Abstract
The 2014 Ebola outbreak, the largest recorded, took us largely unprepared, with no available vaccine or specific treatment. In this context, the World Health Organization (WHO) declared that the humanitarian use of experimental therapies against Ebola Virus (EBOV) is ethical. In particular, an experimental treatment consisting of a cocktail of three monoclonal antibodies (mAbs) produced in tobacco plants and specifically directed to the Ebola virus glycoprotein (GP) was tested in humans, apparently with good results. Several mAbs with high affinity to the GP have been described. This review discusses our current knowledge on this topic. Particular emphasis is devoted to those mAbs that have been assayed in animal models or humans as possible therapies against Ebola. Engineering aspects and challenges for the production of anti-Ebola mAbs are also briefly discussed; current platforms for the design and production of full-length mAbs are cumbersome and costly.
Keywords: Ebola, mAbs, monoclonal antibodies, therapeutic, epidemic, GP
The Ebola virus in brief: Epidemiology, and genetic variability
The Ebola virus (more formally called EBOV, formerly known as Zaire ebolavirus) is one of the most aggressive and feared pathogens known to humans. Its first documented outbreak occurred in 1976 (WHO, 1978; Leroy et al., 2005). Yet no vaccine or specific treatment against Ebola infection is commercially available.
The Ebola virus is an enveloped, non-segmented, RNA virus. EBOV, together with the Marburg virus, belongs to the Filoviridae family. The ecology and epidemiology aspects of Ebola Virus Disease (EVD) have been reviewed elsewhere (Feldmann and Geisbert, 2011). In brief, experimental evidence suggests that fruit bats are its main natural reservoir (Leroy et al., 2005; Leroy et al., 2004). Human outbreaks have been associated with previous occurrences of nonhuman primate outbreaks. The patient-zero cases have been mainly hunters, infected when manipulating dead nonhuman primates (gorillas, chimpanzees) or duikers. Other zero patients include subjects that accidentally came in contact with bats (i.e., workers in bat-infested cotton factories or mines) (Chiappelli et al., 2015; Saenz et al., 2015). Indeed, the Guinean two-year old kid, believed to be the patient zero of the current West Africa outbreak, most probably became infected while playing in a hollow tree infested by insectivorous bats (Baize et al., 2014; Saenz et al., 2015). Subsequent dissemination often occurred by direct contact amongst individuals living together (Leroy et al., 2004), through patient care or through ritual burial practices (Chiappelli et al., 2014; Pandei et al., 2014; Richards et al., 2015).
The genome of the Ebola virus was elucidated in 1993 (Sanchez et al., 1993). The Ebola virus has been diversified into five different species: Zaire, Sudan, Ivory Coast, Reston, and Bundibugyo ebolavirus. All species originated in Africa, with the exception of Reston, which was discovered in Reston Virginia, from a macaque imported from the Philippines (Feldmann and Geisbert, 2011; Caroll et al., 2013). The Zaire species, the protagonist of the current outbreak, is the most virulent. The genetic differences among species are relatively high; a 35% genetic divergence among all species has been reported, based on sequences available up to 2011 (Grard et al., 2011). Until 2013, only 22 complete genome sequences for the EBOV Zaire species were available in international repositories. Most of these were collected during the outbreaks of 1976, 1990, and 2007–2008 in the Democratic Republic of Congo. Gire et al. (2014) have studied 99 EBOV genome sequences from 78 confirmed EVD patients during the current outbreak, providing new and valuable information on the genetic identity of the Zaire EBOV. The authors found 341 fixed substitutions (35 non-synonymous, 173 synonymous, and 133 noncoding) between the 2014 EBOV and all previously published EBOV sequences. With all these new sequences included, the Ebolavirus resource database at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/genome/viruses/variation/ebola) has 149 complete genome sequences of the Zaire EBOV available. Considering these sequences, the EBOV genome variability increases from 35% to 40%–45%. It should be noted that the advances achieved so far in the understanding of the infection mechanisms and in the design of an experimental vaccine and therapies against EBOV are based on the genome information available before the current outbreak. The implications of new genomic variations might be important for Ebola diagnostics and therapy.
The recent Ebola outbreak that began in West Africa in December, 2013, made evident that we are unprepared to effectively control this disease (Leroy et al., 2014; Enserink, 2014; Brad, 2014). As of July 12, 2015, there are more than 27670 documented cases of the infection (including more than 11250 deaths) in six different African countries: Guinea, Liberia, Nigeria, Mali, Sierra Leone and Senegal (WHO, 2015). Additionally, there have been eight cases outside West Africa, five in the USA, one in Spain, one in the United Kingdom and one in Italy (WHO, 2015).
Current therapeutic strategies to treat Ebola
The clinical manifestations of EVD have been reviewed elsewhere (Feldmann and Geisbert, 2011; Bah et al., 2015). As the virus reproduces and spreads in the body, it interferes with blood clotting and disrupts electrolyte balance. Based on the current treatment for EVD, which consists of supportive care, patients are frequently dehydrated and need intravenous or oral fluids with solutions that contain electrolytes (Bah et al., 2015; Lyon et al., 2014; Zhong et al., 2014). Maintaining oxygen levels and modulating coagulation (Feldmann and Geisbert, 2011; Geisbert et al., 2003) are important parts of the treatment scheme for Ebola patients (Bah et al., 2015; Lyon et al., 2014; Zhong et al., 2014). Such interventions can help sustain some patients and allow them to recover. An adequate level of support care might improve survival significantly (Bah et al., 2015; Lyon et al., 2014), but even in such conditions patients can progress toward multiorgan failure, shock, and death. Based on WHO reports, the overall fatality rate for the current Ebola outbreak is 48–49% (WHO, 2015) with a span that goes from 0 to 90% as a strong function of the quality of the supportive care received (Lyon et al., 2014).
As stated before, no commercial vaccines or specific therapies are currently available to combat Ebola. Several experimental vaccines and drugs have been tested in animal models with promising results, and some of them are currently in clinical trials (Kuehn, 2015). A comprehensive analysis of the state of the art in vaccine development strategies against EBOV can be found elsewhere (Marzi et al., 2014). Regarding therapeutic approaches against EBOV, the main strategies tested in animal models include the use of phosphorodiamidate morpholino oligomers (PMOs) (Warfield et al., 2006; Iversen et al., 2012; Warren et al., 2015) small interference RNA molecules (siRNA) (Geisbert et al., 2010; Thi et al., 2014), small-molecule antiviral drugs (Oestereich et al., 2014; Smither et al., 2014; Warren et al., 2014), and full-length mAbs.
Amongst the experimental therapies that have been proposed and tested against EVD, passive immunization using full-length mAbs is arguably the most promising strategy. Several anti-Ebola mAb have been identified and studied by a number of research groups (Wilson et al., 2000; Takada et al., 2003; Lee et al., 2008; Shedlock et al., 2010; Qiu et al., 2011; Marceau et al., 2014) and several mAb cocktails have been developed by different research groups and companies (Olinger et al., 2012; Pettitt et al., 2013; Qiu et al., 2013; Qiu et al., 2014), mainly based on knowledge derived from the study of viral species or strains isolated between 1976 and 1995.
Exceptional humanitarian use of Anti-Ebola mAb-based therapies
In the context of the largest Ebola outbreak ever registered, the WHO has declared that the use of experimental drugs for the humanitarian treatment of Ebola patients is ethical. An experimental treatment (ZMapp™, from Mapp Biopharmaceutical, Inc., San Diego, CA) consisting of a cocktail of three mAbs, produced in tobacco leaves, has been administered to several patients (Hampton, 2014; Goodman, 2014; Qiu et al., 2014) under this humanitarian exception. The evolution of two of the three patients first treated with ZMapp™, two American health workers infected in Liberia and treated at the Emory Hospital, in Atlanta USA, was recently documented by Lyon et al. (2015). Both patients improved their conditions shortly after receiving a first dose of ZMapp™. Since this improvement occurred in the context of aggressive rehydration, electrolyte balancing, and other care measures, the significance of the effect of the mAb cocktail cannot be conclusively established from this application (Lyon et al., 2015). ZMapp™ targets different epitopes of the Ebola virus GP. Before the current outbreak, this mAb cocktail had not been used in humans. Its predecessors had been tested only in murine and nonhuman primate models (Olinger et al., 2012; Pettitt et al., 2013; Qiu et al., 2013).
Interfering with viral attachment or (viral entry) into mammalian cells is a common therapeutic approach to fight envelope viruses (Wisskirchen et al., 2014). The therapeutic use of full-length antibodies against viral infection has been proposed to obstruct viral entry in the context of West Nile (Oliphant et al., 2005), herpes simplex (Highlander et al., 1988), dengue (Crill et al., 2001), influenza infection (Vanderlinden and Naesens, 2014; DiLillo et al., 2014), and SARS (Sui et al., 2004), among others. Several reports on antibodies with a high affinity for EBOV proteins are available (Lee et al., 2008, Olinger et al., 2012; Pettitt et al., 2013; Qiu et al., 2013, Becquart et al., 2014). Most of these studies refer to antibodies that specifically bind to different epitopes of the GP. Indeed, all mAbs proposed to be used as anti-Ebola therapeutics have the virus transmembrane GP as their target. This is logical as transmembrane GP is key to initiate virus attachment and fusion to host membranes (Nanbo et al., 2010; Sakurai et al., 2015; see Figure 1).
Figure 1.
The GP of EBOV. (A) The cylindrical EBOV capsid contains genetic information encoded in single strand, negative sense RNA enclosed in an inner structure composed of NP (indicated in green). (B) GP is expressed as a 676-residue protein (GP0) from the fourth of a total of seven RNA protein genes present in EBOV. (C) GP0 undergoes a post-transductional furin cleavage at the 501–502 site to render two GP units (GP1 and GP2) that bridge together through a disulfide bond to generate GP monomers. Different GP forms originate through transcriptional editing, mainly secreted dimers (sGP), small secreted monomers (small ssGP), and transmembrane GP. (D) Transmembrane GP is a trimer that is anchored in a bilipid layer and structurally supported by VP40. Each of the monomers conforming to the trimer contains a chalice-shaped domain (GP1; indicated in yellow) and a basal/transmembrane domain (GP2; indicated as an orange stalk). The mucin like domain (MLD), a highly glycosylayed domain in GP1 (indicated in lighter shade of yellow), covers the receptor binding domain of GP. (E) Secreted GP (sGP) is believe to act as a distractor of the host immune system, serving also as a target for neutralizing antibodies, diminishing the number of mAb units effectively available for viral entry interference.
GP: The therapeutic target of anti-Ebola mAb-based therapies
GP is its only surface capsid protein, i.e., a transmembrane protein with spiked protrusions on the surface of the virus (see Figure 1) and plays a key role in many important EBOV functions, including the interaction with host cell receptors to activate viral attachment and/or entry. GP is also the most antigenic of the EBOV proteins. For instance, serum from EVD survivors collected a few days after the end of symptoms react mainly with GP peptides (Becquart et al., 2014). Currently, GP is believed to be required, but it is not sufficient for in vivo virulence (Groseth, et al., 2012). Here we provide a brief summary of the current knowledge on the structure and functions of GP, particularly those relevant to the design and/or efficacy of anti-GP mAb therapies.
Several glycoproteins (forms of GP) originate from the GP-encoding RNA sequence: a transmembrane form of GP (normally referred to in literature simply as GP), secreted GP (sGP), and a smaller version of sGP (named small sGP or ssGP) are among the most relevant (Lee et al., 2009; Mehedi et al., 2011; Figure 1). The sGP may act as a distractor to the host immune system (de la Vega et al., 2014; Misasi and Sullivan, 2014; Mohan et al., 2012). It is highly present as a dimer in solution in the serum of infected patients, and serves as a binding target for some anti-GP antibodies produced by the host, perhaps effectively diminishing the number of antibodies available for virus neutralization (Mohan et al., 2012; Ramanan et al., 2011). The transmembrane GP of EBOV is a protein containing a high number of both N-linked and O-linked carbohydrates (Takada et al., 1997). Mature transmembrane GP is a trimer of GP1-GP2 subunits linked by disulfide bonds. Each of these subunits is generated by the proteolytic cleavage of GP0, a precursor polypeptide, during virus assembly. GP1, the membrane-distal subunit, is responsible of viral adhesion to host cells and regulates GP2, the transmembrane subunit, which participates in membrane fusion (White and Schornberg, 2015; Malashkevich et al., 1999).
The most accurate information on the three-dimensional (3D) structure of transmembrane GP has been derived from a small number of well-executed studies (Lee et al., 2008; Beniac et al., 2012; Tran et al., 2014). The structure of the trimeric GP ectodomain (Figure 2) has been more graphically referred as a “chalice,” consisting of a base, a head, and a glycan cap (Lee et al., 2008; Lee et al., 2009). The base projects a transmembrane anchor of 22 residues (651–672 in GP) that attaches GP2 to the viral membrane (Malashkevich et al., 1999), which is structurally composed of protein VP40 (Beniac et al., 2012) and covered by a lipid bilayer originating from the cells of the host upon viral budding (Figure 1(A), (B)).
Figure 2.
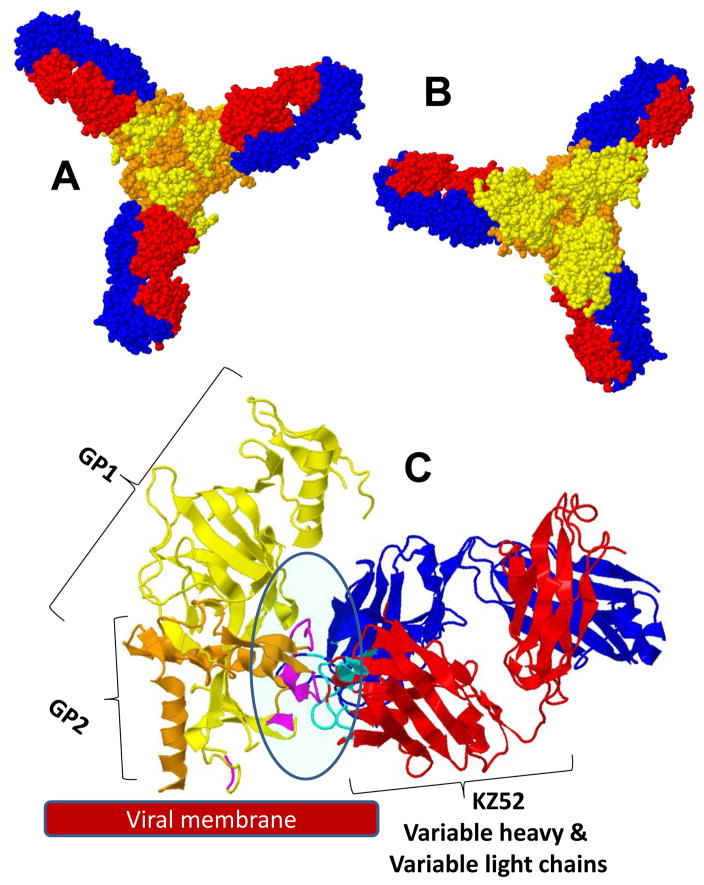
3-D view of the complexity of three monoclonal antibodies binding to GP-EBOV, as resolved using X-ray crystallography (Lee et al., 2008; Lee et al., 2009): (A) bottom view (as seen from the viral surface). The GP1 subunit is colored in yellow; GP2 is colored in orange. In the case of mAb KZ52, only the FAB region is presented (variable light chains in blue; variable heavy chains in red). (B) Top view (the mucin-like domain is not presented).
Within the GP1 subunit, three regions have been frequently referred to in the literature as key to the binding and immune-evasion functions of EBOV: the glycan cap, the mucin-like domain (MLD), and the receptor-binding domain (RBD). The glycan cap and the MLD are highly glycosylated GP1 regions. The MLD, containing both N- and O-linked glycans (Lennenmann et al., 2014), spans from residues 313 to 501 in GP (Figure 3). Several neutralizing antibodies, including two comprised in MB-003 (Olinger et al., 2012), are directed against the MLD (Tran et al., 2014). Recently, Tran et al. (2014) used cryoelectron tomography of EBOV virus-like particles to show the exact 3D location of MLD with respect to the rest of the GP molecule. Functions attributed to MLD include: enhancing viral attachment to target cell surfaces (Marzi et al., 2007; Matsuno et al., 2010), protecting conserved regions of GP from antibody recognition, and sterically masking important immune regulatory molecules, such as MHC1 (major histocompatibility complex 1) or β1 integrin, on the surfaces of infected cells (Lennemann et al., 2014; Francica et al., 2009; Reynard et al., 2009).
Figure 3.
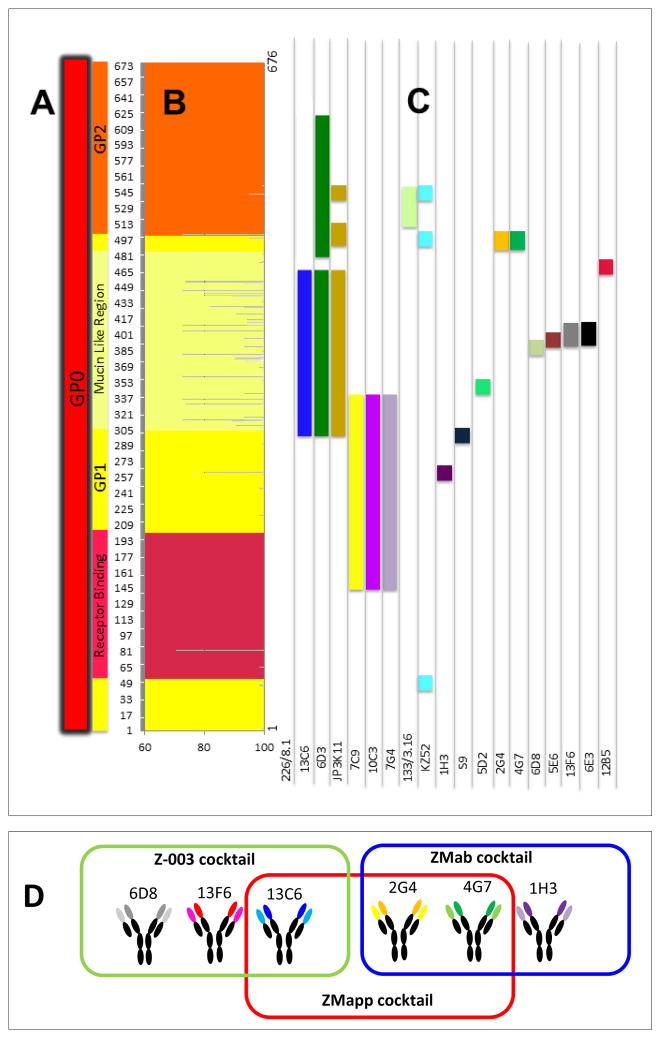
Different anti-GP mAbs bind to different epitopes in EBOV-GP. (A) The different subregions on GP1 (yellow section) and GP2 (orange section) are represented. The receptor-binding domain (RBD) in GP1 is indicated in red; the mucin-like domain (MLD) in GP1 is indicated in light yellow. (B) The percentage of genetic conservation among Zaire EBOV variants ranges from 100% to 76%. The MLD is the least-conserved domain in GP1. The RBD in the GP1 is a well-conserved region. (C) Mapping of the epitopes of 19 anti-GP mAbs reported in the literature. Different epitopes are coded with different colors (one epitope per column). In most cases, the epitope location was derived from studies in which truncated GP proteins were exposed to mAb binding. Therefore, all the residues within each color segment do not necessarily interact effectively with the corresponding binding mAb. (D) Zoom of the conformational epitope for KZ52 in GP1-GP2. (F) ZMapp™ is composed of mAbs included in predecessor formulations (MB-003 and ZMAb, produced by Mapp Biopharmaceutical, Inc, and Dephyrus Inc., respectively)
The glycan cap is the other highly glycosylated region within GP1 as it contains 6 N-linked glycosylation sites. The hyper-glycosylated character of GP1 has a major role in this steric immune shielding/masking. In an elegant set of site-directed mutagenesis experiments by Lennemann et al. (2014), the N-linked glycan sites on EBOV GP1 (a total of 15) were systematically disrupted to better understand their role in GP function. The loss of EBOV GP1 glycosylation sites enhanced pseudo-virion infection in Vero cells. Results also indicated that the glycan cap/MLD domains mask the GP1 RBD residues required for binding. EBOV entry into murine macrophages still occurred independently on the presence of GP1 N-glycans, suggesting that N-glycan interactions are not required for entry, (at least) into this cell type, one of the main primary EBOV targets. Also, the removal of all non-MLD GP1 N-glycans enhanced antibody sensitivity. All together, these observations suggest that N-linked glycans on the EBOV GP1 core protect GP from antibody neutralization despite the effect that these glycans might have diminishing infection efficiency (Lennemann et al., 2014).
The role of the glycan cap as a protector of the receptor-binding domain is well understood. However, there is still incomplete knowledge on the mechanisms of EBOV-host cell fusion and subsequent viral entry. A good summary of the current knowledge in this particular area has been recently provided by Gehring et al., (2014). Possibly a number of regions in the GP glycan cap interact with cell receptors to mediate/trigger the fusion of the viral and host membranes (White and Schornberg, 2015; Lee et al., 2009). In vivo GP appears to interact with and infect a wide variety of cells in different tissues. Monocytes, macrophages, and dendritic cells are considered EBOV primary targets (Feldmann and Geisbert, 2011; Gehring et al., 2014). Infection is then distributed through the lymphatic and vascular system to other tissues (Feldmann and Geisbert, 2011; Martines et al., 2014). Affected cells in these tissues include: alveolar macrophages, endothelial cells, fibroblasts, and other interstitial cells (in the lung); Kupffer cells and hepatocytes (in the liver); epidermal dendritic cells, endothelial cells, connective tissue fibroblasts, epithelium cells of the sweat and sebaceous glands (in the skin); cells of the mononuclear phagocytic system, dendritic cells, and fibroblasts (in the spleen and lymph nodes); mononuclear cells within the lamina propria (in the mucosa of the GI tract); endothelial cells (in renal tissue); and monocytes, interstitial cells, and endothelium cells (in testes) (Martines et al., 2014). Several molecules have been suggested as GP1 binding receptors/attachment factors (Gehring et al., 2014) in different cell types, including T-cell Ig and mucin domain 1 (TIM-1) (Kondratowicz et al., 2011) and C-Type Lectins (i.e., L-SIGN and DC-SIGN) in dendritic cells (Alvarez et al., 2000; Simmons et al., 2003). There is a dispute on the role of folate receptors as facilitators of EBOV entry (Simmons et al., 2003; Chan et al., 2001).
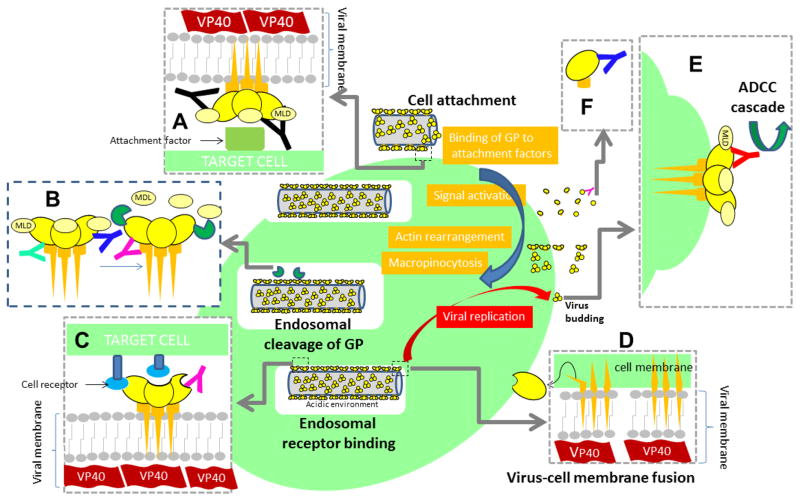
Experimental evidence indicates that cell binding triggers a chain of biochemical signals that lead to viral entry into the cell through macropinocytosis (Nanbo et al., 2010; Figure 4). Endosomal proteolysis of the GP, apparently mediated by low pH-dependent cysteine proteases (i.e., Cathepsin B and L), removes the glycan cap and exposes the receptor binding domain, facilitating further membrane fusion between the virus and the cell (at the endosome). This proteolitic cleavage of the glycan cap appears to be required for infection (Misasi et al., 2012; Chandran et al., 2005) and the endosomal cholesterol transporter Niemann–Pick C1 (NPC1) is believed to be an important intracellular receptor (White and Schornberg, 2015; Carette et al., 2011; Côté et al., 2011). Recently, Sakurai et al. (2015) demonstrated that the endosomal calcium channels, called two-pore channels (TPCs), are required for EBOV entry into host cells. The binding of mAbs to GP interferes at least partially with the virus’s first interaction with host cells, its later entry and infection at the endosome (Figure 5). Precisely, the main rationale for the use of a mAb cocktail (instead of a single mAb) is to extend the breath of protection by binding to multiple GP regions and to more efficiently mitigate immune escape (Both et al., 2013; Ter Meulen et al., 2006).
Figure 4.
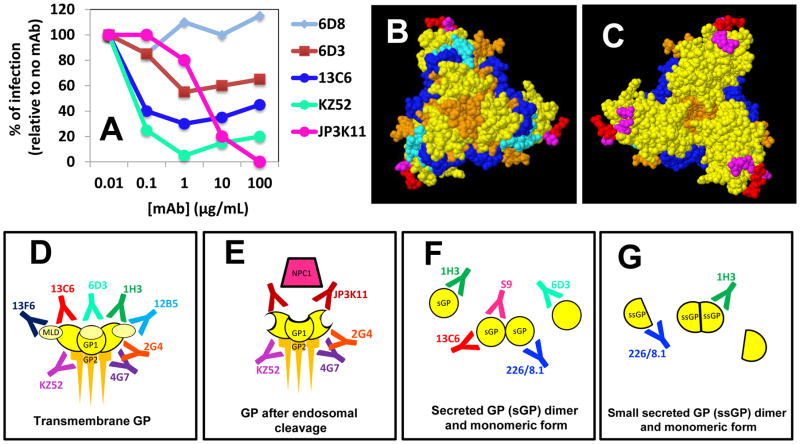
Different mAbs bind to different GP epitopes and interfere at different functional levels. (A) A plot of the percentages of infection inhibition (in vitro) at different mAb concentrations for five different anti-GP mAbs (data modified from Sheldock et al. 2010); (B) Bottom and (C) top view of the chalice of the GP trimer. The epitopes for selected anti-GP mAbs have been indicated with different colors: S9 (red); 1H3 (magenta); JP3K11 (light blue); 133/3.16 (blue). (D) Some mAbs bind to transmembrane GP (E), and/or the enzymatically cleaved form of GP; (F) and/or the monomeric or dimmer versions of sGP (the secreted form of GP) and (G) ssGP.
Figure 5.
Known anti-GP mAbs interfere with key GP functions at different stages of the progression of EBOV infection. (A) Upon interaction with the host cell through attachment factors (not precisely receptors), a complex series of biochemical signals are triggered, eventually leading to EBOV entry through macropinocytosis and endosome formation. Several mAbs are known to interfere with virus-cell attachment (black mAb). (B) Transmembrane GP is cleaved by proteases (dark green symbols) within endosomes. This enzymatic cleavage removes the MLD region (indicated in a lighter shade of yellow) and the glycan cap exposing the RBD at GP1. Several mAbs bind cleaved forms of GP and, by doing so, interfere with GP binding to cell receptors (pink antibody) and (C) further enabling the interaction of viral GP with cell receptors (blue ovals) through the RBD. The interaction of cleaved GP with cell receptors (v.gr. NCP1) triggers virus-cell membrane fusion. After GP binding to receptors, (D) GP is further cleaved, and a significant portion of GP1 is lost, the remaining GP1-GP2 peptide undergoes a geometrical rearrangement to initiate fusion. Some antibodies simultaneously bind epitopes at G1 and G2 interfering with the series of structural arrangements required for virus-cell membrane fusion. (E) Anti-GP mAbs may also bind to the GP molecules exposed at the surface of infected cells, marking them for further host immune response and attacking through mechanisms including antibody-dependent cell-mediated cytotoxicity (ADCC). (F) Binding to sGP conceivably decreases the number of mAb units available to interfere with transmembrane or cleaved GP.
Anti-GP mAb-based therapies: From animal models to a potential clinical use
The first set of experiments that documented a successful use of passive immunization for the EBOV infection in nonhuman primates was published in 2012 (Olinger et al., 2012; Dye et al., 2012), after 15 years of research and multiple failed attempts to prove the therapeutic potential of anti-EBOV antibodies (Qiu and Kobinger, 2014). Dye et al. (2012) used polyclonal antibodies that were directly recovered and concentrated from nonhuman primates (NHP) that survived EBOV infection to treat NHP that were lethally challenged with EBOV. The polyclonal mix provided full protection to the animals even when the first dose was administered 48 hours after the EBOV challenge. During the current outbreak, treatment with plasma or whole blood from convalescent patients has been used. In particular, the massive implementation of EVD treatment using convalescent plasma has been evaluated as a cost-effective countermeasure against EBOV (Gutfraind and Meyers, 2015; Kreil, 2015). The first successful case of the protective use of an anti-GP (EBOV) mAb cocktail in nonhuman primates, that were lethally challenged with EBOV, was reported by Olinger et al. (2012).
Currently, mAb-based therapies have proven to be the most efficient strategy to reverse the progression of a lethal EBOV challenge in nonhuman primates, and there is very limited but promising therapeutic evidence in humans (Qiu and Kobinger, 2014). Nearly 20 anti-GP full-length mAbs have been described in the recent literature, including 1H3, 2G4, 4G7, 5D2, 5E6, 7C9, 7G4, 10C8, KZ52, 13F6, 6D8, 12B5(14G7), 13C6, 6D3, 133/3.16, 226/8.1, 6E3, JP3K11, and S9. At least four different groups have led research efforts on the development of these anti-Ebola therapeutic candidates (see Table 1–3). All of these mAbs target different epitopes (some linear, but most of them conformational) of the GP Zaire EBOV protein.
Table 1.
More than 15 anti-GP (EBOV) full-length mAbs have been described in the literature. The level of conservation among different Zaire EBOV strains varies from 76% to 100%. Among them, examples of linear and conformational epitopes can be found. Z(Zaire), S(Sudan), IC(Ivory Coast).
| mAb | Target AA sequence | Specific strain: GP region | Percentage of conservation | Epitope type (linear or conformational) | Reference |
|---|---|---|---|---|---|
| 1H3 | 267–281 | Z: GP1, sGP | 100% | Conformational | (Qiu et al., 2011) |
| 2G4 | 501–515 | Z: GP1, GP2 | 93.3% | Conformational | (Qiu et al., 2011) |
| 4G7 | 501–515 | Z: GP1, GP2 | 93.3% | Linear | (Qiu et al., 2011) |
| 5D2 | 329–343 | Z: GP1 | 73.3% | Linear | (Qiu et al., 2011) |
| 5E6 | 401–415 | Z: GP1 | 80% | Linear | (Qiu et al., 2011) |
| 7C9 | 157–369 | Z: GP1 | 91.5% | Conformational | (Qiu et al., 2011) |
| 7G4 | 157–369 | Z: GP1 | 91.5% | Conformational | (Qiu et al., 2011) |
| 10C8 | 157–369 | Z: GP1 | 91.5% | Conformational | (Qiu et al., 2011) |
| KZ52 | 51–52 505–513 549–556 |
Z: GP1, GP2 | 100% | Conformational | (Lee et al., 2008) |
| 13F6 | 401–417 | Z: GP1 | 76.4% | Linear | (Wilson et al., 2000) |
| 6D8 | 389–405 | Z: GP1 | 82.4% | Linear | (Wilson et al., 2000) |
| 12B5 | 477–493 | Z: GP1 | 94.1% | Linear | (Wilson et al., 2000) |
| 6E3 | 401–417 | Z: GP1 | 76.4% | Linear | (Wilson et al., 2000) |
| 13C6 | 302–479 494–635 |
Z, S, IC: GP1, sGP | 93.6% | Conformational | (Wilson et al., 2000) |
| 6D3 | 302–479 494–635 |
Z, S, IC: GP1, GP2 | 93.6% | Conformational | (Wilson et al., 2000) |
| 133/3.16 | 521–560 | Z: GP1, GP2 | 97.5% | Conformational | (Takada et al., 2003) |
| 226/8.1 | 1–232 | Z: GP1, sGP, ssGP | 97.4% | Conformational | (Takada et al., 2003) |
| JP3K11 | 302–479 505–514 549–556 |
Z: Cleaved GP1 | 90% | Conformational | (Shedlock et al., 2010) |
| S9 | 293–307 | Z: GP1, sGP | 100% | Linear | (Marceau et al., 2014) |
Table 3.
Subtype (IgGx), nature (neutralizing or non-neutralizing), and animal model used to asses therapeutic efficacy of different anti-Ebola mAbs. Relevant references are included.
| mAb | Subtype | Neutralizing/non-neutralizing | Animal models used | References |
|---|---|---|---|---|
| 1H3 | IgG2a | Neutralizing | Mice, guinea pigs, cynomolgus, rhesus macaques | (Qiu et al., 2012;Qiu et al., 2013) |
| 2G4 | IgG2b | Neutralizing | Mice, guinea pigs, cynomolgus, rhesus macaques | (Qiu et al., 2012;Qiu et al., 2013) |
| 4G7 | IgG2a | Neutralizing | Mice, guinea pigs, cynomolgus, rhesus macaques | (Qiu et al., 2012;Qiu et al., 2013) |
| 5D2 | IgG2a | Non-neutralizing (38%) | Mice, guinea pigs | (Qiu et al., 2012) |
| 5E6 | IgG2a | Non-neutralizing | Mice, guinea pigs | (Qiu et al., 2012) |
| 7C9 | IgG2a | Non-neutralizing | Mice, guinea pigs | (Qiu et al., 2012) |
| 7G4 | IgG1 | Non-neutralizing | Mice, guinea pigs | (Qiu et al., 2012) |
| 10C8 | IgG2a | Non-neutralizing | Mice, guinea pigs | (Qiu et al., 2012) |
| KZ52 | IgG1 | Neutralizing | Mice, guinea pigs, rhesus macaques | (Lee et al., 2008; Parren et al., 2002;) |
| 13F6 | IgG2a | Non-neutralizing | Mice, rhesus macaques | (Wilson et al., 2000; Olinger et al., 2012; Zeitlin et al., 2011)) |
| 6D8 | IgG2a | Non-neutralizing/neutralizing with complement/neutralizing | Mice, rhesus macaques | (Wilson et al., 2000; Shedlock et al., 2010; Olinger et al., 2012; Pettitt et al., 2013) |
| 12B5 | IgG1 | Non-neutralizing | Mice | (Wilson et al., 2000) |
| 6E3 | IgG1 | Non-neutralizing | Mice | (Wilson et al., 2000) |
| 13C6 | IgG2a | Non-neutralizing/neutralizing with complement/neutralizing 65% | Mice, rhesus macaques | (Wilson et al., 2000; Shedlock et al., 2010; Olinger et al., 2012) |
| 6D3 | IgG2a | Non-neutralizing/neutralizing40% | Mice | (Wilson et al., 2000; Shedlock et al., 2010) |
| 133/3.16 | IgG1 | Neutralizing | Mice | (Takada et al., 2003) |
| 226/8.1 | IgG1 | Neutralizing | Mice | (Takada et al., 2003) |
| JP3K11 | IgG1 | Neutralizing | ----- | (Shedlock et al., 2010) |
| S9 | Neutralizing | Mice, guinea pigs | (Marceau et al., 2014) |
While antibody cross-reactivity has been reported among VP40s and NPs of Ebola viruses, antibody binding to GPs is very specific (Kamata et al., 2014). However, some of the mAbs in Tables 1 to 3 also recognize GPs from other EBOV species. For example, mAbs 13C6 and 6D3 bind to epitopes that are highly conserved among Zaire, Sudan, and Ivory Coast EBOVs. Remarkably, a recent paper by Flyak et al. (2015) reported that several anti-GP Marbug mAbs also bind EBOV GP. Figure 3 illustrates, in a graphical way, the GP regions targeted by each of these mAbs. This information is very relevant from the perspective of drug design or drug prescription/personalization. Note that even among the Zaire EBOV strains, some mAbs may not recognize the same epitope with equal affinity in different genetic variants. For instance, the GP region between AA 305 and 510 comprehends the most genetic variations in the Zaire EBOV strains. Therefore, therapeutic mAbs that would target epitopes within this region might not bind with the same GP from different patients or geographic locations. An extreme case for Zaire EBOV is mAb 6E3, which binds to an epitope located in a region with relatively low conservation (76%). On the other hand, mAb 226/8.1 targets an epitope (Takada et al., 2003; Ponomarenko et al., 2014) (Table 1 and 2), which is highly conserved in Zaire EBOV (Figure 3). Next, we review a body of published research that documents the potential therapeutic use of the most promising anti-GP EBOV mAbs among the 20 referred to in Tables 1 to 3. In particular, we refer to experimental evidence on the use of one mAb, named KZ52 (Lee et al., 2008), (see Figure 2), and six mAbs used in three different cocktails (named MB-003, ZMAb, and ZMapp™) that have proven to be protective against EBOV lethal challenges in nonhuman primates (Qiu et al., 2014; Olinger et al., 2012; Pettitt et al., 2013; Qiu et al., 2013). The genetic sequences of mAb KZ52, and all mAbs contained in MB-003, ZMAb, and ZMapp™, have been disclosed (Table 4).
Table 2.
Amino acid sequence of GP epitopes targeted by different anti-Ebola mAbs. The conserved amino acids among different Zaire EBOV strains are marked in black; amino acid positions with differences among Zaire EBOV strains are indicated in gray.
Table 4.
Genetic sequence of the variable heavy (VH) and light (VL) chains of the antibodies including in mAb cocktails proven to be protective in non-human primates in EBOV lethal challenge experiments. The VH and VL sequence of mAb KZ52, protective in guinea pigs but not in human-primates are also included.
| mAb | Chain Heavy | Chain Light | Ref |
|---|---|---|---|
| 13C6 | MGRLTSSFLLLIVPAYVLXQLTLKESGPGI LKPSQTLSLTCSLSGFSLSTSGVGVGWFR QPSGKGLEWLALIWWDDDKYYNPSLKS QLSISKDFSRNQVFLKISNVDIADTATYYC ARRDPFGYDNAMGYWGQGTSVTVSSAK TTAPPVYPLVPGSL |
MGIKMKSQTQAFVFAFLWLSGVDGDIV MTQSQKFMSTSVGDRVSLTCKASQNVG TAVAWYQQKPGQSPKLLIYSASNRYTGV PDRFTGSGSGTDFTLTISNMQSEDLADYF CQQYSSYPLTFGAGTKLELRRADAAPTV SIFPPS |
Hart et al., 2005 |
| 2G4 | GGGLMQPGGSMKLSCVASGFTFSNYWM NWVRQSPEKGLEWVAEIRLKSNNYATH YAESVKGRFTISRDDSKRSVYLQMNTLR AEDTGIYYCTRGNGNYRAMDYWGQGTS VTVSSAKTTPPS |
ASLSVSVGETVSITCRASENIYSSLAWYQ QKQGKSPQLLVYSATILADGVPSRFSGSG SGTQYSLKINSLQSEDFGTYYCQHFWGT PYTFGGGTKLEIKRAD |
Hart et al., 2005 |
| 4G7 | GPELEMPGASVKISCKASGSSFTGFSMNW VKQSNGKSLEWIGNIDTYYGGTTYNQKF KGKATLTVDKSSSTAYMQLKSLTSEDSA VYYCARSAYYGSTFAYWGQGTLVTVSA AKTTAPS |
ASLSASVGETVTITCRASENIYSYLAWYQ QKQGKSPQLLVYNAKTLIEGVPSRFSGS GSGTQFSLKINSLQPEDFGSYFCQHHFGT PFTFGSGTELEIKRAD |
Hart et al., 2005 |
| 1H3 | GAELVKPGASVKLSCTASGFNIKDTYIHW VKQGPEQGLEWIGRIDPANGNTKYDPKF QGKATITADTSSNTAYLQLSGLTSEDTAV YYCARESRISTMLTTGYFDYWGQGTTLT VSSAKTTAPS |
AIMSASPGEKVTMTCSASSSVSYMYWY QQKPGSSPRLLIYDTSNLASGVPVRFSGS GSGTSYSLTISRMEAEDAATYYCQQWSS YPYTFGGGTKLEIKRAD |
Jones et al., 2013 |
| 6D8 | MDFGLIFFIVALLKGVQCDVKLLESGGGL VQPGGSLKLSCAASGFDFSRYWMSWVR QAPGKGLEWIGEINPDSSTINYTPSLKDKF IISRDNAKNTLYLQMSKVRSEDTALYYCT RQGYGYNYWGQGTTLIVSSAKTTAPPVY PLVPGSL |
MKLPVRLLVLMFWIPASSSDVLLTQIPLS LPVSLGDQASISCRSSQSIVHSNGNTYLE WYLQKPGQSPKLLIYKASNRFSGVPDRF SGSGSGTDFTLKINRVEAEDLGVYYCLQ GSHVPSTFGGGTKLEIKRADAAPTVSIFP PSSKLG |
Jones et al., 2013 |
| 13F6 | MELGLSWIFLVLTLKGVKCEVQVVESGG GLVKPGGSLKLSCAASGFAFSSYDMSWV RQTPEKRLEWVAYISRGGGYTYYPDTVK GRFTISRDNAKNTLYLQMSSLKSEDTAM YYCSRHIYYGSSHYYAMDYWGQGTSVT VSSAKTTAPPVYPLAPGSL |
MAWIXLIFFVLHCSGSFSQLVLTQSSSAS FSLGASAKLTCTLSRQHSTYTIEWYQQQ PLKPPRYVMELKKDGSHSTGDGIPDRFS GSSSGADRYLSISNIQPEDEAIYICGVGDT IKEQFVYVFGGGTKVTVLGQPKSTPTLT VFPPSSEELKENKATLVCLISNFSPSGVTV AWKANGTPITQGVDTSNPTKEGNKFMA SSFLHLTSDQWRSHNSFTCQVTHEGDTV EKSLSPAECL |
Jones et al., 2013 |
| 10C8 | GAELVRSGASVKLSCTSSGFNIKDYFLHW VKQRPEQGLEWIGWIDPENGDTEYAPKF QDKATMTADTSSNTAYLHLSSLTSEDTG VYYCNADGNYGKNYWGQGTTLTVSSAK TTAPS |
LSLPVSLGDQASISCRSSQSLVHSNGNTF LHWYLQKPGQSPKLLIYRVSNRFSGVPD RFSGSGSGTDFTLKISRVEAEDLGVYFCS QSTHVPPYTFGGGTKLEIKRAD |
Jones et al., 2013 |
| 7C9 | GAELVKPGASVKLSCTASGFNIKDTYMH WVKERPDKGLEWIGRIDPANGNTKCDSR FQGKATITADTSSNTAYLQLSSLTSEDTA VYYCARRIYFGKGFDFWGQGTTLTVSSA KTTAPS |
SSLSVSAGEKVTMSCKSSQSLFNSGDQK NYLAWYQQKPGQPPKLLIYGASTRESGV PDRFTGSGSGTDFTLTISSVQAEDLAVYY CQNDQFYPPTFGDGTKLDLKRAD |
Jones et al., 2013 |
| 5E6 | GGGLVKPGGSLKLSCAASGSAFSRYDMS WVRQTPEKRLEWVAYISRGGGFIYYPDT VKGRFTISRDNAKNTLYLQMSSLKSDDT AMYYCARHVYYGSSPLYAMDYWGQGT SVTVSSAKTTAPS |
SASFSLGASAKLTCTLSSQHSTFTIEWYQ QQPLKPPKYVMELKKDGSHSTGDGIPDR FSGSSSGADRYLSISNIQPEDEAIYICGVG DTINEQFVYVFGGGTKVTVLG |
Jones et al., 2013 |
| 5D2 | GPGLVRPSQSLSLTCTVTGYSITSDYAWN WIRQFPGNKLEWLGYITNTGSTGFNPSLK SRISITRDTSKNQFFLQLISVTTEDTATYH CARGLAYWGQGTLVTVSAAKTTAPS |
LTLSVTIGQPASISCKSSQSLLDSDGKTYL NWLLQRPGQSPKRLIYLVSKLDSGVTDR FTGSGSGTDFTLKISRVEAEDLGVYYCW QGTHSPFTFGSGTKLEIKRAD |
Jones et al., 2013 |
| KZ52 | EVQLLESGGGLVKPGGSLRLSCAASGFTL INYRXNWVRQAPGKGLEWVSSISSSSSYI HYADSVKGRFTISRDNAENSLYLQXNSL RAEDTAVYYCVREGPRATGYSXADVFDI WGQGTXVTVSSASTKGPSVFPLAPSSKST SGGTAALGCLVKDYFPEPVTVSWNSGAL TSGVHTFPAVLQSSGLYSLSSVVTVPSSSL GTQTYICNVNHKPSNTKVDKKVEPK |
ELVXTQSPDSLAVSLGERATINCKSSQSV LYSSNNKSYLAWYQQKPGQPPKLLIYW ASTRESGVPDRFSGSGSGTDFTLTISSLQA EDVAVYYCQQYYSAPLTFGGGTKVEIKR TVAAPSVFIFPPSDEQLKSGTASVVCLLN NFYPREAKVQWKVDNALQSGNSQESVT EQDSKDSTYSLSSTLTLSKADYEKHKVY ACEVTHQGLRSPVTKSFNR |
Lee et al., 2008 |
MAb KZ52 is one of the first and best described neutralizing EBOV mAbs. Lee et al. (2008) analyzed the 3D structure of the GP bound to antibody KZ52, originally isolated from a 1995 Kikwit outbreak human survivor (Figure 2). The authors used X-ray crystallography to resolve, at the interaction site, the structural details between residues at the surface of GPs and the variable regions (Table 4) of KZ52. The antibody recognizes a relatively small (~20 GP residues), glycan-unprotected region of the protein neighboring the viral membrane surface (see Figure 2(A), (B), (E)). The KZ52-conformational epitope contains residues of GP1 and GP2 (Figure 2(C)) (Lee et al., 2008, 2009). The KZ52 mAb proved to be protective against lethal EBOV challenge in guinea pigs when administered before and immediately after infection (Parren et al., 2002). In these experiments, the medium to high levels of viremia in survivor animals suggested that other mechanisms (i.e., tagging, to activate the elimination of infected cells) besides virus neutralization were responsible for protection. Interestingly, KZ52 was unable to provide protection against lethal challenge in nonhuman primates (Oswald et al., 2007) even at doses of 50 mg/kg, which have been shown to be sufficient in other disease/mAb cases.
Olinger et al.(2012) reported the expression of three anti-GP virus mAbs (named c13C6, h-13F6, and c6D8) in whole plant (Nicotiana benthamiana) cells and in Chinese hamster ovary (CHO). The authors tested these three mAbs (MB-003, a predecessor of ZMapp™) and found them to be protective against lethal Ebola challenge in rhesus macaques when administered 1 h postinfection. The tobacco version of MB-003 was approximately three times more effective than its CHO analog in murine models, presumably due to their non-mammalian (lacking core fucose) glycosylation pattern. In a pilot study on primate models, three 16 mg/kg doses of each tobacco-derived mAb protected three out of three Ebola-infected macaques when MB-003 was administered within the first 48 h after virus exposure. In a more refined experiment (Pettitt et al., 2013), a group of macaques received a lethal Ebola virus challenge and were treated with three doses of 50 mg/kg of MB-003 several hours after they presented Ebola symptoms (four to five days after exposure). Three out of seven macaques survived the challenge. A combination therapy with ZMAb—another cocktail composed of the murine mAbs 1H3, 2G4, and 4G7—and Ad-IFN (adenovirus-vectored interferon-α) was 100% protective in rhesus macaques when administered three days after positive Ebola diagnosis (Qiu et al., 2013).
In a recent report, the different mAbs contained in MB-003 and ZMAb were tested in lethal challenge EBOV experiments in guinea pigs and nonhuman primates (Qiu et al., 2014). In a first round of experiments, the authors tested a single dose of individual mAbs or combinations of them. In these experiments, no individual mAb was able to provide protection levels above 33% of survival; c13C6 proved to be the most effective mAb in single-dose experiments. In contrast, some mAb cocktails provided between 50% and 67% protection levels in guinea pigs. In a second round of lethal challenge experiments, this time using a scheme of three doses of mAb cocktails, some mAb combinations provided full protection. Promisingly, the administration of the mAb therapy started three, four, or five days after the lethal challenge, when viremia, as measured by RT-PCR, was high. The mAb cocktail that rendered the best results was a combination of the mouse/human chimeric mAbs c13C6 (from MB-003), c2G4 (from ZMAb), and the murine mAb m4G7 (from ZMAb). This mAb formulation (see Figure 3D) was selected by Mapp Biopharmaceuticals, Inc. to be further tested under the brand name of ZMapp™ (produced by Mapp Biopharmaceuticals, Inc.), which was recently approved by WHO for humanitarian use on Ebola patients. Clearly, mAb therapy using the ZMapp™ cocktail has proven to be helpful as an emergency resource to treat Ebola patients, although the evidence that exists at this point is insufficient to anticipate high proficiency in large-scale clinical intervention (Goodman et al., 2014).
The limited availability of robust and reliable in vitro assays/platforms that are capable of predicting the therapeutic value of an anti-EBOV mAb in animal models is an important challenge in anti-EBOV mAb research and development; the ability of a monoclonal antibody to neutralize EBOV in cell culture assays does not necessarily mean that this mAb will be protective in animal models. Moreover, the characteristics and conditions that make an antibody protective against EBOV in animal models and/or in humans are not fully understood. Furthermore, the protective ability of a specific mAb in one animal model does not necessarily imply that it provides protection in another animal model.
Nacayama and Saijo (2013) have comprehensively reviewed the different animal models used to study EBOV infection, mainly rodents and non-human primates (NHP), and have summarized their strengths and weaknesses. NHP, particularly rhesus and cynomolgus macaques, better mimic EBOV infection in humans and more closely reproduce the symptoms of the disease than do rodents. However, the use of NHP presents researchers with more significant practical and ethical hurdles (Nacayama and Saijo, 2013; Geisbert et al., 2015) than does the use of rodents. Different rodent models have been developed to do EBOV research, including some knockout variants (Brannan et al., 2015). The practicality of using rodents in the laboratory and the possibility of using genetically modified animals has made them an attractive model with which to study the protective effect of anti-EBOV therapies and vaccines. However, the EBOV has to be adapted to cause lethal infection in rodents, which represents a highly restrictive situation given the high risk presented by handling a BSL-4 pathogen, such as EBOV. Moreover, some widely used rodents, such as mice and guinea pigs, do not exhibit some of the distinctive symptoms of EBOV in primates, such as fever and rash, which further suggests dissimilarities between the anti-EBOV immune response in rodents and primates. In general, anti-EBOV protection in rodents is not a conclusive indicator of protection in humans. Probably the best example of this is the case of mAb KZ52, which is neutralizing and protective in rodents but not in NHPs. In the particular case of anti-EBOV mAb therapies, NHP studies appear to be a mandatory step in predicting efficacy in humans. However, no clear correlation can be established between therapeutic effectiveness in NHPs and humans due to the limited amount of clinical data in humans.
Anti-Ebola mAb-based therapies: Incomplete knowledge on mechanisms of action
As previously stated, the use of anti-GP mAb cocktails is mainly intended to interfere with the functions of EBOV-GP by binding GP at different epitopes (Figure 4). In brief, anti-EBOV mAbs are believed to directly interfere with EBOV infection at four different stages: (a) during host cell attachment, (b) during endosomal protease cleavage, (c) at endosomal NPC1 receptor binding, (d) and during virus-host cell membrane fusion (Figure 5). However, our knowledge on the mechanisms by which the anti-EBOV mAb therapy may work remains incomplete. A summary of the current knowledge on the effects and mechanisms of action of mAb-based therapies follows.
In experiments where MB-003 was administered to nonhuman primates (Olinger et al., 2012; Pettitt et al., 2013), the number of viral gene copies in surviving monkeys, as measured by RT-PCR, was significantly reduced by two to four orders of magnitude after the first two dosages, suggesting that the therapy effectively interferes with viremia progression. In addition, in these experiments, the titer of specific anti-GP IgGs was much higher in survivors. A plausible interpretation of this observation is that mAb therapy retards the infection progress long enough to allow the host immune system to mount a sufficiently efficient immune response.
High anti-GP titers appear to be correlated with EBOV infection survival. On the other hand, a considerable body of experimental evidence indicates that EBOV has multiple ways to evade and interfere with the host immune response. Some (but not all) of these interference strategies are mediated by transmembrane or sGPs (see Figures 1 and 4).
Wong et al. (2012) evaluated the correlation between immune responses and survival in rodents lethally challenged or vaccinated with EBOV using knockout mice with an impaired ability to generate normal B and/or T cell responses. In particular, vaccinated animals with impaired B cell response were unable to survive the challenge, while their wild type counterpart did. Impaired CD4+ animals were capable of mounting a certain level of protection. Results suggested that protection in mice was mainly mediated by B cells and CD4+ T cells. A high correlation between GP-specific total immunoglobulin G (IgG) levels and survival was found in both vaccinated guinea pigs and nonhuman primates. Although the mechanisms for protection after vaccination are not necessarily the same as those for infection intervention, these results clearly suggest that GP plays a relevant role in the infection propagation.
Figure 3 and 4 illustrates the GP regions where each of the mAbs listed in Tables 1 and 2 bind to GP. This information might be useful in the design of mAb cocktail therapies. Note that mAbs may inhibit different GP roles by binding to different epitopes (Figure 3,4B–C)) in different GP forms (Figure 4D–G). Since some epitopes are not accessible to mAb binding in some GP forms (i.e. MDL in transmembrane GP protects the epitopes located nearby the receptor binding domain), some mAbs can only bind the cleaved forms of GP. As stated before, anti-EBOV directly interfere with the progression of EBOV infection at different stages. During host cell attachment, mAbs with binding affinity for the exposed epitopes of transmembrane GP (Figure 4B–D) might sterically impede GP binding to different cell attachment factors (Figure 5A). Other mAbs might obstruct the enzymatic cleavage conducted by proteases at the endosomes (Figure 5B) or block the binding of the cleaved forms of GP (Figure 4E) to cell receptors also present at endosomes, (i.e. NPC1) hence obstructing the activation of the process of endosomal virus-cell fusion (see White and Schornberg, 2015). Moreover, GP also undergoes a GP1-GP2 endosomal cleavage to release GP1 and trigger the complex structural rearrangement that mediates virus-cell membrane fusion. Experimental evidence suggests that some mAbs may bind simultaneously to GP1 and GP2 regions obstructing cleavage or rearrangement (Dias et al., 2011).
The recognition of different epitopes at different stages of viral infection results in differences in observable binding affinities and neutralization kinetics of each mAb. Shedlock et al. (2010) published the most comprehensive comparative study of anti-GP binding affinities published so far (see Figure 4A). In experiments designed to measure the extent of infection inhibition by using five different anti-GP EBOV mAbs (6D3, 13C6, KZ52, 6D8, and JP3K11), the authors exposed endothelial cell cultures to GP-pseudoviral particles in the presence of different concentrations of each mAb. The observed inhibition profiles differed significantly from mAb to mAb (Figure 4(A)). The most drastic infection inhibition was obtained when KZ52 was used, closely followed by 13C6. Interestingly, in both cases, results suggested maximum inhibition at an in vitro concentration of ≈1 μg/mL. JP3K1 was also capable of interfering with infection efficiently in this model, but at much higher concentrations. The authors also report and discuss other important differences in binding/neutralizing behavior among these mAbs. Both KZ52 and JP3K11 bind to conformational epitopes that contain residues in GP1 and GP2. However, the mechanisms by which these two mAbs inhibit infection could be different: JP3K1 can bind to both trimeric GP and cleaved GP. Some controversy remains on the ability of KZ52 to bind effectively to cleaved GP1. Shedlock et al. (2010) found that KZ52 is unable to bind GP-pseudoviral particles (not actual EBOV particles or the isolated GP molecule), which were treated with Cathepsin to cleave GP. Other reports have demonstrated that KZ52 is capable of binding to the purified cleaved GP trimmer (Bale et al., 2011; Hood et al., 2010). It is believed that KZ52 binds cleaved GP restraining conformational changes required for membrane fusion (Dias et al., 2011). On the other hand, 13C6 and 6D3 recognized epitopes in GP and sGP, exhibiting a greater affinity for sGP than for transmembrane GP, probably due to the increased exposure of the corresponding epitope on sGP compared to GP (Shedlock et al., 2010; Murin et al., 2014). The 6D8, a mAb that recognizes a linear epitope in MLD, was unable to interfere with infection and did not exhibit affinity for sGP or cleaved GP (GP loses the MLD during cleavage) (Shedlock et al., 2010). Some other mAbs that also target the MLD have been proven to be protective in animal models. In particular, mAbs 13F6 and 12B5 (12B5 is most probably equivalent to mAb 14G7, referred in Wilson et al., 2000) recognize linear non-glycosylated epitopes of the MLD (Olal et al., 2012. The precise binding site for these two mAbs (13F6 and 12B5) has been described in detail (Olal et al., 2012). In a recent paper, Murin et al. (2014), using single particle EM, attempted to precisely identify the binding sites of each conformational antibody contained in MB-003, ZMAb and ZMapp™ cocktails. Their results confirmed that mAb 13C6, constituent of MB-003 and ZMapp™, binds perpendicularly to the expected plane of the membrane, straight down onto the surface of the GP, in the region of the glycan cap. Similarly, 1H3 binds the glycan cap of GP partially interfering with 13C6. The authors also showed that MAb 13F6 and 12B5 bind the MLD without interfering with each other. MAbs 2G4, 4G7, and 16F6 simultaneously target epitopes at the base of GP. The epitopes of c4G7 and c2G4 overlap extensively. These antibodies differ mainly in their angle of approach to the overlapping binding sites. While c4G7 most likely simultaneously binds GP1 and GP2, c2G4 appears to bind almost exclusively to GP2. mAb 4G7 binds slightly lower on GP, encompassing some of the GP1 base, similar to KZ52. The footprints of both c2G4 and c4G7 identified by Murin et al. (2014), as well as the footprint of KZ52 determined crystallographically (Lee et al., 2008), all include residue Q508 of GP2. A point mutation at Q508 abolishes the binding of c2G4 and c4G7 (Qiu et al., 2013), and also abolishes binding of KZ52 (Murin et al., 2014).
Some experimental evidence suggests that glycosylation (and differences in glycosylation patterns) might play a role in the design and selection of the recombinant platform for the production of anti-Ebola mAbs (Zeitlin et al., 2011). There was a three-fold difference in effectiveness between MB-003 formulations produced in tobacco versus those expressed in CHO cell lines. This suggests that the differences in glycosylation (e.g., the absence of fucose) attached at the constant region portion of the full-length antibody might play a role on the observed therapeutic effect. The absence of core fucose on full-length antibodies increases the binding of mAb to FcγRIII, a well-characterized cell receptor in macrophages (Guilliams et al., 2014). This results in a significant enhancement in antibody-dependent cell-mediated cytotoxicity (ADCC) activity, as compared with a fucosylated antibody, such as those produced by most CHO cell lines (Figure 5E). In conclusion, this observation suggests that full-length antibodies without fucose are better infected-cell markers than full-length antibodies with fucose, but it does not indicate the relative importance of ADCC activation versus simple virus entry interference. On the other hand, CHO-derived MB-003 formulations still protected nonhuman primates from lethal Ebola challenge when administered at a higher dose, suggesting that ADCC stimulation is not an absolute requirement for protection (Olinger et al., 2012).
In summary, there are two main mechanisms that appear to be involved in the therapeutic effect of anti-GP mAbs: interference with viral functions (namely attachment, cleavage or entry) and tagging for immune system attack. We do not know the relative importance of these two effects; no peer-reviewed published research is yet available on this particular topic.
Bottlenecks in anti-Ebola mAb production: Scaling up, cost, and development time
Anti-Ebola mAb therapy appears to be a promising resource to combat EVD. One severe problem remains: the production of mAbs is a complex process from a biopharmaceutical engineering perspective, and currently available production platforms are not sufficiently effective to respond quickly in an emergency. The current Ebola outbreak is unprecedented in terms of the number of people infected, the rapidness of progression, and the broadness of geographical extent (WHO, 2014b). On the other hand, mAb doses required for each patient might be high. Based on the doses that have proven to be effective in preclinical studies in nonhuman primates (i.e., 50 mg kg−1 mAb−1 (Olinger et al., 2012)), a simple extrapolation to humans (average weight of 70 kg) suggests that approximately 10.0 g of mAbs would be required to treat each Ebola patient.
In a scenario in which 5,000 people require ZMapp™ treatment per month (as it has happened in October, 2014), 50,000 g will be required monthly. So far, ZMapp™ has been produced in tobacco plants by a high-yield transient expression strategy in which tobacco leaves from plants that are six to eight weeks old are co-transfected with constructs containing the genetic information for the synchronous production of both heavy and light chains of each antibody (Giritch et al., 2006; Castilho et al., 2011). This strategy is labor intensive but definitely useful for producing sufficient amounts of each mAb rapidly for pilot studies. However, to produce 50 kg of mAbs, approximately 150 tons of leaves would have to be transfected. The separation processes required to recover mAbs from tobacco biomass is not straightforward (Fulton et al., 2015) and has yet to be optimized and scale-up to be suitable for handling tons of biomass. Stable expression in mature tobacco plants is another option that is both less labor intensive and more scalable, but developing a tobacco plant with stable expression requires an investment of several months (Hood et al., 2002; Ma et al., 2003). A more practical option is expression in mammalian cells. Therapeutic mAbs are commercially produced by recombinant technology in suspended mammalian cell cultures in stirred tanks (Li et al., 2010). In an optimized commercial process, at least 2 g mAb L−1 (after purification) can be produced after two weeks of fed-batch culture in a standard 10–13 m3 stirred-tank bioreactor using CHO cells, the warhorse for the production of glycosylated biopharmaceuticals (Li et al., 2010; Garza-García et al., 2014). Producing 100 kg of mAbs would require processing 50000 L of culture media monthly. This proposition is feasible, but the high complexity and cost of mammalian cell culture are serious drawbacks of this technological alternative; the operational cost for a single 10 m3-CHO cell culture is approximately US$10M. Most importantly, to construct and isolate a stable, high-producing, CHO cell clone would demand no less than 90 days of development work; the optimization and scale-up of a CHO cell culture to a 10 m3 bioreactor would demand three or four additional months (Li et al., 2010). Even assuming that an optimized process is in place to produce mAb cocktails in CHO cells, this technological path could still be compromised due to the relatively low productivity of this production platform and the high dose required to treat an Ebola patient. Illustratively, to treat 24,000 patients, approximately the total number of confirmed cases reported up to March 10st 2014, 252 kg of mAbs would be required. That is approximately 3.0% of the yearly worldwide installed capacity for mAb production (Ecker et al., 2014). The use of properly engineered mAb fragments, instead of full-length mAbs, is a therapeutic alternative that has yet to be investigated for Ebola. The use of antibody fragments to neutralize viruses or prevent virus infection is not novel; proof of concept experiments have shown their potential applications in the context of different viral infections such us HIV (Lülf et al., 2014), Influenza A H1N5 (Bal et al., 2015), SARS (Sui et al., 2004), HPV (Culp et al., 2007), and West Nile virus (Gould et al., 2005). A recent contribution by Rodríguez-Martínez et al. (2015) offers proof-of-principle of the application of three anti-EBOV mAb fragments, containing the variable regions the mAbs KZ52, 13C6, and 13F6, in immunological assays to specifically detect recombinant GP.
The production of mAb fragments provides with several important technological (and possibly even therapeutic) advantages. MAb fragments have been suggested before to interfere with other viral infections. It can be easily and massively produced in simpler bioreactors at 1/50 of the cost required to produce full-length mAbs from CHO cells (Figure 5).
The road ahead
MAb-based anti-EBOV therapies must be further investigated to assure their safety and effectiveness at large-scale clinical interventions. No mAb-based cocktail has yet been the subject of a formal clinical trial yet. The ZMapp™ cocktail might enter into clinical trials in the USA this year.
Even preclinical data in non-human primates is still limited; only a handful of mAbs have been tested in NHPs, and only three mAbs, as a cocktail, have been tested in humans. Ideally, the set of mAbs tested in animal models should be expanded. More research is needed to fully understand the mechanisms by which different mAbs (and mAb cocktails) interfere with the progression of EVD in NHP and other more widely available animal models. A rational extrapolation of preclinical data to humans can only be accomplished when the first sets of clinical data becomes available.
Recently, the Viral Hemorrhagic Fever Immunotherapeutic Consortium (VIC), a worldwide research consortium headed by Erika Saphire (known as VIC), initiated a massive screening of anti-GP antibodies from laboratories across the globe in order to identify the best anti-EBOV therapeutic candidates for further preclinical testing. A deeper understanding of the mechanisms by which different mAbs interfere with EBOV infection will provide elements for a more rational design of anti-EBOV mAb cocktails. Relevant details of the process of EBOV entry and infection propagation are still unknown. For example, the importance of the MLD cleavage in exposing the RBD for further interaction of the virus with inner and outer cell surfaces needs to the clarified. In addition, the relative importance of mAb interference with relevant GP functions versus mAb tagging to induce ADCC activity has not yet been investigated. Finally, there are technological issues to be resolved to make feasible the massive production of anti-EBOV mAbs. Currently available platforms have yet to be adapted to produce sufficient mAb quantities fast enough to respond to epidemic outbreaks.
Figure 6.
Comparison of different platforms to produce anti-EBOV immune therapeutics. While transient expression in tobacco leaves is not an easily scalable solution, and CHO cell culture exhibits limitations in capacity and cost, other alternatives such as the production of mAb fragments in bacterial cultures could be a cost-effective alternative to face EBOV epidemics.
Acknowledgments
MMA gratefully acknowledge the institutional funding received from Tecnológico de Monterrey (seed funding to Strategic Research Groups, 2014) and funding provided by CONACyT (Consejo Nacional de Ciencia y Tecnología, México) in the form of a sabbatical scholarship (262130) and student scholarships for ARMI and EGG. GTdS gratefully acknowledge the funding received by CONACyT (scholarship 234713) and Fundación México in Harvard in the form of postdoctoral scholarships. AK and NA acknowledge funding from the National Science Foundation (EFRI-1240443), IMMODGEL (602694), and the National Institutes of Health (EB012597, AR057837, DE021468, HL099073, AI105024, AR063745).
Footnotes
Declaration of conflicts of interest
The authors report no declarations of interest.
References
- 1.Alvarez CP, Lasala F, Carrillo J, Muñiz O, Corbí AL, Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J Virol. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audet J, Wong G, Wang H, Lu G, Gao GF, Kobinger G, Qiu X. Molecular characterization of the monoclonal antibodies composing ZMAb: A protective cocktail against Ebola virus. Sci Rep. 2014;4:6881. doi: 10.1038/srep06881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bah EI, Lamah M-C, Fletcher T, Jacob ST, Brett-Major DM, Sall AA, Shindo N, Fischer WA, Lamontagne F, Saliou SM, Bausch DG, Moumié B, Sprecher A, Lawler JV, Mayet T, Jacquerioz FA, Méndez MF, Vallenas C, Clement C, Mardel S, Faye O, Faye O, Soropogui B, Magassouba N, Koivogui L, Pinto R, Fowler RA. Clinical presentation of patients with Ebola virus disease in Conakry, Guinea. New Engl J Med. 2015;372:40–47. doi: 10.1056/NEJMoa1411249. [DOI] [PubMed] [Google Scholar]
- 4.Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keïta S, De Clerck H, et al. Emergence of Zaire Ebola virus disease in Guinea - preliminary report. New Eng J Med. 2014;371:1418–25. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 5.Bal C, Herbreteau CH, Buchy P, Rith S, Zaid M, Kristanto W, Han V, Reynaud C, Granjard P, Lépine B, Durand C, Tambyah PA. Safety, potential efficacy, and pharmacokinetics of specific polyclonal immunoglobulin F (ab′) 2 fragments against avian influenza A (H5N1) in healthy volunteers: a single-centre, randomised, double-blind, placebo-controlled, phase 1 study. Lancet Infect Dis. 2015;15:285–292. doi: 10.1016/S1473-3099(14)71072-2. [DOI] [PubMed] [Google Scholar]
- 6.Bale S, Liu T, Li S, Wang Y, Abelson D, Fusco M, Woods VL, Jr, Saphire EO. Ebola virus glycoprotein needs an additional trigger, beyond proteolytic priming for membrane fusion. PLoS Neglected Trop Dis. 2011;5:e1395. doi: 10.1371/journal.pntd.0001395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becquart P, Mahlakõiv T, Nkoghe D, Leroy EM. Identification of continuous human B-cell epitopes in the VP35, VP40, nucleoprotein and glycoprotein of Ebola virus. PLoS One. 2014;9:e96360. doi: 10.1371/journal.pone.0096360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beniac DR, Melito PL, deVarennes SL, Hiebert SL, Rabb MJ, Lamboo LL, Jones SM, Booth TF. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS One. 2012;7:e29608. doi: 10.1371/journal.pone.0029608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Both L, Banyard AC, Dolleweerd C, Wright E, Ma JK, Fooks AR. Monoclonal antibodies for prophylactic and therapeutic use against viral infections. Vaccine. 2013;31:1553–1559. doi: 10.1016/j.vaccine.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brady O. Scale up the supply of experimental Ebola drugs. Nature. 2014;512:233. doi: 10.1038/512233a. [DOI] [PubMed] [Google Scholar]
- 11.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brannan JM, Froude JW, Prugar LI, Bakken RR, Zak SE, Daye SP, Wilhelmsen CE, Dye JM. Interferon α/β receptor–deficient mice as a model for Ebola virus disease. J Infect Dis. 2015:jiv215. doi: 10.1093/infdis/jiv215. [DOI] [PubMed] [Google Scholar]
- 13.Carroll SA, Towner JS, Sealy TK, McMullan LK, Khristova ML, Burt FJ, Swanepoel R, Rollin PE, Nichol ST. Molecular evolution of viruses of the family Filoviridae based on 97 whole-genome sequences. J Virol. 2013;87:2608–16. doi: 10.1128/JVI.03118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castilho A, Bohorova N, Grass J, Bohorov O, Zeitlin L, Whaley K, Altmann F, Steinkellner H. Rapid high yield production of different glycoforms of Ebola virus monoclonal antibody. PLoS One. 2011;6:e26040. doi: 10.1371/journal.pone.0026040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan SY, Empig CJ, Welte FJ, Speck RF, Schmaljohn A, Kreisberg JF, Goldsmith MA. Folate receptor-α is a cofactor for cellular entry by Marburg and Ebola viruses. Cell. 2001;106:117–126. doi: 10.1016/s0092-8674(01)00418-4. [DOI] [PubMed] [Google Scholar]
- 16.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiappelli F, Bakhordarian A, Thames AD, Du AM, Jan AL, Nahcivan M, Nguyen MT, Sama N, Manfrini E, Piva F, Rocha R, Maida CA. Ebola: translational science considerations. J Transl Med. 2015;13:11. doi: 10.1186/s12967-014-0362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Côté M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. 2001;75:7769–7773. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Culp TD, Spatz CM, Reed CA, Christensen ND. Binding and neutralization efficiencies of monoclonal antibodies, Fab fragments, and scFv specific for L1 epitopes on the capsid of infectious HPV particles. Virology. 2007;361:435–446. doi: 10.1016/j.virol.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de La Vega MA, Wong G, Kobinger GP, Qiu X. The multiple roles of sGP in Ebola pathogenesis. Viral Immunol. 2015;28:3–9. doi: 10.1089/vim.2014.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, Kawaoka Y, Chandran K, Dye JM, Saphire EO. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dye JM, Herbert AS, Kuehne AI, Barth JF, Muhammad MA, Zak SE, Ortiz RA, Prugar LI, Pratt WD. Postexposure antibody prophylaxis protects nonhuman primates from filovirus disease. Proc Natl Acad Sci USA. 2012;109:5034–5039. doi: 10.1073/pnas.1200409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ecker DM, Ransohoff TC. Mammalian cell culture capacity for biopharmaceutical manufacturing. In: Zhou W, Kantardjieff A, editors. Mammalian cell cultures for biologics manufacturing. Springer; Berlin Heidelberg: 2014. pp. 185–225. [DOI] [PubMed] [Google Scholar]
- 26.Enserink M. Infectious diseases. Ebola drugs still stuck in lab. Science. 2014;345:364–655. doi: 10.1126/science.345.6195.364. [DOI] [PubMed] [Google Scholar]
- 27.Feldmann H, Geisbert T. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flyak AI, Ilinykh PA, Murin CD, Garron T, Shen X, Fusco ML, Hashiguchi T, Bornholdt ZA, Slaughter JC, Sapparapu G, Klages C, Ksiazek TG, Ward AB, Saphire EO, Bukreyev A, Crowe JE., Jr Mechanism of human antibody-mediated neutralization of marburg virus. Cell. 2015;160:893–903. doi: 10.1016/j.cell.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francica JR, Matukonis MK, Bates P. Requirements for cell rounding and surface protein down-regulation by Ebola virus glycoprotein. Virology. 2009;383:237–247. doi: 10.1016/j.virol.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fulton A, Lai H, Chen Q, Zhang C. Purification of Monoclonal Antibody against Ebola GP1 Protein Expressed in Nicotiana benthamiana. Journal of Chromatography A. 2015 doi: 10.1016/j.chroma.2015.02.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garza-García LD, García-López E, Camacho-León S, del Rocha-Pizaña MR, López-Pacheco F, López-Meza J, Araiz-Hernández D, Tapia-Mejía EJ, Trujillo-de Santiago G, Rodríguez-González CA, Alvarez MM. Continuous flow micro-bioreactors for the production of biopharmaceuticals: the effect of geometry, surface texture, and flow rate. Lab Chip. 2014;14:1320–1329. doi: 10.1039/c3lc51301g. [DOI] [PubMed] [Google Scholar]
- 32.Gehring G, Rohrmann K, Atenchong N, Mittler E, Becker S, Dahlmann F, Pöhlmann S, Vondran FWR, David S, Manns MP, Ciesek S, von Hahn T. The clinically approved drugs amiodarone, dronedarone and verapamil inhibit filovirus cell entry. J Antimicrob Chemother. 2014;69:2123–2131. doi: 10.1093/jac/dku091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geisbert TW, Hensley LE, Jahrling PB, Larsen T, Geisbert JB, Paragas J, Young HA, Fredeking TM, Rote WE, Vlasuk GP. Treatment of Ebola virus infection with a recombinant inhibitor of factor VIIa/tissue factor: a study in rhesus monkeys. Lancet. 2003;362:1953–1958. doi: 10.1016/S0140-6736(03)15012-X. [DOI] [PubMed] [Google Scholar]
- 34.Geisbert TW, Lee AC, Robbins M, Geisbert JB, Honko AN, Sood V, Johnson JC, de Jong S, Tavakoli I, Judge A, Hensley LE, Maclachlan I. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet. 2010;375:1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geisbert TW, Strong JE, Feldmann H. Considerations in the use of nonhuman primate models of Ebola virus and Marburg virus infection. J Infect Dis. 2015:jiv284. doi: 10.1093/infdis/jiv284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gire SK, Goba A, Andersen KG, Sealfon RSG, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, Dudas G, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–72. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giritch A, Marillonnet S, Engler C, van Eldik G, Botterman J, Klimyuk V, Gleba Y. Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc Natl Acad Sci USA. 2006;103:14701–14706. doi: 10.1073/pnas.0606631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodman JL. Studying “secret serums”--toward safe, effective Ebola treatments. N Engl J Med. 2014;371:1086–1089. doi: 10.1056/NEJMp1409817. [DOI] [PubMed] [Google Scholar]
- 39.Gould LH, Sui J, Foellmer H, Oliphant T, Wang T, Ledizet M, Murakami A, Noonan K, Lambeth C, Kar K, Anderson JF, de Silva AM, Diamond MS, Koski RA, Marasco WA, Fikrig E. Protective and therapeutic capacity of human single-chain Fv-Fc fusion proteins against West Nile virus. J Virol. 2005;79:14606–14613. doi: 10.1128/JVI.79.23.14606-14613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grard G, Biek R, Tamfum J-JM, Fair J, Wolfe N, Formenty P, Paweska J, Leroy E. Emergence of divergent Zaire Ebola virus strains in Democratic Republic of the Congo in 2007 and 2008. J Infect Dis. 2011;204(Suppl):S776–84. doi: 10.1093/infdis/jir364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groseth A, Marzi A, Hoenen T, Herwig A, Gardner D, Becker S, Ebihara H, Feldmann H. The Ebola virus glycoprotein contributes to but is not sufficient for virulence in vivo. PLoS Pathog. 2012;8:e1002847. doi: 10.1371/journal.ppat.1002847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcγ receptors in dendritic cells and macrophages. Nat Rev Immunol. 2014;14:94–108. doi: 10.1038/nri3582. [DOI] [PubMed] [Google Scholar]
- 43.Gutfraind A, Meyers LA. Evaluating large-scale blood transfusion therapy for the current Ebola epidemic in Liberia. J Infect Dis. 2015:jiv042. doi: 10.1093/infdis/jiv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hampton T. Largest-ever outbreak of Ebola virus disease thrusts experimental therapies, vaccines into spotlight. JAMA. 2014;312:987–989. doi: 10.1001/jama.2014.11170. [DOI] [PubMed] [Google Scholar]
- 45.Hart MK, Wilson J, inventors. The United States of America as represented by the Secretary of the Army, assignee. Monoclonal antibodies and complementarity-determining regions binding to Ebola glycoprotein. 6,875,433. United States patent US. 2005 Apr 5;5
- 46.Highlander SL, Cai WH, Person S, Levine M, Glorioso JC. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J Virol. 1988;62:1881–1888. doi: 10.1128/jvi.62.6.1881-1888.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hood CL, Abraham J, Boyington JC, Leung K, Kwong PD, Nabel GJ. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J Virol. 2010;84:2972–2982. doi: 10.1128/JVI.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hood EE, Woodard SL, Horn ME. Monoclonal antibody manufacturing in transgenic plants--myths and realities. Curr Opin Biotechnol. 2002;13:630–635. doi: 10.1016/s0958-1669(02)00351-8. [DOI] [PubMed] [Google Scholar]
- 49.Iversen PL, Warren TK, Wells JB, Garza NL, Mourich DV, Welch LS, Panchal RG, Bavari S. Discovery and early development of AVI-7537 and AVI-7288 for the treatment of Ebola virus and Marburg virus infections. Viruses. 2012;4:2806–2830. doi: 10.3390/v4112806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones S, Qui X, Feldmann H, Stroeher U, inventors; Jones S, Qiu X, Feldmann H, Stroeher U, assignees. Monoclonal antibodies for Ebola and Marburg viruses. 8,513,391. United States patent US. 2013 Ago;
- 51.Kamata T, Natesan M, Warfield K, Aman MJ, Ulrich RG. Determination of specific antibody responses to the six species of Ebola and Marburg viruses by multiplexed protein microarrays. Clin Vaccine Immunol. 2014;21:1605–1612. doi: 10.1128/CVI.00484-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, Sandersfeld LM, Quinn K, Weller M, McCray PB, Jr, Chiorini J, Maury W. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc Natl Acad Sci USA. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kreil TR. Treatment of Ebola Virus Infection with Antibodies from Reconvalescent Donors. Emerg Infect Dis. 2015;21:521–523. doi: 10.3201/eid2103.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuehn BM. As Ebola Epidemic Begins to Slow, Trials of Drugs and Vaccines Speed Up. JAMA. 2015;313:1000–1002. doi: 10.1001/jama.2015.0942. [DOI] [PubMed] [Google Scholar]
- 55.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee JE, Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lennemann NJ, Rhein BA, Ndungo E, Chandran K, Qiu X, Maury W. Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. mBio. 2014;5:e00862–13. doi: 10.1128/mBio.00862-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez JP, Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–6. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 59.Leroy EM, Labouba I, Maganga GD, Berthet N. Ebola in West Africa: the outbreak able to change many things. Clin Microbiol Infect. 2014;20:O597–9. doi: 10.1111/1469-0691.12781. [DOI] [PubMed] [Google Scholar]
- 60.Leroy EM, Rouquet P, Formenty P, Souquière S, Kilbourne A, Froment JM, Bermejo M, Smit S, Karesh W, Swanepoel R, Zaki SR, Rollin PE. Multiple Ebola virus transmission events and rapid decline of central African wildlife. Science. 2004;303:387–90. doi: 10.1126/science.1092528. [DOI] [PubMed] [Google Scholar]
- 61.Li F, Vijayasankaran N, Shen AY, Kiss R, Amanullah A. Cell culture processes in monoclonal antibody production. mAbs. 2010;2:466–479. doi: 10.4161/mabs.2.5.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lülf S, Matz J, Rouyez MC, Järviluoma A, Saksela K, Benichou S, Geyer M. Structural basis for the inhibition of HIV-1 Nef by a high-affinity binding single-domain antibody. Retrovirology. 2014;11:24. doi: 10.1186/1742-4690-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lyon GM, Mehta AK, Varkey JB, Brantly K, Plyler L, McElroy AK, Kraft CS, Towner JS, Spiropoulou C, Ströher U, Uyeki TM, Ribner BS. Clinical care of two patients with Ebola virus disease in the United States. N Engl J Med. 2014;371:2402–2409. doi: 10.1056/NEJMoa1409838. [DOI] [PubMed] [Google Scholar]
- 64.Ma JKC, Drake PMW, Christou P. The production of recombinant pharmaceutical proteins in plants. Nat Rev Genet. 2003;4:794–805. doi: 10.1038/nrg1177. [DOI] [PubMed] [Google Scholar]
- 65.Malashkevich VN, Schneider BJ, McNally ML, Milhollen MA, Pang JX, Kim PS. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marceau CD, Negi SS, Hernandez H, Callison J, Marzi A, Borisevich V, Braun W, Berry J, Feldmann H, Rockx B. Novel neutralizing monoclonal antibodies protect rodents against lethal filovirus challenges. Trials Vaccinol. 2014;3:89–94. [Google Scholar]
- 67.Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg Viruses. J Pathol. 2015;235:153–174. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- 68.Marzi A, Feldmann H. Ebola virus vaccines: an overview of current approaches. Expert Rev Vaccines. 2014;13:521–531. doi: 10.1586/14760584.2014.885841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marzi A, Möller P, Hanna SL, Harrer T, Eisemann J, Steinkasserer A, Becker S, Baribaud F, Pöhlmann S. Analysis of the interaction of Ebola virus glycoprotein with DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin) and its homologue DC-SIGNR. J Infect Dis. 2007;196(Suppl 2):S237–46. doi: 10.1086/520607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsuno K, Nakayama E, Noyori O, Marzi A, Ebihara H, Irimura T, Feldmann H, Takada A. C-type lectins do not act as functional receptors for filovirus entry into cells. Biochem Biophys Res Commun. 2010;403:144–148. doi: 10.1016/j.bbrc.2010.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mehedi M, Falzarano D, Seebach J, Hu X, Carpenter MS, Schnittler HJ, Feldmann H. A new Ebola virus nonstructural glycoprotein expressed through RNA editing. J Virol. 2011;85:5406–5414. doi: 10.1128/JVI.02190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Misasi J, Chandran K, Yang J-Y, Considine B, Filone CM, Côté M, Sullivan N, Fabozzi G, Hensley L, Cunningham J. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J Virol. 2012;86:3284–3292. doi: 10.1128/JVI.06346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Misasi J, Sullivan NJ. Camouflage and Misdirection: The full-on assault of Ebola virus disease. Cell. 2014;159:477–486. doi: 10.1016/j.cell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohan GS, Li W, Ye L, Compans RW, Yang C. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 2012;8:e1003065. doi: 10.1371/journal.ppat.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murin CD, Fusco ML, Bornholdt ZA, Qiu X, Olinger GG, Zeitlin L, Kobinger GP, Ward AB, Saphire EO. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. P Natl Acad Sci USA. 2014;111:17182–17187. doi: 10.1073/pnas.1414164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakayama E, Saijo M. Animal models for Ebola and Marburg virus infections. Front microbiol. 2013;4:267. doi: 10.3389/fmicb.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oestereich L, Lüdtke A, Wurr S, Rieger T, Muñoz-Fontela C, Günther S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res. 2014;105:17–21. doi: 10.1016/j.antiviral.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 79.Olal D, Kuehne AI, Bale S, Halfmann P, Hashiguchi T, Fusco ML, Lee JE, King LB, Kawaoka Y, Dye JM, Saphire EO. Structure of an antibody in complex with its mucin domain linear epitope that is protective against Ebola virus. J Virol. 2012;86:2809–2816. doi: 10.1128/JVI.05549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Olinger GG, Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, Pauly M, Whaley KJ, Lear CM, Biggins JE, Scully C, Hensley L, Zeitlin L. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci USA. 2012;109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oswald WB, Geisbert TW, Davis KJ, Geisbert JB, Sullivan NJ, Jahrling PB, Parren PWHI, Burton DR. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 2007;3:e9. doi: 10.1371/journal.ppat.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pandey A, Atkins KE, Medlock J, Wenzel N, Townsend JP, Childs JE, Nyenswah TG, Ndeffo-Mbah ML, Galvani AP. Strategies for containing Ebola in West Africa. Science. 2014;346:991–995. doi: 10.1126/science.1260612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parren PWHI, Geisbert TW, Maruyama T, Jahrling PB, Burton DR. Pre- and postexposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J Virol. 2002;76:6408–6412. doi: 10.1128/JVI.76.12.6408-6412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pettitt J, Zeitlin L, Kim DH, Working C, Johnson JC, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, Pauly MH, Whaley KJ, Ingram MF, Zovanyi A, Heinrich M, Piper A, Zelko J, Olinger GG. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci Transl Med. 2013;5:199ra113. doi: 10.1126/scitranslmed.3006608. [DOI] [PubMed] [Google Scholar]
- 86.Ponomarenko J, Vaughan K, Sette A, Maurer-Stroh S. Conservancy of mAb epitopes in Ebolavirus glycoproteins of previous and 2014 outbreaks. PLOS Curr. 2014;3:6. doi: 10.1371/currents.outbreaks.f1a7028a13ce1c5f0bdbb4b0cc0b919b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qiu X, Alimonti JB, Melito PL, Fernando L, Ströher U, Jones SM. Characterization of Zaire Ebolavirus glycoprotein-specific monoclonal antibodies. Clin Immunol. 2011;141:218–227. doi: 10.1016/j.clim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 88.Qiu X, Kobinger GP. Antibody therapy for Ebola: is the tide turning around? Hum Vaccines Immunother. 2014;10:964–967. doi: 10.4161/hv.27813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qiu X, Wong G, Fernando L, Audet J, Bello A, Strong J, Alimonti JB, Kobinger GP. mAbs and Ad-vectored IFN-α therapy rescue Ebola-infected nonhuman primates when administered after the detection of viremia and symptoms. Sci Transl Med. 2013;5:207ra143. doi: 10.1126/scitranslmed.3006605. [DOI] [PubMed] [Google Scholar]
- 91.Ramanan P, Shabman RS, Brown CS, Amarasinghe GK, Basler CF, Leung DW. Filoviral immune evasion mechanisms. Viruses. 2011;3:1634–1649. doi: 10.3390/v3091634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reynard O, Borowiak M, Volchkova VA, Delpeut S, Mateo M, Volchkov VE. Ebolavirus glycoprotein GP masks both its own epitopes and the presence of cellular surface proteins. J Virol. 2009;83:9596–9601. doi: 10.1128/JVI.00784-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Richards P, Amara J, Ferme MC, Kamara P, Mokuwa E, Sheriff AI, Suluku R, Voors M. Social pathways for Ebola virus disease in rural Sierra Leone, and some implications for containment. PLoS Neglected Trop Dis. 2014 doi: 10.1371/journal.pntd.0003567. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rodríguez-Martínez LM, Marquez-Ipiña AR, López-Pacheco F, Pérez-Chavarría R, González-Vázquez JC, González-González E, Trujillo-de Santiago G, Ponce-Ponce de León CA, Zhang YS, Dokmeci MR, Khademhosseini A, Alvarez MM. Antibody derived peptides for detection of Ebola virus glycoprotein. PLoS One. 2015 doi: 10.1371/journal.pone.0135859. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sabalza M, Christou P, Capell T. Recombinant plant-derived pharmaceutical proteins: current technical and economic bottlenecks. Biotechnol Lett. 2014;36:2367–2379. doi: 10.1007/s10529-014-1621-3. [DOI] [PubMed] [Google Scholar]
- 96.Saéz AM, Weiss S, Nowak K, Lapeyre V, Zimmermann F, Düx A, Kühl HS, Kaba M, Regnaut S, Merkel K, Sachse A, Thiesen U, Villányi L, Boesch C, Dabrowski PW, Radonić A, Nitsche A, Leendertz SA, Petterson S, Becker S, Krähling V, Couacy-Hymann E, Akoua-Koffi C, Weber N, Schaade L, Fahr J, Borchert M, Gogarten JF, Calvignac-Spencer S, Leendertz FH. Investigating the zoonotic origin of the West African Ebola epidemic. EMBO Mol Med. 2014;7:17–23. doi: 10.15252/emmm.201404792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sakurai Y, Kolokoltsov AA, Chen CC, Tidwell MW, Bauta WE, Klugbauer N, Grimm C, Wahl-Schott C, Biel M, Davey RA. Two-pore channels control Ebola virus host cell entry and are drug targets for disease treatment. Science. 2015;347:995–998. doi: 10.1126/science.1258758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanchez A, Kiley MP, Holloway BP, Auperin DD. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993;29:215–40. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 99.Shedlock DJ, Bailey MA, Popernack PM, Cunningham JM, Burton DR, Sullivan NJ. Antibody-mediated neutralization of Ebola virus can occur by two distinct mechanisms. Virology. 2010;401:228–235. doi: 10.1016/j.virol.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, Burke E, Buchmeier MJ, Soilleux EJ, Riley JL, Doms RW, Bates P, Pöhlmann S. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology. 2003;305:115–123. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- 101.Smither SJ, Eastaugh LS, Steward JA, Nelson M, Lenk RP, Lever MS. Post-exposure efficacy of oral T-705 (Favipiravir) against inhalational Ebola virus infection in a mouse model. Antiviral Res. 2014;104:153–155. doi: 10.1016/j.antiviral.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 102.Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, Moore MJ, Tallarico AS, Olurinde M, Choe H, Anderson LJ, Bellini WJ, Farzan M, Marasco WA. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci USA. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, Moore MJ, Tallarico ASC, Olurinde M, Choe H, Anderson LJ, Bellini WJ, Farzan M, Marasco WA. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci USA. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takada A, Ebihara H, Feldmann H, Geisbert TW, Kawaoka Y. Epitopes required for antibody-dependent enhancement of Ebola virus infection. J Infect Dis. 2007;196:S347–56. doi: 10.1086/520581. [DOI] [PubMed] [Google Scholar]
- 105.Takada A, Feldmann H, Stroeher U, Bray M, Watanabe S, Ito H, McGregor M, Kawaoka Y. Identification of protective epitopes on Ebola virus glycoprotein at the single amino acid level by using recombinant vesicular stomatitis viruses. J Virol. 2003;77:1069–1074. doi: 10.1128/JVI.77.2.1069-1074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ter Meulen J, Van Den Brink EN, Poon LL, Marissen WE, Leung CS, Cox F, Cheung CY, Bakker AQ, Bogaards JA, van Deventer E, Preiser W, Doerr HW, Chow VT, de Kruif J, Peiris JS, Goudsmit J. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thi EP, Mire CE, Ursic-Bedoya R, Geisbert JB, Lee ACH, Agans KN, Robbins M, Deer DJ, Fenton KA, MacLachlan I, Geisbert TW. Marburg virus infection in nonhuman primates: Therapeutic treatment by lipid-encapsulated siRNA. Sci Transl Med. 2014;6:250ra116. doi: 10.1126/scitranslmed.3009706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tran EEH, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S. Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. J Virol. 2014;88:10958–10962. doi: 10.1128/JVI.00870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vanderlinden E, Naesens L. Emerging antiviral strategies to interfere with influenza virus entry. Med Res Rev. 2014;34:301–339. doi: 10.1002/med.21289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Warfield KL, Swenson DL, Olinger GG, Nichols DK, Pratt WD, Blouch R, Stein DA, Aman MJ, Iversen PL, Bavari S. Gene-specific countermeasures against Ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS Pathog. 2006;2:e1. doi: 10.1371/journal.ppat.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Warren TK, Wells J, Panchal RG, Stuthman KS, Garza NL, Van Tongeren SA, Dong L, Retterer CJ, Eaton BP, Pegoraro G, Honnold S, Bantia S, Kotian P, Chen X, Taubenheim BR, Welch LS, Minning DM, Babu YS, Sheridan WP, Bavari S. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Warren TK, Whitehouse CA, Wells J, Welch L, Heald AE, Charleston JS, Sazani P, Reid SP, Iversen PL, Bavari S. A single phosphorodiamidate morpholino oligomer targeting VP24 protects rhesus monkeys against lethal Ebola virus infection. mBio. 2015;6:e02344–14. doi: 10.1128/mBio.02344-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.White JM, Shornberg KL. A new player in the puzzle of filovirus entry. Nat Rev. 2012;10:317–322. doi: 10.1038/nrmicro2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.WHO. Report of an International Convention. Bull World Health Organ 1978. 1978;56:271–293. [Google Scholar]
- 116.WHO; Media Centre. Fact sheet No. 33. 2014 http://www.who.int/mediacentre/en/. downloaded in October 31, 2014.
- 117.WHO. Regional Office for Africa. Situation reports (from September 2014 to March 2014) Epidemic and pandemic alert and response (EPR) 2014 http://www.afro.who.int/en/clusters-a-programmes/dpc/epidemic-a-pandemic-alert-and-response/sitreps.html. Retrieved March. 12, 2014.
- 118.Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 119.Wisskirchen K, Lucifora J, Michler T, Protzer U. New pharmacological strategies to fight enveloped viruses. Trends Pharmacol Sci. 2014;35:470–478. doi: 10.1016/j.tips.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wong G, Richardson JS, Pillet S, Patel A, Qiu X, Alimonti J, Hogan J, Zhang Y, Takada A, Feldmann H, Kobinger GP. Immune parameters correlate with protection against Ebola virus infection in rodents and nonhuman primates. Sci Transl Med. 2012;4:158ra146. doi: 10.1126/scitranslmed.3004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zeitlin L, Pettitt J, Scully C, Bohorova N, Kim D, Pauly M, Hiatt A, Ngo L, Steinkellner H, Whaley KJ, Olinger GG. Enhanced potency of a fucose-free monoclonal antibody being developed as an Ebola virus immunoprotectant. Proc Natl Acad Sci USA. 2011;108:20690–20694. doi: 10.1073/pnas.1108360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhong Y, Xu J, Li T, Yu X, Sheng M. Potential clinical treatment for Ebola pandemic. Sci China Life Sci. 2014;57:982–984. doi: 10.1007/s11427-014-4756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]