The spore-forming obligate anaerobe Clostridium difficile is the leading cause of antibiotic-associated diarrheal disease in the United States. When C. difficile spores are ingested by susceptible individuals, they germinate within the gut and transform into vegetative, toxin-secreting cells. During infection, C. difficile must also induce spore formation to survive exit from the host. Since spore formation is essential for transmission, understanding the basic mechanisms underlying sporulation in C. difficile could inform the development of therapeutic strategies targeting spores. In this study, we determine the requirement of the C. difficile homolog of SpoVM, a protein that is essential for spore formation in Bacillus subtilis due to its regulation of coat and cortex formation. We observed that SpoVM plays a minor role in C. difficile spore formation, in contrast with B. subtilis, indicating that this protein would not be a good target for inhibiting spore formation.
KEYWORDS: Clostridium difficile, SpoIVA, SpoVM, coat, morphogenesis, spore
ABSTRACT
The spore-forming bacterial pathogen Clostridium difficile is a leading cause of health care-associated infections in the United States. In order for this obligate anaerobe to transmit infection, it must form metabolically dormant spores prior to exiting the host. A key step during this process is the assembly of a protective, multilayered proteinaceous coat around the spore. Coat assembly depends on coat morphogenetic proteins recruiting distinct subsets of coat proteins to the developing spore. While 10 coat morphogenetic proteins have been identified in Bacillus subtilis, only two of these morphogenetic proteins have homologs in the Clostridia: SpoIVA and SpoVM. C. difficile SpoIVA is critical for proper coat assembly and functional spore formation, but the requirement for SpoVM during this process was unknown. Here, we show that SpoVM is largely dispensable for C. difficile spore formation, in contrast with B. subtilis. Loss of C. difficile SpoVM resulted in modest decreases (~3-fold) in heat- and chloroform-resistant spore formation, while morphological defects such as coat detachment from the forespore and abnormal cortex thickness were observed in ~30% of spoVM mutant cells. Biochemical analyses revealed that C. difficile SpoIVA and SpoVM directly interact, similarly to their B. subtilis counterparts. However, in contrast with B. subtilis, C. difficile SpoVM was not essential for SpoIVA to encase the forespore. Since C. difficile coat morphogenesis requires SpoIVA-interacting protein L (SipL), which is conserved exclusively in the Clostridia, but not the more broadly conserved SpoVM, our results reveal another key difference between C. difficile and B. subtilis spore assembly pathways.
IMPORTANCE The spore-forming obligate anaerobe Clostridium difficile is the leading cause of antibiotic-associated diarrheal disease in the United States. When C. difficile spores are ingested by susceptible individuals, they germinate within the gut and transform into vegetative, toxin-secreting cells. During infection, C. difficile must also induce spore formation to survive exit from the host. Since spore formation is essential for transmission, understanding the basic mechanisms underlying sporulation in C. difficile could inform the development of therapeutic strategies targeting spores. In this study, we determine the requirement of the C. difficile homolog of SpoVM, a protein that is essential for spore formation in Bacillus subtilis due to its regulation of coat and cortex formation. We observed that SpoVM plays a minor role in C. difficile spore formation, in contrast with B. subtilis, indicating that this protein would not be a good target for inhibiting spore formation.
INTRODUCTION
The Gram-positive, endospore-forming, gastrointestinal pathogen Clostridioides difficile (1, 2), more commonly known as Clostridium difficile, is a leading cause of health care-associated infections in the United States (3). C. difficile-associated disease ranges from gastroenteritis to more severe conditions like pseudomembranous colitis and toxic megacolon (4). The vegetative form of C. difficile causes these disease symptoms through its secretion of inflammation-inducing toxins (4), while its spore form is essential for C. difficile to transmit disease because C. difficile is an obligate anaerobe (5). Thus, spore formation allows C. difficile to survive exit from the host, and spore germination is essential for C. difficile to initiate infection (6).
Endospore formation is an ancient, highly coordinated developmental process (7, 8) that generates a metabolically dormant spore from the asymmetric division of a vegetative cell. Asymmetric division creates a larger mother cell and a smaller forespore. The mother cell engulfs the forespore in a process analogous to phagocytosis, leaving the double-membrane bound forespore suspended within the mother cell cytosol. A thick layer of modified peptidoglycan known as the cortex is built upon the germ cell wall present between the two membranes surrounding the forespore. The cortex helps maintain metabolic dormancy and confers heat resistance (9, 10). A series of concentric proteinaceous shells known as the coat is also assembled around the outermost forespore membrane. This external layer functions to protect spores against oxidative, chemical, and enzymatic insults (11) and can protect spores following phagocytosis by eukaryotic organisms (12).
In Bacillus subtilis, the coat is made of ~80 different proteins that are assembled in a hierarchal manner around the forespore (11). Landmark proteins recognize the forespore and subsequently recruit additional proteins to create four discrete layers: the basement layer, inner coat, outer coat, and crust (13). The basement layer is the most important of these layers, since mutants deficient in this layer fail to make heat-resistant spores (14–16). A key component of the basement layer is SpoVM (here referred to as VM [14]), a 26-amino-acid small protein (17) that anchors the basement layer to the forespore membrane. VM function depends on its ability to recognize the positive curvature of the forespore and embed itself within the mother cell-derived membrane (18, 19). VM directly binds to and recruits SpoIVA (here referred to as IVA), an ATPase that forms highly stable polymers (20) that encase the forespore (21). This interaction (22) is required for both proteins to preferentially surround the forespore (22–24).
Both IVA and VM are essential morphogenetic proteins in B. subtilis, since IVA and VM mutants fail to properly assemble the coat and make cortex (14, 15). The defect in cortex synthesis presumably underlies their failure to make heat- and chloroform-resistant spores (15, 25, 26). If IVA fails to encase the forespore, a quality control mechanism prevents cortex formation and eventually leads to lysis of the sporulating cell (26). This mechanism is dependent on CmpA (cortex morphogenetic protein A [27]), a ClpXP adaptor protein that targets improperly localized IVA for degradation (26).
IVA also binds to and recruits another coat morphogenetic protein, SpoVID (28) (here referred to as VID). This interaction depends on region A in VID’s C terminus, although it is independent of VID’s C-terminal LysM domain (23). VID then recruits additional coat morphogenetic proteins, SafA and CotE (29–31), that direct assembly of the inner and outer coat layers, respectively (11).
Notably, of the 10 B. subtilis coat morphogenetic proteins that have been identified to date (11), only VM and IVA are widely conserved in the Bacilli and Clostridia, with IVA being strictly conserved (7, 32) and CmpA, VID, SafA, and CotE being conserved exclusively in the Bacillales (7, 33). As a result, relatively little is known about spore coat assembly in the Clostridia. Preliminary analyses in C. difficile of this process have identified SipL as a clostridium-specific functional homolog of VID (34), although unlike B. subtilis VID, SipL is essential for heat-resistant spore formation in C. difficile. In recombinant coaffinity purification analyses, SipL directly binds to IVA through SipL’s C-terminal LysM domain (34). Given that VID binding to IVA in B. subtilis occurs independently of VID’s LysM domain (23), the mechanism by which C. difficile IVA recognizes SipL fundamentally differs from how B. subtilis IVA recognizes VID.
Interestingly, although C. difficile IVA and SipL are dispensable for cortex formation, both coat morphogenetic proteins are required for heat-resistant spore formation (34). These observations contrast with B. subtilis (14, 15), where IVA mutants fail to make cortex and heat-resistant spores and VID mutants make cortex and exhibit modest defects in heat-resistant spore formation. Since these findings are consistent with the absence of CmpA homologs in the Clostridia (27, 33), cortex and coat syntheses appear to be less tightly linked in C. difficile than they are in B. subtilis.
Given that VM is the only other major coat morphogenetic protein that has a homolog in C. difficile (7), in this study we sought to determine whether VM was similarly critical for C. difficile spore formation using genetic, cytological, and biochemical analyses.
RESULTS
Key structural features of VM are conserved between C. difficile and B. subtilis.
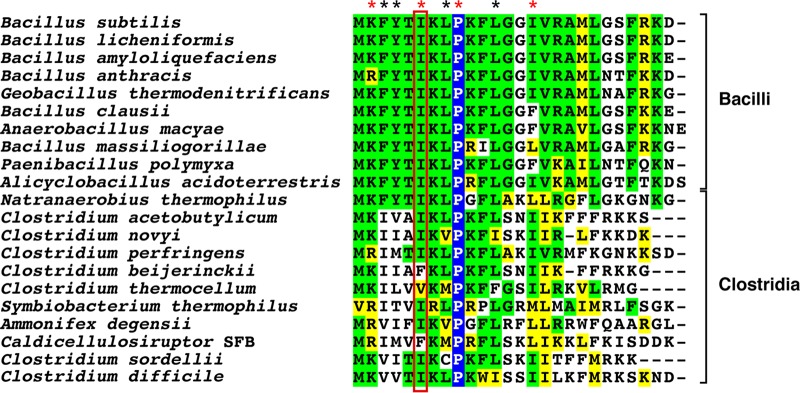
Before assessing the requirement for C. difficile VM during spore formation, we first compared its primary sequence to B. subtilis VM. Extensive mutagenesis analyses have been performed on B. subtilis VM (35), and specific functions have been identified for a subset of its 26 residues (22, 26, 27). This sequence alignment revealed that many of the residues previously identified to be critical for B. subtilis VM function are conserved in C. difficile VM (Fig. 1) (22, 26, 27, 35). Indeed, most of these functionally important residues are conserved across VM homologs in the Bacilli and Clostridia, suggesting that VM may be under negative selection.
FIG 1 .
Sequence alignment of VM homologs in the Bacilli and Clostridia. Pro9 is the only residue that is completely conserved (boxed in blue with white font). This residue introduces a kink into the helical structure of VM (18) and is essential for it to specifically recognize positively curved membranes (19, 24). Conserved identical residues are boxed in green, and conserved similar residues are boxed in yellow. Sequences were derived from the work of Galperin et al. (7). Residues whose mutation to alanine causes severe defects in B. subtilis VM function (35) are indicated with an asterisk, while red asterisks highlight residues that have been shown or are presumed to activate the CmpA pathway in B. subtilis. K2A and I15A mutations prevent cortex production and activate the CmpA-dependent checkpoint pathway (26, 27), while I6A (highlighted by the red box) prevents binding to IVA and IVA encasement of the forespore (22) and thus presumably also activates CmpA.
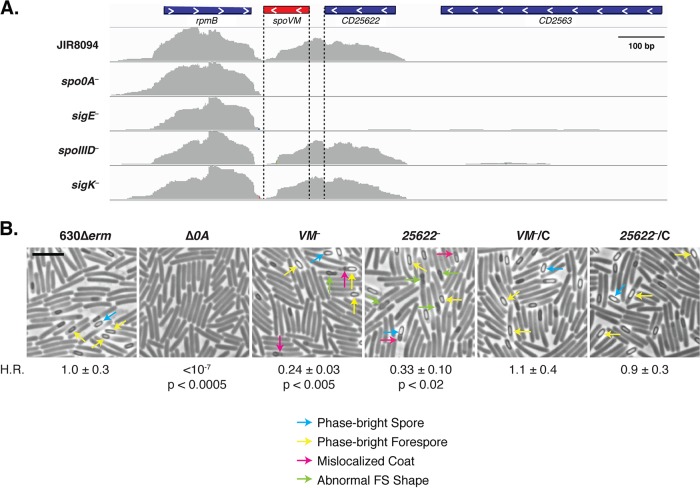
We also analyzed whether C. difficile VM is expressed during sporulation and compared its regulation to that of B. subtilis VM. Analysis of transcriptome sequencing (RNA-Seq) data previously generated in the JIR8094 strain background (36) revealed that VM is transcribed during sporulation under the control of the early-stage mother cell-specific sigma factor σE. This regulation was confirmed using quantitative reverse transcription PCR (qRT-PCR) analyses on a separate set of RNA samples (see Fig. S1B in the supplemental material). This expression pattern differs slightly from B. subtilis VM, which for its expression requires SpoIIID (14), a transcription factor that acts downstream of σE (37). Notably, while σE controls the transcription of IVA, sipL, and VM in C. difficile, IVA and sipL are expressed at 50- to 100-fold-higher levels than VM in the RNA-Seq analyses (Table S3 in Data Set S1).
Transcriptional analyses of the CD25622-VM locus. (A) Representative image of Integrative Genomics Viewer software (65) used to visualize CD25622-VM in RNA-Seq data of sporulating JIR8094 and associated sigma factor mutants from reference 36. The reads mapped to this locus are shown. The direction of transcription is indicated by the angled bracket. (B) qRT-PCR analyses of VM transcript levels in wild-type JIR8094, spo0A mutant, sigE mutant, spoIIID mutant, and sigK mutant strains relative to spo0A mutant. spoIIID and sigK expression depend on sigE, which encodes the mother cell-specific sigma factor σE. ***, P < 0.005. Download FIG S1, TIFF file, 0.1 MB (118.7KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Table S1 shows strains and plasmids used in this study. Table S2 shows primers used in this study. Table S3 shows expression levels of select sporulation genes as measured by RNA-Seq. Table S4 shows sporulation efficiency as measured by chloroform resistance. Table S5 shows frequency of forespore morphological abnormalities detected by phase-contrast microscopy. Download DATA SET S1, PDF file, 0.1 MB (124.8KB, pdf) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
C. difficile VM is largely dispensable for functional spore formation.
Interestingly, during the visualization of the direct reads generated by RNA-Seq we noticed that VM is actually cotranscribed downstream of CD25622 (Fig. 2A and S1B), a gene that encodes a 50-amino-acid protein of unknown function that appears to be primarily conserved among the Peptostreptococcaceae family. This raised the possibility that both VM and CD25622 may play important roles during C. difficile spore formation. To assess their functional contribution during sporulation, we constructed TargeTron insertions in VM and CD25622 in both the 630ΔermΔpyrE and JIR8094 strain backgrounds (Fig. S2). The 630ΔermΔpyrE strain was used to facilitate chromosomal complementation of the VM mutants at the pyrE locus using allele-coupled exchange (ACE) (38). Nevertheless, in both strain backgrounds, the TargeTron insertions in CD25622 likely have polar effects on VM expression such that they are effectively CD25622-VM double mutants.
FIG 2 .
The CD25622-VM locus is largely dispensable for heat-resistant spore formation. (A) RNA-Seq transcript data of the CD25622-VM locus in wild-type JIR8094 and spo0A, sigE, spoIIID, and sigK mutant TargeTron insertion strains (36) visualized using the Integrative Genomics Viewer software (65). spo0A encodes the master transcriptional regulator necessary for induction of sporulation. sigE encodes σE, a mother cell-specific sigma factor that activates spoIIID transcription (59, 66, 67). spoIIID encodes a transcription factor that coordinately activates sigK transcription along with σE (36, 67). The proximity of C. difficile VM to rpmB, which encodes a ribosomal subunit protein, is conserved relative to other VM-carrying organisms (7). CD25622 appears to be unique to the Peptostreptococcaceae (41). The angled brackets indicate the direction of transcription. (B) Phase-contrast microscopy of wild-type 630Δerm, Δ0A, and VM mutant and CD25622-TargeTron mutants and their complements (VM::ermB/C and 25622::ermB/C) sporulating cultures at 20 h. Examples of phase-bright forespores and spores are marked using yellow and blue arrows, respectively. Select forespores with abnormal morphologies are delineated by green arrows, while regions that may correspond to mislocalized coat are highlighted in pink. H.R. refers to the heat resistance of each strain relative to the wild type. The means and standard deviations shown are based on four biological replicates. Statistical significance relative to the wild type was determined using a one-way ANOVA and Tukey’s test. Bars, 5 µm.
Construction of VM, CD25622, and ΔIVA mutants. (A) Schematic of the TargeTron-based insertions in VM (VM::ermB) and CD25622 (CD25622::ermB). The TargeTron insertion in CD25622 is predicted to have polar effects on VM expression. (B) Colony PCR of VM::ermB (VM−) and CD25622::ermB (CD25622−) mutants in the 630ΔermΔpyrE and JIR8094 strain backgrounds. The group II intron is ~2 kb. (C) Schematic and colony PCR of the spoIVA deletion in 630ΔermΔpyrE. Download FIG S2, TIFF file, 0.3 MB (267.6KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To assess the functional requirement for VM and CD25622 during sporulation, we compared the heat resistance of sporulating cultures of VM mutant and CD25622 mutant strains to that of the wild type. C. difficile 630Δerm VM::ermB and 25622::ermB exhibited a relatively modest ~3- to 4-fold heat resistance defect relative to the wild type (P < 0.02, Fig. 2B) in contrast with the >6-log defects of B. subtilis VM mutants relative to the wild type (25). Importantly, the heat resistance defects of 630Δerm VM mutants could be fully complemented by expressing CD25622-VM from the pyrE locus (Fig. 2B). Since VM::ermB and CD25622::ermB mutants in the JIR8094 background exhibited a similar ~5-fold heat resistance defect relative to the wild type (Fig. S3A), loss of VM leads to minor but reproducible decreases in heat-resistant spore formation. Furthermore, CD25622 would appear to have little impact on C. difficile sporulation given that the heat resistance defects of VM::ermB and CD25622::ermB mutants in both strain backgrounds were essentially identical.
The CD25622-VM locus is largely dispensable for heat-resistant spore formation in JIR8094. (A) Phase-contrast microscopy of sporulating cultures of wild-type JIR8094, spo0A mutant, and VM and CD25622-VM TargeTron mutants at 21 h. Examples of phase-bright forespores and spores are marked using yellow and blue arrows, respectively. Forespores with abnormal morphologies are denoted by green arrows, while regions that likely correspond to mislocalized coat are marked in pink. HR refers to the heat resistance of each strain relative to the wild type. The means and standard deviations shown are based on three biological replicates. Statistical significance relative to the wild type was determined using one-way ANOVA and Tukey’s test. Bars, 5 µm. (B) Transmission electron microscopy (TEM) analyses of sporulating cells of wild-type JIR8094 and VM and CD25622-VM TargeTron mutants and their complements (VM−/C and 25622−/C). Bars, 500 nm. Blue arrows mark coat layers that surround the forespore, while pink arrows indicate detached coat layers termed “bearding.” The green arrows mark abnormal forespore shape, where the forespore appears to be pinched at the mother cell-proximal side; the yellow arrows denote areas of abnormal cortex thickness. (C) TEM analyses of sporulating JIR8094 strains. Percentage of cells with the observed phenotype based on analyses of a minimum of 50 TEM images. Download FIG S3, TIFF file, 0.6 MB (579.1KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We also measured the impact of VM on C. difficile sporulating cell chloroform resistance, since a B. subtilis VM mutant is sensitive to chloroform (>6-log defect relative to wild type [25]). An ~4-fold reduction in chloroform resistance was observed in C. difficile VM mutants relative to the wild type (P < 0.05, Table S4 in Data Set S1), with expression of CD25622-VM from the pyrE locus again restoring chloroform resistance to wild-type levels. Taken together, C. difficile VM is necessary for optimal heat- and chloroform-resistant spore formation, but the requirement for VM during spore formation differs markedly between C. difficile and B. subtilis.
Loss of VM causes morphological abnormalities in a subset of C. difficile sporulating cells.
Consistent with the relatively minor role that VM plays during C. difficile spore formation, phase-contrast microscopy revealed that CD25622-VM::ermB and VM::ermB mutants produced wild-type-like, phase-bright forespores and free spores (Fig. 2B, yellow and blue arrows, respectively). Nevertheless, these strains also produced forespores with clear morphological abnormalities. Phase-dark extensions of the phase-bright forespores were frequently observed in ~15% of sporulating VM mutant cells (Fig. 2B, pink arrows, and Table S5 in Data Set S1) similar to the mislocalized, polymerized coat observed in engulfment mutants lacking components of the SpoIIQ-SpoIIIA channel complex (39). VM mutants also produced phase-gray and phase-dark forespores with abnormal shapes (~13%) in which the forespores appeared to be pinched at the mother cell-proximal side (Fig. 2B, green arrow, and Table S5 in Data Set S1). Both these abnormal phenotypes were observed at a higher frequency in VM mutants than in the wild type and the complementation strains (<4%, Table S5 in Data Set S1). Given that similar morphological abnormalities were observed in VM::ermB and CD25622-VM::ermB mutants in the JIR8094 strain background (Fig. S3A), these morphological defects are likely due to loss of VM. These morphological defects would appear to differ from the phase-dark sporelets produced by B. subtilis VM mutants (14), but direct comparisons cannot be made because images of these sporelets have not been published.
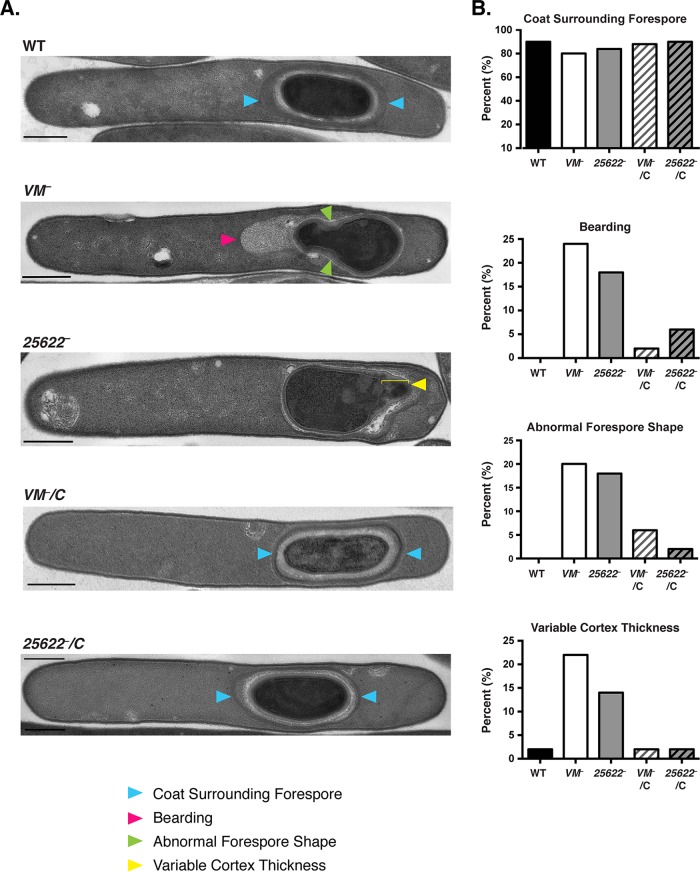
To assess whether polymerized coat is mislocalized in sporulating C. difficile VM mutant cells as predicted by the phase-contrast analyses, we used transmission electron microscopy (TEM) to visualize the multilayered coat. These analyses confirmed that polymerized coat failed to adhere to the forespore-mother cell interface in C. difficile VM mutants (termed “bearding” [Fig. 3A]), similar to the coat localization defects previously reported in SpoIIQ-SpoIIIA channel mutants (39). Bearding was observed in ~20% of sporulating CD25622-VM and VM mutant cells based on analyses of ≥50 images of sporulating cells that had completed engulfment; in contrast, bearding was observed in less than 6% of the complementation strains, and no bearding was observed in wild-type cells (Fig. 3B). Approximately 20% of sporulating VM mutant cells produced forespores that appeared to be “pinched” at one end, whereas only ~5% of forespores in the VM complementation strains, and none of the wild-type cells, exhibited this phenotype (Fig. 3). The frequencies of these phenotypes are slightly higher than the frequency measured using phase-contrast microscopy (Table S5 in Data Set S1), which has considerably lower resolution. TEM analyses also revealed that the VM mutants were more likely to produce cortex of variable thickness than the wild-type and complementation strains (Fig. 3). In total, 35% of 630Δerm VM mutant cells displayed at least one of the three aberrant phenotypes (bearding, abnormal forespore shape, and variable cortex thickness). In contrast, only 2% of wild-type and 10% of the complementation strains’ sporulating cells displayed one or more of these phenotypes.
FIG 3 .
Morphological defects are visible in sporulating VM mutant cells. (A) Transmission electron microscopy (TEM) analyses of sporulating cultures at 23 h of wild-type (WT) 630Δerm, Δ0A, and VM and CD25622-VM TargeTron mutants and their complements (VM::ermB/C and 25622::ermB/C). Bars, 500 nm. Blue arrowheads mark coat layers that encase the forespore, while the pink arrowhead indicates detached coat layers termed “bearding.” The green arrowheads highlight abnormal forespore shape, where the forespore appears to be pinched at the mother cell-proximal side, while the yellow arrowhead denotes areas of abnormal cortex thickness. (B) Percentage of cells with the observed phenotype based on analyses of a minimum of 50 TEM images.
Similar morphological abnormalities were also observed in JIR8094 VM mutants at similar frequencies. Approximately 20% of sporulating JIR8094 VM mutant cells had forespores with coat bearding and/or variable cortex thickness (Fig. S3), phenotypes that were rarely observed in wild-type cells. A smaller proportion of JIR8094 VM mutant cells produced abnormally shaped forespores (6% of VM::ermB and 12% of CD25622-VM::ermB) relative to 630Δerm VM mutant cells. However, wild-type JIR8094 also produced abnormally shaped forespores in 4% of cells (Fig. S3C), whereas wild-type 630Δerm did not. Overall, 32% and 42% of sporulating JIR8094 spoVM::ermB and CD25622-VM:ermB cells displayed at least one of the three morphological defects compared to 11% of wild-type JIR8094 cells. Taken together, these analyses indicate that loss of VM causes some form of morphological defect during spore formation in about one-third of the population.
While cortex thickness varied within a subpopulation of sporulating C. difficile VM mutant cells (Fig. 3), cortex was nevertheless visible in almost all sporulating cells of this mutant. While these observations indicate that VM is dispensable for cortex production in C. difficile, unlike B. subtilis (14, 25, 27), it remains possible that C. difficile VM modulates cortex thickness and/or its composition.
Loss of VM leads to a minor defect in spore purification efficiency.
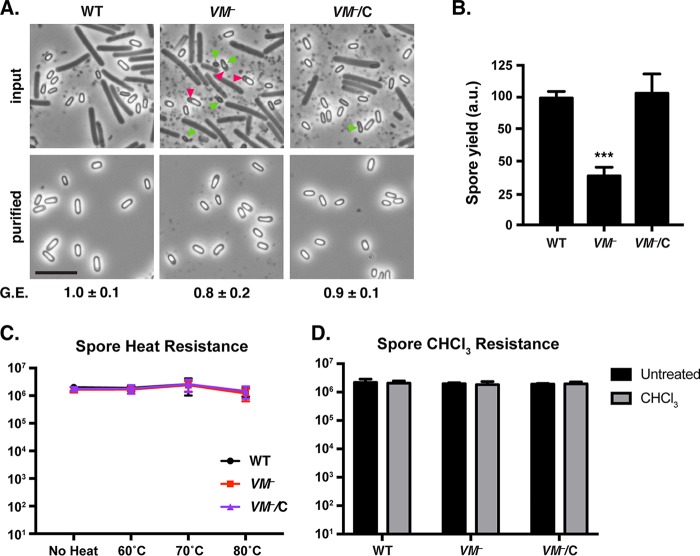
Given that approximately one-third of C. difficile VM mutant cells exhibited morphological defects by phase-contrast microscopy and TEM and approximately two-thirds exhibited heat and chloroform resistance defects (Fig. 2, 3, and S3 and Table S4 in Data Set S1), we wondered whether C. difficile VM mutant spores would be purified less efficiently than wild-type spores. To test this possibility, we compared the spore purification yields of wild-type, VM mutant, and the VM mutant complementation strain (VM/C). Loss of C. difficile VM resulted in an ~3-fold decrease in spore yield relative to the wild type and the VM complementation strain (P < 0.001, Fig. 4A). Importantly, similar levels of sporulation were observed in the input samples, although aberrantly shaped free spores were more frequently visible in the VM mutant than in the wild type and the complementation strain (Fig. 4B). However, once the spores were purified on a density gradient, VM mutant spores were largely indistinguishable from wild-type spores by phase-contrast microscopy (Fig. 4B) and TEM analyses (Fig. S4).
FIG 4 .
VM mutant spores are purified less efficiently than wild type (WT), but purified VM mutant spores have resistance properties similar to those of the wild type. (A) Phase-contrast microscopy images of wild-type, VM mutant, and VM mutant complementation cultures isolated 2 days after sporulation was induced (input) and spores following density gradient isolation (purified). G.E. refers to the germination efficiency of each strain relative to that of the wild type. The means and standard deviations shown are based on three biological replicates. Statistical significance relative to the wild type was determined using a one-way ANOVA and Tukey’s test. Bars, 5 µm. Examples of spores with abnormal forespore shape are marked with green arrowheads, and areas of putative mislocalized coat are marked with pink arrowheads. (B) Spore yields based on purifications from the indicated strains from four biological replicates. Yields were determined by measuring the optical density of spore purifications at 600 nm; yields are expressed in arbitrary units (a.u.). Statistical significance relative to the wild type was determined using one-way ANOVA and Tukey’s test. ***, P < 0.0005. (C and D) Heat (C) and chloroform (D) resistance of spores purified from the indicated strains based on three independent experiments. Numbers at left are numbers of CFUs produced by spores plated on BHIS containing taurocholate germinant.
Transmission electron microscopy of wild-type and VM mutant spores. Presumed cortex layers are marked (Cx). Bars, 100 nm. Download FIG S4, TIFF file, 1 MB (1.1MB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Since the VM mutant exhibited an ~3-fold reduction in spore purification yield, heat resistance, and chloroform resistance relative to the wild type, we tested whether the VM mutant spores that survived the spore purification process would have heat and/or chloroform resistance defects. Loss of VM did not affect heat (Fig. 4C) or chloroform (Fig. 4D) resistance in purified spores, suggesting that VM mutant spores with morphological and functional defects fail to be purified by the density gradient.
VM modulates, but is not essential for, IVA encasement of the C. difficile forespore.
Having established that approximately two-thirds of sporulating C. difficile VM mutant cells have functional defects, we wondered whether the localization of VM’s putative binding partner, IVA, would be disrupted in VM mutant cells. To address this question, we generated a construct encoding an N-terminal fusion of mCherry to C. difficile IVA because a C-terminally green fluorescent protein (GFP)-tagged B. subtilis IVA fails to localize properly, in contrast with N-terminally GFP-tagged B. subtilis (21), and C-terminally His6-tagged C. difficile IVA is produced at low levels in Escherichia coli, in contrast with N-terminally His6-tagged IVA (data not shown). When mCherry-IVA was coproduced with native IVA (WT/mCherry-IVA), the fusion protein (i) localized to the mother cell-forespore interface of cells that had initiated engulfment (Fig. 5, pink arrows) and (ii) encased the forespore of cells that had completed engulfment (Fig. 5, yellow and orange arrows). Taken together, C. difficile IVA preferentially localizes to the forespore in a manner similar to B. subtilis IVA (21).
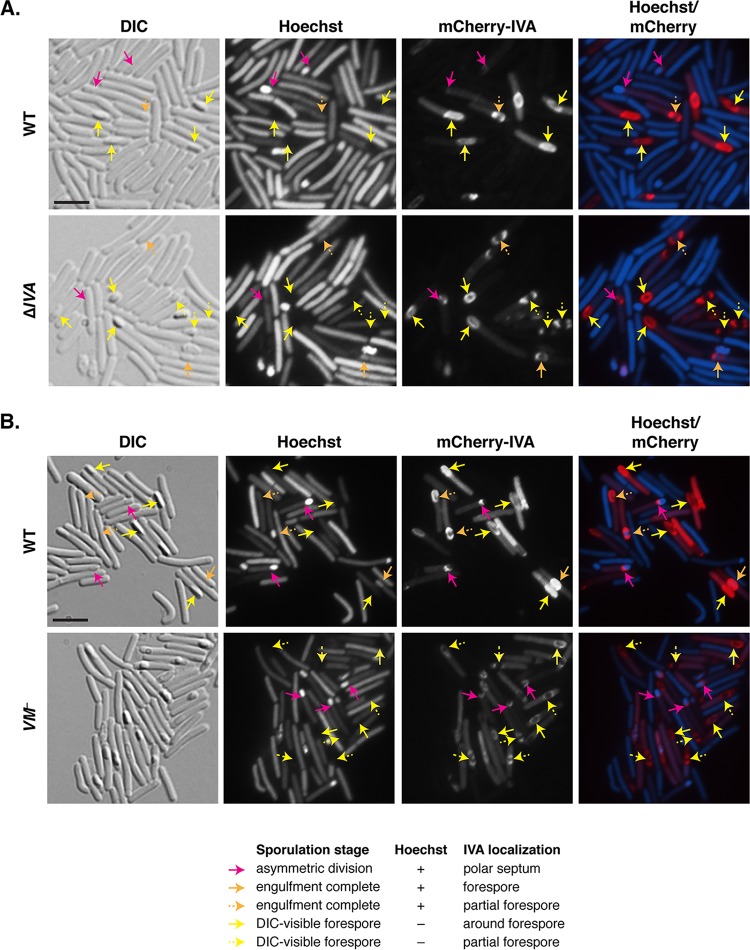
FIG 5 .
IVA can localize to and encase the forespore in the absence of VM. Fluorescence microscopy analyses of wild-type (WT) and ΔIVA sporulating cells (A) and wild-type and VM mutant sporulating cells (B) producing mCherry-IVA at 23 h post-sporulation induction. Differential interference contrast (DIC) microscopy was used to visualize forespores and free spores. The nucleoid was stained with Hoechst stain (blue), and mCherry-IVA fluorescence is shown in red. The merge of Hoechst stain and mCherry is also shown. Pink arrows highlight cells that have undergone asymmetric division but have not completed engulfment; these forespores stain with Hoechst stain (+), and mCherry-IVA localizes to the mother cell-forespore interface. Orange arrows highlight cells that appear to have completed engulfment but whose forespores retain the Hoechst dye (+). Yellow arrows mark forespores that are visible by DIC and no longer retain the Hoechst dye. Orange and yellow solid arrows indicate that mCherry-IVA fully encases the forespore, while dashed arrows indicate partial encasement of the forespore. Bars, 5 µm.
When the mCherry-IVA construct was used to complement ΔIVA, the mCherry-IVA signal was concentrated mostly at the forespore, although a higher proportion of the cells appeared to cap, rather than fully encase, the forespore (Fig. 5A, dashed arrows). Since these observations implied that the mCherry-IVA fusion protein was not fully functional, we measured its ability to complement the heat resistance phenotype of a ΔIVA strain. The resulting ΔIVA/mCherry-IVA strain produced heat-resistant spores ~5-fold less frequently than the wild type (P ≤ 0.05, one-way analysis of variance [ANOVA]), whereas WT/mCherry-IVA produced heat-resistant spores ~2-fold less frequently than the wild type (this difference was not statistically significant [Fig. S5A]). Given that mCherry-IVA was less capable of encasing the forespore in the absence of wild-type IVA and that B. subtilis IVA ATPase mutants exhibit similar defects in encasing the forespore (40), C. difficile mCherry-IVA is likely less efficient at polymerizing than wild-type IVA.
Complementation of ΔIVA by mCherry-IVA and N491G IVA. (A) Phase-contrast microscopy of wild-type 630Δerm and ΔIVA strains complemented with either IVA, mCherryIVA, or N491G IVA, and Δ0A after 22 h of sporulation. Examples of phase-bright forespores and spores are marked using yellow and blue arrows, respectively. Mislocalized coat regions are marked by pink arrows. HR refers to the heat resistance of each strain relative to the wild type. The means and standard deviations shown are based on three biological replicates. Statistical significance relative to the wild type was determined using one-way ANOVA and Tukey’s test. Bars, 5 µm. (B) Western blot analyses of the strains in panel A using anti-IVA and anti-mCherry antibodies. Nonspecific bands are marked with asterisks. A small amount of mCherry-IVA undergoes cleavage during sporulation, appearing to liberate IVA and mCherry. Download FIG S5, TIFF file, 0.6 MB (592.9KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Although most of the mCherry-IVA signal localized to the forespores of WT/mCherry-IVA and ΔIVA/mCherry-IVA, cytosolic mCherry signal was also observed (Fig. 5A). This cytosolic signal likely results from the small amount of mCherry that is liberated upon processing of mCherry-IVA (Fig. S5B). Taken together, these results confirm that C. difficile IVA encases the forespore and that this localization pattern correlates with functional spore formation.
With the functional mCherry-IVA reagent in hand, we next assessed the impact of VM on the localization of mCherry-IVA. The fusion protein localized primarily around the forespore in the absence of VM (Fig. 5B), although 29% of VM mutant cells exhibited partial encasement by mCherry-IVA around the forespore. In contrast, 14% of wild-type cells exhibited this phenotype based on analyses of 100 sporulating cells that had completed engulfment. These results strongly suggest that C. difficile IVA can still localize to and surround the forespore in the absence of C. difficile VM, albeit with somewhat lower efficiency than the wild type. They also contrast with the localization pattern of B. subtilis IVA, which forms foci on the forespore but fails to encase it in ~30% of VM mutant cells (22, 23), relative to ~5% of wild-type cells. Unfortunately, attempts to localize VM during sporulation using fluorescent protein fusions have thus far been unsuccessful.
C. difficile VM binds IVA in vitro.
Since these data indicated that IVA can localize to and surround the forespore even in the absence of VM in the majority of sporulating cells, we wondered whether C. difficile VM even binds C. difficile IVA. In B. subtilis, VM binding to IVA depends on Ile6 in VM and Gly486 in IVA, as a G486V mutation in IVA can rescue the sporulation defect of the I6A VM mutant (22). Interestingly, while Ile6 is conserved in C. difficile VM (Fig. 1), Gly486 is an asparagine in C. difficile IVA (Asn491), other Peptostreptococcaceae spp. (41), and Clostridium perfringens IVA (Fig. S6).
Sequence alignment of the C-terminal domain (68) of IVA homologs in the Bacilli and Clostridia. Residues that are completely conserved are boxed in blue with white text. Conserved identical residues are boxed in green, and conserved similar residues are boxed in yellow. The residues corresponding to Gly486 in B. subtilis IVA are outlined in red. Mutation of Gly486 to valine suppressed the sporulation defect of an I6A VM mutant and restored IVA’s ability to encase the forespore in the I6A VM mutant (22). SpoIVA sequence accession numbers: B. subtilis, NP_390161; Bacillus licheniformis, AAU23942; Paenibacillus sp., GYMC10_2192; Bacillus clausii, WP_011246725; Clostridium acetobutylicum, NP_348339; Clostridium perfringens, ABG84677; Clostridium beijerinckii, WP_026889165; Clostridium butyricum, EDT74816; Clostridium novyi, KEH85260; Clostridium sporogenes, EHN14425; Clostridium bartlettii, CDA10929; Clostridium bifermentans, WP_021433867; Clostridium mangenotii, WP_024621862; Peptostreptococcaceae sp., WP_026900372; Clostridium sordellii, CEQ01088; C. difficile, CAJ69515. Download FIG S6, TIFF file, 0.4 MB (380.3KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
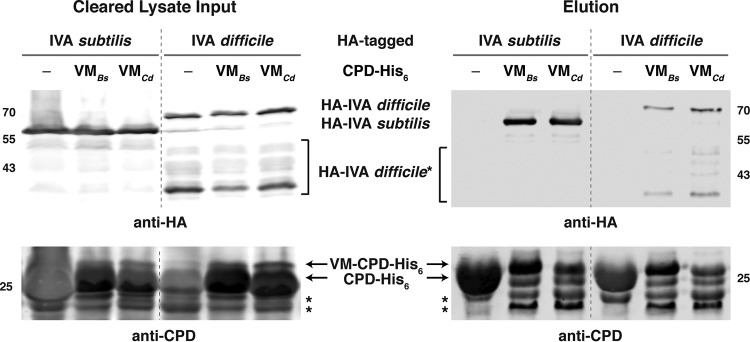
To test whether C. difficile VM can bind IVA, we coproduced His6-tagged C. difficile VM (VMCd) with hemagglutinin (HA)-tagged C. difficile IVA in E. coli and measured the ability of HA-tagged IVA to copurify with His6-tagged VMCd using Ni2+-affinity chromatography. To facilitate the production and detection of VMCd, we fused it to CPD-His6, an inducible, self-cleaving protease domain (42) that can promote the production and purification of small amphipathic helices (43, 44). As a positive control, we measured the ability of HA-tagged B. subtilis IVA to copurify with B. subtilis VM-CPD-His6. As negative controls, we tested whether the HA-tagged IVA variants could copurify with CPD-His6. Last, we also tested whether HA-tagged C. difficile IVA could bind B. subtilis VM-CPD-His6 and vice versa. When VMCd-CPD-His6 was pulled down on nickel-nitrilotriacetic acid (Ni-NTA) beads in the presence of either B. subtilis or C. difficile HA-tagged IVA, both HA-tagged IVA variants were detected in the imidazole elution fraction (Fig. 6). Since VMBs-CPD-His6 also pulled down both HA-tagged IVA variants, whereas CPD-His6 failed to pull down HA-tagged B. subtilis and C. difficile IVA from the cleared lysates, these results indicate that C. difficile IVA can interact specifically with both B. subtilis and C. difficile VM in this heterologous system.
FIG 6 .
C. difficile VM directly binds IVA in coaffinity purification analyses. HA-tagged B. subtilis and C. difficile IVA were coproduced in E. coli with either VM-CPD-His6 fusions, where VM was from either B. subtilis (Bs) or C. difficile (Cd), or CPD-His6 alone. Cleared (soluble) lysate samples were prepared from the cultures following lysis by sonication and centrifugation. CPD-His6 variants were affinity purified, and high imidazole was used to elute the His-tagged proteins and any interacting partners. Cleared lysate and elution samples were analyzed by Western blotting using anti-HA and anti-CPD (42) antibodies. Degradation products of HA-tagged C. difficile IVA are highlighted by the square brackets, while degradation products of VM-CPD-His6 are highlighted by asterisks. Numbers at left and right of the gels indicate molecular masses in kilodaltons.
Despite these observations, both VM-CPD-His6 fusions pulled down more B. subtilis IVA than C. difficile IVA, so we wondered whether the bulkier Asn491 residue in C. difficile IVA diminished its ability to bind VM relative to B. subtilis IVA, which carries a glycine (G486) at this position. To test this possibility, we coproduced HA-tagged C. difficile N491G IVA with VM-CPD-His6. Unfortunately, N491G IVA was degraded more readily than wild-type C. difficile IVA in E. coli (data not shown), so we could not assess whether the N491G mutation enhanced binding to VM. However, when we produced N491G IVA in the ΔIVA background, N491G IVA was detected at wild-type levels and no change in the heat resistance of ΔIVA/N491G IVA was observed (Fig. S5). The latter observation appears to be consistent with the finding that a G486V IVA mutation in B. subtilis does not alter heat-resistant spore formation (22). Taken together, our results indicate that, despite C. difficile VM’s ability to bind IVA, disrupting this interaction in C. difficile has a relatively minor impact on functional spore formation.
DISCUSSION
In B. subtilis, the proper assembly of the coat basement layer is essential for both coat and cortex assembly and thus functional spore formation (8, 11). While extensive analyses have identified key regulators of this process in B. subtilis, namely, VM, IVA, CmpA, and VID (11, 33, 45), major differences in basement layer coat morphogenesis exist between C. difficile and B. subtilis. These include the lack of CmpA and VID homologs in the Clostridia (26, 33), the lack of SipL homologs in the Bacilli (34), the dispensability of IVA for cortex formation in C. difficile (34), and the ability of C. difficile IVA but not B. subtilis IVA to bind LysM domains (23, 34). In the present study, we identified another major difference in spore assembly between C. difficile and B. subtilis: VM is essential for B. subtilis, but not C. difficile, spore formation. Whereas B. subtilis VM mutants exhibit ~6-log reductions in spore purification yields (26) and heat and chloroform resistance (25), only ~3-fold decreases in these properties were observed in C. difficile (Fig. 2 and 3; see also Table S4 in Data Set S1). Thus, this study contributes to a growing body of work indicating that C. difficile and B. subtilis can have differential requirements for conserved factors during sporulation (39, 46, 47).
Our finding that loss of VM does not abolish functional spore formation in C. difficile was somewhat surprising, since (i) key functional residues of B. subtilis VM are conserved in C. difficile (Fig. 1); (ii) C. difficile VM and C. difficile IVA directly interact (Fig. 6), similarly to B. subtilis VM and IVA (22); and (iii) VM is transcribed in the mother cell early during sporulation in both B. subtilis (37) and C. difficile (Fig. 2 and S1). A potential reason why VM may be differentially required between these two organisms is that B. subtilis IVA, but not C. difficile IVA, depends on VM for forespore encasement (Fig. 5B). In B. subtilis, sporulating cells that fail to encase IVA around the forespore (due to loss of VM [23] or IVA ATPase activity [40]) are lysed by a CmpA-dependent checkpoint mechanism (26). While IVA fails to surround the forespore in almost all B. subtilis VM mutant cells, only ~30% of C. difficile VM mutant cells exhibit IVA encasement defects (Fig. 5B). Thus, even if C. difficile employed a checkpoint mechanism analogous to the one activated by CmpA, it would not be strongly induced in a C. difficile VM mutant.
Clearly, a major difference between B. subtilis and C. difficile IVA is their dependence on VM for IVA forespore encasement. C. difficile IVA may be able to independently interact with the forespore membrane largely independently of VM, such that VM is largely dispensable in C. difficile. Alternatively, a redundant factor could help IVA surround the forespore in the absence of VM. While additional possibilities exist, we posit that C. difficile has more redundant factors and fewer checkpoint mechanisms than B. subtilis to maximize spore production prior to exiting the host (33, 48). Whereas spore formation is essential for C. difficile to survive exit from the host (5), this process is only one of several alternative lifestyles that can be adopted by B. subtilis (49). As a result, mechanisms may have evolved to ensure the fidelity of this developmental process (33, 48).
While addressing these questions awaits further analysis, another question raised by this study is why ~70% of VM mutant spores are sensitive to heat and chloroform (Fig. 2; Table S4 in Data Set S1) when only ~30% of VM mutant cells exhibit gross morphological defects (Fig. 3 and 5). Since the cortex is critical for spores to resist heat and chloroform (9, 10), heat- and chloroform-sensitive C. difficile VM mutant cells may have cortex defects that are not visible by TEM. Indeed, we found that ~20% of VM mutant cells produce cortex with various thicknesses (Fig. 3 and S3), suggesting that C. difficile VM alters the activities of the cortex synthesis and/or modification machinery. However, little is known about the factors controlling cortex production and/or modification in C. difficile, although homologs of the B. subtilis cortex synthesis machinery (SpoVB, SpoVD, and SpoVE [50]) and cortex-modifying enzymes (CwlD and PdaA [51, 52]) are conserved in C. difficile. Future studies of cortex production and modification in C. difficile could identify mechanisms by which VM modulates spore formation and may also provide insight into the mechanisms underlying the forespore pinching observed in ~20% of VM mutant cells (Fig. 3).
Whether such mechanisms are related to how B. subtilis links basement layer formation to cortex assembly (27, 33), which is also poorly defined, awaits further investigation. Regardless, our study further highlights that the coat assembly pathway in C. difficile exhibits substantial differences relative to B. subtilis and raises questions as to how these functional differences evolved for the shared coat morphogenetic proteins IVA and VM. Analyses of coat assembly in other clostridial organisms may provide insight into this question.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
JIR8094 (630E [53]) and 630ΔermΔpyrE (38) were used as the parental strains for TargeTron-based gene disruption (54). 630ΔermΔpyrE was also used for pyrE-based allele-coupled exchange (ACE [38]). C. difficile strains are listed in Table S1 in Data Set S1 and were grown on BHIS agar (55) supplemented with taurocholate (TA; 0.1% [wt/vol]; 1.9 mM), kanamycin (50 µg/ml), cefoxitin (8 µg/ml), FeSO4 (50 µM), and/or erythromycin (10 µg/ml) as needed. C. difficile defined minimal medium (CDMM [56]) was supplemented with 5-fluoroorotic acid (5-FOA) at 2 mg/ml and uracil at 5 µg/ml as needed for ACE. Cultures were grown under anaerobic conditions using a gas mixture containing 85% N2, 5% CO2, and 10% H2.
Escherichia coli strains for HB101/pRK24-based conjugations and BL21(DE3)-based protein production are listed in Table S1 in Data Set S1. E. coli strains were grown at 37°C with shaking at 225 rpm in Luria-Bertani broth (LB). The medium was supplemented with chloramphenicol (20 µg/ml), ampicillin (50 µg/ml), or kanamycin (30 µg/ml) as indicated.
E. coli strain construction.
All primers are listed in Table S2 in Data Set S1. For constructing targeting sequences for spoVM and CD25622-VM, a modified plasmid containing the retargeting group II intron, pCE245 (a gift from C. Ellermeier, University of Iowa), was used as a template. For targeting spoVM, primers 1094, 1095, 1096, and 532 (EBS universal primer; Sigma-Aldrich) were used; for targeting CD25622, primers 2186, 2187, 2188, and 532 were used. The resulting sequences were digested with BsrGI and HindIII and cloned into pJS107 (6), a derivative of pJIR750ai (Sigma-Aldrich). Ligations were transformed into DH5α, and the resulting plasmids were transformed into HB101/pRK24.
To construct the CD25622-VM complementation plasmid for ACE, primer pair 2148 and 2149 was used to amplify a region 77 bp upstream of CD25622 and 9 bp downstream of spoVM. To construct the spoIVA complementation constructs, primer pair 2036 and 2037 was used to amplify 80 bp upstream and 159 bp downstream of spoIVA. To construct the N491G spoIVA complementation construct, splicing-by-overlap-extension (SOE) primer pair 2177 and 2178 was used in conjunction with 2036 and 2037. To construct the mCherry-spoIVA construct, primer pair 2202 and 2203 was used to amplify the 80-bp upstream region of spoIVA, while primer pair 2204 and 2205 was used to amplify a codon-optimized mCherry construct (a kind gift of D. Weiss and C. Ellermeier [57]). The resulting PCR products fuse the spoIVA promoter region to codon-optimized mCherry (57) and the spoIVA gene when assembled into pMTL-YN1C digested with NotI and XhoI using Gibson assembly (58).
To construct the pMTL-YN3 ΔspoIVA allelic exchange construct, primer pair 2272 and 2273 was used to amplify regions ~1 kb upstream and downstream of spoIVA. This primer pair was used in conjunction with the SOE primer pair 1746 and 1747, which fuses the first 12 codons of spoIVA to its last 13 codons. The resulting PCR products were ligated into AscI-SbfI-digested pMTL-YN3 using Gibson assembly.
To create the recombinant protein expression constructs for the coaffinity purifications, primer pair 1086 and 1609 was used to amplify spoVM lacking the stop codon using C. difficile genomic DNA as the template. The resulting PCR product was digested with NdeI/SalI and ligated to pET22b-Δ50-CPD (43) to construct a SpoVM-CPD-His6 fusion construct. The B. subtilis SpoVM-CPD expression construct was cloned in a similar manner except that the spoVM gene was digested from pUC57-Kan-spoVM subtilis (synthesized by GenScript) using NdeI/SalI. To construct the HA-tagged B. subtilis spoIVA expression construct, primer pair 1318 and 1237 was used to amplify spoIVA encoding an N-terminal HA tag using B. subtilis DNA as a template. The resulting PCR product was digested with NheI and XhoI and ligated into pET28a digested with the same enzymes. DNA sequencing was used to confirm all the constructs described.
C. difficile strain construction.
TargeTron-based gene disruption using pJS107-spoVM and pJS107-CD25622 inserted into either JIR8094 or 630ΔermΔpyrE was employed as previously described (54, 59). Primer pair 2148 and 2149 was used to screen isolated erythromycin-resistant colonies for TargeTron (2-kb) insertions in spoVM and CD25622. Allele-coupled exchange (ACE [38]) was used as previously described (46) to construct the clean spoIVA deletion in 630ΔermΔpyrE. Primer pair 1743 and 1744 was used to screen isolated FOA-resistant, uracil auxotroph colonies for the ΔspoIVA deletion. At least two clones of each mutant strain were phenotypically characterized.
C. difficile complementation.
The pyrE locus was restored using pMTL-YN1C and pMTL-YN1C-based complementation constructs as previously described (46). At least two independent clones from each complementation strain were phenotypically characterized.
Sporulation.
C. difficile strains were grown from glycerol stocks on BHIS plates containing TA (0.1% [wt/vol], 1.9 mM). Colonies from these plates were used to inoculate liquid BHIS cultures, which were grown to stationary phase either for several hours or overnight. The liquid cultures were back-diluted 1:50 into BHIS, and cultures were grown until they reached an optical density at 600 nm (OD600) between 0.35 and 0.7. One hundred to 150 µl of this culture was used to inoculate 70:30 agar plates (40 ml [34]), and sporulation was induced on this medium for 20 to 24 h. The constant thickness of the 70:30 plates combined with the broth culture inoculation reduced the variability in heat resistance efficiencies between replicates.
Phase-contrast microscopy.
Sporulating cells were enumerated for apparent mislocalized coat and misshapen forespores using phase-contrast microscopy. A minimum of 400 cells (includes sporulating cells and free spores) from at least two independent replicates was counted, and the percentages of phase-bright forespores, phase-dark forespores, phase-dark extensions, and misshapen forespores were determined relative to the total number of sporulating cells. Free spores were also enumerated.
qRT-PCR.
Transcript levels for spoVM and rpoB were determined using cDNA templates prepared from five biological replicates of wild-type JIR8094, spo0A mutant, sigE mutant, spoIIID mutant, and sigK mutant strains (36, 59). Primer pair 1086 and 1087 was used to amplify the spoVM coding sequence, while the housekeeping gene rpoB-specific primers have been previously described (36). qRT-PCR was performed as previously described (36) using SYBR green to quantify transcript levels for both spoVM and rpoB. Transcript levels were normalized to rpoB using the standard curve method and calculated relative to spo0A mutant.
Heat resistance assay on sporulating cells.
C. difficile strains were induced to sporulate for 20 to 24 h as described above. Heat-resistant spore formation was measured as previously described (60). Heat resistance efficiencies represent the average ratio of heat-resistant cells for a given strain to the average ratio determined for the wild type based on a minimum of three biological replicates.
Chloroform resistance assay.
Following the induction of sporulation for 20 to 24 h, a quarter of the 70:30 plate was harvested into phosphate-buffered saline (PBS). Sporulating cells were either mock treated or exposed to 10% chloroform for 15 min with periodic mixing, after which the sample was serially diluted and plated on BHIS-TA plates. Chloroform resistance was calculated by determining the ratio of chloroform-resistant cells to untreated cells and averaging this ratio across four biological replicates. Chloroform resistance efficiencies were determined by comparing the average chloroform resistance for a given strain to that of the wild type.
TEM analyses.
Sporulating cultures (24 h) were fixed and processed for electron microscopy by the University of Vermont Microscopy Center as previously described (34). A minimum of 50 full-length sporulating cells were used for phenotype counting.
Spore purification.
Sporulation was induced on four 70:30 plates as described above for 2 to 3 days. Sporulating cells were scraped into ice-cold water, and a sample was removed to analyze the spore input by phase-contrast microscopy. Spores were purified as previously described (61). Ice-cold water washes were used to lyse sporulating cells, after which released DNA was removed by DNase, and spores were purified on a HistoDenz density gradient and resuspended in equal volumes of water. Purified spores were analyzed by phase-contrast microscopy to ensure that they were >95% pure, and spore yields were determined by measuring the optical density (600 nm) of the spore purifications. Spores were stored in water at 4°C.
Germination assay.
Purified spores (~1 × 107 spores, equivalent to 0.35 OD600 units) were resuspended in 100 µl of water, and 10 µl was removed for serial dilutions and plating on BHIS-TA. Colonies arising from germinated spores were counted between 20 and 24 h. Germination efficiency represents the average number of CFU produced by spores of a given strain on BHIS-TA relative to the average number produced by wild-type spores based on three biological replicates of at least two independent spore preparations.
Spore heat titration.
Approximately 4 × 107 spores (1.5 OD600 units) were resuspended in 420 µl of water. One-hundred-microliter aliquots were removed for the untreated sample and three temperature treatments. Spores were incubated at either 60°C, 70°C, or 80°C for 15 min, and 10-µl aliquots were removed for serial dilution and plating onto BHIS-TA plates similarly to previous studies (46).
Spore chloroform resistance.
Approximately 2 × 107 spores (0.75 OD600 units) were resuspended in 190 µl water. Ninety microliters was then added to tubes containing either 10 µl of water or chloroform. Spores were incubated for 15 min, after which 10 µl of the sample was serially diluted and plated on BHIS-TA plates similarly to previous studies (62).
Fluorescence microscopy.
Live cell fluorescence microscopy was performed using Hoechst 33342 (Molecular Probes; 15 µg/ml) and mCherry protein fusions to localize IVA. Samples were prepared on agarose pads as previously described (39) except that the samples were not imaged until 30 min after harvesting, since this time frame allowed for reconstitution of mCherry fluorescence signal in the anaerobically growing bacteria. Sporulating cells were exposed to ambient oxygen for a maximum of 80 min to minimize DNA fragmentation; no cell lysis was observed under these conditions. Differential interference contrast (DIC) and fluorescence microscopy were performed using a Nikon PlanApo Vc 100× oil immersion objective (1.4 numerical aperture [NA]) on a Nikon Eclipse Ti2000 epifluorescence microscope. An EXi Blue Mono camera (QImaging) with a hardware gain setting of 2.0 was used to acquire multiple fields for each sample in 14-bit format with 2-by-2 binning using NIS-Elements software (Nikon). The Texas Red channel was used to acquire images after a 300- to 400-ms exposure, 75- to 90-ms exposures were used to visualize the Hoechst stain, and ~10- to 20-ms exposures were used for DIC microscopy. Twenty-megahertz images were subsequently imported into Adobe Photoshop CS6 for minimal adjustments in brightness/contrast levels and pseudocoloring.
Western blot analyses.
Samples for immunoblotting were prepared as previously described (61). Briefly, pelleted sporulating cells were resuspended in 100 µl of PBS, and 50-µl samples were freeze-thawed for three cycles and then resuspended in 100 µl EBB buffer (8 M urea, 2 M thiourea, 4% [wt/vol] SDS, 2% [vol/vol] β-mercaptoethanol). The samples were boiled for 20 min, pelleted at high speed, resuspended in the same buffer to maximize protein solubilization, boiled for another 5 min, and then pelleted at maximum speed. Samples were resolved on 12% SDS-PAGE gels, transferred to an Immobilon-FL polyvinylidene difluoride (PVDF) membrane, and blocked in Odyssey Blocking buffer with 0.1% (vol/vol) Tween 20. Mouse anti-SpoIVA (63) and rabbit anti-mCherry (Abcam, Inc.) were used at 1:2,500 and 1:2,000 dilutions. IRDye 680CW and 800CW infrared dye-conjugated secondary antibodies were used at a 1:20,000 dilution, and blots were imaged on an Odyssey LiCor CLx imaging system.
Coaffinity purification assays.
VM-CPD-His6 variants and associated IVA variants were purified on Ni2+-affinity resin from 500 ml of 2YT (0.55% NaCl, 1.0% yeast extract, 1.5% [wt/vol] tryptone) culture as previously described (64). Input samples were taken from cultures following isopropyl-β-d-thiogalactopyranoside (IPTG) induction at 18°C for 15 h. Culture pellets were resuspended in ~25 ml lysis buffer (500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 15 mM imidazole, 10% [vol/vol] glycerol), flash frozen in liquid nitrogen, thawed, sonicated to lyse the sample, and centrifuged to clear insoluble material. Ni-NTA agarose beads (0.5 ml; 5 Prime) were added to the cleared lysate for 3 h. The resin was washed extensively in lysis buffer, and His6-tagged proteins were eluted in 175 µl of high-imidazole buffer (500 mM NaCl, 50 mM Tris, pH 7.5, 175 mM imidazole, 10% glycerol) after nutating the sample for 5 to 10 min.
ACKNOWLEDGMENTS
We thank N. Bishop, M. Von Turkovich, and J. Schwarz for excellent assistance in preparing samples for transmission electron microscopy throughout this study; K. Schutz for excellent technical assistance in conducting the qRT-PCR analyses; D. Weiss and C. Ellermeier for providing a codon-optimized mCherry construct; C. Huston for access to the Nikon microscope; N. Minton (University of Nottingham) for generously providing us with access to the 630ΔermΔpyrE strain and pMTL-YN1C and pMTL-YN3 plasmids for allele-coupled exchange (ACE); and Marcin Dembek for directly providing these materials to us and sharing his specific protocols on ACE with us.
Research in this work was funded by award no. R01AI22232 from the National Institute of Allergy and Infectious Diseases (NIAID) to A.S. A.S. was a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. J.W.R. was a recipient of the American Society for Microbiology Undergraduate Research Fellowship in 2016. The content is solely the responsibility of the author(s) and does not necessarily reflect the views of the Pew Charitable Trusts, NIAID, or the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. 2016. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prevot 1938. Anaerobe 40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Oren A, Garrity GM. 2017. Notification that new names of prokaryotes, new combinations, and new taxonomic opinions have appeared in volume 67, part 4, of the IJSEM. Int J Syst Evol Microbiol 67:2079–2080. doi: 10.1099/ijsem.0.002098. [DOI] [PubMed] [Google Scholar]
- 3.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis MB, Allen CA, Shrestha R, Sorg JA. 2013. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, Rigden DJ. 2012. Genomic determinants of sporulation in Bacilli and Clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol 14:2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan IS, Ramamurthi KS. 2014. Spore formation in Bacillus subtilis. Environ Microbiol Rep 6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atrih A, Foster SJ. 1999. The role of peptidoglycan structure and structural dynamics during endospore dormancy and germination. Antonie Leeuwenhoek 75:299–307. doi: 10.1023/A:1001800507443. [DOI] [PubMed] [Google Scholar]
- 10.Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol 101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 11.Driks A, Eichenberger P. 2016. The spore coat. Microbiol Spectr 4(2). doi: 10.1128/microbiolspec.TBS-0023-2016. [DOI] [PubMed] [Google Scholar]
- 12.Klobutcher LA, Ragkousi K, Setlow P. 2006. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc Natl Acad Sci U S A 103:165–170. doi: 10.1073/pnas.0507121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenney PT, Eichenberger P. 2012. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol Microbiol 83:245–260. doi: 10.1111/j.1365-2958.2011.07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levin PA, Fan N, Ricca E, Driks A, Losick R, Cutting S. 1993. An unusually small gene required for sporulation by Bacillus subtilis. Mol Microbiol 9:761–771. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- 15.Roels S, Driks A, Losick R. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol 174:575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens CM, Daniel R, Illing N, Errington J. 1992. Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J Bacteriol 174:586–594. doi: 10.1128/jb.174.2.586-594.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobbs EC, Fontaine F, Yin X, Storz G. 2011. An expanding universe of small proteins. Curr Opin Microbiol 14:167–173. doi: 10.1016/j.mib.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill RL Jr, Castaing JP, Hsin J, Tan IS, Wang X, Huang KC, Tian F, Ramamurthi KS. 2015. Structural basis for the geometry-driven localization of a small protein. Proc Natl Acad Sci U S A 112:E1908–E1915. doi: 10.1073/pnas.1423868112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. 2009. Geometric cue for protein localization in a bacterium. Science 323:1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramamurthi KS, Losick R. 2008. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell 31:406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price KD, Losick R. 1999. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J Bacteriol 181:781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramamurthi KS, Clapham KR, Losick R. 2006. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol 62:1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang KH, Isidro AL, Domingues L, Eskandarian HA, McKenney PT, Drew K, Grabowski P, Chua MH, Barry SN, Guan M, Bonneau R, Henriques AO, Eichenberger P. 2009. The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol Microbiol 74:634–649. doi: 10.1111/j.1365-2958.2009.06886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu IL, Narayan K, Castaing JP, Tian F, Subramaniam S, Ramamurthi KS. 2015. A versatile nano display platform from bacterial spore coat proteins. Nat Commun 6:6777. doi: 10.1038/ncomms7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cutting S, Anderson M, Lysenko E, Page A, Tomoyasu T, Tatematsu K, Tatsuta T, Kroos L, Ogura T. 1997. SpoVM, a small protein essential to development in Bacillus subtilis, interacts with the ATP-dependent protease FtsH. J Bacteriol 179:5534–5542. doi: 10.1128/jb.179.17.5534-5542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan IS, Weiss CA, Popham DL, Ramamurthi KS. 2015. A quality-control mechanism removes unfit cells from a population of sporulating bacteria. Dev Cell 34:682–693. doi: 10.1016/j.devcel.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebmeier SE, Tan IS, Clapham KR, Ramamurthi KS. 2012. Small proteins link coat and cortex assembly during sporulation in Bacillus subtilis. Mol Microbiol 84:682–696. doi: 10.1111/j.1365-2958.2012.08052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beall B, Driks A, Losick R, Moran CP Jr. 1993. Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J Bacteriol 175:1705–1716. doi: 10.1128/jb.175.6.1705-1716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Francesco M, Jacobs JZ, Nunes F, Serrano M, McKenney PT, Chua MH, Henriques AO, Eichenberger P. 2012. Physical interaction between coat morphogenetic proteins SpoVID and CotE is necessary for spore encasement in Bacillus subtilis. J Bacteriol 194:4941–4950. doi: 10.1128/JB.00914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozin AJ, Samford CS, Henriques AO, Moran CP. 2001. SpoVID guides SafA to the spore coat in Bacillus subtilis. J Bacteriol 183:3041–3049. doi: 10.1128/JB.183.10.3041-3049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozin AJ, Henriques AO, Yi H, Moran CP Jr. 2000. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J Bacteriol 182:1828–1833. doi: 10.1128/JB.182.7.1828-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abecasis AB, Serrano M, Alves R, Quintais L, Pereira-Leal JB, Henriques AO. 2013. A genomic signature and the identification of new sporulation genes. J Bacteriol 195:2101–2115. doi: 10.1128/JB.02110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Decker AR, Ramamurthi KS. 2017. Cell death pathway that monitors spore morphogenesis. Trends Microbiol 25:637–647. doi: 10.1016/j.tim.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putnam EE, Nock AM, Lawley TD, Shen A. 2013. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J Bacteriol 195:1214–1225. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Ooij C, Eichenberger P, Losick R. 2004. Dynamic patterns of subcellular protein localization during spore coat morphogenesis in Bacillus subtilis. J Bacteriol 186:4441–4448. doi: 10.1128/JB.186.14.4441-4448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pishdadian K, Fimlaid KA, Shen A. 2015. SpoIIID-mediated regulation of σK function during Clostridium difficile sporulation. Mol Microbiol 95:189–208. doi: 10.1111/mmi.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel B, Kroos L, Poth H, Youngman P, Losick R. 1989. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev 3:1735–1744. doi: 10.1101/gad.3.11.1735. [DOI] [PubMed] [Google Scholar]
- 38.Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, Collignon A, Cartman ST, Minton NP. 2013. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One 8:e56051. doi: 10.1371/journal.pone.0056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fimlaid KA, Jensen O, Donnelly ML, Siegrist MS, Shen A. 2015. Regulation of Clostridium difficile spore formation by the SpoIIQ and SpoIIIA proteins. PLoS Genet 11:e1005562. doi: 10.1371/journal.pgen.1005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castaing JP, Nagy A, Anantharaman V, Aravind L, Ramamurthi KS. 2013. ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein. Proc Natl Acad Sci U S A 110:E151–E160. doi: 10.1073/pnas.1210554110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yutin N, Galperin MY. 2013. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupardus PJ, Shen A, Bogyo M, Garcia KC. 2008. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science 322:265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen A, Lupardus PJ, Morell M, Ponder EL, Sadaghiani AM, Garcia KC, Bogyo M. 2009. Simplified, enhanced protein purification using an inducible, autoprocessing enzyme tag. PLoS One 4:e8119. doi: 10.1371/journal.pone.0008119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright O, Yoshimi T, Tunnacliffe A. 2012. Recombinant production of cathelicidin-derived antimicrobial peptides in Escherichia coli using an inducible autocleaving enzyme tag. N Biotechnol 29:352–358. doi: 10.1016/j.nbt.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 45.McKenney PT, Driks A, Eichenberger P. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donnelly ML, Fimlaid KA, Shen A. 2016. Characterization of Clostridium difficile spores lacking either SpoVAC or dipicolinic acid synthetase. J Bacteriol 198:1694–1707. doi: 10.1128/JB.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serrano M, Crawshaw AD, Dembek M, Monteiro JM, Pereira FC, Pinho MG, Fairweather NF, Salgado PS, Henriques AO. 2016. The SpoIIQ-SpoIIIAH complex of Clostridium difficile controls forespore engulfment and late stages of gene expression and spore morphogenesis. Mol Microbiol 100:204–228. doi: 10.1111/mmi.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramírez-Guadiana FH, Meeske AJ, Wang X, Rodrigues CDA, Rudner DZ. 2017. The Bacillus subtilis germinant receptor GerA triggers premature germination in response to morphological defects during sporulation. Mol Microbiol 105:689–704. doi: 10.1111/mmi.13728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasudevan P, Weaver A, Reichert ED, Linnstaedt SD, Popham DL. 2007. Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol Microbiol 65:1582–1594. doi: 10.1111/j.1365-2958.2007.05896.x. [DOI] [PubMed] [Google Scholar]
- 51.Fukushima T, Yamamoto H, Atrih A, Foster SJ, Sekiguchi J. 2002. A polysaccharide deacetylase gene (pdaA) is required for germination and for production of muramic delta-lactam residues in the spore cortex of Bacillus subtilis. J Bacteriol 184:6007–6015. doi: 10.1128/JB.184.21.6007-6015.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Popham DL, Helin J, Costello CE, Setlow P. 1996. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc Natl Acad Sci U S A 93:15405–15410. doi: 10.1073/pnas.93.26.15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol 66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- 54.Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. 2010. The ClosTron: mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 80:49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 55.Sorg JA, Dineen SS. 2009. Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol Chapter 9:Unit 9A.1. doi: 10.1002/9780471729259.mc09a01s12. [DOI] [PubMed] [Google Scholar]
- 56.Cartman ST, Minton NP. 2010. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl Environ Microbiol 76:1103–1109. doi: 10.1128/AEM.02525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ransom EM, Ellermeier CD, Weiss DS. 2015. Use of mCherry red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl Environ Microbiol 81:1652–1660. doi: 10.1128/AEM.03446-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 59.Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, Shen A. 2013. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet 9:e1003660. doi: 10.1371/journal.pgen.1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen A, Fimlaid KA, Pishdadian K. 2016. Inducing and quantifying Clostridium difficile spore formation. Methods Mol Biol 1476:129–142. doi: 10.1007/978-1-4939-6361-4_10. [DOI] [PubMed] [Google Scholar]
- 61.Fimlaid KA, Jensen O, Donnelly ML, Francis MB, Sorg JA, Shen A. 2015. Identification of a novel lipoprotein regulator of Clostridium difficile spore germination. PLoS Pathog 11:e1005239. doi: 10.1371/journal.ppat.1005239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edwards AN, Karim ST, Pascual RA, Jowhar LM, Anderson SE, McBride SM. 2016. Chemical and stress resistances of Clostridium difficile spores and vegetative cells. Front Microbiol 7:1698. doi: 10.3389/fmicb.2016.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kevorkian Y, Shirley DJ, Shen A. 2016. Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie 122:243–254. doi: 10.1016/j.biochi.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams CM, Eckenroth BE, Putnam EE, Doublié S, Shen A. 2013. Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog 9:e1003165. doi: 10.1371/journal.ppat.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pereira FC, Saujet L, Tomé AR, Serrano M, Monot M, Couture-Tosi E, Martin-Verstraete I, Dupuy B, Henriques AO. 2013. The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet 9:e1003782. doi: 10.1371/journal.pgen.1003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, Gelfand MS, Dupuy B, Henriques AO, Martin-Verstraete I. 2013. Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet 9:e1003756. doi: 10.1371/journal.pgen.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castaing JP, Lee S, Anantharaman V, Ravilious GE, Aravind L, Ramamurthi KS. 2014. An autoinhibitory conformation of the Bacillus subtilis spore coat protein SpoIVA prevents its premature ATP-independent aggregation. FEMS Microbiol Lett 358:145–153. doi: 10.1111/1574-6968.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptional analyses of the CD25622-VM locus. (A) Representative image of Integrative Genomics Viewer software (65) used to visualize CD25622-VM in RNA-Seq data of sporulating JIR8094 and associated sigma factor mutants from reference 36. The reads mapped to this locus are shown. The direction of transcription is indicated by the angled bracket. (B) qRT-PCR analyses of VM transcript levels in wild-type JIR8094, spo0A mutant, sigE mutant, spoIIID mutant, and sigK mutant strains relative to spo0A mutant. spoIIID and sigK expression depend on sigE, which encodes the mother cell-specific sigma factor σE. ***, P < 0.005. Download FIG S1, TIFF file, 0.1 MB (118.7KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Table S1 shows strains and plasmids used in this study. Table S2 shows primers used in this study. Table S3 shows expression levels of select sporulation genes as measured by RNA-Seq. Table S4 shows sporulation efficiency as measured by chloroform resistance. Table S5 shows frequency of forespore morphological abnormalities detected by phase-contrast microscopy. Download DATA SET S1, PDF file, 0.1 MB (124.8KB, pdf) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of VM, CD25622, and ΔIVA mutants. (A) Schematic of the TargeTron-based insertions in VM (VM::ermB) and CD25622 (CD25622::ermB). The TargeTron insertion in CD25622 is predicted to have polar effects on VM expression. (B) Colony PCR of VM::ermB (VM−) and CD25622::ermB (CD25622−) mutants in the 630ΔermΔpyrE and JIR8094 strain backgrounds. The group II intron is ~2 kb. (C) Schematic and colony PCR of the spoIVA deletion in 630ΔermΔpyrE. Download FIG S2, TIFF file, 0.3 MB (267.6KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The CD25622-VM locus is largely dispensable for heat-resistant spore formation in JIR8094. (A) Phase-contrast microscopy of sporulating cultures of wild-type JIR8094, spo0A mutant, and VM and CD25622-VM TargeTron mutants at 21 h. Examples of phase-bright forespores and spores are marked using yellow and blue arrows, respectively. Forespores with abnormal morphologies are denoted by green arrows, while regions that likely correspond to mislocalized coat are marked in pink. HR refers to the heat resistance of each strain relative to the wild type. The means and standard deviations shown are based on three biological replicates. Statistical significance relative to the wild type was determined using one-way ANOVA and Tukey’s test. Bars, 5 µm. (B) Transmission electron microscopy (TEM) analyses of sporulating cells of wild-type JIR8094 and VM and CD25622-VM TargeTron mutants and their complements (VM−/C and 25622−/C). Bars, 500 nm. Blue arrows mark coat layers that surround the forespore, while pink arrows indicate detached coat layers termed “bearding.” The green arrows mark abnormal forespore shape, where the forespore appears to be pinched at the mother cell-proximal side; the yellow arrows denote areas of abnormal cortex thickness. (C) TEM analyses of sporulating JIR8094 strains. Percentage of cells with the observed phenotype based on analyses of a minimum of 50 TEM images. Download FIG S3, TIFF file, 0.6 MB (579.1KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transmission electron microscopy of wild-type and VM mutant spores. Presumed cortex layers are marked (Cx). Bars, 100 nm. Download FIG S4, TIFF file, 1 MB (1.1MB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Complementation of ΔIVA by mCherry-IVA and N491G IVA. (A) Phase-contrast microscopy of wild-type 630Δerm and ΔIVA strains complemented with either IVA, mCherryIVA, or N491G IVA, and Δ0A after 22 h of sporulation. Examples of phase-bright forespores and spores are marked using yellow and blue arrows, respectively. Mislocalized coat regions are marked by pink arrows. HR refers to the heat resistance of each strain relative to the wild type. The means and standard deviations shown are based on three biological replicates. Statistical significance relative to the wild type was determined using one-way ANOVA and Tukey’s test. Bars, 5 µm. (B) Western blot analyses of the strains in panel A using anti-IVA and anti-mCherry antibodies. Nonspecific bands are marked with asterisks. A small amount of mCherry-IVA undergoes cleavage during sporulation, appearing to liberate IVA and mCherry. Download FIG S5, TIFF file, 0.6 MB (592.9KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence alignment of the C-terminal domain (68) of IVA homologs in the Bacilli and Clostridia. Residues that are completely conserved are boxed in blue with white text. Conserved identical residues are boxed in green, and conserved similar residues are boxed in yellow. The residues corresponding to Gly486 in B. subtilis IVA are outlined in red. Mutation of Gly486 to valine suppressed the sporulation defect of an I6A VM mutant and restored IVA’s ability to encase the forespore in the I6A VM mutant (22). SpoIVA sequence accession numbers: B. subtilis, NP_390161; Bacillus licheniformis, AAU23942; Paenibacillus sp., GYMC10_2192; Bacillus clausii, WP_011246725; Clostridium acetobutylicum, NP_348339; Clostridium perfringens, ABG84677; Clostridium beijerinckii, WP_026889165; Clostridium butyricum, EDT74816; Clostridium novyi, KEH85260; Clostridium sporogenes, EHN14425; Clostridium bartlettii, CDA10929; Clostridium bifermentans, WP_021433867; Clostridium mangenotii, WP_024621862; Peptostreptococcaceae sp., WP_026900372; Clostridium sordellii, CEQ01088; C. difficile, CAJ69515. Download FIG S6, TIFF file, 0.4 MB (380.3KB, tiff) .
Copyright © 2017 Ribis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.