Abstract
The Antarctic bathydraconid dragonfish, Parachaenichthys charcoti, is an Antarctic notothenioid teleost endemic to the Southern Ocean. The Southern Ocean has cooled to −1.8ºC over the past 30 million years, and the seawater had retained this cold temperature and isolated oceanic environment because of the Antarctic Circumpolar Current. Notothenioids dominate Antarctic fish, making up 90% of the biomass, and all notothenioids have undergone molecular and ecological diversification to survive in this cold environment. Therefore, they are considered an attractive Antarctic fish model for evolutionary and ancestral genomic studies. Bathydraconidae is a speciose family of the Notothenioidei, the dominant taxonomic component of Antarctic teleosts. To understand the process of evolution of Antarctic fish, we select a typical Antarctic bathydraconid dragonfish, P. charcoti. Here, we have sequenced, de novo assembled, and annotated a comprehensive genome from P. charcoti. The draft genome of P. charcoti is 709 Mb in size. The N50 contig length is 6145 bp, and its N50 scaffold length 178 362 kb. The genome of P. charcoti is predicted to contain 32 712 genes, 18 455 of which have been assigned preliminary functions. A total of 8951 orthologous groups common to 7 species of fish were identified, while 333 genes were identified in P. charcoti only; 2519 orthologous groups were also identified in both P. charcoti and N. coriiceps, another Antarctic fish. Four gene ontology terms were statistically overrepresented among the 333 genes unique to P. charcoti, according to gene ontology enrichment analysis. The draft P. charcoti genome will broaden our understanding of the evolution of Antarctic fish in their extreme environment. It will provide a basis for further investigating the unusual characteristics of Antarctic fishes.
Keywords: Parachaenichthys charcoti, antarctic dragonfish, notothenioid, de novo genome assembly; genome annotation
Data description
Introduction
The fish fauna of the Southern Ocean is dominated by a single lineage belonging to the perciform suborder Notothenioidei, consisting of 132 species and 8 families. All Antarctic notothenioids have evolved to adapt to the extreme Antarctic marine environment, which includes large seasonal changes in food availability and stably cold water temperature. Notothenioids dominate Antarctic fish, making up 90% of the biomass, and all notothenioids have undergone molecular and ecological diversification to survive in this cold environment. Therefore, they are considered an attractive Antarctic fish model for evolutionary and ancestral genomic studies. Bathydraconidae is a speciose family of the Notothenioidei, the dominant taxonomic component of Antarctic teleosts [1–4]. Parachaenichthys charcoti, the Antarctic bathydraconid dragonfish, was first described by Vaillant in 1906 (Notothenioidei: Bathydraconidae; AphiaID: 234687; Fishbase ID: 7102). They are found in localities around Potter Cove, South Shetland Islands. P. charcoti remain almost exclusively on the inner shelves throughout their ontogeny [5]. Several studies have investigated their ecology and ethology, but there has been no genomic study [5–8]. A comprehensive genetic study is needed to identify the distinguishing characteristics of this Antarctic fish and to provide useful data for understanding Antarctic teleost divergence and evolution.
Library construction and sequencing
P. charcoti (length: ∼45 cm) were collected in nets at depths of 20–30 m in Marian Cove, near King Sejong Station, on the Northern Antarctic Peninsula (62°14'S, 58°47'W) in January 2012 using the hook-and-line method (Fig. 1). High–molecular weight genomic DNA was extracted from P. charcoti using the Gentra Puregene Blood Kit (Qiagen, Valencia, CA, USA). For genomic DNA sequencing, 3 paired-end libraries (PE300, PE400, and PE450) were constructed from sheared genomic DNA (consisting of 300-, 400-, and 450-bp fragments) and subsequently prepared using standard Illumina sample preparation methods. Mate-pair libraries (MP3K, MP5K, MP8K, and MP20K) were prepared for scaffolding, and sequencing was performed according to the manufacturer's instructions (consisting of 3-kb, 5-kb, 8-kb, and 20-kb fragments; Illumina, San Diego, CA, USA).
Figure 1:
Photograph of Antarctic dragonfish, P. charcoti.
Because expressed sequence tags are essential for gene annotation in draft genomes, a transcriptome library was conducted using TruSeq® Sample Preparation v. 2 (Illumina) with total RNA. Total RNA was extracted from liver tissue and purified using the RNeasy Mini Kit (Qiagen) with the RNase-Free DNaseI Kit (Qiagen). Extracted sample quality and concentration were determined with 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). mRNA was isolated from 2 μg of the total RNA for double-stranded cDNA library construction with poly-A selection. For transcriptome sequencing, paired-end libraries (PE500) were constructed from sheared cDNA consisting of 500-bp fragments and subsequently prepared using standard Illumina sample preparation methods. Final transcriptome libraries’ length and concentration were determined with 2100 Bioanalyzer. Transcriptome libraries were sequenced using runs of 300 × 2 paired-end reads (Table 1).
Table 1:
P. charcoti sequencing statistics
| Library | Mode | Insert size (bp) | Library type | Trimmed reads | Trimmed sequence (bp) | Source |
|---|---|---|---|---|---|---|
| PE300 | 2 × 300 | 300 | Paired-end | 28 776 064 | 4 964 428 226 | Genomic DNA |
| PE400 | 2 × 300 | 400 | Paired-end | 139 126 700 | 29 538 419 473 | Genomic DNA |
| PE450 | 2 × 300 | 450 | Paired-end | 85 834 292 | 16 644 575 781 | Genomic DNA |
| MP3K | 2 × 300 | 3000 | Mate-pair | 70 517 546 | 4 925 657 177 | Genomic DNA |
| MP5K | 2 × 300 | 5000 | Mate-pair | 66 623 428 | 4 626 486 038 | Genomic DNA |
| MP8K | 2 × 300 | 8000 | Mate-pair | 61 240 982 | 4 212 744 363 | Genomic DNA |
| MP20K | 2 × 300 | 20 000 | Mate-pair | 86 575 644 | 5 387 730 972 | Genomic DNA |
| PE500 | 2 × 300 | 500 | Paired-end | 25 940 404 | 5 571 197 784 | Liver RNA |
All resulting Illumina reads were trimmed using the FASTX-Toolkit (v. 0.0.11) [9] with the parameters -t 20, -l 70, and -Q 33, after which a paired sequence from the trimmed Illumina reads was selected. All sequencing processes for 3 paired-end libraries (genomic DNA), 4 mate-pair libraries (genomic DNA), and 1 paired-end library (transcriptome) were performed by Korea Polar Research Institutes (data statistics provided in Table 1).
Genome assembly
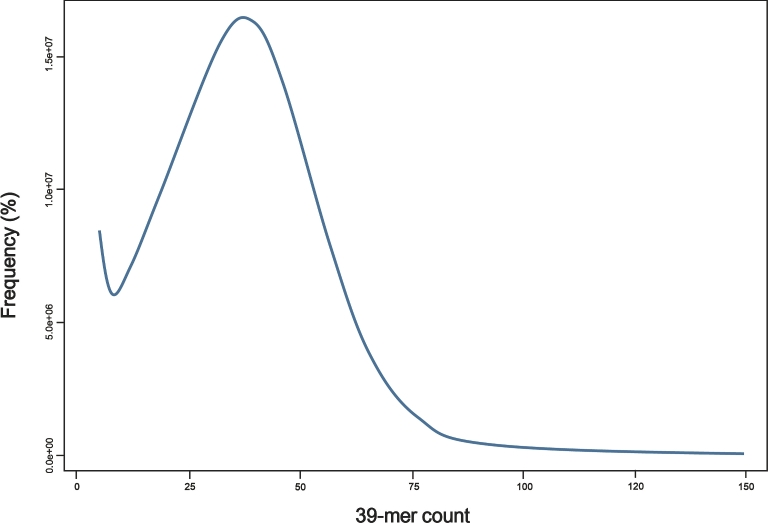
K-mer analysis was conducted using Jellyfish 2.2.5 (Jellyfish, RRID:SCR_005491) [10] to estimate the genome size from DNA paired-end libraries. The estimated genome size is 805 Mb, with the main peak observed at a coverage depth of ∼×39 (Fig. 2). Initial assemblies were performed using the Celera Assembler v. 8.3 (Celera Assembler, RRID:SCR_010750) with trimmed paired-end reads [11]. For the Celera Assembler, paired-end read data were converted into FRG file format using FastqToCA, which is a utility included in the Celera Assembler. Assembly was performed on an 80-processor workstation using Intel Xeon X7460 2.66 GHz processors and 1 Tb of RAM with the following parameters: overlapper = ovl, unitigger = bogart, utgErrorRate = 0.03, utgErrorLimit = 2.5, utgGraphErrorRate = 0.030, utgGraphErrorLimit = 3.25, ovlErrorRate = 0.06, cnsErrorRate = 0.06, cgwErrorRate = 0.1, merSize = 28, doOverlapBasedTrimming = 1, merylMemory = 500 000, merylThreads = 40, ovlMemory = 8 Gb, ovlThreads = 2, ovlConcurrency = 40, ovlHashBlockLength = 300 000 000, ovlRefBlockSize = 7 630 000, and ovlHashBits = 24. The initial assembly had a total size of 709 Mb, N50 contig length of 5039 bp, and N50 scaffold length of 6135 kb, with a GC content of 40.66%. The assembled contig revealed a contig coverage of approximately ×36.57 from the Celera Assembler. Contigs from the initial assembly were used for scaffolding using the stand-alone scaffolding tool SSPACE v. 2.0 (SSPACE, RRID:SCR_005056) with the following parameters: -x 0, -k 3, -a 0.8, and -T 60 [12].
Figure 2:
Estimation of the P. charcoti genome size based on 39-mer analysis. X-axis represents the depth (peak at ×39), and the y-axis represents the proportion. Genome size was estimated to be 805 Mb (total k-mer number/volume peak).
Trimmed mate-pair reads created using the FASTX-Toolkit were used in the scaffolding process. After scaffolding, the number of scaffolds decreased from 153 398 to 12 381, and the N50 scaffold length increased from 6135 to 166 726 bp (Table 2). The total size of the final scaffolds (∼795 Mb) was consistent with the estimated genome size (805 Mb).
Table 2:
Global statistics of the P. charcoti genome assembly
| P. charcoti | ||
|---|---|---|
| Scaffold | Total scaffold length (bases) | 794 596 176 |
| Gap size (bases) | 86 840 902 | |
| Scaffolds (n) | 12 602 | |
| N50 scaffold length (bases) | 178 362 | |
| Max scaffold length (bases) | 1 318 127 | |
| Contig | Total contig length (bases) | 709 540 340 |
| Contigs (n) | 153 398 | |
| N50 contig length (bases) | 6145 | |
| Max contig length (bases) | 65 864 | |
| Annotation | Gene number (n) | 32 712 |
| An average mRNA length (bases) | 1412 | |
| An average CDS length (bases) | 1291 | |
| An average of exons (n) | 8 | |
| Repeat content (% of genome) | 19.4 | |
Gene annotation
MAKER2 annotation pipeline (MAKER, RRID:SCR_005309) was used for genome annotation with default parameters [13]. It first identified repetitive elements using RepeatMasker v. 3.3.0 (Repeat Masker, RRID:SCR_012954) with a de novo repeat library [14], which was constructed using RepeatModeler v. 1.0.3 (RepeatModeler, RRID:SCR_015027) [15] with the Repbase library (Ver. 20 140 131). The SNAP gene finder [16] was selected to perform ab initio gene prediction from this masked genome sequence. Alignment of transcriptome assembly results using BLASTn and homologous protein information from tBLASTx were considered for gene annotation as RNA and protein evidence, respectively. Transcriptome assembly was performed by using the program CLC Genomics Workbench 8.0 with default parameters, and sequencing reads from PE500 (Table 1) were used. Proteins from 6 species were used in the analysis: Notothenia coriiceps (NCBI reference sequence NC_015653.1) and Danio rerio, Gasterosteus aculeatus, Takifugu rubripes, Tetraodon nigroviridis, and Gadus morhua (all from Ensembl release 69). MAKER2 includes integration of the annotation edit distance (AED) metric for controlling the quality of annotation [17]. AED values are bounded between 0 and 1; an AED value of 0 indicated that its aligned evidence and annotated gene showed an exact match. Conversely, a value of 1 indicated no evidence support. But the AED cut-off was not applied for these gene predictions. Instead, AED values were denoted in gene annotation and were considered for orthologous gene analysis and gene gain and loss.
MAKER2 was used to select and revise the final gene model based on all inputs. A total of 32 712 genes were predicted in P. charcoti using MAKER2 (Table 2). The annotated genes contained an average of 8 exons, with an average mRNA length of 1412 bp and coding DNA sequences (CDS) length of 1291 bp. The repeat prediction from MAKER2 showed that repeat sequences accounted for 19.41% of the assembled P. charcoti genome.
To estimate genome assembly and annotation completeness, we performed Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis (BUSCO, RRID:SCR_015008) [18], an approach used for lineage-specific profile libraries, such as those of actinopterygii, and identified 88.6% complete and 5.7% partial eukaryote orthologous gene sets in our assembly (Table 3).
Table 3:
Summarized benchmarks of the BUSCO assessment
| Actinopterygii (%) | |
|---|---|
| Total BUSCO groups searched | 4062a |
| Complete BUSCOs | 88.6 |
| Complete and single-copy | 86.3 |
| Complete and duplicated | 2.3 |
| Partial | 5.7 |
| Missing | 5.7 |
aNumber of total BUSCO groups searched.
To assign preliminary functions for 32 712 genes, we used Blast2GO v. 2.6.0 (Blast2GO, RRID:SCR_005828) [19]. We classified functions for 18 455 (56.42%) predicted genes, which were annotated using BLASTp results and InterproScan (RRID:SCR_005829). Gene ontology annotation terms included “biological process” (20 126, 61.52%), “molecular function” (20 514, 62.71%), and “cellular component” (15 452, 47.23%). Enzyme commission numbers were obtained for 3846 proteins.
Ortholog analysis
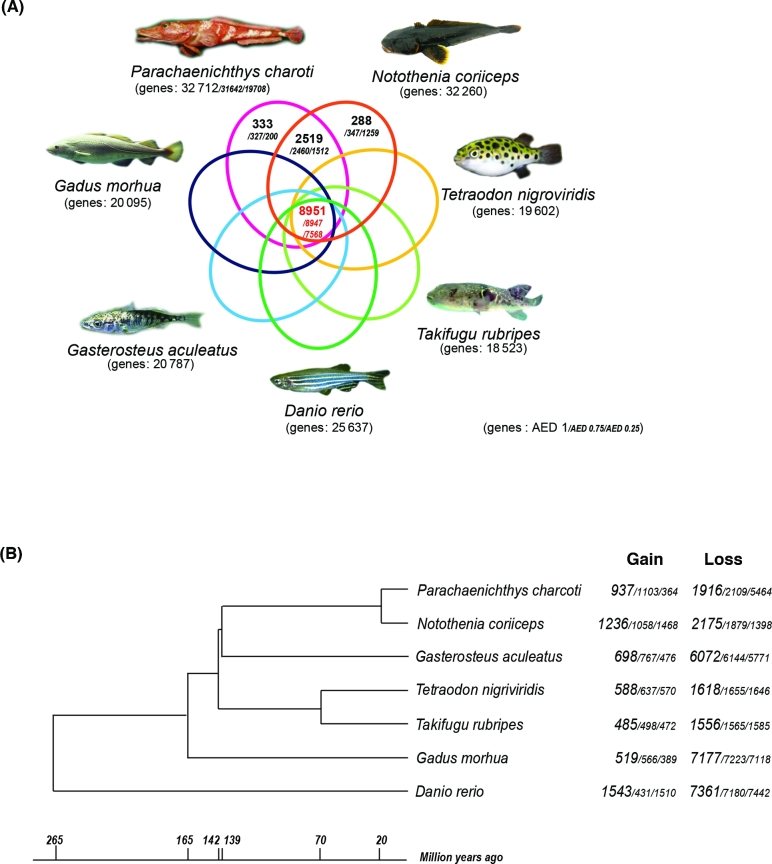
We identified orthologous groups using OrthoMCL (v. 2.0.5) [20], which generated a graphical representation of the sequence relationships, which were then presented in subgraphs using the Markov Clustering Algorithm based on multiple eukaryotic genomes. We used the standard parameters (percentMatchCutoff = 50 and evalueExponentCutoff = –5) and options within OrthoMCL for all steps. We used 7 fish genomes for this analysis (D. rerio, G. aculeatus, T. rubripes, T. nigroviridis, G. morhua, N. coriiceps, and P. charcoti). The coding sequences of 5 genomes were collected from Ensembl release 69, and 1 coding sequence was selected among multiple proteins corresponding to 1 gene. We used the coding sequence from the NCBI reference sequence (NC_015653.1) of N. coriiceps and 3 groups of the coding sequence of P. charcoti from MAKER annotation with different AED thresholds (1, 0.75, and 0.25). In the case of a AED cut-off value of 1, we identified 8951 orthologous groups common to all 7 fish; 288 of 32 636 N. coriiceps genes and 333 of 32 712 P. charcoti genes were not identified in any other species, and 2519 groups were identified only in the 2 Antarctic fish (Fig. 3A). When we applied an AED threshold of 0.25 against gene prediction of P. charcoti, 7568 orthologous groups were identified.
Figure 3:
Comparative genome analyses of the P. charcoti genome. (A) Venn diagram of orthologous gene clusters between 4 arthropod lineages. (B) Gene family gain-and-loss analysis. The number of gained gene families and lost gene families are indicated for each species. Time lines specify divergence times between the lineages.
Likelihood analysis of gene gain and loss
We estimated differences in the size of orthologs to identify gene families that have undergone significant size changes through evolution [21, 22]. We used the program CAFE3.0 [23] and performed analyses against 3 groups including the coding sequence of P. charcoti with different AED thresholds separately. We performed phylogenetic analyses among 7 representative fishes with the protein-coding gene in the orthologous groups to obtain the Newick description of a rooted and bifurcating phylogenetic tree. A total of 8951 orthologous gene sets were selected using the criterion of reciprocal best BLASTP hit and were aligned using PRANK (v. 130820) under a codon model with the “-dna -codon” option [24]; poor alignment sites were eliminated using Gblock (v. 0.91) under a codon model with the “-t = c” option [25]. The remaining alignment regions were concatenated and used in the construction of the phylogenetic tree by using the neighbor-joining method in the MEGA (v. 6) program (MEGA, RRID:SCR_000667) [26]. The ultrametric tree of the species with branch lengths in units of time was prepared by referring TimeTree [27] for CAFE3.0 (Fig. 3B). The program was performed using P < 0.05, and estimated rates of birth (λ) and death (μ) were calculated using the program LambdaMu with the “-s” option. The numbers of gene gains and losses were calculated on each branch of the tree with the “-t” option. P. charcoti gained 937 and lost 1916 gene families (Fig. 3B).
The Antarctic dragonfish P. charcoti is a species in the sister lineage of icefishes [28–30]; it is the only hemoglobinless vertebrate. The dragonfish (Bathydraconidae) and the icefish (Channichthyidae) were generally considered to have evolved from a common notothenioid ancestor, which was characterized by decreased hematocrit and blood hemoglobin concentrations [31–35]. The dragonfish showed the most similar patterns in these trends among red-blooded notothenioid taxa [35]. The globin complex of the dragonfish P. charcoti was hypothesized to be similar in length and organization to that of ancestral icefish prior to loss of functionality [36]. Along with the recently published N. coriiceps genome [37], the genome of P. charcoti will broaden our understanding of how Antarctic fish have evolved to survive in sub-zero temperatures and might provide an important clue to understand the process of evolution to the hemoglobinless Antarctic fish and their distinct phenotypes (an increase of blood volume, low blood viscosity, large bore capillaries, increased vascularity with great capacitance, cardiomegaly, and high blood flow).
Availability of supporting data
The data for the P. charcoti genome and transcriptome have been deposited in the Sequence Read Archive as BioProject PRJNA330735. Other supporting data, including annotations, alignments, and BUSCO results, are available in the GigaScience repository, GigaDB [38].
Abbreviations
AED: annotation edit distance; BUSCO: Benchmarking Universal Single-Copy Orthologs; CDS: coding DNA sequences.
Competing interests
The authors declare no competing interests.
Funding
This work was supported by a grant entitled “Polar Genomics 101 Project (PE17080),” funded by the Korea Polar Research Institute.
Author contributions
H.P. and J.P. conceived and designed experiments and analyses. D.H.A., S.C.S., B.K., S.K., J.K., I.A., and J.P. performed experiments and conducted bioinformatics. D.H.A., S.C.S., B.K., and H.P. wrote the paper.
Supplementary Material
References
- 1. Eastman JT, Pratt D, Winn W. Antarctic Fish Biology: Evolution in a Unique Environment. San Diego: Academic Press; 1993. [Google Scholar]
- 2. Eastman JT, Clarke A. A comparison of adaptive radiations of Antarctic fish with those of nonAntarctic fish. In: Fishes of Antarctica. Milan: Springer; 1998:3–26. [Google Scholar]
- 3. Eastman JT. Antarctic notothenioid fishes as subjects for research in evolutionary biology. Antarct Sci 2000;12(03):276–87. [Google Scholar]
- 4. Eakin RR, Eastman JT, Near TJ. A new species and a molecular phylogenetic analysis of the Antarctic fish genus Pogonophryne (Notothenioidei: Artedidraconidae). Copeia 2009;4(4):705–13. [Google Scholar]
- 5. Casaux RJ, Mazzotta AS, Barrera-Oro ER. Seasonal aspects of the biology and diet of nearshore nototheniid fish at Potter Cove, South Shetland Islands, Antarctica. Polar Biol 1990;11(1):63–72. [Google Scholar]
- 6. Barrera-Oro E. The role of fish in the Antarctic marine food web: differences between inshore and offshore waters in the southern Scotia Arc and west Antarctic Peninsula. Ant Sci 2002;14(4):293–309. [Google Scholar]
- 7. Barrera-Oro ER, Lagger C. Egg-guarding behaviour in the Antarctic bathydraconid dragonfish Parachaenichthys charcoti. Polar Biol 2010;33(11):1585–7. [Google Scholar]
- 8. Eastman JT, Sidell BD. Measurements of buoyancy for some Antarctic notothenioid fishes from the South Shetland Islands. Polar Biol 2002;25(10):753–60. [Google Scholar]
- 9. Gordon A, Hannon GJ. Fast-toolkit. FASTQ/A short-reads pre-processing tools. http://hannonlab.cshl.edu/fastx_toolkit. Accessed 27 Feb 2017.
- 10. Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011;27(6):764–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Myers EW, Sutton GG, Delcher AL et al. . A whole-genome assembly of Drosophila. Science 2000;287(5461):2196–204. [DOI] [PubMed] [Google Scholar]
- 12. Boetzer M, Henkel CV, Jansen HJ et al. . Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011;27(4):578–9. [DOI] [PubMed] [Google Scholar]
- 13. Cantarel BL, Korf I, Robb SMC et al. . MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 2008;18(1):188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tarailo‐Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protocols Bioinform 2009:4–10. [DOI] [PubMed] [Google Scholar]
- 15. Bao Z. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res 2002;12(8):1269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korf I. Gene finding in novel genomes. BMC Bioinformatics 2004;5(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 2011;12(1):491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simão FA, Waterhouse RM, Ioannidis P et al. . BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015:31(19):3210–2. [DOI] [PubMed] [Google Scholar]
- 19. Conesa A, Gotz S, Garcia-Gomez JM et al. . Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005;21(18):3674–6. [DOI] [PubMed] [Google Scholar]
- 20. Li L. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 2003;13(9):2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hahn MW, De Bie T, Stajich JE et al. . Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res 2005;15(8):1153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hahn MW, Han MV, Han S. Gene family evolution across 12 drosophila genomes. PLoS Genet 2007;3(11):e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Bie T, Cristianini N, Demuth JP et al. . CAFE: a computational tool for the study of gene family evolution. Bioinformatics 2006;22(10):1269–71. [DOI] [PubMed] [Google Scholar]
- 24. Loytynoja A, Goldman N. From the cover: an algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A 2005;102(30):10557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000;17(4):540–52. [DOI] [PubMed] [Google Scholar]
- 26. Tamura K, Stecher G, Peterson D et al. . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013;30(12):2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 2006;22(23):2971–2. [DOI] [PubMed] [Google Scholar]
- 28. Balushkin A, Voskoboynikova O. Systematics and phylogeny of antarctic dragonfishes (Bathydraconidae, Notothenioidei, Perciformes). J Ichthyol 1995;35(5):89–104. [Google Scholar]
- 29. Derome N, Chen W, Dettai A et al. . Phylogeny of Antarctic dragonfishes (Bathydraconidae, Notothenioidei, Teleostei) and related families based on their anatomy and two mitochondrial genes. Mol Phylogenet Evol 2002;24(1):139–52. [DOI] [PubMed] [Google Scholar]
- 30. Near TJ. A genomic fossil reveals key steps in hemoglobin loss by the Antarctic icefishes. Mol Biol Evol 2006;23(11):2008–16. [DOI] [PubMed] [Google Scholar]
- 31. Bargelloni L, Marcato S, Patarnello T. Antarctic fish hemoglobins: evidence for adaptive evolution at subzero temperature. Proc Natl Acad Sci U S A 1998;95(15):8670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beers JM, Borley KA, Sidell BD. Relationship among circulating hemoglobin, nitric oxide synthase activities and angiogenic poise in red- and white-blooded Antarctic notothenioid fishes. Comp Biochem Physiol A Mol Integr Physiol 2010;156(4):422–9. [DOI] [PubMed] [Google Scholar]
- 33. D’avino R, Di Prisco G. Antarctic fish hemoglobin: an outline of the molecular structure and oxygen binding properties—I. Molecular structure. Comp Biochem Physiol B Comp Biochem 1988;90(3):579–84. [Google Scholar]
- 34. Di Prisco G. Molecular adaptations of Antarctic fish hemoglobins. In: Fishes of Antarctica. Milan: Springer; 1998:339–53. [Google Scholar]
- 35. Kunzmann A, Caruso C, Prisco GD. Haematological studies on a high-Antarctic fish: Bathydraco marri Norman. J Exp Mar Biol Ecol 1991;152(2):243–55. [Google Scholar]
- 36. Lau Y, Parker SK, Near TJ et al. . Evolution and function of the globin intergenic regulatory regions of the Antarctic dragonfishes (Notothenioidei: Bathydraconidae). Mol Biol Evol 2012;29(3):1071–80. [DOI] [PubMed] [Google Scholar]
- 37. Shin SC, Ahn DH, Kim SJ et al. . The genome sequence of the Antarctic bullhead notothen reveals evolutionary adaptations to a cold environment. Genome Biol 2014;15(9):468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ahn DH, Shin SC, Kim BM et al. . Supporting data for “Draft genome of the Antarctic dragonfish, Parachaenichthys charcoti.” GigaScience Database 2017. 10.5524/100321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.