Abstract
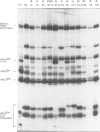
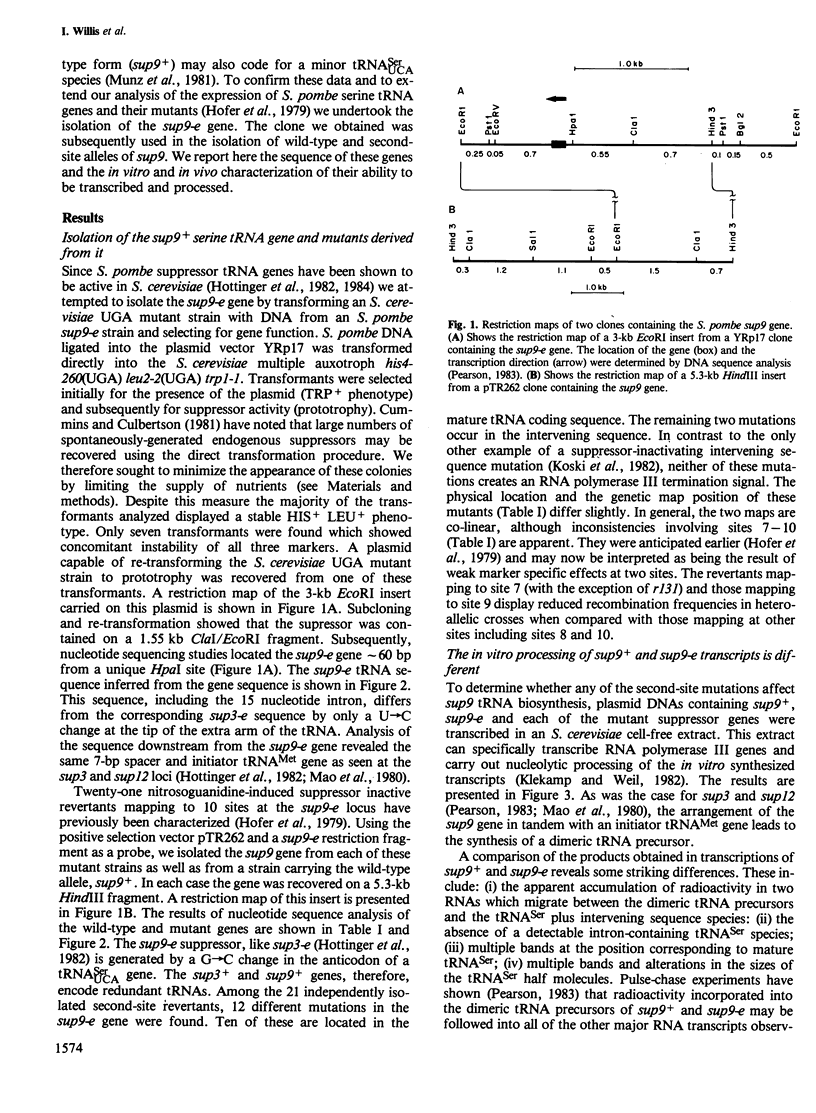
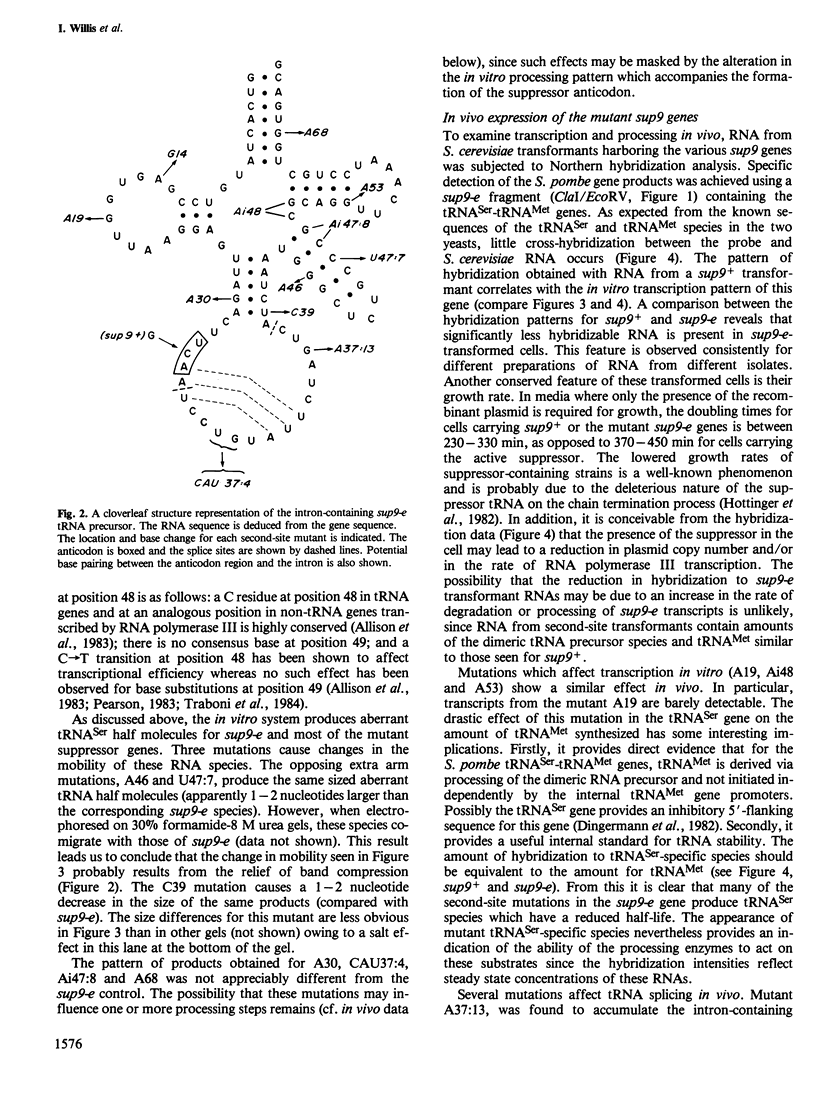
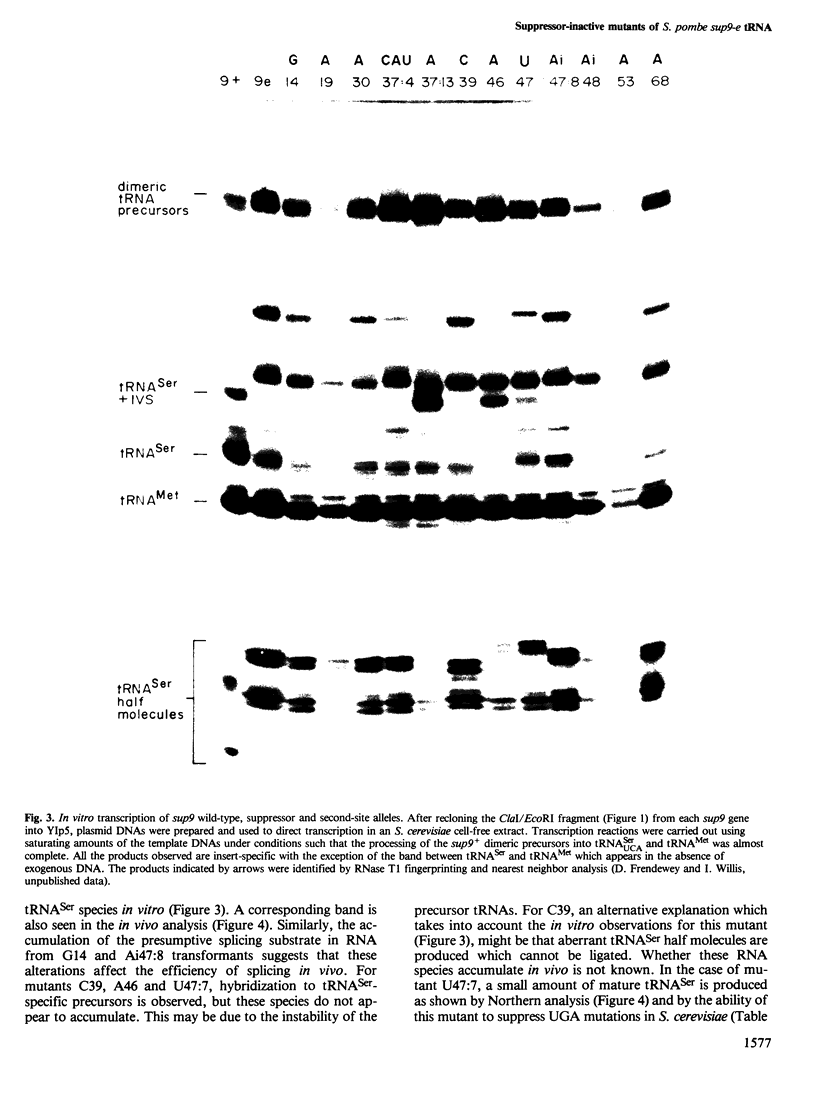
The nucleotide sequences of a Schizosaccharomyces pombe opal suppressor serine tRNA gene (sup9-e) and of 12 in vivo-generated mutant genes, which have lost the ability to suppress UGA mutations, have been determined. Analysis of the expression of these genes in Saccharomyces cerevisiae in vitro and in vivo systems has revealed defects in tRNA gene transcription and precursor tRNA processing. Single base changes in the D-loop, the intron and the extra arm affect the efficiency of splicing of the tRNA precursors while an anti-codon stem mutation may affect the accuracy of this process. Two mutations which occur in the intervening sequence of the sup9-e gene allow an alternate tRNA base pairing configuration. Transcription of the sup9-e gene and of the adjacent tRNAMet gene (located 7 bp downstream) is essentially abolished in vivo by a G----A19 mutation in the tRNASer gene, suggesting that tRNAMet may be derived solely via processing of the tRNASer-tRNAMet dimeric precursor.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison D. S., Goh S. H., Hall B. D. The promoter sequence of a yeast tRNAtyr gene. Cell. 1983 Sep;34(2):655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi M. S., Melton D. A., Cortese R. Site-directed mutagenesis of a tRNA gene: base alterations in the coding region affect transcription. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1388–1392. doi: 10.1073/pnas.79.5.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Melton D. A., Cortese R. Promoter of a eukaryotic tRNAPro gene is composed of three noncontiguous regions. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1195–1199. doi: 10.1073/pnas.79.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby D., Leboy P. S., Guthrie C. Yeast tRNA precursor mutated at a splice junction is correctly processed in vivo. Proc Natl Acad Sci U S A. 1981 Jan;78(1):415–419. doi: 10.1073/pnas.78.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryer D. R., Eccleshall R., Marmur J. Isolation of yeast DNA. Methods Cell Biol. 1975;12:39–44. doi: 10.1016/s0091-679x(08)60950-4. [DOI] [PubMed] [Google Scholar]
- Cummins C. M., Culbertson M. R. Molecular cloning of the SUF2 frameshift suppressor gene from Saccharomyces cerevisiae. Gene. 1981 Sep;14(4):263–278. doi: 10.1016/0378-1119(81)90159-1. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Burke D. J., Sharp S., Schaack J., Söll D. The 5- flanking sequences of Drosophila tRNAArg genes control their in vitro transcription in a Drosophila cell extract. J Biol Chem. 1982 Dec 25;257(24):14738–14744. [PubMed] [Google Scholar]
- Folk W. R., Hofstetter H. A detailed mutational analysis of the eucaryotic tRNAmet1 gene promoter. Cell. 1983 Jun;33(2):585–593. doi: 10.1016/0092-8674(83)90439-7. [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Kurjan J. tRNA synthesis: identification of in vivo precursor tRNAs from parental and mutant yeast strains. Nucleic Acids Res. 1981 Feb 25;9(4):1019–1029. doi: 10.1093/nar/9.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottinger H., Pearson D., Yamao F., Gamulin V., Cooley L., Cooper T., Söll D. Nonsense suppression in Schizosaccharomyces pombe: the S. pombe Sup3-e tRNASerUGA gene is active in S. cerevisiae. Mol Gen Genet. 1982;188(2):219–224. doi: 10.1007/BF00332678. [DOI] [PubMed] [Google Scholar]
- Hottinger H., Stadelmann B., Pearson D., Frendewey D., Kohli J., Söll D. The Schizosaccharomyces pombe sup3-i suppressor recognizes ochre, but not amber codons in vitro and in vivo. EMBO J. 1984 Feb;3(2):423–428. doi: 10.1002/j.1460-2075.1984.tb01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Specific transcription of homologous class III genes in yeast-soluble cell-free extracts. J Biol Chem. 1982 Jul 25;257(14):8432–8441. [PubMed] [Google Scholar]
- Klemenz R., Stillman D. J., Geiduschek E. P. Specific interactions of Saccharomyces cerevisiae proteins with a promoter region of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6191–6195. doi: 10.1073/pnas.79.20.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J., Hottinger H., Munz P., Strauss A., Thuriaux P. Genetic Mapping in SCHIZOSACCHAROMYCES POMBE by Mitotic and Meiotic Analysis and Induced Haploidization. Genetics. 1977 Nov;87(3):471–489. doi: 10.1093/genetics/87.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli J., Kwong T., Altruda F., Söll D., Wahl G. Characterization of a UGA-suppressing serine tRNA from Schizosaccharomyces pombe with the help of a new in vitro assay system for eukaryotic suppressor tRNAs. J Biol Chem. 1979 Mar 10;254(5):1546–1551. [PubMed] [Google Scholar]
- Koski R. A., Allison D. S., Worthington M., Hall B. D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4 gene transcription. Nucleic Acids Res. 1982 Dec 20;10(24):8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski R. A., Clarkson S. G., Kurjan J., Hall B. D., Smith M. Mutations of the yeast SUP4 tRNATyr locus: transcription of the mutant genes in vitro. Cell. 1980 Nov;22(2 Pt 2):415–425. doi: 10.1016/0092-8674(80)90352-9. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D., Gillam S., Smith M. Mutations at the yeast SUP4 tRNATyr locus: DNA sequence changes in mutants lacking suppressor activity. Cell. 1980 Jul;20(3):701–709. doi: 10.1016/0092-8674(80)90316-5. [DOI] [PubMed] [Google Scholar]
- Mao J., Schmidt O., Söll D. Dimeric transfer RNA precursors in S. pombe. Cell. 1980 Sep;21(2):509–516. doi: 10.1016/0092-8674(80)90488-2. [DOI] [PubMed] [Google Scholar]
- Mattoccia E., Baldi M. I., Pande G., Ogden R., Tocchini-Valentini G. P. Mutation in the a block of the yeast tRNAleu3 gene that allows transcription but abolishes splicing and 5'-end maturation. Cell. 1983 Jan;32(1):67–76. doi: 10.1016/0092-8674(83)90497-x. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Munz P., Leupold U., Agris P., Kohli J. In vivo decoding rules in Schizosaccharomyces pombe are at variance with in vitro data. Nature. 1981 Nov 12;294(5837):187–188. doi: 10.1038/294187a0. [DOI] [PubMed] [Google Scholar]
- Newman A. J., Ogden R. C., Abelson J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983 Nov;35(1):117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Nishikura K., Kurjan J., Hall B. D., De Robertis E. M. Genetic analysis of the processing of a spliced tRNA. EMBO J. 1982;1(2):263–268. doi: 10.1002/j.1460-2075.1982.tb01157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Gegenheimer P., Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983 Feb;32(2):525–536. doi: 10.1016/0092-8674(83)90472-5. [DOI] [PubMed] [Google Scholar]
- Rafalski A., Kohli J., Agris P., Söll D. The nucleotide sequence of a UGA suppressor serine tRNA from Schizosaccharomyces pombe. Nucleic Acids Res. 1979 Jun 25;6(8):2683–2695. doi: 10.1093/nar/6.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 1975;12:45–64. doi: 10.1016/s0091-679x(08)60951-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S., DeFranco D., Dingermann T., Farrell P., Söll D. Internal control regions for transcription of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6657–6661. doi: 10.1073/pnas.78.11.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traboni C., Ciliberto G., Cortese R. Mutations in Box B of the promoter of a eucaryotic tRNAPro gene affect rate of transcription, processing, and stability of the transcripts. Cell. 1984 Jan;36(1):179–187. doi: 10.1016/0092-8674(84)90087-4. [DOI] [PubMed] [Google Scholar]