Abstract
CSL is the primary target of the Notch signaling pathway in mammalian cells. It is a DNA binding protein that generally represses transcription in the absence of Notch signaling and activates transcription upon formation of a ternary complex with NICD, the protease-generated intracellular domain of Notch. Previous mapping experiments identified the central third of CSL as both necessary and sufficient for DNA binding and activation by Notch. Here we show that CSL promotes transcription in 293T cells in the absence of added NICD and that this activity requires both the central domain plus the C-terminal third of the protein. Evidence is presented that argues against a contribution of endogenous NICD and instead supports the possibility that distinct coactivators may directly stimulate the activity of CSL in a cell type-specific manner. This conclusion supports a recent finding that Drosophila CSL (Suppressor of Hairless) can also mediate transcriptional activation in the absence of Notch.
INTRODUCTION
Notch proteins define a family of large transmembrane receptors that control cell fate decisions in many developmental systems (1–6). The extracellular domain of Notch contains multiple epidermal growth factor-like repeats while the intracellular domain is distinguished by six cdc10/ankyrin repeats, an OPA and a PEST domain and a functional region N-terminal to the ankyrin domains designated the RAM (or RAM23) domain. The extracellular domain of Notch is cleaved at a site adjacent to the transmembrane domain by a furin-type protease prior to its presentation on the cell surface; however, the two portions of the protein remain associated with one another (7–9). Upon interaction with ligand, two additional proteolytic cleavage events occur which liberate the Notch intracellular domain (NICD) from the membrane, allowing its subsequent entry into the nucleus (10–13). Once in the nucleus, NICD interacts with a DNA binding protein denoted CSL (also known as CBF1/RBP-J in mammals, Suppressor of Hairless [Su(H)] in Drosophila and Xenopus, and Lag-1 in Caenorhabditis elegans) and the resulting complex, which also includes the protein Mastermind and/or SKIP, activates transcription of target genes (14–19).
CSL is a DNA binding protein expressed in a wide array of mammalian tissues and cell lines (20,21). Ablation of CSL in mice results in death prior to 10.5 days of gestation. The mutant mice exhibit severe growth retardation and defects in somite and neural tube development (22). CSL is a pioneer protein that binds DNA as a monomer and recognizes the consensus sequence (C/T)GTGGGAA (23). In the absence of Notch, CSL typically functions as a transcriptional repressor. The repression domain maps to the central third of the protein, which also corresponds to the DNA binding and Notch interaction domains (24). Transcriptional repression can be mediated in two ways. First, CSL can interact directly with components of the basal transcription machinery (TFIIA and dTAFII110), which disrupts activated transcription in vitro (25). Secondly, CSL can interact with corepressor proteins that recruit histone deacetylase enzymes such as HDAC1 and HDAC2. The transcriptional corepressors identified thus far are SMRT and a novel protein denoted CIR (26,27). In the presence of NICD, the corepressors are displaced from CSL and the resulting NICD–CSL complex activates transcription by virtue of an activation domain carried on NICD that recruits the histone acetyltransferase PCAF (28,29). A recent report by Posakony and colleagues (30) provides evidence that Drosophila CSL [Su(H)] responds similarly, but that it can also mediate Notch-independent transcriptional activation in the socket cells of the mechanosensory bristle.
Until recently, no activities had been described for the N- or C-terminal portions of CSL. In a recent report, Honjo and co-workers (31) found that the N- and C-terminal portions of CSL can mediate interactions with the ankyrin repeats of Notch and aid transcriptional activation by NICD. We show here that in 293T cells (an adenovirus-transformed human kidney cell line), CSL is active in the absence of added NICD, and that this activity depends on the Notch interaction domain plus the C-terminal third of the protein. We present evidence that activity of CSL in 293T cells is not dependent on Notch, but is a consequence of distinct cell type-specific coactivators working directly on CSL. This conclusion is consistent with the recent model proposed for Notch-independent activation by Su(H) in Drosophila socket cells.
MATERIALS AND METHODS
Plasmids and transfection assays
Gal4-CBF1 (referred to herein as Gal4-CSL; 16) was the gift of Diane Hayward (Johns Hopkins University). Gal4-CSL(1-233) and Gal4-CSL(233-500) were generated by EcoR1 digestion of the parental plasmid. Other CSL deletion mutants were generated as BamH1–Sac1 fragments by the polymerase chain reaction and inserted downstream of the Gal4 coding region. Expression levels were assessed by western analysis using an anti-Gal4 antibody (Santa Cruz). The Gal4 reporter plasmids 5×Gal4TK-CAT, 5×Gal4TK200-luc and (Gal4)5E1B-luc were obtained from Diane Hayward, Kathryn Calame (Columbia University) and Antonio Giordano (Thomas Jefferson University), respectively. The Notch-dependent reporter 4×wtCBF1Luc (referred to herein as (CSL)4-luc; 16) was the gift of Diane Hayward and the parental vector lacking CSL binding sites, pGL2pro, was obtained from Promega. Expression vectors for NICD (N1-IC; 32), CSL (Flag-CBF1; 16) and SMRT (26) have been described elsewhere. 293T and NIH 3T3 cells were maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum. Transfections were carried out using calcium phosphate coimmunoprecipitation using 7 µg of total DNA per transfection of a 60 mm plate. CAT assays and luciferase assays were performed as described previously (32) and values were normalized to levels of β-galactosidase generated from CMV-β-galactosidase (100 ng). Values represent average results obtained from at least three separate experiments.
Electrophoretic mobility shift assay (EMSA)
The CSL binding site probe was generated with the oligonucleotides 5′-GATCGGAAACACGCCGTGGGAAAAAATTTGGC-3′ and 5′-GATCGCCAAATTTTTTCCCACGGCGTGTTTCC-3′ (complementary strand). Annealed double-stranded probe was labeled by filling 5′ overhangs with Klenow fragment in the presence of [32P]dATP and [32P]dCTP. Labeled probes were purified with a G50 spin column (Boehringer Manheim). Whole cell extracts were made according to Damm et al. (33) and each binding reaction contained 1–4 µl cell extract, 1 µl probe (4 × 104 c.p.m.), 2 µg poly(dI-dC) as non-specific competitor, 1× binding buffer (20 mM HEPES pH 7.9, 60 mM KCl, 1.33 mM MgCl2, 1.0 mM DTT, 4% glycerol) in a final volume of 20 µl. Binding was carried out at 4°C for 1 h and then reactions were loaded onto a 5% polyacrylamide gel using 0.25× TBE. For super shifts, the cell extracts and serum were incubated for 1 h prior to the addition of DNA. CSL antiserum was kindly provided by J.Coligan (NIAID, NIH).
RESULTS
CSL is activated in trans in 293T cells
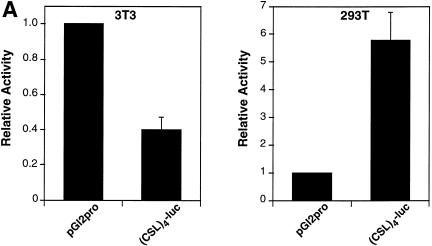
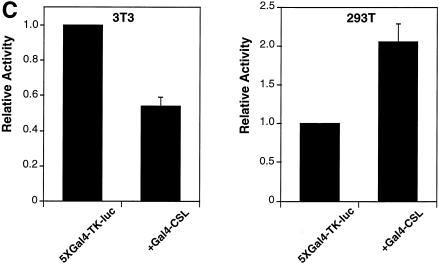
Vertebrate CSL (CBF1/RBP-J) is generally a transcriptional repressor in the absence of Notch. We and others have demonstrated that this is likely to be a consequence of CSL interacting with corepressor proteins (26,27). However, in the course of our studies we noted that promoters containing CSL binding sites were active in the 293T cell line. 293T cells were derived initially from an adenovirus-mediated transformation of primary human kidney cells (they express low levels of E1A and E1B proteins) and then transduced further with a plasmid that expresses a temperature-sensitive SV40 large T antigen. The effect of linking CSL binding sites to the SV40 promoter led, as expected, to a decrease in reporter activity in NIH 3T3 cells (Fig. 1A, left). In contrast, the same sites stimulated expression from the SV40 promoter in 293T cells (Fig. 1A, right). Examination of endogenous CSL DNA binding activity by EMSA failed to reveal any significant differences between NIH 3T3 cells and 293T cells (Fig. 1B). The possibility that these cells harbor a mutant, constitutively-active CSL was ruled out by our observation that plasmid-encoded Gal4-CSL activated transcription in 293T cells in the absence of added NICD (Fig. 1C). We determined that the activity is not a consequence of the resident 12S E1A proteins (see below) and exogenously added T antigen had no appreciable effect on CSL activity (data not shown).
Figure 1.
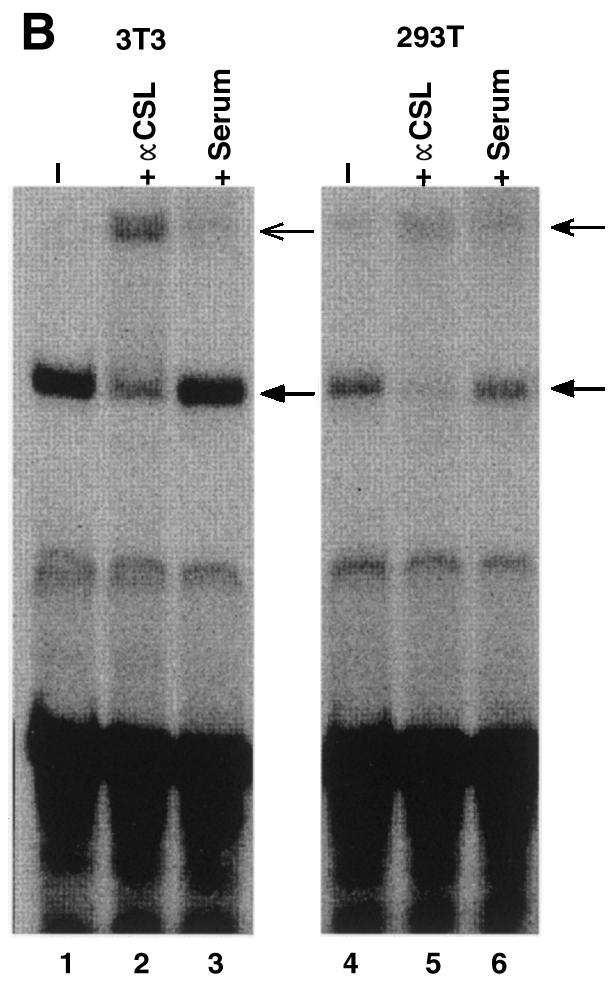
CSL activates transcription in 293T cells. (A) Activity of a CSL-responsive reporter, (CSL)4-luc, compared to the reporter lacking CSL binding sites, pGl2pro, upon transfection of NIH 3T3 and 293T cells. (B) EMSAs using nuclear extracts of NIH 3T3 and 293T cells in conjunction with an oligonucleotide bearing CSL binding sites. The major band is due to CSL by virtue of the change of its mobility in the presence of CSL antiserum (+ αCSL), but not in the presence of control serum (+ Serum). (C) Activity of a Gal4-CSL fusion protein transfected into NIH 3T3 and 293T cells. Cells were transfected with either the Gal4 site-containing reporter alone (5×Gal4-TK-luc) or along with an expression plasmid expressing Gal4-CSL, as indicated. Luciferase activities were determined 2 days after transfection.
Activity of CSL in 293T cells requires a novel functional domain
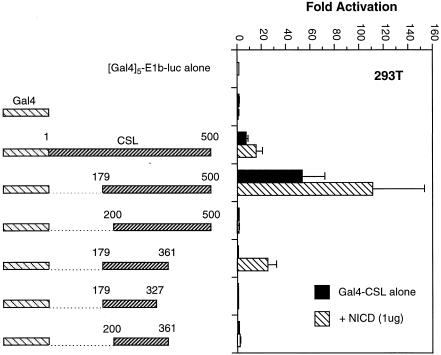
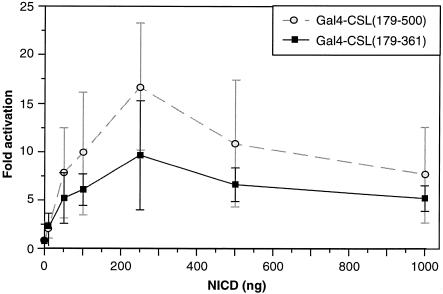
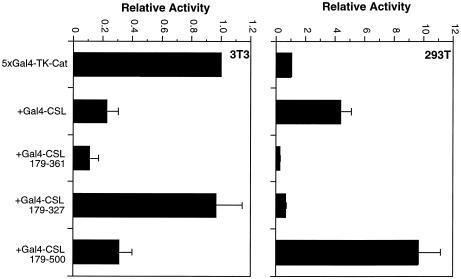
To explore further the activity of CSL in 293T cells, we mapped the domain of CSL required for its activity. We constructed a series of deletion mutants of CSL, linked these to the Gal4 DNA binding domain and tested the ability of the various proteins to activate a TATA box-containing reporter in 293T cells in the presence or absence of NICD. Western analyses confirmed that the various fusion proteins were expressed at comparable levels (data not shown). The data (Fig. 2) can be summarized as follows. First, we confirmed previously published results by showing that the minimum Notch interaction domain maps between CSL amino acids 179 and 361. Secondly, and most importantly, the activation domain of CSL in 293T cells does not coincide precisely with the Notch interaction domain. Although the two domains overlap, sharing an N-terminal boundary [amino acid 179; constructs containing CSL amino acids 200–500 and 233–500 failed to activate transcription (data not shown)], amino acids necessary for CSL activity in the absence of added NICD extend beyond position 361, requiring perhaps the entire C-terminal two-thirds of the CSL protein. We considered the possibility that activity of CSL in 293T cells might still be due to a small amount of activated Notch, but that such low concentrations may require a larger interaction domain on CSL than that mapped in overexpression studies. In fact, it was recently shown that both the N- and C-terminal regions of CSL can bind in vitro to the ankyrin repeats of Notch (31). To address this possibility we varied the concentration of NICD in transfections of NIH 3T3 cells and measured the relative responses of Gal4-CSL fusion proteins carrying either the minimum Notch interaction domain (179–361) or the larger domain (179–500). As shown in Figure 3, although the maximum response of the larger domain was roughly 2-fold greater, consistent with a role for the C-terminal domain in overall transcriptional activation by NICD (31), the relative activities of the larger and smaller domains at low NICD concentrations was not significantly different. These data argue that the preferential activity of the larger CSL domain in 293T cells is not due to an enhanced ability to recruit low concentrations of endogenous activated Notch.
Figure 2.
The activation domain within CSL does not correspond to the Notch interaction domain. The (Gal4)5-E1b-luc reporter was transfected into 293T cells along with the indicated Gal4-CSL fusion proteins, + or – NICD, as indicated.
Figure 3.
The relative responses of Gal4-CSL(179-361) and Gal4-CSL(179-500) to various NICD concentrations. NIH 3T3 cells were transfected with the (Gal4)5-E1b-luc reporter along with either the Gal4-CSL(179-361) or Gal4-CSL(179-500) expression vectors in the presence of increasing amounts of NICD expression plasmid, as indicated.
CSL binds both corepressors and coactivators in the absence of added NICD
CSL recruits transcriptional corepressors to DNA in the absence of NICD and coactivators in the presence of NICD. However, the transition from repression to activation represents a dynamic equilibrium since high levels of the corepressor SMRT can reverse activation by NICD. Activation or repression by CSL should therefore be determined by the relative concentrations of coactivators and corepressors in a given cell. Although CSL may have the potential to directly recruit both, binding of corepressors may prevail in most cells. NICD may serve simply to alter the balance in favor of coactivators.
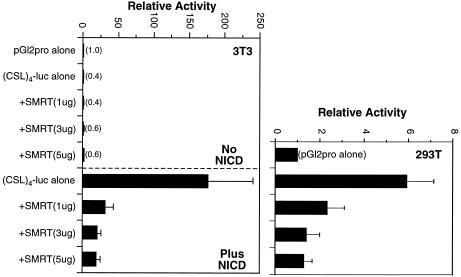
To investigate whether CSL can recruit corepressors in 293T cells, we employed various Gal4-CSL fusion proteins and assessed their ability to repress transcription when linked to a TK promoter (Fig. 4). As expected, Gal4-CSL repressed transcription in NIH 3T3 cells (Fig. 4, left) and the minimum repression domain coincided with amino acids 179–361, similar to the Notch-interaction domain. In 293T cells (Fig. 4, right), repression was not evident for either the full-length CSL protein or the fragment containing the novel activation domain. However, repression was observed in 293T cells when the minimum repression domain was tested, suggesting that CSL has the ability to recruit corepressors in 293T cells, but that this is not apparent due to the preferential recruitment of coactivators. Consistent with this view, overexpression of SMRT led to the inhibition of both NICD-mediated activation of CSL in NIH 3T3 cells and NICD-independent activity of CSL in 293T cells (Fig. 5). That our 293T cells contain normal levels of corepressor proteins was confirmed with a fusion protein containing Gal4 linked to the thyroid hormone receptor (Gal4-TR), which repressed transcription ∼3-fold (data not shown).
Figure 4.
Comparison of activities of Gal4-CSL deletions in NIH 3T3 and 293T cells. NIH 3T3 cells (left) and 293T cells (right) were transfected with the 5×Gal4-TK-luc reporter along with the various Gal4-CSL deletion mutants, as indicated. Activities are represented relative to the reporter alone.
Figure 5.
CSL can bind corepressors in both NIH 3T3 and 293T cells. SMRT overexpression results in repression of both Notch-mediated activation of CSL in NIH 3T3 cells (left) and CSL activity in 293T cells (right). Cells were transfected with the indicated reporter and expression plasmids along with increasing amounts of an SMRT expression plasmid, as indicated. Activities are expressed relative to those obtained with the pGl2pro reporter.
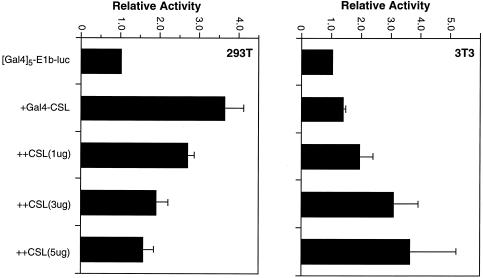
We considered the possibility that a small fraction of the CSL pool in NIH 3T3 cells may recruit coactivators in the absence of added NICD. To address this we overexpressed CSL in an attempt to disrupt the normal balance of available coactivators and corepressors, and examined the effect on Gal4-CSL activity in NIH 3T3 and 293T cells. We used a reporter, Gal4-E1b-luc, whose promoter consists of Gal4 sites linked to only a TATA box. Corepressor proteins recruited to such a promoter will result in neither transcriptional activation nor measurable transcriptional repression. By contrast, even low levels of coactivators recruited to such a promoter will result in a positive signal. Increasing doses of CSL inhibited activity of Gal4-CSL in 293T cells (Fig. 6, left), consistent with CSL binding a limited number of coactivator proteins. By contrast, increasing doses of CSL promoted activity of Gal4-CSL in NIH 3T3 cells to levels that were comparable to those obtained in 293T cells (Fig. 6, right). We conclude that while CSL binds primarily corepressors in NIH 3T3 cells, it can also bind coactivators if the concentration of corepressors is reduced. Thus, both NIH 3T3 and 293T cells contain CSL transcriptional coactivators, but the relative balance is such that the corepressors dominate in NIH 3T3 cells while the coactivators dominate in 293T cells.
Figure 6.
Effect of CSL overexpression on activities of Gal4-CSL in NIH 3T3 and 293T cells. 293T cells (left) and NIH 3T3 cells (right) were transfected with the (Gal4)5-E1b-luc reporter either alone, with the Gal4-CSL expression vector or with the Gal4-CSL expression vector and increasing amounts of a CSL expression vector, as indicated. Activities are expressed relative to those obtained with the reporter alone.
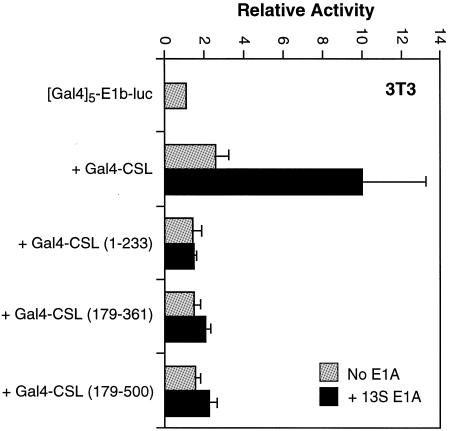
One defining feature of 293T cells is their expression of adenovirus proteins E1A and E1B. Since E1A is a known transcription factor, we considered the possibility that it affects activity of CSL in 293T cells. Of particular interest to us was the 13S form of E1A since it has been shown to be a transcriptional activator (34) and is expressed in 293T cells (35). Indeed, when we cotransfected NIH 3T3 cells with plasmids expressing a Gal4-CSL fusion protein and 13S E1A, we observed transcriptional activation (Fig. 7). This result is consistent with a recent report that describes the ability of 13S E1A to serve as a coactivator for CSL (36). Although this observation is interesting, we found that it cannot easily explain the activity of CSL in 293T cells. None of the individual domains of CSL we tested could support transcriptional activation by 13S E1A (Fig. 7), including the one sufficient for transcriptional activation in 293T cells (amino acids 179–500; see Discussion).
Figure 7.
13S E1A activates full-length CSL, but not the 179–500 deletion mutant. NIH 3T3 cells were transfected with the indicated reporter and expression plasmids with or without a 13S E1A expression plasmid (1 µg), as indicated. Values are expressed relative to the (Gal4)5-E1b-luc reporter alone.
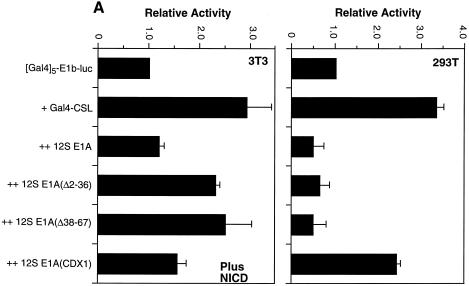
In contrast to 13S E1A, the 12S form of E1A often represses transcription due to its ability to bind the coactivators p300/CBP and PCAF (37). It is expressed in 293T cells at a level comparable to that of 13S (35). Indeed, Notch has been shown previously to bind PCAF and to be inhibited by 12S E1A (29). We found that NICD-mediated activation of Gal4-CSL in NIH 3T3 cells was almost completely inhibited in the presence of transfected 12S E1A (Fig. 8A, left). As expected, N-terminal deletion mutants of E1A that fail to bind PCAF (Δ2–36 and Δ38–67) (37,38) were less effective. We also tested a mutant E1A (CDx1) (39) that cannot bind the retinoblastoma protein and found that it can also inhibit Notch, but not to the same extent as wild-type 12S E1A (Fig. 8A, left).
Figure 8.
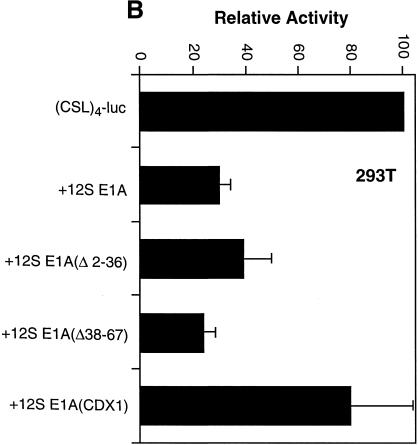
The domains of 12S E1A required for repression of Notch-mediated activation of CSL are different from those required for repression of CSL activity in 293T cells. (A) NIH 3T3 cells (left) and 293T cells (right) were transfected with the indicated reporter and Gal4-CSL expression vector with or without the indicated versions of 12S E1A (1 µg). Activities are expressed relative to the (Gal4)5-E1b-luc reporter alone. (B) 293T cells were transfected with the (CSL)4-luc expression vector along with the indicated 12S E1A (1 µg) expression vectors. Activities are expressed relative to those obtained with the reporter alone.
12S E1A also inhibited activity of Gal4-CSL in 293T cells (Fig. 8A, right). However, unlike what was observed for Notch-mediated activation, the N-terminal deletion mutants of 12S E1A were equally effective in inhibiting Gal4-CSL while the E1A mutant that cannot bind Rb was much less inhibitory (Fig. 8A, right). Similar results were obtained with a reporter that measures the activity of endogenous CSL in 293T cells (Fig. 8B). These data argue that while PCAF (and possibly p300/CBP) is important for Notch-mediated activation in NIH 3T3 cells, it is either not important or not used in the same way for activation of CSL activity in 293T cells. We conclude that activity of CSL in 293T cells relies on distinct coactivators.
DISCUSSION
CSL is a central player in the Notch signaling pathway in vertebrates. In the cases examined thus far it functions as a transcriptional repressor in the absence of the NICD and as a transcriptional activator in its presence. NICD binds CSL directly, displaces corepressor proteins and recruits coactivators by virtue of its own activation domain. Here we provide evidence that CSL has the potential to activate transcription in the absence of added NICD. Our data raise the possibility that transcriptional events generally considered to be Notch-dependent may be activated by other signals as well. Our data are consistent with a recent report that draws a similar conclusion for Drosophila Su(H) (30).
The possibility that CSL is activated in trans in 293T cells independently of Notch is supported by two observations. First, activity of CSL in 293T cells requires a domain comprising amino acids 179–500, whereas that needed for NICD-mediated activation is smaller, consisting of amino acids 179–361. We have verified that the larger domain is not simply more effective at recruiting low levels of NICD in vivo, despite a recent report that shows that the C-terminal region of CSL can bind the Notch ankyrin repeats in vitro (31). Secondly, the effects mediated by mutant E1A proteins indicate that the coactivators utilized by NICD (e.g. PCAF) are distinct from those utilized by CSL in 293T cells. This latter conclusion is particularly relevant since 293T cells express low levels of E1A proteins. Although 13S E1A did stimulate activity of full-length Gal4-CSL fusion protein in NIH 3T3 cells, it did not stimulate a Gal4-CSL fusion protein containing the domain sufficient for activity in 293T cells. It has been reported recently that 13S E1A binds to the N-terminal region of CSL in vitro (36), yet we observed no stimulation by 13S E1A of a Gal4 fusion protein containing only this region in vivo. Although further experiments will be required to map the region of CSL that is necessary and sufficient for in vivo activation by 13S E1A, it does not correspond to our activation domain. Combined with our observation that the 12S E1A inhibited both Notch-mediated and 293T cell-mediated activation of CSL, we conclude that the E1A proteins are not responsible for activating CSL in 293T cells.
The activity of CSL in a particular cell is determined by its relative recruitment of coactivators and corepressors. Although we have not undertaken an extensive analysis, it would appear that in most cell lines CSL recruits corepressor proteins predominantly. NICD shifts this balance in favor of coactivators first by displacing bound corepressors and secondly by directly recruiting coactivators such as PCAF. Our results suggest that even in the absence of NICD, CSL can bind coactivators. This can be observed when the effective concentration of corepressors is artificially reduced (e.g. in NIH 3T3 cells expressing high concentrations of CSL; Fig. 6). 293T cells represent a natural situation in which coactivator recruitment by CSL happens to be dominant, despite the presence of corepressors.
It is clear that the activity of coactivators and corepressors can be regulated at many levels. Modifications of DNA binding proteins can dictate the recruitment of coactivators as exemplified by CREB (40) or the relative recruitment of coactivators and corepressors as exemplified by nuclear hormone receptors (41). It has been shown that components of certain corepressor complexes, specifically class II histone deacetylases, can be regulated at the level of nuclear export (42,43). The manner in which such regulatory networks impinge on CSL activity remains to be determined. Indeed, future studies will need to focus on identifying the coactivators used by CSL. Our experiments argue against an important role for PCAF and p300/CBP since inhibition by 12S E1A did not require domains at the N-terminus known to bind PCAF and p300/CBP (37). In contrast, E1A-mediated inhibition of NICD correlated with PCAF and p300/CBP binding, consistent with published reports (29).
It is also possible that cell type-dependent modifications of CSL may affect the recruitment of these coactivators. Support for cell type-specific modifications of CSL comes from Coligan and colleagues (44) who purified a DNA binding activity by virtue of its ability to bind an NF-kB site and found CSL instead of NF-kB. They went on to show that CSL from other tissue sources was unable to bind their NF-kB probe. This was a property unique to the thymus-derived CSL (in both crude and highly purified forms) and it was postulated to be due to a modification of the CSL protein that relaxed its DNA binding specificity. The nature of the proposed modification is not known. While these data are intriguing, our experiments indicated that CSL proteins from both NIH 3T3 and 293T cells display similar DNA binding specificities, binding well to the CSL consensus and weakly to the NF-kB site (data not shown).
One coactivator known to mimic Notch activity is the EBNA2 protein of Epstein–Barr virus (16,24). Although not a coactivator in the strictest sense, EBNA2 binds CSL and is also likely to recruit cellular coactivators. It is possible that 293T cells harbor a similar bridging protein that recruits coactivators to CSL. Interestingly, we found that the 13S form of E1A stimulated activity of a Gal4-CSL fusion protein and may thus possess the properties of such a bridging protein. While interesting, 13S E1A did not activate through the minimal CSL activation domain and, thus, its presence cannot easily explain the activity of CSL in 293T cells.
Our findings that CSL can be activated by Notch-independent means may have broad implications. Notch is known to exert profound effects on a variety of developmental processes and our results provide evidence that certain cellular environments may mimic activated Notch. Results from Ziff and colleagues may provide clues as to what these environments might be (45,46). They have shown that wnt-mediated transformation of PC12 cells is associated with downregulation of the proneural gene MASH-1 and upregulation of HES-1, two events that normally reside downstream of Notch activation. Furthermore, they have shown that activation of HES-1 transcription is correlated with stimulation of the HES-1 promoter, largely through two of its three CSL binding elements. Whether wnt-transformation mimics the effects we observe in 293T cells remains to be determined. Results with Drosophila indicate that Su(H) activates transcription in the absence of Notch only in socket cells of the bristle organ and is therefore not a consequence of widely used signaling pathways such as wingless (30). Accordingly, even though 293T cells are highly transformed and therefore not representative of normal cells, it may not be surprising if the effects we see are ultimately limited to a small number of select cell types.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Prakash Rao and David Ross for critical reading of the manuscript and members of the Kadesch laboratory (especially Fang Zhao and Peter Ordentlich) for reagents, support and encouragement. This work was supported by funds from the National Institutes of Health (RO1 GM58228).
References
- 1.Artavanis-Tsakonas S., Matsuno,K. and Fortini,M.E. (1995) Notch signaling. Science, 268, 225–232. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S., Rand,M.D. and Lake,R.J. (1999) Notch signaling: cell fate control and signal integration in development. Science, 284, 770–776. [DOI] [PubMed] [Google Scholar]
- 3.Osborne B. and Miele,L. (1999) Notch and the immune system. Immunity, 11, 653–663. [DOI] [PubMed] [Google Scholar]
- 4.Robey E. (1999) Regulation of T cell fate by Notch. Annu. Rev. Immunol., 17, 283–295. [DOI] [PubMed] [Google Scholar]
- 5.Beatus P. and Lendahl,U. (1998) Notch and neurogenesis. J. Neurosci. Res., 54, 125–136. [DOI] [PubMed] [Google Scholar]
- 6.Lewis J. (1998) Notch signalling and the control of cell fate choices in vertebrates. Semin. Cell. Dev. Biol., 9, 583–589. [DOI] [PubMed] [Google Scholar]
- 7.Blaumueller C.M., Qi,H., Zagouras,P. and Artavanis-Tsakonas,S. (1997) Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell, 90, 281–291. [DOI] [PubMed] [Google Scholar]
- 8.Logeat F., Bessia,C., Brou,C., LeBail,O., Jarriault,S., Seidah,N.G. and Israel,A. (1998) The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl Acad. Sci. USA, 95, 8108–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rand M.D., Grimm,L.M., Artavanis-Tsakonas,S., Patriub,V., Blacklow,S.C., Sklar,J. and Aster,J.C. (2000) Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell. Biol., 20, 1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Struhl G. and Adachi,A. (1998) Nuclear access and action of notch in vivo. Cell, 93, 649–660. [DOI] [PubMed] [Google Scholar]
- 11.Schroeter E.H., Kisslinger,J.A. and Kopan,R. (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature, 393, 382–386. [DOI] [PubMed] [Google Scholar]
- 12.Brou C., Logeat,F., Gupta,N., Bessia,C., LeBail,O., Doedens,J.R., Cumano,A., Roux,P., Black,R.A. and Israel,A. (2000) A novel proteolytic cleavage involved in notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell, 5, 207–216. [DOI] [PubMed] [Google Scholar]
- 13.Mumm J.S., Schroeter,E.H., Saxena,M.T., Griesemer,A., Tian,X., Pan,D.J., Ray,W.J. and Kopan,R. (2000) A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell, 5, 197–206. [DOI] [PubMed] [Google Scholar]
- 14.Jarriault S., Brou,C., Logeat,F., Schroeter,E.H., Kopan,R. and Israel,A. (1995) Signalling downstream of activated mammalian Notch. Nature, 377, 355–358. [DOI] [PubMed] [Google Scholar]
- 15.Tamura K., Taniguchi,Y., Minoguchi,S., Sakai,T., Tun,T., Furukawa,T. and Honjo,T. (1995) Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J κ/Su(H). Curr. Biol., 5, 1416–1423. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh J.J., Henkel,T., Salmon,P., Robey,E., Peterson,M.G. and Hayward,S.D. (1996) Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein–Barr virus EBNA2. Mol. Cell. Biol., 16, 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou S., Fujimuro,M., Hsieh,J.J., Chen,L. and Hayward,S.D. (2000) A role for SKIP in EBNA2 activation of CBF1-repressed promoters. J. Virol., 74, 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petcherski A.G. and Kimble,J. (2000) Mastermind is a putative activator for Notch. Curr. Biol., 10, R471–R473. [DOI] [PubMed] [Google Scholar]
- 19.Wu L., Aster,J.C., Blacklow,S.C., Lake,R., Artavanis-Tsakonas,S. and Griffin,J.D. (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet., 26, 484–489. [DOI] [PubMed] [Google Scholar]
- 20.Matsunami N., Hamaguchi,Y., Yamamoto,Y., Kuze,K., Kangawa,K., Matsuo,H., Kawaichi,M. and Honjo,T. (1989) A protein binding to the J κ recombination sequence of immunoglobulin genes contains a sequence related to the integrase motif. Nature, 342, 934–937. [DOI] [PubMed] [Google Scholar]
- 21.Hamaguchi Y., Yamamoto,Y., Iwanari,H., Maruyama,S., Furukawa,T., Matsunami,N. and Honjo,T. (1992) Biochemical and immunological characterization of the DNA binding protein (RBP-J κ) to mouse J κ recombination signal sequence. J. Biochem. (Tokyo), 112, 314–320. [DOI] [PubMed] [Google Scholar]
- 22.Oka C., Nakano,T., Wakeham,A., de la Pompa,J.L., Mori,C., Sakai,T., Okazaki,S., Kawaichi,M., Shiota,K., Mak,T.W. and Honjo,T. (1995) Disruption of the mouse RBP-J κ gene results in early embryonic death. Development, 121, 3291–3301. [DOI] [PubMed] [Google Scholar]
- 23.Tun T., Hamaguchi,Y., Matsunami,N., Furukawa,T., Honjo,T. and Kawaichi,M. (1994) Recognition sequence of a highly conserved DNA binding protein RBP-J κ. Nucleic Acids Res., 22, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh J.J.-D. and Hayward,S.D. (1995) Masking of the CBF1/RBPjk transcriptional repression domain by Epstein–Barr virus EBNA2. Science, 268, 560–563. [DOI] [PubMed] [Google Scholar]
- 25.Olave I., Reinberg,D. and Vales,L.D. (1998) The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev., 12, 1621–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao H.Y., Ordentlich,P., Koyano-Nakagawa,N., Tang,Z., Downes,M., Kintner,C.R., Evans,R.M. and Kadesch,T. (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev., 12, 2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh J.J., Zhou,S., Chen,L., Young,D.B. and Hayward,S.D. (1999) CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl Acad. Sci. USA, 96, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurooka H., Kuroda,K. and Honjo,T. (1998) Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res., 26, 5448–5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurooka H. and Honjo,T. (2000) Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem., 275, 17211–17220. [DOI] [PubMed] [Google Scholar]
- 30.Barolo S., Walker,R.G., Polyanovsky,A.D., Freschi,G., Keil,T. and Posakony,J.W. (2000) A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell, 103, 957–969. [DOI] [PubMed] [Google Scholar]
- 31.Tani S., Kurooka,H., Aoki,T., Hashimoto,N. and Honjo,T. (2001) The N- and C-terminal regions of RBP-J interact with the ankyrin repeats of Notch1 RAMIC to activate transcription. Nucleic Acids Res., 29, 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ordentlich P., Lin,A., Shen,C.P., Blaumueller,C., Matsuno,K., Artavanis-Tsakonas,S. and Kadesch,T. (1998) Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol. Cell. Biol., 18, 2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damm K., Thompson,C.C. and Evans,R.M. (1989) Protein encoded by v-erbA functions as a thyroid-hormone receptor antagonist. Nature, 339, 593–597. [DOI] [PubMed] [Google Scholar]
- 34.Flint J. and Shenk,T. (1997) Viral transactivating proteins. Annu. Rev. Genet., 31, 177–212. [DOI] [PubMed] [Google Scholar]
- 35.Spindler K.R. and Berk,A.J. (1984) Rapid intracellular turnover of adenovirus 5 early region 1A proteins. J. Virol., 52, 706–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansieau S., Strobl,L.J. and Leutz,A. (2001) Activation of the Notch-regulated transcription factor CBF1/RBP-Jκ through the 13SE1A oncoprotein. Genes Dev., 15, 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakravarti D., Ogryzko,V., Kao,H.Y., Nash,A., Chen,H., Nakatani,Y. and Evans,R.M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell, 96, 393–403. [DOI] [PubMed] [Google Scholar]
- 38.Wang H.-G.H., Rikitake,Y., Corrigan Carter,M., Yaciuk,P., Abraham,S.E., Zerler,B. and Moran,E. (1993) Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J. Virol., 67, 476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H.-G.H., Draetta,G. and Moran,E. (1991) E1A induces phosphorylation of the retinoblastoma protein independently of direct physical association between the E1A and retinoblastoma products. Mol. Cell. Biol., 11, 4253–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwok R.P., Lundblad,J.R., Chrivia,J.C., Richards,J.P., Bachinger,H.P., Brennan,R.G., Roberts,S.G., Green,M.R. and Goodman,R.H. (1994) Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature, 370, 223–226. [DOI] [PubMed] [Google Scholar]
- 41.Glass C.K. and Rosenfeld,M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- 42.Grozinger C.M. and Schreiber,S.L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl Acad. Sci. USA, 97, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKinsey T.A., Zhang,C.L., Lu,J. and Olson,E.N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature, 408, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirakata Y., Shuman,J.D. and Coligan,J.E. (1996) Purification of a novel MHC class I element binding activity from thymus nuclear extracts reveals that thymic RBP-Jκ/CBF1 binds to NF-κB-like elements. J. Immunol ., 156, 4672–4679. [PubMed] [Google Scholar]
- 45.Issack P.S. and Ziff,E.B. (1998) Altered expression of helix–loop–helix transcriptional regulators and cyclin D1 in Wnt-1-transformed PC12 cells. Cell Growth Differ ., 9, 837–845. [PubMed] [Google Scholar]
- 46.Issack P.S. and Ziff,E.B. (1998) Genetic elements regulating HES-1 induction in Wnt-1-transformed PC12 cells. Cell Growth Differ ., 9, 827–836. [PubMed] [Google Scholar]