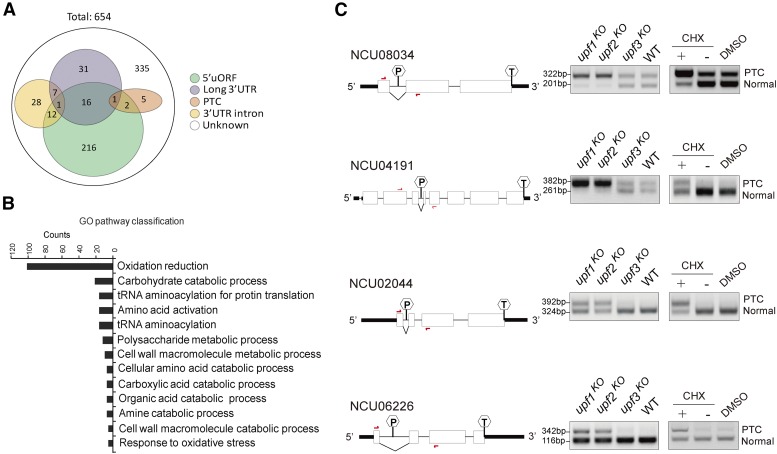
One important role of NMD factors is to eliminate aberrant PTC-containing transcripts. To identify potential NMD substrates, we conducted RNA-seq of upf1KO and WT control strains, which identified ∼654 genes showing upregulation in the upf1KO strain (Figure 2A). Among the total genes (654) upregulated by depletion of UPF1, near half (319) are potential NMD targets as they contain at least one of the key features: premature termination codons, 5′-uORFs, long 3′-UTRs, or introns located in 3′-UTR region. Gene ontology classification identified a variety of pathways that are influenced by the depletion of UPF1 (Figure 2B, Figures S3 and S4 and Table S2 in File S1).
Figure 2.
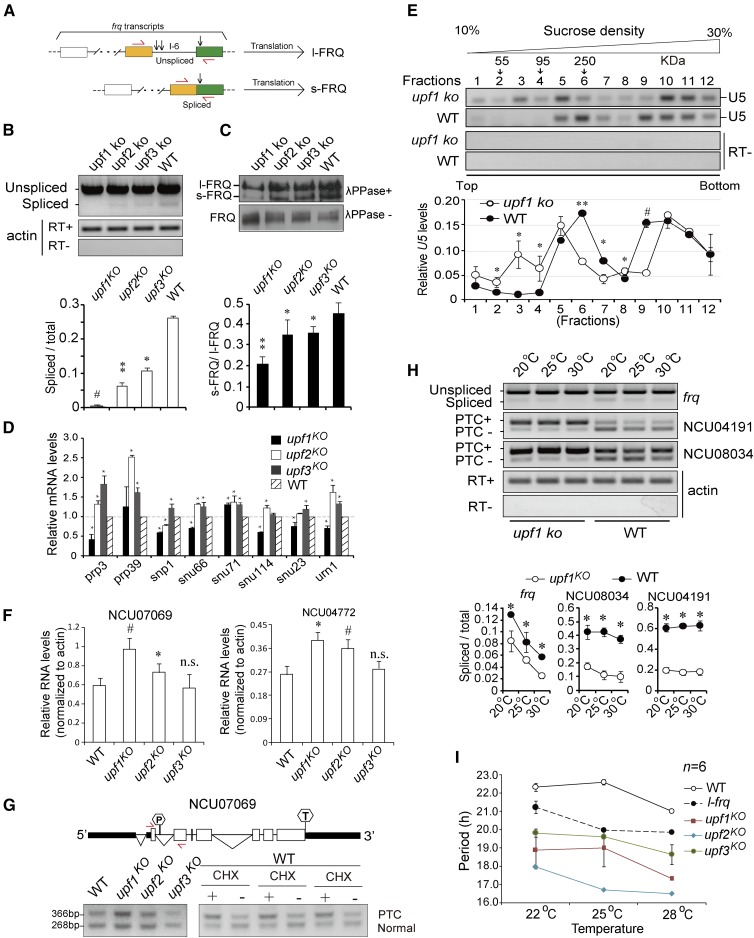
Characterization of genes regulated by Neurospora UPF-1 (A) Distribution of genes with different features upregulated (fold change ≥ 2) in the upf1KO strain. The results were obtained from RNA-seq data. (B) Distribution of the GO categories of genes upregulated in the upf1KO strain revealed by RNA-seq analysis. The genes showing expression with fold change ≥ 2 were analyzed. (C) RT-PCR of spliced and unspliced transcripts of representative PTC-containing genes. The unspliced transcripts contain typical PTCs. The diagrams of these genes are shown. The hexagons with “P” indicate PTCs and the hexagons with “T” indicate normal termination codons. Red arrows denote the primer locations. CHX, cycloheximide; GO, gene ontology; ko, knockout; PTC, premature termination codon; RNA-seq, RNA sequencing; WT, wild-type.
Several genes containing potential PTCs identified from RNA-seq results, including NCU08034, NCU04191, NCU02044 and NCU06226, were subjected to RT-PCR analysis. The results showed that the unspliced species harboring PTCs were significantly elevated in upf1KO and upf2KO but not upf3KO strains. As expected, CHX treatment in the WT strain also resulted in an increase in the levels of PTC-containing transcripts (Figure 2C and Figure S3 in File S1). Together, these data demonstrate that UPF1 and UPF2 are required for the surveillance and decay of PTC-containing transcripts.
UPF1 mediates circadian and diurnal rhythms
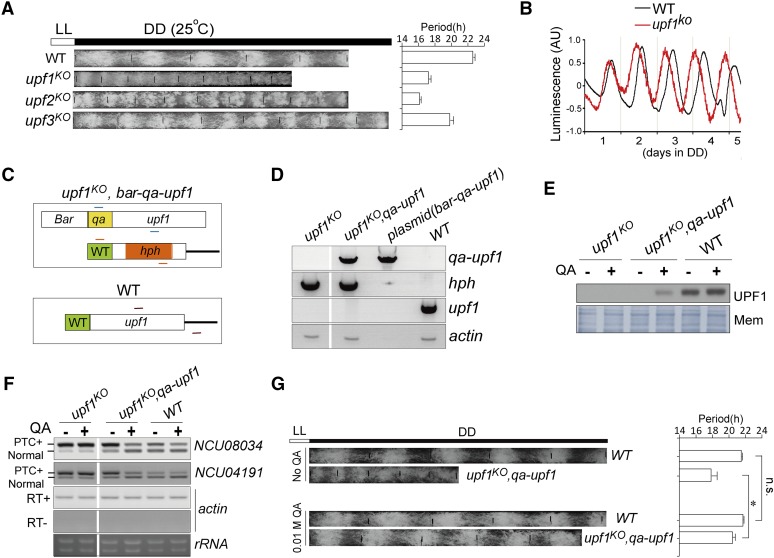
To probe the effects of NMD factors on the circadian clock, these knockout strains were grown in race tubes to compare the circadian periods of conidia banding rhythms in constant dark (DD), and the race tube assay results showed that all the upf1–3 knockout strains displayed shorter circadian periods compared to WT. Among these strains, upf2KO showed the shortest period, followed by upf1KO (Figure 3A).
Figure 3.
Depletion of UPF1 leads to altered circadian rhythms. (A) Conidiation banding rhythms of WT and upf1–3 ko strains revealed by race tube assay. The temperature was 25°. The white bar on the top denotes the LL condition and the black bar denotes the DD condition. (B) Representative results of luciferase reporter assays showing the frq promoter activity of the indicated strains in constant darkness. The measurement of luciferase activity was normalized to subtract the baseline luciferase signal. AU denotes arbitrary unit. Data are mean ± SD. (C) Diagram of strategy to generate constructs and transformants to express upf1 in the upf1KO strain. In the upf1KO strain, the locus of upf1 gene has been replaced with the hph gene. The Upf1 ORF was linked to qa promoter so that the expression of upf1 can be driven by the addition of QA, to generate the upf1KO, bar-qa-upf1 transformants. Transformants were selected by bialaphos resistance conferred by the bar gene. The short lines in different colors denote three primer sets for validation of the obtained transformants. (D) PCR validation of the transformants. The gDNA was isolated and subject to PCR analysis. Upf1KO strain and the bar-qa-upf1 plasmid serve as controls. Actin serves as the PCR control. (E) Western blot results showing the expression of UPF1 in the upf1KO, bar-qa-upf1 strain in the presence or absence of QA (0.01 M). Membrane stained with amido black serves as loading control. (F) Expression of qa-upf1 in upf1KO leads to increased elimination of PTC-containing transcripts. RT-PCR results of NCU008034 and NCU04191 are shown. Data are mean ± SD. (G) Left: race tube assay of upf1KO, qa-upf1. WT and upf1KO served as controls. Right: quantification of the circadian periods. Data are mean ± SD. Significance was detected by Student’s t-test. DD, continuous dark; gDNA, genomic DNA; ko, knockout; LL, continuous light; Mem, membrane; n.s., not significant; PTC, premature termination codon; QA, quinic acid; WT, wild-type.
We introduced a luciferase reporter construct that is under the control of the frq promoter into the upf1KO and WT strains, respectively. As shown in Figure 3B, upf1KO consistently showed a period that was much shorter than that of WT. The short periodicity of upf1KO is consistent with the prd-6 strain, as previously described, which bears a mutation in the upf1 locus (Morgan and Feldman 1997, 2001).
To preclude that our upf1KO strain contains additional mutations affecting circadian rhythms, we generated a strain that constantly expresses upf1 in the background of the upf1KO strain upon addition of qa. The obtained upf1KO, bar-qa-upf1 strain was validated by PCR amplification of the hph gene, endogenous upf1 gene, and transformed cassette-containing upf1 gene with the qa-2 promoter, respectively (Figure 3, C and D). The expression of UPF1 in upf1KO, qa-upf1 was lower than that in WT, as revealed by western blot (Figure 3E); however, such a level of UPF1 was able to overtly reduce the ratio of PTC-containing transcripts of NCU08034 and NCU04191 (Figure 3F), suggesting that the expressed UPF1 driven by qa has normal functions. The upf1KO, qa-upf1 strain exhibits a circadian period of conidiation that is comparable to that of WT despite growing more slowly (Figure 3G). These data demonstrate that the loss of upf1 accounts for the severe disruption of the circadian phenotype in upf1KO strain.
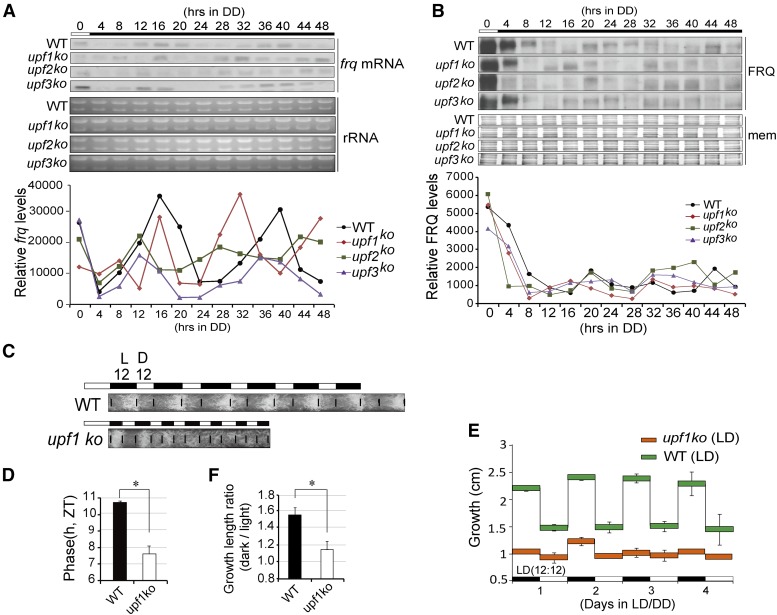
We measured the expression of clock genes in upf1–3 knockout strains, and the results showed that the levels of clock genes were affected in these strains (data not shown). To further assess the impacts of UPF1 on the circadian clock, we performed northern blots and western blots to analyze the expression of frq mRNA and FRQ protein over the course of 48 hr. The results showed that both the RNA and protein rhythms were significantly altered, and the upf1KO and upf2KO strains showed short periods (Figure 4, A and B).
Figure 4.
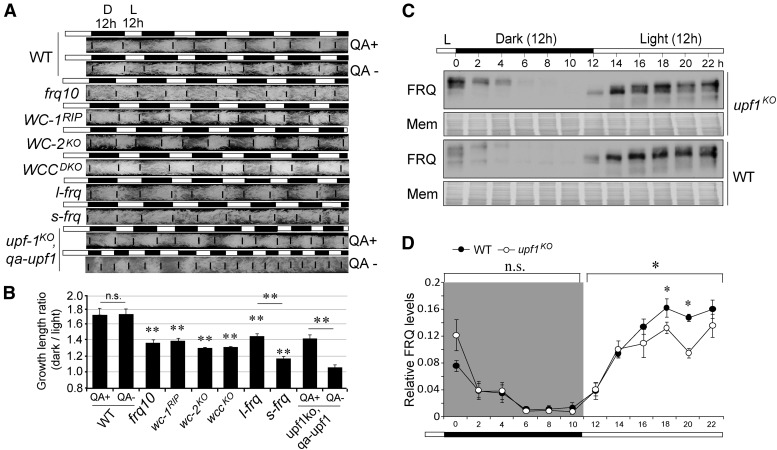
UPF1 regulates Neurospora circadian rhythm. (A) Northern blot results of frq mRNA over 48 hr in WT and upf1–3 ko strains. (B) Western blotting results of FRQ protein over 48 hr in WT and upf1–3 ko strains. (C) Race tube results of WT and upf1KO in LD12:12. (D) Quantification of phase difference between WT and upf1KO in LD12:12. Data are mean ± SD. (E) Quantification of growth length of WT and upf1KO strains in LD12:12 over 4 days. Black and white bars denote the LD conditions. Data are mean ± SD. (F) Quantification of growth ratios of WT and upf1KO of strains in LD. The ratios represent the growth lengths in dark vs. that in the light. Data are mean ± SD. Significance was detected by Student’s t-test. DD, continuous dark; ko, knockout; mem, membrane; WT, wild-type.
We next grew the upf1KO and WT strains in LD12:12 condition and compared their phases, and the results reveal a ∼3 hr advanced phase in the upf1KO strain (Figure 4, C and D). We also measured the growth length of WT, which grew faster in the dark than in the light. In contrast, the growth difference of upf1KO between in the dark and in the light is flat and shows marginal rhythmicity (Figure 4, C, E, and F).
UPF genes regulate the splicing of clock gene frq
Since regulation of the splicing of PTC-containing transcripts of spliceosome-associated factors by NMD factors has been observed in a variety of organisms including Neurospora (Lareau et al. 2007; Saltzman et al. 2008; Weischenfeldt et al. 2012; Lareau and Brenner 2015), it is possible that NMD factors also regulate the splicing of clock genes in this fashion. In Neurospora, the core negative component frq gene does not contain a PTC but is instead comprised of eight splicing variants, and splicing of the sixth intron (intron 6, I-6) determines the synthesis ratio of two FRQ isoforms: large FRQ (l-FRQ) and small FRQ (s-FRQ) (Liu et al. 1997). frq also contains uORFs in the 5′-UTR (Liu et al. 1997), suggesting that frq might be an NMD target, though the turnover of frq mRNA is not significantly affected by the knockout of upf genes (Figure S5 in File S1).
We measured the expression of frq I-6 splicing in the upf knockout strains by RT-PCR, and the results showed that the ratios of spliced variants vs. total transcripts containing I-6 were significantly decreased in the knockout strains. The ratio of spliced/total transcripts was the lowest in upf1KO (Figure 5, A and B). We also extracted proteins from the knockout strains and treated them with λ phosphatase to dephosphorylate FRQ, which allows for the observation of the expression of s-FRQ and l-FRQ (Liu et al. 1997). The western blot results showed that, consistent with frq I-6 splicing, the ratios of s-FRQ/total were significantly attenuated in the knockout strains (Figure 5C). Because s-FRQ supports a longer period in Neurospora (Liu et al. 1997), these data suggest that decreased splicing of frq I-6 might account for the short periods of upf knockout strains.
Figure 5.
UPF1 regulates the splicing of frq transcripts. (A) Schematic representation of frq I-6 splicing and the corresponding protein products l-FRQ and s-FRQ. Arrows in red denote the relative location of PCR primers flanking frq I-6. Vertical arrows denote the locations of the three initiation codons. (B) Top: RT-PCR results of frq I-6 splicing in upf1–3 knockout and WT strains. Actin amplified in the presence/absence of reverse transcription served as control. Below: densitometric quantification of the results. Data are mean ± SD. (C) Top: western blot results showing the ratio of s-FRQ protein in upf1–3 knockout and WT strains. The proteins were either treated or untreated with λ phosphatase before running electrophoresis. Below: densitometric quantification of the results. Data are mean ± SD. (D) qRT-PCR results showing the expression of snRNP genes in upf1–3 knockout strains. The levels were normalized to that in WT. The expression data were normalized to actin. Data are mean ± SD. (E) Top: RT-PCR results of the levels of U5 snRNA in the fractions from sucrose gradients of upf1KO and WT strains. Upper panel: representative U5 snRNA profiles. Locations of the protein markers (55, 95, and 250 kDa) from parallel gradients are indicated above the panel. Below: densitometric quantification of U5 distribution. Data are mean ± SD. The total value from 12 densitometric analyses of each experiment was normalized to be 1. Data are mean ± SD. (F) Quantification of relative RNA levels of NCU07069 and NCU04772 in indicated strains. The data were normalized to the expression of actin. Data are mean ± SD. (G) RT-PCR of spliced and unspliced transcripts of NCU07069. Top: schematic representation of exon/intron structure of NCU07069. Arrows in red denote the relative location of PCR primers. Below: RT-PCR results of NCU07069 expression in indicated strains or with/without CHX treatment. (H) Top: splicing of frq and two PTC-containing genes is repressed in upf1KO strain at three temperatures. Upper panel: RT-PCR results of frq I-6, NCU04191, and NCU08034. Below: densitometric quantification of the spliced and unspliced transcripts. Data are mean ± SD. (I) Quantification of circadian periods of upf1–3 KO and WT strains at various temperatures. Data are mean ± SD. Significance was detected by Student’s t-test. CHX, cycloheximide; ko, knockout; PTC, premature termination codon; qRT-PCR, quantitative RT-PCR; WT, wild-type.
To validate this possibility, we first investigated the expression of a number of spliceosome genes in upf1–3 knockout strains, which include prp3, prp39, snp1, snu66, snu71, snu114, snu23 and urn1. The quantitative RT-PCR (qRT-PCR) results showed that, compared to WT, the levels of all of the tested genes except prp39 were decreased in upf1KO, while they were mostly upregulated in upf2KO and upf3KO strains (Figure 5D), suggesting that NMD factors are involved in the regulation of spliceosome components, though the roles of UPF1 might differ from those of UPF2 and UPF3. We next conducted sucrose density gradient sedimentation to fractionate the total protein extracts from the upf1KO and WT strains. The small nuclear RNA (snRNA) U5 is critical for cell viability and pre-mRNA splicing (O’Keefe and Newman 1998). The distribution of U5 in the sucrose density gradient fractions was evaluated by RT-PCR, and the results revealed a different distribution pattern of U5 snRNA in the fractionated samples from that in WT (Figure 5E).
Arginine/serine-rich splicing factors (SR proteins) control splicing specificity, and in species of different kingdoms including Neurospora, the transcripts of some SR genes, e.g., SRSF5, harbor PTCs that are NMD targets (Lareau and Brenner 2015). In humans, all of the SR family members have splice forms and contain PTCs or alternative introns in the 3′-UTR (Lareau et al. 2007). We identified several SR homologs in Neurospora, including NCU07069, NCU03491 and NCU04772, and sequence analysis indicates that NCU07069, a homolog of SRSF4, bears a potential PTC (Figure S6 in File S1). RT-PCR results showed that the expression of NCU07069 and NCU04772 was significantly higher in upf1KO and upf2KO strains compared to that in WT (Figure 5F and Figure S7 in File S1). With primers spanning the PTC-containing intron, RT-PCR results showed that the inclusion of this intron was induced in the upf1KO strain. In addition, CHX treatment also led to induced inclusion of this intron (Figure 5G). Together, these data suggest that UPF1 may affect splicing, through regulating the expression of spliceosome components and related factors.
We next measured the splicing of frq I-6 and two representative PTC-containing genes, NCU04191 and NCU08034, and the results showed that the ratios of the spliced species of these three gene transcripts decreased at different temperatures in upf1KO (Figure 5H). At three different temperatures, all of the upf1–3KO strains exhibit shorter periods than the WT control (Figure 5I). A previous study reported that the prd-6 strain showed conidiation rhythms even at very low or high temperatures (Morgan and Feldman 1997, 2001). However, we failed to observe discernable rhythms at temperatures lower than 22° and higher than 28° in the upf1KO strain. Even at 25°, upf1KO showed conidiation rhythmicity for only the first few days on the race tube and became arrhythmic afterward (Figure 3A). Moreover, as shown in Figure 5I, the circadian period of upf1KO was significantly shorter than that of the l-frq strain, which exclusively expresses l-FRQ but not s-FRQ.
UPF1 regulates LD growth rhythm in LD conditions
As shown in Figure 4, the upf1KO strain displayed an altered growth rhythm in LD conditions. To verify whether UPF1 mediates growth through the circadian clock or not, we first looked at the growth rhythms of a series of clock mutants including frq10 (frq knockout strain), wc-1RIP (a strain with the wc-1 gene silenced), wc-2KO (wc-2 knockout strain), and wccDKO (a strain in which both wc-1 and wc-2 are depleted) in LD12:12 conditions by calculating the growth length during night vs. that during daytime. According to the race tube results, the LD growth ratio in the night vs. daytime in the WT strain was ∼1.7, while the LD growth ratios in the clock mutants were significantly lower (Figure 6, A and B). It is noteworthy to mention that, even in these mutant strains in which the circadian oscillator was disrupted, the growth rates at night were still higher than in the day. These data suggest that the circadian clock participates in the regulation of the daily diurnal growth rhythm in Neurospora.
Figure 6.
upf1KO strain displays a changed growth rhythm. (A) Clock gene mutants exhibit altered diurnal growth rhythms in LD12:12. The results were obtained from race tube assay. The growth front of mycelia growth was labeled at the L/D transition time points, twice a day. QA+ denotes the presence and QA− denotes absence of qa in the culture, respectively. (B) Quantification of the data from (A). Data are mean ± SD. (C) Representative western blot results of FRQ in upf1KO and WT strains in LD12:12. Black and white bars denote the LD conditions. (D) Quantification of FRQ levels in upf1KO and WT strains in LD12:12 cycles. The total FRQ amount of each strain was normalized to be 1. Black and white bars denote the LD conditions. Data are mean ± SE. Significance was detected by two-way repeated-measures ANOVA. D, dark; ko, knockout; mem, membrane; n.s., not significant; QA, quinic acid; WT, wild-type.
In addition, we analyzed the diurnal growth rhythms of the l-frq and s-frq strains, and the results showed that a lack of either l-FRQ in the s-frq strain or s-FRQ in the l-frq strain led to a decreased LD growth ratio, and the s-frq strain displayed a lower growth ratio than l-frq (Figure 6, A and B). We also compared the growth rhythms in the upf1KO, qa-upf1 strain, and the results showed that, upon the addition of QA, the LD growth ratio was significantly increased (Figure 6, A and B), which confirms the idea that UPF1 controls the diurnal growth rhythm.
We next conducted western blots to analyze the changes in FRQ levels in LD12:12, and the results showed that during light, the FRQ protein levels of upf1KO are significantly lower than that in WT. In contrast, the FRQ levels in the dark are comparable between these two strains (Figure 6, C and D). These results suggest that the changes in the amount of FRQ in light might be associated with the altered growth rhythms.
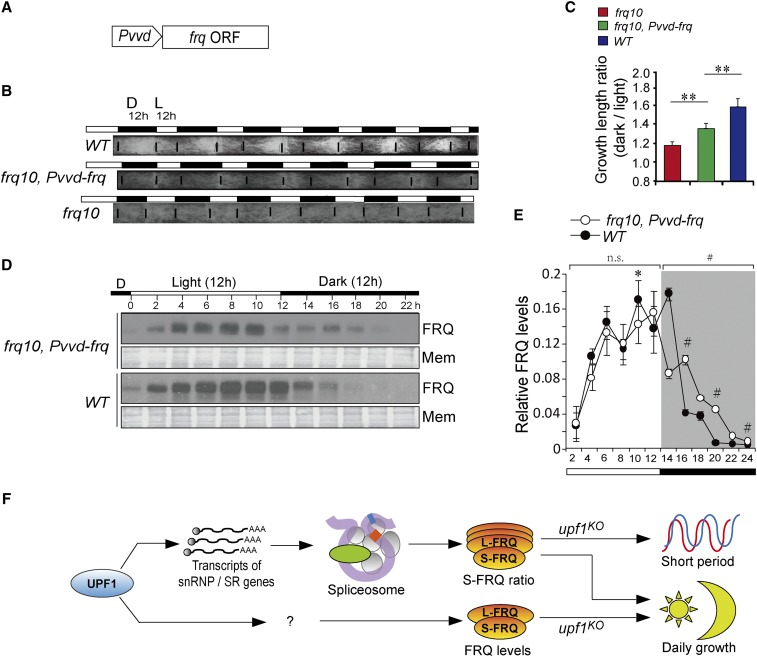
VIVID (VVD) is a blue light receptor that controls the entrainment and adaptation to light change in the Neurospora circadian clock (Heintzen et al. 2001; Schwerdtfeger and Linden 2003; Elvin et al. 2005; Hunt et al. 2007; Malzahn et al. 2010). The expression of vvd can be rapidly induced by the light, but it is only weakly expressed in the dark (Heintzen et al. 2001; Elvin et al. 2005). To verify whether the lower FRQ levels in the light can explain the changed growth rhythm in the upf1KO strain, we generated a strain frq10, Pvvd-frq, which expresses frq under the control of the vvd promoter (Figure 7A). As the expression of vvd is predominantly occurs in the light (Hurley et al. 2012), the FRQ profiles would be expected to be different from that of WT. As expected, the race tube assays in LD12:12 showed that the LD growth ratio of frq10, Pvvd-frq was significantly increased compared to frq10 (Figure 7, B and C). In LD12:12, the frq10, Pvvd-frq strain showed an altered FRQ profile in which FRQ was relatively higher in the dark (Figure 7, D and E).
Figure 7.
Effects of diurnal levels of FRQ on Neurospora growth rhythm. (A) Schematic diagrams showing the cassette to overexpress the frq gene in frq10. The transcription of frq is controlled by the vvd promoter. (B) Race tube results of WT and frq10, Pvvd-frq strains. (C) Quantification of growth ratios of WT, Pvvd-frq, frq10, and WT strains in LD. The ratios represent the growth lengths in dark vs. that in the light. (D) Diurnal profile of FRQ protein is altered in frq10, Pvvd-frq, and WT strains. Representative western blot results of FRQ in upf1KO and WT strains in LD12:12. (E) Quantification of FRQ levels in upf1KO and WT strains in an LD12:12 cycle. The total FRQ amount of each strain was normalized to be 1. Data are mean ± SE. Significance detected by two-way repeated-measures ANOVA. (F) Model for UPF1 in the regulation of circadian and diurnal growth rhythms. UPF1 mediates the ratio of l-FRQ/s-FRQ through the spliceosome, which further contributes to the shorter period in the upf1KO strain. UPF1 also regulates the day/night FRQ levels through unidentified factors, which account for the altered diurnal growth rhythms in the upf1KO strain. Mem, membrane; n.s., not significant; SR gene, arginine/serine-rich splicing factor gene; WT, wild-type.
Discussion
NMD is a quality control mechanism that degrades a variety of aberrant mRNAs, including harboring premature termination (nonsense) codons and certain other types of aberrant transcripts. In addition, NMD also regulates the expression of a substantial set of normal transcripts (Kervestin and Jacobson 2012; Schweingruber et al. 2013). When translated, these aberrant mRNAs can produce truncated proteins with dominant-negative or deleterious gain-of-function activities. In humans, ∼30% of all known diseases result from mutations in the production of mRNAs with PTCs (Khajavi et al. 2006).
It has been shown that EJC-mediated NMD and NMD factors participate in the regulation of mRNA stability of target genes in Neurospora (Zhang and Sachs 2015). In this work, we first showed that UPF1 can bind to UPF2 and that this process is compromised if the translation is repressed (Figure 1, C–F). In addition, for several representative genes, the ratio of unspliced to spliced transcripts containing PTCs is significantly increased in the upf1KO and upf2KO strains (Figure 2C). These findings together suggest that these factors play conserved roles in regulating the NMD in Neurospora and, according to RNA-seq data, the predominant NMD target genes are 5′-uORF-containing transcripts in Neurospora.
In Neurospora, the depletion of UPF1 led to a very short circadian period in the first few days (Figure 3, A and B). The depletion of UPF1 resulted in a change in the spliceosome gene expression and assembly. As a consequence, a decrease in the proportion of spliced frq transcripts containing frq I-6 was observed in the depletion strains of UPFs (Figure 5B). In Neurospora, s-FRQ supports a longer circadian period while l-FRQ supports a shorter circadian period (Liu et al. 1997). As such, the decreased s-FRQ ratio may account for the shortened periods of the upfs deletion strains. The expression of upf1 driven by the qa promoter restored the circadian period (Figure 3G), which is consistent with the model that UPF1 mediates the circadian clock through the alternative splicing of frq. NMD factors may affect splicing events of frq and other genes through targeting the transcripts of spliceosomal components including SR proteins. As it has been shown that frq transcribes eight splicing variants including the unspliced pre-mRNA, splicing and turnover of which are differentially regulated (Zhang et al. 2015), it is possible that in addition to the spliceosome, NMD might also affect the ratio of frq transcripts with I-6 to those lacking I-6 by regulating the turnover of frq splicing variants.
Despite the increase in l-FRQ through alternative splicing accounting for the short period, the period of the upf1KO strain was still much shorter than that in l-frq, suggesting that the alteration of frq I-6 splicing only partially explains the short period in the upf1KO strain. Together with the fact that the rhythmicity of upf1KO is disrupted after the first few days, these data suggest that NMD also regulates the circadian oscillator in Neurospora through additional pathways other than splicing. As UPF1 mediates a number of downstream pathways (Figure 2B), it is possible that it regulates the gene expression of oscillator components in both direct and indirect ways. For instance, among the influenced gene upon the depletion of upf genes, several protein kinases and protein phosphatases that are known regulators of the circadian clock are under NMD control and their levels are altered in the knockout strains of upf genes. Moreover, as Neurospora NMD factors control mRNA turnover (Zhang and Sachs 2015), certain clock regulators could be mediated in this fashion.
The prd-6 strain showed conidiation rhythms under temperatures ranging from 17 to 34°, and the temperature compensation was aberrant compared to the WT strain (Morgan and Feldman 1997). In contrast, in this work no discernable rhythms in the upf1KO strain were observed at temperatures beyond 22–28°. According to unpublished data, the original prd-6 strain bears a spontaneous frameshift mutation in upf1 resulting in a premature termination codon in the UPF1 coding region (Zhang and Sachs 2015). As such, gain-of-function mutation might be an explanation accounting for the difference in conidiation rhythmicity at lower and higher temperatures and the temperature compensation in the prd-6 strain from those in the upf1KO strain.
The circadian clocks efficiently coordinate growth and development with respect to time of day in various species, and light interacts with the circadian systems, which results in diurnal growth rhythms (Dowson-Day and Millar 1999). In lower organisms, the dominant growth at night might be explained by the “escape of light” hypothesis (Parker and Rossman 1971; Matos et al. 2014). Arabidopsis hypocotyl elongation is under the control of both light and the circadian clock, and mutations in clock genes caused impairment in the growth rhythms (Dowson-Day and Millar 1999; Nozue et al. 2007). In Brachypodium distachyon, the daily temperature change plays an important role in regulating the growth rhythm instead of the circadian clock (Matos et al. 2014). At the hormonal level, the amounts of some hormones closely oscillate in a circadian fashion. In plants, the response to auxin is under circadian control (Nozue and Maloof 2006; Covington and Harmer 2007). In animals, it is known that the growth hormones undergo a diurnal control that oscillates with a period near 24 hr (Leatherland et al. 1974; Brandenberger and Weibel 2004). Overall, the growth rhythms seem to represent a crucial adaptation to the environment, the disruption of which may undermine their fitness in the environment. However, the underlying mechanisms are not yet clearly understood.
The diurnal vs. nocturnal growth of Neurospora shows obvious rhythmicity, shown by faster growth in dark (Figure 4C). Depletion of UPF1 resulted in dramatic abolishment of this growth rhythmicity, suggesting that UPF1 may have a role in the establishment of the diurnal growth rhythm. In a series of clock gene mutants, the growth ratio (LD) of all these strains was significantly decreased (Figure 6), suggesting the involvement of clock components in regulating growth rhythmicity. In frq10, Pvvd-frq strain, the LD growth ratios were significantly altered, suggesting that the FRQ levels affect the diurnal growth rhythm. In addition to its influence on the circadian period and phase, the deletion of upf1 caused altered diurnal levels of FRQ protein and a flattened diurnal growth rhythm. Together, these data demonstrate that UPF1 may control the diurnal growth rhythm by regulating the expression of clock genes including FRQ (Figure 7F). As in those strains in which the circadian system is disrupted, including frq10, wc-1RIP, wc-2KO and wccDKO, the diurnal growth ratios are significantly higher than that in upf1KO, suggesting that certain other photoreceptors or light-interacting factors are implicated in the regulation of diurnal growth rhythmicity, and that such potential factors should be under NMD control. In Neurospora, established and putative photoreceptors comprise WC-1, VVD, Phytochrome orthologs PHY-1/2, the CRYPTOCHROME homolog CRY, and NEW EUKARYOTIC OPSIN-1 (NOP-1) (Chen et al. 2010). Whether these factors regulate Neurospora growth rhythmicity as well as the mechanisms by which they may do this remains to be investigated.
Supplementary Material
Supplemental material is available online at http://www.genetics.org/content/suppl/2017/06/08/genetics.117.202788.DC1.
Acknowledgments
We thank Yi Liu (University of Texas Southwestern Medical Center) for generous help and comments, and Deborah Bell-Pedersen (Texas A&M University) for generously providing the pBARKS1 plasmid. This work was supported by the National 973 Program of China (grant numbers 2011CB711000 and 2012CB947600) and the National Natural Science Foundation of China (grant numbers 31071122, 31571205, and 157120578) to J.G.
Footnotes
Communicating editor: M. Freitag
Literature Cited
- Adhvaryu K., Firoozi G., Motavaze K., Lakinthomas P. L., 2016. PRD-1, a component of the circadian system of Neurospora crassa, is a member of the DEAD-box RNA helicase family. J. Biol. Rhythms 31: 258–271. [DOI] [PubMed] [Google Scholar]
- Aronson B. D., Johnson K. A., Dunlap J. C., 1994. Circadian clock locus frequency: protein encoded by a single open reading frame defines period length and temperature compensation. Proc. Natl. Acad. Sci. USA 91: 7683–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L., Loros J. J., Dunlap J. C., 2012. The circadian clock of Neurospora crassa. FEMS Microbiol. Rev. 36: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellpedersen D., Cassone V. M., Earnest D. J., Golden S. S., Hardin P. E., et al. , 2005. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet. 6: 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger G., Weibel L., 2004. The 24-h growth hormone rhythm in men: sleep and circadian influences questioned. J. Sleep Res. 13: 251–255. [DOI] [PubMed] [Google Scholar]
- Chamieh H., Ballut L., Bonneau F., Le Hir H., 2008. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 15: 85–93. [DOI] [PubMed] [Google Scholar]
- Chang Y., Imam J. S., Wilkinson M. F., 2007. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76: 51–74. [DOI] [PubMed] [Google Scholar]
- Chen C., Dunlap J. C., Loros J. J., 2010. Neurospora illuminates fungal photoreception. Fungal Genet. Biol. 47: 922–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington M. F., Harmer S. L., 2007. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 5: e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Underbrink K. M., Fink G. R., 1980. Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics 95: 833–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson-Day M. J., Millar A. J., 1999. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 17: 63–71. [DOI] [PubMed] [Google Scholar]
- Ebbole D., Sachs M. S., 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37: 17–18. [Google Scholar]
- Elvin M., Loros J. J., Dunlap J. C., Heintzen C., 2005. The PAS/LOV protein VIVID supports a rapidly dampened daytime oscillator that facilitates entrainment of the Neurospora circadian clock. Genes Dev. 19: 2593–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatscher T., Boehm V., Gehring N. H., 2015. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell. Mol. Life Sci. 72: 4523–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S. A., Mockler T. C., 2012. Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes. Biol. Direct 7: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin S. A., Priest H. D., Givan S. A., Shen R., Bryant D. W., et al. , 2010. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fury M. G., Zieve G. W., 1996. U6 snRNA maturation and stability. Exp. Cell Res. 228: 160–163. [DOI] [PubMed] [Google Scholar]
- Görl M., Merrow M., Huttner B., Johnson J., Roenneberg T., et al. , 2001. A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J. 20: 7074–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geever R. F., Giles N. H., 1983. Point mutations and DNA rearrangements 5′ to the inducible qa-2 gene of Neurospora allow activator protein-independent transcription. Proc. Natl. Acad. Sci. USA 80: 7298–7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooch V. D., Mehra A., Larrondo L. F., Fox J., Touroutoutoudis M., et al. , 2008. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot. Cell 7: 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Cheng P., Yuan H., Liu Y., 2009. The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell 138: 1236–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Cheng P., Liu Y., 2010. Functional significance of FRH in regulating the phosphorylation and stability of Neurospora circadian clock protein FRQ. J. Biol. Chem. 285: 11508–11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Brown A. H., Jacobson A., 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17: 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C., Nater M., Apel K., Staiger D., 1997. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94: 8515–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C., Loros J. J., Dunlap J. C., 2001. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104: 453–464. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Papp A., Pulak R., Ambros V., Anderson P., 1989. A new kind of informational suppression in the Nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S., Thompson S., Elvin M., Heintzen C., 2010. VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc. Natl. Acad. Sci. USA 107: 16709–16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt S. M., Elvin M., Crosthwaite S. K., Heintzen C., 2007. The PAS/LOV protein VIVID controls temperature compensation of circadian clock phase and development in Neurospora crassa. Genes Dev. 21: 1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. M., Chen C., Loros J. J., Dunlap J. C., 2012. Light-inducible system for tunable protein expression in Neurospora crassa. G3 2: 1207–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J. M., Larrondo L. F., Loros J. J., Dunlap J. C., 2013. Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered Neurospora clock protein FRQ. Mol. Cell 52: 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Li X., Serin G., Maquat L. E., 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L. E., 2008. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 9: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kervestin S., Jacobson A., 2012. NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajavi M., Inoue J. R., Lupski K., 2006. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 14: 1074–1081. [DOI] [PubMed] [Google Scholar]
- Kojima S., Shingle D. L., Green C. B., 2011. Post-transcriptional control of circadian rhythms. J. Cell Sci. 124: 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T., Li W., Hoque M., Popp M. W., Ermolenko D. N., et al. , 2014. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 28: 1900–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L. F., Brenner S. E., 2015. Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol. Biol. Evol. 32: 1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L. F., Brooks A. N., Soergel D. A., Meng Q., Brenner S. E., 2007. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv. Exp. Med. Biol. 623: 190–211. [DOI] [PubMed] [Google Scholar]
- Larrondo L. F., Loros J. J., Dunlap J. C., 2012. High-resolution spatiotemporal analysis of gene expression in real time: in vivo analysis of circadian rhythms in Neurospora crassa using a FREQUENCY-luciferase translational reporter. Fungal Genet. Biol. 49: 681–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauinger L., Diernfellner A., Falk S., Brunner M., 2014. The RNA helicase FRH is an ATP-dependent regulator of CK1a in the circadian clock of Neurospora crassa. Nat. Commun. 5: 3598. [DOI] [PubMed] [Google Scholar]
- Leatherland J. F., McKeown B. A., John T. M., 1974. Circadian rhythm of plasma prolactin, growth hormone, glucose and free fatty acid in juvenile kokanee salmon, Oncorhynchus nerka. Comp. Biochem. Physiol. A Comp. Physiol. 47: 821–828. [DOI] [PubMed] [Google Scholar]
- Lim C., Allada R., 2013. Emerging roles for post-transcriptional regulation in circadian clocks. Nat. Neurosci. 16: 1544–1550. [DOI] [PubMed] [Google Scholar]
- Linde L., Boelz S., Neuyilik G., Kulozik A. E., Kerem B., 2007. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur. J. Hum. Genet. 15: 1156–1162. [DOI] [PubMed] [Google Scholar]
- Liu Y., Garceau N. Y., Loros J. J., Dunlap J. C., 1997. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the Neurospora circadian clock. Cell 89: 477–486. [DOI] [PubMed] [Google Scholar]
- Lombardi L., Schneider K., Tsukamoto M., Brody S., 2007. Circadian rhythms in Neurospora crassa: clock mutant effects in the absence of a frq-based oscillator. Genetics 175: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzahn E., Ciprianidis S., Káldi K., Schafmeier T., Brunner M., 2010. Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell 142: 762–772. [DOI] [PubMed] [Google Scholar]
- Matos D. A., Cole B. J., Whitney I. P., Mackinnon K. J., Kay S. A., et al. , 2014. Daily changes in temperature, not the circadian clock, regulate growth rate in Brachypodium distachyon. PLoS One 9: e100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L. E., Tarn W. Y., Isken O., 2010. The pioneer round of translation: features and functions. Cell 142: 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan L. W., Feldman J. F., 1997. Isolation and characterization of a temperature-sensitive circadian clock mutant of Neurospora crassa. Genetics 146: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan L. W., Feldman J. F., 2001. Epistatic and synergistic interactions between circadian clock mutations in Neurospora crassa. Genetics 159: 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan L. W., Greene A. V., Bellpedersen D., 2003. Circadian and light-induced expression of luciferase in Neurospora crassa. Fungal Genet. Biol. 38: 327–332. [DOI] [PubMed] [Google Scholar]
- Nolte C., Staiger D., 2015. RNA around the clock - regulation at the RNA level in biological timing. Front. Plant Sci. 6: 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Maloof J. N., 2006. Diurnal regulation of plant growth. Plant Cell Environ. 29: 396–408. [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M. F., Duek P. D., Lorrain S., Fankhauser C., et al. , 2007. Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- O’Keefe R. T., Newman A. J., 1998. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 17: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D. C., Rossman L. G., 1971. Human growth hormone release in sleep: nonsuppression by acute hyperglycemia. J. Clin. Endocrinol. Metab. 32: 65–69. [DOI] [PubMed] [Google Scholar]
- Rehman S., Shawl A. S., Kour A., Andrabi R., Sudan P., et al. , 2008. An endophytic Neurospora sp. from Nothapodytes foetida producing camptothecin. Appl. Biochem. Microbiol. 44: 203–209. [PubMed] [Google Scholar]
- Rehwinkel J., Raes J., Izaurralde E., 2006. Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem. Sci. 31: 639–646. [DOI] [PubMed] [Google Scholar]
- Saltzman A. L., Kim Y. K., Pan Q., Fagnani M. M., Maquat L. E., et al. , 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 28: 4320–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoning J. C., Streitner C., Page D. R., Hennig S., Uchida K., et al. , 2007. Auto‐regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 52: 1119–1130. [DOI] [PubMed] [Google Scholar]
- Schweingruber C., Rufener S. C., Zund D., Yamashita A., Muhlemann O., 2013. Nonsense-mediated mRNA decay — mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta 1829: 612–623. [DOI] [PubMed] [Google Scholar]
- Schwerdtfeger C., Linden H., 2003. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22: 4846–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D., Zecca L., Kirk D. A. W., Apel K., Eckstein L., 2003. The circadian clock regulated RNA‐binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre‐mRNA. Plant J. 33: 361–371. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J., Waage J., Tian G., Zhao J., Damgaard I., et al. , 2012. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 13: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wan Y., Huang G., Wang D., Yu X., et al. , 2015. The exosome controls alternative splicing by mediating the gene expression and assembly of the spliceosome complex. Sci. Rep. 5: 13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sachs M. S., 2015. Control of mRNA stability in fungi by NMD, EJC and CBC factors through 3′UTR introns. Genetics 200: 1133–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Shen Y., Yang S., Wang J., Hu Q., et al. , 2009. Ubiquitin ligase components cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J. Biol. Chem. 285: 4355–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Guo J., Cha J., Chae M., Chen S., et al. , 2013. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495: 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains used are available upon request. The RNA-seq data sets are available at the GEO database (GSE97157).