Abstract
Twenty AdoMet-dependent methyltransferases (MTases) have been characterized structurally by X-ray crystallography and NMR. These include seven DNA MTases, five RNA MTases, four protein MTases and four small molecule MTases acting on the carbon, oxygen or nitrogen atoms of their substrates. The MTases share a common core structure of a mixed seven-stranded β-sheet (6↓ 7↑ 5↓ 4↓ 1↓ 2↓ 3↓) referred to as an ‘AdoMet-dependent MTase fold’, with the exception of a protein arginine MTase which contains a compact consensus fold lacking the antiparallel hairpin strands (6↓ 7↑). The consensus fold is useful to identify hypothetical MTases during structural proteomics efforts on unannotated proteins. The same core structure works for very different classes of MTase including those that act on substrates differing in size from small molecules (catechol or glycine) to macromolecules (DNA, RNA and protein). DNA MTases use a ‘base flipping’ mechanism to deliver a specific base within a DNA molecule into a typically concave catalytic pocket. Base flipping involves rotation of backbone bonds in double-stranded DNA to expose an out-of-stack nucleotide, which can then be a substrate for an enzyme-catalyzed chemical reaction. The phenomenon is fully established for DNA MTases and for DNA base excision repair enzymes, and is likely to prove general for enzymes that require access to unpaired, mismatched or damaged nucleotides within base-paired regions in DNA and RNA. Several newly discovered MTase families in eukaryotes (DNA 5mC MTases and protein arginine and lysine MTases) offer new challenges in the MTase field.
INTRODUCTION
Methyl transfers are alkylation reactions central to cellular biochemistry, and S-adenosyl-l-methionine (AdoMet) is by far the most commonly used methyl donor molecule. The AdoMet methyl group is bound to a charged sulfur atom, which thermodynamically destabilizes the molecule and makes the relatively inert methylthiol of the methionine moiety very reactive (1) toward polarizable nucleophiles (N, O and S) and activated C atoms (carbanions). The AdoMet-dependent methyltransferases (MTases) act on a wide variety of target molecules, including DNA, RNA, proteins, polysaccharides, lipids and a range of small molecules. Since 1993, 20 of these enzymes have been characterized structurally, almost all with bound AdoMet or the reaction product S-adenosyl-l-homocysteine (AdoHcy) and some with bound substrates.
In prokaryotes, over 3000 restriction–modification systems have been discovered so far, and they have been found across the full spectrum of known bacterial species, including both eubacteria and archaea (see http://rebase.neb.com/) (2). While the functions and various types of restriction–modification systems are discussed in this issue and elsewhere (for example, reviewed in 3), structures for six type II MTases are currently available: two 5-methylcytosine (5mC), one N4-methylcytosine (N4mC) and three N6-methyladenine (N6mA) MTases (Table 1).
Table 1. Structurally characterized AdoMet-dependent MTases.
| Enzyme |
Source organism |
Target atom |
Enzyme E.C. no. |
PDB |
Reference |
| DNA MTases | |||||
| M.HhaI | Haemophilus haemolyticus | Cytosine-C5 | 2.1.1.73 | 2hmy | 91 |
| M.HaeIII | Haemophilus aegyptius | Cytosine-C5 | 2.1.1.73 | 1dct | 64 |
| M.PvuII | Proteus vulgaris | Cytosine-N4 | 2.1.1.113 | 1boo | 43 |
| M.DpnII | Streptococcus pneumoniae | Adenine-N6 | 2.1.1.72 | 2dpm | 46 |
| M.RsrI | Rhodobacter sphaeroides | Adenine-N6 | 2.1.1.72 | 1eg2 | 44 |
| M.TaqI | Thermus aquaticus | Adenine-N6 | 2.1.2.72 | 2adm | 97,119 |
| DNMT2 | Human | Cytosine-C5 (?) | 2.1.1.37 (?) | 1g55 | 120 |
| Protein MTases | |||||
| CheR | Salmonella typhimurium | Glutamate-O | 2.1.1.80 | 1af7,1bc5 | 121,122 |
| PRMT3 | Rat | Arginine-N | 2.1.1.125 | 1f3l | 25 |
| Hmt1 | Saccharomyces cerevisiae | Arginine-N | 2.1.1.125 | 1g6q | 38 |
| PIMT | Thermus maritima | Isoaspartate-O | 2.1.1.77 | 1dl5 | 26 |
| RNA MTases | |||||
| VP39 | Vaccinia orthopox virus | mRNA nucleoside-2′-O | 2.1.1.57 | 1v39 | 123–126 |
| ErmAM | S.pneumoniae | rRNA adenine-N6 | 2.1.1.48 | 1yub | 127 |
| ErmC′ | Bacillus subtilis | rRNA adenine-N6 | 2.1.1.48 | 2erc | 128,129 |
| Fibrillarin homolog | M.jannaschii | rRNA nucleoside-2′-O (?) | 2.1.1.66 (?) | 1fbn | 29 |
| FTSJ | E.coli | RNA | 2.1.1.? | 1eiz,1ej0 | 130 |
| Small molecule MTases | |||||
| COMT | Rat | Catechol-O | 2.1.1.6 | 1vid | 131 |
| GNMT | Rat | Glycine-N | 2.1.1.20 | 1xva,1d2c | 132,133 |
| ChOMT | Medicago sativa L | Chalcone-O | 2.1.1.? | 1fpq, 1fp1 | 134 |
| IOMT | Medicago sativa L | Isoflavone-O | 2.1.1.? | 1fpx, 1fp2 | 134 |
| Other MTases | |||||
| Mj0882 | M.jannaschii | ? | ? | 1dus | 31 |
| HI0319 | H.influenzae | Small molecule (?) | ? | 1im8 | 32 |
| HI0766 | H.influenzae | tRNA (?) | 2.1.1.34 (?) | 1j85 | 32 |
In mammals, recent publications clearly define, in addition to Dnmt1 (4), two additional distinct phylogenetic 5mC MTase families, Dnmt2 (5–7) and Dnmt3a and Dnmt3b (8). All of the enzymes preserve the same basic organization of the conserved MTase motifs in a C-terminal catalytic domain that resembles the prokaryotic 5mC MTases (reviewed in 9–11). Less is known about the enzymology and structure of the recently described enzymes, but considering that methylation in eukaryotes is found predominantly in the sequence CpG or CpNpG, it is likely that the eukaryotic enzymes have similar sequence specificities. However, embryonic stem cells have significant 5mC residues at CpA and, to a lesser extent, at CpT (12). The unique features of the individual 5mC MTase families are found in the large N-terminal domain that varies in size between the different MTase families and is likely responsible for the regulation of their diverse biological functions—in the normal development of animal (13,14) and plant species (15–17), gene repression (18), X chromosome inactivation (19), genome imprinting (20) and replication timing (21). The tasks of dissecting the functional roles of the N-terminal region of the different 5mC MTase families are just beginning. How genome methylation patterns are established during development and how the altered function of these enzymes contributes to the aberrant methylation changes that accompany embryonic development (13,14) and disease (14,22,23) is presently unknown.
While some proteins exert their effects simply through binding interactions, other proteins both bind to DNA and catalyze chemical reactions. These include polymerases, nucleases, glycosylases, MTases and various integrases and recombinases that rearrange DNA segments. DNA binding frequently deforms the usual B-helix and bending and kinking of DNA is common. Proteins that perform chemistry on the DNA bases have a difficult accessibility problem. This problem was resolved in 1994 by the discovery of ‘base flipping’—when a structure was reported for the ternary complex of the 5mC DNA MTase, M.HhaI, its DNA substrate and the methyl donor AdoMet (24). It was proposed that other classes of DNA MTases and some DNA glycosylases might also use base flipping to gain access to DNA bases (24). Both predictions proved accurate.
The purpose of this review is to provide a brief overview of the known structures of AdoMet-dependent MTases, particularly DNA MTases, and summarize current knowledge about base flipping, including all systems in which it is proven to occur. Finally, we will focus on the available structures of the HhaI MTase that has served as a structural paradigm for DNA MTases and DNA base flipping enzymes.
AN AdoMet-DEPENDENT MTase FOLD
At this time, the structural characterization of 20 AdoMet-dependent MTases has been reported (Table 1). These include DNA MTases (two generating 5mC, one generating N4mC and three generating N6mA), RNA MTases (two closely related enzymes generating N6mA in rRNA, one generating mRNA cap-specific 2′-O-methylribose), protein MTases generating glutamyl carboxymethyl ester, l-isoaspartate methyl ester and methylarginine, and small molecule MTases that act on catechol, glycine, chalcone and isoflavone. Of the known MTase structures, two transfer the methyl group to a carbon atom, six to oxygen atoms and nine to nitrogen atoms (Table 1). The substrates for human DNMT2, Methanococcus jannaschii fibrillarin homolog and Escherichia coli FtsJ are unknown.
A striking feature of all the structures is that they share a common core structure referred to as an ‘AdoMet-dependent MTase fold’ (Figs 1–4). Many of these proteins have domains outside the core structure that play a role in substrate recognition or in separate functions. Central to this shared core structure is a mixed seven-stranded β-sheet (green in Figs 1–4). Strand 7 is antiparallel to the other six strands, and is inserted into the sheet between strands 5 and 6 (6↓ 7↑ 5↓ 4↓ 1↓ 2↓ 3↓); this feature is thus far characteristic of the AdoMet-dependent MTases, with one exception. The protein arginine (R) MTase (PRMT) lacks the anti-parallel β-hairpin (6↓ 7↑) (Fig. 3) (25). In addition, the protein isoaspartyl MTase (PIMT) has the order of strands 6 and 7 reversed in the primary sequence (26). It is possible that the insertion of the antiparallel strand 7 between strands 5 and 6 plays an important functional role in some MTases. In HhaI DNA MTase the loop between strands 6 and 7 stabilizes both the substrate (cytosine) and the DNA-recognition domain. In a simplified version, strands 1–3 form the part of the MTase that interacts with the AdoMet, while strands 4–7, particularly the loop after strand 4, which usually contains the catalytic amino acids, are in the region binding the diverse substrates methylated by these enzymes.
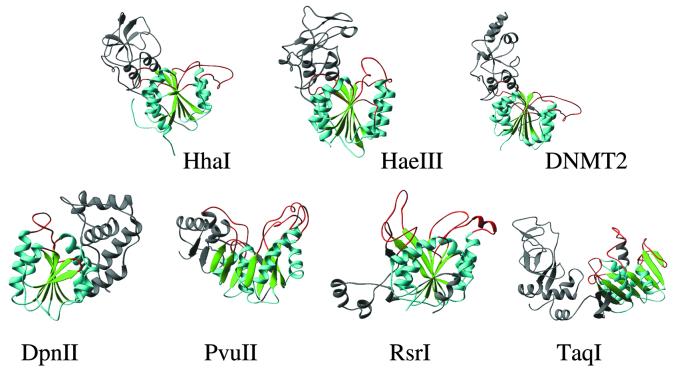
Figure 1.
Examples of DNA MTases (see Table 1). M.HhaI (5mC), M.HaeIII (5mC), human DNMT2, M.DpnII (N6mA, α), M.PvuII (N4mC, β), M.RsrI (N6mA, β) and M.TaqI (N6mA, γ). The AdoMet-dependent MTase fold is colored in green (β strands), cyan (α helices) and red (the loops after the carboxyl ends of β strands). The region(s) outside the MTase fold is colored in gray.
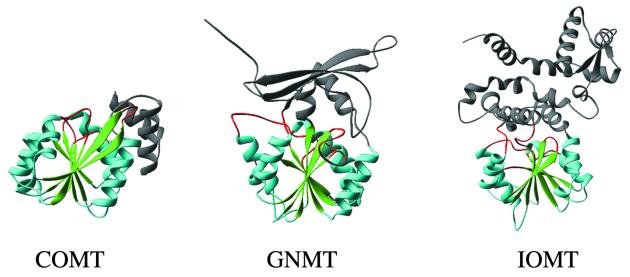
Figure 4.
Examples of small molecule MTases: COMT (catechol-O), GNMT (glycine-N), and IOMT (isoflavone-O) or closely related ChOMT (chalcone-O).
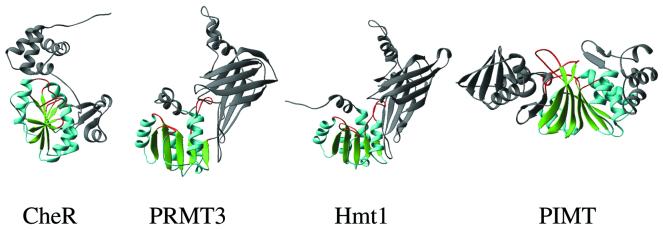
Figure 3.
Examples of protein MTases. CheR (glutamate-O), PRMT3 (arginine-N), Hmt1 (arginine-N) and PIMT (isoaspartate-O).
The high degree of structural similarity among AdoMet-dependent MTases is not reflected by a corresponding degree of sequence conservation (27). In fact, only three positions are highly conserved among the structurally-characterized enzymes (27): a Gly (motif I) in the loop after strand 1, a negatively-charged Asp/Glu (motif II) in the carboxyl end of strand 2 and a hydrophobic Val/Ile/Leu (motif IV) within strand 4. Only one of these (motif I) is obvious from the sequence alone without structural guidance (28), though substantial conservation can exist within a particular subfamily of MTase (defined according to their reactions and substrates, such as the DNA 5mC MTases).
Structural proteomics
While the lack of sequence-level conservation has made it difficult to determine relationships among the various AdoMet-dependent MTases, the high degree of conservation of the MTase fold has served as a prototype to identify hypothetical MTases of unannotated proteins. In one of the first structural proteomics efforts (http://www-kimgrp/lbl.gov/genomics/proteinlist.html), the crystal structure of the fibrillarin homolog from M.jannaschii (Mj0697) revealed a MTase-like C-terminal domain (Fig. 2) (29). Fibrillarin is a phylogenetically conserved protein essential for efficient processing of pre-rRNA through its association with a class of small nucleolar RNAs (snoRNAs) during ribosomal RNA biogenesis (30). A majority of fibrillarin-associated snoRNAs function in rRNA 2′-O-methylation within a base-paired duplex region. In addition, a structure of Mj0882 deposited in PDB (1dus) has the MTase fold (31).
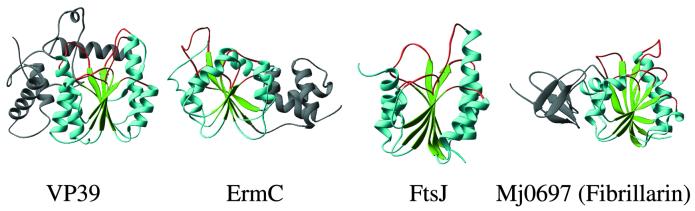
Figure 2.
Examples of RNA MTases. VP39 (mRNA nucleoside-2′-O), ErmC′ or closely related ErmAM (rRNA N6mA), FtsJ and fibrillarin homolog.
Other examples are the two new structures of proteins (HI0319 and HI0766) from Haemophilus influenzae (http://s2f.umbi.umd.edu). HI0319 is homologous to some MTases acting on small molecules and has a core structure with a MTase fold and a bound AdoHcy (32). HI0766 is a truncated version of SpoU, a MTase acting on tRNA (33). It is worth noting that AdoMet concentrations in E.coli were reported to be in the 300–500 µM range (34) and copurification of AdoMet has been noted for many DNA MTases expressed in E.coli (35–37). Bound AdoMet or AdoHcy has been found in almost all (except yeast Hmt1; 38) structurally characterized known MTases, no matter whether extra cofactor was added or not during crystallization.
Circular permutation of DNA MTases
An analysis of the family of DNA 5mC MTases revealed 10 conserved amino acid sequence motifs (39) and a target-recognizing domain(s) (TRD) (40). Nine of these motifs were also found to occur in the three different linear orders (families α, β and γ) in the DNA amino MTases (41). Currently, we have examples for each family (Table 2). The 5mC (M.HhaI and M.HaeIII) and the γ (M.TaqI) MTases differ only in the placement of one helix and its associated conserved motif X (respectively at the C- and N-termini of the protein) (42). In the β MTases M.PvuII and M.RsrI, the N-terminus is just upstream of strand 3 (43,44), which means that the conserved motifs are in a simple permuted order relative to the γ MTases (45). The α MTase M.DpnII has a large insertion (TRD) between strands 2 and 3 (46). Despite the altered linear order of the sequence motifs and secondary structure elements, they are still able to yield the same tertiary structure and maintain the core MTase fold.
Table 2. Circular permutation of DNA MTases.
| Family |
Motif order (linear) |
Strand order (linear) |
Examples |
| α | I (AdoMet)-TRD-IV (DPPY) | 1-2-TRD-3-4-5–6-7 | M.DpnII (N6mA) |
| β | IV (D/SPPY)-TRD-I (AdoMet) | 3-4-5-6-7-TRD-1-2 | M.RsrI (N6mA) and M.PvuII (N4mC) |
| γ | I (AdoMet)-IV (NPPY)-TRD | 1-2-3-4-5-6-7-TRD | M.TaqI (N6mA) |
| 5mC | I (AdoMet)-IV (PC)-TRD | 1-2-3-4-5-6-7-TRD | M.HhaI and M.HaeIII (5mC) |
DNA BASE FLIPPING
It might seem surprising that the same core structure works for a very different class of MTases that act on substrates differing in size from small molecules (catechol or glycine) to protein and DNA. The solution to this apparent puzzle is that macromolecule MTases do not methylate protein or DNA per se, but methylate a specific base within a DNA molecule or a specific amino acid within a protein. This raises the problem of substrate accessibility. How can the MTase deliver the target to its catalytic center? One elegant solution found by the DNA MTases is shown in Figure 5A, and was originally discovered for HhaI MTase (24). In the protein–DNA complex, the target cytosine is no longer buried within the double helix, but has been rotated on its flanking sugar–phosphate bonds so that it projects out into a typically concave catalytic pocket. No covalent bonds were broken to carry out the base flipping process, however, the base pairing hydrogen bonds were broken and the stacking π interactions with the adjacent base pairs were lost.
Figure 5.
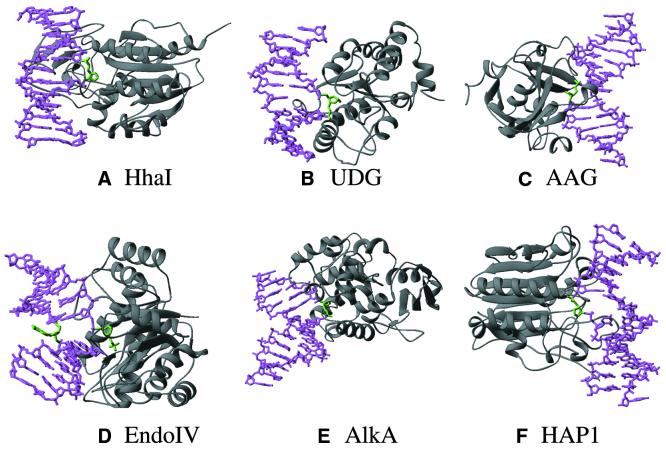
Examples of DNA base flipping proteins (see Table 3) in complex with oligonucleotide containing an abasic site. (A) M.HhaI (a DNA 5mC MTase), (B) human UDG, (C) human AAG, (D) E.coli endonuclease IV, (E) E.coli AlkA and (F) human HAP1. The protein is colored gray, the DNA is represented as a magenta stick model with the flipped abasic site in green, usually buried in a surface pocket in the protein.
There are still a number of things we do not understand about base flipping, including how it is initiated and how (or if) it is related to the recognition of the substrate sequence. Two interesting features are known, however. First, in the structure of M.HhaI with a DNA substrate having an abasic (apurinic/apyrimidinic or AP) site at the position of the target cytosine, the enzyme still moves the sugar–phosphate backbone to the ‘flipped out’ position (Fig. 5A) (47). A similar conformation is also observed for the flipped-out abasic nucleotide in four glycosylase–DNA complexes: uracil DNA glycosylase (UDG) (48) (Fig. 5B), mismatch-specific uracil glycosylase (MUG) (49,50), alkyladenine glycosylase (AAG) (51) (Fig. 5C) and alkylation glycosylase (AlkA) (52) (Fig. 5E). It thus appears that the base per se is not the target for the structural change in the DNA. Since an abasic sugar is flipped by M.HhaI, AAG and AlkA, UDG and MUG, there is clearly nothing special about the cytosine, 3-methyladenine, uracil, thymine or any of the bases that is required for flipping. Thus, we conclude that it is the backbone that is targeted for rotation by the enzyme and the base is merely carried along with it (53).
Secondly, base flipping by M.HhaI will work with guanine and uracil (47) or probably any base at the target position (54,55). The methylation reaction is sensitive to the base at the target position, but the base flipping step is not. This property has been used to measure base flipping because 2-aminopurine fluorescence increases dramatically when it is removed from the stacking environment in double helical DNA (56). Examples include M.EcoRI (57,58), M.TaqI and M.HhaI (59), M.EcoP15I (60) and E.coli UDG (61). However, the practice of this technique needs some precautions. Large changes in 2-aminopurine fluorescence have been observed in M.EcoP15I (60) and M.EcoRV (62) for substrates which do not carry the substitution at the site of methylation/flipping. On the other hand, small or no changes have been observed in M.EcoRV (62) and M.RsrI (63) for substrates which do carry the substitution at the site of flipping/methylation. Therefore, changes in 2-aminopurine fluorescence should not be regarded as definitive proof of base flipping but as evidence for conformational or environmental changes of the DNA which lead to unstacking of the 2-aminopurine base. Base flipping is merely one example of such a change.
Several other examples of base flipping have now been observed in structures of protein–DNA complexes (Table 3). In the structure of M.HaeIII bound to DNA, the substrate cytosine is flipped out from the DNA helix (64), as observed for M.HhaI. The M.HaeIII structure is, so far, unique in having some rearrangement of the bases adjacent to the flipped cytosine. Recently, the structure of a ternary complex between M.TaqI, an adenine-N6-specific DNA MTase, and its two substrates, the specific DNA and a non-reactive AdoMet analog, revealed a flipped adenine (65). However, the three available MTase–DNA complexes use three different means of stabilizing DNA structure with a flipped base (66). The latest and most amazing observations on base flipping came from structure of the 30S ribosomal subunit complexed with the antibiotics paromomycin (67) or initiation factor IF1 (68). Paromomycin binds in the major groove of helix H44 of 16S RNA and flips out the functionally important bases A1492 and A1493; the flipped-out bases point directly into the ribosomal A site and interact with the minor groove of the codon–anticodon duplex (Fig. 6, left). Binding of IF1 occludes the A site and flips out A1492 and A1493 and buries them in pockets in IF1 (Fig. 6, right).
Table 3. Known base-flipping systems.
| Specific protein |
Catalytic reaction |
PDB |
Reference |
| DNA MTases | |||
| M.HhaI | Forms 5-methylcytosine in DNA | 9mht | 24,47 |
| M.HaeIII | Forms 5-methylcytosine in DNA | 1dct | 64 |
| M.TaqI | Forms N6-methyladenine in DNA | 1G38 | 65 |
| DNA glycosylases | |||
| T4 endonuclease V | Removes pyrimidine dimers from DNA | 1vas | 78 |
| Human UDG | Removes uracil from DNA | 2ssp | 48,71,135 |
| E.coli MUG | Removes uracil or thymine from DNA containing G:T or G:U | a | 49,50 |
| Human AAG | Removes 3-methyladenine from DNA | 1bnk | 51 |
| E.coli AlkA | Removes 3-methyladenine from DNA | 1diz | 52 |
| hOGG1 | Remove 8-oxoguanine from DNA | 1ebm | 74 |
| AP endonucleases | |||
| E.coli endonuclease IV | Cleaves the DNA backbone 5′ of AP sites | 1qum | 79 |
| Human AP endonuclease (HAP1 or APE1) | Cleaves 5′ to AP sites | 1dew | 80 |
aThe coordinates of MUG–DNA complexes are currently not available in PDB.
Figure 6.
30S ribosomal subunit flipped-out A1492 and A1493 from helix 44 of 16S RNA by binding of (left) paromomycin (seen in the difference electron density) and (right) initiation factor IF1 (in purple). The protein S12 is in orange, helix H44 in cyan.
DNA repair enzymes
Besides DNA MTases, base flipping has since been found in a variety of DNA repair enzymes (53,69) that function through either the base excision repair pathway (see references in Table 3) or a direct reversal mechanism (70). Some authors refer to this phenomenon as ‘nucleotide flipping’ (51,71). The enzymes in the base excision repair pathway, proved to use base flipping, encompass several lesion-specific DNA N-glycosylases that remove the damaged base by cleaving the glycosylic bond between the base and deoxyribose of the DNA backbone and AP endonucleases that cleaves the phosphodiester backbone 5′ to an AP site.
DNA glycosylases
Human UDG. UDG removes uracil residues from DNA. Base flipping was confirmed by the description of a structure for human UDG complexed with a uracil-containing double-stranded DNA (71): the uracil and deoxyribose are rotated 180° from their starting structure within DNA. Even though the glycosidic bond was cleaved the uracil remains in the binding pocket, while Leu272 penetrates into the DNA helix and occupies the space left by the flipped base. A ‘push and pull’ mechanism for base flipping was thus suggested for human UDG (71). However, Parikh et al. (48) suggested neither ‘push’ (the missing leucine side chain of the L272A enzyme does not affect flipping) nor ‘pull’ (UDG can flip an AP site) is essential for flipping. Stivers et al. (72) suggested an enzyme-assisted active mechanism for uracil flipping by E.coli UDG. Panayotou et al. (73) suggested a passive mechanism for viral UDG in which the enzyme traps a transient extrahelical uracil in the free substrate. Further work is needed to settle the issue.
Escherichia coli MUG. MUG removes pyrimidines from mismatches opposite guanine arising from deamination of cytosine (to uracil) or 5-methylcytosine (to thymine). Structures of E.coli MUG have been obtained for an abasic–DNA product complex (49) and a non-hydrolysable deoxyuridine analog (50). The crystal structures show great similarity to UDG, especially around the active site. In the structure of the native DNA complex, the glycosidic bond of the uracil base has been hydrolyzed and a flipped-out abasic site is left in the enzyme’s active site. In the structure of the modified DNA complex, the non-hydrolyzable substrate analog is flipped out and bound in the base-binding pocket.
HhH DNA glycosylase. The crystal structures of two helix–hairpin–helix DNA glycosylase–DNA complexes have revealed a common structural feature of base flipping. Escherichia coli 3-methyladenine DNA glycosylase II (AlkA) induces a 66° bend in the DNA with a marked widening of the minor groove and flips a 1-azaribose abasic nucleotide out of the DNA (52) (Fig. 5E). Human 8-oxoguanine DNA glycosylase (hOGG1) bends the DNA (∼70°) at the oxoG:C base pair and flips the oxoG into the enzyme active site (74). Other known members of the helix–hairpin–helix DNA glycosylase family—E.coli endonuclease III (75) that removes pyrimidine radiolysis products from DNA, MutY (76) that removes adenine from mispairs with 8-oxoguanine and guanine, and MutM (Fpg) (77) that removes a wide range of oxidatively damaged bases—will likely flip the base in a similar manner to AlkA (52).
Human 3-methyladenine DNA glycosylase. Human 3-methyladenine DNA glycosylase is another structurally unrelated alkylation glycosylase, alternatively named alkyladenine glycosylase (AAG) that removes 3-methyladenine and a wide variety of other damaged bases from DNA. Lau et al. (51) reported a structure for AAG complexed to DNA containing a pyrrolidine abasic nucleotide, a potent inhibitor of excision repair glycosylases, which is flipped into the enzyme’s active site (Fig. 5C).
T4 endonuclease V. T4 endonuclease V is a DNA glycosylase/AP lyase that can initiate repair of cis–syn cyclobutane pyrimidine dimers in DNA by cleaving the glycosidic bond of the 5′ pyrimidine and then cleaving the phosphodiester backbone. T4 endonuclease V does not flip out the damaged bases, but rather it kinks the dimer-containing DNA, at an angle of ∼60°, and moves the nucleotide opposite the 5′ pyrimidine of the dimer into a binding pocket on the surface of the enzyme (78). The key feature associated with endonuclease V base flipping is that the hole in DNA, created by movement of the nucleotide, is filled by the enzyme inserting its active site amino acids into that hole. Thus, through a change in the structure of the DNA the enzyme is correctly positioned to carry out a nucleophilic attack on the 5′ pyrimidine of the dimer.
AP endonucleases
Escherichia coli endonuclease IV is the hydrolytic AP endonuclease. The structure of the enzyme complexed with AP-containing DNA revealed that the protein loops intercalate side chains at the abasic site, compress the DNA backbone, bend the DNA, and promote base flipping of both the AP site and its mismatched guanine (79). The extrahelical AP site is bound in an enzyme pocket and the target scissile phosphate bond is cleaved (Fig. 5D). A flipped abasic nucleotide also occurs in another structurally unrelated AP endonuclease, human APE1 (80) (Fig. 5F). In addition, E.coli exonuclease III (81) has a structure similar to human APE1 and will very likely bind to DNA and flip the abasic nucleotide in a manner similar to APE1.
HHAI DNA MTase
HhaI MTase, a 327-residue protein, methylates the internal cytosine of its recognition sequence 5′-GCGC-3′/3′-CGCG-5′ (82,83). Crystal structures of M.HhaI are available in various combinations with cofactor and DNA (Table 4), and represent a rich structural paradigm for AdoMet-dependent MTases and DNA base flipping enzymes. The structural information also provides the basis for studies of this enzyme using other techniques: NMR (84), 2-aminopurine fluorescence (59), a chemical probe for thymine in DNA (85), molecular dynamics (86), DNA π-electron transfer (87), biochemistry and kinetics (88), and mutagenesis (89,90).
Table 4. Structures of HhaI MTase.
| PDB |
Protein |
Cofactor |
DNA |
Reference |
| 1hmy | WT | AdoMet | – | 91 |
| 2hmy | WT | AdoMet | – | 92 |
| 3mht | WT | AdoHcy | Unmethylated DNA | 93 |
| 4mht | WT | AdoHcy | Fully methylated DNA | 93 |
| 5mht | WT | AdoHcy | Hemimethylated DNA | 94 |
| 1fjx | T250G | AdoHcy | Unmethylated DNA | 90 |
| 7mht | WT | AdoHcy | G:A mismatch | 47 |
| 8mht | WT | AdoHcy | G:U mismatch | 47 |
| 9mht | WT | AdoHcy | G:AP mismatch | 47 |
| 1mht | WT | AdoHcy | 5-Fluoro-5-methyl-2′-deoxycytosine | 24 |
| 6mht | WT | AdoHcy | 4′-Thio-2′-deoxycytosine | 95 |
| 10mh | WT | AdoHcy | 5,6-Dihydro-5-azacytosine | 96 |
Structures are available for M.HhaI bound to AdoMet (91,92), non-covalently bound to all three states of DNA methylation (unmethylated, hemimethylated and fully-methylated) (93,94), base pair mismatch-containing DNA (47) and nucleoside analog-containing DNA (24,95,96). The protein has two lobes, a conserved AdoMet-dependent catalytic core domain and a DNA-recognition domain (91), which is common to all structurally-characterized DNA MTases, M.HaeIII (64), M.TaqI (97), M.PvuII (43), M.DpnII (46) and M.RsrI (44).
Three states of DNA methylation
An important aspect of DNA methylation is the ability of DNA MTases to distinguish DNA substrates with methyl groups on one strand (hemimethylated DNA) from those which carry no methyl groups. In bacteria, many of the type II enzymes such as M.HhaI are equally active on unmethylated and hemimethylated DNA, but with an increased affinity for asymmetrically methylated DNA [M.EcoRI (98), M.MspI (99), M.HaeIII (100), M.EcoRV (37), M.RsrI (63)]. In mammals, human ‘maintenance’ MTase Dnmt1 shows a preference for hemimethylated DNA in vitro (101). Although mouse Dnmt3 may represent ‘the long sought after de novo MTase’ (8,14), recombinant Dnmt3 in vitro methylates unmethylated and hemimethylated DNA at equal rates (8).
The three M.HhaI structures (3mht contains unmodified C, 4mht contains symmetrically methylated C and 5mht contains a hemimethylated substrate) are rather similar, with one of the target nucleotides (only the unmethylated target C in 5mht) flipped out of the DNA helix and fitting snugly into the active site of the enzyme. On the complementary strand, the target C (methylated or unmethylated) remains stacked in the DNA helix. Both the N4 (NH2) group and the methyl group of 5mC on the complementary strand interact with the protein (see figure 1b of ref. 94); these interactions may enable the MTase to recognize both unmodified cytosine (de novo) and methylated cytosine in the hemimethylated substrate (maintenance).
In 4mht the 5mC residue in the fully methylated oligonucleotide, which is the reaction product, is flipped out of the DNA helix in the same manner as with the unmodified target C in 3mht and 5mht. This is surprising but consistent with biochemical data, which suggest that the binding specificity for M.HhaI is asymmetric 5′-GXGC-3′/3′-CGCG-5′ and determined by the nucleotides neighboring the target (X) nucleotide (54). In other words, the MTase does not depend on the flippable base for its binding specificity.
Nucleoside analogs
The fact that HhaI MTase does not show much binding specificity for the flippable base may reflect a need to leave that base unencumbered by recognition contacts. This provides an opportunity to probe the structural and chemical interactions involved in sequence-specific recognition and catalysis using nucleotide analogs incorporated into synthetic oligonucleotides in the position of the target nucleotide. One prominent analog is 2-aminopurine, which has been used extensively to probe base flipping (see above).
There is also potential clinical significance in designing novel DNA MTase inhibitors, which may be used to reverse the effects of DNA methylation (102). The effects include mutagenicity caused by the spontaneous deamination of 5mC to T and promoter hypermethylation which causes gene silencing and is believed to be a problem in many cancers. Three nucleotide analogs/inhibitors have been used in structural studies including a 4′-thionucleoside (95) and two mechanism-based inhibitors of 5mC MTases, namely 5-fluoro-2′-deoxycytidine (103,104) and 5,6-dihydro-5-azacytosine (a hydrolytically stable replacement for 5-azacytosine) (105).
A complex containing 5-fluoro-2′-deoxycytosine as the target base was first crystallized in the presence of AdoMet, which resulted in the formation of a covalent linkage between the S atom of the enzyme nucleophile Cys81 and the C6 atom of cytosine generating 5-methyl-5-fluoro-2′-deoxydihydrocytosine and AdoHcy (24). Interestingly, 5mC RNA MTases contain sequence motifs that are very similar to those found in the DNA 5mC MTases, including the Pro–Cys, but use a different Cys as a catalytic nucleophile (106).
When 4′-thio-2′-deoxycytosine is incorporated as the target base in the recognition sequence for M.HhaI, binding to the 4′-thio-modified DNA is almost identical to that of the unmodified DNA under equilibrium conditions (95). In contrast, methyl transfer was strongly inhibited in solution. Surprisingly, the flipped 4′-thio-2′-deoxycytosine in the 6mht crystal structure was partially methylated, the result of a much slowed reaction that allowed direct visualization of the methyl transfer between the donor atom (the AdoMet S) and the acceptor atom (the activated cytosine C5) (see figure 9 of ref. 95).
5,6-Dihydro-5-azacytosine (DHAC) contains a cytosine-like ring lacking aromatic character with an sp3-hybridized carbon (CH2 group) at position 6 and an NH group at position 5, thus resembling the transition state of the dihydrocytosine intermediate in the reaction pathway of the 5mC DNA MTases (see figure 2 of ref. 107). The 10mh structure, containing DHAC as the target, showed that DHAC occupies the active site of M.HhaI but with no covalent bond formed between the sulfur atom of nucleophile Cys81 and the pyrimidine C6 carbon (96). This result indicates that the DHAC-containing DNA, behaving as a transition-state mimic, is sufficient to produce strong inhibition of DNA MTases.
Mismatched bases
Following the discovery of base flipping by M.HhaI, the effects of replacing the target cytosine by mismatched bases, including adenine, guanine, thymine and uracil, were investigated (54,55). Many DNA MTases were found to bind even more tightly to such mismatched substrates, including human Dnmt1 (108), M.HpaII (55), M.EcoRV (109), M.EcoRII (110), T4 Dam (111), M.EcoRI (58), M.EcoR124I (112) and M.EcoP15I (60). Furthermore, the uracil can be enzymatically methylated and converted to thymine at low efficiencies (54,55); and the binding of these MTases at the G:U mismatch prevented its repair by uracil DNA glycosylase in vitro (55). The highest affinity of M.HhaI was for a gap formed by removal of the target nucleotide and both phosphodiester linkages (54).
Three ternary structures (7mht, 8mht and 9mht) of M.HhaI complexed with AdoHcy and a non-palindromic oligonucleotide containing a G:A, G:U or G:AP mismatch at the target base pair, respectively, have been determined (47). The mismatched adenine, uracil and abasic site are flipped out and located in the enzyme’s active site, respectively. It seems likely that this active site pocket is non-specific for binding, but specific for methylation. In the light of the non-specific binding pocket, the DNA MTase may be more related to the repair enzymes such as AAG that have broad substrate specificity. On the other hand, the methylation reaction is specific in that catalysis occurs only when the flipped base is cytosine or uracil (at low efficiency), more closely resembling the behavior of MUG.
M.HhaI–AdoMet binary complexes
Two distinct binding orientations of AdoMet have been observed in the binary M.HhaI–AdoMet (91,92), suggesting that the enzyme–AdoMet complex does form but lacks catalytic competence in the absence of DNA. These findings were supported by pre-steady-state partitioning analysis of M.HhaI (88). This reconciles the proposed ordered mechanism of binding in which the DNA is bound first (107) and explains the stable association between AdoMet and M.HhaI (36). It is interesting to note the differences in the order of substrate binding between the 5mC MTases and the amino MTases examined so far. 5mC MTases M.HhaI (88,107), M.MspI (113) and mouse Dnmt1 (114) follow a sequential mechanism with the DNA binding first. In Wu and Santi’s ordered mechanism (107), after methyl transfer, AdoHcy dissociates followed by methylated DNA. In support of this ordered mechanism, the structure of M.HaeIII (another 5mC MTase) does not have AdoMet or AdoHcy bound, but the C5 position of the target 5-fluoro-2′-deoxycytosine carries both the fluorine and the methyl group (64), indicating that methyl transfer has taken place and AdoHcy had diffused out of the enzyme–DNA complex. The amino MTases have all three possibilities: M.EcoRV (α), M.RsrI (β) and M.EcoRI (γ) all bind AdoMet first (62,63,98,115), CcrM (an orphan MTase) binds DNA first (116), and M.EcoP15I (a type III MTase) and M.EcaI (β) both have a random equilibrium mechanism (117,118).
M.HhaI–DNA binary complex
The binary M.HhaI–DNA complex has been studied by NMR (84). The solution study showed that the initial product of binding is a complex containing normal B-DNA. Only later does a conformational change take place that results in the flipped base in an intermediate position (or a series of intermediates). The intermediate position is neither in its original stacked position in the DNA helix nor in its final position in the catalytic pocket of the enzyme. The addition of AdoHcy greatly enhances the locking of the flipped-out base into the enzyme’s active site pocket (the methyl transfer turnover would take place with the addition of AdoMet). The two-state model of flipping has been used to fit the kinetic data for M.EcoRI (N6mA), M.HhaI (5mC) and M.EcoRV (N6mA) (58,62,90).
PROSPECTIVES
As we move into the new millenium, we are in a fast moving world of genomics and proteomics. There is a tremendous amount of work to be done to understand the binding, catalytic and kinetic mechanisms of both existing and newly identified families of AdoMet-dependent MTases. Computer searches of newly completed genomes show an abundance of new, putative MTases including many for which substrates cannot even be guessed. There is a dire need for more biochemistry to characterize this large family of enzymes. Even in the cases of MTases for which structures are known, only a few have the target of methylation bound.
The phenomenon of base flipping is appearing in a number of DNA repair enzymes following its initial discovery in the DNA MTases, and many additional enzymes are likely candidates for use of this novel mechanism. It remains to be proven whether base flipping is an active process in which the protein rotates the sugar–phosphate backbone and brings the base out of the helix or a passive one in which the protein binds to a transiently flipped base.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Alan M. Friedman for providing PIMT coordinates, Drs Mark A. Saper and Ursula Jakob for FTSJ coordinates, Drs Dick Gumport and Mair Churchill for M.RsrI coordinates, Dr Osnat Herzberg for communicating results before publication, and Drs Venki Ramakrishnan and Andrew Carter for providing images for Figure 6. Work in our laboratories is supported by grants from the National Institutes of Health (GM46127 to R.J.R. and GM49245 and GM61355 to X.C.).
References
- 1.Walsh C. (1979) Enzymatic Reaction Mechanisms. W.H. Freeman, San Francisco, CA, pp. 851–859.
- 2.Roberts R.J. and Macelis,D. (2000) REBASE – restriction enzymes and methylases. Nucleic Acids Res., 28, 306–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dryden D.T.F. (1999) Bacterial DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific Publishing, NJ, pp. 283–340.
- 4.Bestor T.H. (1988) Cloning of a mammalian DNA methyltransferase. Gene, 74, 9–12. [DOI] [PubMed] [Google Scholar]
- 5.Yoder J.A. and Bestor,T.H. (1998) A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet., 7, 279–284. [DOI] [PubMed] [Google Scholar]
- 6.Okano M., Xie,S. and Li,E. (1998) Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res., 26, 2536–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van den Wyngaert I., Sprengel,J., Kass,S.U. and Luyten,W.H.M.L. (1998) Cloning and analysis of a novel human putative DNA methyltransferase. FEBS Lett., 426, 283–289. [DOI] [PubMed] [Google Scholar]
- 8.Okano M., Xie,S. and Li,E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet., 19, 219–220. [DOI] [PubMed] [Google Scholar]
- 9.Vertino P.M. (1999) Eukaryotic DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), AdoMet-Dependent Methyltransferases: Structures and Functions. World Scientific Publishing, NJ, pp. 341–372.
- 10.Bird A.P. and Wolffe,A.P. (1999) Methylation-induced repression – belts, braces, and chromatin. Cell, 99, 451–454. [DOI] [PubMed] [Google Scholar]
- 11.Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402. [DOI] [PubMed] [Google Scholar]
- 12.Ramsahoye B.H., Biniszkiewicz,D., Lyko,F., Clark,V., Bird,A.P. and Jaenisch,R. (2000) Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA, 97, 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed] [Google Scholar]
- 14.Okano M., Bell,D.W., Haber,D.A. and Li,E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- 15.Finnegan E.J., Peacock,W.J. and Dennis,E.S. (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl Acad. Sci USA, 93, 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakutani T., Jeddeloh,J.A., Flowers,S.K., Munakata,K. and Richards,E.J. (1996) Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl Acad. Sci. USA, 93, 12406–12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronemus M.J., Galbiati,M., Ticknor,C., Chen,J. and Dellaporta,S.L. (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science, 273, 654–657. [DOI] [PubMed] [Google Scholar]
- 18.Bird A.P. (1993) Functions for DNA methylation in vertebrates. Cold Spring Harb. Symp. Quant. Biol., 58, 281–285. [DOI] [PubMed] [Google Scholar]
- 19.Riggs A.D. and Pfeifer,G.P. (1992) X-chromosome inactivation and cell memory. Trends Genet., 8, 169–174. [DOI] [PubMed] [Google Scholar]
- 20.Shemer R. and Razin,A. (1996) Establishment of imprinted methylation patterns during development. In Russo,V.E.A., Martienssen,R.A. and Riggs,A.D. (eds), Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press, New York, NY, pp. 215–229.
- 21.Siegfried Z. and Cedar,H. (1997) DNA methylation: a molecular lock. Curr. Biol., 7, R305–R307. [DOI] [PubMed] [Google Scholar]
- 22.Xu G.-L., Bestor,T.H., Bourc’his,D., Hsieh,C.-L., Tommerup,N., Bugge,M., Hulten,M., Qu,X., Russo,J.J. and Viegas-Pequignot,E. (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature, 402, 187–191. [DOI] [PubMed] [Google Scholar]
- 23.Hansen R.S., Wijmenga,C., Luo,P., Stanek,A.M., Canfield,T., Weemaes,C.M.R. and Gartler,S.M. (1999) The Dnmt3b DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl Acad. Sci. USA, 96, 14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klimaauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) HhaI methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Zhou,L. and Cheng,X. (2000) Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J., 19, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skinner M.M., Puvathingal,J.M., Walter,R.L. and Friedman,A.M. (2000) Crystal structure of protein isoaspartyl methyltransferase: a catalyst for protein repair. Structure, 8, 1189–1201. [DOI] [PubMed] [Google Scholar]
- 27.Fauman E.B., Blumenthal,R.M. and Cheng,X. (1999) Structure and evolution of AdoMet-dependent methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), AdoMet-Dependent Methyltransferases: Structures and Functions. World Scientific Publishing, NJ, pp. 1–38.
- 28.Niewmierzycka S., A. and Clarke, (1999) S-adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem., 274, 814–824. [DOI] [PubMed] [Google Scholar]
- 29.Wang H., Boisvert,D., Kim,K.-K., Kim,R. and Kim,S.-H. (2000) Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J., 19, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 31.Hung L.W., Huang,L., Kim,R. and Kim,S.-H. (2000) Crystal structure and functional analysis of a hypothetical protein, Mj0882, from Methanococcus jannaschii. http://www.rcsb.org/pdb/cgi/explore.cgi?pid=22617983827596&pdbId=1DUS.
- 32.Lim K., Zhang,H., Tempczyk,A., Bonander,N., Toedt,J., Howard,A.J., Eisenstein,E. and Herzberg,O. (2000) Hypothetical proteins from Haemophilus influenzae: two new structures implying methyltransferase function. Abstract of American Crystallographic Association annual meeting, p. 36 (July 22–27, 2000, St Paul, MN).
- 33.Holmes W.M. (1999) tRNA Methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), AdoMet-Dependent Methyltransferases: Structures and Functions. World Scientific Publishing, NJ, pp. 185–198.
- 34.Javor G.T. (1993) Depression of adenosylmethionine content of Escherichia coli by thioglycerol. Antimicrob. Agents Chemother., 24, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piekarowicz A. and Brzezinski,R. (1980) Cleavage and methylation of DNA by the restriction endonuclease HinfIII isolated from Haemophilus influenzae Rf. J. Mol. Biol., 144, 415–429. [DOI] [PubMed] [Google Scholar]
- 36.Kumar S., Cheng,X., Pflugrath,J.W. and Roberts,R.J. (1992) Purification, crystallization, and preliminary X-ray diffraction analysis of an M.HhaI-AdoMet complex. Biochemistry, 31, 8648–8653. [DOI] [PubMed] [Google Scholar]
- 37.Szczelkun M.D. and Connolly,B.A. (1995) Sequence-specific binding of DNA by the EcoRV restriction and modification enzymes with nucleic acids and cofactor analogues. Biochemistry, 34, 10724–10733. [DOI] [PubMed] [Google Scholar]
- 38.Weiss V.H., McBride,A.E., Soriano,M.A., Filman,D.J., Silver,P.A. and Hogle,J.M. (2000) The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nature Struct. Biol., 7, 1165–1171. [DOI] [PubMed] [Google Scholar]
- 39.Posfai J., Bhagwat,A.S., Posfai,G. and Roberts,R.J. (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res., 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauster R., Trautner,T.A. and Noyer-Weidner,M. (1989) Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J. Mol. Biol., 206, 305–312. [DOI] [PubMed] [Google Scholar]
- 41.Malone T., Blumenthal,R.M. and Cheng,X. (1995) Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol., 253, 618–632. [DOI] [PubMed] [Google Scholar]
- 42.Schluckebier G., O’Gara,M., Saenger,W. and Cheng,X. (1995) Universal catalytic domain structure of AdoMet-dependent methyltransferases. J. Mol. Biol., 247, 16–20. [DOI] [PubMed] [Google Scholar]
- 43.Gong W., O’Gara,M., Blumenthal,R.M. and Cheng,X. (1997) Structure of PvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res., 25, 2702–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scavetta R.D., Thomas,C.B., Walsh,M.A., Szegedi,S., Joachimiak,A., Gumport,R.I. and Churchill,M. (2000) Structure of RsrI methyltransferase, a member of the N6-adenine β class of DNA methyltransferases. Nucleic Acids Res., 28, 3950–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeltsch A. (1999) Circular permutations in the molecular evolution of DNA methyltransferase. J. Mol. Evol., 49, 161–164. [DOI] [PubMed] [Google Scholar]
- 46.Tran P.H., Korszun,Z.R., Cerritelli,S., Springhorn,S.S. and Lacks,S.A. (1998) Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of Streptococcus pneumoniae bound to S-adenosylmethionine. Structure, 6, 1563–1575. [DOI] [PubMed] [Google Scholar]
- 47.O’Gara M., Horton,R.J., Roberts,R.J. and Cheng,X. (1998) Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nature Struct. Biol., 5, 872–877. [DOI] [PubMed] [Google Scholar]
- 48.Parikh S.S., Mol,C.D., Slupphaug,G., Bharati,S., Krokan,H.E. and Tainer,J.A. (1998) Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J., 17, 5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barrett T.E., Savva,R., Panayotou,G., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. [DOI] [PubMed] [Google Scholar]
- 50.Barrett T.E., Scharer,O.D., Savva,R., Brown,T., Jiricny,J., Verdine,G.L. and Pearl,L.H. (1999) Crystal structure of a thwarted mismatch glycosylase DNA repair complex. EMBO J., 18, 6599–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau A.Y., Scharet,O.D., Samson,L., Verdine,G.L. and Ellenberger,T. (1998) Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell, 95, 249–258. [DOI] [PubMed] [Google Scholar]
- 52.Hollis T., Ichikawa,Y. and Ellenberger,T. (2000) DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J., 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts R.J. and Cheng,X. (1998) Base flipping. Annu. Rev. Biochem., 67, 181–198. [DOI] [PubMed] [Google Scholar]
- 54.Klimaauskas S. and Roberts,R.J. (1995) M.HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Res., 23, 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang A.S., Shen,J.-C., Zingg,J.-M., Mi,S. and Jones,P.A. (1995) HhaI and HpaII DNA methyltransferases bind DNA mismatches, methylate uracil and block DNA repair. Nucleic Acids Res., 23, 1380–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward D.C., Reich,E. and Stryer,L. (1969) Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem., 244, 1228–1237. [PubMed] [Google Scholar]
- 57.Allan B.W. and Reich,N.O. (1996) Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry, 35, 14757–14762. [DOI] [PubMed] [Google Scholar]
- 58.Allan B.W., Beechem,J.M., Lindstrom,W.M. and Reich,N.O. (1998) Direct real time observation of base flipping by the EcoRI DNA methyltransferase. J. Biol. Chem., 273, 2368–2373. [DOI] [PubMed] [Google Scholar]
- 59.Holz B., Klimaauskas,S., Serva,S. and Weinhold,E. (1998) 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res., 26, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reddy Y.V. and Rao,D.N. (2000) Binding of EcoP15I DNA methyltransferase to DNA reveals a large structural distortion within the recognition sequence. J. Mol. Biol., 298, 597–610. [DOI] [PubMed] [Google Scholar]
- 61.Stivers J.T. (1998) 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res., 26, 3837–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gowher H. and Jeltsch,A. (2000) Molecular enzymology of the EcoRV DNA-(adenine-N6)-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J. Mol. Biol., 303, 93–110. [DOI] [PubMed] [Google Scholar]
- 63.Szegedi S.S., Reich,N.O. and Gumport,R.I. (2000) Substrate binding in vitro and kinetics of RsrI [N6-adenine] DNA methyltransferase. Nucleic Acids Res., 28, 3962–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reinisch K.M., Chen,L., Verdine,G.L. and Lipscomb,W.N. (1995) The crystal structure of HaeIII methyltransferase covalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell, 82, 143–153. [DOI] [PubMed] [Google Scholar]
- 65.Goedecke K., Pignot,M., Goody,R.S., Scheidig,A.J. and Weinhold,E. (2001) Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with a cofactor analog. Nature Struct. Biol., 8, 121–125. [DOI] [PubMed] [Google Scholar]
- 66.Blumenthal R.M. and Cheng,X. (2001) A Taq attack displaces bases. Nature Struct. Biol., 8, 101–103. [DOI] [PubMed] [Google Scholar]
- 67.Carter A.P., Clemons,W.M., Brodersen,D.E., Morgan-Warren,R.J., Wimberly,B.T. and Ramakrishnan,V. (2000) Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature, 407, 340–348. [DOI] [PubMed] [Google Scholar]
- 68.Carter A.P., Clemons,W.M., Brodersen,D.E., Morgan-Warren,R.J., Hartsch,T., Wimberly,B.T. and Ramakrishnan,V. (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science, 291, 498–501. [DOI] [PubMed] [Google Scholar]
- 69.Lloyd R.S. and Cheng,X. (1997) Mechanistic link between DNA methyltransferases and DNA repair enzymes by base flipping. Biopolymers, 44, 139–151. [DOI] [PubMed] [Google Scholar]
- 70.Daniels D.S., Mol,C.D., Arvai,A.S., Kanugula,A., Pegg,A.E. and Tainer,J.A. (2000) Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding. EMBO J., 19, 1719–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Slupphaug G., Mol,C.D., Kavli,B., Arvai,A.S., Krokan,H.E. and Tainer,J.A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- 72.Stivers J.T., Pankiewicz,K.W. and Watanabe,K.A. (1999) Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry, 38, 952–963. [DOI] [PubMed] [Google Scholar]
- 73.Panayotou G., Brown,T., Barlow,T., Pearl,L.H. and Savva,R. (1998) Direct measurement of the substrate preference of uracil-DNA glycosylase. J. Biol. Chem., 273, 45–50. [DOI] [PubMed] [Google Scholar]
- 74.Bruner S.D., Norman,D.P.G. and Verdine,G.L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- 75.Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guan Y., Manuel,R.C., Arvai,A.S., Parikh,S.S., Mol,C.D., Miller,J.H., Lloyd,R.S. and Tainer,J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Struct. Biol., 5, 1058–1063. [DOI] [PubMed] [Google Scholar]
- 77.Sugahara M., Mikawa,T., Kumasaka,T., Yamamoto,M., Kato,R., Fukuyama,K., Inoue,Y. and Kuramitsu,S. (2000) Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J., 19, 3857–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vassylyev D.G., Kashiwagi,T., Mikami,Y., Ariyoshi,M., Iwai,S., Ohtsuka,E. and Morikawa,K. (1995) Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell, 83, 773–782. [DOI] [PubMed] [Google Scholar]
- 79.Hosfield D.J., Guan,Y., Haas,B.J., Cunningham,R.P. and Tainer,J.A. (1999) Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell, 98, 397–408. [DOI] [PubMed] [Google Scholar]
- 80.Mol C.D., Izumi,T., Mitra S. and Tainer,J.A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature, 403, 451–456. [DOI] [PubMed] [Google Scholar]
- 81.Mol C.D., Kuo,C.-F., Thayer,M.M., Cunningham,R.P. and Tainer,J.A. (1995) Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature, 374, 381–386. [DOI] [PubMed] [Google Scholar]
- 82.Roberts R.J., Myers,P.A., Morrison,A. and Murray,K. (1976) A specific endonuclease from Haemophilus haemolyticus. J. Mol. Biol., 103, 199–208. [DOI] [PubMed] [Google Scholar]
- 83.Mann M.B. and Smith,H.O. (1979) In Usdin,E., Borchardt,R.T. and Creveling,C.R. (eds), Proceedings of the Conference on Transmethylation. Elsevier, New York, NY, pp. 483–492.
- 84.Klimaauskas S., Szyperski,T., Serva,S. and Wuthrich,K. (1998) Dynamic modes of the flipped-out cytosine during HhaI methyltransferase-DNA interactions in solution. EMBO J., 17, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Serva S., Weinhold,E., Roberts,R.J. and Klimaauskas,S. (1998) Chemical display of thymine residues flipped out by DNA methyltransferases. Nucleic Acids Res., 26, 3473–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lau E.Y. and Bruice,T.C. (1999) Active site dynamics of HhaI methyltransferase: insights from computer simulation. J. Mol. Biol., 293, 9–18. [DOI] [PubMed] [Google Scholar]
- 87.Rajski S.R., Kumar,S., Roberts,R.J. and Barton,J.K. (1999) Protein-modulated DNA electron transfer. J. Am. Chem. Soc., 121, 5615–5616. [Google Scholar]
- 88.Lindstrom W.M., Flynn,J. and Reich,N.O. (2000) Reconciling structure and function in HhaI DNA cytosine-C-5 methyltransferase. J. Biol. Chem., 275, 4912–4919. [DOI] [PubMed] [Google Scholar]
- 89.Mi S., Alonso,D. and Roberts,R.J. (1995). Functional analysis of Gln-237 mutants of HhaI methyltransferase. Nucleic Acids Res., 23, 620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vilkaitis G., Dong,A., Weinhold,E., Cheng,X. and Klimaauskas,S. (2000) Functional roles of the conserved Threonine-250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem., 275, 38722–38730. [DOI] [PubMed] [Google Scholar]
- 91.Cheng X., Kumar,S., Posfai,J., Pflugrath,J.W. and Roberts,R.J. (1993) Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell, 74, 299–307. [DOI] [PubMed] [Google Scholar]
- 92.O’Gara M., Zhang,X., Roberts,R.J. and Cheng,X. (1999) Structure of a binary complex of HhaI methyltransferase with S-adenosyl-l-methionine formed in the presence of a short non-specific DNA oligonucleotide. J. Mol. Biol., 287, 201–209. [DOI] [PubMed] [Google Scholar]
- 93.O’Gara M., Klimaauskas,S., Roberts,R.J. and Cheng,X. (1996) Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaI methyltransferase-DNA-AdoHcy complexes. J. Mol. Biol., 261, 634–645. [DOI] [PubMed] [Google Scholar]
- 94.O’Gara M., Roberts,R.J. and Cheng,X. (1996) A structural basis for the preferential binding of hemimethylated DNA by HhaI DNA methyltransferase. J. Mol. Biol., 263, 597–606. [DOI] [PubMed] [Google Scholar]
- 95.Kumar S., Horton,J.R., Jones,G.D., Walker,R.T., Roberts,R.J. and Cheng,X. (1997) DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res., 25, 2773–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sheikhnejad G., Brank,A., Christman,J.K., Goddard,A., Alvarez,E., Ford,H.,Jr, Marquez,V.E., Marasco,C.J., Sufrin,J.R., O’Gara,M. and Cheng,X. (1999) Mechanism of inhibition of DNA-(cytosine C5) methyltransferases by oligodeoxyribonucleotides containing 5, 6-dihydro-5-azacytosine. J. Mol. Biol., 285, 2021–2034. [DOI] [PubMed] [Google Scholar]
- 97.Labahn J., Granzin,J., Schluckebier,G., Robinson,D.P., Jack,W.E., Schildkraut,I. and Saenger,W. (1994) Three-dimensional structure of the adenine-specific DNA methyltransferase M.TaqI in complex with the cofactor S-adenosylmethionine. Proc. Natl Acad. Sci. USA, 91, 10957–10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reich N.O. and Mashhoon,N. (1990) Inhibition of EcoRI DNA methylase with cofactor analogs. J. Biol. Chem., 265, 8966–8970. [PubMed] [Google Scholar]
- 99.Dubey A.K. and Roberts,R.J. (1992) Sequence-specific DNA binding by the MspI DNA methyltransferase. Nucleic Acids Res., 20, 3167–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen L., MacMillan,A.M. and Verdine,G.L. (1993) Mutational separation of DNA binding from catalysis in a DNA cytosine methyltransferase. J. Am. Chem. Soc., 115, 5318–5319. [Google Scholar]
- 101.Pradhan S., Bacolla,A., Wells,R.D. and Roberts,R.J. (1999) Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem., 274, 33002–33010. [DOI] [PubMed] [Google Scholar]
- 102.Bender C.M., Zingg,J.-M. and Jones,P.A. (1998) DNA methylation as a target for drug design. Pharm. Res., 15, 175–187. [DOI] [PubMed] [Google Scholar]
- 103.Santi D.V., Garrett,C.E. and Barr,P.J. (1983) On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell, 33, 9–10. [DOI] [PubMed] [Google Scholar]
- 104.Osterman D.G., DePillis,G.D., Wu,J.C., Matsuda,A. and Santi,D.V. (1988) 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry, 27, 5204–5210. [DOI] [PubMed] [Google Scholar]
- 105.Santi D.V., Norment,A. and Garrett,C.E. (1984) Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc. Natl Acad. Sci. USA, 81, 6993–6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y. and Santi,D.V. (2000) m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl Acad. Sci. USA, 97, 8263–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu J.C. and Santi,D.V. (1987) Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem., 262, 4778–4786. [PubMed] [Google Scholar]
- 108.Smith S.S. (1994) Biological implications of the mechanism of action of human DNA (cytosine-5) methyltransferase. Prog. Nucleic Acid Res. Mol. Biol., 49, 65–111. [DOI] [PubMed] [Google Scholar]
- 109.Cal S. and Connolly,B.A. (1997) DNA distortion and base flipping by the EcoRV DNA methyltransferase. J. Biol. Chem., 272, 490–496. [DOI] [PubMed] [Google Scholar]
- 110.Sheluho D., Yebra,M.J., Khariwala,S.S. and Bhagwat,A.S. (1997) Lack of correlation between binding of EcoRII methylase to DNA duplexes containing mismatches and the promotion of C to T mutations. Mol. Gen. Genet., 255, 54–59. [DOI] [PubMed] [Google Scholar]
- 111.Malygin E.G., Petrov,N.A., Gorbunov,Y.A., Kossykh,V.G. and Hattman,S. (1997) Interaction of the phage T4 Dam DNA-(N6-adenine) methyltransferase with oligonucleotides containing native or modified (defective) recognition sites. Nucleic Acids Res., 25, 4393–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mernagh D.R., Taylor,I.A. and Kneale,G.G. (1998) Interaction of the type I methyltransferase M.EcoR124I with modified DNA substrates: sequence discrimination and base flipping. Biochem. J., 336, 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bhattacharya S.K. and Dubey,A.K. (1999) Kinetic mechanism of cytosine DNA methyltransferase MspI. J. Biol. Chem., 274, 14743–14749. [DOI] [PubMed] [Google Scholar]
- 114.Flynn J. and Reich,N.O. (1998) Murine DNA (cytosine-5)-methyltransferase: steady-state and substrate trapping analyses of the kinetic mechanism. Biochemistry, 37, 15162–15169. [DOI] [PubMed] [Google Scholar]
- 115.Surby M.A. and Reich,N.O. (1996) Contribution of facilitated diffusion and processive catalysis to enzyme efficiency: Implications for the EcoRI restriction-modification system. Biochemistry, 35, 2201–2208. [DOI] [PubMed] [Google Scholar]
- 116.Berdis A.J., Lee,I., Coward,J.K., Stephens,C., Wright,R., Shapiro,L. and Benkovic,S.J. (1998) A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA. Proc. Natl Acad. Sci. USA, 95, 2874–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rao D.N., Page,M.G.P. and Bickle,T.A. (1989) Cloning, over-expression and the catalytic properties of the EcoP15 modification methylase from Escherichia coli. J. Mol. Biol., 209, 599–606. [DOI] [PubMed] [Google Scholar]
- 118.Szilak L., Der,A., Deak,F. and Venetianer,P. (1993) Kinetic characterization of the EcaI methyltransferase. Eur. J. Biochem., 218, 727–733. [DOI] [PubMed] [Google Scholar]
- 119.Schluckebier G., Kozak,M., Bleimling,N., Weinhold,E. and Saenger,W. (1997) Differential binding of S-adenosylmethionine, S-adenosylhomocysteine and sinefungin to the adenine-specific DNA methyltransferase M.TaqI. J. Mol. Biol., 265, 56–67. [DOI] [PubMed] [Google Scholar]
- 120.Dong A., Yoder,J.A., Zhang,X., Zhou,L., Bestor,T.H. and Cheng,X. (2001) Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res., 29, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Djordjevic S. and Stock,A.M. (1997) Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure, 5, 545–558. [DOI] [PubMed] [Google Scholar]
- 122.Djordjevic S. and Stock,A.M. (1998) Chemotaxis receptor recognition by methyltransferase CheR. Nature Struct. Biol., 5, 446–450. [DOI] [PubMed] [Google Scholar]
- 123.Hodel A.E., Gershon,P.D., Shi,X. and Quiocho,F.A. (1996) The 1.85 Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell, 85, 247–256. [DOI] [PubMed] [Google Scholar]
- 124.Hodel A.E., Gershon,P.D., Shi,X., Wang,S.M. and Quiocho,F.A. (1997) Specific protein recognition of an mRNA cap through its alkylated base. Nature Struct. Biol., 4, 350–354. [DOI] [PubMed] [Google Scholar]
- 125.Hodel A.E., Gershon,P.D. and Quiocho,F.A. (1998) Structural basis for sequence non-specific recognition of 5′-capped mRNA by a cap modifying enzyme. Mol. Cell, 1, 443–447. [DOI] [PubMed] [Google Scholar]
- 126.Hu G., Hodel,A.E., Gershon,P.D. and Quiocho,F.A. (1999) mRNA cap recognition: dominant role of enhanced stacking interactions between methylated bases and protein aromatic side chains. Proc. Natl Acad. Sci. USA, 96, 7149–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu L., Petros,A.M., Schnuchel,A., Zhong,P., Severin,J.M., Walter,K., Holzman,T.F. and Fesik,S.W. (1997) Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide-streptogramin antibiotic resistance. Nature Struct. Biol., 4, 483–489 [published erratum appears in Nature Struct. Biol., (1997) 4, 592]. [DOI] [PubMed] [Google Scholar]
- 128.Bussiere D.E., Muchmore,S.W., Dealwis,C.G., Schluckebier,G., Nienaber,V.L., Edalji,R.P., Walter,K.A., Ladror,U.S., Holzman,T.F. and Abad-Zapatero,C. (1998) Crystal structure of ErmC′, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry, 37, 7103–7112. [DOI] [PubMed] [Google Scholar]
- 129.Schluckebier G., Zhong,P., Stewart,K.D., Kavanaugh,T.J. and Abad-Zapatero,C. (1999) The 2.2 Å structure of the rRNA methyltransferase ermC’ and its complexes with cofactor and cofactor analogs: implications for the reation mechanism. J. Mol. Biol., 289, 277–291. [DOI] [PubMed] [Google Scholar]
- 130.Buegl H., Fauman,E.B., Staker,B.L., Zheng,F., Kushner,S.R., Saper,M.A., Bardwell,J.C.A. and Jakob,U. (2000) RNA methylation under heat shock control. Mol. Cell, 6, 349–360. [DOI] [PubMed] [Google Scholar]
- 131.Vidgren J., Svensson,L.A. and Liljas,A. (1994) Crystal structure of catechol O-methyltransferase. Nature, 368, 354–358. [DOI] [PubMed] [Google Scholar]
- 132.Fu Z., Hu,Y., Konnishi,K., Takata,Y., Ogawa,H., Gomi,T., Fujioka,M. and Takusagawa,F. (1996) Crystal structure of glycine N-methyltransferase from rat liver. Biochemistry, 35, 11985–11993. [DOI] [PubMed] [Google Scholar]
- 133.Huang Y., Komoto,J., Konishi,K., Takata,Y., Ogawa,H., Gomi,T., Fujioka,M. and Takusagawa,F. (2000) Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes. J. Mol. Biol., 298, 149–162. [DOI] [PubMed] [Google Scholar]
- 134.Zubieta C., He,X.-Z., Dixon,R.A. and Noel,J.P. (2000) Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nature Struct. Biol., 8, 271–279. [DOI] [PubMed] [Google Scholar]
- 135.Parikh S.S., Walcher,G., Jones,G.D., Slupphaug,G., Krokan,H.E., Blackburn,G.M. and Tainer,J.A. (2000) Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl Acad. Sci. USA, 97, 5083–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]