Abstract
Holliday junction (HJ) is a hallmark intermediate in DNA recombination and must be processed by dissolution (for double HJ) or resolution to ensure genome stability. Although HJ resolvases have been identified in all domains of life, there is a long-standing effort to search in prokaryotes and eukarya for proteins promoting HJ migration. Here, we report the structural and functional characterization of a novel ATPase, Sulfolobus islandicus PilT N-terminal-domain-containing ATPase (SisPINA), encoded by the gene adjacent to the resolvase Hjc coding gene. PINA is conserved in archaea and vital for S. islandicus viability. Purified SisPINA forms hexameric rings in the crystalline state and in solution, similar to the HJ migration helicase RuvB in Gram-negative bacteria. Structural analysis suggests that ATP binding and hydrolysis cause conformational changes in SisPINA to drive branch migration. Further studies reveal that SisPINA interacts with SisHjc and coordinates HJ migration and cleavage.
Keywords: ATPase, DNA recombinational repair, Holliday junction migration, archaea, Sulfolobus islandicus
Introduction
DNA recombination is one of the fundamental pathways for all domains of life and is essential for genome stability and genetic diversity. This process generates a four-way DNA intermediate, also called a Holliday junction (HJ) [1]. HJs are required to be dissolved or resolved for proper DNA repair and chromosome segregation [2,3]. Whereas the dissolution of a double HJ is accomplished in eukarya by a complex composed of BLM/Sgs1 RecQ helicase, type IA topoisomerase TOP3/TopoIIIa, and RMI1-RMI2/Rmi1 [2,3], the resolution reaction is achieved by a class of nucleases, the HJ resolvases, which catalyze the formation of paired nicks in the phosphodiester backbones of two opposing strands of HJDNA leading to two heteroduplex DNA products [4].
The prototypical HJ resolvase, Escherichia coli RuvC, is a dimeric endonuclease [5,6]. Although HJ resolvases have been identified in all domains of life including archaeal Hjc/Hje [7,8], GEN1 in mammals [9,10], and Yen1 in yeast [9], these nucleases have no sequence or structural homology with RuvC at the protein level [11]. It is well documented that RuvC accomplishes HJ cleavage in association with RuvA and RuvB [12–14], a tetrameric DNA binding protein [15,16] and a hexameric ATPase [17,18], respectively. RuvA interacts directly with HJ DNA and RuvB to regulate branch migration [19,20]. RuvB forms two hexameric rings on two opposite DNA arms of a HJ [21,22]. The RuvAB complex drives the migration of HJs to the preferred DNA sequence for incision by the RuvC dimer. In addition, RuvAB can target stalled replication forks in vivo and directly convert them into HJs, which can be processed by either fork reversal or recombination repair [23]. In vitro, the RuvAB complex can also generate a completely unwound product consisting of the paired nascent leading and lagging strands from stalled replication forks [24,25]. Apart from the RuvAB complex, E. coli RecG promotes the formation and migration of HJs by fork regression in vitro [26]. In eukarya, several proteins, including BLM [27,28], WRN [29], RecQ1 [30], RecQ5 [31], Rad54 [32], Rad5 [33], and FancM [34], have been reported to promote HJ migration. In archaea, Hjm (Hel308A) was reported to catalyze HJ migration in vitro [35] and is important for the restart of stalled replication forks [36,37]. However, the equivalents of the E. coli RuvAB migration motor have not been reported in eukarya or in archaea.
Here, we report the identification and characterization of a novel ATPase, the PilT N-terminal-(PIN)-domain-containing ATPase (PINA) from Sulfolobus islandicus (SisPINA). The PINA-coding gene is vital for cell viability in S. islandicus, and its homologs exist in all archaea species. SisPINA is able to mediate HJ migration and interact physically and functionally with SisHjc resolvase. The crystal structure of SisPINA reveals a hexameric ring assembly similar to that of RuvB. Together, these results suggest that PINA is a RuvB-like motor driving HJ migration.
The PIN domain was originally named for its similarity to the N-terminal domain of a type IV pili (PilT) ATPase from Myxococcus xanthus [38]. However, as indicated by Satyshur et al. [39], “the PIN domain is a misleading annotation because the authentic PilT N terminus domain has neither sequence nor structural homology with the PIN domain”. In fact, the PIN domain is a protein module consisting of about 130 aa residues present in all three domains of life. Some PIN domains have conserved acidic residues and a serine/threonine residue, forming a catalytic triad similar to those of FEN-1 family proteins such as FEN-1, T4 RNase H, and Taq polymerase I exonucease domain [38,40]. The majority of PIN-domain-containing proteins studied so far are nucleases and ribonucleases, which function as toxins of toxin/antitoxin system (VapBC) [41,42], translational inhibitors of growth regulation in bacteria [43,44], or enzymes in nonsense-mediated mRNA decay and surveillance [45,46], rRNA maturation [47,48], and down regulation of immune response [49] in eukarya. Some PIN-domain-containing proteins lack the canonical triad and are inactive as nucleases [45,50], but they may have other functions such as Poly (ADP-ribose) binding [50]. Our results suggest that the PIN-domain-containing ATPase PINA plays a key role in DNA recombination and repair.
Results
Identification of a novel ATPase associated with the resolvase Hjc
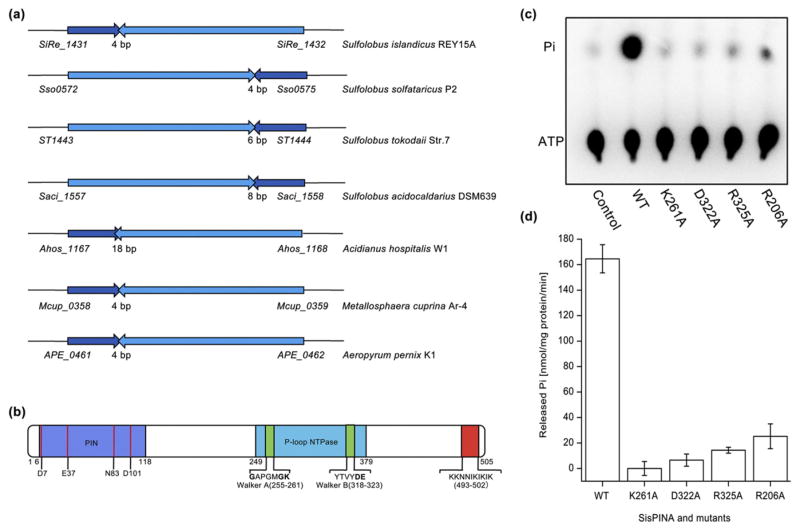
The hyperthermophilic archaeon S. islandicus Rey15A has two HJ resolvases, Hjc and Hje. Hjc is conserved in all archaea, whereas Hje is present only in some species of Sulfolobus. We previously showed that either gene can be deleted, but double knockout is lethal [51], in agreement with the observation that the DNA recombination pathway is essential in hyperthermophilic archaea [52,53]. To search for proteins that promote HJ migration and coordinate cleavage by Hjc, we overexpressed a C-terminal His-tagged Hjc in S. islandicus. A protein was co-purified from the cells with the His-tagged Hjc (Fig. S1). Interestingly, this protein was found to be the product of SiRe_1432, the gene located at the same locus with that encoding Hjc (SiRe_1431) in the S. islandicus Rey15A genome [54]. Gene SiRe_1432 has a 4-bp overlap with SiRe_1431 and is oriented in the opposite direction to SiRe_1431 in the genome (Fig. 1a). Bioinformatics and biochemical (see below) analysis indicated that SiRe_1432 encodes a P-loop ATPase with a PIN domain [38,55]; therefore, we named the protein SisPINA (Fig. 1b). In addition, there is a lysine-rich segment at the C terminus of SisPINA (Fig. 1b).
Fig. 1.
Genome context and domain organization of a PINA in Sulfolobus and its close relatives. (a) The genes encoding SisPINA and SisHjc and their homologs overlap over 4–18 bp and are oriented in opposite directions in the genomes of Sulfolobus and its close relatives. (b) Schematic domain organization of SisPINA. The PIN domain, P-loop NTPase domain, and the positively charged cluster at the C terminus are shown in blue, cyan, and red, respectively. The Walker A and Walker B motifs are shown in green. (c) Representative thin layer chromatographic profile of ATP hydrolysis by SisPINA and mutants. Control, no protein; WT, SisPINA; K261A, mutant at Walker A; D322A, mutant at Walker B; R325A, mutant at the highly conserved residue next to Walker B motif; R206A, mutant at a predicted arginine finger residue. (d) Quantification of the ATPase activity of SisPINA and the mutants in (c). The values were based on at least three independent replicates. The data were quantified using Image J [National Institutes of Health (NIH)].
SisPINA contains a central P-loop ATPase domain with typical Walker A (GAPGMGK) and Walker B (YTVYDE) motifs [56] (the conserved residues are shown in bold; Fig. 1b). To test if SisPINA has ATPase activity, we expressed SisPINA in E. coli and purified the protein by butyl, heparin, and gel-filtration chromatography (Fig. S2a). SisPINA displayed higher ATPase activity at basic pHs (pH 8.0 and 9.0) than at neutral (pH 7.0) or acidic pH (pH 6.0; Fig. S3a). In the presence of Ca2+, 168 nmol/mg protein/min ATP was hydrolyzed, while in the presence of magnesium and manganese, the ATPase activity was reduced to 104 and 97 nmol/mg protein/min (Fig. S3b). However, NaCl (0–1.0 M) had no apparent effect under standard assay conditions (Fig. S3c). In addition, a time course of the ATP hydrolysis by SisPINA was determined. As shown in Fig. S4a, around 32% of ATP was hydrolyzed at 12 min, and the hydrolysis rate was approximately 111 nmol/mg protein/min (Fig. S4a). We also examined the effect of various DNA substrates on the ATPase activity of SisPINA. As shown in Fig. S4b, the ATPase activity of SisPINA was not stimulated by ssDNA, dsDNA, 3′-overhang, 5′-overhang, Y-shaped DNA, HJ, or Replication fork.
To clarify the role of Walker A, Walker B, and other conserved residues in the ATPase activity of SisPINA, we performed ATP hydrolysis assays on one Walker A mutant (SisPINAK261A), one Walker B mutant (SisPINAD322A), one conserved-arginine mutant (SisPINAR325A), and one conserved putative arginine-finger mutant (SisPINAR206A; Figs. 1c, d, and S2b). In the presence of calcium, 45.3% ATP was hydrolyzed by the wild-type SisPINA in a reaction time of 10 min, whereas no more than 6.6% ATP was hydrolyzed by any of the four PINA mutants. These results indicate that the Walker A and Walker B motifs, and the specific conserved arginine residues, are important for the ATPase activity of SisPINA.
To verify the interaction between SisPINA and SisHjc, we performed a pull-down assay using individually purified SisPINA and SisHjc proteins (Fig. S5). Histagged SisHjc was able to pull down non-tagged SisPINA by Ni-resins (Fig. S5), confirming protein– protein interactions between them.
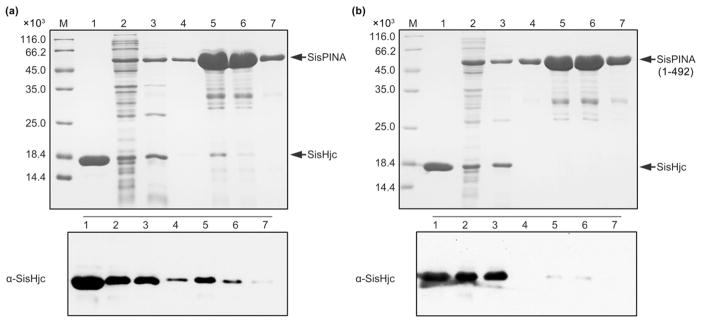
To further investigate the interaction between SisPINA and SisHjc, we co-expressed an N-terminal His-tagged PINA and untagged Hjc in bacteria. When the His-tagged SisPINA was purified from the bacterial extract by His-tag affinity chromatography, the results of SDS-PAGE and Western blotting showed that the non-tagged SisHjc was co-purified with the His-tagged SisPINA (Fig. 2a). As a control, we expressed the non-tagged SisHjc alone and applied the protein extract to nickel affinity chromatographic analysis. The result showed that non-tagged SisHjc could not bind to the Ni-resins (not shown). However, when SisHjc was co-expressed with a His-tagged truncated mutant (containing residues 1–492) lacking the C-terminal lysine-rich region, the non-tagged SisHjc was barely detectable (Fig. 2b), showing a dramatic decrease in the level of SisHjc co-purified with SisPINA. These results demonstrate that there is a protein–protein interaction between SisPINA and SisHjc, and the C-terminal region of SisPINA plays a key role in this interaction.
Fig. 2.
SisPINA interacts with SisHjc through its C terminus. (a) SisHjc was co-purified with His-tagged SisPINA from E. coli cells harboring co-expression vector pRSFDuet-1-SisPINA-SisHjc. Top: SDS-PAGE of the protein samples from the indicated steps of purification by His-tag affinity chromatography. The gels were stained with Coomassie blue. Bottom: western blot to monitor the presence of Hjc in the samples using polyclonal antibody against SisHjc. (b) Same experiments as in (a) except that SisPINA was replaced with a C-terminal truncation mutant, SisPINA(1–492). M, molecular size marker; 1, purified SisHjcE12A as a control; 2, whole extract; 3, heat-resistant supernatant; 4–7, elution fractions.
The gene encoding PINA is conserved across archaea and is essential for cell viability in S. islandicus
To understand the distribution of SisPINA homologs in the three domains of life, we performed a BLAST homology search. The results showed that all archaea species, including major and minor phyla, possess a SisPINA homolog (Figs. S6b and S7, and Table S5), whereas no PINA homolog was found in bacteria or eukarya. Consistent with the prevalence of SisPINA homologs across archaea, the phylogenetic tree based on SisPINA homologs (Fig. S6b) is similar to that generated using 16S rDNA sequences of corresponding species (Fig. S6a). Interestingly, in all Sulfolobus species and close relatives Acidianus, Metallosphaera, and Aeropyrum, the two genes encoding the resolvase Hjc and SisPINA homologs are located at the same locus, with a 4- to 18-bp-overlap and transcribed in opposite directions (Fig. 1a). The results suggest that PINA is a conserved ATPase in archaea and has an association with the HJ resolvase Hjc.
To understand the functional importance of PINA, we utilized a previously established S. islandicus Rey15A (E233S) gene deletion and complementation system [57,58] to test the impact of SisPINA gene knockout on cell viability (Fig. S8). According to this method [57], if the protein is encoded by a gene that is nonessential for cell viability, cells will grow and form white colonies on a medium containing uracil and 5-fluoroorotic acid (5-FOA). If the cells do not grow on this medium, the gene is likely essential (Fig. S8a). After several rounds of counterselection, we obtained no colonies from cells with the SisPINA gene deleted, implying that the cell cannot survive without SisPINA.
To confirm the biological importance of SisPINA, we tested if the chromosomal deletion of SisPINA can be complemented by the ectopic expression of SisPINA from a plasmid, pSSRlacS-SisPINA, which has a simvastatin selection marker [58]. Strain pMID-SisPINA-T, harboring plasmid pSSRlacS-SisPINA, was prepared, and the cells of this strain were counterselected on a medium consisting of mineral salts, sucrose, casamino acids, and a mixed vitamin solution (MSCV) plus uracil, 5-FOA, and simvastatin. Cells formed colonies, and the colonies were transferred into a liquid medium for further culture. The chromosomal PINA-coding gene was not amplified by PCR using gene-flanking primers (Table S2), providing the evidence of the gene deletion, while plasmid pSSRlacS-SisPINA was present in the cells (Fig. S8b and c). Even in the absence of simvastatin, the plasmid was maintained in the cells. These results together indicate that SisPINA is essential for the viability of S. islandicus.
SisPINA mediates branch migration in HJ DNA and unwinds Y-shaped DNA in vitro
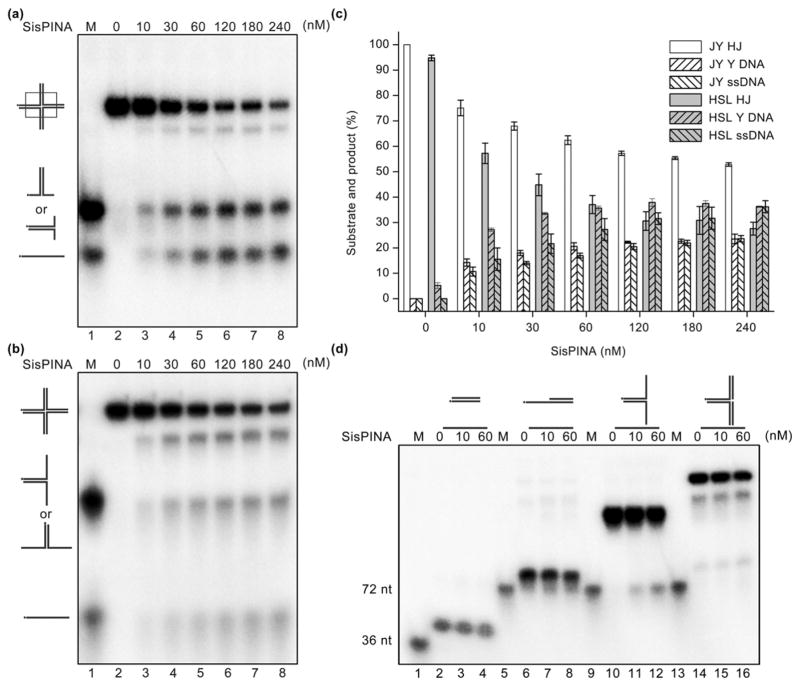
After observing that SisPINA exhibits ATPase activity and is associated with resolvase SisHjc, we further explored whether SisPINA can catalyze branch migration in HJ DNA (Fig. 3). We utilized two synthetic HJ substrates, mobile HSL [35] and fixed JY [59], for branch migration and unwinding analysis. HSL was made from four 70 nts forming four-way junction with 30 bps at the junction center, while JY was made from four 72-nt oligonucleotides with no homologous region (Table S1). Y-shaped DNA was produced by SisPINA for both HJ substrates, suggesting that SisPINA was able to drive branch migration (Fig. 3a,b). Notably, a slow migration band was observed immediately ahead of the HJ substrate band when SisPINA was mixed with either HJ substrate. We confirmed that this band is the product that one arm has released from HJ DNA (Fig. S9). Under the same conditions, SisPINA is more active on the mobile HJHSL than the fixed HJ JY (Fig. 3a–c). For example, at 60 nM SisPINA, 60% HSL HJ was processed, whereas only about 40% JY HJ was processed. To our surprise, SisPINA further processed both HJ DNA substrates into single-stranded DNA (ssDNA; Fig. 3a and b), showing the ability to unwind Y-shaped DNA (Fig. 3d). However, two ATPase-deficient mutants, SisPINAK261A and SisPINAR206A (Fig. 1c and d), lost the ability to promote branch migration or unwind HJ DNA (and likely the unwinding activity of Y-shaped DNA as well; Fig. S10), indicating that these activities of SisPINA are dependent upon ATP hydrolysis. Based on these results, we conclude that PINA is able to promote branch migration and unwind Y-shaped DNA powered by ATP hydrolysis.
Fig. 3.
SisPINA catalyzes HJ branch migration and unwinds Y-shaped DNA. (a and b) Representative gel electrophoretic profiles of SisPINA processing mobile (HSL) and fixed (JY) HJ DNA, respectively. (c) Quantification of the results in (a) and (b). Data were quantified using Image J (NIH), and each value was based on data from at least three independent experiments. (d) Analysis of the unwinding activity of SisPINA toward various DNA substrates: dsDNA (lanes 2–4), 5′-overhang (lanes 6–8), Y-shaped DNA (lanes 10–12), and replication fork DNA (lanes 14–16). Lane 1, 32P-labeled 36-nt ssDNA; lanes 5, 9, and 13, 32P-labeled 72-nt and 36-nt ssDNA as markers (M). The concentrations of SisPINA (as hexamer) were 0, 10, and 60 nM, as indicated above the gel. All reactions were carried out in a 20-μl mixture at 55 °C for 30 min, as described in Materials and Methods.
To further characterize the DNA-unwinding activity of SisPINA, various DNA substrates were tested, including blunt-ended double-stranded DNA (dsDNA; 36-bp), 5′-overhang (72-nt with 36-bp), Y-shaped (72-nt with 36-bp matches), and replication fork DNA (72-nt parental strands and 36-nt nascent strands; Table S1). Intriguingly, SisPINA can only unwind Y-shaped DNA among these substrates (Fig. 3d). However, SisPINA binds to most of these DNA substrates as shown by electrophoretic mobility shift assays. SisPINA formed stable protein–DNA complexes with replication fork and HJ DNA, as indicated by the shifted bands above the corresponding free DNA bands (Fig. S11). In addition, SisPINA reduced the level of free DNA in reactions with a blunt-ended DNA or a 72-nt ssDNA (Fig. S11b), suggesting that there are unstable protein–DNA interactions of SisPINA with these DNA substrates as well. Based on electrophoretic mobility shift assay results with 40 nM SisPINA, the affinity of SisPINA for these various DNA substrates ranges from highest to lowest in such order as HJ DNA ~ replication fork DNA ≫ Y-shaped DNA > 5′-overhang DNA ≫ dsDNA~ssDNA (Fig. S11).
Binding of SisHjc to HJ inhibits branch migration by SisPINA
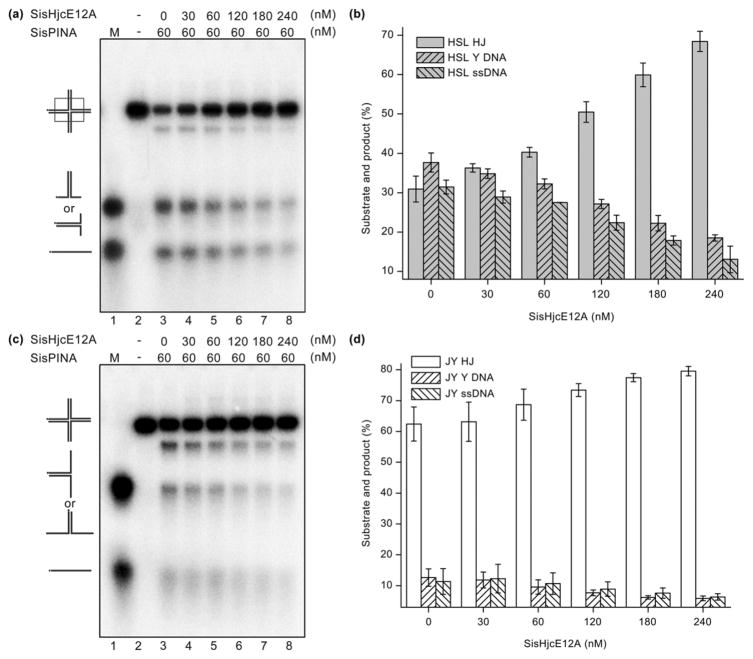
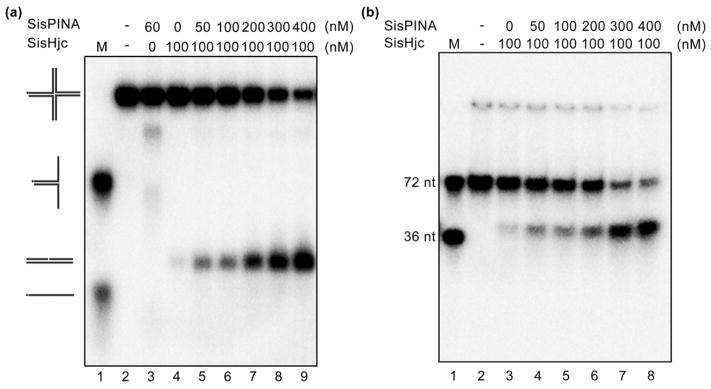
We have shown that SisPINA and SisHjc interact with each other (Figs. 2 and S5). To investigate if this interaction has any impact on their individual biochemical activities, we repeated the SisPINA HJ-processing experiments in the presence of the resolvase SisHjc. To avoid HJ cleavage by SisHjc during the process, mutant SisHjcE12A, which has a deficient DNA-cutting but normal DNA-binding activity, was used [60]. In the absence of SisHjcE12A, around 65% HSL DNA was processed by SisPINA into Y-shaped DNA (branch-migration product) and ssDNA (Fig. 4a and b). When the concentration of added SisHjcE12A was increased, the level of branch migration by SisPINA decreased (Fig. 4a and b), indicating that the binding of SisHjcE12A to HJ inhibits branch migration by SisPINA. For example, when the concentration of SisHjcE12A reached 240 nM, only 33% HSL DNA was processed by SisPINA compared to 65% processed in the absence of SisHjcE12A (Fig. 4a and b). However, the ratio of Y-shaped DNA to ssDNA was almost the same, indicating that SisHjcE12A does not inhibit Y-shaped DNA unwinding by SisPINA (Fig. 3a versus Fig. 4a). Similar results were obtained for the fixed HJ substrate JY (Fig. 4c and d).
Fig. 4.
HJ branch migration by SisPINA is inhibited by the nuclease-deficient mutant SisHjcE12A. Representative results of gel electrophoresis of (a) HSL HJ and (c) JY HJ processed by SisPINA in the presence of SisHjcE12A. Increasing concentrations of SisHjcE12A (as dimer) were added to a reaction mixture containing 60 nM SisPINA (as hexamer). (b and d) Quantification of the results in (a) and (c), respectively. Quantification of data using Image J (NIH) was based on values from at least three independent experiments.
SisPINA enhances the cleavage of the preferred strand by SisHjc during HJ resolution
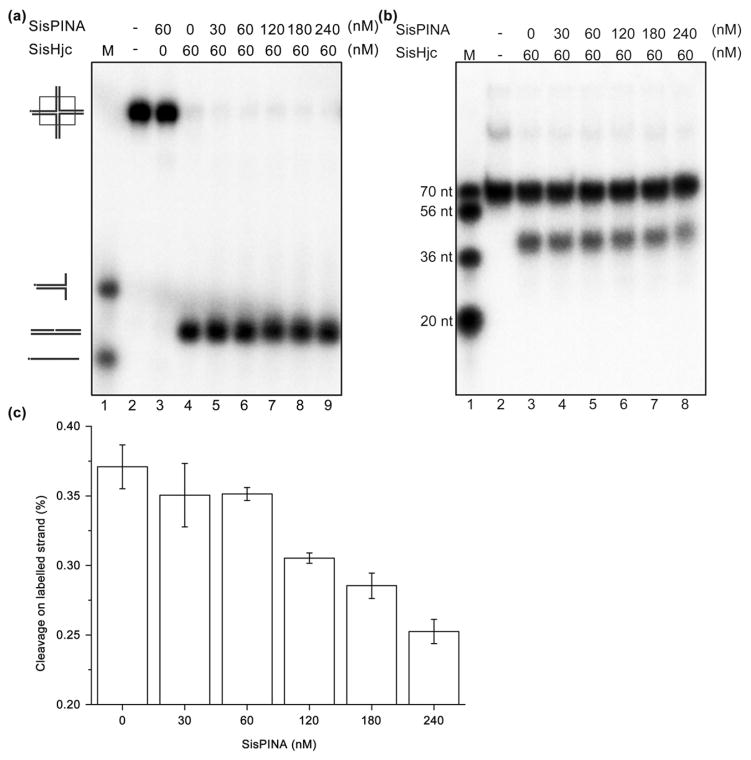
We next examined the effect of SisPINA on the HJ-cleavage activity of SisHjc. In the absence of SisPINA, SisHjc is able to resolve JY HJ by itself, but the cleavage was not efficient (Fig. 5). When SisPINA was added to the mixture, the cleavage of JY by SisHjc was enhanced and increased with increasing concentration of SisPINA (Fig. 5). The cleaved oligonucleotides are a little longer than the 36-nt marker (Fig. 5b), in agreement with previous observations that archaeal Hjc resolvase cuts the phosphodiester bonds between the third and fourth nucleotides on the 3′ side of the junction [7,60,61]. By comparing the level of ssDNA products in the denaturing gel with that of dsDNA products in the native gel (Fig. 5), the fixed HJ substrate JY was resolved with cleavage by SisHjc almost exclusively on the labeled strand 1 and strand 3 (Fig. S12).
Fig. 5.
SisPINA stimulates the cleavage activity of SisHjc on a fixed HJ substrate. The cleavage of the JY substrate by SisHjc in the presence of SisPINA at the indicated concentrations was analyzed by (a) native and (b) denaturing gel electrophoresis. The reaction was carried out at 55 °C for 30 min for native gel electrophoresis or at 65 °C for 30 min for denaturing gel electrophoresis. The use of 55 °C for the native gel electrophoresis is to prevent the product from denaturing at higher temperatures.
In contrast to the relatively low efficiency of cleavage of the fixed HJ substrate JY, SisHjc displayed much higher cleavage activity on the mobile HJ substrate HSL (compare lane 4 in Fig. 6a with lane 4 in Fig. 5a). According to the results of native gel electrophoresis, SisHjc (60 nM) was able to cleave all of the HSL HJ, whereas less than 5% of the fixed HJ substrate JY was resolved under the same conditions. When the HJ substrates are completely cleaved, the upper band generated during denaturing gel electrophoresis (uncut labeled strand; Fig. 6b) represents the dsDNA product formed by cleavage of unlabeled strands (Fig. S12), whereas the lower band represents cleavage of the labeled strand. The results showed that only about 35% of the labeled strand in the mobile HJ substrate HSL was cleaved (lower band in lane 4 of Fig. 6b), although 100% HSL was resolved (lane 4 in Fig. 6a), indicating that 65% HSL was resolved by cleavage of unlabeled strands. Interestingly, when an increasing amount of SisPINA was added to the reaction, the amount of cleaved 32P-labeled ssDNA decreased, indicating that the preference for cleavage of unlabeled strands of the mobile HJ substrate HSL was enhanced by SisPINA, whereas cleavage of the labeled strand was either unchanged or inhibited.
Fig. 6.
SisPINA enhances the cleavage of the unlabeled strand by SisHic on a mobile HJ substrate. Analysis of the cleavage activity of SisHjc on HSL by (a) native and (b) denaturing gel electrophoresis in the presence of SisPINA at the indicated concentrations. The reaction was carried out at 55 °C for 30 min. (c) Amount of labeled strand cleaved by Hjc relative to total HSL HJ. The density of the substrate or product bands was quantified using Image J (NIH). Each value was based on at least three independent experiments.
Crystal structure of SisPINA reveals a hexameric-ring assembly like that of bacterial RuvB helicase
In order to further characterize SisPINA at the molecular level, we determined the crystal structure of the Walker A mutant SisPINAK261A after many trials failed to obtain high-quality crystals of the wild-type SisPINA protein. The crystal structure was solved at 2.8 Å resolution by the multiple-wavelength anomalous diffraction (MAD) method (Table 1) using seleno-methionine (SeMet)-substituted SisPINAK261A. SisPINAK261A was crystallized in space group P1211 with six molecules per asymmetric unit. The six SisPINA molecules assemble into a hexameric ring (Fig. 7a), which is in agreement with the results of size-exclusion chromatography (Fig. S13). The elution profile of SisPINAK261A showed a major peak corresponding to a hexamer and a minor one presumably to be the monomer of the protein. In crystals, as shown in Fig. S14, each PINA hexamer (green) is surrounded by nine PINA hexamers: three hexmers (red) in the same level forming a triangle, three hexamers (yellow, magenta, cyan) half way up, and three hexamers (yellow, magenta, cyan) half way down to fill the gaps among the three red hexamers.
Table 1.
Statistics for data collection and refinement
| Data collection | BL12-2 | BL14-1 | BL12-2 | BL12-2 | BL12-2 |
| Crystal | Native | SeMet | SeMet | SeMet | SeMet |
| Space group | P 1 21 1 | P 1 21 1 | P 1 21 1 | P 1 21 1 | P 1 21 1 |
| Cell dimensions | |||||
| a, b, c (Å) | 101.72, 148.98, 122.13 | 102.04, 149.32, 122.49 | 102.03, 149.32, 122.48 | 102.08, 149.33, 122.50 | 101.93, 149.07, 122.07 |
| α, β, γ (°) | 90.00, 104.07, 90.00 | 90.00, 104.14, 90.00 | 90.00, 104.10, 90.00 | 90.00, 104.13, 90.00 | 90.00, 104.15, 90.00 |
| Peak | Inflection | High | Low | ||
| Wavelength (Å) | 0.9795 | 0.9725 | 0.9797 | 0.9184 | 1.0332 |
| Resolution (Å) | 39.49–2.82 (2.87–2.82)* | 39.59–3.20 (3.28–3.20) | 39.59–3.50 (3.59–3.50) | 39.60–3.20 (3.28–3.20) | 39.45–3.42 (3.51–3.42) |
| Rmerge(%) | 0.145 (2.453) | 0.557 (5.520) | 0.303 (1.970) | 0.281 (2.810) | 0.319 (4.499) |
| I/σI | 16.1 (4.3) | 9.9 (0.7) | 9.3 (1.2) | 9.0 (0.6) | 9.7 (0.6) |
| Completeness (%) | 97.9 (94.8) | 99.5 (95.6) | 98.1 (94.0) | 98.0 (93.1) | 97.6 (93.3) |
| Redundancy | 7.0 (6.6) | 29.4 (22.3) | 7.1 (6.6) | 6.3 (3.4) | 14.0 (12.7) |
| Refinement | |||||
| Resolution (Å) | 39.49–2.82 | ||||
| No. of reflections | 83,650 | ||||
| Rwork/Rfree | 23.52/26.07 | ||||
| No. of atoms | |||||
| Protein | 20,068 | ||||
| Glycerol | 132 | ||||
| 1,2-Ethanediol | 52 | ||||
| Water | 275 | ||||
| B-factors | |||||
| Protein | 82.73 | ||||
| Glycerol | 94.19 | ||||
| 1,2-Ethanediol | 89.67 | ||||
| Water | 72.47 | ||||
| R.m.s.d. | |||||
| Bond lengths (Å) | 0.008 | ||||
| Bond angles (°) | 1.421 | ||||
| Ramachandran | |||||
| Core | 95.4% | ||||
| Allowed | 4.5% | ||||
| Gen. Allowed | 0.0% | ||||
| Disallowed | 0.0% | ||||
Values in parentheses are for highest-resolution shell.
Fig. 7.
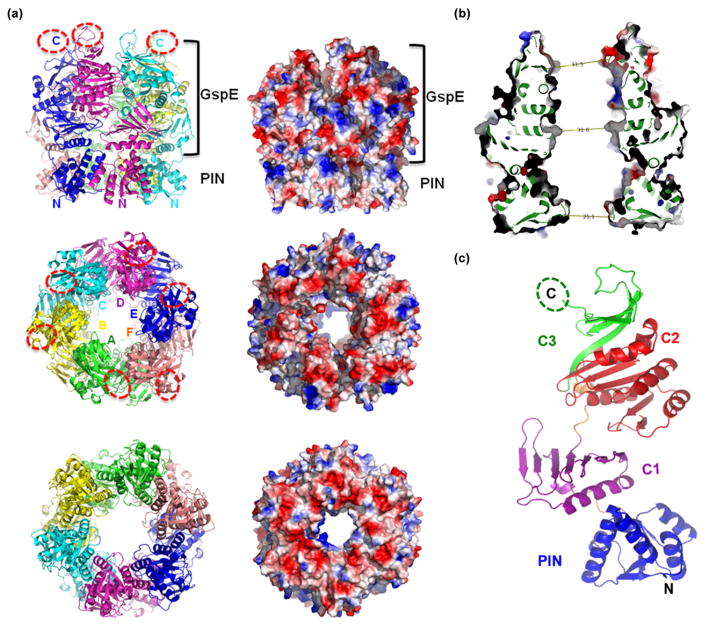
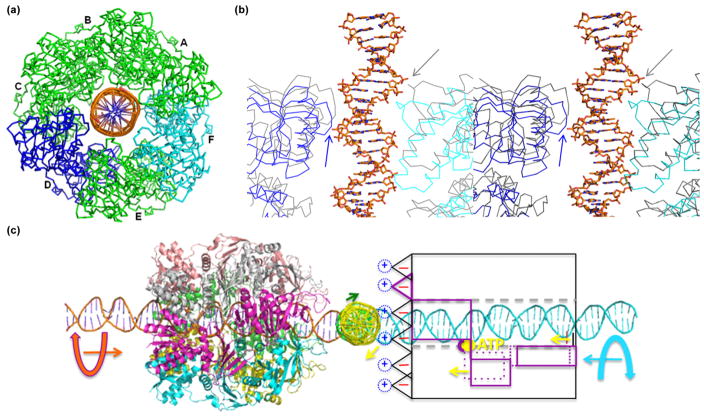
Crystal structure of SisPINA. (a) SisPINA forms a hexameric crown in crystals. The ribbon (left) and electrostatic potential surface (right) presentations of the side view (top), the crown points of the C-terminal domains (middle), and the crown ring of the PIN domains (bottom) are shown. The six subunits are displayed in different colors in a ribbon model. Dashed oval rings represent the missing C-terminal tails. Positive and negative electrostatic potential surfaces are shown in blue and red, respectively. Sections with structural homology to PIN or secretion ATPase (GspE) are indicated. (b) Dimensions of the central channel in the PINA hexamer. Cross-section of the hexamer shown in electron potential surfaces and green ribbons. The distances at three locations along the channel are measured. The N-terminal entrance has the smallest diameter of ~25 Å by the clamp loops of PINA subunits. (c) The structure of a SisPINA monomer in a ribbon model. The four domains, PIN, C1, C2, and C3, are shown in blue, magenta, red, and green, respectively. The linkers between the domains are in orange. The missing C-terminal tail is presented by a dashed oval ring.
The overall shape of the SisPINA hexamer looks like a crown (Fig. 7a, top), with the PIN domains forming the crown ring and with the C-terminal domains forming the crown points. The SisPINA crown has outer dimensions of 92 Å (diameter) × 100 Å (height). The central hole is a tunnel of about 31 Å in diameter (Fig. 7b). Therefore, the hexameric crown is big enough to hold a normal dsDNA in its central tunnel. Interestingly, the loops formed by residues 74–82 (GDDSKKGEA) of the PIN domains narrow the central tunnel to its smallest diameter (~25 Å) at the N-terminal entrance (Fig. 7b), implying that these loops (referred to as the clamp loop from here on) could potentially have the strongest interactions with the dsDNA in the tunnel of the SisPINA crown. In agreement with this hypothesis, these loops provide positive electrostatic potential to the N-terminal entrance of the tunnel, whereas the crown ring has predominantly negative electrostatic potential surface (Fig. 7a, bottom). The positively charged residues K78K79 at the tip of this clamp loop likely interact directly with the negatively charged phosphodiester backbones of the dsDNA.
Structure of SisPINA
Each SisPINA molecule consists of four domains, with the overall shape resembling that of a sea horse, with the PIN domain as the head and the three other domains (C1, C2, and C3) as the body (Fig. 7c). Although the full-length SisPINAK261A was crystallized, the 69 residues (437–505) at the C terminus are not detectable for any of the 6 SisPINA molecules in the crystal due to the lack of electron density, indicating a flexible C-terminal tail that can adopt different secondary structures and/or positions to interact with the Hjc resolvase, as shown in Fig. 2.
The PIN domain (residues 1–118) consists of a central sheet of five parallel β-strands surrounded by four α-helices on one side and two α-helices on the other side (Fig. 7c). As expected, the PIN domain of SisPINA shares structural similarities with PIN domains of other proteins, including the nuclease PAE2754 (PDB code 1V8P, Dali Z-score 7.2, RMSD of 3.4 Å over 96 residues) from Pyrobaculum aerophilum. PAE2754, the first reported structure of a PIN-domain family protein in archaea [55], is a structural homolog of T4RNase Hand other exonucleases, which usually contain three or four conserved acidic residues for binding to one or two divalent metal ions required for catalysis [62]. However, we did not detect any nuclease activity in the mutant SisPINA (1–120) containing only the PIN domain (data not shown). Structural comparison revealed that the negatively charged residue aspartate (D92) at the active site of PAE2754 is replaced by asparagine (N83) in SisPINA (Fig. S15). This replacement may eliminate the ability of the PIN domain in SisPINA to bind two divalent metal ions required for the nuclease activity. Therefore, the PIN domain of SisPINA has no nuclease activity but likely retains its ability to interact with DNA, an important property for SisPINA to catalyze branch migration.
The PIN domain of SisPINA is connected to the C1 domain by a short loop (residues 119–124). The C1 domain (residues 125–225) contains a central sheet of seven antiparallel strands with two helices on one side adjacent to the PIN domain (Fig. 7c). The C1 domain is connected to the C2 domain through a long linker containing a one-turn helix (residues 226–236). The C2 domain (residues 237–380) is the conserved P-loop ATPase domain consisting of a central sheet of six parallel strands flanked by two helices on one side and four helices on the other, next to the C3 domain. The central sheet of the C2 domain is extended by a long seventh antiparallel strand. The C-terminal half of this seventh strand is also part of the C3 domain (residues 381–436) formed by four antiparallel strands with a large loop (residues 397–404) forming the points of the crown.
Although SisPINA shows structural homology in the P-loop ATPase (C2) domain with the RuvB helicase (PDB code 3PFI, Dali Z-score 7.8, RMSD of 5.8 Å over 129 residues), structural analysis and comparison by the Dali server [63] indicate that the seahorse body (residues 120–436) of SisPINA shares the highest structural similarity to the archaeal secretion ATPase GspE (PDB code 2OAP, Dali Z-score 20.2, RMSD of 2.9 Å over 267 residues), twitching motility protein PilT (PDB code 2GSZ, Dali Z-score 19.4, RMSD of 3.0 Å over 258 residues), and Flal ATPase (PDB code 4IHQ, Dali Z-score 19.4, RMSD of 3.8 Å over 269 residues), all of which are type II/IV secretion hexameric ATPases. One of the structural features conserved in these ATPases is the conformational dynamics revealed by multiple conformations observed for the subunits within the hexameric assembly [39,64–66]. Similarly, two different subunit conformations are identified in the PINA hexamer.
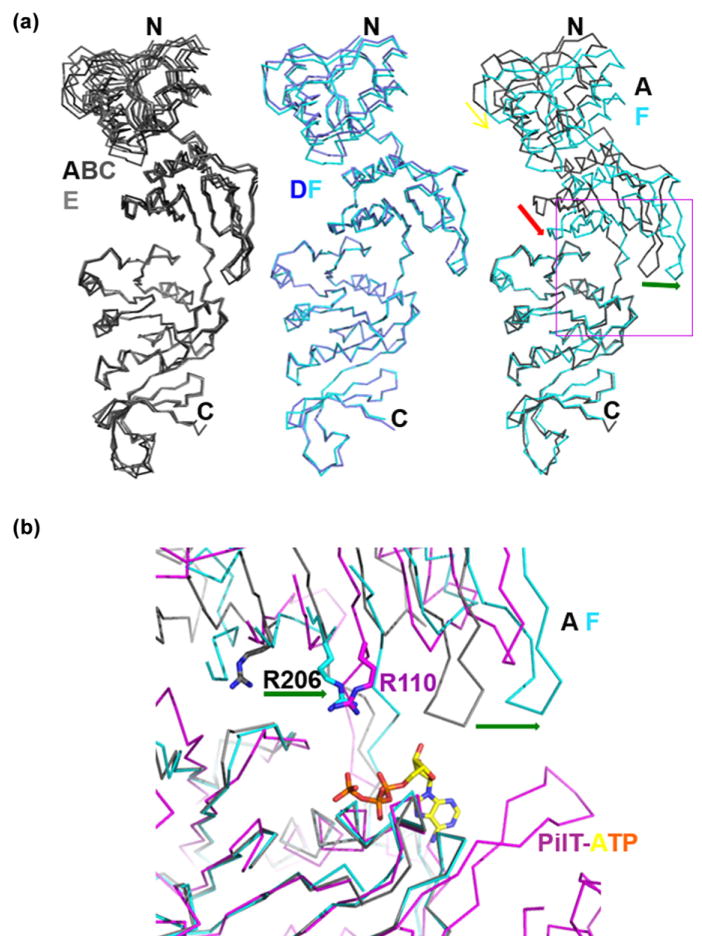
Two different subunit conformations within the PINA hexamer
Of the six SisPINA molecules in the hexameric assembly, subunits A, B, C, and E have the same conformation, while subunits D and F share a different conformation caused by the shift of the linker between the C1 and C2 domains (Fig. 8a). There are three noticeable structural differences between these two subunit conformations (Fig. 8a): (1) the central sheet of the C1 domain moves closer to the C2 domain (red arrow), (2) the loop between the second and third strands of the C1 domain shifts away from the C2 domain (green arrow), and (3) the clamp loop (residues 74–82) of the PIN domain closes down (yellow arrow). The result of these conformational changes is two different ATP-binding pockets between subunit A (the same as subunit B, C, and E) and subunit F (the same as subunit D; Fig. 8b). Compared to the structure (magenta wires in Fig. 8b) of the ATP-bound PilT ATPase (PDB code 2EWW), the ATP-binding pocket of subunit F (and D) represents the ATP-binding conformation: the ATP (in sticks) bound to the P-loop motif and arginine residue R206 is approximately in the same position as arginine residue R110 of the PilT ATPase. Residue R110 was reported to be conserved among members of the type II/IV secretion ATPase superfamily and is important for ATP hydrolysis by virtue of stabilizing the leaving phosphate group [39]. In subunit A (and B, C, E), however, arginine residue R206 is too far away to make any contact with the leaving phosphate group (Fig. 8b), suggesting that subunit A (and subunit B, C, and E with the same conformation) represents the SisPINA conformation without ATP. In agreement with this structural analysis, substitution of R206 with alanine in SisPINA dramatically reduced the ATPase activity (Fig. 1c and d).
Fig. 8.
Two subunit conformations observed in the PINA hexamer. (a) Superimposition of the six SisPINA subunits over the ATPase domain. The six subunits are shown in a wire model. Four subunits (A, B, C, and E) in gray have the same conformation (left), whereas two subunits D (blue) and F (cyan) have a different conformation (middle) caused by the shift of the linker between the C1 and C2 domains (right). The linker shift results in three changes (right): (1) the lower level (red arrow) of the central-sheet of the C1 domain, (2) the shift (green arrow) of the loop between the second and third strands of the sheet in the C1 domain, and (3) the closing down (yellow arrow) of the clamp loop of the PIN domain. (b) Different ATP-binding pockets between the two subunit conformations. The A (gray) and F (cyan) subunits of SisPINA are superimposed on the ATP-bound PilT ATPase (PDB code 2EWW, magenta) over the ATPase domain, ensuring the best superimposition in the Walker A motif (phosphate-binding loop). ATP from PilT is shown in colored sticks. The “arginine finger” residue R110 of PilT is displayed in sticks (N atoms in blue) together with the corresponding arginine residue R206 of SisPINA. The shifts of both R206 and the loop between the subunits A and F are indicated by the green arrows. Only the interface between domains C1 and C2 framed in the magenta square on the right panel of (a) is shown.
Discussion
Here, we have reported the identification and characterization of a novel ATPase SisPINA. Previously the PINA homolog in S. solfataricus, the Sso0572 protein, was studied together with other four predicted secretion ATPases based on their sequence homology to bacterial PilT secretion ATPase [67]. Sso0572 was active as an ATPase in the presence of divalent cations including Ca2+, Mn2+, and Mg2+, in agreement with our results. However, unlike other four candidates, the upstream and downstream genes of sso0572 gene are unrelated to secretion system, strongly suggesting that the Sso0572 protein is not a secretion ATPase. In contrast, our results suggest that SisPINA is involved in DNA metabolism. In S. islandicus, like all the genes encoding proteins involved in DNA recombination such as RadA, Mre11, Rad50, HerA, NurA, and Hjm [53,68], SisPINA is essential. It is highly likely that PINA is a protein that participates in DNA recombinational repair in this archaeaon.
Our results indicate that SisPINA preferably binds to HJ and replication fork DNA over other forms of DNA. Based on the structural and electrostatic potential surface features of the SisPINA hexamer, we propose that the SisPINA hexameric crown binds to HJ DNA with the central tunnel (25–31 Å in diameter) large enough to hold one dsDNA arm (20 Å in diameter; Fig. 9a). Because the C terminus of SisPINA is important for its interaction with SisHjc, the crown points likely face the junction when SisPINA binds to the arms of HJ so that it not only allows SisPINA to interact with SisHjc bound at the junction but also has the positively charged C-terminal tails (dotted small circles in Fig. 9c, right cartoon) to reduce the repulsive force between the DNA backbones (central yellow and green DNA in Fig. 9c) and negatively charged surfaces of the crown points (triangles in Fig. 9c, right cartoon).
Fig. 9.
Models of SisPINA bound to HJ DNA and branch migration. (a) Model of the SisPINA hexamer with dsDNA (as one arm of an HJ DNA) bound through its central tunnel. Subunits A, B, C, and E are shown as green wires, whereas subunits D and F are displayed in blue and cyan, respectively. The dsDNA was adapted from the DNA–UvrD complex [72] (PDB code 2IS4) with the phosphodiester backbones shown as orange ribbons. (b) Stereo view of the DNA-SisPINA model at the cross-section of DNA and the clamp loops from subunits D (blue) and F (cyan). In order to show that the closing down (indicated by blue and gray arrows) of the clamp loop possibly causes the 2-bp translocation of the dsDNA by ATP binding and hydrolysis, subunit B (light gray, representing conformation without ATP) is superimposed on subunit D and F (ATP-bound conformation), respectively. Other subunits are not shown for visibility, whereas dsDNA is shown in sticks. (c) Model of two SisPINA hexamers bound to the open X-HJ DNA. The open X-HJ DNA was made by dsDNA from the DNA–UrvD complex (PDB code 2IS4) and the DNA–RuvA complex (PDB code 1C7Y) with modifications. The four arms are shown in green, cyan, yellow, and orange, respectively. One SisPINA hexamer bound to one arm (orange) of the HJ DNA is shown as ribbons with the six subunits in different colors. The other SisPINA hexameric crown bound to the opposite DNA arm (cyan) is shown in the scheme to illustrate the movement possibly induced by ATP binding and hydrolysis to drive the translocation of dsDNA. The direction of DNA translocation is indicated by arrows in colors matching those of the DNA arms. One subunit is shown in magenta boxes representing different domains: solid lines for empty SisPINA and dashed lines for ATP (yellow ball)-bound SisPINA. Two small yellow arrows indicate the conformational changes induced by ATP binding. The clamp loop is indicated by black and magenta bars. The dashed gray lines indicate the central tunnel of the SisPINA hexameric crown. The orientations of the two SisPINA hexamers are based on the results of SisPIN–Hjc interactions. The missing C-terminal tails are indicated by dashed blue circles, with “+” representing positive charge. The negative electrostatic potential surfaces of the crown points are indicated by “−” within the triangles of the crown points.
The central tunnel of the SisPINA hexamer is narrowed down to its smallest diameter of ~25 Å at the N-terminal entrance by the clamp loops (Figs. 9b and 7b), suggesting an important role for the clamp loop in DNA interaction and translocation. The six subunits of the SisPINA crown have two different conformations representing ATP-free and ATP-bound states, respectively, when compared to the complex structure of the PilT ATPase with bound ATP (Fig. 8b). As shown in Fig. 9b, compared to the conformation without ATP (gray wires), the clamp loop of SisPINA closes down about 2 bps in subunits D (blue wires) and F (cyan wires), which represents the ATP-bound conformation. Based on these structural analyses, we propose that ATP hydrolysis by subunit D (and F) would release the clamp loop and propel the 2-bp translocation of the dsDNA arm encircled by the central tunnel of the SisPINA crown (Fig. 9c, right cartoon). Because subunits D and F (120° or 240° away) instead of subunits D and A (180° away) within the SisPINA hexameric ring have the ATP-bound conformation (Fig. 9a), the dsDNA arm will likely be forced to rotate 60° when the SisPINA hexamer propels the dsDNA by 2 bps through the central tunnel (Fig. 9c). In order to maintain the overall subunit interfaces within the SisPINA hexamer, the next two subunits A and E become ATP-bound and ready for the next 2-bp translocation powered by the energy released by ATP hydrolysis. When two SisPINA hexamers bind separately on the opposite DNA arms of the HJ DNA (Fig. 9c), the two SisPINA hexamers propel the two DNA arms toward the junction, and these two dsDNA arms rotate in opposite directions to neutralize the supercoil stress. Driven by these two rotations, the other two dsDNA arms without SisPINA will rotate automatically and drive the junction to migrate along the DNA until the Hjc resolvase is recruited to the junction for resolution.
Archaeal Hjc resolvase functions as a dimer. Crystal structures of some Hjc resolvases have been published [69,70]. In addition, the crystal structure (3.1-Å resolution) of Archaeoglobus fulgidus Hjc resolvase in complex with an HJ DNA is also available in the Protein Data Bank (PDB code 2WJ0); unfortunately, no publication is available to illustrate the structural details. As shown in Fig. S16a, two Hjc dimers bind to the HJ DNA, with one dimer interacting with one side of the junction. Each dimer has only one Hjc monomer (red or green ribbons in Fig. S16) to approach the phosphodiester backbone at its active site (residues D38, E51, and K53 in spheres). The other Hjc monomer (yellow in Fig. S16) has limited contact with DNA. The active Hjc monomer (red or green) also has interactions with the adjacent DNA arm, therefore leading to the formation of a stacked X-HJ conformation (Fig. S16), consistent with the previous observation that Hjc manipulated a synthetic HJ substrate into an X shape in solution [60]. The stacked X-HJ DNA should be difficult for branch migration compared to open HJ junctions, supporting our observation that the binding of SisHjc to the HJ junction inhibited the branch migration by SisPINA (Fig. 4). In addition, the binding of SisHjc to the junction may sterically hinder branch migration by SisPINA as well. For these reasons, we propose that SisPINA binds to HJ before Hjc and catalyzes branch migration until Hjc is recruited. The binding of Hjc to the HJ DNA stops the branch migration and initiates the cleavage at the junction. The binding of SisPINA to the HJ DNA may facilitate the recruitment of Hjc to the HJ DNA, therefore enhancing the HJ cleavage by Hjc. Since no RuvA homologs have so far been identified in archaea, we think that PINA possibly has both HJ opening and migration functions because PINA alone can bind to HJ and replication fork DNA with intrinsic ATPase activity. We are trying to identify other proteins that may play a role like RuvA and coordinate with PINA in HJ processing.
The AfuHjc–HJ complex structure may provide some hints to strand preference by Hjc incision during HJ resolution. Hjc binds to HJ as a dimer, but the two monomers interact with the HJ DNA asymmetrically as shown by the crystal structure with one monomer engaged in the active form with the HJ DNA, while the other monomer interacts with limited contacts with DNA. Therefore, how the two Hjc dimers bind to the DNA strands of the HJDNA may provide the structural basis for strand preference of Hjc on HJ cleavage. As shown in Fig. S16, the red and green DNA strands are the preferred cutting strand for the red or green Hjc monomer, respectively. We have shown that SisHjc preferably cut the labeled strand of the fixed HJ substrate and the unlabeled strand of the mobile HJ substrate as described in the Results. We also showed that SisPINA enhanced the preferred cleavage for both HJ substrates (Figs. 5 and 6). Since two SisPINA hexamers bind to only two of the four arms in the HJ DNA, the interactions of SisPINA with SisHjc may enhance the asymmetric distribution of the two monomers of each Hjc dimer at the DNA junction and facilitate the interaction of the active Hjc monomer with the preferred DNA strand in the HJ DNA, therefore enhancing the preferred DNA cleavage as well. This is consistent with the model proposed in Fig. S16b, showing that the magenta PINA hexamer interacts with the red Hjc monomer, while the cyan PINA hexamer interacts with the green Hjc monomer, but neither PINA hexamer has interactions with the non-active Hjc mononers (yellow). Further studies, particularly the structure of the complex consisting of PINA, Hjc, and HJ DNA, are required to fully understand this process.
Materials and Methods
Phylogenetic analysis
The amino acid sequences of PINA homologs and the 16S rDNA sequences of corresponding species were obtained from the non-redundant protein sequence database (National Center for Biotechnology Information). Neighbor-joining trees were constructed using the bootstrap method and Mega 5.5 software. The number of bootstrap replications was 1000.
Construction of the SisPINA knockout plasmid and transformation of S. islandicus E233S
The plasmid pMID-SisPINA for the knockout of the gene encoding SisPINA (SiRe_1432) was constructed according to a method described previously [57,71]. Briefly, the genomic DNA fragments containing gene SiRe_1432 (G-arm, 841 bp), the upstream flanking sequences (L-arm, 895 bp), and the downstream flanking sequence (R-arm, 845 bp) were amplified by PCR using S. islandicus E233S genomic DNA as the template and the corresponding primers (Table S2). The primers contained restriction sites SphI/XhoI, XhoI/NcoI, and MluI/SalI for the L-, R-, and G-arms, respectively. The amplified fragments were digested with the corresponding enzymes and ligated sequentially into pMID. All inserted sequences were confirmed by sequencing (BGI, Beijing, China). pMID-SisPINA was linearized by digestion with SphI and SalI, and the fragment containing the marker pyrEF-lacS (1 μg) was utilized to transform S. islandicus E233S cells by electroporation as previously described [57,71]. Cell suspension (100 μl) was spread onto a plate of a medium consisting of mineral salts (M), 0.2% (wt/vol) sucrose (S), 0.2% (wt/vol) casamino acids (C), and a mixed vitamin solution (V) (MSCV) solidified with 1% Gelrite (Sigma). The plate was incubated at 75 °C for 7 days, and the colonies were stained with X-gal. The blue colonies were selected and transferred into MSCV liquid medium and cultured at 75 °C for 7 days. The cells from 3-ml culture were collected by centrifugation, and the genomic DNA was extracted. The genotype of the transformant was identified by PCR using the SisPINA flanking primers (Table S2). The purified strain was named pMID-SisPINA-T (having functional SisPINA and the inserted pyrEF-lacS marker). For counterselection, the cells were further cultured in MSCV liquid medium for 3 days and transferred into fresh liquid medium. After two more rounds of subculture, the transformed cells were spread on a MSCV plate containing uracil and 5-FOA. The genotype of the clones appearing on the plate was determined by PCR.
Genetic complementation
The simvastatin selection system [58] was used for the genetic complementation of SisPINA. The DNA fragment containing the gene encoding SisPINA was obtained by the digestion of pET15bm-SisPINA using NdeI and SalI restriction enzymes. The gene fragment was then cloned into pSeSD [72] at the NdeI/SalI site, and the resultant plasmid was named pSeSD-SisPINA. The fragment containing the arabinose promoter and the SisPINA gene in pSeSD-SisPINA was amplified using primers pSSRlacs-araP-XmaI-Forward and SisPINA-SalI-Reverse (Table S2). The amplified fragment was digested with XmaI and SalI and ligated into pSSRlacS [58], generating plasmid pSSRlacS-SisPINA. Competent pMID-SisPINA-T cells were prepared according to the method described previously [58]. The cells were transformed with pSSRlacS-SisPINA by electroporation and transferred into a 30-ml MTSV liquid medium{0.2%(wt/vol) tryptone [T]} containing 12 μM simvastatin. After incubation for about 10 days, when the OD600 of the culture reached 0.6, 5-ml cell culture was transferred into fresh MTSV liquid medium containing 12 μM simvastatin. Cells harboring pSSRlacS-SisPINA were enriched by repeating this process four times. Cells (50 μl; at OD600 of 0.4) were then spread on MCSV plates containing uracil, 5-FOA, and 12 μM simvastatin and incubated at 75 °C for about 10 days. The colonies were transferred onto fresh plates and cultured for 10 days. Cells were then picked and transferred into liquid MCSV medium containing uracil, 5-FOA, and 12 μM simvastatin. Cells from 3-ml culture were collected, and the genomic DNA was extracted. The genotype was determined by PCR using primers flanking the gene encoding SisPINA (Table S2). The plasmid was extracted and confirmed by digestion with NdeI and SalI followed by sequencing.
Gene cloning and construction of plasmid for protein expression in E. coli
The gene encoding PINA from S. islandicus REY15A (SisPINA, SiRe_1432) was amplified by PCR using the genomic DNA as the template and SisPINA-NdeI-Forward and SisPINA-SalI-Reverse as the primers (Table S2). The PCR product was digested with NdeI and SalI, and the gene fragment was cloned into the NdeI/SalI site of pET15bm, a modified pET15b. The resultant plasmid was named pET15bm-SisPINA and used for the expression of untagged SisPINA without any tag. The plasmids used to express site-directed mutants SisPINAK261A and SisPINAD322A were constructed by overlap PCR using the above primers and site-specific primers (Table S2). The amplified fragments were digested with NdeI and SalI and inserted into the NdeI/SalI site of the pET15bm vector. To construct the plasmids for the expression of site-directed mutants SisPINAR206A and SisPINAR325A, we amplified fragments by PCR with the primers listed in Table S2. The fragment was digested with NdeI and SalI and inserted into the pET22b vector.
The gene encoding SisHjc (SiRe_1431) was amplified from S. islandicus REY15A genomic DNA, using SisHjc-NdeI-Forward and SisHjc-XhoI-Reverse (Table S2) as the primers. The fragment was digested with NdeI and SalI and cloned into the NdeI/XhoI site of pRSFDuet-1 (Novagen). The resultant plasmid was named pRSFDuet-1-SisHjc and used for the expression of SisHjc in E. coli. Similar procedures were used to construct the plasmid pRSFDuet-1-SisHjcE12A using SisHjcE12A-NdeI-Forward and SisHjc-XhoI-Reverse (Table S2) as the primers.
Expression and purification of recombinant proteins in E. coli
E. coli BL21-Condonplus (DE3)-RIL cells harboring pET15bm-SisPINA were grown in 1000 ml LB broth supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). The cells were cultured at 37 °C until the OD600 reached 0.6 when IPTG was added to a final concentration of 0.2 mM to induce protein expression. The cells were further cultured at 16 °C for 16 h, harvested by centrifugation at 6300g for 10 min, and disrupted by sonication in 30-ml buffer A [25 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 1 mM DTT, and 5% glycerol]. The disrupted cells were centrifuged at 13,500g for 20 min. Then, the supernatant was incubated at 70 °C for 25 min and centrifuged again at 13,500g for 20 min. The heat-resistant supernatant was precipitated by 50% saturated ammonium sulfate. The precipitated proteins were resuspended in buffer A containing 0.8 M (NH4)2SO4 and loaded onto a HiTrap-Butyl FF column. The proteins eluted at 0.12–0.0 M (NH4)2SO4 were fractionated on a HiTrap-Heparin column pre-equilibrated with buffer containing 25 mM Tris–HCl (pH 8.0), 1 mM EDTA, 1 mM DTT, and 5% glycerol. The fractions eluted at 0.8–1.0 M NaCl were pooled and loaded onto a Superdex 200 gel-filtration column pre-equilibrated in buffer containing 25 mM Tris–HCl (pH 8.0), 500 mM NaCl, 1 mM DTT, and 5% glycerol. Similar procedures were used to express and purify the site-directed mutants of SisPINA.
The procedures for the culture and expression of SisHjc were similar to those for SisPINA except that ampicillin was replaced with kanamycin in the medium. The cells were disrupted by sonication in a buffer containing 25 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, and 5% glycerol. The sample was centrifuged at 13,500g for 20 min. The supernatant was heat-treated at 70 °C for 25 min and centrifuged at 13,500g for 20 min. The supernatant was loaded onto a HiTrap-Heparin column pre-equilibrated in buffer containing 25 mM Tris–HCl (pH 8.0), 1 mM EDTA, and 5% glycerol. The eluted proteins were pooled, concentrated, and loaded onto a Superdex 200 gel-filtration column equilibrated in buffer containing 25 mM Tris–HCl (pH 8.0), 100 mM NaCl, and 5% glycerol. Similar procedures were used to purify SisHjcE12A and N-terminal His-tagged SisHjc.
Aliquots of the purified proteins were immediately frozen in liquid nitrogen and stored at −80 °C. The protein concentration was determined by the Bradford method.
Co-expression and purification of SisPINA and SisHjc in E. coli
pRSFDuet-1 was used to construct the vector for the co-expression of SisPINA and SisHjc in E. coli. The fragment containing the gene encoding SisPINA was amplified by PCR using primers listed in Table S2, digested with BamHI and SalI, and cloned into the vector at the BamHI/SalI site. The fragment containing the gene encoding SisHjc was amplified by PCR with primers SisHjc-NdeI Forward and SisHjc-XhoI Reverse (Table S2), digested with NdeI and XhoI, and inserted into the vector at the NdeI/XhoI site. The resultant plasmid, pRSFDuet-1-SisPINA-SisHjc, was used for the co-expression of SisPINA with an N-terminal 6× His tag and untagged SisHjc in E. coli BL21 (DE3)-CodonPlus-RIL cells. The procedures for culture and co-expression of SisPINA and SisHjc were similar to those for SisHjc. The cell pellet was resuspended in 30-ml buffer A [25 mM Tris–HCl (pH 8.0), 100 mM NaCl, and 5% glycerol] and disrupted by sonication. The sample was centrifuged at 13,500g for 20 min, and the supernatant was treated at 70 °C for 25 min. The sample was centrifuged again at 13,500g for 20 min, and the supernatant was loaded onto the nickel affinity column pre-equilibrated in buffer A. The column was washed with wash buffer (buffer A containing 40 mM imidazole), and SisPINA and its associated proteins were eluted with elution buffer (buffer A containing 400 mM imidazole). Purified proteins were analyzed by SDS-PAGE. The same procedure was applied to construct the pRSFDuet-1-N-SisPINA (1–492)-SisHjc plasmid and analyze the co-expression and purification of SisPINA (1–492) and SisHjc.
Western blotting
Protein samples were mixed with 5× loading buffer [250 mM Tris–HCl (pH 6.8), 10% SDS, 0.5% BPB, 50% glycerol, and 5% 2-mercaptoethanol] and loaded onto 15% gel for SDS-PAGE analysis. After electrophoresis, the gel was transferred onto a PVDF membrane. First, the membrane was blocked in skim milk, washed with TBST, and incubated with a rabbit primary antibody and anti-rabbit HRP-conjugated secondary antibody (HuaAn Biotechnology Limited) following the standard protocol. The primary rabbit polyclonal anti-SisHjc antibody was made by HuaAn Biotechnology Limited with the 15-aa synthetic peptide NAKKPKGSAVERNIC. The band was visualized with Immobilon™ Western Chemiluminescent HRP Substrate (Millipore), and the images were obtained by Imagequant™ 400 (GE Healthcare).
Preparation of DNA substrates
Oligonucleotides (Table S1) were labeled at the 5′-end by T4 polynucleotide kinase. The reaction was conducted in a 30-μl mixture containing 2 μM oligonucleotide, 1× T4 polynucleotide kinase buffer, 15 units T4 polynucleotide kinase (Promega), and 15 μCi [γ-32P] ATP. The mixture was incubated at 37 °C for 30 min, and the reaction was stopped by heating at 95 °C for 10 min. The labeled oligonucleotide was purified using a MicroSpinTMG-25 column (GE Healthcare) according to the manufacturer’s instructions.
DNA substrates were prepared by annealing in a 50-μl mixture containing 7 mM Tris–HCl (pH 8.0), 7 mM MgCl2, 1 μM unlabeled oligonucleotide, and 0.4 μM labeled oligonucleotide. The mixture was incubated at 95 °C for 10 min and then gradually cooled down to room temperature. The annealed substrates were purified by gel electrophoresis (6% polyacrylamide) in 1 × TBE buffer [89 mM Tris–borate (pH 8.3) and 2 mM EDTA]. The bands corresponding to the substrates were excised and eluted from the gel by diffusion at room temperature for 18–36 h into a buffer containing 25 mM Tris–HCl (pH 8.0) and 50 mM NaCl.
ATPase assay
The standard assay for ATPase activity was performed in a 20-μl mixture containing 25 mM Tris–HCl (pH 8.0), 50 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM ATP, 0.2 μCi labeled [γ-32P]-ATP, and 84 nM SisPINA (as a hexamer). The reactions were performed at 65 °C for 10 min. Aliquots (0.8 μl) were spotted onto a polyethylenimine-cellulose plate (Merck). ATP and released Pi were separated by thin layer chromatography in 1 M acetic acid and 0.5 M lithium chloride. The plate was exposed to a phosphorimager. The phosphorimager was then scanned with the Cyclone Plus Storage Phosphor System (PerkinElmer).
DNA binding assay
Standard DNA binding assay was carried out in a 20-μl mixture containing 25 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 0.1 mg/ml bovine serumalbumin (BSA), 12% glycerol, 1 mM DTT, 5 nM of 32P-labeled synthetic DNA molecule, and the indicated amount of SisPINA. The samples were incubated at 55 °C for 30 min, and aliquots (10 μl) were loaded onto a 6% polyacrylamide gel. The electrophoresis was run at 205 V in 0.5× TBE buffer [44.5 mM Tris-borate (pH 8.3) and 1.0 mM EDTA] for 45 min. The gels were exposed to a phosphorimager. The phosphorimager was then scanned with the Cyclone Plus Storage Phosphor System (PerkinElmer).
DNA helicase assay
The DNA helicase activity was carried out as previously described with slight modification [73]. Briefly, the reaction was performed in a 20-μl mixture containing 20 mM Tris–HCl (pH 8.0) 0.1 mg/ml BSA, 1 mM DTT, 10 mM ATP, 5 mM MgCl2, 5 nM DNA substrate, and the indicated amount of SisPINA at 55 °C for 30 min and stopped by the addition of 10 μl stop buffer (20 mM EDTA, 1%SDS, 2 mg/ml proteinase K [Takara], and 0.02% bromophenol blue). The samples were further incubated at 55 °C for 10 min. The samples (20 μl) were loaded onto an 8% native polyacrylamide gel. Electrophoresis was run at 130 V for 90 min in 1× TBE buffer [89 mM Tris-borate (pH 8.3) and 1 mM EDTA]. The gels were exposed to a phosphorimager. The phosphorimager was then scanned with the Cyclone Plus Storage Phosphor System (PerkinElmer).
HJ cleavage assay
The HJ cleavage assay was performed in a 20-μl mixture containing 25 mM Tris–HCl (pH 8.0) 50 mM NaCl, 0.1 mg/ml BSA, 1 mM DTT, 2 mM ATP, 5 mM MgCl2, JY HJ DNA or HSL HJ DNA, SisHjc, and the indicated amount of SisPINA. The reaction was incubated at 55 °C for 30 min unless otherwise indicated. After 30 min, the reaction products were analyzed by native or denaturing gel electrophoresis. For native gel electrophoresis, 10-μl stop buffer (same as for helicase assay) was added and incubated at 55 °C for another 10 min, and then, 20-μl samples were loaded onto an 8% native gel. For denaturing gel electrophoresis, 2 μl proteinase K (20 mg/ml) and 2 μl 10% SDS were added and incubated at 55 °C for 5 min. Then, 20-μl denaturing buffer (95% formamide, 10 mM EDTA, and 0.02% bromophenol blue) was added and incubated at 95 °C for another 5 min. The products were immediately transferred to 4 °C for 5 min. Reaction mixture (30 μl) was loaded onto a 15% denaturing polyacrylamide gel containing 8 M urea. The gels were exposed to a phosphorimager. The phosphorimager was then scanned with the Cyclone Plus Storage Phosphor System (PerkinElmer).
Crystallization of SisPINAK261A
Original crystals of SisPINAK261A were observed in a buffer of 0.1 M sodium malonate (pH 5.0) and 12% (wt/vol) polyethylene glycol (PEG) 3350. Good-quality crystals were obtained in a 5-μl sitting drop containing 3 μl protein and 2 μl 0.1 M sodium malonate (pH 5.0), 12% (wt/vol) PEG3350, and 3% glycerol. Using a microseeding method, SeMetsubstituted SisPINAK261A crystals were obtained under the same conditions. Crystals were cryoprotected by addition of 20% PEG200 for data collection.
Structure determination and refinement
The crystal structure was determined by 4 wavelengths (4w) SeMet MAD phasing, and the phases extended to the best native dataset at 2.82-Å resolution. Low resolution of the crystal diffraction and the high number of expected Se sites (90, 6 × 15) required high multiplicity from multiple anomalous datasets. The majority of the data were obtained from a single crystal, which was collected initially at the Se “peak” wavelength at BL14-1 with a 200 × 90 μm beam. The three 360° Se-peak runs had different doses (0.43, 3.3, 1.3 MGy) and ISa values (16.7, 22.2, 24.1).
Two 3-wavelength SeMet MAD datasets were collected on the same crystal at BL12-2 with a 150 × 100 μm beam with the following total doses in parenthesis (0.71, 3.60 MGy; a fivefold difference; and with beam attenuated 98% and 90%) with “peak” ISa values of 30.3 and 40.2; “high” wavelength ISa values of 44.2 and 48.8; and “inflection” wavelength ISa values of 33.6 and 41.9. The Friedel pairs were collected close together in time (5-degree inverse-beam protocol).
These two runs were scaled together for “high” and “inflection”, whereas a total of five “peak” datasets from BL14-1 and BL12-2 were also scaled with XSCALE (version March 1, 2015) [74] with the zero-dose and strict-absorption corrections. The total absorbed dose for the main crystal was 9.3 MGy. A single dataset from different crystal’s 4w MAD experiment with similar processing schemes (corrections) was used for the “low” dataset (ISa = 22.6; absorbed dose = 2.63 MGy). The SeMet positions were located with a successful SHELXCDE [75] run performed by HKL2MAP graphical interface [76] containing merged data from five “peak” datasets collected at both beamlines with low doses and two merged datasets for each “inflection” and “high” wavelengths datasets. Single datasets were used for “low” wavelength and the “native” (sulfur containing methionine versus SeMet in the other 4w datasets). Despite strong anomalous signal to ~3.6 Å, a heavy-atom search for 90 sites (6monomers × 15 SeMet residues per chain) in SHELXD was successful only within the 40–5.3 Å range. It produced reasonable solution with correlation coefficient close to the considered “solved” structure 25% (24.8%). Of the 126 initially identified sites, only 59 passed the internal significance test. Subsequent BUCCANNER [77] runs were used to automatically build a protein model, alternatively with PHASER_EP[78] runs to complete the Se anomalous substructure. The final substructure solution from PHASER_EP had 68 unique SeMet sites, indicating the disorder of some of the Se atoms.
The initial model was autobuilt from combined SeMet MAD phases and BUCCANEER autobuilt model with sequence tracing, resulting in a nice-looking model with Rfree of ~44%. During visual inspection, the well-established SeMet positions and aromatic residues were used to confirm the proper sequence placement. In addition, alternative domain connections from the automated BUCCANEER model were established, allowing the proper definition of the unique subunit and reducing the large number of chains to only six. Applying NCS relationships or superimposed structures led to the establishment of the unique sixfold model.
Structure refinement was carried out against the native data set at resolution of 2.82 Å using PHENIX.REFINE [79] and REFMAC [80]. The structural model was completed with manual structure manipulation via Coot [81] using σ-weighted (Fo − Fc) and (2Fo − Fc) electron density maps modified with density modification (DM) [82]. The final model geometry was validated by MOLPROBITY [83].
Accession number
The atomic coordinates and structural factors for SisPINAK261A are deposited to the Protein Data Bank with the accession number 5F4H.
Supplementary Material
Acknowledgments
We thank Mingzhu Chu and Yan Wang for the assistance in the experiments and all lab members for helpful discussion. We also thank the staff at beamlines BL-14-1 and BL-12-2 at SSRL for help with data collection. The study was supported by grants from Natural Science Foundation of China [30930002, 31470184, and 31670061 to Y.S.]; China Scholarship Council for supporting B.Z.’s study in L. Fan’s lab at University of California, Riverside; UC Regents Faculty Development Award to L.F. The license fee for Pymol software used for structural analysis and figure preparation was funded by NIH R01GM108893 to L.F.
Abbreviations used
- HJ
Holliday junction
- PIN
PilT N-terminal
- PINA
PIN-domain-containing ATPase
- SisPINA
PINA from Sulfolobus islandicus
- PilT
type IV pili
- 5-FOA
5-fluoroorotic acid
- MSCV
a medium consisting of mineral salts (M), 0.2% (wt/vol) sucrose (S), 0.2% (wt/vol) casamino acids (C), and a mixed vitamin solution (V)
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
- MAD
multiple-wavelength anomalous diffraction
- BSA
bovine serum albumin
- PEG
polyethylene glycol
- 4w
4 wavelengths
Appendix A. Supplementary Data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2017.02.016.
References
- 1.Holliday R. A mechanism for gene conversion in fungi (reprinted) Genet Res. 2007;89:285–307. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 2.Bizard AH, Hickson ID. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol. 2014;6:1–14. doi: 10.1101/cshperspect.a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarbajna S, West SC. Holliday junction processing enzymes as guardians of genome stability. Trends Biochem Sci. 2014;39:409–419. doi: 10.1016/j.tibs.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 4.West SC, Blanco MG, Chan YW, Matos J, Sarbajna S, Wyatt HD. Resolution of recombination intermediates: mechanisms and regulation. Cold Spring Harb Symp Quant Biol. 2015;80:103–109. doi: 10.1101/sqb.2015.80.027649. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett RJ, Dunderdale HJ, West SC. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- 7.Komori K, Sakae S, Shinagawa H, Morikawa K, Ishino Y. A Holliday junction resolvase from Pyrococcus furiosus: functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in bacteria, eukarya, and archaea. Proc Natl Acad Sci U S A. 1999;96:8873–8878. doi: 10.1073/pnas.96.16.8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kvaratskhelia M, White MF. An archaeal Holliday junction resolving enzyme from Sulfolobus solfataricus exhibits unique properties. J Mol Biol. 2000;295:193–202. doi: 10.1006/jmbi.1999.3363. [DOI] [PubMed] [Google Scholar]
- 9.Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Freeman AD, Declais AC, Wilson TJ, Gartner A, Lilley DM. Crystal structure of a eukaryotic GEN1 resolving enzyme bound to DNA. Cell Rep. 2015;13:2565–2575. doi: 10.1016/j.celrep.2015.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Declais AC, Lilley DMJ. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr Opin Struct Biol. 2008;18:86–95. doi: 10.1016/j.sbi.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Eggleston AK, West SC. Cleavage of Holliday junctions by the Escherichia coli RuvABC complex. J Biol Chem. 2000;275:26,467–26,476. doi: 10.1074/jbc.M001496200. [DOI] [PubMed] [Google Scholar]
- 13.Eggleston AK, Mitchell AH, West SC. In vitro reconstitution of the late steps of genetic recombination in E. coli. Cell. 1997;89:607–617. doi: 10.1016/s0092-8674(00)80242-1. [DOI] [PubMed] [Google Scholar]
- 14.Davies AA, West SC. Formation of RuvABC–Holliday junction complexes in vitro. Curr Biol. 1998;8:725–727. doi: 10.1016/s0960-9822(98)70282-9. [DOI] [PubMed] [Google Scholar]
- 15.Rafferty JB, Sedelnikova SE, Hargreaves D, Artymiuk PJ, Baker PJ, Sharples GJ, et al. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science. 1996;274:415–421. doi: 10.1126/science.274.5286.415. [DOI] [PubMed] [Google Scholar]
- 16.Ariyoshi M, Nishino T, Iwasaki H, Shinagawa H, Morikawa K. Crystal structure of the Holliday junction DNA in complex with a single RuvA tetramer. Proc Natl Acad Sci U S A. 2000;97:8257–8262. doi: 10.1073/pnas.140212997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada K, Kunishima N, Mayanagi K, Ohnishi T, Nishino T, Iwasaki H, et al. Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8. Proc Natl Acad Sci U S A. 2001;98:1442–1447. doi: 10.1073/pnas.031470598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Putnam CD, Clancy SB, Tsuruta H, Gonzalez S, Wetmur JG, Tainer JA. Structure and mechanism of the RuvB Holliday junction branch migration motor. J Mol Biol. 2001;311:297–310. doi: 10.1006/jmbi.2001.4852. [DOI] [PubMed] [Google Scholar]
- 19.Hargreaves D, Rice DW, Sedelnikova SE, Artymiuk PJ, Lloyd RG, Rafferty JB. Crystal structure of E. coli RuvA with bound DNA Holliday junction at 6 A resolution. Nat Struct Mol Biol. 1998;5:441–446. doi: 10.1038/nsb0698-441. [DOI] [PubMed] [Google Scholar]
- 20.Nishino T, Iwasaki H, Kataoka M, Ariyoshi M, Fujita T, Shinagawa H, et al. Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA. J Mol Biol. 2000;298:407–416. doi: 10.1006/jmbi.2000.3675. [DOI] [PubMed] [Google Scholar]
- 21.Parsons CA, Stasiak A, Bennett RJ, West SC. Structure of a multisubunit complex that promotes DNA branch migration. Nature. 1995;374:375–378. doi: 10.1038/374375a0. [DOI] [PubMed] [Google Scholar]
- 22.Yamada K, Miyata T, Tsuchiya D, Oyama T, Fujiwara Y, Ohnishi T, et al. Crystal structure of the RuvA–RuvB complex: a structural basis for the Holliday junction migrating motor machinery. Mol Cell. 2002;10:671–681. doi: 10.1016/s1097-2765(02)00641-x. [DOI] [PubMed] [Google Scholar]
- 23.Baharoglu Z, Petranovic M, Flores MJ, Michel B. RuvAB is essential for replication forks reversal in certain replication mutants. EMBO J. 2006;25:596–604. doi: 10.1038/sj.emboj.7600941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGlynn P, Lloyd RG, Marians KJ. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc Natl Acad Sci U S A. 2001;98:8235–8240. doi: 10.1073/pnas.121007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta S, Yeeles JTP, Marians KJ. Regression of replication forks stalled by leading-strand template damage. J Biol Chem. 2014;289:28,376–28,387. doi: 10.1074/jbc.M114.587907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bianco PR. I came to a fork in the DNA and there was RecG. Prog Biophys Mol Biol. 2015;117:166–173. doi: 10.1016/j.pbiomolbio.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci U S A. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manthei KA, Keck JL. The BLM dissolvasome in DNA replication and repair. Cell Mol Life Sci CMLS. 2013;70:4067–4084. doi: 10.1007/s00018-013-1325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, et al. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bugreev DV, Brosh RM, Jr, Mazin AV. RECQ1 possesses DNA branch migration activity. J Biol Chem. 2008;283:20,231–20,242. doi: 10.1074/jbc.M801582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5 beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 33.Unk I, Hajdu I, Blastyak A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair. 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 34.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 35.Fujikane R, Komori K, Shinagawa H, Ishino Y. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J Biol Chem. 2005;280:12,351–12,358. doi: 10.1074/jbc.M413417200. [DOI] [PubMed] [Google Scholar]
- 36.Guy CP, Bolt EL. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 2005;33:3678–3690. doi: 10.1093/nar/gki685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujikane R, Shinagawa H, Ishino Y. The archaeal Hjm helicase has recQ-like functions, and may be involved in repair of stalled replication fork. Genes Cells. 2006;11:99–110. doi: 10.1111/j.1365-2443.2006.00925.x. [DOI] [PubMed] [Google Scholar]
- 38.Arcus VL, McKenzie JL, Robson J, Cook GM. The PIN-domain ribonucleases and the prokaryotic VapBC toxin– antitoxin array. Protein Eng Des Sel. 2011;24:33–40. doi: 10.1093/protein/gzq081. [DOI] [PubMed] [Google Scholar]
- 39.Satyshur KA, Worzalla GA, Meyer LS, Heiniger EK, Aukema KG, Misic AM, et al. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol. 2000;10:R888–R890. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 41.Mate MJ, Vincentelli R, Foos N, Raoult D, Cambillau C, Ortiz-Lombardia M. Crystal structure of the DNA-bound VapBC2 antitoxin/toxin pair from Rickettsia felis. Nucleic Acids Res. 2012;40:3245–3258. doi: 10.1093/nar/gkr1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das U, Pogenberg V, Subhramanyam UK, Wilmanns M, Gourinath S, Srinivasan A. Crystal structure of the VapBC-15 complex from Mycobacterium tuberculosis reveals a two-metal ion dependent PIN-domain ribonuclease and a variable mode of toxin–antitoxin assembly. J Struct Biol. 2014;188:249–258. doi: 10.1016/j.jsb.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robson J, McKenzie JL, Cursons R, Cook GM, Arcus VL. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J Mol Biol. 2009;390:353–367. doi: 10.1016/j.jmb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, et al. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc Natl Acad Sci U S A. 2006;103:9464–9469. doi: 10.1073/pnas.0603673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fatica A, Tollervey D, Dlakic M. PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA. 2004;10:1698–1701. doi: 10.1261/rna.7123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu J, Peng W, Sun Y, Wang X, Xu Y, Li X, et al. Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase. Nucleic Acids Res. 2012;40:6957–6965. doi: 10.1093/nar/gks359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang F, Shi J, Chen SH, Bian C, Yu X. The PIN domain of EXO1 recognizes poly(ADP-ribose) in DNA damage response. Nucleic Acids Res. 2015;43:10,782–10,794. doi: 10.1093/nar/gkv939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Q, Li Y, Zeng C, Song T, Yan Z, Ni J, et al. Genetic analysis of the Holliday junction resolvases Hje and Hjc in Sulfolobus islandicus. Extremophiles. 2015;19:505–514. doi: 10.1007/s00792-015-0734-5. [DOI] [PubMed] [Google Scholar]
- 52.Fujikane R, Ishino S, Ishino Y, Forterre P. Genetic analysis of DNA repair in the hyperthermophilic archaeon, Thermococcus kodakaraensis. Genes Genet Syst. 2010;85:243–257. doi: 10.1266/ggs.85.243. [DOI] [PubMed] [Google Scholar]
- 53.Zhang C, Tian B, Li S, Ao X, Dalgaard K, Gokce S, et al. Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem Soc Trans. 2013;41:405–410. doi: 10.1042/BST20120285. [DOI] [PubMed] [Google Scholar]
- 54.Guo L, Brugger K, Liu C, Shah SA, Zheng H, Zhu Y, et al. Genome analyses of Icelandic strains of Sulfolobus islandicus, model organisms for genetic and virus–host interaction studies. J Bacteriol. 2011;193:1672–1680. doi: 10.1128/JB.01487-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arcus VL, Backbro K, Roos A, Daniel EL, Baker EN. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J Biol Chem. 2004;279:16,471–16,478. doi: 10.1074/jbc.M313833200. [DOI] [PubMed] [Google Scholar]
- 56.Aravind L, Iyer LM, Leipe DD, Koonin EV. A novel family of P-loop NTPases with an unusual phyletic distribution and transmembrane segments inserted within the NTPase domain. Genome Biol. 2004;5:R30.1–R30.10. doi: 10.1186/gb-2004-5-5-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng L, Zhu H, Chen Z, Liang YX, She Q. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009;13:735–746. doi: 10.1007/s00792-009-0254-2. [DOI] [PubMed] [Google Scholar]
- 58.Zheng T, Huang Q, Zhang C, Ni J, She Q, Shen Y. Development of a simvastatin selection marker for a hyperthermophilic acidophile, Sulfolobus islandicus. Appl Environ Microbiol. 2012;78:568–574. doi: 10.1128/AEM.06095-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hishida T, Iwasaki H, Yagi T, Shinagawa H. Role of walker motif A of RuvB protein in promoting branch migration of Holliday junctions. J Biol Chem. 1999;274:25,335–25,342. doi: 10.1074/jbc.274.36.25335. [DOI] [PubMed] [Google Scholar]
- 60.Kvaratskhelia M, Wardleworth BN, Norman DG, White MF. A conserved nuclease domain in the archaeal Holliday junction resolving enzyme Hjc. J Biol Chem. 2000;275:25,540–25,546. doi: 10.1074/jbc.M003420200. [DOI] [PubMed] [Google Scholar]
- 61.Daiyasu H, Komori K, Sakae S, Ishino Y, Toh H. Hjc resolvase is a distantly related member of the type II restriction endonuclease family. Nucleic Acids Res. 2000;28:4540–4543. doi: 10.1093/nar/28.22.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 64.Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, et al. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBOJ. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reindl S, Ghosh A, Williams GJ, Lassak K, Neiner T, Henche AL, et al. Insights into Flal functions in archaeal motor assembly and motility from structures, conformations, and genetics. Mol Cell. 2013;49:1069–1082. doi: 10.1016/j.molcel.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Albers SV, Driessen AJM. Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology. 2005;151:763–773. doi: 10.1099/mic.0.27699-0. [DOI] [PubMed] [Google Scholar]
- 68.Huang Q, Liu L, Liu J, Ni J, She Q, Shen Y. Efficient 5′–3′ DNA end resection by HerA and NurA is essential for cell viability in the crenarchaeon Sulfolobus islandicus. BMC Mol Biol. 2015;16:2. doi: 10.1186/s12867-015-0030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bond CS, Kvaratskhelia M, Richard D, White MF, Hunter WN. Structure of Hjc, a Holliday junction resolvase, from Sulfolobus solfataricus. Proc Natl Acad Sci U S A. 2000;98:5509–5514. doi: 10.1073/pnas.091613398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K. Crystal structure of the archaeal Holliday junction resolvase Hjc and implications for DNA recognition. Structure. 2001;9:197–204. doi: 10.1016/s0969-2126(01)00576-7. [DOI] [PubMed] [Google Scholar]
- 71.Zhang C, Guo L, Deng L, Wu Y, Liang Y, Huang L, et al. Revealing the essentiality of multiple archaeal pcna genes using a mutant propagation assay based on an improved knockout method. Microbiology. 2010;156:3386–3397. doi: 10.1099/mic.0.042523-0. [DOI] [PubMed] [Google Scholar]
- 72.Peng N, Deng L, Mei Y, Jiang D, Hu Y, Awayez M, et al. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl Environ Microbiol. 2012;78:5630–5637. doi: 10.1128/AEM.00855-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang S, Wei T, Hou G, Zhang C, Liang P, Ni J, et al. Archaeal DNA helicase HerA interacts with Mre11 homologue and unwinds blunt-ended double-stranded DNA and recombination intermediates. DNA Repair. 2008;7:380–391. doi: 10.1016/j.dnarep.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 74.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- 77.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 78.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 80.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 81.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 82.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.