Abstract
CTP synthetases catalyze the last step of pyrimidine biosynthesis and provide the sole de novo source of cytosine-containing nucleotides. As a central regulatory hub, they are regulated by ribonucleotide and enzyme concentration through ATP and UTP substrate availability, CTP product inhibition, GTP allosteric modification, and quaternary structural changes including the formation of CTPinhibited linear polymers (filaments). Here, we demonstrate that nicotinamide redox cofactors are moderate inhibitors of Escherichia coli CTP synthetase (EcCTPS). NADH and NADPH are the most potent, and the primary inhibitory determinant is the reduced nicotinamide ring. Although nicotinamide inhibition is noncompetitive with substrates, it apparently enhances CTP product feedback inhibition and GTP allosteric regulation. Further, CTP and GTP also enhance each other’s effects, consistent with the idea that NADH, CTP, and GTP interact with a common intermediate enzyme state. A filamentblocking mutation that reduces CTP inhibitory effects also reduced inhibition by GTP but not NADH. Protein-concentration effects on GTP inhibition suggest that, like CTP, GTP preferentially binds to the filament. All three compounds display nearly linear dose-dependent inhibition, indicating a complex pattern of cooperative interactions between binding sites. The apparent synergy between inhibitors, in consideration with physiological nucleotide concentrations, points to metabolically relevant inhibition by nicotinamides, and implicates cellular redox state as a regulator of pyrimidine biosynthesis.
Graphical abstract
CTP synthetases (CTPSs) are ubiquitous enzymes that produce CTP by amination of UTP1 (reviewed in ref 2). CTP concentrations are the lowest of the four ribonucleotides in many cells.3-5 Catalyzing the last step of the pyrimidine biosynthetic pathway and as the only de novo source of cytosine nucleotides, CTPSs are critical regulatory hubs with a number of inputs that modulate their activity including ribonucleotide concentrations and phosphorylation.2,6-10 CTPSs have been suggested as drug targets for cancer, sleeping sickness, and recently, Mycobacterium tuberculosis infection.11-13
Structural analyses of CTPSs depict an active tetramer composed of two-domain monomers (Figure 1;2,7,14,15 Welin et al., unpublished, PDB 2VKT, 2W7T). ATP hydrolysis drives this unfavorable reaction by phosphorylating the unreactive UTP O4 atom, and the resulting phosphate is displaced by ammonia derived from glutamine hydrolysis.16,17 The Nterminal kinase-like amidoligase domain promotes UTP phosphorylation and ammonia transfer reactions, while the C-terminal glutamine amidotransferase domain produces ammonia and coordinates its delivery to the amidoligase active site.
Figure 1.
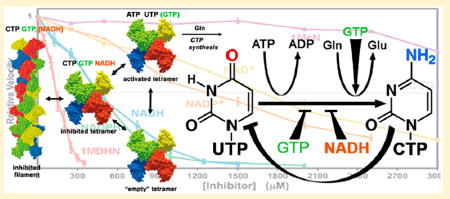
Nucleotide regulation of EcCTPS. EcCTPS is in a dimer–tetramer equilibrium favoring the tetramer in the presence of nucleotides ATP and UTP, since their binding sites are completed upon tetramerization (see refs 2 and 23). GTP (<250 μM) activates glutamine hydrolysis. Product feedback inhibitor CTP binds the tetramer, competitively preventing UTP binding. CTP-bound tetramers are also in equilibrium with an inhibited filament (see ref 31). It is unknown whether EcCTPS dimers bind GTP. Mutual enhancements of each other’s inhibitory potencies by CTP, NADH, and GTP and the lack of competition of GTP and NADH with ATP and UTP substrates suggest that they preferentially bind an inhibited tetramer conformation distinct from the “active” one. CTP and NADH can bind this conformation in the absence of other ligands. Responses to a filament-disrupting mutation suggest that CTP and GTP also preferentially interact with the filament (indicated by an asterisk) while NADH does not.
The structural mechanisms of CTPS regulation are only partially understood (Figure 1). All four nucleotides contribute activating or inhibiting inputs. Active tetramer assembly from inactive dimers is promoted by ATP and UTP substrates,18-22 which bind at an interfacial active site formed by the N-terminal amidoligase domains.2,7,23 Feedback inhibition is effected by the product CTP, which competes with UTP by binding at a distinct but overlapping site.23 Allosteric regulator GTP is required for efficient CTP synthesis but also can inhibit it; low concentrations (0–200 μM) activate glutamine hydrolysis up to 50-fold, while higher concentrations (IC50 ≈ 500 μM) inhibit CTP synthesis without affecting glutamine hydrolysis.10 However, its binding sites are still speculative.2 In many eukaryotes, two independent CTPS isoforms are further regulated by multiple protein phosphorylations.6,8,9,24-26 More recently, bacterial, human, yeast, and Drosophila CTPSs were demonstrated to dynamically aggregate into micrometerscale linear filaments.27-30 Barry et al. demonstrated that Escherichia coli filaments are a regulatory assembly that is induced by CTP binding.31 They also determined the filament cryoEM structure, which revealed a superhelix of tetramers. Analyses of human, Drosophila, and yeast enzymes suggest that different factors might control eukaryotic filament formation and structure.32-34
While ribonucleotide influences on CTPS activity are relatively well characterized, there has been no evidence for its regulation by other metabolites. Serendipitously, we discovered that the redox cofactor NADH inhibits E. coli CTP synthetase (EcCTPS). Here, we describe inhibition by nicotinamides and their interactions with other known EcCTPS regulators. Although by themselves nicotinamides are moderate inhibitors, their apparent synergy with other natural inhibitors suggests that their effects on EcCTPS may be physiologically relevant.
MATERIALS AND METHODS
Enzyme Production
EcCTPS was expressed as an N- or C-terminal His6 fusion protein. The EcCTPS reading frame was amplified from E. coli K-12 and ligated into pET28b, fused in frame to the N-terminal His6 tag sequence at the NdeI site.23 The C-terminal His-tagged protein was created by replacing the N-terminal region XbaI–NdeI fragment with that of pET41 to yield the native N-terminus, while deletion mutagenesis fused the C-terminal lysine residue to a His6 tag via a Val–Glu linker.
EcCTPS enzymes were overexpressed and purified using metal chelate chromatography. Proteins were produced in BL21(DE3)* cells, which express rare tRNAs (Life Technologies), enhancing yields 1.5–2-fold over BL21(DE3). One liter of culture was inoculated with a single fresh colony and grown on LB broth at 37 °C until A600nm reached 0.6–0.8 and then induced with 100 μM IPTG. After 3–5 h growth, cells were centrifuged (2000g, 10 min, 4 °C), and the pellets were stored at −80 °C. Cell pellets were thawed on ice, resuspended in lysis/wash buffer (20 mM Tris-Cl, 5 mM imidazole, 500 mM NaCl, pH 7.9) containing 1 mM PMSF, and lysed using a microfluidizer. The cleared lysates (15 000g, 45 min, 4 °C) were loaded onto 3–5 mL beds of Ni-NTA resin equilibrated with lysis/wash buffer. The columns were washed with 200 mL of lysis/wash buffer, followed by 200 mL of lysis/wash buffer containing 2 M NaCl, and then again with 200 mL of lysis/wash. The proteins were eluted with minimal elution buffer (20 mM Tris-Cl, 500 mM imidazole, 500 mM NaCl, pH 7.9), and immediately dialyzed against 20 mM Tris-Cl, 10 mM EDTA, 500 mM NaCl, pH 8.1 for 4 h, followed by overnight dialysis against 20 mM Tris-Cl, 0.1 mM EDTA, 500 mM NaCl, 2 mM DTT, pH 8.1. The dialyzed eluates were concentrated using Amicon Y-100 spin concentrators to 10–15 A280 units/mL, then flash-frozen in 50 μL aliquots and stored at −80 °C. Typical recoveries were 20–30 mg of purified EcCTPS per liter of cell culture. For assays, storage tubes were rapidly thawed under running cold water, immediately mixed, and stored at 4°C. Maximum activity was achieved within 12 h after thawing and diminished with extended storage at 4 °C. Freezing and storage in 500 mM NaCl-containing buffer extended the halflife of thawed enzyme to more than 6 weeks, compared to about 3 weeks using a buffer with no added salt.
Quantification of Protein, Nucleotide, and Nicotinamides
Enzyme concentrations were determined from the 280 nm absorbance. To make our data comparable to previous work, we used the historical extinction coefficient 0.89A280 mg–1-mL–1 (5.5 × 103 M–1).35 It should be noted that the EXPASY PROTPARAM calculator (http://web.expasy.org/protparam/) yields values of 0.67A280 mg–1-mL–1 and 4.0 × 103 M–1, which would result in a 1.33-fold higher protein concentrations than are indicated.
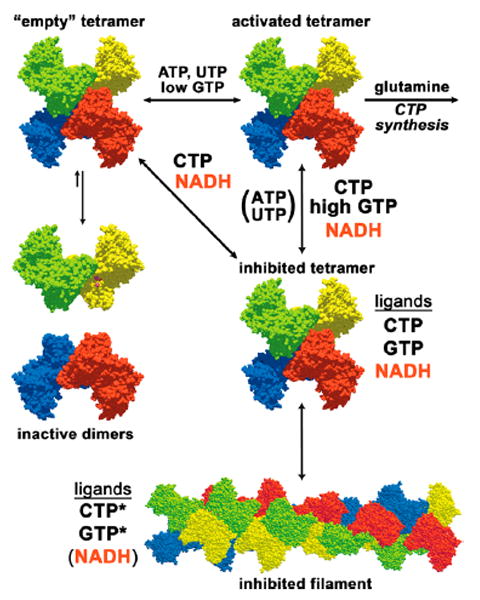
Solid nucleotides were obtained from Sigma-Aldrich unless otherwise noted. The structures of the nicotinamides used in this study are depicted in Figure 2. Solutions were prepared by dissolving solids in equimolar Na-HEPES pH 8.0 at twice the desired final concentration: 100 mM for ATP (no. A3377), UTP (no. U6750 or no. U6875), CTP (no. C1506), NADH (no. N6505), NADPH (no. N7505), NAD+ (no. N0632), and NADP+ (no. N0505); 0.02 mM for GTP (no. G8877); 100 mM for 1-methyl dihydronicotinamide (1MDHN, Santa Cruz Biotechnology, no. sc-213351); and 250 mM for 1-methylnicotinamide (1MeN, Santa Cruz Biotechnology, no. sc-237583). Monitored with pH paper, the pH was adjusted to 7.5–8.0 using 1 M NaOH. Nucleotide concentrations were determined from UV absorbance using the appropriate extinction coefficient (ATP, ɛ(259 nm) = 1.54 × 104 M–1; UTP, ɛ(262 nm) = 1.00 × 104 M–1; GTP, ɛ(253 nm) = 1.37 × 104 M–1; CTP, ɛ(271 nm) = 9.0 × 103 M–1; NADH/NADPH, ɛ(339 nm) = 6.22 × 103 M–1; NAD+/NADP+, ɛ(259 nm) = 1.7 × 104 M–1; 1MDHN, ɛ(360 nm) = 7.06 × 103 M–1;36 1MeN, ɛ(265 nm) = 3.94 × 103 M–1).37 The mixtures were diluted to their final storage volumes with water and requantified by UV absorbance. Stocks were apportioned into 50–150 uL aliquots in screw-capped tubes prior to freezing at −80 °C. Concentrations of further dilutions were redetermined prior to use. Solutions of 1MDHN oxidized rapidly at 4 °C, and aliquots were only allowed to stand for less than 2 h before use. Glutamine stocks (Sigma/Aldrich no. G3126, 100 mM in 1 mL aliquots) were treated similarly except that no pH adjustment was necessary and concentrations were solely based on weight.
Figure 2.
Structures of nicotinamide compounds used in this study. (a) Structural differences between reduced and oxidized nicotinamide rings. Oxidized forms are aromatic, charged, and planar, while reduced forms are neutral and in a “boat” conformation. (b) Structural differences between the different R groups of the nicotinamides. Phosphorylation occurs on the 2′-OH of the adenylate portion of NAD+/NADH (−OX) to give NADP+/NADH.
CTP Synthesis Assay
Assays were carried out using an HP-6853 spectrophotometer outfitted with a water-jacketed 8-cell cuvette holder. The cuvettes were pre-equilibrated at 37 °C (liquid temperature in cuvette). Absorbance at 291 nm versus time data were acquired using the UV–visible Chemstation software package in “Kinetics” mode. Sampling times were 0.5–8 s depending on the duration of the experiment (100–500 s).
Reaction buffer contained 60 mM Na-HEPES, pH 8.0, 10 mM MgCl2, 0.5 mM EGTA, and 2 mM Na-azide. Standard saturating substrate concentrations were 600 μM UTP, 1.5 mM ATP, and 10 mM glutamine. The S0.5 values are given in Table 1. Unless otherwise stated, 200 μM GTP and 200 nM EcCTPS were used. Both N-terminal and C-terminal His-tagged enzymes yielded indistinguishable kinetic values and proteinconcentration dependencies on specific activity. The majority of the inhibition data reported here were obtained using the Nterminal His-tagged form.
Table 1.
Kinetic Constants for EcCTPS at 37°Ca
| limiting substrate | conditiona | S0.5 (μM) | Hill coeff, nH |
|---|---|---|---|
| UTP | 59 ± 15 | 1.4 ± 0.3 (23)b | |
| 150 μM ATP | 200 ± 40 | 1.8 ± 0.1 (6) | |
| ATP | 130 ± 10 | 1.7 ± 0.2 (18)c | |
| 60 μM UTP | 310 ± 20 | 1.9 ± 0.4 (5) | |
| glutamined | 360 ± 20e | 1.1 ± 0.1 (11) | |
|
| |||
| effector | conditiona | IC50/EC50 (μM) | |
|
| |||
| CTP | IC50 | 370 ± 60f (6) | |
| 50 μM UTP 100 nM EcCTPS | IC50 | 160 ± 10g (4) | |
| GTPh | EC50 | 38 ± 3 (4) | |
| IC50 | 540 ± 60 | ||
Unless noted, substrates were present at saturation with optimal GTP. ([UTP] = 600 μM, [ATP] = 1500 μM, [GTP] = 200 μM, [glutamine] = 10 mM). S0.5 and nH values were determined by fitting data to the Hill function. IC50 values were estimated by linear extrapolation of data points straddling 50% inhibition values (see Materials and Methods). The number of experiments used for each determination is given in parentheses.
The S0.5 and nH values did not significantly vary from 10 to 2000 nM enzyme (at 100 nM, S0.5 = 57 ± 8, nH = 1.3 ± 0.2, from seven experiments).
The S0.5 and nH values did not significantly vary from 50 to 400 nM enzyme (at 100 nM, S0.5 = 126 ± 13, nH = 1.7 ± 0.3, from six experiments).
These values were determined at 100 nM enzyme and include four values determined in the presence of 500 μM NADH, which had no discernible effect.
Lineweaver–Burke analysis gave 411 ± 44 μM as the Km value. Bearne and Iyengar reported a Km of 320 μM using HPLC.58
The limiting slopes of the biphasic Hill plots were −1.22 ± 0.3 and −4.8 ± 1.9, at the low and high concentration ranges, respectively.
The limiting slopes of the biphasic Hill plots were −0.6 ± 0.2 and −1.9 ± 0.3, at the low and high concentration ranges, respectively.
GTP EC50 and IC50 values were estimated by linear extrapolation of data points straddling 50% activation and inhibition values, respectively. Nonlinear Hill plots yielded limiting slopes of 1.8 ± 0.2 and −4.6 ± 0.3, at the low and high concentration ranges, respectively. Fitting the data from individual experiments to a two-site model,10 where nact = 1, gave the following averaged parameters: kact = 10.0 ± 0.6 s−1, KA = 57 ± 7 μM, Ki = 308 ± 24 μM, and ninh = 5.0 ± 0.4. Previously reported values were, respectively, 10.3, 23, 190, and 3.8.10
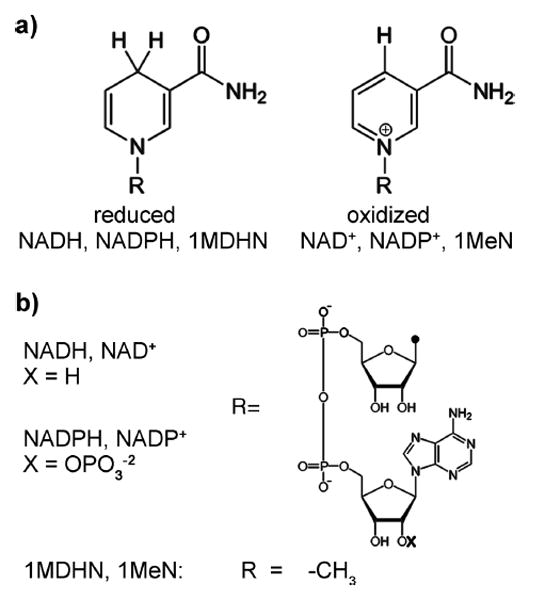
The reaction procedure is illustrated in Figure 3a. For each reaction, three tubes were prepared: “A” tubes contained nucleotides, inhibitors, and reaction buffer, “B” tubes contained 2 μM EcCTPS in reaction buffer, and “C” tubes contained 100 mM glutamine. Component concentrations were adjusted to achieve the desired final levels when the contents of all three tubes are combined. A and C tubes were pre-equilibrated at 37 °C. At time = 0, B tubes, stored on ice, were transferred to a room temperature water bath (21–22 °C) and incubated for 3 min (“annealing”). The contents of tube A were then added, and the mixture was further incubated at 37 °C for 4 min. CTP synthesis was initiated by transferring the entire contents to tube C, mixing, and then immediately transferring the mixture to a prewarmed cuvette.
Figure 3.
In vitro EcCTPS reactions. (a) Scheme for performing EcCTPS reactions. EcCTPS enzyme (2–40 μM) was annealed for 3 min at 21 °C in reaction buffer then mixed with prewarmed nucleotide substrates and effectors and incubated for an additional 4 min at 37 °C. To initiate the synthesis reaction, the enzyme–nucleotide solutions were mixed with 10-fold concentrated 37 °C glutamine solution and immediately transferred to 37 °C cuvettes to measure the change in UV absorbance at 291 nm. (b) Concentration dependence of the annealing reaction on specific activity (apparent kcat). Different amounts of EcCTPS were annealed at either 4 μM (black circles) or at 10-fold final concentration (100–4000 nM, gray squares) EcCTPS prior to dilution to 10–400 nM into nucleotides and glutamine as described in panel a and Materials and Methods. Note that the apparent kcat values for the two curves converge between 2 and 2.4 μM. (c) Dependence of apparent kcat values on final [EcCTPS] in the reaction. In a separate experiment from panel b, 4 μM EcCTPS was preincubated at 21 °C, then diluted to the final concentrations indicated on the horizontal axis (5–200 nM) upon addition of nucleotides and glutamine as described in panel a and Materials and Methods. Under these conditions, kcat increases approximately 5–9% going from 100 to 500 nM final EcCTPS (Figure S8).
In protein-dependent experiments in which high (> μM) protein concentrations were used (Figure 7c), EcCTPS was annealed at 40 μM, and the reactions were carried out as previously described.31 At the highest final EcCTP concentration used (8 μM, Figure 7c), the NaCl contribution from enzyme stocks did not exceed 20 mM, which has a negligible effect on rate (Figure S1). The maximum additional sodium ion contributions from triphosphates and nicotinamides ranged 11–18.5 mM. This amount represents a significant increase above the reaction buffer contribution (46 mM). However, based on the effects of 0.1 M NaCl (Figure S1) or 0.1 M NaOAc (Figure S6) on apparent kcat, the velocity reduction due to 20 mM additional sodium ion is 4.4–6% (0.22–0.30%/mM Na+). The sodium ion concentration differences in comparisons between inhibitor titrations in the absence or presence of another inhibitor were 1–1.9 mM.
Figure 7.
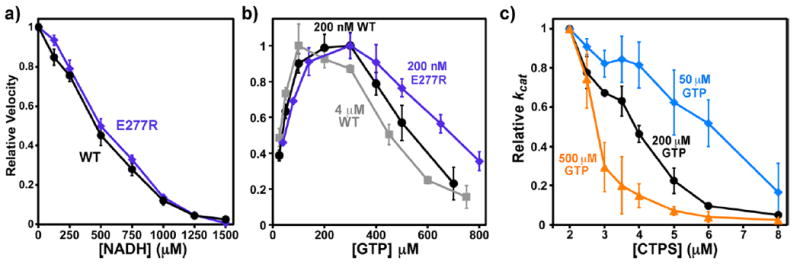
The role of inhibitory filament formation in NADH and GTP inhibition. (a) Relative CTP synthesis velocities of 200 nM WT (black circles) and E277R (see ref 31, in digo diamonds) in the presence of 0–1500 μM NADH normalizing the V0 equal to one (V0 (WT) = 1.24 ± 0.05 μM s−1, V0 (E277R) = 1.07 ± 0.03 μM s−1). Error bars indicate the standard deviation from the average values of three experiments after normalizing V0 equal to one. The filament blocking mutation E277R does not significantly affect NADH inhibition. (b) Relative velocities of 200 nM WT (black circles), 4 μM WT (gray squares), or 200 nM E277R (indigo diamonds) as a function of 25–800 μM GTP. Error bars indicate the standard deviation from the average of four experiments. For comparison purposes, the maximum velocities of each experiment were normalized to one prior, to averaging. The average Vmax values were 1.57 ± 0.11 s−1 (200 nM WT), 14.0 ± 1.7 s−1 (4 μM WT), and 1.60 ± 0.11 μM s−1 (200 nM 277R). The E277R mutation reduced both the activating and inhibitory effects of GTP, while high protein concentrations increased them. The EC50 and IC50 values, respectively, were 38 ± 3 and 540 ± 60 μM (200 nM WT), 27 ± 7 and 455 ± 46 μM (4 μM WT), and 46 ± 6 and 697 ± 4 μM (200 nM E277R). Fitting the data to a two-site activation/inhibition model (fits not shown) yielded the following parameters: for 200 nM WT, kact = 10.7 s−1, KA = 58 μM, Ki = 301 μM, and ninh = 4.7 (R2 = 0.993); for 4 μM WT, kact = 4.7 s−1, KA = 41 μM, Ki = 221 μM, and ninh = 4.1 (R2 = 0.984); and 200 nM E277R, kact = 10.7 s−1, KA = 90 μM, Ki = 342 μM, and ninh = 3.8 (R2 = 0.996). (c) Relative kcat values from at 2–8 μM final EcCTPS in the presence of 50 μM (blue diamonds), 200 μM (black circles), and 500 μM (orange triangles) GTP. Reactions were performed as previously described 31 with annealing carried out at 40 μM EcCTPS. Error bars indicate the standard deviation from the average kcat of three experiments, after normalizing the maximum kcat of each to one. GTP enhances protein-dependent inhibition. The apparent kcat values at 2 μM EcCTPS are 2.9 s−1(50 μM), 5.0 ± 0.1 s−1(200 μM), and 2.7 ± 1.0 s−1(500 μM). The enzyme concentrations for 50% autoinhibition are 6.1 μM (50 μM), 3.9 μM (200 μM), and 2.8 μM (500 μM).
EcCTPS requires at least 2 mM free magnesium ion for maximal activity.38 The maximum triphosphate concentrations used here were 2.3–3.9 mM, allowing for greater than 6 mM minimum free magnesium ion.
Data Analysis
CTP production velocities were calculated using the extinction coefficient difference between UTP and CTP at 291 nm, 1338 M–1.39 The delay between glutamine mixing and the first spectrophotometric data point was 5–10 s. Linear rates persisted for at least 10–20 s unless substrates were limiting. Initial rates were extracted from the earliest linear regions (6–20 data points) of the A291 versus time data using linear regression.
Descriptive substrate kinetic parameters were obtained by nonlinear fitting of velocity data to the Hill equation
using the Solver Add-in function in Excel to minimize the value of the sum of the squared differences between calculated and observed rates. Substrate titrations yielded linear Hill plots over the concentrations ranges used (ATP, 35–1500 μM; UTP, 15–600 μM; glutamine 100–5000 μM). All inhibition data yielded nonlinear Hill plots, apparently biphasic (for CTP, see Figure 4a) so we reported descriptive IC50 values, which were determined by linear extrapolation between the two data points that straddled the 50% activity value (Tables 2 and 3). Mean values and standard deviations for all kinetic constants were calculated from individual experimental values using Excel. In an attempt to understand the Hill plot curvature and the nature of inhibitor binding cooperativity, we fit NADH and CTP v vs [I] inhibition data to a generalized sequential binding model for a tetramer:40
where each Kn corresponds to the dissociation constant between binding n – 1 and n inhibitors. The model is highly simplified, and low data/parameter ratios limited fitting accuracy. Boundary restraints were applied to K1 and the ratios K2/K1, K3/K2 and K4/K3 (>0, 0.03–1000, 0.03–1000, 0.00001–30, respectively). For some data sets, one additional low inhibitor concentration value between 0 and the first measured concentration point was added due to lack of data coverage, estimated from other experiments outside the fit data sets. These points were required to prevent K1 from becoming unrealistically small, K2 from becoming concomitantly unrealistically large, and the calculated binding curve from unusual developing extreme downward concavity at the low concentration range during fitting cycles. Even without these points, the patterns of apparent cooperativity were consistent (see Discussion).
Figure 4.
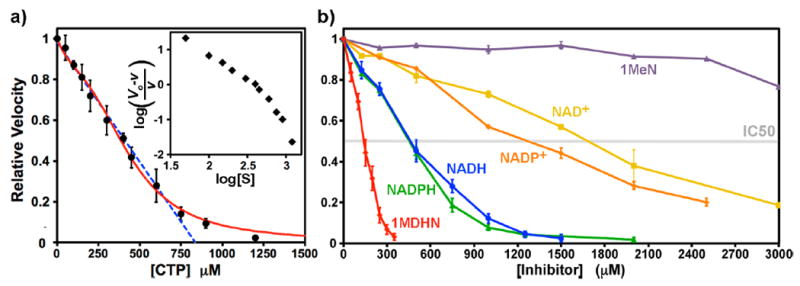
(a) Dose–response curve for CTP inhibition. Data points are indicated (black circles) as the averages of three, six, or nine separate determinations. The error bars indicate ±SD. The dose response is nearly linear up to 750 μM CTP (blue dashed line, R = 0.993). The best-fit curve to a four-site sequential binding model is also shown (red curve, see Materials and Methods). The parameters K1, K2, K3, and K4 were 446, 2120, 1173, and 7.5 μM, respectively. The fit IC50 value is 385 μM. A Hill plot is shown in the inset. The limiting slopes at low and high CTP concentrations are −1.5 and −4.0, respectively, and the average slope is −2.0. (b) Dose-dependent EcCTPS inhibition by various reduced and oxidized nicotinamide compounds. Each line is labeled and color-coded for the particular compound (see Figure 2). Reactions were performed under standard conditions (see Materials and Methods), with inhibitors present during the nucleotide preincubation period (see Figure 3). Numeric IC50 values are found in Table 2.
Table 2.
Inhibition Constants for Nicotinamide EcCTPS Inhibitorsa
| inhibitor | IC50,b μM | IC50ox/IC50red |
|---|---|---|
| NADH | 470 ± 40 (6) | 3.7 |
| NAD+ | 1700 ± 70 (2) | |
| NADPH | 450 ± 20 (3) | 2.8 |
| NADP+ | 1280 ± 50 (3) | |
| 1MDHN | 140 ± 10 (7) | >28 |
| 1MeN | 4000(est) (2) | |
| ADP | 4500(est) (2) |
All reactions contained 200 nM EcCTPS, 1.5 mM ATP, 0.6 mM UTP, 0.2 mM GTP, and 10 mM glutamine.
IC50 values were estimated by linear extrapolation of data points straddling 50% inhibition values. All nicotinamide inhibitors yielded nonlinear Hill plots, with limiting slopes of −0.89 ± 0.16 and −4.0 ± 1.0, at the low and high concentration ranges, respectively. The numbers of independent determinations used in the calculations are given in parentheses.
Table 3.
Interactions between NADH, CTP, and GTPa
| noneb | 400 μM CTP | 500 μM NADH | 50 μM GTP | 500 μM GTP | 140 μM 1MeDHN | 1200 μM NAD+ | |
|---|---|---|---|---|---|---|---|
| CTP IC50 | 370 ± 60 | 160 ± 40 (4) | 620 ± 30 (3) | 220 ± 60 (3) | 200 ± 20 (3) | 160 ± 30 (2) | |
| NADH IC50 | 470 ± 40 | 220 ± 90 (4) | 770 ± 30 (3) | 240 ± 10 (3) | |||
| GTP EC50 | 38 ± 3 | 21 ± 2 (4) | 24 ± 2 (4) | 20 (1) | |||
| GTP IC50 | 540 ± 60 | 410 ± 50 (4) | 350 ± 40 (4) | 330 (1) | |||
| 1MDHN IC50 | 140 ± 10 | 73 ± 6 (3) |
Initial values of K1, K2, K3, and K4 were set to the experimental IC50 values. Although the fitted values were sensitive to initial estimates, the trends over a 10-fold range of initial values were robust and suggested complex cooperativity patterns (see Discussion).
Descriptive values EC50 and IC50 were also reported for the activation and inhibition portions of the GTP dose–response curves and were calculated analogously to the S0.5 values (Tables 2 and 3). For comparison purposes, we also fit the data to a two-site activation/inhibition model from Bearne and coworkers,10
with the assumptions that EcCTPS is inactive in the absence of GTP and is fully inhibited by it. GTP parameters and data are reported in the legend for Figure 6.
Figure 6.
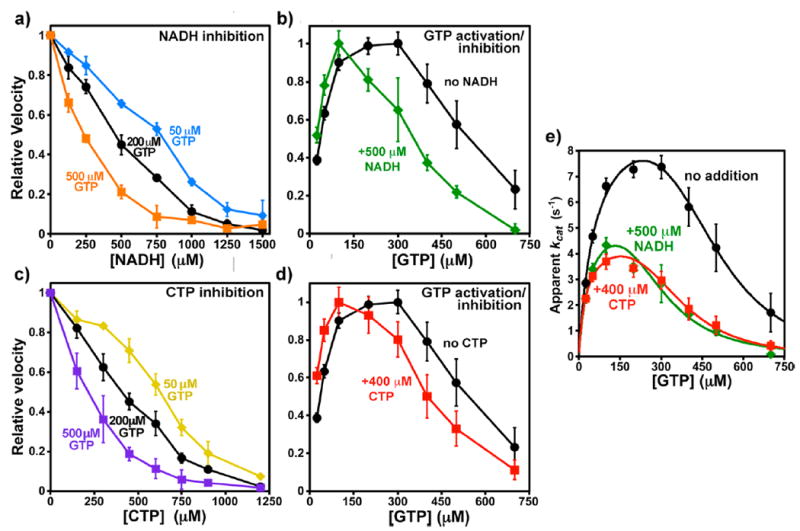
Interactions of CTP or NADH with GTP. Apparent EC50 and IC50 values are given in Table 3. (a) Relative CTP synthesis velocities with 0–1500 μM NADH in the presence of GTP at 50 μM (blue diamonds), 200 μM (black circles), and 500 μM (orange squares). Error bars indicate the standard deviation for three experiments after normalizing to V0 equal to one. The V0 values were 0.64 ± 0.01, 1.21 ± 0.05, and 0.70 ± 0.04 μM s−1, respectively. (b) Relative velocities as a function of 25–700 μM GTP in the absence (black circles) or presence (green squares) of 500 μM NADH. Error bars indicate the standard deviation for four experiments after averaging and then normalizing to Vmax equal to one. The Vmax values were 1.47 ± 0.09 and 0.86 ± 0.06 μM s−1, respectively. (c) Relative velocities with 0–1200 μM CTP in the presence of GTP at 50 μM (yellow diamonds), 200 μM (black circles), and 500 μM (purple squares). Error bars indicate the standard deviation for three experiments after normalizing to V0 equal to one. The V0 values were 0.65 ± 0.01, 1.16 ± 0.10, and 0.77 ± 0.09 μM s−1, respectively. (d) Relative velocities as a function of 25–700 μM GTP in the absence (black circles) or presence (green squares) of 500 μM NADH. Error bars indicate the standard deviation for four experiments after averaging and then normalizing to Vmax equal to one. The Vmax values were 1.47 ± 0.09 and 0.74 ± 0.06 μM s−1, respectively. (e) Fit of GTP titration data to a two-site inhibition model 10 (see Materials and Methods). Unnormalized kcat values with data from experiments with no inhibitor (black circles), 500 μM NADH (green diamonds), or 400 μM CTP (red squares) are shown (see Table 3), along with curves, calculated from the following parameters fit to the averaged data: (i) uninhibited (black circles), kact = 10.0 s−1, KA = 57 μM, Ki = 301 μM, and ninh = 4.8 (R2 = 0.993); (ii) NADH inhibited (green diamonds) kact = 6.0 s−1, KA = 39 μM, Ki = 175 μM, and ninh = 4.1 (R2 = 0.966); (iii) CTP inhibited (red squares) kact = 4.8 s−1, KA = 27 μM, Ki = 202 μM, and ninh = 4.5 (R2 = 0.962).
RESULTS
Activity Assay Optimization
EcCTPS kinetic constants were reported previously,16,19,22,35,39,41,42 but the collective results did not agree well. Our own initial attempts also yielded inconsistent values. We traced these problems to enzyme storage, determination of nucleotide concentrations, assay enzyme concentrations, and preincubation conditions. Our present assay procedure incorporates additional features that greatly improve reproducibility: careful spectrophotometric determination of nucleotide concentrations, enzyme storage in high salt and room-temperature preincubation (“annealing”) at high enzyme concentration prior to exposure to substrates (See Materials and Methods, Figure 3a). At 4 °C, EcCTPS reportedly dissociates into monomers but reforms dimers when shifted to higher temperature.19,43 The annealing procedure, which is aimed at maximizing dimer reassembly, improved assay reproducibility and increased the specific activity by 20–40% (Figure S2). The protein concentration during annealing was also critical, with maximum specific activity achieved between 2 and 4 μM EcCTPS. Annealing at a fixed high concentration prior to enzyme dilution into substrate/buffer solution lead to a smaller apparent kcat dependence on final enzyme concentration compared to annealing at 10-fold final concentrations (annealing concentrations of 50 nM to 2 μM, compare solid black and dashed gray lines, Figure 3b). Even when annealing was performed at 4 μM enzyme, the specific activity increased 50% between 5 and 50 nM final EcCTPS (Figure 3c), which we attribute to the CTPS dimer–tetramer equilibrium favoring the active tetramer at higher CTPS concentrations. At assay concentrations higher than 2 μM, the specific activity decreased due to inhibitory filament formation.31 The apparent kcat values, typically 5.5–6.5 s−1, ranging up to 8 s−1, at 200 nM enzyme, are comparable to those reported elsewhere.16,19,22,42
Our kinetic constants were robust over a number of individual experimentalists and enzyme preparations. For the data reported here, baseline characterizations of UTP, ATP, CTP, and GTP concentration dependences, data from two or three individual experimentalists and two to four distinct enzyme preparations were utilized. Subsequently, seven additional experimentalists have obtained average S0.5, EC50, or IC50 values within 1 SD of those reported here (data not shown). The kcat values vary the most (ranging 40–60%), in part because of different ages of the thawed enzyme. N-values are given in Tables 1 and 2 as well as the figure legends.
Our ATP and UTP S0.5 values, 60 and 130 μM, respectively, at saturating concentrations of other substrates (Table 1 and Figure S3), agreed well with some reported values16 but differed from others.19,22,39,44 As previously observed,19 the S0.5 values of ATP and UTP are interdependent (Table 1). At subsaturating ATP concentration (150 μM), the S0.5 of UTP increased to 200 μM, while at 60 μM UTP, the ATP S0.5 value increased to 330 μM. Interestingly, we found that lowering the GTP concentration from the optimal (200 μM) to its EC50 (50 μM) also reduced the ATP S0.5 value to 81 ± 5 μM (n = 5). However, at 500 μM GTP, the ATP S0.5 value was similar to that measured at 200 μM GTP (155 ± 25 μM, n = 5).
Protein concentration did not significantly affect the UTP S0.5 value at saturating ATP, between 10 and 2000 nM EcCTPS, and had no significant effect on the ATP S0.5 value at saturating UTP, from 50 to 400 nM EcCTPS (Table 1, footnotes b and c, and Figure S3). This result is curious since the tetramerization equilibrium is thought to be coupled to UTP binding18,19,22 and the reduced kcat at low EcCTPS concentrations suggested incomplete tetramer formation (Figure 3c). Therefore, in addition to tetramer formation, the UTP–ATP interdependence may be exerted via on-enzyme site–site interactions.
The GTP concentration dependence of specific activity resembled that previously measured10 and when fit to the same two-site activation/inhibition model yielded parameter values that agreed within a factor of 2 (Table 1, footnote h).
From these data, we codified “standard assay conditions” of 200 nM final enzyme annealed at 2 μM with saturating substrates 600 μM UTP, 1500 μM ATP, and 10 mM glutamine and 200 μM GTP effector.
Under standard conditions, the IC50 of feedback inhibitor CTP is ~370 μM, whereas at 50 μM UTP (near the UTP S0.5 value), the CTP IC50 is 160 μM. These values are comparable to those measured under similar conditions.39,44 Thus, at 50 μM UTP, the CTP IC50 is 2.5-fold higher than the UTP concentration, while at saturating UTP (600 μM), the IC50 is 1.6-fold lower (Table 1). This increase in relative inhibitory potency when the competitive substrate concentration increases suggests cooperativity between UTP and CTP binding and underscores the complexity of intersite interactions in CTPSs. In fact, the dose-dependence curve for CTP inhibition is atypical (Figure 4a) in that it was nearly linear up to 85% inhibition at 750 μM CTP (R = 0.993) and yielded an apparently biphasic Hill plot (See Materials and Methods and Figure 4a, inset). This behavior has not been previously described and suggests that cooperative interactions between CTP binding sites in the tetramer follow a complex pattern (see Discussion). This behavior was also observed for nicotinamide and GTP inhibition.
EcCTPS Is Inhibited by Nicotinamides
EcCTPS is inhibited by nicotinamide-containing compounds. Dosedependent inhibition data for six nicotinamides, determined under standard assay conditions, is shown in Figure 4b and Table 2.
NADH inhibits CTP synthesis activity with an IC50 of 470 μM, with greater than 95% inhibition at 1250 μM. Inhibition potency was independent of enzyme concentration from 50 to 400 nM (Figure S4). Like CTP, the dose dependence was complex, being nearly linear from 0 to 750 μM, and yielded nonlinear Hill plots. Potent inhibition required the reduced cofactor. The oxidized NAD+ cofactor inhibited ~3-fold less effectively (IC50 = 1700 μM). NADPH had similar inhibitory potency to NADH. Curiously, NADP+ was a somewhat better inhibitor than NAD+ (IC50 = 1280 μM) and concentration dependence for both 2′-phosphorylated cofactors exhibited slightly more curvature.
The reduced nicotinamide ring is a critical recognition element. The NADH analog 1-methyl 1,4-dihydronicotinamide (1MDHN, Figure 2) is more effective than NADH (IC50 = 140 μM), whereas the oxidized analog 1-methylnicotinamide (1MeN) was a very poor inhibitor (<25% inhibition at 3000 μM).
The inhibitory properties of NADH/NADPH and NAD+/NADP+ do not apparently involve critical interaction of the adenine nucleotide portion with the ATP site, since ATP had the same S0.5 value in the presence or absence of these cofactors. In agreement with this idea, ADP was a poor inhibitor with only 35% inhibition at 3000 μM (est. IC50 ≈ 4.5 mM, Table 2). The v vs [S] curves obtained varying UTP or glutamine were also unaffected by NADH or 1MDHN. Conversely, when titrated in the presence of ATP, UTP, or glutamine substrates at their S0.5 values, the NADH dose–response curves were identical to those with saturating substrates (Figure S5). These data demonstrate that nicotinamide inhibition is noncompetitive with ATP, UTP, and glutamine.
NADH, CTP, and GTP Mutually Increase Each Other’s Potency
To probe potential on-enzyme interactions between inhibitors, we titrated CTP synthesis reactions with each inhibitor in the presence of the others near their IC50 values (Table 3). In the presence of 400 μM CTP, the shape of the NADH dose–response curve became more concave and the apparent NADH inhibition potency increased ~2.1-fold with an IC50 of 220 μM (Figure 5a). Similarly, in the presence of 500 μM NADH, CTP is a 2.3-fold more potent feedback inhibitor (IC50 = 160 μM) (Table 3, Figure 5b). Thus, the two apparently increase each other’s inhibitory activity in a mutual fashion. Similar behavior was observed with 1MDHN and CTP: 140 μM 1MDHN increased CTP inhibition potency 1.8- fold, and 400 μM CTP increased 1MDHN potency more than 2-fold (Table 3). Although NAD+ was a weaker inhibitor, at its IC50, CTP efficacy increased similarly as with NADH (Table 3), suggesting that both oxidized and reduced forms inhibit by similar mechanisms.
Figure 5.
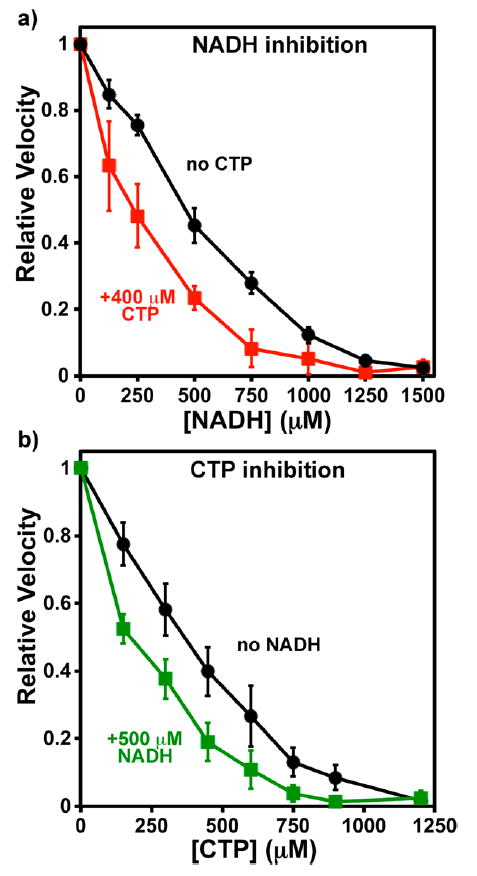
Interactions between NADH and CTP. Error bars indicate the standard deviation of the indicated number of experiments after normalizing to V0 equal to one. IC50 values are given in Table 3. (a) Relative CTP synthesis velocities with 0–1500 μM NADH in the absence (black circles) or presence (red squares) of 400 μM CTP. The average V0 values were 1.23 ± 0.05 (n = 6) and 0.61 ± 0.05 μM s−1 (n = 4), respectively. (b) Relative velocities with 0–1200 μM CTP in the absence (black circles) or presence (green squares) of 500 μM NADH. The average V0 values were 1.31 ± 0.30 (n = 6) and 0.66 ± 0.01 μM s−1 (n = 4), respectively.
GTP is an activator at low concentrations (EC50 ≈ 38 μM, maximum activation at ~200 μM) and an inhibitor at concentrations above 250 μM, with an IC50 of 540 μM (Table 1, Figure 6b). As with CTP, NADH inhibition was enhanced by GTP (Figure 6a). NADH inhibitory strength increased with increasing GTP over both activating and inhibiting concentrations. From 50 to 500 μM GTP, the NADH IC50 value decreased from 770 to 240 μM. Complementarily, 500 μM NADH altered the GTP concentration dependence, steepening both the activation and inhibition portions of the curve (EC50 = 24 μM, IC50 = 350 μM) (Figure 6b) and shifting the optimal GTP concentration to approximately 100 μM. At its IC50, 1MDHN had a similar effect on the GTP dose–response of EcCTPS activity as NADH (Table 3). Therefore, GTP and NADH also apparently increase each other’s activities.
Since NADH amplified the effects of both CTP and GTP, we investigated whether these two inhibitors also act collaboratively. Indeed, the presence of one modifies the concentration dependence of the other. As GTP concentration increased from 50 to 500 μM, the CTP IC50 decreased from 620 to 220 μM (Figure 6c). In turn, and similar to NADH, 400 μM CTP shifted the GTP EC50 and IC50 values to lower concentrations, from 38 μM and 540 μM to 21 μM and 410 μM, respectively (Figure 6d). The data suggest that NADH, CTP, and GTP are mutually synergistic (See Discussion). Plots of 1/IC50 versus GTP concentration yielded a straight lines with nonzero intercepts at 0 μM GTP corresponding to 1080 and 860 μM for NADH and CTP, respectively (R = 0.995). These results imply that both inhibitors can bind to EcCTPS without GTP, albeit with lower affinity.
Testing the Role of the Inhibited Filament in Inhibitor Interactions
This mutual enhancement of CTP, GTP, and NADH effects suggests that these molecules interact with the same conformation of the enzyme. One possibility was that NADH and GTP stabilize the CTP-bound inhibitory filament described by Barry et al.31 In that work, we demonstrated that the presence of CTP increased the self-inhibition of the enzyme at high concentrations, presumably by selectively binding the filament conformation over the free tetramer. Likewise, CTP was a more effective inhibitor at high enzyme concentrations that favor filament formation. We also demonstrated that a mutation that compromised filament formation, E277R, also reduced CTP inhibitory potency.
To test the filament role in NADH and GTP inhibition, we compared the activities of wild-type and E277R EcCTPS at different NADH and GTP concentrations. The NADH dose–response showed no significant differences between wild-type and mutant (Figure 7a). For GTP, the E277R dose–response curve was shifted toward higher concentrations, particularly for the inhibitory portion (Figure 7b). Furthermore, at 4 μM EcCTPS, a concentration at which filament formation reduced activity by ~50%,31 the dose–response was shifted toward lower concentrations (Figure 7b). Interestingly, EcCTPS selfinhibition from filament formation was magnified by increasing GTP from 50 to 500 μM. (Figure 7c). Taken together, these data suggest that some GTP interactions with EcCTPS, unlike those of NADH, are linked to protein–protein interactions within the inhibitory filament.
DISCUSSION
Central metabolic pathways are commonly regulated by intermediates related to their products or precursors; for example, EcCTPS is inhibited by its product CTP (Figure 1) and activated by ATP and UTP substrates. Metabolites may also regulate multiple pathways through “cross-talk”; for example, GTP allosteric regulation of EcCTPS. “Reporter metabolites” globally communicate cellular metabolite and energy status to the metabolic network, which integrates their inputs to optimize pathway outputs over a wide range of conditions.45 Indeed, nicotinamides epitomize this role. In bacteria, conserved Rex repressors link carbon and energy metabolism to redox state,46,47 as reported by NADH/NAD+ levels and ratio, thereby controlling operons involved in anaerobic respiration, fermentation, and amino acid metabolism. Nicotinamides also modulate some enzyme activities, either directly through substrate availability (oxidases, dehydrogenases, histone deacetylases) or much less commonly through allosteric regulatory interactions48 (citrate synthase, phosphorylase, phosphoribulokinase, and pyruvate kinase49-52). Nicotinamide regulatory targets control both shortterm cellular responses, such as metabolic pathway and operon outputs, and long-term ones, like epigenetic modifications. Our data point to pyrimidine biosynthesis as a potential nicotinamide-regulated pathway.
Nicotinamides inhibit EcCTPS with moderate affinities, with reduced forms having higher inhibitory potency than oxidized forms. Inhibition is noncompetitive with substrates but increases the effectiveness of both product inhibitor CTP and allosteric modifier GTP. We further showed that CTP and GTP also amplify each other’s potencies. Together, the data suggest that all three molecules inhibit EcCTPS via a common enzyme conformation. The mutual binding enhancements by these metabolites may shift nicotinamide dose–responses into concentration ranges that can contribute to cellular EcCTPS regulation.
The primary inhibitory determinant is the dihydronicotinamide moiety, since the cofactor analog 1MDHN is more potent than the complete cofactor. The dihydronicotiamide ring may provide a framework for discovery of novel EcCTPS inhibitors. Although the affinity is modest, the N1 atom is readily derivatized allowing for the synthesis and evaluation of a number of analogs. Unlike 1MDHN, the oxidized analog 1MeN binds even less tightly than NAD+ and NADP+ suggesting that the ribo-dinucleotide moiety contributes significantly to their binding. The ADP portion is only a moderate contributor, with ADP inhibiting less well than even 1MeN.
Where are the nicotinamide binding sites? NAD+ and 1MDHN enhance EcCTPS inhibition by CTP similarly to NADH, supporting the idea that NADH, NAD+, and 1MDHN induce similar changes in EcCTPS structure, likely by binding to the same site. While the most obvious candidate is the ATP binding site, lack of comparable inhibition by ADP, unchanged v vs [ATP] behavior in the presence of NAD+ and NADH, and ATP-insensitive NADH dose–response all argue against this idea. The UTP site is also ruled out since NADH inhibition is noncompetitive with UTP and subsaturating UTP does not alter the NADH dose–response.
Another obvious candidate is the GTP inhibitory site. Indeed, NADH inhibition resembles that of guanosine and other purines.44,53 Like guanosine, NADH inhibits ammoniadependent synthesis (Figure S6) and similarly shifts both GTP activation and inhibition curves10,44 (Figure 6e, compare to Figure 4 in ref 44). Potentiation of GTP dual effects is diagnostic for mutual and nonexclusive binding in noncooperative systems. Although an explicit model is difficult to construct, apparent cooperative binding of NADH and GTP could be exerted allosterically through binding to the same sites on different subunits. A complication is that the locations of the GTP activation and inhibition sites and whether they are distinct are unknown. Although structural comparisons with GTP-binding proteins are suggestive,2 we have been unable to locate the bona fide GTP binding site(s) using crystallography. Soaking EcCTPS crystals in 100 mM GTP loads GTP into the ATP site (James Endrizzi and E.B., unpublished data, Figure S7). However, 500 μM GTP does not significantly modify the UTP or ATP S0.5 values, arguing against GTP occupying substrate sites in solution.
The apparent synergism between NADH, CTP, and GTP suggest potential coupling between their binding sites. While mild enhancement of inhibitory potency can result simply from coresidency of noninteracting ligands on the same enzyme particle,54 the more extensively altered dose–response curve shapes and increased potencies provide clear evidence that binding by one molecule exerts positive cooperativity for the binding the others. Interestingly, the effects of GTP at suboptimal (50 μM), optimal (200 μM), and inhibitory (500 μM) concentrations are remarkably similar on both CTP and NADH (compare Figure 6, panels a and c), where the IC50 values show a similar proportional dependence on GTP concentration (see Results). In turn, CTP and NADH have very similar effects on GTP concentration dependences (Figure 6b,d,e).
The mutual effects of NADH, CTP, and GTP are most straightforwardly rationalized by all three inhibitors binding preferentially to the same inhibited enzyme conformation (Figure 1). This conformation is likely accessed by the free enzyme since none of the effectors seem to require the others in order to inhibit. In aggregate, the data implicate separate binding sites for the three inhibitors: lack of CTP site binding by GTP, inability of GTP and NADH to promote filament formation, and CTP and NADH enhancement of both GTP activation and inhibition provide some evidence for “synergy” that is nonexclusive, cooperative binding.54 However, the complexity and cooperativity of the individual binding isotherms (see next paragraph) make it challenging to distinguish between true synergy, which by definition excludes synergists from binding to the same site even on different subunits,54 and cooperative additivity (see above) using the data at hand. Practically, this would require extensive analysis of second inhibitor effects at multiple degrees of inhibition by the first, which is beyond the scope of this work.
EcCTPS exhibits uncommon inhibitor dose–responses, hinting at an unusual regulatory strategy and complex patterns of interactions between binding sites. Unlike more typical hyperbolic or sigmoidal responses to ligands, CTP, nicotinamide, and GTP inhibition curves were very linear, were not fit well by typical logit binding models, and yielded nonlinear Hill plots. Although much has been written concerning the deficiencies of Hill plots,40,55,56 they can be useful for phenomenological characterization, such as in our velocity titrations with UTP and ATP (Table 1). Hill plot curvature can result when there is not a smooth change in binding interactions when successive ligands are bound, that is, uneven cooperativity.40 Although testing explicit models for ligand binding site–site interactions within EcCTPS tetramers is beyond the resolution of our data, we applied the sequential interaction model of Koshland and co-workers to gain more insight into the unusual dose–response behavior.40 This model treats all four sites as spatially equivalent and is specified by four independent equilibrium dissociation constants that represent transitions between species with different numbers of bound ligands (see Materials and Methods and Figure 4a).
While low data/parameter ratios precluded convergent solutions in the sequential binding model, the four dissociation constants for CTP, NADH, NADPH, and 1MDHN inhibitions at activating amounts of GTP (50 or 200 μM, Figures 4, 5a,b, and 6a,c) displayed similar trends. Dissociation constant ratios, which indicate cooperativities for each successive binding step, exhibited a consistent but uneven pattern. Binding the second ligand was anticooperative, binding the third was somewhat easier, and binding the final one is essentially concerted with the third (K2/K1 ≈ 4–40, K3/K2 ≈ 0.2–0.8, and K4/K3 ≈ 10−2–10−5). When two ligands were present, as for dose–responses of CTP inhibition with NADH, 1MDHN, NAD+, or GTP (Figures 5b and 6c, Table 3) and NADH inhibition with CTP or GTP (Figures 5a and 6a), the ratios shifted so that the third binding step became highly anticooperative (K2/K1 ≈ 0.4–0.8, K3/K2 ≈ 40–250, and K4/K3 ≈ 10−4–10−5). These results suggest that to attain a nearly linear dose–response, EcCTPS has evolved a complex pattern of intersubunit interactions, precluding straightforward and detailed mechanistic explanations for apparent inhibitor synergy and binding cooperativity. Nonetheless, the similar dissociation constant ratio patterns for both inhibitors alone or in the presence of another inhibitor strongly support the idea that all interact with the same conformation of the enzyme and induce similar functional and structural changes.
The recently described CTP-inhibited EcCTPS filaments31 might have provided an intriguing global explanation for apparent synergism: that the inhibited filaments preferentially bind not only CTP but also NADH and GTP. GTP is a less effective inhibitor of E277R, while increased protein concentration, which favors filament formation, enhances GTP inhibition (Figure 7b). Protein-dependent inhibition is potentiated by increasing GTP (Figure 7c). GTP behavior mirrors that of CTP31 and suggests that it also preferentially binds the filament. However, GTP has not been observed in high-resolution cryoEM reconstructions of EcCTPS filaments (Justin Kollman, personal communication). In contrast to GTP, NADH inhibition is not affected by the filament-disrupting E277R mutation (Figure 7a) indicating it does not bind preferentially to the filament.
The minimal scheme for EcCTPS regulation is depicted in Figure 1. Active or free enzymes are sequestered from the CTP synthesis pathway by inhibitor binding. The inhibited tetramer is in equilibrium with the inhibited filament. CTP and GTP preferentially bind the filament over the inhibited tetramer, while nicotinamides bind the inhibited tetramer preferentially or equally to the filament and do not significantly affect this conformational equilibrium.
Our experiments uncovered previously undocumented inhibitory interactions between CTP and GTP. This cooperation provides a further mechanism by which EcCTPS can finetune its responses to the outputs of both the GTP and CTP synthesis pathways. In human cells, the key enzyme in GTP synthesis, IMP dehydrogenase, colocalizes with CTPS1, implying that the intracellular regulatory balance between these two nucleotides is fundamentally important.57
What is the physiological relevance of EcCTPS nicotinamide inhibition? It might be advantageous to minimize CTPS activity under conditions where CTP, GTP, or NADH/NADPH accumulate, that is, high anabolite levels and low energy and biosynthesis expenditures. However, uncertainties in free intracellular concentrations and the complex network of interactions between inhibitors make it difficult to assess the contribution of nicotinamide levels and redox state to EcCTPS output in vivo. The inhibitory concentrations determined here in vitro are high compared to some previous estimates of absolute intracellular log-phase concentrations.4,5 However, such measurements are variable and their accuracy is difficult to assess. A recent estimate for total (free and bound) metabolites in glucose-grown, log phase E. coli for NADH, NADPH, CTP, and GTP are 83 μM, 120 μM, 2 mM, and 6 mM, respectively.3 Using respiratory substrates, glycerol and acetate, the average concentrations of NADH and NADPH are even higher (130 μM and 290 μM, respectively), ranging up to 850 μM and 1.7 mM (Supplementary Table 3 from ref 3). These nicotinamide levels in conjunction with saturating concentrations of CTP, GTP or both, may exert significant influence on EcCTPS activity. The higher intracellular NADPH concentrations over NADH could indicate that the phosphorylated form may be the primary effector. Further, while reduced cofactors are more potent inhibitors, we should take care in understating the impact of weaker inhibition by oxidized cofactor. Total NAD+ cellular concentrations are high in glucose, glycerol, or acetate grown cells (average 2.6 mM, 4.1 mM, and 2.4 mM, respectively, ranging 1.3–13 mM), well within the range expected to effect significant inhibition. On the other hand, NADP+ concentrations are very low (2 μM), much lower than its IC50.
We described inhibition of EcCTPS by redox cofactors that hints of crosstalk between the cellular redox state and pyrimidine biosynthesis. This work further contributes to the emerging view of CTPSs as a regulatory node, subject to a diverse set of inputs that influence complex conformational equilibria in order to precisely balance intracellular nucleotide levels, much like the regulation of nitrogen metabolism by glutamine synthetases.
Supplementary Material
Acknowledgments
The authors thank Chris Fraser and Irwin Segel (U.C. Davis) for helpful discussions.
Funding
This work was funded in part by NIH Grants R01GM63109 and R01GM97073 (E.P.B), as well as 5RO1GM107384 (Z.G), the Human Frontier Science Program (Z.G. and J.K.) and the Department of Molecular and Cellular Biology at UC Davis (E.B.).
ABBREVIATIONS
- EcCTPS
Escherichia coli CTP synthetase
- ATP
adenosine 5’-triphosphate
- ADP
adenosine 5’-diphosphate
- UTP
uridine-5’- triphosphate
- CTP
cytidine-5’-triphosphate
- GTP
guanosine-5’-triphosphate
- NADH
1,4 dihydronicotinamide adenine dinucleotide (reduced)
- NAD+
nicotinamide adenine dinucleotide (oxidized)
- NADPH
1,4 dihydronicotinamide adenine dinucleotide 2’-phosphate (reduced)
- NADP+
nicotinamide adenine dinucleotide 2’-phosphate (oxidized)
- 1MDHN
1- methyl 1,4-dihydronicotinamide
- 1MeN
1-methyl nicotinamide
- IPTG
isopropylthiogalactoside
- EDTA
ethylene diamine tetraacetic acid
- NTA
nitrilo-triacetic acid
- Vmax
maximum enzyme velocity under given conditions
- V0
uninhibited enzyme velocity
- kcat
apparent rate constant for the enzyme catalyzed reaction (V/[E])
- S0.5
substrate concentration required to achieve one-half Vmax
- IC50
inhibitor concentration that reduces enzyme velocity to one-half Vmax
- EC50
activator concentration required to achieve one-half Vmax
- nH
Hill number
Footnotes
The authors declare no competing financial interest.
Supporting Information
- Dependence of EcCTPS activity on NaCl concentration, time course of EcCTPS activity with preincubation at 21 °C, representative kinetic data for UTP and ATP concentration dependence of EcCTPS velocity and apparent kcat, independence of NADH inhibition on enzyme activity, independence of NADH inhibition on ATP, UTP and glutamine concentrations, NADH inhibits ammonia-dependent CTP synthesis, high GTP binds to the ATP site in EcCTPS crystals, and small increase in EcCTPS specific activity at concentrations greater than 2 μM in 21 °C preincubation step (PDF)
References
- 1.Lieberman I. Enzymatic amination of uridine triphosphate to cytidine triphosphate. J Biol Chem. 1956;222:765–775. [PubMed] [Google Scholar]
- 2.Endrizzi JA, Kim H, Anderson PM, Baldwin EP. Crystal structure of Escherichia coli cytidine triphosphate synthetase, a nucleotide-regulated glutamine amidotransferase/ATP-dependent amidoligase fusion protein and homologue of anticancer and antiparasitic drug targets. Biochemistry. 2004;43:6447–6463. doi: 10.1021/bi0496945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 6.Chang YF, Carman GM. CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae. Prog Lipid Res. 2008;47:333–339. doi: 10.1016/j.plipres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto M, Omi R, Nakagawa N, Miyahara I, Hirotsu K. Crystal structures of CTP synthetase reveal ATP, UTP, and glutamine binding sites. Structure. 2004;12:1413–1423. doi: 10.1016/j.str.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Higgins MJ, Graves PR, Graves LM. Regulation of human cytidine triphosphate synthetase 1 by glycogen synthase kinase 3. J Biol Chem. 2007;282:29493–29503. doi: 10.1074/jbc.M703948200. [DOI] [PubMed] [Google Scholar]
- 9.Kassel KM, Au DR, Higgins MJ, Hines M, Graves LM. Regulation of human cytidine triphosphate synthetase 2 by phosphorylation. J Biol Chem. 2010;285:33727–33736. doi: 10.1074/jbc.M110.178566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacDonnell JE, Lunn FA, Bearne SL. Inhibition of E. coli CTP synthase by the “positive” allosteric effector GTP. Biochim Biophys Acta, Proteins Proteomics. 2004;1699:213–220. doi: 10.1016/j.bbapap.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Mori G, Chiarelli LR, Esposito M, Makarov V, Bellinzoni M, Hartkoorn M, Degiacomi G, Boldrin F, Ekins S, de Jesus Lopes Ribeiro AL, Marino LB, Centarova I, Svetlikova Z, Blasko J, Kazakova E, Lepioshkin A, Barilone N, Zanoni G, Porta A, Fondi M, Fani R, Baulard AR, Mikusova K, Alzari PM, Manganelli R, de Carvalho LP, Riccardi G, Cole ST, Pasca MR. Thiophenecarboxamide Derivatives Activated by EthA Kill Mycobacterium tuberculosis by Inhibiting the CTP Synthetase PyrG. Chem Biol (Oxford, U K) 2015;22:917–927. doi: 10.1016/j.chembiol.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofer A, Steverding D, Chabes A, Brun R, Thelander L. Trypanosoma brucei CTP synthetase: a target for the treatment of African sleeping sickness. Proc Natl Acad Sci U S A. 2001;98:6412–6416. doi: 10.1073/pnas.111139498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams JC, Kizaki H, Weber G, Morris HP. Increased CTP synthetase activity in cancer cells. Nature. 1978;271:71–73. doi: 10.1038/271071a0. [DOI] [PubMed] [Google Scholar]
- 14.Kursula P, Flodin S, Ehn M, Hammarstrom M, Schuler H, Nordlund P, Stenmark P. Structure of the synthetase domain of human CTP synthetase, a target for anticancer therapy. Acta Crystallogr, Sect F: Struct Biol Cryst Commun. 2006;62:613–617. doi: 10.1107/S1744309106018136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauritsen I, Willemoes M, Jensen KF, Johansson E, Harris P. Structure of the dimeric form of CTP synthase from Sulfolobus solfataricus. Acta Crystallogr, Sect F: Struct Biol Cryst Commun. 2011;67:201–208. doi: 10.1107/S1744309110052334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis DA, Villafranca JJ. Investigation of the mechanism of CTP synthetase using rapid quench and isotope partitioning methods. Biochemistry. 1989;28:8454–8459. doi: 10.1021/bi00447a027. [DOI] [PubMed] [Google Scholar]
- 17.von der Saal W, Anderson PM, Villafranca JJ. Mechanistic investigations of Escherichia coli cytidine-5’-triphosphate synthetase. Detection of an intermediate by positional isotope exchange experiments. J Biol Chem. 1985;260:14993–14997. [PubMed] [Google Scholar]
- 18.Long CW, Levitzki A, Koshland DE., Jr The subunit structure and subunit interactions of cytidine triphosphate synthetase. J Biol Chem. 1970;245:80–87. [PubMed] [Google Scholar]
- 19.Anderson PM. CTP synthetase from Escherichia coli: an improved purification procedure and characterization of hysteretic and enzyme concentration effects on kinetic properties. Biochemistry. 1983;22:3285–3292. doi: 10.1021/bi00282a038. [DOI] [PubMed] [Google Scholar]
- 20.Thomas PE, Lamb BJ, Chu EH. Purification of cytidine-triphosphate synthetase from rat liver, and demonstration of monomer, dimer and tetramer. Biochim Biophys Acta, Protein Struct Mol Enzymol. 1988;953:334–344. doi: 10.1016/0167-4838(88)90042-8. [DOI] [PubMed] [Google Scholar]
- 21.Pappas A, Yang WL, Park TS, Carman GM. Nucleotide-dependent tetramerization of CTP synthetase from Saccharomyces cerevisiae. J Biol Chem. 1998;273:15954–15960. doi: 10.1074/jbc.273.26.15954. [DOI] [PubMed] [Google Scholar]
- 22.Lunn FA, Macleod TJ, Bearne SL. Mutational analysis of conserved glycine residues 142, 143 and 146 reveals Gly(142) is critical for tetramerization of CTP synthase from Escherichia coli. Biochem J. 2008;412:113–121. doi: 10.1042/BJ20071163. [DOI] [PubMed] [Google Scholar]
- 23.Endrizzi JA, Kim H, Anderson PM, Baldwin EP. Mechanisms of product feedback regulation and drug resistance in cytidine triphosphate synthetases from the structure of a CTP-inhibited complex. Biochemistry. 2005;44:13491–13499. doi: 10.1021/bi051282o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi MG, Park TS, Carman GM. Phosphorylation of Saccharomyces cerevisiae CTP synthetase at Ser424 by protein kinases A and C regulates phosphatidylcholine synthesis by the CDP-choline pathway. J Biol Chem. 2003;278:23610–23616. doi: 10.1074/jbc.M303337200. [DOI] [PubMed] [Google Scholar]
- 25.Han GS, Sreenivas A, Choi MG, Chang YF, Martin SS, Baldwin EP, Carman GM. Expression of Human CTP synthetase in Saccharomyces cerevisiae reveals phosphorylation by protein kinase A. J Biol Chem. 2005;280:38328–38336. doi: 10.1074/jbc.M509622200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park TS, O’Brien DJ, Carman GM. Phosphorylation of CTP synthetase on Ser36, Ser330, Ser354, and Ser454 regulates the levels of CTP and phosphatidylcholine synthesis in Saccharomyces cerevisiae. J Biol Chem. 2003;278:20785–20794. doi: 10.1074/jbc.M301394200. [DOI] [PubMed] [Google Scholar]
- 27.Carcamo WC, Satoh M, Kasahara H, Terada N, Hamazaki T, Chan JY, Yao B, Tamayo S, Covini G, von Muhlen CA, Chan EK. Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS One. 2011;6:e29690. doi: 10.1371/journal.pone.0029690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu JL. Intracellular compartmentation of CTP synthase in Drosophila. J Genet Genomics. 2010;37:281–296. doi: 10.1016/S1673-8527(09)60046-1. [DOI] [PubMed] [Google Scholar]
- 30.Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010;190:541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barry RM, Bitbol AF, Lorestani A, Charles EJ, Habrian CH, Hansen JM, Li HJ, Baldwin EP, Wingreen NS, Kollman JM, Gitai Z. Large-scale filament formation inhibits the activity of CTP synthetase. eLife. 2014;3:e03638. doi: 10.7554/eLife.03638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calise SJ, Carcamo WC, Krueger C, Yin JD, Purich DL, Chan EK. Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cell Mol Life Sci. 2014;71:2963–2973. doi: 10.1007/s00018-014-1567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aughey GN, Grice SJ, Shen QJ, Xu Y, Chang CC, Azzam G, Wang PY, Freeman-Mills L, Pai LM, Sung LY, Yan J, Liu JL. Nucleotide synthesis is regulated by cytoophidium formation during neurodevelopment and adaptive metabolism. Biol Open. 2014;3:1045–1056. doi: 10.1242/bio.201410165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noree C, Monfort E, Shiau AK, Wilhelm JE. Common regulatory control of CTP synthase enzyme activity and filament formation. Mol Biol Cell. 2014;25:2282–2290. doi: 10.1091/mbc.E14-04-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levitzki A, Koshland DE., Jr Ligand-induced dimer-to-tetramer transformation in cytosine triphosphate synthetase. Biochemistry. 1972;11:247–253. doi: 10.1021/bi00752a016. [DOI] [PubMed] [Google Scholar]
- 36.Ortiz-Maldonado M, Gatti D, Ballou DP, Massey V. Structure-function correlations of the reaction of reduced nicotinamide analogues with p-hydroxybenzoate hydroxylase substituted with a series of 8-substituted flavins. Biochemistry. 1999;38:16636–16647. doi: 10.1021/bi991603u. [DOI] [PubMed] [Google Scholar]
- 37.Wang SY. Photochemistry of N1-methylnicotinamide salts. Biochemistry. 1968;7:3740–3744. doi: 10.1021/bi00850a055. [DOI] [PubMed] [Google Scholar]
- 38.Robertson JG, Villafranca JJ. Characterization of metal ion activation and inhibition of CTP synthetase. Biochemistry. 1993;32:3769–3777. doi: 10.1021/bi00065a032. [DOI] [PubMed] [Google Scholar]
- 39.Long CW, Pardee AB. Cytidine triphosphate synthetase of Escherichia coli B. I. Purification and kinetics. J Biol Chem. 1967;242:4715–4721. [PubMed] [Google Scholar]
- 40.Cornish-Bowden A, Koshland DE., Jr Diagnostic uses of the Hill (Logit and Nernst) plots. J Mol Biol. 1975;95:201–212. doi: 10.1016/0022-2836(75)90390-3. [DOI] [PubMed] [Google Scholar]
- 41.Levitzki A, Stallcup WB, Koshland DE., Jr Half-of-the-sites reactivity and the conformational states of cytidine triphosphate synthetase. Biochemistry. 1971;10:3371–3378. doi: 10.1021/bi00794a009. [DOI] [PubMed] [Google Scholar]
- 42.MacLeod TJ, Lunn FA, Bearne SL. The role of lysine residues 297 and 306 in nucleoside triphosphate regulation of E. coli CTP synthase: inactivation by 2’,3’-dialdehyde ATP and mutational analyses. Biochim Biophys Acta, Proteins Proteomics. 2006;1764:199–210. doi: 10.1016/j.bbapap.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 43.Robertson JG. Determination of subunit dissociation constants in native and inactivated CTP synthetase by sedimentation equilibrium. Biochemistry. 1995;34:7533–7541. doi: 10.1021/bi00022a029. [DOI] [PubMed] [Google Scholar]
- 44.Lunn FA, MacDonnell JE, Bearne SL. Structural requirements for the activation of Escherichia coli CTP synthase by the allosteric effector GTP are stringent, but requirements for inhibition are lax. J Biol Chem. 2008;283:2010–2020. doi: 10.1074/jbc.M707803200. [DOI] [PubMed] [Google Scholar]
- 45.Gruning NM, Lehrach H, Ralser M. Regulatory crosstalk of the metabolic network. Trends Biochem Sci. 2010;35:220–227. doi: 10.1016/j.tibs.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Brekasis D, Paget MS. A novel sensor of NADH/ NAD+ redox poise in Streptomyces coelicolor A3(2) EMBO J. 2003;22:4856–4865. doi: 10.1093/emboj/cdg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravcheev DA, Li X, Latif H, Zengler K, Leyn SA, Korostelev YD, Kazakov AE, Novichkov PS, Osterman AL, Rodionov DA. Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex. J Bacteriol. 2012;194:1145–1157. doi: 10.1128/JB.06412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Sohngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duckworth HW, Anderson DH, Bell AW, Donald LJ, Chu AL, Brayer GD. Structural basis for regulation in gram-negative bacterial citrate synthases. Biochem Soc Symp. 1987;54:83–92. [PubMed] [Google Scholar]
- 50.Jenkins JA, Johnson LN, Stuart DI, Stura EA, Wilson KS, Zanotti G, Wilkie DR. Phosphorylase: control and activity. Philos Trans R Soc, B. 1981;293:23–41. doi: 10.1098/rstb.1981.0057. [DOI] [PubMed] [Google Scholar]
- 51.Runquist JA, Narasimhan C, Wolff CE, Koteiche HA, Miziorko HM. Rhodobacter sphaeroides phosphor-ibulokinase: binary and ternary complexes with nucleotide substrate analogs and effectors. Biochemistry. 1996;35:15049–15056. doi: 10.1021/bi9619334. [DOI] [PubMed] [Google Scholar]
- 52.Devin A, Nogueira V, Leverve X, Guerin B, Rigoulet M. Allosteric activation of pyruvate kinase via NAD+ in rat liver cells. Eur J Biochem. 2001;268:3943–3949. doi: 10.1046/j.1432-1327.2001.02306.x. [DOI] [PubMed] [Google Scholar]
- 53.Roy AC, Lunn FA, Bearne SL. Inhibition of CTP synthase from Escherichia coli by xanthines and uric acids. Bioorg Med Chem Lett. 2010;20:141–144. doi: 10.1016/j.bmcl.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 54.Breitinger H-G. Drug Synergy – Mechanisms and Methods of Analysis. In: Acree W, editor. Toxicity and Drug Testing. InTech; Rijeka, Croatia: 2012. pp. 143–166. [DOI] [Google Scholar]
- 55.Bindslev N. Drug-Acceptor Interactions: Modeling Theoretical Tools to Test and Evaluate Experimental Equilibrium Effects. Co-Action Publishing; Järfälla, Sweden: 2008. [Google Scholar]
- 56.Ricard J, Cornish-Bowden A. Co-operative and allosteric enzymes: 20 years on. Eur J Biochem. 1987;166:255–272. doi: 10.1111/j.1432-1033.1987.tb13510.x. [DOI] [PubMed] [Google Scholar]
- 57.Carcamo WC, Calise SJ, von Muhlen CA, Satoh M, Chan EK. Molecular cell biology and immunobiology of mammalian rod/ring structures. Int Rev Cell Mol Biol. 2014;308:35–74. doi: 10.1016/B978-0-12-800097-7.00002-6. [DOI] [PubMed] [Google Scholar]
- 58.Iyengar A, Bearne SL. An assay for cytidine 5(’)-triphosphate synthetase glutaminase activity using high performance liquid chromatography. Anal Biochem. 2002;308:396–400. doi: 10.1016/s0003-2697(02)00240-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


