Abstract
The contribution of lncRNAs to tumor progression and regulatory mechanisms driving their expression are areas of intense investigation. Here, we characterize the binding of heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) to a nucleic acid structural element located in exon 12 of PNUTS (also known as PPP1R10) pre-RNA that regulates its alternative splicing. HnRNP E1 release from this structural element, following its silencing, nucleo-cytoplasmic translocation or in response to TGFβ, allows alternative splicing and generates a non-coding isoform of PNUTS. Functionally the lncRNA-PNUTS serves as a competitive sponge for miR-205 during epithelial-mesenchymal transition. In mesenchymal breast tumor cells and in breast tumor samples, the expression of lncRNA-PNUTS is elevated and correlates with levels of ZEB mRNAs. Thus, PNUTS is a bifunctional RNA encoding both PNUTS-mRNA and lncRNA-PNUTS each eliciting distinct biological functions. While PNUTS-mRNA is ubiquitously expressed, lncRNA-PNUTS appears to be tightly regulated dependent on hnRNP E1’s status and tumor context.
Keywords: Serine/threonine-protein phosphatase 1 regulatory subunit 10 (PPP1R10, also known as PNUTS), heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1), epithelial-mesenchymal transition (EMT), miR-205, alternative splicing, long noncoding RNA (lncRNA)
Introduction
Breast cancer in females and lung cancer in males are the most frequently diagnosed cancers and the leading cause of cancer death worldwide1. Although metastasis is the overwhelming cause of mortality in patients with solid tumors, the molecular and cellular mechanisms that drive tumor cells to become metastatic remain largely unknown2–4.
Noncoding RNAs have recently emerged as key mediators of tumor progression through their regulation of both oncogenic and tumor suppressive pathways5,6. LncRNAs have been implicated in cellular processes such as proliferation, apoptosis, migration and cell invasion and their dysregulated expression has been observed in various human cancers7,8. Despite these recent findings, the regulatory role of lncRNAs in mediating these cellular processes and in cancer development remains an area of active investigation and the subject of controversy9,10.
Epithelial-mesenchymal transition (EMT) is a developmental process aberrantly reactivated during tumor progression of epithelial cells and contributes to resistance of both conventional and targeted therapies. We have previously demonstrated that post-transcriptional regulation of gene expression plays an important role in EMT, especially during TGFβ-mediated EMT14,15. We described a transcript-selective translational regulatory pathway involving the binding of hnRNP E1 protein to a BAT structural element (for TGF-beta-activated translational element) located in the 3'-UTR of transcripts involved in EMT-related tumor progression14,16. In addition, hnRNP E1 protein was previously described to regulate other critical cellular processes such as transcription, mRNA stability, transport and splicing17.
Alternative splicing regulates over 90% of multi-exon protein-coding genes in humans18 and hnRNP E1 is well documented for its repressive role in this process. HnRNP E1 represses tumor cell invasion by inhibiting the alternative splicing of CD4419 and binds to the growth hormone receptor (GHR) pseudoexon and prevent its usage, thus allowing expression of a functional protein20.
Here we report the binding of hnRNP E1 to an alternative splicing site in the pre-RNA of PNUTS to control the generation of an alternative spliced isoform of PNUTS that we describe as a lncRNA involved in EMT-related tumor progression. The study reveals that the PNUTS pre-RNA transcript serves as a bifunctional RNA capable of generating PNUTS-mRNA or lncRNA-PNUTS in an hnRNP E1 dependent and cell context-dependent manner.
Results
The predicted lncRNA-PNUTS is upregulated upon hnRNP E1 loss and during tumor cell progression
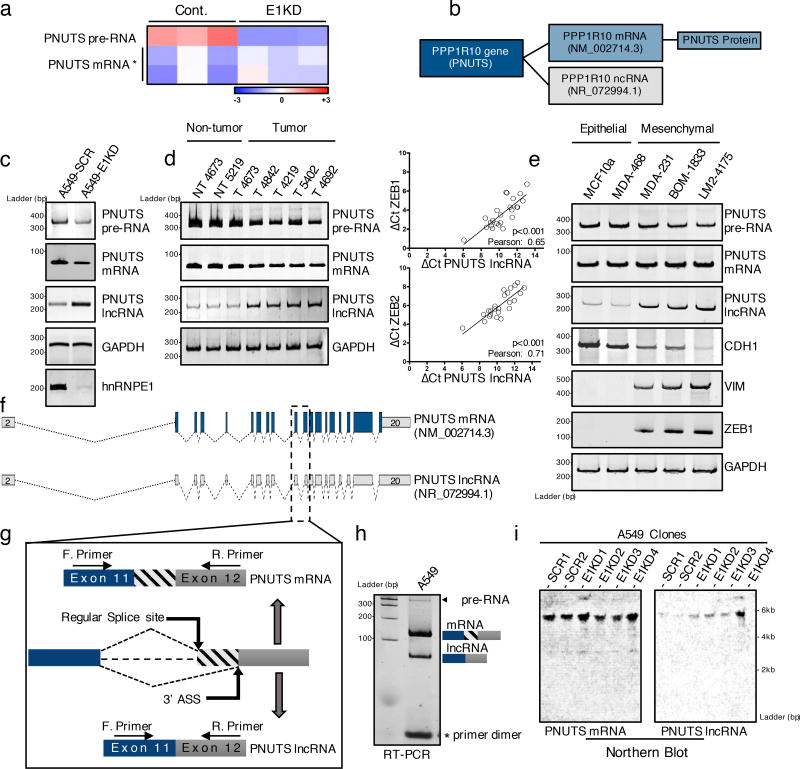
Using an hnRNP E1 knockdown-induced EMT model in NMuMG cells (Supplementary figure 1a) we performed an Affymetrix array analysis and identified PNUTS pre-RNA as downregulated upon hnRNP E1 knockdown while the associated PNUTS-mRNA remained relatively unaffected (Figure 1a) suggestive of a differential processing of PNUTS pre-RNA. Interestingly, the human PNUTS gene is described to encode two sequenced variants. While variant 1 encodes the well-characterized PNUTS-mRNA, variant 2 has not been investigated and is predicted to be a lncRNA (Figure 1b). We validated the differential processing of PNUTS pre-RNA by RT-PCR analysis with primers specific to PNUTS isoforms (Figure 1c, Supplementary figure 6).
Figure 1. PNUTS alternative splicing occurs upon hnRNP E1 loss and is increased in mesenchymal tumor cells.
(a) Heat map of Affymetrix array data showing expression levels (log2 fold) of either PNUTS pre-RNA or PNUTS mRNA in control (CTRL) or hnRNP E1 knockdown (E1KD) NMuMG cells. The data were generated from triplicates samples. * Two distinct probes were used to target the spliced PNUTS RNA.
(b) NCBI database accession numbers of PNUTS mRNA and PNUTS predicted lncRNA isoform in human.
(c) Validation by RT-PCR analysis with primers specific to PNUTS isoforms of alternative PNUTS gene processing upon hnRNP E1 knockdown in human A549 cell line.
(d) (Left) PNUTS isoform expression levels analyzed by RT-PCR in human breast tumor samples (T) or non-tumor counterparts (NT). (Right) Quantitative RT-PCR analysis of lncRNA-PNUTS, ZEB-1 and ZEB-2 expression in 24 human breast tumor samples. Relative expression levels of transcripts were calculated using the ΔCt method normalizing to GAPDH. Correlations between transcript expression levels were evaluated using Pearson correlation coefficient test. (Linear regression, df=24-2, a Pearson score > 0.515 and p<0.05 was considered as significant). Source data are available in Supplementary table 2.
(e) PNUTS isoform expression screening by RT-PCR analysis in MCF10a mammary gland epithelial cells and MDA-MB-468 breast cancer epithelial cells, or in the metastasis progression model of MDA-MB-231 mesenchymal cell line (MDA-231, BOM-1833, LM2-4175). E-Cadherin (CDH1) was used as epithelial marker while vimentin (VIM) and ZEB1 were used as mesenchymal cell specific markers.
(f) Map of PNUTS isoforms acquired by sequence alignment and drawn by using fancyGene online software.
(g) Schematic representation of the alternative splicing region of the PNUTS variants (ASS: Alternative Splicing Site).
(h) RT-PCR amplification of exon11–exon12 junction encompassing the predicted alternative splicing site using intron-flanking PCR primers as indicated in (g).
(i) Northern-blot analysis of both PNUTS mRNA and lncRNA isoforms expression in control (SCR) or hnRNP E1 knockdown (E1KD) A549 cell clones.
The biological significance of PNUTS pre-RNA differential processing is demonstrated in human breast tumor samples (Figure 1d) and in breast cancer cell lines (Figure 1e). We observed upregulation of the predicted lncRNA-PNUTS in breast tumor samples and a correlation between ZEB1/ZEB2 mesenchymal markers expression and lncRNA-PNUTS (Figure 1d) but not with the PNUTS-mRNA (Supplementary figure 1b). We also observed a correlation between lncRNA-PNUTS expression and the epithelial/mesenchymal status of breast cancer cells. In the more mesenchymal MDA-MB-231 cell line, and its metastatic bone (BOM-1833) and metastatic lung (LM2-4175) derivatives21, we observed increased expression of the predicted lncRNA-PNUTS correlating with the expression of the mesenchymal markers vimentin and Zeb1; whereas, in the more epithelial MCF10a and MDA-MB-468 cell lines expressing the epithelial marker E-cadherin, there was less expression of the predicted lncRNA (Figure 1e).
The NCBI database predicts the generation of lncRNA-PNUTS as a result of the removal of 61 bases in the 5'-region of the exon 12 leading to a break in the open reading frame (ORF) of the transcript (Figure 1f, 1g). By RT-PCR, using flanking primers (Supplementary figure 6), we demonstrated the existence of an alternative splice product of the expected size (Figure 1h) and validated the alternative splicing model by sequencing (Supplementary figure 1c). The alternative splice site is also identified in mouse (Supplementary figure 1d, 1e), and northern-blot validated the size of the full-length lncRNA-PNUTS and its upregulation following hnRNP E1 knockdown (Figure 1i).
hnRNP E1 prevents the splicing of the lncRNA-PNUTS isoform by binding to a BAT structural element located at the alternative splice site
The alternative splice site in exon 12 of the PNUTS gene is also predicted in silico by the HSF finder22 (Supplementary figure 2a). Interestingly, this alternative site has a higher consensus splice site value (91.74) than the regular splice site (79.38) used to generate the PNUTS-mRNA (Supplementary figure 2a), suggesting the existence of an inhibitory mechanism of alternative splicing site utilization. Since hnRNP E1 is a known repressor of alternative splicing19,20 and its knockdown results in upregulation of lncRNA-PNUTS, we postulated that it is an endogenous repressor of PNUTS pre-RNA splicing.
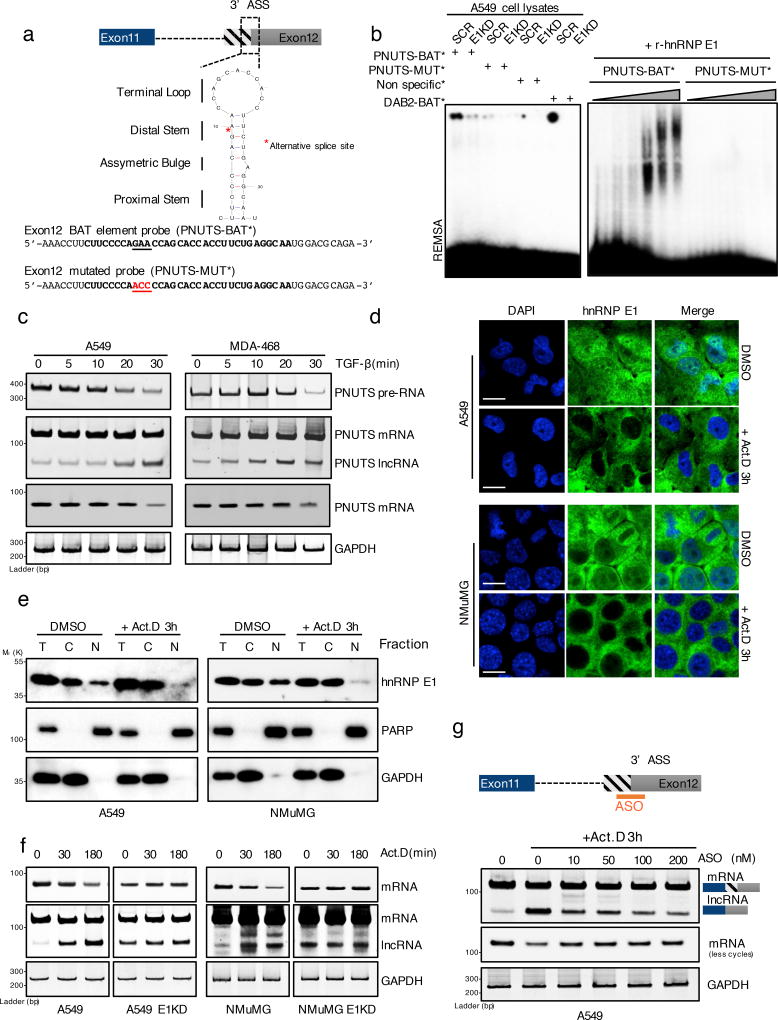
We previously described that hnRNP E1 binds to a consensus BAT element, consisting of a stem-loop structure with an asymmetric bulge located in the 3’-UTR of RNAs14,23. An analysis of the secondary structure of the PNUTS alternative splicing site in human and mouse sequences revealed the existence of a similar evolutionary conserved BAT-like element encompassing the alternative splicing site (Figure 2a ; Supplementary figure 1f). We thus designed PNUTS BAT alternative splicing site RNA probes, wild-type or mutated, to perform RNA electromobility shift assays (REMSA) and validated the direct and specific binding of hnRNP E1 to the structural element. Combinations of WCLs from A549 cells with the wild-type probe show a significant gel shift that is abolished by using either the mutant probe or WCLs prepared from A549 cells silenced for hnRNP E1 (Figure 2b). Direct binding was further validated using recombinant hnRNP E1 protein (Figure 2b). To test whether hnRNP E1 removal from the BAT alternative splicing site can mediate alternative splicing, we used two methods to induce its dissociation from RNA. First, based on our earlier demonstration that TGFβ-induced Akt2 phosphorylation of hnRNP E1 leads to its loss of binding and release from the BAT element14,16, we treated cells with TGFβ and observed alternative splicing occurring in both A549 and MDA-468 cells, generating the lncRNA-PNUTS isoform within 30 minutes and persisting for ~ 3 h (Figure 2c, Supplementary figure 1g). Second, since inhibition of transcription with RNA polymerase inhibitors results in cytoplasmic accumulation of many splicing factors24, we treated cells with actinomycin D (Act.D) and performed both immunofluorescence analysis and cell fractionation to investigate hnRNP E1 localization upon transcriptional inhibition (Figure 2 d,e). We observed that Act.D induced the release of hnRNP E1 from pre-RNAs and its nuclear (N)/cytoplasmic (C) shuttling, resulting in cytoplasmic accumulation (Figure 2 d,e). Concomitantly, cytoplasmic accumulation of hnRNP E1, in response to Act.D, results in a strong induction of PNUTS alternative splicing which is not observed in hnRNP E1 silenced cells (Figure 2f). These results suggest that hnRNP E1 binding to the PNUTS pre-RNA alternative splicing site mediates an inhibitory effect on alternative splicing. To further validate this splicing model, we designed an antisense oligo (ASO) to the alternative splicing site to prevent its utilization. As shown in A549 cells (Figure 2g), the ASO prevents, in a concentration-dependent manner, the alternative splicing induced by hnRNP E1 release following Act.D treatment.
Figure 2. hnRNP E1 protein prevents PNUTS alternative splicing by its specific binding to a BAT structural element.
(a) Secondary structure of the human PNUTS alternative splicing site as predicted by the Mfold algorithm (ΔG= −3.90 kcal.mol−1). The underlined nucleotides colored in red represent the mutant probe used for the REMSA experiment in (b) and (c). The red asterisk represents the exact alternative splicing site leading to the lncRNA-PNUTS isoform generation. (ASS: Alternative Splicing Site)
(b) (Left) RNA electromobility shift assay (REMSA) experiment using either wild-type [PNUTS-BAT] or mutated [PNUTS-MUT] α-32P-labelled PNUTS alternative splicing site probes combined with control (SCR) or hnRNP E1 knockdown (E1KD) A549 cell lysates. The [PNUTS-MUT] probe was mutated by a nucleotide substitution to destroy its secondary structure. [Non specific] and [DAB2-BAT] α-32P-labelled probes were used as negative and positive controls respectively. [DAB2-BAT] corresponds to the BAT-sequence located on the Dab2–3'UTR already described to bind to hnRNP E1. (Right) REMSA using a combination of [PNUTS-BAT] or mutated [PNUTS-MUT] α-32P-labelled probes with increasing concentration of recombinant hnRNP E1 protein purified from e. coli bacteria.
(c) Time course experiment using RT-PCR analysis of PNUTS gene processing after addition of 5ng.mL−1 of TGFβ.
(d) Confocal microscopy imaging of the hnRNP E1 nucleocytoplasmic shuttling by addition of 5µg.mL−1 of Actinomycin D for 3h in A549 and NMuMG cells cultures. Scale bar: 10µM
(e) Characterization of the nucleocytoplasmic transportation of hnRNP E1 following Act.D treatment by using cell fractionation and subsequent western-blot analysis of hnRNP E1 expression. To check the fractions purity, GAPDH and PARP were used as cytoplasmic and nuclear compartment markers respectively.
(f) Time course experiment using RT-PCR analysis of PNUTS predicted-lncRNA alternative splicing activation upon addition of 5µg.mL−1 of Act.D in control (CTRL) or hnRNP E1 silenced (E1KD) A549 and NMuMG cells.
(g) Inhibition of alternative splicing induced by Act.D in A549 cells using and antisense oligonucleotide (ASO) targeting the alternative splicing site of PNUTS.
GAPDH was used as a loading control. Unprocessed original scans of blots are shown in Supplementary Fig. 7
PNUTS alternative splicing product is a cytosolic and nuclear lncRNA
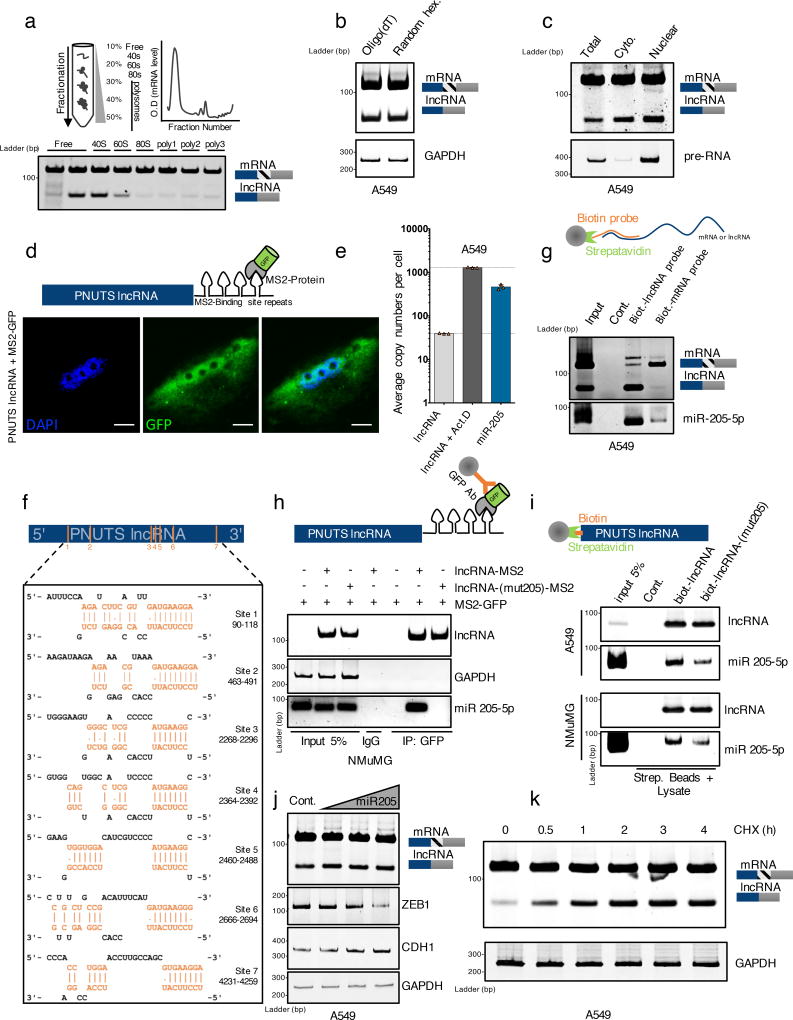
PNUTS mRNA encodes the 99 kDa PNUTS protein. However, PNUTS alternative splicing leads to a break at position K318 generating a downstream premature stop codon, potentially allowing the generation of a 41 kDa truncated protein. By immunoblot analysis, using an N-terminal generated PNUTS antibody, we failed to detect a truncated expression product even upon lncRNA-PNUTS overexpression in various cell lines (Supplementary figure 2b). Further, as analyzed by polysome fractionation, lncRNA-PNUTS is observed only in the non-translating, monosomal fractions and not in the actively translating, polysomal fractions compared to PNUTS mRNA (Figure 3a), underlining its non-translatability. Finally, endogenous lncRNA-PNUTS has a poly(A)+ tail (Figure 3b) and is located in both the cytoplasmic and nuclear compartments as observed by cell fractionation, GFP tracking microscopy employing the MS2-Tag strategy and FISH analysis (Figure 3c, 3d, Supplementary figure 2c).
Figure 3. PNUTS alternative splicing product is non-coding and interacts with miR-205.
(a) Polysome fractionation experiment of A549 cells followed by RT-PCR analysis of PNUTS mRNA and lncRNA-PNUTS expression in each fraction.
(b) RT-PCR analysis of PNUTS mRNA and lncRNA-PNUTS expression after the use of oligo-(dT) or random hexanucleotides as primers for initial reverse transcription reaction.
(c) RT-PCR analysis of lncRNA-PNUTS expression in A549 cells. The total, cytoplasmic (Cyto.) and nuclear fractions are shown. PNUTS pre-RNA and PNUTS mRNA were used as endogenous controls to monitor the fractions purity.
(d) Confocal microscopy imaging of subcellular localization of lncRNA-PNUTS using co-transfection of a MS2-tagged-RNA construct of lncRNA-PNUTS and a fused MS2-GFP protein construct. Scale bar: 5µM.
(e) The exact copy numbers of lncRNA-PNUTS (basal levels or levels following activation by Actinomycin D treatment for 3h) and miR-205 were quantified with limiting-dilution qRT-PCR. Data are shown as mean ± s.d., n= 3 independent experiments per condition. Source data are available in Supplementary table 2.
(f) In silico prediction of MiR-205 binding sites located on lncRNA-PNUTS, obtained using the DIANA-microT web server.
(g) Selective pull-down of either endogenous lncRNA-PNUTS or PNUTS-mRNA isoforms by using antisense biotinylated probes followed by miRNA-specific RT-PCR analysis to detect endogenously associated miR-205 with lncRNA-PNUTS in A549 cells.
(h) MS2-RIP followed by miRNA-specific RT-PCR analysis to detect the association of miR-205 with lncRNA-PNUTS in NMuMG cells. LncRNA-PNUTS and GAPDH expression were used as internal controls.
(i) A549 and NMUMG cell lysates incubated with in vitro transcribed biotin-labeled lncRNA-PNUTS were subjected to pull-down followed by miRNA extraction and analysis by RT-PCR.
(j) A549 cells overexpressing lncRNA-PNUTS were transfected with an increasing concentration of a synthetic miR-205 mimic and the lncRNA expression was assessed by RT-PCR. ZEB-1 and CDH1 were used to monitor the efficiency of miR-205 overexpression on mesenchymal-epithelial transition (MET) process.
(k) Time course experiment by using RT-PCR analysis of lncRNA-PNUTS levels upon addition of 10µg.mL−1 cycloheximide in A549 cells.
GAPDH was used as a loading control.
lncRNA-PNUTS interacts with miR-205
Given the subcellular localization of lncRNA-PNUTS, we next explored its biological function as a presumed competing-endogenous RNA (ceRNA). By in silico analysis, we predicted 21 miRNAs targeting at least 5 sites with a score higher than 0.6 (Supplementary table 1). We focused on miRNA binding sites most represented in the CDS of the cognate mRNA rather than in its 3'-UTR region to explore the intrinsic properties of the full-length lncRNA-PNUTS as a ceRNA. Among the 10 miRNAs meeting this criterion, miR-205 was an obvious candidate due to its critical role in EMT and its high conservation among species25. We used quantitative real-time PCR to quantify the copy numbers of both miR-205 and lncRNA-PNUTS per cell since comparable levels are suggestive of ceRNA function (Figure 3e). FISH analysis (Supplementary figure 2c) demonstrates co-localization of miR-205 with lncRNA-PNUTS and not PNUTS-mRNA. This is suggesting of a preferential interaction of miR-205 with the lncRNA isoform which was further confirmed by the use of biotinylated antisense probes (Figure 3g).
LncRNA-PNUTS harbors seven miR-205 sites, including one located in the 3'-UTR of the cognate PNUTS mRNA (Figure 3f). To ensure that the part including the first six miR-205 binding sites is functionally active, we cloned this portion, either wild-type or mutated for the miRNA-205 binding sites, into the MS2-TRAP vector and validated the specific binding by MS2-tagged RNA affinity purification strategy and by avidin-affinity pull-down of cellular lysates (Figure 3h, 3i). To investigate the decay mechanism(s) of lncRNA-PNUTS, we treated A549 cells with increasing concentrations of miR-205 which results in an expected decrease in ZEB1 and increase in E-cadherin expression levels, respectively; however, miR-205 levels have no significant impact on the level of lncRNA-PNUTS (Figure 3j). Moreover, cycloheximide treatment of cells for up to 4h considerably increases the expression of lncRNA-PNUTS, suggesting its sensitivity to nonsense-mediated mRNA decay (NMD) (Figure 3k).
lncRNA-PNUTS regulates EMT migration and invasion in vitro through its miR-205 interaction
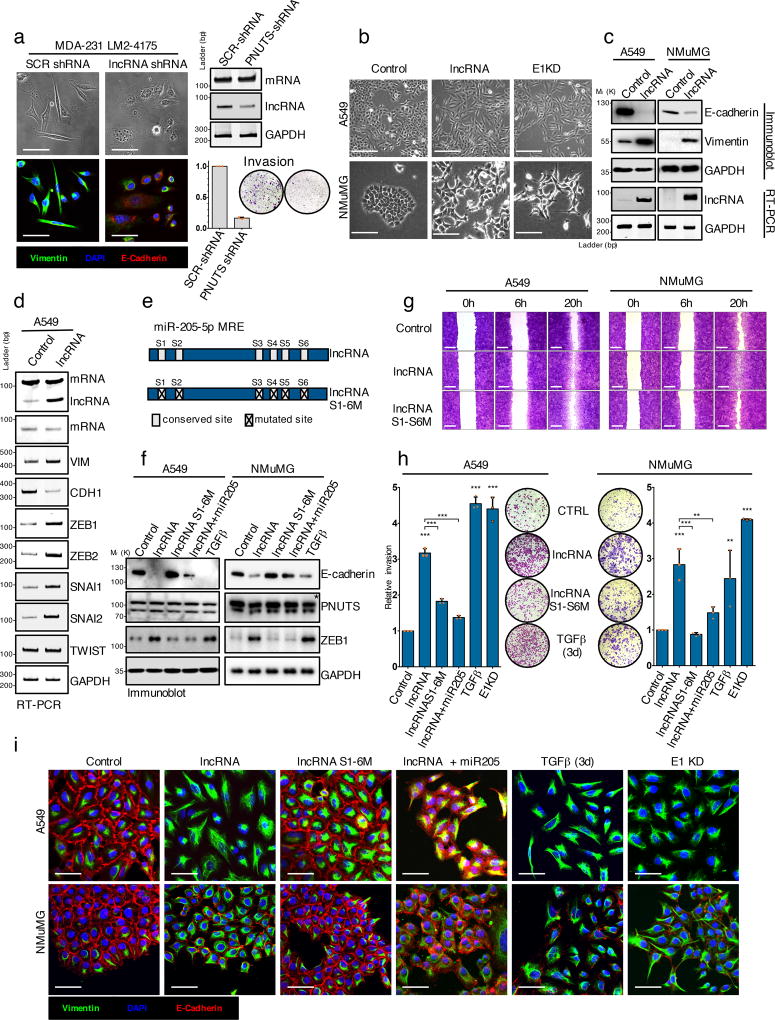
Since lncRNA-PNUTS interacts with miRNA-205, a well-established regulator of ZEB proteins and of epithelial cell maintenance, we investigated its effects on ZEB expression and cell plasticity. We silenced endogenous lncRNA-PNUTS in mesenchymal and invasive MDA231-LM2-4175 cells that express high levels of the lncRNA (Figure 1e) to test whether lncRNA-PNUTS could modulate cell plasticity. LncRNA-PNUTS silencing led to a significant decrease in cell invasion correlating with reduced vimentin expression (mesenchymal marker) and re-expression of epithelial marker E-cadherin concomitant to morphological changes (Figure 4a). To assess whether lncRNA-PNUTS generation is a prerequisite for TGFβ-mediated EMT, we treated A549 cells with ASO to the alternative splice site (Figure 2g) and showed that it significantly impaired TGFβ-mediated EMT (Supplementary figure 3a, 3d, 3e). This was confirmed using siRNA specifically targeting lncRNA-PNUTS to prevent either TGFβ-mediated or hnRNP E1 knockdown-mediated EMT (Supplementary figure 3b, 3c). Moreover, the overexpression of lncRNA-PNUTS induced an EMT in both A549 and NMuMG as characterized by a morphological change from an epithelial-like, cobblestone phenotype to a more spindle-shaped mesenchymal phenotype, an E-cadherin/vimentin switch (Figure 4 b,c), and an accompanying increase in the levels of the EMT-TFs, ZEB1, ZEB2, SNAI1 and SNAI2 (Figure 4d). While the wild-type lncRNA-PNUTS induced an EMT associated to a downregulation of E-cadherin and upregulation of ZEB1, co-transfection with miRNA-205 as well as the overexpression of the miR-205-mutant form of the lncRNA-PNUTS abolished this effect (Figure 4 e,f). The lncRNA-PNUTS controls both migration and invasion (Figure 4 g,h) of A549 and NMuMG cells in a miR-205 binding site-dependent manner and miR-205 overexpression is able to abolish this effect (Figure 4h). These observations were further validated by immunofluorescence (Figure 4i).
Figure 4. LncRNA-PNUTS regulates EMT and cell migration/invasion in vitro.
(a) MDA-231-LM2-4175 cells stably silenced for lncRNA-PNUTS were analyzed by immunofluorescence (left) using antibodies against vimentin (green), E-cadherin (red) and merged with DAPI (blue). Scale bar: 50µM. lncRNA-PNUTS silencing was monitored by RT-PCR (right, top). Invasive capacities of control (SCR-shRNA) or lncRNA-PNUTS silenced (PNUTS shRNA) cells were monitored in modified Boyden chamber assay (right, bottom). (Mean ± s.d., n= 3 independent experiments per condition). Source data are available in Supplementary table 2.
(b) A549 and NMuMG cells stably overexpressing lncRNA-PNUTS were analyzed using bright-field microscopy. hnRNP E1 knockdown (E1KD) cells were used as controls. Scale bar: 100µM.
(c) Western-blot (top) and RT-PCR (bottom) analysis of E-cadherin, vimentin and lncRNA-PNUTS in A549 and NMuMG cells overexpressing lncRNA-PNUTS.
(d) RT-PCR analysis of several EMT-related transcription factors in A549 cells stably overexpressing lncRNA-PNUTS.
(e) Schematic outlining the constructs used in this study for wild-type (lncRNA) or mutated (lncRNAS1-6M) lncRNA.
(f) Western-blot analysis of E-cadherin, PNUTS and ZEB1 protein expression in A549 and NMuMG cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS and treated or not with synthetic miR-205 mimic or TGFβ for 3 days. TGFβ was used as a positive control. * PNUTS protein band.
(g) Wound-healing migration assay of control (CTRL), lncRNA-PNUTS (lncRNA) or mutated lncRNA-PNUTS (lncRNAS1-6M) A549 and NMuMG cell models. Scale bar: 400µM
(h) Modified Boyden chamber invasion assay of wild-type (lncRNA) or mutated (lncRNAS1-6M) lncRNA-PNUTS overexpressing A549 and NMuMG cells pre-treated +/− synthetic miR-205 mimic or TGFβ for 3 days. hnRNP E1 knockdown (E1KD) and TGFβ treated cells were used as a positive control. (Mean ± s.d., n= 3 independent experiments per condition, ANOVA followed by post-hoc Tukey's multiple comparisons test, *p<0.05; **p<0.01; ***p<0.001, NS, not significant). Source data are available in Supplementary table 2.
(i) Confocal microscopy imaging of co-immunostaining of vimentin (green), E-cadherin (red) and merged with DAPI (blue) in A549 and NMuMG cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS and treated +/− synthetic miR-205 mimic or TGFβ for 3 days. Scale bar: 50µM.
For all western-blots and RT-PCRs GAPDH was used as a loading control. Unprocessed original scans of blots are shown in Supplementary Fig7
lncRNA-PNUTS controls the miR-205/ZEB/E-cadherin axis
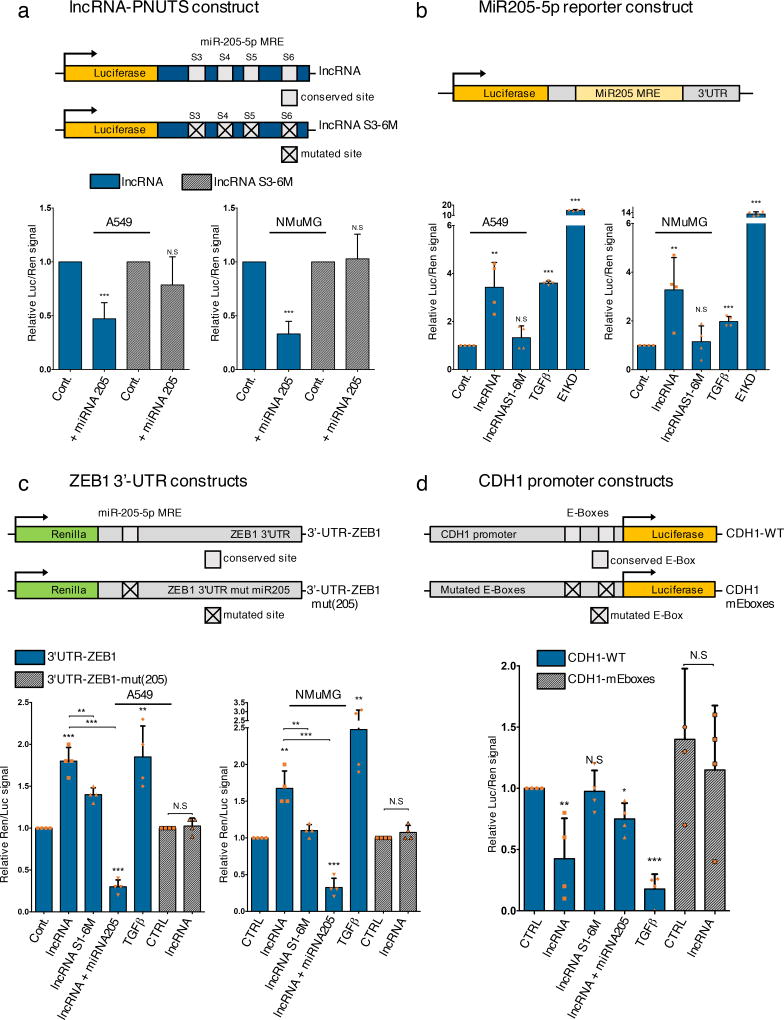
Using luciferase reporter assays we next validated the regulation of the miR-205/ZEB/E-cadherin axis by lncRNA-PNUTS. We first confirmed the binding of miR-205 to lncRNA-PNUTS by cloning the S3-S6-miR-205 binding sites of lncRNA-PNUTS as a 3'-UTR of the luciferase CDS (Figure 5a) and demonstrated that co-transfection with miR-205 in A549 and NMUMG cells reduces luciferase expression with the wild-type (WT), but not the mutated S3-S6-miR205 construct (Figure 5a). Second, using a luciferase reporter construct whose stability is dependent on miR-205 binding (miR-205-MRE), we demonstrated that WT, but not the miR-205-mutated-lncRNA-PNUTS decreases miR-205 bioavailability (Figure 5b). Third, using constructs of renilla fused to the ZEBs 3’-UTR (Figure 5c; Supplementary figure 4), we observed that WT lncRNA-PNUTS stabilized ZEBs 3'-UTR and this effect was partially abolished using the miR-205-mutated lncRNA or reversed by co-transfection with miR-205 mimics (Figure 5c; Supplementary figure 4). Importantly, no effects were observed on the ZEB1 3’-UTR mutated for its miR-205 binding site (Figure 5c). Last, using an E-cadherin (CDH1) promoter luciferase construct that contains either wild-type or mutant ZEBs binding sites (E boxes), we observed an ~ 50% decrease of luciferase activity upon lncRNA-PNUTS overexpression. This downregulation was partially rescued by co-transfection with miR-205 or abolished by using either the mutated lncRNA-PNUTS or the CDH1 promoter mutated for its E-boxes (Figure 5d).
Figure 5. LncRNA-PNUTS controls the miR-205/ZEB/E-Cadherin axis.
(a) Dual luciferase reporter assays to test the interaction between miR-205 and lncRNA-PNUTS (S3 to S6 region) by using a synthetic miR-205 mimic (+ miRNA 205) co-transfected with wild-type (LUC-lncRNA) or mutated (LUC-lncRNA-S3-S6M) constructs of lncRNA-PNUTS cloned into the 3'-UTR of the luciferase reporter gene. For each condition, assays were normalized to Renilla reporter gene expression. (mean ± s.d., n= 7 independent experiments per condition, two-tailed Student t test, ***p<0.001, NS, not significant).
(b) Dual-Luciferase reporter assay of miR-205 bioavailability in A549 and NMuMG cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS. TGFβ treatment and hnRNP E1 knockdown (E1KD) were used as internal controls. For each condition, assays were normalized to Renilla reporter gene expression. (mean ± s.d., n= 4 independent experiments per condition, two-tailed Student t test, **p<0.01; ***p<0.001, NS, not significant).
(c) The wild-type (REN-3'-UTR-ZEB1) 3'-UTR of ZEB1 cloned into the 3'-UTR of the Renilla gene was transfected in A549 and NMuMG cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS and treated +/− synthetic miR-205 mimic or TGFβ for 3 days. Mutated construct (REN-3'-UTR-ZEB1-mut(205)) for the miR-205 binding site located in the 3'-UTR of ZEB1 was also used. TGFβ was used as a positive control. For each condition, assays were normalized to Luciferase reporter gene expression. (mean ± s.d., n= 4 independent experiments per condition, two-tailed Student t test, **p<0.01; ***p<0.001, NS, not significant).
(d) The wild-type (prom-CDH1-WT) proximal promoter of E-Cadherin driving the luciferase reporter gene expression was transfected in A549 cells overexpressing wild-type (lncRNA) or mutated (lncRNAS1-6M) constructs of lncRNA-PNUTS and treated or not with synthetic miR-205 mimics or TGFβ for 3 days. Mutated construct for both E2 Boxes 1 and 3 (Prom-CDH1-mEboxes) located on the promoter was also used. TGFβ was used as a positive control. For each condition, assays were normalized to Renilla reporter gene expression. (mean ± s.d., n= 4 independent experiments per condition, two-tailed Student t test, *p<0.05; **p<0.01; ***p<0.001, NS, not significant).
LncRNA-PNUTS regulates tumor implantation, growth and metastasis
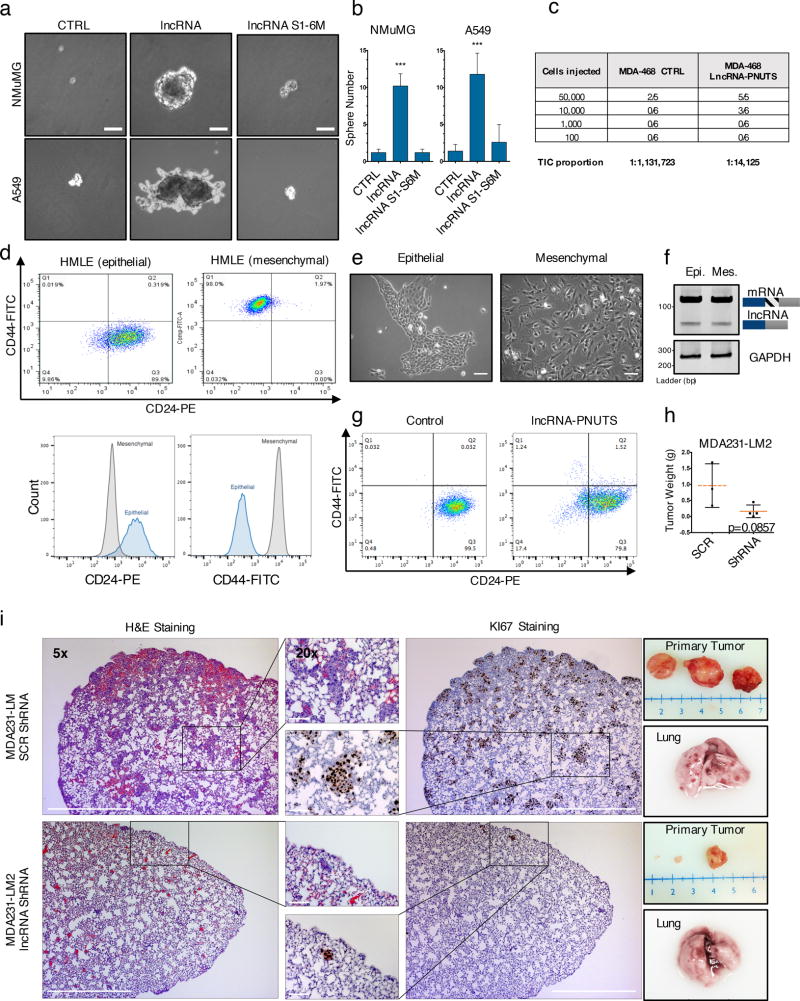
Given the role of miR-205 in regulating mammary stem cell fate and tumorigenesis through EMT26, we investigated whether lncRNA-PNUTS contributes to these phenotypes. Utilizing the tridimensional, sphere formation assay, we showed a significant increase in sphere-formation of A549 and NMuMG cells induced by lncRNA-PNUTS dependent on its miR-205 binding sites (Figure 6a & 6b). Using limiting dilutions of MDA-468 cells in an in vivo fat pad injection assay, we observed lncRNA overexpression resulted in an ~80-fold increase of tumor initiating cell number compared to control (Figure 6c). Next, we FACS sorted HMLE cells (Figure 6 d, e) using CD24/CD44 to determine whether lncRNA-PNUTS is upregulated in stem cells. Although the CD24−/CD44+ subpopulation (mesenchymal phenotype) is known to be highly enriched for tumor-initiating cells27,28, we did not observe an upregulation of the lncRNA-PNUTS in this subpopulation compared to the CD24+/CD44− subpopulation (epithelial phenotype; Figure 6f). Nevertheless, overexpression of lncRNA-PNUTS in the epithelial subpopulation induced a significant decrease in the number of CD24+/CD44− cells and revealed a minor subpopulation of cells harboring the CD24−/CD44+ phenotype (Figure 6g ; Supplementary figure 5a). Additionally, silencing of lncRNA-PNUTS in MDA231-LM2 cells led to an increased expression of the CD24 epithelial marker (Supplementary figure 5b).
Figure 6. LncRNA-PNUTS promotes tumor initiation/growth and metastasis in vivo.
(a) Bright-field microscopy pictures of in vitro mammosphere/oncosphere formation assay in NMuMG and A549 cells overexpressing empty vector (CTRL), lncRNA-PNUTS (lncRNA) or mutated lncRNA-PNUTS (lncRNAS1-6M). Scale bar: 20µM
(b) Absolute quantification of the sphere numbers obtained in (a). (mean ± s.d., n=5 independent experiments, two-tailed Student t test, ***p<0.001 NS, not significant). Source data are available in Supplementary table 2.
(c) Number of tumor formed upon limiting dilution injection of control and lncRNA-PNUTS overexpressing MDA-468 cells. MDA-MB-468 cells were injected into the mammary fat pads of 6–8 week-old female mice in limiting dilution. TIC, tumor-initiating cells number was determined using ELDA software46. Number of mice used for each condition is indicated.
(d) Flow cytometry analysis of CD24/CD44 cell surface expression levels in the epithelial (CD44−/CD24+ sorted cells) and mesenchymal (CD44+/CD24− sorted cells) HMLE subpopulations.
(e) Cell morphology observed by phase-contrast microscopy. Scale bar: 50µM.
(f) RT-PCR analysis of lncRNA-PNUTS expression level in mesenchymal and epithelial sorted HMLE cells.
(g) Flow cytometry analysis of the CD24/CD44 cell surface expression levels in the epithelial (CD44−/CD24+ sorted cells) HMLE subpopulation expressing empty vector (control) or overexpressing lncRNA-PNUTS.
(h)Tumor weight of primary tumors obtained following mammary fat pad injection of MDA-231-LM2 expressing scrambled control (SCR) or lncRNA-PNUTS targeting shRNA (shRNA) in NOD/SCID mice. (mean ± s.d., n= 4 mice per condition, two-tailed Mann-Whitney test, p=0.08570). Source data are available in Supplementary table 2.
(i) (Left) Histopathological analysis of paraffin-embedded lung serial sections of mice injected in the mammary fat pad with MDA-231-LM2 expressing scrambled or lncRNA-PNUTS targeting shRNA. Haematoxylin and Eosin (H&E) staining and immunostaining of Ki67 protein was performed in serial lung sections to identify macro- and micro-metastases. (Right) Photographs of primary tumors and of a representative lung collected for each condition. Scale bar: 500 µM
We next tested the effects of lncRNA-PNUTS on tumor progression in vivo and demonstrated that silencing of lncRNA-PNUTS in MDA-231-LM2 cells impairs tumor formation when injected orthotopically into mammary fat pads (Figure 6h). Moreover, lncRNA-PNUTS also contributes to in vivo metastasis as we observed a significant decrease of lung colonization in MDA-231-LM2 cells silenced for lncRNA-PNUTS compared to their scrambled-control counterparts (Figure 6i).
Discussion
The discovery of lncRNAs as biologically relevant molecules has led to a rethinking of the central dogma of biology and unraveled new layers of cellular and molecular complexities. It is now well established that many genes can encode both mRNA and ncRNA. For instance, a large number of miRNAs and most of the snoRNAs are processed from spliced introns. Furthermore, circular RNAs can be processed from introns or back-spliced exons29. Moreover, the existence of large numbers of bifunctional RNAs whose isoforms are regulated by alternative splicing, as described herein, were previously theorized and predicted based on genome-wide data-mining of alternative splicing events30. Here, we describe how under different cellular contexts, a gene, through alternative splicing, can encode for either an mRNA or a lncRNA and demonstrate the biological relevance of the generated lncRNA in targeting and sequestering miR-205 to ultimately regulate EMT (Model Figure 6e).
Since alternative splicing and generation of lncRNA-PNUTS is an early event in TGFβ-mediated EMT, we postulate that lncRNA-PNUTS operates as a transient inhibitor of miR-205 to allow for the temporal upregulation of ZEBs and subsequent regulation of downstream EMT events. Indeed ZEBs proteins are reciprocally linked in a feedback loop with the miR-200 family, each strictly controlling the expression of the other31. In this way, a transient, but nevertheless, strong decrease in miR-205 bioavailability, sufficient to activate the ZEB proteins, would allow for transcriptional repression of the miR-200 family or other miRNAs such as miR-183 or miR-203 thereby further stabilizing ZEB proteins and reinforcing the EMT process. Furthermore, the transient nature of the lncRNA-PNUTS allows for its early regulation of the miR-205/ZEB axis during EMT but is not sufficient to sustain a decrease in PNUTS-mRNA and protein expression, thus allowing independent functions of the isoforms. Moreover, the fact that the lncRNA-PNUTS is upregulated in tumor samples compared to their non-tumor counterpart despite its transient functional role might be the result of tumor heterogeneity with regard to TGFβ signaling32,33 and that at any given time lncRNA-PNUTS is elevated in certain tumor cells. Finally, the alternative splicing of PNUTS is consistently accompanied by a decrease in the expression of the PNUTS pre-RNA. Since RNA splicing occurs co-transcriptionally, this decrease could be explained by the influence of transcriptional regulators pausing the RNAPII elongation complex to allow splicing to proceed on the alternative site34.
LncRNA-PNUTS is localized in both the cytoplasm and nucleus (Figure 3 c,d ; Supplementary figure 2c) and while its function as a ceRNA could be attributed to its cytoplasmic localization, its role in the nuclear compartment was not investigated herein. Nuclear biogenesis of lncRNA-PNUTS might explain its localization in the nucleus although we speculate that it could also be involved in nuclear processes such as transcription or epigenetic regulation as other previously described lncRNAs35–37. Moreover, we demonstrate that the transcriptional inhibitor Actinomycin D is a potent activator of the PNUTS alternative splicing through its effects on hnRNP E1 translocation from the nucleus to cytoplasm. Furthermore, lncRNA-PNUTS is upregulated by cycloheximide, a compound widely used to inhibit translation but also to test the sensitivity of RNAs to NMD by inhibiting the first round of translation38 and which is also known to induce transport of hnRNPs into the cytoplasm39. Based on these results, we expect that any anti-tumor agents whose pharmacological properties block transcription, might activate the alternative splicing of PNUTS. Given the role of EMT in drug resistance and the contribution of miR-205 in chemotherapy sensitivity11,40–42, it will be of interest to evaluate the contribution of lncRNA-PNUTS in EMT-mediated drug resistance mechanisms.
Although it is of note that both PNUTS isoforms share miR-205 sites raising the obvious question as to why the PNUTS mRNA does not itself serve to sequester miR-205 and regulate EMT. We postulate that the location of miRNA binding sites in the CDS of the cognate mRNA, relative to those in the lncRNA, may affect miRNA binding. As previously established, ribosomal hindrance could interfere with the ability of the miRNA to attach to its target site if he is located in the CDS43 and this is supported by our data demonstrating preferential co-localization and binding of miR-205 with lncRNA-PNUTS. Additionally, the two PNUTS isoforms could have distinct secondary structures who might also explain the preferential binding of miR-205 to the lncRNA-isoform. The fact that miR-205 binds weakly to the PNUTS-mRNA, presumably to its site in the 3'-UTR, could also contribute to PNUTS protein stability during alternative splicing since the sponge activity of the lncRNA-PNUTS could counteract the inhibitory effect of the miR-205 initially occurring on the 3'-UTR of the PNUTS-mRNA.
Collectively, our work confirms the key roles of lncRNAs and RNA-binding proteins in biological processes and human diseases. The study describes the generation and function of a lncRNA, and of an RNA-binding protein with which it associates, as key regulators of EMT and of the mesenchymal properties of tumor cells. Our in vivo data also demonstrate that modulation of lncRNA-PNUTS regulates the metastatic potential of tumor cells. Since most cancer-related mortalities result from metastatic disease and no mutations that are selective for metastases have been identified3,44,45, it is imperative to identify potential metastatic mediators for prognostic and therapeutic benefit. As such, the identification of both hnRNP E1 and lncRNA-PNUTS provide two additional targets that could potentially serve as predictive markers of metastasis and/or chemoresistance, as well as effective targets for anti-metastatic therapies.
Methods
Cell culture, antibodies, primers and reagents
NMuMG, A549, MCF7, CaCo-2, HMLE, MCF10a and MDA-MB-468 cells were obtained from the American Type Culture Collection (ATCC), and the MDA231 progression model was graciously provided by Dr. Joan Massagué. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Cat. No. SH30081.01, GE Healthcare Life Sciences) high glucose supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic solution (penicillin G, streptomycin, amphotericin B). MCF10a cells were obtained from ATCC and cultured in DMEM:F12 supplemented with 5% horse serum, 0.5 µg/ml hydrocortisone, 10 µg/ml insulin, 20 ng/ml epidermal growth factor, 100ng/mL Cholera toxin and 1% antibiotic/antimycotic. All cells were cultured in a 37 °C, 5% CO2 incubator. TGFβ2 was a generous gift from Genzyme Corporation. Antibody dilutions, company names, catalogue numbers and clone numbers and their respective dilutions are listed below. Puromycin and G418 were purchased from InvivoGen. Cycloheximide and Actinomycin D were purchased from Sigma. Site-directed mutagenesis was performed using a site-directed mutagenesis kit from Life Technologies as described by the manufacturer. Mouse monoclonal anti-E-cadherin (Clone [4A2], Cat. No. #14472, 1:2,000 dilution), Rabbit monoclonal anti-E-cadherin (Clone [24E10], Cat. No #3195, 1:2,000 dilution), Rabbit monoclonal anti-vimentin (Clone [D21H3], Cat. No. #5741, 1:2,000 dilution), Rabbit monoclonal anti-ZEB1 (Clone [D80D3], Cat. No. #3396, 1:1,000 dilution), Rabbit monoclonal anti-Ki67 (Clone [D2H10], Cat. No. #9027, 1:1,600 dilution) and Rabbit polyclonal anti-PARP (Cat. No. #9542, 1:2,000 dilution) were purchased from Cell signaling technology company. Mouse monoclonal anti-GAPDH (Clone [6C5], Cat. No. sc32233, 1:5,000 dilution) and Mouse monoclonal anti-HSP90 (Clone [F-8], Cat. No. sc-13119, 1:2,000 dilution) were purchased from Santa Cruz Biotechnology company. Rabbit monoclonal anti-vimentin (Clone [EPR3776], Cat. No. Ab92547, 1:2,000 dilution) was purchased from Abcam company. Rabbit polyclonal anti-PNUTS (Cat. No. 24450-1-AP, 1:1,000 dilution) was purchased from Proteintech company. Mouse monoclonal anti-hnRNPE1 (Clone [1G2] Cat. No. H00005093-M01, 1:1,000 dilution) was purchased from Abnova company. Mouse monoclonal anti-GFP (clones [7.1 & 13.1], Cat. No. 11 814 460 001, 1:500 dilution) was purchased from Roche company. The primers used in PCR analysis were purchased from IDT company and are listed as follows: hCDH1-F TGCCGCCATCGCTTACACCA ; hCDH1-R CCACGCTGGGGTATTGGGGG ; hGAPDH-F TGATGACATCAAGAAGGTGGTGAAG ; hGAPDH-RTCCTTGGAGGCCATGTGGGCCAT ; hPNUTS-v1-F CCAAGCCCCTTTGAAGGGAAA ; hPNUTS-v1-R CTGGGGAAGAAGGTTTGGCTGT ; hPNUTS-v1-v2-flanking-F AAGTACTGTCACCTACGGCTGCC ; hPNUTS-v1-v2-flanking-R GGACGGTCTGCGTCCATTGC ; hPNUTS-v2-boundary-F GTACTGTCACCTACGGCTGCCAAGAAC ; hPNUTS-v2-boundary-R TGCCTTCCTCAGGCCATGTCA ; hPNUTS-v2-boundary2-F TGCCTTCCTCAGGCCATGTCA ; hPNUTS-v2-boundary2-R TGCTGGTTCTTGGCAGCCGT ; hSNAI1-F CCTCAAGATGCACATCCGAAG ; hSNAI1-R ACATGGCCTTGTAGCAGCCA ; hSNAI2-F CCCACACATTACCTTGTGTTTGCAA ; hSNAI2-R CAAATGCTCTGTTGCAGTGAGG ; hTWIST-F GGACAAGCTGAGCAAGATTCAGA ; hTWIST-R GTGAGCCACATAGCTGCAG ; hVIM-F CAACGACAAAGCCCGCGTCG ; hVIM-R GCGCAGGGCGTCATTGTTCC ; hZEB1-F GGCAGAGAATGAGGGAGAAG ; hZEB1-R CTTCAGACACTTGCTCACTACTC ; hZEB2-F TCTCGCCCGAGTGAAGCCTT ; hZEB2-R GGGAGAATTGCTTGATGGAGC ; mPNUTS-v1-v2-flanking-F AGGTACTATCGCCGACTGCT ; mPNUTS-v1-v2-flanking-R GGGCGGTCCGTGTCCATGGG.
Transfections
All cell transfections were carried out using 5 µg DNA (or specified amount) per 8 ml of medium with cells at 70% confluence cultured in 100 mm plates. The transfection reagent Lipofectamine (ThermoFisher Scientific) was used according to the protocol provided by the manufacturer.
Transfection of small interfering RNA
Two specific sequences were designed across the new exon-exon junction generating the lncRNA-PNUTS. The sequences were submitted to a BLAST search against the human genome to ensure the specificity of the siRNA to the targeted sequence. Two corresponding scramble duplexes, which do not recognize any sequence in the human genome, were used as controls. The sense and antisense strands were then annealed to obtain duplexes with identical 3′ overhangs. For transfection of the siRNA duplexes, 75,000 cells were seeded in a six-well plates containing 2 mL of culture medium. Twenty-four hours after the seeding, the cells were transfected by phosphate calcium precipitation by adding in each well 200µL of a mixture containing the siRNA duplexes, 140 mM NaCl, 0.75 nM Na2HPO4, 6 nM glucose, 5 mM KCl, 25 mM HEPES, and 125 mM CaCl2. Twenty-four hours after transfection, the cells were extensively washed with PBS and incubated for 48 hours in culture medium before they were harvested for reverse transcription-PCR (RT-PCR) analyses and Western blotting analyses.
ASO oligo design and usage
Antisense oligonucleotide (IDT) was designed against the splicing site used to generate the lncRNA-PNUTS in order to prevent its usage. The oligo consists of modified 2′ O methyl phosphothioates oligonucleotide where each ribose and each phosphate group was modified by a 2′ O methyl modification or a single sulfur, respectively (sequence: mG*mU*mG*mG* mU*mG*mC* mU*mG*mG* mU*mU*mC* mU*mG). Cells were transfected with the indicated amount of the oligo 2 hours prior to treatment of the cells. For reverse transcription step, the RNAs were pre-heated in the reaction mix (65°C/5min, 75°C/2min, 35°C/30sec) prior to addition of the reverse transcriptase and RT reaction.
Polysome profiling
Cells were extracted in TMK100 buffer (10mM Tris-HCl, pH 7.4, 5mM MgCl2, 1% (v/v) Triton X-100, 0.5% deoxycholate, 2mM DTT, 100ug.mL−1 cycloheximide) and then supernatant were collected by centrifugation (14000 rpm, 10min). Cell extracts were layered onto sucrose gradients (10–50%) and centrifuged at 35,000 r.p.m. in a SW40Ti rotor for 3 h at 4 °C. Fractions were collected using a density gradient fractionation system (Teledyne Isco) and then RNA was isolated using Trizol. Monosomal and polysomal fractions were determined by analysis of 18 S and 28 S rRNA levels using denaturing agarose gel electrophoresis.
Modified Boyden chamber invasion assay
Invasion across a basement membrane was performed using BD BioCoat™ Matrigel™ Invasion Chambers (BD Biosciences) as per manufacturer’s instructions. Briefly, a total of 105 cells were placed in the upper compartment of the invasion chamber (BD BioCoat Matrigel Invasion Chamber, BD Biosciences) for 24 h at 37 °C. Non-invading cells were removed with a swab and the filters were then fixed in methanol and stained with crystal violet. Quantification of the invasion assay was performed by spectrophotometry after resuspension of the stain.
Western Blot Analysis
Western blot analysis was performed by standard SDS-PAGE. Whole cell lysates were prepared from 2–5 × 106 cells in 300 µl of lysis buffer (20 mM Tris, pH 7.4, 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 1 mM Na3VO4, and protease inhibitors). Lysates were sonicated and clarified by centrifugation at 4 °C for 10 min in a Beckman tabletop microcentrifuge at maximum speed. Typically, 5–20 µg of whole cell lysates were separated on 10 or 12% acrylamide minigels and transferred to Immobilon-P membrane (Millipore). The membrane was blocked for 1 h in wash buffer (PBS containing 0.1% Tween 20) containing 5% nonfat dry milk followed by an overnight incubation with primary antibody diluted in the same blocking buffer. After extensive washing, the blot was incubated with secondary antibody for 1 h in blocking buffer, washed, and processed using the ECL+ Western blotting detection system (Amersham Biosciences).
Immunofluorescence, FISH and imaging
For immunofluorescence, cells were fixed for 15 min in PBS containing 3.7% (w/v) paraformaldehyde, followed by permeabilization with 0.2% (w/v) Triton X-100. Cells were then incubated 1h in 3% BSA and incubated overnight with primary antibody at 4°C. Then cells were incubated with secondary antibodies conjugated with Alexa Fluor (Life Technologies) at room temperature for 1 h followed by three washes with PBS before analysis with the FV10i confocal laser scanning microscope (Olympus).
For FISH analysis, cells were fixed for 15 min in PBS containing 3.7% (w/v) paraformaldehyde, then slides were incubated overnight at 37°C in hybridization solution (10% formamide, 2X SSC, 10% dextran sulfate (w/v), 10 µM each probe, labelled with ATTO-488,590 and 649 respectively, IDT). Cells were then washed twice for 30 min at 37°C with 10% formamide in 2X SSC. DAPI was applied during the second wash. Cells were then rinsed twice with 2X SSC before imaging in 2X SSC buffer.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections were deparaffinized in xylene, rehydrated in alcohol, and processed as follows: Sections were incubated with target retrieval solution (Dako) in a steamer for 45 min followed by 3% hydrogen peroxide solution for 10 min and protein block (Dako) for 20 min at room temperature. Sections were incubated overnight in a humid chamber at 4°C with antibody against Ki-67 purchased from Cell signaling technology company (Clone [D2H10], Cat. #9027, 1:1600 dilution) followed by biotinylated secondary antibody (Vector laboratories) for 30 min and ABC reagent for 30 min. Immunocomplexes of horseradish peroxidase were visualized by DAB reaction (Dako), and sections were counterstained with hematoxylin before mounting. Micrographs of stained sections were taken using a Leica DMIL LED microscope with Amscope camera and acquisition software.
Immunoprecipitation assays and biotin-pulldown
MS2-TRAP immunoprecipitation assays were performed as described previously46. Immunoprecipitated RNA was isolated from beads by addition of Trizol, followed by RT-PCR as described above. LncRNA-PNUTS biotinylation was performed using the biotin RNA Labeling mix (Sigma) and the T7 RNA polymerase (Promega) after PCR amplification of lncRNA-PNUTS vectors. Biotin-pull-down were performed by using antisense biotinylated probes (IDT) specific to PNUTS isoforms. RNA was isolated from beads by addition of Trizol, followed by mRNA or miRNA-specific RT-PCR analysis (Quantimir, System Biosciences).
RNA electromobility shift assays
Recombinant hnRNP E1 protein was prepared as previously described26 and allowed to incubate for 30 min at 4°C in RNA–protein binding buffer (40 mM Tris–HCl pH = 7.5, 30 mM KCl, 1 mM MgCl2, 0.01% NP40, 1 mM dithiothreitol). After binding, a loading buffer composed of 50% glycerol and bromophenol blue/xylene cyanol was added to samples. Samples were loaded into non-denaturing polyacrylamide gel, electrophoresed and autoradiographed.
Dual-luciferase reporter assays
Transient transfections were performed using XtremeGENE 9 DNA transfection reagent on 50,000 cells plated in a 24-well plate. At 24 hours after transfection, the cells were lysed in 100µL of passive lysis buffer and the firefly luciferase activity as well as the renilla activity were determined with a luminometer using the Dual Luciferase Assay System (Promega) on 20 µL of lysate. For each experiment, either the firefly luciferase or renilla activity was normalized to either the activity of the renilla or firefly luciferase used as internal control. The results were expressed as fold induction determined by normalizing each firefly luciferase or Renilla value to the internal control value and by dividing these normalized values with the mean normalized value of the corresponding reporter construct transfected with the empty expression vector.
Flow Cytometry
Single-cell suspensions of cells were washed three times in PBS containing 1% BSA followed by incubation in 100 µl PBS/1% BSA containing anti-CD24 (PE) and anti-CD44 (FITC) antibody (BD Biosciences) for 2 hours at room temperature. Cells were then washed three times in PBS containing 1% BSA and resuspended in 500 µl PBS. Samples were analyzed using the BD LSRFortessa Analytic Flow Cytometer. FACS sorting of CD44+/CD24− and CD44−/CD24+ HMLE populations was performed using the FACSAria II Cell Sorter and FACSDiva™ 6 software (BD Biosciences).
Microarray processing and analysis
Conversion of total RNA into labelled material, mouse genome 430 2.0 GeneChip hybridization, and post hybridization washing, staining, and scanning were performed in accordance with Affymetrix protocols by the MUSC Proteogenomics Core Facility. Hybridization data were processed with Affymetrix Expression Console software to obtain normalized hybridization data (RMA algorithm) and detection scores (MAS5 algorithm). This data was imported into dChip software for hierarchical clustering and comparative analysis where a combination of fold change and Student’s t-test (unpaired) was utilized to identify genes changing significantly for pairwise relationships. Pathway analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and Molecular Signature Database (MSigDB) platforms. Raw data files were deposited in the NCBI Gene Expression Omnibus (GEO) repository as series GSE94637.
Statistics and reproducibility
Invasion assay were subjected to ANOVA statistical analysis followed by post-hoc Tukey's multiple comparisons test. For Luciferase reporter assays, statistical analysis was performed by two-tailed Student t test. Human tumor samples analysis were subjected to Pearson correlation score analysis (df=24-2, a Pearson score > 0.515 and p<0.05 was considered as significant). No statistical method was used to predetermine sample size and experiments were not randomized, and we were not blinded to allocation during experiments and outcome assessment. The representative images shown in Figures 1, 2, 3, 4, 6, S1, S2, S3, S5 are representative of at least 2 independent experiments performed with similar results, excepted for in vivo experiments and Supplementary figure 3d with only one repeat. All other experiments were repeated 2 or more times, as indicated in the legends. All the results are expressed as mean ± SD. * p < 0.05; ** p < 0.01; *** p < 0.001.
Human samples
The use of human breast tumor tissues and database were approved by the Institutional Review Board for Human Research of the Medical University of South Carolina. Written informed consent from the donors for research use of tissue in this study was obtained prior to acquisition of the specimen. Samples were confirmed to be tumor or normal based on pathological assessment.
Ethics statement
Animals were kept on a 12:12 h light–dark cycle and provided with food and water ad libitum. All experiments were performed according to approved protocols of the Institutional Animal Care and Use Committee (IACUC), Medical University of South Carolina.
Data availability
Microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE94637. Source data for Figure 1e, 3e, 4a, 4h, 6b, S1b, S3d, S3e, and S4 have been provided as Supplementary Table 2. Unprocessed original scans of blots are shown in Supplementary Figure 7. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
Authors are grateful to Dr. Monika Gooz for help with the microscopic analysis. We also thank Dr. Yuan Shao for histopathological analysis and Dr. Kiwana Gibbs for providing the human tumor samples. We thank Dr. Mohanty Bidyut for his assistance with the cloning and Ken Noguchi for his help with hnRNP E1 plasmid constructs and Dr. Jennifer Isaacs for her precious help concerning hypoxia experiments. This work was supported in part by the Cell & Molecular Imaging, Small Animal Imaging, and the Biorepository & Tissue Analysis Shared Resources, Hollings Cancer Center, Medical University of South Carolina (P30 CA138313), and the Shared Instrumentation Grant S10 OD018113. This work was also supported by grants CA555536 and CA154664 from the National Cancer Institute to P.H.H.
Footnotes
Author Contributions
The study was supervised by P.H.H. The conception and design were made by S.G and P.H.H. The methodology was developed by S.G. and P.H.H. Acquisition and interpretation of the data was made by S.G. and P.H.H. The Affymetrix experiment and care of the mice was provided by B.H. Mice injections and dissection were made by S.G., L.L. and C.O. The manuscript was written by S.G. and P.H.H. and all the authors contributed to its reviewing. V.P., V.K.G., J.A.D. provided critical discussion.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Jemal A, et al. Global cancer statistics. CA. Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 3.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Weigelt B, Peterse JL, van’t Veer LJ. Breast cancer metastasis: markers and models. Nat. Rev. Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 5.Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int. J. Cancer. 2016;139:269–280. doi: 10.1002/ijc.30039. [DOI] [PubMed] [Google Scholar]
- 6.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batista PJ, Chang HY. Long Noncoding RNAs: Cellular Address Codes in Development and Disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan J, et al. A Long Noncoding RNA Activated by TGF-β Promotes the Invasion-Metastasis Cascade in Hepatocellular Carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Leone S, Santoro R. Challenges in the analysis of long noncoding RNA functionality. FEBS Lett. 2016;590:2342–2353. doi: 10.1002/1873-3468.12308. [DOI] [PubMed] [Google Scholar]
- 10.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 11.Fischer KR, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith BN, Bhowmick NA. Role of EMT in Metastasis and Therapy Resistance. J. Clin. Med. 2016;5 doi: 10.3390/jcm5020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye X, Weinberg RA. Epithelial–Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhury A, et al. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grelet S, et al. The human NANOS3 gene contributes to lung tumour invasion by inducing epithelial-mesenchymal transition. J. Pathol. 2015;237:25–37. doi: 10.1002/path.4549. [DOI] [PubMed] [Google Scholar]
- 16.Hussey GS, et al. Identification of an mRNP Complex Regulating Tumorigenesis at the Translational Elongation Step. Mol. Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhury A, Chander P, Howe PH. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1’s multifunctional regulatory roles. RNA N.Y.N. 2010;16:1449–1462. doi: 10.1261/rna.2254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T, et al. PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Mol. Cancer. 2010;9:72. doi: 10.1186/1476-4598-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akker SA, et al. Pre-spliceosomal binding of U1 small nuclear ribonucleoprotein (RNP) and heterogenous nuclear RNP E1 is associated with suppression of a growth hormone receptor pseudoexon. Mol. Endocrinol. Baltim. Md. 2007;21:2529–2540. doi: 10.1210/me.2007-0038. [DOI] [PubMed] [Google Scholar]
- 21.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desmet F-O, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown AS, Mohanty BK, Howe PH. Identification and characterization of an hnRNP E1 translational silencing motif. Nucleic Acids Res. 2016;44:5892–5907. doi: 10.1093/nar/gkw241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tennyson CN, Klamut HJ, Worton RG. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat. Genet. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 25.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 26.Chao C-H, et al. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J. Clin. Invest. 2014;124:3093–3106. doi: 10.1172/JCI73351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L. Splicing noncoding RNAs from the inside out. Wiley Interdiscip. Rev. RNA. 2015;6:651–660. doi: 10.1002/wrna.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulveling D, Francastel C, Hubé F. Identification of potentially new bifunctional RNA based on genome-wide data-mining of alternative splicing events. Biochimie. 2011;93:2024–2027. doi: 10.1016/j.biochi.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 31.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, et al. Metastatic heterogeneity of breast cancer cells is associated with expression of a heterogeneous TGFβ-activating miR424–503 gene cluster. Cancer Res. 2014;74:6107–6118. doi: 10.1158/0008-5472.CAN-14-0389. [DOI] [PubMed] [Google Scholar]
- 33.Oshimori N, Oristian D, Fuchs E. TGF-β Promotes Heterogeneity and Drug Resistance in Squamous Cell Carcinoma. Cell. 2015;160:963–976. doi: 10.1016/j.cell.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornblihtt AR, et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013;14:153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 35.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West JA, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xing Z, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter MS, et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 39.Laury-Kleintop LD, Tresini M, Hammond O. Compartmentalization of hnRNP-K during cell cycle progression and its interaction with calponin in the cytoplasm. J. Cell. Biochem. 2005;95:1042–1056. doi: 10.1002/jcb.20486. [DOI] [PubMed] [Google Scholar]
- 40.Hu Y, et al. miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016;7:e2291. doi: 10.1038/cddis.2016.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Liu H, Yu J, Yu H. Chemoresistance to doxorubicin induces epithelial-mesenchymal transition via upregulation of transforming growth factor β signaling in HCT116 colon cancer cells. Mol. Med. Rep. 2015;12:192–198. doi: 10.3892/mmr.2015.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q-Q, et al. Twist1-mediated adriamycin-induced epithelial-mesenchymal transition relates to multidrug resistance and invasive potential in breast cancer cells. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009;15:2657–2665. doi: 10.1158/1078-0432.CCR-08-2372. [DOI] [PubMed] [Google Scholar]
- 43.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanharanta S, Massagué J. Origins of metastatic traits. Cancer Cell. 2013;24:410–421. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat. Med. 2013;19:1450–1464. doi: 10.1038/nm.3391. [DOI] [PubMed] [Google Scholar]
- 46.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE94637. Source data for Figure 1e, 3e, 4a, 4h, 6b, S1b, S3d, S3e, and S4 have been provided as Supplementary Table 2. Unprocessed original scans of blots are shown in Supplementary Figure 7. All other data supporting the findings of this study are available from the corresponding author on reasonable request.