Abstract
Osteogenesis Imperfecta (OI) is a genetic disorder characterized by various clinical features including bone deformities, low bone mass, brittle bones, and connective tissue manifestations. The predominant cause of OI is due to mutations in the two genes that encode type I collagen. However, recent advances in sequencing technology has led to the discovery of novel genes that are implicated in recessive and dominant OI. These include genes that regulate the post-translational modification, secretion and processing of type I collagen as well as those required for osteoblast differentiation and bone mineralization. As such, OI has become a spectrum of genetic disorders informing about the determinants of both bone quantity and quality. Here we summarize the known genetic causes of OI, animal models that recapitulate the human disease and mechanisms that underlie disease pathogenesis. Additionally, we discuss the effects of disrupted collagen networks on extracellular matrix signaling and its impact on disease progression.
Keywords: Osteogenesis imperfecta, collagen type I, osteoblast, extracellular matrix, TGF-β signaling
1. INTRODUCTION
Osteogenesis Imperfecta (OI) is a genetically heterogeneous skeletal dysplasia that affects approximately 1 in 10,000–20,000 births [1,2]. Patients with OI feature a prominent skeletal phenotype with a wide clinical spectrum of severities ranging from low bone mass (OI type I), to progressive bone deformities with increased incidence of fractures (OI type III/IV) and perinatal lethality (OI type II) [3,4]. Additionally, OI patients may exhibit Dentinogenesis Imperfecta (abnormal tooth development), craniofacial abnormalities and joint hypermobility, as well as extra-skeletal manifestations including blue sclerae, hearing impairment, and intrinsic and extrinsic lung abnormalities [2–4]. The majority of OI cases occur as a result of autosomal dominant mutations in the genes encoding type I collagen (COL1A1 and COL1A2) [3]. In addition, approximately 10% of all OI cases are caused by recessive mutations in genes that regulate the post-translational modification, secretion, and processing of type I collagen, as well as those that modulate osteoblast differentiation and bone mineralization [2]. For example, defects in the prolyl 3-hydroxylase complex, which serves to convert a single proline residue at position 986 of the proα1(I) chain into 3-hydroxyproline, leads to abnormal collagen fibrillogenesis and severe OI [5]. Abnormalities in other genes that regulate the proper folding of type I procollagen molecules in the endoplasmic reticulum (ER) can delay secretion [6–8]. Additionally, recent studies have shown that defects in genes that are essential for osteoblast differentiation and function can result in recessive OI [9,10].
Anti-resorptive treatments using bisphosphonates are one of the pharmacological mainstays of OI therapy. Whereas bisphosphonates have been shown to increase bone mineral density and reduce fracture risk in pediatric OI patients, they may be less effective in treating adult patients with OI [11]. Thus, understanding the biochemical and molecular mechanisms underlying the pathogenesis of OI and OI-related diseases could significantly impact novel drug discovery for targeted-mechanism based treatments. This review summarizes the genetic causes of dominant and recessive OI, animal models, and current thinking on the biochemical and molecular mechanisms that drive disease progression. Reviews with greater emphasis on the biochemistry of collagen modifications and cross-linking [12–14] and human disease and therapeutic interventions [2,15] are discussed elsewhere.
2. GENETIC CAUSES AND MECHANISMS OF OSTEOGENESIS IMPERFECTA
2.1 Collagen genes
The fibrillar type I collagen are trimeric molecules composed of two α1(I) chains and one α2(I) chain. The helical domain type I collagen primarily consists of Gly-X-Y repeats with the X and Y frequently being occupied by proline and hydroxyproline residues, respectively [12]. The helical domain is flanked by two globular domains, the N- and C-propeptides, that are eventually removed during collagen assembly [13]. The most frequent cause of classic OI is due to glycine substitutions in the helical domain of type I collagen, which may affect helical assembly. Glycine substitutions in the α1(I) helical domains are associated with more severe phenotypes, including lethality, whereas mutations in the α2(I) helical domain are less severe. Because procollagen assembly begins from the C-propeptide, glycine substitutions in this domain can interfere with chain association and folding and cause OI [2]. Here we describe several mouse models of OI that recapitulate key features of human mutations (Table 1).
Table 1.
Summary of mouse genetic models of dominant OI with type I collagen mutations.
| Gene | Mouse model | Skeletal Phenotype | Type | Reference |
|---|---|---|---|---|
| Col1a1 | Col1a1Mov13/+ | Reduced bone mass but recovers by 15 weeks | I | [16,17,23] |
| Aga2+/− | Reduced bone mass with fractures | III | [26] | |
| Brtl+/− | Reduced bone mass and strength but recovers by 12 months | IV | [28,143] | |
| Col1a2 | Oim−/− | Reduced bone mass and strength with fractures | III-like | [31,32,34–38] |
| Col1a2tm1.1Mcbr | Reduced bone mass and strength | IV | [43] |
Animal models
2.1.1 Col1a1Mov13
Col1a1Mov13 mice were generated by insertion of the Moloney murine leukemia virus (MoMLV) in the first intron of Col1a1 [16,17]. Mice with homozygous insertion (Col1a1Mov13/Mov13) were embryonic lethal due to the complete absence of Col1a1 mRNA despite intact Col1a2 expression [18,19]. Col1a1Mov13/+ mice produced less but structurally normal type I collagen, with increased bone brittleness due to reduced post-yield displacement [20–22]. By 15-weeks, however, Col1a1Mov13/+ mice increased periosteal growth without significant abnormalities in biomechanical strength [23]. Thus, this model closely mimics the mild, non-deforming OI type I, which in humans is due to a heterozygous null COL1A1 allele, resulting in a quantitative reduction of type I collagen without affecting collagen structure [24]. Also, this mouse model captures the natural history of these patients, who exhibit reduced fracture rates and a milder phenotype after puberty [4].
2.1.2 Aga2+/−
The Aga2+/− mice were generated by N-ethyl-N-nitrosourea (ENU) mutagenesis that caused a C-terminal frameshift mutation in Col1a1. While a significant number of Aga2+/− mice died after birth, likely due to cardiorespiratory defects [25], non-lethal mutants displayed reduced size and low bone mass, owing to increased bone turnover [26]. In the same study, an increase in endoplasmic recitulum (ER)-retention of procollagen was observed in Aga2+/− primary dermal fibroblasts. Furthermore, Aga2+/− osteoblasts significantly increased apoptosis, both in vitro and in vivo, which was associated with elevated levels of unfolded protein response (UPR) genes such as BiP, Hsp47 and Ddit3 [26] that may be induced by ER stress (See “Collagen secretion and ER stress”). Aga2+/− mice also manifests with frequent fractures, and kyphosis/scoliosis, which, together with the variation in phenotypic severity, provides a model of OI type III [26,27].
2.1.3 Brtl+/− (Brittle IV)
The Brtl+/− knockin mouse carries a glycine to cysteine substitution at amino acid position 349 of the Col1a1 allele, which was previously characterized in a patient with a moderately severe OI type IV. 40–60% of Brtl+/− mice died shortly after birth due to respiratory distress [17]. The remaining non-lethal Brtl+/− mice were significantly smaller in size and showed reduced bone mineral density, cortical thickness, cross-sectional area and biomechanical properties until 6-months of age, which were largely corrected by 12-months of age [28]. Consistent with these observations, histomorphometric analyses revealed a significant reduction in osteoblast function with a concomitant increase in osteoclast numbers in Brtl+/− mice at 6-months of age [29]. Taken together, Brtl+/− mice are phenotypically heterogeneous in severity, ranging from moderate to perinatal lethality, and show the classic OI symptoms of low bone mass, bone deformities and fragility. Hence, this mouse model recapitulates the phenotype of moderate to severe forms of human OI type IV, in which there is variable expressivity as is seen in patients with the same mutation [30].
2.1.4 Oim−/−
The oim−/− mouse model has a recessively inherited mutation in the C-terminal propeptide of Col1a2, which prevents its incorporation into the collagen triple helix and generates α1(I) homotrimers [31]. Oim−/− mice were smaller in size and had reduced bone mineral density when compared with either wild type or oim+/− mice [32]. The reduction in bone mass is likely due to high bone turnover, as oim−/− mice show increased osteoblast and osteoclast numbers by histomorphometry [33]. Bones from oim−/− mice have altered mineral content with dense and disordered apatite crystals, which likely contribute to increased brittleness [32,34–38]. Furthermore, collagen cross-linking is altered in oim−/− mice. Specifically, stabilizing enzymatic lysyl oxidase-mediated cross-links between fibrils are decreased whereas non-enzymatic glycation-induced cross-links are increased in oim−/− mice, both of which may contribute to the increase in bone fragility [39]. Despite the lack of change in bone mineral density, oim+/− mice also showed abnormalities in biomechanical properties including reduced ductility, which measures the degree of bone to deform preceding a fracture [39,40]. Thus, these results suggest that oim+/− mice have qualitative bone defects independent of bone mass. Because oim−/− mice develop low bone mass, bent bones, and frequent and early fractures, this model phenotypically resembles the moderate to severe OI type III [31,33,41] while oim+/− represents a less severe form of OI [40]. Oim−/− mice displayed increased numbers and branching of canaliculi concomitant with increased numbers and greater spherical morphology of osteocyte lacunae compared to controls [42], which indicates that the alterations in type I collagen may affect osteoblast and osteocyte differentiation.
2.1.5 Col1a2tm1.1Mcbr
Col1a2tm1.1Mcbr mice were generated by insertion of a knockin allele carrying a G-to-T substitution that converts the triple-helical codon for glycine-610 to cysteine, which is identical to a COL1A2 variant that was identified in an Amish population [43]. Col1a2tm1.1Mcbr mice showed reduced bone mass and cortical thickness as well as decreased biomechanical properties [43,44]. In the same study, an increase in mineral-to-collagen ratio was observed, indicating more brittleness in Col1a2tm1.1Mcbr bones [43,44]. Similar to the Aga2+/− model, increased expression of Ddit3 and Hsp47 was observed in Col1a2tm1.1Mcbr mice, indicating that high ER stress and UPR activation may potentially account for osteoblast dysfunction in mutant mice [45]. Taken together, the Col1a2tm1.1Mcbr mouse model recapitulates the moderately deforming OI type IV [43].
2.2 Post-translational modification and cross-linking
During collagen synthesis, nascent type I procollagen molecules are translocated into the endoplasmic reticulum where they are subject to various post-translational modifications (e.g. lysyl and prolyl-hydroxylation), which are important for proper collagen synthesis, transport and stability. These modifications are subsequently used as substrates for lysyl oxidases to convert specific lysine residues to lysyl-pyridinoline (LP) or hydroxylysine residues to hydroxylysyl-pyridinoline (HP) to generate inter-collagen cross-links [14,46]. Overall, post-translational modification of collagen can influence the formation of covalent cross-links between the collagen telopeptide and helical domains, which can govern its tensile properties.
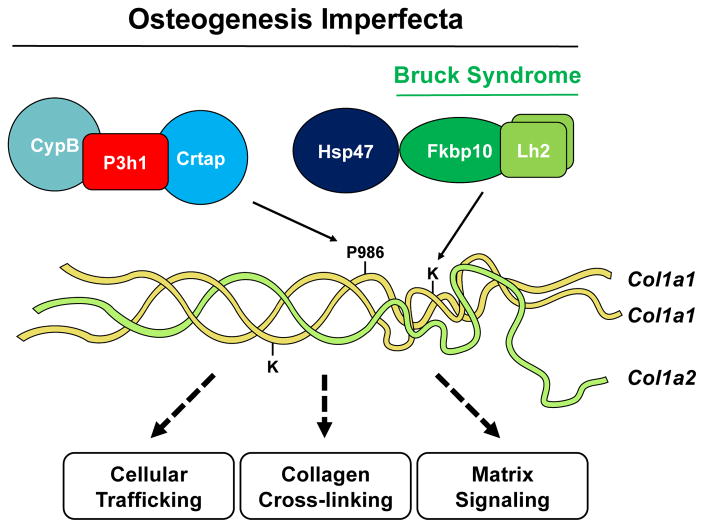
2.2.1 Prolyl 3-hydroxylase complex (P3h1/Crtap/CypB)
Prolyl 3-hydroxylase 1 (P3H1, encoded by LEPRE1) belongs to a group of prolyl 3-hydroxylases (P3H) which makes up the larger 2-oxoglutarate dioxygenase domain containing family of enzymes. Initially purified from chick embryos, P3h1 was observed to co-purify with cartilage associated protein (Crtap) and Cyclophilin B (CypB, encoded by Ppib) [47]. The P3h1 complex converts a single proline in the helical region of type I procollagen (Pro986 of chain α1(I) and Pro707 of chain α2(I)) to 3-hydroxyproline (3Hyp) [48]. However, the biological significance of this modification was not completely understood. Interestingly, homozygous deletion of Crtap downregulates 3Hyp and results in reduced bone mass, increased bone brittleness and impaired biomechanical parameters in mice. Consistent with this observation, loss of function mutations in CRTAP, P3H1 and PPIB have been identified in patients with severe recessive OI [47,49–52]. Similarly, Lepre1−/− mice displayed skeletal abnormalities, including reduced growth, osteopenia and decreased bone strength, indicating that loss of the P3h1 complex and 3Hyp may account for the skeletal phenotypes in these mouse models [53]. Interestingly, knockin mice containing a single amino acid substitution of the P3h1 catalytic site, which abolished enzymatic activity while retaining the ability to bind with Crtap and form the P3h1 complex, caused osteopenia without discernible effect on mouse growth [54]. These observations support the notion that P3h1 enzymatic activity and 3Hyp may be an essential requirement for fibrillar collagen in mineralized tissues, and that defects in either Crtap or P3h1, may cause recessive OI. In addition, homozygous deletion of the peptidyl-prolyl isomerase, Ppib, resulted in mice of smaller size, kyphosis and decreased bone mass [55]. In the same study, Ppib deletion reduced P3h1 but not Crtap levels, indicating that Ppib is essential for P3h1 stability [55]. Whereas the lack of 3Hyp does not affect collagen stability, it may allow prolonged access to other collagen-modifying enzymes, which leads to overmodification of the helical domain of type I procollagen and increased cross-linking [47]. Interestingly, overmodification is observed in Lepre1−/− and Crtap−/− mice but not in Ppib−/− mice, which may be in part due to the mutual requirement of P3h1 and Crtap to maintain the trimeric complex [5,56].
2.2.2 Lysyl-hydroxylase complex (Plod2/Fkbp10)
Lysyl hydroxylase 1-3 (LH1-3) are encoded by procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1-3 (PLOD1-3), respectively, and serve to convert lysine to hydroxylysine (Hyl) residues, which are subsequently used as substrates by the family of lysyl oxidases (LOX) to generate pyridinoline cross-links. Hyl residues are generated by specific enzymes depending on the location of the lysine residue within the procollagen molecule. For example, PLOD1 regulates helical Hyl formation whereas PLOD2 governs Hyl formation in the telopeptide regions [57]. In humans, loss of function mutations in PLOD2 results in OI combined with congenital joint contractures, known as Bruck syndrome [58,59]. Similarly, mutations in FKBP10 causes OI and Bruck syndrome that is accompanied by a significant decrease in telopeptide lysyl-hydroxylation and subsequent inter-chain cross-linking [60–63]. Due to the similarities in phenotypic features, cellular localization and the lack of intrinsic lysyl-hydroxylase function of FKBP65, it was speculated that FKBP65 and LH2 may need to form a complex to exert enzymatic function [2,12]. Indeed, a recent study has shown that physical interaction between FKBP65 and LH2 promotes dimerization and activation of LH2 in cell culture [64]. This interaction was specific to LH2 as FKBP65 did not bind with either LH1 or LH3, corroborating the observation that loss of Fkbp10 does not reduce helical lysyl hydroxylation [63,64]. Similar observations have been made by an independent group (D. Krakow submitted). Still, whether the biochemical interaction between FKBP65 and LH2 is biologically important calls for further investigation. Future studies warrant genetic evaluation between Fkbp10 and Plod2 to determine the physiological relevance of its interaction. Unlike the human mutation, Fkbp10−/− mice died before birth due to embryonic lethality, possibly due to vasculature defects, generalized tissue fragility or poor lung function [65]. In this regard, a conditional knockout mouse model for Fkbp10 would be useful to determine the bone and tendon functions of Fkbp10 removal in postnatal mice.
2.2.3 Serpinh1/Hsp47
Hsp47 is an ER-chaperone that contributes to proper assembly of the collagen triple helix. Hsp47−/− mice are embryonic lethal at E11.5 due to defects in collagen synthesis [66]. Mechanistically, Hsp47 regulates triple-helix stability via direct binding to specific arginine residues that lie at the interface between Hsp47 and collagen [66–68]. Loss of function mutations in HSP47 causes recessive OI due to aggregation and delayed secretion of procollagen molecules [69,70]. While Hsp47 is not directly involved in the post-translational modification of collagen, loss of function mutations of HSP47 (p.L326P) in a dog OI model caused overmodification of type I collagen and increased cross-linking without affecting 3Hyp (Pro986) [71]. This indicates that structural defects of collagen may contribute to abnormalities in post-translational modification. Given that FKBP65 and HSP47 co-eluted in a velocity sedimentation experiment [47] and were also shown to interact by proximity ligation assay, it is possible that HSP47 associates with the FKBP65/LH2 complex, albeit in a weak or transient fashion [69].
2.3 Collagen secretion and ER stress
Newly synthesized proteins must undergo proper conformational change prior to secretion. This process occurs in the endoplasmic reticulum (ER), a eukaryotic organelle that is important for calcium homeostasis, protein folding and secretion. Protein misfolding or excess protein synthesis can cause ER stress and elicit the unfolded protein response (UPR), which activates specific transcriptional programs to cope with ER stress. The most well studied UPR mechanisms include the PERK-eIF2α-ATF4, IRE1α-Xbp1 and ATF6α pathways [72]. Novel recessive OI genes for which the pathogenic mechanisms may involve ER stress and UPR have now recently been identified.
2.3.1 Creb3l1/OASIS
The cAMP response element-binding protein 3-like 1 (Creb3l1) is a basic leucine zipper (bZIP) transcription factor belonging to the CREB/ATF family. Creb3l1, also known as old astrocyte specifically induced substance (OASIS), shares high structural similarities with ATF6. Creb3l1 is highly expressed in osteoblasts and homozygous deletion in mice causes severe osteopenia due to reduced osteoblast function [73]. The molecular function of Creb3l1 known thus far is two-fold: Creb3l1 directly binds to a UPRE-like sequence in the Col1a1 promoter region to drive its expression, and it also appears to regulate the secretion of matrix proteins [6]. Loss of function mutations in CREB3L1 have been shown to cause recessively inherited severe OI with spontaneous fractures in human [74].
2.3.2 Mbtps2
Membrane-bound transcription factor protease, site 2 (Mbtps2) is localized in the Golgi membrane where it cleaves substrates involved in the ER stress response, including Creb3l1/OASIS, ATF6 and sterol regulatory element binding protein (SREBP). A novel missense mutation in MPTBS2 which affected a motif that is important for protease catalytic function caused moderate/severe X-linked recessive form of OI in two independent families [7]. In the same study, OI patient osteoblasts showed reduced cleavage of Creb3l1/OASIS and decreased LH1 levels concomitant with lower levels of hydroxylation of helical lysine (K87) and higher LP/HP ratio [7].
2.3.3 Tric-b
Trimeric intracellular cation channel subtype B (Tric-b, also known as TMEM38B in human) is expressed ubiquitously at low levels and functions to regulate intracellular calcium release [75]. Tric-b−/− mice die soon after birth due to lung defects. At birth, Tric-b−/− mice display significant loss of bone mineralization [8]. Primary calvarial osteoblasts from Tric-b−/− mice showed reduced mineralization despite an increase in collagen protein accumulation in the ER upon recombinant BMP-2 treatment, indicating defects in collagen secretion [8]. Loss of function mutations in TRIC-B cause moderate to severe recessive OI [76–80]. Similar to what is observed in mice, human fibroblasts from OI patients with TRIC-B mutations showed decreased synthesis, secretion and deposition of type I collagen [79]. Further studies are needed to determine how loss of TRIC-B causes defects in collagen biogenesis.
2.4 Collagen processing
Procollagen molecules must undergo proteolytic processing of the N- and C-terminal domains preceding fibril formation [13]. Removal of the C-propeptides is particularly important as it is necessary and sufficient for collagen fibril formation [13,81]. Key cross-linking evens then occur via oxidation of telopeptide hydroxyl-lysines by the family of lysyl oxidases (LOX) [12].
2.4.1 Bmp1
Bone morphogenetic protein 1 (Bmp1) encodes a secreted procollagen C-proteinase that is closely related to the tolloid family of proteases and is functionally distinct from other bone-inducing BMPs [82]. In addition to procollagen, BMP1 has also been shown to exhibit protease activity on LOX and other extracellular matrix (ECM) proteins [83,84]. Bmp1−/− embryos showed reduced ossification of the frontal, parietal and interparietal bones of the skull but no discernible abnormalities in the axial or appendicular skeleton [82]. The lack of a robust skeletal phenotype in Bmp1−/− mice are likely due to residual C-proteinase activity of tolloid-like 1 (Tll1) as simultaneous deletion of Bmp1 and Tll1 in postnatal mice significantly reduced bone mass, length and biomechanical properties owing to increased bone turnover [84]. In human OI patients, mutations in BMP1 had variable effects on bone mass but all cases reported frequent fractures that are associated with decreased cleavage of the collagen C-terminal domain [85–89]. Interestingly, the cleaved C-terminal carboxy propeptide has been reported to influence cellular behavior [90], but further studies are needed to determine whether these functions can contribute to OI pathogenesis.
2.5 Osteoblast differentiation and mineralization
Osteoblasts are derived from mesenchymal lineage cells and are the primary source of collagen deposition in bone [91]. Abnormalities in osteoblast proliferation, differentiation and function significantly impacts the quality and quantity of bone. Once mature osteoblasts secrete collagen into the ECM, multiple factors regulate the mineralization process. Emerging evidence has shed light on the relevance of genes that are important for osteoblast differentiation and mineralization in the context of OI in human patients.
2.5.1 Wnt1
The WNT signaling pathway has critical roles in regulating osteoblast differentiation and function. Upon binding of WNT ligands to Frizzled receptors and co-receptors including the low-density lipoprotein receptor-related protein 5 or 6 (LRP5, LRP6), β-catenin is localized to the nucleus and activates downstream target genes [91]. The first evidence of the importance of WNT signliang in human was demonstrated by reports on loss of function mutations in LRP5, which causes osteoporosis-pseudoglioma syndrome (OPPG) [92]. In contrast, activating mutations in LRP5/6 or mutations in molecules that block ligand-receptor interaction, including Dickkopf-related 1 (DKK1) and sclerostin (SOST), lead to high bone mass [93–97]. In addition, WNT signaling regulates β-catenin-independent pathways in bone [98–100]. While the mechanism by which WNT signaling regulates bone formation is well documented, less is known about which WNT ligands are most critical for regulating bone formation. Interestingly, recent studies have shown that loss of function mutations in WNT1 cause OI, whereas heterozygous WNT1 mutations can lead to early onset osteoporosis [9,101,102]. Here, the semidominant inheritance (i.e. homozygous mutations causing a more severe phenotype than the heterozygous mutation), supports a critical time and dose dependent requirement for this ligand. In mice, the Swaying mouse model (Wnt1sw/sw) which has a spontaneous loss of function mutation in Wnt1, showed frequent fractures, low bone mass and decreased bone strength, recapitulating the phenotypes of human patients [103].
2.5.2 Sp7/Osx
Sp7/Osterix (Osx) is a zinc finger-containing transcription factor that is essential for osteoblast differentiation [104]. Sp7/Osx-null mice completely lack bone formation due to defects in osteoblast differentiation [104] whereas postnatal deletion mildly reduces bone mass due to impairment of osteoblast numbers and function [105]. Mechanistically, Sp7/Osx is recruited to AT-rich enhancers of osteoblast target genes by regulatory complexes containing Distal-less homeobox (Dlx) genes [106]. Interestingly, a frameshift mutation in OSX caused recessive OI in human with mild bone deformities and recurrent fractures [10].
2.5.3 Serpinf1/Pedf
Serpinf1 encodes pigment epithelium-derived factor (PEDF), a secreted glycoprotein originally known for its neurotropic and antiangiogenic features. Loss of function mutations in SERPINF1 causes recessive OI type VI, which features long bone fractures and deformities due to impaired mineralization (i.e., osteomalacia) [107,108]. Serpinf1−/− mice display a mild decrease in bone mass and reduced biomechanical strength accompanied by increased brittleness [109]. Because PEDF is a secreted protein that is primarily produced in the liver and at lower levels in bone, it was hypothesized that restoration of circulating PEDF levels may correct the bone phenotype in OI type VI. In one study, which used a helper-dependent adenovirus (HDAd) to drive human SERPINF1 expression in mouse liver, overexpression of PEDF did not improve the bone phenotype in Serpinf1−/− mice despite restoring biologically active PEDF serum levels [110]. In a separate study, however, intraperitoneal injection of PEDF-containing microspheres markedly increased bone mass and partially improved biomechanical parameters [111]. One possible explanation for this apparent difference in efficacy between the two methods may be the ability of PEDF to inhibit Wnt signaling at high levels [111], but this warrants further investigation.
2.5.4 Ifitm5/Bril
The interferon inducible transmembrane protein family 5 (Ifitm5), also known as Bone restricted Ifitm-like protein (Bril), is a member of the Ifitm family of proteins. Unlike other Ifitm genes that were initially known to regulate germ cell specification [112], Ifitm5 expression is most prominent in osteoblasts [113,114]. Interestingly, Ifitm5−/− mice displayed bent bones in newborn mice (which was later corrected in adulthood) and shorter appendicular elements but no apparent effects on bone mass [114]. The majority of human mutations in IFITM5 are caused by a unique heterozygous mutation (c.-14C>T) in the 5′-untranslated region, which results in autosomal dominant OI type V featuring hyperplastic callus formation [115–119]. Transgenic mice overexpressing the mutant form of Ifitm5 showed delayed mineralization, in utero fractures and severe skeletal malformation, but postnatal bone mass could not be determined due to perinatal lethality [120]. In the same study, transgenic mice overexpressing the wildtype form of Ifitm5 showed normal growth and development, indicating that the human mutation (c.-14C>T) likely has a neomorphic effect in bone [120]. It is worth noting that another heterozygous mutation (c.119C>T) which causes S40L substitution in BRIL protein, showed phenotypic features of type VI OI but not that of type V OI [121]. In this study, serum PEDF levels were significantly reduced, indicating that the S40L substitution in SERPINF1 affects PEDF stability [121]. Thus, IFITM5 may regulate bone formation through interaction with PEDF in a context dependent manner.
2.6 OSTEOGENESIS IMPERFECTA AND TGF-β SIGNALING
The collagen network in skeletal elements serves as a scaffold for its growth and houses a diverse number of proteoglycans, growth factors and cytokines that regulate tissue organization and function. Thus, it is conceivable that defects in collagen structure may adversely affect the extracellular microenvironment leading to dysregulation of cell-matrix interactions and cell signaling.
The transforming growth factor beta (TGF-β) signaling modulates cell proliferation, lineage determination and differentiation in a variety of tissues [122]. The TGF-β ligand is secreted as an inactive latent form, whereas active TGF-β is non-covalently associated with its propeptide LAP (latency-associated peptide) to form the small latent complex (SLC) [123]. The SLC can bind to the latent TGF-β binding protein (LTBP) that can modulate sequestration of the TGF-β complex in the ECM [124,125]. Activated TGF-β signaling is primarily transduced via kinase cascades. Upon ligand binding, TGF-β receptors form a heterotetrameric receptor complex that induces phosphorylation of Smad2/3, which subsequently localizes into the nucleus and interacts with co-activators and co-repressors to modulate gene expression [126].
In bone, TGF-β is secreted by osteoblasts and stored in the bone matrix, primarily in association with SLC, while it often binds with LTBPs in non-skeletal tissues [124,127]. Additionally, TGF-β bioactivity can be modulated by small leucine-rich proteoglycans (SLPRs) such as decorin, which binds to active TGF-β and collagen fibrils [128–130]. TGF-β can be released from the bone matrix and activated during bone resorption by osteoclasts [131], upon which it can locally attract osteoblast precursor cells and stimulate their proliferation and differentiation to facilitate new bone formation [132]. Hence, TGF-β is an important factor that locally couples bone resorption with bone formation to properly maintain bone mass [132]. However, continuous activation of TGF-β signaling has been shown to inhibit terminal osteoblast differentiation and function, leading to low bone mass while increasing osteocyte density [122,133,134].
Interestingly, bones of Crtap−/− and Col1a2tm1.1Mcbr mice show a phenotypic overlap with a genetic model of increased TGF-β signaling, including increased numbers of osteoblasts and osteoclasts with impaired osteoblast function, low bone mass and increased osteocyte density [132,133,135–137]. Furthermore, Crtap−/− mice demonstrate lung abnormalities similar to those observed in Marfan-syndrome, where increased TGF-β activity has been identified as a contributing pathogenic mechanism [138,139]. Interestingly, Smad2 phosphorylation and TGF-β target gene expression were increased in both Col1a2tm1.1Mcbr and Crtap−/− mice [135]. Furthermore, TGF-β-reporter activity was elevated in Crtap−/− mice, in vivo [135]. Importantly, TGF-β neutralizing antibody (1D11) treatment reduced osteoblast and osteoclast numbers, normalized the osteocyte density and improved bone biomechanical strength in both Col1a2tm1.1Mcbr and Crtap−/− mice to wildtype levels [135]. Furthermore, 1D11 partially corrected the lung defects in Crtap−/− mice. Collectively, these findings indicate that increased TGF-β signaling may be a common mechanism that contributes to the skeletal and extraskeletal phenotypes in dominant and recessive forms of OI.
The mechanisms leading to abnormal TGF-β signaling in OI are incompletely understood. In vitro, binding of recombinant decorin to type I collagen of Crtap−/− mice is reduced compared with wildtype collagen, which raises the possibility that the abnormal collagen in OI impairs the ability of decorin, and potentially other SLRPs, to modulate TGF-β signaling [135]. Abnormalities in TGF-β signaling were also observed in Brtl+/− mice, which exhibit phenotypic variability with either a moderately severe or lethal phenotype [140]. Bones from lethal Brtl+/− mice showed increased expression of TGF-β, but no change in the phosphorylation of Smad2/3, whereas bone from non-lethal Brtl+/− mice demonstrated no increased TGF-β expression, but increased Smad2/3 activation. Furthermore, lethal Brtl+/− mice showed abnormal cytoskeletal structure and function, which affected intracellular trafficking and integrin-mediated signaling. Because integrins have been shown to activate TGF-β in the ECM, it is possible that defects in integrin function may be a contributing factor to abnormal TGF-β signaling in OI [140,141]. Together, these findings indicate roles for dysregulated matrix-cell signaling in the pathogenesis of OI. Future clinical studies warrant evaluation of the therapeutic safety and value of targeting altered signaling pathways for the treatment of OI patients.
The potential contributions of another important class of ECM proteins, such as proteoglycans may be underscored by these abnormalities in extracellular-matrix signaling. Specifically, SLRPs interact with collagens as a class and may mediate complex protein-protein interactions which are redundant in nature. In support of this notion, loss of function models of single SLRP mutations often have minimal phenotypes, while more severe bone and connective tissue phenotypes are revealed when they are intercrossed in combination [142]. Hence, the future understanding of how classes of SLRPs and proteoglycans can regulate collagen function and matrix-cell signaling will be critical in delineating the mechanistic basis of these disorders.
3. CONCLUSIONS AND FUTURE DIRECTIONS
The past decade has seen explosive growth in the discovery of novel genes that cause OI, and our understanding of the underlying molecular mechanisms. Beginning with the identification of CRTAP, 14 new genes that cause OI have been discovered, primarily owing to significant advances in genomic technology. In this review, we described genetic causes of OI, mouse models that recapitulate the human disease to varying degrees (Table 1, 2), as well as common mechanisms that contribute to OI pathophysiology (Figure 1).
Table 2.
Summary of mouse genetic models of dominant and recessive OI.
| Gene | Mouse model | Modification | Cross-linking | Skeletal Phenotype | Type | Reference |
|---|---|---|---|---|---|---|
| POST-TRANSLATIONAL MODIFICATION AND CROSS-LINKING | ||||||
| Crtap | Crtap−/− | 3Hyp↓ Overmodification |
HP/LP↑ | Reduced bone mass and strength | VII | [47] |
| Lepre1 | Lepre1−/− | 3Hyp↓ Hyl↑ Overmodification |
HP/LP↑ | Reduced bone mass and strength | VIII | [53] |
| CypB | CypB−/− | 3Hyp↓ Hyl (K87)↓ |
HP/LP↓ HP+LP↑ |
Reduced bone mass and strength | IX | [55,56] |
| Fkbp10 | Fkbp10−/− | Hyl (telo) ↓ | HP/LP↓h HP+LP↓h |
Embryonic lethal | XI, Bruck syndrome | [63,65] |
| Plod2 | n.a. | Hyl (telo) ↓h Hyp↑h |
HP+LP↓h | No mouse model available | Bruck syndrome | [59] |
| Serpinh1 | Hsp47−/− | Overmodification | HP/LP↑d | Reduced bone mass | X | [66,71] |
| COLLAGEN SECRETION AND ER STRESS | ||||||
| Creb3l1 | Creb3l1−/− | n.d. | n.d. | Reduced bone mass with fractures | XVI | [6] |
| Mbtps2 | n.a. | Hyl (K87) ↓ | HP/LP↓h | No mouse model available | n.a. | [7] |
| Tric-b | Tric-b−/− | 3Hyp↓h Hyl (helical) ↓h Hyl (telo)↑h |
n.d. | Reduced bone mass | XIV | [8,79] |
| COLLAGEN PROCESSING | ||||||
| Bmp1 | Bmp1−/− | n.d. | n.d. | Skull defects | XIII | [82] |
| Bmp1−/−;; Tll1−/− | n.d. | n.d. | Reduced bone mass and strength | [84] | ||
| OSTEOBLAST DIFFERENTIATION AND MINERALIZATION | ||||||
| Wnt1 | sw/sw | n.d. | n.d. | Reduced bone mass and strength with fractures and increased ductility | XV | [103] |
| Sp7/Osx | Sp7/Osx−/− | n.d. | n.d. | No bone formation | XII | [10,104] |
| Serpinf1 | Pedf−/− | n.d. | n.d. | Reduced bone mass | VI | [109] |
| Ifitm5 | Ifitm5−/− | n.d. | n.d. | Bone mass unchanged | V | [114] |
| Tg(Col1a1-Ifitm5c.-14C>T) | Unchanged | Unchanged | Embryonic lethal | [120] | ||
3Hyp 3-hydroxyproline, Hyl hydroxylysine, telo telopeptide, HP hydroxylysylpyridinoline, LP lysylpyridinoline,
data from human patients.
data from dog model.
Figure 1.
Genes involved in the post-translational modification of type I collagen. The P3h1 complex serves to generate 3Hyp at P986 of the α(I) chain and the Lh2 complex lysyl hydroxylates telopeptide lysine residues. Defects in members of the P3h1 complex causes OI, whereas Lh2/Fkbp65 mutations cause Bruck syndrome in conjunction with OI. In addition to dominant mutations in Col1a1 or Col1a2, common causes of OI encompass altered collagen trafficking, cross-linking and extracellular matrix signaling.
The fundamental understanding of the mechanisms that underlie disease pathogenesis is essential to devise specific treatment options for the various OI symptoms. Still, investigating both general and specific mechanisms of OI pathogenesis is important to reach this goal. In this regard, the identification of altered TGF-β signaling in both dominant and recessive OI matrix has emerged as a targetable pathway for therapy. It will be of interest to determine whether other matrix signaling pathways are altered in unique OI cases. To this end, using modern technology to massively screen altered gene expression and cell signaling pathways may provide a useful platform to discover pharmacological targets for treatment of OI.
Recent studies have shown that OI has tissue-specific effects. For example, mutations in FKBP10 causes joint contractures in conjunction with deforming OI while other forms of OI cause skin abnormalities. Thus, future studies warrant further investigation in the pathogenic mechanisms and consequence of OI in extraskeletal tissue, including tendons, muscles and skin. Given that human fibroblasts in OI patients do not completely reflect the molecular changes in bones or the affected tissue, generation of animal models that recapitulate the human phenotype is crucial to overcome the limitations studying patient samples originating from unaffected tissues.
Lastly, in OI, alterations in the complex post-translational modifications and residue-specific cross-linking is associated with changes in bone mass and mineral-to-matrix content (Table 1). However, the regulatory mechanisms by which they control bone formation is not completely understood. Further studies will be critical for identifying the specific mechanisms and to develop new treatment options to improve bone quantity and quality and pave way for the next generation of therapies to treat OI and OI-related connective tissue disorders.
As a collection of disorders, the study of OI has not only impacted our understanding of the genetic determinants of bone mass and bone quality in the context of disease but also in physiological bone homeostasis. Moreover, the disorders currently identified to date point to important common pathophysiological mechanisms that contribute differentially to the integration of bone mass and quality. They include broadly, defects in intracellular trafficking and UPR in response to ER stress, abnormalities that affect collagen cross-linking, and finally, disorders that affect cellular-matrix signaling (Figure 1). The future challenge will be to develop mechanistic therapies that will deliver on genotype specific treatments in the form of personalized medicine.
Highlights.
Osteogenesis Imperfecta is a genetic disorder of skeletal and connective tissues
Defects in collagen assembly and matrix quality can affect extracellular signaling
Development of mechanistic therapies can benefit genotype-specific treatments
Acknowledgments
We thank members of the Lee lab for critical reading of the manuscript and Brian C. Dawson for help with figure illustrations. This work was supported by the BCM Intellectual and Developmental Disabilities Research Center (HD024064) from the Eunice Kennedy Shriver National Institute of Child Health & Human Development, the BCM Advanced Technology Cores with funding from the NIH (AI036211, CA125123, and RR024574), the Rolanette and Berdon Lawrence Bone Disease Program of Texas, and the BCM Center for Skeletal Medicine and Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Folketstad L, Hald JD, Ersbøll AK, Gram J, Hermann AP, Langdahl B, et al. Fracture Rates and Fracture Sites in Patients with Osteogenesis Imperfecta - A Nationwide Register-Based Cohort Study. J Bone Miner Res. 2016 doi: 10.1002/jbmr.2920. [DOI] [PubMed] [Google Scholar]
- 2.Forlino A, Marini JC. Osteogenesis imperfecta. Lancet. 2016;387:1657–1671. doi: 10.1016/S0140-6736(15)00728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rauch F, Glorieux FH. Osteogenesis imperfecta. The Lancet. 2004 doi: 10.1016/S0140-6736(04)16051-0. [DOI] [PubMed] [Google Scholar]
- 4.Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. Journal of Medical Genetics. 1979;16:101–116. doi: 10.1136/jmg.16.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldridge D, Schwarze U, Morello R, Lennington J, Bertin TK, Pace JM, et al. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum Mutat. 2008;29:1435–1442. doi: 10.1002/humu.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murakami T, Saito A, Hino S-I, Kondo S, Kanemoto S, Chihara K, et al. Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nature Cell Biology. 2009;11:1205–1211. doi: 10.1038/ncb1963. [DOI] [PubMed] [Google Scholar]
- 7.Lindert U, Cabral WA, Ausavarat S, Tongkobpetch S, Ludin K, Barnes AM, et al. MBTPS2 mutations cause defective regulated intramembrane proteolysis in X-linked osteogenesis imperfecta. Nature Communications. 2016;7:11920. doi: 10.1038/ncomms11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao C, Ichimura A, Qian N, Iida T, Yamazaki D, Noma N, et al. Mice lacking the intracellular cation channel TRIC-B have compromised collagen production and impaired bone mineralization. Science Signaling. 2016;9:ra49–ra49. doi: 10.1126/scisignal.aad9055. [DOI] [PubMed] [Google Scholar]
- 9.Laine CM, Joeng KS, Campeau PM, Kiviranta R, Tarkkonen K, Grover M, et al. WNT1 mutations in early-onset osteoporosis and osteogenesis imperfecta. N Engl J Med. 2013;368:1809–1816. doi: 10.1056/NEJMoa1215458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lapunzina P, Aglan M, Temtamy S, Caparrós-Martín JA, Valencia M, Letón R, et al. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am J Hum Genet. 2010;87:110–114. doi: 10.1016/j.ajhg.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi CG, Zhang Y, Yuan W. Efficacy of Bisphosphonates on Bone Mineral Density and Fracture Rate in Patients With Osteogenesis Imperfecta: A Systematic Review and Meta-analysis. Am J Ther. 2016;23:e894–904. doi: 10.1097/MJT.0000000000000236. [DOI] [PubMed] [Google Scholar]
- 12.Eyre DR, Weis MA. Bone collagen: new clues to its mineralization mechanism from recessive osteogenesis imperfecta. Calcif Tissue Int. 2013;93:338–347. doi: 10.1007/s00223-013-9723-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canty EG, Kadler KE. Procollagen trafficking, processing and fibrillogenesis. Journal of Cell Science. 2005;118:1341–1353. doi: 10.1242/jcs.01731. [DOI] [PubMed] [Google Scholar]
- 14.Bateman JF, Boot-Handford RP, Lamandé SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nature Reviews Genetics. 2009;10:173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 15.Tosi LL, Warman ML. Mechanistic and therapeutic insights gained from studying rare skeletal diseases. Bone. 2015;76:67–75. doi: 10.1016/j.bone.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Harbers K, Kuehn M, Delius H, Jaenisch R. Insertion of retrovirus into the first intron of alpha 1(I) collagen gene to embryonic lethal mutation in mice. Proc Natl Acad Sci USa. 1984;81:1504–1508. doi: 10.1073/pnas.81.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnieke A, Harbers K, Jaenisch R. Embryonic lethal mutation in mice induced by retrovirus insertion into the α1(I) collagen gene. Nature. 1983;304:315–320. doi: 10.1038/304315a0. [DOI] [PubMed] [Google Scholar]
- 18.Löhler J, Timpl R, Jaenisch R. Embryonic lethal mutation in mouse collagen I gene causes rupture of blood vessels and is associated with erythropoietic and mesenchymal cell death. Cell. 1984;38:597–607. doi: 10.1016/0092-8674(84)90514-2. [DOI] [PubMed] [Google Scholar]
- 19.Jaenisch R, Harbers K, Schnieke A, Löhler J, Chumakov I, Jähner D, et al. Germline integration of moloney murine leukemia virus at the Mov13 locus leads to recessive lethal mutation and early embryonic death. Cell. 1983;32:209–216. doi: 10.1016/0092-8674(83)90511-1. [DOI] [PubMed] [Google Scholar]
- 20.Bonadio J, Saunders TL, Tsai E, Goldstein SA, Morris-Wiman J, Brinkley L, et al. Transgenic mouse model of the mild dominant form of osteogenesis imperfecta. Proc Natl Acad Sci USa. 1990;87:7145–7149. doi: 10.1073/pnas.87.18.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobsen CM, Schwartz MA, Roberts HJ, Lim K-E, Spevak L, Boskey AL, et al. Enhanced Wnt signaling improves bone mass and strength, but not brittleness, in the Col1a1(+/mov13) mouse model of type I Osteogenesis Imperfecta. Bone. 2016;90:127–132. doi: 10.1016/j.bone.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jepsen KJ, Goldstein SA, Kuhn JL, Schaffler MB, Bonadio J. Type-I collagen mutation compromises the post-yield behavior of Mov13 long bone. J Orthop Res. 1996;14:493–499. doi: 10.1002/jor.1100140320. [DOI] [PubMed] [Google Scholar]
- 23.Bonadio J, Jepsen KJ, Mansoura MK, Jaenisch R, Kuhn JL, Goldstein SA. A murine skeletal adaptation that significantly increases cortical bone mechanical properties. Implications for human skeletal fragility. J Clin Invest. 1993;92:1697–1705. doi: 10.1172/JCI116756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willing MC, Deschenes SP, Slayton RL, Roberts EJ. Premature chain termination is a unifying mechanism for COL1A1 null alleles in osteogenesis imperfecta type I cell strains. The American Journal of Human Genetics. 1996;59:799–809. [PMC free article] [PubMed] [Google Scholar]
- 25.Thiele F, Cohrs CM, Flor A, Lisse TS, Przemeck GKH, Horsch M, et al. Cardiopulmonary dysfunction in the Osteogenesis imperfecta mouse model Aga2 and human patients are caused by bone-independent mechanisms. Human Molecular Genetics. 2012;21:3535–3545. doi: 10.1093/hmg/dds183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisse TS, Thiele F, Fuchs H, Hans W, Przemeck GKH, Abe K, et al. ER stress-mediated apoptosis in a new mouse model of osteogenesis imperfecta. PLoS Genet. 2008;4:e7. doi: 10.1371/journal.pgen.0040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forlino A, Cabral WA, Barnes AM, Marini JC. New perspectives on osteogenesis imperfecta. Nature Publishing Group. 2011;7:540–557. doi: 10.1038/nrendo.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozloff KM, Carden A, Bergwitz C, Forlino A, Uveges TE, Morris MD, et al. Brittle IV mouse model for osteogenesis imperfecta IV demonstrates postpubertal adaptations to improve whole bone strength. J Bone Miner Res. 2004;19:614–622. doi: 10.1359/JBMR.040111. [DOI] [PubMed] [Google Scholar]
- 29.Uveges TE, Collin-Osdoby P, Cabral WA, Ledgard F, Goldberg L, Bergwitz C, et al. Cellular mechanism of decreased bone in Brtl mouse model of OI: imbalance of decreased osteoblast function and increased osteoclasts and their precursors. J Bone Miner Res. 2008;23:1983–1994. doi: 10.1359/jbmr.080804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Dijk FS, Sillence DO. Osteogenesis imperfecta: clinical diagnosis, nomenclature and severity assessment. Am J Med Genet. 2014;164A:1470–1481. doi: 10.1002/ajmg.a.36545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chipman SD, Sweet HO, McBride DJ, Davisson MT, Marks SC, Shuldiner AR, et al. Defective pro alpha 2(I) collagen synthesis in a recessive mutation in mice: a model of human osteogenesis imperfecta. Proc Natl Acad Sci USa. 1993;90:1701–1705. doi: 10.1073/pnas.90.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips CL, Bradley DA, Schlotzhauer CL, Bergfeld M, Libreros-Minotta C, Gawenis LR, et al. Oim mice exhibit altered femur and incisor mineral composition and decreased bone mineral density. Bone. 2000;27:219–226. doi: 10.1016/s8756-3282(00)00311-2. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=10913914&retmode=ref&cmd=prlinks. [DOI] [PubMed] [Google Scholar]
- 33.Kalajzic I, Terzic J, Rumboldt Z, Mack K, Naprta A, Ledgard F, et al. Osteoblastic response to the defective matrix in the osteogenesis imperfecta murine (oim) mouse. Endocrinology. 2002;143:1594–1601. doi: 10.1210/endo.143.5.8807. [DOI] [PubMed] [Google Scholar]
- 34.Camacho NP, Hou L, Toledano TR, Ilg WA, Brayton CF, Raggio CL, et al. The material basis for reduced mechanical properties in oim mice bones. J Bone Miner Res. 1999;14:264–272. doi: 10.1359/jbmr.1999.14.2.264. [DOI] [PubMed] [Google Scholar]
- 35.Fratzl P, Paris O, Klaushofer K, Landis WJ. Bone mineralization in an osteogenesis imperfecta mouse model studied by small-angle x-ray scattering. J Clin Invest. 1996;97:396–402. doi: 10.1172/JCI118428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camacho NP, Landis WJ, Boskey AL. Mineral changes in a mouse model of osteogenesis imperfecta detected by Fourier transform infrared microscopy. Connect Tissue Res. 1996;35:259–265. doi: 10.3109/03008209609029199. [DOI] [PubMed] [Google Scholar]
- 37.Vanleene M, Porter A, Guillot P-V, Boyde A, Oyen M, Shefelbine S. Ultra-structural defects cause low bone matrix stiffness despite high mineralization in osteogenesis imperfecta mice. Bone. 2012;50:1317–1323. doi: 10.1016/j.bone.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-Florez N, Garcia-Tunon E, Mukadam Q, Saiz E, Oldknow KJ, Farquharson C, et al. An investigation of the mineral in ductile and brittle cortical mouse bone. J Bone Miner Res. 2015;30:786–795. doi: 10.1002/jbmr.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carriero A, Zimmermann EA, Paluszny A, Tang SY, Bale H, Busse B, et al. How tough is brittle bone? Investigating osteogenesis imperfecta in mouse bone. J Bone Miner Res. 2014;29:1392–1401. doi: 10.1002/jbmr.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saban J, Zussman MA, Havey R, Patwardhan AG, Schneider GB, King D. Heterozygous oim mice exhibit a mild form of osteogenesis imperfecta. Bone. 1996;19:575–579. doi: 10.1016/s8756-3282(96)00305-5. [DOI] [PubMed] [Google Scholar]
- 41.Misof BM, Roschger P, Baldini T, Raggio CL, Zraick V, Root L, et al. Differential effects of alendronate treatment on bone from growing osteogenesis imperfecta and wild-type mouse. Bone. 2005;36:150–158. doi: 10.1016/j.bone.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Carriero A, Doube M, Vogt M, Busse B, Zustin J, Levchuk A, et al. Altered lacunar and vascular porosity in osteogenesis imperfecta mouse bone as revealed by synchrotron tomography contributes to bone fragility. Bone. 2014;61:116–124. doi: 10.1016/j.bone.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Daley E, Streeten EA, Sorkin JD, Kuznetsova N, Shapses SA, Carleton SM, et al. Variable bone fragility associated with an Amish COL1A2 variant and a knock-in mouse model. J Bone Miner Res. 2010;25:247–261. doi: 10.1359/jbmr.090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masci M, Wang M, Imbert L, Barnes AM, Spevak L, Lukashova L, et al. Bone mineral properties in growing Col1a2(+/G610C) mice, an animal model of osteogenesis imperfecta. Bone. 2016;87:120–129. doi: 10.1016/j.bone.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirigian LS, Makareeva E, Mertz EL, Omari S, Roberts Pilgrim AM, Oestreich AK, et al. Osteoblast Malfunction Caused by Cell Stress Response to Procollagen Misfolding in α2(I)-G610C Mouse Model of Osteogenesis Imperfecta. J Bone Miner Res. 2016;31:1608–1616. doi: 10.1002/jbmr.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eyre D. Collagen cross-linking amino acids. Meth Enzymol. 1987;144:115–139. doi: 10.1016/0076-6879(87)44176-1. [DOI] [PubMed] [Google Scholar]
- 47.Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, Monticone M, et al. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127:291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 48.Hudson DM, Eyre DR. Collagen prolyl 3-hydroxylation: a major role for a minor post-translational modification? Connect Tissue Res. 2013;54:245–251. doi: 10.3109/03008207.2013.800867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grafe I, Alexander S, Yang T, Lietman C, Homan EP, Munivez E, et al. Sclerostin Antibody Treatment Improves the Bone Phenotype of Crtap(−/−) Mice, a Model of Recessive Osteogenesis Imperfecta. J Bone Miner Res. 2016;31:1030–1040. doi: 10.1002/jbmr.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnes AM, Chang W, Morello R, Cabral WA, Weis M, Eyre DR, et al. Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. N Engl J Med. 2006;355:2757–2764. doi: 10.1056/NEJMoa063804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cabral WA, Chang W, Barnes AM, Weis M, Scott MA, Leikin S, et al. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet. 2007;39:359–365. doi: 10.1038/ng1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Dijk FS, Nesbitt IM, Zwikstra EH, Nikkels PGJ, Piersma SR, Fratantoni SA, et al. PPIB mutations cause severe osteogenesis imperfecta. Am J Hum Genet. 2009;85:521–527. doi: 10.1016/j.ajhg.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vranka JA, Pokidysheva E, Hayashi L, Zientek K, Mizuno K, Ishikawa Y, et al. Prolyl 3-hydroxylase 1 null mice display abnormalities in fibrillar collagen-rich tissues such as tendons, skin, and bones. Journal of Biological Chemistry. 2010;285:17253–17262. doi: 10.1074/jbc.M110.102228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Homan EP, Lietman C, Grafe I, Lennington J, Morello R, Napierala D, et al. Differential Effects of Collagen Prolyl 3-Hydroxylation on Skeletal Tissues. PLoS Genet. 2014;10:e1004121. doi: 10.1371/journal.pgen.1004121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi JW, Sutor SL, Lindquist L, Evans GL, Madden BJ, Bergen HR, et al. Severe osteogenesis imperfecta in cyclophilin B-deficient mice. PLoS Genet. 2009;5:e1000750. doi: 10.1371/journal.pgen.1000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cabral WA, Perdivara I, Weis M, Terajima M, Blissett AR, Chang W, et al. Abnormal type I collagen post-translational modification and crosslinking in a cyclophilin B KO mouse model of recessive osteogenesis imperfecta. PLoS Genet. 2014;10:e1004465. doi: 10.1371/journal.pgen.1004465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Slot AJ, Zuurmond A-M, Bardoel AFJ, Wijmenga C, Pruijs HEH, Sillence DO, et al. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J Biol Chem. 2003;278:40967–40972. doi: 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- 58.Puig-Hervás MT, Temtamy S, Aglan M, Valencia M, Martínez-Glez V, Ballesta-Martínez MJ, et al. Mutations in PLOD2 cause autosomal-recessive connective tissue disorders within the Bruck syndrome--osteogenesis imperfecta phenotypic spectrum. Hum Mutat. 2012;33:1444–1449. doi: 10.1002/humu.22133. [DOI] [PubMed] [Google Scholar]
- 59.Ha-Vinh R, Alanay Y, Bank RA, Campos-Xavier AB, Zankl A, Superti-Furga A, et al. Phenotypic and molecular characterization of Bruck syndrome (osteogenesis imperfecta with contractures of the large joints) caused by a recessive mutation in PLOD2. Am J Med Genet. 2004;131:115–120. doi: 10.1002/ajmg.a.30231. [DOI] [PubMed] [Google Scholar]
- 60.Barnes AM, Cabral WA, Weis M, Makareeva E, Mertz EL, Leikin S, et al. Absence of FKBP10 in recessive type XI osteogenesis imperfecta leads to diminished collagen cross-linking and reduced collagen deposition in extracellular matrix. Hum Mutat. 2012;33:1589–1598. doi: 10.1002/humu.22139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alanay Y, Avaygan H, Camacho N, Utine GE, Boduroglu K, Aktas D, et al. Mutations in the gene encoding the RER protein FKBP65 cause autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86:551–559. doi: 10.1016/j.ajhg.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kelley BP, Malfait F, Bonafe L, Baldridge D, Homan E, Symoens S, et al. Mutations in FKBP10 cause recessive osteogenesis imperfecta and Bruck syndrome. J Bone Miner Res. 2011;26:666–672. doi: 10.1002/jbmr.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwarze U, Cundy T, Pyott SM, Christiansen HE, Hegde MR, Bank RA, et al. Mutations in FKBP10, which result in Bruck syndrome and recessive forms of osteogenesis imperfecta, inhibit the hydroxylation of telopeptide lysines in bone collagen. Human Molecular Genetics. 2012;22:1–17. doi: 10.1093/hmg/dds371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gjaltema RAF, van der Stoel MM, Boersema M, Bank RA. Disentangling mechanisms involved in collagen pyridinoline cross-linking: The immunophilin FKBP65 is critical for dimerization of lysyl hydroxylase 2. Proceedings of the National Academy of Sciences. 2016;113:7142–7147. doi: 10.1073/pnas.1600074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lietman CD, Rajagopal A, Homan EP, Munivez E, Jiang MM, Bertin TK, et al. Connective tissue alterations in Fkbp10−/− mice. Human Molecular Genetics. 2014;23:4822–4831. doi: 10.1093/hmg/ddu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nagai N, Hosokawa M, Itohara S, Adachi E, Matsushita T, Hosokawa N, et al. Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis. The Journal of Cell Biology. 2000;150:1499–1506. doi: 10.1083/jcb.150.6.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Widmer C, Gebauer JM, Brunstein E, Rosenbaum S, Zaucke F, Drögemüller C, et al. Molecular basis for the action of the collagen-specific chaperone Hsp47/SERPINH1 and its structure-specific client recognition. Proceedings of the National Academy of Sciences. 2012;109:13243–13247. doi: 10.1073/pnas.1208072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ishida Y, Kubota H, Yamamoto A, Kitamura A, Bächinger HP, Nagata K. Type I collagen in Hsp47-null cells is aggregated in endoplasmic reticulum and deficient in N-propeptide processing and fibrillogenesis. Mol Biol Cell. 2006;17:2346–2355. doi: 10.1091/mbc.E05-11-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duran I, Nevarez L, Sarukhanov A, Wu S, Lee K, Krejci P, et al. HSP47 and FKBP65 cooperate in the synthesis of type I procollagen. Human Molecular Genetics. 2015;24:1918–1928. doi: 10.1093/hmg/ddu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Christiansen HE, Schwarze U, Pyott SM, AlSwaid A, Al Balwi M, Alrasheed S, et al. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86:389–398. doi: 10.1016/j.ajhg.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lindert U, Weis MA, Rai J, Seeliger F, Hausser I, Leeb T, et al. Molecular Consequences of the SERPINH1/HSP47 Mutation in the Dachshund Natural Model of Osteogenesis Imperfecta. Journal of Biological Chemistry. 2015;290:17679–17689. doi: 10.1074/jbc.M115.661025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nature Reviews Cancer. 2014;14:581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- 73.Murakami T, Hino S-I, Nishimura R, Yoneda T, Wanaka A, Imaizumi K. Distinct mechanisms are responsible for osteopenia and growth retardation in OASIS-deficient mice. Bone. 2011;48:514–523. doi: 10.1016/j.bone.2010.10.176. [DOI] [PubMed] [Google Scholar]
- 74.Symoens S, Malfait F, D’hondt S, Callewaert B, Dheedene A, Steyaert W, et al. Deficiency for the ER-stress transducer OASIS causes severe recessive osteogenesis imperfecta in humans. Orphanet Journal of Rare Diseases. 2013;8:154. doi: 10.1186/1750-1172-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Venturi E, Sitsapesan R, Yamazaki D, Takeshima H. TRIC channels supporting efficient Ca(2+) release from intracellular stores. Pflugers Arch. 2013;465:187–195. doi: 10.1007/s00424-012-1197-5. [DOI] [PubMed] [Google Scholar]
- 76.Rubinato E, Morgan A, D’Eustacchio A, Pecile V, Gortani G, Gasparini P, et al. A novel deletion mutation involving TMEM38B in a patient with autosomal recessive osteogenesis imperfecta. Gene. 2014;545:290–292. doi: 10.1016/j.gene.2014.05.028. [DOI] [PubMed] [Google Scholar]
- 77.Shaheen R, Alazami AM, Alshammari MJ, Faqeih E, Alhashmi N, Mousa N, et al. Study of autosomal recessive osteogenesis imperfecta in Arabia reveals a novel locus defined by TMEM38B mutation. Journal of Medical Genetics. 2012;49:630–635. doi: 10.1136/jmedgenet-2012-101142. [DOI] [PubMed] [Google Scholar]
- 78.Volodarsky M, Markus B, Cohen I, Staretz-Chacham O, Flusser H, Landau D, et al. A Deletion Mutation in TMEM38B Associated with Autosomal Recessive Osteogenesis Imperfecta. Hum Mutat. 2013 doi: 10.1002/humu.22274. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 79.Cabral WA, Ishikawa M, Garten M, Makareeva EN, Sargent BM, Weis M, et al. Absence of the ER Cation Channel TMEM38B/TRIC-B Disrupts Intracellular Calcium Homeostasis and Dysregulates Collagen Synthesis in Recessive Osteogenesis Imperfecta. PLoS Genet. 2016;12:e1006156. doi: 10.1371/journal.pgen.1006156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lv F, Xu X-J, Wang J-Y, Liu Y, Asan, Wang J-W, et al. Two novel mutations in TMEM38B result in rare autosomal recessive osteogenesis imperfecta. J Hum Genet. 2016;61:539–545. doi: 10.1038/jhg.2016.11. [DOI] [PubMed] [Google Scholar]
- 81.Kadler KE, Hojima Y, Prockop DJ. Assembly of collagen fibrils de novo by cleavage of the type I pC-collagen with procollagen C-proteinase. Assay of critical concentration demonstrates that collagen self-assembly is a classical example of an entropy-driven process. J Biol Chem. 1987;262:15696–15701. [PubMed] [Google Scholar]
- 82.Suzuki N, Labosky PA, Furuta Y, Hargett L, Dunn R, Fogo AB, et al. Failure of ventral body wall closure in mouse embryos lacking a procollagen C-proteinase encoded by Bmp1, a mammalian gene related to Drosophila tolloid. Development. 1996;122:3587–3595. doi: 10.1242/dev.122.11.3587. [DOI] [PubMed] [Google Scholar]
- 83.Uzel MI, Scott IC, Babakhanlou-Chase H, Palamakumbura AH, Pappano WN, Hong HH, et al. Multiple bone morphogenetic protein 1-related mammalian metalloproteinases process pro-lysyl oxidase at the correct physiological site and control lysyl oxidase activation in mouse embryo fibroblast cultures. J Biol Chem. 2001;276:22537–22543. doi: 10.1074/jbc.M102352200. [DOI] [PubMed] [Google Scholar]
- 84.Muir AM, Ren Y, Butz DH, Davis NA, Blank RD, Birk DE, et al. Induced ablation of Bmp1 and Tll1 produces osteogenesis imperfecta in mice. Human Molecular Genetics. 2014;23:3085–3101. doi: 10.1093/hmg/ddu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martínez-Glez V, Valencia M, Caparrós-Martín JA, Aglan M, Temtamy S, Tenorio J, et al. Identification of a mutation causing deficient BMP1/mTLD proteolytic activity in autosomal recessive osteogenesis imperfecta. Hum Mutat. 2011;33:343–350. doi: 10.1002/humu.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Asharani PV, Keupp K, Semler O, Wang W, Li Y, Thiele H, et al. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and zebrafish. Am J Hum Genet. 2012;90:661–674. doi: 10.1016/j.ajhg.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fahiminiya S, Al-Jallad H, Majewski J, Palomo T, Moffatt P, Roschger P, et al. A polyadenylation site variant causes transcript-specific BMP1 deficiency and frequent fractures in children. Human Molecular Genetics. 2014;24:516–524. doi: 10.1093/hmg/ddu471. [DOI] [PubMed] [Google Scholar]
- 88.Valencia M, Caparrós-Martín JA, Sirerol-Piquer MS, García-Verdugo JM, Martínez-Glez V, Lapunzina P, et al. Report of a newly indentified patient with mutations in BMP1 and underlying pathogenetic aspects. Am J Med Genet. 2014;164A:1143–1150. doi: 10.1002/ajmg.a.36427. [DOI] [PubMed] [Google Scholar]
- 89.Cho SY, Asharani PV, Kim O-H, Iida A, Miyake N, Matsumoto N, et al. Identification and in vivo functional characterization of novel compound heterozygous BMP1 variants in osteogenesis imperfecta. Hum Mutat. 2015;36:191–195. doi: 10.1002/humu.22731. [DOI] [PubMed] [Google Scholar]
- 90.Vincourt J-B, Etienne S, Cottet J, Delaunay C, Malanda CB, Malanda B, et al. C-propeptides of procollagens I alpha 1 and II that differentially accumulate in enchondromas versus chondrosarcomas regulate tumor cell survival and migration. Cancer Res. 2010;70:4739–4748. doi: 10.1158/0008-5472.CAN-10-0046. [DOI] [PubMed] [Google Scholar]
- 91.Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nature Reviews Molecular Cell Biology. 2012;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- 92.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 93.Babij P, Zhao W, Small C, Kharode Y, Yaworsky PJ, Bouxsein ML, et al. High bone mass in mice expressing a mutant LRP5 gene. J Bone Miner Res. 2003;18:960–974. doi: 10.1359/jbmr.2003.18.6.960. [DOI] [PubMed] [Google Scholar]
- 94.Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, et al. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. The American Journal of Human Genetics. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ai M, Holmen SL, Van Hul W, Williams BO, Warman ML. Reduced affinity to and inhibition by DKK1 form a common mechanism by which high bone mass-associated missense mutations in LRP5 affect canonical Wnt signaling. Molecular and Cellular Biology. 2005;25:4946–4955. doi: 10.1128/MCB.25.12.4946-4955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Semenov MV, He X. LRP5 mutations linked to high bone mass diseases cause reduced LRP5 binding and inhibition by SOST. J Biol Chem. 2006;281:38276–38284. doi: 10.1074/jbc.M609509200. [DOI] [PubMed] [Google Scholar]
- 97.Ellies DL, Viviano B, McCarthy J, Rey J-P, Itasaki N, Saunders S, et al. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res. 2006;21:1738–1749. doi: 10.1359/jbmr.060810. [DOI] [PubMed] [Google Scholar]
- 98.Chen J, Tu X, Esen E, Joeng KS, Lin C, Arbeit JM, et al. WNT7B promotes bone formation in part through mTORC1. PLoS Genet. 2014;10:e1004145. doi: 10.1371/journal.pgen.1004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Esen E, Chen J, Karner CM, Okunade AL, Patterson BW, Long F. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metabolism. 2013;17:745–755. doi: 10.1016/j.cmet.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karner CM, Esen E, Okunade AL, Patterson BW, Long F. Increased glutamine catabolism mediates bone anabolism in response to WNT signaling. J Clin Invest. 2014;125:551–562. doi: 10.1172/jci78470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Keupp K, Beleggia F, Kayserili H, Barnes AM, Steiner M, Semler O, et al. Mutations in WNT1 cause different forms of bone fragility. Am J Hum Genet. 2013;92:565–574. doi: 10.1016/j.ajhg.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pyott SM, Tran TT, Leistritz DF, Pepin MG, Mendelsohn NJ, Temme RT, et al. WNT1 mutations in families affected by moderately severe and progressive recessive osteogenesis imperfecta. Am J Hum Genet. 2013;92:590–597. doi: 10.1016/j.ajhg.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Joeng KS, Lee Y-C, Jiang MM, Bertin TK, Chen Y, Abraham AM, et al. The swaying mouse as a model of osteogenesis imperfecta caused by WNT1 mutations. Human Molecular Genetics. 2014;23:4035–4042. doi: 10.1093/hmg/ddu117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 105.Baek W-Y, de Crombrugghe B, Kim J-E. Postnatally induced inactivation of Osterix in osteoblasts results in the reduction of bone formation and maintenance. Bone. 2010;46:920–928. doi: 10.1016/j.bone.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hojo H, Ohba S, He X, Lai LP, McMahon AP. Sp7/Osterix Is Restricted to Bone-Forming Vertebrates where It Acts as a Dlx Co-factor in Osteoblast Specification. Developmental Cell. 2016;37:238–253. doi: 10.1016/j.devcel.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Homan EP, Rauch F, Grafe I, Lietman C, Doll JA, Dawson B, et al. Mutations in SERPINF1 cause osteogenesis imperfecta type VI. J Bone Miner Res. 2011;26:2798–2803. doi: 10.1002/jbmr.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Becker J, Semler O, Gilissen C, Li Y, Bolz HJ, Giunta C, et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am J Hum Genet. 2011;88:362–371. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bogan R, Riddle RC, Li Z, Kumar S, Nandal A, Faugere M-C, et al. A mouse model for human osteogenesis imperfecta type VI. J Bone Miner Res. 2013;28:1531–1536. doi: 10.1002/jbmr.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rajagopal A, Homan EP, Joeng KS, Suzuki M, Bertin T, Cela R, et al. Restoration of the serum level of SERPINF1 does not correct the bone phenotype in Serpinf1 null mice. Mol Genet Metab. 2016;117:378–382. doi: 10.1016/j.ymgme.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Belinsky GS, Sreekumar B, Andrejecsk JW, Saltzman WM, Gong J, Herzog RI, et al. Pigment epithelium-derived factor restoration increases bone mass and improves bone plasticity in a model of osteogenesis imperfecta type VI via Wnt3a blockade. The FASEB Journal. 2016;30:2837–2848. doi: 10.1096/fj.201500027r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lange UC, Saitou M, Western PS, Barton SC, Surani MA. The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol. 2003;3:1. doi: 10.1186/1471-213x-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moffatt P, Gaumond M-H, Salois P, Sellin K, Bessette M-C, Godin E, et al. Bril: a novel bone-specific modulator of mineralization. J Bone Miner Res. 2008;23:1497–1508. doi: 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- 114.Hanagata N, Li X, Morita H, Takemura T, Li J, Minowa T. Characterization of the osteoblast-specific transmembrane protein IFITM5 and analysis of IFITM5-deficient mice. J Bone Miner Metab. 2010;29:279–290. doi: 10.1007/s00774-010-0221-0. [DOI] [PubMed] [Google Scholar]
- 115.Cho T-J, Lee K-E, Lee S-K, Song SJ, Kim KJ, Jeon D, et al. A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. Am J Hum Genet. 2012;91:343–348. doi: 10.1016/j.ajhg.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Semler O, Garbes L, Keupp K, Swan D, Zimmermann K, Becker J, et al. A mutation in the 5′-UTR of IFITM5 creates an in-frame start codon and causes autosomal-dominant osteogenesis imperfecta type V with hyperplastic callus. Am J Hum Genet. 2012;91:349–357. doi: 10.1016/j.ajhg.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rauch F, Moffatt P, Cheung M, Roughley P, Lalic L, Lund AM, et al. Osteogenesis imperfecta type V: marked phenotypic variability despite the presence of the IFITM5 c.-14C>T mutation in all patients. Journal of Medical Genetics. 2013;50:21–24. doi: 10.1136/jmedgenet-2012-101307. [DOI] [PubMed] [Google Scholar]
- 118.Guillén-Navarro E, Ballesta-Martínez MJ, Valencia M, Bueno AM, Martínez-Glez V, López-González V, et al. Two mutations in IFITM5 causing distinct forms of osteogenesis imperfecta. Am J Med Genet. 2014;164A:1136–1142. doi: 10.1002/ajmg.a.36409. [DOI] [PubMed] [Google Scholar]
- 119.Grover M, Campeau PM, Lietman CD, Lu JT, Gibbs RA, Schlesinger AE, et al. Osteogenesis imperfecta without features of type V caused by a mutation in the IFITM5 gene. J Bone Miner Res. 2013;28:2333–2337. doi: 10.1002/jbmr.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lietman CD, Marom R, Munivez E, Bertin TK, Jiang MM, Chen Y, et al. A transgenic mouse model of OI type V supports a neomorphic mechanism of the IFITM5 mutation. J Bone Miner Res. 2015;30:489–498. doi: 10.1002/jbmr.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Farber CR, Reich A, Barnes AM, Becerra P, Rauch F, Cabral WA, et al. A novel IFITM5 mutation in severe atypical osteogenesis imperfecta type VI impairs osteoblast production of pigment epithelium-derived factor. J Bone Miner Res. 2014;29:1402–1411. doi: 10.1002/jbmr.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Janssens K, Gershoni-Baruch R, Guañabens N, Migone N, Ralston S, Bonduelle M, et al. Mutations in the gene encoding the latency-associated peptide of TGF-beta 1 cause Camurati-Engelmann disease. Nat Genet. 2000;26:273–275. doi: 10.1038/81563. [DOI] [PubMed] [Google Scholar]
- 123.Gentry LE, Lioubin MN, Purchio AF, Marquardt H. Molecular events in the processing of recombinant type 1 pre-pro-transforming growth factor beta to the mature polypeptide. Molecular and Cellular Biology. 1988;8:4162–4168. doi: 10.1128/mcb.8.10.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dallas SL, Park-Snyder S, Miyazono K, Twardzik D, Mundy GR, Bonewald LF. Characterization and autoregulation of latent transforming growth factor beta (TGF beta) complexes in osteoblast-like cell lines. Production of a latent complex lacking the latent TGF beta-binding protein. J Biol Chem. 1994;269:6815–6821. [PubMed] [Google Scholar]
- 125.Nunes I, Gleizes PE, Metz CN, Rifkin DB. Latent transforming growth factor-beta binding protein domains involved in activation and transglutaminase-dependent cross-linking of latent transforming growth factor-beta. The Journal of Cell Biology. 1997;136:1151–1163. doi: 10.1083/jcb.136.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Massague J. TGFβ signalling in context. Nature Reviews Molecular Cell Biology. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hering S, Isken E, Knabbe C, Janott J, Jost C, Pommer A, et al. TGFbeta1 and TGFbeta2 mRNA and protein expression in human bone samples. Exp Clin Endocrinol Diabetes. 2001;109:217–226. doi: 10.1055/s-2001-15109. [DOI] [PubMed] [Google Scholar]
- 128.Hildebrand A, Romaris M, Rasmussen LM, Heinegård D, Twardzik DR, Border WA, et al. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor β. Biochem J. 1994;302:527–534. doi: 10.1042/bj3020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takeuchi Y, Kodama Y, Matsumoto T. Bone matrix decorin binds transforming growth factor-beta and enhances its bioactivity. J Biol Chem. 1994;269:32634–32638. [PubMed] [Google Scholar]
- 130.Markmann A, Hausser H, Schönherr E, Kresse H. Influence of decorin expression on transforming growth factor-beta-mediated collagen gel retraction and biglycan induction. Matrix Biology. 2000;19:631–636. doi: 10.1016/s0945-053x(00)00097-4. [DOI] [PubMed] [Google Scholar]
- 131.Oreffo RO, Mundy GR, Seyedin SM, Bonewald LF. Activation of the bone-derived latent TGF beta complex by isolated osteoclasts. Biochemical and Biophysical Research Communications. 1989;158:817–823. doi: 10.1016/0006-291X(89)92795-2. [DOI] [PubMed] [Google Scholar]
- 132.Tang Y, Wu X, Lei W, Pang L, Wan C, Shi Z, et al. TGF-β1–induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nature Medicine. 2009;15:757–765. doi: 10.1038/nm.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Erlebacher A, Derynck R. Increased expression of TGF-beta 2 in osteoblasts results in an osteoporosis-like phenotype. The Journal of Cell Biology. 1996;132:195–210. doi: 10.1083/jcb.132.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. The EMBO Journal. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grafe I, Yang T, Alexander S, Homan EP, Lietman C, Jiang MM, et al. Excessive transforming growth factor-β signaling is a common mechanism in osteogenesis imperfecta. Nature Medicine. 2014;20:670–675. doi: 10.1038/nm.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rauch F, Travers R, Parfitt AM, Glorieux FH. Static and dynamic bone histomorphometry in children with osteogenesis imperfecta. Bone. 2000;26:581–589. doi: 10.1016/s8756-3282(00)00269-6. [DOI] [PubMed] [Google Scholar]
- 137.Sarathchandra P, Pope FM, Kayser MV, Ali SY. A light and electron microscopic study of osteogenesis imperfecta bone samples, with reference to collagen chemistry and clinical phenotype. J Pathol. 2000;192:385–395. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH704>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 138.Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 139.Baldridge D, Lennington J, Weis M, Homan EP, Jiang MM, Munivez E, et al. Generalized connective tissue disease in Crtap−/− mouse. PLoS ONE. 2010;5:e10560. doi: 10.1371/journal.pone.0010560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bianchi L, Gagliardi A, Maruelli S, Besio R, Landi C, Gioia R, et al. Altered cytoskeletal organization characterized lethal but not surviving Brtl+/− mice: insight on phenotypic variability in osteogenesis imperfecta. Human Molecular Genetics. 2015;24:6118–6133. doi: 10.1093/hmg/ddv328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wipff P-J, Hinz B. Integrins and the activation of latent transforming growth factor beta1 - an intimate relationship. Eur J Cell Biol. 2008;87:601–615. doi: 10.1016/j.ejcb.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 142.Corsi A, Xu T, Chen XD, Boyde A, Liang J, Mankani M, et al. Phenotypic effects of biglycan deficiency are linked to collagen fibril abnormalities, are synergized by decorin deficiency, and mimic Ehlers-Danlos-like changes in bone and other connective tissues. J Bone Miner Res. 2002;17:1180–1189. doi: 10.1359/jbmr.2002.17.7.1180. [DOI] [PubMed] [Google Scholar]
- 143.Forlino A, Porter FD, Lee EJ, Westphal H, Marini JC. Use of the Cre/lox recombination system to develop a non-lethal knock-in murine model for osteogenesis imperfecta with an alpha1(I) G349C substitution. Variability in phenotype in BrtlIV mice. J Biol Chem. 1999;274:37923–37931. doi: 10.1074/jbc.274.53.37923. [DOI] [PubMed] [Google Scholar]