Abstract
Background Recent application of molecular-based technologies has considerably advanced our understanding of complex processes in plant–pathogen interactions and their key components such as PAMPs, PRRs, effectors and R-genes. To develop novel control strategies for disease prevention in citrus, it is essential to expand and consolidate our knowledge of the molecular interaction of citrus plants with their pathogens.
Scope This review provides an overview of our understanding of citrus plant immunity, focusing on the molecular mechanisms involved in the interactions with viruses, bacteria, fungi, oomycetes and vectors related to the following diseases: tristeza, psorosis, citrus variegated chlorosis, citrus canker, huanglongbing, brown spot, post-bloom, anthracnose, gummosis and citrus root rot.
Keywords: Citrus immunity, citrus psorosis, tristeza of citrus, citrus variegated chlorosis (CVC), citrus canker, huanglongbing (HLB), brown spot, post-bloom, anthracnose, gummosis, and citrus root rot
INTRODUCTION
Citrus trees are grown in more than 140 countries in tropical and subtropical areas. The long life span of a citrus tree (in some cases more than a century) leads to complex interactions with micro-organisms throughout the soil and above-ground areas (Wang et al., 2015). Citrus production is heavily affected by diseases caused by a diverse range of viruses, bacteria, fungi, oomycetes and nematodes. These diseases increase the cost of production, are responsible for low productivity and involve annual losses of millions of dollars (Neves et al., 2014). In this context, it is essential to expand and consolidate knowledge of citrus interactions with their pathogens to implement new control and/or management approaches. The advancement of molecular biology technologies has greatly expanded our understanding of the complex processes of host–pathogen interactions, both in model herbaceous as well as in woody and perennial plants such as citrus species. The complexity of citrus pathosystems, associated with extensive monospecific or monoclonal plantations, indicates that the chemical control model for disease, pests or vectors, when feasible, cannot be sustainable in the medium and long term.
Knowledge of the biological processes that lead to disease and of how organisms behave during the interactions with their hosts is crucial for proposing new control strategy models. This review provides an update on important aspects of the interaction of citrus with several of their pathogens. Whilst considerable advances have been made, it is also clear that many stages of disease development processes are still not well understood and behave differently than observed in model plant systems.
GENERAL ASPECTS OF PLANT INNATE IMMUNITY
Plant cells have a large number of receptors anchored on the cell surface, which are crucial to sense extracellular signals and for cell-to-cell communication. Pattern recognition receptors (PRRs) act as cellular ‘antenna’ and allow plants to detect a wide range of danger signals including non-self (PAMPs, MAMPs, HAMPs and VAMPs – pathogen, microbe, herbivore and virus-associated molecular patterns) and even self-derived compounds (damage-associated molecular patters or DAMPs), which are released upon herbivore and pathogen attack. The presence of PRRs represents a critical step in host perception and self-defence against attackers by triggering innate immune responses (Jones and Dangl, 2006).
The structures perceived by PRRs are conserved across certain microbe classes and are related to primary functions for their fitness (Medzhitov and Janeway, 1997; Nürnberger and Brunner, 2002). Thus, the genetic factors coding for the recognized pattern molecules are less likely to be mutated or lost during the microbe evolutionary processes. The recognition by PRRs of this set of conserved molecules confers broad-spectrum resistance against microbes sharing the same PAMP. Despite this conservation, some PAMPs are still subject to selection pressure during co-evolution with host plants. The modification of key amino acid residues at the recognition sites allows adapted pathogens to evade perception by PRRs (Boller and Felix, 2009; Monaghan and Zipfel, 2012).
Pattern recognition receptors
Although only a few PRR–PAMP pairs have been identified, all the known PRRs are modular transmembrane proteins and they are either receptor-like kinases (RLKs) or receptor-like proteins (RLPs) containing ligand-binding ectodomains (Goff and Ramonell, 2007; Monaghan and Zipfel, 2012). In general, plant RLKs have a single pass transmembrane (TM) domain for anchorage, a variable N-terminal extracellular domain (ECD) for ligand binding and a C-terminal intracellular kinase domain (KD) that relays downstream signalling (Shiu and Bleecker, 2001). RLKs represent a large and diverse gene family with more than 600 and 1100 members identified in Arabidopsis and rice, respectively (Shiu and Bleecker, 2001; Shiu et al., 2004). The RLKs with leucine-rich repeats as ECDs (LRR–RLKs) constitute the largest subfamily and contain most of the identified PRRs (Goff and Ramonell, 2007). The LRR–RLKs Flagellin Sensing 2 (FLS2) and EF-Tu Receptor (EFR) from Arabidopsis and XA21 from rice represent the best-studied plant PRRs. FSL2 and EFR activate PAMP-triggered immunity (PTI) responses by sensing elicitor epitopes from bacterial flagellin (flg22), elongation factor Tu (elf18), whereas XA21 is elicited by Ax21 (sulfated RaxX) (Boller and Felix, 2009; Pruitt et al., 2015). The Arabidopsis PEPR1 and PEPR2 are LRR–RLKs that trigger PTI defence responses by perceiving as DAMPs the conserved Pep epitopes produced by cleavage of propeptides (PROPEPs) (Yamaguchi and Huffaker, 2011). Although RNA-silencing represents the main resistance strategy against viruses, Arabidopsis NIK1 and NIK2 are LRR–RLKs with an important role in antiviral immunity responses. Instead of LRRs, some RLKs perceive PAMPs by LysM motifs in the ECD (LysM–RLKs), such as CERK1 (Chitin Elicitor Receptor Kinase 1) that has three extracellular LysM domains and triggers PTI by recognizing fungal chitin oligosaccharides (Miya et al., 2007). Plant RLPs also have a TM domain and an ECD but do not have a KD, except for a short cytoplasmic tail lacking any obvious signalling domain. Thus, it must complex with KD proteins to transduce the signals in the cytoplasm after PAMP recognition by the ECD (Shiu and Bleecker, 2003). The Arabidopsis RLPs LYM1 and LYM3 and the rice orthologues LYP4 and LYP6 recognize peptidoglycans as PAMPs but complexes with LysM RLKs are necessary to trigger immunity responses (Zipfel, 2014). Other important PRR RLPs identified are the rice chitin elicitor-binding protein (CEBiP), which recognizes fungal chitin as an elicitor, and the tomato LeEIX1 and LeEIX2, which are able to detect fungal ethylene inducing xylanase EIX as PAMP (Kaku et al., 2006).
PAMP-triggered immunity (PTI)
To activate the PTI response, pathogen structures must be perceived by the PRR ECD, with subsequent signal transduction in the cytoplasm. Several molecules are used by plants to encode signals acquired by pathogen recognition for delivery of information downstream of PRRs to proteins related to signal interpretation and activation of defence response genes (Zipfel et al., 2004; Denoux et al., 2008). PAMP recognition leads to numerous plant signals, including an oxidative burst by the generation of reactive oxygen species (ROS), calcium influx, activation of the mitogen-activated protein kinase (MAPK) cascade, nitric oxide (NO) burst, ethylene production, callose deposition at the cell wall, and expression of defence-related genes involved in immunity responses (Boller and Felix, 2009).
GENERAL ASPECTS OF EFFECTOR-TRIGGERED IMMUNITY
Effectors
Many definitions of effectors are available in the literature. For this review, the following definition will be used: effectors are molecules released/associated with an organism that alters the physiology, structure or function of another organism. Specifically, effectors are pathogen molecules that can modify host cell structures and manipulate function, facilitating infection and/or triggering defence responses. Unlike the terms ‘avirulence’, ‘elicitor’, ‘toxin’ and ‘virulence’, the term effector is neutral and does not imply a negative or positive impact on the outcome of the host–pathogen interaction. Effectors are responsible for promoting pathogen penetration and persistence inside the host tissue, as well as suppression of immune responses, allowing access to nutrients, proliferation and growth (Göhre and Robatzek, 2008).
Common features from well-characterized effectors are used by plant pathologists to search for possible candidate molecules from new and old pathogens. These candidates are usually small secreted proteins, which are rich in cysteine and show no obvious homology to other known proteins (Göhre and Robatzek, 2008). Secreted effectors reach their cellular target either at the intercellular interface of the host and pathogen cells (apoplastic effectors) or inside the host cells (cytoplasmic effectors) (Kamoun, 2006; Djamei et al., 2011).
R-genes
Plant defence through effector-triggered immunity (ETI) is based on the highly specific interaction between products from pathogen avirulence genes (Avr) and products from host resistance genes (R), according to the gene-for-gene hypothesis (Flor, 1971). R proteins can recognize pathogen effectors directly or indirectly through their effects on host cells (Win et al., 2012). Indirect recognition occurs through R protein-mediated monitoring of effector disturbances in distinct host cellular targets of an effector, consistent with the so-called ‘guard hypothesis’ (Dangl and Jones, 2001). Currently, two variations of this model are recognized. In one, the R receptor is constitutively associated with the host intermediate factor, whereas in the other, the pathogen effector first associates with a host target and the complex formed is then recognized by the immune receptor (Caplan et al., 2008; Elmore et al., 2011). The major evidence for the guard hypothesis was obtained in the R/Avr system between Arabidopsis thaliana and Pseudomonas syringae pv. tomato, where the modification of the host factor RIN4 by the bacterial Avr gene product activates the R protein RPM1, resulting in plant resistance (Mackey et al., 2002).
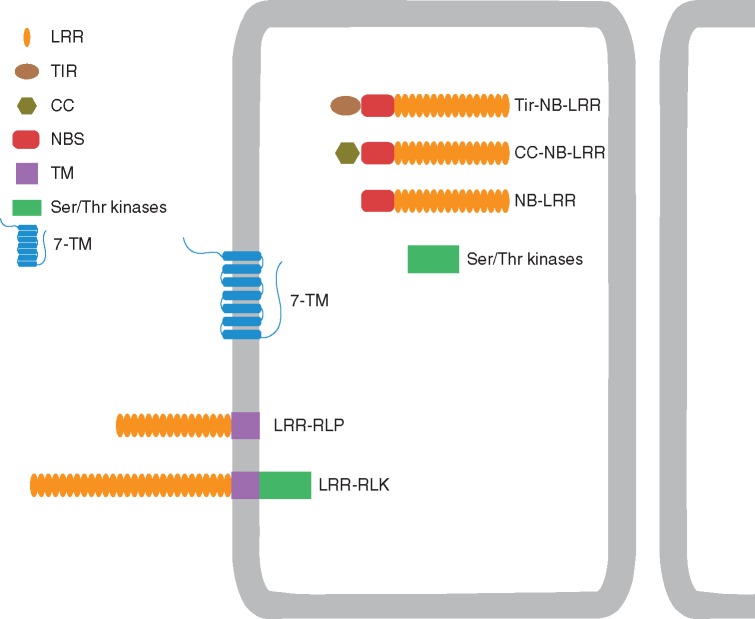
Structurally, R-genes commonly present a central nucleotide-binding site (NBS) domain, a C-terminal LRR region to mediate pathogen recognition and an N-terminal variable domain mainly identified as TIR (Toll/Interleucina-1) or CC (Coiled-coil) (Elmore et al., 2011; Gururani et al., 2012). Besides TIR-NBS-LRR and CC-NBS-LRR, other major classes of R-genes include the RLKs (containing an extracellular LRR, a transmembrane domain and a cytoplasmic kinase domain), RLPs (which are similar to the RLKs but lack the kinase domain) and cytoplasmic enzymatic R-genes that contain neither LRR nor NBS groups (Gururani et al., 2012).
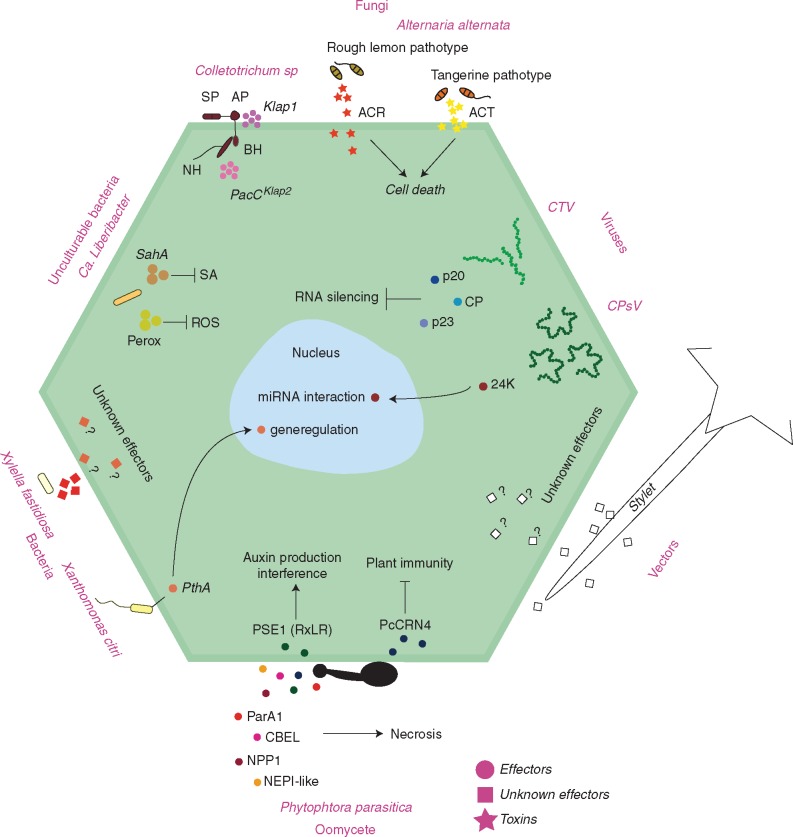
CITRUS–PATHOGEN INTERACTIONS
Citrus overview
The Brazilian citrus industry is one of the most important in the world. It accounts for over a third of the world’s sweet orange production and more than 50 % of the orange juice production, both frozen-concentrated orange juice (FCOJ) and not from concentrate (NFC) juice. São Paulo State is the main producer, processor and exporter of orange juice, and accounts for 80 % of the orange, lime and lemon production and 45 % of the mandarin production in Brazil. From 2005 to 2015, São Paulo was responsible for the production of 97 % of FCOJ, 99·5 % of NFC juice, 95 % of essential oils, and 99·5 % of dried or fresh orange fruit exports in Brazil (Neves et al., 2014).
Although Brazil is the world’s largest producer of fresh oranges and orange juice, productivity of the Brazilian citrus industry is considered very low (approximately two boxes per tree per year, each box with 40·8 kg). Low yield is associated with high incidence of pests and diseases, reduced genetic diversity of scions and rootstocks, and production in non-irrigated areas.
It has been estimated that more than 60 % of the costs of citrus production in Brazil are associated with control of pests and diseases. Diseases such as citrus variegated chlorosis (CVC), leprosis, black and brown spot (CBS), sudden death, citrus canker, gummosis, root rot, tristeza and huanglongbing (HLB) are the more important diseases seen in orchards (Machado et al., 2011).
When a disease is associated with rootstock, its replacement by more resistant material has been the most effective method of control. However, when the disease mainly affects the canopy, replacement with a more resistant cultivar is not always feasible, either because there is no available resistance source or the resistant cultivar is not acceptable to the market (Machado et al., 2011). Moreover, the complexity of citrus pathosystems is very high and often involves pathogens that invade plants systemically and with highly efficient vectors. Genetic improvement-based control is possible, but involves long and costly selection and evaluation programmes, with genetic barriers that can hinder breeding.
Pattern recognition receptors in citrus
Despite the large number of disease-causing pathogens in citrus, no PRR has yet been functionally well characterized. However, some RLKs and RLPs have been identified with roles in pathogen perception in innate immunity responses (Fig. 1). Rodrigues et al. (2013) analysed RNA-Seq data for CVC in resistant mandarin (Citrus reticulata), infected with Xylella fastidiosa. They identified, in addition to a leucine-rich repeat receptor-like protein (RLP12), two other up-regulated genes (Ciclev10004108m and Ciclev10014130m) similar to LRR-RLKs, which might be related to PTI responses activated by X. fastidiosa PAMP recognition. Moreover, a novel RLK with a lectin domain (Lec) in the ECD was isolated from C. limon in response to the fungus Capnodium citri, the causal agent of sooty mould. Investigations of this interaction suggest a defence mechanism against pathogens mediated via a signal transduction pathway which can be modulated by a PRR (De Felice and Wilson, 2009).
Fig. 1.
Domain structures of Citrus PRRs and R-genes.
Transient expression of the flagellin- and hook-associated protein (Fla) from the bacterium Candidatus Liberibacter asiaticus (CaLas), which causes HLB citrus disease, induced cell death, callose deposition and up-regulation of BAK1 transcripts in Nicotiana benthamiana. The conserved domain flg22Las also triggered different degrees of PAMP activity in citrus plants, suggesting that FlaLas acts as a PAMP and can be recognized in citrus in addition to N. benthamiana (Zou et al., 2012). The conserved flg22 derived from Xanthomonas citri subsp. citri, the bacterial agent of citrus canker disease, also triggered rapid ROS production and induction of PTI marker genes, mainly in the group of more resistant citrus genotypes (Shi et al., 2015). These results suggest a citrus PRR that is able to recognize flagellin exists and might be important in triggering resistance against HLB and X. citri subsp. citri. Functional orthologues of FLS2 with different perception specificities were previously characterized in other plants such as tomato, rice, grapevine and N. benthamiana (Robatzek et al., 2007; Takai et al., 2008; Chakravarthy et al., 2010). Genetically engineered plants with overexpression of PRRs provide a promising strategy to increase plant immunity. Transgenic C. sinensis overexpressing rice Xa21 showed increased resistance to citrus canker. Recently, a reduced susceptibility to X. citri was also demonstrated in citrus plants expressing the FLS2 receptor from N. benthamiana (Hao et al., 2016).
The Citrus EST project (CitEST) consists of an expressed sequence tag (EST) database obtained from citrus species under a diverse range of conditions, including stresses caused by the main pathogens (Targon et al., 2007). This database represents a useful source of genomic information for further understanding of host defence mechanisms such as those associated with innate immunity responses. Guidetti-Gonzalez and Carrer (2007) conducted in silico analyses with CitEST and identified genes with similarity to the RLP ethylene-inducing xylanase EIX1 in C. sinensis infected with X. fastidiosa, suggesting a specific role against bacterial pathogens rather than associated with fungal response. Moreover, contigs and singletons similar to Xa21 and Xa26 rice receptors were identified in C. sinensis, C. reticulata and Poncirus trifoliata in response to X. fastidiosa and Citrus tristeza virus (CTV). Functional characterization of the available data from CitEST is necessary to advance our understanding of the mechanisms associated with citrus innate immunity responses.
R-genes in citrus
A recent genome-wide comparative analysis identified NBS genes in the genomes of C. sinensis and C. clementine (Y. Wang et al., 2015). The authors found 618, 650 and 508 NBS genes in C. clementina, C sinensis China and C. sinensis USA, respectively. However, the typical TIR- and CC-NBS-LRR classes of R-genes correspond to only 32 % of the total NBS genes in C. clementina, 29 % in C. sinensis China and 18 % in C. sinensis USA genomes. Phylogenetic analysis discriminated Citrus NBS genes into three groups with highly variable C-terminal LRR motifs, responsible for recognizing pathogen effectors, which implies different roles for the groups in the citrus immune system. The majority of Citrus NBS genes are physically clustered in the genome. Most clusters contain genes from the same phylogenetic group; genes in the same cluster tend to be on the same strand, which indicates that the expansion of NBS genes in Citrus is primarily due to tandem duplication. Furthermore, both the hybrid C. sinensis and the original C. clementina have similar numbers and types of NBS genes, consistent with their derivation from a common ancestor.
Besides this screening at the genome level, a first attempt to identify genes coding for resistance proteins in the citrus transcriptome was accomplished using CitEST. In the CitEST database, 259 contigs and 332 singletons related to R-genes were identified, wherein a total of 137 R-genes showed similarities to different categories including NBS-LRR, CC-NBS-LRR, TIR-NBS-LRR, cytoplasmic Ser/Thr kinases and the seven-transmembrane (7-TM) family of resistance proteins. Although some of those sequences were present in citrus libraries from healthy and infected samples, most came from plants challenged with pathogens. The large number of expressed putative R-like genes found in the CitEST database, mainly in pathogen-infected libraries, suggests that they function as resistance genes in citrus (Guidetti-Gonzalez and Carrer, 2007). The structures of these R-genes are shown in Fig. 1.
Other evidence for the involvement of R-genes in the citrus defence response to pathogens comes from global gene expression analyses. In CVC-resistant mandarins (C. reticulata Blanco), one gene encoding an NBS-LRR-like disease resistance protein was up-regulated 30 d after inoculation with X. fastidiosa, indicating that some bacterial signals are recognized by the plant, triggering defence mechanisms to prevent disease (Souza et al., 2007). Furthermore, the CC-NBS-LRR gene was up-regulated in mandarin 1 d after infection with X. fastidiosa (Rodrigues et al., 2013). Likewise, in citrus hybrids resistant to Phytophthora parasitica infection, both the TIR-NBS-LRR RPS4 gene and another R-gene of the same class were up-regulated, indicating that these genes may be involved in the recognition of effectors produced by P. parasitica, thus inducing the plant defence system (Boava et al., 2011).
Finally, some citrus R-genes were also functionally validated, such as the Ctv-R, a CC-NBS-LRR gene, which confers resistance to CTV in P. trifoliata. Ctv-R incorporation into susceptible plants results in different levels of resistance to CTV infection, confirming its role as a disease resistance protein (Rai, 2006). It remains to be demonstrated whether the large number of putative Citrus R-genes and their allelic variants identified in both genome and transcriptome studies effectively promote defence in resistant plants against the wide range of citrus pathogens.
CITRUS–VIRUS INTERACTIONS
In contrast to other plant–pathogen systems, the primary plant immune strategy against viruses is RNA silencing (Incarbone and Dunoyer, 2013), a mechanism for control of both gene expression and viral infection mediated by the action of small interfering RNAs (siRNAs). Antiviral RNA silencing is triggered by double-stranded RNA (dsRNA) replication intermediates or structures within RNA viral genomes. These viral dsRNAs are recognized by the RNAseIII endonuclease Dicer (DCL) that processes them into virus-derived siRNAs. A strand of these molecules is incorporated into an RNA-induced silencing complex (RISC) containing an Argonaute (AGO) protein, which finally guides sequence-specific silencing of the homologous viral genome. This mechanism of antiviral control is highly efficient, since the target sequence is dictated by the virus itself, which in turn cannot evolve to avoid sequence-based recognition (Obbard et al., 2009). However, viruses have developed a means to counteract the RNA silencing: viral suppressors of RNA silencing (VSRs). The VSRs are present in most, if not all, plant viruses, presenting different modes of action to target many steps of the RNA silencing pathway (Incarbone and Dunoyer, 2013).
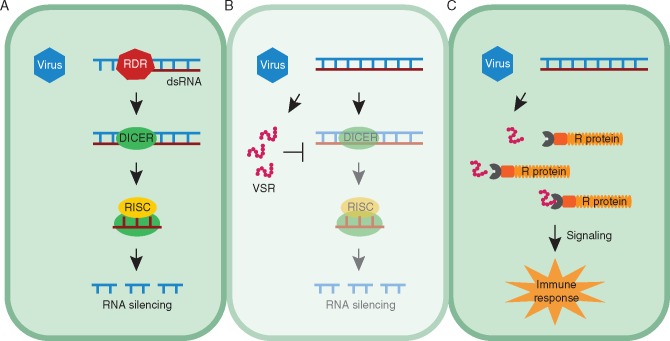
A remarkable parallel can be observed between the activation and suppression of RNA silencing on the one hand and the classic zig-zag scheme for PTI/ETI resistance on the other (Fig. 2). In fact, it has been accepted that these processes are manifestations of the same phenomenon (Incarbone and Dunoyer, 2013). In this regard, virus-derived dsRNA can be considered a VAMP, because it constitutes a mandatory pattern in RNA virus replication. The silencing machinery, composed of DCL and RISC, forms the first line of defence that recognizes those patterns, similar to PTI. VSRs are the virulence effectors that overcome RNA silencing, triggering ETS. As expected for pathogen effectors, VSRs are highly diverse, involving many different strategies to overcome plant defence, and are under strong selection, evolving much faster than other viral genes (Obbard et al., 2009). As a consequence, plants could present R-proteins capable of perceiving VSR effects and triggering typical outputs of ETI, such as the hypersensitive response (HR) (Incarbone and Dunoyer, 2013).
Fig. 2.
Parallel between the classical pattern-triggered immunity (PTI)/effector-triggered immunity (ETI) response and the framework of RNA silencing activation and suppression. (A) Upon virus attack, RNA-dependent RNA polymerases (RDR) produce dsRNA, a virus-associated molecular pattern (VAMP). Similar to PTI, the RNA silencing machinery coordinated by Dicer and RNA induced silencing complex (RISC) recognize and process viral PAMPs, forming the first layer of defence. (B) Viruses have acquired viral supressors of RNA silencing (VSRs) as effectors that suppress host defence, resulting in effector-triggered susceptibility (ETS). (C) In turn, plants developed resistance (R) proteins that recognize viral effectors and activate ETI.
Citrus–Citrus tristeza virus interaction
Our current understanding of the mechanisms involved in Citrus–virus interactions is mainly focused on CTV, a positive-sense single-stranded RNA (ssRNA) virus member of the genus Closterovirus. Depending on the virus isolate and the variety/rootstock combination, CTV can cause any of four distinct syndromes in citrus plants: decline (plant death), stem pitting (aberrant phloem development, resulting in pits in the wood), seedling yellows (stunting and leaf chlorosis) and, most commonly, a complete lack of symptoms, even when the virus multiplies to high titres (Dawson et al., 2013). In the last century, due to the massive use of the decline-sensitive sweet orange/sour graft orange combination, CTV caused losses of over 100 million trees worldwide, becoming the most economically important virus affecting the citrus industry (Moreno et al., 2008).
CTV infection is restricted to phloem-associated cells, resulting in limited cell-to-cell and long-distance movement. Host interference in both virus infection mechanisms is dependent on the citrus genotype (Dawson et al., 2013). The virus systemically infects its hosts using only the long-distance movement from source-to-sink, with cell-to-cell movement absent or limited to only small clusters of adjacent cells even in the more susceptible citrus species (Folimonova et al., 2008). This distribution within the host is thought to be related to the interaction of virus gene products with specific hosts (Dawson et al., 2013).
To reduce CTV titre and systemic infection, citrus species employ RNA silencing. In turn, CTV has developed three effectors that exhibit VSR activities: p20, p23 and coat protein (CP) (Lu et al., 2004). The p20 and CP proteins suppress intercellular silencing, preventing the spread of the silencing signal and probably the activation of host defences, while p20 and p23 suppress intracellular silencing, reducing viral degradation. Given that RNA silencing is a central host defence to contain viral replication and even restrict the virus to phloem cells, the constitutive expression of p23 has been reported to increase the CTV titre in sour orange and to allow CTV to escape from the confines of the phloem in both sour and sweet orange (Fagoaga et al., 2011). However, even with the establishment of a successful systemic infection, some degree of CTV genome silencing still occurs, suggesting that RNA silencing cannot completely inhibit viral replication and infection and that the three effectors cannot completely block RNA silencing. The constant arms race between the virus and its hosts (Obbard et al., 2009; Dawson et al., 2013) may have led to this balanced co-evolutionary process, so that the virus remains in the host without causing severe symptoms or plant death.
Besides the three effectors with VSR activities required for CTV to overcome host resistance, the virus has other non-conserved genes – p33, p18 and p13 – that are not needed for infection of most CTV hosts, but are necessary in different combinations for infection of certain citrus species (Tatineni et al., 2011). It has been suggested that CTV acquired these non-conserved genes for movement and overcoming host resistance, further extending its host range (Dawson et al., 2013).
Notably, the CTV effectors identified (Table 1) are not only required to suppress host defences and establish infection, but can also be involved in induction of disease symptoms. For example, the balance between expression of p33, p13 and p18 determines the severity of the stem-pitting symptom: deletion of different combinations of these genes can induce large increases in stem pits (Tatineni and Dawson, 2012). Similarly, ectopic expression of the VSR p23 induces virus-like symptoms (Flores et al., 2013). For other described pathosystems, it has been shown that ectopic expression of VSRs alters the plant small RNA regulatory pathway, inducing symptoms (Pacheco et al., 2012). Changes in the accumulation patterns of miRNAs have also been reported in CTV-infected citrus plants (Ruiz-Ruiz et al., 2011), suggesting that suppression of host RNA silencing defences by CTV also affects the plant small RNA regulatory pathway, resulting in symptom expression.
Table 1.
Effectors from citrus viruses and cognate R proteins from citrus species
| Virus | Effector | R protein | Reference |
|---|---|---|---|
| Citrus tristeza virus | p20 | – | Lu et al. (2004) |
| p23 | – | ||
| Coat protein (CP) | – | ||
| p33 | – | Tatineni and Dawson (2012) | |
| p18 | – | ||
| p13 | – | ||
| – | Ctv-R (Poncirus trifoliata) | Rai (2006) | |
| Citrus psorosis virus | 24K | – | Reyes et al. (2016) |
Although CTV effectors are known, no corresponding plant R-gene has been identified. However, a CC-NB-LRR R protein with an unknown corresponding Avr CTV gene has been characterized (Rai, 2006) (Table 1). The locus, Ctv-R, is a single dominant gene from P. trifoliata, which confers broad-spectrum resistance to the majority of CTV isolates. Sequence analysis of the Ctv genomic region located the locus in a 121-kb region comprising ten genes. Susceptible grapefruit plants transformed with some of these ten candidate Ctv-R genes result in different levels of resistance, such as an absence of initiation of infection, its slow spread or an initial appearance of infection followed by its subsequent eradication (Rai, 2006). However, some of the viral proteins recognized by NB-LRR are not VSRs (de Ronde et al., 2014). Therefore, the CTV protein recognized by CTV-R may not necessarily be analogous to other effector proteins that suppress PTI.
Citrus–Citrus psorosis virus interaction
Besides CTV, information on citrus molecular interactions with other infecting viruses is still scarce. However, the RNA silencing mechanism has been suggested to be involved in the citrus response to Citrus psorosis virus (CPsV), a negative-stranded RNA virus from the genus Ophiovirus and causal agent of psorosis disease (Achachi et al., 2014). In CPsV-infected plants, higher temperatures promote attenuated symptoms, reduce levels of viral RNA and increase virus-derived siRNA. Previous work revealed that RNA silencing is weaker at low temperatures and stronger at high temperatures (Chellappan et al., 2005). Thus, the impairment of CPsV infection may be due to the temperature-induced enhancement of RNA silencing (Velázquez et al., 2010).
To date, a VSR from CPsV to counteract viral silencing has not been characterized. However, a recent study demonstrated that infection by CPsV promotes a down-regulation of C. sinensis endogenous micro-RNAs (miRNAs) (mainly miR156 and miR171) and a consequent up-regulation of its target genes (the transcription factors Squamosa promoter-binding protein-like, SLP, and Scarecrow-like, SCL) (Reyes et al., 2016). Modulation of the miRNA pathway by plant viruses as a result of VSR activities has already been described and is thought to be a viral strategy to bypass host defences and induce symptoms (Jay et al., 2011; Padmanabhan et al., 2013). The up-regulated targets, SPL and SCL, are involved in the activation of programmed cell death and in the decrease of chlorophyll biosynthesis, respectively; thus, large amounts of both transcripts may contribute to the necrosis and chlorosis symptoms manifested by citrus plants infected by CPsV (Reyes et al., 2016). Furthermore, the authors demonstrated that the CPsV 24K protein physically interacts with pre-miR156 and pre-miR171, suggesting the protein is responsible for altering precursor processing and subsequent biogenesis of miRNAs (Reyes et al., 2016). Once the 24K protein affects the miRNA pathway, whose components are shared by antiviral silencing, and induces the expression of genes probably involved in disease symptom development, the protein may be considered a potential VSR and effector of CPsV (Table 1).
CITRUS–BACTERIA INTERACTIONS
Citrus–Xylella fastidiosa interaction
Xylella fastidiosa is a Gram-negative bacterium that causes CVC disease in sweet orange and Pierce’s disease (PD) in grapevine, and infects other economically important crops (Hopkins and Purcell, 2002; Bové and Ayres, 2007). Xylella fastidiosa is limited to xylem vessels and its transmission under field conditions occurs via insect vectors (sharpshooters). Once in the susceptible citrus plant, X. fastidiosa systemically colonizes the xylem vessels forming a biofilm. The resultant sap flow blockage in vessels by biofilm has been suggested as the main factor associated with X. fastidiosa pathogenicity. In C. sinensis, leaf symptoms are described as yellow spots on the adaxial surface that can develop into necrosis as the disease progresses (Souza et al., 2009).
In fruits, the disease promotes size reduction and premature ripening, which has been responsible for losses of millions of dollars in citrus agribusiness (Bové and Ayres, 2007).
Given the economic importance of this bacterium, several studies have been conducted to understand the biology of X. fastidiosa, including functional gene studies, and investigation of the mechanisms of tolerance to antimicrobial compounds and bacterial colonization (Rodrigues et al., 2008; Caserta et al., 2010; Muranaka et al., 2012). However, few studies have focused on the interaction of X. fastidiosa and citrus, especially regarding the role of effectors in disease development.
In C. reticulata (resistant to X. fastidiosa), Coletta-Filho et al. (2007) demonstrated that X. fastidiosa can survive in the initial stages of infection in this host, suggesting that the resistant plant recognizes X. fastidiosa in some way, and triggers the plant defence response. To better understand this interaction, EST libraries were created using sweet orange with and without CVC symptoms and mandarin inoculated with X. fastidiosa (Gmitter et al., 2012). Data analysis showed that genes associated with a defence response are also up-regulated in susceptible plants, but primarily when the bacteria have already colonized the plant, showing that these genes are induced but at later stages of infection. In contrast, in resistant plants, different sets of genes are up-regulated at different time points during the interaction. At initial stages of infection, the induced genes were related to pathogen recognition, signal transduction and defence. At the second time point, the induced genes were associated with signal transduction (MAPK cascade) and with a defence response, including ethylene-related transcription factor, LOX gene associated with the jasmonic acid pathway, and S-adenosyl-l-methionine: salicylic acid methyltransferase (Gmitter et al., 2012). These findings reinforce the hypothesis that the resistant host triggers defence genes after recognition of X. fastidiosa. Indeed, Rodrigues et al. (2013) verified by RNA-seq analysis that genes associated with PTI are induced in C. reticulata 1 d after X. fastidiosa inoculation. Such genes are putative PAMP receptors, genes associated among others with the formation of secondary xylem, cell-wall synthesis and ROS, suggesting that this plant was able to recognize unknown PAMPs of X. fastidiosa and, consequently, activated defence responses. Although no PAMPs for X. fastidiosa have yet been characterized, Kunze et al. (2004) demonstrated elicitation activity of EF-Tu peptide from X. fastidiosa in an alkalinization assay. However, the elicitation was much lower when compared with other peptides from different bacteria. Additional studies for verification that EF-Tu is a bona fide PAMP for X. fastidiosa are warranted.
Another gene induced identified following RNA-seq and EST analyses is a CC-NBS-LRR gene (Gmitter et al., 2012; Rodrigues et al., 2013). The CC-NBS-LRR genes have been reported as cytoplasmic receptor proteins, which usually recognize effector proteins triggering ETI. Although no effector has been described so far in the X. fastidiosa CVC strain, in the PD strain some putative effectors were recently reported (Zhang et al., 2015; Nascimento et al., 2016). Among them, the LipA/LesA gene (PD1703) (Zhang et al., 2015; Nascimento et al., 2016) was characterized as a lipase/esterase and was identified as a key gene for pathogenesis of X. fastidiosa in grapevine (Nascimento et al., 2016). A loss-of-function lesA mutant produced far fewer symptomatic leaves when compared with the wild-type infection. In the CVC strain there is a homologue of the LipA/LesA gene in its genome (XF0357) (Nascimento et al., 2016); however, whether this gene has a role in the pathogenesis of X. fastidiosa in citrus needs to be verified. These results suggest that X. fastidiosa might be secreting effectors by alternative systems. The type III secretion system (T3SS), which is classically associated with secretion of effectors in other bacteria, is lacking in X. fastidiosa strains. Thus, the CC-NBS-LRR protein could be recognizing some yet unknown effector and, consequently, leading to plant defence responses (Rodrigues et al., 2013).
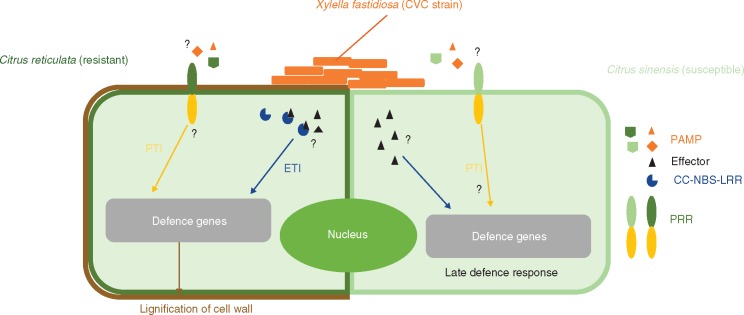
Rodrigues et al. (2013) reported that the resistance mechanism of C. reticulata could be associated with reinforcement of xylem cell walls, since expression of auxin-related genes associated with cell-wall modification was up-regulated in the initial stage of infection. Curiously, this kind of resistance response is similar to that occurring with necrotrophic pathogens in the early stages of infection, where PAMPs, mediated by cell-wall degradation, can be recognized by the plant host, triggering an immunity response. Even though X. fastidiosa is not a necrotrophic organism, this bacterium can degrade plant cell walls in xylem vessels (Pérez-Donoso et al., 2010) and be recognized by the plant host through some as yet unknown mechanism. Consistent with this hypothesis, Niza et al. (2015) recently showed that X. fastidiosa remains trapped in primary xylem of resistant plants due to lignin accumulation that coincides with the initial stage of infection (Fig. 3). In susceptible hosts, the bacterial strains were able to colonize primary and secondary xylem vessels, with lignification of primary xylem cells delayed and occurring later after infection, with no impairment of bacterial spread in the plant. Thus, the induction of lignification is suggested to be a physical defence response to X. fastidiosa infection in resistant plants, preventing the movement of this bacterium in the plant.
Fig. 3.
Schematic representation of X. fastidiosa interaction with resistant and susceptible genotypes.
Citrus–Xanthomonas interactions
Xanthomonas is a genus of phytopathogenic Gram-negative bacteria that are known to cause disease in more than 200 plant families, including many economically important crops, such as rice, tomato, and citrus. Xanthomonas citri is the etiological agent of citrus canker, one of the most devastating diseases that affects citrus orchards worldwide. Initially, X. citri grows on leaf surfaces in structured biofilms (Rigano et al., 2007) and it then enters the plant through stomata or injuries, and colonizes the mesophyll parenchyma.
Citrus canker infection is characterized by raised water-soaked lesions, which further progress to form the cankers. When trees are severely affected, infections cause premature fruit drop and defoliation, resulting in significant yield losses (Brunings and Gabriel, 2003). To date, there is no cure for citrus canker, and preventive copper spray application is one of the unique control measures that can be adopted, other than plant eradication (Behlau et al., 2014).
There are three main types of citrus canker described, based on the strains that cause disease and the aggressiveness of the symptoms. Type A is the most aggressive cancrosis and is caused by X. citri (syn. X. axonopodis pv. citri). It originated in Asia and is able to infect all citrus varieties without any true resistance being recognized, despite the field tolerance described for some citrus genotypes such as ‘Muscia’ (C. reticulata) (Brunings and Gabriel, 2003; Carvalho et al., 2015). Two other variants of Type A citrus canker are known: A*, which was first described in southern Asia in 1998, and Aw (Wellington strain), which was isolated in Florida in 2003. Both Aw and A* have a narrow host range, infecting only Mexican lime (C. aurantifolia) and alemow (C. macrophylla) (Vernière et al., 1998; Sun et al., 2004). Type B is also known as ‘false canker’ and was first described in Argentina in 1923 but has also been detected in Uruguay and Paraguay. It is virtually restricted to C. limon, although it can cause mild infections in all citrus varieties. Type C was isolated in São Paulo (Brazil) and, like Aw and A* strains, it is restricted to C. aurantifolia (Moreira et al., 2010). Cancrosis of both Types B and C are caused by Xanthomonas fuscans subsp. aurantifolli strains.
Lipopolysaccharide (LPS) and adhesin are virulence factors that can be recognized as PAMPs. LPS has a role in the activation of basal defences in both host (C. sinensis ‘Valencia late’) and non-host (Nicotiana tabacum ‘Petit Havana’) plants (Petrocelli et al., 2012). Similarly, the adhesin XacFhaB was also involved in the triggering of plant defence responses. The role of the three XacFhaB regions as PAMPs was investigated and all adhesin regions were able to induce basal immune responses in host and non-host plants. When citrus leaves were pre-infiltrated with XacFhaB regions, an inhibition of X. citri growth was observed, confirming the induction of defence responses and control of citrus canker (Garavaglia et al., 2016).
Another molecule recognized as a PAMP is flagellin. Studies using flg22 of X. citri (flg22Xcc) 306 have shown that, in susceptible citrus genotypes ‘Duncan’ grapefruit and ‘Navel’ orange, no significant ROS production or PTI/ETI was associated with flg22Xcc treatment. However, resistant genotypes ‘Nagami’ kumquat and ‘Sun Chu Sha’ showed higher ROS production following flg22Xcc treatment (Shi et al., 2015), indicating that these plants are able to recognize this PAMP (Fig. 4).
Fig. 4.
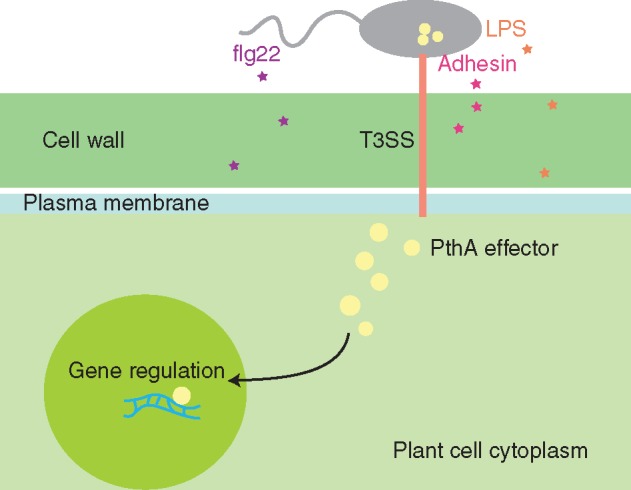
PAMPs and effectors in X. citri. The known PAMPs for X. citri flagellin (flg22), adhesion and lipopolysaccharides (LPS) are present and enter the host cell. The T3SS-delivered effectors, such as PthA4 and its homologues, are injected into the host cell and travel to the nucleus, where they can act as transcriptional regulators.
Most Xanthomonas genomes sequenced to date have the hrp/hrc gene cluster, which render these cells capable of assembling a fully functional T3SS (Ryan et al., 2011). The needle-like structure formed by the T3SS proteins is responsible for the delivery of effectors directly inside the host cell (Fig. 4). All citrus canker-causing strains have similar T3SS gene clusters (Moreira et al., 2010; Neha Jalan et al., 2013) but there are differences among their effector pool (Moreira et al., 2010). The presence of the T3SS is necessary for full virulence of X. citri, which has been demonstrated in many studies (e.g. Laia et al., 2009). Therefore, the T3SS secreted effectors will be presented in more detail below.
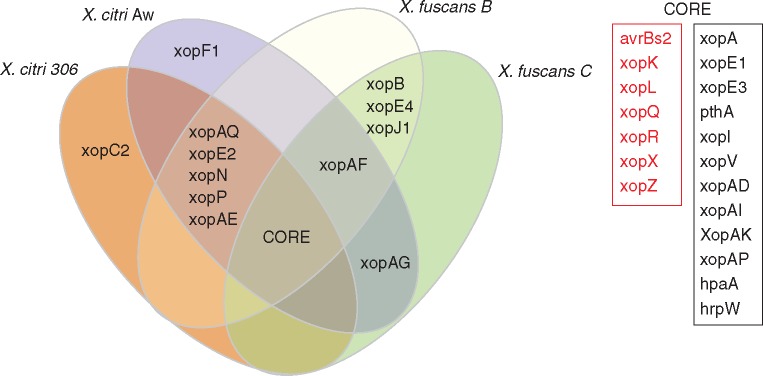
Comparative analysis of the genomic sequence of four citrus canker-causing Xanthomonas [X. citri 306 (da Silva et al., 2002) X. citri Aw (Jalan et al., 2013) and X. fuscans B and C (Moreira et al., 2010)] reveals that they have 19 common effectors, which are therefore considered as the ‘core’ citrus canker effector pool. Among these, seven are also found in all other Xanthomonas genomes (avrBs2, xopK, xopL, xopQ, xopR, xopX and xopZ), and are considered the core effector set for this genus. The other 12 genes (xopA, xopE1, xopE3, pthA4 and/or its functional homologues, xopI, xopV, xopAD, xopAI, xopAK, xopAP, hpaA and hrpW) were found in the four citrus canker strains mentioned above (Neha Jalan et al., 2013).
The genes xopAF and avrGf1 (=xopAG) are completely absent only in X. citri 306 (Neha Jalan et al., 2013). XopAF seems to promote the growth of X. citri Aw in Mexican lime (C. aurantifolia), contributing to virulence, while the presence of the AvrGf1 effector (= XopAG) is responsible for the HR observed in grapefruit (C. paradisi), restricting the host range of X. citri Aw (Rybak et al., 2009; Escalon et al., 2013; Jalan et al., 2013). Deletion of avrGf1 enabled X. citri Aw to colonize grapefruit even though the symptoms were less severe than those caused by X. citri 306 (Rybak et al., 2009). In contrast, for sweet orange, symptoms were not visible in deletion mutants infecting this cultivar, suggesting that other factors may be acting in this host range restriction (Neha Jalan et al., 2013). A similar effector (AvrGf2) is found in X. fuscans C, also inducing ETI in grapefruit (Gochez et al., 2015). Interestingly, X. fuscans B has an almost identical gene coding for avrGf2, but its sequence is interrupted by a transposon, indicating that its ability to infect grapefruit may be due to the absence of a fully functional protein (Moreira et al., 2010).
Besides XopAG, Moreira et al. (2010) found other gene-coding effectors that occur in X. fuscans strains but are absent in the X. citri 306 genome, such as xopB, xopE4, xopJ (avrXccB) and xopAF (avrXv3). The gene sequence of xopE4 is similar to avrXopE3, but due to its low amino acid sequence identity (31 %), it was considered a different effector. Another effector gene (avrXccA2) was found only in a few strains of X. fuscans, but it was not found in the two sequenced strains (X. fuscans B and C). In addition, X. fuscans C is uniquely depleted in effectors such as xopE2, xopN, xopP, xopAE (Moreira et al., 2010) and XopAQ (Neha Jalan et al., 2013). A general overview of the T3SS-delivered effectors is shown in Fig. 5.
Fig. 5.
General overview of T3SS-delivered effectors found in citrus-canker causing Xanthomonas species. The centre of the Venn diagram represents the core effectors found among these genomes (see box on the right), with these Xanthomonas core effectors shown in red.
One of the most important T3SS-delivered effectors found among the citrus canker strains is PthA4 and its homologues. This protein is a T3SS effector delivered inside the plant cell and is a key effector responsible for canker development. Its presence alone is capable of inducing canker formation, while its absence suppresses the appearance of cankers (Al-Saadi et al., 2007). PthA and its homologues are members of the transcription activator-like (TAL) effector family of proteins (formerly known as the AvrBs3/PthA family).
The TAL effectors are proteins that can control gene expression of the host cell they are delivered into, where they can enter the cell nucleus and act as transcriptional regulators favouring pathogen development. Therefore, TAL effectors must target specific DNA sequences which can be accomplished due to the presence of conserved (almost identical) repeats that include a repeat variable di-residue (RVD). Each of the RVDs recognize one nucleotide and the juxtaposed RVDs target a given DNA sequence (Streubel et al., 2012).
Xanthomonas citri 306 has four nearly identical copies of the TAL-effector PthA4, all present on plasmids, but only one of them, PthA4, is the main effector involved in canker formation (Swarup et al., 1992). This effector does not determine host range in citrus species and varieties, but does restrict this pathogen to citrus. It is recognized by non-host plants, triggering ETI in all non-citrus plants tested to date (Swarup et al., 1992).
PthA homologues are known to elicit the classical symptoms of citrus canker, which are hyperplastic and hypertrophic water-soaked lesions that become thicker and darker, ultimately causing the epidermis to rupture, spreading the bacteria (Brunings and Gabriel, 2003). Recently, among other possibilities, one of the targets of PthA4 was found to be the plant gene CsLOB1 (a member of the lateral organ boundaries family of transcription factors), which forms pustules when over expressed. Interestingly, the activation of this gene was observed with all the PthA4 variants found in citrus canker-causing Xanthomonas (Li et al., 2014). However, not only CsLOB seems to be controlled by PthA effectors, and the regulation of many genes is possibly required, with the dioxygenase gene (DIOX) being another possible candidate (Abe and Benedetti, 2016). Although PthA4 is indeed the eliciting factor needed for canker formation, the other PthA copies present in X. citri 306 seem to have an important role in canker development, especially in some citrus varieties (Abe and Benedetti, 2016).
Citrus–HLB interaction
HLB is a century-old disease that has emerged as the most destructive citrus disease worldwide in the past few decades (Wang and Trivedi, 2013). HLB is associated with three phloem-limited Gram-negative bacteria belonging to the genus Liberibacter (Bové, 2014). No efficient method for cultivation has been found to isolate these bacteria to date, and thus they remain in the ‘Candidatus’ status (Fleites et al., 2014). HLB-associated ‘Candidatus Liberibacter’ are named according to the place they were first detected: ‘Candidatus Liberibacter asiaticus’ (CaLas – Asia), ‘Candidatus Liberibacter africanus’ (CaLaf – Africa) and ‘Candidatus Liberibacter americanus’ (CaLam – America) (Garnier et al., 2000; Wang and Trivedi, 2013). Owing to its wide distribution, ‘CaLas’ has been the most studied bacteria of the genus ‘Ca. Liberibacter’ related to citrus HLB. Diaphorina citri and Tryoza eritrea are the main insect vectors of the bacteria associated with HLB disease; the first one is responsible for transmission of ‘CaLas’ and ‘CaLam’ in Asia and America, and T. eritrea transmits ‘CaLaf’ in Africa (Bové, 2014).
Due to its negative effects on citrus yield, the disease is studied using different approaches with the aim of developing HLB-resistant citrus varieties. Several genome sequences were obtained from isolates of Liberibacter species (da Graça et al., 2016). These sequences have been extensively explored and have led to new studies on the biology of the pathogen.
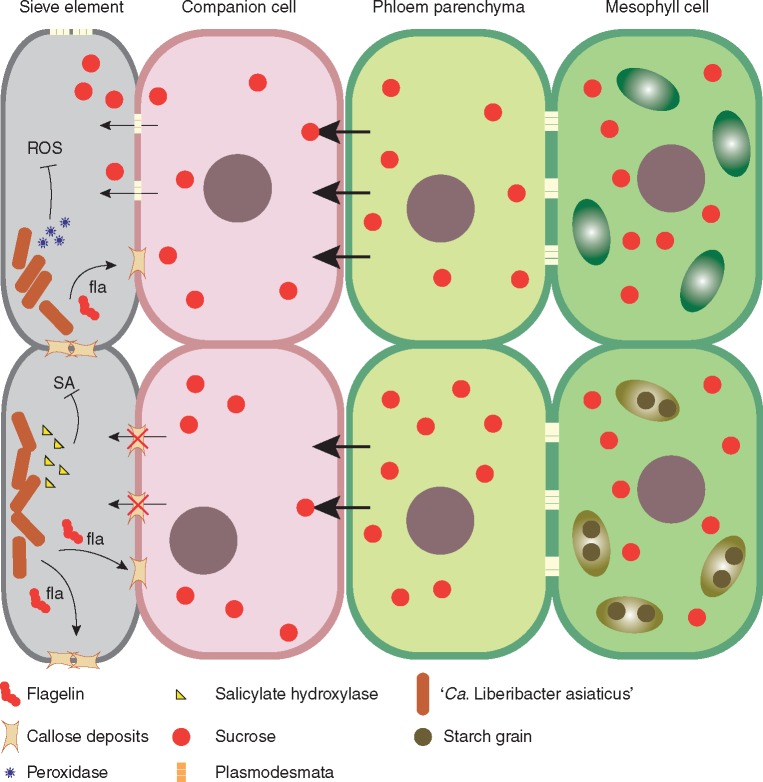
Thus far, no conclusive pathogenicity mechanism of the ‘Ca. Liberibacter spp.’ has been identified (da Graça et al., 2016). Phloem dysfunction appears to be the primary alteration that might determine the emergence of other symptoms (Koh et al., 2012). When studying this alteration, Zou et al. (2012) showed that ‘CaLas’ encodes a flagellin and a hook-associated protein (fla) with PAMP activity. In this research, a synthetic flg22Las peptide induced callose deposition in N. benthamiana, although with a weaker response than observed in other well-characterized plant–pathogen systems. In microscopic analysis of ‘CaLas’-infected citrus, accumulation of callose was observed in sieve plates (Koh et al., 2012). Excessive callose deposition in phloem plasmodesmata may interrupt photoassimilate distribution along the source and sink system, causing starch over-accumulation in leaf chloroplasts (Koh et al., 2012). Other factors such as phloem protein (PP2) accumulation in sieve plates and collapse of phloem cells might contribute to phloem dysfunction (J.-S. Kim et al., 2009) (Fig. 6).
Fig. 6.
An interaction model between citrus and ‘Candidatus Liberibacter asiaticus’. Reported components of the liberibacter possibly associated with its virulence mechanisms regarding suppression of host immunity and manipulation of its physiology. Flagellin components induce blockage of phloem plasmodesmata and impair sap flow between cells. Starch grains accumulate in chloroplasts in response to several changes in enzymatic activities. Salicylic acid might be broken down into catechol by hydroxylases and reactive oxygen species are suppressed by the activity of peroxidases of the bacteria.
Besides anatomical alterations, several metabolic imbalances and genetic reprogramming are observed in HLB-infected plants (Aritua et al., 2013; Mafra et al., 2013; Chin et al., 2014). A protein with potential salicylate hydroxylase activity was identified in the ‘CaLas’ genome, which might convert salicylic acid into catechol (Wang and Trivedi, 2013). Salicylic acid is a hormone that plays an important role in induction of plant defence systems against biotrophic pathogens (Yusuf et al., 2013). Therefore, ‘CaLas’ might evade plant defences by modulating overall defence of its hosts. This hypothesis is supported by studies demonstrating depression of the salicylic acid pathway in susceptible citrus plants (Xu et al., 2015) (Fig. 6).
Another interesting characteristic found in ‘Ca. Liberibacter spp.’ is the presence of prophages integrated in their genomes (Zhang et al., 2011). Many bacterial pathogens contain prophages or phage remnants integrated in their genomes that encode virulence factors (Menouni et al., 2014). ‘CaLas’, for example, carries two predicted prophages: an excision prophage (SC2) and a chromosomally integrated prophage (SC1) (Fleites et al., 2014). SC1 replicates and forms phage particles in the phloem of ‘CaLas’-infected periwinkle and in citrus, but not in the psyllids (Fu et al., 2014). Both phages encode two proteins with peroxidase activity (Zhang et al., 2011). These peroxidases might protect the bacteria against ROS produced by the plant during infection (Jain et al., 2015). The SC1 prophage also encodes functional holin (SC1_gp110) and endolysin (SC1_gp035) proteins that might be implicated in bacterial membrane lysis and cell-wall degradation during bacteriophage egress (Fleites et al., 2014). An incomplete phage/prophage variant (iFP3) derived from recombination of FP1 (SC1) and FP2 (SC2) was reported in ‘CaLas’ (Zhou et al., 2013). iFP3 is absent or detected at low levels in psyllids but it is abundant in host plants. Furthermore, it might be associated with blotchy mottle symptoms and disease development (Zhou et al., 2013; Pitino et al., 2014).
Given that ‘Ca. Liberibacter spp.’ are intracellular pathogens, there is a conjecture that the bacteria secrete effector proteins directly into host cell cytoplasm and modulate its physiology (Puttamuk et al., 2014). ‘Ca. Liberibacter spp.’ contain a general secretory pathway, which might contribute to the secretion of these molecules (Hao et al., 2013). A few studies have reported findings about potential effectors of ‘Ca. Liberibacter spp.’. A putative serralysin was predicted in the ‘CaLas’ genome (Cong et al., 2012). This protein is a metalloprotease associated with the type I secretion system (T1SS) and might act as a virulence factor. It is very likely that serralysin plays an important role in infection as an anti-immune mechanism by degrading host proteins, as already described in other pathosystems (Felfoldi et al., 2009). A protein containing a von Willebrand factor type A domain (vWA) was also identified in the ‘CaLas’ genome (Cong et al., 2012). This protein is predicted to be associated with cell adhesion, migration and signal transduction, although it has yet to be well characterized in ‘CaLas’. Other candidates for virulence factors are reported in the analyses carried out by Cong et al. (2012).
A putative effector (CLIBASIA_05315) was identified in the ‘CaLas’ genome (Pitino et al., 2016). This protein fused to GFP was localized in chloroplasts of Nicotiana benthamiana after transient expression in leaf tissue and induced cell death associated with H2O2 accumulation, electrolyte leakage and callose deposition. This protein was also localized in chloroplasts of transgenic citrus and resulted in leaf chlorosis and plant growth retardation. Another potential effector candidate named LasAI was also identified, which induced an increase in the number of root hairs when expressed in A. thaliana and an increased number of trichomes when transiently expressed in N. benthamiana (Pitino et al., 2015).
Despite the recent advances in identification of effectors, a model that explains the pathogenicity mechanism of the bacteria associated with HLB has not been established. Furthermore, the group of liberibacters might behave as an obligate ‘energy parasite’ rather than a pathogen (Haapalainen, 2014). Several research groups are currently focusing on the identification and characterization of effector proteins of ‘Ca. Liberibacter spp.’, and it is expected that in a few years we will have an improved view of this pathosystem evolution (Table 2).
Table 2.
PAMPs and putative effectors of ‘Ca. Liberibacter asiaticus’
| Gene | Description | Reference | |
|---|---|---|---|
| PAMPs | Flagellin | flagellin | Zou et al. (2012) |
| Peroxidase | hypothetical protein SC2_gp095 | Jain et al. (2015) | |
| Las5315 | hypothetical protein | Pitino et al. (2016) | |
| LasAI | hypothetical protein | Pitino et al. (2015) | |
| Putative effectors | Protein serine/tyrosine phosphatase | hypothetical protein | Cong et al. (2012) |
| Serralysin | Serralysin | Wang and Trivedi (2013) | |
| Haemolysin | Haemolysin | ||
| Salicylate hydroxylase | Monooxygenase FAD-binding protein |
CITRUS–FUNGI INTERACTIONS
Citrus post-bloom fruit drop and key lime anthracnose
The genus Colletotrichum comprises at least 600 species and includes a number of important pathogens that cause economically significant losses on various crops worldwide. Disease caused by this group of fungi is known as anthracnose (O’Connell et al., 2012). Colletotrichum species are characterized by a distinctive hemibiotrophic lifestyle (O’Connell et al., 2012). Infection occurs through a brief biotrophic phase followed by a necrotrophic phase, in which secondary hyphae spread throughout the host tissue. Production of orange–brown lesions on petals of open citrus flowers, induction of abscission of young fruit and formation of persistent calyces are characteristic symptoms of post-bloom fruit drop (PFD) (de Goes et al., 2008). In key lime, C. acutatum infects all parts of the plant, causing anthracnose symptoms associated with key lime anthracnose (KLA) (You and Chung, 2007).
Damm et al., (2012) described species of the C. acutatum complex and reported that citrus is attacked by more than one species of this complex. Pinho and Pereira (2015) identified a new species, C. abscissum, as being the agent responsible for PFD (Crous et al., 2015). Colletotrichum gloeosporioides was reported as the casual agent of PDF, although it exhibits lower pathogenicity levels in comparison with the C. acutatum complex (Lima et al., 2011).
The high variability and ability of this fungus to infect all sweet orange varieties and key lime must be related to a large arsenal of effector proteins in its genome. Similarly, in the genomes and transcriptomes of C. higginsianum infecting A. thaliana and C. graminicola infecting maize, 365 and 177 candidate secreted effectors were found, respectively (O’Connell et al., 2012).
Currently, studies focusing on understanding plant–pathogen interactions have shown that activation of these effectors is orchestrated and in some cases it follows specific patterns, such as expression in waves during infection (Giraldo and Valent, 2013). In hemibiotrophic fungi, gene expression studies have increasingly shown highly controlled gene regulation specific for each infection phase (Giraldo and Valent, 2013).
Colletotrichum spp. proteins related to pathogenicity have not yet been fully characterized and the identification of effector proteins in citrus is underway. The restriction enzyme-mediated integration (REMI) technique, which was used by Chen et al. (2005) for transformation of C. acutatum, resulted in six mutants not pathogenic in key lime. The KLAP1 gene was responsible for the loss of C. acutatum pathogenicity (Table 3). KLAP1-null mutants were unable to develop the penetration stage on leaves of key lime, but were able to cause orange–brown lesions similar to the wild-type on flower petals. The actual function of KLAP1 remains uncertain but this gene may encode a putative transcription activator necessary for penetration of the hyphae on key lime leaves, suggesting that these proteins may be effectors.
Table 3.
Candidate effector genes identified in Colletotrichum acutatum of citrus pathogen
| Gene | Description | Reference | |
|---|---|---|---|
| Putative effectors | KLAP1 | Hypothetical transcription activator | Chen et al. (2005) |
| PacCKLAP2 | Hypothetical PH regulation | You et al. (2007) |
Unlike KLAP1, mutants of the PacCKLAP2 gene characterized by You et al. (2007) were less effective in causing anthracnose in key lime and sweet oranges, demonstrating that PacCKLAP2 is a common gene for virulence of C. acutatum in both pathosystems (Table 3). Colletotrichum acutatum transformants not expressing transcripts of the PacCKLAP2 gene were unable to grow at high pH. However, they are capable of forming appressoria on the host leaves and flowers, but fail to colonize the surrounding tissue. Enzymatic tests showed a decrease in alkaline phosphatase activity, proteases and cellulases in PacCKLAP2 null mutants (You et al., 2007). Absence of these enzymes prevents fungal development at high pH and should be required for C. acutatum pathogenesis. CUT1 gene transcripts decreased indicating that PacCKLAP2 also regulates the expression of cutinases (You and Chung, 2007). If the gene expression pattern of C. higginsianum were similar to C. acutatum when infecting citrus species, this would suggest that the protein coded by the PacCKLAP2 gene is a ‘candidate effector’. This protein is secreted by the appressorium and it seems to be important for the development of pathogenicity in the host tissue.
Alternaria brown spot in citrus
Alternaria brown spot, caused by Alternaria alternata, is an important disease of tangerines and their hybrids, affecting leaves, twigs and immature fruit (Canihos et al., 1999). This fungus is also responsible for rough lemon brown spot (Timmer et al., 2003). There are two different pathotypes that produce host-specific toxins (HSTs), which are responsible for causing the disease (Tsuge et al., 2013).
One of the main barriers to prevent the fungal penetration of plant pathogens is the cell wall, and therefore many fungi secrete extracellular enzymes that can degrade cell-wall polymers. One of these enzymes produced by the genus Alternaria is endopolygalacturonase (endoPG) (Isshiki et al., 1997). An A. citri mutant for this gene resulted in a significant reduction in its ability to cause black rot symptoms in citrus (Isshiki et al., 2001). There is no Alternaria endoPG receptor described in Citrus; however, in A. thaliana, a receptor was identified as a leucine-rich repeat receptor-like protein (RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1, RBPG1), which recognizes fungal endoPGs (Zhang et al., 2014).
The laccase enzyme was suggested as an elicitor of the genus Alternaria and could be involved in the pathogenesis of A. alternata in Citrus. After challenging ‘Fortune’ mandarin, C. limon and C. paradise with Alternaria, the flavonoid degradation pathway was activated in the host plants in association with the de novo synthesis of the phytoalexin scoparone. This metabolism of flavonoids is caused by an extracellular laccase, which utilizes Citrus flavonoids as a substrate (Díaz et al., 2015). Another elicitor from Alternaria is β-1,3-,1,6-oligoglucan, which is a fungal cell-wall component. This glucan, when applied to a tobacco plant model (BY-2), induced chitinase activity (Shinya et al., 2006). In soybean, the PRR recognizing Phytophthora megasperma β-glucan was identified as the β-glucan binding protein (GBP) (Umemoto et al., 1997). However, this MAMP and its corresponding PRR have not been studied in citrus species or in any other organisms.
Effectors of necrotrophic pathogens include phytotoxins and proteinaceous effectors (Wang et al., 2014). Phytotoxins can be divided into general toxins (non-HSTs), to which many plant species are sensitive, and HSTs, where sensitivity is restricted to specific host genotypes (Oliver and Solomon, 2010). Based on their chemical structure, phytotoxins are classified as polyketides, non-ribosomal peptides, alkaloids, terpenes or metabolites of mixed biosynthetic origin (Stergiopoulos et al., 2012).
Recent studies have shown that several necrotrophs secrete HSTs, which play a crucial role in disease outcome (Izumi et al., 2012). Therefore, the toxins may be considered as the major group of effectors of necrotrophic fungi, since they have characteristics of avirulence genes (Stergiopoulos et al., 2012). These toxins secreted by necrotrophic fungi are detected by R-genes of susceptible plants in order to trigger the HR and initiate cell death. This phenomenon is known as dominant susceptibility (Liu et al., 2009). Thus, interactions between necrotrophic effectors and genes for susceptibility of plants are called inverse interactions of the gene-for-gene theory (Oliver and Solomon, 2010).
In Citrus, the two pathotypes of A. alternata are identified according to the production of HSTs. One is the tangerine pathotype, which produces ACT-toxin specific to tangerine (C. reticulata ‘Blanco’) and their hybrids. The other is the rough lemon pathotype, which affects rough lemon (C. jambhiri ‘Lush’) and Rangpur lemon (C. limonia ‘Osbeck’), and produces ACR-toxin (Tsuge et al., 2013). These HSTs are essential for host-selective infection and disease development (Tsuge et al., 2013).
Due to the lifestyle of Alternaria, cell death is the main mechanism for success of the pathogen. HSTs produced by this species have a central role in the host cell-death induction mechanism and are critical for successful pathogenesis. Other classic effectors, which evade recognition or hinder the defence, do not necessarily cause disease, given that the toxins are always identified as the central component in pathogenesis.
The chemical structure of the ACT-toxin consist of three parts: 9,10-epoxy-8-hydroxy-9-methyl-decatrienoic acid (EDA), valine and a polyketide (Kohmoto, 1993). One putative target site of ACT-toxins in tangerine is the plasma membrane (Kohmoto, 1993). These toxins are quickly translocated through the vascular system, causing rapid electrolyte leakage and necrotic lesions along the veins (Chung, 2012). After infection of citrus leaves by A. alternata, the induction of fast lipid peroxidation and accumulation of hydrogen peroxide (H2O2) occur (Lin et al., 2011) (Fig. 7).
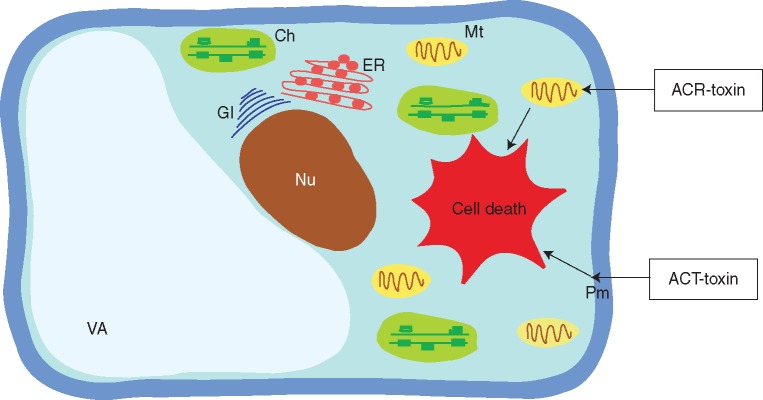
Fig. 7.
Schematic representation of target sites of toxins produced by A. alternata. The putative target site of ACT-toxins in tangerine is the plasma membrane. After the infection of citrus leaves, induction of fast lipid peroxidation and accumulation of hydrogen peroxide (H2O2) occurs, resulting in cell death. The target of ACR-toxin is the mitochondrion, which after contact with the host plant causes a rapid increase in electrolyte leakage and consequent cell death. Ch, chloroplast; ER, endoplasmic reticulum; Gl, Golgi apparatus; Mt, mitochondrion; Nu, nucleus; Pm, plasma membrane; Va, vacuole.
The rough lemon pathotype of A. alternata produces a host-selective ACR-toxin and causes Alternaria leaf spot disease of rootstock species such as rough lemon (C. jambhiri), Rangpur lime (C. limonia) and a hybrid of rough lemon and acid mandarin, Rangpur lime (C. limonia ‘Osbeck’) (Akimitsu et al., 2003). The structure of ACR-toxin I consists of a polyketide with an α-dihydropyrone ring in a 19-carbon polyalcohol (Akimitsu et al., 2003). ACR-toxins are polyketide secondary metabolites and the ACRTS2 gene that encodes a polyketide synthase (PKS) essential for biosynthesis of these toxins. This gene was identified in the rough lemon pathotype (Izumi et al., 2012). The target of ACR-toxin is the mitochondrion (Kohmoto et al., 1984). After coming into contact with the host plant, ACR-toxins cause water congestion and veinal necrosis, and induce a rapid increase of electrolyte leakage (Kohmoto, 1979) (Fig. 7).
The effectors, PAMPs, DAMPs as well as toxins involved in the pathogenesis of A. alternata in Citrus are shown in Table 4.
Table 4.
The effectors involved in the pathogenesis of A. alternata in Citrus
| Gene | Description | Reference | |
|---|---|---|---|
| PAMP/DAMP | Endopolygalacturonase (endoPG) | enzyme | Isshiki et al. (1997) |
| laccase enzyme | enzyme | Díaz et al. (2015) | |
| β-1,3-, 1,6-oligoglucans | glucan | Shinya et al. (2006, 2007) | |
| Putative effectors | ACT-toxin | toxin | Kohmoto (1993) |
| ACR-toxin | toxin | Akimitsu et al. (2003) |
The involvement of chitinases (Ch) and β-1,3-glucanases (Glu) in the defence response against A. alternata was investigated in C. limon seedlings. Following inoculation of the fungus, increased activity of these enzymes was observed with the detection of a new Ch isoenzyme and of three new Glu enzymes (Fanta et al., 2003).
Enhancement of the citrus plant immune system was observed after treatment of ‘Fortune’ mandarin with hexanoic acid (Hx). The diameter of the lesions was significantly reduced in plants treated with Hx and challenged with A. alternata compared with non-treated plants. Furthermore, treated plants showed an increase in callose deposition and activation of the jasmonic acid pathway (Llorens et al., 2013).
Other defence mechanisms were reported in citrus plants exposed to toxins that do not cause disease. After inoculation of rough lemon with the tangerine pathotype, which produces an ACT-toxin that is not toxic to this plant, the induction of several defence-related genes such as chitinases (Gomi et al., 2002a), lipoxygenase (Gomi et al., 2002b), epoxide hydrolase (Gomi et al., 2003b), hydroperoxide lyase (Gomi et al., 2003a), chalcone synthase (Gotoh et al., 2002), miraculin-like protein (Tsukuda et al., 2006) and thaumatin-like protein (B.-G. Kim et al., 2009) was observed.
CITRUS–OOMYCETES INTERACTIONS
Commonly mistaken as fungi because of morphological similarities, the oomycetes are a group of eukaryotic micro-organisms that include pathogens of insects, crustaceans, fish, vertebrates, micro-organisms and plants. Modern molecular phylogenies based on rRNA sequences, amino acid data for mitochondrial proteins and four protein-encoding chromosomal genes have been used to identify the oomycetes as a unique lineage of stramenopile eukaryotes, unrelated to true fungi but closely related to heterokont photosynthetic algae (ADL et al., 2005; Kamoun et al., 2015).
The genus Phytophthora consists of more than 100 plant-pathogenic species with worldwide distribution. As pathogens they infect more than 250 plant families and damaging crops and natural ecosystems (Kroon et al., 2012). Several Phytophthora species have been associated with disease in citrus plants. The predominant species in citrus orchards and nurseries worldwide include: P. boehmeriae Saw., P. cactorum (Lebert & Cohn) Srhöter, P. capsici Leonian, P. cinnamomi Rands, P. citricola Saw., P. citrophthora (Sm. & Sm.) Leonian, P. drechsleri Tucker, P. hibernalis Carne, P. megasperma Drechsler, P. palmivora (Butler) Butler, P. nicotiane (=P. parasitica) Dastur., P. parasitica and P. citrophthora (Luz, 2001). Phytophthora parasitica is the main citrus pathogen due to its geographical distribution and severity (Panabieres et al., 2016).
Phytophthora species are able to secrete two types of effectors related to their localization in plant tissues: the apoplastic or extracellular effectors, such as elicitins and NPP-like effectors; and cytoplasmic effectors, such RxRL and Crinkler effectors (CRNs), which possess special amino acid motifs in their structure enabling their entry inside cells independent of the presence of the pathogen (Fig. 8) (Hogenhout et al., 2009; Kamoun, 2009).
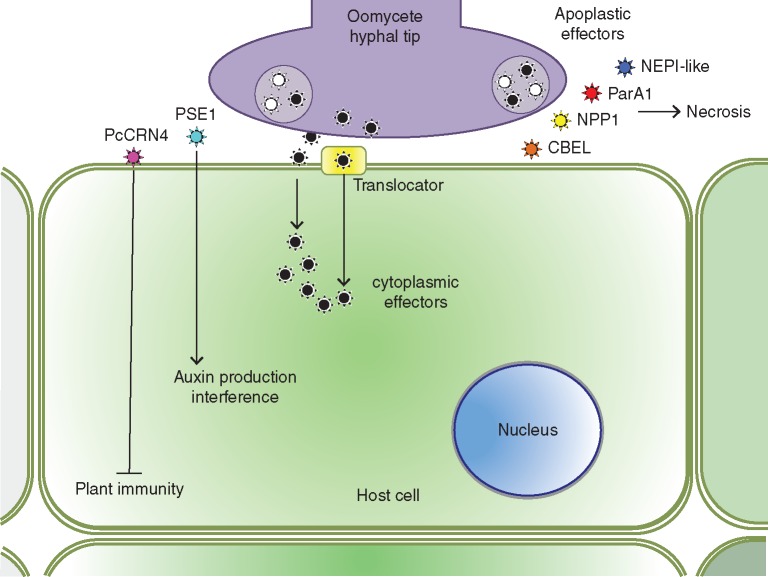
Fig. 8.
Oomycete pathogens of citrus release apoplastic effectors such as NEP-like, ParA1, NPP1 and CBEL, which can elicit plant responses and necrosis and/or cytoplasmic effectors, such as PSE1 (RxLR effector) and PcCRN4 (Crinkler effector), which use the plant machinery (a translocator) to invade the cytoplasm and interfere with auxin production or suppress plant immunity, respectively.
Apoplastic Phytophthora effectors
Elicitins
Elicitins are extracellular proteins with low molecular weight (about 10 kDa) which are secreted by most members of the genus Phytophthora (Oßwald et al., 2014). The first characterized elicitin was INF-1 from P. infestans. INF-1 induces a strong HR in tobacco plants (Kamoun et al., 1998). Sharing features of PAMPs, elicitins are widely used to induce HR and to study defence in plants. Studies on the crystallography and functional characterization of elicitins in model plants such as A. thaliana and N. benthamiana have been extensive. Usually, elicitins lead to local and systemic responses in plants after inducing an oxidative burst in cells through efflux of K+ and Cl− and influx of Ca2− (Fellbrich et al., 2002). This phenomenon is found not only in members of the family Solanacea but also in Brassicacea plants. However, little is known about the role of these proteins in compatible interactions (Bhattacharjee et al., 2006; Svozilová et al., 2009).
Among the oomycetes pathogens of citrus plants, species such as P. parasitica, P. citrophthora, P. citricola, P. capsici, P. drechsleri, P. palmivora and P. megasperma are highlighted since they have several types of elicitins and elicitin-like proteins organized as multigenes. Phytophthora parasitica, for instance, secretes the elicitin ParA1 which induces a very strong HR in tobacco (Kamoun, 1993). A simple search for elicitin proteins in public databanks such as NCBI yields more than 500 elicitin-related proteins, 19 of which are characterized as elicitins and 489 are hypothetical elicitins (Geer et al., 2010). The EST sequencing project for P. parasitica led to the identification of ten different elicitin classes (Panabières et al., 2005). Most abundantly expressed are the class 1 proteins of parasiticein and encoded by at least four genes (ParA1.1–ParA1.4) (Panabières et al., 2005). Parasiticein genes from classes 5 and 6 (PAR5 and PAR6) have N-terminal sequence similarities with a phospholipase from P. capsici, suggesting an involvement of PAR5 and PAR6 in membrane remodelling (Nespoulous et al., 1999). For the P. parasitica–citrus interaction, it was found that elicitins were up-regulated at the later stages of infection, indicating elicitins are correlated with the late necrosis in tissues of susceptible varieties of citrus (Boava et al., 2011).
Other Phytophthora effectors
Other reported effectors of P. parasitica that might have important roles in the interaction with citrus plants include: NEPl-like protein (necrosis and ethylene-inducing peptide), NPP1 (necrosis-inducing Phytophthora protein 1) (Fellbrich et al., 2002), the gene family encoding apoplastic polygalacturonases (Yan and Liou, 2005; Wu et al., 2008) and CBEL (cellulose-binding, elicitor and lectin activity) apoplastic effectors, which are purified from cell walls and, importantly, induce HR when infiltrated in leaves of tobacco and A. thaliana (Khatib et al., 2004; Oßwald et al., 2014). It is also suggested that the structure of the hyphal cell wall and attachment to cellulosic substrates, such as plant surfaces, depend on CBEL effectors (Gaulin et al., 2002). The main role of the aforementioned effectors during infection of citrus plants by P. parasitica is still obscure.
Cytoplasmic Phytophthora effectors
RxLR effectors
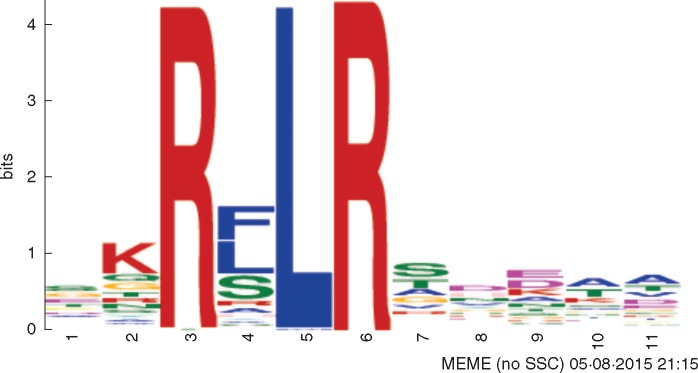
The proteins from the RxLR family are cytoplasmic modular effectors carrying a conserved amino acid motif in their N-terminal structure: RxLR (R: arginine; x: any amino acid; L: leucine; R: arginine) (Fig. 9) (Win et al., 2007). The RxLR motif is particularly interesting because it enables the delivery of these proteins into the interior of cells using the plant machinery (Grouffaud et al., 2008). One of the most studied RxLR effectors is the P. infestans AVR3a, which suppresses cell death induced by the elicitin INF-1 (Bos et al., 2009).
Fig. 9.
Conserved RxRL domain in P. parasitica effectors.
RxLR effectors from P. parasitica were shown to be differentially expressed in the necrotrophic phase of infection of A. thaliana (Attard et al., 2014). Through localization studies, GFP-labelled RxLRs were observed in hyphae and appressoria. Another study demonstrated that the P. parasitica RxLR effector PSE1 (penetrating specific effector 1) promotes infection of A. thaliana by interfering with auxin physiology (Evangelisti et al., 2013).
Searches for RxLR effectors in the genome of P. parasitica INRA-310 (originally isolated from tobacco in Australia, but also pathogenic to citrus) at the Fungidb platform (fungidb.org) rendered 179 hits. An additional eight genomes of different P. parasitica isolates are now also available at the Broad Institute (olive.broadinstitute.org/projects/phytophthora_parasitica), each showing a repertoire of RxRL effectors. To our knowledge, no P. parasitica RxLR effector has yet been characterized functionally.
Crinkler effectors (CRN)
Crinklers are cytoplasmic effectors originally described in P. infestans. Today, it is accepted that CRN effectors are secreted by most Phytophthora species as well as other plant-pathogenic micro-organisms such as Hyaloperonospora arabidopsidis (Baxter et al., 2010), Bremia lactucae (Stassen and Van den Ackerveken, 2011) and Pythium ultimum (Lévesque et al., 2010). The name Crinkler was originally used because of the crinkling phenotype of leaves infected with P. infestans (Torto et al., 2003).
The structure of CRN effectors presents a highly conserved N-terminal amino acid domain: Leu-Xaa-Leu-Phe-Leu-Ala-Lys (LxLFLAK; Fig. 10) (Haas et al., 2009). Functional characterization of CRN (PsCRN70) of P. sojae in N. benthamiana showed that the effector suppressed cell death induced by the INF-1 elicitin. INF-1 would act as a PAMP inducing cell death (Schornack et al., 2010), while PsCRN70 would suppress the responses contributing to pathogen virulence (Rajput et al., 2014).
Fig. 10.
Conserved LxLFLAK domain in P. parasitica CRN effectors.
The CRN family shows extensive expansion in all sequenced Phytophthora species, including P. parasitica (Tyler et al., 2006; Haas et al., 2009). Phytophthora capsici secretes PcCRN4, which is an effector essential to pathogen virulence, since it suppresses plant-immunity responses (Mafurah et al., 2015). Searching for CRN effectors in the genome of P. parasitica INRA-310 at the Fungidb platform (fungidb.org) renders 26 hits. However, most of the CRN effectors related to the Phytophthora–citrus interaction remain without any functional characterization.
INTERACTIONS OF CITRUS AND PATHOGEN VECTORS
Phytophagous insects and mites play an important role in agriculture as pests or vectors of diverse pathogens. Similar to many microbes, arthropods are also able to manipulate the physiological mechanisms of host plants (Stuart, 2015).
Plants recognize arthropod attacks by perception of HAMPs arising from wounds, contact, oral (regurgitate and saliva) and oviposition secretions. These then trigger signalling cascades similar to those characterized in a large range of plant–microbe interactions. The best known insect HAMPs are elicitors present in oral secretions, such as β-glucosidase, fatty acid-amino acid conjugates (FACs), glucose oxidase and inceptins (Sharma et al., 2014; Acevedo et al., 2015). These molecules promote activation of several plant defence responses, such as induction of plant calcium fluxes, activation of MAPK pathways, production of ROS, volatile emissions, and biosynthesis and signalling of plant defence hormones, such as jasmonic acid (JA), ethylene (ET) and salicylic acid (SA) (Wu and Baldwin, 2010).
During feeding, insects deliver their effectors through saliva, avoiding HAMP recognition. Insect saliva is composed of several important proteins, which facilitate entry and movement of the stylet of sap-sucking insects such as hydrolases, and perform detoxification of plant defence substances (superoxide dismutases, peroxidases and phosphatases) (Nicholson et al., 2012; Sharma et al., 2014). Calmodulin and/or other Ca2+ binding proteins are also present in insect saliva, preventing the occlusion of sieve-tubes caused by callose deposition or other innate plant responses (Will et al., 2007; Carolan et al., 2009; Hattori et al., 2012). Glucose oxidase (GOX), an effector present in the oral secretions of several insects, manipulates SA- and JA-related genes in different host plants (Zhu-Salzman et al., 2005; Hogenhout and Bos, 2011). Additionally, some effectors are able to induce chlorosis or repression of microbe elicitors (Bos et al., 2010; Elzinga and Jander, 2013), suppress plant protease inhibitors and other defence genes, or promote post-translational modifications (Acevedo et al., 2015), as well as repress wound-inducing responses (Consales et al., 2012).
Conversely, plant-associated microbes can manipulate plants and the physiology of their vectors to facilitate their transmission and dispersion (Felton and Tumlinson, 2008; Clark et al., 2010; Frago et al., 2012; Junker, 2014; Acevedo et al., 2015).
Since the transmission of many citrus diseases is facilitated by arthropods, here we provide some aspects of insect–microbe interactions for the major vectors associated with citrus crops.
Huanglongbing (HLB)
Diaphorina citri (Asian citrus psyllid, ACP) is the most important vector of HLB (Fig. 11), responsible for transmission of ‘CaLas’ bacteria across the Asian and American continents and transmission of ‘CaLam’ in South America (Bové, 2014; Haapalainen, 2014).
Fig. 11.
(A) Nymphs and (B) adults of Diaphorina citri, the vector of Candidatus Liberibacter spp., the causal agent of HLB.
Modifications of citrus volatile profiles caused by attack of this insect were reported (Hijaz et al., 2013), although until recently no candidate effectors were proposed for D. citri. However, in the genome of this insect there is a glucose oxidase-like gene and a putative secreted β-glucosidase without functional characterization, which are putative effectors of D. citri. This interference is based upon the characterization of these molecules as effectors in other insect species (Hogenhout and Bos, 2011; Consales et al., 2012).
Additionally, 86 miRNA sequences were found in the D. citri EST database. These sequences are phylogenetically related to 15 biotic stress-associated miRNA sequences of the Hessian fly (Mayetiola destructor), which are expressed in specific plant host genotypes. These observations led to the suggestion that these miRNAs may act on plant–insect interactions by regulating virulence factors (Khalfallah et al., 2015).
In contrast, ‘CaLas’ strongly manipulate insect physiology. The presence of ‘CaLas’ increased mortality of infected adult insects, delayed development of infected ACPs and increased fecundity (infected females laid more eggs than healthy females) (Pelz-Stelinski and Killiny, 2016). ‘CaLas’-infected ACP insects have decreased expression of detoxification genes such cytochrome P450, esterases and glutathione transferases (Tiwari et al., 2011b). Repression of these genes promotes loss of fitness and increases susceptibility to insecticides (Tiwari et al., 2011a, b, c). ‘CaLas’ infection also interferes with the metabolism and immune system of D. citri, manipulating free amino acid availability, iron transport and cytoskeleton networks, in addition to suppressing 90 % of the immune genes at immature stages (Fisher et al., 2014; Vyas et al., 2015). Down-regulation of insect immune genes may be associated with expression of an as yet uncharacterized gene ‘CaLas’, containing an imelysin-like domain. In other bacterial species, homologues of this gene act to suppress the immune systems of insects (Yan et al., 2013). In addition, ‘CaLas’ induces modification of plant volatiles making them more attractive to D. citri and allowing bacterial dispersal (Mann et al., 2012; Hijaz et al., 2013).
Citrus variegated chlorosis
Xylem sap-feeding insects belonging to the families Cicadellidae and Cercopidae (Fig. 12) are the major vectors of the bacteria X. fastidiosa (Almeida et al., 2013). Until recently, no putative effector molecules were described for these insects during interactions with X. fastidiosa. However, some genes were characterized as being important for this interaction.
Fig. 12.
Macugonalia leucomelas, one of the sharpshooter vectors of CVC disease.
Gene expression analysis revealed that Homalodisca coagulata promotes lower activation of stress-related genes than mechanical wounds in citrus plants, suggesting the presence of an insect elicitor which modulates plant response (Mozoruk et al., 2006).
Salivary β-glucosidase of Homalodisca vitripennis is important for transmission of X. fastidiosa, digesting bacterial biofilm in the Pierce’s disease pathosystem (Backus et al., 2012, 2015). The action of this enzyme may also be important for CVC disease, since biofilm formation is critical for establishment and transmission of X. fastidiosa through its host.
Many studies of Pierce’s disease of grapevines have demonstrated that initial adhesion and retention of X. fastidiosa of its insect vector are related to fimbrial and afimbrial adhesins (such Hxf, XadA, and fimA genes), as well as LPS (Killiny and Almeida, 2009, 2014; Killiny et al., 2012; Orlovskis et al., 2015; Rapicavoli et al., 2015). This colonization is dependent on a quorum-sensing mechanism composed of components of the rfp cluster (regulation of pathogenicity factors), which senses a diffusible signalling factor (DFS) and alters the expression of many genes involved with attachment and biofilm formation, including hxfA, hxfB and fimA (Baccari et al., 2014; Ryan et al., 2015).
The extracellular polysaccharides (EPS) and outer membrane vesicles of X. fastidiosa also play a role in retention and transmission of this pathogen by insect vectors (Killiny et al., 2013; Ionescu et al., 2014).
Citrus tristeza disease
Citrus tristeza disease is vectored by the brown citrus aphid (Aphis citricidus). The majority of insect effectors characterized so far belong to aphid family members, but only two of these molecules were identified from A. citricidus. The first is a protein, C002, which has no functional characterization, but plays an important role in the feeding behaviour of pea aphid (Acyrthosiphon pisum), reducing the time of contact of this insect with plant sieve elements when this gene is suppressed (Mutti et al., 2008). Expression of C002 homologues of Myzus persicae in N. benthamiana enhances aphid fecundity (Bos et al., 2010). These data support the hypothesis that A. citricidus C002 may be important for the interaction of this aphid with its plant host. The second A. citricidus putative effector is a cysteine-rich protein that was first identified in A. pisum and is present across many aphid species (Guo et al., 2014). This protein is highly expressed in salivary glands and in the first instar of A. pisum; however, knockdown of this gene does not interfere with survival or feeding (Guo et al., 2014).
Other important arthropods
Mites, along with insects, are the most important agricultural pests in the arthropod group. In citrus crops, the mite Brevipalpus yothersi is notable for being a vector of citrus leprosis virus (CiLV) but no effector molecules have yet been characterized. Evidence of plant volatile and defence manipulation by mites has been reported in the literature (Albarouki and Deising, 2013; Zhurov et al., 2014; Alba et al., 2015; Martel et al., 2015; Godinho et al., 2016). In a recent study, three salivary effectors were identified for two mite species: Tetranychus urticae and Tetranychus evansi (Villarroel et al., 2016). These proteins were described acting on modulation of JA and SA responses and increasing mite performance.
OVERVIEW, CONCLUSIONS AND PERSPECTIVES
As perennial woody plants, citrus are in constant interaction with various abiotic and biotic factors, including viroids, viruses, mollicutes, bacteria, oomycetes and fungi. Many of the pathosystems are highly complex and require a deeper understanding to enable development of suitable disease control strategies.
The present review summarizes important information available regarding PAMPs, PRRs, effectors and R-genes associated with the main interactions of citrus species and their pathogens and insect vectors (Fig. 13). Whilst gaps still remain, available information is potentially useful in the development of disease-resistant plants through both conventional breeding programmes and biotechnology-based approaches, which include the development of transgenic or cisgenic citrus, genome editing and host induced gene silencing (HIGS). We have not included in this review those citrus pathogens that still lack robust information regarding molecular interaction with their hosts; however such information will probably be available in the near future.
Fig. 13.
Schematic representation of a citrus cell and its general molecular interactions with the main fungi, viruses, vectors, oomycetes, bacteria and unculturable bacteria. Fungi: The infection process of C. acutatum requires proteins Klap 1 for appressorium development and infection on leaves of key lime. Proteins PacCKLAP2 are required for pH adjustment in the transition from biotrophic hyphae to necrotrophic. Spore (SP), appressorium (AP), biotrophic hyphae (BH), necrotrophy hyphae (NH). The conidia produced by both pathotypes of A. alternata germinate quickly and begin to produce toxins. The putative target site of ACT-toxins in tangerine is the plasma membrane and of ACR-toxin is the mitochondrion, both leading to cell death. Virus:Citrus tristeza virus (CTV) deploys three effectors – p20, p23 and coat protein (CP) – with activities of viral suppressors of RNA silencing (VSR) to overcome host resistance. Citrus psorosis virus (CPsV) 24K protein physically interacts with pre-miRNA in the nucleus and induces the expression of genes involved in disease symptom development, suggesting a putative VSR function. Vectors: During feeding, vectors deliver putative effectors through saliva in stylets into the plant intercellular space which can modulate host plant recognition, volatile emission and defence. Oomycetes: Oomycete pathogens of citrus can secrete two types of proteins: apoplastic and cytoplasmic effectors. The apoplastic effectors, such as NEP-like, ParA1, NPP1 and CBEL, are frequently related to plant responses and necrosis elicitation. The cytoplasmic effectors, such as PSE1 (RxLR effector) and PcCRN4 (Crinkler effector), may interfere with the physiology of plants (auxin production) or suppress plant immunity, respectively. Bacteria:X. citri is able to inject into the plant host cell effectors such as PthA, which heads towards the cell nucleus where it controls gene regulation. Xylella fastidiosa may have effector molecules that are secreted into the plant, but none have been characterized to date. Unculturable bacteria: ‘Candidatus Liberibacter asiaticus’ might quench H2O2 accumulation and signalling events by secretion of peroxidase enzyme during early stages of infection. A functional salicylate hydroxylase (SahA) predicted in the ‘CaLas’ genome converts salicylic acid (SA) into catechol and might suppress SA-mediated defence.
ACKNOWLEDGEMENTS
This work was supported by Fundação de Amparo a Pesquisa no Estado de São Paulo (FAPESP) (grant nos.: 2015/14498-6 to R.J.D.D., 2014/19375-7 to C.M.R., 2013/21924-6 to R.R.S.N., 2013/01395-9 to S.C.P., and 2014/0036-8 to G.D.A.) and the Conselho Nacional de Ciência e Tecnologia (CNPQ), grant no.: 164342/2015-0 to P.M.M.M. and 573848/2008-4 to M.A.M.
LITERATURE CITED
- Abe VY, Benedetti CE.. 2016. Additive roles of PthAs in bacterial growth and pathogenicity associated with nucleotide polymorphisms in effector-binding elements of citrus canker susceptibility genes. Molecular Plant Pathology 17: 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo FE, Rivera-Vega LJ, Chung SH, Ray S, Felton GW.. 2015. Cues from chewing insects – the intersection of DAMPs, HAMPs, MAMPs and effectors. Current Opinion in Plant Biology 26: 80–86. [DOI] [PubMed] [Google Scholar]
- Adl SM, Simpson AGB, Farmer MA, et al. 2005. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. The Journal of Eukaryotic Microbiology 52: 399–451. [DOI] [PubMed] [Google Scholar]
- Achachi A, Barka EA, Ibriz M. 2014. Recent advances in Citrus psorosis virus. Virusdisease 24: 261–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimitsu K, Peever TL, Timmer LW.. 2003. Molecular, ecological and evolutionary approaches to understanding Alternaria diseases of citrus. Molecular Plant Pathology 4: 435–446. [DOI] [PubMed] [Google Scholar]
- Alba JM, Schimmel BC, Glas JJ, et al. 2015. Spider mites suppress tomato defenses downstream of jasmonate and salicylate independently of hormonal crosstalk. New Phytologist 205: 828–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarouki E, Deising HB. 2013. Infection structure-specific reductive iron assimilation is required for cell wall integrity and full virulence of the maize pathogen Colletotrichum graminicola. Molecular Plant Microbe Interactions 26: 695–708. [DOI] [PubMed] [Google Scholar]
- Almeida RPP, Coletta-Filho HD, Lopes JRS.. 2013. Xylella fastidiosa. Manual of Security Sensitive Microbes and Toxins. CRC Press, 841–850. [Google Scholar]
- Al-Saadi A, Reddy JD, Duan YP, Brunings AM, Yuan Q, Gabriel DW.. 2007. All five host-range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host-range variation. Molecular Plant-Microbe Interactions 20: 934–943. [DOI] [PubMed] [Google Scholar]
- Aritua V, Achor D, Gmitter FG, Albrigo G, Wang N.. 2013. Transcriptional and microscopic analyses of citrus stem and root responses to Candidatus liberibacter asiaticus infection. PLoS One 8: 4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong MR, Whisson SC, Pritchard L, et al. 2005. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proceedings of the National Academy of Sciences of the United States of America 102: 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard A, Evangelisti E, Kebdani-Minet N, et al. 2014. Transcriptome dynamics of Arabidopsis thaliana root penetration by the oomycete pathogen Phytophthora parasitica. BMC Genomics 15: 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccari C, Killiny N, Ionescu M, Almeida RPP, Lindow SE.. 2014. Diffusible signal factor-repressed extracellular traits enable attachment of Xylella fastidiosa to insect vectors and transmission. Phytopathology 104: 27–33. [DOI] [PubMed] [Google Scholar]
- Backus EA, Andrews KB, Shugart HJ, Greve CL, Labavitch JM, Alhaddad H.. 2012. Salivary enzymes are injected into xylem by the glassy-winged sharpshooter, a vector of Xylella fastidiosa. Journal of Insect Physiology 58: 949–959. [DOI] [PubMed] [Google Scholar]
- Backus EA, Shugart HJ, Rogers EE, Morgan JK, Shatters R.. 2015. Direct evidence of egestion and salivation of Xylella fastidiosa suggests sharpshoosters can be ‘flying syringes’. Phytopathology 105: 608–620. [DOI] [PubMed] [Google Scholar]
- Baroncelli R, Zapparata A, Sarrocco S, et al. 2015. Molecular diversity of anthracnose pathogen populations associated with UK strawberry production suggests multiple introductions of three different Colletotrichum species. PLoS One 10: e0129140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter L, Tripathy S, Ishaque N, et al. 2010. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science (New York, N.Y.) 330: 1549–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlau F, Barelli NL, Belasque J. Jr, 2014. Lessons from a case of successful eradication of citrus canker in a citrus-producing farm in São Paulo State, Brazil. Journal of Plant Pathology. 96: 561–568. [Google Scholar]
- Bhadauria V, Banniza S, Vandenberg A, Selvaraj G, Wei Y.. 2011. EST mining identifies proteins putatively secreted by the anthracnose pathogen Colletotrichum truncatum. BMC Genomics 12: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee S, Hiller NL, Liolios K, et al. 2006. The malarial host-targeting signal is conserved in the irish potato famine pathogen. PLoS Pathogens 2: 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch PRJ, Rehmany AP, Pritchard L, Kamoun S, Beynon JL.. 2006. Trafficking arms: oomycete effectors enter host plant cells. Trends in Microbiology 14: 8–11. [DOI] [PubMed] [Google Scholar]
- Boava LP, Cristofani-Yaly M, Mafra VS, et al. 2011. Global gene expression of Poncirus trifoliata, Citrus sunki and their hybrids under infection of Phytophthora parasitica. BMC Genomics 12: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, et al. 2009. Breaking the code of DNA binding specificity of TAL-type III effectors. Science (New York, N.Y.) 326: 1509–12. [DOI] [PubMed] [Google Scholar]
- Boissy G, de La Fortelle E, Kahn R, et al. 1996. Crystal structure of a fungal elicitor secreted by Phytophthora cryptogea, a member of a novel class of plant necrotic proteins. Structure (London, England: 1993) 4: 1429–1439. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G.. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annual Review of Plant Biology 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Bos JIB, Chaparro-Garcia A, Quesada-Ocampo LM, McSpadden Gardener BB, Kamoun S.. 2009. Distinct amino acids of the Phytophthora infestans effector AVR3a condition activation of R3a hypersensitivity and suppression of cell death. Molecular Plant-Microbe Interactions 22: 269–281. [DOI] [PubMed] [Google Scholar]
- Bos JIB, Prince D, Pitino M, Maffei ME, Win J, Hogenhout SA. 2010. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genetics 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bové JM. 2014. Huanglongbing or yellow shoot, a disease of Gondwanan origin: will it destroy citrus worldwide? Phytoparasitica 42: 579–583. [Google Scholar]
- Bové JM, Ayres AJ.. 2007. Etiology of three recent diseases of citrus in São Paulo State: sudden death, variegated chlorosis and huanglongbing. IUBMB Life 59: 346–354. [DOI] [PubMed] [Google Scholar]
- Brunings AM, Gabriel DW.. 2003. Xanthomonas citri: Breaking the surface. Molecular Plant Pathology 4: 141–157. [DOI] [PubMed] [Google Scholar]
- Canchaya C, Fournous G, Brüssow H.. 2004. The impact of prophages on bacterial chromosomes. Molecular Microbiology 53: 9–18. [DOI] [PubMed] [Google Scholar]
- Canihos Y, Peever TL, Timmer LW.. 1999. Temperature, leaf wetness, and isolate effects on infection of Minneola tangelo leaves by Alternaria sp. Plant Disease 83: 429–433. [DOI] [PubMed] [Google Scholar]
- Caplan J, Padmanabhan M, Dinesh-Kumar SP.. 2008. Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host and Microbe 3: 126–135. [DOI] [PubMed] [Google Scholar]
- Carazzolle MF, Formighieri EF, Digiampietri LA, Araujo MRR, Costa GGL, Pereira GAG.. 2007. Gene projects: a genome web tool for ongoing mining and annotation applied to CitEST. Genetics and Molecular Biology 30: 1030–1036. [Google Scholar]
- Carolan JC, Fitzroy CIJ, Ashton PD, Douglas AE, Wilkinson TL.. 2009. The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics 9: 2457–2467. [DOI] [PubMed] [Google Scholar]
- Carvalho SA, de Carvalho Nunes WM, Belasque J, et al. 2015. Comparison of resistance to asiatic citrus canker among different genotypes of citrus in a long-term canker-resistance field screening experiment in Brazil. Plant Disease 99: 207–218 [DOI] [PubMed] [Google Scholar]
- Caserta R, Takita MA, Targon ML, et al. 2010. Expression of Xylella fastidiosa fimbrial and afimbrial proteins durine biofilm formation. Applied and Environmental Microbiology 76: 4250–4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Molecular Microbiology 49: 277–300. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Velásquez AC, Ekengren SK, Collmer A, Martin GB.. 2010. Identification of Nicotiana benthamiana genes involved in pathogen-associated molecular pattern-triggered immunity. Molecular Plant-Microbe Interactions 23: 715–726. [DOI] [PubMed] [Google Scholar]
- Chellappan P, Vanitharani R, Ogbe F, Fauquet CM.. 2005. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiology 138: 1828–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-Q, Cao L, Dekkers KL, et al. 2005. A gene with domains related to transcription regulation is required for pathogenicity in Colletotrichum acutatum causing key lime anthracnose. Molecular Plant Pathology 6: 513–525. [DOI] [PubMed] [Google Scholar]
- Chin EL, Mishchuk DO, Breksa AP, Slupsky CM.. 2014. Metabolite signature of Candidatus liberibacter asiaticus infection in two citrus varieties. Journal of Agricultural and Food Chemistry 62: 6585–6591. [DOI] [PubMed] [Google Scholar]
- Chung K-R. 2012. Stress response and pathogenicity of the necrotrophic fungal pathogen Alternaria alternata. Scientifica 2012: 635431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EL, Karley AJ, Hubbard SF.. 2010. Insect endosymbionts: manipulators of insect herbivore trophic interactions? Protoplasma 244: 25–51. [DOI] [PubMed] [Google Scholar]
- Coletta-Filho HD, Pereira EO, Souza AA, Takita MA, Cristofani-Yale M, Machado MA. 2007. Analysis of resistance to Xylella fastidiosa within a hybrid population of Pera sweet orange × Murcott tangor. Plant Pathology 56: 661–668. [Google Scholar]
- Cong Q, Kinch LN, Kim B-H, Grishin N V.. 2012. Predictive sequence analysis of the Candidatus Liberibacter asiaticus proteome. PloS one 7: e41071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consales F, Schweizer F, Erb M, et al. 2012. Insect oral secretions suppress wound-induced responses in Arabidopsis. Journal of Experimental Botany 63: 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, et al. 2015. Fungal Planet description sheets: 320–370. Persoonia 34: 167–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, Crous PW.. 2012. The Colletotrichum acutatum species complex. Studies in Mycology 73: 37–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG.. 2001. Defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Dawson WO, Garnsey SM, Tatineni S, Folimonova SY, Harper SJ, Gowda S.. 2013. Citrus tristeza virus-host interactions. Frontiers in Microbiology 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice B, Wilson RR.. 2009. Molecular characterization of a novel pathogen-responsive receptor kinase-like in Citrus limon. Tree Genetics & Genomes 6: 47–56. [Google Scholar]
- Denoux C, Galletti R, Mammarella N, et al. 2008. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Molecular Plant 1: 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz L, Del Río JA, Pérez-Gilabert M, Ortuño A.. 2015. Involvement of an extracellular fungus laccase in the flavonoid metabolism in Citrus fruits inoculated with Alternaria alternata. Plant Physiology and Biochemistry 89: 11–17. [DOI] [PubMed] [Google Scholar]
- Djamei A, Schipper K, Rabe F, et al. 2011. Metabolic priming by a secreted fungal effector. Nature 478: 395–398. [DOI] [PubMed] [Google Scholar]
- Dou D, Kale SD, Wang X, et al. 2008. Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. The Plant Cell 20: 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JM, Lin ZJD, Coaker G.. 2011. Plant NB-LRR signaling: upstreams and downstreams. Current Opinion in Plant Biology 14: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga D, Jander G.. 2013. The role of protein effectors in plant-aphid interactions. Current Opinion in Plant Biology 16: 451–456. [DOI] [PubMed] [Google Scholar]
- Escalon A, Javegny S, Vernière C, et al. 2013. Variations in type III effector repertoires, pathological phenotypes and host range of Xanthomonas citri pv. citri pathotypes. Molecular Plant Pathology 14: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti E, Govetto B, Minet-Kebdani N, et al. 2013. The Phytophthora parasitica RXLR effector Penetration-Specific Effector 1 favours Arabidopsis thaliana infection by interfering with auxin physiology. New Phytologist 199: 476–489. [DOI] [PubMed] [Google Scholar]
- Fagoaga C, Pensabene-Bellavia G, Moreno P, Navarro L, Flores R, Peña L.. 2011. Ectopic expression of the p23 silencing suppressor of Citrus tristeza virus differentially modifies viral accumulation and tropism in two transgenic woody hosts. Molecular Plant Pathology 12: 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanta N, Ortega X, Pérez LM.. 2003. The development of Alternaria alternata is prevented by chitinases and β-1,3-glucanases from Citrus limon seedlings. Biological Research 36: 411–20. [DOI] [PubMed] [Google Scholar]
- Felfoldi G, Marokhazi J, Kepiro M, Venekei I.. 2009. Identification of natural target proteins indicates functions of a serralysin-type metalloprotease, PrtA, in anti-immune mechanisms. Applied and Environmental Microbiology 75: 3120–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellbrich G, Romanski A, Varet A, et al. 2002. NPP1, a Phytophthora-associated trigger of plant defence in parsley and Arabidopsis. The Plant Journal 32: 375–390. [DOI] [PubMed] [Google Scholar]
- Felton GW, Tumlinson JH.. 2008. Plant-insect dialogs: complex interactions at the plant-insect interface. Current Opinion in Plant Biology 11: 457–63. [DOI] [PubMed] [Google Scholar]
- Fisher T, He R, Nelson W.. 2014. A comparative transcriptomic approach to elucidate Psyllid-Ca. Liberibacter interactions. Journal of Citrus Pathology. [Google Scholar]
- Fleites LA, Jain M, Zhang S, Gabriel DW.. 2014. ‘ Ca. Liberibacter asiaticus’ prophage late genes may limit host range and culturability. Applied Environmental Microbiology 80: AEM.01958-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH. 1971. Current status of the gene-for-gene concept. Annual Review of Phytopathology 9: 275–296. [Google Scholar]
- Flores R, Ruiz-Ruiz S, Soler N, et al. 2013. Citrus tristeza virus p23: a unique protein mediating key virus-host interactions. Frontiers in Microbiology 4: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folimonova SY, Folimonov AS, Tatineni S, Dawson WO.. 2008. Citrus tristeza virus: survival at the edge of the movement continuum. Journal of Virology 82: 6546–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frago E, Dicke M, Godfray HCJ.. 2012. Insect symbionts as hidden players in insect-plant interactions. Trends in Ecology & Evolution 27: 705–711. [DOI] [PubMed] [Google Scholar]
- Fu S, Li Z, Tan J, Su H, Hartung J, Zhou CY.. 2014. Ultrastructural changes and putative phage particles observed in sweet orange leaves infected with ‘Candidatus Liberibacter asiaticus.’ Plant Disease 141010072153003. [DOI] [PubMed] [Google Scholar]
- Garavaglia BS, Zimaro T, Abriata LA, Ottado J, Gottig N.. 2016. XacFhaB adhesin, an important Xanthomonas citri subsp. citri virulence factor, is recognized as a pathogen-associated molecular pattern. Molecular Plant Pathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier M, Jagoueix-Eveillard S, Cronje PR, Le Roux HF, Bové JM.. 2000. Genomic characterization of a liberibacter present in an ornamental rutaceous tree, Calodendrum capense, in the Western Cape province of South Africa. Proposal of ‘Candidatus liberibacter africanus subsp. capensis.’ International Journal of Systematic and Evolutionary Microbiology 50: 2119–2125. [DOI] [PubMed] [Google Scholar]
- Geer LY, Marchler-Bauer A, Geer RC, et al. 2010. The NCBI BioSystems database. Nucleic Acids Research 38: D492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo MC, Valent B.. 2013. Filamentous plant pathogen effectors in action. Nature Reviews Microbiology 11: 800–814. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. 2005. Contrasting mechanisms of defence against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Gmitter FG, Chen C, Machado MA, et al. 2012. Citrus genomics. Tree Genetics & Genomes 8: 611–626. [Google Scholar]
- Gochez AM, Minsavage G V., Potnis N, Canteros BI, Stall RE, Jones JB.. 2015. A functional XopAG homologue in Xanthomonas fuscans pv. aurantifolii strain C limits host range. Plant Pathology 64: 1207–1214. [Google Scholar]
- Godinho DP, Janssen A, Dias T, et al. 2016. Down-regulation of plant defence in a resident spider mite species and its effect upon con- and heterospecifics. Oecologia 180: 161–7. [DOI] [PubMed] [Google Scholar]
- de Goes A, Garrido RBO, Reis RF, Baldassari RB, Soares MA.. 2008. Evaluation of fungicide applications to sweet orange at different flowering stages for control of postbloom fruit drop caused by Colletotrichum acutatum. Crop Protection 27: 71–76. [Google Scholar]
- Goff KE, Ramonell KM.. 2007. The role and regulation of receptor-like kinases in plant defence. Gene Regulation and Systems Biology 1: 167–175. [PMC free article] [PubMed] [Google Scholar]
- Göhre V, Robatzek S.. 2008. Breaking the barriers: microbial effector molecules subvert plant immunity. Annual Review of Phytopathology 46: 189–215. [DOI] [PubMed] [Google Scholar]
- Gomi K, Itoh N, Yamamoto H, Akimitsu K.. 2002a. Characterization and functional analysis of Class I and II acidic chitinase cDNA from rough lemon. Journal of General Plant Pathology 68: 191–199. [Google Scholar]
- Gomi K, Yamamoto H, Akimitsu K.. 2002b. Characterization of a lipoxygenase gene in rough lemon induced by Alternaria alternata. Journal of General Plant Pathology 68: 21–30. [Google Scholar]
- Gomi K, Yamamato H, Akimitsu K.. 2003a. Epoxide hydrolase: a mRNA induced by the fungal pathogen Alternaria alternata on rough lemon (Citrus jambhiri Lush). Plant Molecular Biology 53: 189–199. [DOI] [PubMed] [Google Scholar]
- Gomi K, Yamasaki Y, Yamamoto H, Akimitsu K.. 2003b. Characterization of a hydroperoxide lyase gene and effect of C6-volatiles on expression of genes of the oxylipin metabolism in Citrus. Journal of plant physiology 160: 1219–31. [DOI] [PubMed] [Google Scholar]
- Gotoh Y, Nalumpang S, Isshiki A, et al. 2002. A cDNA encoding polygalacturonase-inhibiting protein induced in citrus leaves by polygalacturonase of Alternaria citri. Journal of General Plant Pathology 68: 57–61. [Google Scholar]
- da Graça JV, Douhan GW, Halbert SE, et al. 2016. Huanglongbing: an overview of a complex pathosystem ravaging the world’s citrus. Journal of Integrative Plant Biology 58: 373–387. [DOI] [PubMed] [Google Scholar]
- Grouffaud S, van West P, Avrova AO, Birch PRJ, Whisson SC.. 2008. Plasmodium falciparum and Hyaloperonospora parasitica effector translocation motifs are functional in Phytophthora infestans. Microbiology 154: 3743–3751. [DOI] [PubMed] [Google Scholar]
- Guidetti-Gonzalez S, Carrer H.. 2007. Putative resistance genes in the CitEST database. Genetics and Molecular Biology 30: 931–942. [Google Scholar]
- Guo K, Wang W, Luo L, Chen J, Guo Y, Cui F.. 2014. Characterization of an aphid-specific, cysteine-rich protein enriched in salivary glands. Biophysical Chemistry 189: 25–32. [DOI] [PubMed] [Google Scholar]
- Gururani MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, Park SW.. 2012. Plant disease resistance genes: current status and future directions. Physiological and Molecular Plant Pathology 78: 51–65. [Google Scholar]
- Haapalainen M. 2014. Biology and epidemics of Candidatus Liberibacter species, psyllid-transmitted plant-pathogenic bacteria. Annals of Applied Biology 165: 172–198. [Google Scholar]
- Haas BJ, Kamoun S, Zody MC, et al. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398. [DOI] [PubMed] [Google Scholar]
- Hao G, Boyle M, Zhou L, Duan Y.. 2013. The intracellular Citrus Huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ encodes two novel autotransporters. PLoS One 8: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao G, Pitino M, Duan Y, Stover E.. 2016. Reduced susceptibility to Xanthomonas citri in transgenic citrus expressing the FLS2 receptor from Nicotiana benthamiana. Molecular Plant-Microbe Interactions 29: 132–142. [DOI] [PubMed] [Google Scholar]
- Hattori M, Nakamura M, Komatsu S, Tsuchihara K, Tamura Y, Hasegawa T.. 2012. Molecular cloning of a novel calcium-binding protein in the secreted saliva of the green rice leafhopper Nephotettix cincticeps. Insect Biochemistry and Molecular Biology 42: 1–9. [DOI] [PubMed] [Google Scholar]
- Hijaz F, El-Shesheny I, Killiny N.. 2013. Herbivory by the insect Diaphorina citri induces greater change in citrus plant volatile profile than does infection by the bacterium, Candidatus Liberibacter asiaticus. Plant Signaling & Behavior doi:10.4161/psb.25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenhout SA, Van der Hoorn R a L, Terauchi R, Kamoun S.. 2009. Emerging concepts in effector biology of plant-associated organisms. Molecular Plant-Microbe Interactions 22: 115–122. [DOI] [PubMed] [Google Scholar]
- Hopkins DL, Purcell AH.. 2002. Xylella fastidiosa: cause of Pierce’s disease of grapevines and other emergent diseases. Plant Disease 86: 1056–1066. [DOI] [PubMed] [Google Scholar]
- Huitema E, Vleeshouwers VG, Cakir C, Kamoun S, Govers F.. 2005. Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans. Molecular Plant-Microbe Interactions 18: 183–193. [DOI] [PubMed] [Google Scholar]
- Incarbone M, Dunoyer P.. 2013. RNA silencing and its suppression: novel insights from in planta analyses. Trends in Plant Science 18: 382–392. [DOI] [PubMed] [Google Scholar]
- Ionescu M, Zaini PA, Baccari C, Tran S, da Silva AM, Lindow SE.. 2014. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proceedings of the National Academy of Sciences of the United States of America 111: E3910–E3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki A, Akimitsu K, Nishio K, Tsukamoto M, Yamamoto H.. 1997. Purification and characterization of an endopolygalacturonase from the rough lemon pathotype ofAlternaria alternata, the cause of citrus brown spot disease. Physiological and Molecular Plant Pathology 51: 155–167. [Google Scholar]
- Isshiki A, Akimitsu K, Yamamoto M, Yamamoto H.. 2001. Endopolygalacturonase is essential for citrus black rot caused by Alternaria citri but not brown spot caused by Alternaria alternata. Molecular Plant-Microbe Interactions 14: 749–57. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohtani K, Miyamoto Y, et al. 2012. A polyketide synthase gene, ACRTS2, is responsible for biosynthesis of host-selective ACR-toxin in the rough lemon pathotype of Alternaria alternata. Molecular Plant-Microbe Interactions 25: 1419–1429. [DOI] [PubMed] [Google Scholar]
- Jain M, Fleites L, Gabriel DW.. 2015. Prophage encoded peroxidase in ‘Candidatus Liberibacter asiaticus’ is a secreted effector that suppresses plant defenses. Molecular Plant-Microbe Interactions 3: 150827055927002. [DOI] [PubMed] [Google Scholar]
- Jalan N, Kumar D, Andrade MO, et al. 2013. Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp. citri provide insights into mechanisms of bacterial virulence and host range. BMC Genomics 14: 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay F, Wang Y, Yu A, et al. 2011. Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis. PLoS Pathogens 7: e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL.. 2006. The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Junker RR. 2014. New synthesis: a holobiontic view on plant-insect interactions. Journal of Chemical Ecology 40: 521. [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, et al. 2006. Plant cells recognize chitin fragments for defence signaling through a plasma membrane receptor. Proceedings of the National Academy of Sciences of the United States of America 103: 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S. 1993. A gene encoding a host-specific elicitor protein of Phytophthora parasitica. Molecular Plant-Microbe Interactions 6: 573. [DOI] [PubMed] [Google Scholar]
- Kamoun S. 2006. A catalogue of the effector secretome of plant pathogenic oomycetes. Annual Review of Phytopathology 44: 41–60. [DOI] [PubMed] [Google Scholar]
- Kamoun S. 2009. The secretome of plant-associated fungi and oomycetes In: Deising HB, ed. The Mycota. Berlin: Springer, 173–180. [Google Scholar]
- Kamoun S, van West P, Vleeshouwers V, de Groot KE, Govers F.. 1998. Resistance of nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. The Plant cell 10: 1413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S, Furzer O, Jones JDG, et al. 2015. The top 10 oomycete pathogens in molecular plant pathology. Molecular Plant Pathology 16: 413–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki H, Saitoh H, Ito A, et al. 2003. Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Molecular Plant Pathology 4: 383–391. [DOI] [PubMed] [Google Scholar]
- Khalfallah Y, Bouktila D, Makni M, Makni H.. 2015. Tracking microRNAs with a potential for virulence regulation in the pea aphid, Acyrthosiphon pisum Harris (Hemiptera: Aphidae), and the Asian citrus psyllid, Diaphorina citri Kuwayama (Hemiptera: Psyllidae). African Entomology 23: 502–509. [Google Scholar]
- Khatib M, Lafitte C, Esquerre-Tugaye M-T, Bottin A, Rickauer M.. 2004. The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytologist 162: 501–510. [Google Scholar]
- Killiny N, Almeida RPP.. 2009. Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Applied and Environmental Microbiology 75: 521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killiny N, Almeida RPP.. 2014. Factors affecting the initial adhesion and retention of the plant pathogen Xylella fastidiosa in the foregut of an insect vector. Applied and Environmental Microbiology 80: 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killiny N, Rashed A, Almeida RPP.. 2012. Disrupting the transmission of a vector-borne plant pathogen. Applied and Environmental Microbiology 78: 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killiny N, Martinez RH, Dumenyo CK, Cooksey DA, Almeida RPP.. 2013. The Exopolysaccharide of Xylella fastidiosa is essential for biofilm formation, plant virulence, and vector transmission. Molecular Plant-Microbe Interactions 26: 1044–1053. [DOI] [PubMed] [Google Scholar]
- Kim B-G, Fukumoto T, Tatano S, et al. 2009. Molecular cloning and characterization of a thaumatin-like protein-encoding cDNA from rough lemon. Physiological and Molecular Plant Pathology 74: 3–10. [Google Scholar]
- Kim J-S, Sagaram US, Burns JK, Li J-L, Wang N.. 2009. Response of sweet orange (Citrus sinensis) to ‘Candidatus Liberibacter asiaticus’ infection: microscopy and microarray analyses. Phytopathology 99: 50–57. [DOI] [PubMed] [Google Scholar]
- Kleemann J, Rincon-Rivera LJ, Takahara H, et al. 2012. Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathogens 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh EJ, Zhou L, Williams DS, et al. 2012. Callose deposition in the phloem plasmodesmata and inhibition of phloem transport in citrus leaves infected with ‘Candidatus Liberibacter asiaticus.’ Protoplasma 249: 687–697. [DOI] [PubMed] [Google Scholar]
- Kohmoto K. 1979. Host-selective toxins from Alternaria citri. Phytopathology 69: 667. [Google Scholar]
- Kohmoto K. 1993. Isolation and biological activities of two host-specific toxins from the tangerine pathotype of Alternaria alternata. Phytopathology 83: 495. [Google Scholar]
- Kohmoto K, Kondoh Y, Kohguchi T, Otani H, Nishimura S, Scheffer RP.. 1984. Ultrastructural changes in host leaf cells caused by host-selective toxin of Alternaria alternata from rough lemon. Canadian Journal of Botany 62: 2485–2492. [Google Scholar]
- Kroon LPNM, Brouwer H, de Cock AWA M, Govers F.. 2012. The genus Phytophthora anno 2012. Phytopathology 102: 348–364. [DOI] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. 2004. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. The Plant Cell 16: 3496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laia ML, Moreira LM, Dezajacomo J, et al. 2009. New genes of Xanthomonas citri subsp. citri involved in pathogenesis and adaptation revealed by a transposon-based mutant library. BMC microbiology 9: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque CA, Brouwer H, Cano L, et al. 2010. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biology 11: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lherminier J, Benhamou N, Larrue J, et al. 2003. Cytological characterization of elicitin-induced protection in tobacco plants infected by Phytophthora parasitica or Phytoplasma. Phytopathology 93: 1308–1319. [DOI] [PubMed] [Google Scholar]
- Li Z, Zou L, Ye G, et al. 2014. A potential disease susceptibility gene CsLOB of citrus is targeted by a major virulence effector PthA of Xanthomonas citri subsp. citri. Molecular Plant 7: 912–915. [DOI] [PubMed] [Google Scholar]
- Lima WG, Spósito MB, Amorim L, Gonçalves FP, de Filho PAM.. 2011. Colletotrichum gloeosporioides, a new causal agent of citrus post-bloom fruit drop. European Journal of Plant Pathology 131: 157–165. [Google Scholar]
- Lin C-H, Yang SL, Chung K-R.. 2011. Cellular responses required for oxidative stress tolerance, colonization, and lesion formation by the necrotrophic fungus Alternaria alternata in Citrus. Current Microbiology 62: 807–815. [DOI] [PubMed] [Google Scholar]
- Liu Z, Faris JD, Oliver RP, et al. 2009. SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathogens 5: e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens E, Fernández-Crespo E, Vicedo B, Lapeña L, García-Agustín P.. 2013. Enhancement of the citrus immune system provides effective resistance against Alternaria brown spot disease. Journal of Plant Physiology 170: 146–154. [DOI] [PubMed] [Google Scholar]
- Lochman J, Kasparovsky T, Damborsky J, et al. 2005. Construction of cryptogein mutants, a proteinaceous elicitor from Phytophthora, with altered abilities to induce a defence reaction in tobacco cells. Biochemistry 44: 6565–6572. [DOI] [PubMed] [Google Scholar]
- Lu R, Folimonov A, Shintaku M, et al. 2004. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proceedings of the National Academy of Sciences of the United States of America 101: 15742–15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz E. 2001. Doenças causadas por Phytophthora no Brasil. Campinas: Editora Rural1: 1–22.
- Machado MA, Cristofani-Yaly M, Bastianel M.. 2011. Breeding, genetic and genomic of citrus for disease resistance. Revista Brasileira de Fruticultura 33: 158–172. [Google Scholar]
- Mackey D, Holt BF, Wiig A, Dangl JL.. 2002. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- Mafra V, Martins PK, Francisco CS, Ribeiro-Alves M, Freitas-Astúa J, Machado MA. 2013. Candidatus Liberibacter americanus induces significant reprogramming of the transcriptome of the susceptible citrus genotype. BMC Genomics 14: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafurah JJ, Ma H, Zhang M, et al. 2015. A virulence essential CRN effector of Phytophthora capsici suppresses host defence and induces cell death in plant nucleus. PLoS One 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Ali JG, Hermann SL, et al. 2012. Induced release of a plant-defence volatile ‘deceptively’ attracts insect vectors to plants infected with a bacterial pathogen. PLoS Pathogens 8: e1002610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Zhurov V, Navarro M, et al. 2015. Tomato whole genome transcriptional response to Tetranychus urticae identifies divergence of spider mite-induced responses between tomato and arabidopsis. Molecular Plant Microbe Interactions 28: 343–61. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CAJ.. 1997. Innate immunity: Minireview the virtues of a nonclonal system of recognition. Cell 91: 295–298. [DOI] [PubMed] [Google Scholar]
- Menouni R, Hutinet G, Petit M-A, Ansaldi M.. 2014. Bacterial genome remodeling through bacteriophage recombination. FEMS Microbiology Letters 362: 1–10. [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, et al. 2007. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 104: 19613–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C.. 2012. Plant pattern recognition receptor complexes at the plasma membrane. Current Opinion in Plant Biology 15: 349–357. [DOI] [PubMed] [Google Scholar]
- Moreira LM, Almeida NF, Potnis N, et al. 2010. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics 11: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno P, Ambros S, Albia-Marti MR, Guerri J, Pena L.. 2008. Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Molecular Plant Pathology 9: 251–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan W, Kamoun S.. 2007. RXLR effectors of plant pathogenic oomycetes. Current Opinion in Microbiology 10: 332–338. [DOI] [PubMed] [Google Scholar]
- Mozoruk J, Hunnicutt LE, Cave RD, Hunter WB, Bausher MG.. 2006. Profiling transcriptional changes in Citrus sinensis (L.) Osbeck challenged by herbivory from the xylem-feeding leafhopper Homalodisca coagulata (Say) by cDNA macroarray analysis. Plant Science 170: 1068–1080. [Google Scholar]
- Muranaka LS, Takita MA, Olivato JC, Kishi LT, de Souza AA. 2012. Global expression profile of biofilm resistance to antimicrobial compounds in the plant-pathogenic bacterium Xylella fastidiosa reveals evidence of persister cells. Journal of Bacteriology 194: 4561–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutti NS, Louis J, Pappan LK, et al. 2008. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proceedings of the National Academy of Sciences of the United States of America 105: 9965–9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento R, Gouran H, Chakraborty S, et al. 2016. The type II secreted lipase/esterase LesA is a key virulence factor required for Xylella fastidiosa pathogenesis in grapevines. Scientific Reports 6: 18598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nespoulous C, Gaudemer O, Huet J-C, Pernollet J-C.. 1999. Characterization of elicitin-like phospholipases isolated from Phytophthora capsici culture filtrate. FEBS Letters 452: 400–406. [DOI] [PubMed] [Google Scholar]
- Neves MF, Trombin VG, Milan P, Lopes FF, Cressoni F,Kalaki R.. 2014. O Retrato da citricultura Brasileira, In: FM Neves, ed. São Paulo: CitrusBR, p138. [Google Scholar]
- Nicholson SJ, Hartson SD, Puterka GJ.. 2012. Proteomic analysis of secreted saliva from Russian wheat aphid (Diuraphis noxia Kurd.) biotypes that differ in virulence to wheat. Journal of Proteomics 75: 2252–2268. [DOI] [PubMed] [Google Scholar]
- Niza B, Coletta-Filho HD, Merfa MV, Takita MA, de Souza AA.. 2015. Differential colonization patterns of Xylella fastidiosa infecting citrus genotypes. Plant Pathology 64: 1259–1269. [Google Scholar]
- Nürnberger T, Brunner F.. 2002. Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Current Opinion in Plant Biology 5: 318–324. [DOI] [PubMed] [Google Scholar]
- O’Connell RJ, Thon MR, Hacquard S, et al. 2012. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nature Genetics 44: 1060–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM.. 2009. The evolution of RNAi as a defence against viruses and transposable elements. Philosophical transactions of the Royal Society of London. Series B, Biological Sciences 364: 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver RP, Solomon PS.. 2010. New developments in pathogenicity and virulence of necrotrophs. Current Opinion in Plant Biology 13: 415–419. [PubMed] [Google Scholar]
- Orlovskis Z, Canale MC, Thole V, Pecher P, Lopes JR, Hogenhout SA.. 2015. Insect-borne plant pathogenic bacteria: getting a ride goes beyond physical contact. Current Opinion in Insect Science 9: 16–23. [DOI] [PubMed] [Google Scholar]
- Oßwald W, Fleischmann F, Rigling D, et al. 2014. Strategies of attack and defence in woody plant – Phytophthora interactions. Forest Pathology 44: 169–190. [Google Scholar]
- Pacheco R, García-Marcos A, Barajas D, Martiáñez J, Tenllado F.. 2012. PVX–potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana. Virus Research 165: 231–235. [DOI] [PubMed] [Google Scholar]
- Padmanabhan MS, Ma S, Burch-Smith TM, Czymmek K, Huijser P, Dinesh-Kumar SP.. 2013. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathogens 9: e1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panabières F, Amselem J, Galiana E, Le Berre J-Y.. 2005. Gene identification in the oomycete pathogen Phytophthora parasitica during in vitro vegetative growth through expressed sequence tags. Fungal Genetics and Biology 42: 611–23. [DOI] [PubMed] [Google Scholar]
- Panabieres F, Ali GS, Allagui MB, et al. 2016. Phytophthora nicotianae diseases worldwide: new knowledge of a long-recognised pathogen. Phytopathologia Mediterranea 55: 20–40. [Google Scholar]
- Peever TL, Su G, Carpenter-Boggs L, Timmer LW.. 2004. Molecular systematics of Citrus-associated Alternaria species. Mycologia 96: 119. [PubMed] [Google Scholar]
- Pelz-Stelinski KS, Killiny N.. 2016. Better together: association with ‘ Candidatus Liberibacter asiaticus’ increases the reproductive fitness of its insect vector, Diaphorina citri (Hemiptera: Liviidae). Annals of the Entomological Society of America 109: 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Donoso AG, Sun Q, Roper MC, Greve LC, Kirkpatrick B, Labavitch JM.. 2010. Cell wall-degrading enzymes enlarge the pore size of intervessel pit membranes in healthy and Xylella fastidiosa-infected grapevines. Plant Physiology 152: 1748–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrocelli S, Tondo ML, Daurelio LD, Orellano EG.. 2012. Modifications of Xanthomonas axonopodis pv. citri lipopolysaccharide affect the basal response and the virulence process during citrus canker. PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino M, Hoffman MT, Zhou L, Hall DG, Stocks IC, Duan Y.. 2014. The phloem-sap feeding mealybug (Ferrisia virgata) carries ‘Candidatus Liberibacter asiaticus’ populations that do not cause disease in host plants. PLoS One 9: e85503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino M, Hao G, Duan Y.. 2015. Expression of the LasAI effector of ‘ Candidatus Liberibacter asiaticus’ induced proliferation of root hair and trichome in transformed plants. In Abstracts from the 4th International Research Conference on Huanglongbing: 1. [Google Scholar]
- Pitino M, Armstrong CM, Cano LM, Duan Y.. 2016. Transient expression of Candidatus Liberibacter asiaticus effector induces cell death in Nicotiana benthamiana. Frontiers in Plant Science 7: 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Schwessinger B, Joe A, et al. 2015. The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Science Advances 1: e1500245–e1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Voinnet O.. 2013. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nature Reviews Microbiology 11: 745–760. [DOI] [PubMed] [Google Scholar]
- Puttamuk T, Zhou L, Thaveechai N, Zhang S, Armstrong CM, Duan Y.. 2014. Genetic diversity of Candidatus Liberibacter asiaticus based on two hypervariable effector genes in Thailand. PLoS One 9: e112968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai M. 2006. Refinement of the Citrus Tristeza Virus resistance gene (Ctv) positional map in Poncirus trifoliata and generation of transgenic grapefruit (Citrus paradisi) plant lines with candidate resistance genes in this region. Plant Molecular Biology 61: 399–414. [DOI] [PubMed] [Google Scholar]
- Rajput NA, Zhang M, Ru Y, Liu T, Xu J, Liu L, Mafurah JJ, Dou D.. 2014. Phytophthora sojae effector PsCRN70 suppresses plant defences in Nicotiana benthamiana. PLoS One 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli JN, Kinsinger N, Perring TM, et al. 2015. O antigen modulates insect vector acquisition of the bacterial plant pathogen Xylella fastidiosa. Applied and Environmental Microbiology 81: 8145–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis MS, Takita MA, Palmieri DA, Machado MA.. 2007. Bioinformatics for the Citrus EST project (CitEST). Genetics and Molecular Biology 30: 1024–1029. [Google Scholar]
- Reyes CA, Ocolotobiche EE, Marmisollé FE, et al. 2016. Citrus psorosis virus 24K protein interacts with citrus miRNA precursors, affects their processing and subsequent miRNA accumulation and target expression. Molecular Plant Pathology 17: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigano L, Siciliano F, Enrique R, et al. 2007. Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Molecular Plant-Microbe Interactions 20: 1222–1230. [DOI] [PubMed] [Google Scholar]
- Robatzek S, Bittel P, Chinchilla D, et al. 2007. Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Molecular Biology 64: 539–547. [DOI] [PubMed] [Google Scholar]
- Rodrigues CM, Takita M, Coletta-Filho HD, et al. 2008. Copper resistance of biofilm cells of the plant pathogen Xylella fastidiosa. Applied Microbiology and Biotechnology 77: 1145–1157. [DOI] [PubMed] [Google Scholar]
- Rodrigues CM, de Souza AA, Takita MA, Kishi LT, Machado MA.. 2013. RNA-Seq analysis of Citrus reticulata in the early stages of Xylella fastidiosa infection reveals auxin-related genes as a defence response. BMC Genomics 14: 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ronde D, Butterbach P, Kormelink R.. 2014. Dominant resistance against plant viruses. Frontiers in Plant Science 5: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ruiz S, Navarro B, Gisel A, et al. 2011. Citrus tristeza virus infection induces the accumulation of viral small RNAs (21-24-nt) mapping preferentially at the 3’-terminal region of the genomic RNA and affects the host small RNA profile. Plant Molecular Biology 75: 607–19. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Vorhölter F-J, Potnis N, et al. 2011. Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nature Reviews Microbiology 9: 344–355. [DOI] [PubMed] [Google Scholar]
- Ryan RP, An S-Q, Allan JH, McCarthy Y, Dow JM.. 2015. The DSF family of cell-cell signals: an expanding class of bacterial virulence regulators. PLoS Pathogens 11: e1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak M, Minsavage GV, Stall RE, Jones JB.. 2009. Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host. Molecular Plant Pathology 10: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S, van Damme M, Bozkurt TO, et al. 2010. Ancient class of translocated oomycete effectors targets the host nucleus. Proceedings of the National Academy of Sciences of the United States of America 107: 17421–17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Cao M, Leung D, Tyler BM.. 2004. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Molecular Plant-Microbe Interactions 17: 394–403. [DOI] [PubMed] [Google Scholar]
- Sharma A, Khan AN, Subrahmanyam S, Raman A, Taylor GS, Fletcher MJ.. 2014. Salivary proteins of plant-feeding hemipteroids – implication in phytophagy. Bulletin of Entomological Research 104: 117–136. [DOI] [PubMed] [Google Scholar]
- Shi Q, Febres VJ, Jones JB, Moore GA.. 2015. Responsiveness of different citrus genotypes to the Xanthomonas citri ssp. citri-derived pathogen-associated molecular pattern (PAMP) flg22 correlates with resistance to citrus canker. Molecular Plant Pathology 16: 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya T, Ménard R, Kozone I, et al. 2006. Novel β-1,3-, 1,6-oligoglucan elicitor from Alternaria alternata 102 for defence responses in tobacco. The FEBS Journal 273: 2421–2431. [DOI] [PubMed] [Google Scholar]
- Shinya T, Hanai K, Gális I, et al. 2007. Characterization of NtChitIV, a class IV chitinase induced by β-1,3-, 1,6-glucan elicitor from Alternaria alternata 102: antagonistic effect of salicylic acid and methyl jasmonate on the induction of NtChitIV. Biochemical and Biophysical Research Communications 353: 311–317. [DOI] [PubMed] [Google Scholar]
- Shiu S-H, Bleecker AB.. 2001. Plant receptor-like kinase gene family: diversity, function, and signaling. Science Signaling 2001: re22–re22. [DOI] [PubMed] [Google Scholar]
- Shiu S, Bleecker AB.. 2003. Expansion of the receptor-like kinase / pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiology 132: 530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S, Karlowski W, Pan R.. 2004. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. The Plant Cell 16: 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva CR, Ferro J, Reinach FC, et al. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417: 459–463. [DOI] [PubMed] [Google Scholar]
- Souza AA De, Takita MA, Coletta-Filho HD, et al. 2007. Comparative analysis of differentially expressed sequence tags of sweet orange and mandarin infected with Xylella fastidiosa. Genetics and Molecular Biology 30: 965–971. [Google Scholar]
- Souza AA, Takita MA, Amaral AM, Coletta-Filho HD, Machado MA.. 2009. Citrus responses to Xylella fastidiosa infection, the causal agent de citrus variegated chlorosis. Tree and Forestry Science and Biotechnology 3: 957–964. [Google Scholar]
- Stassen JHM, Van den Ackerveken G.. 2011. How do oomycete effectors interfere with plant life? Current Opinion in Plant Biology 14: 407–414. [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I, Collemare J, Mehrabi R, De Wit PJGM.. 2012. Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiology Reviews 37: 67–93. [DOI] [PubMed] [Google Scholar]
- Streubel J, Blücher C, Landgraf A, Boch J.. 2012. TAL effector RVD specificities and efficiencies. Nature Biotechnology 30: 593–595. [DOI] [PubMed] [Google Scholar]
- Stuart J. 2015. Insect effectors and gene-for-gene interactions with host plants. Current Opinion in Insect Science 9: 56–61. [DOI] [PubMed] [Google Scholar]
- Sun X, Stall RE, Jones JB, et al. 2004. Detection and characterization of a new strain of citrus canker bacteria from key/Mexican lime and alemow in south Florida. Plant Disease 88: 1179–1188. [DOI] [PubMed] [Google Scholar]
- Svozilová Z, Kasparovský T, Skládal P, Lochman J.. 2009. Interaction of cryptogein with its binding sites in tobacco plasma membrane studied using the piezoelectric biosensor. Analytical Biochemistry 390: 115–20. [DOI] [PubMed] [Google Scholar]
- Swarup S, Yang Y, Kingsley MT, Gabriel DW.. 1992. An Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Molecular Plant-Microbe Interactions 5: 204–213. [DOI] [PubMed] [Google Scholar]
- Takai R, Isogai A, Takayama S, Che F-S.. 2008. Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Molecular Plant-Microbe Interactions 21: 1635–1642. [DOI] [PubMed] [Google Scholar]
- Targon MLPN, Takita MA, Amaral AM do, et al. 2007. CitEST libraries. Genetics and Molecular Biology 30: 1019–1023. [Google Scholar]
- Tatineni S, Dawson WO.. 2012. Enhancement or attenuation of disease by deletion of genes from Citrus Tristeza Virus. Journal of Virology 86: 7850–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatineni S, Robertson CJ, Garnsey SM, Dawson WO.. 2011. A plant virus evolved by acquiring multiple nonconserved genes to extend its host range. Proceedings of the National Academy of Sciences, USA 108: 17366–17371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer LW, Peever TL, Solel Z, Akimitsu K.. 2003. Alternaria diseases of citrus – Novel pathosystems. Phytopathologia Mediterranea 42:3–16. [Google Scholar]
- Tiwari S, Gondhalekar a D, Mann RS, Scharf ME, Stelinski LL.. 2011a. Characterization of five CYP4 genes from Asian citrus psyllid and their expression levels in Candidatus Liberibacter asiaticus-infected and uninfected psyllids. Insect Molecular Biology 20: 733–44. [DOI] [PubMed] [Google Scholar]
- Tiwari S, Pelz-Stelinski K, Mann RS, Stelinski LL.. 2011b. Glutathione transferase and cytochrome P450 (general oxidase) activity levels in Candidatus Liberibacter asiaticus-infected and uninfected asian citrus psyllid (Hemiptera: Psyllidae). Annals of the Entomological Society of America 104: 297–305. [Google Scholar]
- Tiwari S, Pelz-Stelinski K, Stelinski LL.. 2011c. Effect of Candidatus Liberibacter asiaticus infection on susceptibility of Asian citrus psyllid, Diaphorina citri, to selected insecticides. Pest Management Science 67: 94–99. [DOI] [PubMed] [Google Scholar]
- Torto TA, Li S, Styer A, et al. 2003. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Research 13: 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T, Harimoto Y, Akimitsu K, et al. 2013. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiology Reviews 37: 44–66. [DOI] [PubMed] [Google Scholar]
- Tyler BM, Tripathy S, Zhang X, et al. 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science (New York, N.Y.) 313: 1261–1266. [DOI] [PubMed] [Google Scholar]
- Umemoto N, Kakitani M, Iwamatsu A, Yoshikawa M, Yamaoka N, Ishida I.. 1997. The structure and function of a soybean beta-glucan-elicitor-binding protein. Proceedings of the National Academy of Sciences of the United States of America 94: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez K, Renovell A, Comellas M, et al. 2010. Effect of temperature on RNA silencing of a negative-stranded RNA plant virus: Citrus psorosis virus. Plant Pathology 59: 982–990. [Google Scholar]
- Vernière C, Hartung JS, Pruvost OP, et al. 1998. Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from Southwest Asia. European Journal of Plant Pathology 104: 477–487. [Google Scholar]
- Villarroel CA, Jonckheere W, Alba JM, et al. 2016. Salivary proteins of spider mites suppress defences in Nicotiana benthamiana and promote mite reproduction. The Plant Journal 86: 119–131. [DOI] [PubMed] [Google Scholar]
- Vyas M, Fisher TW, He R, et al. 2015. Asian citrus psyllid expression profiles suggest Candidatus Liberibacter asiaticus-mediated alteration of adult nutrition and metabolism, and of nymphal development and immunity. PLoS One 10: e0130328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Trivedi P.. 2013. Citrus huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology 103: 652–65. [DOI] [PubMed] [Google Scholar]
- Wang N, Jin T, Trivedi P, et al. 2015. Announcement of the International Citrus Microbiome (Phytobiome) Consortium. Journal of Citrus Pathology 2. [Google Scholar]
- Wang X, Jiang N, Liu J, Liu W, Wang G-L.. 2014. The role of effectors and host immunity in plant-necrotrophic fungal interactions. Virulence 5: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou L, Li D, et al. 2015. Genome-wide comparative analysis reveals similar types of NBS genes in hybrid Citrus sinensis genome and original Citrus clementine genome and provides new insights into non-TIR NBS genes. PLoS One 10: e0121893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, et al. 2007. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450: 115–118. [DOI] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thönnessen A, van Bel AJE.. 2007. Molecular sabotage of plant defence by aphid saliva. Proceedings of the National Academy of Sciences of the United States of America 104: 10536–10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win J, Morgan W, Bos J, et al. 2007. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. The Plant Cell 19: 2349–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win J, Chaparro-Garcia A, Belhaj K, et al. 2012. Effector biology of plant-associated organisms: concepts and perspectives. Cold Spring Harbor Symposia on Quantitative Biology 77: 235–47. [DOI] [PubMed] [Google Scholar]
- Wu C-H, Yan H-Z, Liu L-F, Liou R-F.. 2008. Functional characterization of a gene family encoding polygalacturonases in Phytophthora parasitica. Molecular Plant-Microbe Interactions 21: 480–489. [DOI] [PubMed] [Google Scholar]
- Wu J, Baldwin IT.. 2010. New insights into plant responses to the attack from insect herbivores. Annual Review of Genetics 44: 1–24. [DOI] [PubMed] [Google Scholar]
- Xu M, Li Y, Zheng Z, Dai Z, Tao Y, Deng X.. 2015. Transcriptional analyses of mandarins seriously infected by ‘Candidatus Liberibacter asiaticus.’ PLoS One 10: e0133652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A.. 2011. Endogenous peptide elicitors in higher plants. Current Opinion in Plant Biology 14: 351–357. [DOI] [PubMed] [Google Scholar]
- Yan H-Z, Liou R-F.. 2005. Cloning and analysis of pppg1, an inducible endopolygalacturonase gene from the oomycete plant pathogen Phytophthora parasitica. Fungal Genetics and Biology 42: 339–350. [DOI] [PubMed] [Google Scholar]
- Yan Q, Sreedharan A, Wei S, et al. 2013. Global gene expression changes in Candidatus Liberibacter asiaticus during the transmission in distinct hosts between plant and insect. Molecular Plant Pathology 14: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You B-J, Chung K-R.. 2007. Phenotypic characterization of mutants of the citrus pathogen Colletotrichum acutatum defective in a PacC-mediated pH regulatory pathway. FEMS Microbiology Letters 277: 107–114. [DOI] [PubMed] [Google Scholar]
- You B-J, Choquer M, Chung K-R.. 2007. The Colletotrichum acutatum gene encoding a putative pH-responsive transcription regulator is a key virulence determinant during fungal pathogenesis on citrus. Molecular Plant-Microbe Interactions 20: 1149–1160. [DOI] [PubMed] [Google Scholar]
- Yusuf M, Hayat S, Alyemeni MN, Fariduddin Q, Ahmad A.. 2013. Salicylic acid: physiological roles in plants In: Hayat S, Ahmad A, Alyemeni NM, eds. Salicylic Acid. Dordrecht: Springer, 15–30. [Google Scholar]
- Zhang L, Kars I, Essenstam B, et al. 2014. Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiology 164: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Flores-Cruz Z, Zhou L, et al. 2011. ‘ Ca. Liberibacter asiaticus’ carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant infections. Molecular Plant-Microbe Interactions 24: 458–468. [DOI] [PubMed] [Google Scholar]
- Zhang S, Chakrabarty PK, Fleites LA, Rayside PA, Hopkins DL, Gabriel DW.. 2015. Three new Pierce’s disease pathogenicity effectors identified using Xylella fastidiosa biocontrol strain EB92-1. PLoS One 10: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z-L, Zhao Y.. 2013. Transcriptome comparison and gene coexpression network analysis provide a systems view of citrus response to ‘Candidatus Liberibacter asiaticus’ infection. BMC Genomics 14: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Powell C a, Li W, Irey M, Duan Y.. 2013. Prophage-mediated dynamics of ‘Candidatus Liberibacter asiaticus’ populations, the destructive bacterial pathogens of citrus huanglongbing. PLoS One 8: e82248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurov V, Navarro M, Bruinsma KA, et al. 2014. Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiology 164: 384–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu-Salzman K, Zhu-Salzman K, Bi I-L, Bi I-L, Liu T-X, Liu T-X.. 2005. Molecular strategies of plant defence and insect counter-defence. Insect Science 12: 3–15. [Google Scholar]
- Zipfel C. 2014. Plant pattern-recognition receptors. Trends in Immunology 35: 345–351. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, et al. 2004. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
- Zou H, Gowda S, Zhou L, Hajeri S, Chen G, Duan Y.. 2012. The destructive citrus pathogen, ‘Candidatus Liberibacter asiaticus’ encodes a functional flagellin characteristic of a pathogen-associated molecular pattern. PloS One 7: e46447. [DOI] [PMC free article] [PubMed] [Google Scholar]