Abstract
In this study, we investigated whether intrinsic glial dysfunction contributes to the pathogenesis of schizophrenia (SCZ). Our approach was to establish humanized glial chimeric mice using glial progenitor cells (GPCs) produced from induced pluripotent stem cells derived from patients with childhood-onset SCZ. After neonatal implantation into myelin-deficient shiverer mice, SCZ GPCs showed premature migration into the cortex, leading to reduced white matter expansion and hypomyelination relative to controls. The SCZ glial chimeras also showed delayed astrocytic differentiation and abnormal astrocytic morphologies. When established in myelin wild-type hosts, SCZ glial mice showed reduced prepulse inhibition and abnormal behavior, including excessive anxiety, antisocial traits and disturbed sleep. RNAseq of cultured SCZ hGPCs revealed disrupted glial differentiation-associated and synaptic gene expression, indicating that glial pathology was cell-autonomous. Our data therefore suggest a causal role for impaired glial maturation in the development of schizophrenia, and provide a humanized model for its in vivo assessment.
Keywords: induced pluripotent stem cell, iPSC, glia, glial differentiation, myelin, dysmyelination, astrocyte, schizophrenia, childhood-onset schizophrenia, mouse models
eTOC summary
Goldman and colleagues use mice chimerized with human patient-derived glial progenitor cells to ask whether glia contribute to childhood-onset schizophrenia. The defects in cell differentiation, myelination, and behavior that they see strongly suggest that glial cells do in fact have a previously unappreciated role in the pathogenesis of this disease.
INTRODUCTION
Schizophrenia is a uniquely human disorder, whose phylogenetic appearance parallels that of glial evolution, which accelerated with the appearance of hominids (Horrobin, 1998; Oberheim et al., 2006; Oberheim et al., 2009). In particular, astroglial complexity and pleomorphism increased significantly with hominid evolution, which suggests an association between human glial evolution and the development of human-selective neurological disorders, the neuropsychiatric disorders as a case in point. Indeed, a number of both genome-wide association and differential expression studies have highlighted the frequent dysregulation of glial-selective genes, both astrocytic and oligodendrocytic, in schizophrenia (Aberg et al., 2006; Georgieva et al., 2006; Hakak et al., 2001; Hof et al., 2002; Roy et al., 2007; Takahashi et al., 2011; Walsh et al., 2008). At the same time, a number of investigators have highlighted the marked differences between humans and rodents in glial gene expression, in contrast to the relatively conserved nature of neuronal gene expression across mammals (Miller et al., 2010; Sim et al., 2009; Zhang et al., 2016). Together these studies suggest the correlative association of human glial evolution with the phylogenetic appearance of schizophrenia.
Patients with schizophrenia are typically characterized by a relative paucity of white matter and often frank hypomyelination (Connor et al., 2011; Fields, 2008; Gogtay et al., 2008; McIntosh et al., 2008; Munoz Maniega et al., 2008; Takahashi et al., 2011), and a number of both pathological and neuroimaging studies have highlighted deficiencies in both oligodendroglial density and myelin structure in affected patients (Fields, 2008; Langmead and Salzberg, 2012; Rapoport et al., 2005; Xia et al., 2014), including at the ultrastructural level (Pruitt et al., 2007; Uranova et al., 2011; Uranova et al., 2007). Furthermore, recent studies have emphasized the role of oligodendrocytes in the metabolic support of neurons, suggesting myelin-independent mechanisms whereby oligodendrocytic dysfunction might yield neuronal pathology (Lee et al., 2012; Simons and Nave, 2015). Yet despite compelling genetic, cellular, pathological, and radiological studies that have correlated glial and myelin pathology with schizophrenia, many have assumed that clinical hypomyelination among schizophrenics is secondary to neuronal pathology, so that the contribution of cell-autonomous glial dysfunction to schizophrenia has not been well studied.
This lapse has in large part been due to the lack of animal models of human glial pathophysiology: mouse brains have mouse glia. We therefore asked whether this limitation might be addressed using a novel model of human glial chimeric mice (Goldman et al., 2015; Han et al., 2013; Windrem et al., 2008) paired with the development of protocols for generating bipotential astrocyte-oligodendrocyte glial progenitor cells (GPCs) from patient-specific human induced pluripotent stem cells (hiPSCs) (Wang et al., 2013). In these human glial chimeric mouse brains, the majority of resident glia are replaced by human glia and their progenitors (Windrem et al., 2014), allowing human glial physiology, gene expression, and effects on neurophysiological function to be assessed in vivo, in live adult mice (Han et al., 2013). In this study, we used this glial chimeric model to assess the contribution of human glia to schizophrenic disease phenotype. To this end, we prepared hGPCs from iPSCs derived from fibroblasts taken from either juvenile-onset schizophrenic (SCZ) patients or their normal controls, assessed the differential gene expression of SCZ hGPCs relative to those of normal subjects, and transplanted these cells into immunodeficient neonatal mice to produce patient-specific human glial chimeric mice. The glial chimeric mice were then analyzed in regards to the effects of SCZ derivation on astrocytic and oligodendrocytic differentiation in vivo, as well as on behavioral phenotype, and the data thereby obtained correlated to disease-associated gene expression (Figure 1).
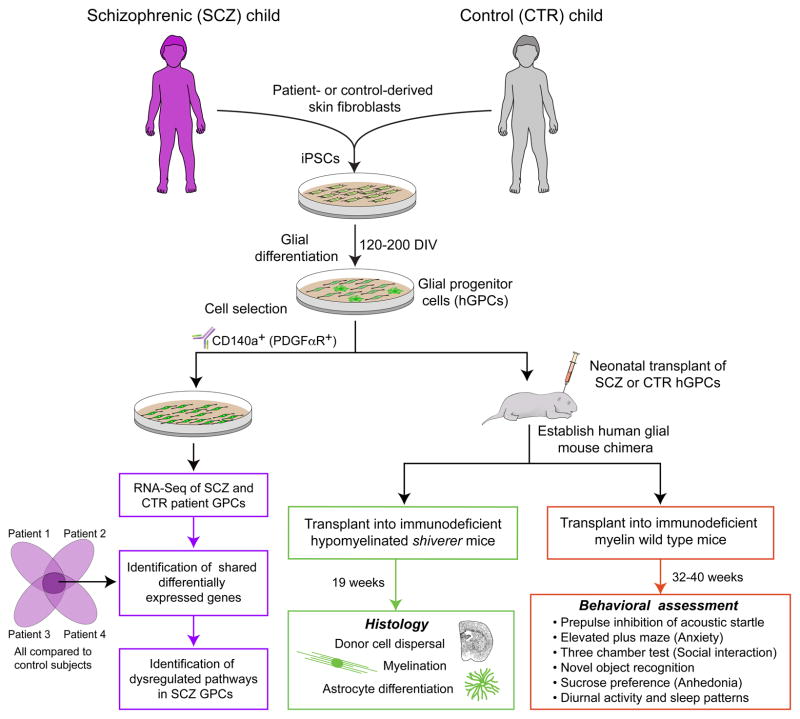
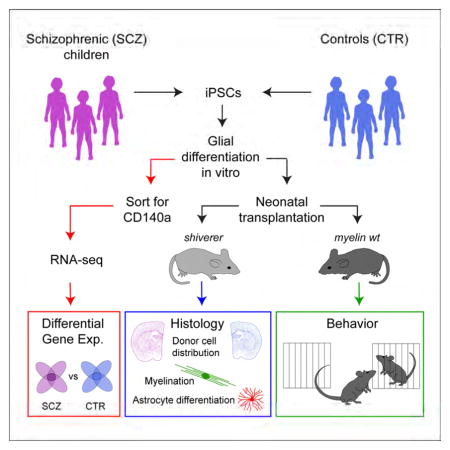
Figure 1. Functional and genomic assessment of schizophrenia-derived glial progenitor cells.
This schematic summarizes the steps involved in our analysis of glial progenitor cells derived from individuals with juvenile-onset schizophrenia, compared to GPCs derived from behaviorally-normal controls. The major output data include effects of SCZ origin on in vivo oligodendrocyte maturation and myelination (Figure 2); in vivo astrocyte differentiation and phenotype (Figure 3); in vitro differential gene expression (Figures 4 and 5); and behavioral phenotype of the human glial chimeric host animals (Figure 6).
RESULTS
Patients with juvenile-onset schizophrenia, as well as healthy young adult controls free of known mental illness, were recruited and skin biopsies obtained from each. Patient identifiers were not available to investigators besides the treating psychiatrist, although age, gender, race, diagnosis and medication history accompanied cell line identifiers. Fibroblasts were isolated from each sample; from these, 11 new independent hiPSC lines were derived from 8 patient samples (5 juvenile-onset schizophrenia patients and 3 healthy gender-matched and age-analogous controls (Table S1). iPSCs were generated using excisable floxed polycistronic hSTEMCCA lentivirus (Somers et al., 2010; Zou et al., 2012) encoding Oct4, Sox2, Klf4 and c-Myc (Takahashi et al., 2007; Welstead et al., 2008). All lines were initially characterized and validated as pluripotent using global transcriptome profiling by RNA sequencing to assess pluripotent gene expression, as well as immunostaining for Oct4, Nanog, and SSEA4. The identity of each iPSC line was confirmed to match the parental donor fibroblasts using short tandem repeat (STR)-based DNA fingerprinting. iPSC line isolates were also karyotyped concurrently with these experiments to confirm genomic integrity. An additional well-characterized hiPSC control line, C27(Chambers et al., 2009), was also used, to ensure that our control engraftment and differentiation data were consistent with prior studies in our lab (Wang et al., 2013). Altogether, we evaluated hGPC preparations from 7 iPSC lines derived from 5 SCZ patients, and 5 iPSC lines derived from 4 control subjects (Table S1). We instructed these iPSC cells to GPC fate as previously described (Wang et al., 2013), and after >105 days in vitro (DIV) under glial differentiation conditions validated the predominant GPC phenotype of each cell population using flow cytometry for CD140a/PDGFαR (Figure S1) (Sim et al., 2011). To optimize glial differentiation in vivo, we limited transplants to those preparations in which most cells were CD140a+ GPCs, with the remainder astroglial.
We first asked whether SCZ hGPCs differed from wild-type hGPCs in their myelination competence. To this end, we implanted SCZ hGPCs into neonatal immunodeficient shiverer mice (rag2−/− × MBPshi/shi), a congenitally hypomyelinated mutant lacking myelin basic protein (MBP)(Roach et al., 1983; Rosenbluth, 1980). As these otherwise myelin-deficient mice matured, their engrafted hGPCs differentiated into both astrocytes and myelinogenic oligodendrocytes, yielding mice chimeric for individual patient-derived glia(Windrem et al., 2008; Windrem et al., 2014). By this means, we established mice with patient-specific, largely humanized forebrain white matter, derived from SCZ or control subjects (Figures 2A–D).
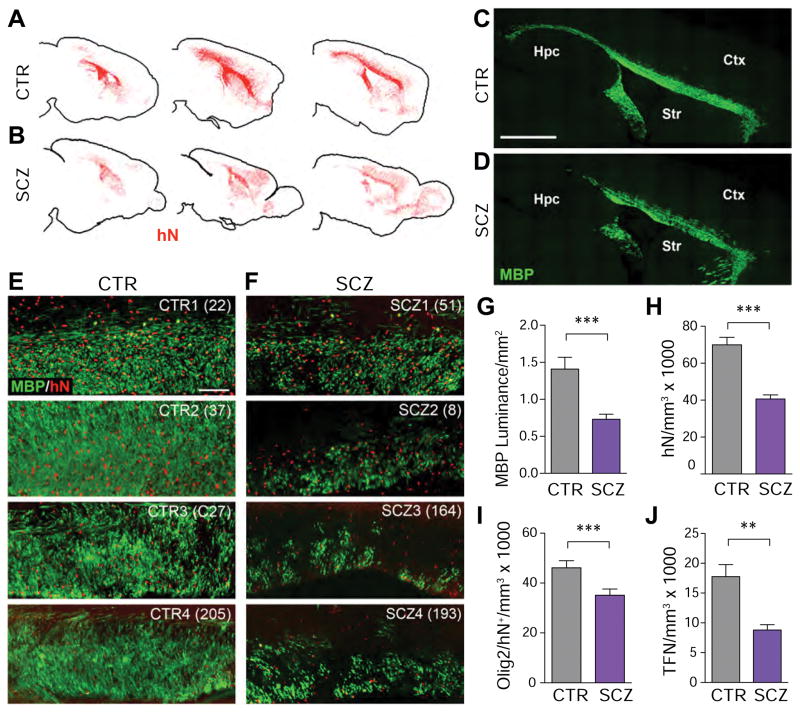
Figure 2. Schizophrenia-derived hGPCs exhibit aberrant dispersal and relative hypomyelination.
Human iPSC GPC chimeras were established by neonatal hGPC injection into shiverer hosts and sacrificed at 19 weeks. GPCs derived from a control subject (A) dispersed primarily in the major white matter tracts, whereas (B) SCZ-derived GPCs (15 yo male) showed less white matter residence and more rapid cortical infilitration. C–D, Sagittal sections reveal that callosal myelination by SCZ GPCs (D) was less dense than that by control hGPCs (C). E–F, Higher power images from chimeric mice engrafted with hGPCs from 4 control patients (E) vs. chimeric mice engrafted with hGPCs from 4 different SCZ patients (F). G, MBP luminance confirmed the greater callosal myelination of CTRL GPC-engrafted vs. SCZ GPC-engrafted mice at 19 weeks (means of 4 different SCZ and CTRL patients each, n>3 mice/patient) (p=0.0002, t-test). H, Absolute donor cell densities were lower in SCZ than control hGPC-engrafted corpus callosum (p<0.0001, t-test), as were the densities of olig2+ hGPCs and oligodendroglia (I) (p=0.0064, t-test) and transferrin (TFN)+ oligodendroglia (J) (p<0.0001, t-test).
See also Figure S2.
SCZ glial chimeric mice were uniformly hypomyelinated
We first noted that the SCZ hGPCs manifested an aberrant pattern of migration upon neonatal transplantation. Normal control hGPCs invariably expanded through the white matter before colonizing the cortical gray matter (Figure 2A), as we have previously noted in both fetal tissue- and hiPSC GPC-engrafted shiverer mice (Wang et al., 2013; Windrem et al., 2008). In contrast, SCZ GPCs preferentially migrated earlier into the gray matter in shiverer mice, with large numbers traversing without stopping in the callosal white matter (n=4 lines from 4 different patients, each with >3 mice/patient, each vs. paired controls) (Figure 2B; Figure S2). This resulted in significantly fewer donor hGPCs in the white matter of shiverers engrafted with SCZ GPCs (Figures 2H–2I; Figure S2). Importantly, this was associated with substantially diminished central myelination in these mice, as reflected by both MBP immunostaining (Figures 2C–D and 2E–F) and myelin luminance (Figure 2G).
Since the SCZ hGPC-engrafted shiverers manifested deficient myelination, we asked whether this was due to a relative failure of SCZ hGPCs to remain within white matter, or rather to a cell-intrinsic failure in myelinogenesis. Examining 19 wk-old SCZ and control hGPC-engrafted shiverer mice, we found significantly fewer human nuclear antigen (hNA)-defined donor-derived cells in SCZ hGPC-engrafted shiverer white matter (40,615 ± 2,189 × 103 hNA+ cells/mm3, n=18) than in mice identically transplanted with control hGPCs (69,970 ± 4,091/mm3; n=32; p<0.0001 by 2-tailed t test (Fagerland and Sandvik, 2009; Zimmerman, 2004)) (Figure 2H). Moreover, the numbers of hNA+ donor cells co-expressing the oligodendroglial lineage marker Olig2 were similarly depressed in the SCZ hGPC-engrafted mice (33,619 ± 2,435/mm3, n=26), relative to control hGPC-engrafted mice (46,139 ± 2,858/mm3, n=17; p<0.002) (Figure 2I). On that basis, we next found that the density of transferrin-defined human oligodendroglia was similarly lower in the callosal white matter of SCZ hGPC chimeras, than in control hGPC chimeras (8,778±892.2/mm3, n=25; vs. 17,754±2,023/mm3, n=17, respectively; p=0.0006, Mann-Whitney) (Figure 2J). These data indicate that SCZ GPCs are deficient not only in their colonization of the forebrain white matter, but also in their oligodendrocytic differentiation, with a resultant suppression of central myelinogenesis. Together, these findings suggest that SCZ hGPCs migrate aberrantly, traversing rather than homing to developing white matter, thus yielding relatively poor white matter engraftment, deficient myelin formation, and premature cortical entry relative to normal GPCs.
SCZ glial chimeric mice manifested developmentally-delayed astrocytic maturation
We next asked whether the SCZ hGPCs that prematurely entered the gray matter differentiated instead into astrocytes in that environment, or whether they rather manifested an impairment in lineage progression that prevented their astrocytic differentiation as well. Both SCZ and control hGPC-engrafted shiverer brains were immunostained for astrocytic glial fibrillary acidic protein (GFAP) at 19 weeks after neonatal graft, using a species-specific anti-human GFAP antibody. We found that astrocytic maturation from engrafted hGPCs was markedly deficient in the SCZ hGPC-engrafted brains (n=19, derived from 3 SCZ patient lines, and n=12 control mice, from 3 control patients) (Figures 3A–B). In the callosal white matter, as well as in both the striatal and cortical gray matter, astrocytic differentiation by SCZ hGPCs was significantly less than that of control GPCs, such that whereas all control hGPC forebrains showed dense human GFAP+ astrocytic maturation, far fewer SCZ hGPCs manifested hGFAP expression and astrocytic phenotype (controls: 6,616 ± 672.3 GFAP+ cells/mm3 in callosum, n=12; SCZ: 1,177 ± 276.6 GFAP+ callosal cells/mm3, n=19; p<0.0001 by 2-way t-test (Figure 3C). This defect in astrocytic differentiation was consistently observed in all mice (n=19) derived from the 3 SCZ patients assessed, compared to the control GPC-engrafted mice (n=12) derived from 3 normal subjects (Figure 3D), and reflected in part the lower proportion of GFAP+ astrocytes that developed among engrafted human cells in the SCZ HGPC-engrafted brains (Figure 3E). Furthermore, Sholl analysis of individual astroglial morphologies (Sholl, 1953), as imaged in 150 μm sections and reconstructed in Neurolucida (Figure 3J), revealed that astrocytes in SCZ hGPC chimeras differed significantly from their control hGPC-derived counterparts, with fewer primary processes (Figure 3F), less proximal branching (Figure 3G), longer distal fibers (Figure 3H), and less coherent domain structure (Figure 3I). Thus, SCZ hGPCs derived from multiple patients exhibited a common defect in phenotypic maturation, and hence proved deficient in astrocytic differentiation as well as myelination.
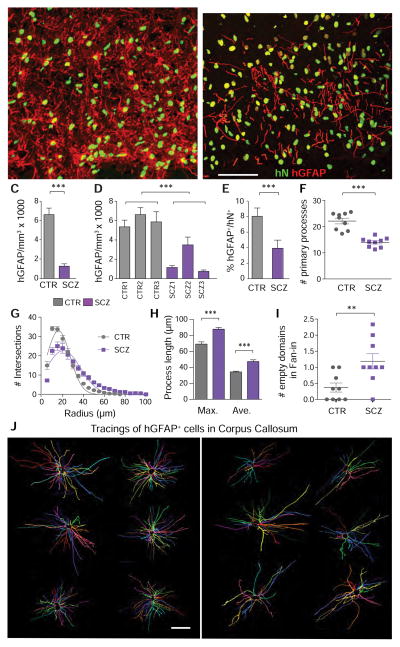
Figure 3. Astrocytic differentiation is impaired in schizophrenia hGPC chimeric brain.
Human iPSC GPC chimeras were established in immunodeficient shiverer hosts and sacrificed at 19 weeks, and astrocytic diffeerntiation assessed. A–B, representative images of the corpus callosum of mice neonatally injected with iPSC GPCs derived from either control (A, line 22) or schizophrenic (B, line 164) subjects (human nuclear antigen, green; glial fibrillary acidic protein, red). A, Control hiPSC GPCs from all tested patients rapidly differentiated as GFAP+ astrocytes with dense fiber arrays in both callosal white and cortical gray matter. B, In contrast, SCZ GPCs were slow to develop mature GFAP expression. At 19 weeks, GFAP+ astrocyte densities were significantly greater in mice chimerized with control than SCZ-derived GPCs, both as a group (C), and when analyzed line-by-line (D). This was not just a function of less callosal engraftment, as the proportion of human donor cells that developed GFAP and astrocytic phenotype was significantly lower in SCZ- than control GPC-engrafted mice (E). Sholl analysis of individual astroglial morphologies(Sholl, 1953), as imaged in 150 μm sections and reconstructed in 3D by Neurolucida (J), revealed that astrocytes in SCZ hGPC chimeras differed significantly from their control hGPC-derived counterparts, with fewer primary processes (F), less proximal branching (G), and longer distal fibers (H). When the 3-D tracings (J) were assessed by Fan-in radial analysis (MBF Biosciences)(Dang et al., 2014), control astrocytic processes were noted to extended uniformly in all directions, but SCZ astrocyte processes left empty spaces, indicative of a discontiguous domain structure (I). ***p<0.0001, by t-test (C, E, F, H; by 2-way ANOVA in D; **p<0.002 in I; p<0.0001 by non-linear comparison in G.
Scale, A–B = 50 μm, J = 25 μm.
SCZ hGPCs showed cell-autonomous misexpression of differentiation-associated genes
To better define the molecular basis for the apparent impediment to terminal glial differentiation in SCZ GPC-engrafted mice, and to define which aspects of that deficit might be cell-autonomous, we next used RNA-seq analysis to identify the differentially expressed genes of SCZ iPSC-derived GPCs, relative to those of control-derived glia. We used sequencing data to reconstruct the transcriptional patterns of hGPCs derived from 4 different SCZ and 3 control patients. hGPCs were derived at time points ranging from 154 to 242 days in vitro, and sorted for hGPCs using CD140a-targeted FACS. Using a 5% FDR and a fold-change threshold of 2, we identified a total of 118 mRNAs that were differentially expressed by CD140a-sorted SCZ hGPCs relative to their control iPSC hGPCs (Figures 4A–B). Among those genes most differentially expressed by CD140a-sorted SCZ hGPCs were a host of glial differentiation-associated genes, in particular those associated with early oligodendroglial and astroglial lineage progression, which were uniformly down-regulated in the SCZ hGPCs relative to their normal controls (Figures 4C and 4F). These included a coherent set of the key GPC lineage transcription factors OLIG1, OLIG2, SOX10, and ZNF488, as well as genes encoding stage-regulated proteins involved in myelination such as GPR17, UGT8, OMG, and FA2H (Figure 4G; see Table S2 and Figures S3 and S4 for detailed gene expression data).
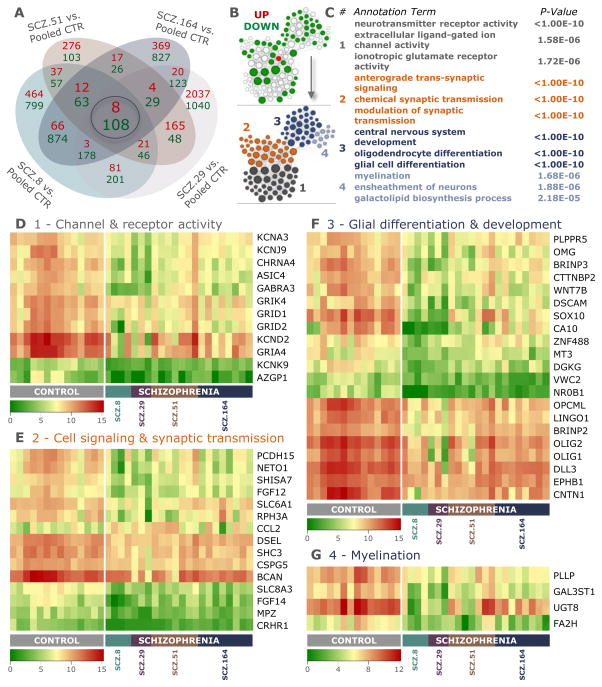
Figure 4. Schizophrenia-derived hGPCs suppress glial differentiation-associated gene expression.
RNA sequence analysis reveals differential gene expression by SCZ hGPCs. A, Intersection of lists of differentially expressed genes (DEGs) (log2-fold change >1.00, FDR 5%) obtained by comparison of hGPCs derived from 4 different schizophrenia patients, compared to pooled control hGPCs. B, Network representation of functional annotations for the intersection gene list shown in A. In the upper network, green and red nodes represent down- and up-regulated genes, respectively, and white nodes represent significantly associated annotation terms (FDR-corrected p< 0.01; annotation terms include GO:BP, GO:MF, pathways, and gene families, and nodes are sized by degree). Lower network highlights 4 highly interconnected modules identified by community detection. (C) Top annotation terms identified for each module in B. D, Heat map representation of 12 conserved differentially expressed genes that are associated to module 1 (grey in B, 32.4%), which includes annotations related to neurotransmitter receptor and gated channel activity. E, Heat map representation of 15 conserved differentially expressed genes associated to module 2 (orange in B, 28.7%), which comprises annotations related to cell-to-cell signaling and synaptic transmission. F, Heat map representation of 21 conserved differentially expressed genes associated to module 3 (dark blue in B, 28.7%); annotations related to CNS and glial differentiation. G, Heat map representation of 4 conserved differentially expressed genes that are associated to module 4 (light blue in B, 10.2%), with annotations related to myelination and lipid biosynthesis. The absolute expression in heat maps is shown in UQ-normalized, log2-transformed counts (Li et al., 2015).
See also Figures S3 and S4, and Table S2.
These expression data suggest that the diminished myelination of SCZ hGPC-transplanted shiverer brains reflected aberrant oligodendrocytic differentiation from the engrafted SCZ hGPCs. Similarly, since hGPCs give rise to astrocytes as well as oligodendrocytes, the RNA expression data suggest an analogous impediment to astrocytic differentiation. The functional consequences of the latter are especially profound, given the critical role for astrocytes in synaptic development and function; indeed, the relative suppression of astrocytic differentiation by SCZ hGPCs suggests a glial contribution to the impaired synaptic function noted in schizophrenia. In that regard, further functional analysis of SCZ-associated dysregulated hGPC genes identified channel and receptor activity, as well as synaptic transmission, as the most differentially affected functions besides glial differentiation (Figures 4D–E). These disease-linked channel and synapse-associated genes were largely down-regulated in the SCZ hGPCs, and included a number of potassium channel genes (Figure 4D), including KCND2, KCNJ9, KCNK9 and KCNA3, as well as a number of transcripts associated with synaptic development and function (Figure 4E and Table S2). The latter included NXPH1, NLGN3, and LINGO1, among others (Table S3, and Figures S3 and S4), synaptic genes whose dysregulation has been previously linked to both SCZ and the autism spectrum disorders (Andrews and Fernandez-Enright, 2015; Fernandez-Enright et al., 2014; Mackowiak et al., 2014; Salyakina et al., 2011; Sudhof, 2008). Whereas the expression of these latter genes was suppressed in hGPCs derived from all 4 SCZ patients, other synapse-associated genes, such as NRXN1, NLGN1, DSCAML1, and the SLITRKs 2–5, were sharply down-regulated in hGPCs derived from 3 of the 4 patients, but not in the fourth (Table S3). Yet other synapse-associated transcripts, like NXPH3 and NTRNG2, were similarly down-regulated in some patients, but not others. TaqMan low density arrays were used for quantitative real-time PCR validation of these and other dysregulated transcripts of interest, and confirmed the significant differential down-regulation of these differentiation and synaptic function-associated genes (Figure 5).
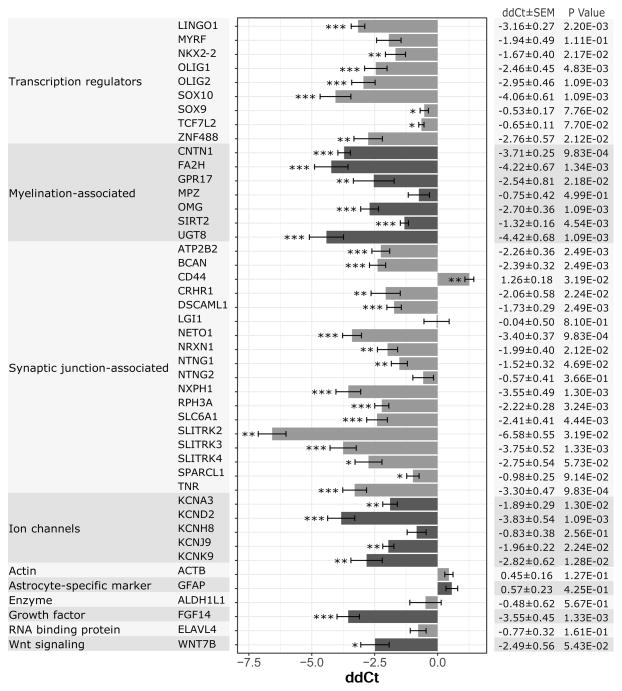
Figure 5. Impaired glial differentiation-associated gene expression by SCZ hGPCs.
The expression of dysregulated genes in SCZ-derived GPCs, as identified by RNA-seq analysis, was assessed by TaqMan Low Density Array (TLDA) RT-qPCR, then compared to that of control hGPCs. Expression data were normalized to GAPDH endogenous control. Mean ddCt values and standard error ranges, calculated from 4 pooled SCZ GPC lines (n = 19) that were individually compared to 3 pooled control GPC lines (n = 10), are shown. The difference in gene expression by SCZ and control hGPCs was assessed by paired t-tests, followed by multiple testing correction by Benjamini-Hochberg (BH) procedure (*** = p <0.01, ** = p < 0.05, * = p <0.1). 48 genes were assessed. 45 genes are shown, excluding the endogenous control and genes that had high proportions of undetermined or unreliable reactions, LRFN1 and NEUROD6. The vast majority of genes were confirmed as dysregulated in SCZ-derived GPCs. Analysis of TLDA data was performed in ExpressionSuite Software version 1.1, supplied by Applied Biosciences.
See also Table S3.
Together, these data suggest the importance of glial-associated synaptic gene expression in schizophrenia, while emphasizing the heterogeneity of pathways that might be mechanistically complicit in its dysregulation. These data also highlight the point that while the neuronal localization of these synaptic proteins has long been recognized, their synthesis by glia and synaptic contributions thereof have not been specifically discussed, although cell type-specific transcriptional databases have noted significant glial expression of these genes (Zhang et al., 2014). Since NRXN1, a synapse-associated transcript closely linked to schizophrenia (Sudhof, 2008), was one of the most strongly and consistently down-regulated glial genes across our patients, we verified the down-regulation of its expression by SCZ glia, by immunoblotting CD140a-sorted, neuron-free isolates of SCZ and control hGPCs. Western blots revealed that neurexin-1 was indeed abundantly expressed by human GPCs, and that neurexin-1 protein levels were sharply lower in otherwise matched SCZ hGPCs (Figure S5).
SCZ glial chimerization yielded disease-specific behavioral phenotype
We next asked whether the alterations in glial distribution and differentiation observed in mice engrafted with SCZ hGPCs might alter the behavioral phenotype of the host mice. In particular, we postulated that the aberrant infiltration of hGPCs and their derived astroglia into the developing cortex might influence information processing within the cortex once mature. As noted, past studies have reported both the influence of astrocytic networks on synaptic efficacy and plasticity, and the differential competence of hominid glia in this respect (Han et al., 2013; Oberheim et al., 2009). Human glial chimeric mice manifest a lower threshold for hippocampal long-term potentiation (LTP) and learn more rapidly, with superior performance in a variety of learning tasks, which include auditory fear conditioning, novel object and place recognition, and Barnes maze navigation. In each of these tests - but not in any test of social interactivity or primary perception – human glial chimeras acquire new causal associations more quickly than do allografted or untransplanted controls (Han et al., 2013). Thus, engrafted human GPCs and their daughter glia can integrate into, and substantially modify, developing neural networks (Franklin and Bussey, 2013). On that basis, we postulated that the disruption in normal glial development noted in our SCZ glial chimeras might yield disease-associated changes in learning and behavior. To address this question, we assessed the behavioral phenotypes of immunodeficient but otherwise wild-type mice neonatally engrafted with SCZ GPCs, relative to matched hosts engrafted with control-derived GPCs. For these experiments, we used normally-myelinated hosts rather than shiverer mice, so as to produce mice chimeric only for human GPCs and astrocytes, and not for oligodendroglia, thus isolating any observed behavioral effects to SCZ hGPCs and astrocytes.
We first asked whether schizophrenic derivation of engrafted glia affected prepulse inhibition (PPI), a behavioral hallmark of both clinical schizophrenics and animal models thereof (Ewing and Grace, 2013). PPI reflects the coordination of sensorimotor gating in the CNS, and its diminution may predict aspects of schizophrenic phenotype (Ivleva et al., 2013; Kohl et al., 2013). We found that when assessed at 6 months of age – the latest time-point at which the C57Bl/6 background strain of our rag1−/− mice can be reliably assessed, since these mice suffer premature auditory loss which might otherwise diminish auditory PPI – that mice engrafted with SCZ hGPCs exhibited significantly diminished auditory prepulse inhibition (Figure 6A), and did so at all volumes of pre-pulse. Given the strong effect of SCZ glial chimerization on PPI, we next asked if SCZ glial chimerization might be associated with changes in behavior on cognitive and socialization tests. To that end, we compared SCZ and control chimeras on a battery of behavioral tests that included: 1) the elevated plus maze, a measure of anxiety (Walf and Frye, 2007); 2) the 3-chamber social challenge (Yang et al., 2011); 3) novel object recognition, a focused measure of executive memory (Bevins and Besheer, 2006), and 4) the preference for sucrose water, a test for anhedonia (Barnes et al., 2014; Willner et al., 1987). In each, mice chimerized with one of 3 SCZ or 3 control patient-derived lines were compared; each line was derived from a different patient. Between 6–12 recipient mice were engrafted and tested per cell line, or 17–36 mice per group for each behavioral comparison, with a typically equal balance of male and female recipients. These animals were tested beginning between 30–36 weeks of age, and testing typically lasted 3 weeks. Over the tested age range, the SCZ GPC chimeric mice exhibited a number of significant differences in behavior relative to their control hGPC-engrafted counterparts. Normal control-engrafted mice are significantly more likely to explore the open arms (horizontal segments), whereas SCZ mice spent most of their time in the closed maze arms (vertical segments), consistent with greater anxiety (p=0.036, 2-tailed t test). The SCZ hGPC mice exhibited greater avoidance of the open arms in the elevated plus maze than did their normal hGPC-engrafted controls (n=36 mice/group, each including 12 mice engrafted with hGPCs from each of 3 patients; p=0.036, 2-tailed t test), suggesting that the SCZ hGPC mice were prone to higher anxiety when challenged (Figure 6B). In addition, the SCZ hGPC mice showed less preference for sucrose water), consistent with relative anhedonia (Figure 6C), less interest in stranger mice in the 3-chamber social test (Figure 6D), and relatively poor novel object recognition (Figure 6E), reflecting relative impairment in executive memory.
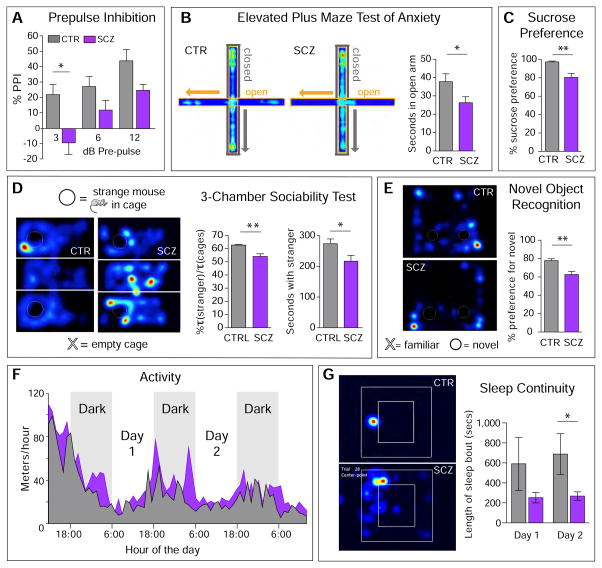
Figure 6. Schizophrenia-derived human glial chimeras have significant behavioral abnormalities.
A–E, Behavioral tests were performed in mice chimerized with one of 3 SCZ or 3 control hGPC lines, each line from a different patient. 7–20 recipient mice were tested per cell line, males and females equally. A, Prepulse inhibition Normally-myelinated rag1−/− mice engrafted with SCZ hGPCs had reduced auditory pre-pulse inhibition (PPI) at all volumes of pre-pulse (A). The extent of PPI differed significantly between control (n=13) and SCZ (n=27) hGPC-engrafted animals (p=0.0008 by ANOVA, F=11.76 [1,114]). B. Elevated Plus Maze Left panel, representative heat maps of the cumulated movement of a mouse engrafted with SCZ hGPCs, relative to its matched normal hGPC-engrafted control, in the elevated plus maze, a test designed to assess anxiety, in which preference for enclosed space and avoidance of open height suggests greater anxiety. Right panel, Mice engrafted with hGPCs from 3 SCZ patients (12 implanted mice each, for n=36 mice total) spent more time in the closed maze arms than did control-engrafted mice (n=36, also derived from 3 patients) (p=0.036, 2-tailed t test). C. Sucrose Preference SCZ GPC-engrafted mice were less likely to prefer sweetened water, suggesting relative anhedonia (p=0.02, Mann-Whitney t-test; n=30 mice derived from 3 SCZ lines; n=17 mice from 3 control lines). D. 3-Chamber Socialization Test Mice engrafted with hGPCs were placed into the middle chamber of a box divided into 3 compartments, one holding an empty cage (bottom, “X” in D) and one containing an unfamiliar mouse (top, filled white circle), then video-tracked for 10 minutes. Mice engrafted with SCZ hGPCs (right heat-map) avoided strangers more than did control mice (left heat-map), spending less time with strangers whether analyzed as the proportion of time spent with the stranger mouse relative to the empty cage (left bar graph; p=0.005) or the net amount of time spent with the stranger mouse (right bar graph; p=0.02); 3 SCZ lines, 39 mice; 4 control lines, 52 mice). E. Novel Object Recognition Mice engrafted with SCZ hGPCs showed significantly poorer novel object recognition (p=0.0006; 3 SCZ lines, 19 mice; 3 control lines, 28 mice).
F–G. The diurnal activity and sleep patterns of adult mice (70–80 weeks old) engrafted neonatally with either SCZ or CTRL hGPCs were assessed for 72 hrs in closed chambers (Noldus Ethovision), under continuous video recording. F. The average distance traveled in meters/hr over a 72 hr period was calculated and compared between CTRL mice (gray fill, n=8 mice; lines 22 and 17) and SCZ mice (purple fill; n=10, line 52). Time of day is shown as a 24-hour cycle, with the dark phase indicated by gray background shading. The SCZ mice were significantly more active throughout the observation period than CTRL-engrafted mice (p<0.0001, ANOVA, F=19.32 [1,851]. G. Left, Sample heat-maps of one hour of activity during the light phase (16:00 hrs, 2nd day in box), the normal period of sleep for mice. The control mouse (left), remains inactive for the entire hour, while the SCZ mouse moves about the cage during much of the hour. Right, The SCZ mice exhibited sleep patterns that were fragmented into bouts of shorter duration than their normal hGPC- chimeric controls (p=0.0026 by ANOVA, F=12.08 [1,24]. Means ± SEM; unpaired, two-tailed Welch-corrected t-tests.
As an additional metric of SCZ-associated behavior, we then assessed sleep and diurnal activity patterns of human SCZ and CTRL glial chimeras, directly comparing mice engrafted with either SCZ (line 52) or control (line 22) hGPCs. We found that mice engrafted with SCZ GPCs were significantly more active than control mice engrafted with normal hGPCs As measured by meters moved per hour, over the course of a 72-hour video-recording (Noldus Ethovision), the SCZ hGPC chimeric mice moved significantly more than their normal hGPC-engrafted controls (2-way ANOVA, F=48.35; p<0.0001) (Figure 6F). Interestingly, while the SCZ-associated increment in activity largely occurred during night-time periods of wakefulness, the SCZ mice also manifested disrupted sleep patterns, as measured by the duration of bouts of inactivity, a surrogate for EEG-validated sleep (McShane et al., 2010; Pack et al., 2007) (Figure 6G). Within the half-hour following the phase transition from dark to light (when mice normally sleep), the CTRL mice had more continuous, uninterrupted patterns of sleep, with an average sleep bout of 511.5 ± 36.4 seconds (8.53 minutes), whereas SCZ mice were asleep for 306.2 ± 43.7 seconds, or 5.1 minutes per bout (p<0.01 by 2-way ANOVA, with Bonferroni post hoc t tests). The shorter average periods of inactivity manifested by SCZ hGPC mice during the normal daytime transition to sleep suggests that SCZ hGPC chimerization disrupted normal daytime sleep patterns, while increasing night-time activity. Together, these results suggest that SCZ glial chimerization was sufficient to yield heightened anxiety and fear in engrafted recipients, as well as disease-associated deficits in socialization, cognition, and sleep patterning, all features associated with human schizophrenia.
DISCUSSION
These data suggest a significant contribution of cell-autonomous glial pathology to the genesis and development of juvenile-onset schizophrenia. In these human glial chimeric mice, schizophrenia-derived iPSC hGPCs exhibited aberrant migration with deficient engraftment in the central white matter, relative to age-analogous and gender-matched control iPSC hGPCs. Although a fraction of those SCZ hGPCs that did remain within the white matter differentiated as normal myelinogenic oligodendroglia, the premature cortical influx and hence lower density of donor-derived cells in the white matter of SCZ hGPC-engrafted mice resulted in the latter’s overt hypomyelination, relative to mice engrafted with control GPCs. Thus, SCZ hGPCs appeared to traverse rather than home in to the nascent white matter, resulting in sparse hGPC colonization and hence deficient forebrain myelination. The aberrant dispersal pattern of SCZ hGPCs suggests that SCZ GPCs may not recognize developmental stop signals that permit progenitors to dwell and expand within the presumptive white matter before colonizing the cortical mantle, and may instead be biased towards rapid entry into the cortical gray matter. These observations in human SCZ glial chimeric mice are especially intriguing given the well-described hypomyelination of schizophrenic patients (Davis et al., 2003; Najjar and Pearlman, 2015; Sigmundsson et al., 2001; Voineskos et al., 2013), particularly so in early onset disease (Gogtay et al., 2008; Gogtay and Rapoport, 2008; Samartzis et al., 2014).
These anatomic observations were especially intriguing in light of the differential gene expression pattern of the SCZ hGPCs, which revealed that the cells were deficient not only in early glial differentiation–associated transcripts, but also in genes that encode for synaptic proteins typically associated with transducing activity-dependent signals (Sudhof, 2008). Together, these anatomic and transcriptional data suggest that SCZ hiPSC-derived GPCs might be subject to impaired phenotypic differentiation, that might result in their neglect of the local neuronal signals that typically regulate the expansion and maturation of GPCs(Barres and Raff, 1993); this might account for their rapid transit through the white matter into the overlying cortex, and hence the diminished callosal GPC density and hypomyelination of SCZ chimeric shiverer mice (Figure 2). Thus, the myelination defect in SCZ hGPC chimeras appeared due to both deficient oligodendrocytic differentiation and the relative dearth of SCZ hGPCs remaining within the white matter. Moreover, astrocytic differentiation from SCZ hGPCs was also impaired, and may have contributed further to hypomyelination in the SCZ glial chimeras, given the metabolic dependence of mature oligodendrocytes upon local astrocytes (Amaral et al., 2013; John, 2012).
Importantly, the defective astrocytic maturation of SCZ hGPCs might also have profound effects on developmental synaptogenesis and circuit formation, as well as on myelinogenesis. Neural connectivity and synaptic development are both intimately dependent upon astrocytic guidance (Clarke and Barres, 2013; Ullian et al., 2001), and hence upon the appropriate timing of astrocytic appearance and maturation. As a result, any disruption in astrocytic maturation by SCZ hGPCs, as observed in each of the SCZ lines we studied, might be expected to significantly confound the construction and functional architecture of those neural networks in which SCZ hGPCs are resident. Moreover, glial progenitors themselves may have significant interactions with local neurons (Sakry et al., 2014), such that their dysfunction might disrupt local neuronal response thresholds and circuit formation.
Besides the anatomic observation of deficient astrocytic maturation in SCZ hGPC chimeras, our genomic analysis of SCZ-derived hGPCs revealed the significant down-regulation in hGPCs derived from all 4 SCZ patients of a number of synaptic genes, including neuroligin-3, neuroexophilin-1, and LINGO1 relative to their normal controls (Table S3; Figure 5). Other synapse-associated genes, such as neurexin-1 and DSCAML1 were significantly and sharply down-regulated in GPCs derived from 3 patients (lines 8, 29, and 51) but not in the fourth (line 164). Similarly, SLITRKs 2–5 were significantly and sharply down-regulated in GPCs derived from 3 patients (lines 8, 51, and 164), but not in a fourth (line 29), which was instead associated with sharp down-regulation of LINGO1, DSCAML1, and several neurexins and neuroexophilins; these data suggesting the heterogeneity of transcriptional dysfunction that may lead to a final common pathway of glial-involved synaptic dysfunction in SCZ (Tables S2 and S3). These transcripts are critical contributors to synaptic stabilization and function (Sudhof, 2008), but while typically considered neuronal, may be produced significantly by glial cells as well (Zhang et al., 2014). The relative down-regulation of these genes by SCZ hGPCs may reflect the suppression of mature glial transcripts in these cells, coincident with their relative block in glial differentiation. This in turn may lead to a relative failure of SCZ hGPCs and their derived astrocytes to provide these key proteins to their neuronal partners, as well as a potential failure on the part of glial progenitors receiving synaptic inputs to respond to afferent stimulation (De Biase et al., 2010; Lin and Bergles, 2004). Thus, besides the structural havoc that might be expected of a cortical connectome formed without normal astrocytic support, the synaptic structure of the resultant networks might be expected to be destabilized by poor SCZ glial provision to the synaptic cleft of key astrocytic proteins required for normal synaptic maintenance and function.
Schizophrenia is genetically heterogeneous, so that anatomic and behavioral pathology may vary significantly among animals chimerized with GPCs derived from different patients. It is thus critical that the results obtained from chimeras established with control hiPSC GPCs be stable across both distinct lines of donor cells, and among recipient mice. The chimeric brains established from the hGPCs of 3 different SCZ patients were thus compared anatomically to those established from GPCs derived from 3 control patients. None of the controls manifested the white matter-avoidant dispersal pattern of the SCZ hGPC chimeras. Similarly, we had never noted this pattern of SCZ hGPC avoidance of the white matter in any of several hundred human glial chimeras engrafted in other studies with either fetal tissue-derived (Windrem et al., 2008; Windrem et al., 2014) or normal iPSC-derived (Wang et al., 2013) hGPCs.
Besides their clear anatomic phenotype, the SCZ hGPC-chimeric mice manifested robust behavioral phenotypes. They exhibited significantly attenuated prepulse inhibition relative to control-engrafted mice, relative anhedonia, excessive anxiety, deficient socialization with avoidance of conspecifics, and disrupted patterns of diurnal activity and sleep. These data establish that SCZ glial engraftment may yield an abnormal behavioral phenotype in recipient mice, along behavioral axes that typify selected aspects of schizophrenic behavioral pathology in humans. In that regard, while an extensive literature has implicated GPCs (Bergles et al., 2010; De Biase et al., 2010) as well as astroglia (Araque et al., 1998; Kang et al., 1998) in the modulation of synaptic plasticity and learning (Han et al., 2013), our data do not implicate one phenotype over the other in the modulation of behavior by SCZ glial chimerization; our chimeric mice are colonized by both donor-derived human GPCs and their derived astrocytes. That said, our observations of significant defects in SCZ glial maturation shared by hGPCs derived from multiple independent patients, associated in each with hypomyelination and disrupted astrocytic differentiation, as well as with abnormal behavioral phenotypes in the resultant SCZ GPC chimeras, together suggest a strong causal contribution of glial pathology to schizophrenia. In addition, these data highlight the potential of disease-specific humanized chimeras in defining the respective contributions of glial and neuronal dysfunction in the genesis and course of neurological disease.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to, and will be fulfilled by, the lead and corresponding author, Steve Goldman (goldman@sund.ku.dk or steven_goldman@urmc.rochester.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Patient identification, protection and sampling
Patients from which these lines were derived were diagnosed with disabling degrees of schizophrenia with onset in early adolescence; informed consent was obtained and all patients and their guardians consented by a child psychiatrist (RLF) under an approved protocol of Case Western School of Medicine, blinded as to subsequent line designations, and no study investigators had access to patient identifiers.
iPSC line derivation and production of GPCs
Punch biopsies of the skin were obtained from patients with juvenile onset schizophrenia (ages 10 to 17 years old) and controls (ages 24 to 32 years old). The ages, genders, and ethnicities of each line used in this study are individually identified in Supplemental Table 1. In brief, 4 control subjects were used: CTRL 1 (lines 19, 22; 26 year-old male); CTRL 2 (line 37; 32 year-old female); CTRL 3 (line 205; 25 year-old male); CTRL 4 (line C27, fully de-identified), while 5 schizophrenic patients were used: SCZ 1 (line 8; 10 year-old female); SCZ 2 (lines 51, 52; 16 year-old male); SCZ 3 (lines 29, 31; 12 year-old male); SCZ 4 (line 164; 14 year old female); SCZ 5 (line 193, 15 year-old female).
Induced pluripotent stem cells (iPSC) lines were derived from these patient samples using an excisable floxed polycistronic hSTEMCCA lentiviral vector. Short tandem repeat (STR)-based DNA fingerprinting was used to confirm iPSC identity, as a match to original patient or control donor. Additional genotyping was performed using Illumina Omni5 SNP arrays; these data are available in dbGAP (http://www.ncbi.nlm.nih.gov/gap). The iPSCs were then driven toward a glial progenitor cell (GPC) fate using previously described protocols (Wang et al., 2013). Cells were harvested between 160–240 DIV, by which time most typically expressed the bipotential GPC marker PDGFαR/CD140a, while the remainder were A2B5+/CD140a− astrocytes. The karyotypes of all iPSC lines were assessed during glial differentiation to ensure genotypic stability of the cells utilized in all experiments presented here (karyotyping by WiCell, Madison, WI). All iPSCs showed a normal karyotype, except for line 51, which was found to have a balanced Robertsonian translocation of chromosome 13, an anomaly previously associated with juvenile-onset schizophrenia (Graw et al., 2012).
Host transplantation
Homozygous shiverer mice (The Jackson Laboratory, Bar Harbor, ME) were crossed with homozygous rag2-null immunodeficient mice (Shinkai et al., 1992) on the C3h background (Taconic, Germantown, NY, USA) to generate shi/shi × rag2−/− myelin-deficient, immunodeficient mice (Windrem et al., 2008). In addition, rag1−/− normally-myelinated immunodeficient mice (B6.129S7-Rag1tm1Mom/J), were obtained from the Jackson Laboratory and bred in our lab. Suspensions of single-cells or small clusters of hiPSC-derived GPCs were spun down to 100,000 cells/μl. Neonates were anesthetized by cooling, and transplanted bilaterally in the corpus callosum with a total of 200,000 cells, as described (Windrem et al., 2004); see Method Details section for transplant procedure. At 3 months of age (shi/shi × rag2−/−) or after completion of behavioral testing at 6–9 months (rag1−/− only), transplanted mice were anesthetized with pentobarbital, then perfusion fixed with cold HBSS+/+ followed by 4% paraformaldehyde (PF) with a 2 hour post-fixation in cold PF. All animal procedures were approved by the University of Rochester’s Committee on Animal Resources.
METHOD DETAILS
Transplantation
Shiverer x Rag2-null and Rag1-null neonatal mice were transplanted on postnatal day 1 or 2. Half of the litter was removed from the dam and placed in a humidified warming chamber. For this we used a sterilized plastic box, lined with sterile gauze dampened with Hanks balanced salt solution, and warmed on a heating block. The pups to be injected were then wiped with Povidone-Iodine and wrapped in sterile gauze to prevent direct contact with ice, then cryo-anesthetized for 2 to 6 minutes, depending on size. The pups were then removed from ice and cleaned with an alcohol prep pad, then laid in a customized neonatal mouse holder made of baked molded clay. The pups were injected directly through the skin and skull osteoid into both the rostral (at Bregma, ML ± 1.0 mm, ventral 1.0 mm) and caudal (AP −1.0, ML ± 1.0 mm, ventral 0.9 mm) corpus callosum. Following injections, pups were cleaned with alcohol prep pads and returned to the warming chamber for recovery. Upon recovery, the first half of the litter was returned to the dam, and the second half put in the humidified chamber. Pups were weaned between 21 and 28 days, then group housed.
Immunolabeling of tissue sections
Brains were cryopreserved, embedded in OCT (Tissue-Tek OCT, Sakura Finetek, Torrance, CA) and sectioned at 20 μm, either sagittally or coronally, on a cryostat. Human cells were identified with mouse antihuman nuclei, clone 235-1 at 1:800 (MAB1281, EMD Millipore, Billerica, MA). Myelin basic protein was labeled with rat anti-MBP at 1:25 (Ab7349, Abcam, Cambridge, MA), oligodendrocyte progenitors with anti-human-specific PDGFRa (D13C6, XP® rabbit mAb 5241, 1:300, Cell Signaling Tech), oligodendrocytes with mouse anti-human-specific transferrin (clone HT1/13.6.3, 08691231, MP Biomedicals), astrocytes with anti-human-specific GFAP (SMI 21 at 1:1000, Covance, Princeton, NJ). Alexa Fluor secondary antibodies, goat anti- mouse, rat, and rabbit 488, 568, 594, and 647 were used at 1:400 (Life Technologies, Carlsbad, CA).
Antibodies and dilutions used
See Resource Table.
Western blots
GPCs derived from CWRU22 and CWRU51 were sorted by FACS for CD140a at DIV160-200, directly into cell lysis buffer (NP40, Invitrogen, FNN0021) with protease inhibitor (Roche, 183617025) on ice. The insoluble fraction was removed by centrifugation at 12,000 g for 5 min at 4°C, and the supernatant analyzed for total protein with BCATM Protein Assay Kit (Thermo, 23227). 10 μg sample aliquots were separated on 4–12% gradient gels by SDS-PAGE electrophoresis (XCell SureLock, Invitrogen, 071210). Separated protein was transferred to PVDF membranes, which were blocked with 5% dry milk and incubated sequentially with a rabbit polyclonal anti-neurexin-1 antisera (Millipore, ABN161-1, 1:1000) at 4°C overnight, then washed and followed serially by a mouse monoclonal anti-β actin (Abcam, ab173838, 1:5000) at RT for 1h, and anti-mouse and anti-rabbit secondary antibodies (GE Healthcare, 95107-322 and 95107-328, 1:10000) at RT for 1h. Membranes were visualized by chemiluminescence (Mix ECLTM Reagent, GE Healthcare, RPN2236) through exposure of X-ray film. Experiments were repeated 3 times, with 3 different sets of cells.
Behavior
Behavioral tests were scored using either ANY-maze (Stoelting, Wood Dale, IL) or EthoVision (Noldus). Behavioral testing began at either 25 weeks (for pre-pulse inhibition) or 30–36 weeks (all other tests), and typically lasted 3 weeks; starting age was matched between experimentals and controls. A total of 6–12 recipient mice were engrafted and tested per cell line, or 17–36 mice per group for each behavioral comparison, with a roughly equal balance of male (M) and female (F) recipients. All behavioral tests were done by experimenters blinded as to the treatment condition and implanted cell line.
Tests were performed in the same sequence for all mice, and included: 1) Elevated Plus Maze Each test mouse was placed in the center of a raised, plus-shaped apparatus, consisting of 2 enclosed arms and 2 open arms, facing an open arm (Walf and Frye, 2007). Each tested mouse was videotaped and scored for time spent in the open vs. closed arms. 2) Three chamber social choice The test apparatus is a plexiglass enclosure divided into thirds with connecting doors (Ugo Basile, Italy) (Yang et al., 2011). Each test mouse was first acclimated to the central chamber or 5 min. The doors to the outer chambers were then removed, and the test mouse allowed to explore all three chambers for 10 min. The test mouse was then guided back to the central chamber, and a same sex and age stranger mouse was placed in a cylindrical container in one side chamber, while an empty cylindrical container was placed in the opposite side chamber. The mouse was then recorded for 10 min, and scored with respect to the amount of time it spent with the stranger mouse vs. the empty compartment. 3) Novel Object Recognition Each test mouse was placed in an empty 1 ft2 testing chamber for 5 min to acclimate, then removed, and two identical objects were placed in the chamber. The mouse was returned to the chamber with the objects, placed facing directly away from them, recorded for 10 min and scored for time spent in proximity to each object (Bevins and Besheer, 2006). After one hour, the experiment was repeated, with one of the two objects replaced by a novel object. 4) Pre-pulse inhibition Each mouse was placed in a restraint chamber inside a larger isolation cabinet, equipped with sound, light, and air puff generators (SR-LAB, San Diego Instruments), and auditory PPI assessed as described (Geyer and Swerdlow, 2001). 5) Sucrose preference This experiment was always performed last, as mice were individually housed in order to measure liquid consumption. Sucrose preference was determined by the percentage of sucrose water consumed as a proportion of all water consumed (Willner et al., 1987). Water is delivered in our colony by Hydropac (Lab Products, Inc.), so an additional Hydropac containing sucrose water was added to the cage and the two packs were weighed daily.
Activity and sleep assessment
Individually-housed mice were video recorded in 12″ × 12″ × 13.5″ acrylic chambers, using infra-red cameras during the dark phase, for 72 continuous hours under 12/12 light/dark conditions. The distance traveled in meters per hour was calculated by Noldus Ethovision software, and averaged across 8 CTRL mice (gray fill, lines 22 and 17) and 10 SCZ mice (purple fill, line 52). In addition, transitions between phases of the light cycle (measured 30 minutes before and 30 minutes after light changes) were analyzed in terms of the number of consecutive seconds of immobility as a percentage of total immobility (AnyMaze, Stoelting), per 30 min measurement block as described (McShane et al., 2010; Pack et al., 2007).
RNA-seq
hGPCs assessed for gene expression were first sorted by fluorescence-activated cell sorting on the basis of the cell surface marker CD140a (BD Pharmingen) as described (Sim et al., 2011) (Figure S2). Using polyA-selection, mRNA was isolated from these PDGFRα+ hGPCs, which were derived from iPSCs made from 4 patients with juvenile-onset schizophrenia (SCZ line numbers 8 [n=4 independent cell preparations], 29 [n=3], 51 [n=7], and 164 [n=8]); and 3 demographically similar healthy controls (CTR lines 22 [n=3], 37 [n=4], and 205 [n=7]). Sequencing libraries were prepared using the TruSeq RNA v2 kit, and sequenced on an Illumina HiSeq 2500 platform for approximately 45 million 1×100 bp reads per sample. The sequencing reads were pre-processed by trimming off adapter and low-quality sequences from the 3’ end using Trimmomatic (Bolger et al., 2014). The quality of reads before and after pre-processing was assessed with FastQC (D’Antonio et al., 2015), and the pre-processed reads were then aligned to the RefSeq NCBI reference human genome version GRCh38 (Pruitt et al., 2007) with Subread read aligner (Liao et al., 2013) using Hamming distance to break ties between more than one optimal mapping locations. Raw gene counts were obtained from BAM alignment files with the featureCounts tool (Liao et al., 2014).
For bioinformatic analysis, see Quantification and Statistical Analysis section below
QUANTIFICATION AND STATISTICAL ANALYSIS
Imaging and quantitative histology
For mapping the distribution of human nuclei, or photographing gross distribution of myelin at low power, whole brain sections were imaged on a Leica LMD 6500. Imaging for phenotypic counts was performed on an Olympus BX51 driven by Stereo Investigator software (MBF, Williston, VT).
Astrocyte morphometrics
Shiverer x rag2-null mice were sacrificed at 4.5 months of age and their white matter astrocyte morphologies assessed. 150 μm thick coronal slices were taken by Vibratome at Bregma -1.0 mm from control (22, 37 and C27) or SCZ (51, 164, 193) hGPC-engrafted mice, incubated in mouse anti-hGFAP for 1 week, then 4 hrs in Alexa 568 goat anti-mouse. The slices were mounted on slides and imaged at 100× by confocal (Leica SP8). The images were traced using Neurolucida 360 (MicroBrightfield, Inc.); all tracings were done by experimenters blinded as to the treatment condition.
Individual astrocytes were selected from the middle of the corpus callosum at mid-depth so as to capture cells and their processes in their entirety. Nine cells/brain were analyzed by Neurolucida with Sholl analysis, as 3 cells/slice and 3 slices/brain, taken at 500, 1000, and 1500 μm lateral of the midline. Two or three brains were assessed for each of three lines produced from separate patients, for a total of 8 brains and 72 traced cells/condition, for both CTRL and SCZ-engrafted groups. For Sholl analysis, concentric shells placed at successively increasing diameters of 5 μm were centered on the cell body, and the number of intersections between cell processes and shells counted (Sholl, 1953). For the assessment and quantitative description of astrocytic fiber 3D architecture, Fan-in analysis (MBF Biosciences) was used as previously described for studies of dendritic topology (Dang et al., 2014).
Myelin luminance analysis
To measure forebrain myelination, we used luminance analysis based on measurement of myelin basic protein (MBP) immunofluorescence. Evenly-spaced and uniformly sampled coronal sections were stained for MBP as described, and images taken at 10× using a Nikon Ni-E and Nikon DS-Fi1 camera. The corpus callosum was selected as region of interest, and mean intensity values were obtained using NIS Elements v.4.5.
Statistical analysis
Unless otherwise noted, all analyses were done in GraphPad Prism v.7. Individual tests were performed as noted for each experiment; all data shown as Means ± SEM.
Bioinformatics
Raw RNAseq gene counts were obtained as noted above. After eliminating lowly expressed transcripts with <5 reads in more than 5 samples across the dataset, the count data was normalized using the RUVSeq (Risso et al., 2014) R Bioconductor package (Gentleman et al., 2004) to account for variance. As described in the RUVSeq manual, the normalization was accomplished in the following three-step procedure: 1) in silico negative control genes were determined by first-pass differential expression analysis by the edgeR (Robinson et al., 2010) and DESeq2 (Love et al., 2014) R Bioconductor packages, taking genes with FDR-adjusted P values >0.75, as calculated by both methods (approximately 7000 genes were unaffected by the condition of interest); 2) the negative control genes were then used in the RUVs function of the RUVSeq package, for calculation of variance factors; and, 3) the second-pass differential expression analysis (5% FDR and log2 fold change >1) for determining disease-dysregulated genes was performed using the original counts, adjusting for RUVs-calculated variance factors by multi-factor GLM models implemented in the edgeR and DESeq2 packages.
This three-step analysis, with filtering for low-expressed transcripts, was used to compare each SCZ-derived hGPC cell line to the pooled CTR-derived hGPCs. The intersection of the resulting 4 individual lists of differentially expressed genes was taken as the conserved representative list of SCZ-dysregulated genes. In the normalization procedure for each comparison, the number of RUVs-calculated variance factors was limited to 1 for line 29, 3 for lines 8 and 164, and 7 for line 51, as determined by principal component and hierarchical clustering analyses performed with native R functions (https://www.R-project.org). To obtain average fold changes and P values for dysregulated genes in all 4 SCZ hGPC lines, a differential expression comparison of pooled SCZ to pooled CTR lines was performed by the same filtering and analysis workflow with the number of variance factors limited to 9.
For all differential expression comparisons, only the significant results that agreed between edgeR and DESeq2 methods were used in downstream analysis. Once individual fold changes and P values for dysregulated genes in all 4 SCZ hGPC lines were established relative to control lines, the differential expression of pooled SCZ to pooled CTR lines was performed. For each SCZ cell line, separate DE comparisons were performed against each control line and the intersection of the DE genes was taken as a representative list for that SCZ line against the control population. Fold changes and FDR-adjusted P values reported were calculated by edgeR. Functional annotation of the conserved set of SCZ-dysregulated genes was done using ToppCluster (Kaimal et al., 2010) and Ingenuity Pathway Analysis (IPA) (http://www.qiagen.com/ingenuity). Network visualization and analysis of the results of functional annotation were performed in Gephi (Jacomy et al., 2014) visualization software.
Real-Time PCR
Expression levels in SCZ- and control derived GPCs of selected target genes identified by RNA-seq were assayed by TaqMan Low Density Array (TLDA) Real-Time PCR. The raw data were analyzed in EspressionSuite Software version 1.1 supplied by Applied Biosystems and exported into HTqPCR R package (27) for relative quantification analysis.
DATA AND SOFTWARE AVAILABILITY
For streamlined execution of the above data processing and analysis routines, a set of Python and R scripts was developed. The code and the data files needed to reproduce our normalization and differential expression pipeline are available from https://github.com/cbtncph/GoldmanetalSCZ2016.
All genomic data have been deposited to GEO, accession number GSE86906.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies used in tissue | ||
| Rat monoclonal anti-myelin basic protein, 1:25 | Abcam | Cat#ab7349; RRID: AB_305869 |
| Rabbit polyclonal anti-transferrin, 1:800 | Abcam | Cat#ab9538; RRID: AB_307325 |
| Mouse monoclonal anti-human GFAP, clone SMI 21R, 1:500 | Covance Research Products Inc | Cat#SMI-21R-500; RRID: AB_509979 |
| Rabbit monoclonal anti-Ki-67, clone SP6 | ThermoFisher Scientific | Cat#RM-9106-S |
| Mouse monoclonal anti-human cytoplasmic marker | Takara Bio Inc | Cat#aB-121-U-050; RRID: AB_2632385 |
| Rat monoclonal anti-BrdU clone BU1/75 (ICR1) | Bio-Rad | Cat#MCA2060; RRID: AB_323427 |
| Rabbit polyclonal anti-olig2, 1:500 | Neuromics | Cat#RA25017, 25081 |
| Mouse anti-NG2, clone 9.2.27, 1:200 | Millipore | Cat#MAB2029 |
| Rabbit monoclonal anti-PDGF Receptor alpha, clone D13C6, 1:300 | Cell Signaling Technology | Cat#5241S; RRID: AB_10692773 |
| Goat anti-mouse IgG (H+L) Alexa Fluor 647, 1:400 | ThermoFisher Scientific | Cat#A-21235; RRID: AB_2535804 |
| Goat anti-mouse IgG1 Alexa Fluor 488, 1:400 | ThermoFisher Scientific | Cat#A-21121; RRID: AB_2535764 |
| Goat anti-mouse IgG (H+L) Alexa Fluor 488, 1:400 | ThermoFisher Scientific | Cat#A-11029; RRID: AB_2534088 |
| Goat anti-mouse IgG (H+L) Alexa Fluor 568, 1:400 | ThermoFisher Scientific | Cat#A-11031; RRID: AB_144696 |
| Goat anti-mouse IgG1 Alexa Fluor 568, 1:400 | ThermoFisher Scientific | Cat#A-21124; RRID: AB_2535766 |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor 647, 1:400 | ThermoFisher Scientific | Cat#A-21245; RRID: AB_2535813 |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor 568, 1:400 | ThermoFisher Scientific | Cat#A-11036; RRID: AB_2534094 |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor 488, 1:400 | ThermoFisher Scientific | Cat#A-11034; RRID: AB_2576217 |
| Goat anti-Rat IgG (H+L) Alexa Fluor 647, 1:400 | ThermoFisher Scientific | Cat#A-21247; RRID: AB_141778 |
| Goat anti-Rat IgG (H+L) Alexa Fluor 568 | ThermoFisher Scientific | Cat#A-11077; RRID: AB_2534121 |
| Goat anti-Rat IgG (H+L) Alexa Fluor 488 | ThermoFisher Scientific | Cat#A-11006; RRID: AB_2534074 |
| Bacterial and Virus Strains | ||
| NA | ||
| Experimental Models: Organisms/Strains | ||
| Mouse: C3Fe.SWV-Mbpshi/J | Jackson Laboratory | Cat#001428 |
| Mouse: B6.129S7-Rag1tm1Mom/J | Jackson Laboratory | Cat#002216 |
| Mouse: C3H.129S6(B6)-Rag2tm1FwaN12 | Taconic | Cat#000602-M |
| Software and Algorithms | ||
| Photoshop CS6 | Adobe | |
| Illustrator CS6 | Adobe | |
| StereoInvestigator v11 | MBF Bioscience | |
| Neurolucida 360 v2 | MBF Bioscience | |
| Neurolucida Explorer v11 | MBF Bioscience | |
| Leica Metamorph AF v2 | Leica Biosystems | |
| Leica Application Suite X | Leica Biosystems | |
| Equipment: Histological analysis | ||
| Cryostat | Hacker Instruments | Model OTF |
| Cryostat | Leica Biosystems | Cat#CM3050S |
| Vibratome | Vibratome | 1000 Plus |
| Disposable microtome blades | C.L. Sturkey | Cat#DT315G50 |
| Double edge PTFE coated stainless steel blades | Ted Pella Inc | Cat#121-6 |
| Surgipath X-tra precleaned micro slides | Leica Biosystems | Cat#38002002 |
| DMi8 | Leica Biosystems | |
| DM6000B | Leica Biosystems | |
| DFC 360 FX camera | Leica Biosystems | |
| BX51 | Olympus | |
| DP30BW camera | Olympus | |
| Orca-R2 Digital CCD Camera | Hamamatsu | Cat#C10600-10B |
| MAC 5000 | Ludl Electronic Prods. | Cat#73005001 |
| Focus DR Linear encoder | Ludl Electronic Prods. | Cat#99A420 |
| STG 4″×3″ Stepper | Ludl Electronic Prods. | Cat#99S100LE2MBF |
| Antibodies used in vitro | ||
| Alexa Fluor 488-SSEA4 | Invitrogen | Cat#A14810 |
| APC-conjugated mouse IgG1, Isotype Control | Miltenyi Biotec | Cat#130-092-214 |
| APC-mouse IgM, Isotype Control | Miltenyi Biotec | Cat#130-093-176 |
| APC-conjugated mAb A2B5 | Miltenyi Biotec | Cat#130-093-582 |
| APC-conjugated anti-CD44 | Miltenyi Biotec | Cat#130-095-177 |
| APC-conjugated anti-CD133/1 | Miltenyi Biotec | Cat#130-090-826 |
| PE-conjugated anti-CD140a | BD Pharmingen | Cat#556002 |
| Anti-olig2 | R&D Systems | Cat#AF2418 |
| PE-conjugated mouse IgG2a, Isotype Control | BD Pharmingen | Cat#555574 |
| Anti-PDGFα receptor | Cell Signaling Tech. | Cat#5241S |
| Antibodies used for Western Blot | ||
| Neurexin 1, 1:1000 | EMD Millipore | ABN161-1 |
| Chemicals, Peptides, and Recombinant Proteins used in vitro | ||
| bFGF | Sigma | Cat#F0291 |
| Biotin | Sigma | Cat#B4639 |
| dibutyryl cAMP | Sigma | Cat#D0260 |
| Heparin | Fisher | Cat#NC9484621 |
| IGF-1 | R&D Systems | Cat#291-G1-050 |
| Laminin | Corning | Cat#354232 |
| NT3 | R&D Systems | Cat#267-N3-025 |
| PDGFaa | R&D Systems | Cat#221-AA-50 |
| Purmorphamine | Calbiochem | Cat#80603-730 |
| Retinoic acid | Sigma | Cat#R2625 |
| T3 | Sigma | Cat#T5516-1MG |
| Critical Commercial Assays | ||
| Custom TaqMan Array Card | Applied Biosystems | N/A |
| Ovation PicoSL WTA System V2 | NuGEN | Cat#3312 |
| RNeasy mini kit | QIAGEN | Cat#74104 |
| Taqman Universal master mix | Applied Biosystems | Cat #4304437 |
| TruSeq RNA library Prep Kit V2 | Illumina | Cat#RS-122-2001 |
| Deposited Data | ||
| Raw RNA-seq data | GEO datasets: https://www.ncbi.nlm.nih.gov/gds/ | GEO accession number: GSE86906 |
| Processed RNA-seq data (count matrix) and R scripts for data analysis | This paper | GitHub repository: https://github.com/cbtncph/GoldmanetalSCZ2016 |
| Human reference genome NCBI build 38, GRCh38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/human |
| Experimental Models: Cell Lines | ||
| C27 | L. Studer, SKI | N/A |
| CWRU8 | P.Tesar, Case Western | N/A |
| CWRU19 | P.Tesar, Case Western | N/A |
| CWRU22 | P.Tesar, Case Western | N/A |
| CWRU29 | P.Tesar, Case Western | N/A |
| CWRU31 | P.Tesar, Case Western | N/A |
| CWRU37 | P.Tesar, Case Western | N/A |
| CWRU51 | P.Tesar, Case Western | N/A |
| CWRU52 | P.Tesar, Case Western | N/A |
| CWRU164 | P.Tesar, Case Western | N/A |
| CWRU193 | P.Tesar, Case Western | N/A |
| CWRU205 | P.Tesar, Case Western | N/A |
| Software and Algorithms used in cell culture | ||
| FlowJo | TreeStar Inc. | N/A |
| Software and Algorithms used in RNA-seq | ||
| Trimmomatic (version 0.32) | Bolger et al. (2014) | http://www.usadellab.org/cms/?page=trimmomatic |
| Subread (version 1.4.6-p3) | Liao et al. (2013) | http://subread.sourceforge.net/ |
| featureCounts (version 1.4.6-p3) | Liao et al. (2014) | http://subread.sourceforge.net/ |
| R | R Core Team (2016) | https://www.R-project.org/ |
| RUVSeq (version 1.6.2) | Risso et al. (2014) | http://www.bioconductor.org/packages/release/bioc/html/RUVSeq.html |
| EDASeq (version 2.6.2) | Risso et al. (2011) | http://www.bioconductor.org/packages/release/bioc/html/EDASeq.html |
| edgeR (version 3.14.0) | Robinson et al. (2010) | http://www.bioconductor.org/packages/release/bioc/html/edgeR.html |
| DESeq2 (version 1.12.4) | Love et al. (2014) | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ToppCluster | Kaimal et al. (2010) | https://toppcluster.cchmc.org/ |
| Gephi (version 0.9.1) | https://gephi.org/ | https://gephi.org/ |
| Ingenuity Pathway Analysis | QIAGEN | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ |
| Software and Algorithms used in RT-qPCR | ||
| ExpressionSuite Software (version 1.1) | Applied Biosystems | https://www.thermofisher.com/dk/en/home/technical-resources/software-downloads/expressionsuite-software.html |
| Other | ||
| Agilent Bioanalyzer | Agilent | N/A |
| BD FACS Aria IIIU | BD Biosciences | N/A |
| HiSeq 2500 | Illumina Inc. | N/A |
| Nanodrop 1000 spectrophotometer | Nanodrop | N/A |
| Olympus IX71 Inverted Research Microscope | Olympus | N/A |
| QuantStudio 12K Flex Real-Time PCR system | Applied Biosystems | N/A |
Supplementary Material
Highlights.
Human glial chimeric mice were made using iPSCs derived from schizophrenic subjects
SCZ glial chimeras develop abnormal astrocytic morphology and hypomyelination
Differentiation-associated gene expression is impaired in SCZ glial progenitors
SCZ glial chimeric mice have broad behavioral and sleep abnormalities
Acknowledgments
Supported by NIMH, NINDS, the G. Harold and Leila Y. Mathers Charitable Foundation, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and the Novo Nordisk Foundation. We thank Alexis Yagielski and Simrat Dhaliwal for histological assistance, and Jennifer Wintermute for assistance in cell culture, Lorenz Studer (Memorial Sloan-Kettering) for the C27 hiPSC cell line, Nora McNamara (Case Western) for assistance in patient identification, Leslie Cooperman and Elizabeth Shick (Case Western) for assistance in producing the iPSC lines, and Chiara Cirelli (U. Wisconsin) for her comments on our sleep and activity data. This paper is dedicated to the memory of Mr. James T. Handelman of the Mathers Charitable Foundation, whose early and ardent support made this work possible.
All genomic data have been deposited to GEO, accession number GSE86906.
Footnotes
Author contributions: MO did the genomic analysis; DCM and ZL prepared RNA and validated both RNA and protein expression; JB, DCM, and ZL raised cells, under the direction of SW; JM, SS and LZ assessed myelination and astrocytic morphologies, under the direction of MSW; RHM and PT produced the iPSCs from patient samples provided by RLF; LZ and KM performed the blinded behavioral analyses, while MN and MSW analyzed the behavioral and sleep data; MSW organized the histological and behavioral figures, while MO organized the genomics figures and tables; PT, MSW, MN and SAG conceived the project; SAG directed the overall effort and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral AI, Meisingset TW, Kotter MR, Sonnewald U. Metabolic aspects of neuron-oligodendrocyte-astrocyte interactions. Front Endocrinol (Lausanne) 2013;4:54. doi: 10.3389/fendo.2013.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JL, Fernandez-Enright F. A decade from discovery to therapy: Lingo-1, the dark horse in neurological and psychiatric disorders. Neurosci Biobehav Rev. 2015;56:97–114. doi: 10.1016/j.neubiorev.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Araque A, Parpura V, Sanzgiri R, Haydon P. Glutamate-dependent astrocyte modulation of synaptic transmission between cultured hippocampal neurons. European J Neurosci. 1998:10. doi: 10.1046/j.1460-9568.1998.00221.x. [DOI] [PubMed] [Google Scholar]
- Barnes SA, Der-Avakian A, Markou A. Anhedonia, avolition, and anticipatory deficits: assessments in animals with relevance to the negative symptoms of schizophrenia. Eur Neuropsychopharmacol. 2014;24:744–758. doi: 10.1016/j.euroneuro.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA, Raff MC. Proliferation of oligodendrocyte precursor cells depends on electrical activity in axons. Nature. 1993;361:258–260. doi: 10.1038/361258a0. [DOI] [PubMed] [Google Scholar]
- Bergles DE, Jabs R, Steinhauser C. Neuron-glia synapses in the brain. Brain Res Rev. 2010;63:130–137. doi: 10.1016/j.brainresrev.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins RA, Besheer J. Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study ‘recognition memory’. Nat Protoc. 2006;1:1306–1311. doi: 10.1038/nprot.2006.205. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature biotechnology. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LE, Barres BA. Glia keep synapse distribution under wraps. Cell. 2013;154:267–268. doi: 10.1016/j.cell.2013.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor CM, Crawford BC, Akbarian S. White matter neuron alterations in schizophrenia and related disorders. International journal of developmental neuroscience : the official journal of the International Society for Developmental Neuroscience. 2011;29:325–334. doi: 10.1016/j.ijdevneu.2010.07.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Antonio M, D’Onorio De Meo P, Pallocca M, Picardi E, D’Erchia AM, Calogero RA, Castrignano T, Pesole G. RAP: RNA-Seq Analysis Pipeline, a new cloud-based NGS web application. BMC Genomics. 2015;16:S3. doi: 10.1186/1471-2164-16-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang V, Medina B, Das D, Moghadam S, Martin KJ, Lin B, Naik P, Patel D, Nosheny R, Wesson Ashford J, et al. Formoterol, a long-acting beta2 adrenergic agonist, improves cognitive function and promotes dendritic complexity in a mouse model of Down syndrome. Biol Psychiatry. 2014;75:179–188. doi: 10.1016/j.biopsych.2013.05.024. [DOI] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Archives of general psychiatry. 2003;60:443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- De Biase LM, Nishiyama A, Bergles DE. Excitability and synaptic communication within the oligodendrocyte lineage. J Neurosci. 2010;30:3600–3611. doi: 10.1523/JNEUROSCI.6000-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing SG, Grace AA. Evidence for impaired sound intensity processing during prepulse inhibition of the startle response in a rodent developmental disruption model of schizophrenia. Journal of psychiatric research. 2013 doi: 10.1016/j.jpsychires.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerland MW, Sandvik L. Performance of five two-sample location tests for skewed distributions with unequal variances. Contemp Clin Trials. 2009;30:490–496. doi: 10.1016/j.cct.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Fernandez-Enright F, Andrews JL, Newell KA, Pantelis C, Huang XF. Novel implications of Lingo-1 and its signaling partners in schizophrenia. Translational psychiatry. 2014;4:e348. doi: 10.1038/tp.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD. White matter in learning, cognition and psychiatric disorders. Trends in neurosciences. 2008;31:361–370. doi: 10.1016/j.tins.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin RJ, Bussey TJ. Do your glial cells make you clever? Cell stem cell. 2013;12:265–266. doi: 10.1016/j.stem.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva L, Moskvina V, Peirce T, Norton N, Bray NJ, Jones L, Holmans P, Macgregor S, Zammit S, Wilkinson J, et al. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12469–12474. doi: 10.1073/pnas.0603029103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer MA, Swerdlow NR. Measurement of startle response, prepulse inhibition, and habituation. Curr Protoc Neurosci. 2001;Chapter 8(Unit 8):7. doi: 10.1002/0471142301.ns0807s03. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Lu A, Leow AD, Klunder AD, Lee AD, Chavez A, Greenstein D, Giedd JN, Toga AW, Rapoport JL, et al. Three-dimensional brain growth abnormalities in childhood-onset schizophrenia visualized by using tensor-based morphometry. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15979–15984. doi: 10.1073/pnas.0806485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N, Rapoport JL. Childhood-onset schizophrenia: insights from neuroimaging studies. Journal of the American Academy of Child and Adolescent Psychiatry. 2008;47:1120–1124. doi: 10.1097/CHI.0b013e31817eed7a. [DOI] [PubMed] [Google Scholar]
- Goldman SA, Nedergaard M, Windrem MS. Modeling cognition and disease using human glial chimeric mice. Glia. 2015;63:1483–1493. doi: 10.1002/glia.22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graw SL, Swisshelm K, Floyd K, Carstens BJ, Wamboldt MZ, Ross RG, Leonard S. Isochromosome 13 in a patient with childhood-onset schizophrenia, ADHD, and motor tic disorder. Mol Cytogenet. 2012;5:2. doi: 10.1186/1755-8166-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Chen M, Wang F, Windrem M, Wang S, Shanz S, Xu Q, Oberheim NA, Bekar L, Betstadt S, et al. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell stem cell. 2013;12:342–353. doi: 10.1016/j.stem.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof PR, Haroutunian V, Copland C, Davis KL, Buxbaum JD. Molecular and cellular evidence for an oligodendrocyte abnormality in schizophrenia. Neurochem Res. 2002;27:1193–1200. doi: 10.1023/a:1020981510759. [DOI] [PubMed] [Google Scholar]
- Horrobin DF. Schizophrenia: the illness that made us human. Medical hypotheses. 1998;50:269–288. doi: 10.1016/s0306-9877(98)90000-7. [DOI] [PubMed] [Google Scholar]
- Ivleva EI, Moates AF, Hamm JP, Bernstein IH, O’Neill HB, Cole D, Clementz BA, Thaker GK, Tamminga CA. Smooth pursuit eye movement, prepulse inhibition, and auditory paired stimuli processing endophenotypes across the schizophrenia-bipolar disorder dimension. Schizophrenia bulletin. 2013 doi: 10.1093/schbul/sbt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacomy M, Venturini T, Heymann S, Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PloS one. 2014;9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GR. Investigation of astrocyte - oligodendrocyte interactions in human cultures. Methods Mol Biol. 2012;814:401–414. doi: 10.1007/978-1-61779-452-0_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaimal V, Bardes EE, Tabar SC, Jegga AG, Aronow BJ. ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic acids research. 2010;38:W96–102. doi: 10.1093/nar/gkq418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Jiang L, Goldman SA, Nedergaard M. Astrocyte-mediated potentiation of inhibitory synaptic transmission. Nature neuroscience. 1998;1:683–692. doi: 10.1038/3684. [DOI] [PubMed] [Google Scholar]
- Kohl S, Heekeren K, Klosterkotter J, Kuhn J. Prepulse inhibition in psychiatric disorders--apart from schizophrenia. Journal of psychiatric research. 2013;47:445–452. doi: 10.1016/j.jpsychires.2012.11.018. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Morrison BM, Li Y, Lengacher S, Farah MH, Hoffman PN, Liu Y, Tsingalia A, Jin L, Zhang PW, et al. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature. 2012;487:443–448. doi: 10.1038/nature11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Piao Y, Shon HS, Ryu KH. Comparing the normalization methods for the differential analysis of Illumina high-throughput RNA-Seq data. BMC Bioinformatics. 2015;16:347. doi: 10.1186/s12859-015-0778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic acids research. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin SC, Bergles DE. Synaptic signaling between GABAergic interneurons and oligodendrocyte precursor cells in the hippocampus. NatNeurosci. 2004;7:24–32. doi: 10.1038/nn1162. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackowiak M, Mordalska P, Wedzony K. Neuroligins, synapse balance and neuropsychiatric disorders. Pharmacol Rep. 2014;66:830–835. doi: 10.1016/j.pharep.2014.04.011. [DOI] [PubMed] [Google Scholar]
- McIntosh AM, Munoz Maniega S, Lymer GK, McKirdy J, Hall J, Sussmann JE, Bastin ME, Clayden JD, Johnstone EC, Lawrie SM. White matter tractography in bipolar disorder and schizophrenia. Biological psychiatry. 2008;64:1088–1092. doi: 10.1016/j.biopsych.2008.07.026. [DOI] [PubMed] [Google Scholar]
- McShane BB, Galante RJ, Jensen ST, Naidoo N, Pack AI, Wyner A. Characterization of the bout durations of sleep and wakefulness. J Neurosci Methods. 2010;193:321–333. doi: 10.1016/j.jneumeth.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz Maniega S, Lymer GK, Bastin ME, Marjoram D, Job DE, Moorhead TW, Owens DG, Johnstone EC, McIntosh AM, Lawrie SM. A diffusion tensor MRI study of white matter integrity in subjects at high genetic risk of schizophrenia. Schizophrenia research. 2008;106:132–139. doi: 10.1016/j.schres.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Najjar S, Pearlman DM. Neuroinflammation and white matter pathology in schizophrenia: systematic review. Schizophrenia research. 2015;161:102–112. doi: 10.1016/j.schres.2014.04.041. [DOI] [PubMed] [Google Scholar]
- Oberheim N, Wang X, Goldman SA, Nedergaard M. Astrocytic complexity distinguishes the human brain. Trends in Neurosciences. 2006;29:1–10. doi: 10.1016/j.tins.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Oberheim NA, Takano T, Han X, He W, Lin JH, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, et al. Uniquely hominid features of adult human astrocytes. J Neurosci. 2009;29:3276–3287. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack AI, Galante RJ, Maislin G, Cater J, Metaxas D, Lu S, Zhang L, Von Smith R, Kay T, Lian J, et al. Novel method for high-throughput phenotyping of sleep in mice. Physiol Genomics. 2007;28:232–238. doi: 10.1152/physiolgenomics.00139.2006. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic acids research. 2007;35:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport JL, Addington AM, Frangou S, Psych MR. The neurodevelopmental model of schizophrenia: update 2005. Molecular psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach A, Boylan K, Horvath S, Prusiner SB, Hood LE. Characterization of cloned cDNA representing rat myelin basic protein: absence of expression in brain of shiverer mutant mice. Cell. 1983;34:799–806. doi: 10.1016/0092-8674(83)90536-6. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth J. Central myelin in the mouse mutant shiverer. J Comp Neurol. 1980;194:639–648. doi: 10.1002/cne.901940310. [DOI] [PubMed] [Google Scholar]
- Roy K, Murtie JC, El-Khodor BF, Edgar N, Sardi SP, Hooks BM, Benoit-Marand M, Chen C, Moore H, O’Donnell P, et al. Loss of erbB signaling in oligodendrocytes alters myelin and dopaminergic function, a potential mechanism for neuropsychiatric disorders. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8131–8136. doi: 10.1073/pnas.0702157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakry D, Neitz A, Singh J, Frischknecht R, Marongiu D, Biname F, Perera SS, Endres K, Lutz B, Radyushkin K, et al. Oligodendrocyte precursor cells modulate the neuronal network by activity-dependent ectodomain cleavage of glial NG2. PLoS Biol. 2014;12:e1001993. doi: 10.1371/journal.pbio.1001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyakina D, Cukier HN, Lee JM, Sacharow S, Nations LD, Ma D, Jaworski JM, Konidari I, Whitehead PL, Wright HH, et al. Copy number variants in extended autism spectrum disorder families reveal candidates potentially involved in autism risk. PloS one. 2011;6:e26049. doi: 10.1371/journal.pone.0026049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samartzis L, Dima D, Fusar-Poli P, Kyriakopoulos M. White matter alterations in early stages of schizophrenia: a systematic review of diffusion tensor imaging studies. J Neuroimaging. 2014;24:101–110. doi: 10.1111/j.1552-6569.2012.00779.x. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, lam K, Oltz E, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall A, et al. RAG2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87:387–406. [PMC free article] [PubMed] [Google Scholar]
- Sigmundsson T, Suckling J, Maier M, Williams S, Bullmore E, Greenwood K, Fukuda R, Ron M, Toone B. Structural abnormalities in frontal, temporal, and limbic regions and interconnecting white matter tracts in schizophrenic patients with prominent negative symptoms. Am J Psychiatry. 2001;158:234–243. doi: 10.1176/appi.ajp.158.2.234. [DOI] [PubMed] [Google Scholar]
- Sim FJ, McClain CR, Schanz SJ, Protack TL, Windrem MS, Goldman SA. CD140a identifies a population of highly myelinogenic, migration-competent and efficiently engrafting human oligodendrocyte progenitor cells. Nature biotechnology. 2011;29:934–941. doi: 10.1038/nbt.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim FJ, Windrem MS, Goldman SA. Fate determination of adult human glial progenitor cells. Neuron Glia Biol. 2009;5:45–55. doi: 10.1017/S1740925X09990317. [DOI] [PubMed] [Google Scholar]
- Simons M, Nave KA. Oligodendrocytes: Myelination and Axonal Support. Cold Spring Harb Perspect Biol. 2015 doi: 10.1101/cshperspect.a020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW, et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Sakurai T, Davis KL, Buxbaum JD. Linking oligodendrocyte and myelin dysfunction to neurocircuitry abnormalities in schizophrenia. Progress in neurobiology. 2011;93:13–24. doi: 10.1016/j.pneurobio.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, Barres BA. Control of synapse number by glia. Science (New York, NY) 2001;291:657–661. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- Uranova NA, Vikhreva OV, Rachmanova VI, Orlovskaya DD. Ultrastructural alterations of myelinated fibers and oligodendrocytes in the prefrontal cortex in schizophrenia: a postmortem morphometric study. Schizophrenia research and treatment. 2011;2011:325789. doi: 10.1155/2011/325789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uranova NA, Vostrikov VM, Vikhreva OV, Zimina IS, Kolomeets NS, Orlovskaya DD. The role of oligodendrocyte pathology in schizophrenia. Int J Neuropsychopharmacol. 2007;10:537–545. doi: 10.1017/S1461145707007626. [DOI] [PubMed] [Google Scholar]
- Voineskos AN, Felsky D, Kovacevic N, Tiwari AK, Zai C, Chakravarty MM, Lobaugh NJ, Shenton ME, Rajji TK, Miranda D, et al. Oligodendrocyte genes, white matter tract integrity, and cognition in schizophrenia. Cereb Cortex. 2013;23:2044–2057. doi: 10.1093/cercor/bhs188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walf AA, Frye CA. The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat Protoc. 2007;2:322–328. doi: 10.1038/nprot.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science (New York, NY) 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wang S, Bates J, Li X, Schanz S, Chandler-Militello D, Levine C, Maherali N, Studer L, Hochedlinger K, Windrem M, et al. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12:252–264. doi: 10.1016/j.stem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welstead GG, Brambrink T, Jaenisch R. Generating iPS cells from MEFS through forced expression of Sox-2, Oct-4, c-Myc, and Klf4. J Vis Exp. 2008 doi: 10.3791/734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner P, Towell A, Sampson D, Sophokleous S, Muscat R. Reduction of sucrose preference by chronic unpredictable mild stress, and its restoration by a tricyclic antidepressant. Psychopharmacology (Berl) 1987;93:358–364. doi: 10.1007/BF00187257. [DOI] [PubMed] [Google Scholar]
- Windrem MS, Nunes MC, Rashbaum WK, Schwartz TH, Goodman RA, McKhann G, Roy NS, Goldman SA. Fetal and adult human oligodendrocyte progenitor cell isolates myelinate the congenitally dysmyelinated brain. Nature medicine. 2004;10:93–97. doi: 10.1038/nm974. [DOI] [PubMed] [Google Scholar]
- Windrem MS, Schanz SJ, Guo M, Tian GF, Washco V, Stanwood N, Rasband M, Roy NS, Nedergaard M, Havton LA, et al. Neonatal chimerization with human glial progenitor cells can both remyelinate and rescue the otherwise lethally hypomyelinated shiverer mouse. Cell Stem Cell. 2008;2:553–565. doi: 10.1016/j.stem.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windrem MS, Schanz SJ, Morrow C, Munir J, Chandler-Militello D, Wang S, Goldman SA. A competitive advantage by neonatally engrafted human glial progenitors yields mice whose brains are chimeric for human glia. J Neurosci. 2014;34:16153–16161. doi: 10.1523/JNEUROSCI.1510-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Abazyan S, Jouroukhin Y, Pletnikov M. Behavioral sequelae of astrocyte dysfunction: focus on animal models of schizophrenia. Schizophrenia research. 2014 doi: 10.1016/j.schres.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Silverman JL, Crawley JN. Automated three-chambered social approach task for mice. Curr Protoc Neurosci. 2011;Chapter 8(Unit 8):26. doi: 10.1002/0471142301.ns0826s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman DW. A note on preliminary tests of equality of variances. Br J Math Stat Psychol. 2004;57:173–181. doi: 10.1348/000711004849222. [DOI] [PubMed] [Google Scholar]
- Zou XY, Yang HY, Yu Z, Tan XB, Yan X, Huang GT. Establishment of transgene-free induced pluripotent stem cells reprogrammed from human stem cells of apical papilla for neural differentiation. Stem Cell Res Ther. 2012;3:43. doi: 10.1186/scrt134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.