Abstract
Proteins with amino acid homorepeats have the potential to be detrimental to cells and are often associated with human diseases. Why then are homorepeats prevalent in eukaryotic proteomes? In yeast, we find that homorepeats are enriched in proteins that are essential, pleiotropic and buffer environmental insults. Their presence increases functional versatility of proteins by mediating protein interactions and facilitating spatial organization in a repeat-dependent manner. During evolution, homorepeats are preferentially retained in proteins with stringent proteostasis, which might minimize repeat-associated detrimental effects such as unregulated phase separation and protein aggregation. Their presence facilitates rapid protein divergence through accumulation of amino acid substitutions, which often affect linear motifs and post-translational modification sites. This may result in rewiring protein interaction and signalling networks. Thus, homorepeats are distinct modules that are often retained in stringently regulated proteins. Their presence facilitates rapid exploration of the genotype-phenotype landscape of a population, thereby contributing to fitness.
Repetitive sequences are an important source of genetic variation and are prevalent across all eukaryotic genomes. Amino acid homorepeats (HRs) of various lengths have been implicated in diverse human diseases (e.g., abnormal polyQ expansion in huntingtin leads to Huntington’s disease1). HR mediated pathogenicity can arise due to diverse molecular reasons. For instance, anomalous polyA length in the transcription factor Foxl2 alters its subcellular localization2, whereas abnormal polyQ length in the androgen receptor affects its protein abundance, stability and its interactions in the cell3–5. Importantly, over-expression of wild type homorepeat containing proteins (HRPs) such as ataxin-1 and androgen receptor with non-pathogenic HR lengths tends to phenocopy the detrimental effects of HRPs with abnormally long HRs4,6. This effect is primarily mediated by the potential of repeats to form aggregates and sequester soluble proteins into deposits, resulting in loss of protein activity, or gain of toxicity due to the aggregates1,7.
Although homorepeats can have negative consequences, the presence of HRs could also be beneficial8. PolyQ in the fungal RNA binding protein Whi3 is required for cell-cycle control, whereas polyA in the fly transcription factor Exd, polyS in the human lysyl hydroxylase Jmdj6 and polyH in diverse human transcription factors influence subcellular localization9–12. While polyQ and polyP have been linked to transcriptional activation, polyA in insect Hox genes aids in transcriptional repression13,14. PolyQ length variation in White Collar-1 tunes the circadian rhythm in Neurospora15, and variation in the polyQ:polyA length ratio in the transcription factor Runx2 is linked to variation in canine skull morphology16. Thus amino acid HR can drive varied molecular effects, depending on their length, amino acid type, and the biochemical/molecular function of the protein that harbors them, ultimately leading to either detrimental or beneficial outcomes for organismal phenotypes17.
Amino acid HRs are prevalent in eukaryotic proteins, comprising ~15% of any eukaryotic proteome (Supplementary Notes 1)18–20. Such HRPs are involved in a wide variety of functions including DNA and RNA binding, signalling and adhesion19–21. In this study we investigate the following questions: Why are HRPs prevalent in eukaryotic proteomes, although altered expression or abnormal HR length is often detrimental? Why are HRs found only in certain proteins? If HRs are important, what benefits do they provide to an organism?
Results
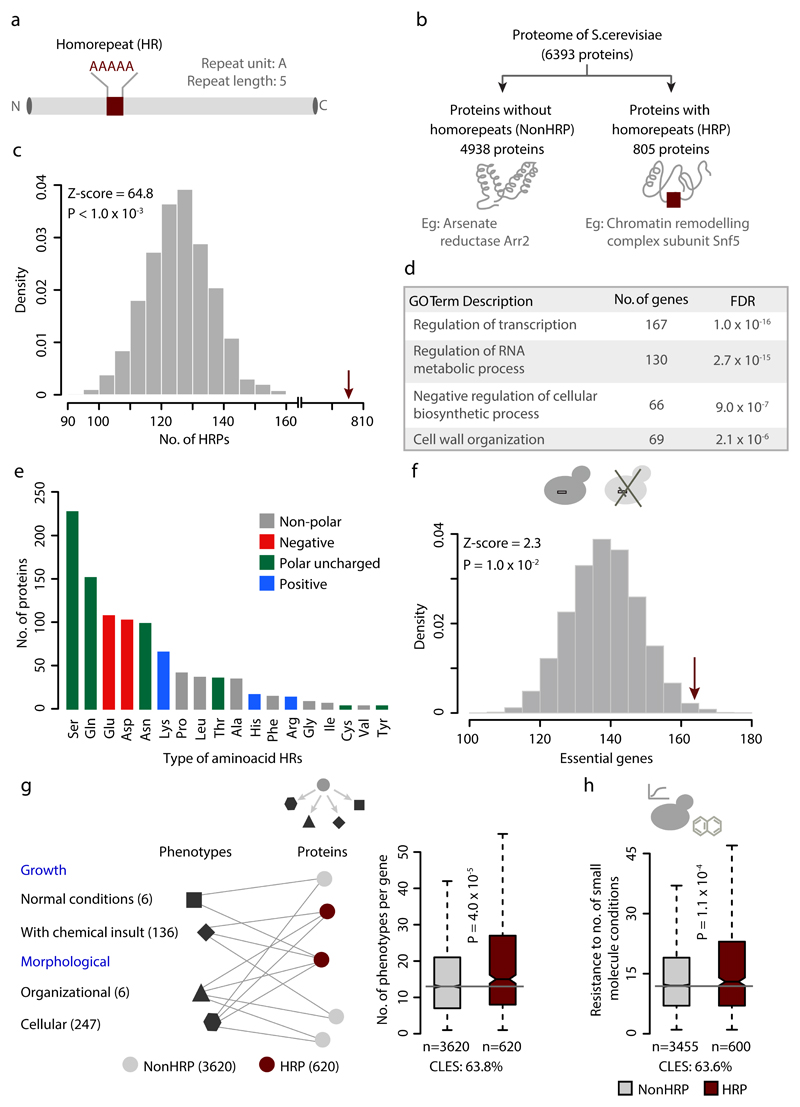
We carried out a comparative genomics study investigating multiple large-scale datasets using budding yeast as a model organism. We first identified 805 yeast proteins with amino acid HRs encompassing at least one continuous stretch of ≥ 5 identical amino acids (Fig. 1a-b). This cut-off was based on the observations that (i) the occurrence of identical amino acid stretches of length five or more is statistically significant compared to randomized sequences with similar amino acid composition (Fig. 1c) and (ii) the smallest HR length, whose length variation is implicated in human diseases, is five (polyD in the cartilage oligomeric complex protein COMP22). HRPs in yeast are involved in vital regulatory processes such as transcription regulation and nucleic acid metabolism, similar to HRPs in other organisms19 (Fig. 1d). Repeats of polar amino acids are prevalent in yeast HRPs (Fig. 1e). Using multiple genome-scale datasets, we compared yeast HRPs with a set of 4938 proteins that lacked HRs (NonHRPs) for their phenotypes upon perturbation, adaptability to different environmental insults, regulatory, and evolutionary properties (Supplementary Fig. 1). To ensure that the NonHRPs do not contain any repetitive sequences, we did not consider proteins with imperfect repeats (n=650). To control for enrichments in the trends that were found to be associated with HRPs, we also compared them to different groups of relevant NonHRPs such as highly disordered proteins, proteins with similar functions, essential and highly pleiotropic proteins, length-matched proteins and proteins with non-repetitive amino acid bias (discussed below and Online Methods).
Figure 1. Proteins with homorepeats in yeast and their physiological importance.
Yeast proteins with HRs (HRPs) and without HRs (NonHRPs; panels a-b). (c) Enrichment of homorepeats of length ≥5 in yeast was tested by generating 1000 shuffled proteomes, using permutation test. Each protein in the yeast proteome was shuffled for the sequence order, by keeping the length and amino acid composition constant. The null expectation in the shuffled proteomes (grey histogram) and the actual observation (red arrow) of HRPs are shown. The Z-score indicates the distance of the actual observation to the mean of random expectation in terms of number of standard deviation. P-value was estimated as the ratio of random observations greater than or equal to the number of actually observed HRPs to the total number of random samples. (d) Gene Ontology (GO) biological processes enriched in HRPs. (e) Distribution of proteins with different amino acid repeat types. (f) Enrichment of HRPs among essential genes, tested using permutation test by performing 10,000 iterations. The random expectation (grey histogram) and the actual observation (red arrow) of essential HRPs are shown. Boxplots of distributions of (g) pleiotropic effects, (h) the number of small molecules that a gene confers resistance to, among HRPs and NonHRPs. The black line within the box represents the median and the boxes represent the first and third quartile. The notches correspond to ~95% confidence intervals for the median. Whiskers (dashed lines) show the data points up to 1.5 times the interquartile range from the box. Values lying beyond the whiskers are considered outliers and not shown to improve visualization. Effect size is indicated by common language effect size (CLES), which describes the probability that a randomly sampled data point from a distribtion A will be greater than a data point sampled from distribution B.
HRs are prevalent in essential and highly pleiotropic genes that buffer environmental perturbations
Genes that are essential for growth in rich medium show an enrichment to contain HRs (Fig. 1f), suggesting that HRPs are important for cell fitness. We constructed a yeast gene to phenotype (G2P) network consisting of 4220 genes and 395 phenotypes describing growth (in normal conditions and upon chemical insults) and morphological (organizational and cellular) features upon genetic alteration (deletion and over-expression). HRPs are associated with a higher number of phenotypes than NonHRPs (Fig. 1g), even upon minimizing the redundancy in the G2P network by merging phenotypes that share ≥50% of genes (Supplementary Fig. 2a). Investigation of the fitness data shows that deletion of non-essential HRPs make the cells sensitive to a higher number of chemical insults and stresses compared to NonHRPs (Fig. 1h). Therefore, HRs are prevalent in proteins whose presence tends to buffer environmental perturbations and chemical insults. Thus, proteins with HRs tend to be essential, influence a range of growth and morphological phenotypes (pleiotropic), and tend to confer adaptability to diverse environmental insults.
HRPs affect multiple biological processes through molecular interactions
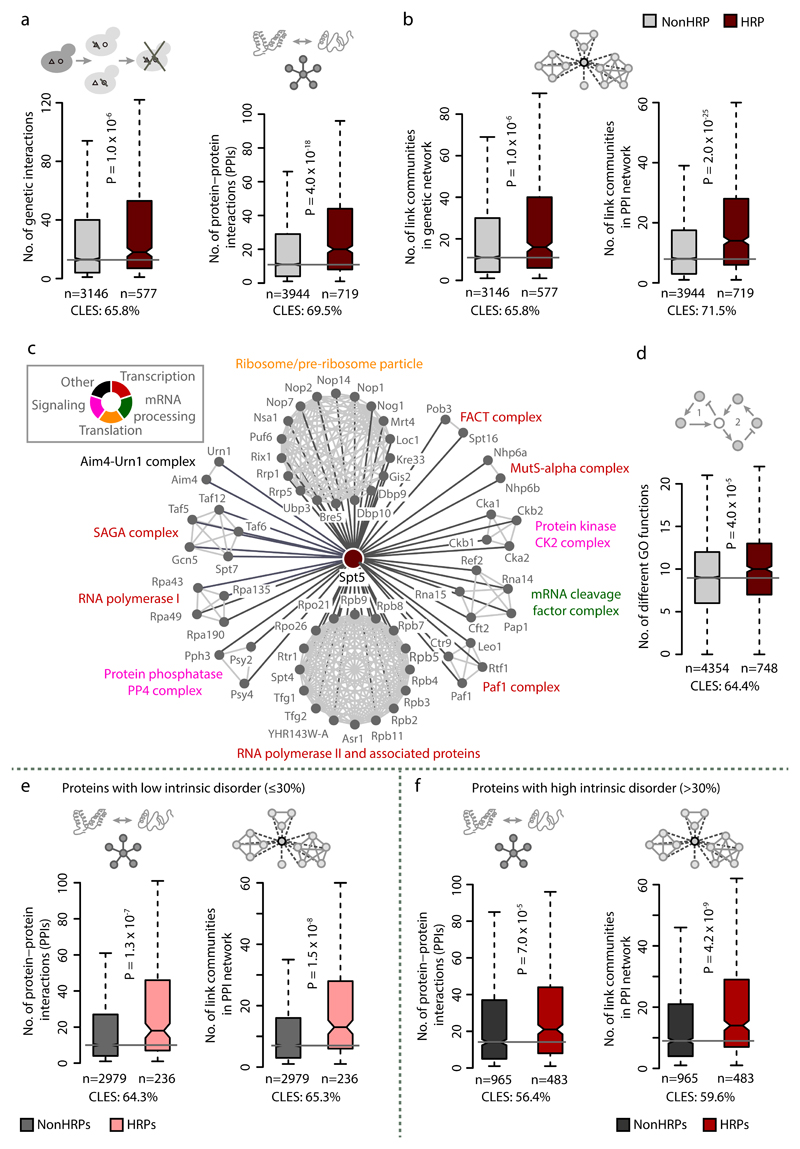
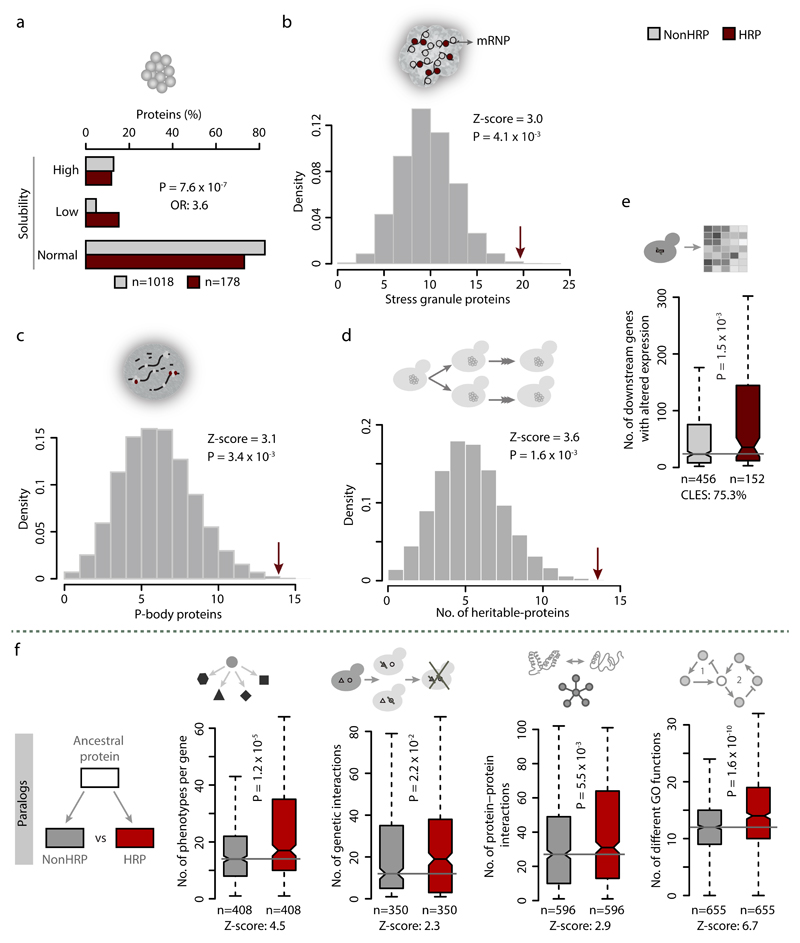
To understand the molecular basis of the observed pleiotropy in proteins with HRs, we examined both genetic and biochemical interaction networks. HRPs tend to have a higher number of genetic and protein interactions (Fig. 2a) than NonHRPs, which could explain their influence on multiple phenotypes. A network analysis of link communities, which represent interconnected sets of genes that participate in similar biological processes23, revealed that proteins with HRs participate in more communities than NonHRPs (Fig. 2b). Thus HRPs often connect diverse biological processes by acting as inter-community hubs, as exemplified by the polyD containing transcriptional regulator Spt5 (Fig. 2c). These observations are consistent with the higher multifunctionality index of HRPs, defined by the number of Gene Ontology (GO) terms associated with each gene24 (Fig. 2d). These trends are not confounded by the extent of intrinsic protein disorder (Fig. 2e-f), protein length (Supplementary Fig. 2b) or differences in biological functions between HRPs and NonHRPs (Supplementary Fig. 2c). Importantly, these trends are also stronger compared to proteins with non-repetitive amino acid bias (Supplementary Fig. 2d). HRPs are overrepresented among proteins with low solubility and those that form intracellular phase-separated structures such as stress granules, P-bodies and heritable proteins (Fig. 3a-d). These observations collectively suggest that HRPs tend to spatially organize protein-protein interactions within cells.
Figure 2. HRPs are functionally versatile and mediate interactions.
Boxplots of distributions of (a) genetic interactions and protein-protein interactions, (b) link communities in genetic and protein-protein interaction network and (d) different GO terms a gene is associated with (functional versatility). Statistical significance was assessed using Wilcoxon rank sum test and false discovery rate (FDR) corrected for multiple testing. (c) Network illustrating link communities in the protein interaction network of polyD containing transcriptional regulator Spt5, affecting diverse biological processes. Proportion of intrinsic disorder in a protein can influence the propensity of proteins to interact with multiple partners and hence its functionality60. To investigate the effect of protein disorder on functional versatility of HRPs, we classified NonHRPs and HRPs based on intrinsic disorder content, as those with (i) low disorder (≤30%) and (ii) high disorder fraction (>30%) and drew comparisons. Boxplots of distribution of number of protein-protein interactions and link communities in PPI network among HRPs and NonHRPs with low (panel e) and high (panel f) disorder content. Statistical significance was assessed using Wilcoxon rank sum test and FDR corrected for multiple testing, with effect sizes displayed as CLES. Irrespective of the extent of protein disorder, HRPs tend to have more interactions and participate in diverse processes.
Figure 3. HRPs form higher-order assemblies and homorepeats might contribute to the functional versatility of HRPs.
(a) Percentage of proteins with high, low and normal solubility among HRPs and NonHRPs. Statistical signficance was assessed using Chi-squared test and effect sizes displayed as odds ratios (OR). Enrichment of HRPs among (b) stress granule proteins, (c) P-body proteins and (d) heritable proteins was tested using permutation test, by performing 10,000 randomizations. The Z-score indicates the distance of the actual observation to the mean of random expectation in terms of number of standard deviation. P-values were estimated as the ratio of randomly observed proteins greater than or equal to the number of actually observed HRPs to the total number of random samples (10,000). (e) Box plot of distribution of genes whose expression is altered upon deletion of HRP and NonHRP regulators (from gene-perturbation network).Statistical significance was estimated using Wilcoxon rank sum test and effect size provided as CLES. (f) Distribution of pleiotropic effects, number of genetic interactions, protein-protein interactions and different GO terms among divergent paralog pairs in which one is a HRP and the other a NonHRP. P-values was estimated using Wilcoxon matched pairs test and corrected for mutliple testing (FDR). Effect sizes are indicated by Z-scores.
While HRPs have a comparable number of target genes in terms of the protein-DNA and protein-RNA interaction network (Supplementary Fig. 2e-f), an analysis of the Gene Perturbation Network25 revealed that deletion of HRP regulators affects expression of a larger number of targets than their NonHRP counterparts (Fig. 3e). Thus proteins with HRs regulate a larger part of the genome/transcriptome than NonHRPs.
HRs might contribute to the trends observed in HRPs
To assess the role and contribution of HRs to the observed trends, we computed likelihood ratios based on conditional probabilities. We find that HRPs have higher likelihood ratios than NonHRPs (Supplementary Fig. 3). This suggests that the presence of HRs might contribute to the feature in HRPs and that the emergence of HRs within proteins might facilitate the acquisition of these features. To investigate this, we examined paralog pairs that arose by gene-duplication and studied divergent pairs, wherein one paralog contained the HR and the other did not (HRP-NonHRP pairs; Fig. 3f). Among such paralog pairs, the one with a HR tends to have a higher number of (i) phenotypes, (ii) genetic and protein interactions and (iii) GO functions compared to the ones that did not contain a HR (Fig. 3f). Thus, our observations collectively suggest that the presence of HRs is associated with, and may influence, a large number of phenotypes and biological processes. Compared to the length of the repeat, the type of amino acid repeat might have a bigger influence on the molecular effects of HRs and its impact on fitness (suggested by likelihood ratio estimates; Supplementary Fig. 3-4).
PolyQ repeat in Snf5 affects fitness and confers adaptability to diverse environmental insults
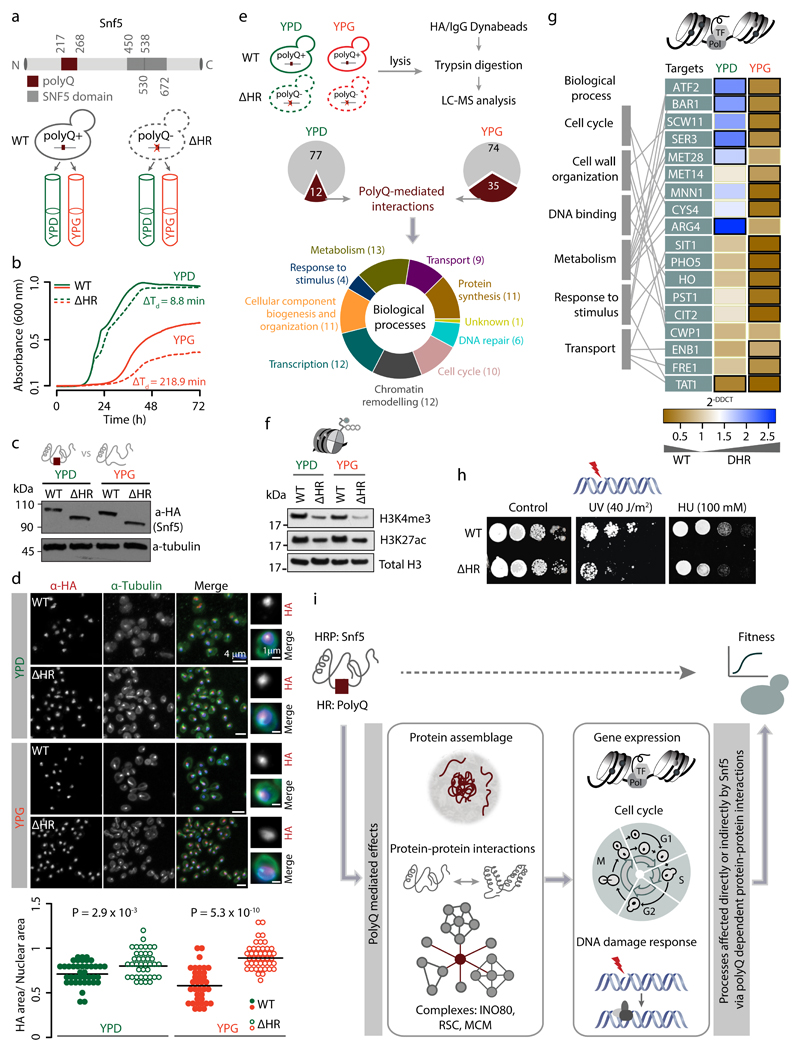
To establish the endogenous roles of amino acid homorepeat in a protein, we investigated a pleiotropic hub in the protein interaction network, Snf5, which is a core component of the Swi/Snf chromatin-remodeling complex and contains a polyQ HR. We generated a haploid mutant strain by deleting the longest stretch of HR in the protein (polyQ; amino acids 217–270; ΔHR) and compared it with the wild type (WT) strain (Fig. 4a). This ensures that SNF5 ΔHR is expressed from its endogenous promoter and regulated in its natural context. We examined the effect of the HR on growth in two different carbon sources, namely glucose (YPD) and glycerol (YPG). The ΔHR mutant displays a minor delay in growth kinetics in YPD, while the delay is more pronounced in YPG (Fig. 4b). Consistently, we observe an increase in budding index (Supplementary Fig. 5a), highlighting an increase in the doubling time. Thus, polyQ in Snf5 might influence growth rates by either directly or indirectly affecting the cell cycle. Deletion of polyQ did not affect the protein abundance (Fig. 4c) or sub-cellular localization of Snf5. However, the WT protein forms punctate structures whereas the ΔHR protein tends to be diffuse (Fig. 4d). This is consistent with our genome-scale analysis (Fig. 3a-d) and suggests that the polyQ in Snf5 is involved in forming protein assemblies in the cell that could facilitate protein-protein interactions.
Figure 4. Homorepeat (PolyQ) in Snf5 affects fitness and adaptability by mediating protein-protein interactions and influencing diverse biological processes.
(a) Yeast strains with wildtype (WT) or polyQ HR deleted (ΔHR) Snf5 tagged with HA were grown in YP+2% glucose (YPD) and YP+4% glycerol (YPG). (b) Mean growth curves of WT or ΔHR (n=4). Doubling time (Td) was derived from the slope of the best fit obtained by linear regression. (c) Immunoblot analysis of HA-tagged Snf5 with loading control (Tubulin). (d) Immunofluorescence analysis of Snf5 using anti-HA (red) and anti-tubulin (green). Magnified images of HA staining (right panel) and the ratio of HA area/Nuclear area for ~40 cells in each condition (bottom panel) are provided. Statistical significance was assessed using Wilcoxon rank sum test. (e) The pie chart depicts polyQ-dependent interactions of Snf5 (observed in WT but absent in ΔHR) in YPD and YPG. The bottom chart shows the diverse biological processes affected by polyQ dependent interactors (numbers in parenthesis). Snf5 interactors were identified with a minimum of 2 unique peptides in atleast 2 of the 3 experiments. (f) Immunoblot analysis of histone modifications from Snf5 WT or ΔHR strains, with total histone H3 as loading control. (g) Heatmap showing differences in the transcript levels of Snf5 targets (WT and ΔHR), quantified by qPCR. Dark black borders represent statistically significant alteration in gene-expression relative to WT (student’s t-test; FDR corrected for multiple testing). Each target is linked to its biological process on the left. (h) Colony formation assay of WT or ΔHR SNF5 yeast strains in serial dilution assay, following exposure to genotoxic stress. (i) Schema illustrating that polyQ HR in Snf5 contributes to fitness by mediating interactions with proteins belonging to diverse biological processes and formation of higher order assemblages.
To test this, we experimentally determined the interactome of WT and ΔHR Snf5, in YPD and YPG using affinity capture followed by mass spectrometry (Fig. 4e). We identified a total of 146 interactions for WT Snf5, of which 130 were not reported before (Supplementary Dataset 1; Supplementary Fig. 5b-c). We detected all Swi/Snf complex members in both strains and both growth conditions, suggesting that polyQ in Snf5 does not affect formation of the Swi/Snf complex. However, 12/89 (~13.5%) interactions in YPD, and a striking 35/109 (~32%) interactions in YPG are mediated in a polyQ dependent manner (Supplementary Fig. 5d). These polyQ dependent interactions involve proteins with diverse functions such as chromatin remodeling, transcription, cell cycle and DNA repair (Fig. 4e). Therefore, Snf5 might influence different cellular processes through its polyQ dependent interactions with the different complexes/proteins in the cell (Supplementary Notes 2). In other words, the repeat dependent membership of Snf5 in different assemblies might provide distinct molecular contexts (e.g. transcription, DNA repair, cell cycle etc.) for realizing the biochemical function/activity of Snf5.
In line with this possibility and consistent with the loss of interactions with members of other chromatin remodeling complexes and transcription factors, deletion of polyQ in Snf5 resulted in a global reduction of active histone marks such as H3K4 trimethylation and H3K27 acetylation (Fig. 4f). A number of known Snf5 targets display significantly altered expression levels between WT and ΔHR in both carbon sources, with more pronounced effects in YPG (Fig. 4g). The WT strains were able to grow after exposure to UV or in the presence of hydroxyurea but the ΔHR mutants were susceptible in both conditions (Fig. 4h) suggesting that polyQ in Snf5 confers adaptability and fitness in response to genotoxic insults.
Thus, the presence of polyQ expands the functional versatility of Snf5 beyond that of the Snf5 domain, by facilitating interactions with proteins from diverse biological processes in a polyQ-dependent manner (Fig. 4i). In this way, the polyQ HR within Snf5 influences different phenotypes and determines fitness and adaptability in diverse environments.
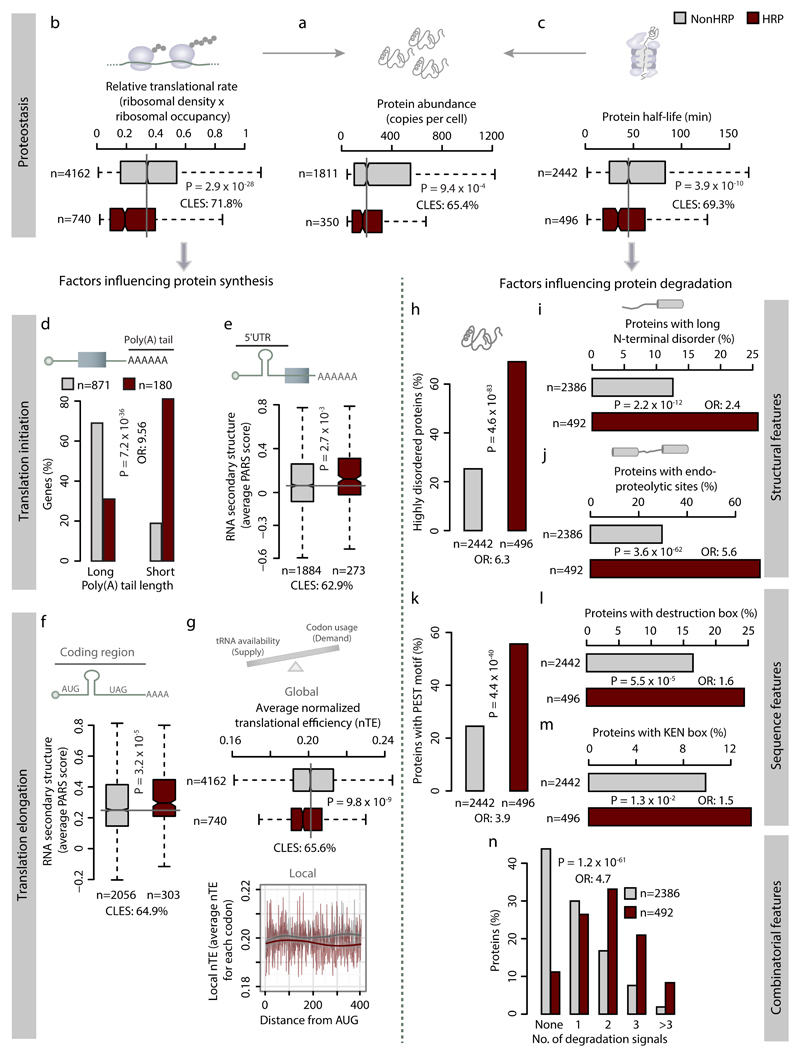
The activity of HRPs is highly regulated in cells
Given the functional versatility, pleiotropic effects and the ability of proteins with HRs to spatially organize proteomes, we investigated how their activity is regulated in cells. Protein activity could be regulated by (i) controlling steady state abundance (coarse-tuning) and/or (ii) altering chemical states by post-translational modifications (fine-tuning). Proteins with HRs are generally less abundant, have lower synthesis rates and are turned over more rapidly than NonHRPs (Fig. 5a-c). Such differences collectively affect the abundance and the amount of time that HRPs spend in the cell. Importantly, this stringent control of HRPs is more pronounced compared to highly disordered NonHRPs and NonHRPs with non-repetitive amino acid bias, as discerned from synthesis and degradation rates (Supplementary Fig. 6a-c). Such a tight regulation is critical for fitness, because over-expression of HRPs is more often toxic (Supplementary Fig. 6d).
Figure 5. HRPs are stringently regulated at multiple levels during synthesis and degradation.
Boxplots showing distribution of (a) protein abundance, (b) translational rate, (c) protein half-life, (e) 5' UTR and (f) coding region mRNA secondary structures, and (g) translational efficiency among HRPs and NonHRPs (left panel). Statistical significance was assessed using Wilcoxon rank sum test, with FDR corrected P-values and effect size as CLES provided in each panel. The right panel of (g) shows the local translational efficiency (TE) represented as the average TE for each codon from AUG to 400 codons for HRPs and NonHRPs. Barplots showing percentages of HRPs and NonHRPs with (d) long and short poly(A) tails, (i) long N-terminal disorder, (j) endo-proteolytic sites, (k) PEST motifs, (l) Destruction box, (m) KEN box and (n) Combinatorial degradation signals. Percentage of proteins that are highly disordered (protein disorder >30%) among HRPs and NonHRPs is shown in panel h. Statistical significance was assessed using Fisher’s exact test and corrected for multiple testing and effect sizes displayed as OR. For panel (n) OR was estimated for the presence of more than one degradation signal.
Transcripts of HRPs are enriched for shorter poly(A) tails and have more complex 5' UTR structures (Fig. 5d-e), both of which reduce translation initiation rates26,27. They more often harbor secondary structures in the coding region (Fig. 5f), and contain sub-optimal codons, even after accounting for the codon ramp (Fig. 5g). These features might slow both translation elongation and global translation efficiency28,29. At the protein level, HRPs tend to be enriched for high disorder content (Fig. 5h), long disordered segments at N-terminus and internal regions (Fig. 5i-j), putative PEST, D-box and KEN box motifs compared to NonHRPs (Fig. 5k-m), all of which facilitate rapid protein turnover30–34. The presence of more than one molecular feature within HRPs (Fig. 5n) suggests that their activity could be regulated through rapid degradation in diverse conditions by different mechanisms. The ability to more stringently regulate the abundance of HRPs could control the equilibrium between the soluble and phase-separated structures, and ensure that the spatially organized proteome remains dynamic and under control32,35.
In terms of fine-tuning protein activity, HRPs have more post-translational modifications (Supplementary Fig. 6e) than NonHRPs, suggesting that they are regulated by a variety of signaling proteins that may be active in different conditions. The distribution of PTM sites show that they occur within and immediately around homorepeats (Supplementary Fig. 6f). These PTMs might act as interaction switches and determine the membership of HRPs among specific complexes36. Diverse PTMs, such as phosphorylation and acetylation could not only promote protein degradation37–39, but also regulate the dynamic and reversible assembly of phase separated structures40. We note that our observations do not rule out the direct involvement of HRs in proteostasis. For instance, mRNA secondary structures formed by tri-nucleotide repeats (encoding amino acid HRs) might contribute to slow translation elongation. Additionally, HRs of certain amino acid types and/or lengths can directly influence protein solubility41 and stability42.
Although the reported trends may be applicable to a large number of HRPs, they are unlikely to apply to every single HRP. Nevertheless, features such as pleiotropic effects, adaptability, functional versatility and proteostasis can collectively differentiate HRPs and NonHRPs through a random forest machine learning based approach (Supplementary Fig. 7).
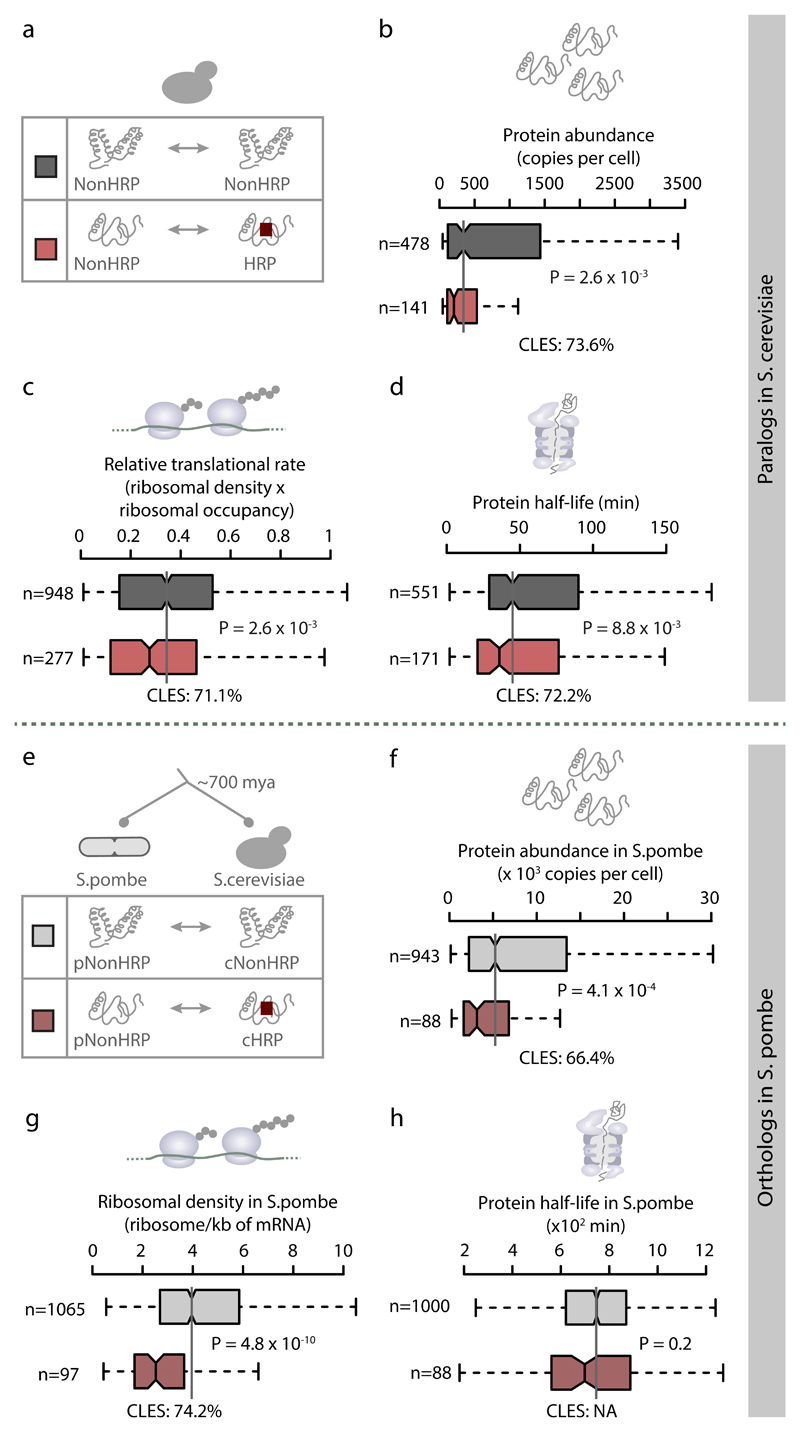
Stringent proteostasis is a pre-requisite for homorepeat retention during evolution
During evolution, does the emergence of HRs in proteins leads to their stringent proteostasis, or are HRs retained in proteins that are already stringently regulated? By comparing groups of NonHRPs that do or do not have HRP paralogs (Fig. 6a), we find that NonHRPs with HRP paralogs are low in abundance, have slower translational rates and display shorter half-lives (Fig. 6b-d) compared to the group of NonHRPs that do not have HRP paralogs. This suggests that although HRs might emerge in any protein, they might be preferentially retained in proteins that are already stringently regulated. We then investigated the one-to-one orthologs of S.cerevisiae in Schizosaccharomyces pombe (estimated divergence time of ~700 million years). We classified the NonHRP orthologs of S.pombe (pNonHRPs) into those that are still NonHRPs in S.cerevisiae (pNonHRP-cNonHRP) and those that have HRs in S.cerevisiae (pNonHRP-cHRP; Fig. 6e) and compared their regulatory properties. The pNonHRP-cHRP orthologs are significantly less abundant and have lower translational rates compared to pNonHRP-cNonHRP orthologs in S.pombe (Fig. 6f-g). However, the half-lives were comparable (Fig. 6h). This suggests that during evolution, HRs were most likely retained in already stringently regulated proteins. This is consistent with the idea that pre-existing proteins that are stringently regulated are better poised to tolerate the emergence of HRs as their regulation readily minimizes their negative effects (e.g., uncontrolled aggregation, leading to protein sequestration and loss of function). This may also explain why the presence of HRs is tolerated in only certain proteins.
Figure 6. Homorepeats tend to be retained in proteins that are stringently regulated.
Distribution of (b) protein abundance, (c) translation rate and (d) protein half-life among NonHRPs with HRP paralogs and NonHRPs whose paralogs never acquired a HR in S.cerevisiae (panel a). (e) Classification of NonHRPs in S.pombe (pNonHRPs) into those whose one-to-one orthologs in S.cerevisiae are either NonHRPs (pNonHRP-cNonHRP) or HRPs (pNonHRP-cHRP). Boxplots of distributions of (f) protein abundance, (g) ribosomal density for protein synthesis rates and (h) protein half-life for the two classes of pNonHRPs in S.pombe. Statistical significance was assessed using Wilcoxon rank sum test with a false discovery rate correction for multiple testing and effect sizes are displayed as CLES.
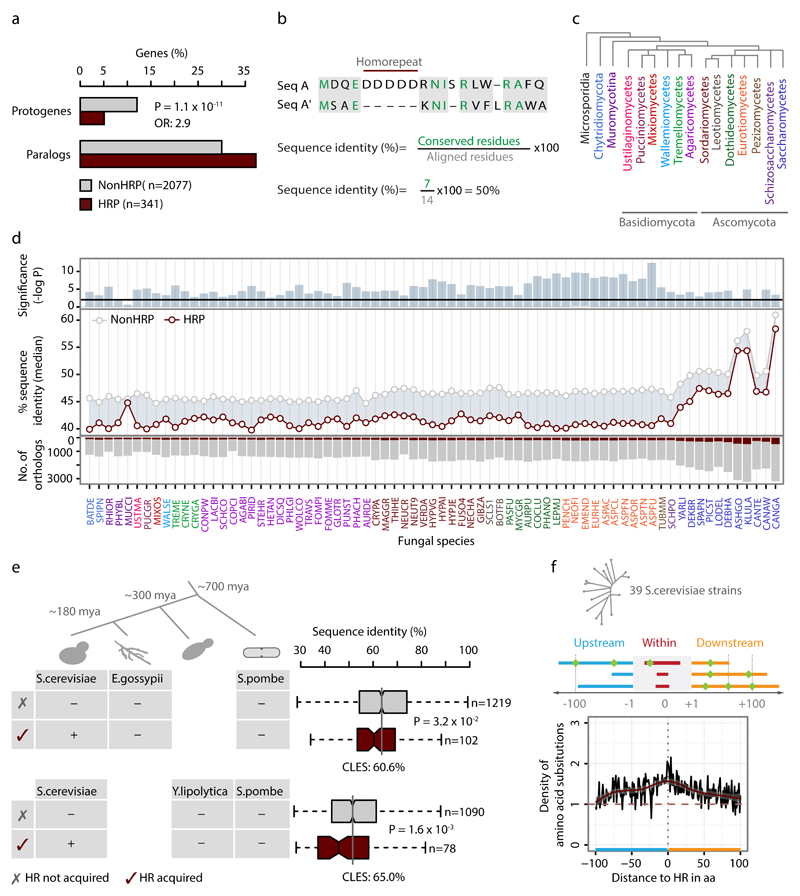
Proteins that contain a HR diverge rapidly across different time-scales
Proteins that contain a HR are more represented among paralogs (gene duplicates) than among protogenes, which originate de novo (Fig. 7a). This is in line with the observation that HRs tends to be retained in pre-existing genes that are under stringent proteostasis. To investigate if proteins that contain a HR evolve differently from those without a HR, we computed the sequence identity of one-to-one orthologs of yeast HRPs and NonHRPs across different evolutionary time scales (74 fungal species; ~1 billion years of evolutionary distance; Supplementary Notes 3). Only the alignable regions were considered and gaps were not considered (Fig. 7b). HRPs in S.cerevisiae show lower sequence identity among their corresponding orthologs across almost all the fungal species compared to NonHRPs (Fig. 7c-d), suggesting that HRPs diverge faster than NonHRPs across species. The average median difference between NonHRPs and HRPs is 4.6% which corresponds to ~14 amino acid substitutions for a 300 amino acid long protein. The saturation effect (i.e. same position can be mutated several times given sufficient time in evolution) resulting from the several variable sites could be one of the reasons for observing the linear shift in the rate of change of sequence identity of HRPs compared to NonHRPs, over long evolutionary time-scales (Fig. 7d, middle panel). To investigate whether HRs contribute to the rapid divergence of HRPs, we first identified S.cerevisiae proteins that acquired a HR at two different time points: from the time of speciation from the common ancestor of S.cerevisiae and (a) Eremothecium gossypii (~180Mya), and (b) that of Yarrowia lipolytica (~300Mya) using S.pombe as an out-group. In both situations, orthologs that acquired HRs in S.cerevisiae diverged more than those that did not acquire a HR (Fig. 7e). Similarly, paralogs that have acquired a HR diverged faster than their NonHRP counterparts (Supplementary Fig. 8a). These results suggest that the presence of HRs is associated with and may drive the rapid divergence of proteins harboring them.
Figure 7. HRPs preferentially arise by gene-duplication and rapidly diverge across different time-scales.
(a) Proportion of HRPs and NonHRPs that arise by de novo gene birth (protogenes) or gene duplication (paralogs). Statistical signficance was assessed using Fisher’s Exact test, corrected for multiple testing, with effect size represented using OR. (b) Estimation of sequence identity among yeast orthologs and paralogs by considering only aligned positions and disregarding gaps which might arise due to HRs. (c) Fungal phylogenetic classes studied here. (d) Sequence divergence among yeast HRPs and NonHRPs with their one-to-one orthologs in 73 of 74 fungal species (with at least 100 orthologs for HRPs in each species). The color of the abbreviated species names corresponds to phylogenetic classes in panel c. Taxonomic and phylogenetic details of the fungal species are provided in Supplementary Notes. Median divergence of HRPs and NonHRPs with their orthologs in each species is shown (middle panel). Statistical significance was assessed by comparing the distribution of divergence of yeast HRPs and NonHRPs with their orthologs in each species using Wilcoxon rank sum test (upper panel). The black line shows P-value cut-off corrected for multiple testing. The bottom panel shows the number of orthologs of yeast HRP (red) and NonHRP (grey) in each species. (e) Classification of S.cerevisiae proteins into those that have or have not acquired HRs based on the HR status of their corresponding orthologs in E.gossypii and Y.lipolytica, using S.pombe as an out-group (left panel). Boxplots showing the distributions of sequence identity of yeast proteins that have or have not acquired HRs (right panel). Statistical significance was assessed using Wilcoxon rank sum test and effect size displayed as CLES (f) Density of amino acid substitutions within (0 in the X-axis) and 100 amino acids on either side of the HR, among yeast strains.
At shorter time-scales of divergence (~thousands of years), i.e. among the 39 different strains of S.cerevisiae (~thousands of years), the regions around HRs tend to accumulate more amino acid substitutions compared to the rest of the protein (Fig. 7f), although the density of such substitutions between HRPs and NonHRPs over the entire protein was comparable. This observation supports the emerging view that there is a higher tendency to find substitutions in and around repeat containing regions in the genome43–46, and highlight the potential impact of HRs in proteome evolution. Collectively, these findings suggest that proteins containing a HR tend to acquire more amino acid substitutions and evolve rapidly, irrespective of the time-scale considered.
HR-associated variation might rewire interactions and facilitate rapid adaptation
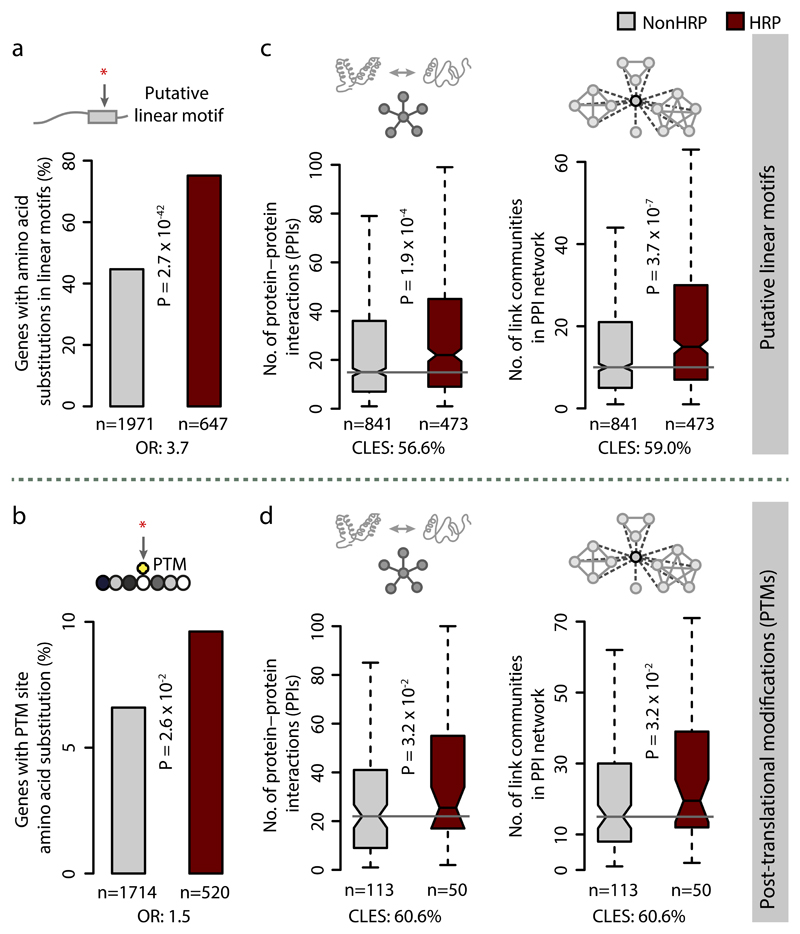
To assess whether the HR associated substitutions affect functional sites in a protein, we integrated the amino acid substitution data for the different the yeast strains with functional site information. Compared to NonHRPs, amino acid substitutions within HRPs more often map to putative linear motifs and PTM sites that mediate protein-peptide interaction (Fig. 8a-b). HRPs with substitutions in these functionally relevant sites have higher number of interactions and link communities in the protein interaction network, compared to NonHRPs that also have substitutions in such sites (Fig. 8c-d). The differences remain significant even after controlling for the number of protein interactions or the density of linear motif residues or PTM sites (Supplementary Fig. 8b-e). Furthermore, the conditional probability of finding (i) amino acid substitutions and (ii) those that affect functionally relevant sites is higher in polypeptide segments that contain HRs than that of finding a HR in polypeptide segments with amino acid substitutions (Online Methods and Supplementary Fig. 8f-g). These observations collectively suggest that HR-associated variation has the potential to alter protein interaction networks and rewire the targets of signaling pathways. Thus, HRs act as distinct modules that increase the genetic diversity of a population (i.e. standing genetic variation) by affecting functionally relevant sites around a HR in key proteins.
Figure 8. Amino acid substitutions in HRPs often affecting functionally relevant sites.
Distribution of HRPs and NonHRPs with substitutions within (a) putative linear motifs, identified using ANCHOR 61 and (b) experimentally determined PTM sites. Boxplots showing distributions of protein-protein interactions and link communities in the protein interaction network among HRPs and NonHRPs with amino acid substitutions in (c) putative linear motifs and (d) PTM sites. Statistical signficance was assessed using Wilcoxon rank sum test and effect size displayed as CLES (c and d) and Fisher’s exact test with ORs indicating the effect size (a and b), corrected for multiple testing.
A more detailed investigation of essential and highly pleiotropic HRPs reveals that they might form a rapidly evolvable part of the proteome (Supplementary Fig. 9-10). Thus, HR-associated variation of pleiotropic and critical hub proteins among individuals in a population may facilitate the adaptation of an organism to diverse environments by permitting rapid exploration of the genotype-phenotype landscape.
Discussion
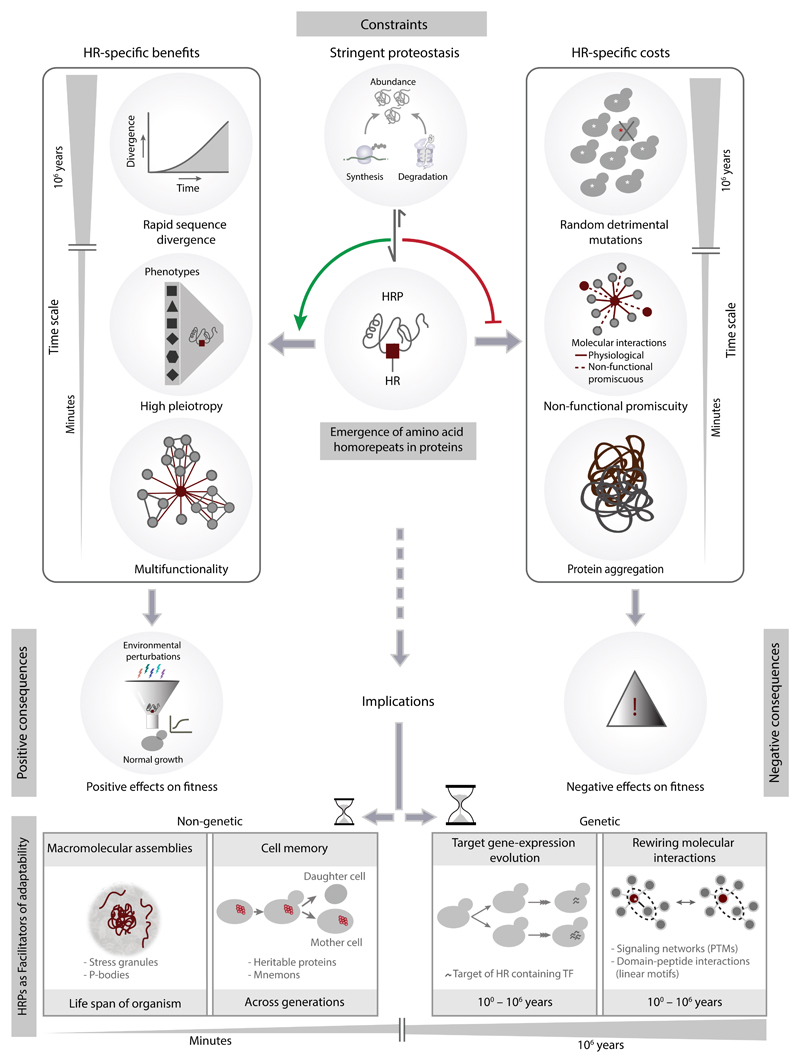
From a biochemical and cell biological perspective, our observations reveal that the presence of HRs has the potential to increase the functional versatility of proteins by facilitating repeat-dependent interactions and their spatial organisation in cells by forming assemblies. From a genetics and evolutionary perspective, HRs facilitate rapid protein divergence, might rewire interactions and favour the emergence of standing genetic variation upon which selection could operate. The emergence of homorepeats in a protein can have both positive and negative consequences at different time-scales (Fig. 9).
Figure 9. Constraints, consequences and implications of the emergence of homorepeats in proteins.
The top panel illustrates that stringent proteostasis facilitates retention of HRs in proteins, favoring HR-specific benefits on fitness and alleviating their negative effects on fitness, at different time-scales. The bottom panel highlights how HRPs facilitate evolvability and adaptability at different time-scales through homorepeat dependent non-genetic and genetic effects.
Our findings show that HRs are often present in proteins that influence diverse phenotypes and buffer environmental insults. The existence of repeat dependent interactions highlights an underappreciated component of our understanding of protein interaction networks, and opens up new directions for future research in the area of repeat edgetics, similar to the concept of single amino acid edgetics47. HR dependent interactions can provide new molecular contexts for a protein at different times and in different conditions where the biochemical function of the HRP can be realized in a spatially localized and regulated manner in a cell. However, in certain situations such as altered protein abundance or abnormal repeat length variation, their high interaction potential can lead to negative consequences such as sequestration of other proteins via non-functional promiscuous interactions and protein aggregation48.
At longer time-scales, the presence of HRs leads to rapid divergence of proteins due to variation in repeat length8 and accumulation of more amino acid substitutions. Diverse mechanisms such as recurrent repair upon replication stalling at nucleotide repeats, unequal crossing over following recombination, transcription coupled repair and the higher error rates of non-replicative DNA polymerases that are recruited at such sites during repair might all collectively contribute to nucleotide repeat-induced mutagenesis and lead to the rapid sequence divergence of HRPs43,46,49–52. In addition, the lower expression levels of HRPs might also influence its rapid evolutionary rate53. The association of homorepeats with disordered regions might also influence the rate of divergence. Detailed analysis of the nature of amino acid substitutions may help uncouple the influence of disordered regions and homorepeats on the rapid divergence of proteins with HRs. Thus the presence of a HR results in standing genetic variation of a population. Although a deleterious consequence of the HR-associated amino acid substitution might be detrimental for an individual, it does not affect the entire population. However, if one of the individuals carrying a specific HR-associated mutation has better fitness when environments change, that specific mutation may be positively selected and benefit the species. Thus, HRs can be considered as an evolutionary module that confers evolvability and adaptability to organisms in a population.
The beneficial effects of HRPs raise the evolutionary conundrum of why their presence is restricted to certain proteins. Our results suggest that HRs tend to be retained in proteins whose abundance and activity are under stringent control, possibly to minimize HR-associated deleterious effects such as the sequestration of proteins through non-functional promiscuous interactions and protein aggregation. This suggests that stringent proteostasis might be a requirement for the tolerance of HRs in a protein.
HRs in a protein might facilitate adaptability at different time-scales (Fig. 9, bottom panel). At shorter time-scales HRPs facilitate adaptability through non-genetic mechanisms. For instance, they facilitate formation of assemblies, such as stress granules, that permit adaptation under diverse conditions54. They also act as heritable proteins55 and contribute to trans-generational adaptability, by forming physical entities that encode molecular memory as in the case of polyQ/polyN containing mnemons of yeast Whi3p56. At longer time-scales they might facilitate adaptability through genetic mechanisms. Recently, we showed that HR length variation in transcription factors facilitates adaptation through rapid expression divergence of their targets41. Here, we find that HR associated amino acid substitutions more often affect linear motifs and PTMs. Such variation might fuel evolution of interaction sites and permit rapid evolution of new substrates for kinases or modifying enzymes of signaling pathways. Thus, the presence of a HR might lead to the rewiring of molecular networks such as signaling (through PTM site variation) and domain-peptide interaction networks (through linear motif variation)57,58. In this manner, HRPs can generate genotypic and phenotypic diversity and facilitate organismal adaptation to new environmental niches.
While we report general trends for proteins with HRs, and provide experimental validation for specific cases, every trend observed here might not be applicable to all proteins with homorepeats. Furthermore, in addition to effects of repeats at the protein level (i.e. proteome), they could also have various consequences at the nucleic acid level (i.e. genome and transcriptome). Although the distribution of the type and length of HRs vary across eukaryotes, our findings in yeast provide important insights for understanding their relevance in other species including humans.
The current understanding of the role of proteostasis in conferring robustness and evolvability primarily focuses on molecular chaperones, such as Hsp90, involved in protein folding and enabling accumulation of cryptic variation in a population59. The findings presented here points to the existence of another strategy, wherein stringent proteostasis facilitates the retention of homorepeats, which in turn leads to rapid protein divergence that may fuel rewiring of interaction networks and signaling pathways. Given that HRPs are more prevalent among paralogs (i.e. gene duplicates), it suggests that the genetic redundancy coupled with the rapid mutation accumulation of HRPs provides a ‘low risk-high gain’ for accelerating evolution and adaptation to diverse environments. The synergy between stringent proteostasis, increased evolvability and functional versatility make HRPs important for fitness and vital contributors to the functioning of cells.
Online Methods
Genome-level computational investigations
Classification of yeast proteome based on
(i). The presence of homorepeats
Yeast proteins were classified into proteins with homorepeats (HRPs) if they harbored at least one continuous stretch of ≥5 identical amino acids. The proteins that lacked HRs (NonHRPs) were used as the control group for all genome-scale comparisons. To ensure that we used the most appropriate control set, we disregarded all NonHRPs, which contained imperfect repeats (n=650). Imperfect repeats included all repeats of unit sizes two and longer, with similarity score of 70% or higher between repeating units. These were identified using TREKS62. A compendium of different genome-scale datasets analyzed in this study is provided in Supplementary Table 1.
(ii). Disorder fraction
The disorder status of every residue in the yeast proteome was inferred using DISOPRED263 and the disorder content averaged over the entire protein length, assigned as the disorder fraction of a protein. Repeats are known to overlap with disordered regions64. To examine if protein disorder confounded our observations on the functional attributes and proteostasis of HRPs, we compared distributions of diverse features of HRPs with low (≤30%) and high intrinsic disorder (>30%) with NonHRPs with similar intrinsic disorder content (Fig. 2e-f, Supplementary Figs. 6 and 7).
(iii). Protein length
To examine if the functional attributes of HRPs are independent of protein length, we selected HRPs and NonHRPs with similar lengths and compared distributions of connectivity in protein interaction network and the number of link communities each protein participates in (Supplementary Fig. 2b).
(iv). Non-repetitive amino acid bias
We compared distribution of different features among HRPs and NonHRPs containing non-repetitive amino acid bias (Supplementary Figs. 2d and 6), to investigate if the observed trends for physiological importance, functional versatility and stringent proteostasis are similar between these two groups. Sequences with compositional amino acid bias in NonHRPs were identified using LPS-annotate employing default parameters65,66, but employing a stringent detection P value cutoff of <1 × 10−7, (~ double the default detection threshold).
(vi). Functional similarity
To rule out the confounding effect of differences in functional versatility of HRPs resulting from the differences in biological functions between HRPs and NonHRPs, we randomly selected NonHRPs with similar biological functions as HRPs and drew comparisons. We first assigned Gene Ontology biological processes to HRPs using GOSlim annotation retrieved from The Saccharomyces Genome Database (SGD)67. About 97 GO terms could be assigned to HRPs. For each GO term we counted the number of HRPs. We then obtained at random, equivalent numbers of NonHRPs for each GO term and compiled them into a single list of “functionally similar NonHRPs”. We compared the differences in distributions for number of protein-protein interactions, genetic interactions, link communities in protein-protein interaction network and genetic network between the two sets of functionally similar groups of HRPs and NonHRPs, for each of the 100 randomizations (Supplementary Fig. 2c).
(vi). Essentiality for growth
To examine if the physiological and functional impact, regulation and evolution of HRPs are influenced by enrichment of HRPs for essential genes, we undertook comparisons of distributions of different features between essential HRPs and essential NonHRPs (Supplementary Fig. 9).
(vii). High pleiotropy
We classified yeast proteins into those with low, medium and high pleiotropic effects based on the number of phenotypes they were linked to in the yeast gene-to-phenotype network using tertile cutoffs. To examine if high pleiotropic effects of HRPs influence the physiological and functional impact, regulation and evolution of HRPs, we compared distributions of different features between highly pleiotropic HRPs and highly pleiotropic NonHRPs (Supplementary Fig. 10).
Classification of yeast paralogs
Paralog protein pairs were identified as described in the Supplementary Table 1. Based on the presence/absence of homorepeats, paralog pairs were classified as similar pairs, where both the paralogs lacked HRs (NonHRP:NonHRP) or divergent pairs, where one paralog contained the HR while the other did not (HRP:NonHRP). To examine if the emergence of HR within a protein facilitated acquisition of physiological and functional attributes of HRPs, we compared members of divergent pairs (Fig. 3f). To test if HRs emerge in proteins that are already stringently regulated during evolution, we compared NonHRPs of similar pairs with NonHRPs belonging to divergent pairs (Fig. 6a-d). To test if HRs facilitate rapid evolvability, we tested for differences in the distribution of sequence identities among similar (NonHRP:NonHRP) and divergent (HRP:NonHRP) paralog pairs (Supplementary Fig. 8a).
Classification of yeast proteins and their orthologs
One-to-one orthologs of yeast proteins across different fungal proteomes were obtained from OMA browser68 (Supplementary Table 1). To test if, during evolution, HRs tend to emerge and are retained in already stringently regulated proteins across species, we classified S.pombe NonHRP (pNonHRPs) orthologs of S.cerevisiae proteins into those whose S.cerevisiae orthologs have not acquired HRs (pNonHRP-cNonHRP) and those that have acquired HRs (pNonHRP-cHRP). Protein regulatory features measured in S.pombe were compared for these two classes of pNonHRPs (Fig. 6e-h).
To test if HRs facilitate rapid divergence of HRPs across evolutionary time-scales, we identified S.cerevisiae proteins that (a) acquired a HR from the time of the common ancestor of S.cerevisiae and E.gossypii (~180Mya), and that of S.cerevisiae and Y.lipolytica (~300Mya) using S.pombe as an out-group and (b) did not acquire a HR. We then compared the distribution of protein sequence identity between these two groups (Fig. 7e).
Gene Ontology enrichment analysis
Gene Ontology (GO) biological process enrichment among HRPs, essential HRPs and NonHRPs, highly pleiotropic HRPs and NonHRPs was obtained using DAVID server69. FDR values, corrected for multiple testing highlight the significance of the enrichment of a GO term.
Calculation of sequence identity
We obtained one-to-one orthologs of yeast proteins in 74 fungal species from OMA browser68. For each pairwise alignment, we computed percentage sequence identity using Biostrings package in R by considering only the aligned regions, ignoring gaps, to ensure that gaps arising from emergence of repeats do not confound the calculation of sequence identity.
Statistical analysis
Differences in the distribution of continuous variables among different classes of proteins were assessed using the non-parametric tests- Wilcoxon rank sum test or Wilcoxon matched pairs test, as appropriate. Median and confidence intervals (C.I. = 1.58(IQR/√n), where IQR is the interquatile range and ‘n’ is the sample size of each group) were computed. Median differences between the compared groups and Common Language Effect Size (CLES)70 were calculated as estimates of the effect size. CLES describes the probability that a randomly sampled data point from a distribtion A, will be greater than a data point sampled from distribution B and was computed as previously described71. Briefly, based on the recommendation from 72, we first computed U-statistic:
where W is the Wilcoxon test statistic, ns is the smaller of na and nb (the sample size of each dataset). CLES is then given by:
For comparisons done using Wilcoxon matched pairs test (Fig. 3f), Z-scores were estimated using the R coin package. Differences in distributions of categorical variables were tested using Fisher’s exact test or Chi-squared test. Effect sizes were estimated using odds ratio (OR). Enrichment of HRPs in different classes of proteins (e.g. stress granule proteins) was assessed using permutation test. In each permutation, every HRP was replaced with a random gene. The number of such randomly obtained genes that overlapped with a specific class of proteins was noted for 10,000 randomizations. From this we estimated the Z-score, which indicates the distance of the actual observation to the mean of random expectation in terms of the number of standard deviations. P-values were estimated as the ratio of the randomly observed proteins greater than or equal to the number of actually observed HRPs to the total number of random samples (10,000). Null expectation of homorepeats of length ≥5 in yeast proteome, Z-score and P-value was obtained by permutation test using 1000 shuffled proteomes by shuffling each protein for sequence order, keeping the length and amino acid composition constant. Test for association between the length of homorepeats and different features were done using Pearson’s correlation. Extent and the type (positive/negative) of correlation were estimated by Pearson’s correlation coefficient. Correction for multiple testing was done by Benjamini-Hochberg method (FDR) to control for false discovery rate for comparisons between similar classes (such as HRPs versus NonHRPs), using the same test (such as Wilcoxon rank sum test or Fisher’s Exact test) but different datasets pertaining similar biological attribute such as physiological and functional relevance and proteostasis (Supplementary Table 1).
For Bayesian inferences, features were classified as either categorical (e.g., essentiality) or quantitative (e.g., no. of phenotypes per gene) depending on the data type (Supplementary Fig. 3). Using tertile-cutoffs quantitative features were classified into three bins as low, medium and high. For features where HRPs had significantly higher values than NonHRPs (e.g., no. of phenotypes per gene) we selected the ‘high’ bin, whereas for features in which HRPs had significantly low values than NonHRPs (e.g., protein abundance) we selected the ‘low’ bin. Conditional probabilities for finding a feature given that the protein contains a HR and for finding a protein with HR given a feature were estimated. Based on the conditional probabilities, we obtained likelihood ratio (LR) estimates as the ratio of probability of finding a feature given that the protein contains a HR divided by the probability of finding a feature given that the protein does not contain a HR. All statistical analyses were performed using R.
The ability of the features associated with HRPs, pertaining to (i) pleiotropy (no. of phenotypes/gene and essentiality), (ii) adaptability (resistance to small molecules), (iii) functional versatility (disorder fraction, no. of protein-protein interactions, no. of link communities in PPI network, no. of GO functions) and (iv) proteostasis (protein abundance, relative translational rate and protein half-life) to distinguish between HRPs and non-HRPs was assessed using a random forest (RF) model (For details see legend of Supplementary Fig. 7).
Trained models were evaluated on the test set and performance summary metrics (precision, recall, and F1 scores) were calculated. Precision reflects that ability of the classifier not to label a NonHRP as a HRP or vice versa and is computed as follows:
Recall reflects the ability of the classifier to find all the positive samples and is computed as follows:
F1 is the weighted harmonic mean of the precision and recall. Features were ranked in relative importance by their mean decrease in impurity, weighted by the probability of reaching the associated node. Feature importance values were normalized to the maximum value. Modeling was performed using the scikit-learn Python library73. Since disorder fraction was one of the important features that distinguished HRPs and NonHRPs, an additional RF model was trained to distinguish between HRPs (n=611) and highly disordered proteins (HDPs; NonHRPs with disorder fraction >30%; n=770). The dataset was first balanced by subsampling HDPs to reach n=650. RF hyperparameters were tuned as before, using 10-fold cross-validation and grid search (yielding optima: criterion="entropy”, max_features="log2”, max_depth=50).
Gene-level experimental investigations
Strains and culture conditions
Yeast genetic manipulations were performed using standard methods74 in the endogenous Snf5 locus in S288c strain. Primers used for amplifications and generating strains are listed in Supplementary Table 2. Deletion of the repeat region in the mutant (ΔHR) was verified by sequencing with primers flanking the repeat region. Additionally, we created 6xHA-tagged versions of the SNF5 WT and ΔHR, from plasmid pYM14 (Euroscarf). Fresh colonies, grown at 30°C for 3 days in solid YPD, were picked and incubated over-night in liquid YPD at 30°C. These cells were employed as start-up for cultures that were grown over night in YPD (YP with 2% glucose) or YPG (YP with 4% glycerol) and 30°C, for different experiments.
Cell growth rate measurement
Using an overnight culture, we achieved an initial OD600 of 0.001 in YPD or YPG. The growth of the cells was followed in a Tecan 2000 pro plate reader measuring the absorbance at 600nm every 10min for 3 days. For budding index measurements, 0.5x106 cells in stationary phase were reinoculated into 5ml of YPD or YPG and samples collected after indicated time points. Samples were then analysed by Vi-cell, images acquired and mitotic cells were quantified by using Fiji (Image J) software. Results depicted are mean of 3 independent experiments for 50 fields each.
Immunofluorescence analysis
Cells in exponential phase were fixed in 4% formaldehyde for an hour. Spheroplasts of fixed cells (prepared in 3.2μl of β-Mercaptoethanol and 5μl of 5mg/ml zymolase) were permeabilised in 100μl PBS+0.05% Tween20. About 25μl of this suspension was then loaded onto 96 well plates and settled for 5min. Cells were blocked in 1mg/ml BSA in PBS for 30min in a humid chamber. After 3 washes with PBS, primary antibody cocktail (1:500 of HA and tubulin) was added and incubated for 1h at room temperature. Subsequently, cells were washed and secondary antibody mix containing rabbit Alexa Flour 555 and mouse Alexa 488 was added (1:1000 in blocking solution) and incubated in the dark for 1h. Samples were finally washed in PBS and mounted in propyl gallate solution with DAPI. Samples were stored in 4°C until visualization. Images were acquired using LSM710 microscope (Zeiss) and images rendered and quantified using Imaris software (Bitplane, UK).
Immunoblotting and Immunoprecipitation analysis
Yeast cells expressing HA tagged WT or ΔHR SNF5 grown in YPD or YPG were lysed in HEPES buffer containing 100mM KCl, 150mM KOac, Protease inhibitor cocktail and 0.1% Triton X-100 using a glass bead beater. Samples were then centrifuged at 4°C, at 10000rpm for 10min and the lysate was collected. For immunoblot analysis, 30μg of total protein from each condition was loaded on a 4-12% NuPAGE gel and transfer was performed with iblot 1.0 as per manufacturer’s instruction (Invitrogen). Primary antibodies used were HA-tag (Millipore 1:1000 in milk), and Tubulin (Sigma, 1:2000 in milk). For immunoprecipitations, 10mg of protein from each condition was precleared with 20μl of Agarose beads for 30min. The precleared lysate was incubated with 50μl of HA tagged Dynabeads and incubated at 4°C for 4h washed 4 times with lysis buffer and three times with 25mM Ammonium bicarbonate. Yeast histones were extracted using the established protocol 75 and immunoblotted in a 4-20% gel.
NanoLC-MS/MS analysis
Bead bound proteins were tryspin digested twice by the addition Trypsin. Peptides were acidified with Formic Acid to a final concentration of 5% and desalted before injection. Separation was achieved using an Acclaim PepMap 100 column (C18, 3μl, 100 Å, ThermoScientific) with an internal diameter of 75μm and capillary length of 25cm. A flow rate of 300nl/minute was used with a solvent gradient of 5% B to 45% in 50min followed by an increase to 95% in 25 min. Solvent A was 0.1% Formic Acid, 2% MeCN and 5% DMSO v/v in water and Solvent B with similar composition containing 80% MeCN.
The mass spectrometer was operated in positive ion mode using an Nth order double-play method to automatically switch between Orbitrap-MS and LTQ Velos-MS/MS acquisition. Survey full-scan MS spectra (from 400 to 1,600 m/z) were acquired in the Orbitrap with resolution (R) 60,000 at 400 m/z (after accumulation to a target of 1,000,000 charges in the LTQ). The method used allowed sequential isolation of the 20 most intense ions for fragmentation in the linear ion trap. Charge state screening was enabled, and precursors with unknown charge state or a charge state of 1 were excluded. Acquired RAW files were analysed using the Sequest 76 and Mascot 77 search engines running under Proteome Discoverer version 1.4. The Saccharomyces cerevisiae protein database used for searches was retrieved in canonical form from the Uniprot Knowledge Base (as on July 27, 2014). Variable modifications included oxidized methionine residues and de-amidated glutamine and asparagine. Search conditions allowed for a single missed cleavage with a 40ppm MS mode fragment tolerance coupled with 0.5 Da for MS/MS ions. A false discovery rate of 1% was applied in all cases. IgG was used as negative control. Proteins identified in IgG even with a single peptide were not considered. A minimum of 3 unique peptide counts in 2 independent experiments was required to consider as hits (Supplementary Dataset 1). Interactions that were present in WT but absent in ΔHR were classified as ‘polyQ dependent interactions’.
Target gene expression analysis
To examine the effect of Snf5 polyQ deletion on target gene-expression, we selected 18 functionally diverse non-essential targets with high fold changes in expression upon SNF5 gene deletion compared to wild type in one or both of the previous studies78,79. Yeast cells were lysed in Lithium acetate -SDS solution and RNA was extracted with TRIzol® Reagent following manufacturers instructions (LifeTechnologies). Reverse transcription was performed using the RevertAid H Minus First Strand cDNA Synthesis Kit (ThermoScientific). The concentration of the cDNA generated was adjusted and qPCR was performed using the SYBR® Green PCR Master Mix (Lifetechnologies). The reactions (5 replicates per gene) were performed in an Eco-illumina qPCR thermocycler (Illumina). The measured geometric mean of 3 different reference genes (ALG9, TAF10 and TFC180) were employed to normalise the data. The mRNA quantification was measured by the ΔCt method. The variation between WT and ΔHR in YPD and YPG was calculated as 2–ΔΔCt.
Statistical analysis
Doubling time (Td) of WT or ΔHR SNF5 strains in YPD and YPG was obtained from growth curves of the slopes of the best fit obtained by linear regression. Statistical significance for differences in budding index of cells after re-inoculation of stationary phase cultures from WT or ΔHR in YPD or YPG after 1, 3 and 4h was assessed using ANOVA. Statistical significance for differences in the distribution of HA area/nuclear area between the WT and ΔHR Snf5 in YPD and YPG was assessed using Wilcoxon rank sum test. Statistical significance for expression changes of Snf5 targets between WT and ΔHR was assessed by Students’s T-test and the P-values were corrected for multiple testing using the false discovery rate method designed by Benjamini, Hochberg and Yekutieli.
Source Data of computational and experimental studies are available with paper online (Supplementary tables 1-2, Supplementary dataset 1-2 and Supplementary Notes 4). Other data are available upon request.
Supplementary Material
Acknowledgements
We thank B. Lenhard, C. Semple, M. Torrent, A.J. Venkatakrishnan, B. Lang and members of our group for stimulating discussions and their comments on this study. This work was supported by the Medical Research Council (MC_U105185859; S.C., N.S.L., S.B., M.M.B.), Human Frontier Science Program (RGY0073/2010; M.M.B.), the European Molecular Biology Organization Long term fellowship (S.C.), Young Investigator Program (M.M.B.; K.J.V.), ERASysBio+ (GRAPPLE; S.C., S.B., and M.M.B.), Marie Curie actions (FP7-PEOPLE-2011-IEF-299105; N.S.G.) and Cancer Research UK (P.L.C). M.M.B. is a Lister Institute Prize Fellow. We thank C. Taylor and C. d’Santos from the CRUK-Cambridge Institute proteomics core facility.
Footnotes
Author Contributions
S.C and M.M.B. conceived the project and wrote the manuscript with inputs from all authors. S.C., K.J.V., S.B., and M.M.B. designed the study; S.C., G.C., N.S.L., E.I., and S.B. collected the datasets and undertook computational investigations; S.C., P.L.C., N.S.G., and R.G. designed and performed the experiments. All authors participated in interpreting the results. S.C led the study and M.M.B. supervised the project.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moumne L, et al. Differential aggregation and functional impairment induced by polyalanine expansions in FOXL2, a transcription factor involved in cranio-facial and ovarian development. Hum Mol Genet. 2008;17:1010–9. doi: 10.1093/hmg/ddm373. [DOI] [PubMed] [Google Scholar]
- 3.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–55. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 4.Tsuda H, et al. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell. 2005;122:633–44. doi: 10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Cortes CJ, et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat Neurosci. 2014;17:1180–9. doi: 10.1038/nn.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monks DA, et al. Overexpression of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proc Natl Acad Sci U S A. 2007;104:18259–64. doi: 10.1073/pnas.0705501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nasrallah IM, Minarcik JC, Golden JA. A polyalanine tract expansion in Arx forms intranuclear inclusions and results in increased cell death. J Cell Biol. 2004;167:411–6. doi: 10.1083/jcb.200408091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 2010;44:445–77. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 9.Stevens KE, Mann RS. A balance between two nuclear localization sequences and a nuclear export sequence governs extradenticle subcellular localization. Genetics. 2007;175:1625–36. doi: 10.1534/genetics.106.066449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolf A, et al. The polyserine domain of the lysyl-5 hydroxylase Jmjd6 mediates subnuclear localization. Biochem J. 2013;453:357–70. doi: 10.1042/BJ20130529. [DOI] [PubMed] [Google Scholar]
- 11.Salichs E, Ledda A, Mularoni L, Alba MM, de la Luna S. Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. 2009;5:e1000397. doi: 10.1371/journal.pgen.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C, et al. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell. 2013;25:572–84. doi: 10.1016/j.devcel.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galant R, Carroll SB. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–3. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- 14.Gerber HP, et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–11. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 15.Michael TP, et al. Simple sequence repeats provide a substrate for phenotypic variation in the Neurospora crassa circadian clock. PLoS One. 2007;2:e795. doi: 10.1371/journal.pone.0000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fondon JW, 3rd, Garner HR. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci U S A. 2004;101:18058–63. doi: 10.1073/pnas.0408118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–4. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 18.Karlin S, Brocchieri L, Bergman A, Mrazek J, Gentles AJ. Amino acid runs in eukaryotic proteomes and disease associations. Proc Natl Acad Sci U S A. 2002;99:333–8. doi: 10.1073/pnas.012608599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alba MM, Guigo R. Comparative analysis of amino acid repeats in rodents and humans. Genome Res. 2004;14:549–54. doi: 10.1101/gr.1925704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faux NG, et al. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15:537–51. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faux NG, et al. RCPdb: An evolutionary classification and codon usage database for repeat-containing proteins. Genome Res. 2007;17:1118–27. doi: 10.1101/gr.6255407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delot E, King LM, Briggs MD, Wilcox WR, Cohn DH. Trinucleotide expansion mutations in the cartilage oligomeric matrix protein (COMP) gene. Hum Mol Genet. 1999;8:123–8. doi: 10.1093/hmg/8.1.123. [DOI] [PubMed] [Google Scholar]
- 23.Ahn YY, Bagrow JP, Lehmann S. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–4. doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- 24.Koch EN, et al. Conserved rules govern genetic interaction degree across species. Genome Biol. 2012;13:R57. doi: 10.1186/gb-2012-13-7-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kemmeren P, et al. Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell. 2014;157:740–52. doi: 10.1016/j.cell.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 26.Munroe D, Jacobson A. mRNA poly(A) tail, a 3' enhancer of translational initiation. Molecular and cellular biology. 1990;10:3441–55. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature reviews Molecular cell biology. 2010;11:113–27. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen JD, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gingold H, Pilpel Y. Determinants of translation efficiency and accuracy. Molecular systems biology. 2011;7:481. doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Lee R, et al. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 2014;8:1832–44. doi: 10.1016/j.celrep.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–8. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 32.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–8. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes & development. 2000;14:655–65. [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–8. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 35.Gsponer J, Babu MM. Cellular strategies for regulating functional and nonfunctional protein aggregation. Cell Rep. 2012;2:1425–37. doi: 10.1016/j.celrep.2012.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woodsmith J, Kamburov A, Stelzl U. Dual coordination of post translational modifications in human protein networks. PLoS Comput Biol. 2013;9:e1002933. doi: 10.1371/journal.pcbi.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mateo F, et al. Degradation of cyclin A is regulated by acetylation. Oncogene. 2009;28:2654–66. doi: 10.1038/onc.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian MX, et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153:1012–24. doi: 10.1016/j.cell.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. The EMBO journal. 1992;11:1773–84. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergeron-Sandoval LP, Safaee N, Michnick SW. Mechanisms and Consequences of Macromolecular Phase Separation. Cell. 2016;165:1067–79. doi: 10.1016/j.cell.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 41.Gemayel R, et al. Variable Glutamine-Rich Repeats Modulate Transcription Factor Activity. Mol Cell. 2015;59:615–27. doi: 10.1016/j.molcel.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fishbain S, et al. Sequence composition of disordered regions fine-tunes protein half-life. Nat Struct Mol Biol. 2015;22:214–21. doi: 10.1038/nsmb.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonald MJ, Wang WC, Huang HD, Leu JY. Clusters of nucleotide substitutions and insertion/deletion mutations are associated with repeat sequences. PLoS Biol. 2011;9:e1000622. doi: 10.1371/journal.pbio.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenz C, Haerty W, Golding GB. Increased substitution rates surrounding low-complexity regions within primate proteins. Genome Biol Evol. 2014;6:655–65. doi: 10.1093/gbe/evu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huntley MA, Clark AG. Evolutionary analysis of amino acid repeats across the genomes of 12 Drosophila species. Mol Biol Evol. 2007;24:2598–609. doi: 10.1093/molbev/msm129. [DOI] [PubMed] [Google Scholar]
- 46.McDonald MJ, et al. Mutation at a distance caused by homopolymeric guanine repeats in Saccharomyces cerevisiae. Sci Adv. 2016;2:e1501033. doi: 10.1126/sciadv.1501033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dreze M, et al. 'Edgetic' perturbation of a C. elegans BCL2 ortholog. Nat Methods. 2009;6:843–9. doi: 10.1038/nmeth.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woerner AC, et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–6. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- 49.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol. 2005;12:654–62. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 50.Mar Alba M, Santibanez-Koref MF, Hancock JM. Amino acid reiterations in yeast are overrepresented in particular classes of proteins and show evidence of a slippage-like mutational process. J Mol Evol. 1999;49:789–97. doi: 10.1007/pl00006601. [DOI] [PubMed] [Google Scholar]
- 51.Shah KA, Mirkin SM. The hidden side of unstable DNA repeats: Mutagenesis at a distance. DNA Repair (Amst) 2015;32:106–12. doi: 10.1016/j.dnarep.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah KA, et al. Role of DNA polymerases in repeat-mediated genome instability. Cell Rep. 2012;2:1088–95. doi: 10.1016/j.celrep.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J, Yang JR. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 2015;16:409–20. doi: 10.1038/nrg3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narayanaswamy R, et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci U S A. 2009;106:10147–52. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chakrabortee S, et al. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell. 2016;167:369–381 e12. doi: 10.1016/j.cell.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caudron F, Barral Y. A super-assembly of Whi3 encodes memory of deceptive encounters by single cells during yeast courtship. Cell. 2013;155:1244–57. doi: 10.1016/j.cell.2013.10.046. [DOI] [PubMed] [Google Scholar]
- 57.Levy ED, Landry CR, Michnick SW. How perfect can protein interactomes be? Sci Signal. 2009;2:pe11. doi: 10.1126/scisignal.260pe11. [DOI] [PubMed] [Google Scholar]
- 58.Hancock JM, Simon M. Simple sequence repeats in proteins and their significance for network evolution. Gene. 2005;345:113–8. doi: 10.1016/j.gene.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 59.Jarosz DF, Taipale M, Lindquist S. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu Rev Genet. 2010;44:189–216. doi: 10.1146/annurev.genet.40.110405.090412. [DOI] [PubMed] [Google Scholar]
- 60.Ekman D, Light S, Bjorklund AK, Elofsson A. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol. 2006;7:R45. doi: 10.1186/gb-2006-7-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dosztanyi Z, Meszaros B, Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–6. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jorda J, Kajava AV. T-REKS: identification of Tandem REpeats in sequences with a K-meanS based algorithm. Bioinformatics. 2009;25:2632–8. doi: 10.1093/bioinformatics/btp482. [DOI] [PubMed] [Google Scholar]
- 63.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–9. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 64.Simon M, Hancock JM. Tandem and cryptic amino acid repeats accumulate in disordered regions of proteins. Genome Biol. 2009;10:R59. doi: 10.1186/gb-2009-10-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harbi D, Kumar M, Harrison PM. LPS-annotate: complete annotation of compositionally biased regions in the protein knowledgebase. Database (Oxford) 2011;2011:baq031. doi: 10.1093/database/baq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 2011;39:D289–94. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 70.McGraw KO, Wong SP. A common language effect size statistic. Psychol Bull. 1992;111:361–5. [Google Scholar]
- 71.Weatheritt RJ, Gibson TJ, Babu MM. Asymmetric mRNA localization contributes to fidelity and sensitivity of spatially localized systems. Nat Struct Mol Biol. 2014;21:833–9. doi: 10.1038/nsmb.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grissom RJ, Kim JJ. Effect Sizes for Research: Univariate and Multivariate Applications. Routledge; 2012. [Google Scholar]
- 73.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–30. [Google Scholar]
- 74.Amberd DC, Burke D, Strathern JN. Methods in Yeast Genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 75.Rossmann MP, Stillman B. Immunoblotting histones from yeast whole-cell protein extracts. Cold Spring Harb Protoc. 2013;2013:625–30. doi: 10.1101/pdb.prot067116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 77.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 78.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39:683–7. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 79.Kemmeren P, et al. Large-scale genetic perturbations reveal regulatory networks and an abundance of gene-specific repressors. Cell. 2014;157:740–52. doi: 10.1016/j.cell.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 80.Teste MA, Duquenne M, Francois JM, Parrou JL. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol. 2009;10:99. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.