Abstract
Anti-influenza drugs play major roles in the management of severe influenza infections. Neuraminidase inhibitors (NAIs), which are active against all influenza A subtypes and the 2 major influenza B lineages, constitute the only class of antivirals recommended for the control of influenza epidemics and eventual pandemics. Thus, the emergence of NAI resistance could be a major clinical concern. Although most currently circulating influenza A and B strains are susceptible to NAIs, clinical cases of influenza viruses harboring single or multiple NA substitutions or deletions conferring a cross-resistance phenotype to the 2 main NAIs (oseltamivir and zanamivir) have been reported, mostly in immunocompromised individuals. Moreover, such events seem to be more frequent in A(H1N1)pdm09 viruses containing the H274Y substitution together with other NA changes (I222R, E119D/G). This review summarizes the therapeutic regimens leading to the emergence of NAI cross-resistant influenza A and B viruses as well as the virologic properties of such variants.
Keywords: influenza, neuraminidase, oseltamivir, resistance, zanamivir
Seasonal influenza epidemics, currently involving influenza A(H3N2), A(H1N1) and B strains, affect 10% to 20% of the human population each year [1]. Although most people infected with a seasonal influenza strain recover in less than 2 weeks, young children, adults ≥65 years old, and subjects with chronic medical conditions are at higher risk of developing serious influenza complications requiring hospitalization and potentially leading to death [2].
Influenza infections are usually more devastating in immunocompromised patients who tend to develop longer influenza illness with prolonged viral shedding [3]. For instance, seasonal influenza infections have been associated with increased mortality, higher rate of viral pneumonia, and greater risk of developing bacterial/fungal superinfections in stem cell transplant recipients [4], as well as solid organ transplant patients [3]. Consequently, the immunocompromised population could significantly benefit from timely and effective antiviral interventions.
Neuraminidase inhibitors (NAIs), which are active against all influenza A subtypes and the 2 major influenza B lineages, constitute the main class of antivirals for the control of influenza epidemics and pandemics. Four different NAIs have been approved in various countries. The 2 older and most widely prescribed agents are oseltamivir and zanamivir, with the remaining 2 (peramivir and laninamivir) approved in a few countries [5]. As for other antivirals, emergence of NAI-resistant infections remains a serious clinical concern.
The prevalence of NAI resistance was low before the emergence and worldwide dissemination of the influenza A/Brisbane/59/2007-like (H1N1)-H274Y (N2 numbering here and throughout the text) NA variant during the 2008–2009 season [6, 7]. Resistance to oseltamivir became subsequently infrequent (approximately 1%) in A(H1N1)pdm09 viruses and mainly involved the H274Y NA substitution. The prevalence of resistance to zanamivir has consistently remained low, and this was attributed to its infrequent use and its unique structure that is closer to the natural sialic acid substrate [8]. In addition, inhaled zanamivir results in higher drug levels (up to 5000-fold > mean IC50) at the site of viral replication (epithelial cells of the respiratory tract) [8]. Of note, zanamivir has been considered as a suitable alternative for oseltamivir-resistant A(H1N1) viruses harboring the H274Y substitution. Nevertheless, since 2009, there seems to be increased reports of oseltamivir/zanamivir-resistant A(H1N1)pdm09 isolates.
In this article, we review clinical cases involving influenza variants that exhibited concurrent reduced susceptibility to the 2 main NAIs: oseltamivir and zanamivir. Also of note, we summarize drug treatments that led to the emergence of those multidrug-resistant viruses and the related clinical outcome. In addition, we describe the in vitro and in vivo properties of such NA variants and discuss potential management options.
Phenotypic susceptibility to NAIs is categorized according to the World Health Organization NA inhibition assay-based phenotypes as follows: normal inhibition or susceptibility (S) (≤10-fold increase in IC50 for influenza A; ≤5-fold increase for influenza B); reduced inhibition (RI) (between 10- and 100-fold increase for influenza A; between 5- and 50-fold increase for influenza B); and highly reduced inhibition (HRI) (>100-fold increase for influenza A; >50-fold increase for influenza B). Obviously, whether some or all NA mutations described in this review could result in clinically relevant resistance phenotype remains questionable because there is no existing laboratory criteria for unequivocally defining such association. Nevertheless, as for the H274Y substitution and its clearly established resistance to oseltamivir, some mutations associated with an HRI phenotype are likely to be of clinical importance.
STRUCTURAL PROPERTIES AND MECHANISMS OF ACTION OF NAIS
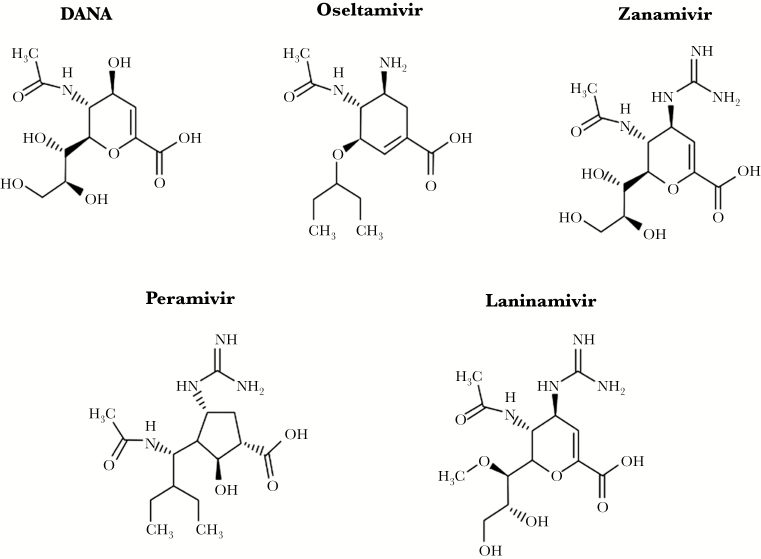
Neuraminidase inhibitors are based on the structure of 2,3- dehydro-2-deoxy-N-acetylneuraminic acid (DANA) (Figure 1) [9]. Oseltamivir is an ethyl ester prodrug that requires ester hydrolysis to be converted to the active form (oseltamivir carboxylate). A bulky lipophilic side chain is included in the compound to improve bioavailability [10]. Zanamivir is a 4-deoxy-4- guanidino analogue of DANA administered by inhalation. Peramivir is a cyclopentane with a negatively charged carboxylate group, a positively charged guanidino group, and a lipophilic side chain [11]. Laninamivir is a long-lasting NAI containing a 4-guanidino group in addition to a 7-methoxy group [12]. Neuraminidase inhibitors target the highly conserved active site of the NA enzyme, which is constituted of 8 functional (R-118, D-151, R-152, R-224, E-276, R-292, R-371, and Y-406) and 11 framework (E-119, R-156, W-178, S-179, D-198, I-222, E-227, H-274, E-277, N-294, and E-425) residues [13].
Figure 1.
Chemical structure of neuraminidase inhibitors (NAIs). All of these NAIs are based on the structure of the 2,3-didehydro analogue of the N-acetyl-neuraminic acid (DANA). The bioavailable prodrug of oseltamivir is an ethyl ester that is converted into the active carboxylate by hepatic esterases. Zanamivir is a 4-deoxy-4-guanidino analogue of DANA. Peramivir is a cyclopentane derivative with a guanidinyl group and a lipophilic chain. Laninamivir is the active product of the esterified octanoate CS-8958. These molecules interact differently within the enzyme active site, which may influence their antiviral activity.
Important structural differences between NAIs may influence the development of resistance. Clinical studies showed that NA substitutions associated with drug resistance involve framework or catalytic residues that are generally subtype- and drug-specific [14]. The most commonly reported mutations that have been selected by oseltamivir therapy are E119V and R292K in H3N2 viruses and H274Y in H1N1 viruses [5].
Clinical Cases of Influenza Variants Exhibiting Reduced Susceptibility to Both Oseltamivir and Zanamivir
A total of 14 cases of influenza A and B infections involving viruses with RI/HRI phenotypes to both oseltamivir and zanamivir have been published as of February 2017 (Table 1). The patients (7 males and 7 females) were aged from 8 months to 88 years. Most cases (12 of 14, 86%) were infected with influenza A viruses, which included 7 A(H1N1)pdm09, 4 A(H3N2), and 1 A(H7N9) strains, whereas the remaining 2 cases (14%) were caused by influenza B viruses.
Table 1.
Summary of Neuraminidase Substitutions Identified in Clinical Influenza A and B Viruses Exhibiting RI/HRI Phenotypes to Both Oseltamivir and Zanamivir
| Gender/Age (Case No.) |
Year of Isolation | Viral Subtype City/Country | NA Mutation(s)a | NA Inhibitor Treatmentb | Immnunological Status/ Underlying Conditions | Clinical Outcome | Ref. |
|---|---|---|---|---|---|---|---|
| M/63 y (1) |
2014 | A(H1N1)pdm09 Geneva, Switzerland |
E119D/ H274Y | OSV (p.o. 150 mg twice daily) for 6 days and ZMV (IV, 600 mg twice daily) for 8 days. (The first mutation [H274Y] appeared after 1 day of OSV therapy.) | Immunocompromised: relapsing acute myeloid leukemia; 7 days post-SCT | Death, 61 days after the onset of illness | [16] |
| M/8 mo (2) |
2014 | A(H1N1)pdm09 District of Columbia, USA |
E119G/ H274Y | OSV for 6 days and ZMV IV for 47 days. (The first mutation [H274Y] appeared after 5 days of OSV therapy.) | Immunocompromised: familial hemophagocytic lymphohistiocytosis, 32 days before CBT | Death, 79 days after the onset of illness | [39] |
| F/55 y (3) |
2010 | A(H1N1)pdm09 Düsseldorf, Germany |
I222R/ H274Y | OSV (p.o. 75 mg once daily) for 14 days and ZMV (inhaled 10 mg twice daily) for 40 days. (The H274Y mutation was present before OSV therapy.) | Immunocompromised: myelodysplastic syndrome; GVHD | Recovery, respiratory samples became negative 154 days after the onset of illness | [40] |
| M/24 y (4) |
2009 | A(H1N1)pdm09 Paris, France |
I222R/ H274Y | OSV (p.o. 75 mg once daily for 6 days). (The 2 mutations appeared after 6 days of OSV therapy.) | Immunocompromised: acute myelogeneous leukemia; CBT (10 months before the onset of illness) | Death, 140 days after the onset of illness | [23] |
| F/14 y (5) |
2009 | A(H1N1)pdm09 Pensylvania, USA |
I222R/ H274Y | OSV (p.o. 60 mg twice daily for 4 days and 150 mg twice daily for 8 days). (The first mutation [H274Y] appeared after 11 days of OSV therapy.) | Immunocompromised: systemic lupus erythematosus, systemic vasculitis, and chronc pancreatis | Death, 74 days after the onset of illness | [41] |
| M/5 y (6) |
2009 | A(H1N1)pdm09 The Netherlands |
I222R | ZMV (IV 20 mg/kg twice daily for 20 day) | Immunocompromised: high- risk acute lymphoblastic leukemia (undergoing preparation for SCT) | Death, 118 days after the onset of illness | [42] |
| F/15 y (7) |
2009 | A(H1N1)pdm09 Toronto, Canada |
I222R | Untreated | Immunocompetent: history of asthma; admitted for respiratory symptoms | Uneventful recovery | [43] |
| F/39 y (8) |
2012 | A(H3N2) Toronto, Canada |
E119V/ Q136K | OSV (p.o. 75 mg twice daily for 10 days and 150 mg twice daily for 20 days) and ZMV (inhalation, 10 mg twice daily for 8 days and IV, 600 mg twice daily for 15 days). (The first mutation [E119V] appeared after 23 days of OSV and 8 days of ZMV therapies). NOTE: The del245-248 mutation was transiently detected |
Immunocompromised: Philadelphia chromosome-positive acute B cell lymphoblastic leukemia; GVHD | Death, 7 months after the onset of illness | [17] |
| F/3 y (9) |
2005 | A(H3N2) Montréal, Canada |
Del245-248 N146K/ S219T/ A272V | ZMV (inhalation, 10–20 mg twice daily for 72 days; 107 days after cessation of a 3-month course of OSV therapy) | Immunocompromised: SCID | After 5 months of ZMV therapy, viral culture became negative for influenza | [44] |
| M/43 y (10) |
2008 | A(H3N2) Bethesda, USA |
Del 245-248 | OSV (p.o. 75 mg twice daily for 5 days) | Immunocompromised: mantle cell lymphoma; HSCT 6 months before diagnosis of influenza | After 12 days of OSV therapy, rapid test and shell vial culture became negative for influenza | [18] |
| M/2.5 y (11) |
2007 | A(H3N2) Texas, USA |
E119I | OSV (p.o. 45 mg twice daily for 20 days) | Immunocompromised: X-linked lymphopro- liferative disorder | Death, 172 days after the onset of illness | [26] |
| M/88 y (12) |
2013 | A(H7N9) Shanghai, China |
R292K | OSV (p.o. 150 mg twice daily for 5 days) and PMV (IV, 600 mg once daily for 1 day) | Immuncompetent, COPD, hypertension and diabetes as underlying diseases | Death, 19 days after the onset of illness | [45] |
| F/2 y (13) |
2001 | B Rochester, USA |
D198N | OSV (p.o. 10 mg twice daily for 4 days [prophylactically], 30 mg twice daily for 14 days, and 20 mg twice daily for 9 days) | Immunocompromised: myelomonocytic leukemia, CBT before the onset of illness | Death, 43 days after the onset of illness | [46] |
| F/18 mo (14) |
1996 | B Memphis, USA |
R152K | ZMV (inhalation, 16–32 mg every 6 h for 14 days) | Immunocompromised: juvenile chronic myelocytic leukemia | Death, 17 days after the onset of illness | [20] |
Abbreviations: CBT, cord blood transplantation; COPD, chronic obstructive pulmonary disease; F, female; GVHD, graft-versus-host disease; HRI, highly reduced inhibition; HSCT, hematopoietic stem cell translplantation; IV, intravenous; M, male; mo, month; NA, neuraminidase; OSV, oseltamivir; PMV, peramivir; RI, reduced inhibition; Ref., reference; SCID, severe combined immunodeficiency disease; SCT, stem cell transplant; y, year; ZMV, zanamivir.
aN2 numbering is used for NA mutations.
bTreatment received before detection of the cross-resistant.
Except for 2 patients who were immunocompetent (case 7, with an history of asthma, and case 12, who had chronic obstructive pulmonary disease, hypertension, and diabetes), all remaining subjects were immunocompromised due to various types of underlying conditions, most notably acute leukemia (8 cases). Fatal outcomes were recorded for 10 cases (71%).
All patients had received NAI treatment before emergence of the resistance mutation except the Canadian A(H1N1)pdm09-I222R patient (case 7). Among the remaining 13 patients, 5 received oral oseltamivir alone; 2 received oseltamivir that was subsequently replaced by inhaled or intravenous (IV) zanamivir, and 3 were simultaneously treated with oseltamivir and zanamivir. Two patients received zanamivir only (cases 6 and 14). A single patient (case 12) received oseltamivir for 5 days in addition to 1 day of peramivir treatment before emergence of the A(H7N9)-R292K substitution.
The mean number of days of NAI treatment before detection of the first mutation among the 13 treated patients was 12.76 (range, 0–72). Patient 3 was infected with the H274Y mutant before starting oseltamivir therapy, which was followed by zanamivir treatment inducing the I222R mutation. In many cases, the isolate with reduced susceptibility to both drugs was detected after only a few days of NAI therapy as shown in cases 1 (6 days of oseltamivir and 8 days of zanamivir), 4 (6 days of oseltamivir), 10 (5 days of oseltamivir), and 12 (5 days of oseltamivir and 1 day of peramivir) (Table 1).
A variety of NA genetic changes consisting of single or double substitutions as well as a 4-amino acid deletion were identified in influenza variants exhibiting RI/HRI to oseltamivir and zanamivir. The 6 variants containing 2 NA substitutions [5 A(H1N1)pdm09 and 1 A(H3N2)] had a subtype-specific hallmark oseltamivir-resistant substitution [H274Y in A(H1N1) and E119V in A(H3N2) viruses] in addition to another substitution [E119D/G or I222R for A(H1N1)pdm09 and Q136K for A(H3N2)]. Single mutants consisted of A(H1N1)pdm09-I222R (2 cases), A(H3N2)-E119I, A(H7N9)-R292K, as well as B-D198N and B-R152K variants. Two A(H3N2) variants had a 4-amino acid NA deletion (del245-248), either alone (case 10) or combined with other NA substitutions with no impact on NAI susceptibility (case 9). The same deletion was also transiently present in samples from case 8. Besides these major NA changes associated with a cross-resistance phenotype to oseltamivir and zanamivir, several additional mutations were detected as minor populations in most cases. This highlights the potential for strong quasi-species events targeting the NA gene during NAI therapy. For instance, E119G/A substitutions were detected at low proportions in case 1. In case 4, an H274Y variant was detected before oseltamivir therapy. The same substitution was detected after 12 days of oseltamivir treatment in a variant from case 6. R292K, R292K/E119V, and E119V/Q136K/del245-248 variants were transiently detected in case 8.
Mecanisms of Resistance to Oseltamivir and Zanamivir Mediated by Neuraminidase Mutations Identified in Clinical Influenza A and B Variants
Influenza viruses with RI/HRI phenotypes to NAIs contained substitutions in the NA protein that directly or indirectly alter the shape of the NA catalytic site, thus reducing inhibitor binding.
A(H1N1) Viruses
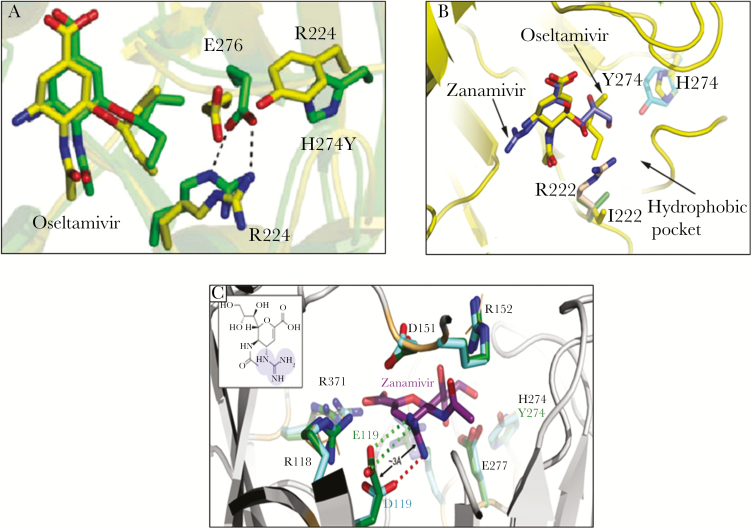
Because oseltamivir has a large hydrophobic side chain, the amino acid E276 must rotate and bond with R224 to create a pocket necessary for accommodating the drug. The H274Y substitution (present in cases 1–5) inhibits the rotation of the E276 residue resulting in HRI phenotype against oseltamivir (and peramivir) but not zanamivir (Figure 2A).
Figure 2.
Modeling of A(H1N1) neuraminidase (NA) substitutions. (A) Superimposition of the molecular dynamic structures of the oseltamivir carboxylate bound to the wild-type (green) and H274Y mutant (yellow) influenza N1 NA. The amino acid E276 of the NA enzyme must rotate and bond with R224 to accomodate oseltamivir binding. The H274Y mutation alters such rearrangement. (B) Schematic representation of the active site of the NA with oseltamivir (yellow) and zanamivir (blue). It could be inferred that the I222R mutation is likely to cause its effect through steric clashes. The extended side chain of arginine would come very close to the hydrophobic moiety of oseltamivir, thus disrupting its binding. For zanamivir, however, the corresponding glycerol moiety sits slightly higher up in the binding site and thus will be less disrupted [15]. (C) Molecular modeling for the A(H1N1)pdm09 E119D/H274Y variant. The positively charged guanidino group of zanamivir forms 2 salt bridges with E119 (green dotted lines) that stabilize zanamivir binding. The shorter side chain of 119D (cyan) allows only 1 salt bridge to this guanidino group (red dotted line). This reduced ability to form salt bridges with zanamivir likely destabilizes inhibitor binding [16]. Adapted from Pizzorno et al. Impact of mutations at residue I223 of the neuraminidase protein on the resistance profile, replication level, and virulence of the 2009 pandemic influenza virus. Antimicrob Agents Chemother. 2012;56:1208–14 and L’Huillier et al. E119D neuraminidase mutation conferring pan-resistance to neuraminidase inhibitors in an A(H1N1)pdm09 isolate from a stem-cell transplant recipient. J Infect Dis 2015;212:1726–34.
Molecular modeling suggested that the I222R framework substitution (cases 3–7) likely induces steric clashes. The extended side chain of an arginine would come very close to the hydrophobic moiety of oseltamivir or peramivir disrupting their binding to the NA enzyme (Figure 2B) [15]. By contrast, for zanamivir, the corresponding glycerol moiety sits slightly higher up in the binding site and thus is less impacted [15].
The E119 residue is involved in the interaction between the NA enzyme and either the guanidine group (present in zanamivir, laninamivir, and peramivir) or the C4 hydroxyl group (present in oseltamivir). Replacement of glutamic acid by amino acids with a shorter side chain such as aspartic acid (case 1) or glycine (case 2) could significantly alter these interactions (Figure 2C) [16].
A(H3N2) Viruses
The E119V substitution in the H3N2 background (case 8) induces resistance to oseltamivir without altering the susceptibility to zanamivir (Figure 3A–B). Therefore, the observed resistance to zanamivir in the double E119V/Q136K mutant is likely due to the Q136K change [17]. The Q136K substitution introduces a larger side chain at this position that is expected to affect the binding of NAIs due to conformational changes in the enzyme-inhibitor complex (Figure 3C–D). Structural modeling suggested the presence of a charge repulsion between the guanidino group of zanamivir and peramivir and lysine at position 136. In contrast, the Q136K substitution is unlikely to cause resistance to oseltamivir because this NAI lacks a functional group close to K136 [17].
Figure 3.

Modeling of A(H3N2) neuraminidase (NA) susbstitutions. Impact of the E119V NA substitution in A(H3N2) viruses. (A) E119V causes oseltamivir resistance by losing the hydrogen bond between side chain of residue 119 and oseltamivir caboxylate amino group. The wild-type (WT) NA-oseltamivir and E119V mutant-oseltamivir are shown as light green color and light blue sticks, respectively. The interactions between water molecules (displayed as spheres) and oseltamivir amino group are stronger in the WT than in the E119V mutant. (B) The guanidino group of zanamivir forms more interactions with environmental residues (E227, W178, and D151), which compensate the loss of interaction due to E119V. The WT-zanamivir and E119V-zanamivir are shown as orange and silver sticks, respectively. (C) Overlay of crystal structure of WT NA from H3N2 (PDB accession no. 4GZP) (blue-green) in complex with oseltamivir (cyan), zanamivir (yellow), and peramivir (pink). Dashes indicate electrostatic interactions. Arrows indicate the distance range between Q136 or A246 and the NAIs. (D) Overlay of NA from H3N2 (Protein Data Bank accession no. 4GZP) with NAIs, as described in the legend to C, with substitutions introduced and colored in brown. Δ245-248 is circled [17]. Adapted from Eshaghi et al. Multiple influenza A (H3N2) mutations conferring resistance to neuraminidase inhibitors in a bone marrow transplant recipient. Antimicrob Agents Chemother 2014;58:7188–9.
The impact of the 4-amino acid deletion (del245-248) (cases 8–10) on the resistance phenotype to oseltamivir and zanamivir seems to rely on residue A246, which, along with R224 and I222 residues, is directly involved in the formation of the hydrophobic pocket adjacent to the NA cavity (Figure 3C–D) [18]. The conformational changes that occur at the surface level due to the del245-248 mutation could disturb the hydrophobic acid pocket. As this pocket accomodates the glycerol side chain of sialic acid and NAI, it may potentially alter NAI-binding.
A(H7N9) Viruses
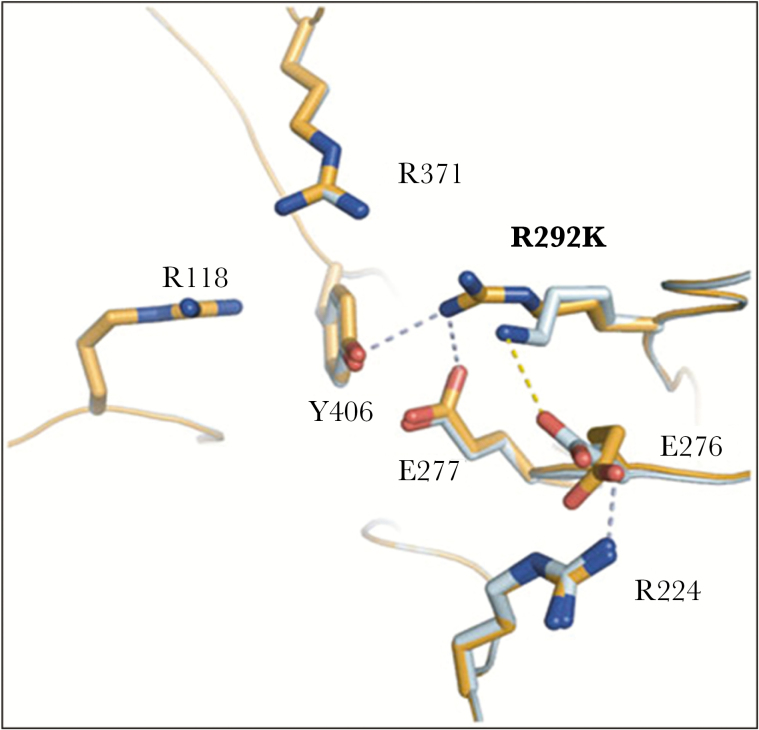
The R292K NA substitution reported in H7N9 variants resistant to oseltamivir, zanamivir, and peramivir (case 12) was shown to inhibit the rotation of E276, preventing the formation of a pocket required to accommodate drug binding (Figure 4) [19].
Figure 4.
Modeling of A(H7N9) neuraminidase (NA) substitution. Comparison of wild-type ([WT] R292) and mutant (K292) A(H7N9) NA uncomplexed active sites. In the WT NA (bright orange), R292 is able to make hydrogen bond with key catalytic residues, Y406 and E277. However, R292 is out of range to interact with E276, which is oriented toward R224. Such conformation is favorable for oseltamivir carboxylate binding. E276 forms a salt bridge with R224. In the mutant NA (pale cyan), K292 is out of range to form any typical hydrogen bonds with Y406 or E277. Instead, K292 is able to form a salt bridge with E276, which is oriented toward R224, a conformation unfavorable for oseltamivir carboxylate binding [19]. Adapted from Wu et al. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res 2013;23:1347–55.
B Viruses
Structural studies on influenza B NA showed that substitutions at the framework residue (D198) (case 13) may affect the interaction of the functional R152 residue with the N-acetyl group on the ligand, resulting in reduced susceptibility to all NAIs [20]. The arginine to lysine change at the functional residue (R152) (case 14) directly affects NA-NAI binding.
Impact of Neuraminidase Mutations Identified in Clinical Influenza A and B Variants With Reduced Susceptibility to Oseltamivir and Zanamivir on Enzyme Properties and Viral Fitness
(Table 2) summarizes the in vitro and in vivo characteristics of NAI-resistant clinical variants or their closely related recombinant viruses. Most A(H1N1)pdm09 variants with cross resistance to oseltamivir and zanamivir (cases 1–5) additionally showed RI/HRI to peramivir and laninamivir. Influenza A(H3N2) isolates from cases 8 and 11 also exhibited HRI to peramivir with no available laninamivir IC50 value. Of note, the del245-248 mutation identified in cases 8–10 had RI phenotype to oseltamivir but did not significantly alter zanamivir suceptibility in NA inhibition assays (only 4-fold change in IC50); however, this variant had a cross-resistance phenotype to oseltamivir and zanamivir in cell culture-based assays [18]. The H7N9-R292K variant (case 11) had HRI, RI, and HRI phenotypes to oseltamivir, zanamivir, and peramivir, respectively.
Table 2.
Impact of Neuraminidase Mutations Identified in Clinical Influenza A and B Variants With Reduced Susceptibility to Oseltamivir/Zanamivir on Drug Resistance Levels, NA Enzyme Properties, and Viral Fitness
| Subtype | NA Mutation(s)a | Fold Increase in IC50 Values vs WT/ Resistance Phenotypeb OSV ZMV PMV LMV |
NA Enzyme Properties vs WT |
Viral Fitness In Vitro vs WT |
Viral Fitness In Vivo vs WT |
Ref. |
|---|---|---|---|---|---|---|
| A(H1N1)pdm09 (RV) |
E119D/H274Y | 790/HRI 903/HRI 5958/HRI 366/ HRI | Reduced NA activity and surface NA expression | Reduced viral titers in MDCK cells; instability of E119D (D119E reversion) | Reduced infectivity in mice; instability of E119D (D119E reversion) | [16, 21] |
| A(H1N1)pdm09 (RV) |
E119D | 25/RI 827/HRI 286/ HRI 702/HRI | Reduced NA activity and surface NA expression | Reduced viral titers in MDCK cells | Reduced infectivity in mice; instability (D119E reversion) | [16, 21] |
| A(H1N1)pdm09 (isolate) |
E119G/H274Y | 2483/HRI 1546/HRI 93433/HRI 605/ HRI | ND | ND | ND | [39] |
| (RV) | E119G | 3/S 832/HRI 51/ RI ND | Reduced NA affinity but comparable velocity | Reduced plaque size and viral titers in MDCK cells | Reduced infectivity in mice; instability of the mutation (G119E reversion in ferrets) | [22] |
| A(H1N1)pdm09 (RV) |
I222R/H274Y | 16300/HRI 17/RI 8975/HRI 17/RI | Reduced NA affinity | Reduced viral titers in MDCK cells | ND | [23] |
| A(H1N1)pdm09 (isolate/RV) |
I222R | 45/RI 10/RI 7/S ND | Reduced NA affinity | Increased viral titers in MDCK cellls | Comparable infectivity and aerosol-transmissibility in ferrets | [23, 25, 42] |
| A(H3N2) (isolate) |
E119V/Q136K | 325/HRI 220/HRI 101/HRI ND | ND | ND | ND | [17] |
| A(H3N2) (isolate) |
Del 245-248 | 55/RI 4/S ND ND | Slight reduction (3-fold) in NA activity | Comparable viral titers in MDCK cells | Comparable fitness and contact transmissibility in ferrets | [18] |
| A(H3N2) (isolate) |
E119I | 977/HRI 11/RI 428/ HRI ND | ND | ND | ND | [26] |
| A(H7N9) (isolate) |
R292K | 4610/HRI 11/RI 563/ HRI ND | Reduced NA affinity and activity | Comparable replication in human tracheobronchial epithelial cells | Comparable virulence in mice and transmissibility in guinea pigs | [27, 28] |
| B (isolate/RV) |
D198N | 8/RI 11/RI 5/S ND | Reduced NA activity | ND | Comparable fitness in ferrets | [29, 46] |
| B (isolate/RV) |
R152K | 100/HRI 70/HRI 400/HRI ND | Reduced NA activity and stability | Reduced viral titers in MDCK cells | Reduced virulence in ferrets, but the mutant had a growth advantaged over the WT in presence of ZMV | [20, 24] |
Abbreviations: HRI, highly reduced inhibition; LMV, laninamivir; MDCK, Madin-Darby Canine Kidney; NA, neuraminidase; ND, not determined; OSV, oseltamivir; PMV, peramivir; Ref., reference; RI, reduced inhibition; RV, recombinant virus; S, susceptibility; WHO, World Health Organization; WT, wild type; ZMV, zanamivir.
aN2 numbering system was used for NA mutations.
bThe phenotype of resistance was determined by NA inhibition assays against OSV, PMV, ZMV, and LMV. Resistance phenotype is determined in accordance with WHO criteria: S (≤10-fold increase for influenza A; ≤5-fold increase for influenza B); RI (between 10- and 100-fold increase for influenza A; between 5- and 50-fold increase for influenza B); HRI (>100-fold increase for influenza A; >50-fold increase for influenza B).
Almost all NA mutations identified in clinical isolates exhibiting cross-resistance to NAIs altered the NA enzyme activity and/or affinity, which likely affected viral fitness (Table 2). The A(H1N1)pdm09 E119D/H274Y and E119G/H274Y isolates (cases 1 and 2) could not be recovered in cell culture. Using reverse genetics, we could rescue the E119D and E119D/H274Y A(H1N1)pdm09 recombinants and demonstrated the genetic instability (reversion) of the E119D mutation in vitro and in animal models [21]. The E119D and E119D/H274Y recombinants had reduced replicative capacity in vitro and decreased infectivity/transmissibility in experimentally infected mice and guinea pigs [21]. Similar findings were observed for the recombinant A(H1N1)pdm09-E119G [22]. The recombinant H274Y/I222R mutant also had reduced viral fitness in Madin-Darby Canine Kidney (MDCK) cells [23].
The R152K mutation significantly altered the NA stability of recombinant influenza B/Beijing/1/87 virus and resulted in poor replication in MDCK cells [24]. Nasal wash viral titers from ferrets infected with the influenza B/Memphis/12/97-R152K variant were also significantly lower versus the wild-type (WT) group [20]. When ferrets were coinfected with different ratios of the 2 viruses, the R152K mutant was less virulent, but it had a growth advantage over the WT in presence of zanamivir [20].
Viral fitness was found to be maintained for some cross-RI/HRI variants. The recombinant A(H1N1)pdm09-I222R mutant (cases 6 and 7) replicated and transmitted as well as the recombinant WT in ferrets [25]. The influenza A/Bethesda/NIH12-0/2008 (H3N2) isolate containing the del245-248 NA mutation had similar viral replicative capacity compared with the WT isolate in MDCK cells [18]. In experimentally infected ferrets, virus could be detected at comparable levels for the WT and the del245-248 NA mutant groups. In addition, the 2 viruses had the same rate of direct-contact transmission in this model [18]. The viral fitness of the H3N2-E119I variant was not formally assessed; however, it was suggested that, similar to the H3N2-E119V variant, which was transmissible in a ferret model, the E119I substitution is unlikely to significantly affect viral infectivity and transmissibility [26]. The identification of H3N2-E119I variants from untreated persons supports this hypothesis [26].
We were surprised to find that the R292K functional substitution had no significant impact on viral fitness of the A(H7N9) virus. Indeed, although the influenza A/Shanghai/1/2013 (H7N9) R292K mutant exhibited lower NA enzyme activity/affinity than the WT virus, this variant replicated at comparable titers versus the WT in MDCK cells [27]. In another report, Hai et al [28] demonstrated that the H7N9-R292K mutant had comparable viral replication in human tracheobronchial epithelial cells, virulence in mice and transmissibility in guinea pigs when compared with the WT virus. The recombinant influenza B/Beijing/1/87 virus containing the D198N substitution was not outcompeted by the WT in ferrets inoculated with a 50:50 mixture of the 2 recombinants [29], suggesting a retained viral fitness for this B variant.
CONCLUSIONS AND PERSPECTIVES
Oseltamivir and zanamivir have been frequently used to treat severe cases of influenza infection. This review demonstrates that, despite some important structural differences between NAIs, mutations conferring RI/HRI phenotypes to both oseltamivir and zanamivir (and also to peramivir and laninamivir for some variants) may emerge in clinic. Such events have occurred more frequently in the immunocompromised population, causing high rates of fatal outcomes despite prolonged antiviral treatments. Such findings are predictable because prolonged viral shedding and lengthy therapy constitute important risk factors for the emergence of viral drug resistance. The 2 pathways leading to dual NAI resistance include the selection of hallmark oseltamivir resistance mutations [A(H1N1)-H274Y and A(H3N2)-E119V] followed by emergence of a second mutation upon zanamivir treatment or the emergence of a single substitution/deletion conferring RI/HRI to various NAIs.
Most notably, all NAI-resistant variants reviewed here were also resistant to the adamantanes. Moreover, although NAI-resistant NA mutations were found to alter, at different levels, the activity/affinity of the NA enzyme, the viral fitness of some variants such as A(H1N1)pdm09-I222R, A(H7N9)-R292K, A(H3N2)-del245-248, and B-D198N could be maintained. This highlights the serious need for additional antiviral strategies against such multidrug-resistant viruses.
This review also suggests a higher propensity of influenza A(H1N1)pdm09 viruses to develop NAI cross-resistance, which is in agreement with a previous study reported before the 2009 pandemic on oseltamivir-treated children showing that oseltamivir resistance was more frequent in influenza A(H1N1) than in A(H3N2) viruses (27% vs 3%, respectively) [30]. More importantly, only the A(H1N1)-H274Y mutation has been unequivocally associated with clinical failure, so one needs to be very cautious in extrapolating IC50 levels and clinical outcomes.
Oseltamivir contributed to most resistant cases reported in this review, reflecting the fact that this is the most widely used agent. Moreover, variants with the most frequent oseltamivir-induced NA mutations [H274Y in A(H1N1)pdm09 and E119V in A(H3N2) viruses] are likely to maintain a significant level of viral fitness and transmissibility [5]. Zanamivir could be potentially more beneficial as first-line option in immunocompromised patients because it has a structure closer to the natural NA substrate and its use provides higher drug concentrations in the respiratory tract versus oseltamivir [8]. On the other hand, inhaled administration is not practical in severe cases, and the IV formulation has not been approved yet. To avoid the sequential use of oseltamivir and zanamivir, the zanamivir-oseltamivir hybrid inhibitor (MS-257), containing both the guanidino group of zanamivir and the hydrophobic pentyloxy group of oseltamivir, appears to be a potentially attractive antiviral option [31].
Antiviral combinations could constitute suitable anti-influenza alternatives because of their potential for increasing antiviral potency and reducing the emergence of resistance [32]. Combined therapies could be particularly helpful in immunocompromised hosts in the presence of high and sustained viral load. Nevertheless, combinations between different NAIs, such as the one between oseltamivir and zanamivir, resulted in a competitive antagonism and failure to improve clinical outcomes [33]. By contrast, combinations of antivirals with different viral targets, such as the amantadine-oseltamivir-ribavirin triple therapy, showed promise despite resistance to 1 agent [34].
Other antivirals with activity against susceptible and NAI-resistant influenza variants are at different stages of preclinical or clinical evaluation. The 2,3-difluorosialic acid compound demonstrated high affinity and slow dissociation from the viral NA, resulting in prolonged NA inactivation [35]. DAS181, a sialidase fusion protein that prevents viral binding on host cells, demonstrated efficacy in vitro as well as in mice infected with the NAI-resistant A(H7N9)-R292K variant [36]. Favipiravir, a viral RNA-dependent ribonucleic acid polymerase inhibitor, demonstrated activity against pan NAI-resistant variants [16] and showed a synergistic effect when combined with oseltamivir or peramivir. Other PA and PB1 inhibitors are also evaluated in preclinical and clinical studies [37]. Nitazoxanide is an antiparasitic agent with a proven anti-influenza activity that is mediated by blocking HA maturation/trafficking, and this compound may also act as an interferon inducer [38]. A potential synergy between this antiviral and zanamivir or oseltamivir was also suggested based on in vitro studies.
CONCLUSIONS
In conclusion, reduced susceptibilty to existing NAIs may develop among immunocompromised patients infected with seasonal influenza A and B viruses as well as with avian influenza strains during NAI therapy, with some variants retaining viral fitness and transmissibility. This has important clinical and public health implications because existing NAIs constitute the only antiviral class currently approved against influenza infections. The management of NAI-resistant influenza infections remains a challenging task and requires a better understanding of the mechanisms of resistance and active evaluation of novel antivirals targeting different viral functions.
Acknowledgments
Financial support. This work was funded by a Canadian Institutes of Health Research grant (no. 229733; to G. B.) for a research program on the pathogenesis, treatment, and prevention of respiratory and herpes viruses.
Potential conflicts of interest. G. B. has received institutional grant support from BioCryst. Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Health topic, Influenza Available at: http://www.who.int/topics/influenza/en/.
- 2. Memoli MJ, Athota R, Reed S et al. . The natural history of influenza infection in the severely immunocompromised vs nonimmunocompromised hosts. Clin Infect Dis 2014; 58:214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ison MG, Hirsch HH. Influenza: a recurrent challenge to transplantation. Transpl Infect Dis 2010; 12:95–7. [DOI] [PubMed] [Google Scholar]
- 4. Choi SM, Boudreault AA, Xie H et al. . Differences in clinical outcomes after 2009 influenza A/H1N1 and seasonal influenza among hematopoietic cell transplant recipients. Blood 2011; 117:5050–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Samson M, Pizzorno A, Abed Y, Boivin G. Influenza virus resistance to neuraminidase inhibitors. Antiviral Res 2013; 98:174–85. [DOI] [PubMed] [Google Scholar]
- 6. Meijer A, Lackenby A, Hungnes O et al. . Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerg Infect Dis 2009; 15:552–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hurt AC, Ernest J, Deng YM et al. . Emergence and spread of oseltamivir- resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res 2009; 83:90–3. [DOI] [PubMed] [Google Scholar]
- 8. Moscona A. Neuraminidase inhibitors for influenza. N Engl J Med 2005; 353:1363–73. [DOI] [PubMed] [Google Scholar]
- 9. McKimm-Breschkin JL. Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza Other Respir Viruses 2013; 7(Suppl 1):25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim CU, Lew W, Williams MA et al. . Structure-activity relationship studies of novel carbocyclic influenza neuraminidase inhibitors. J Med Chem 1998; 41:2451–60. [DOI] [PubMed] [Google Scholar]
- 11. Babu YS, Chand P, Bantia S et al. . BCX-1812 (RWJ-270201): discovery of a novel, highly potent, orally active, and selective influenza neuraminidase inhibitor through structure-based drug design. J Med Chem 2000; 43:3482–6. [DOI] [PubMed] [Google Scholar]
- 12. Kubo S, Tomozawa T, Kakuta M et al. . Laninamivir prodrug CS-8958, a long- acting neuraminidase inhibitor, shows superior anti-influenza virus activity after a single administration. Antimicrob Agents Chemother 2010; 54:1256–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Colman PM, Hoyne PA, Lawrence MC. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J Virol 1993; 67:2972–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McKimm-Breschkin JL. Resistance of influenza viruses to neuraminidase inhibitors—a review. Antiviral Res 2000; 47:1–17. [DOI] [PubMed] [Google Scholar]
- 15. Pizzorno A, Abed Y, Bouhy X et al. . Impact of mutations at residue I223 of the neuraminidase protein on the resistance profile, replication level, and virulence of the 2009 pandemic influenza virus. Antimicrob Agents Chemother 2012; 56:1208–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. L’Huillier AG, Abed Y, Petty TJ et al. . E119D neuraminidase mutation conferring pan-resistance to neuraminidase inhibitors in an A(H1N1)pdm09 isolate from a stem-cell transplant recipient. J Infect Dis 2015; 212:1726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eshaghi A, Shalhoub S, Rosenfeld P et al. . Multiple influenza A (H3N2) mutations conferring resistance to neuraminidase inhibitors in a bone marrow transplant recipient. Antimicrob Agents Chemother 2014; 58:7188–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Memoli MJ, Hrabal RJ, Hassantoufighi A et al. . Rapid selection of a transmissible multidrug-resistant influenza A/H3N2 virus in an immunocompromised host. J Infect Dis 2010; 201:1397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu Y, Bi Y, Vavricka CJ et al. . Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res 2013; 23:1347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gubareva LV, Matrosovich MN, Brenner MK et al. . Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J Infect Dis 1998; 178:1257–62. [DOI] [PubMed] [Google Scholar]
- 21. Abed Y, Bouhy X, L’Huillier AG et al. . The E119D neuraminidase mutation identified in a multidrug-resistant influenza A(H1N1)pdm09 isolate severely alters viral fitness in vitro and in animal models. Antiviral Res 2016; 132:6–12. [DOI] [PubMed] [Google Scholar]
- 22. Pizzorno A, Abed Y, Rheaume C et al. . Evaluation of recombinant 2009 pandemic influenza A (H1N1) viruses harboring zanamivir resistance mutations in mice and ferrets. Antimicrob Agents Chemother 2013; 57:1784–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. LeGoff J, Rousset D, Abou-Jaoudé G et al. . I223R mutation in influenza A(H1N1)pdm09 neuraminidase confers reduced susceptibility to oseltamivir and zanamivir and enhanced resistance with H275Y. PLoS One 2012; 7:e37095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jackson D, Barclay W, Zürcher T. Characterization of recombinant influenza B viruses with key neuraminidase inhibitor resistance mutations. J Antimicrob Chemother 2005; 55:162–9. [DOI] [PubMed] [Google Scholar]
- 25. van der Vries E, Veldhuis Kroeze EJ, Stittelaar KJ et al. . Multidrug resistant 2009 A/H1N1 influenza clinical isolate with a neuraminidase I223R mutation retains its virulence and transmissibility in ferrets. PLoS Pathog 2011; 7:e1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okomo-Adhiambo M, Demmler-Harrison GJ, Deyde VM et al. . Detection of E119V and E119I mutations in influenza A (H3N2) viruses isolated from an immunocompromised patient: challenges in diagnosis of oseltamivir resistance. Antimicrob Agents Chemother 2010; 54:1834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yen HL, McKimm-Breschkin JL, Choy KT et al. . Resistance to neuraminidase inhibitors conferred by an R292K mutation in a human influenza virus H7N9 isolate can be masked by a mixed R/K viral population. MBio 2013; 4:pii: e00396-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hai R, Schmolke M, Leyva-Grado VH et al. . Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun 2013; 4:2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mishin VP, Hayden FG, Gubareva LV. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemother 2005; 49:4515–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stephenson I, Democratis J, Lackenby A et al. . Neuraminidase inhibitor resistance after oseltamivir treatment of acute influenza A and B in children. Clin Infect Dis 2009; 48:389–96. [DOI] [PubMed] [Google Scholar]
- 31. Wu Y, Gao F, Qi J et al. . Resistance to mutant group 2 influenza virus neuraminidases of an oseltamivir-zanamivir hybrid inhibitor. J Virol 2016; 90:10693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ison MG. Influenza prevention and treatment in transplant recipients and immunocompromised hosts. Influenza Other Respir Viruses 2013; 7(Suppl 3):60–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duval X, van der Werf S, Blanchon T et al. . Efficacy of oseltamivir-zanamivir combination compared to each monotherapy for seasonal influenza: a randomized placebo-controlled trial. PLoS Med 2010; 7:e1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seo S, Englund JA, Nguyen JT et al. . Combination therapy with amantadine, oseltamivir and ribavirin for influenza A infection: safety and pharmacokinetics. Antivir Ther 2013; 18:377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tai SH, Agafitei O, Gao Z et al. . Difluorosialic acids, potent novel influenza virus neuraminidase inhibitors, induce fewer drug resistance-associated neuraminidase mutations than does oseltamivir. Virus Res 2015; 210:126–32. [DOI] [PubMed] [Google Scholar]
- 36. Marjuki H, Mishin VP, Chesnokov AP et al. . An investigational antiviral drug, DAS181, effectively inhibits replication of zoonotic influenza A virus subtype H7N9 and protects mice from lethality. J Infect Dis 2014; 210:435–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Muratore G, Goracci L, Mercorelli B et al. . Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proc Natl Acad Sci U S A 2012; 109:6247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rossignol JF. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral Res 2014; 110:94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tamura D, DeBiasi RL, Okomo-Adhiambo M et al. . Emergence of multidrug-resistant influenza A(H1N1)pdm09 virus variants in an immunocompromised child treated with oseltamivir and zanamivir. J Infect Dis 2015; 212:1209–13. [DOI] [PubMed] [Google Scholar]
- 40. Grund S, Gkioule C, Termos T et al. . Primarily oseltamivir-resistant influenza A (H1N1pdm09) virus evolving into a multidrug-resistant virus carrying H275Y and I223R neuraminidase substitutions. Antivir Ther 2015; 20:97–100. [DOI] [PubMed] [Google Scholar]
- 41. Nguyen HT, Fry AM, Loveless PA et al. . Recovery of a multidrug-resistant strain of pandemic influenza A 2009 (H1N1) virus carrying a dual H275Y/I223R mutation from a child after prolonged treatment with oseltamivir. Clin Infect Dis 2010; 51:983–4. [DOI] [PubMed] [Google Scholar]
- 42. van der Vries E, Stelma FF, Boucher CA. Emergence of a multidrug-resistant pandemic influenza A (H1N1) virus. N Engl J Med 2010; 363:1381–2. [DOI] [PubMed] [Google Scholar]
- 43. Eshaghi A, Patel SN, Sarabia A et al. . Multidrug-resistant pandemic (H1N1) 2009 infection in immunocompetent child. Emerg Infect Dis 2011; 17:1472–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baz M, Abed Y, McDonald J, Boivin G. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis 2006; 43:1555–61. [DOI] [PubMed] [Google Scholar]
- 45. Hu Y, Lu S, Song Z et al. . Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet 2013; 381:2273–9. [DOI] [PubMed] [Google Scholar]
- 46. Ison MG, Gubareva LV, Atmar RL et al. . Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J Infect Dis 2006; 193:760–4. [DOI] [PubMed] [Google Scholar]